Abstract
Aspergillus flavus is a saprophytic fungus that infects corn, peanuts, tree nuts and other agriculturally important crops. Once the crop is infected the fungus has the potential to secrete one or more mycotoxins, the most carcinogenic of which is aflatoxin. Aflatoxin contaminated crops are deemed unfit for human or animal consumption, which results in both food and economic losses. Within A. flavus, two morphotypes exist: the S strains (small sclerotia) and L strains (large sclerotia). Significant morphological and physiological differences exist between the two morphotypes. For example, the S-morphotypes produces sclerotia that are smaller (< 400 μm), greater in quantity, and contain higher concentrations of aflatoxin than the L-morphotypes (>400 μm). The morphotypes also differ in pigmentation, pH homeostasis in culture and the number of spores produced. Here we report the first full genome sequence of an A. flavus S morphotype, strain AF70. We provide a comprehensive comparison of the A. flavus S-morphotype genome sequence with a previously sequenced genome of an L-morphotype strain (NRRL 3357), including an in-depth analysis of secondary metabolic clusters and the identification SNPs within their aflatoxin gene clusters.
Introduction
Aspergillus flavus is an agriculturally significant fungus, whose pathogenesis can occur in either field or post-harvest conditions [1]. This fungus may also secrete one or more toxic secondary metabolites, termed mycotoxins, the most carcinogenic of which is aflatoxin [2]. Aflatoxin exists naturally in several forms depending on the fungal strain, the most potent of which is aflatoxin B1 (AFB1)[3]. Aflatoxin contamination of food and feed results in commodities that are unfit for consumption, which equates to millions of dollars in economic losses annually [3–6].
Within species A. flavus, two morphological groups (i.e. morphotypes) have been characterized based on sclerotia production: the S strains (small sclerotia) and the L strains (large sclerotia) [7]. The S morphotype typically produces sclerotia less than 400 μm in diameter and produce higher quantities of AFB1 in culture than the L morphotype. These also differ in pigmentation, pH homeostasis in culture, and the number of sclerotia and spores produced [7–10]. Previous sequence analysis of three genes from a collection of A. flavus strains indicated that two distinct phylogenetic groups (groups I and II) exist within the species, however this does not allow the complete discernment between toxigenic/atoxigenic, or between S- and L-morphotypes. Group I consists of both S- and L-strains that produce AFB1, while group II is comprised of S-strains that produce AFB1 and/or G-aflatoxins (AFG) [8, 11] (although producers of the AFG metabolite appear to be found in Group I as well). Because significant morphological and physiological differences exist between the two morphotypes, a comparison of the two is useful for elucidating mechanisms of virulence, toxin production and development.
Both S and L strains of A. flavus are pathogenic to agricultural crops. Horn et al. [12] found, in a survey of peanut fields throughout the southern United States, that nearly all S strains and approximately 70% of L strains produce both AFB1 and another toxic metabolite, cyclopiazonic acid (CPA). A later study by Abbas et al. [9], in the Mississippi Delta, reported that L strains exhibited much higher virulence since they were far more prevalent in corn, peanut, rice and soil samples than S strains, however all crops were susceptible to AFB1 contamination by at least one morphological type.
NRRL 3357 (ATCC 200026, GenBank assembly accession: GCA_000006275.2) is an A. flavus L strain morphotype that has been developed as a model organism over many years, and across multiple studies, evaluating secondary metabolite production [13–15], genome profiling [16, 17], proteome functional analysis [18], contamination of crop supplies [19, 20], the development of biological control agents [17], and many others. The sequencing of strain NRRL 3357 was the first fully sequenced A. flavus genome, initially reported on by Payne et al. [21], and subsequently re-sequenced to produce a less fragmented genome by Nierman et al. [22]. The S strain sequenced for this study (AF70) was originally isolated using a dicloran-amended media from soil obtained in a Pima Cotton field in Yuma Valley, AZ, USA, and first characterized by Cotty et al. in 1989. It produces very high AFB1 levels, especially during infection of maize kernels [7]. Strain AF70 has been used for the characterization of fungal development and growth [23, 24], virulence of crops [25], secondary metabolite production [15], oxidative stress [26], genome profiling [27] and numerous other studies.
Here we report on morphological differences and vegetative compatibility, and provide a genome-wide assessment of the important differences, between strains AF70 and NRRL 3357. Further, we describe the sequencing and methodology used for the annotation of strain AF70, which utilized RNA-seq reads from available experiments to train gene prediction algorithms. Our analysis of the secondary metabolic gene clusters revealed significant differences in the metabolic profiles of these strains, and we identify key single nucleotide polymorphisms (SNPs) in their respective aflatoxin gene clusters.
Materials and methods
Strains and growth conditions
Aspergillus flavus AF70 (ATCC 200026) [7] was sequenced using the techniques described below. Genomic comparisons were made to A. flavus strain NRRL 3357 [22]. For imaging, determining spore production, sclerotia diameter, and AFB1 analysis, fresh spores from both strains were first produced by plating stocks on V8 agar plates (10% V8 juice, 3% ammonium sulfate, 1% uracil, pH 5.2). For AFB1 analysis, Yeast-Glucose-Trace Element (YGT) medium was supplemented with 6.0 g/L casamino acids and 1 ml/mg trace element solution as previously described [28–30]. The YGT consisted of 0.5% yeast extract, 2% glucose, 20 g agar, and 1 ml of trace element solution per liter of medium [28]. Selection media used in vegetative compatibility group (VCG) assays consisted of amended Czapek-dox (CZ; Difco, BD)), Mutation Induction Media (MIT), Nitrate, Nitrite or Hypoxanthine media [31]. The amended CZ included 35 g of CZ broth, 25 g KClO3, 10 ml of 5 mg/ml Rose Bengal stock solution (Sigma-Aldrich, St. Louis, MO) and 2% Bacto agar (Sigma-Aldrich) per liter, pH 7.2. MIT media consisted of 35 g CZ broth, 15 g KClO3 and 2% Bacto agar per liter with a final pH of 6.5. Nitrate medium consisted of (per L) 35g CZ, 2% Bacto Agar, Nitrite media consisted of (per L) 50g Sucrose, 10g KH2PO4, 2g MgSO4-7H2O, 1 ml micronutrients, 10mM NaNO2, and 2% Bacto Agar. Hypoxanthine media was identical to nitrite media except 100 mg/L hypoxanthine replaced NaNO2.
Phenotypic comparison of A. flavus morphotypes
For obtaining images of fungal phenotype, 2x105 fresh spores from strains AF70 and NRRL 3357 were spot plated on YGT agar plates and grown for 6 days at 30 °C. Stereoscope images were taken on a Zeiss Stereoscope Discovery.v20 using an Axiocam MRc5 digital camera system (Zeiss, Oberkochen, Germany). To determine spore production, 1x106 fresh spores were point inoculated on YGT and V8 media plates, and grown for 9 days at 30 °C in 12 hr light/dark cycles. Three representative plugs were removed from each plate and added to 500 μl 0.02% Triton-X 100 (TTX-100). A hemocytometer (Hausser Scientific, Horsham, PA) was used for counting spores, and in every case >100 spores were counted. Three independent plates for each strain and medium were used.
