Abstract
Golgi membrane protein 1 (GOLM1) is a transmembrane glycoprotein of the Golgi cisternae, which is implicated in carcinogenesis of multiple types of cancer. In this study, using data from the Gene Expression Omnibus and The Cancer Genome Atlas, we compared the expression of GOLM1 in lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) and studied its prognostic value in terms of overall survival (OS) and recurrence-free survival (RFS) in these 2 subtypes of non-small cell lung cancer (NSCLC). Results showed that GOLM1 was significantly upregulated in both LUAD and LUSC tissues compared to the normal controls. However, GOLM1 expression was higher in LUAD tissues than in LUSC tissues. More importantly, using over 10 years’ survival data from 502 patients with LUAD and 494 patients with LUSC, we found that high GOLM1 expression was associated with unfavorable OS and RFS in patients with LUAD, but not in patients with LUSC. The following univariate and multivariate analyses confirmed that increased GOLM1 expression was an independent prognostic indicator of poor OS (hazard ratio [HR]: 1.30, 95% confidence interval [CI]: 1.11-1.54, P = .002) and RFS (HR: 1.37, 95% CI: 1.14-1.64, P = .001) in patients with LUAD. Of 511 cases with LUAD, 248 (48.5%) had heterozygous loss (−1), while 28 (5.5%) of 511 cases with LUAD had low-level copy gain (+1). In addition, we also found that the methylation status of 1 CpG site (chr9: 88,694,942-88,694,944) showed a weak negative correlation with GOLM1 expression (Pearson r = −0.25). Based on these findings, we infer that GOLM1 might serve as a valuable prognostic biomarker in LUAD, but not in LUSC. In addition, DNA copy number alterations and methylation might be 2 important mechanisms of dysregulated GOLM1 in LUAD.
Keywords: GOLM1, lung adenocarcinoma, prognosis, copy number, methylation
Introduction
Golgi membrane protein 1 (GOLM1) is also known as Golgi phosphoprotein 2 or Golgi membrane protein GP73. It is a transmembrane glycoprotein of the Golgi cisternae, which is typically expressed in the epithelial cells of normal human tissues.1 Since the Golgi apparatus has critical roles in modification and transportation of the proteins, the dysregulation of its associated proteins may result in significantly altered cellular processes and functions.2
A series of previous studies observed that GOLM1 is upregulated in multiple cancers and modulates their malignant behaviors. In prostate cancer, GOLM1 upregulation can promote cancer cell proliferation, migration, and invasion and inhibit apoptosis via activating the phosphoinositide 3 kinase-Akt-mammalian target of rapamycin (PI3K-AKT-mTOR) signaling3 and has been considered as a biomarker for detecting prostate cancer and aggressiveness of the disease.4,5 In hepatocellular carcinoma (HCC), aberrant GOLM1 expression enhances proliferation and migration of HCC cell lines and growth of xenograft tumors in mice via its regulation of membrane protein trafficking.6 In addition, it also has certain prognostic value in terms of overall survival (OS) and recurrence-free survival (RFS) in HCC.7,8
Recent studies found that GOLM1 was upregulated in non-small cell lung cancer (NSCLC), and its upregulation was associated with unfavorable clinicopathological features and can promote the proliferation and invasion of NSCLC cells.9,10 Actually, NSCLC constitutes of 3 subtypes, including lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), and large cell carcinoma, among which LUAD and LUSC are the dominant subtypes.11 These 2 subtypes have distinct molecular features and also have different prognostic markers.12-14 Patients with LUAD had significantly higher serum GOLM1 compared to healthy individuals.10 Therefore, it is meaningful to explore the prognostic value of GOLM1 expression and the mechanisms of its dysregulation in each subtype of NSCLC.
In this study, by using data from The Cancer Genome Atlas (TCGA), we compared the expression of GOLM1 in LUAD and LUSC and studied its prognostic value in terms of OS and RFS in these 2 subtypes of NSCLC.
Materials and Methods
Data Mining in Gene Expression Omnibus
The microarray data that compared gene expression profiles between LUAD and LUSC were retrieved in Gene Expression Omnibus (GEO) data sets. The expression of GOLM1 in one previous array (GSE10245),15 which examined gene expression in 40 LUAD cases and 18 LUSC cases, was examined using GEO2R.
Retrospective Data Mining in TCGA
The level 3 data in TCGA-LUAD and in TCGA-LUSC were downloaded using the UCSC Xena browser (https://xenabrowser.net/). These cohorts included over 500 cases of primary LUAD or LUSC, respectively. In TCGA-LUAD, tumor tissues from 514 patients and normal tissues from 59 controls were subjected to RNA-seq. In TCGA-LUSC, tumor tissues from 502 patients and normal tissues from 51 controls were subjected to RNA-seq; 502 of the 514 patients with LUAD and 494 of the 502 patients with LUSC had OS data recorded. Their clinicopathological information, including age at initial pathologic diagnosis, gender, smoking history, nodal status, pathologic stage, residual tumors, the history of radiation therapy/targeted molecular therapy, recurrence status, RFS in days, OS status, and OS in days, was downloaded.
The gene-level thresholded GISTIC2-processed copy number alterations (CNAs) and the DNA methylation status (Illumina (San Diego, California) 450k infinium methylation beadchip) of GOLM1 were also downloaded. In the methylation beadchip, the methylation status of 6 CpG islands in GOLM1 DNA was measured; 453 patients with LUAD and 371 patients with LUSC had GOLM1 DNA CNAs and GOLM1 RNA expression quantified at the same time.
Immunohistochemistry Staining of GOLM1
Immunohistochemistry (IHC) staining of GOLM1 in normal respiratory epithelial tissues and in LUAD tissues were reviewed using images in the Human Protein Atlas (HPA) (http://www.proteinatlas.org/),16,17 an online database aiming to map all the human proteins in cells, tissues, and organs.
