Abstract
We observed the assembly of a nucleus-like structure in bacteria during viral infection. Using fluorescence microscopy and cryo-electron tomography, we showed that Pseudomonas chlororaphis phage 201ϕ2-1 assembled a compartment that separated viral DNA from the cytoplasm. The phage compartment was centered by a bipolar tubulin-based spindle, and it segregated phage and bacterial proteins according to function. Proteins involved in DNA replication and transcription localized inside the compartment, whereas proteins involved in translation and nucleotide synthesis localized outside. Later during infection, viral capsids assembled on the cytoplasmic membrane and moved to the surface of the compartment for DNA packaging. Ultimately, viral particles were released from the compartment and the cell lysed. These results demonstrate that phages have evolved a specialized structure to compartmentalize viral replication.
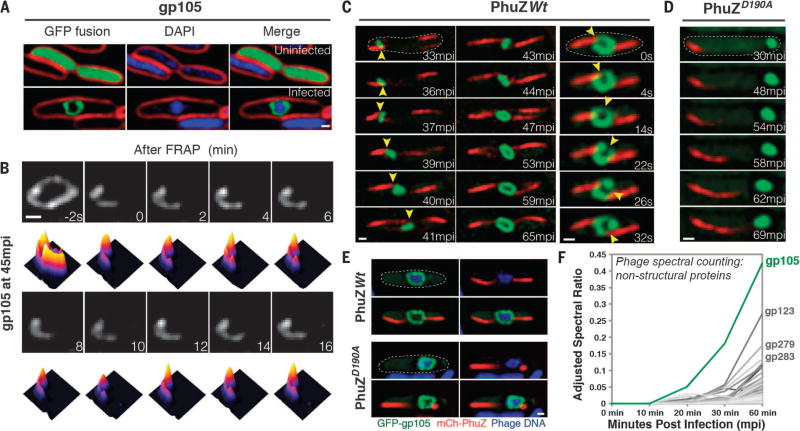
Viruses have been coevolving with cellular life for more than 2 billion years (1). We recently described a bacteriophage reproduction pathway in Pseudomonas species in which a phage-encoded tubulin-like cytoskeletal protein (PhuZ) forms a bipolar spindle that positions replicating phage DNA at the cell midpoint (2–6). To search for other proteins required for this pathway, we used mass spectrometry to identify phage 201ϕ2-1 proteins expressed during lytic growth (7–9) and then created fusions of these proteins to green fluorescent protein (GFP) for localization profiling (table S1). We next examined the localization pattern of each protein in cells before and during phage infection. gp105 is the first and most highly expressed phage protein after infection (Fig. 1F). In uninfected cells, GFP-gp105 was uniformly distributed, but surprisingly, it formed a somewhat spherically shaped structure surrounding phage DNA at the midcell after 45 min of infection (Fig. 1A). This structure was observed in every infected cell, with 87% of cells containing a single structure and the remaining cells containing either two (10%) or three structures (3%) (fig. S1). In photobleaching experiments, fluorescence from GFP-gp105 did not recover after 16 min, which indicates that GFP-gp105 formed a relatively stable structure around the phage DNA (Fig. 1B).
Fig. 1. gp105, an early and highly expressed phage protein, forms a shell around viral DNA during phage infection.
(A) N-terminal fusion of gp105 to GFP (GFP-gp105; green) is diffuse in uninfected P. chlororaphis cells (top) and assembles a shell that encloses the phage DNA at 45 min postinfection (mpi) with phage 201ϕ2-1 (bottom). Cell membranes were stained with FM 4–64 (red), and DNA was stained with DAPI (blue). (B) Fluorescence recovery after photobleaching (FRAP) of the GFP-gp105 shell at 45 mpi. The bleached area of the shell generated at time 0 min does not recover over the course of 16 min. Heat maps show GFP intensity corresponding to the images above. (C and D) Time-lapse imaging (measured in minutes postinfection or seconds) of GFP-gp105 in the presence of mCherry-tagged (red) (C) wild-type PhuZ and (D) mutant PhuZD190A. (C) As indicated by arrowheads, the shell moves to the midcell and rotates (in this case, clockwise) during phage infection in the presence of wild-type PhuZ. See also movies S1 and S3. (D) In the presence of PhuZD190A, the shell remains at the cell pole throughout the experiment and does not rotate. A lack of rotation is sometimes observed in the presence of wild-type PhuZ for larger gp105 shells such as that shown in (B). Dashed circles indicate the border of the cells. (E) Static images of infected host cells expressing GFP-gp105 together with either wild-type mCherry-PhuZ or mutant mCherry-PhuZD190A at 60 mpi. Phage DNA was stained blue with DAPI. (A to E) Scale bars, 0.5 µm. (F) Protein mass spectrometry analysis of phage-infected cells showing putative nonstructural phage proteins until 60 mpi. The gp105 protein (green line) is the most highly expressed phage protein. (See table S1.)
