Abstract
A central mechanism regulating translation initiation in response to environmental stress involves phosphorylation of the α subunit of eukaryotic initiation factor 2 (eIF2α). Phosphorylation of eIF2α causes inhibition of global translation, which conserves energy and facilitates reprogramming of gene expression and signaling pathways that help to restore protein homeostasis. Coincident with repression of protein synthesis, many gene transcripts involved in the stress response are not affected or are even preferentially translated in response to increased eIF2α phosphorylation by mechanisms involving upstream open reading frames (uORFs). This review highlights the mechanisms regulating eIF2α kinases, the role that uORFs play in translational control, and the impact that alteration of eIF2α phosphorylation by gene mutations or small molecule inhibitors can have on health and disease.
Maintenance of protein homeostasis requires appropriate regulation of translation, as well as protein folding, transport, and degradative processes. Environmental stresses and physiological stimuli can rapidly disrupt protein homeostasis, triggering cell-adaptive responses that are critical to restore the integrity of the proteome. However, the functionality of the adaptive responses can decline or be altered with chronic stress or with aging, leading to diseases that can afflict multiple organs, including the neural system and those contributing to metabolic health. This review addresses the role of translational control in adaptive responses to environmental stresses and the processes by which phosphorylation of the α subunit of eukaryotic initiation factor 2 (P-eIF2α) can modulate translation genome wide to restore protein homeostasis. Key themes in the review will be the mechanisms regulating eIF2α kinases, the role that upstream open reading frames (uORFs) play in translational control, and the impact that altered P-eIF2α levels by gene mutations or small molecule inhibitors can have on health and disease.
PHOSPHORYLATION OF eIF2α DIRECTS TRANSLATION CONTROL
A major mechanism regulating the initiation phase of protein synthesis involves P-eIF2α at serine-51. The eIF2, combined with guanosine triphosphate (GTP), is critical for providing initiator methionyl-transfer RNA (tRNA) (Met-tRNAiMet) to the 43S preinitiation complex that contains the small ribosomal subunit and a myriad of additional translation initiation factors. In the predominant pathway, the preinitiation complex then combines with the 5′-7-methylguanosine “cap” of messenger RNA (mRNA) and scans processively 5′- to 3′- along the leader of the transcript in search of an initiation codon. Complementary binding of the Met-tRNAiMet to the start codon in the P site of the 40S ribosomal subunit triggers cessation of scanning and hydrolysis of GTP associated with eIF2. Following release of eIF2•GDP (guanosine diphosphate), the large 60S ribosomal subunit then joins to form the 80S ribosome, which carries out the elongation phase of protein synthesis. To facilitate the next round of translation initiation, GDP associated with eIF2 needs to be exchanged for GTP, a process catalyzed by a guanine nucleotide exchange factor, eIF2B. In response to diverse stresses, P-eIF2α alters this translation factor so that it binds tightly to a regulatory portion of eIF2B, thus inhibiting the recycling of eIF2•GDP to the active GTP-bound form (Fig. 1). As a consequence, there is lowered eIF2•GTP and delivery of the Met-tRNAiMet to ribosomes, culminating in a sharp reduction in global translation initiation.
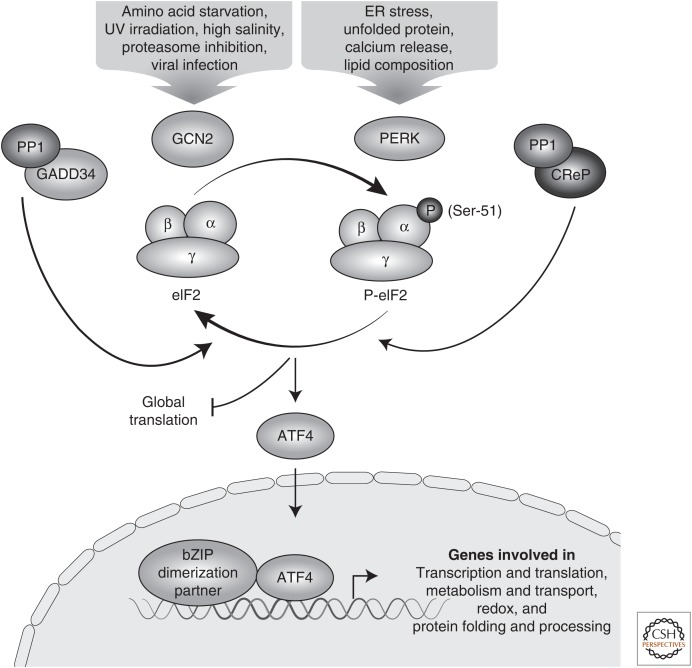
Figure 1.
Phosphorylation of the α subunit of eukaryotic initiation factor 2 (eIF2α) regulates global and gene-specific translation. The eIF2α kinases general control nonderepressible 2 (GCN2) and protein kinase R (PKR)-like endoplasmic reticulum (ER) kinase (PERK) are activated by nutritional stress or perturbations in the ER, respectively. Type 1 protein phosphatase complex (PP1c) combines with CReP to dephosphorylate eIF2α during basal conditions and GADD34 in feedback control of the integrated stress response (ISR). Phosphorylation of eIF2α reduces global translation initiation coincident with preferential translation of ATF4, encoding a basic zipper (bZIP) transcriptional activator that dimerizes with other transcript factors to regulate transcription of ISR genes that function in adaptation to stress.
Repression of translation initiation is an efficient mechanism to conserve energy and nutrients, which are amply consumed by protein synthesis. Furthermore, lowering general translation allow cells to reconfigure gene expression and signaling pathways that optimize stress alleviation. For example, arrest of translational initiation by increased levels of P-eIF2α leads to polysome disassembly that triggers formation of stress granules, which are cytosolic foci of untranslated mRNAs and associated 40S ribosomal subunits and proteins (Kedersha et al. 2013; Ivanov et al. 2017). Stress granules serve as a triage center, sorting incoming messenger ribonucleoproteins for mRNA decay or sequestration for eventual return to the cytoplasm for translation. Therefore, stress granules are critical for reprogramming gene expression. Signaling proteins and enzymes can also be recruited to stress granules, influencing their respective cellular pathways.
Inhibition of global protein synthesis also reshapes the proteome, as proteins that are labile will rapidly be depleted from cells. The biological consequences of these proteomic changes are shown by the activation of nuclear factor κB (NF-κB) in response to accumulation of P-eIF2α and ultraviolet (UV) irradiation (Wu et al. 2004; Jiang and Wek 2005). NF-κB is a transcriptional regulator of genes involved in inflammation, cell proliferation, and apoptosis, and is inhibited by binding to IκBα. Lowered synthesis of IκBα as a consequence of induced P-eIF2α, combined with rapid turnover of IκBα protein, causes a release of IκBα from NF-κB that facilitates NF-κB entry into the nucleus for targeted transcriptional regulation.
FAMILY OF eIF2α KINASES ACTIVATED BY DIFFERENT STRESSES
Coincident with global repression of protein synthesis, select gene transcripts can be resistant or even preferentially translated in response to induced P-eIF2α. An important preferentially translated gene is ATF4, which features uORFs embedded in its mRNA that serve as a “bar code” for scanning ribosomes for selective translation (Harding et al. 2000a; Lu et al. 2004; Vattem and Wek 2004). ATF4 is a basic zipper (bZIP) transcription factor of genes involved in nutrient import, metabolism, and alleviation of oxidative stress (Harding et al. 2003). Because P-eIF2α and ATF4 are induced by diverse stresses, this pathway is referred to as the integrated stress response (ISR) (Harding et al. 2003). In mammals, there are four different eIF2α kinases, each containing distinct regulatory domains that serve to sense the cell stress environment through engagement with regulatory ligands and proteins. This review will focus on two of the eIF2α kinase family members, general control nonderepressible 2 (GCN2 or EIFAK4) and protein kinase R (PKR)-like endoplasmic reticulum (ER) kinase (PERK or EIF2AK3), which respond to perturbations in the cytosol and ER, respectively (Fig. 1). The other eIF2α kinases include HRI (EIF2AK1), which primarily functions to balance globin synthesis with heme availability during erythropoiesis, and PKR (EIF2AK2), which participates in the innate immune response to viral infection.
In the example of GCN2, starvation for amino acids enhance P-eIF2α levels and translational control, which quickly limits incorporation of amino acids into nascent polypeptides. In addition to the protein kinase domain, GCN2 has a regulatory region homologous to histidyl-tRNA synthetase (HARS), which binds to uncharged tRNAs that accumulate during deprivation for nutrients (Wek et al. 1989, 1995; Dong et al. 2000). Binding to uncharged tRNA is thought to lead to conformational changes in GCN2 that trigger autophosphorylation and release of inhibitory interactions between the regulatory regions of GCN2 and the kinase domain, resulting in increased P-eIF2α (Lageix et al. 2014, 2015). It should be emphasized that GCN2 can bind to a range of different uncharged tRNAs to monitor the availability of their respective amino acids. Activation of GCN2 also requires GCN1 protein, which binds to the amino-terminal RWD domain of GCN2 and is thought to facilitate GCN2 access to uncharged tRNAs (Marton et al. 1993, 1997). GCN2 can be inhibited by the regulatory protein IMPACT (YIH1), which competes with this eIF2α kinase for its association with GCN1 (Sattlegger et al. 2004; Pereira et al. 2005). Finally, many other stresses have been reported to activate GCN2, including UV irradiation, high salinity, and glucose deprivation (Yang et al. 2000; Goosens et al. 2001; Deng et al. 2002; Zaborske et al. 2009). These stresses also require the function of the HARS-related domain of GCN2, supporting the idea that this mode of regulation is required at least in part for GCN2 activation by stresses not directly linked with amino acid depletion.