Data Mining in the Kaplan-Meier Plotter
The association between GOLM1 expression and OS or first progression-free survival (FPS) in patients with LUAD was also examined by data mining in the Kaplan-Meier plotter, an online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in NSCLC by integrating 1715 samples of 10 independent data sets.18 The OS and FPS curves were generated by using the optimal cutoff of GOLM1 expression. The hazard ratio (HR) with 95% confidence intervals (CI) and log-rank P value were calculated.
Statistical Analysis
Statistical analysis was conducted using GraphPad Prism 6.0 (GraphPad Inc., La Jolla, California) or SPSS 19.0 software package (SPSS Inc, Chicago, Illinois). Group comparison was performed using Welch unequal variances t test. Receiver operating characteristic (ROC) curve analysis for death and recurrence detection was applied to identify the best cutoff (Youden index) for GOLM1 expression in Kaplan-Meier curves of OS and RFS. Therefore, if no significant difference was observed in OS/RFS under this model, there is no other cutoff that will generate significant results. In brief, the optimal cutoff threshold values were determined at the point on the ROC curve at which Youden index (sensitivity + [100% − specificity]) was maximal. Log-rank test was conducted to examine the significance of the difference between the survival curves. Chi-square tests were used to assess the association between GOLM1 expression and the clinicopathological parameters. Univariate and multivariate Cox regression analysis was performed to examine the independent prognostic value of GOLM1 expression in terms of OS and RFS. The GOLM1 expression was first treated as a continuous variable in univariate and multivariate analysis to avoid the loss of statistical power.19 Univariate and multivariate analyses were also performed by setting its expression as category variables. Linear regression analysis was performed to assess the correlation between GOLM1 expression and the methylation status of the CpG islands. P < .05 was considered statistically significant.
Results
Golgi Membrane Protein 1 Was Significantly Upregulated in LUAD and LUSC Compared to Normal Lung Tissues
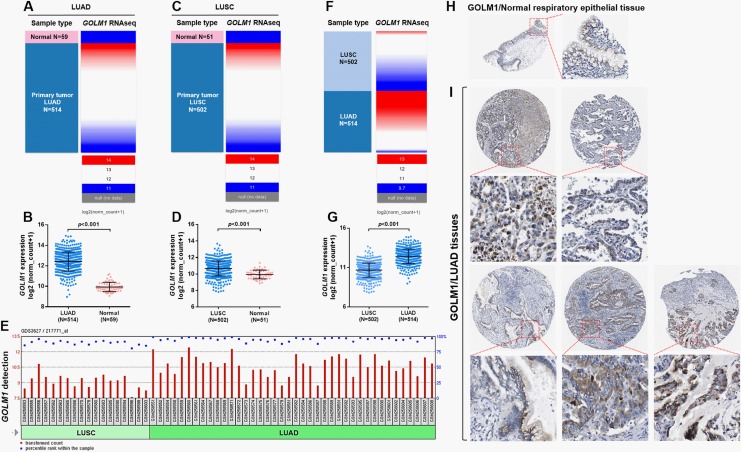
Using RNA-seq data in TCGA, we compared GOLM1 expression between cancerous and normal lung tissues. Results showed that both LUAD and LUSC tissues (N = 514 and 502, respectively) had significantly elevated GOLM1 expression compared to their respective normal controls (N = 59 and 51 respectively; Figure 1A–D). Using data from 1 previous array (GSE10245) that compared gene expression profiles between 40 cases with LUAD and 18 cases with LUSC, we found that GOLM1 expression was significantly higher in LUAD tissues compared to LUSC tissues (Figure 1E). Group comparison using RNA-seq data from TCGA also confirmed substantially increased GOLM1 expression in LUAD compared to LUSC tissues (Figure 1F-G). The IHC staining images in the HPA showed that normal respiratory epithelial tissues had moderate GOLM1 staining in cytoplasm and membrane (Figure 1H). In comparison, in 5 cases of LUAD tissues, nearly all cancer cells had moderate to high GOLM1 staining (Figure 1I).
Figure 1.
Golgi membrane protein 1 (GOLM1) was significantly upregulated in lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) compared to normal lung tissues. A-D, Heatmap (A and C) and plot chart (B and D) of GOLM1 expression in LUAD (A and B) and LUSC (C and D) tissues compared to their respective normal controls. E, The GOLM1 detection call in 40 cases with LUAD and 18 cases with LUSC. Comparison was performed using data from GSE10245,15 with GEO2R. F and G, Heatmap (F) and plot chart (G) comparing GOLM1 expression between LUAD (N = 514) and LUSC (N = 502) cases in The Cancer Genome Atlas (TCGA). H and I, Immunohistochemistry (IHC) staining of GOLM1 expression in normal respiratory epithelial tissue (H) and in 5 cases of normal LUAD tissues. Images were obtained from http://www.proteinatlas.org/ENSG00000135052-GOLM1/pathology/tissue/lung+cancer#ihc.
