Abstract
MicroRNAs are small, noncoding RNAs that regulate gene expression at the post-transcriptional level in essentially all aspects of Caenorhabditis elegans biology. More than 140 genes that encode microRNAs in C. elegans regulate development, behavior, metabolism, and responses to physiological and environmental changes. Genetic analysis of C. elegans microRNA genes continues to enhance our fundamental understanding of how microRNAs are integrated into broader gene regulatory networks to control diverse biological processes, including growth, cell division, cell fate determination, behavior, longevity, and stress responses. As many of these microRNA sequences and the related processing machinery are conserved over nearly a billion years of animal phylogeny, the assignment of their functions via worm genetics may inform the functions of their orthologs in other animals, including humans. In vivo investigations are especially important for microRNAs because in silico extrapolation of their functions using mRNA target prediction programs can easily assign microRNAs to incorrect genetic pathways. At this mezzanine level of microRNA bioinformatic sophistication, genetic analysis continues to be the gold standard for pathway assignments.
Keywords: Caenorhabditis elegans, microRNA, Argonaute, miRISC, mutant phenotypes, WormBook
Overview
HERE, we discuss the current understanding of how microRNAs function in Caenorhabditis elegans. While striving to be as comprehensive as possible, we will emphasize the contexts in which research using C. elegans has provided unique insight into evolutionarily conserved aspects of microRNA biology. We will also highlight where worm microRNA research motivates interesting, unanswered questions and potentially fertile opportunities for future research.
Genetic Analysis of C. elegans MicroRNA Function
Much of what is known about microRNA function in C. elegans is derived from studies of microRNA gene mutants (Table 1, Table 2, and Table 3). Forward genetic screens identified lin-4 and let-7 based on the developmental abnormalities caused by single-gene knockout mutations (Chalfie et al. 1981; Ferguson and Horvitz 1985; Reinhart et al. 2000). Discovery of lin-4 and let-7 mutations with visible phenotypes enabled the identification of the gene products of lin-4 (Lee et al. 1993) and let-7 (Reinhart et al. 2000) as microRNAs: short, 21–22 nt RNAs processed from longer hairpin precursors. Classical genetic analysis (rather than the more promiscuous genome-scale mRNA target prediction programs) was also used to assign these microRNA genes to genetic pathways. Phenotype suppression genetics or epistasis analysis enabled the discovery of protein-coding mRNA targets of these microRNAs (Ambros 1989; Reinhart et al. 2000; Slack et al. 2000). These genetically discovered target mRNAs bore complementarity to the upstream microRNA (Lee et al. 1993; Wightman et al. 1993) and were regulated at the level of translation (Wightman et al. 1993; Olsen and Ambros 1999; Stadler et al. 2012) or mRNA stability (Bagga et al. 2005). Dozens of other microRNA genes in C. elegans were subsequently identified by cDNA cloning (Lau et al. 2001; Lee and Ambros 2001). Their functions were tested by generating strains that were singly or multiply mutant for these microRNAs (Miska et al. 2007). These reverse genetics studies led to the realization that microRNAs, with lin-4 and let-7 being notable exceptions, often function redundantly with members of the same microRNA family (Abbott et al. 2005) or other microRNA families (Brenner et al. 2010).
Table 1. Genetically-defined functions of C. elegans microRNA genes.