To understand the timing of the formation of this structure, we performed time-lapse microscopy in which we expressed GFP-gp105 in P. chlororaphis and then followed its assembly during phage infection. We also expressed mCherry-PhuZ to simultaneously monitor spindle assembly. Upon addition of phage to the cells, GFP-gp105 formed a small focus near the cell pole and mCherry-PhuZ assembled filaments at both cell poles, setting up the bipolar spindle (Fig. 1C). Over time, PhuZ polymers pushed the GFP-gp105 shell to the cell center (Fig. 1C), confirming that these dynamically unstable filaments center phage DNA by a pushing mechanism. The small GFP-gp105 shell increased in size while being pushed to the midcell [Fig. 1C, 33 to 41 min postinfection (mpi), and movie S1], where it continued to grow (Fig. 1C, 43 to 65 mpi, and movie S1) as DNA replication progressed (5). Centrally positioned GFP-gp105 shells continued to be pushed back and forth by opposing filaments of the bipolar PhuZ spindle, causing them to oscillate near the cell center (fig. S2 and movie S2). When opposing PhuZ filaments pushed transversely on opposite sides of the GFP-gp105 shell, the entire structure rotated in position (Fig. 1C, right, and movie S3). In the presence of a mutant PhuZ protein (PhuZD190A) that is unable to hydrolyze GTP (guanosine triphosphate) and forms static filaments, the GFP-gp105 shell remained at the cell pole and exhibited little motion, suggesting that both movement and rotation of the shell were caused by forces exerted by the PhuZ spindle (Fig. 1, D and E).
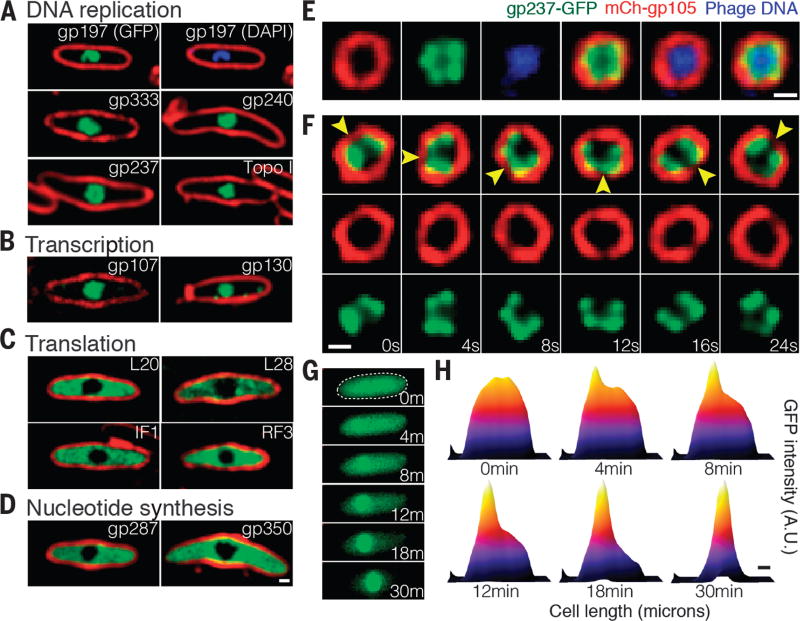
Localization profiling of GFP fusions to 52 phage proteins revealed a high level of organization inside the infected cell in which many proteins were localized inside the gp105 shell along with the phage DNA whereas other proteins were excluded (figs. S3 to S5). Proteins suspected to be involved in DNA replication (Fig. 2A) or transcription (Fig. 2B) colocalized with the DNA inside the shell (fig. S3). This included phage encoded homologs of DNA helicase (gp197), DNA ligase (gp333), RNase H (gp240), RecA (gp237), and two homologs of the β′ subunits of RNA polymerase (gp107 and gp130) (Fig. 2, A and B). In addition, host DNA topoisomerase I was found inside the gp105 shell, indicating that localization to the shell was not limited to phage proteins (Fig. 2A). Three phage proteins of unknown function—gp124, gp125, and gp126—also localized inside the shell, and we thus speculate that they might be involved in DNA processes such as replication, transcription, or recombination (fig. S4). In contrast, GFP fusions to host proteins involved in translation, including the 50S ribosomal proteins L20 and L28, the translation initiation factor 1 (IF1), and the peptide chain release factor 3 (RF3), were located in the cytoplasm outside of the shell (Fig. 2C). Phage homologs of the metabolic enzymes thymidylate kinase (gp287), which catalyzes phosphorylation of thymidine monophosphate to thymidine diphosphate, and thymidylate synthase (gp350), which catalyzes the conversion of deoxyuridine monophosphate to deoxythymidine monophosphate, were also excluded from the shell (Fig. 2D), as was GFP alone (fig. S5). This subcellular compartmentalization of enzymes and DNA resembles the organization of eukaryotic cells containing a nucleus.
