Abstract
EPIGEN-Brazil is one of the largest Latin American initiatives at the interface of human genomics, public health, and computational biology. Here, we present two resources to address two challenges to the global dissemination of precision medicine and the development of the bioinformatics know-how to support it. To address the underrepresentation of non-European individuals in human genome diversity studies, we present the EPIGEN-5M+1KGP imputation panel—the fusion of the public 1000 Genomes Project (1KGP) Phase 3 imputation panel with haplotypes derived from the EPIGEN-5M data set (a product of the genotyping of 4.3 million SNPs in 265 admixed individuals from the EPIGEN-Brazil Initiative). When we imputed a target SNPs data set (6487 admixed individuals genotyped for 2.2 million SNPs from the EPIGEN-Brazil project) with the EPIGEN-5M+1KGP panel, we gained 140,452 more SNPs in total than when using the 1KGP Phase 3 panel alone and 788,873 additional high confidence SNPs (info score ≥ 0.8). Thus, the major effect of the inclusion of the EPIGEN-5M data set in this new imputation panel is not only to gain more SNPs but also to improve the quality of imputation. To address the lack of transparency and reproducibility of bioinformatics protocols, we present a conceptual Scientific Workflow in the form of a website that models the scientific process (by including publications, flowcharts, masterscripts, documents, and bioinformatics protocols), making it accessible and interactive. Its applicability is shown in the context of the development of our EPIGEN-5M+1KGP imputation panel. The Scientific Workflow also serves as a repository of bioinformatics resources.
The EPIGEN-Brazil Initiative (https://epigen.grude.ufmg.br/) is one of the largest Latin American initiatives at the interface of human genomics, public health, and computational biology. Here, we present how we are addressing two challenges to global dissemination of precision medicine and to the development of the bioinformatics know-how to support it. These challenges are (1) the persistent and severe underrepresentation of non-European individuals in human genome diversity studies and well-designed genetic epidemiology studies (Alexander et al. 2009; Bustamante et al. 2011; Check Hayden 2016; Popejoy and Fullerton 2016); and (2) the lack of transparency and reproducibility in the entire scientific process, including bioinformatics protocols (Iqbal et al. 2016).
The underrepresentation of globally diverse individuals in genomic studies is not simply due to lack of their enrollment in these studies. Much more compelling is the need for a more global distribution of research groups with a strong background in genomics and bioinformatics, leading and performing this kind of study. In this context, the overarching goal of the EPIGEN-Brazil Initiative is to study the genomic diversity and its effects on complex phenotypes in Brazil, the most populous Latin American country (Borges et al. 2016; Lima-Costa et al. 2016; Marques et al. 2017). Brazil's more than 200 million inhabitants are the product of admixture that occurred during the last 500 years between Amerindians, Europeans, Africans, and their descendants. Interestingly, Brazil was the largest destiny of the African diaspora, and we have recently shown that Brazilians host on their genomes the diversity of African groups that have not yet been included in population genomics studies, such as Bantu Angola and Mozambique populations, two sources of the slave trade that originated in territories controlled by the Portuguese Crown (Kehdy et al. 2015).
