Drosophila Suppressor of Hairy-wing [Su(Hw)] is a multivalent transcription factor. Although best known for its gypsy retrotransposon insulator function, its functions at non-gypsy genomic binding sites are poorly understood...
Keywords: Drosophila, spermatogenesis, Su(Hw), transcriptional regulation, chromatin insulator, cyst cells
Abstract
Drosophila Suppressor of Hairy-wing [Su(Hw)] protein is an example of a multivalent transcription factor. Although best known for its role in establishing the chromatin insulator of the gypsy retrotransposon, Su(Hw) functions as an activator and repressor at non-gypsy genomic sites. It remains unclear how the different regulatory activities of Su(Hw) are utilized during development. Motivated from observations of spatially restricted expression of Su(Hw) in the testis, we investigated the role of Su(Hw) in spermatogenesis to advance an understanding of its developmental contributions as an insulator, repressor, and activator protein. We discovered that Su(Hw) is required for sustained male fertility. Although dynamics of Su(Hw) expression coincide with changes in nuclear architecture and activation of coregulated testis-specific gene clusters, we show that loss of Su(Hw) does not disrupt meiotic chromosome pairing or transcription of testis-specific genes, suggesting that Su(Hw) has minor architectural or insulator functions in the testis. Instead, Su(Hw) has a prominent role as a repressor of neuronal genes, consistent with suggestions that Su(Hw) is a functional homolog of mammalian REST, a repressor of neuronal genes in non-neuronal tissues. We show that Su(Hw) regulates transcription in both germline and somatic cells. Surprisingly, the essential spermatogenesis function of Su(Hw) resides in somatic cyst cells, implying context-specific consequences due to loss of this transcription factor. Together, our studies highlight that Su(Hw) has a major developmental function as a transcriptional repressor, with the effect of its loss dependent upon the cell-specific factors.
DEVELOPMENT depends upon differential gene expression controlled largely at the transcriptional level. Transcriptional regulation requires the long-range action of regulatory elements, such as enhancers and silencers that modulate the output from gene promoters (Lelli et al. 2012). The limited promoter specificity of enhancers and silencers challenges transcriptional fidelity (Kermekchiev et al. 1991; Schoenherr et al. 1996). However, metazoan genomes carry chromatin insulators that constrain enhancer and silencer action by partitioning chromosomes into physically distinct domains that shield promoters from inappropriate regulatory inputs (Raab and Kamakaka 2010; Ali et al. 2016). Although the transcriptional contributions of enhancers, silencers, and insulators differ, recent studies suggest that individual transcription factors have the capacity to confer more than one of these distinct functions (Nikolaev et al. 2009; Nakahashi et al. 2013; Han et al. 2016; Baxley et al. 2017; Nevil et al. 2017). Mechanisms underlying such regulatory complexity are poorly understood. Defining the individual functions of multivalent transcription factors will advance our understanding of how functional complexity of these factors is achieved.
Drosophila Suppressor of Hairy-wing [Su(Hw)] is a globally expressed transcriptional regulator. Investigations of the regulatory functions of Su(Hw) have largely focused on its role in establishing the insulator within the gypsy retrotransposon. This insulator contains a cluster of 12 tightly spaced Su(Hw) binding sites (SBSs) that confer both position-dependent enhancer blocking and form a barrier to heterochromatin spreading (Holdridge and Dorsett 1991; Geyer and Corces 1992; Roseman et al. 1993; Scott and Geyer 1995; Scott et al. 1999; Parnell and Geyer 2000). Su(Hw) establishes the gypsy insulator through recruitment of two Broad-complex, Tramtrack and Bric-a-brac/Poxvirus and Zinc Finger domain proteins, Modifier of (mdg4)67.2 (Georgiev and Gerasimova 1989) and Centrosomal Protein of 190 kDa (Pai et al. 2004). The effectiveness of gypsy insulator function depends upon the number, orientation, and location of insulators inserted between enhancers and silencers and their target promoters (Scott et al. 1999; Cai and Shen 2001; Muravyova et al. 2001; Kuhn et al. 2003; Kyrchanova et al. 2008). Strikingly, pairs of gypsy insulators interact, such that placing two gypsy insulators in between an enhancer and promoter permits the enhancer to bypass the insulator and activate the promoter (Scott et al. 1999; Cai and Shen 2001; Muravyova et al. 2001; Kuhn et al. 2003; Kyrchanova et al. 2008). Such interactions can be long-range. For example, gypsy insulators have been found to interact when separated by megabase distances and even on separate chromosomes (Kravchenko et al. 2005). Besides gypsy, Su(Hw) binds over 3000 non-gypsy SBSs in the genome. It remains unclear whether non-gypsy SBSs share properties with the gypsy insulator. Support for a genome-wide insulator role for Su(Hw) comes from findings that Su(Hw) contributes to the pairing of interphase homologous chromosomes in Drosophila (Fritsch et al. 2006; Hartl et al. 2008). However, other observations challenge this view. First, the gypsy insulator function of Su(Hw) requires binding to clusters of SBSs, an organization of sites that is rare in the Drosophila genome (Scott et al. 1999; Parnell et al. 2006). Indeed, standalone SBSs fail to confer enhancer blocking (Schwartz et al. 2012). Second, only a third of SBSs are associated with the insulator cofactors Modifier of (mdg4)67.2 and Centrosomal Protein of 190 kDa (Negre et al. 2010; Schwartz et al. 2012). Third, the majority of non-gypsy SBSs differ in sequence composition from gypsy SBSs (Baxley et al. 2017). As the SBS sequence correlates with distinct chromatin features, how Su(Hw) binds DNA might influence its regulatory output (Baxley et al. 2017). Taken together, these observations imply that Su(Hw) at non-gypsy SBSs might have distinct properties from the gypsy insulator.
An understanding of Su(Hw) function at non-gypsy SBSs is emerging through studies of its contributions to development. Investigations of the role of Su(Hw) in oogenesis uncovered activator and repressor functions (Soshnev et al. 2008, 2013), revealing that its transcriptional repressor function is required for egg production (Soshnev et al. 2013). Such findings emphasize that in addition to insulator function, Su(Hw) has a direct role in transcriptional regulation. Here, we study the role of Su(Hw) in spermatogenesis, motivated from findings that Su(Hw) expression is spatially restricted in the testis (Figure 1A). We establish that Su(Hw) is essential for sustained male fertility. Spermatogenesis is a complex developmental process, involving pairing of homologous chromosomes needed for meiosis and execution of a testis-specific transcriptional program that includes activation of coregulated gene clusters for spermiogenesis (Fuller 1998; Shevelyov et al. 2009; White-Cooper 2010, 2012; McKee et al. 2012). These features make spermatogenesis an ideal platform for investigating potential roles for Su(Hw) as an architectural and transcriptional regulator at endogenous, non-gypsy sites. Our studies of su(Hw)−/− testes revealed that Su(Hw) is dispensable for homolog pairing and meiotic chromosome segregation, but is required for transcriptional regulation. Even so, Su(Hw) loss does not affect transcription of testis-specific gene clusters. Instead, we find that the major feature of gene misregulation is derepression of genes that display enriched expression levels in neuronal tissues. These observations reinforce proposals that Su(Hw) functions similarly to mammalian REST (Soshnev et al. 2013), a repressor of neuronal genes in non-neuronal tissues. Notably, genes regulated by Su(Hw) in the testis and ovary differ, highlighting that loss of this transcription factor has context-specific consequences. Using cell-type-specific tools to restrict Su(Hw) expression, we demonstrate that the essential spermatogenesis function of Su(Hw) resides in somatic cyst cells, not in germ cells. Further studies will be needed to identify Su(Hw) regulated genes in these cells. Taken together, our studies show that Su(Hw) is a multifunctional transcriptional regulator during Drosophila development.
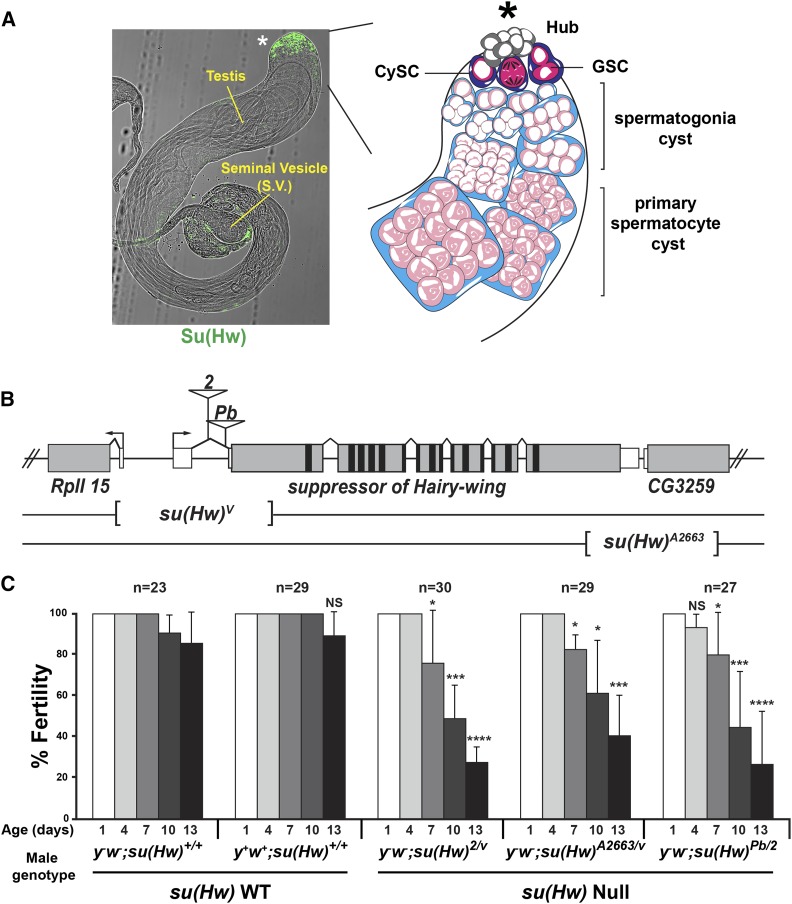
Figure 1.
