Summary
We recently demonstrated that the large Pseudomonas chlororaphis bacteriophage 201φ2-1 assembles a nucleus-like structure that encloses phage DNA and segregates proteins according to function, with DNA processing proteins inside and metabolic enzymes and ribosomes outside the nucleus. Here we investigate the replication pathway of the Pseudomonas aeruginosa bacteriophages φKZ and φPA3. Bacteriophages φKZ and φPA3 encode a proteinaceous shell that assembles a nucleus-like structure that compartmentalizes proteins and DNA during viral infection. We show that the tubulin-like protein PhuZ encoded by each phage assembles a bipolar spindle that displays dynamic instability and positions the nucleus at midcell. Our results suggest that the phage spindle and nucleus play the same functional role in all three phages, 201φ2-1, φKZ and φPA3, demonstrating that these key structures are conserved among large Pseudomonas phages.
Keywords: Spindle, nucleus, phage, tubulin, PhuZ
Graphical abstract
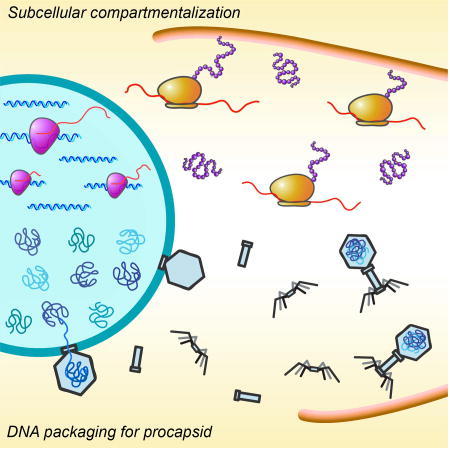
Introduction
Bacteria generally lack a nucleus, the membrane-bound organelle that separates genetic material from the cytoplasm in Eukaryotes. We recently described a nucleus-like structure in bacteria, assembled by phage 201Φ2-1 in Pseudomonas chlororaphis cells during infection (Chaikeeratisak et al., 2017). This “phage nucleus” is bound by a proteinaceous barrier composed of gp105 that encloses viral DNA and separates cellular functions. Like the eukaryotic equivalent, DNA replication and transcription occur inside the phage nucleus, while translation and metabolic enzymes are localized in the cytoplasm. While a membrane bound structure containing chromosomal DNA has been reported in Planctomycetes, it also contains ribosomes, indicating that proteins and DNA are not compartmentalized separately (Boedeker et al., 2017; Fuerst and Sagulenko, 2011; Sagulenko et al., 2017). The subcellular organization observed during phage 201Φ2-1 infection of P. chlororaphis is unique, and prompted us to investigate if it is conserved during replication of other phages.
Positioning of the phage nucleus is controlled by PhuZ, a phage-encoded tubulin which shares only ~18% amino acid sequence identity with the E. coli cell division protein FtsZ (Chaikeeratisak et al., 2017; Kraemer et al., 2012). The crystal structure of PhuZ revealed that it contains a conserved tubulin fold with a long C-terminal aspartic acid rich tail (Kraemer et al., 2012). PhuZ assembles a unique triple stranded polymer in which protofilaments twist around each other and the C-terminal tail plays a key role in making longitudinal contacts with adjacent subunits (Zehr et al., 2014). PhuZ filaments display dynamic instability both in vitro and in vivo (Erb et al., 2014). During phage infection, PhuZ forms a bipolar spindle composed of dynamically unstable filaments that position phage DNA at midcell (Erb et al., 2014). Mutations in the catalytic domain that eliminate PhuZ GTPase activity disrupt both spindle dynamics and phage nucleus positioning and reduce phage burst size by 50% (Erb et al., 2014; Kraemer et al., 2012). The PhuZ spindle is the only known example of a cytoskeletal structure in Bacteria or Archaea that shares three key properties with the eukaryotic spindle: dynamic instability, a bipolar array of filaments, and central positioning of DNA. Phage-encoded tubulins like PhuZ may represent a transitional evolutionary step between simplified bacterial filaments, such as FtsZ and Bacillus plasmid segregation protein TubZ (Aylett et al., 2010; Chen and Erickson, 2008; Larsen et al., 2007; Montabana and Agard, 2014), and more complicated structures like the eukaryotic spindle. Further insight into PhuZ and its relatives may allow us to dissect the evolutionary relationships between the cytoskeletal proteins of phages, bacteria, and eukaryotes.
Here we investigate Pseudomonas aeruginosa phages ΦPA3 (Monson et al., 2011) and ΦKZ (Mesyanzhinov et al., 2002), which encode homologs of PhuZ. Like PhuZ201, both PhuZΦPA3 and PhuZΦKZ assemble filaments in vitro (Aylett et al., 2013; Zehr et al., 2014). The conservation of certain critical interactions in PhuZΦPA3 as well as the pattern produced by a 2D class average of segments of filaments suggest that PhuZΦPA3 also forms a three-stranded polymer like that of PhuZ201 (Zehr et al., 2014). PhuZΦKZ has a tubulin fold and assembles filaments and protofilaments similar to those of PhuZ201 (Aylett et al., 2013). It was previously proposed that despite the high level of conservation seen in the tertiary structures of PhuZΦKZ and PhuZ201, the divergence of the protein sequence might result in different functional roles of these two proteins (Aylett et al., 2013). Here we show that assembly of a nucleus-like structure and the biological function of the phage-encoded tubulin are conserved in the reproduction pathways of these three phages. We demonstrate that the PhuZ tubulins from both phages ΦPA3 and ΦKZ form dynamic bipolar spindles that function to position the phage nucleus at the cell center. ΦPA3 and ΦKZ also encode homologs of the nuclear shell protein gp105 from phage 201Φ2-1. Fluorescence microscopy shows that these homologs similarly form a shell surrounding phage DNA. In addition, we show that each phage encodes a RecA-related protein that localizes inside the nucleus with phage DNA. The major capsid protein is also conserved among these three phages and localizes on the surface of the phage nucleus. Our results suggest that the phage nucleus and tubulin spindle are conserved in the replication pathways of large Pseudomonas phages ΦPA3, ΦKZ, and 201Φ2-1.
