Abstract
N6-Methyladenine (m6dA) has been discovered as a novel form of DNA methylation prevalent in eukaryotes; however, methods for high-resolution mapping of m6dA events are still lacking. Single-molecule real-time (SMRT) sequencing has enabled the detection of m6dA events at single-nucleotide resolution in prokaryotic genomes, but its application to detecting m6dA in eukaryotic genomes has not been rigorously examined. Herein, we identified unique characteristics of eukaryotic m6dA methylomes that fundamentally differ from those of prokaryotes. Based on these differences, we describe the first approach for mapping m6dA events using SMRT sequencing specifically designed for the study of eukaryotic genomes and provide appropriate strategies for designing experiments and carrying out sequencing in future studies. We apply the novel approach to study two eukaryotic genomes. For green algae, we construct the first complete genome-wide map of m6dA at single-nucleotide and single-molecule resolution. For human lymphoblastoid cells (hLCLs), it was necessary to integrate SMRT sequencing data with independent sequencing data. The joint analyses suggest putative m6dA events are enriched in the promoters of young full-length LINE-1 elements (L1s), but call for validation by additional methods. These analyses demonstrate a general method for rigorous mapping and characterization of m6dA events in eukaryotic genomes.
N6-Methyladenine (m6dA) is the most prevalent form of DNA methylation in prokaryotes, most commonly associated with restriction-modification (RM) systems that defend hosts against invading foreign genomes (Casadesús and Low 2006; Wion and Casadesús 2006). In addition, increasing evidence suggests m6dA also plays important roles in the regulation of bacterial gene expression (Fang et al. 2012), cell cycle (Kozdon et al. 2013), virulence (Heithoff et al. 1999), and antibiotic susceptibility (Jen et al. 2014). The prevalence of m6dA in eukaryotes was unclear until recent studies demonstrated its existence in algae (Fu et al. 2015), fungi (Mondo et al. 2017), worm (Greer et al. 2015), and insect (Zhang et al. 2015), as well as recently in vertebrates (Koziol et al. 2016), including mammals (Wu et al. 2016). These recent studies have revealed diverse functions impacted by m6dA events in eukaryotes, including the regulation of gene expression (Fu et al. 2015; Wu et al. 2016; Zhou et al. 2016; Mondo et al. 2017), transposons (Zhang et al. 2015; Wu et al. 2016), and cross-talk with histone modifications (Fu et al. 2015; Wu et al. 2016). The existence of m6dA modifications across a diverse set of eukaryotic genomes opens up an exciting paradigm (Luo et al. 2015) in epigenetics and epigenomics regarding the regulation of biological processes in eukaryotic systems, in addition to the widely studied cytosine methylation.
Several methods have been developed to map m6dA in eukaryotic genomes. DNA immunoprecipitation (DIP) with anti-N6-methyladenine antibodies followed by next-generation sequencing (m6dA-DIP-seq) has identified genomic regions enriched for m6dA events in several species (Fu et al. 2015; Greer et al. 2015; Zhang et al. 2015). The combination of m6dA-DIP-seq and exonuclease digestion (m6dA-CLIP-exo-seq) provides increased resolution (Fu et al. 2015). However, due to the nature of antibody-based methods, both m6dA-DIP-seq and m6dA-CLIP-exo-seq lack the ability to identify m6dA events at single-nucleotide resolution and might be confounded by certain biases (Lentini et al. 2017). Furthermore, the immunoprecipitation process loses information necessary to study cell-to-cell epigenetic heterogeneity (i.e., partial methylation) in the cell population of interest (Fu et al. 2015; Greer et al. 2015; Zhang et al. 2015). Thus, these antibody-based approaches are limited with respect to their ability to elucidate the characteristics of m6dA events at high resolution. A complementary approach was developed using m6dA-sensitive or m6dA-dependent restriction enzymes (REs) (Fu et al. 2015; Luo et al. 2016), both of which provide single-nucleotide resolution and enable estimates of partial methylation at each nucleotide position. However, these RE-based methods have a fundamental limitation in that they can only examine a limited set of motif sites (specific to the REs used) and therefore provide a largely incomplete view of m6dA methylome in any given organism.
Single-molecule real-time (SMRT) sequencing (Eid et al. 2009) by Pacific Biosciences enabled the genome-wide mapping of m6dA in prokaryotes at single-nucleotide resolution (Fang et al. 2012) and at single-molecule level (Beaulaurier et al. 2015). SMRT sequencing monitors not only the pulse fluorescence associated with each incorporated nucleotide but also the time between the incorporation events, termed the inter-pulse duration (IPD). Deviation of an IPD distribution from the expected level, as reflected by the IPD ratio, is highly correlated with the presence of modifications of the nucleotide corresponding to the IPD deviation or its neighboring nucleotides (Flusberg et al. 2010; Schadt et al. 2013). Given this feature of SMRT sequencing, m6dA methylomes have been mapped for hundreds of bacterial and archaeal genomes, revealing many novel insights into m6dA biology in prokaryotes (Sánchez-Romero et al. 2015; Blow et al. 2016). SMRT sequencing has also been used to detect m6dA in some eukaryotic species (Greer et al. 2015; Wu et al. 2016; Mondo et al. 2017). Although promising, the effective use of SMRT sequencing for studying m6dA in eukaryotic genomes has not been rigorously examined.
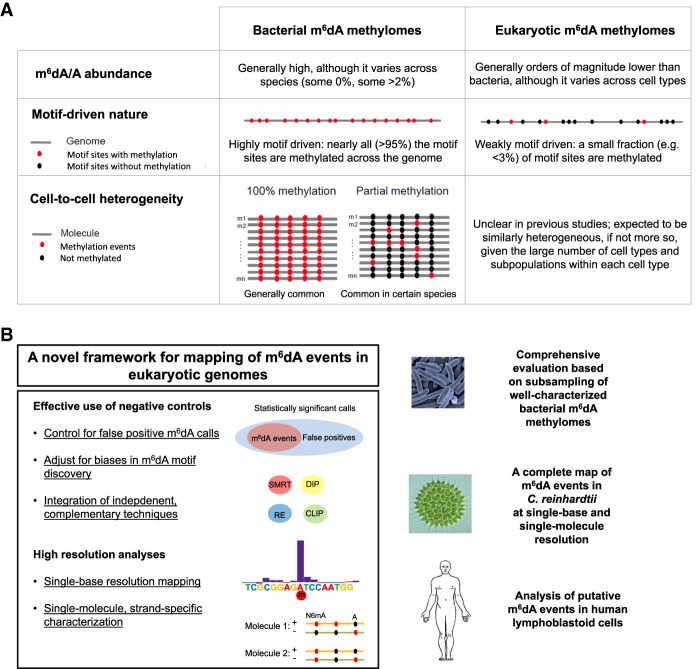
In fact, there are fundamental differences (Fig. 1A) between the prokaryotic and eukaryotic m6dA methylomes that raise a caution in the use of SMRT sequencing, and more generally third-generation sequencing (Manrao et al. 2012; Laszlo et al. 2013; Schreiber et al. 2013), for the detection of m6dA events in eukaryotic genomes. First, m6dA abundance (m6dA/A) is orders of magnitudes lower in eukaryotes than in prokaryotes (Fig. 1A; Casadesús and Low 2006; Fang et al. 2012; Fu et al. 2015; Greer et al. 2015; Luo et al. 2015; Zhang et al. 2015). Given a certain false-positive rate (FPR) associated with IPD-based detection of DNA modifications, m6dA calls from eukaryotes with low m6dA abundance are expected to have high false-discovery rates (FDRs). If the FDR becomes too high, then true m6dA events would be overwhelmed by the large number of false-positive ones. Second, m6dA events in prokaryotes are highly sequence specific due to their involvement in and the nature of RM systems (Casadesús and Low 2006; Fang et al. 2012). Typically, an active methyltransferase methylates nearly all occurrences (often >95%) of its target sequence motif in a prokaryotic genome (Fig. 1A; Fang et al. 2012; Blow et al. 2016). In contrast, m6dA events are much less motif driven in eukaryotes (Fu et al. 2015; Luo et al. 2016; Wu et al. 2016), likely due to their involvement in functional regulation rather than RM systems (Fig. 1A). For example, m6dA motifs have been identified in Chlamydomonas reinhardtii (Fu et al. 2015), Plasmodium falciparum (Luo et al. 2016), and mouse embryonic stem cells (mESCs) (Wu et al. 2016), where very few occurrences (often <3%) of the motif across the genome sites are methylated, i.e., weakly motif driven. Complicating matters further, other types of DNA modifications (DNA damages, m5C and its derivatives in the process of demethylation, etc.) occurring at neighboring bases can disturb the IPD ratios at an adenine site in question (Flusberg et al. 2010; Schadt et al. 2013), leading to false-positive m6dA calls. As a result, the weakly motif-driven nature of m6dA events in eukaryotic genomes poses a critical challenge in differentiating m6dA events from other types of DNA modifications. Finally, cell-to-cell epigenetic heterogeneity of m6dA has been increasingly recognized in prokaryotes (Casadesús and Low 2013; Manso et al. 2014; Beaulaurier et al. 2015), and m6dA in eukaryotes is expected to be similarly heterogeneous, if not more so, considering the large number of cell types and subpopulations of a given cell type (Fig. 1A; Huang et al. 2000; Heintzman et al. 2009; Miller et al. 2012; Shulha et al. 2013). Thus, the ability to study eukaryotic m6dA methylation at single-molecule resolution and to characterize cell-to-cell heterogeneity is desired to achieve a better understanding of m6dA biology in eukaryotes.