To determine sclerotium diameter, 2x105 fresh spores from each strain were point inoculated on YGT plates. After 6 days of growth on YGT agar, sclerotia were harvested with 0.01% TTX-100 and rinsed onto filter paper (Whatman #4, Whatman Ltd.). Three independent plates for each strain were used. Sclerotium diameter was measured by averaging two cross-measurements. Measurements were conducted using Axiovision 4.8 image analysis software (Zeiss).
To determine if both morphotype representatives are of the same VCG, the method of Horn et al. [32] was used with some modification. Spores were inoculated onto 0.5X V8 plates. After 5 to 7 days of growth, five agar plugs containing spores were cut and put into sterilized glass vials containing 2.5 ml deionized water. Twenty microliters of each spore suspension was then inoculated onto the center of multiple plates containing a single type of selection medium. Between days 5–14, mutations were identified as a cloudy, thick growth that outgrew the wild-type. Observed mutants were then transferred to MIT agar plates for further selection. After approximately 3 days of growth, 1 plug from the outer growth was then transferred to a nitrogen-selecting medium, where vegetative compatibility was determined by pairing complementary mutants (nia-D, nir-A or cnx).
Aflatoxin extraction and ultra-performance liquid chromatography (UPLC) analysis
After growth of mycelia on solid supplemented YGT media, 220 ml of 2:1 acetone:water was added and shaken vigorously for 16 hours, then allowed to separate. The supernatant was extracted with 150 ml methylene chloride, which was then evaporated under filtered nitrogen gas. Each dried sample extract was resuspended in 1 ml acetonitrile, then transferred to a 2 ml, 0.45 μm nylon filter centrifuge tube (Spin-X; Corning Inc., Corning, NY) and centrifuged at 14000 rpm for 1 min. Injections (1 μl) of filtered extract were analyzed by UPLC. AFB1 and AFB2analysis was performed with modifications to a procedure previously described [33]. UPLC analyses were performed with a Waters Acquity H-Class System combined with an Acquity fluorescence (FLR) detector (Waters, Milford, MA). Sample extract (1 μl) was injected for separation through an Acquity BEH C18 column (1.7 um, 2.1 x 50 mm) at 30 °C. Run time was 3 min with an elution flow rate of 0.4 ml/min and isocratic mobile phase consisting of methanol:water (40:60). AFB1 detection wavelength was 365 nm (excitation) and 440 nm (emission) and retention time was 2.1 min. A calibration curve with high linearity (R2 = 0.9993) was constructed for AFB1 from a series of diluted standards (Sigma-Aldrich). Statistical analysis conducted using Graphpad Prism 7 (Graphpad Software Inc).
Sequencing and assembly of the A. flavus AF70 genome
DNA was extracted from 10 g of A. flavus AF70 mycelia harvested after 24 h growth in PDB (Becton, Dickinson and Co., Franklin Lakes, NJ) and continuous shaking at 30 °C. Briefly, mycelia were ground to a fine powder in liquid N2, then subjected to CTAB extraction (1% CTAB [mixed alkyltrimethyl-ammonium bromide], 200mM Tris-HCl pH8.0, 0.8M NaCl, 1% B-mercaptoethanol, 10mM EDTA), followed by Proteinase K (500ug/ml) digestion, two rounds of phenol:chloroform extraction and ethanol precipitation [34]. Long strands of visible DNA were removed using a glass rod, and shipped frozen to J. Craig Venter Institute, Bethesda, MD. The sample quality was assayed using an agarose gel and an Agilent 2100 bioanalyzer (Santa Clara, CA). Sequencing of strain AF70 was performed using single end 100 bp reads in the Illumina GA-II (Illumina, San Diego, CA). A total of 20,255,792 reads were obtained equivalent to roughly 70X sequence coverage. The assembly was done using CLC Genomics Workbench de novo assembler (v4.3). Raw Reads are available at the sequence read archive, SRA accession: SRP131888 (www.ncbi.nih.gov/sra).
Genome annotation
Gene predictions were performed using the MAKER pipeline (version 2.31.4) [35]. First, repetitive elements were identified in the initial AF70 genome sequence by MAKER using the RepeatMasker [36] algorithm in conjunction with the RepBase [37] repeat library for fungi and MAKER’s built-in repeat database. To train the gene prediction algorithm Augustus [38], Trinity was used to assemble a transcriptome from in-house generated RNA-seq experiments for A. flavus AF70 [23]. MAKER was then run using the assembled transcriptome, the predicted proteins from A. oryzae, A. flavus NRRL 3357, and A. nidulans (with the “protein2genome” and “est2genome” options set to 1), and fungal protein sequences from the SWISS-Pro database to develop a set of predicted genes. These genes were filtered using maker2zzf based on how well they fit the EST evidence, and narrowed down to about 1,200 genes which were then used to train the first iteration of Augustus. MAKER then produced ab initio gene predictions from the repeat-masked AF70 genomic sequence using the GeneMark [39] and trained Augustus. Gene prediction was also performed on the AF70 sequence using the PASA pipeline [40]. These sets of predictions were included in subsequent runs of MAKER through the “pred_gff” option. Augustus was retrained using results from the second run of MAKER, and MAKER was run a third time. All runs of MAKER were performed with the “single_exon” set to 1 and “correct_est_fusion” set to 1. Gene predictions that had no overlapping EST or protein homology evidence were included in the final set of predictions if they were found to have an InterPro domain when examined using InterProScan. The final annotations were manually edited using WebApollo (version 2013-11-22). The sequence and annotation file was uploaded to NCBI under Genbank Assembly Accession GCA_000952835.1. The genome sequence and predicted proteins for A. oryzae and A. nidulans were obtained from AspGD (www.aspergillusgenome.org, accessed 9/15/14). The sequence for NRRL 3357 was obtained from NCBI (accessed 4/1/14).
Comparative analyses of A. flavus morphotypes
For construction of Venn Diagrams, coding sequences were aligned using blastn. Sequences with coverage > 50%, identity over 70% and an e-value < 1e-10 were considered orthologous. For Gene Ontology Enrichment Analysis, the R package GOSeq was used [41]. Construction of the phylogenetic tree involved orthologous proteins that were detected using Proteinortho [42], aligned with Muscle [43], and concatenated into a 1.1 Mb amino acid alignment using GBLOCKS [44]. The tree was produced using RAxML with A. zonatus as the outgroup taxon. The inset shows closely related species with expanded branch lengths. For secondary metabolic cluster prediction, the Antibiotics-Secondary Metabolite Analysis Shell (anti-SMASH) program was used [45]. Default parameters were used except for the incorporation of the ClusterFinder algorithm. Anti-SMASH is designed to predict 43 categories of gene clusters (e.g. Type 1–3 polyketide synthases [PKS], Non-ribosomal peptide-synthetases [Nrps], terpenes, etc.), thus it generally provides a relatively comprehensive list of cluster predictions. Examination of the clusters within the AF70 and NRRL 3357 genomes was undertaken using MultiGeneBlast [46] in tblastn mode. Each of the known eukaryotic cluster entries from the MIBiG database (accessed 7/1/17) [47] was used as a query against the AF70 and NRRL 3357 genomes. The entry with the most genes was chosen for clusters that had redundant entries. Variant analysis involved creating simulated short reads of the AF70 genome using bbmap [48], mapping those reads to the NRRL 3357 genome using BWA [49], calling variants using Freebayes [50], and annotating variants using SNPeff [51]. Although the total number of orthologous genes will be different than the method described above, the pipeline provides a more accurate estimation of SNP content and is in keeping with best practices [52].