PERK is a transmembrane protein situated in the ER, which functions as part of the unfolded protein response (UPR) (Shi et al. 1998; Harding et al. 1999, 2000b). The UPR features both translational and transcriptional gene expression that serves to expand the processing capacity of the ER (Walter and Ron 2011). Regulation of PERK is complex, in part because there are numerous conditions that readily perturb the ER and because the stresses are typically not measured directly but instead are inferred by assessing activation of PERK and the other UPR sensors IRE1 and ATF6. A prevailing model for the regulation of PERK is that the amino-terminal portion of PERK can bind to the ER-resident chaperone BiP (GRP78/HSPA5), maintaining this eIF2α kinase in a repressed conformation (Bertolotti et al. 2000; Ma et al. 2002). Stressful conditions in the ER that disrupt protein folding can trigger the release of the chaperone BiP from PERK, providing for an activated conformation that induces PERK autophosphorylation and P-eIF2α.
The rationale for BiP release from PERK during ER stress is attributed to accumulating unfolded protein in the ER effectively competing for binding with the ER lumenal portion of this eIF2α kinase. BiP dissociation from PERK would be readily reversed when the ER stress is remedied in the cell (Bertolotti et al. 2000). It was generally assumed that BiP bound with PERK through the peptide-binding portion of this ER chaperone. An alternative model has been put forth that the ATPase domain of BiP binds with PERK and this interaction is released when unfolded protein engages with the canonical peptide-binding domain of BiP (Carrara et al. 2015). Given that BiP is abundant in the ER, it has been argued that the BiP regulatory model of PERK is too coarse for rapid activation of PERK during ER stress (Pincus et al. 2010). For the observed rapid activation of PERK, it has instead been proposed that the lumenal portion of PERK can accommodate direct binding to unfolded protein. This idea is supported by peptide-binding experiments with the UPR sensory protein IRE1 from yeast (Gardner and Walter 2011), but is still unresolved for PERK.
It is noteworthy that PERK has functions independent of its eIF2α kinase activity, as increased cytosolic Ca2+ levels can also trigger oligomerization of PERK in the ER, which is suggested to stabilize PERK interactions with the actin-binding protein filamin A (FLNA) (van Vliet et al. 2017). The PERK/FLNA interaction drives F-actin remodeling, facilitating contacts between the ER and plasma membrane that function in the regulation of Ca2+ fluxes and lipid signaling. These results indicate that the biological effects attributed to loss of PERK do not always involve dysregulation of eIF2α phosphorylation and translational control.
Enhanced P-eIF2α by GCN2 and PERK are balanced by dephosphorylation by type 1 protein phosphatase complex (PP1c) that is directed to eIF2α via scaffolding proteins GADD34 (PPP1R15A) and CReP (PPP1R15B) (Fig. 1) (Connor et al. 2001; Novoa et al. 2001; Jousse et al. 2003). GADD34 and CReP share sequence similarity in their carboxy-terminal PP1c-anchoring motifs, but have dissimilar regions that serve to engage with eIF2α (Choy et al. 2015). Expression of GADD34 is enhanced by stress and elevated levels of P-eIF2α and is central for restoration of translation through feedback control of the ISR. CReP functions to maintain lower levels of P-eIF2α during basal conditions. Although expression of CReP is suggested to be constitutive, there is potential cross regulation between these two PP1c regulatory proteins (Young et al. 2015). It is also noteworthy that both GADD34 and CReP association with PP1c is stabilized by direct binding with monomeric G-actin, and the abundance and activity of the complex and levels of P-eIF2α are responsive to changes in the polymeric status of actin (Chambers et al. 2015; Chen et al. 2015). The involvement of the cytoskeleton as a spatial organizer and regulator of key processes in translation is an emerging theme and may also affect the regulation of GCN2 (Silva et al. 2016). Disruption of the actin cytoskeleton is suggested to facilitate IMPACT release from GCN1, making the activator GCN1 protein more accessible for association with GCN2. GCN2 can then bind uncharged tRNAs more efficiently, leading to enhanced phosphorylation of eIF2α.
Emphasizing the importance of CReP and GADD34 in the implementation and function of the ISR, mice deleted for CReP are growth impaired and deficient for erythropoiesis (Harding et al. 2009). Combined loss of both CReP and GADD34 leads to early embryonic lethality, which can be rescued by expression of a version of eIF2α that that is refractory to phosphorylation. These findings highlight the critical roles that appropriate dephosphorylation of eIF2α plays in regulating translational control in the ISR and in mammalian development.
uORFs IN ISR TRANSLATIONAL CONTROL
In addition to ATF4, a number of key ISR regulatory genes affecting diverse cell functions are preferentially translated by mechanisms involving uORFs (Fig. 2). By definition, a uORF encodes at least two amino acid residues followed by a termination codon, which can be fully upstream or overlapping the primary coding sequence (CDS). About half of human genes encode putative uORFs (Iacono et al. 2005; Calvo et al. 2009; Resch et al. 2009). Whereas the mere presence of a predicted uORF does not necessarily indicate that it is translated, a report by Qian and colleagues used ribosome profiling to identify nearly 8000 translation initiation sites upstream of human CDSs (Lee et al. 2012). Their findings suggest that uORF initiation sites also include non-AUG codons, with CUG being the most prominent. There are technical concerns about potential translation artifacts in profiling studies; nonetheless, the prevalence of uORFs is striking among mammalian transcripts. It is important to note that predicted uORFs are present among mRNAs that are repressed, not affected, or preferentially translated during cellular stress and in the presence of elevated levels of P-eIF2α. Therefore, the presence of a uORF alone is not predictive of whether an mRNA is preferentially translated in response to P-eIF2α induction. Rather, the specific properties of uORFs and their placement and combinations in the 5′-leader of target mRNAs determine translation efficiency in response to P-eIF2α induction (Fig. 3). Additionally, secondary structures in the 5′-leader of mRNAs and RNA-binding proteins can influence the functions of uORFs and translational control.
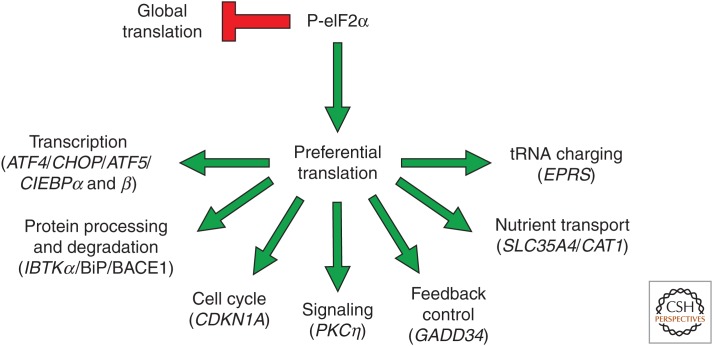
Figure 2.
Phosphorylation of the α subunit of eukaryotic initiation factor 2 (P-eIF2α) enhances translation of multiple integrated stress response (ISR) genes by mechanisms involving upstream open reading frames (uORFs). P-eIF2α reduces global protein synthesis concurrent with preferential translation genes involved in diverse cellular functions. Preferential translation of ATF4, CHOP, GADD34, EPRS, and CDKN1A involves uORFs as described in the text. IBTKα (Baird et al. 2014; Willy et al. 2017), BiP (Starck et al. 2016), BACE1 (O’Connor et al. 2008), PKCη (Raveh-Amit et al. 2009), SLC35A4 (Andreev et al. 2015; Sidrauski et al. 2015), and CAT1 (Yaman et al. 2003) have also been reported to be preferentially translated directly or indirectly by P-eIF2α during stress.