Results
Key features of the phage nucleus are conserved in ΦKZ, ΦPA3, and 201Φ2-1
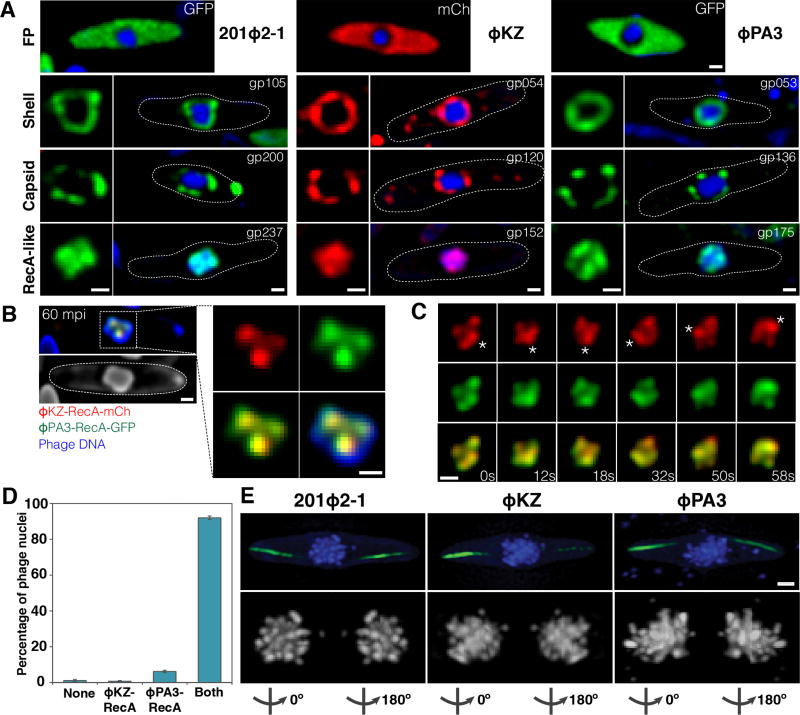
The main component of the phage 201Φ2-1 nuclear shell is gp105, one of the earliest phage proteins expressed after infection. GFP-gp105 fusions assemble a structure that encloses viral DNA, appearing as a ring in cross-sections (Chaikeeratisak et al., 2017). To determine if this nucleus-like structure is conserved in P. aeruginosa phages ΦKZ and ΦPA3, we identified homologs of gp105 in these phages (gp54 in ΦKZ and gp53 in ΦPA3) and tagged them with the fluorescent proteins (FP) GFP or mCherry. When expressed from a plasmid in uninfected cells, these proteins formed small foci that were not specifically localized (Figure S1A), but upon infection, both proteins assembled a spherical structure surrounding phage DNA (Figure 1A, shell). As we previously observed with 201Φ2-1 in P. chlororaphis, the ΦKZ and ΦPA3 shells in P. aeruginosa were centrally positioned at 60 minutes post infection (mpi) (Figure 1A, shell).
Figure 1. 201Φ2-1, ΦKZ, and ΦPA3 assemble a nucleus-like structure that compartmentalizes proteins and DNA during viral replication.
Cells were grown on an agarose pad and the fusion proteins were induced with arabinose at the indicated concentrations. P. chlororaphis was infected with 201Φ2-1 and P. aeruginosa with phages ΦKZ or ΦPA3, and imaged at approximately 60 mpi (A,B,D) or 90 mpi (C,E). Phage DNA is stained with DAPI (blue). These proteins do not assemble in uninfected cells. All scale bars equal 0.5 micron. See also Figure S1.
A) Top row: Fluorescence micrographs of infected P. chlororaphis and infected P. aeruginosa cells expressing fluorescent proteins (GFP, green) or (mCherry, red) alone from the arabinose promoter. Second row: Fluorescence micrographs of infected P. chlororaphis and infected P. aeruginosa cells expressing fluorescent protein fusions to the conserved nuclear shell proteins gp105 of 201Φ2-1 (0.2% arabinose), gp054 of ΦKZ (0.025% arabinose), and gp053 of ΦPA3 (0.025% arabinose). In rows two through four, the square panel on the left is an enlarged image of fluorescent proteins shown within cells whose border is indicated by a dotted line. Third row: Fluorescence micrographs of infected P. chlororaphis and infected P. aeruginosa cells expressing fluorescent protein fusions to the major capsid proteins gp200 of 201Φ2-1 (0.2% arabinose), gp120 of ΦKZ (0.05% arabinose), and gp136 of ΦPA3 (0.05% arabinose). Fourth row: Fluorescence micrographs of infected P. chlororaphis and infected P. aeruginosa cells expressing fluorescent protein fusions to the RecA-like protein gp237 of 201Φ2-1 (0.2% arabinose), gp152 of ΦKZ (0.025% arabinose), and gp175 of ΦPA3 (0.025% arabinose).
B) Co-localization of RecA-related proteins ΦKZ-gp152-mCherry (red) and ΦPA3-gp175-GFP (green) in the 201Φ2-1 nucleus (stained blue by DAPI or gray) during phage 201Φ2-1 infection. P. chlororaphis expressing ΦKZ-gp152-mCherry and ΦPA3-gp175-GFP (0.1% arabinose) was infected with 201Φ2-1 and visualized at 60 mpi. The region indicated by a dashed box is magnified and shown on the right. Scale bar equals 0.5 micron.
C) Time-lapse imaging (seconds) of ΦKZ-gp152-mCherry (red) and ΦPA3-gp175-GFP (green) in the 201Φ2-1 nucleus showing both RecA-related proteins form foci which move together as the 201Φ2-1 nucleus rotates over the course of 58 seconds. The position of the focus (asterisk) was tracked for the duration of the time-lapse. Scale bar equals 0.5 micron. See also Movie S1.
D) Graph showing percentage of 201Φ2-1 nuclei of infected P. chlororaphis cells containing no proteins inside (none), either ΦKZ-gp152 or ΦPA3-gp175 inside, or both RecA-related proteins inside. Data were collected from the infected cells at 60 mpi from at least 3 different fields and are represented as mean ± standard error of the mean (n = 263).