We also utilized nearly 70 kb of genomic sequence from the aflatoxin gene clusters of AF70 (A. flavus S-morphotype) and NRRL 3357 (A. flavus L-morphotype) to compare the quantities and types of single nucleotide polymorphisms (SNPs) that differentiated their biosynthetic pathways. The aflatoxin cluster sequences for both strains were acquired from the NCBI database and uploaded as FASTA files to the sequence analysis program Sequencher (Gene Codes Corporation, Michigan). To accurately identify the start and end of each gene and intergenic region, within the aflatoxin cluster, we also added sequences from individual A. flavus cluster genes to the alignment. Once the cluster sequences were aligned, and the correct orientation of individual cluster genes was verified, the process of scrolling along the cluster sequences began. For each gene and intergenic region, observations were noted for the numbers of transition/transversion SNPs as well as deletions within each strain’s cluster sequence. Another comparison involved multi-locus sequence typing (MLST) for NRRL3357 and AF70. Alignments for each of six genomic regions were prepared using Sequencher and then exported as Nexus files: two aflatoxin cluster intergenic regions (aflM/aflN and aflW/aflX), and four non-cluster regions (amdS, trpC, MAT1-1 and mfs). These loci have proven, in previous MLST studies for A. flavus, to accurately segregate individuals within a population in lieu of vegetative compatibility group (VCG) testing [53]. The Nexus files were then imported into a suite of nucleotide analysis programs (SNAP) that are used for population inferences [54]. Using one of the workbench programs, SNAP: Combine, we concatenated the six different alignments to make a single sequence alignment. Another program (SNAP: Map) was used to collapse the sequences into haplotypes using the parameters of recoded indels and excluded infinite sites violations. The resulting map file indicates whether or not each strain examined is considered an “individual” based on its haplotype designation [55].
Results and discussion
Morphological characterization of A. flavus morphotypes
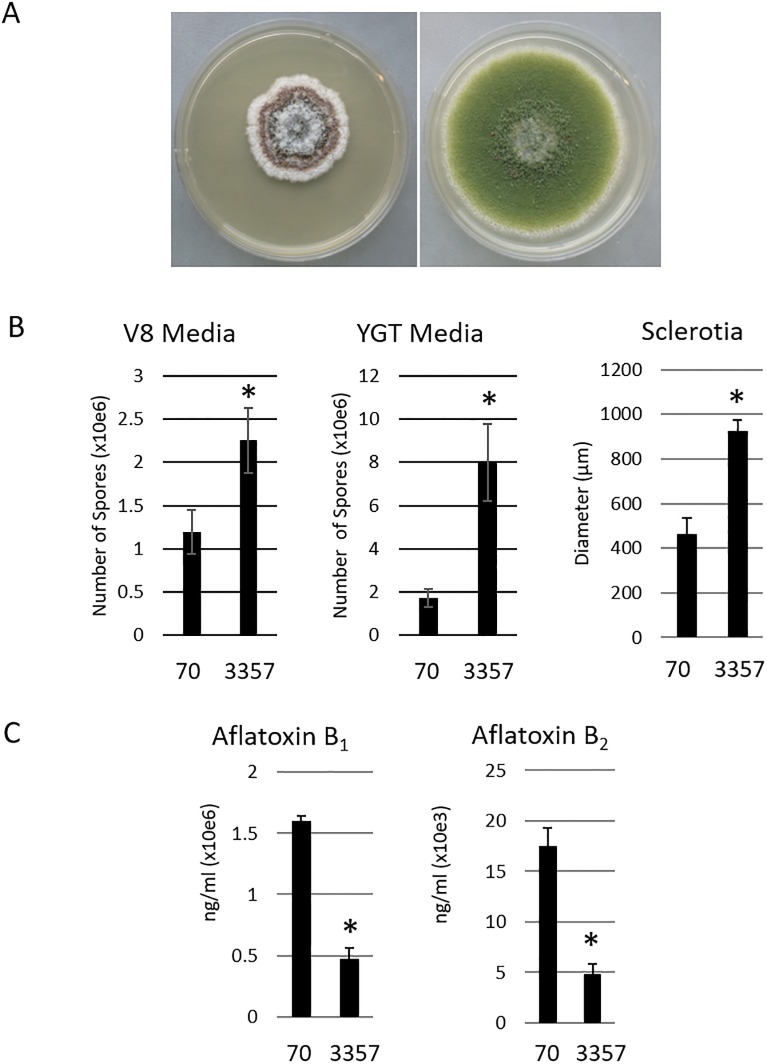
The phenotypic differences exhibited between AF70 and NRRL 3357 are relevant for contextually analyzing a genomic comparison. While some characteristics have been reported previously [7], we wanted a direct and current comparison of their morphological characteristics, aflatoxin production and diameter of sclerotia. Although equal amounts of spore suspension from each strain were plated on YGT, the AF70 (S-morphotype) strain demonstrated slower growth, greater quantities of sclerotia, and a lack of olive-green conidia, compared to the NRRL 3357 L-morphotype strain (Fig 1A). AF70 contained significantly higher spore content (Fig 1B, left and center) and cross-sectional measurements of sclerotia indicated the average size of sclerotia was approximately 400 um, whereas the average diameter for NRRL 3357 sclerotia was just under 1000 um (Fig 1B, right). AFB1 and B2 levels were 4-fold higher and 3.5-fold higher, respectively, for S-morphotype AF70 than for L-morphotype NRRL 3357 (Fig 1C).
Fig 1. Phenotype comparisons for A. flavus strains AF70 and NRRL 3357.
A) After 6 days growth on YGT media, S-morphotype AF70, left, demonstrated several rings of pigmentation correlating with production of numerous small sclerotia, with unpigmented hyphae in the outermost edges of growth. Conversely, L-morphotype NRRL 3357 maintains typical olive-green pigmentation with no sclerotia visible. B) Spore production on two media types, V8 and YGT showed NRRL 3357 consistently producing more spores. C) HPLC quantification of aflatoxin AFB production shows higher concentrations produced by S-morphotype AF70. Student’s t-test where P ≤ 0.05 indicated by asterisk (*).
Collectively, our morphological comparisons of AF70 and NRRL 3357 support general observations relating to both morphotypes. It was also determined, through our VCG assays, that strains NRRL 3357 and AF70 are not vegetatively compatible (S1 Fig), which is consistent with previous findings [56].