Spatially restricted Su(Hw) expression reveals a requirement in male fertility. (A) Location of Su(Hw) in the testis. Left: phase contrast image of a su(Hw)+/+ testis superimposed with a confocal image of the same testis stained with antibodies against Su(Hw) (green). The asterisk marks the position of the somatic hub cells at the anterior tip of the testis. The location of the seminal vesicle (S.V.) is shown. Right: schematic representation of stages of spermatogenesis found at the anterior tip of the testis. Somatic hub cells (asterisk) support germline stem cells (GSCs, dark pink) and somatic cyst stem cells (CySCs, dark blue). Asymmetric division of both stem cells produces a unit of differentiation containing two postmitotic somatic cyst cells (light blue) and one germ cell (light pink). Four mitotic germ cell divisions lead to the formation of a differentiation unit comprised of 16 interconnected spermatogonial cells. Following completion of the mitotic divisions and another S-phase, germ cells within the differentiation unit arrest to form primary spermatocytes. Germ cell nuclei undergo dramatic changes in chromosome (white) organization during spermatogenesis. In GSCs and spermatogonia, chromosomes occupy the entire nuclear space, but in primary spermatocytes, chromosome disperse into chromosomal territories. (B) Diagram of the su(Hw) locus. The 5′ and 3′ UTRs of the su(Hw) gene are shown in white and the coding region in gray, with the location of the zinc fingers shown in black. Structures of the su(Hw) alleles used in this study are shown, including positions of insertions in the su(Hw)2 and su(Hw)Pb alleles, and positions of deletions in the su(Hw)v and su(Hw)A2663 alleles. (C) Sperm depletion assay. Fertility outcomes of males from two su(Hw)+/+ (y1 w1118 and CS, respectively) and three su(Hw)−/− (genotypes shown) mutant backgrounds tested in the sperm exhaustion assay. The total number of males of each genotype tested is shown on top of the graph. The X-axis shows the age of the male when mated. The Y-axis shows the percentage of males that were fertile, with bars indicating the variation of fertility in three biological replicates. Statistical analysis of the fertility of su(Hw)−/− males relative to one su(Hw)+/+ genotype (y1 w1118; NS, not significant, * P < 0.05, *** P < 0.001, **** P < 0.0001, Student’s t-test).
Materials and Methods
Drosophila stocks and culture conditions
Drosophila stocks were raised on standard cornmeal/agar medium. All crosses were carried out at 25°, with 70% humidity. Three su(Hw)+/+ strains were used in this study, including y1w1118, Canton-S (CS; Bloomington Stock Center, BL1), and y1w67c23; P[EPgy2]AmtEY02782 (BL15598; Bloomington Stock Center). Four su(Hw) protein null alleles were used in these studies (Figure 1B and Supplemental Material, Figure S1), including su(Hw)v, which carries a ∼1.7 kb deletion encompassing the su(Hw) and RpII15 promoters (Harrison et al. 1992); su(Hw)2, which carries an insertion of an ∼1.3 kb element into the first intron of the su(Hw) gene (Parkhurst et al. 1988); su(Hw)Pb, which carries an insertion of a white marked piggyBac element into the second exon of the su(Hw) gene [su(Hw)e04061 in FlyBase]; and su(Hw)A2663, which carries a deletion in the last su(Hw) exon, extending to the downstream CG3259 gene (Baxley et al. 2017). In analyses of su(Hw) mutant phenotypes, heteroallelic combinations of su(Hw) mutant were always studied, commonly the su(Hw)v/2 genotype (Figure 2, Figure 3, and Figure 4). Other stocks used were spermatocyte arrest (sa)-GFP, w+/CyO (Chen et al. 2005) and Enhancer of zeste [E(z)]-GFP, w+/CyO (Eun et al. 2014), both generous gifts from Dr. Xin Chen, Johns Hopkins University.
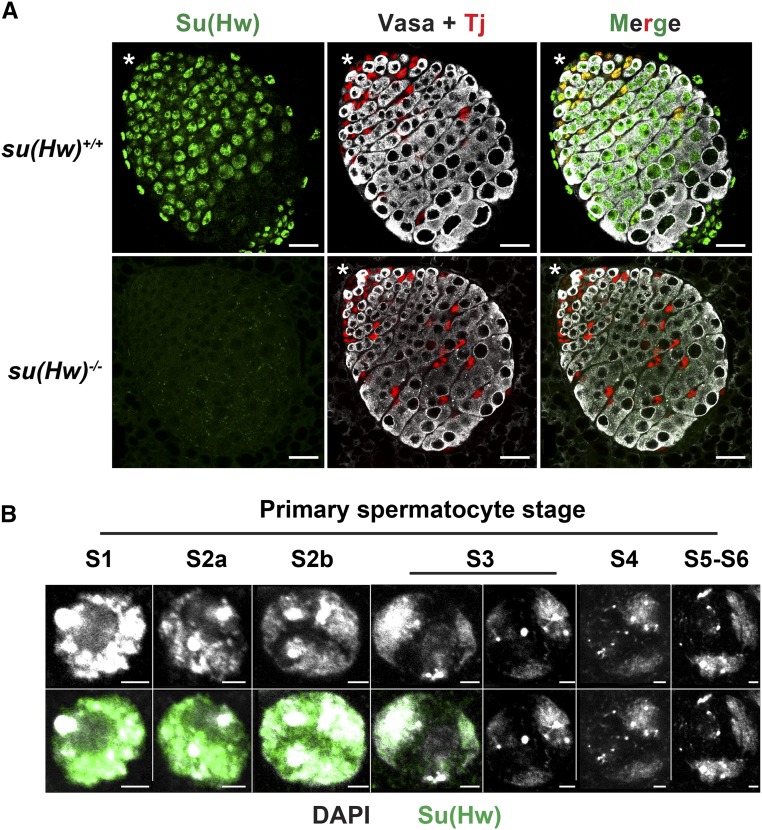
Figure 2.
Su(Hw) is lost during primary spermatocyte maturation. (A) Shown are confocal images of su(Hw)+/+ and su(Hw)2/v second instar larval testes stained with antibodies against Su(Hw) (green), Traffic jam (Tj; red), and Vasa (white). The asterisk indicates the anterior tip of the larval testis. Bar, 20 μm. (B) Shown are confocal images of representative nuclei corresponding to stages of spermatocyte growth (stages 1–6) in su(Hw)+/+ testes stained with DAPI (white) and antibodies against Su(Hw) (green). Top images are DAPI only, bottom images are DAPI and Su(Hw). Bar, 2 μm.
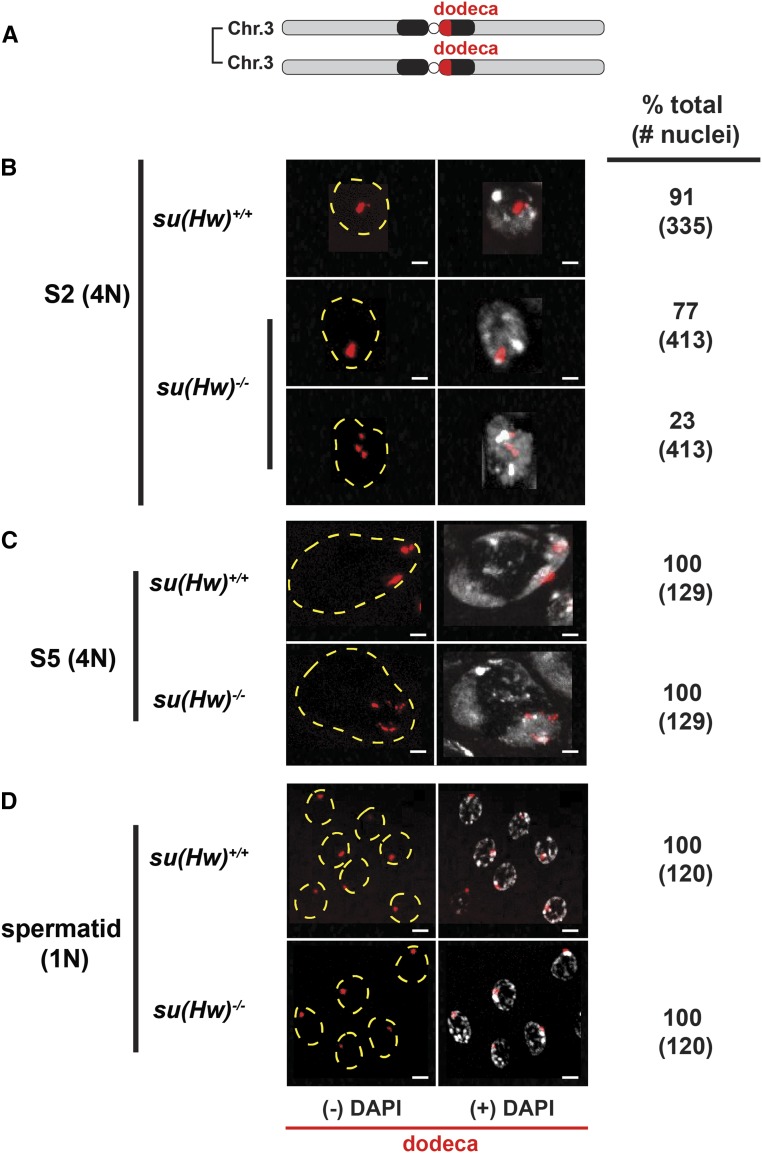
Figure 3.
Su(Hw) is dispensable for chromosome pairing and segregation. (A) Schematic of a pair of third chromosomes showing the location of the pericentric dodeca satellite repeats used in the FISH analysis. (B–D) Shown are confocal images of stage 2, stage 5, and spermatid nuclei in 3-day-old su(Hw)+/+ and su(Hw)2/v testes. The table shows the percentage of total nuclei that demonstrate the represented phenotype, with the total number of nuclei scored indicated in parentheses. Note that for the su(Hw)−/− stage 2 nuclei, two nuclei are shown. The top image represents the majority phenotype, whereas the bottom image represents the minority. FISH signals (red) are shown alone (−DAPI) or with DAPI (+DAPI; white). Yellow dotted lines show the nuclear outline. Bar, 2 μm.
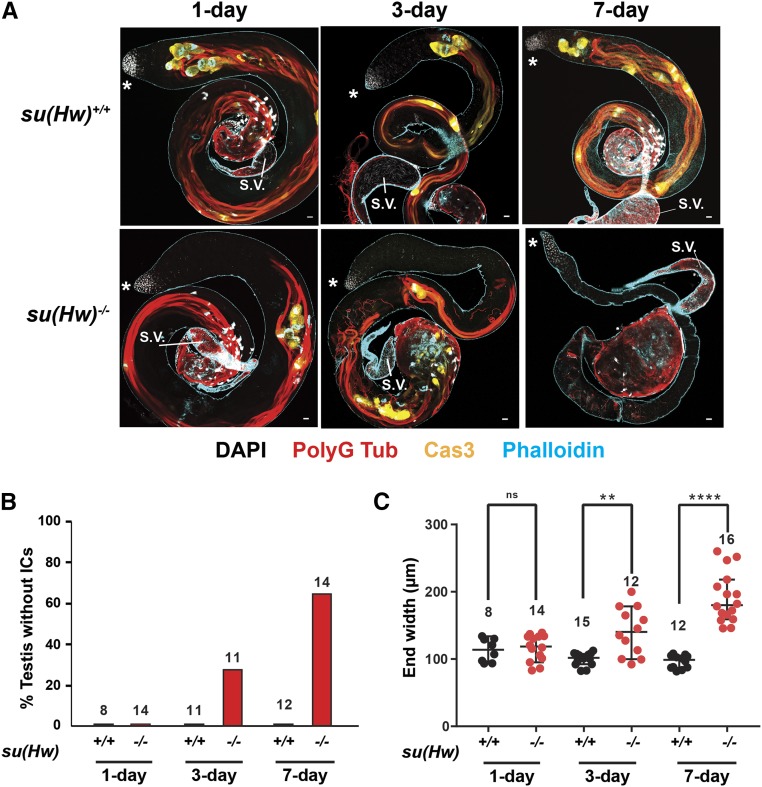
Figure 4.