Comparison of GOLM1 Expression Between the Groups With Different Survival Outcomes
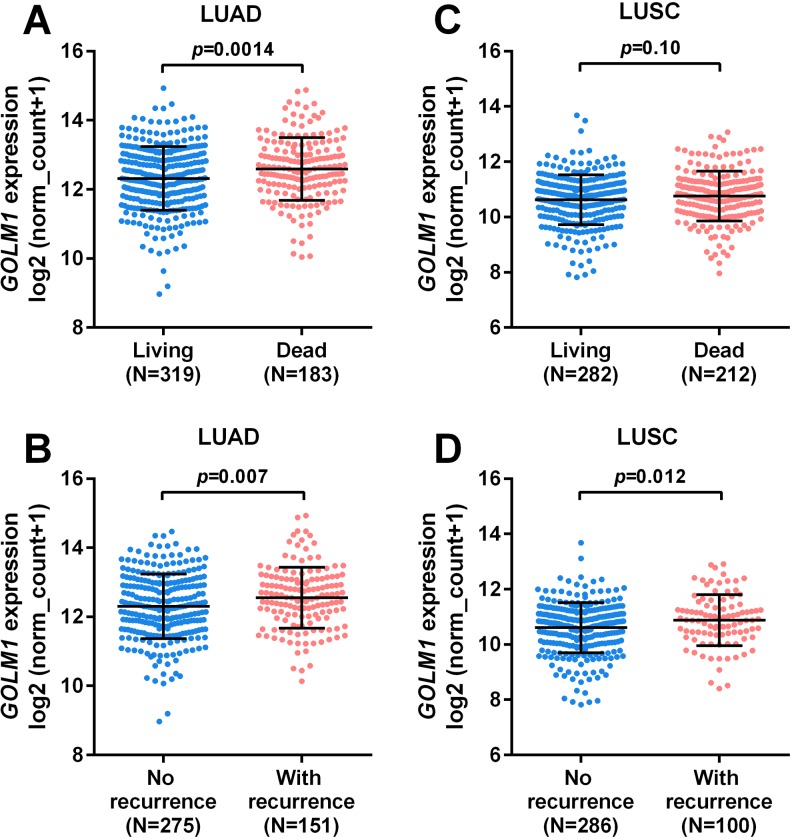
Using OS and RFS as survival indicators, we compared the expression of GOLM1 in patients with LUAD/LUSC with different survival outcomes. Results showed that in patients with LUAD, GOLM1 expression was significantly higher in the deceased and recurrence groups compared to their respective control groups (P = .0014 and .007, respectively; Figure 2A and B). In comparison, no significant difference was observed between the living and dead patients with LUSC (Figure 2C). But the patients with LUSC with disease recurrence had a significantly higher expression of GOLM1 compared to the group without recurrence (P = .012; Figure 2D).
Figure 2.
Comparison of Golgi membrane protein 1 (GOLM1) expression between the groups with different survival outcomes. A-D, Comparison of GOLM1 expression between the deceased and the living groups (A and C) and between the groups with or without recurrence (B and D) in patients with lung adenocarcinoma (LUAD; A-B) and in patients with lung squamous cell carcinoma (LUSC; C-D). Survival data were obtained from The Cancer Genome Atlas (TCGA).
High GOLM1 Expression Was Associated With Poor OS and RFS in Patients With LUAD, but not in Patients With LUSC
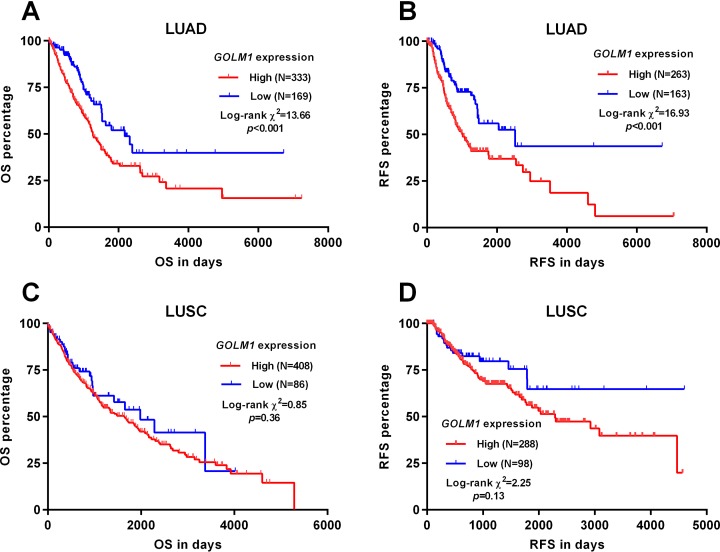
By generating Kaplan-Meier survival curves, we analyzed the association between GOLM1 expression and OS/RFS in patients with LUAD and LUSC, respectively. Both patients with LUAD and LUSC were divided into high/low GOLM1 expression group by using the best cutoff model. Results showed that the high GOLM1 expression group had significantly worse OS (P < .001) and RFS (P < .001) compared to the low GOLM1 expression group (Figure 3A and B). In comparison, there was no significant association between GOLM1 expression and OS or RFS in patients with LUSC (P = .36 and .13, respectively; Figure 3C and D).
Figure 3.
High Golgi membrane protein 1 (GOLM1) expression was associated with poor overall survival (OS) and recurrence-free survival (RFS) in patients with lung adenocarcinoma (LUAD), but not in patients with lung squamous cell carcinoma (LUSC). A-D, Kaplan-Meier curves of OS (A and C) and RFS (B and D) in patients with LUAD (A-B) and LUSC (C-D), respectively. Both patients with LUAD and LUSC were divided into high/low GOLM1 expression group using the best cutoff model. Survival data were obtained from The Cancer Genome Atlas (TCGA).
The association between GOLM1 expression and the clinicopathological features in patients with LUAD is summarized in Table 1. Chi-square analysis showed that the high GOLM1 expression group was associated with a significantly higher rate of recurrence (112/275 vs 39/151, P = .002) and death (140/333 vs 43/169, P < .001), compared to the low GOLM1 expression group (Table 1). No significant difference was observed in other clinicopathological features. Then, we conducted univariate and multivariate analyses to examine the independent prognostic value of GOLM1 expression in terms of OS and RFS. Results showed that advanced pathologic stages (III/IV), nodal invasion, with residual tumors, and increased GOLM1 expression were associated with unfavorable OS (Table 2) and RFS (Table 3) in patients with LUAD. Multivariate analysis showed that increased GOLM1 expression was an independent prognostic indicator of poor OS (HR: 1.30, 95% CI: 1.11-1.54, P = .002, as a continuous variable; HR: 1.87, 95% CI: 1.32-2.66, P < .001, as category variables; Table 2) and RFS (HR: 1.37, 95% CI: 1.14-1.64, P = .001, as a continuous variable; HR: 2.09, 95% CI: 1.45-3.01, P < .001, as category variables; Table 3).