| Conserved family | MicroRNA | Function | Target(s) | References |
|---|---|---|---|---|
| mir-125 | lin-4 | Developmental timing | lin-14; lin-28 | Chalfie et al. (1981)a Ambros (1989)a Lee et al. (1993); Moss et al. (1997) |
| Postdauer developmental timing | hbl-1 | Karp and Ambros (2012)a | ||
| Dauer formation | lin-14 | Liu and Ambros (1989)a | ||
| Vulva fate patterning | lin-14 | Li and Greenwald (2010)a | ||
| HSN axon extension | lin-14; lin-28 | Olsson-Carter and Slack (2010)a | ||
| Axon guidance | lin-14 | Zou et al. (2012)a | ||
| Life span | lin-14 | Boehm and Slack (2005)a | ||
| Energy homeostasis | lin-14 | Dowen et al. (2016)a | ||
| mir-237 | Radiation sensitivity | jun-1 | Metheetrairut et al. (2017)a | |
| let-7 family | let-7 | Developmental timing | lin-41; hbl-1; daf-12 | Reinhart et al. (2000)a, Slack et al. (2000); Abrahante et al. (2003), Lin et al. (2003); Grosshans et al. (2005) |
| Hypodermal cell fate, vulva integrity | opt-2; prmt-1; T27D12.1; lin-41 | Reinhart et al. (2000)a, Hunter et al. (2013); Hunter et al. (2013); Hunter et al. (2013); Slack et al. (2000), Ecsedi et al. (2015) | ||
| Axon regenerative capacity | lin-41 | Zou et al. (2013)a | ||
| Nucleolar size | ncl-1 | Yi et al. (2015)a | ||
| Life span | akt-1/2 | Ren and Ambros (2015)a, D. Wang et al. (2017) | ||
| Survival on P. aeruginosa | sdz-24 | Ren and Ambros (2015)a, Zhi et al. (2017) | ||
| Energy homeostasis | lin-41 | Dowen et al. (2016)a | ||
| mir-84 | Motor neuron connectivity | hbl-1 | Thompson-Peer et al. (2012)a | |
| let-7, mir-84 | Molting cycle exit | nhr-23; nhr-25 | Hayes et al. (2006)a | |
| Vulva integrity | let-60 | Johnson et al. (2005)a | ||
| mir-48, mir-84, mir-241 | Developmental timing | hbl-1; daf-12 | Abbott et al. (2005)a; Hammell et al. (2009a) | |
| Dauer formation | daf-12; hbl-1 | Hammell et al. (2009a)a; Karp and Ambros (2011) | ||
| Life span | Ren and Ambros (2015)a | |||
| Survival on P. aeruginosa | skn-1 | Liu et al. (2013)a, Ren and Ambros (2015) | ||
| lsy-6 | ASE left/right specification | cog-1 | Johnston and Hobert (2003)a |
Where there is more than one target and more than one reference, references are listed in the order of the targets in the preceding column. HSN, hermaphrodite-specific neuron.
Denotes the reference(s) that first reported the function.
Table 2. Genetically-defined functions of C. elegans microRNA genes.
| Conserved family | MicroRNA | Function | Target(s) | References |
|---|---|---|---|---|
| mir-1 | mir-1 | Gonadal morphogenesis | Brenner et al. (2010)a | |
| Synaptic function | unc-29; unc-63; mef-2 | Simon et al. (2008)a | ||
| mir-34 | mir-34 | Dauer formation | daf-16 | Isik et al. (2016)a |
| Gonadal morphogenesis | cdc-42; pat-3 | Burke et al. (2015)a | ||
| Life span | atg-9 | Yang et al. (2013)a | ||
| Heat and oxidative stress resistance | Yang et al. (2013)a | |||
| nc | DNA damage response | Kato et al. (2009)a | ||
| mir-35-42 | Embryonic development | Alvarez-Saavedra and Horvitz (2010)a | ||
| Developmental apoptosis | egl-1 | Sherrard et al. (2017)a | ||
| Fecundity | sup-26 | McJunkin and Ambros (2014)a | ||
| Sex determination | nhl-2; sup-26 | McJunkin and Ambros (2017)a | ||
| Embryonic hypoxic stress resistance | sup-26 | Kagias and Pocock (2015)a | ||
| mir-100 | mir-51-56 | Pharyngeal development | cdh-3 | Shaw et al. (2010)a, Alvarez-Saavedra and Horvitz (2010)a |
| Regulation of microRNA activity | Brenner et al. (2012)a | |||
| nc | mir-57 | Posterior patterning | nob-1 | Zhao et al. (2010)a |
