Summary
Aneuploidy, whole chromosome or chromosome arm imbalance, is a near-universal characteristic of human cancers. In 10,522 cancer genomes from The Cancer Genome Atlas (TCGA), aneuploidy was correlated with TP53 mutation, somatic mutation rate, and expression of proliferation genes. Aneuploidy was anti-correlated with expression of immune signaling genes, due to decreased leukocyte infiltrates in high-aneuploidy samples. Chromosome arm-level alterations show cancer-specific patterns, including loss of chromosome arm 3p in squamous cancers. We applied genome engineering to delete 3p in lung cells, causing decreased proliferation rescued in part by chromosome 3 duplication. This study defines genomic and phenotypic correlates of cancer aneuploidy and provides an experimental approach to study chromosome arm aneuploidy.
Keywords: aneuploidy, cancer genomics, genome engineering, lung squamous cell carcinoma
Graphical abstract
Introduction
Aneuploidy, an unbalanced number of chromosomes, was first observed in tumor cells over 100 years ago (reviewed in Holland and Cleveland, 2009) and is a predominant cancer feature occurring in ~90% of solid tumors (Weaver and Cleveland, 2006). Even though it is the among the oldest described cancer alterations, and even though genomics efforts have allowed high-throughput “karyotyping” of patient cancers, the role of aneuploidy in tumorigenesis is still a mystery.
Aneuploidy and focal copy number alterations represent two classes of somatic copy number alteration (SCNA) (Tang and Amon, 2013). Studies of genomic and phenotypic correlates of aneuploidy and SCNAs have examined copy number based on cytoband (Carter et al., 2006) or based on number of SCNAs, including broad and focal events (Davoli et al., 2017; Buccitelli et al, 2017). By these definitions, increased SCNA levels were reported to correlate with proliferation pathways (Carter et al., 2006; Davoli et al., 2017; Buccitelli et al., 2017) and to anti-correlate with immune signaling within individual tumor types (Davoli et al., 2017; Buccitelli et al., 2017).
We define aneuploidy as SCNAs of whole chromosomes and of chromosome arms. In cancer, aneuploidy affects more of the genome than any other somatic genetic alteration (Beroukhim et al., 2010; Zack et al, 2013; Mitelman, 2000). The most frequent recurrent arm alterations occur in over 30% of tumors, whereas the most frequent recurrent focal copy number alterations occur at a frequency below 0.15 (Beroukhim et al., 2010). Chromosome arm SCNAs are more common than whole chromosome SCNAs, occurring at higher frequencies in 12 of 16 cancer types analyzed (Beroukhim et al., 2010). Cancer subtypes are often characterized by tumor-specific patterns of chromosomal arm-level alterations (Ried et al., 2012), including squamous subtypes of lung, esophageal, and bladder tumors (Hoadley et al., 2014). Specific chromosome arm level alterations have also defined groups of tumors that are responsive to particular therapies, such as low grade gliomas with 1p/19q co-deletions that have been shown to be responsive to specific chemoradiotherapy regimens (Cairncross et al., 2013).
The functional effect of an individual aneuploidy is often thought to be due to the deletion of a tumor suppressor or overexpression of an oncogene (Liu et al., 2016). However, increasing evidence suggests that the effects of broad SCNAs result from the alteration of a combination of genes (Xue et al., 2012; Bonney et al., 2015). Recent SCNA models suggest that the phenotypic effect of chr_17p loss is due to more than TP53 loss of heterozygosity (Liu et al., 2016). Chr_3p loss is an early event in lung squamous cell carcinoma observed in preneoplastic regions in the lung (Hung et al., 1995; Sundaresan et al., 1992), yet studies suggest that its contributions to tumor development are not the result of loss of one gene (Wistuba et al., 2000).
To study the role of aneuploidy in tumor development, models of whole chromosome aneuploidy have been developed. Trisomies and monosomies have been extensively modeled in yeast, where they slow proliferation and induce proteosomal stress (Torres et al., 2007; Sheltzer et al., 2011). Mouse cells with Robertsonian translocations can be used to model one trisomy at a time (Williams et al., 2008), or cells can be compared that differ only by single chromosomes added by microcell-mediated cellular transfer (Sheltzer et al., 2017; Stingele et al., 2012). These studies also show that whole chromosome aneuploidy leads to cellular senescence and decreased proliferation and transformation capabilities; however, karyotype evolution can lead to a rescue of proliferation rates (Sheltzer et al., 2017). There is one characterized model of targeted chromosome arm-level deletion, where chromosome arm 8p was deleted in mammary epithelial cells by targeting TALENs at either end of the chromosome arm and screening for recombination (Cai et al., 2016). In this model, 8p deletion does not lead to an increase in growth rate or tumorigenic potential.
Recent advances in targeting of endonucleases allow approaches to generating broad chromosomal alterations in vitro. Cas9-CRISPR (clustered regularly interspaced short palindromic repeats) systems are particularly advantageous for their high efficiency and design tools available for targeting DNA throughout the genome (Mali et al., 2013; Hsu et al., 2013). CRISPR-targeting, when combined with an artificial telomere-containing plasmid, can be used to truncate a chromosome arm (Uno et al., 2017).
Here, we apply methods that define chromosome arm-level aneuploidy and a global cancer aneuploidy score to 10,522 tumors of 33 types in the Cancer Genome Atlas (TCGA), and develop and analyze an experimental cellular model for chromosome arm-level aneuploidy. By combining analysis of highly annotated cancer genomes and the experimental ability to manipulate chromosomes, we can advance our understanding of the effects of aneuploidy and specific chromosome arm-level alterations in cancer development and progression.
Results
Generation of aneuploidy scores for 10,522 TCGA cancers
To study features associated with aneuploidy, we first generated an aneuploidy score reflecting the total number of chromosome arms with arm-level copy-number alterations in a sample. When identifying these arm-level alterations, we were cognizant that two SCNAs that did not overlap could be misconstrued as a single event if we simply tallied arms with more than 50% of their length altered – for instance, if the SCNAs originated on opposite ends of the chromosome arm. Conversely, a simple length cutoff might exclude smaller SCNAs even if they cover most of the genomic region covered by larger SCNAs called as arm-level events (Figure S1A). We therefore clustered SCNAs on each arm based upon their locations and lengths (using a Gaussian mixture model) to identify events with likely similar consequences (Figure S1B and Method S1). Clusters in which the mean length of that SCNA was greater than 80% of the chromosome arm were considered positive for an arm alteration, samples whose SCNA length was less than 20% of a chromosome arm were considered negative, and clusters whose SCNAs were of intermediate length were not called.
We applied this approach to 10,522 samples spanning 33 cancer types from The Cancer Genome Atlas (TCGA) pan-cancer dataset (Table S1). Somatic DNA copy number was determined from Affymetrix SNP 6.0 arrays profiling of tumor samples. From SNP array data and mutational data, we used the ABSOLUTE algorithm (Carter et al., 2012) to generate segmented absolute copy number and estimate sample purity, ploidy, and number of whole genome doublings. SCNAs, identified by ABSOLUTE to be clonal, were called as deviations from the euploid level of the sample: we considered a purely tetraploid cell (identified in 18 samples in our analysis) as having no arm-level SCNAs. (See STAR Methods and Table S1 for additional information about expression and mutation data.) Using this arm-calling approach, we determined the arm-level SCNA status of more than 400,000 chromosome arms and more than 175,000 non-acrocentric whole chromosomes for the 10,522 cancer genomes (Table S2).
Next, we calculated an aneuploidy score that reflects the total burden, or number, of arm-level events in each sample. This aneuploidy score ranged from 0 to 39, the total number of human autosomal chromosome arms: p and q arms for chromosomes 1-12 and 16-20, together with q arms for the acrocentric chromosomes 13-15 and 21-22 (Figure 1A, Figure S1C). Previous studies have demonstrated that there is less than a two-fold change in frequency of arm alteration due to arm length (Beroukhim et al., 2010), and our aneuploidy score highly correlates with fraction of genome altered by aneuploidy (Spearman’s rank correlation coefficient = 0.975, Figure S1D). For these reasons, we chose the sum of arms altered for subsequent analyses.
Figure 1. Aneuploidy correlates with ploidy, genome doubling status, and tumor type.
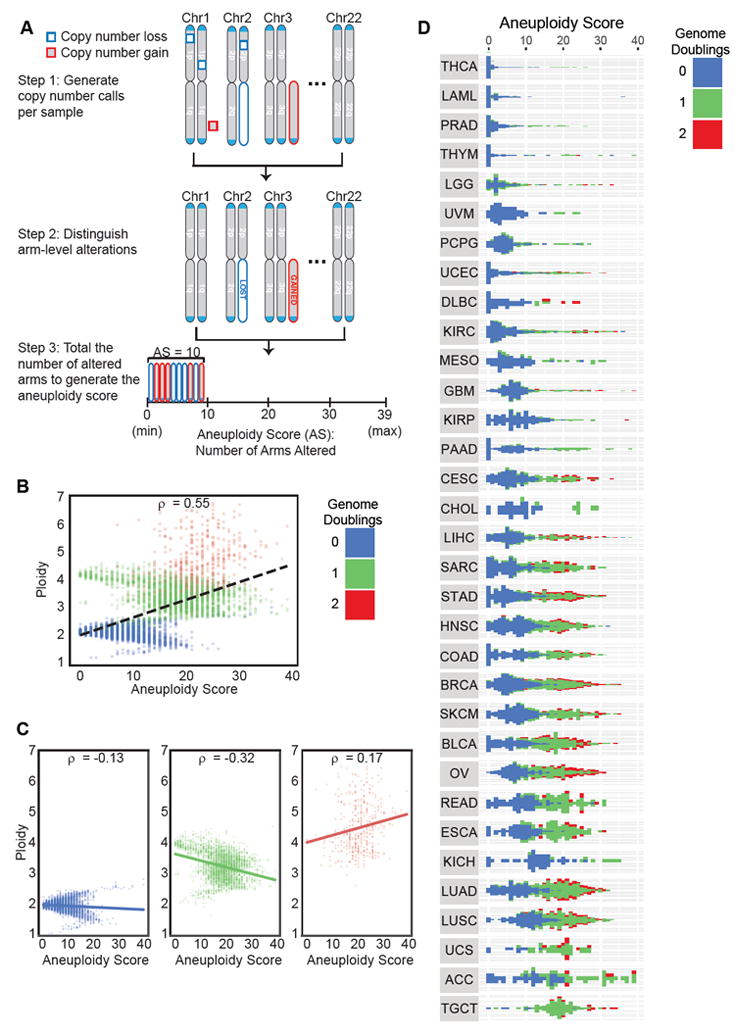
(A) Schematic of aneuploidy score. Step 1 is to generate copy number calls per sample, including somatic copy number alterations (SCNAs) of all sizes. Step 2 is to distinguish arm-level alterations within these SCNAs. Step 3 is to total the number of altered arms to generate the aneuploidy score.
(B) Each tumor sample is organized by genome doubling status (blue = not doubled, green = 1 genome doubling, red = 2 or more genome doublings). X-axis is aneuploidy score, the sum of the number of altered chromosome arms. Y-axis is ploidy as determined by ABSOLUTE. Spearman’s rank correlation coefficient = 0.55.
(C) X-axis is aneuploidy score, the sum of the number of altered chromosome arms. Y-axis is ploidy as determined by ABSOLUTE. Samples are separated by whole genome doubling status: samples without genome doubling (left, blue, Spearman’s rank correlation coefficient = -0.13), samples with one genome doubling (middle, green, Spearman’s rank correlation coefficient = -0.32), and samples with 2 or more genome doublings (right, red, Spearman’s rank correlation coefficient = 0.17).
(D) Each tumor sample is organized by tumor type and genome doubling status (blue = not doubled, green = 1 genome doubling, red = 2 or more genome doublings). Samples are organized by tumor type, and ranked from least to most aneuploid samples within a tumor type. X-axis is aneuploidy score, y-axis is sample number.
See also Figure S1 and Tables S1 and S2.
Samples that have undergone whole genome doubling, as defined by ABSOLUTE, have a higher degree of aneuploidy (Spearman’s rank correlation coefficient = 0.55), suggesting that tumors with increased ploidy are more prone to aneuploidy events (Figure 1B). However, among samples of the same genome doubling status, increased aneuploidy is generally associated with decreasing ploidy, indicating that the arm-level events that contribute to aneuploidy are more often absolute losses than gains (Figure 1C).
Across the entire sample panel, 88% of cancers had at least some detectable aneuploidy (mean aneuploidy score of 10.0); however, this rate varied substantially across cancer types (s.d. between cancer type aneuploidy means = 4.8; Figure 1D). For example, only 26% of thyroid carcinomas have any chromosome arm-level alteration (mean aneuploidy score of 0.87), and less than half of acute myeloid leukemias and thymomas have these alterations (mean aneuploidy scores of 1.6 and 3.8, respectively). In contrast, virtually all glioblastomas (99%, mean aneuploidy score of 8.2), uterine carcinosarcomas (96%, mean aneuploidy score of 17.2), and testicular germ cell tumors (99%, mean aneuploidy score of 18.7) have at least one aneuploidy event (Figure 1D).
Relationship between cancer mutation frequencies and aneuploidy
A variety of studies have assessed the relationship between chromosomal copy number alteration and somatic mutation (Ciriello et al., 2013; Zack et al., 2013; Davoli et al., 2017). We applied the aneuploidy scores of the full pan-cancer cohort of TCGA cases to examine this relationship. To assess whether gene mutations are associated with aneuploidy, we used a multivariable linear regression model that accounts for cancer type and the number of mutations per sample.
In this analysis, TP53 was an outlier, with the highest coefficient in the linear model (towards enrichment of mutations among aneuploid samples) and the highest statistical significance (Figure 2A, Table S3), consistent with previous studies (Ciriello et al., 2013; Zack et al., 2013; Davoli et al., 2017). We also detected significant associations between aneuploidy and mutation rate for 34 other genes. However, the correlation coefficients for these genes were always negative (towards enrichment of mutations in low aneuploidy samples) and never exceeded a magnitude of 0.027, whereas the coefficient for TP53 was positive 0.13.
Figure 2. Aneuploidy score correlates with TP53 mutations and overall mutation rate.
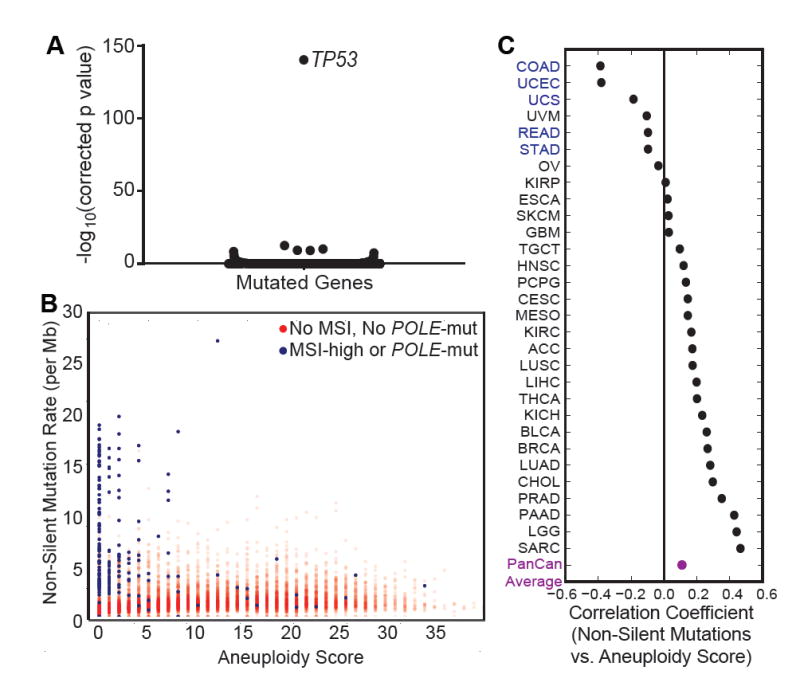
(A) Y-axis is -log10 Bonferroni corrected p value for linear model coefficient of aneuploidy score. Dots represent every mutated gene.
(B) X-axis is aneuploidy score. Y-axis is rate of non-silent mutations per megabase (square root). Blue samples have been called as microsatellite instability (MSI)-high or POLE mutated, whereas red samples do not have these features or have not been called.
(C) Spearman correlation coefficients for aneuploidy score and mutation rate across TCGA tumor types, arranged from smallest to largest value. Tumor types in blue have MSI-high or POLE mutated samples. Average of correlation coefficients across cancer types is in purple.
See also Figure S2 and Tables S3 and S4.
Next, we assessed the relationship between somatic mutation rate and aneuploidy. It has been previously reported that there is an inverse relationship between the frequencies of recurrent copy number alterations and of recurrent somatic mutations in cancer (Ciriello et al., 2013; Figure S2A). While we do also observe that cancers with very high mutation rates have low aneuploidy scores (Figure 2B), these high mutation rate/low aneuploidy cancers largely exhibit high levels of microsatellite instability or POLE mutation, primarily in colon adenocarcinoma and endometrial cancers (Figure 2B).
In contrast, when these hypermutated tumors are excluded, we observed a positive correlation between mutation frequency and aneuploidy score (Spearman’s rank correlation coefficient = 0.38), as well as between recurrent SCNAs and recurrent mutations (Spearman’s rank correlation coefficient = 0.34). The positive correlation between mutation and aneuploidy was found in most cancer types with the notable exceptions of colorectal carcinoma (COAD and READ) and uterine carcinoma (UCEC and UCS) as well as uveal melanoma (UVM) and stomach adenocarcinoma (STAD) (Figure 2C, Table S4). All of these exceptions other than uveal melanoma had some cases of microsatellite instability or POLE mutation.
