Abstract
The relationship between evolutionary genome remodeling and the three-dimensional structure of the genome remain largely unexplored. Here, we use the heavily rearranged gibbon genome to examine how evolutionary chromosomal rearrangements impact genome-wide chromatin interactions, topologically associating domains (TADs), and their epigenetic landscape. We use high-resolution maps of gibbon–human breaks of synteny (BOS), apply Hi-C in gibbon, measure an array of epigenetic features, and perform cross-species comparisons. We find that gibbon rearrangements occur at TAD boundaries, independent of the parameters used to identify TADs. This overlap is supported by a remarkable genetic and epigenetic similarity between BOS and TAD boundaries, namely presence of CpG islands and SINE elements, and enrichment in CTCF and H3K4me3 binding. Cross-species comparisons reveal that regions orthologous to BOS also correspond with boundaries of large (400–600 kb) TADs in human and other mammalian species. The colocalization of rearrangement breakpoints and TAD boundaries may be due to higher chromatin fragility at these locations and/or increased selective pressure against rearrangements that disrupt TAD integrity. We also examine the small portion of BOS that did not overlap with TAD boundaries and gave rise to novel TADs in the gibbon genome. We postulate that these new TADs generally lack deleterious consequences. Last, we show that limited epigenetic homogenization occurs across breakpoints, irrespective of their time of occurrence in the gibbon lineage. Overall, our findings demonstrate remarkable conservation of chromatin interactions and epigenetic landscape in gibbons, in spite of extensive genomic shuffling.
The spatial organization of a genome and its chromatin interactions play a crucial role in mediating vital cellular functions, such as gene regulation (Lieberman-Aiden et al. 2009) and DNA replication (Pope et al. 2014). Intra-genomic interactions organize chromosomes into functional compartments called topologically associating domains (TADs). Within TADs, nearby loci (i.e., enhancers and genes) interact more frequently with each other than with regions located elsewhere in the genome (Dixon et al. 2012). Genes located in the same TAD are often coregulated, coexpressed, and display correlation in epigenetic marks of chromatin activity (Nora et al. 2012). Genes and regulatory elements in neighboring TADs are epigenetically and functionally insulated by TAD boundaries. The zinc-finger CCCTC-binding factor (CTCF) is thought to play a role in TAD boundary formation and mediation of long-range chromatin interactions; however, other factors, including the level of transcriptional activity and presence of retrotransposons, may also contribute to boundary formation (Dixon et al. 2012).
TADs appear to be largely conserved in their structure and organization across tissues and species, as the majority (53.8%) of TADs have been found to be evolutionarily conserved between human and mouse embryonic stem cells (Dixon et al. 2012). Moreover, visual pairwise comparisons of genome-wide chromatin conformation capture (Hi-C) and CTCF binding data between mouse, rhesus, dog, and rabbit show that overall TAD structure is maintained in syntenic regions (Vietri Rudan et al. 2015). Interestingly, conservation of TAD organization and structure appears to strongly relate to TAD size: Large TADs (>1 Mb) show higher conservation across cell types and species (Schmitt et al. 2016), whereas sub-TADs (100 kb–1 Mb) are more variable and contribute to cell- or species-specific gene regulation (Dixon et al. 2012; Phillips-Cremins et al. 2013; Rao et al. 2014). Despite recent advancement in the understanding of TAD structure and function, the mechanisms behind evolutionary TAD conservation have yet to be fully explored.
Chromosomal rearrangements (deletions, duplications, translocations, and inversions) are large-scale events that can drastically influence TAD integrity and organization during species evolution. Until now, studies that have examined the consequences of chromosomal rearrangements on TADs primarily focused on pathological structural variations and therefore were limited to discrete genomic regions. Using genetic engineering in patient cell lines and mouse models, these studies show that rearrangements that remove or misplace TAD boundaries can initiate ectopic interactions between genes and regulatory elements of neighboring TADs, leading to aberrant phenotypes and pathology (e.g., limb malformations, leukemia) (Gröschel et al. 2014; Lupianez et al. 2015; Franke et al. 2016; Hnisz et al. 2016; Dixon et al. 2017). However, there is a paucity of information on the evolution of genome topology including how long-distance DNA interactions and the epigenetic landscape of TADs change following evolutionary rearrangements. With the exception of one study (Vietri Rudan et al. 2015), previous comparative work mainly focused on broad comparisons of TADs between human and mouse and leaves out comparisons between more closely related species (Dixon et al. 2012; Rao et al. 2014). Moreover, these studies lack a detailed and quantitative assessment of the relationship between synteny breakpoints and the epigenetic structure of TADs.
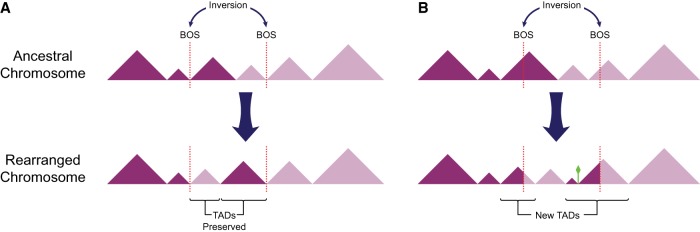
Effects of chromosomal rearrangements on TAD organization and gene regulation during genome evolution depend on the location of rearrangement breakpoints. Rearrangements occurring at domain borders will maintain TAD integrity, likely preserving gene regulation within these units (Fig. 1). However, these events might still modify the physical positioning of loci within the nucleus and perturb long-distance cis and trans interactions between nuclear territories (Harewood and Fraser 2014). Rearrangements that disrupt TADs by breaking the DNA within boundaries might generate new functional interactions and even new TADs (Fig. 1). These rearrangements can cause disease (Redin et al. 2017; Zepeda-Mendoza et al. 2017), making them likely to be eliminated by purifying selection because of their negative effect on fitness. However, a small portion of new TADs might contribute to new traits, giving rise to evolutionary novelties favored by selection.
Figure 1.
Relative position of breaks of synteny differentially affects TAD integrity. Alternative consequences of hypothetical ancestral inversions (top) are demonstrated. TADs are represented with triangles and positions of breaks of synteny (BOS) are depicted with dotted lines. (A) BOS occurring at TAD boundaries in the ancestral genome, rearrange TADs as intact modules. (B) BOS within TAD bodies disrupt ancestral TADs and may give rise to new TADs in the gibbon genome. Moreover, a new TAD boundary (green diamond) can emerge within an ancestral TAD.
Gibbons are particularly appealing experimental models for studying the relationship between TADs and evolutionary chromosomal rearrangements, as this family of apes have experienced an unusually high number of chromosomal rearrangements since their relatively recent divergence from humans (∼17 million years ago). In addition, a high-quality, well-annotated genome is available for the northern white-cheeked gibbon (Nomascus leucogenys, NLE) (Carbone et al. 2014) along with a high-resolution identification of evolutionary breaks of synteny with the human genome, validated by both DNA sequencing and molecular cytogenetics (Carbone et al. 2006, 2009a; Roberto et al. 2007; Girirajan et al. 2009; Capozzi et al. 2012). Finally, the high genetic identity between gibbon and human (∼96%) enables the use of many genetic, epigenetic, and computational tools designed for use in human studies and facilitates the comparison between the two species. In this study, we take advantage of the unique rearrangements that have occurred in the gibbon genome to examine the evolutionary consequences of heavy genome reshuffling on long-distance DNA interactions and integrity of TADs and their overall epigenetic landscape.
Results
Gibbon Hi-C interaction map highlights structural variations and recent genomic rearrangements
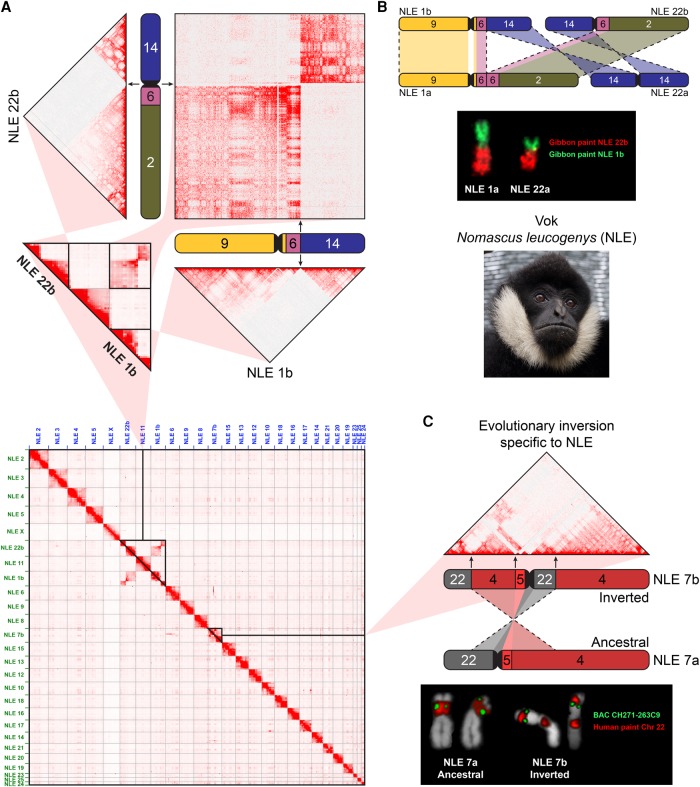
Whole-genome maps of chromatin interactions can be used to identify and study nonreference structural variations in a genome (Dixon et al. 2017; Harewood et al. 2017). To characterize the relationship between gibbon chromosomal rearrangements and TAD organization, we performed genome-wide chromatin conformation capture sequencing (Hi-C) using a lymphoblastoid cell line (LCL) that we established from a male northern white-cheeked gibbon (Nomascus leucogenys, Vok, #NLL600) belonging to the same species used to generate the gibbon genome reference (Supplemental Table S1; Carbone et al. 2014). Our Hi-C data exhibited evidence for a translocation between gibbon Chromosomes 1 and 22, which is a known polymorphic rearrangement in the genus Nomascus (Fig. 2A; Koehler et al. 1995). The individual used to generate the Hi-C data appears to carry the ancestral forms, NLE 1a (corresponding to human Chromosomes 9, 6, and 2) and NLE 22a (corresponding to human Chromosome 14), whereas the reference genome contains sequences for the derivative NLE 1b (corresponding to human Chromosomes 9, 6, and 14) and NLE 22b (corresponding to human Chromosomes 14, 6, and 2). In our Hi-C map, this nonreference translocation appears as strong inter-chromosomal interactions in conjunction with absence of intra-chromosomal interactions (Fig. 2A). We confirmed this translocation by fluorescent in situ hybridization (FISH) using whole-chromosome paints for NLE 1b and NLE 22b on Vok chromosome spreads (Fig. 2B; Supplemental Fig. S1).