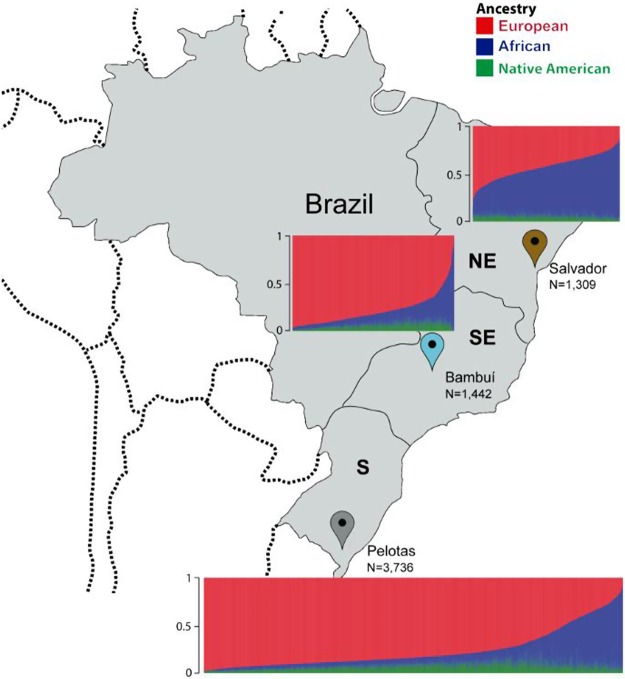
The EPIGEN-Brazil Initiative is studying 6487 Brazilians from the three largest population-based cohorts of the country (Fig. 1; Supplemental Table S1; Supplemental Material Sections 1, 2.1): (1) Salvador-SCAALA in northeast Brazil, with predominant African ancestry (18 years of follow-up) (Barreto et al. 2006); (2) the Bambuí Cohort Study of Aging in Minas Gerais in the southeast of the country (15 years of follow-up) (Lima-Costa et al. 2011); and (3) the 1982 Pelotas Birth-Cohort Study in southern Brazil (30 years of follow-up) (Victora and Barros 2006).
Figure 1.
Continental admixture of the EPIGEN-Brazil population-based cohorts. Ancestry was estimated using the ADMIXTURE software (Alexander et al. 2009), as in Kehdy et al. (2015). European, African, and Native American ancestry are, respectively: 42.8%, 50.8%, and 6.4% in Salvador; 78.5%, 14.8%, and 6.7% in Bambuí; and 76.1%, 15.9%, and 8% in Pelotas. Figure adapted from Kehdy et al. (2015).
The EPIGEN-Brazil Initiative is a strategic project funded by the Brazilian Ministry of Health, and it integrates research areas well established in the country, such as epidemiology, public health, and human genetics (Salzano and Freire-Maia 1967; Barreto 2004; Salzano 2018) with bioinformatics, that is a vigorous emerging area in Brazil. To address the need for more global research groups, one of the main goals of the EPIGEN-Brazil Initiative is to strengthen research capabilities in these research areas in Brazil, and we are training dozens of graduate students and postdoctoral researchers from Brazil and other Latin American countries. In Latin America, we are collaborating with the National Institute of Health from Peru to study the genomic diversity of the Peruvian population (Harris et al. 2017), which differs from the Brazilian population in having a predominant Native American ancestry.
The failing on diversity of human genomics and the EPIGEN-Brazil imputation panel
Imputation is the prediction of missing genotypes based on the pattern of linkage disequilibrium of a reference panel. For GWAS and fine-mapping studies, cosmopolitan public panels for imputation exist, such as the 1000 Genomes Project (1KGP) Phase 3 (Sudmant et al. 2015), based on whole-genome sequencing (WGS) data. In addition to the 1092 individuals from Phase 1, Phase 3 of the 1KGP panel has incorporated 1412 new individuals, including four new populations from Africa, one from admixed Latin America, two from East Asia, and five from South Asia, each with 61–113 individuals (Supplemental Table S3; Supplemental Material Section 2.2.2). Notwithstanding this improvement in the coverage of global genetic diversity, studies continue to show that imputation accuracy may be improved by using WGS or high-density SNP data from individuals with similar genetic background to the target population (Thornton and Bermejo 2014; Ahmad et al. 2017; Mitt et al. 2017). However, for studies performed in non-European populations, WGS or high-density array data are still rare. Next we present a new imputation panel specific for admixed Brazilian and Latin American populations and show that the inclusion of high-density array data from the Brazilian population improve imputation quality in respect to the use of the 1KGP (Phase 3) panel alone.
Addressing lack of transparency and reproducibility of genomic studies
A second challenge faced by global dissemination of bioinformatics and the know-how to support precision medicine is the lack of transparency and reproducibility of the entire scientific process (Iqbal et al. 2016). This limits the worldwide flow of bioinformatics knowledge necessary to build and train research groups with a solid bioinformatics background. Although there are several claims for more transparency and reproducibility of all the scientific process in biomedical literature (Sandve et al. 2013; Kolker et al. 2014; Iqbal et al. 2016), advances from genomic initiatives to share bioinformatics protocols are still rare.