| Embryonic viability | Brenner et al. (2012)a | |||
| mir-58 family (bantam in Drosophila) | mir-58, mir-80-82, mir-1834, mir-2209 | Developmental apoptosis | egl-1 | Sherrard et al. (2017)a |
| Dauer formation | daf-1; daf-4; sta-1 | Alvarez-Saavedra and Horvitz (2010)a, de Lucas et al. (2015); de Lucas et al. (2015); Lozano et al. (2016) | ||
| Body size | dbl-1; sma-6; daf-4; | Alvarez-Saavedra and Horvitz (2010)a; de Lucas et al. (2015); de Lucas et al. (2015) | ||
| Timing of egg laying | Alvarez-Saavedra and Horvitz (2010)a | |||
| Locomotion | Alvarez-Saavedra and Horvitz (2010)a | |||
| Life span | cbp-1 | Vora et al. (2013)a | ||
| Tissue specificity of immune response | pmk-2 | Pagano et al. (2015)a | ||
| nc | mir-59 | Embryonic viability | Brenner et al. (2012)a | |
| Adult viability | Brenner et al. (2012)a | |||
| Gonadal morphogenesis | Brenner et al. (2012)a |
Where there is more than one target and more than one reference, references are listed in the order of the targets in the preceding column.
Denotes the reference(s) that first reported the function.
In the first column, nc denotes microRNAs that are not members of well conserved seed families.
Table 3. Genetically-defined functions of C. elegans microRNA genes.
| Conserved family | MicroRNA | Function | Target(s) | References |
|---|---|---|---|---|
| nc | mir-60 | Oxidative stress | zip-10 | Kato et al. (2016)a |
| nc | mir-61 | Vulva development | vav-1 | Yoo and Greenwald (2005)a |
| mir-64-66, mir-229 | Heat stress | Nehammer et al. (2015)a | ||
| nc | mir-67 | Avoidance of P. aeruginosa | sax-7 | Ma et al. (2017)a |
| mir-70 | Survival on P. aeruginosa | Kudlow et al. (2012)a | ||
| mir-71 | mir-71 | L1 diapause survival | age-1; unc-31 | Zhang et al. (2011)a |
| Post L1 diapause developmental timing | hbl-1; lin-42 | Zhang et al. (2011)a | ||
| AWC left/right specification | tir-1 | Hsieh et al. (2012)a | ||
| Life span | de Lencastre et al. (2010)a, Boulias and Horvitz (2012) | |||
| Heat stress | Nehammer et al. (2015)a | |||
| nc | mir-73-74 | Adult viability | Brenner et al. (2010)a | |
| nc | mir-79 | Neuronal migration | sqv-5; sqv-7 | Pedersen et al. (2013)a |
| mir-29 | mir-83 | Gonadal morphogenesis | cdc-42; pat-3 | Brenner et al. (2010)a, Burke et al. (2015); Burke et al. (2015) |
| mir-124 | mir-124 | Dauer formation | Than et al. (2013)a | |
| Gonadal morphogenesis | Brenner et al. (2010)a | |||
| nc | mir-228 | Embryonic viability | Brenner et al. (2010)a | |
| nc | mir-233 | Survival on P. aeruginosa | sca-1 | Dai et al. (2015)a |
| nc | mir-234 | Dauer formation | Than et al. (2013)a | |
| mir-92 | mir-235 | Adult viability | Brenner et al. (2010)a | |
| L1 diapause arrest | nhr-91 | Kasuga et al. (2013)a | ||
| nc | mir-238 | Nicotine signaling | acr-19 | Rauthan et al. (2017)a |
| nc | Life span | de Lencastre et al. (2010)a | ||
| nc | mir-239 | Life span | de Lencastre et al. (2010)a | |
| nc | mir-246 | Life span | de Lencastre et al. (2010)a | |
| nc | mir-251, mir-252 | Survival on P. aeruginosa | Kudlow et al. (2012)a | |
| nc | mir-259 | Gonadal morphogenesis | Brenner et al. (2010)a | |
| mir-273 | ASE left/right specification | die-1 | Chang et al. (2004)a | |
| mir-365 | mir-786 | Defecation cycle length | elo-2 | Miska et al. (2007)a, Kemp et al. (2012) |
| mir-791 | CO2 sensing | akap-1a; cah-3b | Drexel et al. (2016)a |
Where there is more than one target and more than one reference, references are listed in the order of the targets in the preceding column.