Genomic comparison of AF70 and NRRL 3357
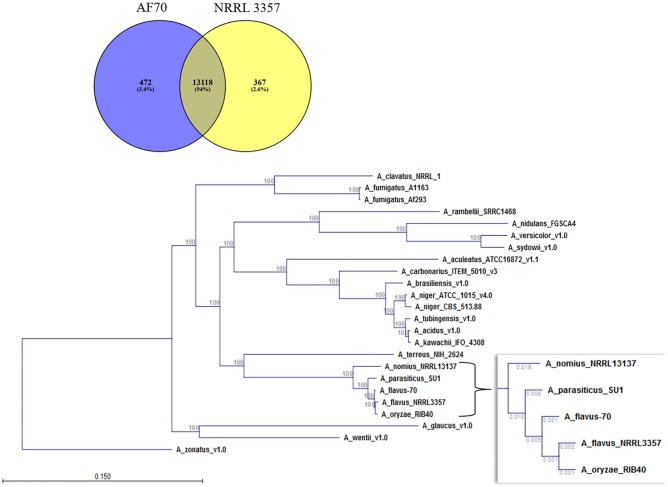
Of the approximately 13,500 genes predicted to exist in NRRL 3357 and AF70, 13,118 genes were predicted to be orthologous based on blastn results of the coding sequences versus the genome based on 70% identity, 50% query coverage, and an E value <1x10-10 (Fig 2). Also based on this criterion, 472 genes were found to be unique to AF70, while 367 genes were found to be unique to NRRL 3357. Using Proteinortho to determine the number of orthologues indicated 2,397 coding sequences unique to NRRL 3357 and 2,039 unique to AF70. However, it should be pointed out that differences in annotation could contribute significantly to this determination. Phylogenetic comparison including multiple Aspergillus species, and using concatenated amino acid sequence alignment, indicates NRRL 3357 shares highest sequence identity with A. oryzae (Fig 2B). Also observed was that both of the examined A. flavus morphotypes share a common ancestor. Gene enrichment analysis of the genes unique to each morphotype was conducted to determine if broad categories of genes are present/absent in either genome. Genes unique to strain AF70 were enriched in cytochrome P450 mono-oxygenases (GO:0016705 oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen) (Table 1 and S1 Table). Three of the identified genes are part of a unique gene cluster (AFLA70_220g001820, AFLA70_220g001850, AFLA70_220g001880).
Fig 2. Genome comparisons of A. flavus strain AF70 and NRRL 3357.
A) Number of genes identified as unique or shared between strains AF70 and NRRL 3357 (S and L strains, respectively) indicate 94% of genes are shared (orthologous). B) Protein sequences were compared and used to illustrate the hierarchal relationship between strains AF70, NRRL 3357 and closely related species, and indicate that NRRL 3357 (“A_flavus_NRRL3357”) is more closely related to A. oryzae (“A_oryzae_RIB40”) than A. flavus AF70 (“A_flavus 70”). Values on main tree indicate bootstrap values. Values on inset indicate branch length.
Table 1. Gene Ontology categories of genes that are significantly enriched in strain AF70 and NRRL 3357.
| Unique to Strain AF70 | Unique to Strain NRRL 3357 | ||||
|---|---|---|---|---|---|
| Biological Processes | E value | Description | Biological Processes | E value | Description |
| GO:0009116 | 0.000 | nucleoside metabolic process | |||
| GO:0055114 | 0.001 | oxidation-reduction process | GO:0016998 | 0.000 | cell wall macromolecule catabolic process |
| GO:0009116 | 0.001 | nucleoside metabolic process | GO:0055114 | 0.001 | oxidation-reduction process |
| GO:0015991 | 0.015 | ATP hydrolysis coupled proton transport | GO:0034551 | 0.013 | mitochondrial respiratory chain complex III assembly |
| GO:0038032 | 0.027 | termination of G-protein coupled receptor signaling pathway | GO:0006098 | 0.044 | pentose-phosphate shunt |
| Molecular Function | E value | Description | Molecular Function | E value | Description |
| GO:0016705 | 0.000 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen | GO:0016491 | 0.000 | oxidoreductase activity |
| GO:0020037 | 0.000 | heme binding | GO:0004497 | 0.002 | monooxygenase activity |
| GO:0005506 | 0.000 | iron ion binding | GO:0004252 | 0.011 | serine-type endopeptidase activity |
| GO:0016831 | 0.008 | carboxy-lyase activity | GO:0003824 | 0.013 | catalytic activity |
| GO:0015078 | 0.011 | hydrogen ion transmembrane transporter activity | GO:0016614 | 0.015 | oxidoreductase activity, acting on CH-OH group of donors |
| GO:0031177 | 0.020 | phosphopantetheine binding | GO:0004616 | 0.016 | phosphogluconate dehydrogenase (decarboxylating) activity |
| GO:0015299 | 0.022 | solute:proton antiporter activity | GO:0050660 | 0.016 | flavin adenine dinucleotide binding |
| GO:0016772 | 0.032 | transferase activity, transferring phosphorus-containing groups | GO:0015124 | 0.020 | allantoate transmembrane transporter activity |
| GO:0016740 | 0.032 | transferase activity | GO:0045482 | 0.020 | trichodiene synthase activity |
| GO:0008270 | 0.035 | zinc ion binding | GO:0008762 | 0.024 | UDP-N-acetylmuramate dehydrogenase activity |
| GO:0016491 | 0.039 | oxidoreductase activity | GO:0016705 | 0.026 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen |
| GO:0004497 | 0.049 | monooxygenase activity | GO:0005506 | 0.031 | iron ion binding |
| GO:0020037 | 0.034 | heme binding | |||
| GO:0000293 | 0.039 | ferric-chelate reductase activity | |||
| GO:0005384 | 0.039 | manganese ion transmembrane transporter activity | |||
| GO:0042936 | 0.039 | dipeptide transporter activity | |||
| GO:0097079 | 0.039 | selenite:proton symporter activity | |||
| GO:0009055 | 0.049 | electron carrier activity | |||
Secondary metabolic gene clusters in A. flavus AF70 and NRRL 3357
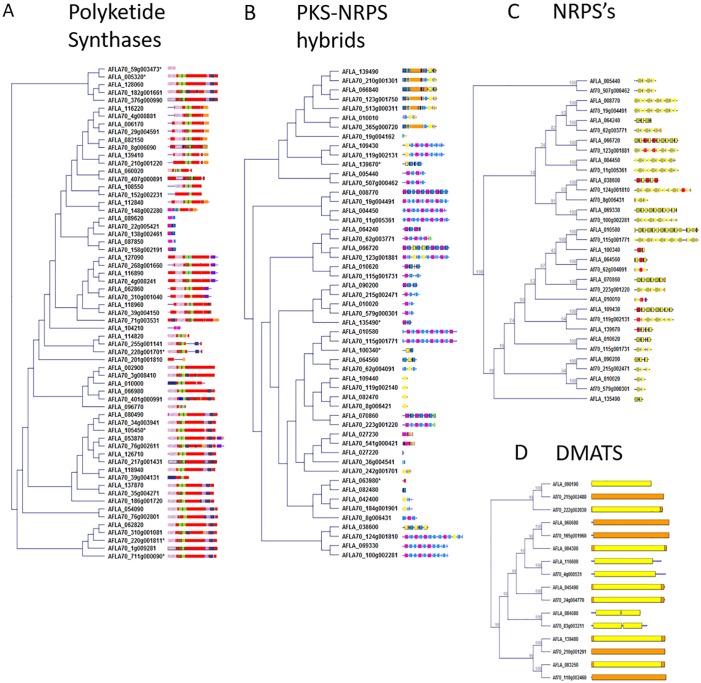
The metabolic clusters within A. flavus strains AF70 and NRRL 3357 were identified by antiSMASH, and alignment of each of the PKS, PKS-NRPS hybrid, NRPS and dimethylallyl tryptophan synthetase (DMAT) enzymes was conducted to illustrate the identity of the domains within each enzyme (Fig 3A–3C). The genomes of AF70 and NRRL 3357 were queried for known eukaryotic biosynthetic clusters using tblastn with MultiGeneBlast, which searches for each gene individually and scores the hits based on their proximity to hits from other genes (see Methods) (Table 2). The criterion for presuming that a cluster was “present” in either strain was that 50% of the genes in the MIBiG cluster were present [57]. The results indicate that 22 characterized clusters are present in both of our examined A. flavus morphotypes, consisting of the well-characterized aflatoxin gene cluster, as well as the aflavarin, aflatrem, and cyclopiazonic acid gene clusters. Nine clusters were predicted to be unique to S-morphotype AF70, including the agriculturally relevant carcinogen ochratoxin. To our knowledge, ochratoxin cluster genes have not been previously identified or characterized in A. flavus strains, thus AF70 may provide such an opportunity. Further study would be necessary to determine if even a minimal amount of ochratoxin production is detectable in AF70, and if not, then identify the cause for its non-production. As well, it may be important to determine if this cluster was inherited through horizontal gene transfer from a closely-related ochratoxigenic species such as A. alliaceus [58]. Six clusters are putatively unique to NRRL 3357.