E) SIM images of infected P. chlororaphis and infected P. aeruginosa at 90 mpi showing encapsidated phage clusters localized at midcell as determined by DAPI staining (blue or gray). GFP-PhuZ filaments (green) extend from both cell poles toward the viral nucleus at midcell. 3D-SIM and rotation of the phage nucleus around Y-axis show that the nucleus is surrounded with encapsidated phages (degree of rotation is indicated below each subset). Scale bar equals 0.5 micron.
One hallmark of the nucleus-like structure established by phage 201Φ2-1 is the selective compartmentalization of proteins. For example, we previously demonstrated that the 201Φ2-1 RecA-related protein (gp237-GFP) accumulated inside the phage nucleus, forming foci on top of viral DNA, while soluble GFP remained excluded in the cytoplasm. To determine if the shells assembled by phages ΦKZ and ΦPA3 have the ability to compartmentalize proteins, we fused GFP or mCherry to the RecA-related proteins of ΦPA3 (gp175-GFP) and ΦKZ (gp152-mCherry) and then compared their localization to the fluorescent proteins expressed alone during infection of P. aeruginosa. While soluble GFP or mCherry expressed during infection were excluded from the phage nucleus (Figure 1A, FP, and S1B), the RecA-related proteins were highly concentrated inside the nucleus where they formed foci coincident with phage DNA (Figure 1A, RecA-like).
In order to determine if proteins from one phage can be recognized and localized properly by another phage, we simultaneously expressed the RecA-related proteins from ΦPA3 (gp175-GFP) and ΦKZ (gp152-mCherry) in P. chlororaphis and then infected the cells with phage 201Φ2-1. Both RecA-related proteins localized together inside 92% (n=263) of the phage nuclei (Figure 1B–D, S1C). Surprisingly, in cases where we could observe individual foci formed by the two RecA-FP fusions, the two foci co-localized (Figure 1B–C). In time-lapse microscopy, as the entire phage 201Φ2-1 nucleus rotated due to forces exerted by the PhuZ spindle (Chaikeeratisak et al., 2017), the two foci rotated together, suggesting that these proteins are localized to specific regions of DNA within the nucleus that are undergoing replication or repair (Figure 1C). These results suggest that the proteinaceous shell and its key property of compartmentalizing DNA and proteins are conserved among the three large Pseudomonas phages.
Progeny phages are clustered around the phage nucleus
The phage nucleus not only serves as a site of DNA replication, but it also provides a surface for docking of capsids in order to package viral DNA. To determine if capsid docking is conserved, we fluorescently tagged the major capsid proteins from ΦKZ (gp120-mCherry) and ΦPA3 (gp136-GFP) and compared their localization to that of the capsid protein (gp200-GFP) of 201Φ2-1. At approximately 60 mpi, when we expect DNA packaging to occur, each protein formed foci that surrounded the phage nucleus (Figure 1A, capsid).
To study the 3-dimensional assembly of phage around the nucleus independently of capsid-GFP fusions, we fixed infected cells and stained them with DAPI. We previously demonstrated that the DNA of the host chromosome is degraded early on during infection, and that only phage DNA is visualized at later time points (Erb et al., 2014; Kraemer et al., 2012). We used super-resolution structured illumination microscopy (SIM) to examine P. chlororaphis cells after infection with 201Φ2-1 and P. aeruginosa cells after infection with ΦKZ or ΦPA3. At early time points before DNA packaging begins, the intensity of DAPI staining is uniform across the nucleus, but after DNA is encapsidated, viral particles appear as bright, fluorescent foci on the surface of the nucleus (Figure S1D, (Kraemer et al., 2012)). At 90 mpi, encapsidated phage particles can be observed as a dense cluster of foci in the cell center in all three infected Pseudomonas cells (Figure 1E and S1E). This arrangement of particles agrees with results obtained using GFP tagged capsids (Figure 1A, capsid) and with tomography of phage 201Φ2-1 (Chaikeeratisak et al., 2017). Taken together, these results suggest that the phage nucleus plays a conserved role in subcellular compartmentalization as the site of DNA replication and progeny phage packaging during the viral life cycle.
PhuZΦKZ and PhuZΦPA3 require a threshold concentration for filament assembly in vivo
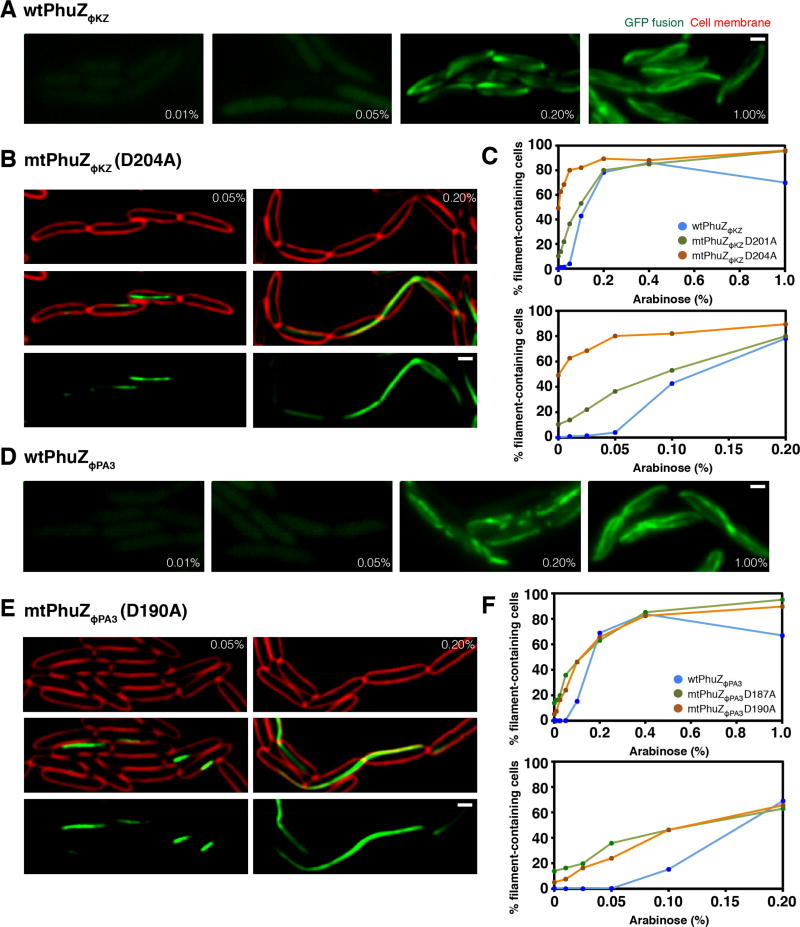