Denotes the reference(s) that first reported the function.
In the first column, nc denotes microRNAs that are not members of well conserved seed families.
The findings from C. elegans genetics studies suggest a classification of microRNAs into two broad functional classes. One class includes lin-4 and let-7, which control developmental switches, where a single major microRNA regulates the expression of a single major target. Single-gene mutations in these microRNAs cause visible phenotypes. The second class encompasses most of the other C. elegans microRNAs and exerts redundant and/or conditional functions in the context of developmental or physiological robustness. These microRNAs generally act in conjunction with other microRNAs and can act on multiple targets.
Heterochronic microRNAs and larval development
The first microRNAs to be identified were the products of the C. elegans genes lin-4 (Lee et al. 1993) and let-7 (Reinhart et al. 2000). These microRNAs emerged from classical Mendelian genetic analysis of strains that had relatively rare recessive mutations, and exhibited visible defects in egg laying or developmental timing (or heterochrony) (Chalfie et al. 1981; Ambros and Horvitz 1984, 1987). For example, lin-4(e912) was identified by its unusual adult morphology and egg-laying defects in homozygous, mutant hermaphrodites. The primary targets of lin-4 and let-7 were identified as the protein-coding genes lin-14 and lin-41, respectively, by genetic epistasis and by examining their roles in developmental timing (Ambros 1989). For example, lin-14 loss-of-function (lf) mutations cause precocious expression of L2 and later cell fates, which is in contrast to the reiterated L1 phenotype of lin-4(lf). Importantly, in double mutants, lin-14(lf) suppresses lin-4(lf) phenotypes, consistent with a role of lin-4 in repression of lin-14 activity to control transitions from L1 to later cell fates. Similarly, lin-41(lf) causes precocious adult fates, while let-7(lf) causes reiteration of the L4 and delay of adult fates. Moreover, lin-41(lf) is epistatic to let-7(lf), consistent with negative regulation of lin-41 by let-7 (Slack et al. 2000).
The identification of lin-14 as the direct target of lin-4 originally emerged from the discovery of evolutionarily conserved base-pairing complementarity between lin-4 and lin-14 3′-UTR sequences (Lee et al. 1993; Wightman et al. 1993). Likewise, there are conserved sites complementary to lin-4 in the 3′-UTR of another heterochronic gene target, lin-28 (Moss et al. 1997), and sites complementary to let-7 in the 3′-UTR of its direct target lin-41 (Slack et al. 2000). The pattern of predicted base pairing of lin-4 and let-7 to their respective targets is characterized by conserved complementarity of the 5′ nucleotides of the microRNA. In particular, nucleotides 2–8, now named the “seed” region, demonstrate significant conservation, with incomplete, variable pairing in the more 3′ regions of the microRNA, especially in the case of let-7 and its mRNA targets. This foreshadowed the principle of seed-pairing that is now recognized as an organizing principle of animal microRNA function and evolution.