Table 1.
Comparison of the Clinicopathological Parameters Between High and Low GOLM1 Expression Groups in Patients With LUAD.a
| Parameters | GOLM1 Expression | χ2 | P Value | ||
|---|---|---|---|---|---|
| High, N = 333 | Low, N = 169 | ||||
| Age, mean ± SD | 64.77 ± 9.82 | 66.42 ± 10.15 | .082 | ||
| Gender | Female | 178 | 93 | 0.11 | .74 |
| Male | 155 | 76 | |||
| Smoking history | 2/3/4/5 | 282 | 134 | 1.80 | .18 |
| 1 | 43 | 29 | |||
| No data | 8 | 6 | |||
| Pathologic stage | III/IV | 76 | 30 | 1.58 | 1.26 |
| I/II | 253 | 135 | |||
| Discrepancy/no data | 4 | 4 | |||
| Nodal invasion | N0 | 208 | 116 | 3.40 | .07 |
| N1/2/3 | 121 | 46 | |||
| NX/no data | 4 | 7 | |||
| Residual tumors | R0 | 228 | 108 | 0.20 | .65 |
| R1/R2 | 10 | 6 | |||
| RX/no data | 95 | 55 | |||
| Radiation therapy | No | 255 | 133 | 0.021 | .89 |
| Yes | 40 | 20 | |||
| No data | 38 | 16 | |||
| Targeted molecular therapy | No | 193 | 103 | 0.35 | .56 |
| Yes | 102 | 48 | |||
| No data | 38 | 18 | |||
| Recurrence status | No | 163 | 112 | 9.46 | .0021 |
| Yes | 112 | 39 | |||
| No data | 58 | 18 | |||
| Living status | Living | 193 | 126 | 13.33 | <.001 |
| Dead | 140 | 43 | |||
Abbreviations: GOLM1, Golgi membrane protein; LUAD, lung adenocarcinoma; R0, No residual tumor; R1, Microscopic residual tumor; R2, Macroscopic residual tumor; RX, The presence of residual tumor cannot be assessed; SD, standard deviation.
a Smoking history: 1, lifelong nonsmoker; 2, current smoker; 3, current reformed smoker (for >15 years); 4, current reformed smoker (for ≤15 years); 5, current reformed smoker (duration not specified).
Table 2.
Univariate and Multivariate Analysis of Overall Survival in Patients With LUAD.a
| Parameters | Univariate Analysis | Multivariate Analysis | ||||||
|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | |||
| Lower | Upper | Lower | Upper | |||||
| GOLM1 (continuous variable) | ||||||||
| Age (continuous) | 1.01 | 0.99 | 1.02 | .33 | ||||
| Gender, female vs male | 0.94 | 0.70 | 1.26 | .67 | ||||
| Smoking history, 2/3/4/5 vs 1 | 0.91 | 0.60 | 1.38 | .66 | ||||
| Pathologic stage III/IV vs I/II | 2.65 | 1.94 | 3.61 | <.001 | 1.74 | 1.20 | 2.52 | .003 |
| Nodal status, positive vs negative | 2.57 | 1.91 | 3.45 | <.001 | 1.87 | 1.32 | 2.65 | <.001 |
| Residual tumors, yes vs no | 3.94 | 2.20 | 7.03 | <.001 | 3.28 | 1.80 | 5.97 | <.001 |
| GOLM1 | 1.30 | 1.11 | 1.53 | .001 | 1.30 | 1.11 | 1.54 | .002 |
| GOLM1 (category variables) | ||||||||
| Age (continuous) | 1.01 | 0.99 | 1.02 | .33 | ||||
| Gender, female vs male | 0.94 | 0.70 | 1.26 | .67 | ||||
| Smoking history, 2/3/4/5 vs 1 | 0.91 | 0.60 | 1.38 | .66 | ||||
| Pathologic stage III/IV vs I/II | 2.65 | 1.94 | 3.61 | <.001 | 1.75 | 1.21 | 2.53 | .003 |
| Nodal status, positive vs negative | 2.57 | 1.91 | 3.45 | <.001 | 1.85 | 1.30 | 2.61 | .001 |
| Residual tumors, yes vs no | 3.94 | 2.20 | 7.03 | <.001 | 3.28 | 1.81 | 5.94 | <.001 |
| GOLM1 | 1.90 | 1.35 | 2.67 | <.001 | 1.87 | 1.32 | 2.66 | <.001 |
Abbreviations: CI, confidence interval; HR, hazard ratio; GOLM1, Golgi membrane protein 1; LUAD, lung adenocarcinoma.
a Pathologic stage, nodal status, residual tumors, and GOLM1 expression were included in multivariable analysis.
Table 3.