Loss of Su(Hw) causes defects in spermiogenesis. (A) Representative confocal images of 1-, 3-, and 7-day-old su(Hw)+/+ and su(Hw)2/v testes stained with antibodies against polyglycylated tubulin (PolyG Tub; red, marks sperm tails), cleaved caspase 3 (Cas3, yellow, marks ICs), and phalloidin (blue, marks actin in ICs and elsewhere in the testis). Bar, 20 μm. Asterisk marks anterior of testis. (B and C) Quantification of su(Hw)2/v phenotypes as males age. End width: width of the posterior end of the testes leading into the seminal vesicle (S.V.). Numbers above data sets correspond to the number of testes examined. ** P < 0.01, **** P < 0.0001, ns, not significant, Student’s t-test.
Sperm exhaustion assay
We performed a sperm exhaustion assay to determine whether su(Hw) males remain fertile as they age, using the procedure described in Barton et al. (2016). For each experiment, 10 1-day-old su(Hw)+/+ or su(Hw)−/− males were individually mated with three y1w1118 virgin females. After 3 days, males were isolated, transferred to a new vial with three fresh virgin females, and allowed to mate for 3 days. This process was repeated four times until males reached an age of 16 days. Males were scored fertile if five or more progeny were produced. Vials containing males that died at any time during an experiment were eliminated from scoring. Three independent biological replicates were performed for each genotype.
Testes RNA isolation and analyses
For each replicate, ∼100 pairs of testes were dissected from 3-day-old males. RNA was extracted using TRIzol (Invitrogen, Carlsbad, CA) followed by purification on RNeasy spin columns (Qiagen, Valencia, CA). Three independent biological replicates were performed for each genotype. Microarray hybridization was conducted by the Iowa Institute of Human Genetics using the Affymetrix GeneChip Drosophila genome 2.0 arrays (#900532). Data were analyzed with Partek Genomics Suite 6.5 Gene Expression pipeline, normalized with Robust Multi-array Average (Irizarry et al. 2003). Differentially expressed genes were determined using one-way ANOVA, using a cutoff of a twofold expression change with 1% false discovery rate. Su(Hw) target genes were identified using the set of SBSs (2932) identified in the ovary (Soshnev et al. 2012), extending it to include additional sites identified in the overlapping set of nonovary SBSs identified in embryos, Kc and Mbn cells (1056), for a total of 3988 SBSs.
RNA levels of individual genes were quantified using reverse transcription quantitative PCR (RT-qPCR). For each replicate, ∼80 pairs of testes from 3-day-old males were dissected and stored at −80° until needed. RNA was extracted with TRIzol (Ambion), treated with DNAse I (Ambion DNA-free kit), and reverse-transcribed with High-Capacity cDNA Archive Kit with random primers (Applied Biosystems, Foster City, CA). qPCR analyses were performed with iQ SYBR green supermix (BioRad, Hercules, CA). A list of primers is found in Table S1. All experiments were performed based on manufacturers’ protocols. Expression levels were all relative to Ribosomal protein L32 (Rpl32).
Immunohistochemical analyses
For each experiment, 5–10 pairs of testes were dissected in cold phosphate buffered saline (PBS) solution. Testes were fixed in 4% EM grade paraformaldehyde (Electron Microscopy Sciences no. 15710) in PBST (PBS with 0.3% v/v Triton X-100), washed in PBST and blocked in 5% w/v BSA at room temperature for 1 hr. Samples were incubated with primary antibody overnight at 4°. Alexa Fluor-conjugated secondary antibodies (Molecular Probes, Eugene, OR) were added at room temperature. Testes were washed in PBST, stained with 1 µg/ml DAPI (Thermo Fisher scientific) and mounted in Vectashield (Vector Laboratories). Images were collected with a Zeiss 710 confocal microscope, and processed using ZEN imaging software. Primary antibodies included goat α-Su(Hw) at 1:300; guinea pig α-Traffic jam at 1:10,000 (Dorothea Godt, University of Toronto); rabbit α-cleaved caspase 3 at 1:400 (Cell Signaling Technology); mouse α-pan polyglycylated tubulin at 1:100 (AXO 49; Millipore); rabbit α-GFP at 1:2000 (Life Technologies); and rabbit α-vasa at 1:300 (Santa Cruz Biotechnology). Actin was stained using Texas Red-X phalloidin at 1:500 (Life Technologies).
Fluorescent in situ hybridization in whole-mount testes
Fluorescent in situ hybridization (FISH) protocols were adapted from an ovary FISH procedure (Joyce et al. 2013). Briefly, ∼20 testes were dissected from 3-day-old males and immediately fixed for 10 min in 200 µl fixative (4% paraformaldehyde, 1 × PBS, and 0.5% NP40) mixed with 600 µl heptane. After fixing, samples were washed and blocked in 1.5% BSA for 1 hr. For hybridization, samples were gradually exchanged into 50% formamide and allowed to prehybridize in a thermocycler at 37° for 4 hr, 92° for 3 min, and 60° for 20 min. Formamide was removed and 36 µl of hybridization buffer (2 × saline-sodium citrate buffer with 0.1% Tween20, 50% formamide, 10% dextran sulfate, 200 pmol probes, 1 µl RNAse A (Thermo Scientific) was added. DNA was denatured in a thermocycler at 91° for 1.5 min, followed by hybridization of probes at 37° for 16–20 hr. After hybridization, samples were washed twice at 37° in 50% formamide, and transferred to room temperature for one wash in 20% formamide, four washes in 2 × saline-sodium citrate buffer with 0.1% Tween20, and then stained with 1 µg/ml DAPI (Thermo Fisher scientific) for 20 min before mounting in Vectashield (Vector Laboratories). FISH images were taken using 1 µm intervals with a Zeiss 710 confocal microscope and processed with ZEN imaging software. Two FISH signals were considered as separate if the distance between centers was at least 0.7 µm and was separated by a black pixel. Fluorophore-conjugated oligonucleotide probes were ordered from Integrated DNA Technologies. These included the following sequences: the 359 repeats (5′-GGGATCGTTAGCACTGGTAATTAGCTGC-3′), the AATAC satellite (5′-AATACAATACAATACAATACAATACAATAC-3′), and the dodeca satellite (5′-ACGGGACCAGTACGG-3′).
Analyses of tissue-restricted Su(Hw) function
To deplete Su(Hw) in a cell-restricted manner, we used the UAST RNA interference (RNAi) line su(Hw)HMS00970 (BL34006; Bloomington Stock Center). To produce Su(Hw) in a cell-restricted manner, we generated flies that carried a P[UASp-su(Hw), w+] transgene. This transgene was made by cloning the su(Hw) gene (+634 to +3296) into a KpnI to BamHI digested UASp vector (gene bank AY831681). Once transgenic flies were obtained, the P[UASp-su(Hw), w+] transgene was crossed into a su(Hw)−/− background. Three Gal4 driver lines were used to generate cell-restricted Su(Hw) production, including w*; eya-Gal4/CyO, w*; C587-gal4 (a generous gift from Dr. Xin Chen, Johns Hopkins University) and w*; vasa-Gal4 (a generous gift from Dr. Leanne Jones, University of California Los Angeles).
Western analyses
Su(Hw) protein levels produced in su(Hw)+/+ and su(Hw)−/− were assessed using an SDS-polyacrylamide gel electrophoresis system of protein extracts from whole males. Briefly, two-male equivalents were loaded in each well, and then electrophoresed on a 4–20% Tris gel, and blotted onto nitrocellulose membranes. Membranes were probed with primary antibodies against Su(Hw) (guinea pig 1:250; Baxley et al. 2011), and α-tubulin (mouse 1:20,000, T5168; Sigma, St. Louis, MO). HRP-conjugated secondary antibodies (donkey anti-guinea pig IgG, 1:20,000; Jackson ImmunoResearch; rabbit anti-mouse IgG, 1:20,000, A9044; Sigma) were added, and detected using WesternBright Quantum kit (K-12042-D10; Advansta).
Data availability
Strains will be provided upon request. Microarray data are submitted to Gene Expression Omnibus under accession number GSE109601. The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6229223.
Results and Discussion
Su(Hw) is required for sustained male fertility
The decline of Su(Hw) in spermatogenesis suggested to us that Su(Hw) might have a role in sperm production. Whereas Su(Hw) has a well-known requirement in oogenesis for egg production (Klug et al. 1968), a requirement in spermatogenesis was unknown. We reasoned that su(Hw) null males might appear fertile because of sex-specific differences in gametogenesis that influence the presentation of sterility. Whereas female germ cells begin differentiation in late larval development (Gilboa and Lehmann 2004; Song et al. 2007), male germ cells begin these processes at the end of embryogenesis (Le Bras and Van Doren 2006; Sheng et al. 2009). Because su(Hw) homozygous mutant embryos are produced from su(Hw) heterozygous mutant females, Su(Hw) protein is present during embryogenesis. This maternal Su(Hw) protein might permit early germ cell functions and fertility of young males. However, developing su(Hw) null animals would not sustain Su(Hw) production, leading to Su(Hw) depletion and emerging sterility in aging males. For this reason, we used a sperm exhaustion assay to test for a requirement for Su(Hw) in male fertility. In this assay, su(Hw)+/+ and su(Hw)−/− males were individually mated to su(Hw)+/+ virgin females that were freshly supplied every 3 days during a 16-day period. The fertility of each male was then measured by assaying progeny output. In total, we analyzed two su(Hw)+/+ and three su(Hw)−/− backgrounds (Figure 1, B and C and Figure S1). The fertility of su(Hw)+/+ males was largely unchanged over the 16-day assay period, with ∼80% of males producing progeny. In contrast, the fertility of su(Hw)−/− males declined, with only ∼20% of 13- to 16-day males producing progeny. These experiments reveal that Su(Hw) is needed for sustained male fertility, demonstrating that its developmental contributions are not restricted to the ovary.
Su(Hw) is lost during primary spermatocyte maturation
To understand how Su(Hw) contributes to spermatogenesis, we defined its temporal and spatial localization in the testis. In the Drosophila testis, spermatogenesis initiates within a single stem cell niche called the hub, located at the anterior tip (Figure 1A). This hub supports two stem cell populations, germline stem cells and cyst stem cells (White-Cooper 2012). Upon asymmetric division of both stem cell populations, differentiating daughters move away from the hub, forming an initial unit of differentiation that consists of two postmitotic somatic cyst cells and one germ cell. A subsequent series of four mitotic germ cell divisions forms a differentiation unit comprised of 16 interconnected spermatogonial cells, which immediately enter the S phase and arrest at the G2-M transition as primary spermatocytes (Figure 1A). Progression toward meiosis requires maturation of spermatocytes, which includes striking nuclear growth that is important for chromosome territory formation and meiotic homolog segregation. Meiosis produces a differentiation unit comprised of 64 spermatids that undergo processes of spermatid elongation and individualization of sperm tails. Through these changes in germ cell number, the two somatic cyst cells continue to enclose the germ cells. Cyst cells codifferentiate with germ cells (Zoller and Schulz 2012), growing in size and displaying morphological differences upon terminal differentiation that distinguishes the head and tail cyst cells. Once formed, mature sperm transfer to the seminal vesicle. The process of spermatogenesis takes ∼10 days, with premeiotic development lasting ∼5 days. As a result, larval testes contain all pre- but not postmeiotic stages of spermatogenesis, whereas adult testes carry all stages of spermatogenesis.