Fig 3. Secondary metabolic gene cluster backbone genes.
The “backbone enzymes” of secondary metabolic gene clusters in A. flavus strains AF70 and NRRL 3357 (S- and L-morphotypes, respectively) were compared and clustered according to amino acid sequence similarity. The colored segments indicate domains identified by InterProScan. A) Polyketide synthases (PKSs), B) Polyketide synthase-nonribosomal peptide synthetase hybrids (PKS-NRPSs), C) nonribosomal peptide synthetases (NRPSs), and D) dimethylallyl tryptophan synthases (DMATs).
Table 2. Percentage of genes identified as being present in A. flavus strains AF70 and NRRL 3357 that are putatively involved in the production of the indicated toxin*.
| Clusters in NRRL 3357 and AF70 | Clusters in AF70 | ||||||
| Cluster Type | MIBiG ID | % in 3357 | % in 70 | Cluster Type | MIBiG ID | % in 3357 | % in 70 |
| 1,8-dihydroxynaphthalene | BGC0001258 | 100.0% | 100.0% | Phytocassane / oryzalides | BGC0000672 | 45.5% | 54.5% |
| 4,4’-piperazine-2,5-diyldimethyl-bis-phenol | BGC0001234 | 100.0% | 100.0% | Radicicol | BGC0000134 | 40.0% | 80.0% |
| Aflavarin | BGC0001304 | 100.0% | 100.0% | Alternapyrone | BGC0000012 | 40.0% | 60.0% |
| Aspirochlorine | BGC0001123 | 100.0% | 100.0% | Grayanic acid | BGC0001266 | 33.3% | 66.7% |
| Ustiloxin B | BGC0000627 | 100.0% | 100.0% | LL-Z1272 beta | BGC0001390 | 33.3% | 66.7% |
| Aflatoxin | BGC0000007 | 100.0% | 72.0% | Ochratoxin A | BGC0001030 | 33.3% | 66.7% |
| Aflatrem | BGC0000629 | 94.1% | 100.0% | Aphidicolin | BGC0000676 | 33.3% | 50.0% |
| Aflatoxin/sterigmatocystin | BGC0000011 | 92.3% | 73.1% | NG-391 | BGC0001026 | 33.3% | 50.0% |
| Sterigmatocystin | BGC0000152 | 88.2% | 58.8% | Marneral | BGC0000669 | 25.0% | 50.0% |
| Cyclopiazonic acid | BGC0000977 | 85.7% | 100.0% | ||||
| Tenellin | BGC0001049 | 80.0% | 80.0% | Clusters in NRRL 3357 | |||
| Chaetoglobosin | BGC0000968 | 71.4% | 71.4% | Cluster Type | MIBiG ID | % in 3357 | % in 70 |
| Ferrichrome | BGC0000901 | 66.7% | 66.7% | Acetylaszonal-enin | BGC0000293 | 66.7% | 0.0% |
| Trans-resorcylide | BGC0001246 | 66.7% | 66.7% | Aspyridone | BGC0000959 | 55.6% | 33.3% |
| Depudecin | BGC0000046 | 66.7% | 50.0% | Monodictyphen-one | BGC0000101 | 50.0% | 0.0% |
| Paxilline | BGC0001082 | 62.5% | 50.0% | T-toxin | BGC0000155 | 50.0% | 0.0% |
| Desmethylbassianin | BGC0001136 | 60.0% | 60.0% | Fujikurins | BGC0001305 | 50.0% | 33.3% |
| Asperthecin | BGC0000684 | 57.1% | 57.1% | Penitrem | BGC0001375 | 40.0% | 0.0% |
| Dehydrocurvularin | BGC0000045 | 50.0% | 50.0% | ||||
| Fumosorinone | BGC0001218 | 50.0% | 50.0% | ||||
| PR toxin | BGC0000667 | 50.0% | 50.0% | ||||
| Stipitatic acid | BGC0000154 | 50.0% | 50.0% | ||||
*Clusters with less than 50% genes present are presumed to be insufficient for production of the metabolite.
Identification of high impact SNPs found in secondary metabolic gene clusters
An analysis of A. flavus strains AF70 and NRRL 3357 genomes allowed us to summarize and identify high impact SNPs. This analysis provides a foundation to describe the phenotypic differences between strains, such as differences in aflatoxin or sclerotium production. For this analysis, 13,403 the genomes were aligned, and SNPs were classified as having either high, moderate or low impact. High impact polymorphisms include exon deletions, premature stop codons and frameshift mutations. Moderate impacts consist of mutations occurring at the 3’ end of the gene, or mutations resulting in a change in amino acid. Low impact mutations are less likely to affect protein function. Our findings indicate that a majority of the genes (11,888) contain between 1 and 10 SNPs (Table 3) in NRRL 3357 The A. flavus S- (AF70) and L-strain (NRRL 3357) morphotypes differ in their production of aflatoxin (Fig 1). To examine if this is potentially due to structural differences between their aflatoxin clusters, we further examined those SNPs identified in their respective coding sequences. Table 4 and S2 Table detail the SNPs present in the aflatoxin gene clusters for both morphotypes examined.
Table 3. Summary of SNPs present in the A. flavus NRRL 3357 genome when queried against the A. flavus AF70 genome.
| Impact | 1 SNP | 1<x<10 SNPs | >10 SNPs |
|---|---|---|---|
| High | 987 | 349 | 0 |
| Moderate | 2,875 | 5,141 | 609 |
| Low | 2,762 | 6,398 | 908 |
Table 4. Genes in the aflatoxin biosynthetic cluster with SNP impacts classified as high, moderate, or low impact.