A still valid and compelling claim and concept were formulated by Bourne (2010), proposing to move away from the classical scientific articles to a more interactive publication of Scientific Workflows. Bourne defined a Scientific Workflow as “part process and part container for content (or pointers to that content), that is significantly broader and more integrated than what is sent for publication today, namely, a manuscript and supplemental information in an essentially computationally unusable form.” Thus, a Scientific Workflow is a more complex concept than, and should not be confused with, a bioinformatics Workflow/Pipeline Management System such as Taverna (Wolstencroft et al. 2013) or Galaxy (Afgan et al. 2016), although the latter may be used to implement Scientific Workflows.
Here, we present the EPIGEN-Brazil Scientific Workflow (http://www.ldgh.com.br/scientificworkflow), a tool for transparent and reproducible bioinformatics analyses, and exemplify it in the context of our EPIGEN-5M+1KGP imputation panel. Our Scientific Workflow includes four self-contained components—scientific publications, flowcharts, masterscripts, and documents—that represent different stages of the scientific process. The scientific publications include both the final research products and the scientific hypotheses. The flowcharts are conceptual visualizations of research tasks performed as part of scientific publications, and the masterscripts are the operational computational execution (programs) of tasks represented by the flowcharts. Documents comprise other information such as technical reports, workshop presentations, and intermediate results.
Results and discussion
Imputation experiments
We genotyped 4.3 million SNPs in 265 admixed individuals from the EPIGEN-Brazil Initiative (90, 88, and 87 individuals randomly selected from the Salvador, Bambuí, and Pelotas cohorts, respectively) (Fig. 1; Supplemental Table S2; Supplemental Material Section 2.2.1). We present a new imputation reference panel (hereafter, the EPIGEN-5M+1KGP panel), which is the fusion of the haplotypes derived from the EPIGEN-5M data set with the public 1KGP Phase 3 imputation panel (Supplemental Table S4; Supplemental Fig. S1; Supplemental Material Sections 2.3, 2.4, 2.5.1). Hereafter, the 1KGP Phase 3 panel will be simply called 1KGP. In the context of GWAS and fine-mapping studies in Brazilian and other Latin American populations with a predominant mix of European and African ancestries, we tested whether using the EPIGEN-5M+1KGP imputation panel improves imputation in respect to the 1KGP imputation panel alone.
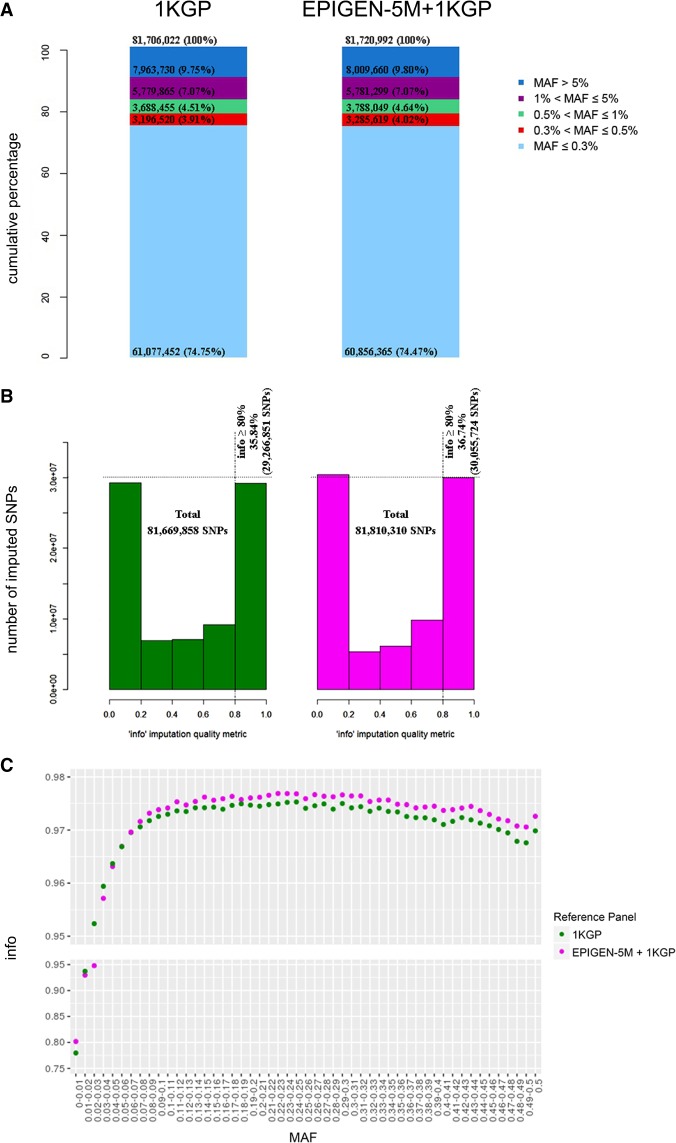