The realization that the let-7 microRNA sequence is deeply conserved across animal phylogeny, including in humans, (Pasquinelli et al. 2000) triggered a search for other microRNAs in C. elegans, (Lau et al. 2001; Lee and Ambros 2001) in Drosophila, and in mammalian cells (Lagos-Quintana et al. 2001). The advent of protocols for the specific prospecting of 20–25-nt RNAs and deep sequencing technologies suited for short-insert libraries enabled the rapid expansion of microRNAs from a C. elegans cottage industry to a global effort, encompassing essentially all plant and animal experimental systems. It soon became clear that, in addition to let-7, many microRNAs are evolutionarily conserved, with highly conserved seed regions that define families of microRNA genes of common evolutionary origin. It also suggested that the seed region comprises a functional domain of microRNAs that is primarily responsible for the specificity of microRNA–target recognition. Although certain microRNAs, exemplified by let-7 (Pasquinelli et al. 2000), are well conserved over their entire ∼22 nt length, other conserved microRNAs, such as lin-4 (mir-125 in other animals) preserve only the seed region. This suggests that certain microRNAs have been under more complex evolutionary constraints than others. However, the nature of these constraints is still not understood.
lin-4 and let-7 regulate a range of stage-specific developmental events across diverse tissues, and the phenotypes of lin-4 or let-7 mutants include altered timing of expression of stage-specific genes (Liu et al. 1995; Slack et al. 2000). GFP reporters driven by the promotor of the adult-specific collagen gene col-19 have been used to screen for heterochronic mutants, and to quantify the expression of precocious and retarded hypodermal adult fates (Abrahante et al. 2003). In addition, stage-specific expression of yolk proteins and other energy carriers by the intestine, and their transport to the germline upon the initiation of adulthood, is of particular significance to reproduction. This program of intertissue transport of energy reserves from the soma to the germline is regulated by lin-4 and let-7, acting via downstream heterochronic genes in the hypodermis (Dowen et al. 2016).
Heterochronic microRNA pathways impact development of the vulva; for example, lin-4 is required for the proper expression of the Vulval Precursor Cell (VPC) fate in the L2 stage (Chalfie et al. 1981; Euling and Ambros 1996), for the specification of certain VPC progeny cell fates (Li and Greenwald 2010), and let-7 is critical for the proper morphogenesis and structural integrity of the vulva (Johnson et al. 2005; Ecsedi et al. 2015).
Additional microRNAs function within the complex signaling networks that regulate vulval cell fate specification; for example, lin-12/Notch signaling in presumptive vulval secondary cells triggers the expression of mir-61, which in turn represses vav-1, a Vav oncogene ortholog that opposes lin-12 activity (Yoo and Greenwald 2005). Thus, mir-61 functions in a feedback loop with lin-12 and vav-1 to reinforce the specification of secondary vulval fates.
Functional redundancy within microRNA seed families
The assignment of mRNA targets to microRNAs identified by deep sequencing of animal small RNAs has been haunted by the hundreds of potential targets predicted by computational approaches (Lewis et al. 2005; Agarwal et al. 2015). The loops and base mismatches characteristic of genetically discovered and validated microRNA–mRNA interactions (Wightman et al. 1993; Ha et al. 1996; Slack et al. 2000; Ecsedi et al. 2015) confound the accurate prediction of animal microRNA targets. By contrast, plant microRNAs, which generally perfectly base pair along their entire 21–24 nt to target mRNAs, can be easily assigned to particular mRNA targets, and hence to particular pathways (Rhoades et al. 2002). The genome-wide identification of C. elegans microRNAs, many of which, like lin-4 and let-7, are also evolutionarily conserved, suggested that the functions of these microRNAs have been under strong selection for the billion-year history of animals. It was assumed that such conserved microRNAs were likely to have conserved functions that could be revealed by genetic analysis in C. elegans. Surprisingly, most microRNA single-gene mutants, including for those that are conserved in phylogeny, displayed no readily evident phenotypes (Miska et al. 2007). Therefore, lin-4 and let-7 were essentially the only C. elegans microRNA genes for which single-gene mutations caused visible phenotypes, which partially explains why only these two microRNA genes had been previously cloned from genetically identified loci [although the nonconserved lsy-6 microRNA and its target mRNA cog-1 did emerge from genetic analysis of neural development (Johnston and Hobert 2003)].