To define the timing of Su(Hw) loss, we stained larval su(Hw)−/− and su(Hw)+/+ testes with antibodies against Su(Hw), the germline-specific helicase Vasa and the somatic transcription factor Tj (Figure 2A). We found that Su(Hw) is present in the somatic cells in the hub, the cyst stem cells, and early-stage postmitotic cyst cells, indicated by colocalization with Tj (Figure 2A). Su(Hw) expression is maintained in late-stage cyst cells, revealed by staining of adult testes for late-stage cyst cell markers (Figure S2A; data not shown). In contrast, Su(Hw) is lost in germ cells during primary spermatocyte maturation, indicated by the presence of Vasa-positive cells that lack Su(Hw) staining (Figure 2, A and B). Using the previously defined criteria of chromosome organization and nuclear size (Cenci et al. 1994), we found that Su(Hw) declines in stage 3 spermatocytes and is absent in all later-stage germ cells (Figure 2B and Figure S2A). The sharpness of the Su(Hw) decline in postmitotic cells suggests that Su(Hw) might be actively degraded during spermatocyte maturation. Consistent with the proposal that Su(Hw) protein levels are regulated, we were unable to produce Su(Hw) protein beyond stage 3 of spermatocyte development using the GAL4-UAS system. We found that levels of Su(Hw) were unchanged in males carrying the P[UASp-su(Hw), w+] responder transgene with either the vasa or Tubulin GAL4 driver (data not shown). Taken together, our data show that Su(Hw) loss is germ-cell-specific, occurring midway through spermatocyte maturation.
Su(Hw) is dispensable for pairing and separation of homologous chromosomes
Based upon the timing of Su(Hw) loss and previous findings that Su(Hw) contributes to somatic homolog pairing (Fritsch et al. 2006; Hartl et al. 2008), we postulated that Su(Hw) might be required for homolog pairing in germ cells. Drosophila spermatogenesis does not require synapsis or recombination for homologous chromosome segregation. Instead homologous chromosomes pair before entrance into the premeiotic prophase (McKee et al. 2012). Pairing of euchromatic and heterochromatic regions is found in stage 1 and 2 spermatocytes, but disappears in stage 3 spermatocytes when chromosomes move into distinct nuclear regions or territories. The hallmark of successful pairing is the separation of chromosomes into distinct territories, with each territory containing one bivalent (Vazquez et al. 2002; Tsai et al. 2011; McKee et al. 2012). Territories are visualized in maturing spermatocytes using DAPI staining. To test whether Su(Hw) has a role in homolog pairing in the male germline, we completed FISH studies using a pericentromeric probe to the dodeca satellite repeat found on the third chromosome (Figure 3A). We found that 91% of su(Hw)+/+ stage 2 spermatocyte nuclei had one or two spots of probe hybridization (Figure 3B), consistent with previous studies (92%; Tsai et al. 2011). In contrast, su(Hw)−/− stage 2 spermatocyte nuclei showed a reduced number of nuclei (77%) with one or two spots, with 23% having more than two spots of hybridization (Figure 3B). These data imply that su(Hw)−/− stage 2 spermatocyte nuclei have incomplete pairing of third chromosomes, suggesting that Su(Hw) might contribute to homolog associations. We reasoned that if this increase in the number of FISH spots represented defects in third chromosome pairing, then chromosome territory formation would be disrupted, resulting in distribution of third chromosomes into different territories. Such defects would be visualized as dodeca spots distributed to more than one DAPI-stained territory. To test this prediction, we analyzed dodeca FISH signals in stage 5 spermatocytes. Of the 129 su(Hw)−/− stage 5 nuclei analyzed, all FISH signals were confined to a single territory. Even so, we noted that FISH signals in su(Hw)−/− stage 5 nuclei remained more dispersed than in su(Hw)+/+ stage 5 nuclei (Figure 3C). Taken together, these data indicate that homolog pairing and assortment occur normally in the absence of Su(Hw).
As a second test of effects of Su(Hw) on homolog pairing, we examined chromosome distribution in spermatids. If homolog pairing was defective, then meiosis would produce aneuploidy, such that some spermatids would contain multiple FISH signals and others would have none. To test this prediction, we quantified dodeca FISH signals in spermatids. Of the 120 su(Hw)−/− spermatids analyzed, all contained a single FISH signal (Figure 3D), indicating that meiotic segregation of third chromosomes occurred normally in su(Hw) mutants. We extended these studies to test whether Su(Hw) loss affected pairing interactions needed for segregation of the X-Y chromosome pair. We used FISH to simultaneously detect the X-chromosome 359 repeats and the Y-chromosome AATAC repeats (Figure S3A). Quantification of FISH signals in fields of over 250 su(Hw)−/− spermatids revealed that all spermatids showed a FISH signal, with each spermatid containing a signal for either the 359 (X-chromosome) or the AATAC (Y-chromosome) repeats. These data indicate that meiotic chromosome segregation of the X-Y and the third chromosome pairs is not affected in su(Hw) mutants. We reason that differences in the increased number of dodeca FISH signals in su(Hw)−/− stage 2 and stage 5 spermatocytes might result from decondensation of pericentric regions, but these changes do not alter associations needed for meiotic chromosome segregation. The absence of a detectable role for Su(Hw) in germline chromosome pairing might result from compensation by other architectural proteins. Even so, our data are consistent with other studies that suggest that the requirements for somatic and germline pairing differ (Tomkiel et al. 2001; Thomas et al. 2005; Yan et al. 2010; Joyce et al. 2012).
Loss of Su(Hw) causes defects in sperm production
To gain a better understanding of the role of Su(Hw) in male fertility, we examined the phenotype of su(Hw)−/− testes in aging males. As premeiotic stages of spermatogenesis appear normal (Figure 2A), adult testes were stained to assess postmeiotic stages of sperm development. A prominent postmeiotic process is sperm individualization, which is characterized by the formation of individualization complexes (ICs). ICs are actin-rich structures that form around sperm nuclei and travel from the posterior to anterior tip of the testis. IC movement results in extrusion of excess cytoplasm and organelles into cyst bulges, and encases each sperm in its own plasma membrane (Fabian and Brill 2012). We used antibodies against polyglycylated tubulin to identify sperm tails during the individualization process, as well as phalloidin and antibodies against cleaved caspase 3 to identify ICs. In 1-day-old su(Hw)+/+ testes, sperm tails and ICs were distributed along the length of testis, extending to the anterior tip, a phenotype that is maintained as males aged (Figure 4 and Figure S2A). However, in 1-day-old su(Hw)−/− testes, ICs form but fail to reach the testis tip, possibly due to tangled sperm tails. As su(Hw)−/− testes age, these phenotypes worsen, and ICs become disorganized and evidentially are lost (Figure 4B). Coincident with these changes, the width of the end of the testis increases (Figure 4C). As a result, the seminal vesicles in su(Hw)−/− testes are small and appear to contain few sperm. Notably, morphological changes of su(Hw)−/− testes precede our detected fertility defects, emphasizing the stringency of our sperm depletion assay. Based on these findings, we conclude that Su(Hw) function is required for spermiogenesis.
Loss of Su(Hw) alters gene expression in the testis
Large scale changes in transcription accompany spermatogenesis. Most transcription occurs in mid to late stages of primary spermatocyte maturation, with only a few genes transcribed in spermatids (White-Cooper 2010). We reasoned that the spermiogenesis defects in su(Hw)−/− testes might result from altered gene expression in spermatocytes. Notably, one-third of testis-specific genes are organized into cotranscribed gene clusters (Boutanaev et al. 2002). These clustered domains are transcriptionally repressed through association with the nuclear lamina and the Drosophila B-type lamin, Lamin Dm0, until the appropriate stage of spermatogenesis (Shevelyov et al. 2009). For several reasons, we postulated that Su(Hw) might be required for establishing repression of the domains encompassing these testis-specific gene clusters. First, Su(Hw) globally localizes to repressive chromatin, found near genes of low transcription (Bushey et al. 2009; Filion et al. 2010; Roy et al. 2010). Second, Su(Hw) binds at lamina-associated domain borders and at specific positions within lamina-associated domains to fine-tune genome-nuclear lamin interactions (van Bemmel et al. 2010). Third, mutation of lamin Dm0 disrupts Su(Hw) insulator function (Capelson and Corces 2005). Together, these observations indicated that Su(Hw) might act as an insulator protein that demarcates chromatin domains required for repression of the testis-specific gene clusters.
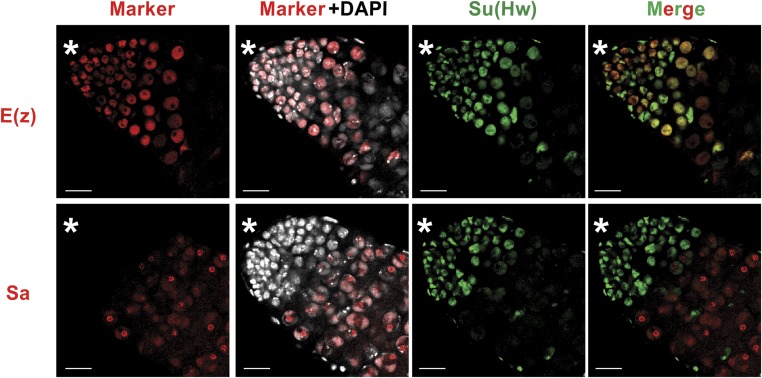
As a first step toward addressing Su(Hw) contributions to the testis-specific transcriptional program, we determined when Su(Hw) was lost relative to activation of testis transcription. The testis transcriptional program depends upon two transcriptional activator complexes, corresponding to a testis-specific Myb-MuvB/dREAM complex (testis meiotic arrest complex; tMAC) and a testis-specific TFIID complex (tTAF) that contains paralogues of TATA-binding proteins (Hiller et al. 2001, 2004). These activator complexes counteract transcriptional repression conferred by the transcriptional repressor complex Polycomb Repressive Complex 2 (PRC2) that carries the H3K27me3 writer E(z), a complex that is lost just before activation of the transcriptional program (Chen et al. 2011). The relative timing of Su(Hw) loss was defined by costaining su(Hw)+/+ testes with antibodies against Su(Hw) and GFP to identify either the GFP-tagged PRC2 component E(z) or the tTAF component Sa (Figure 5). We found that Su(Hw) and E(z) are coexpressed in early spermatocytes and lost midway through primary spermatocyte maturation, with Su(Hw) disappearing at a slightly earlier time (Figure 5). Consistent with this profile, Su(Hw) is absent in spermatocytes expressing Sa. Taken together, these data indicate that Su(Hw) is lost before the testis-specific transcriptional program begins.