The EPIGEN-5M+1KGP and the 1KGP imputation panels have a similar number of variants and allele frequency spectra (Fig. 2A; Supplemental Fig. S2), although the EPIGEN-5M+1KGP has 14,970 more SNPs and 530 (∼10%) more haplotypes than the 1KGP imputation panel (5538 versus 5008 haplotypes, respectively) (Supplemental Table S4). More importantly, after phase inference (Supplemental Tables S5, S6; Supplemental Material Section 2.5.2), when we imputed a target SNPs data set (the 6487 admixed individuals genotyped for 2.2 million SNPs from the EPIGEN-Brazil project) (Fig. 1; Kehdy et al. 2015) with the EPIGEN-5M+1KGP panel, we gained 140,452 more SNPs in total and 788,873 additional high confidence SNPs (info score ≥0.8) than when using the 1KGP panel alone (Fig. 2B; Supplemental Tables S7, S8; Supplemental Material Section 2.5.3). Thus, the major effect of the inclusion of the EPIGEN-5M data set in a new imputation panel is not only to gain more SNPs but also to improve the quality of imputation. Particularly, the EPIGEN-5M+1KGP panel improves imputation quality in respect to 1KGP across a wide range of allele frequencies (Fig. 2C; Supplemental Figs. S3–S6). Therefore, imputation quality (i.e., info score) improves with the inclusion of the EPIGEN-5M data set even if it derives from high-density array data, rather than from WGS (which would be optimal). Imputation quality improves whether we input the entire EPIGEN-Brazil target data set or each of the cohorts separately. This suggests that the assembled EPIGEN-5M+1KGP imputation panel performs better than the 1KGP panel for a variety of study sizes, admixture levels, and post-Columbian demographic histories. Moreover, because high-density array data improve imputation quality, the 2.2 million SNPs data set previously published by Kehdy et al. (2015) may also be used for imputation for GWAS performed in Latin American populations with lower-density arrays.
Figure 2.
Comparison between the 1000 Genomes Project (1KGP) and EPIGEN-5M+1KGP imputation reference panels for autosomal chromosomes. The EPIGEN-5M+1KGP panel is the fusion of the haplotypes derived from the EPIGEN-5M data set (the genotyping of 265 EPIGEN-Brazil individuals for 4.3 million SNPs) with the public 1KGP Phase 3 imputation panel. (A) Allele frequency spectrum of variants by their minor allele frequency (MAF) in each imputation reference panel. The number of SNPs is described in each category, and the percentages are calculated dividing the number of SNPs in each MAF class by the total number of SNPs of each imputation reference panel (top). (B) Distribution of the info score quality metric for imputation results. The dashed vertical line indicates the 0.8 threshold info score value, and the horizontal line indicates the highest number of SNPs info score ≥0.8 achieved by a reference panel. (C) Imputation quality (mean info score) as a function of MAF for the target data set after imputation with each of the tested reference panels (MAF bin sizes of 0.01).