For some single-microRNA gene mutants, the lack of visible phenotypes can be attributed to genetic redundancy among microRNAs of the same seed family. In a systematic genetic analysis of 15 of the 23 microRNA families in C. elegans (Alvarez-Saavedra and Horvitz 2010), mutant strains were generated that lacked most or all members of a given microRNA seed family. For 12 of these families, full family knockout caused no strong observable synthetic phenotypes. For two families, the mir-35 family (mir-35-42) and the mir-51 family (mir-51-56), synthetic embryonic arrest phenotypes resulted from knockout of the entire family, and for the mir-58 family (mir-58.1, -58.2, -80, -81, -82, -1834, -2209a, -2209b, and 2209c), deletion of multiple paralogs caused a complex syndrome of morphological and behavioral defects (Alvarez-Saavedra and Horvitz 2010).
Similarly, animals multiply-mutant for the let-7 paralogs (mir-48, mir-84, and mir-241) exhibit heterochronic phenotypes characterized by repetition of the L2 cell fate programs (Abbott et al. 2005). By examining other combinations of mutations in the let-7 family microRNAs, other developmental timing functions for this family emerged. These functions include the regulation of the timing of exit from the L4-to-adult molt by the action of mir-84 and let-7 on their targets, the nuclear hormone receptor transcription factors (TFs) nhr-23 and nhr-25 (Hayes et al. 2006).
Thus, among a sample of 15 microRNA families in C. elegans, four families (let-7, mir-35, mir-51, and mir-58) are associated with phenotypes resulting from the deletion of multiple members of the family. What about the other 11 of these families, for which complete genetic deletion of all members of the family did not uncover detectable phenotypes (Alvarez-Saavedra and Horvitz 2010)? Perhaps these could represent microRNAs whose functions depend on particular physiological or stress conditions (see Longevity, stress responses, and stress robustness below), and/or they may function redundantly with microRNAs of other families (see Sensitized backgrounds uncover cryptic microRNA functions below).
Sensitized backgrounds uncover cryptic microRNA functions
One explanation for the apparent lack of visible phenotypes for microRNA gene deletion mutants, besides the functional redundancy among microRNAs of the same family discussed above, emerged from studies designed to uncover otherwise cryptic microRNA functions using sensitized genetic backgrounds (Brenner et al. 2010). A significant finding from this study is that many C. elegans microRNAs functionally interact with microRNAs of other seed families. For example, for at least six microRNAs of distinct seed families, single-gene knockout caused gonad migration defects in an alg-1(0) background, where microRNA activity was broadly compromised, owing to loss of one of the two microRNA-specific Argonautes (ALG-1 and ALG-2) (Brenner et al. 2010). This suggests that these microRNAs may functionally interact with each other and/or with other microRNAs in regulating pathways related to the program of gonadal morphogenesis. The roles of microRNAs in gonadal morphogenesis was not previously appreciated. Based on the findings that deletion of either mir-34 or mir-83 (the C. elegans ortholog of mammalian miR-29) could impact this phenotype in the alg-1(0)-sensitized background (Brenner et al. 2010), common targets of mir-34 and mir-83 were identified (Burke et al. 2015). Interestingly, these include conserved components of cell migration and cell adhesion, pat-3/integrin and cdc-42.
Synergy between unrelated microRNA families is perhaps not unexpected, considering that the 3′-UTRs of mRNAs often have multiple microRNA complementary sites. Distinct microRNA families can even interact negatively; mir-52 loss-of-function results in suppression of the phenotypes of let-7 family mutants (Brenner et al. 2012). It is not clear whether the apparent opposition between mir-52 and let-7 microRNAs is direct, for example by competition for overlapping target sites, or indirect, for example via impacting separate but opposing pathways.
Longevity, stress responses, and stress robustness
Another explanation for the apparent lack of visible phenotypes for microRNA gene deletion mutants, besides functional redundancy among microRNAs of the same family or redundancy across families, emerged from experiments designed to stress mutant animals in an effort to uncover conditional functions for the microRNAs. Investigators speculated that some microRNA mutations might yield conditional phenotypes revealed only by subjecting mutant animals to the appropriate stress regimen.