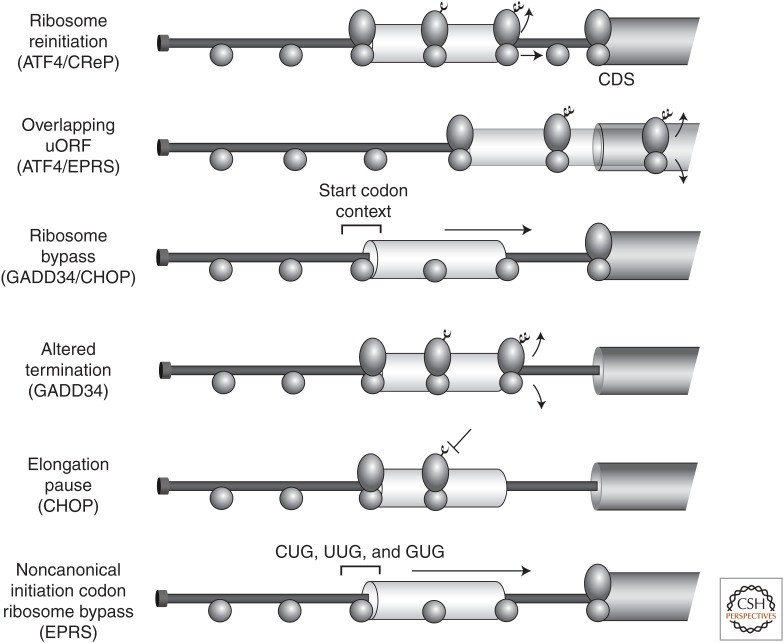
Figure 3.
Upstream open reading frames (uORFs) can have different functions in preferential translation in the integrated stress response (ISR). The uORFs and their function are highlighted for the indicated gene transcripts. The 5′-leader of the messenger RNAs (mRNAs) is indicated as a solid line. The coding sequences (CDSs) are indicated by the bar on the far right of each transcript, with uORFs indicated by the light gray bars. Scanning and elongating ribosomes are indicated by the ovals, with small and large ribosomal subunits. Arrows indicate ribosome bypass, reinitiation or termination, and release.
Typically, uORFs are inhibitory to translation of the downstream CDS. Repression by uORFs can be considerable or more moderate, depending on the degree to which ribosomes initiate translation at the uORF and the ability of the terminating ribosomes to reinitiate translation downstream at a subsequent CDS (Fig. 3). Preferential translation of mRNAs in the ISR involves ribosome bypass or leaky scanning through inhibitory uORFs. How does P-eIF2α allow the ribosomes to proceed through barrier uORFs? The 5′-leader of ATF4 contains a strong inhibitory uORF2, which overlaps out-of-frame with the ATF4 CDS, and a short uORF1 that acts as a positive element in ATF4 translation, promoting downstream reinitiation of translation (Fig. 4A) (Harding et al. 2000a; Lu et al. 2004; Vattem and Wek 2004). Following translation of the 5′-proximal uORF1, 40S ribosomal subunits are thought to be retained on the ATF4 mRNA and resume scanning. The scanning 40S subunits then rapidly reacquire a new eIF2•GTP• Met-tRNAiMet ternary complex that is abundant in nonstressed conditions when P-eIF2α is low. As a result, ribosomes initiate translation at the next available CDS, uORF2. Translation of the overlapping out-of-frame uORF2 results in translation termination and ribosome dissociation 3′ of the initiation codon of the ATF4 CDS. Therefore, ATF4 protein levels are reduced and there is lowered transcription of target ISR genes.
Figure 4.
The integrated stress response (ISR) features different translational control mechanisms with upstream open reading frames (uORFs). (A) Illustration of the mechanism of ATF4 delayed translation reintiation that functions to enhance ATF4 synthesis on phosphorylation of the α subunit of eukaryotic initiation factor 2 (P-eIF2α) and stress. In nonstressed conditions, there are low levels of P-eIF2α and abundant eIF2•GTP (guanosine triphosphate). Following translation of uORF1 (green bar), ribosomes (ovals indicated by large and small subunits) rapidly reacquire new eIF2•GTP•Met-tRNAiMet and reinitiate at the inhibitory uORF2 (red bar), which overlaps out-of-frame with the ATF4 coding sequence (CDS) (blue bar). Therefore, there are low levels of ATF4 and its target genes in the absence of stress. In response to stress, enhanced P-eIF2α and low eIF2•GTP delay reinitiation, allowing ribosomes to proceed through uORF2, and instead translate the ATF4 CDS. (B) Translation of GADD34 involves a fraction of the translating ribosome scanning through an inhibitory uORF2 (red bar) in response to P-eIF2α and stress. The uORF1 (gray bar), which overlaps out-of-frame uORF2, is not well translated and is a modest dampener in the translation of GADD34. (C) Expression of CReP involves a fraction of ribosomes translating uORF2 (red bar) and reinitiating at the CReP CDS independent P-eIF2α and stress. Therefore, synthesis of CReP is largely constitutive regardless of stress conditions. The CReP ORF1 (gray bar) functions to lower translation of the CReP CDS only modestly. (D) Substitution of the Pro-Pro-Gly-stop codons and nine nucleotides 3′- of the GADD34 uORF2 for the corresponding uORF2 regions in the CReP transcript (indicated by red portion of the uORF and messenger RNA [mRNA]) leads to lowered translation of the CReP hybrid that is preferentially translated in response to stress and P-eIF2α induction (Young et al. 2015).
During ER stress or nutrient deprivation, induction of P-eIF2α lowers the levels of eIF2•GTP that are required for delivery of Met-tRNAiMet to the reinitiating ribosomes. As a consequence, after translation of uORF1, the scanning 40S ribosomal subunit requires more time to acquire a new eIF2 ternary complex needed for recognition of the next initiation codon in the ATF4 mRNAs. The delay in the acquisition of eIF2 ternary complex allows the 40S ribosomal subunit to scan through the start codon for the inhibitory uORF2 and instead promotes translation initiation at the next available initiation codon, the ATF4 CDS (Fig. 4A). Increased levels of ATF4 directly enhance adaptive target genes in the ISR (Fig. 1). Translational control by delayed reinitiation was originally described by Hinnebusch and colleagues in budding yeast for the related transcriptional activator GCN4 (Abastado et al. 1991; Hinnebusch 2005).
Given the diverse stress conditions enhancing ATF4 translation, there may be additional modulators of ATF4. For example, another short uORF has been identified upstream of uORF1 in ATF4 that is occupied by ribosomes in profiling studies. Prior experiments using luciferase reporters fused to 5′-segments of the ATF4 transcript did not detect any appreciable changes in the induction of translation on ER stress when this upstream uORF was omitted (RC Wek, unpubl.). However, levels of induced ATF4 translation measured using reporters transfected into cultured cells are typically lower than those determined for endogenous ATF4 so there may be additional regulatory features.
Furthermore, mRNA sequences proximal to the 5′-cap can enhance the recruitment of the eIF4E subunit of the cap-binding eIF4F complex and translation efficiency (Keys 2016; Keys and Sabatini 2017). These so-called “juxtaposed sequences” may influence ATF4 translation and may be critical for loading of the 43S preinitiation complex onto the ATF4 transcript when eIF2•GTP levels are diminished with increased levels of P-eIF2α and stress. Finally, base modifications in RNA, such as N6-methyladenosine, may influence the efficiency of ribosome scanning and reinitiation that can affect ATF4 translation (Meyer et al. 2015; Wang et al. 2015; Zhou et al. 2015).
PREFERENTIAL TRANSLATION BY RIBOSOME BYPASS
A number of transcripts that are preferentially translated in the ISR involve a mechanism featuring a single uORF. One example is CHOP, whose translational and transcriptional expression is enhanced by P-eIF2α. Early in the stress response, CHOP triggers transcription of genes with adaptive functions, including those related to ATF4 (Marciniak et al. 2004; Han et al. 2013). However, with extended stress and sustained P-eIF2α induction, continued CHOP expression can trigger expression of genes that elicit apoptosis (Marciniak et al. 2004; Marciniak and Ron 2006; Oslowski and Urano 2011). Thus, CHOP is central to the balance between the adaptive functions of the ISR during acute stress versus induction of cell death during chronic stress conditions.
Preferential translation of CHOP features a single uORF that serves to stall elongating ribosomes as judged by experiments with translational reporters and in vitro toeprinting analyses, preventing reinitation at the downstream CHOP CDS (Fig. 3) (Jousse et al. 2001; Palam et al. 2011; Young et al. 2016b). In response to stress and accumulating P-eIF2α, a subset of scanning ribosomal subunits proceed through the CHOP uORF and instead initiate at the CDS. Part of the ability of ribosomes to bypass the uORF in response to increased levels of P-eIF2α involves a less-than-optimal context of the uORF start codon. Emphasizing the importance of the uORF in CHOP expression, mutations that prevent translation of the uORF substantially increase the levels of CHOP during both basal and stress conditions and modify the pattern of induction of CHOP expression in the ISR (Young et al. 2016b). Elevated CHOP levels sensitize cells to stress, with accelerated apoptosis on cell exposure to ER stress.