Figure 2.
Gibbon Hi-C map highlights nonreference chromosomal rearrangements. (A, bottom) Whole-genome Hi-C interaction matrix is shown for the gibbon named Vok (photo shown on the right) aligned to the reference gibbon genome (Nleu3.0). (Top) A close-up view of the Hi-C data corresponding to the reciprocal translocation between gibbon Chromosomes 1 and 22 shows a lack of intra-chromosomal interactions in the regions mobilized by the rearrangement, and stronger-than-expected inter-chromosomal interactions. (B, top) A scheme demonstrates the reciprocal translocation that formed NLE1a/22a (present in Vok) from the ancestral NLE 1b/22b (present in reference). (Bottom) FISH validation of the rearrangement on Vok chromosomes with chromosome paints for NLE 1b (green) and NLE 22b (red). (C, top) Hi-C matrix for NLE 7b displays an example of “ghost interactions” between regions corresponding to human Chromosome 22 separated by a pericentromeric inversion of the ancestral NLE 7a. (Bottom) FISH with the chromosome paint for human Chr 22 (red) shows a split signal indicative of the inversion in Vok, but not the sister taxa Nomascus gabriellae. (Gibbon chromosomes are labeled outside of chromosome ideograms, and corresponding human chromosomes are color coded and indicated within ideograms.)
Other interaction signals suggestive of nonreference translocations were also observed. For instance, we identified strong interactions between a 2-Mb “strip” on NLE 7b (Chr 7b: 39,750,000–42,500,000) and the entire NLE 6 (Supplemental Fig. S2A), whereas the same strip barely interacted with other regions in cis. Similar patterns of unusual inter-chromosomal interaction were found in Hi-C data of rhesus, rabbit, and dog (Supplemental Fig. S2B). It should be noted, however, that these putative translocations were not experimentally validated and may simply reflect errors in the genome assemblies.
We observed that genomic regions that were syntenic in the ancestral chromosome sometimes maintained their interaction even after loss of synteny occurred in gibbon due to rearrangement. These “ghost interactions” were mainly detected in recent evolutionary rearrangements with the most prominent case being present in gibbon Chromosome NLE 7b, whose current structure resulted from the most recent rearrangement in the genus Nomascus. This rearrangement, which is a pericentromeric inversion of the ancestral form (NLE 7a) (Fig. 2C) is a known evolutionary fixed rearrangement first identified by karyotyping (Couturier and Lernould 1991) and subsequently tested for presence/absence in about 50 gibbon individuals (Carbone et al. 2009b). This inversion has been found and experimentally validated in all northern white-cheeked gibbons tested so far, including Vok (Carbone et al. 2009b) (Fig. 2C), but appears to be absent in the southern white-cheeked gibbon; hence, its occurrence can be dated to roughly 2 million years ago (mya). Our Hi-C interaction data showed that regions of NLE 7b corresponding to human Chromosome 22 still interact strongly with one another despite being separated on different chromosome arms in the new genomic arrangement. We also observed a few cases of weaker, yet visually evident, inter-chromosomal ghost interactions across other chromosomes. Namely, we observed stronger-than-background interactions between portions of NLE 14 and NLE 19 homologous to human Chromosome 17. Moreover, some regions that were broken up by recent evolutionary rearrangements still display similar patterns of interaction with other chromosomes in trans. For example, the chromosomal segments in NLE 7b that are homologous to human Chromosome 22 all display strong inter-chromosomal interactions with NLE 14 and NLE 6, whereas overall, the rest of NLE 7b exhibits weaker interaction with these chromosomes (Supplemental Fig. S2A).
Gibbon–human breaks of synteny colocalize with gibbon TAD boundaries
We define a break of synteny (BOS) as a locus of the gibbon genome whose 5′ and 3′ ends are homologous to different, nonsyntenic segments of the human genome (Supplemental Table S2). Each gibbon BOS therefore corresponds to two distinct regions in the human genome, either located on the same chromosome (in the case of inversions) or on two different human chromosomes (in the case of translocations, fissions, or fusions). The human genome was selected to represent the ancestral hominoid genome because of its unmatched quality among primate genome assemblies. The rhesus macaque genome was used to discriminate human-specific and great ape-specific from gibbon-specific rearrangements (Carbone et al. 2006). To compensate for the lower quality of the rhesus genome and possible assembly errors, we also extensively relied on cytogenetics data gathered on this species (http://www.biologia.uniba.it/macaque/) to validate our predictions.
We characterized a total of 67 gibbon-specific BOS. Among these, 33 were identified at single base-pair resolution, whereas the remaining 34 BOS contained insertions of repetitive elements of variable sizes that lack synteny with the human genome and were therefore defined as “intervals.” All these BOS regions have been previously described and validated by FISH and Sanger sequencing (Carbone et al. 2009a; Girirajan et al. 2009) and represent a highly curated set that excludes ambiguous mapping or genomic regions with assembly issues.
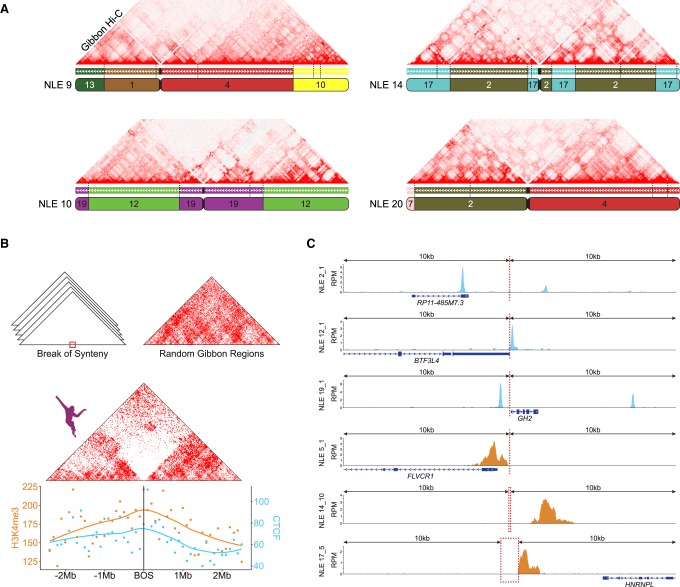
Alignment of gibbon Hi-C contact maps against the UCSC Genome Browser gibbon–human chain allowed us to visually inspect the correspondence between BOS and genomic interactions. We observed an overall reduction of chromatin interactions across gibbon BOS (Fig. 3A). To examine whether this was a general pattern, we averaged Hi-C data surrounding all BOS (±2.5 Mb) into a single two-dimensional contact map using HOMER (Fig. 3B; Heinz et al. 2010). We found a clear reduction in interactions between the two sides of the overlaid BOS, a pattern characteristic of TAD boundaries. Repeating this process with randomly generated regions with the same chromosome distribution and size of the gibbon BOS did not produce a similar signal (Fig. 3B).
Figure 3.
Gibbon–human breaks of synteny display epigenetic signatures of TAD boundaries. (A) As examples, Hi-C matrix for four representative gibbon chromosomes (NLE 9, 10, 14, and 20) are shown along with their corresponding gibbon–human chain from the UCSC Genome Browser. Corresponding human chromosomes are color coded and labeled within each gibbon chromosome ideogram. Positions of all gibbon–human BOS sites are marked on the chain track with vertical dashed lines and demonstrate that chromosomal interactions are often reduced across BOS. (B, center) Averaged interaction maps show juxtaposition of the gibbon Hi-C signal from regions flanking all gibbon–human BOS (±2.5 Mb) and flanking random genomic regions (top right corner). Overall, chromatin contacts are highly depleted across BOS, but not random regions. (Bottom) CTCF and H3K4me3 ChIP-seq peak counts with smoothed Loess curves in 100-kb bins across the BOS (±2.5 Mb) show enrichment of these epigenetic marks at BOS. (C) Examples CTCF (blue) and H3K4me3 peaks (orange) in a 20-kb window around BOS: (RPM) reads per million mapped reads.
TAD boundaries are occupied by CTCF and epigenetic marks of active transcription (i.e., H3K4me3) (Dixon et al. 2012). Therefore, if gibbon BOS colocalize with TAD boundaries, we expect to observe the same epigenetic marks in these regions. To this end, we analyzed newly generated H3K4me3 ChIP-seq data for Vok and the gibbon used for the reference genome (Asia) and used previously published gibbon CTCF ChIP-seq data (Carbone et al. 2014). Although more significant for CTCF, we found significant enrichment for both marks within 20 kb of gibbon BOS (two-sided permutation P-values, CTCF: <0.001; H3K4me3: 0.034) (Fig. 3B,C). We also found that CpG density in the 20 kb surrounding the gibbon BOS is significantly higher than the rest of the genome (two-sided permutation P-value: <0.001) (Supplemental Methods; Supplemental Figs. S3–S6) and that BOS are significantly enriched in CpG islands, CpG shores, and SINE elements (Supplemental Fig. S6). These genomic features were previously described as characteristic of TAD boundaries in human and mouse (Dixon et al. 2012; Sun et al. 2017).