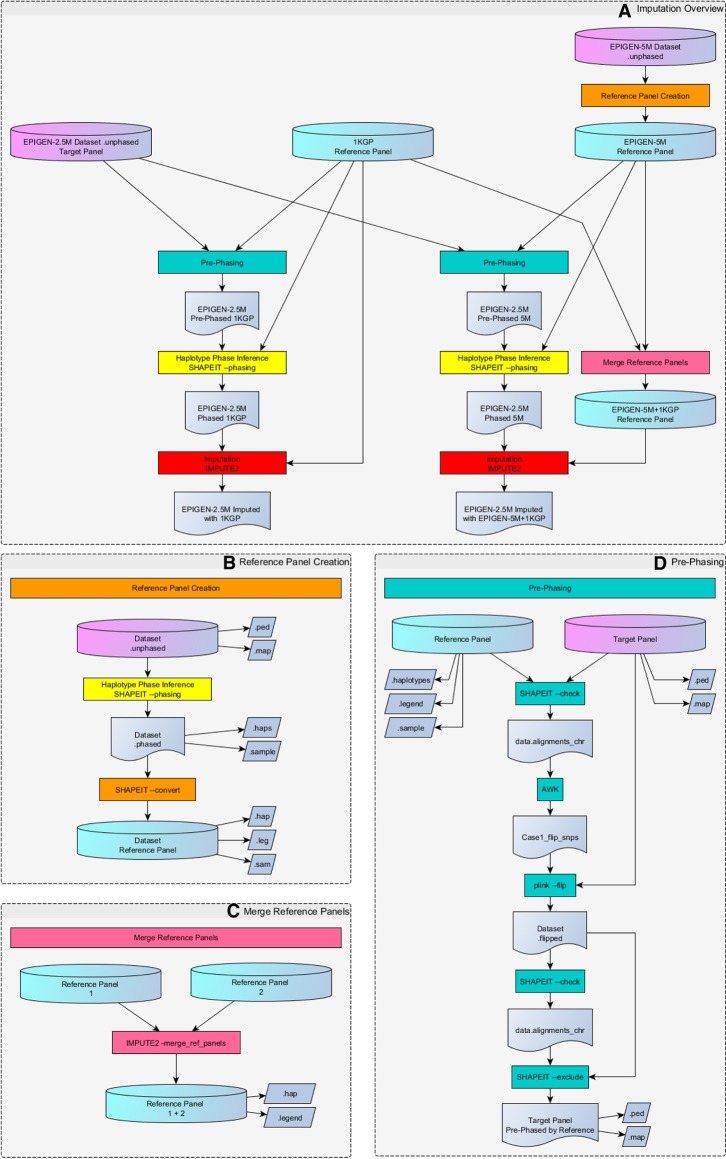
The case of the EPIGEN-5M+1KGP imputation panel exemplifies the applicability of the Scientific Workflow (Supplemental Material Section 3). All methodological steps to obtain the panel are delineated in Methods and are also visualized as a Scientific Workflow flowchart in http://www.ldgh.com.br/scientificworkflow/flowcharts.php (Fig. 3). The corresponding masterscripts that computationally operationalize the flowchart are available at http://www.ldgh.com.br/scientificworkflow/master_scripts.php (Supplemental Material Section 3; Supplemental Figs. S7, S8).
Figure 3.
Flowchart of the whole imputation process (see the EPIGEN-Brazil Scientific Workflow: http://www.ldgh.com.br/scientificworkflow/flowcharts.php). (A) Overview of the complete imputation process. (B,C) Two previous tasks may be required for imputation if it is necessary to create or merge reference panels. The Reference Panel Creation task (B, and orange color process in A) converts a data set of unphased genotypes into a reference panel, producing the EPIGEN-5M Reference Panel of haplotypes from the EPIGEN-5M data set. The Merge Reference Panels task (C, and pink color process in A) produces combinations of two different panels using IMPUTE2 software, generating the EPIGEN-5M+1KGP Reference Panel. The imputation process itself consists of three main tasks: pre-phasing, haplotype phase inference, and imputation. The pre-phasing task (D, and green color processes in A) performs strand alignment between target and reference panel using software SHAPEIT2, PLINK, and the scripting language AWK. Haplotype phase inference task (yellow color processes in A) of the target data set uses the methodology implemented in the software SHAPEIT2, generating .haps and .sample files (target data set aligned and phased with the Reference Panel). The latter files serve as input for the imputation task (red color processes in A) conducted with software IMPUTE2, following the “best practices” guidelines in the software documentation.