Another example of ribosome bypass of a uORF is that of GADD34, which contains two uORFs (Fig. 4B) (Lee et al. 2009; Young et al. 2015). uORF2 is the primary inhibitor of downstream translation at the GADD34 CDS and is sufficient to confer preferential translation in response to P-eIF2α. Translation of the Pro-Pro-Gly codons juxtaposed to the termination codon in uORF2 is suggested to block translation reinitiation at the CDS, lowering levels of GADD34 expression during basal conditions (Young et al. 2015). Preferential translation of GADD34 in response to stress and elevated P-eIF2α levels occurs by a fraction of the scanning ribosomal subunits bypassing uORF2 by a mechanism involving, at least in part, the poor start codon context of this inhibitory uORF. It is interesting to note that translation of CReP is resistant to P-eIF2α induction (Andreev et al. 2015; Young et al. 2015). The CReP transcript also has two uORFs that are frequently bypassed even during nonstressed conditions. Furthermore, uORF2 allows for efficient reinitiation of translation at the CDS (Fig. 4C). If the termination codon and 3′-flanking sequences from the GADD34 uORF2 are substituted for those in the CReP uORF2, translation of CReP becomes induced on stress and with elevated levels of P-eIF2α (Fig. 4D) (Young et al. 2015). This finding emphasizes the importance of precise uORF properties to convey translational control in the ISR.
Preferential translation can also occur via bypass of uORFs with noncanonical initiation codons. Enhanced expression of the bifunctional glutamyl-prolyl tRNA synthetase, EPRS, serves to increase the appropriately charged tRNA pool and prime the cell for resumption of translation once the cellular stress is alleviated (Fig. 3). Two uORFs featuring UUG and CUG initiation codons are considered to be the primary regulators of EPRS preferential translation (Young et al. 2016a). An inhibitory uORF with a CUG initiation codon overlaps out-of-frame with the EPRS CDS. On the other hand, a uORF featuring UUG terminates upstream of the CDS and allows some of the ribosomes to reinitiate at the downstream EPRS CDS (Young et al. 2016a). Both uORFs are bypassed to a moderate extent during basal conditions, with enhanced bypass efficiency during eIF2α-P and stress.
PREFERENTIAL TRANSLATION VARIES BETWEEN STRESSES
There is robust P-eIF2α induction in response to a spectrum of stress conditions, but the pattern of gene-specific translation can be specifically tailored to best adapt to each stress condition. This is noteworthy because the uORFs are embedded in each gene transcript and would not appear to be readily modified for a given stress. Three explanations can be provided for gene-specific translation tailored to a given stress. First, ATF4 and other ISR genes subject to preferential translation also have enhanced transcriptional expression in response to ER or nutrient stress, which would increase the amount of mRNA available for translation during the progression of the stress response. However, following exposure to high physiological doses of UV-B or UV-C, there is repressed transcription of ATF4, sharply lowering the amount of ATF4 mRNA that is available for preferential translation (Dey et al. 2010, 2012; Collier et al. 2015). Lowered ATF4 levels also reduce the transcription and, ultimately, the translation of the downstream target gene CHOP. Repression of global translation is important for cell survival in response to UV stress, and forced expression of ATF4 sensitizes cells. Knockdown of CHOP suppresses this sensitivity, emphasizing the idea that elevated expression of CHOP is detrimental in response to UV irradiation (Collier et al. 2015). The dynamics of changes in both mRNA and translation in response to increased levels of P-eIF2α provide a vehicle to differentially regulate the expression of key ISR genes in response to different stress conditions.
A second explanation features alternative gene promoters and pre-mRNA splicing that can create gene transcripts with different 5′-leaders and uORF configurations, which alter mRNA translation during P-eIF2α induction. For example, the ATF5α variant that is controlled by the mechanism of delayed translation initiation is derived from a different promoter than the ATF5β variant, which has an expanded collection of uORFs that appear to largely dampen translation (Watatani et al. 2008; Zhou et al. 2008). Both variants express the same CDS, with ATF5α participating in the ISR and ATF5β mRNA being expressed predominantly during early development (Hansen et al. 2002). Pre-mRNA splicing can alter translation of CDKN1A (p21/WAF1), which contributes to cell-cycle arrest and increased survival in response to starvation for amino acids. Among the many CDKN1A spliced variants in mice that alter the 5′-leader of the gene transcripts, variant 2 features three uORFs that provide for preferential translation in response to induced P-eIF2α (Lehman et al. 2015).
A final explanation for distinct stress-specific programs of preferential translation involves the notion that mRNA translation is spatially organized and regulated in cells and that a given stress can differentially disrupt cell compartments. Nicchitta and colleagues (Reid et al. 2014; Reid and Nicchitta 2015) found that ER-bound ribosomes synthesize a significant fraction of proteins, both those slated to be retained in the cytosol as well as those destined for the secretory pathway. The ER-associated translation system is suggested to be dynamic and be reorganized in response to physiological cues and cellular stresses. In this way, the ER environment for translation may be quite distinct from the cytosol and the influences of P-eIF2α induction may vary, yielding differences in preferential translation.
ISR AND DISEASE
Emphasizing the broad and diverse impact of the ISR, mutations have been identified in ISR genes that afflict distinct tissues and present with different pathologies. For example, nonsense, frameshift, and missense mutations have been reported in PERK, leading to Wolcott–Rallison syndrome, which is characterized by neonatal diabetes, osteoporosis, digestive dysfunctions, and hepatic complications, culminating in early death (Delepine et al. 2000; Senée et al. 2004). The inability of PERK to induce translational control in Wolcott–Rallison syndrome leads to disruption of protein homeostasis, especially in specialized secretory tissues that require robust ER secretory processes.
Loss of GCN2 function causes pulmonary disorders, including pulmonary arterial hypertension (PAH), pulmonary veno-occlusive disease (PVOD), and pulmonary capillary hemangiomatosis (PCH) (Best et al. 2014, 2017; Eyries et al. 2014). The rationale for why GCN2 deficiency triggers pulmonary disorders in humans is currently not understood. The lungs are challenged by a variety of inhaled stress agents, including smoke, airborne particles, and microbes. Appropriate induction of the ISR may be critical for cell resistance to these insults and for pulmonary vascular remodeling. Supporting this idea, ATF4 plays a central role in antioxidation responses and cysteine sufficiency, along with angiogenesis through enhanced expression of vascular endothelial growth factor (VEGF) (Harding et al. 2003; Roybal et al. 2005; Fusakio et al. 2016). GCN2 can also participate in cell proliferation and differentiation, which may be critical for the health of pulmonary tissues and the immune system (Munn et al. 2005; Collier et al. 2017). Finally, it is suggested that loss of GCN2/ATF4 may disrupt signaling through BMPR2 (Eichstaedt et al. 2016). Mutations in BMPR2 are found in the majority of familial PAH, which segregates as an autosomal dominant with incomplete penetrance.
Missense mutations in CReP that destabilize its association with PP1c have been reported to lead to early-onset diabetes, along with growth retardation and microcephaly and learning disabilities, and liver pathologies (Abdulkarim et al. 2015; Kernohan et al. 2015; Mohammad et al. 2016). In islet β cells of the pancreas, loss of CReP leads to increased levels of P-eIF2α, which lowers insulin synthesis and secretion and sensitizes these cells to enhanced apoptosis in response to ER stress (Abdulkarim et al. 2015).
Mutations in genes encoding one of the five different subunits of eIF2B lead to vanishing white matter (VWM), or childhood ataxia with central nervous system hypomyelination, which features severe white matter abnormalities, including myelin and cystic degeneration (Leegwater et al. 2001; van der Knaap et al. 2002). The resulting lowered exchange of eIF2•GDP to the GTP-bound form is suggested to lead to some activation of the ISR independent of stress. When combined with stress induction of P-eIF2α, the VWM residue substitutions in eIF2B can enhance the amplitude of the ISR and alter the timing of the response, which is suggested to trigger the destructive features of the ISR (Richardson et al. 2004; Pavitt and Proud 2009).
It is important to note that some VWM mutations do not appear to alter eIF2B interactions with eIF2 or its guanine nucleotide exchange activity. Two related functions have been attributed to eIF2B, which could be adversely effected by VWM mutations (Jennings and Pavitt 2010; Jennings et al. 2013, 2017). During translation initiation, GTP associated with eIF2 is hydrolyzed by another translation factor, eIF5, which retains association with eIF2•GDP and thwarts spontaneous release of the nucleotide. eIF2B then promotes release of eIF5 from eIF2 before catalyzing the exchange of eIF2•GDP to the GTP-bound form. Additionally, eIF2B ensures that the phosphorylated version of eIF2 is not included in the eIF2•GTP•Met-tRNAiMet complex. These additional eIF2B functions may help explain the complex decameric structure of this exchange factor and may be additional targets for disruption by VWM mutations.