| NRRL 3357 Gene | AF70 Gene | Gene (function) | High | Moderate | Low |
|---|---|---|---|---|---|
| AFLA_139430 | AFLA70_210g001241 | aflU (P450 monooxygenase) | 4 | 6 | 8 |
| AFLA_139250 | AFLA70_106g003541 | aflL (P450 monooxygenase) | 1 | 16 | 60 |
| AFLA_139420 | AFLA70_210g001231 | aflT (transmembrane protein) | 1 | 8 | 8 |
| AFLA_139140 | AFLA70_106g003640 | aflYa (NADH oxidase) | 1 | 5 | 1 |
| AFLA_139230 | AFLA70_106g003561 | aflI (cytochrome P450 monooxygenase) | 0 | 22 | 26 |
| AFLA_139220 | AFLA70_106g003571 | aflO (O-methyltransferase B) | 0 | 18 | 53 |
| AFLA_139240 | AFLA70_106g003551 | aflLa (hypB, putative oxygenase) | 0 | 15 | 14 |
| AFLA_139210 | AFLA70_106g003581 | aflP (O-methyltransferase) | 0 | 11 | 19 |
| AFLA_139150 | AFLA70_106g003632 | aflY (oxygenase) | 0 | 11 | 9 |
| AFLA_139410 | AFLA70_210g001220 | aflC (polyketide synthase A) | 0 | 11 | 6 |
| AFLA_139160 | AFLA70_106g003561 | aflX (monooxygenase) | 0 | 10 | 23 |
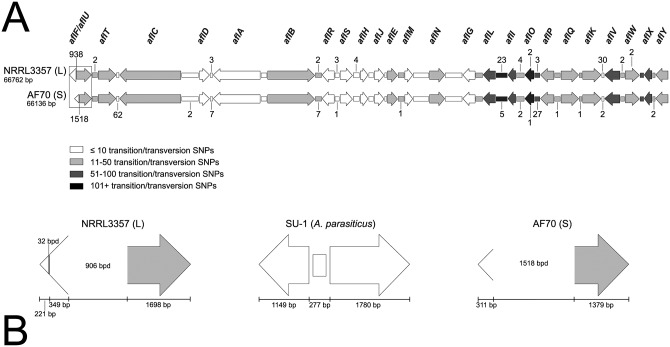
To analyze the entire aflatoxin cluster, including intergenic regions, Fig 4 shows the results of our comparative polymorphism analysis between the aflatoxin gene clusters of NRRL 3357 and AF70, for which we found 1192 transition or transversion mutations (1.78% of the aflatoxin gene cluster) that differentiated the two morphotypes. The cluster of NRRL 3357 alone contained 116 deletions (0.17%), while the cluster of AF70 contained 742 deletions (1.11%). In total, more than 2000 SNPs within the aflatoxin cluster alignment could be used to differentiate these strains based on genotype. The greatest span of cluster alignment (9825 bp) having the lowest percentage of SNPs (0.35%) was found to encompass the start of the aflC/aflD intergenic region through the end of the aflA/aflB intergenic region. The second greatest span and second lowest percentage (7146 bp and 0.39%, respectively) encompassed the aflR gene through the end of the aflJ/aflE intergenic region. The intergenic region with the highest percentage (14.15%) of SNPs per span of genomic sequence (1039 bp) was aflL/aflI, and the gene with the highest percentage of SNPs (9.06%) per span of genomic sequence (1335 bp) was aflO. The highest concentration of deletions for both strains was found to reside in the aflF/aflU (norB/cypA) regions of their aflatoxin clusters. This region in L-morphotype NRRL 3357 exhibited 34 deleted bases not observed in AF70, and the S-morphotype AF70 exhibited 619 deleted bases not observed in NRRL 3357. Since both isolates are of the same mating type (MAT1-1), the presence of SNPs would be required to help differentiate the sequences, but their respective sequences were identical so our MLST analysis was based on five genomic loci.
Fig 4. Schematic of the aflatoxin cluster as a comparison of polymorphisms that differentiate an A. flavus L-morphotype strain (NRRL 3357) from an A. flavus S-morphotype strain (AF70).
The genes are shown as arrows and the intergenic regions are shown as boxes (A and B). Shading for each gene and intergenic region relates to the quantity of transition and transversion SNPs observed (legend). Any number noted above or below a gene or intergenic region for panel A represents the quantity of base pair deletions (bpd) found within the NRRL 3357 or AF70 cluster sequences, respectively. The boxed aflF/aflU regions in panel A are enlarged in panel B, for which the genes in this region (for both A. flavus morphotypes) are compared to the same (complete) genomic region in the SU-1 A. parasiticus strain. Areas noted with bpd indicate large-scale deletions observed.
It has been reported that the inability of A. flavus to produce G-aflatoxins is based on partial or complete deletion of the aflU gene, and that the amount of deletion observed in the aflF/aflU region will correlate with sclerotial morphology [59]. For example, the L-strain genotype for the aflF/aflU region results in an amplicon that is approximately 1 kb in size, while the S-strain genotype results in amplicon size of approximately 300 bp. Therefore, based on the report of Ehrlich and co-workers [59], NRRL 3357 is an L-morphotype strain and AF70 is an S-morphotype strain, which supported these strains being individuals and not sharing a VCG. Our MLST findings further corroborate the VCG results for these strains. VCGs in filamentous fungi are considered to be determined based on specific heterokaryon incompatibility (het) loci [60], and the identities of the relevant loci have not been identified in A. flavus.
In conclusion, the sequence and phenotypic differences quantified here between these A. flavus morphotypes are significant factors to consider in experimental design. In addition to being visibly different morphotypes, these model organisms produce significantly different levels of toxins. Here we describe additional differences in secondary metabolic gene cluster profiles, and identify several high impact SNPs within their aflatoxin gene clusters that could account for their differences in toxin production. This information should contribute to the further use and development of these strains in examining both fungal biology and their pathogenic potential.
Supporting information
The results here illustrate a lack of cleft formation between mycelia of AF70 and NRRL 3357, containing complementary mutations, therefore indicating they are not vegetatively compatible.
(TIF)
GO categories of genes that are unique to strains AF70 and NRRL 3357.
(XLSX)
Comprehensive list of SNPs identified in AF70 and NRRL 3357 aflatoxin gene cluster.
(XLSX)
Acknowledgments
We thank Drs. Heping Cao, Dr. Niranjan Baisakh and Perng Kuang Chang for critical reading of the manuscript; Jonte Ellison and Carol Carter-Wientjes for assistance with phenotype characterization; and Shannon Beltz for assistance with morphological and growth medium information in the Materials and Methods section.
Data Availability
The genome reported here is available at NCBI.gov under Genbank Assembly Accession GCA_000952835.1.
Funding Statement
This work was funded by the United States Department of Agriculture.
References
- 1.Payne GA, Yu J. Ecology, development and gene regulation in Aspergillus flavus In: Machida M, Gomi K, editors. Aspergillus: Molecular Biology and Genomics. Norfolk, U.K.: Caister Academic Press; 2010. p. 157–71. [Google Scholar]
- 2.Amaike S, Keller NP. Aspergillus flavus. Annu Rev Phytopathol. 2011;49:107–33. 10.1146/annurev-phyto-072910-095221 . [DOI] [PubMed] [Google Scholar]
- 3.Baan R, Grosse Y, Straif K, Secretan B, El Ghissassi F, Bouvard V, et al. A review of human carcinogens; Part F: Chemical agents and related occupations. The Lancet Oncology. 2009;10(12):1143–4. 10.1016/S1470-2045(09)70358-4 [DOI] [PubMed] [Google Scholar]
- 4.Wu F. Mycotoxin reduction in Bt corn: potential economic, health, and regulatory impacts. Transgenic Res. 2006;15(3):277–89. 10.1007/s11248-005-5237-1 . [DOI] [PubMed] [Google Scholar]
- 5.Vardon P, McLaughlin C, Nardinelli C. Potential economic costs of mycotoxins in the United States. Council for Agricultural Science and Technology (CAST) Mycotoxins: risks in plant, animal, and human systems. Task Force Report No. 139. Ames, Iowa2003.