Each of the P. aeruginosa phage encodes a PhuZ homolog that is divergent in sequence. PhuZΦKZ shares only 31% identity and PhuZΦPA3 only 46% identity with PhuZ201, with most of the conservation occurring within the nucleotide binding pocket and C-terminal tail (Figure S2). To determine if the divergent PhuZ proteins have similar dynamic polymerization properties in vivo, we made GFP fusions to the N-termini of PhuZΦKZ and PhuZΦPA3 under an inducible arabinose promoter on a plasmid in P. aeruginosa. When the GFP-PhuZ fusions were induced at low levels (≤0.05%) of arabinose, diffuse fluorescence was observed (Figure 2A and 2D). Filaments appeared when arabinose levels were increased to 0.1% (Figure 2C and 2F). At 1% arabinose induction, long filaments were formed that extended the length of the cell (Figure 2A and 2D). Protein induction from this plasmid is approximately linear in P. aeruginosa with arabinose concentrations below 0.5% (Figure S3C) and proteins are expressed at uniform levels among cells throughout the population (Figure S3D). These results suggest that both GFP-PhuZΦKZ/ΦPA3 monomers must reach a threshold critical concentration for assembly to occur, as is the case with PhuZ201, TubZ, and other tubulins. To confirm the presence of the catalytic T7 loops in the PhuZ homologs, we generated mutations in this motif in which the conserved aspartic acid was replaced by alanine: GFP-PhuZΦKZ-D201A or D204A and GFP-PhuZΦPA3-D187A or D190A (Figure 2B, 2E, S3E, and S3F). When these mutant proteins were expressed in P. aeruginosa, filaments appeared at a comparatively lower arabinose concentration, 0.05%, suggesting that the catalytic point mutant requires a lower concentration for stable filament assembly (Figure 2C and 2F), as we previously observed with PhuZ201 catalytic mutants. The mutant proteins formed long polymers that often became trapped in septa during cell division, resulting in extensive chaining of cells (Figure 2B, 2E, S3E, and S3F).
Figure 2. PhuZ requires a critical threshold concentration to polymerize in P. aeruginosa.
Cells were grown on an agarose pad and the fusion proteins were induced at the indicated arabinose concentrations. Cell membranes were stained with FM4-64 (red). All scale bars equal 1 micron. See also Figures S2 and S3.
A) Fluorescence images of cells expressing wild-type GFP-PhuZΦKZ at the indicated arabinose concentrations.
B) P. aeruginosa cells expressing the GTPase mutant GFP-PhuZΦKZD204A at the indicated arabinose concentrations.
C) Graph showing the percentage of cells containing either wild-type or mutant PhuZΦKZ (D201A or D204A) filaments when fusion proteins are expressed at increasing levels of arabinose ranging from 0% to 1%. The graph below replots the same data showing the range of arabinose concentration from 0% to 0.2%. Data represented as mean values were collected from the induced cells at indicated arabinose concentration from at least 3 different fields with at least 200 cells per field.
D) Fluorescence images of cells expressing wild-type GFP-PhuZΦPA3 at the indicated arabinose concentrations.
E) P. aeruginosa cells expressing the GTPase mutant GFP-PhuZΦPA3D190A at the indicated arabinose concentrations.
F) Graph showing the percentage of cells containing either wild-type or mutant PhuZΦPA3 (D187A or D190A) filaments when fusion proteins are expressed at increasing levels of arabinose ranging from 0% to 1%. The graph below replots the same data showing the range of arabinose concentration from 0% to 0.2%. Data represented as mean values were collected from the induced cells at indicated arabinose concentration from at least 3 different fields with at least 200 cells per field.
PhuZΦKZ and PhuZΦPA3 polymers display dynamic instability
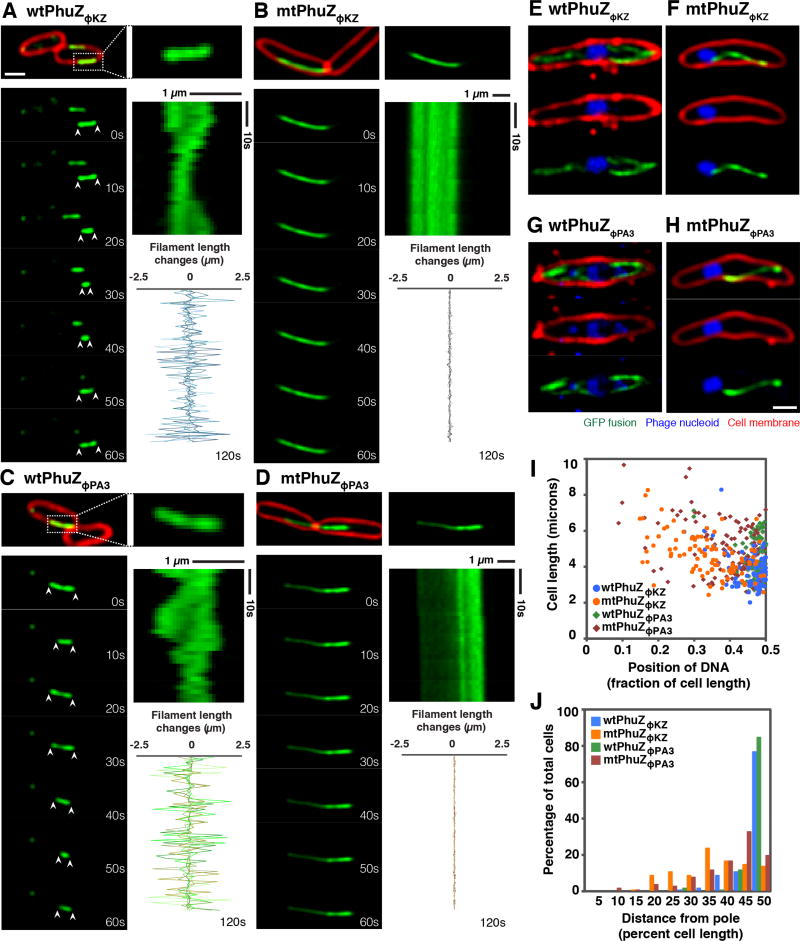
Dynamic instability is a key property of microtubules that allows them to search space and center chromosomal DNA. PhuZ201 forms filaments that undergo dynamic instability, which is essential for its function of centering the phage nucleus. We therefore set out to determine if PhuZΦKZ and PhuZΦPA3 also display dynamic instability in vivo. When we expressed GFP-PhuZΦKZ and GFP-PhuZΦPA3 above the apparent critical threshold concentration for assembly in uninfected P. aeruginosa cells, the filaments displayed evidence of dynamic instability, undergoing cycles of polymerization and depolymerization (Figure 3A and 3C). Figure 3A–D shows time-lapse microscopy of individual filaments over the course of one minute. Quantitation of filament length over time shows that the ends of the wild type PhuZΦPA3 and PhuZΦKZ filaments are dynamically unstable, continuously extending and retracting over a period of 2 minutes (Figure 3A and 3C; graphs). In contrast, time-lapse microscopy and quantitation of filament length of GFP-PhuZΦKZ-D204A and GFP-PhuZΦ PA3-D187A showed that the mutant filaments remained the same length over the course of 2 minutes (Figure 3B and 3D; graphs) suggesting that dynamic instability requires GTPase activity as is the case with PhuZ201 and microtubules.