Next, we used our Hi-C data to computationally predict the position of gibbon TAD boundaries and examine if they significantly overlap with BOS. Since size and position of predicted TAD boundaries can vary depending on the algorithm and parameters used, we predicted TADs with 140 different parameter combinations (seven matrix resolutions, five cutoffs, and four window sizes) (Supplemental Methods) using the directionality index method (Dixon et al. 2012) and TADtool (Kruse et al. 2016). As expected, we observed that parameter choices greatly impacted the location and size of TAD boundaries (Supplemental Fig. S7). To quantify the overlap of TAD boundaries with BOS in an unbiased way, for each set of parameters we performed permutation analyses comparing the number of BOS-TAD boundary overlaps to the number of overlaps of TAD boundaries with 10,000 sets of random regions that had the same size and chromosomal distribution as the BOS regions. Independent of the window size and cutoff, we found a significant correspondence between gibbon BOS and TAD boundaries for all resolution values (permutation P-values <0.001), except the two resolution extremes (10 kb and 1 Mb) (Supplemental Fig. S8).
Regions orthologous to gibbon BOS colocalize with boundaries of larger TADs in other mammalian species
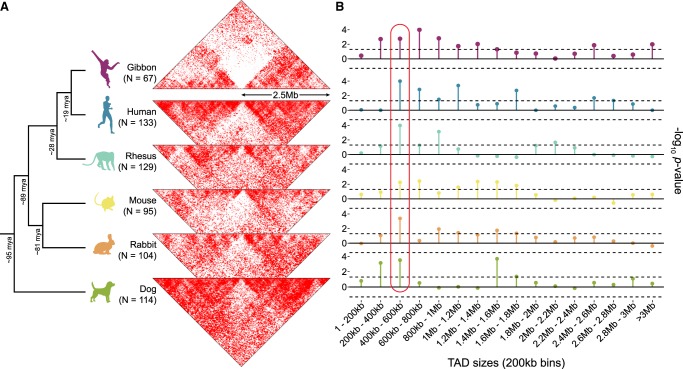
To better understand the evolutionary relationship between gibbon TAD boundaries and chromosomal rearrangements, we examined genomic regions orthologous to gibbon BOS (Supplemental Table S2) and TAD structures in five other species (human, rhesus, mouse, dog, and rabbit) using publicly available, species-specific Hi-C data (Grubert et al. 2015; Vietri Rudan et al. 2015). First, we normalized and overlaid the Hi-C data from these species to examine DNA interactions ±2.5 Mb around loci orthologous to gibbon BOS (Fig. 4A).
Figure 4.
Evolutionary context of the overlap between TAD boundaries and BOS. (A) The two-dimensional gibbon Hi-C histogram (Fig. 3B) is compared with Hi-C histograms for five other mammalian species at loci orthologous to the gibbon BOS (±2.5 Mbp). Decreased contact density across these loci in non-gibbon species suggests that breakpoint regions in gibbon are more likely to be TAD boundaries in other species. (N) Number of breakpoints that successfully lifted over from the gibbon genome to each species. (B) Lollipop plots show −log10 P-values from permutation analyses testing the overlap between gibbon BOS and TADs binned by size. This cross-species comparison points to consistently significant overlap of BOS with boundaries of 400–600 kb TADs (circled in red). Dotted lines mark P = 0.05 significance threshold (no multiple-test correction).
Since each side of a gibbon BOS maps to a disjointed ancestral region, there were roughly twice as many regions composing each map, with some regions missing, because the liftOver tool failed to identify syntenic regions from the gibbon genome. All species showed a distinct reduction in contacts across the sites orthologous to gibbon BOS, suggesting that loci where rearrangements occurred in gibbon are more likely to be TAD boundaries in other species (Fig. 4A).
To predict TAD boundaries in all five species, we used TADtool (Kruse et al. 2016) with the 140 different parameter combinations previously used in gibbons. We detected a significant overlap between TAD boundaries and regions orthologous to gibbon BOS across all species for the majority of parameter combinations (Supplemental Fig. S8). As expected, the association was more consistent in the gibbon genome, which is likely due to independent species-specific remodeling events in each lineage. The weakest overlap was observed in dog, possibly due to the lower Hi-C library complexity for this species (Supplemental Table S1).
Several studies indicate that larger TADs (>1 Mb) show more conservation across species and tissues, whereas smaller subdomains (100 kb–1 Mb) appear more dynamic and change during development (Dixon et al. 2012; Nora et al. 2012; Phillips-Cremins et al. 2013). To facilitate comparison across species and establish the relationship between BOS and boundaries based on TAD size, we repeated our permutation analyses after grouping TADs by size, rather than by parameter combinations (Supplemental Methods). First, TADs were grouped based on size (1 bp–50 kb, 50–100 kb, 100–250 kb, 250–500 kb, 500 kb–1 Mb, 1–2.5 Mb, and >2.5 Mb) in groups that had roughly the same number of TADs. We found boundaries of TADs in the 500 kb–1 Mb bin overlapped with gibbon BOS most often, and this was true for regions orthologous to BOS in all other species, except dog (Supplemental Fig. S9). After increasing the resolution of our comparisons by binning TADs into smaller fixed-size bins (incrementing by 200 kb), we observed the most significant overlap between BOS and boundaries of TADs in the 400–600 kb bin (Fig. 4B; Supplemental Fig. S10).
Additionally, we used permutation tests to ascertain whether the TAD boundaries that overlap with BOS are largely the same across species. We found that TAD boundaries that overlap with BOS were more conserved than would be expected by chance for the two TAD size groups that we investigated (500 kb–1 Mb and 400 kb–600 kb) (Supplemental Methods; Supplemental Fig. S11). Focusing on just 500 kb–1 Mb TADs, we identified 19 loci orthologous to gibbon BOS that overlap with TAD boundaries in all species. Across the non-gibbon species, these loci correspond to 15 regions, since eight of the 19 BOS derive from reciprocal rearrangements in gibbon and thus map to adjacent locations in non-gibbon species. The 211 genes within ±500 kb of these highly conserved TAD boundaries include genes with essential developmental functions (Supplemental Table S3). The misregulation of many of these genes has been implicated in lethality in mouse knockout models and human disorders. Among these is LYPD6, whose product is involved in Wnt/beta catenin signaling and has been found duplicated (microduplication of 2q23.1) or disrupted in autism and other congenital disorders characterized by severe intellectual disabilities (Chung et al. 2012; Nilsson et al. 2017). This gene set also includes GGN (gametogenetin), whose complete loss leads to embryonic lethality at the very early stages of preimplantation due to compromised meiotic double-strand break (DSB) repair (Jamsai et al. 2013). In addition, a recent study reported disruption of three of the highly conserved TAD boundaries, which encompasses 36 of the 211 genes in three patients with congenital disorders, including severe developmental impairment (Redin et al. 2017). Furthermore, 161 of the 211 genes (73.6%) are found in a recently curated list of 3455 disease-associated genes from the Online Mendelian Inheritance of Man (OMIM) (Dickinson et al. 2016), with 34 (21.11%) of them associated with a disease phenotype. Diseases in this list included retinitis pigmentosa (genes IFT172, ZNF513, and RHO), gastrointestinal defects (TTC7A), and Charcot–Marie–Tooth disease, type 1C (PMP22 and LITAF) (Supplemental Table S3).
Large new TADs are rarely generated by evolutionary chromosomal rearrangements
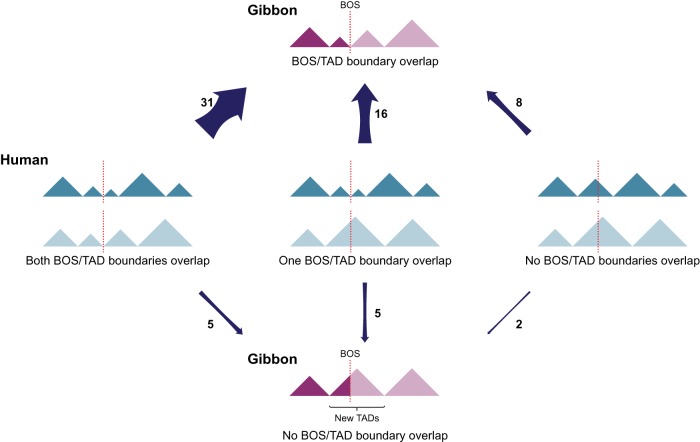
Chromosomal rearrangements have the potential to break existing TADs and generate new ones. Although this phenomenon was recently observed in cancer genomes (Dixon et al. 2017), it has not, to our knowledge, been explored in an evolutionary setting. There are six possible scenarios when the ancestral and current position of rearrangement breakpoints and TAD boundaries are taken into account. In the case in which a BOS colocalizes with a TAD boundary in gibbon, the two sites in the ancestral chromosome could have occurred (1) both in boundaries, (2) both inside TAD bodies, or (3) one in a boundary and the other inside a TAD body. The same three scenarios can apply when a BOS does not overlap with a TAD boundary in gibbon (Fig. 5). When looking at boundaries of TADs sized 500 kb–1 Mb, we find that 55 of 67 BOS overlap with TAD boundaries in gibbon; for 31 of these BOS, both sides colocalize with TAD boundaries in the human genome, which represents the ancestral state. This proportion is significantly higher than any of the other scenarios (two-sided McNemar's test, P = 0.0005) and indicates that evolutionary rearrangements are skewed toward maintaining TADs as intact units.