Figure 5.
Su(Hw) is lost prior to activation of testis-specific transcription. One-day-old su(Hw)+/+, E(z)-GFP or su(Hw)+/+, Sa-GFP testes were costained with DAPI (white) and antibodies against Su(Hw) (green) and GFP (red) to identified either E(z) or Sa (tTAF), respectively. The asterisk indicates the anterior tip of the testis. Bar, 20 μm.
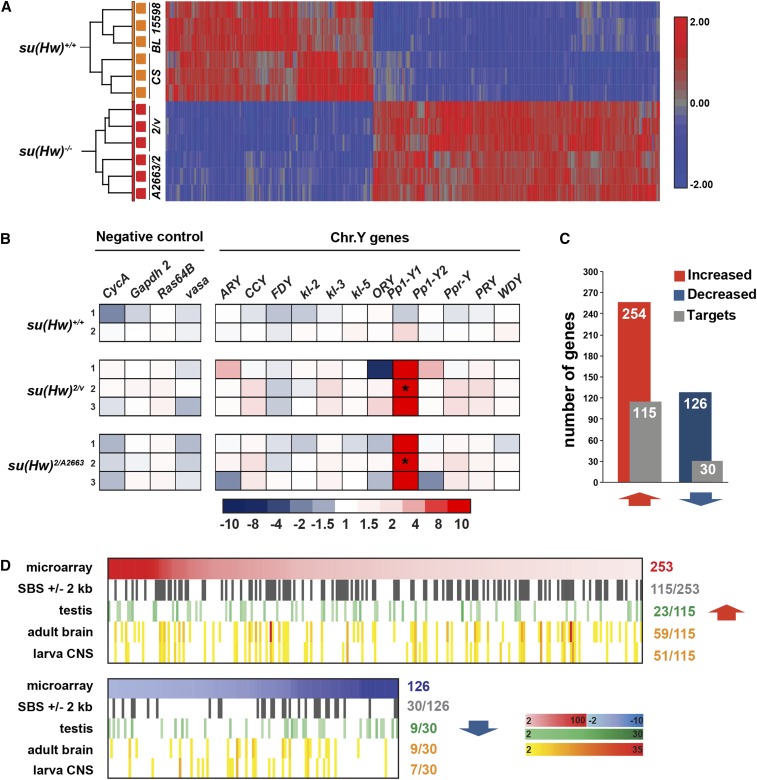
As a second step toward addressing Su(Hw) contribution to testis transcription, we measured RNA levels in 3-day-old testes isolated from two su(Hw)+/+ and two su(Hw)−/− genetic backgrounds, using the Drosophila 2.0 Affymetrix microarray (Figure 6A). We chose 3-day-old testes, because su(Hw) mutant phenotypes are manifest at this age (Figure 4). We complemented our microarray analyses with direct measurements of Y-chromosome gene expression (Figure 6B) because Y chromosome genes are unrepresented on the array. Of the ∼13,000 genes analyzed, we identified 380 genes that changed expression at least twofold in su(Hw)−/− relative to su(Hw)+/+ testes (Figure 6C). Among these genes, we identified possible Su(Hw) target genes by searching for misregulated genes that carried at least one SBS within the transcribed region or within 2 kb of upstream or downstream regulatory region—criteria used previously to identify targets (Soshnev et al. 2013). These analyses identified 145 Su(Hw) target genes (38%; Figure 6C), representing a significant enrichment relative to total number of genes in the genome that carry at least one SBS (n = 2674; 15.7%; P < 0.0001). We randomly selected 21 upregulated [Su(Hw) repressed] and eight downregulated [Su(Hw) activated] target genes and used RT-qPCR analyses to measure gene expression in two su(Hw) mutant backgrounds relative to su(Hw) wild-type controls (Figure 7 and Figure S4A). These studies confirmed misexpression of the majority (86%) of Su(Hw)-repressed genes. However, less than half (38%) of the Su(Hw)-activated genes were validated (Figure S4A), suggesting that the microarray was less effective at identifying this class of gene for an unknown reason. These transcriptional data indicate that Su(Hw) has a primary role as a transcriptional repressor in spermatogenesis, echoing findings in the ovary wherein the majority (71%) of target genes were Su(Hw) repressed (Soshnev et al. 2013). Comparison between testis- and ovary-regulated genes revealed only 21 shared targets, of which 20 correspond to Su(Hw)-repressed genes. Strikingly, the gene responsible for female sterility, RNA-binding protein 9, is not misregulated in the testes (data not shown). These data demonstrate that Su(Hw) loss causes tissue-specific derepression of transcription, emphasizing that the consequences of the loss of a transcriptional repressor depend upon the constellation of transcription factors expressed in the cell.
Figure 6.
Loss of Su(Hw) alters transcription in the testis. (A) Cluster analysis of microarray array data from testes dissected from 3-day-old males of two su(Hw)+/+ and two su(Hw)−/− backgrounds. Three biological replicates were completed for each genotype. (B) Heat map of fold changes of gene expression defined by RT-qPCR of Y-chromosome genes, measuring gene expression levels in RNA isolated from 3-day-old su(Hw)+/+ (CS) and two su(Hw)−/− mutant males. Fold change in expression was determined by normalizing levels to the housekeeping gene RpL32 and is relative to RNA levels in one of the three su(Hw)+/+ (CS) RNA samples. The color key corresponding to fold change is shown below. Asterisks indicate genes that change expression at least twofold with P < 0.05 (Student’s t-test). (C) Bar graph summary of total upregulated (254) and downregulated (126) genes in su(Hw)−/− testes (red and blue bars) alongside the number of Su(Hw) target genes in each class (115 and 30, respectively; gray bars). (D) Graphs of misregulated genes ranked by fold changes (microarray), with red corresponding to upregulated (top) and blue corresponding to downregulated genes (bottom); target genes within the misregulated gene set (genes with SBSs); genes with testis-enriched expression (green scale, testis); and genes with adult brain and larval CNS-enriched expression (orange scale, adult brain larva CNS). Enrichment is defined as a twofold or higher expression in the tissue compared to the whole fly. Numbers to the right of each analyses summarize the number of genes in the category. Bars at the right of the downregulated graph summarize the color key for corresponding fold changes in gene expression.
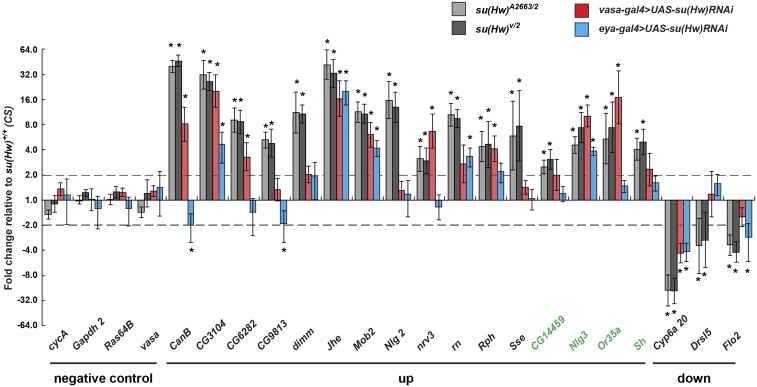
Figure 7.
Su(Hw) is required for transcriptional regulation in the testes. RT-qPCR of four negative controls, 16 upregulated and three downregulated Su(Hw) target genes in RNA isolated from 3-day-old testes dissected from two su(Hw)−/− (gray and black), one vasa-gal4; UAS-su(Hw)RNAi (red), and one eya-gal4; UAS-su(Hw)RNAi (blue) males. Genes indicated in green are Su(Hw) targets that are also derepressed in kmg−/− testes. Fold change in expression was determined by normalizing levels to the housekeeping gene RpL32 and is relative to RNA levels in su(Hw)+/+ (CS). Asterisks indicate genes that change expression at least twofold with P < 0.05 (Student’s t-test).
The Drosophila 2.0 Affymetrix microarray includes probe sets for transposable elements. Among these are probes for the LTR of the gypsy retrotransposon, which displayed a 2.6-fold increase in gypsy RNA in su(Hw) mutants. These data suggested that Su(Hw) might regulate gypsy mobilization through transcriptional repression. However, previous studies showed no role for Su(Hw) in gypsy regulation in the testis (Parkhurst and Corces 1986; Smith and Corces 1995). For this reason, we used three primer sets to measure changes in gypsy RNA, finding that only the LTR primers showed higher mRNA levels in the su(Hw) mutants (Figure S5). As such, transcriptional changes observed in our microarray data are likely to reflect the presence of structural variants of gypsy within the genomes of the tested strains. Taken together, our data imply that Su(Hw) does not regulate transcription of the gypsy retrotransposon transcription in testis.
We took a closer look at the properties of Su(Hw)-regulated testis target genes. Surprisingly, none of the Su(Hw)-repressed target genes lie within testis-specific gene clusters or on the Y-chromosome. Instead, Su(Hw) target genes share the feature that these genes are normally expressed in the adult brain (P < 0.0001) or the larval CNS (P = 0.0004; Figure 6D), properties again shared with Su(Hw) target genes in the ovary. Based on these data, we conclude that Su(Hw) has a prominent developmental role as a repressor of neuronal genes in non-neuronal tissues.
Kmg is a testis-specific transcription factor that is expressed upon commitment to germ cell differentiation and is essential for spermatogenesis (Kim et al. 2017). Kmg loss blocks spermatocyte differentiation, coincident with the upregulation of 500 genes, many of which are not normally expressed in the testis, but expressed in specific differentiated tissues, such as the eye and brain (Kim et al. 2017). Although genes that are derepressed in kmg−/− and su(Hw)−/− testes have similar features, only 13 of these genes are shared between the two mutants, of which only four are Su(Hw) target genes. Although transcriptional derepression upon Kmg loss requires the tMAC component Always Early (Aly) (Kim et al. 2017), the majority (67%) of Su(Hw)-repressed target genes remain derepressed in aly, su(Hw) double mutants (Figure S4B), suggesting that Aly is not required for activation of these target genes. Together, our data indicate that the Su(Hw) and Kmg represent two transcriptional repressors that regulate distinct gene networks in the testis.