In addition to the pathologies resulting from mutations in the ISR genes directly involved in the regulation of P-eIF2α or its effect on eIF2•GTP exchange, there have been reports of genetic disorders that alter stress activation of the eIF2α kinases and the adaptation functions of the ISR. For example, DNAJC3 (P58IPK) is present in the ER lumen and directly aids the chaperone function of BiP by enhancing its ATPase activity and facilitating association of unfolded polypeptides to BiP (Rutkowski et al. 2007; Petrova et al. 2008). Mutations in DNAJC3 were reported to cause diabetes and widespread neurodegeneration (Synofzik et al. 2014). Loss of DNAJC3 is suggested to disrupt BiP function, increasing the levels of unfolded protein in the ER. This disruption in protein homeostasis can chronically induce PERK phosphorylation of eIF2α, triggering apoptosis. IER3IP1 is another ER protein that is linked with regulation of PERK. Mutations that disrupt IER3IP1 lead to pathologies related to Wolcott–Rallison syndrome, including neonatal diabetes, microcephalogy, and developmental delays, along with seizures (Abdel-Salam et al. 2012; Shalev et al. 2014). IER3IP1 has a putative G-patch domain found in RNA-associated proteins, and loss of IER3IP1 lowered activation of PERK and the ISR in cultured β islet cells exposed to ER stress, culminating in increased cell death (Sun and Ren 2017). Finally, elevated levels of P-eIF2α have been reported in the diseased brain tissues from Alzheimer’s patients and from mouse models of Alzheimer’s disease. Genetic deletion of PERK lowered the P-eIF2α induction and translational control, and restored synaptic plasticity and memory in mice that expressed familial Alzheimer’s disease-related mutations (Ma et al. 2013; Sossin and Costa-Mattioli 2017). Similar outcomes were observed for deletion of GCN2 in the Alzheimer’s disease model mice, which further supports the idea that aberrant induction of P-eIF2α is an underlying contributor to the pathophysiology of Alzheimer’s disease.
THERAPEUTIC TARGETS IN THE ISR
Small molecules have been identified that thwart induction of translational control, or alternatively accentuate the ISR pathway. Those that block P-eIF2α induction and the ISR include PERK and GCN2 inhibitors (Robert et al. 2009; Axten et al. 2012; Harding et al. 2012). For example, the PERK inhibitor GSK2606414 blocks induction of P-eIF2α and interrupts translational control in response to ER stress in cultured cells. This is shown by the application of GSK2606414 to cultured islet β cells subjected to high levels of glucose, which sharply interfered with activation of the PERK portion of the UPR, culminating in rapid accumulation of misfolded insulin protein (Harding et al. 2012). A fluorinated analog of this small molecule, GSK2656157, has been optimized for preclinical development with therapeutic applications for cancer and neurodegenerative disorders (Axten et al. 2013). A cautionary note is that extended exposure to these PERK inhibitors alone can induce P-eIF2α, suggesting compensatory mechanisms that are, at least in part, independent of the ISR (Krishnamoorthy et al. 2014). Supporting this idea, these GSK molecules potently inhibit the protein kinase RIPK1, which functions in the tumor necrosis factor α (TNF-α) pathway, affecting inflammation and cell death (Rojas-Rivera et al. 2017).
ISRIB is another small molecule inhibitor of the ISR that was identified for its ability to block induction of ATF4 translation in response to ER stress (Sidrauski et al. 2013). ISR does not block P-eIF2α induction per se, but rather stimulates the guanine nucleotide exchange activity of eIF2B, thus compensating for the inhibitory effect of P-eIF2α (Sekine et al. 2015; Sidrauski et al. 2015). As a consequence, ISRIB allows for retention of global translation and thwarts assembly of stress granules in response to stress. Synthesis of specific proteins and synaptic plasticity in the hippocampus are critical for the formation and maintenance of memory. By diminishing the ISR-dependent translation, treatment with ISRIB or genetic alterations that disrupt P-eIF2α induction in mice improve memory in a learning paradigm that requires long-term potentiation (Sidrauski et al. 2013). In contrast, long-term memory is impaired by the small molecule, salubrinal, which prevents PP1c dephosphorylation of P-eIF2α and sustains the ISR and translational control (Costa-Mattioli et al. 2007). Persistent activation of the ISR also occurs in traumatic brain injury, and treatment with ISRIB reverses the cognitive deficits associated with the hippocampus in two different injury models in mice (Chou et al. 2017).
As noted for salubrinal, some small molecules enhance and sustain the ISR. Salubrinal was first discovered in a screen for chemicals that protect cultured cells from pharmacologically induced ER stress (Boyce et al. 2005). Salubrinal affords protection in neurodegenerative model systems that are associated with the induced UPR (Sokka et al. 2007; Reijonen et al. 2008; Saxena et al. 2009; Colla et al. 2012). However, depending on the disease model, sustained P-eIF2α induction by salubrinal can have deleterious consequences (Moreno et al. 2012; Collier et al. 2015). Another strategy for small molecule activation of the ISR involves provoking a defined stress for targeted activation of an eIF2α kinase. Halofuginone is a potent inhibitor of prolyl-tRNA synthetase and rapidly activates GCN2 and the ISR (Keller et al. 2012). Prior treatment with halofuginone induces expression of stress-resistant proteins that protect against subsequent renal and hepatic ischemic injury in a mouse surgery reperfusion model (Peng et al. 2012).
LOOKING TO THE FUTURE: IMPORTANT UNRESOLVED QUESTIONS
As highlighted in this review, P-eIF2α regulates translation of individual gene transcripts that collectively contribute to global changes in protein synthesis and restoration of protein homeostasis. Given the central role of uORFs in the ISR, it is important to identify their mechanistic contributions to repression, resistance, and preferential mRNA translation. What are the varied mechanisms by which uORFs are bypassed in response to P-eIF2α? Furthermore, it is important to establish accurate predictive rules for uORF regulatory functions in translational expression. Many single-nucleotide polymorphisms (SNPs) have been identified in humans that alter potential uORFs (Calvo et al. 2009). Do these genetic variations alter ISR function in health and disease? Finally, how can our knowledge of the ISR be applied to clinical practice? Certainly, small molecules such as ISRIB have great therapeutic potential, but there are also challenges as disruptions in key elements of the ISR have the potential for altering cell adaptation and triggering death processes.
ACKNOWLEDGMENTS
This laboratory is supported by National Institutes of Health (NIH) Grants GM049164, DK109714, AI124723, and the Ralph W. and Grace M. Showalter Research Trust Fund.
Footnotes
Editors: Michael B. Mathews, Nahum Sonenberg, and John W.B. Hershey
Additional Perspectives on Translation Mechanisms and Control available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Abastado JP, Miller PF, Jackson BM, Hinnebusch AG. 1991. Suppression of ribosomal reinitiation at upstream open reading frames in amino acid-starved cells forms the basis of GCN4 translational control. Mol Cell Biol 11: 486–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdel-Salam GM, Schaffer AE, Zaki MS, Dixon-Salazar T, Mostafa IS, Afifi HH, Gleeson JG. 2012. A homozygous IER3IP1 mutation causes microcephaly with simplified gyral pattern, epilepsy, and permanent neonatal diabetes syndrome (MEDS). Am J Med Genet A 158A: 2788–2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdulkarim B, Nicolino M, Igoillo-Esteve M, Daures M, Romero S, Philippi A, Senee V, Lopes M, Cunha DA, Harding HP, et al. 2015. A missense mutation in PPP1R15B causes a syndrome including diabetes, short stature, and microcephaly. Diabetes 64: 3951–3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreev DE, O’Connor PB, Fahey C, Kenny EM, Terenin IM, Dmitriev SE, Cormican P, Morris DW, Shatsky IN, Baranov PV. 2015. Translation of 5′ leaders is pervasive in genes resistant to eIF2 repression. eLife 4: e03971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axten JM, Medina JR, Feng Y, Shu A, Romeril SP, Grant SW, Li WH, Heerding DA, Minthorn E, Mencken T, et al. 2012. Discovery of 7-methyl-5-(1-{[3-(trifluoromethyl)phenyl]acetyl}-2,3-dihydro-1H-indol-5-yl)-7H-p yrrolo[2,3-d]pyrimidin-4-amine (GSK2606414), a potent and selective first-in-class inhibitor of protein kinase R (PKR)-like endoplasmic reticulum kinase (PERK). J Med Chem 55: 7193–7207. [DOI] [PubMed] [Google Scholar]
- Axten JM, Romeril SP, Shu A, Ralph J, Medina JR, Feng Y, Li WH, Grant SW, Heerding DA, Minthorn E, et al. 2013. Discovery of GSK2656157: An optimized PERK inhibitor selected for preclinical development. ACS Med Chem Lett 4: 964–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird TD, Palam LR, Fusakio ME, Willy JA, Davis CM, McClintick JN, Anthony TG, Wek RC. 2014. Selective mRNA translation during eIF2 phosphorylation induces expression of IBTKα. Mol Biol Cell 25: 1686–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. 2000. Dynamic interaction of BiP and ER stress transducers in the unfolded protein response. Nat Cell Biol 2: 326–332. [DOI] [PubMed] [Google Scholar]