Figure 5.
Alternative evolutionary relationship between BOS and TADs. Schematics show all possible scenarios between BOS (dotted lines) and TADs (triangles) in the gibbon (purple) and human (ancestral, blue) genomes. Arrow width reflects prevalence of the scenario in the gibbon genome, and the number beside the arrow represents the number of occurrences of each scenario.
Of the remaining 24 BOS that overlap with TAD boundaries in gibbon, 16 have evolved from breaking DNA at one ancestral TAD boundary and one TAD body. All other four evolutionary possibilities have occurred much less often (Fig. 5).
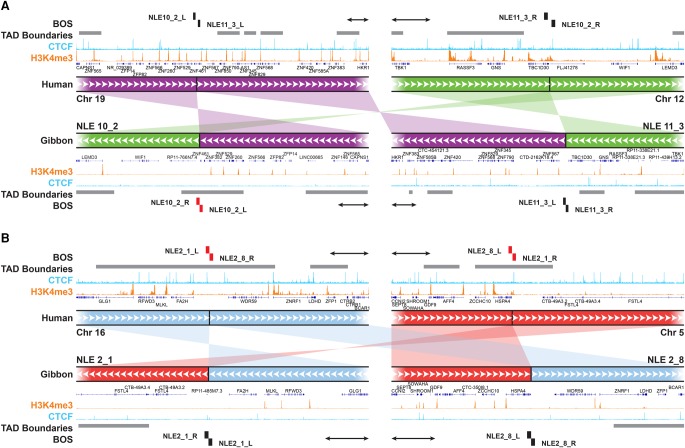
Our findings show that it is quite uncommon for gibbon BOS to occur within TAD bodies, away from boundaries. In total, we only found 12 BOS that did not colocalize with TAD boundaries in the gibbon genome (Fig. 5). Of these, seven BOS were located on three gibbon chromosomes (NLE 8, NLE 11, and NLE 13) and four of these seven are results of reciprocal translocations. We found only two gibbon BOS (NLE 11_3 and NLE 8_3) that did not overlap with boundaries in both gibbon and human. These two rearrangements have broken ancestral TADs and created new ones in the gibbon genome. One of these events involves gibbon Chromosomes NLE 10 and NLE 11, which originated from a reciprocal translocation between ancestral chromosomes corresponding to human Chromosomes 12 and 19 (Fig. 6A). On human Chromosome 19, this rearrangement broke a cluster of highly transcribed Kruppel-associated box (KRAB)-containing zinc-finger (ZNF) protein genes (Dai et al. 2003), bringing a portion of the ZNF cluster together with TBC1D30, GNS, and RASSF3 within a gibbon TAD (Fig. 6A). Because the breakpoint on NLE 11 does not overlap with a boundary, this rearrangement could cause the regulatory landscape of the ZNF cluster to merge with one of the downstream genes (GNS and RASSF3). However, this effect may have been mitigated by an ancestral TAD boundary marked by a CTCF binding event upstream of the BOS (Fig. 6A). Looking at the reciprocal rearrangement formed by this event, our analysis revealed that the breakpoint on NLE 10 (NLE 10_2) corresponded to a new TAD boundary (i.e., present in gibbon, but not in human or rhesus) that insulates the active ZNF cluster from other genes deriving from human Chromosome 12 (WIF1 and LEMD3). In five of the 67 BOS, we found breakpoints that occurred at ancestral (human) TAD boundaries but that are not boundaries in the gibbon genome. By generating new gibbon-specific TADs, these types of rearrangements might lead to functional novelty. When inspecting the Hi-C data, we found that three of these BOS (NLE 2_8, NLE 20_1, and NLE 11_4) have a TAD boundary located nearby. One of these cases is the reciprocal translocation between chromosomes corresponding to human 16 and 5, followed by an inversion that gave rise to NLE 2 (Fig. 6B). The breakpoints of this inversion (NLE 2_1 and NLE 2_8) do not perfectly colocalize with TAD boundaries in gibbon, but NLE 2_8 is very close to the next boundary (∼30 kb) with no genes present in between. In the other two instances (NLE 14_13 and NLE 2_1), a new gibbon TAD has been created. Overall, these observations demonstrate that new TADs have rarely been generated in the gibbon and suggest that new TAD boundaries may have emerged to prevent ectopic interactions and gene misregulation when TAD structures are disrupted (Lupianez et al. 2016; Dixon et al. 2017).
Figure 6.
New TADs and TAD boundaries can emerge from genomic rearrangements. (A) An example of a reciprocal translocation and inversion whose breakpoints (NLE 10_2 and NLE 11_3) do not overlap with TAD boundaries (gray horizontal bars) in human (top tracks). Within the gibbon genome (bottom tracks), breakpoint NLE 11_3 (right) maps within a TAD body, nearby an ancestral boundary, and NLE 10_2 breakpoint (left) corresponds to a new gibbon-specific boundary on NLE 10. ChIP-seq pileups for H3K4me3 (orange) and CTCF (light blue) are shown for human and gibbon. (B) Example of reciprocal translocations in which breakpoints (NLE 2_1 and NLE 2_8) are both within TAD boundaries in human (top tracks) and but not in gibbon (bottom tracks). A new TAD was created by the rearrangement involving NLE 2_1. Gray horizontal bars represent TAD boundaries of every computationally predicted TAD falling into the 500 kb–1 Mb size range. BOS overlapping with boundaries are marked in red. All scale bars represent 100 kb.
The chromatin on the two sides of gibbon BOS maintain their ancestral epigenetic identity
Few studies have explored the extent of epigenetic remodeling that occurs after evolutionary chromosomal rearrangements. We asked whether the two sides of gibbon BOS display different epigenetic states, as may be expected from distal nonrelated chromatin or have undergone epigenetic homogenization after becoming contiguous on the rearranged chromosome. To characterize the epigenetic landscape of gibbon BOS, we generated whole-genome bisulfite sequencing (WGBS) data from whole blood of Vok (Supplemental Methods). To normalize for effects of CpG density, we calculated residual methylation after fitting logistic functions to methylation, given CpG density (Supplemental Fig. S4). We observed large differences in the distribution of CpG methylation, CpG density, and H3K4me3 peaks between the two sides of BOS, often with a sharp switch occurring at the breakpoint (Fig. 7A).
Figure 7.
Gibbon BOS maintain their ancestral epigenetic identity and resemble nonsyntenic regions. (A) Examples of BOS showing a noticeable difference in CpG density (green track) and methylation (black track) between the two sides of the rearrangement with the switch occurring at the BOS (black blocks). Homology with the human chromosomes is shown below each BOS. Gibbon-specific repeats within the breakpoint explain the gap with the human alignment. (NLE) Nomascus leucogenys; (Meth) methylation. (B) Ranked barbell plots show the difference in residual methylation and CpG density between the two sides of each of the gibbon BOS. Each point represents a BOS side, and a line segment joins the two sides from the same BOS. BOS are ordered vertically by magnitude of the difference between sides. Black lines on the left show the rank associated with percentiles of distal permutation regions, whereas blue lines on the right show ranks for percentiles for adjacent permutation regions. Color-coding by age of the rearrangement highlights that old BOS (5–18 mya) are as likely as young ones (<5 mya) to show a large difference between the two sides. (C) Scatterplots of Δ residual methylation (left) and Δ CpG density (right) between gibbon and human BOS regions; each point represents one BOS. The line shows a least-squared linear regression, and the points are color-coded as in B.
To quantify this observation, we calculated the absolute difference in mean residual CpG methylation and CpG density (Δmeth and ΔCpG) between the two sides of each BOS (±10 kb) and used permutation analyses to evaluate whether the Δ values observed in BOS were greater than would be expected by chance. We performed the same analysis for H3K4me3 peaks (ΔH3K4me3), but with a larger window around BOS (±500 kb) to account for the lower density of the peaks (Supplemental Methods; Supplemental Fig. S12). We found that despite being contiguous on gibbon chromosomes, the two sides of BOS are significantly different in their epigenetic characteristics and behave like regions distantly located in the gibbon genome (Supplemental Table S4). Leveraging WGBS data already available for human (Hernando-Herraez et al. 2015), and generated in-house for rhesus, we used regions orthologous to the gibbon BOS to in silico reconstruct the arrangements found in gibbon and repeated the permutation analysis in these species. As expected, in both human and rhesus, CpG density and DNA methylation between the two sides of the constructed BOS look as different as distal regions (Supplemental Table S4). To test whether the epigenetic landscape across BOS homogenizes with time, we split gibbon BOS based on their cytogenetically determined age (Capozzi et al. 2012) into ancestral (5–18 mya old) or Nomascus-specific (<5 mya old) when possible. We found no clear relationship between age of BOS and difference in residual methylation and CpG density across BOS, as older BOS also show large absolute Δmeth and ΔCpG values (Fig. 7B).
Very similar results were obtained for human and rhesus (Supplemental Fig. S13A), and the amount of epigenetic difference measured for each BOS is highly correlated between species (Pearson correlations for gibbon versus human ΔCpG: 0.57, Δmeth: 0.31; gibbon versus rhesus ΔCpG: 0.70, Δmeth: 0.44), suggesting that the epigenetic differences across BOS is inherited from the primate common ancestor (Fig. 7C; Supplemental Fig. S13B). Overall, these results indicate that chromosomal regions largely maintain their ancestral epigenetic landscape after rearrangement, and that physical contiguity at BOS does not lead to epigenetic homogenization over time.