Fig. 2. DNA replication and transcription occur inside the phage compartment, whereas translation occurs outside.
(A to D) Localization profiling of GFP-tagged phage and host proteins involved in various functions at 60 mpi. All proteins were fused to GFP (green), DNA was stained with DAPI (blue), and membranes were stained with FM 4–64 (red). (A) Proteins predicted to be involved in DNA replication, including the phage proteins gp237 (RecA homolog), gp197 (helicase homolog), gp333 (ligase homolog), gp240 (RNase H homolog), and host DNA topoisomerase I. (B) Two phage-encoded proteins predicted to be involved in transcription, gp107 and gp130 (RNA polymerase β′ subunit homologs). (C) Host proteins involved in translation, including ribosomal proteins L20 and L28, translation initiation factor 1 (IF1), and peptide chain release factor 3 (RF3). (D) Phage proteins predicted to be involved in nucleotide synthesis. including gp287 (thymidylate kinase homolog) and gp350 (thymidylate synthase homolog). (E), mCherry-gp105 (red), the RecA homolog gp237-GFP (green), and DAPI-stained DNA (blue), demonstrating localization of gp237 and DNA inside the gp105 shell. (F) Time-lapse imaging (in seconds) showing rotation of the compartment and its contents. As indicated by arrowheads, the shell and gp237 rotate together (in this cell, in a counterclockwise direction), suggesting that the entire structure and its contents rotate. See also movie S5. (G) gp237 moves from the host cell cytoplasm to the compartment during phage infection in this 30-min time lapse (m, minutes). The dashed circle indicates the border of the cell. (H) Heat map of GFP intensity of gp237 in the host cell as corresponding to the fluorescent micrographs reveals that gp237 moves into and accumulates in the gp105 shell. (See also movie S4.) Scale bars in (D) to (F) and (H), 0.5 µm. A.U., arbitrary units.
To gain insight into the mechanism by which proteins might enter the structure, we performed time-lapse microscopy on cells that expressed a GFP-tagged copy of the phage-encoded RecA homolog gp237. Before infection, gp237-GFP was located in the cytoplasm, but after addition of phage, it gradually accumulated inside the growing phage compartment and nearly undetectable levels remained in the cytoplasm after 30 min (Fig. 2, G and H, and movie S4). Thus, proteins can translocate to the phage compartment post-translationally. Once inside, gp237-GFP formed foci (Fig. 2E) that rotated together with the mCherry-gp105 shell, completing one rotation in ~30 s, suggesting that the entire structure rotates as a singular unit (Fig. 2F, movie S5, and fig. S6).