Perhaps nothing is as stressful as aging. The first microRNA found to function in longevity was lin-4, which acts via its major downstream heterochronic gene target lin-14 to promote normal life span, at least in part by engaging the daf-16 and hsf-1 transcriptional programs (Boehm and Slack 2005). Similarly, let-7 family microRNAs seem to be integrated into pathways affecting fertility and longevity (Ren and Ambros 2015; D. Wang et al. 2017).
Evidence that other microRNAs could function in regulating life span came from sensitized genetic backgrounds, including pash-1(ts) mutants (carrying a weak mutation in the microRNA maturation factor gene pash-1 that affects all microRNAs) shifted to the nonpermissive temperature during adulthood (Lehrbach et al. 2012), or from animals depleted for alg-1 specifically during adulthood (Kato et al. 2011), where life span was shortened, presumably due to the compromised microRNA activity in these mutants (it should be noted that a standard caveat applies regarding shortened-life span phenotypes, wherein the genetic lesion may not identify a regulator of longevity per se, but rather could partially disable a pathway essential for robust health.)
Candidates for specific microRNAs that could control adult life span were identified by profiling microRNAs during adulthood to identify those whose levels change with age (Ibáñez-Ventoso et al. 2006; de Lencastre et al. 2010). Examples of specific microRNA genes where deletion mutations impact life span include mir-71, mir-238, mir-239.1, mir-239.2, and mir-246 (de Lencastre et al. 2010). An independent systematic survey of microRNA mutants for life span defects, coupled with mosaic analysis tests for cell autonomy, identified a strong role for mir-71 function in neurons in regulating normal adult life span (Boulias and Horvitz 2012).
A classic mode of regulating longevity is by dietary restriction (DR). One such DR model is the C. elegans mutant eat-2(ad1116), which is defective in eating. Profiling of microRNAs in eat-2(ad1116) adults compared to wild-type uncovered sets of microRNAs whose expression, and hence function, could be linked to DR-regulated longevity (Pandit et al. 2014). In another study, deletion of the microRNA mir-80 induced DR-like phenotypes, including extended longevity via its regulation of cbp-1/CREB-binding protein mRNA translation (Vora et al. 2013).
mir-34 is an evolutionarily conserved microRNA with multiple functions in C. elegans. These functions include regulation of life span (Yang et al. 2013), and conferring robustness against physiological and developmental challenges, including dauer formation (Isik et al. 2016). Roles for mir-34 in dauer formation were revealed by examination of the morphology and measuring the survival capacity of mir-34 mutant larvae. In this context, an interesting DAF-16-mir-34 feedback loop appears to mediate robustness of the dauer larva program (Isik et al. 2016).
mir-34 plays an evolutionarily conserved function in DNA damage responses. Similar to mammalian cells, where mir-34 is upregulated in response to radiation-induced DNA damage, C. elegans mir-34 is induced after irradiation; however, unlike in mammalian cells where irradiation induction of mir-34 requires p53 (Rokavec et al. 2014), C. elegans mir-34 induction is independent of cep-1 (which is considered to be a functional p53 ortholog despite relatively weak sequence homology). Even without p53 involvement, the mir-34 mutant C. elegans displays abnormal survival of somatic and germline cells after irradiation, consistent with mir-34 functioning to regulate apoptotic and nonapoptotic cell death, possibly in parallel to cep-1/p53 (Kato et al. 2009). Another radiation sensitivity phenotype was found for mutants of mir-237, the only other member of the lin-4 family in C. elegans (Metheetrairut et al. 2017).