Figure 1.
Differences between bacterial and eukaryotic m6dA methylomes and a novel approach for mapping m6dA events in eukaryotic organisms. (A) Comparison between bacterial and eukaryotic m6dA methylomes over three aspects. (B) A novel approach for mapping and characterizing m6dA events in eukaryotic genomes. The novel approach, including a set of methods as summarized on the left, is comprehensively evaluated using subsampled bacterial m6dA methylome data and applied to Chlamydomonas reinhardtii (green algae) and human lymphoblastoid cells (LCLs).
Motivated by the above challenges, here we propose the first approach (Fig. 1B) for mapping of m6dA events using SMRT sequencing specifically designed for the study of eukaryotic genomes. By using well-characterized m6dA methylomes, we systematically investigate the factors affecting the sensitivity and specificity of m6dA detection at the levels of single nucleotides, single molecules, and individual motifs. This comprehensive evaluation provides a strategic framework that can help the study design, m6dA detection, and interpretation in future studies of eukaryotic m6dA methylomes using SMRT sequencing, as well as its critical integration with independent and complementary sequencing methods. We applied the approach to examine m6dA in green algae and human genomes. These applications demonstrate a general method and guideline for mapping and rigorous characterization of m6dA events in eukaryotic genomes.
Results
A novel approach and comprehensive evaluations for detecting and characterizing m6dA in eukaryotic genomes
We developed a novel approach with three core components to address challenges posed by the three fundamental differences between the prokaryotic and eukaryotic m6dA methylomes. We will first present each of the core components and associated evaluations, followed by the application of the novel approach to green algae and human genomes (Fig. 1B).
Design of SMRT sequencing and rigorous detection of m6dA events
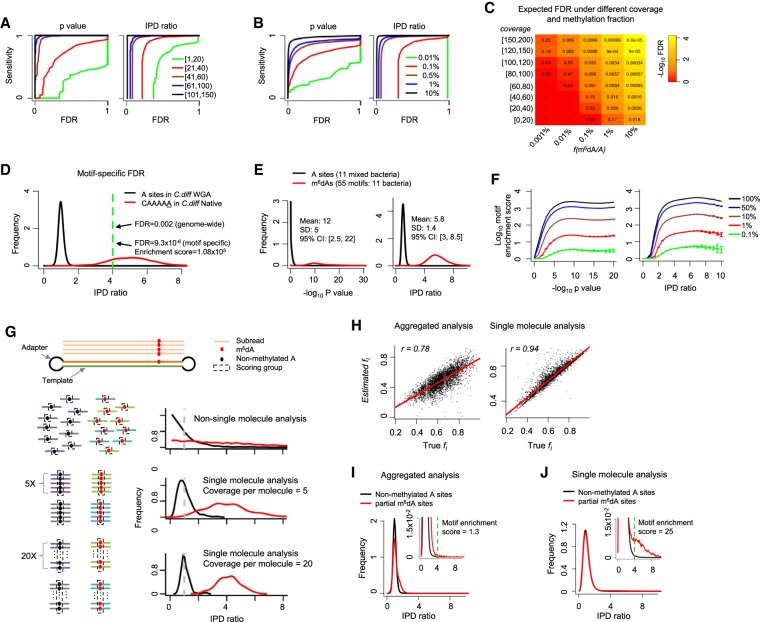
The genome-wide mapping of 5-methylcytosine (m5C) via bisulfite sequencing builds on the accurate base calling of Illumina sequencing (Schuster 2008). In contrast, SMRT sequencing–based detection of DNA modifications is facilitated through statistical tests (Fang et al. 2012; Schadt et al. 2013; Beaulaurier et al. 2015) comparing the observed distribution of IPD values at each nucleotide locus with the expected IPD value of the same base in the same sequence context, but without methylation. The latter IPD value is estimated from whole-genome amplified (WGA; methylation free) samples (Flusberg et al. 2010). For a given genome, millions or billions of nucleotides are tested, making false-positive calls due to multiple hypothesis testing a serious concern (Supplemental Text; Supplemental Fig. S1). To account for the multiple hypothesis testing, we use a FDR (Reiner et al. 2003; Fang et al. 2012) calculated by comparing the global distribution of IPD ratios (or P-values from Student's t-test) (https://www.pacb.com/wp-content/uploads/2015/09/WP_Detecting_DNA_Base_Modifications_Using_SMRT_Sequencing.pdf) in native versus WGA samples (Methods). For a given genome, the FDR of m6dA detection conceptually reflects the fraction of false positives among total m6dA calls and depends on two major factors: (1) the fraction of methylated adenines across the genome, f(m6dA/A), and (2) per-strand sequencing depth, coverage (i.e., average number of IPD values for each strand of the genome reference). f(m6dA/A) can be estimated from high-performance liquid chromatography (HPLC) coupled with m6dA mass spectrometry (MS) (Supplemental Text; Fu et al. 2015; Greer et al. 2015; Zhang et al. 2015), while coverage depends on SMRT sequencing read depth. By using bacteria with well-characterized m6dA methylomes (Supplemental Table S1), we systematically evaluated the variation in FDR over different levels of f(m6dA/A) and coverage (Methods). As expected, for each level of detection sensitivity, lower FDRs can be achieved with higher levels of f(m6dA/A) and coverage (Fig. 2A,B). We further estimated the expected levels of FDRs for genomes with different levels of m6dA f(m6dA/A) at different levels of coverage (Methods; Fig. 2C). Notably, while moderate coverage (e.g., ∼20×) is sufficient to achieve a fairly low FDR (e.g., approximately 0.03) for species with higher f(m6dA/A) levels (e.g., ∼1%), deep coverage (e.g., ∼150×) is necessary for species with low f(m6dA/A) levels (e.g., ∼0.001%) in order to achieve even modest FDRs (e.g., approximately 0.2). This systematic evaluation provides a rational strategy that can help determine the depth of SMRT sequencing in future studies of eukaryotic genomes based on the f(m6dA/A) values estimated from HPLC/MS data. It is worth noting that the coverage required to achieve a certain level of FDR estimated in Figure 2C is specifically for calling fully (∼100%) methylated m6dA events at single-nucleotide resolution. For other types of epigenomic analyses such as motif discovery and consensus analysis across multiple genomic sites (e.g., transcription start sites) (Fu et al. 2015), the requirement on sequencing depth can be lower, depending on specific m6dA patterns in different organisms.
Figure 2.
Comprehensive evaluation of m6dA detection based on SMRT-seq data. (A,B) Sensitivity-FDR curves at different levels of per strand SMRT-seq coverage (A) and fraction of methylated A sites in the genome (B). Curves are estimated based on either P-value or IPD ratio; both are shown. FDR estimation is based on the coverage-matched native (Escherichia coli with m6dA at GATC sites; Methods) and WGA samples. (C) FDRs estimated for different combinations of per strand SMRT-seq coverage and fraction of m6dA sites, f(m6dA/A), in the genome. FDR estimation is based on the coverage-matched native and WGA samples (Methods) at an IPD ratio of four. (D) Motif specific methylation detection leads to more reliable m6dA calls with lower FDRs. (E) Distribution of P-values (−log10) and IPD ratios of m6dA events (red) and nonmethylated A's (black) from 11 well-characterized bacterial m6dA methylomes. (F) Enrichment score for motifs with different fractions of motif sites methylated across the genome fm(m6dA/A), estimated based on P-value (−log10; left) and IPD ratio (right). SMRT-seq data from 11 bacterial species/strains with well-characterized m6dA methylomes are used for this simulation analysis. (G) Schematic illustrating single-molecule-level analysis for the estimation of partial methylation. A single molecule (two DNA strands and two adapters) and the subreads that are produced from the top strand of this molecule in SMRT-seq (top). For a given genomic position, when non-single-molecule analysis is performed, IPD ratios for the methylated and nonmethylated subreads follow two exponential distributions (red and black curves in the second panel). In contrast, when single-molecule analysis was performed, IPD ratios across all molecules follow two normal distributions with smaller variance over increasing coverage per molecule strand (third and fourth panels). (H) Estimation of partial methylation fl(m6dA/A) by aggregate analysis (left) and single-molecule-level analysis (right). x-axis indicates background truth fl based on simulation; y-axis, estimated fl; and dots, 4359 A's with known fraction of m6dA methylation based on subsampling from a well-characterized E. coli m6dA methylome. (I,J) Distribution of IPD ratios for partially methylated m6dA sites and nonmethylated A's based on aggregate analysis (I) and single-molecule level analysis (J). The inset provides an enlarged view. The motif enrichment score for the same, known methylation motif GATC significantly differs between the two types of analyses (1.3 in aggregated analysis vs. 25 in single-molecule analysis).