Univariate and Multivariate Analysis of Recurrence-Free Survival in Patients With LUAD.a
| Parameters | Univariate Analysis | Multivariate Analysis | ||||||
|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | |||
| Lower | Upper | Lower | Upper | |||||
| GOLM1 (continuous variable) | ||||||||
| Age (continuous) | 1.01 | 0.99 | 1.03 | .32 | ||||
| Gender, female vs male | 1.10 | 0.79 | 1.52 | .57 | ||||
| Smoking history, 2/3/4/5 vs. 1 | 1.21 | 0.75 | 1.94 | .44 | ||||
| Pathologic stage III/IV vs I/II | 1.71 | 1.17 | 2.51 | .006 | 1.34 | 0.84 | 2.15 | .22 |
| Nodal status, positive vs negative | 1.63 | 1.18 | 2.26 | .003 | 1.26 | 0.84 | 1.88 | .26 |
| Residual tumors, yes vs no | 3.81 | 1.84 | 7.89 | <.001 | 3.87 | 1.81 | 8.27 | <.001 |
| GOLM1 | 1.37 | 1.15 | 1.63 | .001 | 1.37 | 1.14 | 1.64 | .001 |
| GOLM1 (category variables) | ||||||||
| Age (continuous) | 1.01 | 0.99 | 1.03 | .32 | ||||
| Gender, female vs male | 1.10 | 0.79 | 1.52 | .57 | ||||
| Smoking history, 2/3/4/5 vs 1 | 1.21 | 0.75 | 1.94 | .44 | ||||
| Pathologic stage III/IV vs I/II | 1.71 | 1.17 | 2.51 | .006 | 1.32 | 0.83 | 2.10 | .25 |
| Nodal status, positive vs negative | 1.63 | 1.18 | 2.26 | .003 | 1.26 | 0.84 | 1.89 | .26 |
| Residual tumors, yes vs no | 3.81 | 1.84 | 7.89 | <.001 | 3.99 | 1.87 | 8.48 | <.001 |
| GOLM1 | 2.08 | 1.46 | 2.97 | <.001 | 2.09 | 1.45 | 3.01 | <.001 |
Abbreviations: CI, confidence interval; HR, hazard ratio; GOLM1, Golgi membrane protein 1; LUAD, lung adenocarcinoma.
a Clinical stage, nodal status, residual tumors, and GOLM1 expression were included in multivariable analysis.
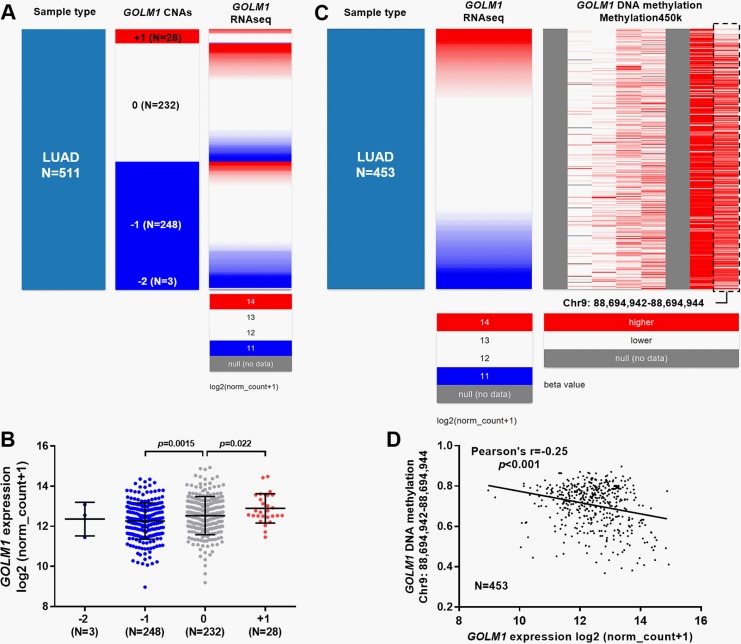
Expression of GOLM1 Was Modulated by DNA CNAs and Methylation Status in LUAD
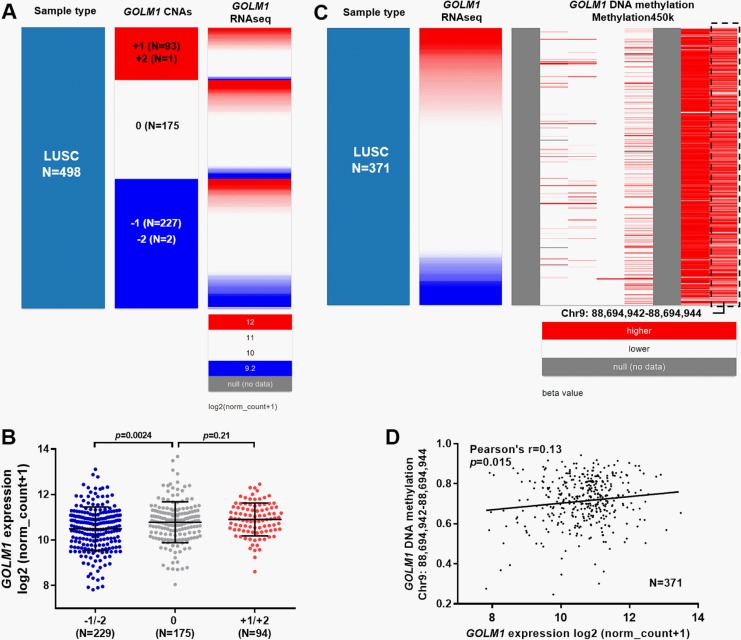
By examining CNAs in 511 cases of LUAD, we found 248 (48.5%) cases had heterozygous loss (−1), which was associated with significantly decreased GOLM1 expression compared to the copy neutral cases (0, N = 232; Figure 4A and B). Besides, 28 (5.5%) cases had low-level copy gain (+1), which was associated with significantly increased GOLM1 expression (Figure 4A and B). Then, we examined the methylation status of 6 CpG sites in GOLM1 DNA in 453 patients with LUAD. Interestingly, we found that the methylation status of 1 CpG site (chr9: 88,694,942-88,694,944) showed a negative correlation with GOLM1 expression (Figure 4C). By performing linear regression analysis, we confirmed a weak negative correlation (Pearson r = −0.25, P < .001; Figure 4D). Then, we examined the association between GOLM1 expression and its DNA CNAs/methylation in patients with LUSC with available data. Results showed that GOLM1 DNA amplification (+1/+2) was more common in patients with LUSC (n = 94, 18.9%; Figure 5A). However, the DNA amplification was not necessarily associated with GOLM1 upregulation, compared to the copy neutral cases (n = 175; Figure 5B, P = .21). Inconsistent with LUAD, DNA deletion was associated with decreased GOLM1 expression in patients with LUSC (Figure 5A and B, P = .0024). Regarding GOLM1 DNA methylation, we failed to identify apparent association between GOLM1 expression and the methylation of the 6 CpG sites (Figure 5C). The CpG (chr9: 88,694,942-88,694,944) site negatively correlated with GOLM1 expression in patients with LUAD and was not relevant to GOLM1 expression in patients with LUSC (Pearson r = 0.13; Figure 5D).