- Best DH, Sumner KL, Austin ED, Chung WK, Brown LM, Borczuk AC, Rosenzweig EB, Bayrak-Toydemir P, Mao R, Cahill BC, et al. 2014. EIF2AK4 mutations in pulmonary capillary hemangiomatosis. Chest 145: 231–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best DH, Sumner KL, Smith BP, Damjanovich-Colmenares K, Nakayama I, Brown LM, Ha Y, Paul E, Morris A, Jama MA, et al. 2017. EIF2AK4 mutations in patients diagnosed with pulmonary arterial hypertension. Chest 151: 821–828. [DOI] [PubMed] [Google Scholar]
- Boyce M, Bryant KF, Jousse C, Long K, Harding HP, Scheuner D, Kaufman RJ, Ma D, Coen DM, Ron D, et al. 2005. A selective inhibitor of eIF2α dephosphorylation protects cells from ER stress. Science 307: 935–939. [DOI] [PubMed] [Google Scholar]
- Calvo SE, Pagliarini DJ, Mootha VK. 2009. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc Natl Acad Sci 106: 7507–7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrara M, Prischi F, Nowak PR, Kopp MC, Ali MM. 2015. Noncanonical binding of BiP ATPase domain to Ire1 and Perk is dissociated by unfolded protein CH1 to initiate ER stress signaling. eLife 4: e03522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers JE, Dalton LE, Clarke HJ, Malzer E, Dominicus CS, Patel V, Moorhead G, Ron D, Marciniak SJ. 2015. Actin dynamics tune the integrated stress response by regulating eukaryotic initiation factor 2α dephosphorylation. eLife 4: e04872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Rato C, Yan Y, Crespillo-Casado A, Clarke HJ, Harding HP, Marciniak SJ, Read RJ, Ron D. 2015. G-actin provides substrate-specificity to eukaryotic initiation factor 2α holophosphatases. eLife 4: e04872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou A, Krukowski K, Jopson T, Zhu PJ, Costa-Mattioli M, Walter P, Rosi S. 2017. Inhibition of the integrated stress response reverses cognitive deficits after traumatic brain injury. Proc Natl Acad Sci 114: E6420–E6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy MS, Yusoff P, Lee IC, Newton JC, Goh CW, Page R, Shenolikar S, Peti W. 2015. Structural and functional analysis of the GADD34:PP1 eIF2α phosphatase. Cell Rep 11: 1885–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colla E, Coune P, Liu Y, Pletnikova O, Troncoso JC, Iwatsubo T, Schneider BL, Lee MK. 2012. Endoplasmic reticulum stress is important for the manifestations of α-synucleinopathy in vivo. J Neurosci 32: 3306–3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier AE, Wek RC, Spandau DF. 2015. Translational repression protects human keratinocytes from UVB-induced apoptosis through a discordant eIF2 kinase stress response. J Invest Dermatol 135: 2502–2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier AE, Wek RC, Spandau DF. 2017. Human keratinocyte differentiation requires translational control by the eIF2α kinase GCN2. J Invest Dermatol 137: 1924–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor JH, Weiser DC, Li S, Hallenbeck JM, Shenolikar S. 2001. Growth arrest and DNA damage-inducible protein GADD34 assembles a novel signaling complex containing protein phosphatase 1 and inhibitor 1. Mol Cell Biol 21: 6841–6850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Mattioli M, Gobert D, Stern E, Gamache K, Colina R, Cuello C, Sossin W, Kaufman R, Pelletier J, Rosenblum K, et al. 2007. eIF2α phosphorylation bidirectionally regulates the switch from short- to long-term synaptic plasticity and memory. Cell 129: 195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delepine M, Nicolino M, Barrett T, Golamaully M, Lathrop GM, Julier C. 2000. EIF2AK3, encoding translation initiation factor 2-a kinase 3, is mutated in patients with Wolcott–Rallison syndrome. Nat Genet 25: 406–409. [DOI] [PubMed] [Google Scholar]
- Deng J, Harding H, Raught B, Gingras A, Berlanga J, Scheuner D, Kaufman R, Ron D, Sonenberg N. 2002. Activation of GCN2 in UV-irradiated cells inhibits translation. Curr Biol 12: 1279–1286. [DOI] [PubMed] [Google Scholar]
- Dey S, Baird TD, Zhou D, Palam LR, Spandau DF, Wek RC. 2010. Both transcriptional regulation and translational control of ATF4 are central to the integrated stress response. J Biol Chem 285: 33165–33174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey S, Savant S, Teske BF, Hatzoglou M, Calkhoven CF, Wek RC. 2012. Transcriptional repression of ATF4 gene by CCAAT/enhancer-binding protein β (C/EBPβ) differentially regulates integrated stress response. J Biol Chem 287: 21936–21949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Qiu H, Garcia-Barrio M, Anderson J, Hinnebusch AG. 2000. Uncharged tRNA activates GCN2 by displacing the protein kinase moiety from a bipartite tRNA-binding domain. Mol Cell 6: 269–279. [DOI] [PubMed] [Google Scholar]
- Eichstaedt CA, Song J, Benjamin N, Harutyunova S, Fischer C, Grunig E, Hinderhofer K. 2016. EIF2AK4 mutation as “second hit” in hereditary pulmonary arterial hypertension. Respir Res 17: 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyries M, Montani D, Girerd B, Perret C, Leroy A, Lonjou C, Chelghoum N, Coulet F, Bonnet D, Dorfmuller P, et al. 2014. EIF2AK4 mutations cause pulmonary veno-occlusive disease, a recessive form of pulmonary hypertension. Nat Genet 46: 65–69. [DOI] [PubMed] [Google Scholar]
- Fusakio ME, Willy JA, Wang Y, Mirek ET, Al Baghdadi RJ, Adams CM, Anthony TG, Wek RC. 2016. Transcription factor ATF4 directs basal and stress-induced gene expression in the unfolded protein response and cholesterol metabolism in the liver. Mol Biol Cell 27: 1536–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner BM, Walter P. 2011. Unfolded proteins are Ire1-activating ligands that directly induce the unfolded protein response. Science 333: 1891–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goosens A, Dever TE, Pascual-Ahuir A, Serrano R. 2001. The protein kinase Gcn2p mediates sodium toxicity in yeast. J Biol Chem 276: 30753–30760. [DOI] [PubMed] [Google Scholar]
- Han J, Back SH, Hur J, Lin YH, Gildersleeve R, Shan J, Yuan CL, Krokowski D, Wang S, Hatzoglou M, et al. 2013. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat Cell Biol 15: 481–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen MB, Mitchelmore C, Kjaerulff KM, Rasmussen TE, Pedersen KM, Jensen NA. 2002. Mouse ATF5: Molecular cloning of two novel mRNAs, genomic organization, and odorant sensory neuron localization. Genomics 80: 344–350. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Ron D. 1999. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397: 271–274. [DOI] [PubMed] [Google Scholar]
- Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. 2000a. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell 6: 1099–1108. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Bertolotti A, Zeng H, Ron D. 2000b. Perk is essential for translation regulation and cell survival during the unfolded protein response. Mol Cell 5: 897–904. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, et al. 2003. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell 11: 619–633. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Scheuner D, Chen JJ, Kaufman RJ, Ron D. 2009. Ppp1r15 gene knockout reveals an essential role for translation initiation factor 2 α (eIF2α) dephosphorylation in mammalian development. Proc Natl Acad Sci 106: 1832–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Zyryanova AF, Ron D. 2012. Uncoupling proteostasis and development in vitro with a small molecule inhibitor of the pancreatic endoplasmic reticulum kinase, PERK. J Biol Chem 287: 44338–44344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG. 2005. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol 59: 407–450. [DOI] [PubMed] [Google Scholar]
- Iacono M, Mignone F, Pesole G. 2005. uAUG and uORFs in human and rodent 5′untranslated mRNAs. Gene 349: 97–105. [DOI] [PubMed] [Google Scholar]