Unbiased discovery of m6dA motifs
In contrast to bacteria, m6dA motifs in eukaryotic species are typically only weakly motif driven; i.e., the motif-specific fraction of methylated motif sites across the genome, fm(m6dA/A), is often very low (<3%) (Fig. 1A). When a motif is only methylated at a low fraction across the genome, it becomes difficult to differentiate between the enrichment of the motif due to m6dA events and methylation-independent enrichment of the motif reflecting the intrinsic sequence composition of a eukaryotic genome or certain regions of interests (Bailey 2011). To address this challenge in SMRT sequencing–based m6dA motif enrichment analysis, we develop a motif enrichment score that is calculated as the odds ratio between the frequency of a motif among putative m6dA sites (IPD ratio > r) and the frequency of the motif among all adenine sites in the genome (Supplemental Fig. S2). The reciprocal of this enrichment score approximates the FDR of the motif sites with m6dA events (Methods). To illustrate the use of the motif enrichment score, we first examine a m6dA motif (CAAAAA; fm(m6dA/A) > 95%) in a strain of the bacterium Clostridium difficile, where the motif has an enrichment score of 1.08 × 105 (r = 4) (Fig. 2D), meaning that CAAAAA is 108,000-fold enriched among A's with an IPD ratio > 4 compared with all the A's sites in the genome (Supplemental Text). Next, we collected 11 bacterial species/strains that contain a total of 55 confident m6dA motifs (Fig. 2E; Supplemental Table S1) to systematically evaluate the use of motif enrichment score for detecting m6dA motifs with low fm(m6dA/A) levels as expected in eukaryotic genomes. With the 55 m6dA motifs as background truth, we calibrated motif enrichment scores over different fm(m6dA/A) levels of abundance (Methods; Fig. 2F).
Single-molecule analysis to estimate partial m6dA methylation
In the above methods and analyses, m6dA calling relies on IPDs pooled from separate molecules for each genomic locus. This aggregated analysis works well when each m6dA locus has ∼100% methylation across all molecules. However, epigenetic heterogeneity is often observed in both bacteria (Casadesús and Low 2013; Manso et al. 2014; Beaulaurier et al. 2015) and eukaryotic species (Heng et al. 2009; Smallwood et al. 2014), where only a fraction of cells are methylated at each genomic locus, i.e., partial methylation (Fig. 1A). Partial methylation is characterized by a locus-specific fraction of methylation fl(m6dA/A). By using an E. coli strain with a well-characterized methylome (Methods; Fang et al. 2012), we found that partial methylation significantly reduces the reliability of m6dA event calling by aggregate analysis (Supplemental Fig. S3). To better estimate partial methylation and study cell-to-cell m6dA heterogeneity in eukaryotic genomes, we developed a method for single-molecule-resolution analysis of SMRT sequencing data. In brief, IPDs are grouped by molecules for each genomic site and compared with the expected IPD values (Methods; Fig. 2G). The IPD ratios of methylated (m6dA) and nonmethylated sites (at single-molecule level) follow two normal distributions with means of one and approximately four and variances that decrease as per-molecule, per-strand sequencing coverage increases (Methods; Fig. 2G; Supplemental Text). By using 4359 m6dA sites with different levels of methylation fraction, ranging from 22% to 97%, subsampled from a well-characterized E. coli m6dA methylome (Methods), we found the single-molecule-level analysis provides a more accurate estimation of partial methylation than existing aggregate methods without single-molecule-level analysis (Fig. 2H). In addition, while aggregate analysis can hardly detect the GATC motif in E. coli methylome with simulated partial methylation (motif enrichment score = 1.3; FDR > 0.75) (Fig. 2I), single-molecule analysis clearly recognizes the GATC motif with a motif enrichment score of 25 (FDR < 0.05) (Fig. 2J).
A comprehensive characterization of m6dA in C. reinhardtii
The first genome-wide detection of m6dA in green algae C. reinhardtii was achieved recently, revealing that m6dA has a periodic pattern of deposition around transcriptional start sites (TSSs) that is inversely correlated with nucleosome positioning (Fu et al. 2015). In this previous study, Fu et al. (2015) developed three complementary sequencing-based methods and found that certain motifs were enriched for m6dA events, of which GATC and CATG were confirmed by m6dA-RE-seq, where the underlined base is the base that may or may not be methylated. In a more recent study, Luo et al. (2016) developed a more sensitive version of m6dA-RE-seq by using a methylation-dependent RE, leading to the discovery of two additional m6dA motifs (CATC and GATG). While these two studies fundamentally enhanced our understanding of the m6dA methylome of C. reinhardtii, the single-nucleotide-resolution m6dA map that they provide remains incomplete, and there are possibly additional m6dA motifs yet to be discovered.
A complete map of m6dA and cross-validation with five independent methods
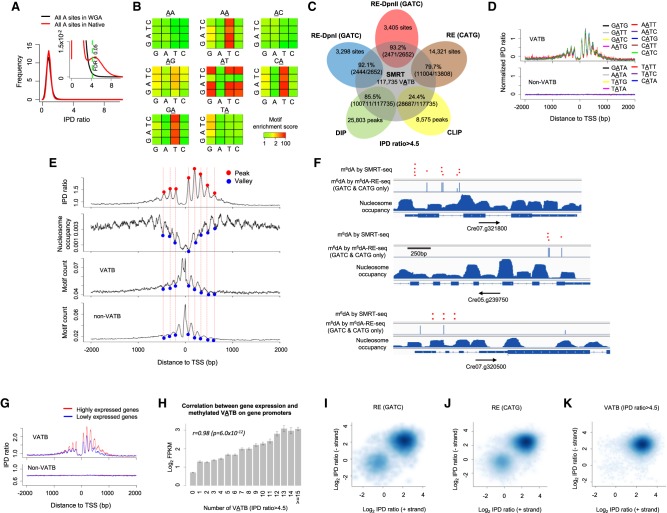
In order to construct a complete m6dA methylome of C. reinhardtii at single-nucleotide resolution, we performed high-coverage SMRT sequencing of both native and WGA samples of the same C. reinhardtii strain used in these recent studies (Supplemental Table S1; Fu et al. 2015; Luo et al. 2016). We used an IPD ratio threshold of >4.5 (FDR < 0.05) (Methods; Fig. 3A; Supplemental Fig. S4a; Supplemental Text) to calculate motif enrichment scores. Among the 16 4-mer motifs centered at AT, nine (VATB, V = A, C, or G and B = C, G, or T) are significantly enriched for the m6dA events in native DNA but not in WGA DNA (Fig. 3B; Supplemental Fig. S4b). In Figure 3B, each 4 × 4 heatmap corresponds to all 16 4-mer motifs, for which second and third bases are fixed at the center/title (e.g., AA). The rows and columns in the heatmaps represent the first and last bases of 4-mer motifs. Each cell in the following 4 × 4 heatmaps shows the motif enrichment score based on native DNA sample. Take the 4 × 4 heatmap with AT on top for example, the upper left corner corresponds to the motif CATG. A red color indicates that CATG has a very high motif enrichment score of approximately 200. For motifs centered at AA, CA, and GA, high motif enrichment scores are also observed when the last base is T (Fig. 3B); this is essentially a trivial consequence of the VATB motifs. It is also worth noting that a small number of additional 4-mer motifs have moderate methylation scores (Fig. 3B, yellow entries) in the native data, to some extent similar to that seen in the WGA data (Supplemental Fig. S4b). This observation suggests that some background noise (Supplemental Text), which may contribute to spurious motif enrichment, should be removed using WGA data as a negative control. Therefore, to further filter the background, we calculated the ratio of the motif scores between the native DNA and the WGA control, demonstrating an even cleaner motif enrichment (Supplemental Fig. S4c). At single-nucleotide level, the 117,735 methylated VATB sites (FDR < 0.05) represent 98.3% of total genomic m6dA calls (Supplemental Table S2) and ∼0.3% of total A sites in the genome. A cross-check among five independent m6dA detection methods shows that single-nucleotide m6dA events called by SMRT sequencing are highly consistent with detections made by m6dA-RE-seq and m6dA-DIP-seq (Fig. 3C; Supplemental Fig. S4d). It is worth nothing that a m6dA event called from SMRT-seq data will be missed by m6dA-RE-seq if the event resides in a motif context not recognized by the RE. m6dA-DIP-seq can miss a m6dA event due to certain bias or lack of sensitivity commonly associated with antibody-based approaches (Supplemental Fig. S4d). Thus, in addition to the four motifs confirmed by RE-based methods (Fu et al. 2015; Luo et al. 2016), SMRT sequencing–based motif analysis revealed five additional m6dA 4-mer motifs and provides a complete motif characterization of the C. reinhardtii m6dA methylome.