Su(Hw) is required in cyst cells for male fertility
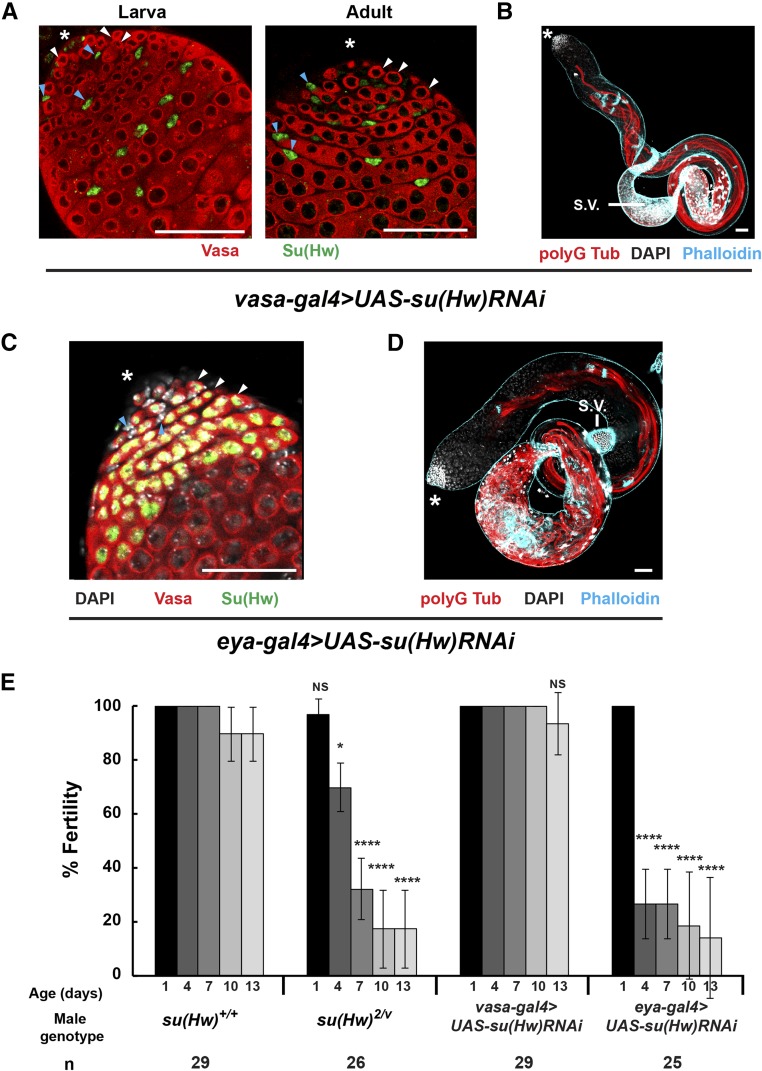
The germ-cell-specific decline of Su(Hw) in spermatogenesis suggested that the Su(Hw) function is required in germ cells for male fertility. Yet, the absence of testis-specific or enriched genes among the targets led us to question this prediction. For this reason, we used the vasa-gal4 driver to direct su(Hw) short hairpin RNA expression to knock down Su(Hw) in germ cells. Immunohistochemical analyses confirmed strong Su(Hw) knockdown in germ cells, as early as second instar larval (Figure 8A). Surprisingly, we also observed lower levels of Su(Hw) staining in early-stage cyst cells near the hub and even occasional Su(Hw) loss (Figure 8A, data not shown). These results indicate low levels of somatic loss using the germline vasa-gal4 driver, possibly due to the early vasa expression in somatic cells of the embryonic male gonad (Renault 2012). Surprisingly, the testes phenotypes in vasa-gal4 > UAS-su(Hw)RNAi males were normal even as the males aged (Figure 8B and Figure S2B). To test whether vasa-gal4 > UAS-su(Hw)RNAi males were fertile, we completed a sperm depletion assay and found that knockdown males retained fertility over the assay period (Figure 8E). These data imply germ cell expression of Su(Hw) is not essential for spermatogenesis. To test the somatic requirements for Su(Hw), we used an eya-gal4 driver to knockdown Su(Hw) in cyst cells. Immunohistochemical analyses confirmed strong and cyst-cell-specific knockdown (Figure 8C). Strikingly, 3-day-old eya-gal4 > UAS-su(Hw) RNAi testes displayed a su(Hw) mutant phenotype, with reduced IC numbers and prominent posterior bulges (Figure 8D). Testing the fertility of eya-gal4 > UAS-su(Hw)RNAi males in a sperm depletion assay showed that knockdown males had a significant reduction in fertility (Figure 8E). We note that fertility of eya-gal4 > UAS-su(Hw)RNAi males declined faster than in su(Hw)−/− males, possibly due to reduced levels of maternally supplied RNA in the RNAi line. Taken together, our cell-type-specific RNAi studies demonstrate that Su(Hw) is required in cyst but not germ cells for sustained male fertility.
Figure 8.
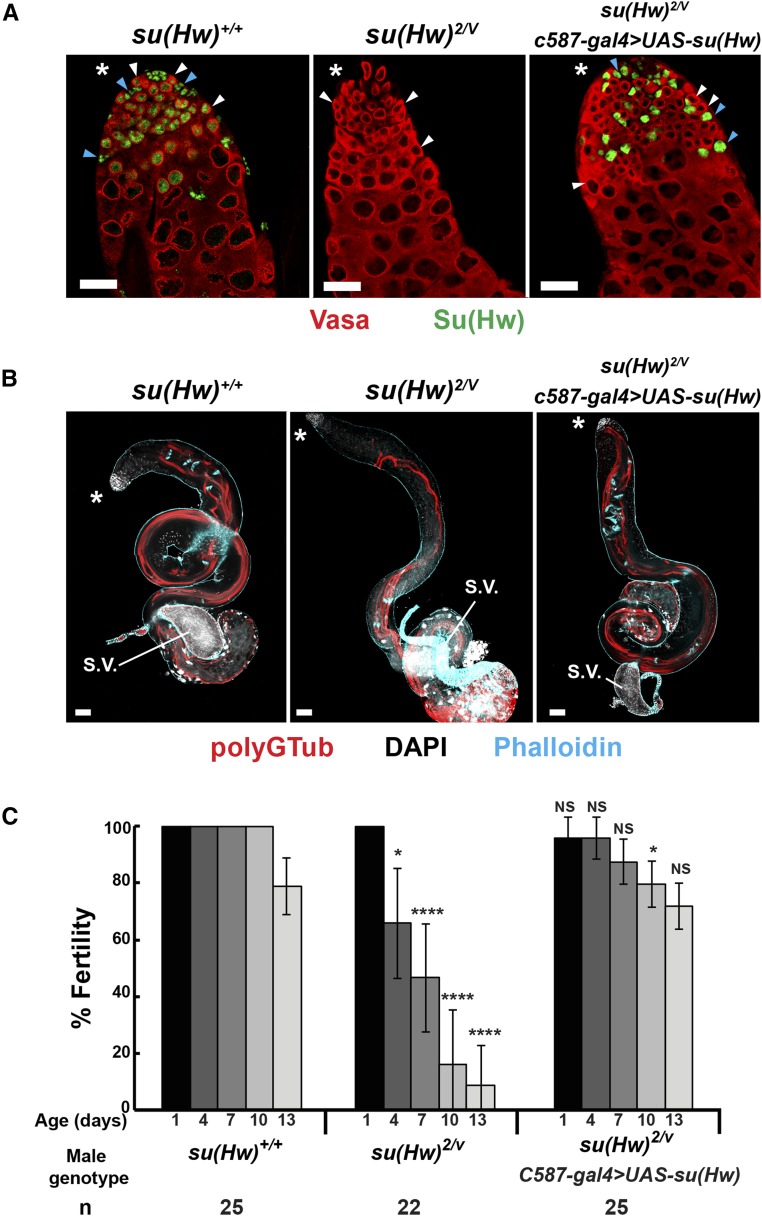
Male fertility requires Su(Hw) expression in cyst cells. (A) Confocal images of a second instar larval and 1-day-old testis isolated from su(Hw)+/+, vasa-gal4 > UAS-su(Hw)RNAi stained with antibodies against Vasa (red), and Su(Hw) (green). Blue arrowheads indicate positions of cyst cells; white arrowheads indicate positions of germ cells. (B) Confocal image of a 3-day-old testis isolated from su(Hw)+/+, vasa-gal4 > UAS-su(Hw)RNAi stained with antibodies against polyG tubulin (red), phalloidin (blue), and DAPI (white). (C) Confocal image of 1-day-old testis isolated from su(Hw)+/+, eya-gal4 > UAS-su(Hw)RNAi stained with antibodies against Vasa and Su(Hw) and DAPI (white). Blue arrowheads indicate positions of cyst cells; white arrowheads indicate positions of germ cells. (D) Confocal image of a 3-day-old testis isolated from su(Hw)+/+, eya-gal4 > UAS-su(Hw)RNAi males stained with antibodies against polyG tubulin (red), phalloidin (blue), and DAPI (white). The asterisks indicate the anterior tip of the testis. (E) Fertility outcomes of males of the indicated genotype tested in the sperm exhaustion assay. Axes are as described in Figure 1. Bars indicate the variation of fertility from three biological replicates (* P < 0.05, **** P < 0.0001, NS, not significant. Student’s t-test). The total number (n) of males tested is shown below the graph. In all panels, bars, 50 μm. S.V., seminal vesicle.
We capitalized on our cell-type-specific knockdown males to address the relative contributions of Su(Hw) to transcriptional regulation in germline or somatic cells of the testis. In these studies, we randomly selected 19 validated Su(Hw) target genes and used RT-qPCR to measure transcriptional levels in RNAs isolated from 3-day-old vasa-gal4 > UAS-su(Hw) RNAi and eya -gal4 > UAS-su(Hw)RNAi testes (Figure 7). We found that 12 (63%) genes were misregulated in at least one of the two knockdown RNAs, including 10 (53%) genes affected upon germ-cell-specific knockdown and seven (37%) affected upon cyst-cell-specific knockdown. Of the 12, five (42%) were shared between cell types. Notably, cell-type-specific knockdown failed to recapitulate a su(Hw) null transcriptional phenotype, even though robust knockdown of Su(Hw) was achieved (Figure 8 and Figure S2B). We predict that even low levels of Su(Hw) might be enough to maintain expression of some genes, based on the previous demonstration that a 95% knock down of Su(Hw) failed to deplete Su(Hw) occupancy at some genomic sites (Vorobyeva et al. 2013). Together, these data demonstrate that Su(Hw) is required for transcriptional regulation in cyst and germ cells.
Our cell-type-specific knockdown studies revealed that somatic Su(Hw) expression is necessary for spermatogenesis. To determine if somatic Su(Hw) expression is sufficient for spermatogenesis, we generated transgenic flies that carried a UAS-su(Hw) expresser transgene and the c587-gal4 > UAS-su(Hw) cyst cell driver in su(Hw)−/− males. Antibody staining of 1-day-old testes revealed restoration of high levels of Su(Hw) protein only in early-stage cyst cells, not germ cells (Figure 9A). In addition, low but detectable levels of Su(Hw) were produced in cyst cells surrounding postmeiotic spermatids (Figure S2C). Strikingly, 3- and 7-day-old su(Hw)−/−, c587-gal4 > UAS-su(Hw) testes had a wild-type appearance (Figure 9B and Figure S2C), with testes carrying many ICs and lacking any posterior bulge. To test whether su(Hw)−/−, c587-gal4 > UAS-su(Hw) males were fertile, we completed a sperm depletion assay (Figure 9C). We found that fertility was restored to wild type levels in 13- to 16-day-old su(Hw)−/−, c587-gal4 > UAS-su(Hw) males (Figure 8C). We were unable to test effects of germ-cell-restricted Su(Hw) expression because the vasa-gal4 driver directed Su(Hw) production in both germ cells and somatic cells (data not shown), as a result of the embryonic expression of vasa in the male gonad (Renault 2012). Based on these studies, we conclude that cyst cell expression of Su(Hw) is both necessary and sufficient for sustained male fertility.