In conclusion, although high-coverage WGS data from populations underrepresented in genomic studies are the optimal source of haplotypes to be used for imputation in genome-wide/fine-mapping association studies, we show here that, in the absence of this kind of data, high-density array data from a few hundreds of individuals from the same populations, used together with the public 1KGP data set, is an alternative to improve imputation quality. Therefore, we expect that the EPIGEN-5M+1KGP imputation panel will allow for better GWAS, admixture mapping/fine-mapping studies in Latin American populations with ancestries that are similar to the Brazilian population studied by the EPIGEN-Brazil Initiative. We also use the EPIGEN-5M+1KGP imputation panel to exemplify our implementation of the concept of Scientific Workflow, in sensu Bourne (2010), which has the goal of making publicly available as much of the scientific process as possible. Since the Scientific Workflow represents different steps of the scientific process, from project development to publication, and with different levels of abstraction and detail, it emerges as a concrete initiative that moves us toward more transparency and reproducibility in bioinformatics analyses.
Methods
Imputation overview
Target data set
The EPIGEN-2.5M data set comprises 2,235,109 SNPs for 6487 Brazilians from three population-based cohorts (1309, 1442, and 3736 individuals from Salvador, Bambuí, and Pelotas, respectively) (Supplemental Table S1, published in Kehdy et al. 2015). EPIGEN-Brazil genome-wide data genotyped for the Illumina Omni 2.5M array are available in the European Nucleotide Archive under EPIGEN Committee Controlled Access mode.
Reference panels
We used two reference panels: (1) the public 1000 Genomes Project Phase 3 haplotypes, version 20130502, (1KGP) (Sudmant et al. 2015); and (2) The EPIGEN-5M+1KGP reference panel, which is the merge of the 1KGP panel and our unpublished EPIGEN-5M panel, bearing 14,970 more SNPs than the public panel solely. The EPIGEN-5M data set was genotyped with the Illumina HumanOmni5-4v1 array. After quality control, the data set comprises 4,102,271 SNPs for 265 Brazilians from the three cohorts (90, 88, and 87 individuals from Salvador, Bambuí, and Pelotas, respectively) (Supplemental Table S2). We used SHAPEIT2 (Delaneau et al. 2013) to infer the chromosome phase of the EPIGEN-5M data set (Supplemental Tables S4–S8).
Pre-phasing between the target and reference panels
We used SHAPEIT2 (Delaneau et al. 2013) to check the consistency of the SNP's strand of the target and the reference panels with the human genome reference sequence (GRCh37/hg19), and PLINK software (Purcell et al. 2007) to flip the strands in case of inconsistencies. Because our data are genotyped with the highest-density array (Omni 5.0) and not NGS-based, a new alignment to GRCh38 would not significantly affect the conclusions.
Haplotype phase inference of the target data set
We phased the target EPIGEN-2.5M data set using (1) the 1KGP haplotypes as phasing references, for the imputation with the 1KGP reference panel; and (2) the EPIGEN-5M data set as phasing reference, for the imputation with the EPIGEN-5M+1KGP reference panel.
Imputation
We performed the imputation using IMPUTE2 v.2.3.2 (Howie et al. 2009) on chromosome chunks of 7 Mb, with additional 250 kb of buffer on both sides (these were used for imputation inference but omitted from the results). We used the effective size parameter (Ne) set to 20,000 and the IMPUTE2 info score as a metric of imputation quality (Supplemental Fig. S1).
Data access
The data generated in this study have been submitted to the European Nucleotide Archive (ENA; https://www.ebi.ac.uk/ena) under accession number PRJEB9080 in EPIGEN Committee Controlled Access mode. All imputation tasks were performed using our Perl masterscript available as Supplemental Material (Supplemental Scripts) and also at our Scientific Workflow website (http://www.ldgh.com.br/scientificworkflow/master_scripts.php). The EPIGEN-5M+1KGP imputation panel in haplotype format is freely available at http://www.ldgh.com.br/scientificworkflow/documents.html.