Figure 3.
Characterization of a complete m6dA methylome of C. reinhardtii reveals novel biological insights. (A) FDR estimation by comparing the IPD ratio distribution of C. reinhardtii native (red) with WGA (black) samples. The inset provides an enlarged view. (B) A rigorous motif enrichment analysis reveals that VATB (V = A, C, or G and B = C, G, or T) is the m6dA motif of in C. reinhardtii. Each 4 × 4 heatmap corresponds to all 16 4-mer motifs, for which the second and third bases are fixed at the center/title (e.g., AA). The rows and columns in the heatmaps represent the first and last bases of 4-mer motifs. Each cell in the following 4 × 4 heatmaps shows the motif enrichment score based on the native DNA sample. (C) Putative m6dA sites called by SMRT-seq are highly consistent with those detected by independent techniques: m6dA-DIP-seq (DIP), m6dA-CLIP-exo-seq (CLIP), and m6dA-RE-seq (RE). (D) VATB, but not non-VATB (i.e., TATN/NATA), motifs have a periodic pattern of IPD ratio distribution around TSSs. Average IPD ratio (normalized by motif frequency) for each of the nine VATB motifs (top) and each of the seven non-VATB motifs (bottom) are plotted around TSSs. (E) Relationship across four different distributions (top to bottom panels): average IPD ratio of VATB sites, nucleosome positioning, and frequency of VATB and non-VATB motif sites. Peaks and valleys of the periodic patterns are indicated by red and blue dots, aligned across the four panels. (F) Illustrative examples showing m6dA sites near the TSSs of three genes. This figure is adapted from Fu et al. (2015), where we project m6dA sites detected by SMRT-seq (red dots; FDR < 0.05; randomly generated heights to ease visualization) on top of GATC and CATG sites detected by m6dA-RE-seq (blue bars; middle) and nucleosome occupancy (bottom). (G) m6dA events at VATB sites are associated with active gene expression. Average IPD ratios are compared between two groups of genes with high (FPKM > 1) and low (FPKM < 1) expression levels. (H) The correlation between the gene expression level in C. reinhardtii and methylated VATB on gene promoters. The x-axis represents the number of methylated VATB sites (IPD ratio > 4.5; FDR = 0.05) within [0, +2000 bp] of TSSs. The y-axis represents the mean log2 FPKM of genes. Error bars, SEs. (I–K) Single-molecule, strand-specific analysis of SMRT-seq data to examine full-, non-, or hemi-methylation status at m6dA sites. Three sets of m6dA sites are analyzed: m6dA in GATC sites (I) and CATG sites (J) based on m6dA-RE-seq (Fu et al. 2015) and (K) VATB sites with high aggregate IPD ratio (IPD ratio > 4.5; Methods) based on SMRT-seq. The x- and y-axes denote the single-molecule, strand-specific IPD ratio of each pair of reverse-complementary VATB sites at the two strands of each single molecule.
High-resolution characterization of m6dA deposition
We next performed a comprehensive characterization of the C. reinhardtii m6dA methylome. We first checked whether the five additional motifs discovered from SMRT sequencing follow a periodic deposition pattern similar to the four previously known motifs (Fu et al. 2015; Luo et al. 2016). The methylated sites of all the nine 4-mers (VATB), but not the other seven 4-mers (non-VATB), are enriched at TSSs with a similar periodic pattern (Fig. 3D) that inversely correlates with nucleosome positioning (Fig. 3E). Next, the completeness and single-nucleotide resolution of this m6dA map allow us to examine the frequency of m6dA events in linker DNAs: An average of one m6dA locus occurs in the linker DNAs between the adjacent nucleosomes near TSSs, with some linkers having approximately 10 m6dA events and some having none (Fig. 3F; Supplemental Fig. S4e). The depletion of m6dA events in the close proximity of TSSs (Fig. 3E) motivated us to check the frequency of VATB motif sites in this region. We found VATB and non-VATB sites have a similar periodic frequency that reaches its peak density near TSSs (Fig. 3E), yet the VATB sites close to TSSs are nonmethylated. Beyond TSSs, regions with high nucleosome occupancy also have high density of VATB sites, yet low levels of m6dA (Fig. 3E). These discrepancies between VATB density and m6dA methylation density suggest the existence of additional factors in the deposition of m6dA events in C. reinhardtii beyond the proximity to TSS and the clearly defined m6dA motif. A further integrative analysis of SMRT sequencing data and the RNA-seq gene expression data from Fu et al. (2015) shows that m6dA events at VATB sites are associated with active gene expression (Fig. 3G,H; Supplemental Fig. S4f), while there is no such correlation between gene expression and the frequency of VATB motif sites (Supplemental Fig. S4g).
Single-molecule strand-specific characterization
A unique advantage of SMRT sequencing is the ability to examine methylation states of the two reverse-complementary VATB sites at the two strands of each molecule. This allows us to further characterize m6dA events at VATB sites in terms of full-, non-, or hemi-methylation states at single-molecule resolution with strand specificity. We examined m6dA calls in GATC (Fig. 3I) and CATG sites (Fig. 3J) detected by m6dA-RE-seq (Fu et al. 2015) and the methylated VATB sites detected by SMRT sequencing (FDR < 0.05) (Fig. 3K; Supplemental Fig. S4h). Consistently, most examined molecules were fully methylated on both strands (Fig. 3I–K, top right corners). We also found that some VATB sites were hemi-methylated (Fig. 3I–K, top left and bottom right corners), which could be right after DNA replication forks and have not been fully methylated yet. Some VATB sites were nonmethylated on both strands of single molecules (Fig. 3I–K; bottom left corners), despite these loci having high-consensus m6dA methylation levels. Collectively, the above comprehensive characterizations reveal the first complete m6dA map of C. reinhardtii and motivate future research toward mechanistic understanding of m6dA deposition.
Integrative analysis of SMRT sequencing data of hLCLs
After interrogating the m6dA distribution in a unicellular eukaryote, we next apply the new method to investigate a more complex genome. The recent discovery of m6dA in mammalian genomes and the enrichment of m6dA in young full-length L1s in mESCs opened new research opportunities (Wu et al. 2016). To date, the deposition patterns of m6dA in human genomes remain unclear. Human lymphoblastoid cells (hLCLs) are transformed from B cells by Epstein-Barr viruses for immortalization and have been widely used in large-scale studies of human genetics and genomics (Reedman and Klein 1973; Young and Rickinson 2004). Recently, whole-genome-wide SMRT sequencing data of hLCLs have been generated to improve human genome assembly (Zook et al. 2016), which also provide a good opportunity to detect putative m6dA events.