Figure 9.
Cyst-cell-restricted Su(Hw) rescues testis morphology and male fertility. (A) Confocal images of 1-day-old testes isolated from su(Hw)+/+, su(Hw)2/v and C587-gal4 >UAS-su(Hw), su(Hw)2/v males stained with antibodies against Vasa (red) and Su(Hw) (green). Blue arrowheads point to cyst cells; white arrowheads point to germ cells. Bar, 20 μm. (B) Confocal images of 3-day-old testes isolated from su(Hw) +/+, su(Hw)2/v and C587-gal4 > UAS-su(Hw), su(Hw)2/v males stained with antibodies against spermiogenesis markers, PolyG tubulin (polyGTub; red), phalloidin (blue), and DAPI (white). Asterisks indicate the anterior tip of the testis. Bar, 50 μm. (C) Fertility outcomes of males of the indicated genotype tested in the sperm exhaustion assay. Axes are as described in Figure 1. Bars indicate the variation of fertility from three biological replicates (* P < 0.05, **** P < 0.0001, NS, not significant, Student’s t-test). The total number (n) of males tested is shown below the graph. S.V., seminal vesicle.
Concluding perspectives
Su(Hw) is a globally expressed multivalent zinc finger transcription factor. Although well-known for its gypsy insulator function, its developmental contributions outside of the gypsy retrotransposon are just beginning to be understood (Baxley et al. 2011; Soshnev et al. 2013). Here, we capitalize on our discovery of a developmental requirement for Su(Hw) in male fertility to investigate the function of Su(Hw) at non-gypsy genomic sites. Spermatogenesis is a complex developmental process involving meiotic chromosome pairing and transcription of lamin-regulated testis-specific gene clusters, both linked to contributions of insulators and architectural proteins such as Su(Hw) (Shevelyov et al. 2009; McKee et al. 2012). Even though Su(Hw) is required for sustained spermatogenesis, it is dispensable for meiotic chromosome pairing and transcription of lamin-regulated testis-specific gene clusters (Figure 3 and Figure 6). These data imply that Su(Hw) has noninsulator roles in the testis. Indeed, we find that loss of Su(Hw) alters transcription, with the majority (79%) of Su(Hw) target genes upregulated in the absence of Su(Hw) (Figure 6). Based on these data, we conclude that Su(Hw) has a prominent repressor role in the testis, a function parallel to its role in the ovary (Soshnev et al. 2013). Strikingly, few Su(Hw) target genes are misexpressed in both the testis and ovary, emphasizing transcriptional changes resulting from loss of a repressor depend on the constellation of transcription factors expressed in a given cell. Most derepressed Su(Hw) testis target genes are normally expressed in neuronal tissues, not the testis (Figure 6), similar to Su(Hw) ovary target genes. Taken together, these data provide additional support for the proposal that Su(Hw) is a functional homolog of mammalian REST proteins that repress neuronal genes in non-neuronal tissues (Soshnev et al. 2013).
Our focus on the testis was motivated by the dynamic change in Su(Hw) levels during the maturation of primary spermatocytes (Figure 1, Figure 2, and Figure 5). However, loss of Su(Hw) in germline cells did not affect male fertility (Figure 8). Instead, we found that Su(Hw) expression in cyst cells is both necessary and sufficient for spermatogenesis (Figure 7, Figure 8, and Figure 9). Dual somatic and germline requirements for Su(Hw) also exist for oogenesis (Soshnev et al. 2013), with gene-specific regulation in both cell types required for fertility. Based on these observations, we predict that the cyst cell requirement for Su(Hw) involves transcriptional repression to prevent ectopic expression of one or more target genes. Currently, the critical target gene(s) of this regulation is unknown. Recent loss of function screens identified genes required in cyst cells for spermiogenesis (Liu et al. 2016; Fairchild et al. 2017). However, none of these genes correspond to Su(Hw) testis target genes. Future studies will be needed to identify how Su(Hw) is integrated into the cyst cell transcriptional regulatory network.
Acknowledgments
We thank Alexey Soshnev and Jaya Guha for technical assistance with these studies, Chantal Allamargot at the University of Iowa Central Microscopy Research Facility for assistance with confocal imaging, and members of the Iowa Institute of Human Genetics for assistance with the microarray experiments. We thank members of the Geyer laboratory for reading the manuscript. We thank the following Drosophila investigators for generously supplying reagents: Xin Chen, Dorothea Godt, and Leanne Jones.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6229223.
Communicating editor: J. Birchler
Literature Cited
- Ali T., Renkawitz R., Bartkuhn M., 2016. Insulators and domains of gene expression. Curr. Opin. Genet. Dev. 37: 17–26. 10.1016/j.gde.2015.11.009 [DOI] [PubMed] [Google Scholar]
- Barton L. J., Lovander K. E., Pinto B. S., Geyer P. K., 2016. Drosophila male and female germline stem cell niches require the nuclear lamina protein Otefin. Dev. Biol. 415: 75–86. 10.1016/j.ydbio.2016.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxley R. M., Soshnev A. A., Koryakov D. E., Zhimulev I. F., Geyer P. K., 2011. The role of the suppressor of Hairy-wing insulator protein in Drosophila oogenesis. Dev. Biol. 356: 398–410. 10.1016/j.ydbio.2011.05.666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxley R. M., Bullard J. D., Klein M. W., Fell A. G., Morales-Rosado J. A., et al. , 2017. Deciphering the DNA code for the function of the Drosophila polydactyl zinc finger protein suppressor of Hairy-wing. Nucleic Acids Res. 45: 4463–4478. 10.1093/nar/gkx040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutanaev A. M., Kalmykova A. I., Shevelyov Y. Y., Nurminsky D. I., 2002. Large clusters of co-expressed genes in the Drosophila genome. Nature 420: 666–669. [DOI] [PubMed] [Google Scholar]
- Bushey A. M., Ramos E., Corces V. G., 2009. Three subclasses of a Drosophila insulator show distinct and cell type-specific genomic distributions. Genes Dev. 23: 1338–1350. 10.1101/gad.1798209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H. N., Shen P., 2001. Effects of cis arrangement of chromatin insulators on enhancer-blocking activity. Science 291: 493–495. [DOI] [PubMed] [Google Scholar]
- Capelson M., Corces V. G., 2005. The ubiquitin ligase dTopors directs the nuclear organization of a chromatin insulator. Mol. Cell 20: 105–116. [DOI] [PubMed] [Google Scholar]
- Cenci G., Bonaccorsi S., Pisano C., Verni F., Gatti M., 1994. Chromatin and microtubule organization during premeiotic, meiotic and early postmeiotic stages of Drosophila melanogaster spermatogenesis. J. Cell Sci. 107: 3521–3534. [DOI] [PubMed] [Google Scholar]
- Chen X., Hiller M., Sancak Y., Fuller M. T., 2005. Tissue-specific TAFs counteract Polycomb to turn on terminal differentiation. Science 310: 869–872. [DOI] [PubMed] [Google Scholar]
- Chen X., Lu C., Morillo Prado J. R., Eun S. H., Fuller M. T., 2011. Sequential changes at differentiation gene promoters as they become active in a stem cell lineage. Development 138: 2441–2450. 10.1242/dev.056572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eun S. H., Shi Z., Cui K., Zhao K., Chen X., 2014. A non-cell autonomous role of E(z) to prevent germ cells from turning on a somatic cell marker. Science 343: 1513–1516. 10.1126/science.1246514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian L., Brill J. A., 2012. Drosophila spermiogenesis: big things come from little packages. Spermatogenesis 2: 197–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairchild M. J., Islam F., Tanentzapf G., 2017. Identification of genetic networks that act in the somatic cells of the testis to mediate the developmental program of spermatogenesis. PLoS Genet. 13: e1007026 10.1371/journal.pgen.1007026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filion G. J., van Bemmel J. G., Braunschweig U., Talhout W., Kind J., et al. , 2010. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell 143: 212–224. 10.1016/j.cell.2010.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsch C., Ploeger G., Arndt-Jovin D. J., 2006. Drosophila under the lens: imaging from chromosomes to whole embryos. Chromosome Res. 14: 451–464. [DOI] [PubMed] [Google Scholar]
- Fuller M. T., 1998. Genetic control of cell proliferation and differentiation in Drosophila spermatogenesis. Semin. Cell Dev. Biol. 9: 433–444. [DOI] [PubMed] [Google Scholar]
- Georgiev P. G., Gerasimova T. I., 1989. Novel genes influencing the expression of the yellow locus and mdg4 (gypsy) in Drosophila melanogaster. Mol. Gen. Genet. 220: 121–126. [DOI] [PubMed] [Google Scholar]
- Geyer P. K., Corces V. G., 1992. DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev. 6: 1865–1873. [DOI] [PubMed] [Google Scholar]
- Gilboa L., Lehmann R., 2004. Repression of primordial germ cell differentiation parallels germ line stem cell maintenance. Curr. Biol. 14: 981–986. [DOI] [PubMed] [Google Scholar]
- Han B. Y., Foo C. S., Wu S., Cyster J. G., 2016. The C2H2-ZF transcription factor Zfp335 recognizes two consensus motifs using separate zinc finger arrays. Genes Dev. 30: 1509–1514. 10.1101/gad.279406.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison D. A., Mortin M. A., Corces V. G., 1992. The RNA polymerase II 15-kilodalton subunit is essential for viability in Drosophila melanogaster. Mol. Cell. Biol. 12: 928–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl T. A., Smith H. F., Bosco G., 2008. Chromosome alignment and transvection are antagonized by condensin II. Science 322: 1384–1387. 10.1126/science.1164216 [DOI] [PubMed] [Google Scholar]
- Hiller M., Chen X., Pringle M. J., Suchorolski M., Sancak Y., et al. , 2004. Testis-specific TAF homologs collaborate to control a tissue-specific transcription program. Development 131: 5297–5308. [DOI] [PubMed] [Google Scholar]
- Hiller M. A., Lin T. Y., Wood C., Fuller M. T., 2001. Developmental regulation of transcription by a tissue-specific TAF homolog. Genes Dev. 15: 1021–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdridge C., Dorsett D., 1991. Repression of hsp70 heat shock gene transcription by the suppressor of hairy-wing protein of Drosophila melanogaster. Mol. Cell. Biol. 11: 1894–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry R. A., Hobbs B., Collin F., Beazer-Barclay Y. D., Antonellis K. J., et al. , 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264. [DOI] [PubMed] [Google Scholar]
- Joyce E. F., Williams B. R., Xie T., Wu C. T., 2012. Identification of genes that promote or antagonize somatic homolog pairing using a high-throughput FISH-based screen. PLoS Genet. 8: e1002667 10.1371/journal.pgen.1002667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce E. F., Apostolopoulos N., Beliveau B. J., Wu C. T., 2013. Germline progenitors escape the widespread phenomenon of homolog pairing during Drosophila development. PLoS Genet. 9: e1004013 10.1371/journal.pgen.1004013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermekchiev M., Pettersson M., Matthias P., Schaffner W., 1991. Every enhancer works with every promoter for all the combinations tested: could new regulatory pathways evolve by enhancer shuffling? Gene Expr. 1: 71–81. [PMC free article] [PubMed] [Google Scholar]