Figure 3. PhuZΦKZ and PhuZΦPA3 show dynamic instability and center the phage nucleus in the cell, while the GTPase mutants fail to position the nucleus at midcell.
Cells were grown on agarose pads and the fusion proteins were induced with 0.05% arabinose (below the critical threshold for filament assembly) for infected cells and with 0.1% arabinose (to study the dynamic instability of filaments) for uninfected cells. Cell membranes were stained red by FM4-64 and phage DNA was stained blue by DAPI. All scale bars equal 1 micron. See also Figures S3 and S4.
(A and C) Fluorescence micrographs of uninfected P. aeruginosa cells induced at 0.1% arabinose to express wild-type GFP-PhuZΦKZ (A) and wild-type GFP-PhuZΦPA3 (C). The filament in the dashed box is magnified and shown on the right. Time-lapse imaging and kymograph for one representative filament for each protein (GFP-PhuZΦKZ (A) and GFP-PhuZΦPA3 (C)) and a graph showing length changes for five representative filaments for each protein show that these filaments display dynamic instability.
(B and D) Fluorescence micrographs of uninfected P. aeruginosa cells induced at 0.1% arabinose to express mutant GFP-PhuZΦKZ (B) and mutant GFP-PhuZΦPA3 (D). Time-lapse imaging and kymograph for one representative filament. The graph shows length changes of five representative mutant filaments.
(E and G) Fluorescence micrographs of ΦKZ-infected (E) and ΦPA3-infected (G) P. aeruginosa cells induced at 0.05% arabinose to express wild-type GFP-PhuZΦKZ (E) or wild-type GFP-PhuZΦPA3 (G). The dynamic GFP-PhuZ spindle (green) and phage nucleus (blue) was centered in the infected cell at 60 mpi.
(F and H) Fluorescence micrographs of ΦKZ-infected (F) and ΦPA3-infected (H) P. aeruginosa cells induced at 0.05% arabinose to express mutant GFP-PhuZΦKZ (F) and mutant GFP-PhuZΦPA3 (H). The mutant PhuZ proteins (green) assembled a single static filament instead of a bipolar spindle resulting in mispositioning of the phage nucleus (blue) at 60 mpi.
(I) Graph showing the position of the phage nucleus in P. aeruginosa cells, expressed as a fraction of cell length. Cells expressed either wild-type GFP-PhuZΦKZ (blue circle), wild-type GFP-PhuZΦPA3 (green diamond), GTPase mutant GFP-PhuZΦKZ (orange circle) and GTPase mutant GFP-PhuZΦPA3 (red diamond). One hundred cells were counted for each group.
(J) Histogram showing position of the phage nucleus in which the data from Figure I is replotted as percentage of total cells. Cells expressed either wild-type GFP-PhuZΦKZ (blue), wild-type GFP-PhuZΦPA3 (green), GTPase mutant GFP-PhuZΦKZ (orange) and GTPase mutant GFP-PhuZΦPA3 (red). One hundred cells were counted for each group.
The PhuZ bipolar spindle centers the phage nucleus after infection with phages ΦKZ and ΦPA3
PhuZ201 is involved in the centering of the phage nucleus during infection of P. chlororaphis. To determine if PhuZΦKZ and PhuZΦPA3 also assemble a bipolar spindle, we expressed GFP-PhuZΦKZ and GFP-PhuZΦPA3 from a plasmid in P. aeruginosa below the critical threshold for filament assembly with very low levels of arabinose induction. We previously established with phage 201Φ2-1 that expressing a small amount of GFP-tagged PhuZ to visualize the spindle does not significantly interfere with the spindle’s ability to display dynamic instability or center the phage nucleus. Based on calculations of the total number of PhuZ monomers in the cell required to form the triple stranded filaments of the bipolar spindle and assuming a critical concentration of assembly of ~2.5 µM based on measurements of PhuZ201 (Kraemer et al., 2012), we expect this level of induction to produce less than 20% of the total PhuZ protein in the cell. Western blot analysis (Figure S3G) verified that plasmid expressed GFP-PhuZ accounted for less than 1/3 of the total PhuZ protein produced at 60 and 90 mpi.