Brazilian EPIGEN Consortium
Isabela O. Alvim,13 Victor Borda,13,14 Mateus H. Gouveia,13,15 Moara Machado,13,16 Rennan G. Moreira,13,17 Fernanda Rodrigues-Soares,13 Hanaisa P. Sant Anna,13 Meddly L. Santolalla,13 Marilia O. Scliar,13 Giordano B. Soares-Souza,13 Roxana Zamudio,13 Camila Zolini13,18
Supplementary Material
Acknowledgments
The EPIGEN-Brazil Initiative is funded by the Brazilian Ministry of Health (Department of Science and Technology from the Secretaria de Ciência, Tecnologia e Insumos Estratégicos) through Financiadora de Estudos e Projetos. The EPIGEN-Brazil investigators received funding from the Brazilian Ministry of Education (CAPES Agency), Brazilian National Research Council (CNPq), the Minas Gerais State Agency for Support of Research (FAPEMIG), and the Minas Gerais Network of Population Genomics and Precision Medicine (FAPEMIG RED00314-16). M.L.S. and V.B. have PhD fellowships from the international Brazilian government programs TWAS-CNPq and CAPES-PEC-PG, respectively. M.R.R. has a São Paulo Research Foundation (FAPESP) fellowship. We used the SAGARANA cluster from the Instituto de Ciências Biológicas from the Federal University of Minas Gerais, and we thank Prof. Miguel Ortega for bioinformatics support.
Footnotes
Departamento de Biologia Geral, Universidade Federal de Minas Gerais, Belo Horizonte, Minas Gerais, 31270-901, Brazil
Instituto Nacional de Salud, Lima, 9, Peru
Instituto de Pesquisa Rene Rachou, Fundação Oswaldo Cruz, Belo Horizonte, Minas Gerais, 30190-009, Brazil
Laboratory of Translational Genomics, National Institute of Health, Bethesda, MD 20877, USA
Laboratório Multiusuário de Genômica, Instituto de Ciências Biológicas, Universidade Federal de Minas Gerais, Belo Horizonte, Minas Gerais, 31270-901, Brazil
Beagle. Belo Horizonte, Minas Gerais, 31710-550, Brazil
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.225458.117.
Contributor Information
Collaborators: Isabela O. Alvim, Victor Borda, Mateus H. Gouveia, Moara Machado, Rennan G. Moreira, Fernanda Rodrigues-Soares, Hanaisa P. Sant Anna, Meddly L. Santolalla, Marilia O. Scliar, Giordano B. Soares-Souza, Roxana Zamudio, and Camila Zolini
References
- Afgan E, Baker D, van den Beek M, Blankenberg D, Bouvier D, Čech M, Chilton J, Clements D, Coraor N, Eberhard C, et al. 2016. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res 44: W3–W10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad M, Sinha A, Ghosh S, Kumar V, Davila S, Yajnik CS, Chandak GR. 2017. Inclusion of population-specific reference panel from India to the 1000 Genomes phase 3 panel improves imputation accuracy. Sci Rep 7: 6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander DH, Novembre J, Lange K. 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res 19: 1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreto ML. 2004. The globalization of epidemiology: critical thoughts from Latin America. Int J Epidemiol 33: 1132–1137. [DOI] [PubMed] [Google Scholar]
- Barreto ML, Cunha SS, Alcântara-Neves N, Carvalho LP, Cruz AA, Stein RT, Genser B, Cooper PJ, Rodrigues LC. 2006. Risk factors and immunological pathways for asthma and other allergic diseases in children: background and methodology of a longitudinal study in a large urban center in Northeastern Brazil (Salvador-SCAALA study). BMC Pulm Med 6: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges MC, Hartwig FP, Oliveira IO, Horta BL. 2016. Is there a causal role for homocysteine concentration in blood pressure? A Mendelian randomization study. Am J Clin Nutr 103: 39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne PE. 2010. What do I want from the publisher of the future? PLoS Comput Biol 6: e1000787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante CD, Burchard EG, De la Vega FM. 2011. Genomics for the world. Nature 475: 163–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Check Hayden E. 2016. A radical revision of human genetics. Nature 538: 154–157. [DOI] [PubMed] [Google Scholar]
- Delaneau O, Zagury JF, Marchini J. 2013. Improved whole-chromosome phasing for disease and population genetic studies. Nat Methods 10: 5–6. [DOI] [PubMed] [Google Scholar]