Discussion
Recent studies demonstrate how structural variations in the genome can alter DNA interaction and topologically associating domains (TADs) in the context of pathology and disease (Lupianez et al. 2015; Franke et al. 2016; Hnisz et al. 2016; Nilsson et al. 2017; Sun et al. 2017). However, the consequences of chromosomal rearrangements during genome evolution have not been extensively explored. In this study, we used a high-resolution map of gibbon–human breaks of synteny (BOS) from the heavily reshuffled gibbon genome and genetic and epigenetic data to thoroughly examine the evolutionary relationship between chromosomal rearrangements and structure of TADs.
Using gibbon Hi-C data, we visualized genome-wide DNA interactions and detected multiple nonreference rearrangements, the most evident of which was experimentally validated and represented a known polymorphic translocation (Fig. 2A; Koehler et al. 1995). We also observed unusual patterns of interactions that we called “ghost interactions.” Ghost interactions broadly refer to interaction patterns that are better explained by the previous chromosomal synteny (before an evolutionary rearrangement), rather than the current chromosomal arrangement. The most prominent case was observed at an experimentally validated and evolutionary recent (∼2 mya) pericentromeric inversion in NLE 7 (Fig. 2C; Supplemental Fig. S2C). Ghost interactions, may simply reflect a passive preservation of the physical conformation of chromatin compartments in 3D, as described in Lieberman-Aiden et al. (2009). In this case, the overall similarity in chromatin state (active versus inactive) may be sufficient to bring the recently separated linear genome together in the nucleus and preserve the 3D structure. Alternatively, we speculate that ghost interactions might, at least partially, represent functional long-distance DNA interactions that are actively preserved as a transitional solution to maintain important regulatory networks after a large-scale genomic rearrangement has occurred. If so, these interactions may later be lost or replaced as novel nearly regulatory interactions evolve. Although the biological and evolutionary implications of ghost interactions are not clear yet, it is a compelling observation, and future analysis of polymorphic rearrangements and recent evolutionary rearrangements in other species may help shed light on this phenomenon and its biological implications.
We next used our Hi-C interaction data to computationally predict TADs in the gibbon genome and compare their location relative to regions of gibbon–human BOS. We found that BOS consistently and significantly colocalize with TAD boundaries in the gibbon genome, and that this association was present independent of the parameters used to predict gibbon TADs (Supplemental Fig. S8). Furthermore, gibbon BOS showed significant enrichment of genetic and epigenetic signatures of TAD boundaries, including higher CpG density than the rest of the genome, enrichment in CTCF binding, H3K4me3, and presence of SINE elements (Fig. 3B; Supplemental Fig. S6). In most cases, the two sides of the gibbon BOS correspond to TAD boundaries in the inferred ancestral hominoid state (Fig. 5), demonstrating that most TADs were maintained as intact modules during and after rearrangement. This overlap was, however, observed more often for larger TADs (500 kb–1 Mb), suggesting that smaller sub-TADs are more variable during evolution (Fig. 4B; Supplemental Fig. S9). Leveraging publicly available data, we also showed that large TADs that overlap with BOS are often conserved across gibbon, human, rhesus macaque, mouse, dog, and rabbit (Fig. 4B; Supplemental Fig. S10); therefore, they preexisted and survived genome remodeling in gibbon. It should be noted that although the human and gibbon Hi-C data both originated from lymphoblastoid cell lines, the rhesus macaque, mouse, dog, and rabbit data were obtained from liver tissue, and tissue-specific differences may exist at fine scales and within some small TADs. However, global interaction patterns and large TADs, which were the main focus of our study, are generally conserved across tissues (Dixon et al. 2012, 2015; Schmitt et al. 2016). Therefore, we do not expect our findings to have been influenced by variation in tissue source.
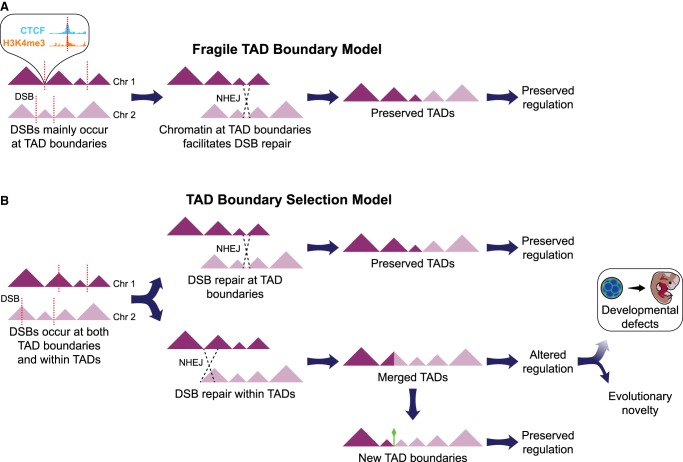
Two non-mutually exclusive models can explain the striking colocalization of BOS and TAD boundaries. The “fragile TAD boundary” model predicts that the distinct genetic and epigenetic properties of TAD boundaries along with their open and transcriptionally active chromatin state, elicit a higher rate of DNA double-strand break (DSB) and repair (Fig. 8; Smerdon 1991). In support of this model, open chromatin and the epigenetic marks found at TAD boundaries (e.g., H3K4me3 and CTCF) have been linked to chromatin fragility in human disease (De and Michor 2011; Li et al. 2012; Tchurikov et al. 2015), species evolution (Carbone et al. 2009a; Lemaitre et al. 2009), and simulation analyses (Berthelot et al. 2015). Moreover, an in vitro study recently showed that chromosome loop anchors bound by CTCF and cohesin are vulnerable to DSBs mediated by topoisomerase 2B (TOP2B) and act as fragile sites for chromosomal rearrangements (Canela et al. 2017). Thus, higher frequency of breakage and repair at TAD boundaries relative to other genomic regions could explain the skewed distribution of rearrangements in gibbon and other species. Under this model, we would also expect to observe a similar trend in somatic (e.g., cancer cell lines) and germline rearrangements in the human population. Indeed, it was recently discovered that nearly all short tandem repeats (STRs) linked to repeat expansion diseases colocalize with TAD boundaries, and their expansion severely compromises those boundaries in Fragile X syndrome and Huntington's disease (Sun et al. 2017). In the “TAD boundary selection” model, DSBs are predicted to occur equally at TAD boundaries and within TAD bodies. However, DSBs that colocalize with boundaries and maintain boundary function are more likely to become evolutionarily fixed, whereas DSBs elsewhere likely reduce fitness and are eliminated by purifying selection (Fig. 8). In agreement with this model, we presented evidence that TAD boundaries that colocalize with BOS have maintained their insulating function and prevented epigenetic homogenization between the two sides of the rearrangements in the gibbon genome (Fig. 7A,B). We also revealed that highly conserved boundaries are located near important developmental genes, suggesting that rearrangements that cause misregulation of these genes were likely negatively selected. Consistent with this observation, a recent study showed that both evolutionary fixed deletions in the great apes and deletions segregating in the human population are depleted at TAD boundaries, whereas deletions implicated in disease, such as neurodevelopmental disorders, do not show this constraint (Fudenberg and Pollard 2018). Overall, our findings suggest that both models may have been at play during the evolution of the gibbon genome, but further studies are required to test these paradigms, for example, by directly examining TAD boundary fragility in vitro (Canela et al. 2017) and testing selection models across different species with rearranged genomes.
Figure 8.
Two models to explain colocalization of BOS and TAD boundaries in genome evolution. (A) Based on the “fragile TAD boundary” model, TAD boundaries carry epigenetic marks associated with DNA double-strand break (DSB, red dotted lines) and repair. DSBs will therefore occur more frequently at TAD boundaries than at other genomic regions and have a higher chance to be repaired and evolutionarily fixed. (B) The “TAD boundary selection” model assumes that DSBs occur equally at TAD boundaries and within TADs. However, rearrangements altering TAD structure by misplacing or deleting TAD boundaries are lost through purifying selection, whereas those maintaining TADs intact are more likely to become evolutionarily fixed. In a small portion of the cases, new TAD boundaries might emerge (green diamond) and survive in the population.
Although the majority of our gibbon BOS colocalize with TAD boundaries, a small subset of the rearrangements created new TADs that were evolutionarily fixed. Many of these new gibbon TADs appear to mitigate ectopic regulation by establishing new boundaries to prevent interactions between the newly joined genomic regions, especially when they present very different regulatory landscapes. For example, on gibbon Chromosome 10, genes WIF1 and LEMD3 were joined by a reciprocal translocation to a transcriptionally active cluster of zinc-finger genes, and a new TAD boundary was formed in gibbon nearby the BOS (Fig. 6A). This new boundary might prevent ectopic activation of LEMD3 whose mutations have been associated with skeletal dysplasia and collagen-type nevi (Buschke-Ollendorff syndrome) and rare mesenchymal dysplasia (Melorheostosis) (Debeer et al. 2003; Hellemans et al. 2004; Fischetto et al. 2017). Alternatively, new TADs might become evolutionarily fixed because they are advantageous. For instance, domain reorganization may bring distant, coregulated genes into the same TAD and facilitate their coordinated expression (Schoenfelder et al. 2010; Harewood and Fraser 2014). We made some anecdotal observations in support of this scenario. On gibbon Chromosome 8, a new TAD originated from a translocation that joined a potassium channel gene (KCNH8) and a vacuolar ATPase (ATP6V1B2), both highly expressed in the human brain (The GTEx Consortium 2013). Mutations in ATP6V1B2 cause a developmental disorder known as Zimmermann-Laband syndrome; the same phenotype is also observed when mutations occur independently in the gene KCNH1 (Kortüm et al. 2015). The overlap in clinical features for these two physically distant genes with apparently unrelated functions was explained by the fact that the two proteins have coordinated action: KCNH1 provides K+ as a counter ion that is needed by ATP6V1B2 to pump protons and achieve acidification of intracellular vacuoles (Kortüm et al. 2015). We therefore speculate that similar coordinated functions exist between KCNH8 (whose function and expression are very similar to KCNH1) and ATP6V1B2, and colocalization of the corresponding two genes in the new TAD in gibbon has facilitated their coexpression. Similarly, we observed joining of two genes of the RAS oncogene GTPases (RAB37 and RAB40B) in a new TAD on NLE 14, and two genes (HMX1 and ADRA2C) implicated in regulating the function of sympathetic neurons (Furlan et al. 2013) in a new TAD on NLE 20 (Supplemental Fig. S14). Overall, these preliminary observations suggest that new TADs that either mitigate ectopic interaction or facilitate coexpression of genes with related function are more likely to be tolerated and fixed in the genome.