- 6.Wu F, Groopman JD, Pestka JJ. Public health impacts of foodborne mycotoxins. Annu Rev Food Sci Technol. 2014;5:351–72. Epub 2014/01/16. 10.1146/annurev-food-030713-092431 . [DOI] [PubMed] [Google Scholar]
- 7.Cotty P. Virulence and cultural characteristics of two Aspergillus flavus strains pathogenic on cotton. Phytopathology. 1989;79(7):808–14. [Google Scholar]
- 8.Geiser DM, Dorner JW, Horn BW, Taylor JW. The Phylogenetics of Mycotoxin and Sclerotium Production in Aspergillus flavus and Aspergillus oryzae. Fungal Genet Biol. 2000;31(3):169–79. 10.1006/fgbi.2000.1215 [DOI] [PubMed] [Google Scholar]
- 9.Abbas HK, Weaver MA, Zablotowicz RM, Horn BW, Shier WT. Relationships between aflatoxin production and sclerotia formation among isolates of Aspergillus section Flavi from the Mississippi Delta. Eur J Plant Pathol. 2005;112(3):283–7. 10.1007/s10658-004-4888-8 [DOI] [Google Scholar]
- 10.Saito M, Tsuruta O. A new variety of Aspergillus flavus from tropical soil in Thailand and its aflatoxin productivity. Proceedings of the japanese association of mycotoxicology. 1993;(37):31–6. [Google Scholar]
- 11.Geiser DM, Pitt JI, Taylor JW. Cryptic speciation and recombination in the aflatoxin-producing fungus Aspergillus flavus. Proc Natl Acad Sci U S A. 1998;95(1):388–93. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horn B, Dorner J. Regional differences in production of aflatoxin B1 and cyclopiazonic acid by soil isolates of Aspergillus flavus along a transect within the United States. Appl Environ Microbiol. 1999;65(4):1444–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rank C, Klejnstrup ML, Petersen LM, Kildgaard S, Frisvad JC, Held Gotfredsen C, et al. Comparative Chemistry of Aspergillus oryzae (RIB40) and A. flavus (NRRL 3357). Metabolites. 2012;2(1):39–56. Epub 2012/01/01. 10.3390/metabo2010039 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim JH, Yu J, Mahoney N, Chan KL, Molyneux RJ, Varga J, et al. Elucidation of the functional genomics of antioxidant-based inhibition of aflatoxin biosynthesis. Int J Food Microbiol. 2008;122(1–2):49–60. Epub 2008/01/02. 10.1016/j.ijfoodmicro.2007.11.058 . [DOI] [PubMed] [Google Scholar]
- 15.Ehrlich KC, Mack BM. Comparison of expression of secondary metabolite biosynthesis cluster genes in Aspergillus flavus, A. parasiticus, and A. oryzae. Toxins. 2014;6(6):1916–28. 10.3390/toxins6061916 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Linz JE, Wee J, Roze LV. Aspergillus parasiticus SU-1 genome sequence, predicted chromosome structure, and comparative gene expression under aflatoxin-inducing conditions: evidence that differential expression contributes to species phenotype. Eukaryot Cell. 2014;13(8):1113–23. Epub 2014/06/22. 10.1128/EC.00108-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donner M, Atehnkeng J, Sikora RA, Bandyopadhyay R, Cotty PJ. Molecular characterization of atoxigenic strains for biological control of aflatoxins in Nigeria. Food Addit Contam Part A Chem Anal Control Expo Risk Assess. 2010;27(5):576–90. Epub 2010/05/11. 10.1080/19440040903551954 . [DOI] [PubMed] [Google Scholar]
- 18.Pechanova O, Pechan T, Rodriguez JM, Williams WP, Brown AE. A two-dimensional proteome map of the aflatoxigenic fungus Aspergillus flavus. Proteomics. 2013;13(9):1513–8. Epub 2013/03/05. 10.1002/pmic.201100659 . [DOI] [PubMed] [Google Scholar]
- 19.Chitarrini G, Nobili C, Pinzari F, Antonini A, De Rossi P, Del Fiore A, et al. Buckwheat achenes antioxidant profile modulates Aspergillus flavus growth and aflatoxin production. Int J Food Microbiol. 2014;189:1–10. 10.1016/j.ijfoodmicro.2014.07.029 [DOI] [PubMed] [Google Scholar]
- 20.Mylroie JE, Ozkan S, Shivaji R, Windham GL, Alpe MN, Williams WP. Identification and Quantification of a Toxigenic and Non-Toxigenic Aspergillus flavus Strain in Contaminated Maize Using Quantitative Real-Time PCR. Toxins. 2016;8(1). Epub 2016/01/08. 10.3390/toxins8010015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Payne GA, Nierman WC, Wortman JR, Pritchard BL, Brown D, Dean RA, et al. Whole genome comparison of Aspergillus flavus and A. oryzae. Med Mycol. 2006;44(Supplement 1):S9–S11. 10.1080/13693780600835716 [DOI] [PubMed] [Google Scholar]
- 22.Nierman WC, Yu J, Fedorova-Abrams ND, Losada L, Cleveland TE, Bhatnagar D, et al. Genome Sequence of Aspergillus flavus NRRL 3357, a Strain That Causes Aflatoxin Contamination of Food and Feed. Genome Announc. 2015;3(2). Epub 2015/04/18. 10.1128/genomeA.00168-15 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilbert MK, Mack BM, Wei Q, Bland JM, Bhatnagar D, Cary JW. RNA sequencing of an nsdC mutant reveals global regulation of secondary metabolic gene clusters in Aspergillus flavus. Microbiol Res. 2016;182:150–61. 10.1016/j.micres.2015.08.007 [DOI] [PubMed] [Google Scholar]
- 24.Cary JW, Han Z, Yin Y, Lohmar JM, Shantappa S, Harris-Coward PY, et al. Transcriptome Analysis of Aspergillus flavus Reveals veA-Dependent Regulation of Secondary Metabolite Gene Clusters, Including the Novel Aflavarin Cluster. Eukaryot Cell. 2015;14(10):983–97. 10.1128/EC.00092-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hruska Z, Rajasekaran K, Yao H, Kincaid R, Darlington D, Brown RL, et al. Co-inoculation of aflatoxigenic and non-aflatoxigenic strains of Aspergillus flavus to study fungal invasion, colonization, and competition in maize kernels. Frontiers in Microbiology. 2014;5:122 10.3389/fmicb.2014.00122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baidya S, Duran RM, Lohmar JM, Harris-Coward PY, Cary JW, Hong S-Y, et al. VeA Is Associated with the Response to Oxidative Stress in the Aflatoxin Producer Aspergillus flavus. Eukaryot Cell. 2014;13(8):1095–103. 10.1128/EC.00099-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moore GG, Mack BM, Beltz SB. Genomic sequence of the aflatoxigenic filamentous fungus Aspergillus nomius. BMC Genomics. 2015;16(1):551 10.1186/s12864-015-1719-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kafer E. The anthranilate synthetase enzyme complex and the trifunctional trpC gene of Aspergillus. Can J Genet Cytol. 1977;19(4):723–38. . [DOI] [PubMed] [Google Scholar]
- 29.Mateles RI, Adye JC. Production of aflatoxins in submerged culture. Appl Microbiol. 1965;13:208–11. Epub 03/01/1965. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Filtenborg O, Frisvad JC, Thrane U. The Significance of Yeast Extract Composition on Metabolite Production in Penicillium In: Samson R, Pitt J, editors. Modern Concepts in Penicillium and Aspergillus Classification. NATO ASI Series. 185: Springer; US; 1990. p. 433–41. [Google Scholar]
- 31.Bayman P, Cotty PJ. Genetic diversity in Aspergillus flavus: association with aflatoxin production and morphology. Canadian Journal of Botany. 1993;71(1):23–31. 10.1139/b93-003 [DOI] [Google Scholar]
- 32.Horn BW, Greene RL. Vegetative compatibility within populations of Aspergillus flavus, A. parasiticus, and A. tamarii from a peanut field. Mycologia. 1995:324–32. [Google Scholar]
- 33.Benvenuti ME, Burgess JA. Rapid Analysis of Aflatoxins in Corn, Cereals, and Almonds Using ACQUITY UPLC H-Class System with Fluorescence Detection. In: Corporation W, editor. Milford, MA2010.