Figure 4.
The correlation between Golgi membrane protein 1 (GOLM1) expression and its DNA copy number alterations (CNAs) or methylation in patients with lung adenocarcinoma (LUAD). A and B, Heatmap (A) and plot chart (B) of GOLM1 expression in groups with different CNAs in patients with LUAD. Homozygous deletion (−2); heterozygous loss (−1), copy neutral (0), and low-level copy gain (+1) groups. C and D, Heatmap (C) and regression analysis (D) of the correlation between GOLM1 expression and its DNA methylation in patients with LUAD.
Figure 5.
The correlation between Golgi membrane protein 1 (GOLM1) expression and its DNA CNAs or methylation in patients with lung squamous cell carcinoma (LUSC). A and B, Heatmap (A) and plot chart (B) of GOLM1 expression in groups with different CNAs in patients with LUSC. Homozygous deletion (−2); heterozygous loss (−1), copy neutral (0), low-level copy gain (+1), and high-level amplification (+2) groups. C and D, Heatmap (C) and regression analysis (D) of the correlation between GOLM1 expression and its DNA methylation in patients with LUSC.
Discussion
The Golgi apparatus is an organelle responsible for posttranslational modification and sorting of protein in mammalian cells. It is polarized in both structure and function, with cis- and trans-side executing different roles. Dysregulated functions of the Golgi apparatus are associated with malignant transformation and pathological development of cancers.20-22 Golgi membrane protein 1 is a cis-Golgi-localized protein which is overexpressed in multiple types of cancer and is closely associated with cancer progression.2,23,24
Some previous studies found that GOLM1 might be a potential diagnostic and prognostic biomarker in some cancers. Prostate tissue GOLM1 RNA might outperform serum prostate-specific antigen in detecting prostate cancer.4 In HCC, GOLM1 expression might be a promising biomarker of chemotherapeutic resistance25 and was associated with poor OS as well as disease-free survival.26 One recent study reported that GOLM1 might also be a valuable biomarker in NSCLC.9 Its upregulation was associated with lymph node metastasis and advanced TNM Classification of Malignant Tumors (TNM) stage of NSCLC and might independently predict poor OS.9 Both LUAD and LUSC are 2 major subtypes of NSCLC but actually are 2 molecularly heterogeneous diseases which have different prognostic markers. The differences in these subtypes are quite complex and far from being fully understood. Expression of PD-L1 is a significant poor prognostic factor in patients with LUAD, but not in those with LUSC.27 SOX2 gene amplification and protein overexpression are associated with better survival outcome in LUSC.13 In comparison, SOX2 upregulation is associated with increased cancer stem cell properties and unfavorable survival in LUAD28,29 (just to name a few). Therefore, it is meaningful to examine the prognostic value of GOLM1 in different subtypes of NSCLC.
In this study, using data from GEO data sets and TCGA-LUAD/LUSC, we found that GOLM1 was significantly upregulated in both LUAD and LUSC tissues compared to the normal controls. However, GOLM1 expression was higher in LUAD tissues than in LUSC tissues. More importantly, using over 10 years’ survival data from 502 patients with LUAD and 494 patients with LUSC, we found that high GOLM1 expression was associated with unfavorable OS and RFS in patients with LUAD, but not in patients with LUSC. The following univariate and multivariate analysis confirmed that increased GOLM1 expression was an independent prognostic indicator of poor OS (HR: 1.30, 95% CI: 1.11-1.54, P = .002, as a continuous variable; HR: 1.87, 95% CI: 1.32-2.66, P < .001, as category variables) and RFS (HR: 1.37, 95% CI: 1.14-1.64, P = .001, as a continuous variable; HR: 2.09, 95% CI: 1.45-3.01, P < .001, as category variables) in patients with LUAD. Using data from the Kaplan-Meier plotter, we confirmed that GOLM1 expression was irrelevant to survival outcomes in patients with LUSC (Supplementary Figure 1A and B). However, we also failed to validate the association between GOLM1 expression and the poor survival in patients with LUAD in this database (Supplementary Figure 1C and D). Actually, the Kaplan-Meier plotter used data from 1715 samples of 10 independent data sets,18 among which the clinicopathological parameters (such as ethnicity, previous chemotherapy, and radiotherapy) of the patients varied significantly. The heterogeneity of the studies might contribute to the different results obtained from TCGA and the Kaplan-Meier plotter. In the future, prospective studies based on a large patient cohort could be performed to validate the findings.