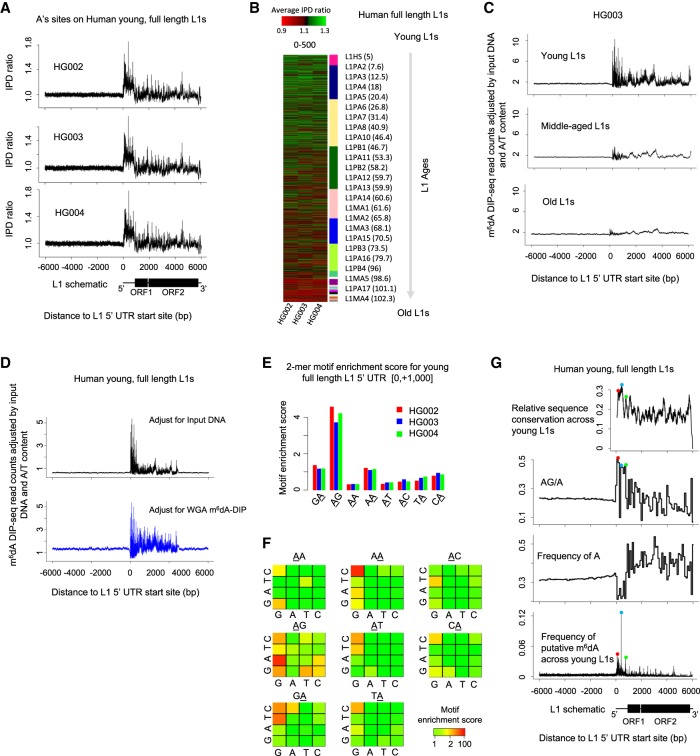
We first used dot blotting to compare hLCLs with negative oligos and mESCs (Wu et al. 2016). The results suggest the existence of m6dA in hLCLs at a m6dA/A level lower than what was observed in mESCs (Supplemental Fig. S5). It is worth noting that, because EBV genome coexists with human genome in hLCLs, the m6dA dot blots reflect m6dA in both EBV genome and human genome. In such cases, sequencing-based study is necessary for a specific genome of interest. We collected the genome-wide SMRT sequencing data (specifically, the subset with P6-C4 chemistry) publicly available for three hLCL samples (HG002, HG003, and HG004) (Zook et al. 2016). Considering the low level of f(m6dA/A) and the sequencing coverage (about 18× per reference strand for aggregate analysis), a genome-wide analysis of m6dA events with current data would probably be associated with a high FDR (Fig. 2C). Therefore, in the current study, we focused on full-length L1s with different ages (Methods) (Castro-Diaz et al. 2014) to test whether putative m6dA is enriched on young full-length L1s in human genome as in mESCs (Wu et al. 2016). A consensus analysis of IPD ratios on adenine (A) sites in the ±6000 bp beyond the 5′ UTRs of L1s across all the 7108 full-length L1s showed that, consistent among the three hLCLs, there is an enrichment of high IPD ratios at young (age < 10 Myr) full-length L1s (Methods; Fig. 4A; Supplemental Fig. S6), but much less enrichment in old L1s (Fig. 4A,B; Supplemental Fig. S6). In addition, this consensus analysis shows that the mean IPD ratio on A sites is relatively higher in the promoter and proximal region of young L1s than in the flanking regions (Fig. 4A). In the mammalian genome, the majority of CG dinucleotides are methylated (m5C), which can confound the m6dA analysis based on SMRT sequencing data because m5C events can affect IPDs at multiple flanking nucleotides (Schadt et al. 2013). To scrutinize this consensus pattern, we next examine multiple factors that may confound SMRT sequencing–based detection of putative m6dA events (Methods; Supplemental Text): effects of m5C events (Hata and Sakaki 1997) on neighboring IPDs (Supplemental Fig. S7; Flusberg et al. 2010; Schadt et al. 2013), outlier IPDs (Fang et al. 2012; Beaulaurier et al. 2015), SNP effects (both homozygous and heterozygous genotypes), and the use of in silico IPD estimation. We found that the consensus IPD ratio pattern in young full-length L1 remains after rigorous filtering of these possible confounding factors (Methods; Supplemental Figs. S8, S9). However, we found that when the same analysis was applied to the other three types of nucleotides (C, G, and T), similar consensus patterns are observed even after the effect of all these confounding factors are filtered out (Supplemental Fig. S10). This unexpected observation suggests the possible coenrichment of other DNA modifications (beyond m5C) in the young L1s together with m6dA, or the possible existence of DNA secondary structure in addition to DNA modifications, which are also expected to affect DNA polymerase kinetics in SMRT sequencing. Without orthogonal validation methods, SMRT sequencing data alone are unable to differentiate among these possibilities.
Figure 4.
m6dA deposition on full-length L1s in hLCLs. (A) Mean IPD ratio of A sites (adjusted by the frequency of A's) across 1274 young (evolutionary age<10 Myr), full-length (>6000 bp) L1s for three hLCL lines, respectively. Consistent across the trio, the IPD ratio is relatively higher in the promoter and proximal region than the flanking regions. (B) The mean IPD ratio of A sites at full-length L1s is inversely correlated with the L1s’ evolutionary ages in hLCLs. The heatmap shows the mean IPD ratio of A's on each L1, [0, +500] from the 5′ UTR start site, for each of the trio. As indicated in the sidebar, L1s (rows) are ordered by their evolutionary ages. Consistently across the trio, the IPD ratio of A sites is higher in younger full-length L1s than in older L1s. (C) Average m6dA-DIP-seq read count (adjusted for the read count in the input DNA sample and the A/T content) on hLCL young (1274), middle-aged (4164), and old L1 elements (1670), respectively. Consistent with SMRT-seq data, m6dA is enriched at the promoter and proximal region of young full-length L1s. (D) Average m6dA-DIP-seq read count adjusted for the A/T content and the read count in two control samples on hLCL young L1 elements, respectively: input DNA as control (black curve in top panel) and m6dA-DIP-seq on WGA as control (blue curve in bottom panel). (E) Motif AG is enriched for putative m6dA events. The barplot represents the motif enrichment score of all dinucleotide motifs in each of the trio. The putative methylated position is underscored. It suggests that motif AG is enriched for high IPD ratios in clear contrast to all the other dinucleotides. (F) Motif enrichment analysis of human young full-length L1s. Each 4 × 4 heatmap corresponds to all 16 4-mer motifs, for which the second and third bases are fixed at the center/title. The rows and columns in the heatmaps represent the first and last bases of 4-mer motifs. Each cell in the following 4 × 4 heatmaps shows the motif enrichment score based on the native DNA sample. (G) Peaks of putative m6dA events across human young full-length L1s occur at loci with certain sequence features. (Top) Level of sequence conservation across young full-length L1 elements based on multiple alignment by Mauve (Darling et al. 2004); (two middle panels) frequency of AG dinucleotides (relative to A's) and A's on young full-length L1s; and (bottom) frequency of putative m6dA events at each locus across all young full-length L1s (averaged among the trio). The peaks of sequence conservation, AG/A frequency, and m6dA frequency across young full-length L1s are colocalized as indicated by the red, blue, and green dots.
We therefore used m6dA-DIP-seq as an independent method to examine the hLCLs derived from the same cell lines (Supplemental Table S3). A consensus analysis of m6dA/A density across all the 7108 full-length L1s shows that m6dA events are enriched in young, but not old, full-length L1s of hLCLs (Methods; Fig. 4C; Supplemental Fig. S11). In addition, we performed a further analysis to examine the possibility that the consensus m6dA pattern across young L1s by m6dA-DIP-seq could be the result of certain biases (Lentini et al. 2017). Specifically, the exact same m6dA-DIP-seq protocol was also performed for hLCL WGA DNA, where essentially no m6dA events are expected, and used as an alternative control to input DNA (Supplemental Table S3). We observed a consistent pattern when two controls are used to compare with m6dA-DIP-seq of native hLCL DNA (Methods; Fig. 4D). These analyses of m6dA-DIP-seq data suggest that m6dA events are enriched in the young full-length L1 of hLCLs and that the m6dA/A level is relatively higher in the promoter and proximal region of young L1s than the downstream region, similar to the observations made from IPD analysis of SMRT sequencing data (Fig. 4A,B). A 2-mer motif analysis of the hLCL SMRT sequencing data (across young L1s) showed that AG is the most enriched for putative m6dA events among all eight 2-mer motifs (Fig. 4E), although it is worth noting the WGA sample also showed modest, weaker enrichment for AG (Methods; Supplemental Fig. S12a; Supplemental Text). In a further analysis of 4-mer motifs, AAGG and CAAG showed the highest motif enrichment scores that are specific to native DNA (Fig. 4F) but not WGA DNA (Supplemental Fig. S12b). We also estimated single-nucleotide sequence conservation in L1s through multiple alignment of young full-length L1s (L1HS and L1PA2; Methods) (Castro-Diaz et al. 2014) and found that the loci with highest frequency of putative m6dA sites (adjusted by the frequency of A's) across young L1s generally occur at the loci that are highly conserved across full-length L1s (Fig. 4G; Darling et al. 2004) and the loci with highest relative frequency of AG (AG/A) (Fig. 4G). These observations suggest the deposition and function of m6dA may be related to sequence conservations in the promoter and proximal regions of young L1s (Goodier and Kazazian 2008); this, however, needs to be validated by future independent methods that have better resolution than m6dA-DIP-seq and less constrained sequence specificity than m6dA-RE-seq.
Discussion
The recent discovery of m6dA in eukaryotic genomes opens up a new and promising dimension of epigenetic research; however, methods for high-resolution, complete mapping of m6dA events are still lacking. Here we presented a novel set of methods and an analytical framework for m6dA characterization in eukaryotic genomes using SMRT sequencing. The key motivation of this study was the characteristics of eukaryotic m6dA methylomes that fundamentally differ from those of prokaryotes, yet all previous computational methods for SMRT sequencing–based detection of m6dA events were designed specifically for the study of prokaryotic methylomes. In addition, we highlighted the importance of tailoring sequencing design and analytical strategy for an organism considering the m6dA/A abundance in its genome as determined by MS and dot blots. For organisms with high m6dA/A abundance, confident (low FDR) m6dA events can be called at both single-nucleotide and single-molecule resolution, allowing a variety of in-depth characterization, as demonstrated in our analysis of the C. reinhardtii m6dA methylome. For organisms with low m6dA/A abundance, however, m6dA events called by SMRT sequencing data are essentially putative events and must be treated with caution, because they are expected to have high FDR as estimated in Figure 2C. In applications that belong to the latter case, consensus analyses, which are more resistant to false-positive calls, should be adopted when applicable, as illustrated in the study of young L1s in hLCLs.