- *.Ivanov P, Kedersha N, Anderson P. 2017. Stress granules and processing bodies in translational control. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings MD, Pavitt GD. 2010. eIF5 has GDI activity necessary for translational control by eIF2 phosphorylation. Nature 465: 378–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings MD, Zhou Y, Mohammad-Qureshi SS, Bennett D, Pavitt GD. 2013. eIF2B promotes eIF5 dissociation from eIF2•GDP to facilitate guanine nucleotide exchange for translation initiation. Genes Dev 27: 2696–2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings MD, Kershaw CJ, Adomavicius T, Pavitt GD. 2017. Fail-safe control of translation initiation by dissociation of eIF2α phosphorylated ternary complexes. eLife 6: e24542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang HY, Wek RC. 2005. Gcn2 phosphorylation of eIF2α activates NF-κB in response to UV irradiation. Biochem J 385: 371–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jousse C, Bruhat A, Carraro V, Urano F, Ferrara M, Ron D, Fafournoux P. 2001. Inhibition of CHOP translation by a peptide encoded by an open reading frame localized in the chop 5′UTR. Nucleic Acids Res 29: 4341–4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jousse, Oyadomari S, Novoa I, Lu PD, Zhang H, Harding HP, Ron D. 2003. Inhibition of a constitutive translation initiation factor 2a phosphatase, CReP, promotes survival of stressed cells. J Cell Biol 163: 767–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Ivanov P, Anderson P. 2013. Stress granules and cell signaling: More than just a passing phase? Trends Biochem Sci 38: 494–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller TL, Zocco D, Sundrud MS, Hendrick M, Edenius M, Yum J, Kim YJ, Lee HK, Cortese JF, Wirth DF, et al. 2012. Halofuginone and other febrifugine derivatives inhibit prolyl-tRNA synthetase. Nat Chem Biol 8: 311–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kernohan KD, Tetreault M, Liwak-Muir U, Geraghty MT, Qin W, Venkateswaran S, Davila J; Care4Rare Canada Consortium, Holcik M, Majewski J, et al. 2015. Homozygous mutation in the eukaryotic translation initiation factor 2α phosphatase gene, PPP1R15B, is associated with severe microcephaly, short stature and intellectual disability. Hum Mol Genet 24: 6293–6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keys HR. 2016. “A multi-level approach to understanding the regulation of translation initiation.” PhD thesis, Massachusetts Institute of Technology, Cambridge, MA. [Google Scholar]
- Keys HR, Sabatini DM. 2017. Juxtacap nucleotide sequence modulates eIF4E binding and translation. bioRxiv 10.1101/165142. [DOI] [Google Scholar]
- Krishnamoorthy J, Rajesh K, Mirzajani F, Kesoglidou P, Papadakis AI, Koromilas AE. 2014. Evidence for eIF2α phosphorylation-independent effects of GSK2656157, a novel catalytic inhibitor of PERK with clinical implications. Cell Cycle 13: 801–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lageix S, Rothenburg S, Dever TE, Hinnebusch AG. 2014. Enhanced interaction between pseudokinase and kinase domains in Gcn2 stimulates eIF2α phosphorylation in starved cells. PLoS Genet 10: e1004326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lageix S, Zhang J, Rothenburg S, Hinnebusch AG. 2015. Interaction between the tRNA-binding and C-terminal domains of yeast Gcn2 regulates kinase activity in vivo. PLoS Genet 11: e1004991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YY, Cevallos RC, Jan E. 2009. An upstream open reading frame regulates translation of GADD34 during cellular stresses that induce eIF2α phosphorylation. J Biol Chem 284: 6661–6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Liu B, Lee S, Huang SX, Shen B, Qian SB. 2012. Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc Natl Acad Sci 109: E2424–E2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leegwater PA, Vermeulen G, Konst AA, Naidu S, Mulders J, Visser A, Kersbergen P, Mobach D, Fonds D, van Berkel CG, et al. 2001. Subunits of the translation initiation factor eIF2B are mutant in leukoencephalopathy with vanishing white matter. Nat Genet 29: 383–388. [DOI] [PubMed] [Google Scholar]
- Lehman SL, Cerniglia GJ, Johannes GJ, Ye J, Ryeom S, Koumenis C. 2015. Translational upregulation of an individual p21Cip1 transcript variant by GCN2 regulates cell proliferation and survival under nutrient stress. PLoS Genet 11: e1005212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu PD, Harding HP, Ron D. 2004. Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J Cell Biol 167: 27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma K, Vattem KM, Wek RC. 2002. Dimerization and release of molecular chaperone inhibition facilitate activation of eukaryotic initiation factor-2 kinase in response to endoplasmic reticulum stress. J Biol Chem 277: 18728–18735. [DOI] [PubMed] [Google Scholar]
- Ma T, Trinh MA, Wexler AJ, Bourbon C, Gatti E, Pierre P, Cavener DR, Klann E. 2013. Suppression of eIF2α kinases alleviates Alzheimer’s disease-related plasticity and memory deficits. Nat Neurosci 16: 1299–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciniak SJ, Ron D. 2006. Endoplasmic reticulum stress signaling in disease. Physiol Rev 86: 1133–1149. [DOI] [PubMed] [Google Scholar]
- Marciniak SJ, Yun CY, Oyadomari S, Novoa I, Zhang Y, Jungreis R, Nagata K, Harding HP, Ron D. 2004. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev 18: 3066–3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marton MJ, Crouch D, Hinnebusch AG. 1993. GCN1, a translational activator of GCN4 in Saccharomyces cerevisiae, is required for phosphorylation of eukaryotic translation initiation factor 2 by protein kinase GCN2. Mol Cell Biol 13: 3541–3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marton MJ, Vazquez de Aldana CR, Qiu H, Chakraburtty K, Hinnebusch AG. 1997. Evidence that GCN1 and GCN20, translational regulators of GCN4, function on elongating ribosomes in activation of eIF2α kinase GCN2. Mol Cell Biol 17: 4474–4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, Pestova TV, Qian SB, Jaffrey SR. 2015. 5′ UTR m6A promotes cap-independent translation. Cell 163: 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad S, Wolfe LA, Stobe P, Biskup S, Wainwright MS, Melin-Aldana H, Malladi P, Muenke M, Gahl WA, Whitington PF. 2016. Infantile cirrhosis, growth impairment, and neurodevelopmental anomalies associated with deficiency of PPP1R15B. J Pediatr 179: 144–149.e2. [DOI] [PubMed] [Google Scholar]
- Moreno JA, Radford H, Peretti D, Steinert JR, Verity N, Martin MG, Halliday M, Morgan J, Dinsdale D, Ortori CA, et al. 2012. Sustained translational repression by eIF2α-P mediates prion neurodegeneration. Nature 485: 507–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munn DH, Sharma MD, Baban B, Harding HP, Zhang Y, Ron D, Mellor AL. 2005. GCN2 kinase in T cells mediates proliferative arrest and anergy induction in response to indoleamine 2,3-dioxygenase. Immunity 22: 633–642. [DOI] [PubMed] [Google Scholar]
- Novoa I, Zeng H, Harding HP, Ron D. 2001. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2α. J Cell Biol 153: 1011–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor T, Sadleir KR, Maus E, Velliquette RA, Zhao J, Cole SL, Eimer WA, Hitt B, Bembinster LA, Lammich S, et al. 2008. Phosphorylation of the translation initiation factor eIF2α increases BACE1 levels and promotes amyloidogenesis. Neuron 60: 988–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oslowski CM, Urano F. 2011. The binary switch that controls the life and death decisions of ER stressed β cells. Curr Opin Cell Biol 23: 207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palam LR, Baird TD, Wek RC. 2011. Phosphorylation of eIF2 facilitates ribosomal bypass of an inhibitory upstream ORF to enhance CHOP translation. J Biol Chem 286: 10939–10949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavitt GD, Proud CG. 2009. Protein synthesis and its control in neuronal cells with a focus on vanishing white matter disease. Biochem Soc Trans 37: 1298–1310. [DOI] [PubMed] [Google Scholar]
- Peng W, Robertson L, Gallinetti J, Mejia P, Vose S, Charlip A, Chu T, Mitchell JR. 2012. Surgical stress resistance induced by single amino acid deprivation requires Gcn2 in mice. Sci Transl Med 4: 118ra111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira CM, Sattlegger E, Jiang HY, Longo BM, Jaqueta CB, Hinnebusch AG, Wek RC, Mello LE, Castilho BA. 2005. IMPACT, a protein preferentially expressed in the mouse brain, binds GCN1 and inhibits GCN2 activation. J Biol Chem 280: 28316–28323. [DOI] [PubMed] [Google Scholar]
- Petrova K, Oyadomari S, Hendershot LM, Ron D. 2008. Regulated association of misfolded endoplasmic reticulum lumenal proteins with P58/DNAJc3. EMBO J 27: 2862–2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pincus D, Chevalier MW, Aragon T, van Anken E, Vidal SE, El-Samad H, Walter P. 2010. BiP binding to the ER-stress sensor Ire1 tunes the homeostatic behavior of the unfolded protein response. PLoS Biol 8: e1000415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raveh-Amit H, Maissel A, Poller J, Marom L, Elroy-Stein O, Shapira M, Livneh E. 2009. Translational control of protein kinase Ceta by two upstream open reading frames. Mol Cell Biol 29: 6140–6148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid DW, Nicchitta CV. 2015. Diversity and selectivity in mRNA translation on the endoplasmic reticulum. Nat Rev Mol Cell Biol 16: 221–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid DW, Chen Q, Tay AS, Shenolikar S, Nicchitta CV. 2014. The unfolded protein response triggers selective mRNA release from the endoplasmic reticulum. Cell 158: 1362–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijonen S, Putkonen N, Norremolle A, Lindholm D, Korhonen L. 2008. Inhibition of endoplasmic reticulum stress counteracts neuronal cell death and protein aggregation caused by N-terminal mutant huntingtin proteins. Exp Cell Res 314: 950–960. [DOI] [PubMed] [Google Scholar]