Golgi membrane protein 1 can regulate several signaling pathways in different cancers. In prostate cancer, it activates the PI3K-AKT-mTOR signaling pathway, thereby promoting cancer cell proliferation, migration, and invasion and inhibiting apoptosis.3 Its expression is suppressed by miR-27b and miR-143/145 cluster in prostate cancer cells.30,31 In HCC, GOLM1 is a critical gene driving cancer cell metastasis. It can selectively interact with epidermal growth factor receptor (EGFR) and supports appropriate anchoring and recycling of EGFR/Receptor tyrosine kinase (RTK) between the trans-Golgi network and the plasma membrane, thereby resulting in prolonged activation of the downstream kinases.23,32 As a crucial signaling transductor, EGFR plays a crucial role in regulating cell proliferation, differentiation, and apoptosis in both normal and cancer cells, via binding with its specific ligands, such as EGF and transforming growth factor (TGF) α.33 In addition, it also promotes the epithelial–mesenchymal transition through the TGF-β1/Smad2 signaling pathway.34 Actually, activating EGFR mutations are quite common in patients with LUAD (approximately 14%-19% of Western patients and 40%-48% of Asian patients) but are far less common in patients with LUSC (around 15% in Asian patients).35,36 Therefore, as an important modulator of EGFR/RTK cell surface recycling, it is possible that GOLM1 upregulation may exaggerate the malignant behaviors of LUAD. This also might help to explain the prognostic difference in GOLM1 between LUAD and LUSC. However, it is necessary to explore the molecular regulative network of GOLM1 in LUAD in future studies.
In this study, we also preliminarily examined the potential mechanisms of GOLM1 dysregulation in NSCLC. By examining GOLM1 DNA CNAs in 511 patients with LUAD and methylation in 453 patients with LUAD, we found that GOLM1 expression was positively correlated with its copy numbers and negatively correlated with its DNA methylation. Therefore, we infer that CNAs and DNA methylation might be 2 important mechanisms of dysregulated GOLM1 in LUAD. In comparison, although GOLM1 expression was significantly decreased in GOLM1 DNA deletion group in patients with LUSC, DNA amplification was not necessarily associated with increased GOLM1 expression. In addition, DNA methylation of the CpG site (chr9: 88,694,942-88,694,944) was irrelevant to GOLM1 expression in patients with LUSC. These findings suggest that GOLM1 expression might be regulated by different mechanisms in the subtypes of NSCLC, which need to be explored by molecular studies in the future.
Conclusion
Golgi membrane protein 1 might serve as a valuable prognostic biomarker in LUAD, but not in LUSC. In addition, DNA CNAs and methylation might be 2 important mechanisms of dysregulated GOLM1 in LUAD.
Supplemental Material
Supplementary_figure_1 for Increased GOLM1 Expression Independently Predicts Unfavorable Overall Survival and Recurrence-Free Survival in Lung Adenocarcinoma by Xi Liu, Lei Chen, and Tao Zhang in Cancer Control
Footnotes
Authors’ Note: Xi Liu and Lei Chen contributed equally to this study.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by a grant from Beijing Natural Science Foundation (7182158).
Supplemental Material: Supplementary material for this article is available online
References
- 1. Kladney RD, Bulla GA, Guo L, et al. GP73, a novel Golgi-localized protein upregulated by viral infection. Gene. 2000;249(1-2):53–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wei S, Dunn TA, Isaacs WB, De Marzo AM, Luo J. GOLPH2 and MYO6: putative prostate cancer markers localized to the Golgi apparatus. Prostate. 2008;68(13):1387–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yan G, Ru Y, Wu K, et al. GOLM1 promotes prostate cancer progression through activating PI3K-AKT-mTOR signaling. Prostate. 2018;78(3):166–177. [DOI] [PubMed] [Google Scholar]
- 4. Varambally S, Laxman B, Mehra R, et al. Golgi protein GOLM1 is a tissue and urine biomarker of prostate cancer. Neoplasia. 2008;10(11):1285–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Li W, Wang X, Li B, Lu J, Chen G. Diagnostic significance of overexpression of Golgi membrane protein 1 in prostate cancer. Urology. 2012;80(4):952.e951–957. [DOI] [PubMed] [Google Scholar]
- 6. Chen X, Wang Y, Tao J, et al. mTORC1 up-regulates GP73 to promote proliferation and migration of hepatocellular carcinoma cells and growth of xenograft tumors in mice. Gastroenterology. 2015;149(3):741–752.e714. [DOI] [PubMed] [Google Scholar]
- 7. Bao YX, Cao Q, Yang Y, et al. Expression and prognostic significance of Golgiglycoprotein73 (GP73) with epithelial-mesenchymal transition (EMT) related molecules in hepatocellular carcinoma (HCC). Diagn Pathol. 2013;8:197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mao Y, Yang H, Xu H, et al. Golgi protein 73 (GOLPH2) is a valuable serum marker for hepatocellular carcinoma. Gut. 2010;59(12):1687–1693. [DOI] [PubMed] [Google Scholar]
- 9. Zhang Y, Hu W, Wang L, Han B, Lin R, Wei N. Association of GOLPH2 expression with survival in non-small-cell lung cancer: clinical implications and biological validation. Biomark Med. 2017;11(11):967–977. [DOI] [PubMed] [Google Scholar]
- 10. Zhang F, Gu Y, Li X, Wang W, He J, Peng T. Up-regulated Golgi phosphoprotein 2 (GOLPH2) expression in lung adenocarcinoma tissue. Clin Biochem. 2010;43(12):983–991. [DOI] [PubMed] [Google Scholar]
- 11. Schmid-Bindert G. Update on antiangiogenic treatment of advanced non-small cell lung cancer (NSCLC). Target Oncol. 2013;8(1):15–26. [DOI] [PubMed] [Google Scholar]