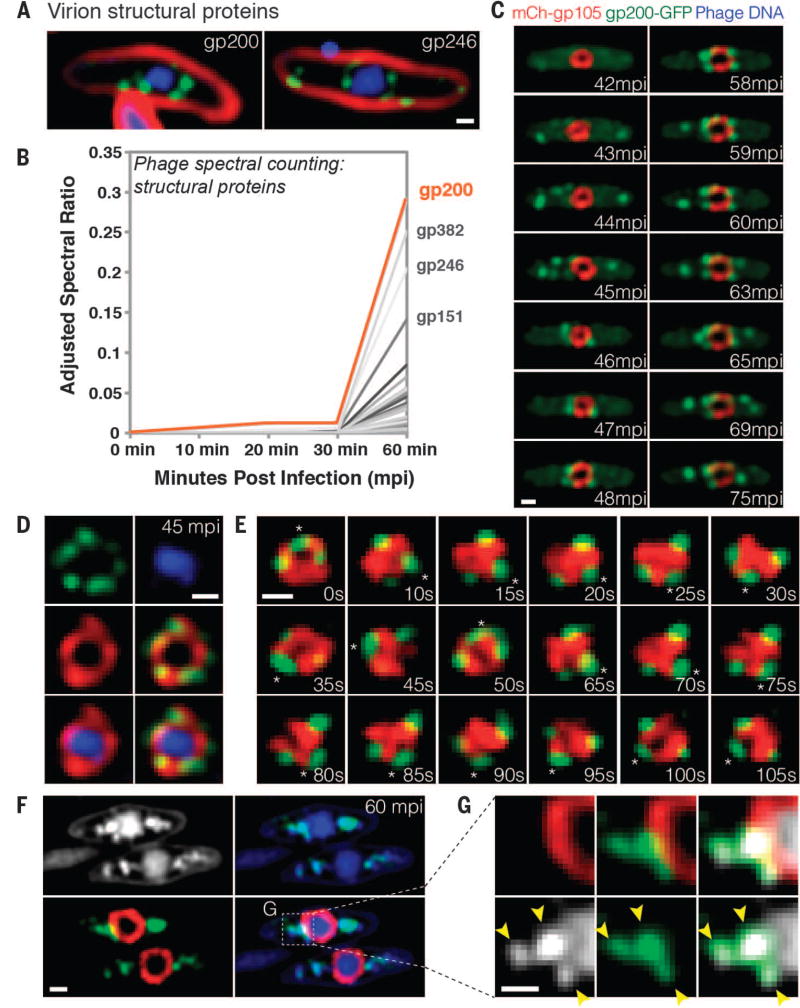
Localization profiling of phage structural proteins allowed us to study phage particle maturation and assembly. GFP fusions to the predicted major capsid protein gp200 and the internal head protein gp246 demonstrated an orchestrated assembly process. Approximately 40 mpi, empty capsids assembled near the cell membrane (Fig. 3C, 43 mpi), forming bright green foci. Capsids (green) then migrated from the membrane to the surface of the phage compartment (red) (Fig. 3C, 46 mpi, and movie S6) and accumulated on the outside of the mCherry-gp105 shell (Fig. 3, C to E). In time-lapse microscopy over shorter time scales, the capsids and the phage compartment rotated together as a single unit (Fig. 3E), undergoing eight successive revolutions in the example shown in movie S7, suggesting that the capsids were tightly docked to the surface. Later, capsids appeared to detach from the phage compartment and migrate away from it (Fig. 3C, 59 to 75 mpi). After 60 min, capsids were costained with 4′,6-diamidino-2-phenylindole (DAPI) (Fig. 3, F and G), showing that DNA packaging occurred while docked at the surface of the phage compartment.
Fig. 3. Capsids migrate to the surface of the phage compartment for DNA encapsidation.