To determine if any epigenetic remodeling and homogenization occurs in the genome after chromosomal rearrangements, we compared aspects of the epigenetic landscape across the gibbon BOS. We measured both residual DNA methylation and H3K4me3 distribution, as indicators of repressed and active chromatin, respectively, and found large differences in chromatin state between the two sides of the rearrangements with the transition occurring exactly at the BOS (Fig. 7C). The overall epigenetic state of chromatin around gibbon BOS was strongly correlated to orthologous regions in human and rhesus (Supplemental Fig. S13A,B); thus, the original epigenetic state remained mostly stable, even in the oldest gibbon BOS (Fig. 7B). Future studies should compare transcription levels and other functional epigenetic marks (e.g., H3K27ac, which marks enhancers), to examine local regulatory and transcriptional changes that might occur even when the overall chromatin state remains unchanged. Nevertheless, our findings demonstrate interesting insight into the preservation of epigenetic landscapes despite extensive evolutionary genomic rearrangements.
In conclusion, this study is the first to show a remarkable correspondence between evolutionary breakpoints and TAD boundaries in the gibbon genome, a critically endangered primate species that recently experienced rapid and heavy chromosome remodeling (Carbone et al. 2014). Our findings provide compelling evidence that this colocalization may be due to both TAD boundaries’ higher fragility and easier repair, as well as purifying selection against rearrangements that disrupt regulation of essential TADs (Fig. 8). Formation of new large-scale TADs does not appear to play a large role in emergence of evolutionary novelty in the gibbon genome. New TADs seem to be more tolerated when ectopic interactions are mitigated through formation of new boundaries, or alternatively, when genes with coordinated expression are placed in the same TAD. Finally, we find that the two sides of evolutionary breakpoints remain epigenetically similar to their ancestral state and do not homogenize even after millions of years. Overall, this study supports a nonrandom mode of chromosome evolution in which functional genomic units remain genetically and epigenetically intact, despite being shuffled around in the genome.
Methods
Annotation of gibbon BOS and orthologous BOS in other species
We compiled a comprehensive list of gibbon–human BOS by incorporating all previous findings from array painting (Carbone et al. 2006), fluorescent in situ hybridization (FISH) experiments, end-sequencing of Bacterial Artificial Chromosomes (BACs) (Roberto et al. 2007; Girirajan et al. 2009), shotgun sequencing, assembly of full BACs (Carbone et al. 2006, 2009a), and the latest gibbon genome assembly, Nleu3.0 (Carbone et al. 2014). We created a naming convention to account for the directionality of the rearrangement and distinguish between the two sides; e.g., NLE 5_2_R_r, indicates the right side (R) of the second BOS on gibbon Chromosome 5, extended 10 kb downstream (r). To enable comparative analyses, we identified regions orthologous to gibbon BOS in human, rhesus macaque, mouse, dog, and rabbit by using BLAT and the UCSC Genome Browser liftOver tool (Kent et al. 2002). From the 67 gibbon BOS, we identified 133 orthologous regions mapped on human (Homo sapiens, hg38), 129 on rhesus (Macaca mulatta, RheMac8), 95 on mouse (Mus musculus, GRC38), 104 on rabbit (Oryctolagus cuniculus, oryCun2), and 114 in dog (Canis familiaris, CanFam3.1) (Supplemental Table S1).
Genome-wide chromatin conformation capture (Hi-C) sequencing and analysis
To prepare the Hi-C library, 2.5 × 107 Vok EBV-transformed cells (Supplemental Methods) were cross-linked with 1% formaldehyde for 5 min on ice and then lysed in Lysis Buffer (0.1% SDS, 0.5% Triton X-100, 20 mM Tris-HCl pH 8.0, 150 mM NaCl, and protease inhibitor cocktail [Roche]). The chromatin was pelleted and washed twice with Hi-C Wash Buffer (HWB; 50 mM Tris-HCl pH 8.0, 50 mM NaCl), then resuspended in 250 µL HWB buffer with 0.6% SDS for 10 min at 68°C. Replicates of 50 µL lysate (<2 µg chromatin per replicate) were bound to 100 µL AMPure XP beads (Beckman Coulter), then washed twice with HWB. Bead-bound chromatin was digested in NEB DpnII Buffer with 5 units of DpnII enzyme for 3 h while shaking at 37°C. After washing beads with HWB, biotinylated dCTP was incorporated by incubating beads in 50 µL of End Fill Mix (NEB Buffer 2, 0.15 mM each dATP, dGTP, dTTP [New England Biolabs], 0.04 mM biotin-14-dCTP [Invitrogen], 3.75 units Klenow [New England Biolabs]) for 30 min while shaking at 25°C. Beads were washed with HWB, and then chromatin proximity ligation was carried out by overnight incubation at 16°C in a 250 µL reaction containing T4 DNA Ligase Buffer (New England Biolabs), 0.1 mg/mL BSA (New England Biolabs), 0.25% Triton X-100 (Thermo Fisher Scientific), and 50 units T4 DNA Ligase (New England Biolabs). Next, 2.5 µL 10 mM dNTPs (New England Biolabs) and 7.5 units T4 DNA polymerase (New England Biolabs) were added to remove biotin-dCTP from unligated ends. The samples were released from the beads in 50 µL cross-link reversal buffer (50 mM Tris pH = 8.0, 1% SDS, 0.25 mM CaCl2, and 0.5 mg/mL Proteinase K) by subsequent incubations at 55°C then 68°C. After cross-link reversal, the samples were cleaned with a 2X AMPure XP bead clean up. DNA was quantified by Qubit fluorometer before Illumina sequencing library preparation using the NEBNext Ultra II DNA Library Preparation Kit (New England Biolabs), following the manufacturer's protocol. The ligation product was pulled down using 25 µL Dynabeads MyOne Streptavidin C1 (Invitrogen) that had been washed with TWB (10 mM Tris-HCl pH 8.0, 0.5 mM EDTA, 0.05% Tween-20) and suspended in NTB (10 mM Tris-HCl pH 8.0, 2M NaCl, 1 mM EDTA). The biotinylated ligation junctions were captured with the prepped Dynabeads and then washed with LWB (10 mM Tris-HCl pH 8.0, 1M LiCl, 1 mM EDTA, 0.05% Tween-20), NWB (10 mM Tris-HCl pH 8.0, 0.1M NaCl, 1 mM EDTA, 0.05% Tween-20), and HWB before continuing on to indexing PCR. Libraries were multiplexed and sequenced on the Illumina NextSeq 500 platform at the OHSU Massive Parallel Sequencing Shared Resources (MPSSR) to generate 75-bp paired-end reads.
Hi-C data sets for the other species: Rhesus, mouse, rabbit, dog, and human were obtained from public sources (Grubert et al. 2015; Vietri Rudan et al. 2015). When the public sources contained multiple read files and replicates, we combined data across replicates. All Hi-C data sets were processed similarly using the Juicer pipeline (Durand et al. 2016). For each species, we used TADtool (Kruse et al. 2016) to call TADs using the directionality index method (Dixon et al. 2012) with 140 parameter combinations: seven resolutions (10, 25, 50, 100, 250, and 500 kb, and 1 Mb), five significance cutoffs (0.1, 0.05, 0.01, 0.005, and 0.001) and four window sizes (1, 2, 5, and 10 Mb). Finally, we used HOMER v4.9 (Heinz et al. 2010) to visualize the Hi-C contact maps overlapped across BOS regions.
Chromatin immunoprecipitation sequencing (ChIP-seq) and data analysis
CTCF ChIP-seq data generation and analysis have been described previously (Carbone et al. 2014). To generate H3K4me3 ChIP-seq libraries, we fixed 5 × 107 cells from EBV-transformed line generated for Vok and Asia (the individual used for the gibbon genome assembly) with 1% formaldehyde on ice for 5 min. Next, cells were lysed with Lysis Buffer (0.1% SDS, 0.5% Triton X-100, 20 mM Tris-HCl pH 8.0, 150 mM NaCl, and protease inhibitor cocktail [Roche]). The lysate was sheared using the Bioruptor Plus sonicator for one cycle of 15 min (30 sec on/off) at high power. A 10-µL aliquot of lysate was taken as “input.” Lysate was split into two preps and each sample was incubated with 1 µg anti-H3K4me3 antibody (abcam Cat. ab8580) overnight at 4°C. Prepared Pierce Protein A/G Magnetic Beads (Thermo Fisher Scientific) were added to lysates, incubated for 2 h at 4°C, and then washed four times with TBST (25 mM Tris-HCl pH 7.6, 250 mM NaCl, 0.05% Tween-20), once with LiCl Wash Buffer (10 mM Tris-HCl pH 8.0, 0.5M LiCl, 1 mM EDTA, 0.05% Tween-20), once with Lysis Buffer, and once with 1X TE. Following washes, samples were eluted with Elution Buffer (1% SDS, 0.1M NaHCO3) and incubated at room temperature for 15 min. Elution reactions were pooled and incubated with NaCl overnight at 65°C to reverse crosslinks. Following RNase A and Proteinase K digests at 37°C and 55°C, respectively, samples were cleaned using standard Phenol:Chloroform cleanup and a 1.6X AMPure XP (Beckman Coulter) bead clean up. Ten nanograms of ChIP and input DNA were used to make next-generation sequencing libraries using the NEBNext Ultra II DNA Library Preparation Kit (New England Biolabs). Libraries were checked with Qubit fluorometer and Bioanalyzer (Agilent) assays and sequenced on the Illumina NextSeq 500 platform to generate single-end 75-bp reads.