- 12. Zhou S, Wang P, Su X, et al. High ECT2 expression is an independent prognostic factor for poor overall survival and recurrence-free survival in non-small cell lung adenocarcinoma. PLoS One. 2017;12(10):e0187356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wilbertz T, Wagner P, Petersen K, et al. SOX2 gene amplification and protein overexpression are associated with better outcome in squamous cell lung cancer. Mod Pathol. 2011;24(7):944–953. [DOI] [PubMed] [Google Scholar]
- 14. Huang ZC, Li H, Sun ZQ, et al. Distinct prognostic roles of HSPB1 expression in non-small cell lung cancer. Neoplasma. 2018;65(1):161–166. [DOI] [PubMed] [Google Scholar]
- 15. Kuner R, Muley T, Meister M, et al. Global gene expression analysis reveals specific patterns of cell junctions in non-small cell lung cancer subtypes. Lung Cancer. 2009;63(1):32–38. [DOI] [PubMed] [Google Scholar]
- 16. Uhlen M, Fagerberg L, Hallstrom BM, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. [DOI] [PubMed] [Google Scholar]
- 17. Uhlen M, Oksvold P, Fagerberg L, et al. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol. 2010;28(12):1248–1250. [DOI] [PubMed] [Google Scholar]
- 18. Gyorffy B, Surowiak P, Budczies J, Lanczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8(12):e82241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mazumdar M, Smith A, Bacik J. Methods for categorizing a prognostic variable in a multivariable setting. Stat Med. 2003;22(4):559–571. [DOI] [PubMed] [Google Scholar]
- 20. Tokuda E, Itoh T, Hasegawa J, et al. Phosphatidylinositol 4-phosphate in the Golgi apparatus regulates cell-cell adhesion and invasive cell migration in human breast cancer. Cancer Res. 2014;74(11):3054–3066. [DOI] [PubMed] [Google Scholar]
- 21. He Y, Wang J, Gou L, et al. Comprehensive analysis of expression profile reveals the ubiquitous distribution of PPPDE peptidase domain 1, a Golgi apparatus component, and its implications in clinical cancer. Biochimie. 2013;95(7):1466–1475. [DOI] [PubMed] [Google Scholar]
- 22. Veldman RJ, Klappe K, Hinrichs J, et al. Altered sphingolipid metabolism in multidrug-resistant ovarian cancer cells is due to uncoupling of glycolipid biosynthesis in the Golgi apparatus. FASEB J. 2002;16(9):1111–1113. [DOI] [PubMed] [Google Scholar]
- 23. Ye QH, Zhu WW, Zhang JB, et al. GOLM1 modulates EGFR/RTK cell-surface recycling to drive hepatocellular carcinoma metastasis. Cancer Cell. 2016;30(3):444–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Donizy P, Kaczorowski M, Biecek P, Halon A, Szkudlarek T, Matkowski R. Golgi-related proteins GOLPH2 (GP73/GOLM1) and GOLPH3 (GOPP1/MIDAS) in cutaneous melanoma: patterns of expression and prognostic significance. Int J Mol Sci. 2016;17(10):1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ye JZ, Yan SM, Yuan CL, et al. GP73 level determines chemotherapeutic resistance in human hepatocellular carcinoma cells. J Cancer. 2018;9(2):415–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen MH, Jan YH, Chang PM, et al. Expression of GOLM1 correlates with prognosis in human hepatocellular carcinoma. Ann Surg Oncol. 2013;(20 suppl 3):S616–S624. [DOI] [PubMed] [Google Scholar]
- 27. Shimoji M, Shimizu S, Sato K, et al. Clinical and pathologic features of lung cancer expressing programmed cell death ligand 1 (PD-L1). Lung Cancer. 2016;98:69–75. [DOI] [PubMed] [Google Scholar]
- 28. Chen S, Xu Y, Chen Y, et al. SOX2 gene regulates the transcriptional network of oncogenes and affects tumorigenesis of human lung cancer cells. PLoS One. 2012;7(5):e36326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Karachaliou N, Rosell R, Viteri S. The role of SOX2 in small cell lung cancer, lung adenocarcinoma and squamous cell carcinoma of the lung. Transl Lung Cancer Res. 2013;2(3):172–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Goto Y, Kojima S, Nishikawa R, et al. The microRNA-23b/27b/24-1 cluster is a disease progression marker and tumor suppressor in prostate cancer. Oncotarget. 2014;5(17):7748–7759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kojima S, Enokida H, Yoshino H, et al. The tumor-suppressive microRNA-143/145 cluster inhibits cell migration and invasion by targeting GOLM1 in prostate cancer. J Hum Genet. 2014;59(2):78–87. [DOI] [PubMed] [Google Scholar]
- 32. Zhu W, Qin L. GOLM1-regulated EGFR/RTK recycling is a novel target for combating HCC metastasis. Sci China Life Sci. 2017;60(1):98–101. [DOI] [PubMed] [Google Scholar]
- 33. Lo HW, Hsu SC, Xia W, et al. Epidermal growth factor receptor cooperates with signal transducer and activator of transcription 3 to induce epithelial-mesenchymal transition in cancer cells via up-regulation of TWIST gene expression. Cancer Res. 2007;67(19):9066–9076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yang HJ, Liu GL, Liu B, Liu T. GP73 promotes invasion and metastasis of bladder cancer by regulating the epithelial-mesenchymal transition through the TGF-beta1/Smad2 signalling pathway. J Cell Mol Med. 2018;22(3):1650–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Han B, Tjulandin S, Hagiwara K, et al. EGFR mutation prevalence in Asia-Pacific and Russian patients with advanced NSCLC of adenocarcinoma and non-adenocarcinoma histology: the IGNITE study. Lung Cancer. 2017;113:37–44. [DOI] [PubMed] [Google Scholar]
- 36. Dearden S, Stevens J, Wu YL, Blowers D. Mutation incidence and coincidence in non small-cell lung cancer: meta-analyses by ethnicity and histology (mutMap). Ann Oncol. 2013;24(9):2371–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary_figure_1 for Increased GOLM1 Expression Independently Predicts Unfavorable Overall Survival and Recurrence-Free Survival in Lung Adenocarcinoma by Xi Liu, Lei Chen, and Tao Zhang in Cancer Control