(A) Localization of the predicted phage structural proteins gp200 (major capsid protein) and gp246 (internal head protein). The proteins were fused to GFP (green), membranes were stained with FM 4–64 (red), and DNA was stained with DAPI (blue) at 60 mpi. (B) Mass spectrometry results showing spectral counting of predicted phage structural proteins in infected host cells until 60 mpi; gp200 (orange line) is the most abundant structural protein. (C) Time-lapse microscopy of mCherry-gp105 (red) and the predicted capsid protein gp200-GFP (green), showing that gp200-GFP was initially diffuse (at 42 mpi) and assembled foci on the cell periphery (at 43 to 45 mpi) that move to the gp105 shell (at 45 to 46 mpi), remain attached for 12 min, and then are released from the shell (at 59 to 75 mpi). Foci translocate to the gp105 shell within 1 to 2 min. See also movie S6. (D) Static images showing gp200-GFP (green) on the surface of the mCherry-gp105 shell (red), with DNA (blue) inside, at 45 mpi. (E) Time lapse showing that gp200-GFP foci rotate together with mCherry-gp105 throughout this 105-s experiment. The position of a single capsid (asterisk) was tracked for the duration of the time lapse. See movie S7. (F and G) Static images of infected cells at 60 mpi to show colocalization of gp200-GFP (green) and phage DNA (blue or white) outside the mCherry-gp105 shell (red). The region indicated by the dashed box in (F) is magnified in (G) to more clearly show the colocalization of DNA (white) within the gp200 foci (green). Arrowheads indicate the colocalization. Scale bars in (A) and (C) to (G), 0.5 µm.
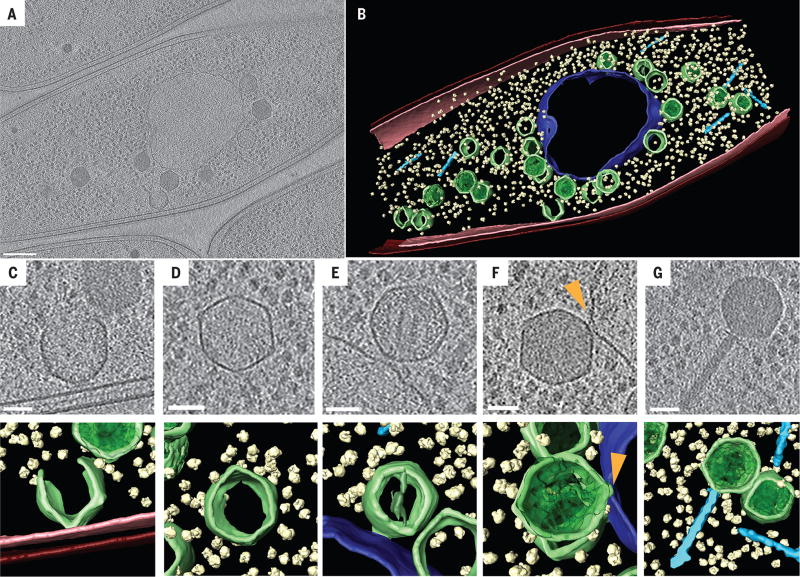
We used cryo-electron tomography (CET) of cryo-focused ion beam (FIB)–milled phage-infected cells to visualize the phage compartment and various steps in phage assembly at high resolution and in a near-native state (10–12). In samples collected at 60 mpi, we observed a central compartment containing a sharply defined but irregularly shaped border (Fig. 4, A and B, and movie S8). The compartment border had an appearance distinct from the cytoplasmic membrane, supporting its composition as a proteinaceous shell with gp105 as a key component. Three-dimensional reconstructions showed a largely continuous envelope with a thickness of ~5 nm forming an apparently enclosed structure. Our fluorescence data indicate that this structure excludes soluble GFP, suggesting that it forms a barrier to protein diffusion (fig. S5). Ribosomes were clearly visible in the cytoplasm but were excluded from the compartment, in agreement with our fluorescence microscopy studies (Fig. 4, A and B). Partially constructed capsids were observed at the bacterial inner membrane (Fig. 4C) where assembly occurs. Capsids containing various amounts of DNA were docked on the surface of the intracellular phage compartment (Fig. 4, B, E, and F) and often a connecting collar was visualized (Fig. 4F, arrow), as well as capsids that appeared to be empty, indicating that this is the site of DNA packaging. Filled capsids and fully assembled phage particles (Fig. 4, B and G) containing tails were observed in the cytoplasm away from the phage compartment. Overall, the CET data were fully consistent with our fluorescence microscopy results and demonstrate that the phage compartment is surrounded by a shell that appears to be contiguous.
Fig. 4. Cryo-electron tomography of the phage compartment during 201ϕ2-1 infection in P. chlororaphis.