- 34.Cummings B, Wood T. A simple and efficient method for isolating genomic DNA from endomycorrhizal spores. Gene analysis techniques. 1989;6(5):89–92. Epub 1989/09/01. . [DOI] [PubMed] [Google Scholar]
- 35.Cantarel BL, Korf I, Robb SM, Parra G, Ross E, Moore B, et al. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 2008;18(1):188–96. Epub 2007/11/21. 10.1101/gr.6743907 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tarailo-Graovac M, Chen N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr Protoc Bioinformatics. 2009;Chapter 4:Unit 4 10 Epub 2009/03/11. 10.1002/0471250953.bi0410s25 . [DOI] [PubMed] [Google Scholar]
- 37.Bao W, Kojima KK, Kohany O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob DNA. 2015;6:11 Epub 2015/06/06. 10.1186/s13100-015-0041-9 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stanke M, Waack S. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics. 2003;19 Suppl 2:ii215–25. Epub 2003/10/10. . [DOI] [PubMed] [Google Scholar]
- 39.Lomsadze A, Ter-Hovhannisyan V, Chernoff YO, Borodovsky M. Gene identification in novel eukaryotic genomes by self-training algorithm. Nucleic Acids Res. 2005;33(20):6494–506. Epub 2005/11/30. 10.1093/nar/gki937 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haas BJ, Salzberg SL, Zhu W, Pertea M, Allen JE, Orvis J, et al. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol. 2008;9(1):R7 10.1186/gb-2008-9-1-r7 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11(2):R14 10.1186/gb-2010-11-2-r14 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lechner M, Findeiss S, Steiner L, Marz M, Stadler PF, Prohaska SJ. Proteinortho: detection of (co-)orthologs in large-scale analysis. BMC Bioinformatics. 2011;12:124 10.1186/1471-2105-12-124 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–7. Epub 2004/03/23. 10.1093/nar/gkh340 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17(4):540–52. Epub 2000/03/31. 10.1093/oxfordjournals.molbev.a026334 . [DOI] [PubMed] [Google Scholar]
- 45.Khaldi N, Seifuddin FT, Turner G, Haft D, Nierman WC, Wolfe KH, et al. SMURF: Genomic mapping of fungal secondary metabolite clusters. Fungal Genet Biol. 2010;47(9):736–41. 10.1016/j.fgb.2010.06.003 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Medema MH, Takano E, Breitling R. Detecting sequence homology at the gene cluster level with MultiGeneBlast. Mol Biol Evol. 2013;30(5):1218–23. Epub 2013/02/16. 10.1093/molbev/mst025 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Medema MH, Kottmann R, Yilmaz P, Cummings M, Biggins JB, Blin K, et al. Minimum Information about a Biosynthetic Gene cluster. Nat Chem Biol. 2015;11(9):625–31. Epub 2015/08/19. 10.1038/nchembio.1890 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bushnell B. BBMap: a fast, accurate, splice-aware aligner. 2014.
- 49.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–60. 10.1093/bioinformatics/btp324 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. arXiv preprint arXiv:12073907. 2012.
- 51.Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012;6(2):80–92. Epub 2012/06/26. 10.4161/fly.19695 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pabinger S, Dander A, Fischer M, Snajder R, Sperk M, Efremova M, et al. A survey of tools for variant analysis of next-generation genome sequencing data. Briefings in Bioinformatics. 2014;15(2):256–78. 10.1093/bib/bbs086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moore GG, Elliott JL, Singh R, Horn BW, Dorner JW, Stone EA, et al. Sexuality generates diversity in the aflatoxin gene cluster: evidence on a global scale. PLoS Pathog. 2013;9(8):e1003574 10.1371/journal.ppat.1003574 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Price EW, Carbone I. SNAP: workbench management tool for evolutionary population genetic analysis. Bioinformatics. 2005;21(3):402–4. 10.1093/bioinformatics/bti003 . [DOI] [PubMed] [Google Scholar]
- 55.Aylor DL, Price EW, Carbone I. SNAP: Combine and Map modules for multilocus population genetic analysis. Bioinformatics. 2006;22(11):1399–401. 10.1093/bioinformatics/btl136 . [DOI] [PubMed] [Google Scholar]
- 56.Abbas HK, Zablotowicz RM, Horn BW, Phillips NA, Johnson BJ, Jin X, et al. Comparison of major biocontrol strains of non-aflatoxigenic Aspergillus flavus for the reduction of aflatoxins and cyclopiazonic acid in maize. Food Addit Contam Part A Chem Anal Control Expo Risk Assess. 2011;28(2):198–208. Epub 2011/01/25. 10.1080/19440049.2010.544680 . [DOI] [PubMed] [Google Scholar]
- 57.Cimermancic P, Medema Marnix H, Claesen J, Kurita K, Wieland Brown Laura C, Mavrommatis K, et al. Insights into Secondary Metabolism from a Global Analysis of Prokaryotic Biosynthetic Gene Clusters. Cell. 2014;158(2):412–21. 10.1016/j.cell.2014.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Varga J, Kevei E, Rinyu E, Teren J, Kozakiewicz Z. Ochratoxin production by Aspergillus species. Appl Environ Microbiol. 1996;62(12):4461–4. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ehrlich KC, Chang PK, Yu J, Cotty PJ. Aflatoxin biosynthesis cluster gene cypA is required for G aflatoxin formation. Appl Environ Microbiol. 2004;70(11):6518–24. 10.1128/AEM.70.11.6518-6524.2004 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Leslie JF. Fungal vegetative compatibility. Annu Rev Phytopathol. 1993;31:127–50. 10.1146/annurev.py.31.090193.001015 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The results here illustrate a lack of cleft formation between mycelia of AF70 and NRRL 3357, containing complementary mutations, therefore indicating they are not vegetatively compatible.
(TIF)
GO categories of genes that are unique to strains AF70 and NRRL 3357.
(XLSX)
Comprehensive list of SNPs identified in AF70 and NRRL 3357 aflatoxin gene cluster.
(XLSX)
Data Availability Statement
The genome reported here is available at NCBI.gov under Genbank Assembly Accession GCA_000952835.1.