Importantly, instead of specifically detecting m6dA events, SMRT sequencing can detect any form of DNA modifications that significantly affect DNA polymerase kinetics as measured by IPD. Different types of DNA modifications at a site of interest or its neighboring sites can lead to similar IPD ratios at the site (Flusberg et al. 2010; Schadt et al. 2013). In a bacterial genome, the forms of DNA methylation are relatively limited (m6dA, m5C, m4C) and highly motif driven, which fundamentally ease the detection and differentiation of m6dA events from other DNA modifications. In contrast, m6dA events in eukaryotic genomes are much less abundant, weakly motif driven, and possibly coexist with other forms of DNA modifications. These differences between bacterial and eukaryotic methylomes call for critical attention in the interpretation of putative m6dA calls based on SMRT sequencing to avoid misinterpretation of false-positive events. The methods and the overall framework we presented in this study highlight the importance of rational design of experiments and SMRT sequencing, as well as rigorous analysis and interpretation of SMRT sequencing data in combination with independent and complementary techniques.
Finally, it is worth noting that the strengths and challenges associated with SMRT sequencing discussed above also largely apply to other third-generation real-time sequencing techniques that also hold promise for the detection of DNA methylation, e.g., Oxford Nanopore (Manrao et al. 2012; Laszlo et al. 2013; Schreiber et al. 2013). Essentially, similar to SMRT sequencing, these other third-generation sequencing methods indirectly detect DNA modifications based on features captured during real-time single-molecule sequencing. Therefore, similar cautions are likely needed in the use of these third-generation sequencing technologies in the mapping and characterization of different forms of DNA modifications in eukaryotic genomes.
Methods
Preprocessing of SMRT sequencing data for IPD analysis
We followed the preprocessing steps as implemented in SMRT portal (https://www.pacb.com/products-and-services/analytical-software/smrt-analysis/). In brief, an initial filtering step removes all subreads with ambiguous alignments (MapQV < 240), low accuracy (<75%), or short-aligned length (fewer than 50 bases). Next, an additional filtering step removes the subread IPD values from the mismatched positions with respect to the reference sequence. Subread IPD normalization corrects for any potential slowing of polymerase kinetics over the course of an entire read (which consists of many subreads) and is done by dividing subread IPD values by their mean.
Estimation of FDR for single-nucleotide-level m6dA calls
The FDR corresponding to a specific threshold on a given statistical measure (e.g., IPD ratio, t-test P-value or identificationQv) is estimated by comparing global distribution of the measure obtained from the native DNA sample with that from a WGA (methylation-free) sample. Specifically, the FDR is calculated as follows:
where m denotes a given statistical measure; denotes the fraction of A's with m > thres out of all A's in WGA, and denotes the fraction of A's with m > thres out of all A's in native DNA sample. There are cases where a WGA sample is not available or some true m6dA motifs are known a priori or discovered based on motif enrichment analysis. In such cases, for each motif, FDRs can be estimated for single-nucleotide-level m6dA calls only among the A sites corresponding to the motif across the genome. Specifically, we describe motif-specific FDR estimation for a specific motif using data from native DNA alone:
where denotes the frequency of motif sites with m > thres among all sites of that putative m6dA motif in native DNA.
Expected FDRs for single-nucleotide-level m6dA calls over different levels of f(m6dA/A) and coverage
We used an E. coli C227 methylome that has been well characterized in the previous study (Supplemental Table S1; Fang et al. 2012). By subsampling both m6dA motif sites and non-m6dA-motif sites, we generated test data sets with different levels of f(m6dA/A) and coverage. For each data set, we estimated the FDR corresponding to the same cutoff on IPD ratio (greater than four).
Methylation enrichment score of a putative m6dA motif
For a real m6dA methylation motif, it is expected that the fraction of A's with high IPD ratios in that motif, i.e., , should be higher than the background in the same native DNA sample, i.e., . So, we define the motif enrichment score as the odds between the two fractions:
The denominator can also be defined as the A sites in native DNA excluding a certain motif. Because m6dA/A level is mostly <5% in both bacteria and eukaryotes, the two alternative definitions are practically the same, and the currently defined one is easier to calculate. It is worth noting that the motif enrichment score is mathematically the reciprocal of motif-specific FDR:
Certain intrinsic biases in SMRT sequencing (e.g., possible biases associated with the in silico control model as described above) can contribute to the small but statistically significant enrichment of certain motifs independent of DNA modifications. These biases can be estimated by calculating methylation enrichment scores for a specific motif using a WGA sample without DNA methylation, i.e., Smotif{WGA}. A motif is enriched for m6dA events if it has a high enrichment score that is specific to the native data but not the WGA data.
Methylation enrichment scores for motifs with different fraction of m6dA methylation
We collected 11 bacterial m6dA methylomes (55 m6dA motifs) that have been well characterized in previous studies (Fang et al. 2012; Beaulaurier et al. 2015; Pak et al. 2015). All the m6dA motif sites are pooled together as true m6dA events. By subsampling from these m6dA sites, we generated test motifs with different fraction of methylation: 100%, 50%, 10%, 1%, and 0.1%. For each of these fractions, we estimated the methylation motif score, Smotif {native}, corresponding to different thresholds of IPD ratios and t-test P-values.
Single-molecule, single-nucleotide-level calculation of IPD ratios
Considering each molecule separately, the IPD values (post-filtering) are grouped by their strand and mapped genomic position, and the mean value is calculated. At each genomic position of a single strand, the mean IPD values for each molecule follow the Gaussian distribution based on the central limit theory.
Methylation fraction calling for each site at single-molecule level
The central limit theorem (CLT) states that, given a sufficiently large sample size, the average of all samples from the same population tends toward a normal distribution, even if the original variables themselves are not normally distributed. Meanwhile, the mean of a sample approximates the mean of the population. In the context of IPD-based DNA modification detection, the IPD values follow an exponential distribution; however, the mean of the IPD values that come from the same molecule and at the same genomic location (referred to as IPD at a single-molecule level) follows normal distributions: either a single normal distribution (fully-methylated or nonmethylated sites) or a mixture of normal distributions (partially methylated sites). Accordingly, given a single site, we can use a Gaussian mixture model (GMM) to estimate the extent of partial methylation. The GMM comprises two normal distributions from methylated and nonmethylated molecules; the mean of IPD for nonmethylated molecules is estimated from either the in silico control model or WGA; the mean of IPD for methylated molecules is learned from the data, and the estimated proportion of two normal distributions reflects the fraction of methylated and nonmethylated molecules. Furthermore, the CLT states that the variance of the sample approximates the variance of the population divided by the sample size. Accordingly, as read coverage increases for each molecule, the variance of normal distributions of IPDs at the single-molecule level decreases, providing a better power for the separation between the methylated and nonmethylated molecules. Therefore, the CLT provides a theoretical foundation to use GMM to call methylation fraction at a single-molecule level.
Simulation of partial m6dA methylation from well-characterized bacterial m6dA methylomes
We use the SMRT sequencing data for E. coli C227 strain (both native and WGA), generated in a recent study (Fang et al. 2012). In E. coli, most GATC sites are ∼100% m6dA methylation (Fang et al. 2012). To simulate partially methylated GATC sites, we randomly select single molecules from both native and WGA data and mix them in different proportions to generate GATC sites with different levels of partial methylation. For each GATC site, the true fraction of m6dA methylation is calculated based on the number of unique molecules from the native and WGA data.
C. reinhardtii DNA extraction
The frozen cell pellet is grounded in liquid nitrogen using a plastic pestle and 1.5-mL LoBind Eppendorf microcentrifuge tubes. We used the NucleoSpin Plant II (Macherey Nagel, catalog no. 740770.50) kit and followed the standard protocol for lysis buffer PL1, using ∼100 mg of tissue/extraction column to extract the DNA. The concentration and quality of the resulting DNA are checked using the Qubit dsDNA high sense kit and 12k DNA BioAnalyzer chip.
hLCLs
Genome-wide SMRT-seq data were from a recent study (Zook et al. 2016). The full human SMRT-seq data contain a mixture of two SMRT-seq chemistries: P5_C3 and P6_C4. Different chemistries are associated with different DNA polymerase kinetics that can significantly impact IPD values, which may lead to false-positive calls. To achieve the most rigorous data analysis, we chose to use P6_C4 SMRT runs only. gDNA is available from Coriell Biorepository: NA24143, NA24149, and NA24385.
Genome references
SMRT-seq data were mapped to the appropriate genomes using BLASR via SMRTportal (https://www.pacb.com/products-and-services/analytical-software/smrt-analysis/). Reads from Illumina sequencing data are mapped using BWA 0.7.8 (Li and Durbin 2009). The C. reinhardtii data were mapped to Chlamydomonas genome (JGI) version 9.1. For hLCL data, we built a customized reference by extracting the [−10,000 nt, +10,000 nt] regions surrounding the 5′ UTR of full-length L1s from the UCSC hg19 and mapped the human data to the faux reference. For the human SMRT sequencing data, we made consistent observations between hg19 and de novo genome assemblies of individual hLCL samples. So, we expect an analysis with GRCh38 would not significantly affect the conclusions.