- Kim J., Lu C., Srinivasan S., Awe S., Brehm A., et al. , 2017. Blocking promiscuous activation at cryptic promoters directs cell type-specific gene expression. Science 356: 717–721. 10.1126/science.aal3096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug W. S., Bodenstein D., King R. C., 1968. Oogenesis in the suppressor of hairy-wing mutant of Drosophila melanogaster. I. Phenotypic characterization and transplantation experiments. J. Exp. Zool. 167: 151–156. [DOI] [PubMed] [Google Scholar]
- Kravchenko E., Savitskaya E., Kravchuk O., Parshikov A., Georgiev P., et al. , 2005. Pairing between gypsy insulators facilitates the enhancer action in trans throughout the Drosophila genome. Mol. Cell. Biol. 25: 9283–9291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn E. J., Viering M. M., Rhodes K. M., Geyer P. K., 2003. A test of insulator interactions in Drosophila. EMBO J. 22: 2463–2471. 10.1093/emboj/cdg241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrchanova O., Chetverina D., Maksimenko O., Kullyev A., Georgiev P., 2008. Orientation-dependent interaction between Drosophila insulators is a property of this class of regulatory elements. Nucleic Acids Res. 36: 7019–7028. 10.1093/nar/gkn781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bras S., Van Doren M., 2006. Development of the male germline stem cell niche in Drosophila. Dev. Biol. 294: 92–103. [DOI] [PubMed] [Google Scholar]
- Lelli K. M., Slattery M., Mann R. S., 2012. Disentangling the many layers of eukaryotic transcriptional regulation. Annu. Rev. Genet. 46: 43–68. 10.1146/annurev-genet-110711-155437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Ge Q., Chan B., Liu H., Singh S. R., et al. , 2016. Whole-animal genome-wide RNAi screen identifies networks regulating male germline stem cells in Drosophila. Nat. Commun. 7: 12149 10.1038/ncomms12149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee B. D., Yan R., Tsai J. H., 2012. Meiosis in male Drosophila. Spermatogenesis 2: 167–184. 10.4161/spmg.21800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muravyova E., Golovnin A., Gracheva E., Parshikov A., Belenkaya T., et al. , 2001. Loss of insulator activity by paired Su(Hw) chromatin insulators. Science 291: 495–498. [DOI] [PubMed] [Google Scholar]
- Nakahashi H., Kwon K. R., Resch W., Vian L., Dose M., et al. , 2013. A genome-wide map of CTCF multivalency redefines the CTCF code. Cell Rep. 3: 1678–1689. 10.1016/j.celrep.2013.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negre N., Brown C. D., Shah P. K., Kheradpour P., Morrison C. A., et al. , 2010. A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet. 6: e1000814 10.1371/journal.pgen.1000814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevil M., Bondra E. R., Schulz K. N., Kaplan T., Harrison M. M., 2017. Stable binding of the conserved transcription factor grainy head to its target genes throughout Drosophila melanogaster. Dev. Genet. 205: 605–620. 10.1534/genetics.116.195685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaev L. G., Akopov S. B., Didych D. A., Sverdlov E. D., 2009. Vertebrate protein CTCF and its multiple roles in a large-scale regulation of genome activity. Curr. Genomics 10: 294–302. 10.2174/138920209788921038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai C. Y., Lei E. P., Ghosh D., Corces V. G., 2004. The centrosomal protein CP190 is a component of the gypsy chromatin insulator. Mol. Cell 16: 737–748. [DOI] [PubMed] [Google Scholar]
- Parkhurst S. M., Corces V. G., 1986. Interactions among the gypsy transposable element and the yellow and the suppressor of hairy-wing loci in Drosophila melanogaster. Mol. Cell. Biol. 6: 47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhurst S. M., Harrison D. A., Remington M. P., Spana C., Kelley R. L., et al. , 1988. The Drosophila su(Hw) gene, which controls the phenotypic effect of the gypsy transposable element, encodes a putative DNA-binding protein. Genes Dev. 2: 1205–1215. [DOI] [PubMed] [Google Scholar]
- Parnell T. J., Geyer P. K., 2000. Differences in insulator properties revealed by enhancer blocking assays on episomes. EMBO J. 19: 5864–5874. 10.1093/emboj/19.21.5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parnell T. J., Kuhn E. J., Gilmore B. L., Helou C., Wold M. S., et al. , 2006. Identification of genomic sites that bind the Drosophila suppressor of Hairy-wing insulator protein. Mol. Cell. Biol. 26: 5983–5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab J. R., Kamakaka R. T., 2010. Insulators and promoters: closer than we think. Nat. Rev. Genet. 11: 439–446. 10.1038/nrg2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault A. D., 2012. Vasa is expressed in somatic cells of the embryonic gonad in a sex-specific manner in Drosophila melanogaster. Biol. Open 1: 1043–1048. 10.1242/bio.20121909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roseman R. R., Pirrotta V., Geyer P. K., 1993. The su(Hw) protein insulates expression of the Drosophila melanogaster white gene from chromosomal position-effects. EMBO J. 12: 435–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S., Ernst J., Kharchenko P. V., Kheradpour P., Negre N., et al. , 2010. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330: 1787–1797. 10.1126/science.1198374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenherr C. J., Paquette A. J., Anderson D. J., 1996. Identification of potential target genes for the neuron-restrictive silencer factor. Proc. Natl. Acad. Sci. USA 93: 9881–9886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz Y. B., Linder-Basso D., Kharchenko P. V., Tolstorukov M. Y., Kim M., et al. , 2012. Nature and function of insulator protein binding sites in the Drosophila genome. Genome Res. 22: 2188–2198. 10.1101/gr.138156.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott K. C., Taubman A. D., Geyer P. K., 1999. Enhancer blocking by the Drosophila gypsy insulator depends upon insulator anatomy and enhancer strength. Genetics 153: 787–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott K. S., Geyer P. K., 1995. Effects of the su(Hw) insulator protein on the expression of the divergently transcribed Drosophila yolk protein genes. EMBO J. 14: 6258–6267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng X. R., Posenau T., Gumulak-Smith J. J., Matunis E., Van Doren M., et al. , 2009. Jak-STAT regulation of male germline stem cell establishment during Drosophila embryogenesis. Dev. Biol. 334: 335–344. 10.1016/j.ydbio.2009.07.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevelyov Y. Y., Lavrov S. A., Mikhaylova L. M., Nurminsky I. D., Kulathinal R. J., et al. , 2009. The B-type lamin is required for somatic repression of testis-specific gene clusters. Proc. Natl. Acad. Sci. USA 106: 3282–3287. 10.1073/pnas.0811933106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith P. A., Corces V. G., 1995. The suppressor of Hairy-wing protein regulates the tissue-specific expression of the Drosophila gypsy retrotransposon. Genetics 139: 215–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X., Call G. B., Kirilly D., Xie T., 2007. Notch signaling controls germline stem cell niche formation in the Drosophila ovary. Development 134: 1071–1080. [DOI] [PubMed] [Google Scholar]
- Soshnev A. A., Li X., Wehling M. D., Geyer P. K., 2008. Context differences reveal insulator and activator functions of a Su(Hw) binding region. PLoS Genet. 4: e1000159 10.1371/journal.pgen.1000159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soshnev A. A., He B., Baxley R. M., Jiang N., Hart C. M., et al. , 2012. Genome-wide studies of the multi-zinc finger Drosophila suppressor of Hairy-wing protein in the ovary. Nucleic Acids Res. 40: 5413–5431. 10.1093/nar/gks225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soshnev A. A., Baxley R. M., Manak J. R., Tan K., Geyer P. K., 2013. The Drosophila suppressor of Hairy-wing insulator protein has an essential role as a transcriptional repressor in the ovary. Development 140: 3613–3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S. E., Soltani-Bejnood M., Roth P., Dorn R., Logsdon J. M., Jr., et al. , 2005. Identification of two proteins required for conjunction and regular segregation of achiasmate homologs in Drosophila male meiosis. Cell 123: 555–568. [DOI] [PubMed] [Google Scholar]
- Tomkiel J. E., Wakimoto B. T., Briscoe A., Jr., 2001. The teflon gene is required for maintenance of autosomal homolog pairing at meiosis I in male Drosophila melanogaster. Genetics 157: 273–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai J. H., Yan R., McKee B. D., 2011. Homolog pairing and sister chromatid cohesion in heterochromatin in Drosophila male meiosis I. Chromosoma 120: 335–351. 10.1007/s00412-011-0314-0 [DOI] [PubMed] [Google Scholar]
- van Bemmel J. G., Pagie L., Braunschweig U., Brugman W., Meuleman W., et al. , 2010. The insulator protein SU(HW) fine-tunes nuclear lamina interactions of the Drosophila genome. PLoS One 5: e15013 10.1371/journal.pone.0015013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez J., Belmont A. S., Sedat J. W., 2002. The dynamics of homologous chromosome pairing during male Drosophila meiosis. Curr. Biol. 12: 1473–1483. [DOI] [PubMed] [Google Scholar]
- Vorobyeva N. E., Mazina M. U., Golovnin A. K., Kopytova D. V., Gurskiy D. Y., et al. , 2013. Insulator protein Su(Hw) recruits SAGA and Brahma complexes and constitutes part of origin recognition complex-binding sites in the Drosophila genome. Nucleic Acids Res. 41: 5717–5730. 10.1093/nar/gkt297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White-Cooper H., 2010. Molecular mechanisms of gene regulation during Drosophila spermatogenesis. Reproduction 139: 11–21. 10.1530/REP-09-0083 [DOI] [PubMed] [Google Scholar]
- White-Cooper H., 2012. Tissue, cell type and stage-specific ectopic gene expression and RNAi induction in the Drosophila testis. Spermatogenesis 2: 11–22. 10.4161/spmg.19088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R., Thomas S. E., Tsai J. H., Yamada Y., McKee B. D., 2010. SOLO: a meiotic protein required for centromere cohesion, coorientation, and SMC1 localization in Drosophila melanogaster. J. Cell Biol. 188: 335–349. 10.1083/jcb.200904040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoller R., Schulz C., 2012. The Drosophila cyst stem cell lineage: partners behind the scenes? Spermatogenesis 2: 145–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains will be provided upon request. Microarray data are submitted to Gene Expression Omnibus under accession number GSE109601. The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6229223.