Relationship between immune infiltrates and aneuploidy
Another important question about the cancer genome is the relationship between genome alterations and the immune response. Tumor mutational burden is known to be associated with response to immune checkpoint inhibition in cancer (Topalian et al., 2016; Rizvi et al., 2015), while aneuploidy has been reported to be associated with decreased immune infiltrate across many tumor types (Davoli et al., 2017). To assess the relationship between aneuploidy and immune infiltrates, we performed analyses of gene expression as a function of aneuploidy scores. The contribution of cells such as fibroblasts, leukocytes, endothelial cells, and other cell types is a confounding factor in the analysis of gene expression in primary tumors, so we also controlled for these features in our analysis. We measured immune and stromal cell populations using computational analyses rather than pathology-based estimates, as sections of the tumor used for histology were different from those used for molecular profiling. For estimating purity, we used ABSOLUTE, an established copy number pipeline used in most TCGA studies that has been benchmarked histologically (Carter et al., 2012; Zack et al., 2013). For leukocyte fraction, estimates based on methylation (TCGA PanCanAtlas immune paper, submitted) and expression (Aran et al., 2017) correlate with a Spearman’s rank correlation coefficient of 0.706 (Figure S2B). Namely, we first identified features reflecting cellular composition that associated with aneuploidy, and included these in our regression model tying gene expression to aneuploidy.
We first found that cancer impurity, as measured by ABSOLUTE (Carter et al., 2012), positively correlated with aneuploidy; in other words, cancers with high aneuploidy were associated with a higher fraction of non-cancerous cells (Figure 3A, Table S4). However, the measurement of leukocyte fractions based on methylation signatures, as previously described (Carter et al., 2012; TCGA Immune Subgroup, submitted), showed a negative correlation with aneuploidy (Figure 3B, Table S4). Most individual tumor types showed a negative correlation between aneuploidy and leukocyte fraction, which was strongest in pancreatic adenocarcinoma and head and neck squamous cell carcinoma (Spearman’s rank correlation coefficients = -0.428 and -0.312, respectively) (Figure 3B, Table S4). This observation is consistent with previous reports that aneuploidy is associated with decreased levels of cancer immune infiltrate (Davoli et al., 2017). Paradoxically, pan-cancer analysis shows a slight positive correlation between aneuploidy score and leukocyte fraction (Spearman’s rank correlation = 0.0568, Figure S2C), but this is driven by differences between tumor types as there is a negative correlation within most tumor types (Figure S2D). Combining the aneuploidy and leukocyte fraction results, the observed positive correlation of aneuploidy with impurity could be related to a non-leukocyte cellular fraction that is associated with aneuploidy (Figure 3C, Table S4).
Figure 3. Aneuploidy score negatively correlates with immune infiltrate, which contributes to decreased expression of immune genes.
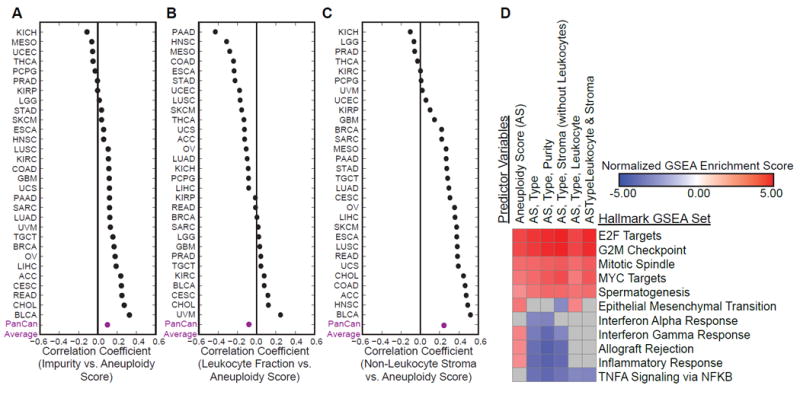
(A) Spearman correlation coefficients for aneuploidy score and impurity across TCGA tumor types, arranged from smallest to largest value. Average of correlation coefficients across tumor types is in purple.
(B) Spearman correlation coefficients for aneuploidy score and leukocyte fraction across TCGA tumor types, arranged from smallest to largest value. Average of correlation coefficients across tumor types is in purple.
(C) Spearman correlation coefficients for aneuploidy score and non-leukocyte stroma across TCGA tumor types, arranged from smallest to largest value. Average of correlation coefficients across tumor types is in purple.
(D) Heatmap of normalized enrichment scores for Hallmark gene sets in GSEA (Gene Set Enrichment Analysis), with FWER (family-wise error rate) p value < 0.01. Gray = not significant or not enriched. Predictor variables describe variables included in linear regression model for gene expression. Pathways are those identified from genes with significant coefficients in linear regression analysis.
See also Figure S2 and Tables S4, S5, and S6.
Another way to assess immune infiltrates is by gene expression rather than methylation data. Using RNA sequencing data from 9,670 TCGA cancers for which there was also aneuploidy data (Table S1, STAR Methods), we generated a linear regression model relating the expression of each gene (measured by RSEM values of RNA sequencing data) in each sample as a function of aneuploidy score, across all cancer types (Table S5). A ranked list of significant aneuploidy score coefficients for each gene was analyzed by gene set enrichment (GSEA) with the MSigDB hallmark gene sets.
In the case of univariate linear regression, we observed a statistically significant (FWER p value < 0.01) enrichment for proliferation pathways (such as E2F targets, mitotic spindle, and G2M checkpoint) and immune pathways (including interferon gamma response, allograft rejection, and immune response) (Figure 3D, column 1, Table S6). At first glance, this result appears contradictory to previously published studies (Davoli et al., 2017; Buccitelli et al., 2017). However, when we added tumor type as an additional variable in the linear regression model, immune gene sets were negatively associated with aneuploidy scores (Figure 3D, column 2, Table S6), consistent with other studies that assessed correlations within specific tumor types (Davoli et al., 2017, Buccitelli et al., 2017). When calculating an immune gene set expression score per sample, we also observed a pan-cancer positive correlation which is driven by tumor type, consistent with the linear regression model (Figure S2E, S2F).
To determine if the purity and immune infiltrate factors contribute to the aneuploidy correlated expression patterns, we added them to our linear model. Adding purity or non-leukocyte stroma as variables did not affect the enrichment of immune signatures (Figure 3D, columns 3 and 4, Table S6), but addition of leukocyte fraction as a variable resulted in the loss of the immune signature enrichment (Figure 3D, column 5, Table S6). These results suggest that the enrichment of the immune expression signature was due to the fraction of leukocyte infiltrate present in that sample. In contrast, pro-proliferative and cell cycle pathways were significantly positively correlated with aneuploidy score, regardless of other predictors in the model (Figure 3D, Table S6).
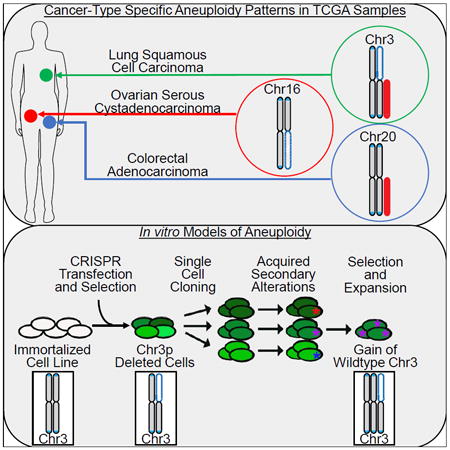
Cancer-type-specific patterns of chromosome arm-level aneuploidies
Next, we assessed the rates of whole chromosome and chromosome arm alteration across human cancer types. Arm or whole chromosome alterations occurred in 88% of cancer samples. Not including acrocentric chromosomes, whole chromosome alterations occurred in 66% of samples and chromosome arm-level alterations occurred in 78% of samples (Table S2). Individual arms and chromosomes were altered at different frequencies (Table S7); 8p and 17p were the most frequently deleted (at 33% and 35%, respectively), 8q was the most commonly gained (33% of samples), and 2p and 2q the least commonly altered (total 18% and 16%, respectively). Chromosome arms 6p, 12q, 17q, and 19q were gained and lost in equal percentages (difference between gain and loss frequency < 0.03), but others were predominantly gained (1q, 7p, 8q, and 20q) or predominantly lost (3p and 17p).
We observed that every cancer type harbors a unique pattern of aneuploidy, with different arms or whole chromosomes altered at different frequencies (Table S7). To compare different cancer types and molecular subtypes, we performed hierarchical clustering of mean arm-level calls (Figure 4A, Table S8), previously performed across 3,000 TCGA samples (Beroukhim et al., 2010). Within an individual cancer type, molecular subtypes often clustered together, such as glioblastoma subtypes and testicular germ cell tumor subtypes (Figure S3A). Consistent with previous analyses, endometrial cancer was a notable exception, as copy number high “serous like tumors” cluster separately from other endometrial subtypes (Figure 4A and TCGA, 2013). Testicular germ cell tumors were characterized by chromosome 12p gain among other aneuploidy events (Taylor-Weiner et al., 2016). Glioblastomas without IDH mutations are characterized by chromosome 7 gain and chromosome 10 loss, consistent with previous studies (Brennan et al., 2013). Low grade gliomas cluster near glioblastomas, but are characterized by distinct alterations of chromosome 1p loss and chromosome 19q gain correlating with IDH mutation (TCGA, 2015), a pattern shown to be therapeutically relevant (Cairncross et al., 2013).
Figure 4. Patterns of arm-level alterations cluster by tumor site, tissue of origin, and squamous morphology.
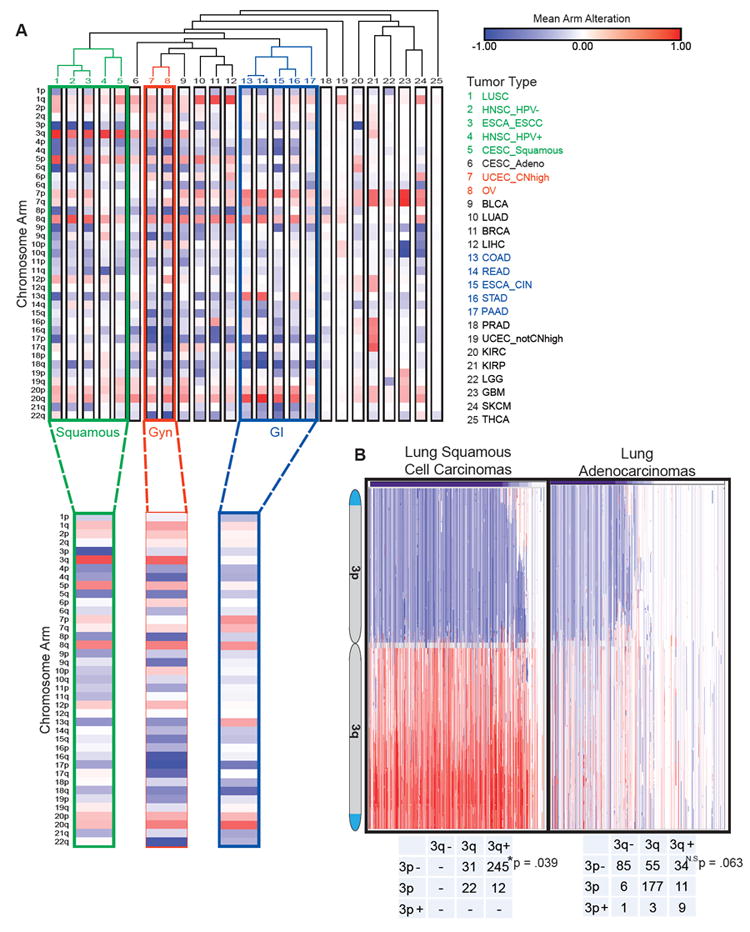
(A) Matrix of mean arm-level alteration within each tumor type/subtype. Hierarchical clustering of tumor type by Pearson’s method.
(B) Integrated genomics viewer (IGV) plots of chromosome 3 copy number alterations in lung squamous cell carcinoma or lung adenocarcinoma. Blue = loss, red = gain. Numbers for lung squamous and lung adenocarcinoma samples that had arm calls for both 3p and 3q. P values represent chi-square test for enrichment of co-occurrence of chromosome 3p loss and chromosome 3q gain.
See also Figure S3 and Tables S7 and S8.
Several cross-tumor clusters emerged from our analysis (Figure 4A, Table S8). Gastrointestinal tumors (colorectal, non-squamous esophageal, stomach, and pancreatic) clustered together with co-occurring gains of 8q, 13q, and chromosome 20, regardless of the status of microsatellite or chromosome instability. We also observed a cluster of some gynecological tumors (ovarian cancer and endometrial cancers with high copy number alterations) and a second cluster of epithelial tumors characterized by 1q gain (lung adenocarcinoma, breast cancers, and liver hepatocellular carcinoma). Neural lineage cancers (low grade glioma, glioblastoma, and melanoma) formed a cluster neighboring additional mesoderm-derived tumors (endometrial cancers with few copy number alterations, renal clear cell carcinoma, and renal papillary cell carcinoma), characterized by recurrent chromosome 7 gain and fewer aneuploidy alterations.
Previous clustering efforts by multiple data types across 12 tumor types showed that squamous cancers from different tissues of origin (lung, esophagus, and bladder) clustered together (Hoadley et al., 2014). Our analyses also separated out squamous cancers based on aneuploidy data alone, suggesting that broad SCNAs are a major determinant of the squamous cluster. A dominant feature of the squamous cancer cluster was chromosome arm 3p loss and chromosome arm 3q gain, which is present in cervical squamous cell carcinomas (CESC) and HPV positive head and neck squamous cell carcinomas (HNSC), and strongest in esophageal squamous cell carcinomas (ESCC), lung squamous cell carcinoma (LUSC) and HPV negative HNSC squamous tumors.
Chromosome 3 alterations are a known feature of lung squamous cell carcinoma (Bass et al., 2009; Hagerstrand et al., 2013; Zabarovsky et al., 2002), with chromosome 3p loss present in preneoplastic lesions in the lung (Hung et al., 1995; Sundaresan et al., 1992). In our lung squamous cell carcinoma dataset, chromosome 3p was deleted in almost 80% of tumors and chromosome arm 3q was gained in over 60% of tumors (Figure 4B). The co-occurrence of chr_3p loss and chr_3q gain was significantly more frequent than would be expected by chance (Figure 4B; chi-square p = 0.0386). Alterations in the reverse direction, chr_3p gain and chr_3q loss, were not observed in our dataset. In contrast, in lung adenocarcinoma, less than half of tumors had 3p loss and only 13% had 3q gain (Campbell et al., 2016), and these did not significantly co-occur (p=0.0626). Chr_3p gains occurred, though rarely, and chr_3q loss occurs at higher rates. As expected, deletion of 3p correlated with a reduction of 3p gene expression across TCGA samples (Figure S3B). Based on the linear modeling of gene expression, chr_3p alteration negatively correlated with hallmark sets of cell cycle (E2F targets/G2M Checkpoint, Figure S3C, FWER p value < 0.01), epithelial mesenchymal transition, interferon gamma response, and TNFa signaling (Figures S3D and S3E, FWER p value < 0.01). These pathways were down-regulated when 3p is gained and up-regulated when 3p is lost.
Genome engineering approach to delete chromosome arm 3p in vitro
Chromosome arm-level aneuploidies occur in almost 80% of cancers, yet have been rarely modeled in human cells (Cai et al., 2016; Uno et al., 2017). Given the continuing scientific mystery regarding the function of cancer aneuploidy, and given that deletion of chromosome 3p occurs in almost 80% of squamous cell lung cancers, we wanted to further understand the effect of chr_3p deletion in lung epithelial cells. We developed a recombination directed approach to remove the 3p chromosome arm. Using the CRISPR-Cas9 system, we generated double strand breaks (DSBs) centromeric to all genes on the 3p arm. We modified a plasmid containing an artificial telomere and puromycin selection cassette (Uno et al., 2017) by addition of one kilobase DNA of sequence homologous to the region centromeric of the chr_3p DSB (Figure 5A). The CRISPR plasmid and telomere-containing plasmid were co-transfected into cells, and the recombination event was selected for with puromycin treatment. We observed successful recombination verified by Sanger sequencing, in a human immortalized lung epithelial cell line (AALE, immortalized by SV40 large T antigen) (Figure S4A). To test for chr_3p hemizygous deletion, we performed quantitative PCR (qPCR) on genomic DNA to compare chr_3p and chr_3q levels. Single cell clones that were positive for recombination were positive for chr_3p hemizygous deletion by qPCR (Figure 5B), as well as whole genome sequencing (Figure 5C and Figure S4B) and karyotyping (Figure 5D).
Figure 5. CRISPR-based approach can delete a chromosome arm in human immortalized cells.
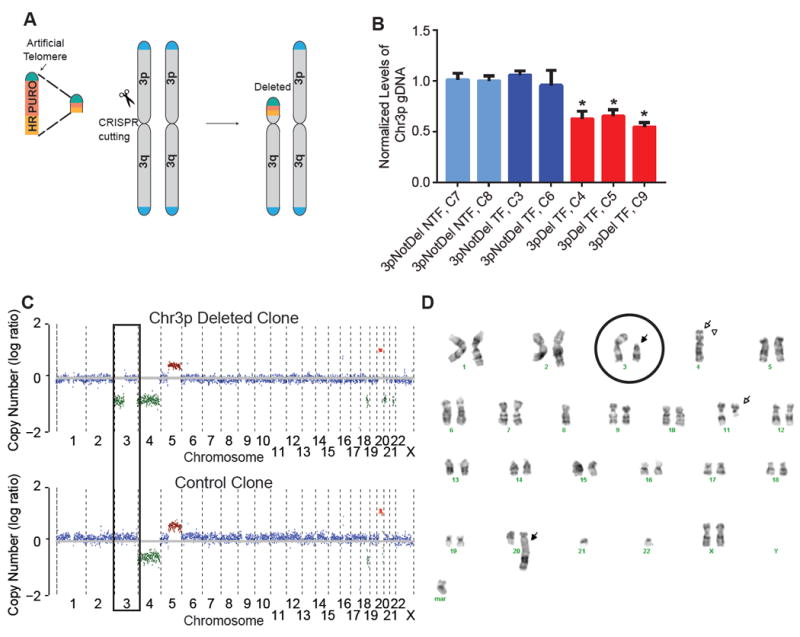
(A) Schematic of CRISPR and recombination based approach to delete chr_3p in vitro. A linearized plasmid containing homologous DNA, a puromycin selection marker, and an artificial telomere is co-transfected with a CRISPR-Cas9 construct to target DNA sequence adjacent to the centromere. Upon transfection, a double-strand break is produced and repaired by homologous directed recombination, removing a chromosome arm and replacing it with an artificial telomere.