- Resch AM, Ogurtsov AY, Rogozin IB, Shabalina SA, Koonin EV. 2009. Evolution of alternative and constitutive regions of mammalian 5′UTRs. BMC Genomics 10: 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson JP, Mohammad SS, Pavitt GD. 2004. Mutations causing childhood ataxia with central nervous system hypomyelination reduce eukaryotic initiation factor 2B complex formation and activity. Mol Cell Biol 24: 2352–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert F, Williams C, Yan Y, Donohue E, Cencic R, Burley SK, Pelletier J. 2009. Blocking UV-induced eIF2α phosphorylation with small molecule inhibitors of GCN2. Chem Biol Drug Des 74: 57–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas-Rivera D, Delvaeye T, Roelandt R, Nerinckx W, Augustyns K, Vandenabeele P, Bertrand MJM. 2017. When PERK inhibitors turn out to be new potent RIPK1 inhibitors: Critical issues on the specificity and use of GSK2606414 and GSK2656157. Cell Death Differ 24: 1100–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roybal CN, Hunsaker LA, Barbash O, Vander Jagt DL, Abcouwer SF. 2005. The oxidative stressor arsenite activates vascular endothelial growth factor mRNA transcription by an ATF4-dependent mechanism. J Biol Chem 280: 20331–20339. [DOI] [PubMed] [Google Scholar]
- Rutkowski DT, Kang SW, Goodman AG, Garrison JL, Taunton J, Katze MG, Kaufman RJ, Hegde RS. 2007. The role of p58IPK in protecting the stressed endoplasmic reticulum. Mol Biol Cell 18: 3681–3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattlegger E, Swanson MJ, Ashcraft EA, Jennings JL, Fekete RA, Link AJ, Hinnebusch AG. 2004. YIH1 is an actin-binding protein that inhibits protein kinase GCN2 and impairs general amino acid control when overexpressed. J Biol Chem 279: 29952–29962. [DOI] [PubMed] [Google Scholar]
- Saxena S, Cabuy E, Caroni P. 2009. A role for motoneuron subtype-selective ER stress in disease manifestations of FALS mice. Nat Neurosci 12: 627–636. [DOI] [PubMed] [Google Scholar]
- Sekine Y, Zyryanova A, Crespillo-Casado A, Fischer PM, Harding HP, Ron D. 2015. Stress responses. Mutations in a translation initiation factor identify the target of a memory-enhancing compound. Science 348: 1027–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senée V, Vattem KM, Delépine M, Rainbow L, Haton C, Lecoq A, Shaw N, Robert J-J, Rooman R, Diatloff-Zito C, et al. 2004. Wolcott–Rallison syndrome: Clinical, genetic, and functional study of EIF2AK3 mutations, and suggestion of genetic heterogeneity. Diabetes 53: 1876–1883. [DOI] [PubMed] [Google Scholar]
- Shalev SA, Tenenbaum-Rakover Y, Horovitz Y, Paz VP, Ye H, Carmody D, Highland HM, Boerwinkle E, Hanis CL, Muzny DM, et al. 2014. Microcephaly, epilepsy, and neonatal diabetes due to compound heterozygous mutations in IER3IP1: Insights into the natural history of a rare disorder. Pediatr Diabetes 15: 252–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Vattem KM, Sood R, An J, Liang J, Stramm L, Wek RC. 1998. Identification and characterization of pancreatic eukaryotic initiation factor 2 α-subunit kinase, PEK, involved in translation control. Mol Cell Biol 18: 7499–7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidrauski C, Acosta-Alvear D, Khoutorsky A, Vedantham P, Hearn BR, Li H, Gamache K, Gallagher CM, Ang KK, Wilson C, et al. 2013. Pharmacological brake-release of mRNA translation enhances cognitive memory. eLife 2: e00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidrauski C, McGeachy AM, Ingolia NT, Walter P. 2015. The small molecule ISRIB reverses the effects of eIF2α phosphorylation on translation and stress granule assembly. eLife 4: e05033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva RC, Sattlegger E, Castilho BA. 2016. Perturbations in actin dynamics reconfigure protein complexes that modulate GCN2 activity and promote an eIF2 response. J Cell Sci 129: 4521–4533. [DOI] [PubMed] [Google Scholar]
- Sokka AL, Putkonen N, Mudo G, Pryazhnikov E, Reijonen S, Khiroug L, Belluardo N, Lindholm D, Korhonen L. 2007. Endoplasmic reticulum stress inhibition protects against excitotoxic neuronal injury in the rat brain. J Neurosci 27: 901–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Sossin W, Costa-Mattioli M. 2017. Translational control in the brain in health and disease. Cold Spring Harb Perspect Biol 10.1101/cshperpsect.a032912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starck SR, Tsai JC, Chen K, Shodiya M, Wang L, Yahiro K, Martins-Green M, Shastri N, Walter P. 2016. Translation from the 5′ untranslated region shapes the integrated stress response. Science 351: aad3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Ren D. 2017. IER3IP1 deficiency leads to increased β-cell death and decreased β-cell proliferation. Oncotarget 8: 56768–56779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Synofzik M, Haack TB, Kopajtich R, Gorza M, Rapaport D, Greiner M, Schonfeld C, Freiberg C, Schorr S, Holl RW, et al. 2014. Absence of BiP co-chaperone DNAJC3 causes diabetes mellitus and multisystemic neurodegeneration. Am J Hum Genet 95: 689–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Knaap MS, Leegwater PA, Konst AA, Visser A, Naidu S, Oudejans CB, Schutgens RB, Pronk JC. 2002. Mutations in each of the five subunits of translation initiation factor eIF2B can cause leukoencephalopathy with vanishing white matter. Ann Neurol 51: 264–270. [DOI] [PubMed] [Google Scholar]
- van Vliet AR, Giordano F, Gerlo S, Segura I, Van Eygen S, Molenberghs G, Rocha S, Houcine A, Derua R, Verfaillie T, et al. 2017. The ER stress sensor PERK coordinates ER-plasma membrane contact site formation through interaction with Filamin-A and F-actin remodeling. Mol Cell 65: 885–899. [DOI] [PubMed] [Google Scholar]
- Vattem KM, Wek RC. 2004. Reinitiation involving upstream open reading frames regulates ATF4 mRNA translation in mammalian cells. Proc Natl Acad Sci 101: 11269–11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Ron D. 2011. The unfolded protein response: From stress pathway to homeostatic regulation. Science 334: 1081–1086. [DOI] [PubMed] [Google Scholar]
- Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C. 2015. N6-methyladenosine modulates messenger RNA translation efficiency. Cell 161: 1388–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watatani Y, Ichikawa K, Nakanishi N, Fujimoto M, Takeda H, Kimura N, Hirose H, Takahashi S, Takahashi Y. 2008. Stress-induced translation of ATF5 mRNA is regulated by the 5′-untranslated region. J Biol Chem 283: 2543–2553. [DOI] [PubMed] [Google Scholar]
- Wek RC, Jackson BM, Hinnebusch AG. 1989. Juxtaposition of domains homologous to protein kinases and histidyl-tRNA synthetases in GCN2 protein suggests a mechanism for coupling GCN4 expression to amino acid availability. Proc Natl Acad Sci 86: 4579–4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek SA, Zhu S, Wek RC. 1995. The histidyl-tRNA synthetase-related sequence in eIF-2 α protein kinase GCN2 interacts with tRNA and is required for activation in response to starvation for different amino acids. Mol Cell Biol 15: 4497–4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willy JA, Young SK, Mosley AL, Gawrieh S, Stevens JL, Masuoka HC, Wek RC. 2017. Function of inhibitor of brutons tyrosine kinase isoform α (IBTKα) in nonalcoholic steatohepatitis links autophagy and the unfolded protein response. J Biol Chem 292: 14050–14065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Tan M, Hu Y, Wang J-L, Scheuner D, Kaufman RJ. 2004. Ultraviolet light activates NFκB through translation inhibition of IκBa synthesis. J Biol Chem 279: 24898–24902. [DOI] [PubMed] [Google Scholar]
- Yaman I, Fernandez J, Liu H, Caprara M, Komar AA, Koromilas AE, Zhou L, Snider MD, Scheuner D, Kaufman RJ, et al. 2003. The zipper model of translational control: A small upstream ORF is the switch that controls structural remodeling of an mRNA leader. Cell 113: 519–531. [DOI] [PubMed] [Google Scholar]
- Yang R, Wek SA, Wek RC. 2000. Glucose limitation induces GCN4 translation by activation of Gcn2 protein kinase. Mol Cell Biol 20: 2706–2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young SK, Willy JA, Wu C, Sachs MS, Wek RC. 2015. Ribosome reinitiation directs gene-specific translation and regulates the integrated stress response. J Biol Chem 290: 28257–28271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young SK, Baird TD, Wek RC. 2016a. Translation regulation of the glutamyl-prolyl-tRNA synthetase gene EPRS through bypass of upstream open reading frames with noncanonical initiation codons. J Biol Chem 291: 10824–10835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young SK, Palam LR, Wu C, Sachs MS, Wek RC. 2016b. Ribosome elongation stall directs gene-specific translation in the integrated stress response. J Biol Chem 291: 6546–6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaborske JM, Narasimhan J, Jiang L, Wek SA, Dittmar KA, Freimoser F, Pan T, Wek RC. 2009. Genome-wide analysis of tRNA charging and activation of the eIF2 kinase Gcn2p. J Biol Chem 284: 25254–25267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Palam LR, Jiang L, Narasimhan J, Staschke KA, Wek RC. 2008. Phosphorylation of eIF2 directs ATF5 translational control in response to diverse stress conditions. J Biol Chem 283: 7064–7073. [DOI] [PubMed] [Google Scholar]
- Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, Qian SB. 2015. Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature 526: 591–594. [DOI] [PMC free article] [PubMed] [Google Scholar]