H3K4me3 ChIP and input sequences (single-end 36-bp reads) for rhesus macaque were obtained from public data (Accession number GSE60269) (Zhao et al. 2006). For both gibbon and rhesus data, we removed adapter sequences using trimmomatic v0.35 (Bolger et al. 2014) and de-duplicated reads using FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/). We then mapped the reads to their corresponding genomes with BWA-MEM v0.7.9a (Li and Durbin 2009) and removed reads with mapping scores below 30. Next, we used phantompeakqualtools v2.0 (Marinov et al. 2014) to run SPP (Kharchenko et al. 2008) and obtain cross-correlation profiles and call a loose set of 300,000 peaks. To determine a cutoff for these peaks, we performed an irreproducibility discovery rate (IDR) analysis (Landt et al. 2012). Using an IDR cutoff of 0.0025 (recommended by ENCODE) (The ENCODE Project Consortium 2012), we obtained 20,828 peaks for gibbon and 21,738 peaks for rhesus.
Whole-genome bisulfite sequencing (WGBS) and data analysis
Whole-genome bisulfite-converted samples were generated for gibbon and rhesus as previously described (Lister et al. 2009). Briefly, 1 µg genomic DNA was extracted from opportunistic whole blood samples from Vok and rhesus and fragmented (Fragmentase Reaction Buffer v2, Fragmentase [New England Biolabs]), end-repaired (T4 DNA Ligase Buffer, dNTP, T4 DNA polymerase, T4 PNK), A-tailed (NEB buffer 2, 1 mM dATP, Klenow exo-), and ligated to 30 µM methylated forked Illumina adapters (Quick Ligase Reaction Buffer, Quick Ligase). Ligated fragments were size-selected (250–500 bp) and bisulfite converted using the Qiagen EpiTect bisulfite kit. Fragments were amplified and sequenced on the Illumina HiSeq 2000 platform to generate 111,876,578 paired-end reads. Human WGBS data was publicly available (Accession SRR2058107 and SRR2058108) (Hernando-Herraez et al. 2015).
Raw reads from human, rhesus, and gibbon were pre-processed using Trim Galore! v0.4.0 (Krueger and Andrews 2011) to remove adapters, bases with Phred score less than 20, and reads shorter than 20 bp. Next, we used bsmooth v0.8.1 (Hansen et al. 2012) to map 250,000 random reads from each data set to their corresponding genomes and generate mbias plots to determine whether the methylation level correlates with the position of the CpG in the reads. Based on these plots and the output of FastQC v0.10.1 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) quality reports, we uniformly trimmed 5 bp from both ends of all reads. We then mapped reads to their respective genomes using Bismark v0.12.5 (Krueger and Andrews 2011) and obtained methylation calls for each read at each CpG locations. In the subsequent analysis, the counts of all CpGs were assessed, but methylation values were only considered for CpG covered by more than four reads. For the human sample, data from two sequencing lanes was merged at the CpG summary level.
Data access
The data generated as part of this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo) under accession number GSE115065. Code is available at https://github.com/nathanlazar/APE_METH_bin and as supplemental file APE_METH_bin.zip. The file ALL_STEPS describes analysis steps for all data sets and the usage of each of our custom scripts.
Supplementary Material
Acknowledgments
We thank the Gibbon Conservation Center (GCC) for providing us with the samples used in this study. In particular, we thank Gabriella Skollar for her inspiring work and passion taking care of these critically endangered species. Illumina sequencing was performed by the OHSU Massively Parallel Sequencing Shared Resource (MPSSR). We also thank the OHSU ExaCloud Cluster Computational Resource and the Advanced Computing Center that allowed us to perform the intensive large-scale data workflows. We thank Brett Davis for mapping the WGBS data. Finally, we thank Drs. Andrew Adey and Eric Jorgenson for insightful discussions about this manuscript and the reviewers for their helpful critiques. N.H.L. and T.J.M. were supported by a fellowship from the National Library of Medicine Biomedical Informatics Research Training Program (NIH) T15 LM007088 during this project; L.C., M.O., C.M., and R.J.O. were supported by National Science Foundation (NSF) award 1613856 and L.C. was supported by NIH/NCRR P51 OD011092 and the Knight Cardiovascular Institute. R.E.G. was supported by NHGRI grant 5U24HG009084.
Author contributions: N.H.L. and L.C. designed the experiments and the analysis plan. N.H.L. performed the analysis. K.A.N. and L.C. performed the experiments. C.M. and R.J.O. performed the FISH experiment. B.O. and R.E.G. contributed to experimental methods. L.C., N.H.L., and M.O. wrote the paper. T.J.M. created the figures. All authors were involved with editing the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.233874.117.
References
- Berthelot C, Muffato M, Abecassis J, Roest Crollius H. 2015. The 3D organization of chromatin explains evolutionary fragile genomic regions. Cell Rep 10: 1913–1924. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canela A, Maman Y, Jung S, Wong N, Callen E, Day A, Kieffer-Kwon KR, Pekowska A, Zhang H, Rao SSP, et al. 2017. Genome organization drives chromosome fragility. Cell 170: 507–521.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capozzi O, Carbone L, Stanyon RR, Marra A, Yang F, Whelan CW, de Jong PJ, Rocchi M, Archidiacono N. 2012. A comprehensive molecular cytogenetic analysis of chromosome rearrangements in gibbons. Genome Res 22: 2520–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone L, Vessere GM, ten Hallers BF, Zhu B, Osoegawa K, Mootnick A, Kofler A, Wienberg J, Rogers J, Humphray S, et al. 2006. A high-resolution map of synteny disruptions in gibbon and human genomes. PLoS Genet 2: e223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone L, Harris RA, Vessere GM, Mootnick AR, Humphray S, Rogers J, Kim SK, Wall JD, Martin D, Jurka J, et al. 2009a. Evolutionary breakpoints in the gibbon suggest association between cytosine methylation and karyotype evolution. PLoS Genet 5: e1000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone L, Mootnick AR, Nadler T, Moisson P, Ryder O, Roos C, de Jong PJ. 2009b. A chromosomal inversion unique to the northern white-cheeked gibbon. PLoS One 4: e4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone L, Harris RA, Gnerre S, Veeramah KR, Lorente-Galdos B, Huddleston J, Meyer TJ, Herrero J, Roos C, Aken B, et al. 2014. Gibbon genome and the fast karyotype evolution of small apes. Nature 513: 195–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung BH, Mullegama S, Marshall CR, Lionel AC, Weksberg R, Dupuis L, Brick L, Li C, Scherer SW, Aradhya S, et al. 2012. Severe intellectual disability and autistic features associated with microduplication 2q23.1. Eur J Hum Genet 20: 398–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couturier J, Lernould JM. 1991. Karyotypic study of four gibbon forms provisionally considered as subspecies of Hylobates (Nomascus) concolor (Primates, Hylobatidae). Folia Primatol (Basel) 56: 95–104. [DOI] [PubMed] [Google Scholar]
- Dai J, Li Y, Ji C, Jin F, Zheng Z, Wang X, Sun X, Xu X, Gu S, Xie Y, et al. 2003. Characterization of two novel KRAB-domain-containing zinc finger genes, ZNF460 and ZNF461, on human chromosome 19q13.1→q13.4. Cytogenet Genome Res 103: 74–78. [DOI] [PubMed] [Google Scholar]
- De S, Michor F. 2011. DNA secondary structures and epigenetic determinants of cancer genome evolution. Nat Struct Mol Biol 18: 950–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeer P, Pykels E, Lammens J, Devriendt K, Fryns JP. 2003. Melorheostosis in a family with autosomal dominant osteopoikilosis: report of a third family. Am J Med Genet A 119A: 188–193. [DOI] [PubMed] [Google Scholar]
- Dickinson ME, Flenniken AM, Ji X, Teboul L, Wong MD, White JK, Meehan TF, Weninger WJ, Westerberg H, Adissu H, et al. 2016. High-throughput discovery of novel developmental phenotypes. Nature 537: 508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485: 376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Jung I, Selvaraj S, Shen Y, Antosiewicz-Bourget JE, Lee AY, Ye Z, Kim A, Rajagopal N, Xie W, et al. 2015. Chromatin architecture reorganization during stem cell differentiation. Nature 518: 331–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon J, Dileep V, Zhan Y, Song F, Le VT, Yardimci GG, Chakraborty A, Bann DV, Wang Y, Clark R, et al. 2017. An integrative framework for detecting structural variations in cancer genomes. bioRxiv 10.1101/119651. [DOI] [Google Scholar]
- Durand NC, Robinson JT, Shamim MS, Machol I, Mesirov JP, Lander ES, Aiden EL. 2016. Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom. Cell Syst 3: 99–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischetto R, Palumbo O, Ortolani F, Palumbo P, Leone MP, Causio FA, Pesce S, Digilio MC, Carella M, Papadia F. 2017. Clinical and molecular characterization of a second family with the 12q14 microdeletion syndrome and review of the literature. Am J Med Genet A 10.1002/ajmg.a.38253. [DOI] [PubMed] [Google Scholar]
- Franke M, Ibrahim DM, Andrey G, Schwarzer W, Heinrich V, Schöpflin R, Kraft K, Kempfer R, Jerković I, Chan WL, et al. 2016. Formation of new chromatin domains determines pathogenicity of genomic duplications. Nature 538: 265–269. [DOI] [PubMed] [Google Scholar]