(B) qPCR measuring chr_3p gDNA normalized to chr_3q gDNA in single cell clones. Bars represent means with error bars +/- standard deviation. Light blue = single cell clones that were not transfected (NTF) and did not have chr_3p deletion, dark blue = single cell clones that were transfected (TF) but did not have chr_3p deletion, and red = single cell clones that were transfected and have chr_3p deletion. (* = p value < 0.05)
(C) Whole genome sequencing output of HMMCopy, for one chr_3p deleted cell clone (top) and a non-deleted control clone (bottom).
(D) Karyotype of one chr_3p deleted cell clone. Chromosome 3 is circled, and arrows point to chromosomal abnormalities. Mar = marker chromosome.
See also Figure S4.
Chromosome arm 3p deleted cells evolve over time
We evaluated the growth of cells with chromosome 3p truncation (Figure 6A). At the first round of characterization, ten passages post-single cell cloning, chromosome 3p deleted cells proliferated more slowly than their non-deleted siblings (Figure 6B, p value < 0.05). The chr_3p hemizygously deleted cells did not undergo increased apoptosis as measured by propidium iodide staining (Figure S4C). However, we observed more cells in G1 of the cell cycle by propidium iodide staining in fixed cells, indicative of cell cycle arrest (Figure S4D, p value < 0.001). AALE parental cells do not form colonies in soft agar (Lundberg et al., 2002), and 3p deleted cells were also negative in this assay for anchorage independent growth (data not shown). RNA sequencing of deleted clones and their non-deleted siblings confirmed down-regulation of chr_3p genes in cis, statistically significant for 64% of genes (FDR < 0.05) (Figure S4E). STAC and ROBO1 were the most down-regulated 3p genes, decreased by more than 15-fold, and chr_3p genes UBA7 and LMCD1 were up-regulated more than two-fold. Chr_3p hemizygously deleted cells also had up-regulation of interferon and immune response pathways by GSEA (FWER < 0.01) (Figure 6C), consistent with the finding that 3p copy number was anti-correlated with these pathways in the pan-cancer analysis (Figures S3D and S3E).
Figure 6. Chromosome 3p lung cells initially have slower proliferation, but normalize over time.
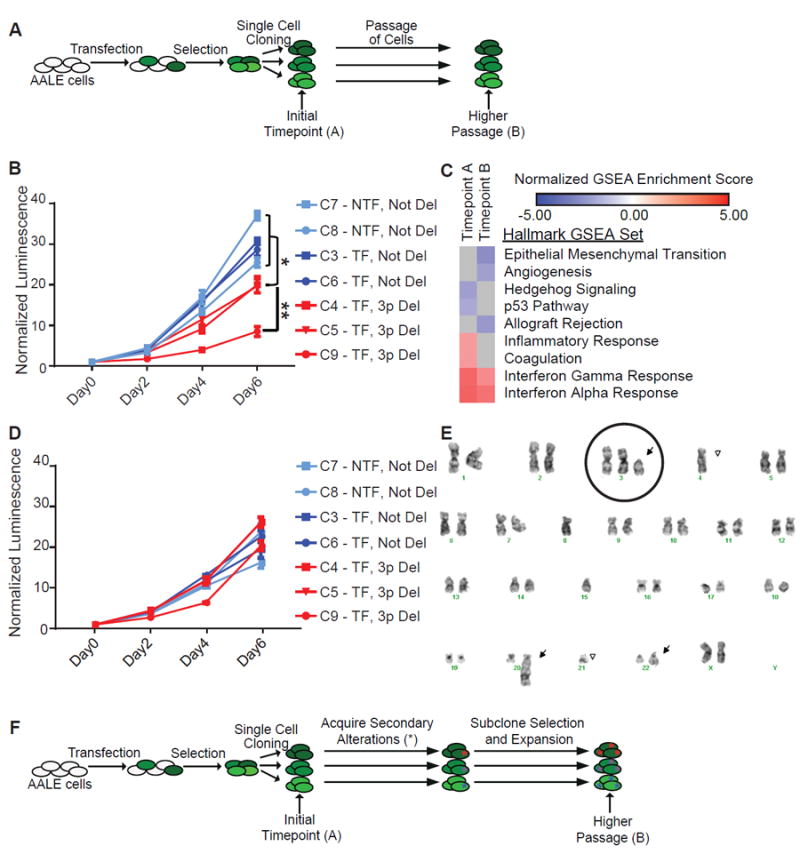
(A) AALE cells were transfected, and one day later cells were selected with puromycin for cells that had incorporated transfected DNA. Cells were single cell cloned to isolate chr_3p deleted clones, and assayed before and after extensive passaging.
(B) Proliferation curves were generated using CellTiter-Glo over 6 days (x-axis). Y-axis is relative luminescence units, normalized to Day 0. Data plotted are means with error bars +/- standard deviation. (* = p value < 0.01, ** = p value < 0.001) Cells were from Timepoint A.
(C) Heatmap of normalized enrichment scores for Hallmark gene sets in GSEA.
(D) Proliferation curves were generated using CellTiter-Glo over 6 days (x-axis). Y-axis is relative luminescence units, normalized to Day 0. Data plotted are means with error bars +/- standard deviation. Cells were from the higher passage population.
(E) Karyotype from one of the chromosome 3p deleted clones. Chromosome 3 is circled, and arrows point to chromosomal abnormalities.
(F) During passaging, cells have gained additional changes (*) to adapt to chr_3p deletion.
See also Figure S4.
After an additional 4-5 passages (approximately 10 population doublings) and one round of freeze-thaw, 3p deleted cells no longer proliferated more slowly (Figure 6D). 53% of chr_3p genes were still significantly down-regulated (FDR < 0.05) (Figure S4F), with STAC and ROBO1 decreased more than 13-fold. Interestingly, UBA7, an ubiquitin enzyme on 3p which targets interferon gene ISG15, was no longer up-regulated. By GSEA, interferon pathways were still up-regulated (Figure 6C), though enrichment scores were lower and other immune signatures were no longer significant. Later stage chr_3p deleted cells had downregulation of genes involved in the epithelial mesenchymal transition and angiogenesis (GSEA FWER < 0.01), but the implications of this remain obscure. We also observed subclones with duplication of the remaining full copy of chromosome 3 in two of the three deleted clones (Figure 6E, Figure S4G). In these subclones with chromosome 3 duplications, 3q is gained and 3p is no longer lost. We conclude that selection of advantageous alterations or expression changes allow chr_3p deleted cells to overcome the negative growth effects of aneuploidy in this model system (Figure 6F). In this section, we have demonstrated that it is possible to model aneuploidy. We have not observed a neutral or positive impact of aneuploidy on cell proliferation. Results are summarized in Table 1.
Table 1.
Summary of results
| Aneuploidy and Ploidy |
| • Across 10,522 pan-cancer samples, the correlation coefficient between ploidy and aneuploidy score is 0.55. |
| • Across 6,800 samples without genome doubling, the correlation coefficient is -0.13. |
| • Across 3,242 samples with one genome doubling, the correlation coefficient is -0.32. |
| • Across 480 samples with two or more genome doublings, the correlation coefficient is 0.17. |
| Aneuploidy and Mutations |
| • Across 9,702 samples (with copy number and mutation data) and corrected by tumor type and mutation rate, TP53 mutation is enriched in samples with higher aneuploidy, with a coefficient of 0.126 and a Bonferroni corrected p value of 5.58*10-141. |
| • Across 9,766 pan-cancer samples in 30 tumor types (with copy number, mutation rate, and leukocyte fraction data), the correlation coefficient between non-silent mutation rate and aneuploidy score is 0.336. |
| • Across 191 samples with high microsatellite instability or POLE mutations, the correlation coefficient between non-silent mutation rate and aneuploidy score is -0.239 |
| • Across 9,575 samples without high microsatellite instability or POLE mutations, the correlation coefficient between non-silent mutation rate and aneuploidy score is 0.379. |
| • Across 9,766 pan-cancer samples, the average tumor-type correlation coefficient between impurity and aneuploidy score is 0.089. |
| • Across 9,766 pan-cancer samples, the average tumor-type correlation coefficient between leukocyte fraction and aneuploidy score is -0.077. |
| • Across 9,766 pan-cancer samples, the average tumor-type correlation coefficient between non-leukocyte stroma and aneuploidy score is 0.241. |
| Chromosome Arm-Alterations and Cross-Tumor Clusters |
| • Clustering 25 tumor subtypes revealed a group of gastrointestinal (endoderm) tumors, including colon adenocarcinoma, rectal adenocarcinoma, esophageal adenocarcinoma, stomach adenocarcinoma, and pancreatic adenocarcinoma. The Pearson’s correlations range from 0.37 to 0.99. |
| • Clustering revealed a group of gynecological (mesoderm) tumors, including endometrial carcinoma with high copy number levels and ovarian cancers. The Pearson’s correlation coefficient is 0.90. |
| • Clustering revealed a group of squamous cell carcinomas (ectoderm), including lung squamous cell carcinoma, head and neck squamous cell carcinoma, esophageal squamous carcinoma, and cervical squamous carcinoma. The Pearson’s correlation coefficient ranges from 0.58 to 0.94. |
| Phenotypes of Chromosome 3p Deletion |
| • Chromosome 3p deletion and chromosome 3q gain are significantly enriched in lung squamous cell carcinoma, with a p value of 0.039. |
| • Chromosome 3p deletion does not induce apoptosis. |
| • Chromosome 3p deletion induces slowed cell cycle (more cells in G1), with a p value of 0.0006. |
| • Chromosome 3p deletion initially induces slower cellular proliferation (p value < 0.0017). |
| • Chromosome 3 duplication occurs in chromosome 3p deleted cells. |
Discussion
Here, we present parallel computational and experimental approaches that provide insight into the largely unexplored role of aneuploidy in cancer. By calculating aneuploidy level in 10,522 tumors, we uncovered the correlation of aneuploidy with TP53 mutations, overall somatic mutation rate, and proliferative signatures, and an inverse correlation with leukocyte fraction. Our analysis revealed expression changes in cell-cycle and immune hallmarks associated with individual chromosome arm-level alterations, independent of aneuploidy level. We also observe tissue specific patterns of aneuploidy, and squamous tumors of different tissue origins clustering together. Using CRISPR technology, we modeled one of the chromosome arm alterations observed in squamous tumors (chr_3p deletion) in human immortalized lung epithelial cells. A decrease in cellular proliferation rate is associated with chr_3p deletion, a phenotype that is reversed by gain of chromosome 3.
Examining pan-cancer aneuploidy analyses
In this study, we defined aneuploidy to include chromosome and arm-level somatic copy number alterations (SCNAs), but not smaller SCNAs. In some cases, computationally derived definitions of aneuploidy and analyses of SCNAs have included both broad and focal SCNAs (Davoli et al., 2017; Buccitelli et al., 2017). Broad and focal events occur by different mechanisms and have different effects; broad events affect a large number of genes by one or few copies, while focal events affect fewer genes with higher amplitudes of gain or with homozygous deletion (Beroukhim et al., 2007). Therefore we chose to restrict our analysis to the broad events, to harmonize the definition of aneuploidy more with the mechanism of generation of the structural and transcriptional consequences.
Aneuploidy level varies greatly across tumor type, impacting the analyses of aneuploidy correlations with mutation rate and immune signatures and potentially leading to conflicting results in the literature (Ciriello et al., 2013; Davoli et al., 2017; Buccitelli et al., 2017). By controlling for tumor type, as well as confounders such as sample purity and cellular composition, our analysis reconciles discrepancies in the literature. Regarding mutation rate and aneuploidy correlations across cancers, mutation rate is inversely proportional to aneuploidy, but this anti-correlation is driven by tumors with high microsatellite instability (MSI, mostly colon adenocarcinoma and endometrial cancers). Once MSI tumors are removed, mutation and aneuploidy are positively correlated (consistent with Davoli et al., 2017 and Buccitelli et al., 2017).
When controlling for tumor type, our results are consistent with a negative correlation between aneuploidy and leukocyte infiltrate that explains a decrease in expression of immune signatures. Interestingly, some studies have shown that abnormal karyotypes or hyperploidy can trigger an immune response (Santaguida et al., 2017; Senovilla et al., 2012). This result suggests the possibility that aneuploid tumor cells may have to overcome or evade this ploidy-related immune response for tumors to progress.
When controlling for tumor type, aneuploidy level, and cellular composition of each sample, we identified immune expression signatures that can independently correlate with individual arm-level copy number alterations. Deletions of some arms, including 3p, 8p, 13q, or 17p, are positively correlated with immune signatures, whereas deletions of other arms, including 4q, 5q, and 14q, are anti-correlated with immune signatures. The distinct correlations of immune signatures with deletion of different arms also suggest that immune signature changes might be due to specific genes or regions within each affected arm rather than to overall aneuploidy.
Genome engineering to model an individual cancer aneuploidy
We have developed a method to generate chromosome arm deletions by initiating centromeric DNA breaks with CRISPR/Cas9 to promote replacement of an arm with a selection cassette and an artificial telomere. The method described here can be applied to deletion of any chromosome arm. In the case of chr_3p deletion, differential expression of chr_3p genes and interferon response genes is consistent with expression changes across TCGA samples, supporting the utility of both our computational and experimental approaches.
Consistent with single chromosome aneuploidy models in human cells (Stingele et al., 2012; Sheltzer et al., 2017), chromosome arm-level deletion of human chr_3p has an anti-proliferative effect that cells can overcome with time. Even though p53 is inhibited in the lung epithelial cells, as they were immortalized with SV40 large T antigen (Lundberg et al., 2002), chr_3p deletion initially caused a decrease in cell proliferation. We initially predicted that deletion of chr_3p would not be as anti-proliferative as other aneuploidies, as it is thought to be an early event in lung tumorigenesis (Hung et al., 1995; Wistuba et al., 2000). However, in our cells, selective pressures have likely induced some secondary changes in the chr_3p deleted cells to promote proliferation.
One previously observed mechanism of selection is the evolution of cell line karyotypes over time (Sheltzer et al., 2017). In two of the three chr_3p deleted clones, we observed subclones with a duplicated wildtype copy of chromosome 3, changing an individual cell from chr_3p deleted to chr_3q gained. Interestingly, chr_3q gain is also a defining feature of squamous cell carcinoma, and contains oncogenes such as SOX2, PIK3CA, and TERC. In the future, we plan to expand our chromosome deletion model to different chromosome arms (Cai et al., 2016; Uno et al., 2017), and thereby to interrogate how specific aneuploidies can induce distinct expression patterns, phenotypes, and selection mechanisms.
Open questions in aneuploidy: Tissue specificity and selection
The tissue specificity we observe in patterns of aneuploidy could be due to the specific transcriptional effects of different aneuploidies. Our work here begins to analyze expression changes induced by each arm alteration while controlling for overall aneuploidy level and other confounders. By focusing our analysis within tumor types and comparing between them, we can uncover whether specific aneuploidies have different transcriptional consequences in different tissues. Determining whether transcriptional consequences have distinct selective advantages and disadvantages in different tissues will require further integration of in vitro models and computational analysis. In addition, specific aneuploidies may trigger unique dependencies; future directions involve interrogating existing in vitro CRISPR and shRNA cell line data to identify potential therapeutic targets that can be validated in the in vitro models.
The major question about cancer aneuploidy remains open: is aneuploidy positively selected for in cancer? We favor the view that aneuploidy is positively selected for, as it is a universal and tissue-consistent feature of epithelial cancer. However, the experimental data on cancer aneuploidy do not currently support this model. There are a few possible explanations for this contradiction. In the case of oncogenic mutations, overexpression in model systems often leads to oncogene-induced senescence (Larsson, 2011); individual aneuploidies may elicit a similar response. In addition, we have not yet identified the exact environment for positive selection of aneuploidy; p53/RB inhibition and TERT activation may not be sufficient to allow selection of aneuploidies. Future work will involve combining models of diverse aneuploidy events, including chromosome arm gains and losses, along with different mutations and different cellular contexts. It will be increasingly important to incorporate models of stromal interaction into prospective studies of aneuploidy and its selection, whether by advanced cellular modeling, single cell sequencing, or use of animal models. In particular, CRISPR manipulation in human lung organoids will be a useful approach to assess interactions of different lung cell types with aneuploidy alterations, as well as potential stromal cell interactions in a 3D tissue model (Barkauskas et al., 2017). Continued integration of computational and experimental approaches will be required to understand how an aneuploidy alteration, affecting hundreds of genes simultaneously, results in an aneuploidy phenotype and contributes to tumor development or, alternatively, is an unselected event.
STAR Methods
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Matthew Meyerson (matthew_meyerson@dfci.harvard.edu).
Experimental Model and Subject Details
All in vitro experiments were performed in XX immortalized lung epithelial cells (AALE cells), derived at Dana-Farber and immortalized by SV40 large-T antigen (Lundberg et al., 2002). Cells were maintained at 37 degrees Celsius and 5% CO2 in Lonza small airway growth medium (CC-3118).
Method Details
Pan-cancer Computational Analyses
Datasets
For 10,522 TCGA samples, somatic DNA copy number was determined from Affymetrix SNP 6.0 arrays. For 9,670 of these 10,522 samples, RSEM (relative standard error of the mean) expression values were determined from Illumina mRNA sequencing data. For 9,756 of these 10,522 samples, mutations were called from whole-exome DNA sequencing data (TCGA MC3 manuscript, in preparation). (See also Table S1.)
Calculating Arm-Level Events and Aneuploidy Score
Using the ABSOLUTE algorithm (Carter et al., 2012) for each sample, we determined the likeliest ploidy and absolute total copy number of each segment in the genome. Each segment was designated as amplified, deleted, or neutral based on whether its copy number was greater than, smaller than, or equal to the sample’s ploidy (rounded to the nearest integer) respectively. For amplifications and deletions separately (collectively “alterations”), segments were joined until either the entire chromosome was considered altered or more than 20% of the genomic length between the start and ends were not altered in the same direction; e.g. >20% deleted or neutral for joining amplification segments. For every combination of arm/chromosome and direction of alteration within each TCGA tumor type, the start coordinates, end coordinates, and percentage length of the longest joined segment were clustered across samples using a Gaussian Mixture Model (Pedregosa et al., 2011, Python package SciKit-Learn). The optimal clustering solution between 2-9 clusters inclusive was chosen based on the lowest BIC (Bayesian information criterion). Tumors in clusters whose mean fraction altered in either specific direction was >=80% were considered “aneuploid.” Tumors altered <20% (in both directions) were considered “non-aneuploid,” and others were designated “other.” Each arm was assigned -1 if lost, +1 if gained, 0 if non-aneuploid, and “NA” if other. Aneuploidy score (number of altered arms) for each tumor is calculated as the sum total of altered arms, for a range of 0 (no arms) to 39 (all arms – long and short arms for each non-acrocentric chromosome, and only long arms for chromosomes 13, 14, 15, 21, and 22).