After induction of GFP-PhuZ at this low level (0.05% arabinose), we infected cells with either ΦKZ or ΦPA3 and observed PhuZ filament formation 60 mpi. In uninfected cells, no filaments were observed, but upon phage infection, GFP-PhuZ assembled dynamic filaments that extended from both poles of the cell toward the nucleus at midcell, forming a bipolar spindle-like structure similar to that observed for PhuZ201 (Figure 1E, 3E, 3G, S4A, and S4C). The phage nucleus was located within 10% of the cell center in approximately 80% of cells after infection with ΦKZ (n=100) or ΦPA3 (n=100) (Figure 3I and 3J). In contrast to wild type, the catalytic mutants GFP-PhuZΦKZ-D204A and GFP-PhuZΦPA3-D187A behaved as dominant negatives that co-assembled with the wild type proteins and formed static polymers that disrupt spindle dynamics and viral DNA positioning. Cells expressing the PhuZ catalytic mutants typically had one long static filament per cell instead of a pair of filaments surrounding the phage nucleus as seen in wild type cells (Figure 3F, 3H, S4B, and S4D). In addition, expression of the mutant proteins led to mispositioning of the phage nucleus (Figure 3F, 3H, 3I, and 3J). Less than 20% of cells expressing GFP-PhuZΦKZ-D204A (n=100) or GFP-PhuZΦPA3-D187A (n=100) had a centered phage nucleus (Figure 3I and 3J). These data suggest that a bipolar spindle formed by dynamic PhuZ polymers is responsible for centering the phage nucleus of ΦKZ and ΦPA3 and therefore, the biological function of PhuZ is conserved in all three phages.
Discussion
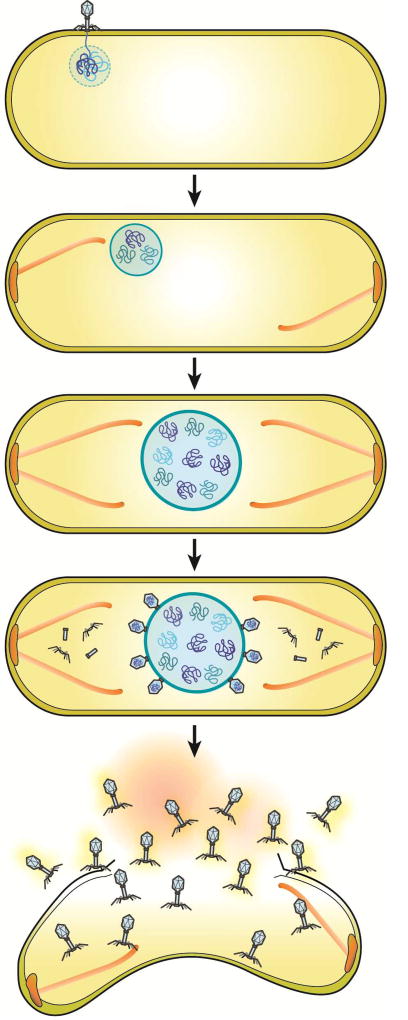
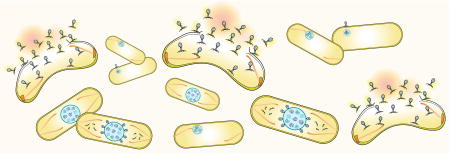
We recently described a nucleus-like structure assembled by phage 201Φ2-1 during P. chlororaphis infection and its role during the lytic life cycle. The phage reorganizes the bacterial cell, forming a proteinaceous shell that encloses viral DNA and enzymes involved in DNA replication and transcription, while excluding metabolic enzymes and ribosomes. The tubulin PhuZ assembles a bipolar spindle that plays a key role in positioning the phage nucleus at midcell (Figure 4). Here, we show that two major components of this replication pathway, the phage nucleus and spindle, are conserved in the related phages ΦKZ and ΦPA3. This remarkable level of organization has not been seen in other bacteriophages or in bacterial or archaeal cells.
Figure 4. Model of phage nucleus assembly and its role in the viral life cycle in large Pseudomonas phages.
As soon as the phage injects DNA into its host, the nuclear shell protein assembles a compartment to protect its genomic DNA from host defenses and to provide a compartment for DNA replication. Dynamically unstable PhuZ filaments set up a spindle with one end anchored at the cell pole while the other end of the spindle pushes the compartment toward midcell. The spindle might rely upon a yet to be discovered factor that organizes its assembly. As DNA replication proceeds inside the compartment, the nucleus grows in size as it is pushed toward the cell center. Late during infection, capsids dock on the surface of the phage nucleus for DNA encapsidation. The mature phage particles are assembled in the cytoplasm and the host cell lyses.
We now show three examples of bacterial viruses with both a nucleus and a tubulin-based spindle, raising the question of how such a complicated system might benefit phage reproduction. One possibility is that the phage nucleus might provide protection for viral DNA from host defenses such as restriction enzymes or CRISPR/CAS systems. In addition, the phage nucleus could also protect against the phage’s own nucleases, which were previously shown to degrade bacterial chromosomal DNA after infection (Erb et al., 2014). A protective barrier might confer a selective advantage, despite the potential added cost of the system. An enclosed structure containing replicating viral DNA requires phage capsids to dock on the surface for DNA encapsidation. If this compartment were located adjacent to the cell membrane near the cell pole, access to a significant portion of its surface would be sterically hindered. Therefore, the spindle might have subsequently evolved to position the nucleus at midcell to provide greater unobstructed surface area for diffusion of molecules in and out of the phage nucleus and for docking of viral capsids. The discovery that viruses evolved both a spindle and a nucleus is in line with previous hypotheses concerning the role of viruses in the evolution of eukaryotic life (Bell, 2001, 2009; Forterre, 2006; Koonin, 2016; Takemura, 2001; Villarreal and DeFilippis, 2000). In particular, the theory of viral eukaryogenesis suggests, in part, that the eukaryotic nucleus evolved when a poxvirus infected an archaeal cell. We speculate that our data support and expand the model of viral eukaryogenesis and suggest that a virus may have provided the origin for the tubulin-based spindle as well as the first step towards nuclear compartmentalization.