Overlap analysis between m6dA calls by different methods
For m6dA-RE-seq, its overlap with SMRT-seq is defined as the ratio between the CATG/GATC sites detected by both SMRT-seq and m6dA-RE-seq, and the CATC/GATG sites detected by SMRT-seq. For m6dA-DIP-seq and m6dA CLIP-seq, their overlap with SMRT-seq is defined as the ratio between putative m6dA sites that are detected by SMRT-seq and covered by at least one peak called from m6dA-DIP-seq/m6dA CLIP-seq, and all the m6dA sites detected by SMRT-seq. The above overlap in a region of interest mainly depends on the ratio between true-positive and false-positive SMRT-seq m6dA detections in that region.
m6dA dot blots
We followed the same protocol as used in the recent study (Wu et al. 2016). Briefly, first, DNA samples were denatured at 95°C for 5 min, cooled down on ice, and neutralized with 10% vol of6.6 M ammonium acetate. Samples were spotted on the membrane (Amersham Hybond-N+, GE), air dried for 5 min, and then UV-crosslinked (2× auto-crosslink, 1800 UV Stratalinker, STRATAGENE). Membranes were blocked in blocking buffer (5% milk, 1% BSA, PBST) for 2 h at room temperature and incubated with m6dA antibodies (202-003, Synaptic Systems, 1:1000) overnight at 4°C. After five washes, membranes were incubated with HRP linked secondary anti-rabbit IgG antibody (1:5,000, Cell Signaling 7074S) for 30 min at room temperature. Signals were detected with ECL Plus Western blotting reagent pack (GE Healthcare).
Full-length L1 elements and their evolutionary ages
We collected the human LINE-1 (L1) transposon annotations from RepeatMasker (Tarailo-Graovac and Chen 2009). Those ∼6-kb-long L1s were treated as full-length L1s (Babushok and Kazazian 2007). The evolutionary age for each L1 subfamily is based on the method of Castro-Diaz et al. (2014).
Consensus analysis of IPD ratios across different L1s
The full-length L1s identified as described above were aligned based on their 5′ UTR sites. At each aligned position, the IPD ratios of a specific base (A/G/C/T) across different L1s were aggregated and normalized to the frequency of that corresponding base (A/G/C/T).
Estimating FPR of m6dA calls for adenines close to m5C sites
E. coli K-12 has m5C at the second cytosine at CC(A/T)GG sites (Kahramanoglou et al. 2012). We used SMRT sequencing data for E. coli from a recent study (Fang et al. 2012) and examined the IPD ratios for A sites within ±10 bp from CC(A/T)GG sites to estimate the false positive, excluding known m6dA events at GATC and AACNNNNNNGTGC/GCACNNNNNNGAA. Based on these selected A sites, we estimate the FPR of m6dA calls due to neighboring m5C sites (Supplemental Fig. S7a)
m6dA DIP sequencing
We followed the same protocol as used in the recent study (Wu et al. 2016). Briefly, genomic DNA from hLCLs derived from a family trio were purified with a DNeasy kit (QIAGEN, 69504). For each sample, 5 µg DNA was sonicated to 200–500 bp with Bioruptor. Then, adapters were ligated to genomic DNA fragments following the Illumina protocol. The ligated DNA fragments were denatured at 95°C for 5 min. Then, the single-stranded DNA fragments were immunoprecipitated with 6 mA antibodies (5 µg for each reaction, 202-003, Synaptic Systems) overnight at 4°C. m6dA-enriched DNA fragments were purified according to the active motif hMeDIP protocol. IP DNA and input DNA were PCR amplified with Illumina indexing primers and were then subjected to multiplexed library construction and sequencing with Illumina HiSeq sequencing.
Analysis of m6dA DIP sequencing data
BWA 0.7.8 (Li and Durbin 2009) was used to align the human m6dA-DIP-seq reads to the UCSC hg19. Peaks called from the green algae genome were obtained from the investigators of the original study.
Consensus analysis of m6dA-DIP-seq reads across different L1s
The putative full-length L1s were aligned based on their 5′ UTR. At each aligned position, the m6dA-DIP-seq read coverage for different L1s were aggregated and normalized to the A/T frequency across all of the full-length L1s. To further rule out the possibility of biased background distribution, we also normalized the average read coverage to the aggregated read coverage from m6dA-DIP-seq of WGA DNA or input DNA.
Software availability
The novel methods presented in the manuscript are implemented in R (R Core Team 2013), and the source codes are available in Supplemental Material and at https://github.com/fanglab/SMRTER.
Data access
The sequencing data from this study have been submitted to the NCBI Sequence Read Archive (SRA; https://www.ncbi.nlm.nih.gov/sra) under the following the accession numbers: SRP102471 (SMRT-seq of Clostridium innocuum native DNA with 1 SMRTcell), SRP102628 (SMRT-seq of C. difficile native DNA with two SMRTcells and WGA with two SMRTcells), SRP105216 (SMRT-seq of Helicobacter pylori native DNA with two SMRTcells), SRP102373 (SMRT-seq of Staphylococcus aureus native DNA with one SMRTcell), SRP105217 (SMRT-seq of C. reinhardtii native DNA with 20 SMRTcells and WGA with 18 SMRTcells), and SRP128153 (SRX3538573: m6dA-DIP-seq of HG002 native DNA; SRX3538574: input DNA of HG002; SRX3538575: m6dA-DIP-seq of HG003 native DNA; SRX3538576: input DNA of HG003n SRX3538577: m6dA-DIP-seq of HG004; SRX3538578: input DNA of HG004; SRX3538579: m6dA-DIP-seq of GM12878 native DNA; SRX3538580: m6dA-DIP-seq of GM12878 WGA; and SRX3538581: input DNA of GM12878).
Competing interest statement
E.E.S. is on the scientific advisory board of Pacific Biosciences.
Supplementary Material
Acknowledgments
We thank the members of the Fang laboratory for critical discussion and the people who contributed to the generation of the publicly available SMRT sequencing data for the human lymphoblastoid cell lines. The work is partially funded by the seed grant (G.F.) from Icahn Institute for Genomics and Multiscale Biology, R01 GM114472 (G.F.) from National Institutes of Health, a Nash Family Research Scholar Award (G.F.) from the Friedman Brain Institute, and a pilot project (G.F.) funded by a P30 center grant ES023515 from the National Institutes of Health. This work was also supported in part through the computational resources and staff expertise provided by the Department of Scientific Computing at the Icahn School of Medicine at Mount Sinai.
Author contributions: G.F. conceived the project and supervised the research. S.Z. and G.F. designed the methods and experiments. S.Z. performed most of the computational analyses. G.D., T.P.W., M.S., Z.H., J.A.G., and R.S. conducted the wet laboratory experiments. G.D. and R.S. designed and conducted SMRT sequencing. S.Z, J.B., T.P.W., Z.H., G.L., A.C., C.H., A.X., R.S., E.E.S., and G.F. contributed to data analysis and interpretation. S.Z. and G.F. wrote the manuscript with input from all coauthors.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.231068.117.