Other scores
Fraction of genome altered by aneuploidy was determined by multiplying each arm altered by its length, and dividing by the length of the genome. Recurrent SCNA scores are the sum of recurrent SCNAs as identified by GISTIC2.0 (output to be available at NCI Genomic Data Commons). Recurrent mutation scores are the sum of recurrent mutations, defined in the TCGA MC3 manuscript (TCGA, in preparation).
Spearman Correlations
Spearman correlation coefficients were calculated using the spearmanr function in the stats package of scipy-0.19.0, which was run using Python-3.5.4 or using cor.test in R (method = “spearman”), which was run using R version 3.2.3.
Linear Modeling
Linear modeling was performed in R version 3.2.3, using lm. The equations used:
P values for each coefficient were calculated by the lm function in R.
Gene Set Enrichment Analysis
Gene set enrichment analysis (GSEA) was performed using Broad GSEA v.3.0 (Subramanian et al., 2005). Ranked lists of genes and coefficients were entered, and enrichment was assessed using Hallmark and Positional Gene Sets.
Clustering
Hierarchical clustering of tissue-means of arm-level alterations was performed using 1-Pearson’s in Morpheus (https://software.broadinstitute.org/morpheus/index.html).
Cell Line Analyses
Karyotype Analysis
Karyotype analysis was performed by the Brigham and Women’s Cytogenomics Core.
RNA Sequencing and Analysis
RNA was isolated from cells using the Qiagen RNAeasy kit (Qiagen 74104), with DNAse treatment. PolyA isolation and sequencing library preparation was performed using the NEBNext Ultra Directional RNA Library Prep Kit. Each set of samples were pooled and sequenced in one lane of HiSeq2500 RR, 100 basepair paired end. Sequencing reads were aligned using STAR (Dobin et al., 2013) and expression level was quantified by RSEM (Li and Dewey, 2011). Differential expression analysis was performed using the edgeR pipeline (Robinson et al., 2010). Ranked lists of genes with fold change were entered into GSEA as described above.
DNA Sequencing and Analysis
DNA was isolated from cells using the QiaAmp Mini DNA kit. Sequencing library preparation was performed using the Nextera DNA Sample Preparation Kit. Samples were pooled and sequencing by miSeq, 300 basepair paired end. Copy number profiles were generated by HMMCopy (Lai et al., 2016). Subclonal analysis was performed using IchorCNA (Adalsteinsson et al., 2017).
Cell Maintenance, Transfection and Proliferation Assays
Transfections were performed using Fugene-6 (Promega E2691) following manufacturer’s instructions at a 3:1 ratio. 24 hours after transfection, cells were selected with 2 μg/mL puromycin through several passages and a round of single cell cloning. For single cell cloning, individual cells were plated in each well of a 96-well plate using cell sorting, with each well containing 100μL of 50% conditioned small airway growth media (SAGM). After single cell cloning and confirmation of telomere recombination, puromycin was no longer added in the media. For cell proliferation assays, 1500 cells per well in a 96-well plate were plated in 100μL of media. Plates were collected at time of plating and days 2, 4, and 6. 50μL of CellTiter-Glo Reagent was added, and plates were incubated for 20 minutes before luminescence readings.
Flow Cytometry
For apoptosis analysis, adherent cells were washed, trypsinized, collected, and stained in propidium iodide solution. Propidium iodide levels were measured on the BD LSRii. For cell cycle analysis, adherent cells were washed, trypsinized, collected, and fixed in 10% formalin buffered solution and 70% ethanol. After fixing, cells were stained with propidium iodide solution for thirty minutes before analysis on the BD LSRii. Cells were assigned to G1, S, and G2 stages of the cell cycle by identifying the G1 peak and 2x G2 peak.
PCR and qPCR
PCR was performed using the Sigma Aldrich AccuTaq enzyme (D8045) with primers listed under key resources. Quantitative PCR (qPCR) was performed using the Power SYBR Green PCR mastermix (ThermoFisher 4367659), with the primers listed above.
Quantification and Statistical Analysis
Quantitative and statistical methods are noted above.
Data and Software Availability
The raw TCGA data, processed data and clinical data can be found at the legacy archive of the GDC (https://portal.gdc.cancer.gov/legacy-archive/search/f) and the PancanAtlas publication page (https://gdc.cancer.gov/about-data/publications/pancanatlas). The mutation data can be found here (https://gdc.cancer.gov/about-data/publications/mc3-2017). TCGA data can also be explored through the Broad Institute FireBrowse portal (http://gdac.broadinstitute.org) and the Memorial Sloan Kettering Cancer Center cBioPortal (http://www.cbioportal.org). For the in vitro assays, RNA-sequencing BAM files are available at the Sequence Read Archive as Study PRJNA436953 (SRP133935), Run Numbers SRR6806553-SRR6806554 and SRR6806559-SRR6806570. Related DNA-sequencing BAM files are available at the Sequence Read Archive as Study PRJNA436953 (SRP133935), Run Numbers SRR6806543-SRR6806552 and SRR6806555-SRR6806558. Details for software availability are in the Key Resource Table.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
| Critical Commercial Assays | ||
| Nextera DNA Sample Preparation Kit | Illumina | Catalog: FC-121-1030 |
| NEBNext Ultra Directional RNA Library Prep Kit | New England Biolabs | Catalog: E7420S |
| QIAamp DNA Mini Kit | Qiagen | Catalog: 51304 |
| Qiagen RNeasy Mini Kit | Qiagen | Catalog: 74104 |
| CellTiter-Glo Luminescence Viability Assay | VWR | Catalog: PAG7572 |
| Deposited Data | ||
| Raw data files for RNA-sequencing of in vitro AALE clones | This paper | https://www.ncbi.nlm.nih.gov/Traces/study/?acc=SRP133935 |
| Raw data files for DNA-sequencing of in vitro AALE clones | This paper | https://www.ncbi.nlm.nih.gov/Traces/study/?acc=SRP133935 |
| Experimental Models: Cell Lines | ||
| Immortalized Lung Epithelial Cells (AALE) | Lundberg et al., 2002 | #N/A |
| Oligonucleotides | ||
| Chr_3p CRISPR (5’ TGATCAGTCAGGTAAGGATG 3’) | This paper | #N/A |
| Recombination PCR Forward (5’CTACCCGCTTCCATTGCTCA 3’) | This paper | #N/A |
| Recombination PCR Reverse (5’TTGGTTGAGCAGTTGGACAT 3’) | This paper | #N/A |
| Chr_3p qPCR Forward (5’ ACAATCCAAACTAGCATGCACA 3’) | This paper | #N/A |
| Chr_3p qPCR Reverse (5’ AGCGTTAGAGGGAGGGGAG 3’) | This paper | #N/A |
| Chr_3q qPCR Forward (5’ CGTGTCCGGGGTAGATCTTG 3’) | This paper | #N/A |
| Chr_3p qPCR Reverse (5’ GCTTACATCCTCGGGCAGAA 3’) | This paper | #N/A |
| Software and Algorithms | ||
| ABSOLUTE | Carter et al., 2012 | Version 1.5 |
| Python Package SciKit-Learn | Pedregosa et al., 2011 | Version 0.16.1 |
| Python Package scipy stats | www.scipy.org | Version 0.19.0 |
| edgeR | Robinson et al., 2010 | |
| STAR | Dobin et al., 2013 | |
| RSEM | Li and Dewey, 2011 | |
| GSEA | Subramanian et al., 2005 | Version 3.0 |
| HMMCopy | Lai et al., 2016 | |
| IchorCNA | Adalsteinsson et al., 2017 | |
| Morpheus | https://software.broadinstitute.org/morpheus/index.html | |
Supplementary Material
Table S2, related to Figure 1. Sample Chromosome Arm Calls and Aneuploidy Scores.
Table S3, related to Figure 2. Aneuploidy Score Coefficients for Gene Mutation Linear Models.
Table S4, related to Figures 2 and 3. Correlation Coefficients with Aneuploidy Score.
Table S5, related to Figure 3. Aneuploidy Score Coefficients for Gene Expression Linear Models.
Table S6, related to Figure 3. Gene Set Enrichment Analysis (GSEA) Enrichment Scores.
Table S7, related to Figure 4. Chromosome Arm Summary Statistics and Chromosome Arm Coefficients for Gene Expression Linear Models.
Table S8, related to Figure 4. Matrix of 1-Pearson's Correlation Coefficients for Tumor Type Clustering.
Highlights.
Aneuploidy, whole chromosome or chromosome-arm imbalance, occurs in 88% of cancers
Aneuploidy correlates with cell cycle genes and anti-correlates with immune levels
Patterns of aneuploidy alterations are tumor-type specific
Engineered chromosome 3p deletion does not promote proliferation in human lung cells
Significance.
Although its universality makes aneuploidy a salient feature of cancer genomes, the functional significance of cancer aneuploidy remains a mystery. Powerful technologies for genome analysis and genome engineering now make the study of aneuploidy more tractable. Here, we look across cancer to identify universal and cancer-type specific characteristics of aneuploidy which further hint at its importance for the process of cancer pathogenesis. Furthermore, the ability to engineer aneuploid chromosome arms will now enable the development of more complex cellular models to generate and investigate the proliferative and survival impact of cancer aneuploidy.
Acknowledgments
All cytogenomics analysis was performed by the CytoGenomics Core at Brigham and Women’s Hospital. A plasmid containing an artificial telomere was kindly shared by Narumi Uno and Mitsuo Oshimura (Uno et al., 2017). A.M.T. was funded by a grant from Uniting Against Lung Cancer and is an American Cancer Society postdoctoral fellow. M.M. acknowledges support from the National Cancer Institute (1R35CA197568) and the Norman R. Seaman Endowment fund. M.M. is an American Cancer Society Research Professor.
Secondary author list
Amy Blum, Samantha J. Caesar-Johnson, John A. Demchok, Ina Felau, Melpomeni Kasapi, Martin L. Ferguson, Carolyn M. Hutter, Heidi J. Sofia, Roy Tarnuzzer, Peggy Wang, Zhining Wang, Liming Yang, Jean C. Zenklusen, Jiashan (Julia) Zhang, Sudha Chudamani, Jia Liu, Laxmi Lolla, Rashi Naresh, Todd Pihl, Qiang Sun, Yunhu Wan, Ye Wu, Juok Cho, Timothy DeFreitas, Scott Frazer, Nils Gehlenborg, Gad Getz, David I. Heiman, Jaegil Kim, Michael S. Lawrence, Pei Lin, Sam Meier, Michael S. Noble, Gordon Saksena, Doug Voet, Hailei Zhang, Brady Bernard, Nyasha Chambwe, Varsha Dhankani, Theo Knijnenburg, Roger Kramer, Kalle Leinonen, Yuexin Liu, Michael Miller, Sheila Reynolds, Ilya Shmulevich, Vesteinn Thorsson, Wei Zhang, Rehan Akbani, Bradley M. Broom, Apurva M. Hegde, Zhenlin Ju, Rupa S. Kanchi, Anil Korkut, Jun Li, Han Liang, Shiyun Ling, Wenbin Liu, Yiling Lu, Gordon B. Mills, Kwok-Shing Ng, Arvind Rao, Michael Ryan, Jing Wang, John N. Weinstein, Jiexin Zhang, Adam Abeshouse, Joshua Armenia, Debyani Chakravarty, Walid K. Chatila, Ino de Bruijn, Jianjiong Gao, Benjamin E. Gross, Zachary J. Heins, Ritika Kundra, Konnor La, Marc Ladanyi, Augustin Luna, Moriah G. Nissan, Angelica Ochoa, Sarah M. Phillips, Ed Reznik, Francisco Sanchez-Vega, Chris Sander, Nikolaus Schultz, Robert Sheridan, S. Onur Sumer, Yichao Sun, Barry S. Taylor, Jioajiao Wang, Hongxin Zhang, Pavana Anur, Myron Peto, Paul Spellman, Christopher Benz, Joshua M. Stuart, Christopher K. Wong, Christina Yau, D. Neil Hayes, Joel S. Parker, Matthew D. Wilkerson, Adrian Ally, Miruna Balasundaram, Reanne Bowlby, Denise Brooks, Rebecca Carlsen, Eric Chuah, Noreen Dhalla, Robert Holt, Steven J.M. Jones, Katayoon Kasaian, Darlene Lee, Yussanne Ma, Marco A. Marra, Michael Mayo, Richard A. Moore, Andrew J. Mungall, Karen Mungall, A. Gordon Robertson, Sara Sadeghi, Jacqueline E. Schein, Payal Sipahimalani, Angela Tam, Nina Thiessen, Kane Tse, Tina Wong, Ashton C. Berger, Rameen Beroukhim, Andrew D. Cherniack, Carrie Cibulskis, Stacey B. Gabriel, Galen F. Gao, Gavin Ha, Matthew Meyerson, Steven E. Schumacher, Juliann Shih, Melanie H. Kucherlapati, Raju S. Kucherlapati, Stephen Baylin, Leslie Cope, Ludmila Danilova, Moiz S. Bootwalla, Phillip H. Lai, Dennis T. Maglinte, David J. Van Den Berg, Daniel J. Weisenberger, J. Todd Auman, Saianand Balu, Tom Bodenheimer, Cheng Fan, Katherine A. Hoadley, Alan P. Hoyle, Stuart R. Jefferys, Corbin D. Jones, Shaowu Meng, Piotr A. Mieczkowski, Lisle E. Mose, Amy H. Perou, Charles M. Perou, Jeffrey Roach, Yan Shi, Janae V. Simons, Tara Skelly, Matthew G. Soloway, Donghui Tan, Umadevi Veluvolu, Huihui Fan, Toshinori Hinoue, Peter W. Laird, Hui Shen, Wanding Zhou, Michelle Bellair, Kyle Chang, Kyle Covington, Chad J. Creighton, Huyen Dinh, HarshaVardhan Doddapaneni, Lawrence A. Donehower, Jennifer Drummond, Richard A. Gibbs, Robert Glenn, Walker Hale, Yi Han, Jianhong Hu, Viktoriya Korchina, Sandra Lee, Lora Lewis, Wei Li, Xiuping Liu, Margaret Morgan, Donna Morton, Donna Muzny, Jireh Santibanez, Margi Sheth, Eve Shinbrot, Linghua Wang, Min Wang, David A. Wheeler, Liu Xi, Fengmei Zhao, Julian Hess, Elizabeth L. Appelbaum, Matthew Bailey, Matthew G. Cordes, Li Ding, Catrina C. Fronick, Lucinda A. Fulton, Robert S. Fulton, Cyriac Kandoth, Elaine R. Mardis, Michael D. McLellan, Christopher A. Miller, Heather K. Schmidt, Richard K. Wilson, Daniel Crain, Erin Curley, Johanna Gardner, Kevin Lau, David Mallery, Scott Morris, Joseph Paulauskis, Robert Penny, Candace Shelton, Troy Shelton, Mark Sherman, Eric Thompson, Peggy Yena, Jay Bowen, Julie M. Gastier-Foster, Mark Gerken, Kristen M. Leraas, Tara M. Lichtenberg, Nilsa C. Ramirez, Lisa Wise, Erik Zmuda, Niall Corcoran, Tony Costello, Christopher Hovens, Andre L. Carvalho, Ana C. de Carvalho, José H. Fregnani, Adhemar Longatto-Filho, Rui M. Reis, Cristovam Scapulatempo-Neto, Henrique C.S. Silveira, Daniel O. Vidal, Andrew Burnette, Jennifer Eschbacher, Beth Hermes, Ardene Noss, Rosy Singh, Matthew L. Anderson, Patricia D. Castro, Michael Ittmann, David Huntsman, Bernard Kohl, Xuan Le, Richard Thorp, Chris Andry, Elizabeth R. Duffy, Vladimir Lyadov, Oxana Paklina, Galiya Setdikova, Alexey Shabunin, Mikhail Tavobilov, Christopher McPherson, Ronald Warnick, Ross Berkowitz, Daniel Cramer, Colleen Feltmate, Neil Horowitz, Adam Kibel, Michael Muto, Chandrajit P. Raut, Andrei Malykh, Jill S. Barnholtz-Sloan, Wendi Barrett, Karen Devine, Jordonna Fulop, Quinn T. Ostrom, Kristen Shimmel, Yingli Wolinsky, Andrew E. Sloan, Agostino De Rose, Felice Giuliante, Marc Goodman, Beth Y. Karlan, Curt H. Hagedorn, John Eckman, Jodi Harr, Jerome Myers, Kelinda Tucker, Leigh Anne Zach, Brenda Deyarmin, Hai Hu, Leonid Kvecher, Caroline Larson, Richard J. Mural, Stella Somiari, Ales Vicha, Tomas Zelinka, Joseph Bennett, Mary Iacocca, Brenda Rabeno, Patricia Swanson, Mathieu Latour, Louis Lacombe, Bernard Têtu, Alain Bergeron, Mary McGraw, Susan M. Staugaitis, John Chabot, Hanina Hibshoosh, Antonia Sepulveda, Tao Su, Timothy Wang, Olga Potapova, Olga Voronina, Laurence Desjardins, Odette Mariani, Sergio Roman-Roman, Xavier Sastre, Marc-Henri Stern, Feixiong Cheng, Sabina Signoretti, Andrew Berchuck, Darell Bigner, Eric Lipp, Jeffrey Marks, Shannon McCall, Roger McLendon, Angeles Secord, Alexis Sharp, Madhusmita Behera, Daniel J. Brat, Amy Chen, Keith Delman, Seth Force, Fadlo Khuri, Kelly Magliocca, Shishir Maithel, Jeffrey J. Olson, Taofeek Owonikoko, Alan Pickens, Suresh Ramalingam, Dong M. Shin, Gabriel Sica, Erwin G. Van Meir, Hongzheng Zhang, Wil Eijckenboom, Ad Gillis, Esther Korpershoek, Leendert Looijenga, Wolter Oosterhuis, Hans Stoop, Kim E. van Kessel, Ellen C. Zwarthoff, Chiara Calatozzolo, Lucia Cuppini, Stefania Cuzzubbo, Francesco DiMeco, Gaetano Finocchiaro, Luca Mattei, Alessandro