These results also provide insight into the plasticity of tubulin and its ability to form a bipolar spindle composed of dynamically unstable filaments despite significant divergence in amino acid sequences. While PhuZΦKZ, and PhuZΦPA3 share only 31% and 46% sequence identity (respectively) with PhuZ201, both formed dynamically unstable bipolar spindles that positioned phage DNA at midcell, and mutations in the catalytic T7 loops of these tubulins resulted in mispositioning of phage DNA. These findings are consistent with those previously outlined for P. chlororaphis phage 201Φ2-1 and show that the centering function is conserved among large bacteriophages infecting different Pseudomonas species. In comparison with eukaryotic alpha/beta tubulin which forms 13 stranded microtubules, PhuZ shares less than 11% amino acid identity and assembles triple-stranded filaments. Despite the large divergence in sequence, PhuZ filaments still display dynamic instability in vitro and in vivo similar to eukaryotic microtubules, suggesting that tubulin sequences can undergo extensive variation and evolution, yet retain the ability to assemble filaments with similar dynamic properties.
EXPERIMENTAL PROCEDURES
Strains, Growth Conditions and Bacteriophage Preparation
P. chlororaphis strain 200-B and P. aeruginosa strain PA01 were grown on Hard Agar (HA) (Serwer et al., 2004) and LB media, respectively. Lysates of each phage (201Φ2-1, ΦPA3, and ΦKZ) were made by infecting saturated bacterial culture with 10 µl of high titer lysate, then incubating for 15 minutes at room temperature. 5 ml of HA or LB top agar (0.35%) was added to the infected cultures, then the top agar mixture was poured over a HA or LB plate. The plates were incubated at 30°C overnight. The following day, the plates that formed web lysis (nearly confluent lysis) were flooded with 5 ml of phage buffer and incubated at room temperature for 6 hours. The phage lysates were then aspirated, clarified by centrifugation at 15,000 rpm for 10 minutes, and stored at 4°C with 0.01% chloroform.
Plasmid Constructions and Bacterial Transformation
Plasmids, as listed in supplemental table 1, were introduced into P. chlororaphis strain 200-B and P. aeruginosa strain PA01 by electroporation. All bacterial cultures were grown at 30°C. For additional details, please see Extended Experimental procedures.
Fluorescence Microscopy
Pseudomonas cells were inoculated on 1.2% agarose pads in concavity slides. Each pad was supplemented with desired arabinose concentrations to induce the protein expression, 1 µg/ml FM4-64 for membrane staining, and 1 µg/ml DAPI for DNA staining (Pogliano et al., 1999). The slides were then incubated in a humid chamber at 30°C for 3 hours for P. chlororaphis strain 200-B and at 37°C for 2 hours for P. aeruginosa strain PA01 prior to fluorescence microscopy. The DeltaVision Spectris Deconvolution microscope (Applied Precision, Issaquah, WA, USA) was used to visualize the cells. For static images, the cells were imaged for at least 8 stacks from the middle focal plane with 0.15 µm increments in the Z-axis and, for time-lapse imaging, the cells were imaged from a single stack at the focal plane with Ultimate Focusing mode. Microscopic images were further processed by the deconvolution algorithm in DeltaVision SoftWoRx Image Analysis Program.
Fixed Cell Imaging
Cells were prepared on 1.2% agarose pads as for live-cell imaging. At desired time point, cells were fixed with paraformaldehyde and glutaraldehyde and washed with 1× PBS, as described by Kraemer et al., 2012. Before imaging, the slides were then covered with a cover slip. Data of static images were collected and processed as stated above.
Single-cell Phage Infection
As described above, Pseudomonas cells were grown on 1.2% agarose pads, supplemented with desired concentration of arabinose to induce fluorescently-tagged protein expression to label wild-type proteins from phage, and then incubated at 30°C for 3 hours without a coverslip in a humid chamber. To begin the phage infection, 3 µl of high-titer lysate (108 pfu/ml) was added to the cells on agarose pads, and then the cells were further incubated to allow the phage infection occurs. At desired time point after phage infection, a coverslip was put on the slide and fluorescence microscopy was then initiated. Data of static images and time-lapse imaging were collected and processed as stated above.
3D-SIM Super-resolution Microscopy Pseudomonas
cells were grown and infected on 1.2% agar pads and then fixed with paraformaldehyde and gluteraldehyde as stated above. Cells were subsequently stained with 1 µg/ml DAPI and then imaged using an Applied Precision/GE OMX V2.2 microscope. Microscopic raw data were sequentially taken by Structured illumination (SI)-super resolution light path to collect 3 µm thickness of samples with 125 nm increments in the Z-axis with compatible immersion oils (Applied Precision). 3D-SIM images were then rendered by standard OMX SI reconstruction parameters in DeltaVision SoftWoRx Image Analysis Program
Statistical Analysis
The number of cells analyzed in the experiments or the number of experimental replicates is indicated in the figure legends. All data are shown as mean values or mean ± standard error of the mean (± SEM). Student’s t test was performed for unpaired data with unequal variance (p value less than 0.05 shows a significant difference).
Supplementary Material
Supplemental Movie 1: Time-lapse imaging, images taken every 2 seconds for 2 minutes, of 201Φ2-1 infected P. chlororaphis cells expressing ΦKZ-gp152-mCherry (Red) and ΦPA3-gp175-GFP (Green) beginning at 90 minutes post infection (mpi). Both RecA-related proteins from ΦKZ and ΦPA3 assemble foci and rotate together with the 201Φ2-1 nucleus suggesting the formation of a singular structure with 201Φ2-1 nucleus. Related to Figure 1.
Acknowledgments
This research was supported by NIH grants GM104556 (J.P.). We acknowledge the use of the UCSD Neuroscience Microscopy Shared Facility, which is financially supported by Grant P30 NS047101.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
V.C., K.N on conception and design, acquisition of data, analysis and interpretation of data, drafting or revising the article; M.E.E., M.L.E., and A.V. on acquisition of data; J.P. on conception and design, analysis and interpretation of data, drafting or revising the article.