(A) Slice through a tomogram of a cryo-focused ion beam–thinned phage-infected cell at 60 mpi. Assembled capsids are docked to an apparently contiguous shell during the process of DNA encapsidation, which produces the darker, filled capsids. Scale bar, 200 nm. (B) Segmentation of the tomogram in (A), showing extracted structures, including the shell (purple), capsids (green), cytoplasmic membrane (pink), outer membrane (red), phage tails (light blue), and ribosomes (yellow). (C to G) Tomographic slices (top) and segmentation images (bottom) of (C) an assembling phage capsid, (D) an empty capsid, (E and F) two capsids docked at the compartment with (E) less or (F) more DNA, and (G) an assembled phage. Scale bars in (C) to (G), 50 nm. The arrowhead in (F) indicates a connecting collar between the capsid and the compartment shell.
Here we describe a nucleus-like structure formed by a virus during infection of bacteria and its role in the phage life cycle (fig. S11). The compartment initially assembles near the cell pole surrounding phage DNA after injection, potentially forming a protective enclosure that shields it from host defense systems. The compartment grows in size as DNA replication proceeds and is centrally positioned by the PhuZ spindle. Previously we showed that phage DNA is centered by PhuZ polymers, but it was unclear how PhuZ filaments might connect to the replicating DNA molecules (4–6). Our results suggest that the nucleus-like compartment described here provides a structure encapsulating the DNA that is pushed by PhuZ filaments. Because phage mRNA transcripts likely originate from inside the compartment, we infer that they must be transported out of the compartment to the cytoplasm to be translated by the ribosomes. Some of the newly expressed proteins, such as those involved in DNA replication and transcription, translocate back into the compartment. How molecules are able to pass in and out of the phage compartment remains unclear. Later during infection, capsids assemble on the cytoplasmic membrane and migrate to the surface of the compartment, where they dock for DNA packaging. After the capsids are filled with DNA, they are released from the surface and coassemble with tails located in the cytoplasm to create mature particles. These results show that phages have evolved a complicated pathway of reproduction in which a bipolar tubulin-based spindle, together with a nucleus-like structure, spatially and temporally organizes DNA replication, transcription, translation, and phage particle assembly. It will be interesting to determine whether this phage-assembly pathway is found in other phages that encode tubulin homologs.
Supplementary Material
Acknowledgments
This research was supported by NIH grants GM031627 (D.A.A.), GM104556 (J.P. and D.A.A.), R01-GM57045 (K.P.), and 1DP2GM123494-01 (E.V.) and by the HHMI (D.A.A.). We acknowledge the use of the UCSD Cryo-Electron Microscopy Facility, which is partially supported by NIH grants to T. S. Baker and a gift from the Agouron Institute to UCSD.
Footnotes
REFERENCES AND NOTES
- 1.Durzyńska J, Goździcka-Józefiak A. Virol. J. 2015;12:169. doi: 10.1186/s12985-015-0400-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Serwer P, et al. Virology. 2004;329:412–424. doi: 10.1016/j.virol.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 3.Thomas JA, et al. Virology. 2008;376:330–338. doi: 10.1016/j.virol.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kraemer JA, et al. Cell. 2012;149:1488–1499. doi: 10.1016/j.cell.2012.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Erb ML, et al. eLife. 2014;3:e03197. [Google Scholar]
- 6.Zehr EA, et al. Structure. 2014;22:539–548. doi: 10.1016/j.str.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guttman M, et al. Proteomics. 2009;9:5016–5028. doi: 10.1002/pmic.200900457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCormack AL, et al. Anal. Chem. 1997;69:767–776. doi: 10.1021/ac960799q. [DOI] [PubMed] [Google Scholar]
- 9.Paoletti AC, et al. Proc. Natl. Acad. Sci. U.S.A. 2006;103:18928–18933. doi: 10.1073/pnas.0606379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Villa E, Schaffer M, Plitzko JM, Baumeister W. Curr. Opin. Struct. Biol. 2013;23:771–777. doi: 10.1016/j.sbi.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 11.Zheng SQ, Palovcak E, Armache J-P, Cheng Y, Agard DA. 2016 http://biorxiv.org/content/early/2016/07/04/061960.
- 12.Oikonomou CM, Jensen GJ. Nat. Rev. Microbiol. 2016;14:205–220. doi: 10.1038/nrmicro.2016.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.