Perin, Bianca Pollo, Chu Chen, John Houck, Pawadee Lohavanichbutr, Arndt Hartmann, Christine Stoehr, Robert Stoehr, Helge Taubert, Sven Wach, Bernd Wullich, Witold Kycler, Dawid Murawa, Maciej Wiznerowicz, Ki Chung, W. Jeffrey Edenfield, Julie Martin, Eric Baudin, Glenn Bubley, Raphael Bueno, Assunta De Rienzo, William G. Richards, Steven Kalkanis, Tom Mikkelsen, Houtan Noushmehr, Lisa Scarpace, Nicolas Girard, Marta Aymerich, Elias Campo, Eva Giné, Armando López Guillermo, Nguyen Van Bang, Phan Thi Hanh, Bui Duc Phu, Yufang Tang, Howard Colman, Kimberley Evason, Peter R. Dottino, John A. Martignetti, Hani Gabra, Hartmut Juhl, Teniola Akeredolu, Serghei Stepa, Dave Hoon, Keunsoo Ahn, Koo Jeong Kang, Felix Beuschlein, Anne Breggia, Michael Birrer, Debra Bell, Mitesh Borad, Alan H. Bryce, Erik Castle, Vishal Chandan, John Cheville, John A. Copland, Michael Farnell, Thomas Flotte, Nasra Giama, Thai Ho, Michael Kendrick, Jean-Pierre Kocher, Karla Kopp, Catherine Moser, David Nagorney, Daniel O’Brien, Brian Patrick O’Neill, Tushar Patel, Gloria Petersen, Florencia Que, Michael Rivera, Lewis Roberts, Robert Smallridge, Thomas Smyrk, Melissa Stanton, R. Houston Thompson, Michael Torbenson, Ju Dong Yang, Lizhi Zhang, Fadi Brimo, Jaffer A. Ajani, Ana Maria Angulo Gonzalez, Carmen Behrens, Jolanta Bondaruk, Russell Broaddus, Bogdan Czerniak, Bita Esmaeli, Junya Fujimoto, Jeffrey Gershenwald, Charles Guo, Alexander J. Lazar, Christopher Logothetis, Funda Meric-Bernstam, Cesar Moran, Lois Ramondetta, David Rice, Anil Sood, Pheroze Tamboli, Timothy Thompson, Patricia Troncoso, Anne Tsao, Ignacio Wistuba, Candace Carter, Lauren Haydu, Peter Hersey, Valerie Jakrot, Hojabr Kakavand, Richard Kefford, Kenneth Lee, Georgina Long, Graham Mann, Michael Quinn, Robyn Saw, Richard Scolyer, Kerwin Shannon, Andrew Spillane, Jonathan Stretch, Maria Synott, John Thompson, James Wilmott, Hikmat Al-Ahmadie, Timothy A. Chan, Ronald Ghossein, Anuradha Gopalan, Douglas A. Levine, Victor Reuter, Samuel Singer, Bhuvanesh Singh, Nguyen Viet Tien, Thomas Broudy, Cyrus Mirsaidi, Praveen Nair, Paul Drwiega, Judy Miller, Jennifer Smith, Howard Zaren, Joong-Won Park, Nguyen Phi Hung, Electron Kebebew, W. Marston Linehan, Adam R. Metwalli, Karel Pacak, Peter A. Pinto, Mark Schiffman, Laura S. Schmidt, Cathy D. Vocke, Nicolas Wentzensen, Robert Worrell, Hannah Yang, Marc Moncrieff, Chandra Goparaju, Jonathan Melamed, Harvey Pass, Natalia Botnariuc, Irina Caraman, Mircea Cernat, Inga Chemencedji, Adrian Clipca, Serghei Doruc, Ghenadie Gorincioi, Sergiu Mura, Maria Pirtac, Irina Stancul, Diana Tcaciuc, Monique Albert, Iakovina Alexopoulou, Angel Arnaout, John Bartlett, Jay Engel, Sebastien Gilbert, Jeremy Parfitt, Harman Sekhon, George Thomas, Doris M. Rassl, Robert C. Rintoul, Carlo Bifulco, Raina Tamakawa, Walter Urba, Nicholas Hayward, Henri Timmers, Anna Antenucci, Francesco Facciolo, Gianluca Grazi, Mirella Marino, Roberta Merola, Ronald de Krijger, Anne-Paule Gimenez-Roqueplo, Alain Piché, Simone Chevalier, Ginette McKercher, Kivanc Birsoy, Gene Barnett, Cathy Brewer, Carol Farver, Theresa Naska, Nathan A. Pennell, Daniel Raymond, Cathy Schilero, Kathy Smolenski, Felicia Williams, Carl Morrison, Jeffrey A. Borgia, Michael J. Liptay, Mark Pool, Christopher W. Seder, Kerstin Junker, Larsson Omberg, Mikhail Dinkin, George Manikhas, Domenico Alvaro, Maria Consiglia Bragazzi, Vincenzo Cardinale, Guido Carpino, Eugenio Gaudio, David Chesla, Sandra Cottingham, Michael Dubina, Fedor Moiseenko, Renumathy Dhanasekaran, Karl-Friedrich Becker, Klaus-Peter Janssen, Julia Slotta-Huspenina, Mohamed H. Abdel-Rahman, Dina Aziz, Sue Bell, Colleen M. Cebulla, Amy Davis, Rebecca Duell, J. Bradley Elder, Joe Hilty, Bahavna Kumar, James Lang, Norman L. Lehman, Randy Mandt, Phuong Nguyen, Robert Pilarski, Karan Rai, Lynn Schoenfield, Kelly Senecal, Paul Wakely, Paul Hansen, Ronald Lechan, James Powers, Arthur Tischler, William E. Grizzle, Katherine C. Sexton, Alison Kastl, Joel Henderson, Sima Porten, Jens Waldmann, Martin Fassnacht, Sylvia L. Asa, Dirk Schadendorf, Marta Couce, Markus Graefen, Hartwig Huland, Guido Sauter, Thorsten Schlomm, Ronald Simon, Pierre Tennstedt, Oluwole Olabode, Mark Nelson, Oliver Bathe, Peter R. Carroll, June M. Chan, Philip Disaia, Pat Glenn, Robin K. Kelley, Charles N. Landen, Joanna Phillips, Michael Prados, Jeff Simko, Jeffry Simko, Karen Smith-McCune, Scott VandenBerg, Kevin Roggin, Ashley Fehrenbach, Ady Kendler, Suzanne Sifri, Ruth Steele, Antonio Jimeno, Francis Carey, Ian Forgie, Massimo Mannelli, Michael Carney, Brenda Hernandez, Benito Campos, Christel Herold-Mende, Christin Jungk, Andreas Unterberg, Andreas von Deimling, Aaron Bossler, Joseph Galbraith, Laura Jacobus, Michael Knudson, Tina Knutson, Deqin Ma, Mohammed Milhem, Rita Sigmund, Andrew K. Godwin, Rashna Madan, Howard G. Rosenthal, Clement Adebamowo, Sally N. Adebamowo, Alex Boussioutas, David Beer, Thomas Giordano, Anne-Marie Mes-Masson, Fred Saad, Therese Bocklage, Lisa Landrum, Robert Mannel, Kathleen Moore, Katherine Moxley, Russel Postier, Joan Walker, Rosemary Zuna, Michael Feldman, Federico Valdivieso, Rajiv Dhir, James Luketich, Edna M. Mora Pinero, Mario Quintero-Aguilo, Carlos Gilberto Carlotti, Jr., Jose Sebastião Dos Santos, Rafael Kemp, Ajith Sankarankuty, Daniela Tirapelli, James Catto, Kathy Agnew, Elizabeth Swisher, Jenette Creaney, Bruce Robinson, Carl Simon Shelley, Eryn M. Godwin, Sara Kendall, Cassaundra Shipman, Carol Bradford, Thomas Carey, Andrea Haddad, Jeffey Moyer, Lisa Peterson, Mark Prince, Laura Rozek, Gregory Wolf, Rayleen Bowman, Kwun M. Fong, Ian Yang, Robert Korst, W. Kimryn Rathmell, J. Leigh Fantacone-Campbell, Jeffrey A. Hooke, Albert J. Kovatich, Craig D. Shriver, John DiPersio, Bettina Drake, Ramaswamy Govindan, Sharon Heath, Timothy Ley, Brian Van Tine, Peter Westervelt, Mark A. Rubin, Jung Il Lee, Natália D. Aredes, Armaz Mariamidze, Anant Agrawal, Jaeil Ahn, Jordan Aissiou, Dimitris Anastassiou, Jesper B. Andersen, Jurandyr M. Andrade, Marco Antoniotti, Jon C. Aster, Donald Ayer, Matthew H. Bailey, Rohan Bareja, Adam J. Bass, Azfar Basunia, Oliver F. Bathe, Rebecca Batiste, Oliver Bear Don’t Walk, Davide Bedognetti, Gloria Bertoli, Denis Bertrand, Bhavneet Bhinder, Gianluca Bontempi, Dante Bortone, Donald P. Bottaro, Paul Boutros, Kevin Brennan, Chaya Brodie, Scott Brown, Susan Bullman, Silvia Buonamici, Tomasz Burzykowski, Lauren Averett Byers, Fernando Camargo, Joshua D. Campbell, Francisco J. Candido dos Reis, Shaolong Cao, Maria Cardenas, Helio H.A. Carrara, Isabella Castiglioni, Anavaleria Castro, Claudia Cava, Michele Ceccarelli, Shengjie Chai, Kridsadakorn Chaichoompu, Matthew T. Chang, Han Chen, Haoran Chen, Hu Chen, Jian Chen, Jianhong Chen, Ken Chen, Ting-Wen Chen, Zhong Chen, Zhongyuan Chen, Hui Cheng, Hua-Sheng Chiu, Cai Chunhui, Giovanni Ciriello, Cristian Coarfa, Antonio Colaprico, Lee Cooper, Daniel Cui Zhou, Aedin C. Culhane, Christina Curtis, Patrycja Czerwi3ska, Aditya Deshpande, Lixia Diao, Michael Dill, Di Du, Charles G. Eberhart, James A. Eddy, Robert N. Eisenman, Mohammed Elanbari, Olivier Elemento, Kyle Ellrott, Manel Esteller, Farshad Farshidfar, Bin Feng, Camila Ferreira de Souza, Esla R. Flores, Steven Foltz, Mitchell T. Frederick, Qingsong Gao, Carl M. Gay, Zhongqi Ge, Andrew J. Gentles, Olivier Gevaert, David L. Gibbs, Adam Godzik, Abel Gonzalez-Perez, Marc T. Goodman, Dmitry A. Gordenin, Carla Grandori, Alex Graudenzi, Casey Greene, Justin Guinney, Margaret L. Gulley, Preethi H. Gunaratne, A. Ari Hakimi, Peter Hammerman, Leng Han, Holger Heyn, Le Hou, Donglei Hu, Kuan-lin Huang, Joerg Huelsken, Scott Huntsman, Peter Hurlin, Matthias Hüser, Antonio Iavarone, Marcin Imielinski, Mirazul Islam, Jacek Jassem, Peilin Jia, Cigall Kadoch, Andre Kahles, Benny Kaipparettu, Bozena Kaminska, Havish Kantheti, Rachel Karchin, Mostafa Karimi, Ekta Khurana, Pora Kim, Leszek J. Klimczak, Jia Yu Koh, Alexander Krasnitz, Nicole Kuderer, Tahsin Kurc, David J. Kwiatkowski, Teresa Laguna, Martin Lang, Anna Lasorella, Thuc D. Le, Adrian V. Lee, Ju-Seog Lee, Steve Lefever, Kjong Lehmann, Jake Leighton, Chunyan Li, Lei Li, Shulin Li, David Liu, Eric Minwei Liu, Jianfang Liu, Rongjie Liu, Yang Liu, William J.R. Longabaugh, Nuria Lopez-Bigas, Li Ma, Wencai Ma, Karen MacKenzie, Andrzej Mackiewicz, Dejan Maglic, Raunaq Malhotra, Tathiane M. Malta, Calena Marchand, R. Jay Mashl, Sylwia Mazurek, Pieter Mestdagh, Chase Miller, Marco Mina, Lopa Mishra, Younes Mokrab, Raymond Monnat, Jr., Nate Moore, Nathanael Moore, Loris Mularoni, Niranjan Nagarajan, Aaron M. Newman, Vu Nguyen, Michael L. Nickerson, Akinyemi I. Ojesina, Catharina Olsen, Sandra Orsulic, Tai-Hsien Ou Yang, James Palacino, Yinghong Pan, Elena Papaleo, Sagar Patil, Chandra Sekhar Pedamallu, Shouyong Peng, Xinxin Peng, Arjun Pennathur, Curtis R. Pickering, Christopher L. Plaisier, Laila Poisson, Eduard Porta-Pardo, Marcos Prunello, John L. Pulice, Charles Rabkin, Janet S. Rader, Kimal Rajapakshe, Aruna Ramachandran, Shuyun Rao, Xiayu Rao, Benjamin J. Raphael, Gunnar Rätsch, Brendan Reardon, Christopher J. Ricketts, Jason Roszik, Carlota Rubio-Perez, Ryan Russell, Anil Rustgi, Russell Ryan, Mohamad Saad, Thais Sabedot, Joel Saltz, Dimitris Samaras, Franz X. Schaub, Barbara G. Schneider, Adam Scott, Michael Seiler, Sara Selitsky, Sohini Sengupta, Jose A. Seoane, Jonathan S. Serody, Reid Shaw, Yang Shen, Tiago Silva, Pankaj Singh, I.K. Ashok Sivakumar, Christof Smith, Artem Sokolov, Junyan Song, Pavel Sumazin, Yutong Sun, Chayaporn Suphavilai, Najeeb Syed, David Tamborero, Alison M. Taylor, Teng Teng, Daniel G. Tiezzi, Collin Tokheim, Nora Toussaint, Mihir Trivedi, Kenneth T. Tsai, Aaron D. Tward, Eliezer Van Allen, John S. Van Arnam, Kristel Van Steen, Carter Van Waes, Christopher P. Vellano, Benjamin Vincent, Nam S. Vo, Vonn Walter, Chen Wang, Fang Wang, Jiayin Wang, Sophia Wang, Wenyi Wang, Yue Wang, Yumeng Wang, Zehua Wang, Zeya Wang, Zixing Wang, Gregory Way, Amila Weerasinghe, Michael Wells, Michael C. Wendl, Cecilia Williams, Joseph Willis, Denise Wolf, Karen Wong, Yonghong Xiao, Lu Xinghua, Bo Yang, Da Yang, Liuqing Yang, Kai Ye, Hiroyuki Yoshida, Lihua Yu, Sobia Zaidi, Huiwen Zhang, Min Zhang, Xiaoyang Zhang, Tianhao Zhao, Wei Zhao, Zhongming Zhao, Tian Zheng, Jane Zhou, Zhicheng Zhou, Hongtu Zhu, Ping Zhu, Michael T. Zimmermann, Elad Ziv, and Patrick A. Zweidler-McKay
The members of The Cancer Genome Atlas Research Network for this project are
NCI/NHGRI Project Team
Samantha J. Caesar-Johnson, John A. Demchok, Ina Felau, Melpomeni Kasapi, Martin L. Ferguson, Carolyn M. Hutter, Heidi J. Sofia, Roy Tarnuzzer, Zhining Wang, Liming Yang, Jean C. Zenklusen, Jiashan (Julia) Zhang
TCGA DCC
Sudha Chudamani, Jia Liu, Laxmi Lolla, Rashi Naresh, Todd Pihl, Qiang Sun, Yunhu Wan, Ye Wu
Genome Data Analysis Centers (GDACs)
The Broad Institute:
Juok Cho, Timothy DeFreitas, Scott Frazer, Nils Gehlenborg, Gad Getz, David I. Heiman, Jaegil Kim, Michael S. Lawrence, Pei Lin, Sam Meier, Michael S. Noble, Gordon Saksena, Doug Voet, Hailei Zhang
Institute for Systems Biology:
Brady Bernard, Nyasha Chambwe, Varsha Dhankani, Theo Knijnenburg, Roger Kramer, Kalle Leinonen, Yuexin Liu, Michael Miller, Sheila Reynolds, Ilya Shmulevich, Vesteinn Thorsson, Wei Zhang
MD Anderson Cancer Center:
Rehan Akbani, Bradley M. Broom, Apurva M. Hegde, Zhenlin Ju, Rupa S. Kanchi, Anil Korkut, Jun Li, Han Liang, Shiyun Ling, Wenbin Liu, Yiling Lu, Gordon B. Mills, Kwok-Shing Ng, Arvind Rao, Michael Ryan, Jing Wang, John N. Weinstein, Jiexin Zhang
Memorial Sloan Kettering Cancer Center:
Adam Abeshouse, Joshua Armenia, Debyani Chakravarty, Walid K. Chatila, Ino de Bruijn, Jianjiong Gao, Benjamin E. Gross, Zachary J. Heins, Ritika Kundra, Konnor La, Marc Ladanyi, Augustin Luna, Moriah G. Nissan, Angelica Ochoa, Sarah M. Phillips, Ed Reznik, Francisco Sanchez-Vega, Chris Sander, Nikolaus Schultz, Robert Sheridan, S. Onur Sumer, Yichao Sun, Yichao Sun, Barry S. Taylor, Jioajiao Wang, Hongxin Zhang
Oregon Health and Science University:
Pavana Anur, Myron Peto, Paul Spellman
University of California Santa Cruz:
Christopher Benz, Joshua M. Stuart, Christopher K. Wong, Christina Yau
University of North Carolina at Chapel Hill:
D. Neil Hayes, Joel S. Parker, Matthew D. Wilkerson
Genome Characterization Centers (GCC)
BC Cancer Agency:
Adrian Ally, Miruna Balasundaram, Reanne Bowlby, Denise Brooks, Rebecca Carlsen, Eric Chuah, Noreen Dhalla, Robert Holt, Steven J.M. Jones, Katayoon Kasaian, Darlene Lee, Yussanne Ma, Marco A. Marra, Michael Mayo, Richard A. Moore, Andrew J. Mungall, Karen Mungall, A. Gordon Robertson, Sara Sadeghi, Jacqueline E. Schein, Payal Sipahimalani, Angela Tam, Nina Thiessen, Kane Tse, Tina Wong
The Broad Institute:
Ashton C. Berger, Rameen Beroukhim, Andrew D. Cherniack, Carrie Cibulskis, Stacey B. Gabriel, Galen F. Gao, Gavin Ha, Matthew Meyerson, Gordon Saksena, Steven E. Schumacher, Juliann Shih
Harvard:
Melanie H. Kucherlapati, Raju S. Kucherlapati
Johns Hopkins:
Stephen Baylin, Leslie Cope, Ludmila Danilova
University of Southern California:
Moiz S. Bootwalla, Phillip H. Lai, Dennis T. Maglinte, David J. Van Den Berg, Daniel J. Weisenberger
University of North Carolina at Chapel Hill:
J. Todd Auman, Saianand Balu, Tom Bodenheimer, Cheng Fan, D. Neil Hayes, Katherine A. Hoadley, Alan P. Hoyle, Stuart R. Jefferys, Corbin D. Jones, Shaowu Meng, Piotr A. Mieczkowski, Lisle E. Mose, Joel S. Parker, Amy H. Perou, Charles M. Perou, Jeffrey Roach, Yan Shi, Janae V. Simons, Tara Skelly, Matthew G. Soloway, Donghui Tan, Umadevi Veluvolu, Matthew D. Wilkerson
Van Andel Research Institute:
Huihui Fan, Toshinori Hinoue, Peter W. Laird, Hui Shen, Wanding Zhou
Genome Sequencing Centers (GSC)
Baylor College of Medicine:
Michelle Bellair, Kyle Chang, Kyle Covington, Chad J. Creighton, Huyen Dinh, HarshaVardhan Doddapaneni, Lawrence A. Donehower, Jennifer Drummond, Richard A. Gibbs, Robert Glenn, Walker Hale, Yi Han, Jianhong Hu, Viktoriya Korchina, Sandra Lee, Lora Lewis, Wei Li, Xiuping Liu, Margaret Morgan, Donna Morton, Donna Muzny, Jireh Santibanez, Margi Sheth, Eve Shinbrot, Linghua Wang, Min Wang, David A. Wheeler, Liu Xi, Fengmei Zhao
The Broad Institute:
Carrie Cibulskis, Stacy B. Gabriel, Julian Hess
Washington University at St. Louis:
Elizabeth L. Appelbaum, Matthew Bailey, Matthew G. Cordes, Li Ding, Catrina C. Fronick, Lucinda A. Fulton, Robert S. Fulton, Cyriac Kandoth, Elaine R. Mardis, Michael D. McLellan, Christopher A. Miller, Heather K. Schmidt, Richard K. Wilson
Bio specimen Core Resource
The International Genomics Consortium:
Daniel Crain, Erin Curley, Johanna Gardner, Kevin Lau, David Mallery, Scott Morris, Joseph Paulauskis, Robert Penny, Candace Shelton, Troy Shelton, Mark Sherman, Eric Thompson, Peggy Yena
Nationwide Children’s Organization:
Jay Bowen, Julie M. Gastier-Foster, Mark Gerken, Kristen M. Leraas, Tara M. Lichtenberg, Nilsa C. Ramirez, Lisa Wise, Erik Zmuda
Tissue Source Sites
Australian Prostate Cancer Research Center:
Niall Corcoran, Tony Costello, Christopher Hovens
Barretos Cancer Hospital:
Andre L. Carvalho, Ana C. de Carvalho, Jose H. Fregnani, Adhemar Longatto-Filho, Rui M. Reis, Cristovam Scapulatempo-Neto, Henrique C. S. Silveira, Daniel O. Vidal
Barrow Neurological Institute:
Andrew Burnette, Jennifer Eschbacher, Beth Hermes, Ardene Noss, Rosy Singh
Baylor College of Medicine:
Matthew L. Anderson, Patricia D. Castro, Michael Ittmann
BC Cancer Agency:
David Huntsman
BioreclamationIVT:
Bernard Kohl, Xuan Le, Richard Thorp
Boston Medical Center:
Chris Andry, Elizabeth R. Duffy
Botkin Hospital:
Vladimir Lyadov, Oxana Paklina, Galiya Setdikova, Alexey Shabunin, Mikhail Tavobilov
Brain Tumor Center at the University of Cincinnati Gardner Neuroscience Institute:
Christopher McPherson, Ronald Warnick
Brigham and Women’s Hospital:
Ross Berkowitz, Daniel Cramer, Colleen Feltmate, Neil Horowitz, Adam Kibel, Michael Muto, Chandrajit P. Raut
Capital Biosciences, Inc.:
Andrei Malykh
Case Comprehensive Cancer Center:
Jill S. Barnholtz-Sloan, Wendi Barrett, Karen Devine, Jordonna Fulop, Quinn T. Ostrom, Kristen Shimmel, Yingli Wolinsky
Case Western Reserve School of Medicine:
Andrew E. Sloan
Catholic University of the Sacred Heart:
Agostino De Rose, Felice Giuliante
Cedars-Sinai Medical Center:
Marc Goodman, Beth Y. Karlan
Central Arkansas Veterans Healthcare System:
Curt H. Hagedorn
Centura Health:
John Eckman, Jodi Harr, Jerome Myers, Kelinda Tucker, Leigh Anne Zach
Chan Soon-Shiong Institute of Molecular Medicine at Windber:
Brenda Deyarmin, Hai Hu, Leonid Kvecher, Caroline Larson, Richard J. Mural, Stella Somiari
Charles University:
Ales Vicha, Tomas Zelinka
Christiana Care Health System:
Joseph Bennett, Mary Iacocca, Brenda Rabeno, Patricia Swanson
CHU of Montreal:
Mathieu Latour
CHU of Quebec:
Louis Lacombe, Bernard Tetu
CHU of Quebec, Laval University Research Center of Chus:
Alain Bergeron
Cleveland Clinic Foundation:
Mary McGraw, Susan M. Staugaitis
Columbia University:
John Chabot, Hanina Hibshoosh, Antonia Sepulveda, Tao Su, Timothy Wang
Cureline, Inc.:
Olga Potapova, Olga Voronina
Curie Institute:
Laurence Desjardins, Odette Mariani, Sergio Roman-Roman, Xavier Sastre, Marc-Henri Stern
Dana-Farber Cancer Institute:
Feixiong Cheng, Sabina Signoretti
Dignity Health Mercy Gilbert Medical Center:
Jennifer Eschbacher
Duke University Medical Center:
Andrew Berchuck, Darell Bigner, Eric Lipp, Jeffrey Marks, Shannon McCall, Roger McLendon, Angeles Secord, Alexis Sharp
Emory University:
Madhusmita Behera, Daniel J. Brat, Amy Chen, Keith Delman, Seth Force, Fadlo Khuri, Fadlo Khuri, Kelly Magliocca, Shishir Maithel, Jeffrey J. Olson, Taofeek Owonikoko, Alan Pickens, Suresh Ramalingam, Dong M. Shin, Gabriel Sica, Gabriel Sica, Erwin G. Van Meir, Erwin G. Van Meir, Hongzheng Zhang
Erasmus Medical Center:
Wil Eijckenboom, Ad Gillis, Esther Korpershoek, Leendert Looijenga, Wolter Oosterhuis, Hans Stoop, Kim E. van Kessel, Ellen C. Zwarthoff
Foundation of the Carlo Besta Neurological Institute, IRCCS:
Chiara Calatozzolo, Lucia Cuppini, Stefania Cuzzubbo, Francesco DiMeco, Gaetano Finocchiaro, Luca Mattei, Alessandro Perin, Bianca Pollo
Fred Hutchinson Cancer Research Center:
Chu Chen, John Houck, Pawadee Lohavanichbutr
Friedrich-Alexander-University:
Arndt Hartmann, Christine Stoehr, Robert Stoehr, Helge Taubert, Sven Wach, Bernd Wullich
Greater Poland Cancer Center:
Witold Kycler, Dawid Murawa, Maciej Wiznerowicz
Greenville Health System Institute for Translational Oncology Research:
Ki Chung, W. Jeffrey Edenfield, Julie Martin
Gustave Roussy institute:
Eric Baudin
Harvard University:
Glenn Bubley, Raphael Bueno, Assunta De Rienzo, William G. Richards
Henry Ford Health System:
Ana deCarvalho, Steven Kalkanis, Tom Mikkelsen, Tom Mikkelsen, Houtan Noushmehr, Lisa Scarpace
Hospices Civils de Lyon:
Nicolas Girard
Hospital Clinic:
Marta Aymerich, Elias Campo, Eva Gine, Armando Lopez Guillermo
Hue Central Hospital:
Nguyen Van Bang, Phan Thi Hanh, Bui Duc Phu
Human Tissue Resource Network:
Yufang Tang
Huntsman Cancer Institute:
Howard Colman, Kimberley Evason
Icahn School of Medicine at Mount Sinai:
Peter R. Dottino, John A. Martignetti
Imperial College London:
Hani Gabra
Indivumed GmbH:
Hartmut Juhl
Institute of Human Virology Nigeria:
Teniola Akeredolu
Institute of Urgent Medicine:
Serghei Stepa
John Wayne Cancer Institute:
Dave Hoon
Keimyung University:
Keunsoo Ahn, Koo Jeong Kang
Ludwich Maximilians University Munich:
Felix Beuschlein
Maine Medical Center:
Anne Breggia
Massachusetts General Hospital:
Michael Birrer
Mayo Clinic:
Debra Bell, Mitesh Borad, Alan H. Bryce, Erik Castle, Vishal Chandan, John Cheville, John A. Copland, Michael Farnell, Thomas Flotte, Nasra Giama, Thai Ho, Michael Kendrick, Jean-Pierre Kocher, Karla Kopp, Catherine Moser, David Nagorney, Daniel O’Brien, Brian Patrick O’Neill, Tushar Patel, Gloria Petersen, Gloria Petersen, Florencia Que, Michael Rivera, Lewis Roberts, Robert Smallridge, Robert Smallridge, Thomas Smyrk, Thomas Smyrk, Melissa Stanton, R. Houston Thompson, Michael Torbenson, Ju Dong Yang, Lizhi Zhang, Lizhi Zhang
McGill University Health Center:
Fadi Brimo
MD Anderson Cancer Center:
Jaffer A. Ajani, Ana Maria Angulo Gonzalez, Carmen Behrens, Jolanta Bondaruk, Russell Broaddus, Bradley Broom, Bogdan Czerniak, Bita Esmaeli, Junya Fujimoto, Jeffrey Gershenwald, Charles Guo, Alexander J. Lazar, Christopher Logothetis, Funda Meric-Bernstam, Funda Meric-Bernstam, Cesar Moran, Lois Ramondetta, David Rice, Anil Sood, Pheroze Tamboli, Timothy Thompson, Patricia Troncoso, Patricia Troncoso, Anne Tsao, Ignacio Wistuba
Melanoma Institute Australia:
Candace Carter, Lauren Haydu, Peter Hersey, Valerie Jakrot, Hojabr Kakavand, Richard Kefford, Kenneth Lee, Georgina Long, Graham Mann, Michael Quinn, Robyn Saw, Richard Scolyer, Kerwin Shannon, Andrew Spillane, Jonathan Stretch, Maria Synott, John Thompson, James Wilmott
Memorial Sloan Kettering Cancer Center:
Hikmat Al-Ahmadie, Timothy A. Chan, Ronald Ghossein, Anuradha Gopalan, Douglas A. Levine, Victor Reuter, Samuel Singer, Bhuvanesh Singh
Ministry of Health of Vietnam:
Nguyen Viet Tien
Molecular Response:
Thomas Broudy, Cyrus Mirsaidi, Praveen Nair
Nancy N. and J.C. Lewis Cancer & Research Pavilion at St. Joseph’s/Candler:
Paul Drwiega, Judy Miller, Jennifer Smith, Howard Zaren
National Cancer Center Korea:
Joong-Won Park
National Cancer Hospital of Vietnam:
Nguyen Phi Hung
National Cancer Institute:
Electron Kebebew, W. Marston Linehan, Adam R. Metwalli, Karel Pacak, Peter A. Pinto, Mark Schiffman, Laura S. Schmidt, Cathy D. Vocke, Nicolas Wentzensen, Robert Worrell, Hannah Yang
Norfolk & Norwich University Hospital:
Marc Moncrieff
NYU Langone Medical Center:
Chandra Goparaju, Jonathan Melamed, Harvey Pass
Oncology Institute:
Natalia Botnariuc, Irina Caraman, Mircea Cernat, Inga Chemencedji, Adrian Clipca, Serghei Doruc, Ghenadie Gorincioi, Sergiu Mura, Maria Pirtac, Irina Stancul, Diana Tcaciuc
Ontario Tumour Bank:
Monique Albert, Iakovina Alexopoulou, Angel Arnaout, John Bartlett, Jay Engel, Sebastien Gilbert, Jeremy Parfitt, Harman Sekhon
Oregon Health & Science University:
George Thomas
Papworth Hospital NHS Foundation Trust:
Doris M. Rassl, Robert C. Rintoul
Providence Health and Services:
Carlo Bifulco, Raina Tamakawa, Walter Urba
QIMR Berghofer Medical Research Institute:
Nicholas Hayward
Radboud Medical University Center:
Henri Timmers
Regina Elena National Cancer Institute:
Anna Antenucci, Francesco Facciolo, Gianluca Grazi, Mirella Marino, Roberta Merola
Reinier de Graaf Hospital:
Ronald de Krijger
René Descartes University:
Anne-Paule Gimenez-Roqueplo
Research Center of Chus Sherbrooke, Québec:
Alain Piche
Research Institute of the McGill University Health Centre:
Simone Chevalier, Ginette McKercher
Rockefeller University:
Kivanc Birsoy
Rose Ella Burkhardt Brain Tumor and Neuro-Oncology Center:
Gene Barnett, Cathy Brewer, Carol Farver, Theresa Naska, Nathan A. Pennell, Daniel Raymond, Cathy Schilero, Kathy Smolenski, Felicia Williams
Roswell Park Cancer Institute:
Carl Morrison
Rush University:
Jeffrey A. Borgia, Michael J. Liptay, Mark Pool, Christopher W. Seder
Saarland University:
Kerstin Junker
Sage Bionetworks:
Larsson Omberg
Saint-Petersburg City Clinical Oncology Hospital:
Mikhail Dinkin, George Manikhas
Sapienza University of Rome:
Domenico Alvaro, Maria Consiglia Bragazzi, Vincenzo Cardinale, Guido Carpino, Eugenio Gaudio
Spectrum Health:
David Chesla, Sandra Cottingham
St. Petersburg Academic University RAS:
Michael Dubina, Fedor Moiseenko
Stanford University:
Renumathy Dhanasekaran
Technical University of Munich:
Karl-Friedrich Becker, Klaus-Peter Janssen, Julia Slotta-Huspenina
The International Genomics Consortium:
Daniel Crain, Erin Curley, Johanna Gardner, David Mallery, Scott Morris, Joseph Paulauskis, Robert Penny, Candace Shelton, Troy Shelton, Eric Thompson
The Ohio State University:
Mohamed H. Abdel-Rahman, Dina Aziz, Sue Bell, Colleen M. Cebulla, Amy Davis, Rebecca Duell, J. Bradley Elder, Joe Hilty, Bahavna Kumar, James Lang, Norman L. Lehman, Randy Mandt, Phuong Nguyen, Robert Pilarski, Karan Rai, Lynn Schoenfield, Kelly Senecal, Paul Wakely
The Oregon Clinic:
Paul Hansen
The Research Institute at Nationwide Children’s Hospital:
Nilsa Ramirez
Tufts Medical Center:
Ronald Lechan, James Powers, Arthur Tischler
University of Alabama at Birmingham Medical Center:
William E. Grizzle, Katherine C. Sexton
UC Cancer Institute:
Alison Kastl
UCSF-Helen Diller Family Comprehensive Cancer Center:
Joel Henderson, Sima Porten
University Hospital of Giessen and Marburg:
Jens Waldmann
University Hospital in Wurzburg, Germany:
Martin Fassnacht
University Health Network:
Sylvia L. Asa
University Hospital Essen:
Dirk Schadendorf
University Hospitals Case Medical Center Hamburg-Eppendorf∷
Marta Couce, Markus Graefen, Hartwig Huland, Guido Sauter, Thorsten Schlomm, Ronald Simon, Pierre Tennstedt
University of Abuja Teaching Hospital:
Oluwole Olabode
University of Arizona:
Mark Nelson
University of Calgary:
Oliver Bathe
University of California:
Peter R. Carroll, June M. Chan, Philip Disaia, Pat Glenn, Robin K. Kelley, Charles N. Landen, Joanna Phillips, Michael Prados, Jeff Simko, Jeffry Simko, Karen Smith-McCune, Scott VandenBerg
University of Chicago Medicine:
Kevin Roggin
University of Cincinnati:
Ashley Fehrenbach, Ady Kendler
University of Cincinnati Cancer Institute:
Suzanne Sifri, Ruth Steele
University of Colorado Cancer Center:
Antonio Jimeno
University of Dundee:
Francis Carey, Ian Forgie
University of Florence:
Massimo Mannelli
University of Hawaii Cancer Center:
Michael Carney, Brenda Hernandez
University of Heidelberg:
Benito Campos, Christel Herold-Mende, Christin Jungk, Andreas Unterberg, Andreas von Deimling
University of Iowa Hospital & Clinics:
Aaron Bossler, Joseph Galbraith, Laura Jacobus, Michael Knudson, Tina Knutson, Deqin Ma, Mohammed Milhem, Rita Sigmund
University of Kansas Medical Center:
Andrew K. Godwin, Rashna Madan, Howard G. Rosenthal
University of Maryland School of Medicine:
Clement Adebamowo, Sally N. Adebamowo
University of Melbourne:
Alex Boussioutas
University of Michigan:
David Beer, Thomas Giordano
University of Montreal:
Anne-Marie Mes-Masson, Fred Saad
University of New Mexico:
Therese Bocklage
University of Oklahoma:
Lisa Landrum, Robert Mannel, Kathleen Moore, Katherine Moxley, Russel Postier, Joan Walker, Rosemary Zuna
University of Pennsylvania:
Michael Feldman, Federico Valdivieso
University of Pittsburgh:
Rajiv Dhir, James Luketich
University of Puerto Rico:
Edna M. Mora Pinero, Mario Quintero-Aguilo
University of São Paulo:
Carlos Gilberto Carlotti Junior, Jose Sebastiao Dos Santos, Rafael Kemp, Ajith Sankarankuty, Daniela Tirapelli
University of Sheffield Western Bank:
James Catto
University of Washington:
Kathy Agnew, Elizabeth Swisher
University of Western Australia:
Jenette Creaney, Bruce Robinson
University of Wisconsin School of Medicine and Public Health:
Carl Simon Shelley
University of Kansas Cancer Center:
Eryn M. Godwin, Sara Kendall, Cassaundra Shipman
University of Michigan:
Carol Bradford, Thomas Carey, Andrea Haddad, Jeffey Moyer, Lisa Peterson, Mark Prince, Laura Rozek, Gregory Wolf
UQ Thoracic Research Centre:
Rayleen Bowman, Kwun M. Fong, Ian Yang
Valley Health System:
Robert Korst
Vanderbilt University Medical Center:
W. Kimryn Rathmell
Walter Reed National Medical Center:
J. Leigh Fantacone-Campbell, Jeffrey A. Hooke, Albert J. Kovatich, Craig D. Shriver
Washington University:
John DiPersio, Bettina Drake, Ramaswamy Govindan, Sharon Heath, Timothy Ley, Brian Van Tine, Peter Westervelt
Weill Cornell Medical College:
Mark A. Rubin
Yonsei University College of Medicine:
Jung Il Lee
Institution Not Provided:
Natalia D. Aredes, Armaz Mariamidze
Institution Addresses
Australian Prostate Cancer Research Center, Epworth Hospital, VIC, Australia
Australian Prostate Cancer Research Center, Epworth Hospital, VIC, Australia
Barretos Cancer Hospital, Av: Antenor Duarte Villela, 1331, Barretos, Sao Paulo, Brazil
Barrow Neurological Institute, St. Joseph’s Hospital and Medical Center, Phoenix, Arizona 85013
Barrow Neurological Institute, St. Joseph’s Hospital and Medical Center, Phoenix, Arizona 85013,
Baylor College of Medicine One Baylor Plaza, Houston, TX 77030
BC Cancer Agency, 675 W 10th Ave, Vancouver, BC V5Z 1L3, Canada
Beth Israel Deaconess Medical Center Harvard University Medical School Boston Mass
BioreclamationIVT, 99 Talbot Blvd Chestertown, MD 21620
Boston Medical Center, Boston MA 02118
Botkin Hospital, 2-y Botkinskiy pr-d, 5, Moskva, Russia, 125284
Brain Tumor and Neuro-oncology Center, Department of Neurosurgery, University Hospitals Case Medical Center, Case Western Reserve School of Medicine, 11100 Euclid Ave, Cleveland, Ohio, 44106
Brain Tumor Center at the University of Cincinnati Gardner Neuroscience Institute, and Department of Neurosurgery, University of Cincinnati College of Medicine, and Mayfield Clinic, 260 Stetson Street, Suite 2200, Cincinnati, Ohio, 45219
Brain Tumor Center at the University of Cincinnati Neuroscience Institute, and Department of Neurosurgery, University of Cincinnati College of Medicine, and Mayfield Clinic, 234 Goodman Street, Cincinnati, Ohio, 45219
Brigham and Women’s Hospital, 75 Francis St, Boston MA 02115
Capital Biosciences, Inc., 900 Clopper Rd, Suite 120, Gaithersburg, MD 20878
Case Comprehensive Cancer Center, 11100 Euclid Ave - Wearn 152, Cleveland, OH 44106-5065
Cedars-Sinai Medical Center, 8700 Beverly Boulevard, Suite 290 West MOT, Los Angeles, CA
Center for Liver Cancer, National Cancer Center Korea, 323 Ilsan-ro, Ilsan dong-gu, Goyang, Gyeonggi 10408, South Korea
Central Arkansas Veterans Healthcare System, Little Rock, AR 72205
CHU of Quebec, Laval University Research Center of Chus 2705, boul. Laurier Bureau TR72
QUÉBEC, Quebec G1V 4G2
Centura Health 9100 E Mineral Cir, Centennial, CO 80112
Chan Soon-Shiong Institute of Molecular Medicine at Windber, Windber, PA 15963
Charles University, Czech Republic
CHU of Quebec, Hôtel-Dieu de Quebec-University Laval, 11 cote du palais, Quebec City, G1R 2J6
CHUM, Montreal, Qc, Canada.