- Harris DN, Song W, Shetty AC, Levano K, Cáceres O, Padilla C, Borda V, Tarazona D, Trujillo O, Sanchez C, et al. 2017. The evolutionary genomic dynamics of Peruvians before, during, and after the Inca Empire. bioRxiv 10.1101/219808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie BN, Donnelly P, Marchini J. 2009. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 5: e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal SA, Wallach JD, Khoury MJ, Schully SD, Ioannidis JP. 2016. Reproducible research practices and transparency across the biomedical literature. PLoS Biol 14: e1002333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehdy FS, Gouveia MH, Machado M, Magalhães WC, Horimoto AR, Horta BL, Moreira RG, Leal TP, Scliar MO, Soares-Souza GB, et al. 2015. Origin and dynamics of admixture in Brazilians and its effect on the pattern of deleterious mutations. Proc Natl Acad Sci 112: 8696–8701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolker E, Ozdemir V, Martens L, Hancock W, Anderson G, Anderson N, Aynacioglu S, Baranova A, Campagna SR, Chen R, et al. 2014. Toward more transparent and reproducible omics studies through a common metadata checklist and data publications. OMICS 18: 10–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima-Costa MF, Firmo JO, Uchoa E. 2011. Cohort profile: the Bambuí (Brazil) Cohort Study of Ageing. Int J Epidemiol 40: 862–867. [DOI] [PubMed] [Google Scholar]
- Lima-Costa MF, Mambrini JV, Leite ML, Peixoto SV, Firmo JO, Loyola Filho AI, Gouveia MH, Leal TP, Pereira AC, Macinko J, et al. 2016. Socioeconomic position, but not African genomic ancestry, is associated with blood pressure in the Bambui-Epigen (Brazil) Cohort Study of Aging. Hypertension 67: 349–355. [DOI] [PubMed] [Google Scholar]
- Marques CR, Costa GN, da Silva TM, Oliveira P, Cruz AA, Alcantara-Neves NM, Fiaccone RL, Horta BL, Hartwig FP, Burchard EG, et al. 2017. Suggestive association between variants in IL1RAPL and asthma symptoms in Latin American children. Eur J Hum Genet 25: 439–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitt M, Kals M, Parn K, Gabriel SB, Lander ES, Palotie A, Ripatti S, Morris AP, Metspalu A, Esko T, et al. 2017. Improved imputation accuracy of rare and low-frequency variants using population-specific high-coverage WGS-based imputation reference panel. Eur J Hum Genet 25: 869–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popejoy AB, Fullerton SM. 2016. Genomics is failing on diversity. Nature 538: 161–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzano FM. 2018. The evolution of science in a Latin-American country: genetics and genomics in Brazil. Genetics 208: 823–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzano FM, Freire-Maia N. 1967. Populações Brasileiras: aspectos demográficos, genéticos e antropológicos. Companhia Editora Nacional, São Paulo, Brazil. [Google Scholar]
- Sandve GK, Nekrutenko A, Taylor J, Hovig E. 2013. Ten simple rules for reproducible computational research. PLoS Comput Biol 9: e1003285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudmant PH, Rausch T, Gardner EJ, Handsaker RE, Abyzov A, Huddleston J, Zhang Y, Ye K, Jun G, Fritz MH, et al. 2015. An integrated map of structural variation in 2,504 human genomes. Nature 526: 75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton TA, Bermejo JL. 2014. Local and global ancestry inference and applications to genetic association analysis for admixed populations. Genet Epidemiol 38: S5–S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora CG, Barros FC. 2006. Cohort profile: the 1982 Pelotas (Brazil) birth cohort study. Int J Epidemiol 35: 237–242. [DOI] [PubMed] [Google Scholar]
- Wolstencroft K, Haines R, Fellows D, Williams A, Withers D, Owen S, Soiland-Reyes S, Dunlop I, Nenadic A, Fisher P, et al. 2013. The Taverna workflow suite: designing and executing workflows of Web Services on the desktop, web or in the cloud. Nucleic Acids Res 41: W557–W561. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.