References
- Babushok DV, Kazazian HH. 2007. Progress in understanding the biology of the human mutagen LINE-1. Hum Mutat 28: 527–539. [DOI] [PubMed] [Google Scholar]
- Bailey TL. 2011. DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics 27: 1653–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulaurier J, Zhu S, Sebra R, Zhang X-S, Rosenbluh C, Deikus G, Shen N, Munera D, Waldor MK, Blaser M, et al. 2015. Single molecule-level detection and long read-based phasing of epigenetic variations in bacterial methylomes. Nat Commun 6: 7438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow MJ, Clark TA, Daum CG, Deutschbauer AM, Fomenkov A, Fries R, Froula J, Kang DD, Malmstrom RR, Morgan RD. 2016. The epigenomic landscape of prokaryotes. PLoS Genet 12: e1005854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadesús J, Low D. 2006. Epigenetic gene regulation in the bacterial world. Microbiol Mol Biol Rev 70: 830–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadesús J, Low DA. 2013. Programmed heterogeneity: epigenetic mechanisms in bacteria. J Biol Chem 288: 13929–13935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Diaz N, Ecco G, Coluccio A, Kapopoulou A, Yazdanpanah B, Friedli M, Duc J, Jang SM, Turelli P, Trono D. 2014. Evolutionally dynamic L1 regulation in embryonic stem cells. Genes Dev 28: 1397–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling AC, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14: 1394–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, Peluso P, Rank D, Baybayan P, Bettman B. 2009. Real-time DNA sequencing from single polymerase molecules. Science 323: 133–138. [DOI] [PubMed] [Google Scholar]
- Fang G, Munera D, Friedman DI, Mandlik A, Chao MC, Banerjee O, Feng Z, Losic B, Mahajan MC, Jabado OJ. 2012. Genome-wide mapping of methylated adenine residues in pathogenic Escherichia coli using single-molecule real-time sequencing. Nat Biotechnol 30: 1232–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flusberg BA, Webster DR, Lee JH, Travers KJ, Olivares EC, Clark TA, Korlach J, Turner SW. 2010. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat Methods 7: 461–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Luo G-Z, Chen K, Deng X, Yu M, Han D, Hao Z, Liu J, Lu X, Doré LC. 2015. N6-Methyldeoxyadenosine marks active transcription start sites in Chlamydomonas. Cell 161: 879–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier JL, Kazazian HH. 2008. Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell 135: 23–35. [DOI] [PubMed] [Google Scholar]
- Greer EL, Blanco MA, Gu L, Sendinc E, Liu J, Aristizábal-Corrales D, Hsu C-H, Aravind L, He C, Shi Y. 2015. DNA methylation on N6-adenine in C. elegans. Cell 161: 868–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata K, Sakaki Y. 1997. Identification of critical CpG sites for repression of L1 transcription by DNA methylation. Gene 189: 227–234. [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW. 2009. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459: 108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heithoff DM, Sinsheimer RL, Low DA, Mahan MJ. 1999. An essential role for DNA adenine methylation in bacterial virulence. Science 284: 967–970. [DOI] [PubMed] [Google Scholar]
- Heng HH, Bremer SW, Stevens JB, Ye KJ, Liu G, Ye CJ. 2009. Genetic and epigenetic heterogeneity in cancer: a genome-centric perspective. J Cell Physiol 220: 538–547. [DOI] [PubMed] [Google Scholar]
- Huang F-P, Platt N, Wykes M, Major JR, Powell TJ, Jenkins CD, MacPherson GG. 2000. A discrete subpopulation of dendritic cells transports apoptotic intestinal epithelial cells to T cell areas of mesenteric lymph nodes. J Exp Med 191: 435–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jen FE-C, Seib KL, Jennings MP. 2014. Phasevarions mediate epigenetic regulation of antimicrobial susceptibility in Neisseria meningitidis. Antimicrob Agents Chemother 58: 4219–4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahramanoglou C, Prieto AI, Khedkar S, Haase B, Gupta A, Benes V, Fraser GM, Luscombe NM, Seshasayee AS. 2012. Genomics of DNA cytosine methylation in Escherichia coli reveals its role in stationary phase transcription. Nat Commun 3: 886. [DOI] [PubMed] [Google Scholar]
- Kozdon JB, Melfi MD, Luong K, Clark TA, Boitano M, Wang S, Zhou B, Gonzalez D, Collier J, Turner SW. 2013. Global methylation state at base-pair resolution of the Caulobacter genome throughout the cell cycle. Proc Natl Acad Sci 110: E4658–E4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koziol MJ, Bradshaw CR, Allen GE, Costa AS, Frezza C, Gurdon JB. 2016. Identification of methylated deoxyadenosines in vertebrates reveals diversity in DNA modifications. Nat Struct Mol Biol 23: 24–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laszlo AH, Derrington IM, Brinkerhoff H, Langford KW, Nova IC, Samson JM, Bartlett JJ, Pavlenok M, Gundlach JH. 2013. Detection and mapping of 5-methylcytosine and 5-hydroxymethylcytosine with nanopore MspA. Proc Natl Acad Sci 110: 18904–18909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lentini A, Lagerwall C, Vikingsson S, Mjoseng HK, Douvlataniotis K, Vogt H, Green H, Meehan RR, Benson M, Nestor CE. 2017. A reassessment of DNA immunoprecipitation-based genomic profiling. bioRxiv 10.1101/224279. [DOI] [PMC free article] [PubMed]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G-Z, Blanco MA, Greer EL, He C, Shi Y. 2015. DNA N6-methyladenine: a new epigenetic mark in eukaryotes? Nat Rev Mol Cell Biol 16: 705–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G-Z, Wang F, Weng X, Chen K, Hao Z, Yu M, Deng X, Liu J, He C. 2016. Characterization of eukaryotic DNA N6-methyladenine by a highly sensitive restriction enzyme-assisted sequencing. Nat Commun 7: 11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manrao EA, Derrington IM, Laszlo AH, Langford KW, Hopper MK, Gillgren N, Pavlenok M, Niederweis M, Gundlach JH. 2012. Reading DNA at single-nucleotide resolution with a mutant MspA nanopore and phi29 DNA polymerase. Nat Biotechnol 30: 349–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manso AS, Chai MH, Atack JM, Furi L, Croix MDS, Haigh R, Trappetti C, Ogunniyi AD, Shewell LK, Boitano M. 2014. A random six-phase switch regulates pneumococcal virulence via global epigenetic changes. Nat Commun 5: 5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Brown BD, Shay T, Gautier EL, Jojic V, Cohain A, Pandey G, Leboeuf M, Elpek KG, Helft J. 2012. Deciphering the transcriptional network of the dendritic cell lineage. Nat Immunol 13: 888–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondo SJ, Dannebaum RO, Kuo RC, Louie KB, Bewick AJ, LaButti K, Haridas S, Kuo A, Salamov A, Ahrendt SR. 2017. Widespread adenine N6-methylation of active genes in fungi. Nat Genet 49: 964–968. [DOI] [PubMed] [Google Scholar]
- Pak TR, Altman DR, Attie O, Sebra R, Hamula CL, Lewis M, Deikus G, Newman LC, Fang G, Hand J. 2015. Whole-genome sequencing identifies emergence of a quinolone resistance mutation in a case of Stenotrophomonas maltophilia bacteremia. Antimicrob Agents Chemother 59: 7117–7120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2013. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org/. [Google Scholar]
- Reedman BM, Klein G. 1973. Cellular localization of an Epstein-Barr virus (EBV)-associated complement-fixing antigen in producer and non-producer lymphoblastoid cell lines. Int J Cancer 11: 499–520. [DOI] [PubMed] [Google Scholar]
- Reiner A, Yekutieli D, Benjamini Y. 2003. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics 19: 368–375. [DOI] [PubMed] [Google Scholar]
- Sánchez-Romero MA, Cota I, Casadesús J. 2015. DNA methylation in bacteria: from the methyl group to the methylome. Curr Opin Microbiol 25: 9–16. [DOI] [PubMed] [Google Scholar]
- Schadt EE, Banerjee O, Fang G, Feng Z, Wong WH, Zhang X, Kislyuk A, Clark TA, Luong K, Keren-Paz A. 2013. Modeling kinetic rate variation in third generation DNA sequencing data to detect putative modifications to DNA bases. Genome Res 23: 129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber J, Wescoe ZL, Abu-Shumays R, Vivian JT, Baatar B, Karplus K, Akeson M. 2013. Error rates for nanopore discrimination among cytosine, methylcytosine, and hydroxymethylcytosine along individual DNA strands. Proc Natl Acad Sci 110: 18910–18915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster SC. 2008. Next-generation sequencing transforms today's biology. Nat Methods 5: 16. [DOI] [PubMed] [Google Scholar]
- Shulha HP, Cheung I, Guo Y, Akbarian S, Weng Z. 2013. Coordinated cell type–specific epigenetic remodeling in prefrontal cortex begins before birth and continues into early adulthood. PLoS Genet 9: e1003433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallwood SA, Lee HJ, Angermueller C, Krueger F, Saadeh H, Peat J, Andrews SR, Stegle O, Reik W, Kelsey G. 2014. Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat Methods 11: 817–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarailo-Graovac M, Chen N. 2009. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr Protoc Bioinformatics Chapter 4: Unit 4.10. [DOI] [PubMed] [Google Scholar]
- Wion D, Casadesús J. 2006. N6-methyl-adenine: an epigenetic signal for DNA–protein interactions. Nat Rev Microbiol 4: 183–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TP, Wang T, Seetin MG, Lai Y, Zhu S, Lin K, Liu Y, Byrum SD, Mackintosh SG, Zhong M. 2016. DNA methylation on N6-adenine in mammalian embryonic stem cells. Nature 532: 329–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young LS, Rickinson AB. 2004. Epstein–Barr virus: 40 years on. Nat Rev Cancer 4: 757–768. [DOI] [PubMed] [Google Scholar]
- Zhang G, Huang H, Liu D, Cheng Y, Liu X, Zhang W, Yin R, Zhang D, Zhang P, Liu J. 2015. N6-Methyladenine DNA modification in Drosophila. Cell 161: 893–906. [DOI] [PubMed] [Google Scholar]
- Zhou C, Liu Y, Li X, Zou J, Zou S. 2016. DNA N6-methyladenine demethylase ALKBH1 enhances osteogenic differentiation of human MSCs. Bone Res 4: 16033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zook JM, Catoe D, McDaniel J, Vang L, Spies N, Sidow A, Weng Z, Liu Y, Mason CE, Alexander N. 2016. Extensive sequencing of seven human genomes to characterize benchmark reference materials. Sci Data 3: 160025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.