Clinic of Urology and Pediatric Urology, Saarland University, Homburg, Germany.
Clinical Breast Care Project, Murtha Cancer Center, Uniformed Services University / Walter Reed National Military Medical Center, Bethesda, MD 20889
Comprehensive Cancer Center Tissue Procurement Shared Resource, Cooperative Human Tissue Network
Midwestern Division, Dept. of Pathology, Human Tissue Resource Network, The Ohio State University, 410 West 10th Ave, Doan Hall, Room E413A, Columbus, OH 43210
Cureline, Inc., 290 Utah Ave, Ste 300, South san Francisco, CA 94080, USA
Dana-Farber Cancer Institute, 450 Brookline Ave, Boston MA, 02215
Dardinger Neuro-Oncology Center, Department of Neurosurgery, James Comprehensive Cancer Center and The Ohio State University Medical Center, 320 W 10th Ave, Columbus, Ohio, 43210
Department of Cardiovascular and Thoracic Surgery. Suite 774 Professional Office Building. 1735 W. Harrison St., Chicago, IL 60612
Department of Epidemiology and Public Health, University of Maryland School of Medicine, Baltimore MD 21201
Department of Genetics & Genomic Sciences, Icahn School of Medicine at Mount Sinai, 1 Gustave L. Levy Place, New York, NY 10029
Department of Hematology and Medical Oncology, Mayo Clinic Arizona, 5779 E. Mayo Blvd, Phoenix AZ 85054
Department of Medicine, University of Wisconsin School of Medicine and Public Health, 1685 Highland Avenue, Madison, WI 53705
Department of Medicine, Washington University in St. Louis, 660 S. Euclid Ave., CB 8066, St. Louis, MO 63110
Department of Medicine, Yonsei University College of Medicine, Seoul, Republic of Korea
Department of Neurological Surgery
Department of Neurosurgery, Emory University School of Medicine, 1365 Clifton Road, NE, Atlanta, GA 30322
Department of Obstetrics and Gynecology, Baylor College of Medicine, One Baylor Plaza, Houston, Texas 77030
Department of Obstetrics/Gynecology and Reproductive Sciences, Icahn School of Medicine at Mount Sinai, 1 Gustave L. Levy Place, New York, NY 10029
Department of Orthopedic Surgery, University of Kansas Medical Center 3901 Rainbow Boulevard, Kansas City, KS 66160
Department of Pathology and Cell Biology, Columbia University, New York, NY10032
Department of Pathology and Immunology, Baylor College of Medicine, One Baylor Plaza, Houston, TX 77030
Department of Pathology and Laboratory Medicine, University of Kansas Medical Center, Kansas City, KS 66206
Department of Pathology, Department of Cell and Molecular Medicine. 570 Jelke South center, 1750 W. Harrison St., Chicago, IL 60612
Department of Pathology, Duke University School of Medicine, Durham, NC 27710
Department of Pathology, Spectrum Health, 35 Michigan NE, Grand Rapids, MI 49503
Department of Pathology, The Ohio State University School of Medicine, N308 Doan Hall, 410 W 10th Ave, Columbus, OH-43210-1267
Department of Pathology, The Ohio State University Wexner Medical Center (Doan Hall N337B, 410 West 10th Ave., Columbus, OH 43210
Department of Pathology. 570 Jelke South center, 1750 W. Harrison St., Chicago, IL 60612
Department of Surgery and Anatomy, Ribeirao Preto Medical School - FMRP, University of Sao Paulo, Brazil, 14049-900
Department of Surgery and Cancer, Imperial College London, Du Cane Road London W12 0NN, UK
Department of Surgery, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA, USA
Department of Surgery, Columbia University, New York, NY 10032
Department of Surgery, University of Michigan, Ann Arbor MI 48109
Department of Urology and Pediatric Urology, University Hospital Erlangen, Friedrich-Alexander-University Erlangen-Nuremberg, 91054 Erlangen, Germany
Department of Urology, Mayo Clinic Arizona, 5779 E. Mayo Blvd, Phoenix AZ 85054
Departments of Neurosurgery and Hematology and Medical Oncology, School of Medicine and Winship Cancer Institute, 1365C Clifton Rd. N.E., Emory University, Atlanta, GA 30322
Departments of Pathology & Translational Molecular Pathology, The University of Texas MD Anderson Cancer Center, 1515 Holcombe Blvd--Unit 85, Houston, Texas, USA
Dept. of Pathology & Laboratory Medicine, University of Cincinnati, UC Health University Hospital, 234 Goodman Street, Cincinnati, OH 45219-0533
Dept. of Pathology, Robert J. Tomsich Pathology & Laboratory Medicine Institute, Lerner Research Inst, Cleveland Clinic Foundation, Cleveland, OH 44195
Dept. of Surgery, Klinikum rechts der Isar, Technical University of Munich, Ismaninger Str. 22, 81675 Munich, Germany
Dignity Health Mercy Gilbert Medical Center 3555 S Val Vista Dr, Gilbert, AZ 85297
Division Molecular Urology, Department of Urology and Pediatric Urology, University Hospital Erlangen, Friedrich-Alexander-University Erlangen-Nuremberg, 91054 Erlangen, Germany
Division of Cancer Epidemiology and Genetics, National Cancer Institute, 9609 Medical Center Dr. Bethesda 20892 USA
Division of Neurosurgical Research, Dpt. Neurosurgery, University of Heidelberg, INF 400, 69120 Heidelberg, Germany
Division of Surgical Oncology, Department of Surgery, Brigham and Women’s Hospital, 75 Francis Street, Boston, MA 02115
Dpt. Neuropathology, University of Heidelberg, INF 224, 69120 Heidelberg, Germany
Dpt. Neurosurgery, University of Heidelberg, INF 400, 69120 Heidelberg, Germany
Duke University
Duke University Medical Center 177 MSRB Box 3156 Durham, NC 27710
Duke University Medical Center, Gynecologic Oncology, Box 3079, Durham, NC USA
Emory University, 1365 Clifton Road, NE Atlanta GA, 30322
Erasmus MC, Wytemaweg 80, 3015 CN, Rotterdam, The Netherlands
Erasmus Medical Center
Erasmus University Medical Center Rotterdam, Cancer Institute, Wytemaweg 80, 3015CN, Rotterdam, the Netherlands
The Foundation of the Carlo Besta Neurological Institute, IRCCS via Celoria 11, 20133
Fred Hutchinson Cancer Research Center, 1100 Fairview Ave N, Seattle, WA 98019
Greater Poland Cancer Center, Garbary 15, 61-866 Poznań Poland
Greenville Health System Institute for Translational Oncology Research 900 West Faris Road Greenville SC 29605 Harvard University Cambridge, MA 02138
Havener Eye Institute, The Ohio State University Wexner Medical Center 915 Olentangy River Rd, Columbus, OH 43212
Henry Ford Hospital 2799 West Grand Blvd Detroit MI USA 48202
Hepatobiliary Surgery Unit, A. Gemelli Hospital, Catholic University of the Sacred Heart, Largo Agostino Gemelli 8, 00168 Rome, Italy
Hermelin Brain Tumor Center, Henry Ford Health System, 2799 W Grand Blvd, Detroit, MI, 48202
Hospices Civils de Lyon, CARDIOBIOTEC, Lyon F-69677, France
Hospital Clinic, Villarroel 180, Barcelona, Spain, 08036
Hue Central Hospital, Hue, Vietnam
Human Tissue Resource Network, Dept. of Pathology, College of Medicine, 1615 Polaris Innovation Ctr, 2001 Polaris, Columbus 43240
Huntsman Cancer Institute, Univ. of Utah, 2000 Circle of Hope, Salt Lake City, UT 84112
Indivumed GmbH, 20251 Hamburg, Germany
René Descartes University, Hospital Europeen Georges Pompidou, 20 rue Leblanc, 75015, Paris, France
Curie Institute, 26 rue Ulm, 75005 Paris, France
Gustave Roussy Institute of Oncology, 39 Rue Camille Desmoulins 94805, Villejuif, France
Institute of Human Virology Nigeria, Abuja, Nigeria
Institute of Pathology, Technical University of Munich, Trogerstr. 18, 83675 Munich, Germany
Institute of Pathology, University Hospital Erlangen, Firedrich-Alexander-University Erlangen-Nuremberg, 91054 Erlangen, Germany
Institute of Urgent Medicine, Republic of Moldova
Regina Elena National Cancer Institute Irccs - Ifo, Via Elio Chianesi 53, 00144, Rome, Italy
John Wayne Cancer Institute, 2200 Santa Monica Blvd, Santa Monica, CA 90404
Keimyung University, Daegu, South Korea
Knight Comprehensive Cancer Institute, Oregon Health & Science University
Ludwich Maximilians University Munich, Ziemssenstrasse 1, D-80336, Munich, Germany
Maine Medical Center, 22 Bramhall St., Portland, ME 04102
Martini-Clinic, Prostate Cancer Center, University Medical Center Hamburg-Eppendorf, Martinistr. 52, D-20246
Hamburg, Germany
Massachusetts General Hospital 55 Fruit Street Boston Ma 02114
Mayo Clinic 5777 E Mayo Blvd, Phoenix, Arizona 85054
Mayo Clinic 4500 San Pablo Road Jacksonville, FL 32224
Mayo Clinic, 200 First St. SW, Rochester, MN 55905
Mayo Clinic, Rochester, MN 55905
McGill University Health Center. 1001 Decarie Blvd, Montreal, QC, Canada H4A 3J1
MD Anderson Cancer Center 1515 Holcombe Blvd. Unit 0085 Houston, TX 77030
MD Anderson Cancer Center, Department of Pathology, Unit 085; 1515
MD Anderson Cancer Center Life Science Plaza Building 2130 W. Holcombe Blvd, Unit 2951 Houston, TX 77030
Office: LSP9.4029
Melanoma Institute Australia, North Sydney, NSW, Australia 2060
Memorial Sloan Kettering Cancer Center Department of Pathology, 1275 York Avenue, New York, NY 10065
Memorial Sloan Kettering Cancer Center, 1275 York Avenue, New York, NY 10065
Memorial Sloan Kettering Cancer Center, Center for Molecular Oncology, 1275 York Avenue, New York, NY 10065
Ministry of Health of Vietnam, Hanoi, Vietnam
Molecular Pathology Shared Resource of Herbert Irving Comprehensive Cancer Center of Columbia University, New York, NY10032
Molecular Response 11011 Torreyana Road San Diego, CA 92121
Murtha Cancer Center, Uniformed Services University / Walter Reed National Military Medical Center, Bethesda, MD 20889
Nancy N. and J.C. Lewis Cancer & Research Pavilion at St. Joseph’s/Candler, 225 Candler Drive, Savannah, GA 31405
National Cancer Hospital of Vietnam
National Cancer Institute, 31 Center Dr, Bethesda, MD 20892
National Cancer Institute, Bethesda, MD 20892
Norfolk & Norwich University Hospital, Norwich, UK. NR4 7UY
NYU Langone Medical Center, Cardiothoracic Surgery, 530 first Avenue, 9V, New York, NY
Oncology Institute, Republic of Moldova
Ontario Tumor Bank - Hamilton site, St. Joseph’s Healthcare Hamilton, Hamilton, Ontario L8N 3Z5, Canada
Ontario Tumor Bank - Kingston site, Kingston General Hospital, Kingston, Ontario K7L 5H6, Canada
Ontario Tumor Bank – Ottawa site, The Ottawa Hospital, Ottawa, Ontario K1H 8L6, Canada.