- Fudenberg G, Pollard KS. 2018. Chromatin features constrain structural variation across evolutionary timescales. bioRxiv 10.1101/285205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlan A, Lubke M, Adameyko I, Lallemend F, Ernfors P. 2013. The transcription factor Hmx1 and growth factor receptor activities control sympathetic neurons diversification. EMBO J 32: 1613–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girirajan S, Chen L, Graves T, Marques-Bonet T, Ventura M, Fronick C, Fulton L, Rocchi M, Fulton RS, Wilson RK, et al. 2009. Sequencing human–gibbon breakpoints of synteny reveals mosaic new insertions at rearrangement sites. Genome Res 19: 178–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gröschel S, Sanders MA, Hoogenboezem R, de Wit E, Bouwman BA, Erpelinck C, van der Velden VH, Havermans M, Avellino R, van Lom K, et al. 2014. A single oncogenic enhancer rearrangement causes concomitant EVI1 and GATA2 deregulation in leukemia. Cell 157: 369–381. [DOI] [PubMed] [Google Scholar]
- Grubert F, Zaugg JB, Kasowski M, Ursu O, Spacek DV, Martin AR, Greenside P, Srivas R, Phanstiel DH, Pekowska A, et al. 2015. Genetic control of chromatin states in humans involves local and distal chromosomal interactions. Cell 162: 1051–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The GTEx Consortium. 2013. The Genotype-Tissue Expression (GTEx) project. Nat Genet 45: 580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KD, Langmead B, Irizarry RA. 2012. BSmooth: from whole genome bisulfite sequencing reads to differentially methylated regions. Genome Biol 13: R83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harewood L, Fraser P. 2014. The impact of chromosomal rearrangements on regulation of gene expression. Hum Mol Genet 23: R76–R82. [DOI] [PubMed] [Google Scholar]
- Harewood L, Kishore K, Eldridge MD, Wingett S, Pearson D, Schoenfelder S, Collins VP, Fraser P. 2017. Hi-C as a tool for precise detection and characterisation of chromosomal rearrangements and copy number variation in human tumours. Genome Biol 18: 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellemans J, Preobrazhenska O, Willaert A, Debeer P, Verdonk PC, Costa T, Janssens K, Menten B, Van Roy N, Vermeulen SJ, et al. 2004. Loss-of-function mutations in LEMD3 result in osteopoikilosis, Buschke-Ollendorff syndrome and melorheostosis. Nat Genet 36: 1213–1218. [DOI] [PubMed] [Google Scholar]
- Hernando-Herraez I, Heyn H, Fernandez-Callejo M, Vidal E, Fernandez-Bellon H, Prado-Martinez J, Sharp AJ, Esteller M, Marques-Bonet T. 2015. The interplay between DNA methylation and sequence divergence in recent human evolution. Nucleic Acids Res 43: 8204–8214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Weintraub AS, Day DS, Valton AL, Bak RO, Li CH, Goldmann J, Lajoie BR, Fan ZP, Sigova AA, et al. 2016. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science 351: 1454–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamsai D, O'Connor AE, Deboer KD, Clark BJ, Smith SJ, Browne CM, Bensley JG, Merriman JA, Yuen WS, Koopman P, et al. 2013. Loss of GGN leads to pre-implantation embryonic lethality and compromised male meiotic DNA double strand break repair in the mouse. PLoS One 8: e56955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. 2002. The human genome browser at UCSC. Genome Res 12: 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV, Tolstorukov MY, Park PJ. 2008. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat Biotechnol 26: 1351–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler U, Bigoni F, Wienberg J, Stanyon R. 1995. Genomic reorganization in the concolor gibbon (Hylobates concolor) revealed by chromosome painting. Genomics 30: 287–292. [DOI] [PubMed] [Google Scholar]
- Kortüm F, Caputo V, Bauer CK, Stella L, Ciolfi A, Alawi M, Bocchinfuso G, Flex E, Paolacci S, Dentici ML, et al. 2015. Mutations in KCNH1 and ATP6V1B2 cause Zimmermann-Laband syndrome. Nat Genet 47: 661–667. [DOI] [PubMed] [Google Scholar]
- Krueger F, Andrews SR. 2011. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27: 1571–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse K, Hug CB, Hernández-Rodríguez B, Vaquerizas JM. 2016. TADtool: visual parameter identification for TAD-calling algorithms. Bioinformatics 32: 3190–3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landt SG, Marinov GK, Kundaje A, Kheradpour P, Pauli F, Batzoglou S, Bernstein BE, Bickel P, Brown JB, Cayting P, et al. 2012. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res 22: 1813–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre C, Zaghloul L, Sagot MF, Gautier C, Arneodo A, Tannier E, Audit B. 2009. Analysis of fine-scale mammalian evolutionary breakpoints provides new insight into their relation to genome organisation. BMC Genomics 10: 335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Harris RA, Cheung SW, Coarfa C, Jeong M, Goodell MA, White LD, Patel A, Kang SH, Shaw C, et al. 2012. Genomic hypomethylation in the human germline associates with selective structural mutability in the human genome. PLoS Genet 8: e1002692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326: 289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. 2009. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462: 315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupianez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, Horn D, Kayserili H, Opitz JM, Laxova R, et al. 2015. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 161: 1012–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupianez DG, Spielmann M, Mundlos S. 2016. Breaking TADs: how alterations of chromatin domains result in disease. Trends Genet 32: 225–237. [DOI] [PubMed] [Google Scholar]
- Marinov GK, Kundaje A, Park PJ, Wold BJ. 2014. Large-scale quality analysis of published ChIP-seq data. G3 (Bethesda) 4: 209–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson D, Pettersson M, Gustavsson P, Förster A, Hofmeister W, Wincent J, Zachariadis V, Anderlid BM, Nordgren A, Mäkitie O, et al. 2017. Whole-genome sequencing of cytogenetically balanced chromosome translocations identifies potentially pathological gene disruptions and highlights the importance of microhomology in the mechanism of formation. Hum Mutat 38: 180–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. 2012. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485: 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips-Cremins JE, Sauria ME, Sanyal A, Gerasimova TI, Lajoie BR, Bell JS, Ong CT, Hookway TA, Guo C, Sun Y, et al. 2013. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell 153: 1281–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope BD, Ryba T, Dileep V, Yue F, Wu W, Denas O, Vera DL, Wang Y, Hansen RS, Canfield TK, et al. 2014. Topologically associating domains are stable units of replication-timing regulation. Nature 515: 402–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. 2014. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159: 1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redin C, Brand H, Collins RL, Kammin T, Mitchell E, Hodge JC, Hanscom C, Pillalamarri V, Seabra CM, Abbott MA, et al. 2017. The genomic landscape of balanced cytogenetic abnormalities associated with human congenital anomalies. Nat Genet 49: 36–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberto R, Capozzi O, Wilson RK, Mardis ER, Lomiento M, Tuzun E, Cheng Z, Mootnick AR, Archidiacono N, Rocchi M, et al. 2007. Molecular refinement of gibbon genome rearrangements. Genome Res 17: 249–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt AD, Hu M, Jung I, Xu Z, Qiu Y, Tan CL, Li Y, Lin S, Lin Y, Barr CL, et al. 2016. A compendium of chromatin contact maps reveals spatially active regions in the human genome. Cell Rep 17: 2042–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, Andrews S, Kurukuti S, Mitchell JA, Umlauf D, Dimitrova DS, et al. 2010. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet 42: 53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smerdon MJ. 1991. DNA repair and the role of chromatin structure. Curr Opin Cell Biol 3: 422–428. [DOI] [PubMed] [Google Scholar]
- Sun J, Zhou L, Emerson DJ, Gilgenast TGG, Titus K, Beagan JA, Phillips-Cremins JE. 2017. Genetic drivers of repeat expansion disorders localize to 3-D chromatin domain boundaries. bioRxiv 10.1101/191213. [DOI] [Google Scholar]
- Tchurikov NA, Fedoseeva DM, Sosin DV, Snezhkina AV, Melnikova NV, Kudryavtseva AV, Kravatsky YV, Kretova OV. 2015. Hot spots of DNA double-strand breaks and genomic contacts of human rDNA units are involved in epigenetic regulation. J Mol Cell Biol 7: 366–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vietri Rudan M, Barrington C, Henderson S, Ernst C, Odom DT, Tanay A, Hadjur S. 2015. Comparative Hi-C reveals that CTCF underlies evolution of chromosomal domain architecture. Cell Rep 10: 1297–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zepeda-Mendoza CJ, Ibn-Salem J, Kammin T, Harris DJ, Rita D, Gripp KW, MacKenzie JJ, Gropman A, Graham B, Shaheen R, et al. 2017. Computational prediction of position effects of apparently balanced human chromosomal rearrangements. Am J Hum Genet 101: 206–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Tavoosidana G, Sjölinder M, Göndör A, Mariano P, Wang S, Kanduri C, Lezcano M, Sandhu KS, Singh U, et al. 2006. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet 38: 1341–1347. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.