References
- Aylett CH, Izoré T, Amos LA, Löwe J. Structure of the tubulin/FtsZ-like protein TubZ from Pseudomonas bacteriophage ΦKZ. J Mol Biol. 2013;425:2164–2173. doi: 10.1016/j.jmb.2013.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylett CH, Wang Q, Michie KA, Amos LA, Lowe J. Filament structure of bacterial tubulin homologue TubZ. Proc Natl Acad Sci U S A. 2010;107:19766–19771. doi: 10.1073/pnas.1010176107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell PJ. Viral eukaryogenesis: was the ancestor of the nucleus a complex DNA virus? J Mol Evol. 2001;53:251–256. doi: 10.1007/s002390010215. [DOI] [PubMed] [Google Scholar]
- Bell PJ. The viral eukaryogenesis hypothesis: a key role for viruses in the emergence of eukaryotes from a prokaryotic world environment. Ann N Y Acad Sci. 2009;1178:91–105. doi: 10.1111/j.1749-6632.2009.04994.x. [DOI] [PubMed] [Google Scholar]
- Boedeker C, Schuler M, Reintjes G, Jeske O, van Teeseling MC, Jogler M, Rast P, Borchert D, Devos DP, Kucklick M, et al. Determining the bacterial cell biology of Planctomycetes. Nat Commun. 2017;8:14853. doi: 10.1038/ncomms14853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaikeeratisak V, Nguyen K, Khanna K, Brilot AF, Erb ML, Coker JK, Vavilina A, Newton GL, Buschauer R, Pogliano K, et al. Assembly of a nucleus-like structure during viral replication in bacteria. Science. 2017;355:194–197. doi: 10.1126/science.aal2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Erickson HP. In vitro assembly studies of FtsZ/tubulin-like proteins (TubZ) from Bacillus plasmids: evidence for a capping mechanism. J Biol Chem. 2008;283:8102–8109. doi: 10.1074/jbc.M709163200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erb ML, Kraemer JA, Coker JK, Chaikeeratisak V, Nonejuie P, Agard DA, Pogliano J. A bacteriophage tubulin harnesses dynamic instability to center DNA in infected cells. Elife. 2014;3 doi: 10.7554/eLife.03197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forterre P. The origin of viruses and their possible roles in major evolutionary transitions. Virus Res. 2006;117:5–16. doi: 10.1016/j.virusres.2006.01.010. [DOI] [PubMed] [Google Scholar]
- Fuerst JA, Sagulenko E. Beyond the bacterium: planctomycetes challenge our concepts of microbial structure and function. Nature Reviews Microbiology. 2011;9:403–413. doi: 10.1038/nrmicro2578. [DOI] [PubMed] [Google Scholar]
- Koonin EV. Viruses and mobile elements as drivers of evolutionary transitions. Philos Trans R Soc Lond B Biol Sci. 2016;371 doi: 10.1098/rstb.2015.0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer JA, Erb ML, Waddling CA, Montabana EA, Zehr EA, Wang H, Nguyen K, Pham DS, Agard DA, Pogliano J. A phage tubulin assembles dynamic filaments by an atypical mechanism to center viral DNA within the host cell. Cell. 2012;149:1488–1499. doi: 10.1016/j.cell.2012.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen RA, Cusumano C, Fujioka A, Lim-Fong G, Patterson P, Pogliano J. Treadmilling of a prokaryotic tubulin-like protein, TubZ, required for plasmid stability in Bacillus thuringiensis. Genes Dev. 2007;21:1340–1352. doi: 10.1101/gad.1546107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesyanzhinov VV, Robben J, Grymonprez B, Kostyuchenko VA, Bourkaltseva MV, Sykilinda NN, Krylov VN, Volckaert G. The genome of bacteriophage phiKZ of Pseudomonas aeruginosa. J Mol Biol. 2002;317:1–19. doi: 10.1006/jmbi.2001.5396. [DOI] [PubMed] [Google Scholar]
- Monson R, Foulds I, Foweraker J, Welch M, Salmond GP. The Pseudomonas aeruginosa generalized transducing phage phiPA3 is a new member of the phiKZ-like group of 'jumbo' phages, and infects model laboratory strains and clinical isolates from cystic fibrosis patients. Microbiology. 2011;157:859–867. doi: 10.1099/mic.0.044701-0. [DOI] [PubMed] [Google Scholar]
- Montabana EA, Agard DA. Bacterial tubulin TubZ-Bt transitions between a two-stranded intermediate and a four-stranded filament upon GTP hydrolysis. Proc Natl Acad Sci U S A. 2014;111:3407–3412. doi: 10.1073/pnas.1318339111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogliano J, Osborne N, Sharp MD, Abanes-De Mello A, Perez A, Sun YL, Pogliano K. A vital stain for studying membrane dynamics in bacteria: a novel mechanism controlling septation during Bacillus subtilis sporulation. Mol Microbiol. 1999;31:1149–1159. doi: 10.1046/j.1365-2958.1999.01255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagulenko E, Nouwens A, Webb RI, Green K, Yee B, Morgan G, Leis A, Lee KC, Butler MK, Chia N, et al. Nuclear Pore-Like Structures in a Compartmentalized Bacterium. PLoS One. 2017;12:e0169432. doi: 10.1371/journal.pone.0169432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serwer P, Hayes SJ, Zaman S, Lieman K, Rolando M, Hardies SC. Improved isolation of undersampled bacteriophages: finding of distant terminase genes. Virology. 2004;329:412–424. doi: 10.1016/j.virol.2004.08.021. [DOI] [PubMed] [Google Scholar]
- Takemura M. Poxviruses and the origin of the eukaryotic nucleus. J Mol Evol. 2001;52:419–425. doi: 10.1007/s002390010171. [DOI] [PubMed] [Google Scholar]
- Villarreal LP, DeFilippis VR. A Hypothesis for DNA Viruses as the Origin of Eukaryotic Replication Proteins. In J Virol. 2000:7079–7084. doi: 10.1128/jvi.74.15.7079-7084.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehr EA, Kraemer JA, Erb ML, Coker JK, Montabana EA, Pogliano J, Agard DA. The structure and assembly mechanism of a novel three-stranded tubulin filament that centers phage DNA. Structure. 2014;22:539–548. doi: 10.1016/j.str.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Movie 1: Time-lapse imaging, images taken every 2 seconds for 2 minutes, of 201Φ2-1 infected P. chlororaphis cells expressing ΦKZ-gp152-mCherry (Red) and ΦPA3-gp175-GFP (Green) beginning at 90 minutes post infection (mpi). Both RecA-related proteins from ΦKZ and ΦPA3 assemble foci and rotate together with the 201Φ2-1 nucleus suggesting the formation of a singular structure with 201Φ2-1 nucleus. Related to Figure 1.