Ontario Tumor Bank, London Health Sciences Centre, London, Ontario N6A 5A5, Canada
Ontario Tumor Bank, Ontario Institute for Cancer Research, Toronto, Ontario M5G 0A3, Canada
Orbital Oncology & Ophthalmic Plastic Surgery Department of Plastic Surgery M.D. Anderson Cancer Center 1515
Holcombe Blvd, Unit 1488 Houston, Texas 77030
Papworth Hospital NHS Foundation Trust, UK
Pathology, St. Joseph’s/Candler, 5353 Reynolds St., Savannah, GA 31405
Professor, Division of Neuropathology, Department of Pathology, University Hospitals Case Medical Center
Program in Epidemiology, Fred Hutchinson Cancer Research Center, Seattle, WA 98109
Providence Health and Services
QIMR Berghofer Medical Research Institute, Herston, QLD, Australia
Radboud Medical University Center, Geert Grooteplein-Zuid 10, Nijmegen, the Netherlands
Regina Elena National Cancer Institute, 00144 Rome, Italy
Reinier de Graaf Hospital, Reinier de Graafweg 5, 2625AD, Delft, the Netherlands
Research Institute of the McGill University Health Centre, McGill University, Montreal, Quebec, Canada
Research Center Of Chus Sherbrooke, Quebec aile 9, porte 6, 3001 12e Avenue Nord, Sherbrooke, QC J1H 5N4, Canada
Rockefeller University 1230 York Ave New York, NY
Rose Ella Burkhardt Brain Tumor and Neuro-Oncology Center ND4-52A, Cleveland Clinic Foundation, 9500 Euclid Ave, Cleveland, OH 44195
Rose Ella Burkhardt Brain Tumor and Neuro-Oncology Center, 9500 Euclid Avenue - CA51, Cleveland, OH 44195
Rose Ella Burkhardt Brain Tumor and Neuro-Oncology Center, Department of Neurosurgery, Neurological and Taussig Cancer Institute, Cleveland Clinic, 9500 Euclid Avenue, Cleveland, Ohio, 44195
Roswell Park Cancer Institute. Elm & Carlton Streets, Buffalo NY 14263
Sage Bionetworks, Seattle, WA 98109
Saint-Petersburg City Clinical Oncology Hospital, 56 Veteranov prospect, Saint-Petersburg, 198255, Russia
Sapienza University of Rome, Piazzale Aldo Moro 5, 00185 Rome, Italy
School of Medicine, National Center for Asbestos Related Research, University of Western Australia, Nedlands, WA, Australia 6009
Sir Peter MacCallum Department of Oncology, University of Melbourne, Parkville, 3050, Victoria, Australia
St. Petersburg Academic University RAS, 8/3 Khlopin Str., St. Petersburg, 194021, Russia
Stanford University, Palo Alto, CA, USA
Stephenson Cancer Center, University of Oklahoma, Oklahoma City, OK USA
Tayside Tissue Bank, University of Dundee, Scotland UK DD1 9SY
The International Genomics Consortium, 445 N. 5th Street, Phoenix, Arizona 85004
The Ohio State University, Columbus, OH 43210
The Ohio State University Comprehensive Cancer Center, 320 W 10th Avenue, Columbus, OH 43210
The Ohio State University Wexner Medical Center (2012 Kenny Rd, Columbus, OH 43221)
The Oregon Clinic 1111 NE 99th Ave, Portland, OR 97220
The Prince Charles Hospital, UQ Thoracic Research Centre, Australia 4032
The Research Institute at Nationwide Children’s Hospital 700 Children’s Drive Columbus Ohio 43205
Tufts Medical center, 800 Washington St. Boston MA 02111
UABMC 401 Beacon Pkwy W Birmingham AL 35209
UC Cancer Institute, 200 Albert Sabin Way, Suite 1012, Cincinnati, OH 45267-0502
UCSF-Helen Diller Family Comprehensive Cancer Center, 550 16th St., Mission Hall WS 6532 Box 3211, San Francisco, CA 94143
University Hospital of Giessen and Marburg, Badingerstrasse 3, 35044, Marburg, Germany
University Hospital in Wurzburg, Germany, Oberdurrbacher Strasse 6, 97080, Wurzburg, Germany
University Health Network, 200 Elizabeth Street, Toronto ON M5G 2C4 Canada
University Hospital Essen, University Duisburg-Essen, German Cancer Consortium, Hufelandstr. 55; 45239 Essen, Germany
University Medical Center Hamburg-Eppendorf, Martinistr. 52, D-20246 Hamburg, Germany
University of Abuja Teaching Hospital, Gwagalada, FCT, Nigeria
University of Arizona Tucson Arizona
University of Calgary, Departments of Surgery and Oncology, 1331 - 29th St NW, Calgary, AB, T2N 4N2
University of California San Francisco, 2340 Sutter St Rm S 229, San Francisco CA 94143
University of California, Irvine 333 City Boulevard West Suite 1400 Orange CA 92868
University of Chicago Medicine 5841 S. Maryland Ave. Room G-216, MC 5094|Chicago, IL 60637
University of Cincinnati Cancer Institute, Brain Tumor Clinical Trials, 200 Albert Sabin Way Suite 1012, Cincinnati, OH 45267
University of Cincinnati Cancer Institute, Holmes Bldg., 200 Albert Sabin Way, Ste 1002, Cincinnati, OH 45267-0502
University of Colorado Cancer Center, Aurora, CO, 80111, USA
University of Dundee, Scotland UK DD1 9SY
University of Florence, Viale Pieraccini 6, 50139 Firenze, Italy
University of Hawaii Cancer Center
University of Iowa Hospital & Clinics, 200 Hawkins Drive, Clinical Trials-Data Management, 11510 PFP, Iowa City, IA 52242
University of Iowa Hospital & Clinics, 200 Hawkins Drive, Hematology/Oncology, C32 GH, Iowa City, IA 52242
University of Iowa Hospital & Clinics, 200 Hawkins Drive, ICTS-Informatics, 272 MRF, Iowa City, IA 52242
University of Iowa Hospital & Clinics, 200 Hawkins Drive, Medicine Administration, 380 MRC, Iowa City, IA 52242
University of Iowa Hospital & Clinics, 200 Hawkins Drive, Molecular Pathology, B606 GH, Iowa City, IA 52242
University of Iowa Hospital & Clinics, 200 Hawkins Drive, Pathology, SW247 GH, Iowa City, IA 52242
University of Kansas Cancer Center, 3901 Rainbow Blvd, Kansas City, KS. 66160
University of Kansas Medical Center Kansas City KS 66160
University of Michigan 500 S State St, Ann Arbor, MI 48109
University of Montreal 2900 Edouard Mont petit Blvd, Montreal, QC H3T 1J4, Canada
University of New Mexico Albuquerque, New Mexico 87131
University of Pennsylvania Philadelphia, PA 19104
University of Pittsburgh, Department of Cardiothoracic Surgery,200 Lothrop St, Suite C-800, Pittsburgh, Pennsylvania 15213
University of Pittsburgh, Department of Pathology, Pittsburgh, Pennsylvania 15213
University of Sheffield Western Bank, Sheffield S10 2TN, UK
University of Washington Seattle, WA 98105
UPR Comprehensive Cancer Center Biobank; University of Puerto Rico Comprehensive Cancer Center, Celso Barbosa St. Medical Center Area, San Juan, PR 00936
Urologic Oncology Branch, Center for Cancer Research, National Cancer Institute, Building 10, Room 1-5940, Bethesda, MD 20892-1107
Valley Health System, 1 Valley Health Plaza, Paramus, NJ 07652
Vanderbilt University Medical Center 1211 Medical Center Dr, Nashville, TN 37232
Washington University School of Medicine, 600 S. Taylor Ave, St. Louis, MO 63110
Weill Cornell Medical College, New York, NY 10065
Footnotes
Declaration of Interests
M.M., A.D.C., A.C.B., and G.F.G. receive research support from Bayer HealthCare Pharmaceuticals. M.M. is a consultant for and equity holder in OrigiMed. R.B. consults for and receives research support from Novartis. Other authors declare no competing interests.
Author Contributions
A.M.T., J.S., G.H., G.F.G., A.C.B., S.S., H.H., and J.L. performed computational analyses; A.M.T. and X.Z. performed in vitro experiments and sequencing; TCGA Research Network provided genomic data of patient samples; A.D.C., R.B., and M.M. supervised the work; C.W. and A.J.L. provided feedback and advice on analyses; A.M.T. and M.M. wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could a3ect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adalsteinsson VA, Ha G, Freeman SS, Choudhury AD, Stover DG, Parsons HA, Gydush G, Reed SC, Rotem D, Rhoades J, et al. Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat Commun. 2017;8:1324. doi: 10.1038/s41467-017-00965-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18:220. doi: 10.1186/s13059-017-1349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkauskas CE, Chung MI, Fioret B, Gao X, Katsura H, Hogan BL. Lung organoids: current uses and future promise. Development. 2017;144:986–997. doi: 10.1242/dev.140103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass AJ, Watanabe H, Mermel CH, Yu S, Perner S, Verhaak RG, Kim SY, Wardwell L, Tamayo P, Gat-Viks I, et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet. 2009;41:1238–1242. doi: 10.1038/ng.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Getz G, Nghiemphu L, Barretina J, Hsueh T, Linhart D, Vivanco I, Lee JC, Huang JH, Alexander S, et al. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc Natl Acad Sci USA. 2007;104:20007–20012. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonney ME, Moriya H, Amon A. Aneuploid proliferation defects in yeast are not driven by copy number changes of a few dosage-sensitive genes. Gene Dev. 2015;29:898–903. doi: 10.1101/gad.261743.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, Zheng S, Chakravarty D, Sanborn JZ, Berman SH, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buccitelli C, Salgueiro L, Rowald K, Sotillo R, Mardin BR, Korbel JO. Pan-cancer analysis distinguishes transcriptional changes of aneuploidy from proliferation. Genome Res. 2017;27:501–511. doi: 10.1101/gr.212225.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Crowther J, Pastor T, Abbasi Asbagh L, Baietti MF, De Troyer M, Vazquez I, Talebi A, Renzi F, Dehairs J, et al. Loss of chromosome 8p governs tumor progression and drug response by altering lipid metabolism. Cancer Cell. 2016;29:751–766. doi: 10.1016/j.ccell.2016.04.003. [DOI] [PubMed] [Google Scholar]
- Cairncross G, Wang M, Shaw E, Jenkins R, Brachman D, Buckner J, Fink K, Souhami L, Laperriere N, Curran W, et al. Phase III trial of chemoradiotherapy for anaplastic oligodendroglioma: long-term results of RTOG 9402. J Clin Oncol. 2013;31:337–343. doi: 10.1200/JCO.2012.43.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell JD, Alexandrov A, Kim J, Wala J, Berger AH, Pedamallu CS, Shukla SA, Guo G, Brooks AN, Murray BA, et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet. 2016;48:607–616. doi: 10.1038/ng.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T, Laird PW, Onofrio RC, Winckler W, Weir BA, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30:413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter SL, Eklund AC, Kohane IS, Harris LN, Szallasi Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat Genet. 2006;38:1043–1048. doi: 10.1038/ng1861. [DOI] [PubMed] [Google Scholar]
- Ciriello G, Miller ML, Aksoy BA, Senbabaoglu Y, Schultz N, Sander C. Emerging landscape of oncogenic signatures across human cancers. Nat Genet. 2013;45:1127–1133. doi: 10.1038/ng.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoli T, Uno H, Wooten EC, Elledge SJ. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science. 2017;355 doi: 10.1126/science.aaf8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerstrand D, Tong A, Schumacher SE, Ilic N, Shen RR, Cheung HW, Vazquez F, Shrestha Y, Kim SY, Giacomelli AO, et al. Systematic interrogation of 3q26 identifies TLOC1 and SKIL as cancer drivers. Cancer Discov. 2013;3:1044–1057. doi: 10.1158/2159-8290.CD-12-0592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoadley KA, Yau C, Wolf DM, Cherniack AD, Tamborero D, Ng S, Leiserson MDM, Niu B, McLellan MD, Uzunangelov V, et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell. 2014;158:929–944. doi: 10.1016/j.cell.2014.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland AJ, Cleveland DW. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol. 2009;10:478–487. doi: 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nature Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung J, Kishimoto Y, Sugio K, Virmani A, McIntire DD, Minna JD, Gazdar AF. Allele-specific chromosome 3p deletions occur at an early stage in the pathogenesis of lung carcinoma. JAMA. 1995;273:558–563. [PubMed] [Google Scholar]
- Lai D, Ha G, Shah S. HMMcopy: Copy number prediction with correction for GC and mappability bias for HTS data. R package version 1.18.0 2016 [Google Scholar]
- Larsson LG. Oncogene- and tumor suppressor gene-mediated suppression of cellular senescence. Semin Cancer Biol. 2011;21:367–376. doi: 10.1016/j.semcancer.2011.10.005. [DOI] [PubMed] [Google Scholar]
- Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Chen C, Xu Z, Scuoppo C, Rillahan CD, Gao J, Spitzer B, Bosbach B, Kastenhuber ER, Baslan T, et al. Deletions linked to TP53 loss drive cancer through p53-independent mechanisms. Nature. 2016;531:471–475. doi: 10.1038/nature17157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg AS, Randell SH, Stewart SA, Elenbaas B, Hartwell KA, Brooks MW, Fleming MD, Olsen JC, Miller SW, Weinberg RA, et al. Immortalization and transformation of primary human airway epithelial cells by gene transfer. Oncogene. 2002;21:4577–4586. doi: 10.1038/sj.onc.1205550. [DOI] [PubMed] [Google Scholar]
- Mali P, Esvelt KM, Church GM. Cas9 as a versatile tool for engineering biology. Nat Methods. 2013;10:957–963. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitelman F. Recurrent chromosome aberrations in cancer. Mutat Res. 2000;462:247–253. doi: 10.1016/s1383-5742(00)00006-5. [DOI] [PubMed] [Google Scholar]
- Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, et al. Scikit-learn: Machine learning in Python. J Mach Learn Res. 2011;12:2825–2830. [Google Scholar]
- Ried T, Hu Y, Difilippantonio MJ, Ghadimi BM, Grade M, Camps J. The consequences of chromosomal aneuploidy on the transcriptome of cancer cells. Biochim Biophys Acta. 2012;1819:784–793. doi: 10.1016/j.bbagrm.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, Lee W, Yuan J, Wong P, Ho TS, et al. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S, Richardson A, Iyer DR, M’Saad O, Zasadil L, Knouse KA, Wong YL, Rhind N, Desai A, Amon A. Chromosome mis-segregation generates cell-cycle-arrested cells with complex karyotypes that are eliminated by the immune system. Dev Cell. 2017;41:638–651. e5. doi: 10.1016/j.devcel.2017.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senovilla L, Vitale I, Martins I, Tailler M, Pailleret C, Michaud M, Galluzzi L, Adjemian S, Kepp O, Niso-Santano M, et al. An immunosurveillance mechanism controls cancer cell ploidy. Science. 2012;337:1678–1684. doi: 10.1126/science.1224922. [DOI] [PubMed] [Google Scholar]
- Sheltzer JM, Blank HM, Pfau SJ, Tange Y, George BM, Humpton TJ, Brito IL, Hiraoka Y, Niwa O, Amon A. Aneuploidy drives genomic instability in yeast. Science. 2011;333:1026–1030. doi: 10.1126/science.1206412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheltzer JM, Ko JH, Replogle JM, Habibe Burgos NC, Chung ES, Meehl CM, Sayles NM, Passerini V, Storchova Z, Amon A. Single-chromosome gains commonly function as tumor suppressors. Cancer Cell. 2017;31:240–255. doi: 10.1016/j.ccell.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheltzer JM, Torres EM, Dunham MJ, Amon A. Transcriptional consequences of aneuploidy. Proc Natl Acad Sci USA. 2012;109:12644–12649. doi: 10.1073/pnas.1209227109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingele S, Stoehr G, Peplowska K, Cox J, Mann M, Storchova Z. Global analysis of genome, transcriptome and proteome reveals the response to aneuploidy in human cells. Mol Syst Biol. 2012;8:608. doi: 10.1038/msb.2012.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaresan V, Ganly P, Hasleton P, Rudd R, Sinha G, Bleehen NM, Rabbitts P. p53 and chromosome 3 abnormalities, characteristic of malignant lung tumours, are detectable in preinvasive lesions of the bronchus. Oncogene. 1992;7:1989–1997. [PubMed] [Google Scholar]
- Tang YC, Amon A. Gene copy-number alterations: A cost-benefit analysis. Cell. 2013;152:394–405. doi: 10.1016/j.cell.2012.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor-Weiner A, Zack T, O’Donnell E, Guerriero JL, Bernard B, Reddy A, Han GC, AlDubayan S, Amin-Mansour A, Schumacher SE, et al. Genomic evolution and chemoresistance in germ-cell tumours. Nature. 2016;540:114–118. doi: 10.1038/nature20596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas (TCGA) Research Network. Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, Shen H, Robertson AG, Pashtan I, Shen R, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas (TCGA) Research Network. Brat DJ, Verhaak RG, Aldape KD, Yung WK, Salama SR, Cooper LA, Rheinbay E, Miller CR, Vitucci M, et al. Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. New Engl J Med. 2015;372:2481–2498. doi: 10.1056/NEJMoa1402121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topalian SL, Taube JM, Anders RA, Pardoll DM. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat Rev Cancer. 2016;16:275–287. doi: 10.1038/nrc.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, Amon A. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science. 2007;317:916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- Uno N, Hiramatsu K, Uno K, Komoto S, Kazuki Y, Oshimura M. CRISPR/Cas9-induced transgene insertion and telomere-associated truncation of a single human chromosome for chromosome engineering in CHO and A9 cells. Sci Rep. 2017;7:12739. doi: 10.1038/s41598-017-10418-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver BA, Cleveland DW. Does aneuploidy cause cancer? Curr Opin Cell Biol. 2006;18:658–667. doi: 10.1016/j.ceb.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Williams BR, Prabhu VR, Hunter KE, Glazier CM, Whittaker CA, Housman DE, Amon A. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science. 2008;322:703–709. doi: 10.1126/science.1160058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wistuba II, Behrens C, Virmani AK, Mele G, Milchgrub S, Girard L, Fondon JW, 3rd, Garner HR, McKay B, Latif F, et al. High resolution chromosome 3p allelotyping of human lung cancer and preneoplastic/preinvasive bronchial epithelium reveals multiple, discontinuous sites of 3p allele loss and three regions of frequent breakpoints. Cancer Res. 2000;60:1949–1960. [PubMed] [Google Scholar]
- Xue W, Kitzing T, Roessler S, Zuber J, Krasnitz A, Schultz N, Revill K, Weissmueller S, Rappaport AR, Simon J, et al. A cluster of cooperating tumor-suppressor gene candidates in chromosomal deletions. Proc Natl Acad Sci USA. 2012;109:8212–8217. doi: 10.1073/pnas.1206062109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabarovsky ER, Lerman MI, Minna JD. Tumor suppressor genes on chromosome 3p involved in the pathogenesis of lung and other cancers. Oncogene. 2002;21:6915–6935. doi: 10.1038/sj.onc.1205835. [DOI] [PubMed] [Google Scholar]
- Zack TI, Schumacher SE, Carter SL, Cherniack AD, Saksena G, Tabak B, Lawrence MS, Zhang CZ, Wala J, Mermel CH, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013;45:1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S2, related to Figure 1. Sample Chromosome Arm Calls and Aneuploidy Scores.
Table S3, related to Figure 2. Aneuploidy Score Coefficients for Gene Mutation Linear Models.
Table S4, related to Figures 2 and 3. Correlation Coefficients with Aneuploidy Score.
Table S5, related to Figure 3. Aneuploidy Score Coefficients for Gene Expression Linear Models.
Table S6, related to Figure 3. Gene Set Enrichment Analysis (GSEA) Enrichment Scores.
Table S7, related to Figure 4. Chromosome Arm Summary Statistics and Chromosome Arm Coefficients for Gene Expression Linear Models.
Table S8, related to Figure 4. Matrix of 1-Pearson's Correlation Coefficients for Tumor Type Clustering.