A role for mir-34 in developmental robustness against stress emerged from studies of genetic interactions between mir-34 and mir-83 (see Sensitized backgrounds uncover cryptic microRNA functions). The relatively mild penetrance of gonad migration defects in mir-34; mir-83 double mutants was dramatically increased by cycling the temperature of developing larvae between temperatures within the worm’s normal temperature range (for example 15 and 25°C). Constant temperature throughout development did not affect the mir-34; mir-83 phenotype, indicating that this mutant appears to be sensitive specifically to changing environmental temperature, suggesting that mir-34 functions with mir-83 to maintain the robustness of gonadal migratory morphogenesis against the stress of unstable temperature (Burke et al. 2015).
Certain C. elegans microRNA mutants were tested in the context of heat stress and functions were identified for several microRNAs, including mir-71, as regulators of the heat stress response (Nehammer et al. 2015). Worms subjected to stress caused by benzo-α-pyrene (Wu et al. 2015) or graphene oxide (Wu et al. 2014) exhibited altered expression of certain sets of microRNAs and, in the latter case, worms with mutations in the genes for some of these microRNAs exhibited altered tolerance to graphene oxide stress. Likewise, mir-35-41 mutant embryos were found to exhibit hypersensitivity to hypoxia stress (Kagias and Pocock 2015) and mir-60 mutants exhibit a dysregulated adaptive response to oxidative stress (Kato et al. 2016).
Studies of the response of C. elegans to pathogen stress have uncovered roles for microRNAs in regulating innate immune pathways. Using a sensitized genetic background, phenotypic evidence emerged for the involvement of microRNAs in regulating the C. elegans antibacterial pathogen response, and the characterization of microRNAs identified by co-immunoprecipitation (co-IP) with the microRNA-Induced Silencing Complex (miRISC) identified candidate pathogen-responsive microRNAs (Kudlow et al. 2012). Mutants of either miR-70 or miR-251/miR-252 showed enhanced survival on Pseudomonas aeruginosa compared to wild-type worms, indicating a negative regulation of immune responses by these microRNAs (Kudlow et al. 2012). mir-233 mutants are more sensitive to infection than wild-type worms, apparently through dysregulation of the unfolded protein response (Dai et al. 2015). mir-67 mutants exhibited reduced pathogen avoidance behavior, apparently from the derepression of the mir-67 target, sax-7 (Ma et al. 2017). Mutations in the microRNA and small interfering RNA (siRNA) maturation factor dcr-1/Dicer confer sensitivity in C. elegans to the Bacillus thuringiensis pathogen, suggesting that microRNAs mediate immunity to the Cry toxins of B. thuringiensis (Iatsenko et al. 2013).
The heterochronic microRNAs, whose functions are primarily the control of developmental cell fates (Heterochronic microRNAs and larval development), have also been found to affect stress responses. let-7 family microRNA mutants were found to exhibit either positive or negative effects on resistance to P. aeruginosa, suggesting a delicate temporal modulation of innate immune pathways in the worm (Ren and Ambros 2015). The activity of let-7 in modulating the innate immune response to P. aeruginosa infection was shown to occur in the intestine, via regulation of SDZ-24-mediated signaling (Zhi et al. 2017). Other candidate targets of let-7 and let-7 family microRNAs for innate immune modulation may include components of the pmk-1/p38 innate immune pathway (Ren and Ambros 2015). No doubt the heterochronic microRNAs will likely be found to have additional roles in the modulation of various aspects of cellular physiology; one example is the regulation of ribosome biogenesis through the repression of ncl-1 by let-7 (Yi et al. 2015).
L1 diapause and dauer larva arrest
L1 larvae that hatch in the absence of food enter a developmentally arrested diapause stage that can survive for many days, and then reinitiate postembryonic development upon encountering food. A screen for microRNA gene mutations that perturb the ability of newly hatched larvae to enter the L1 diapause identified mir-235, the C. elegans homolog of the mammalian oncogenesis-associated microRNA mir-92 (Kasuga et al. 2013). mir-235 mutants fail to properly arrest development when hatched in the absence of food. mir-235 expression is regulated by insulin/IGF signaling, such that mir-235 is elevated during L1 diapause and declines upon feeding. mir-235 seems to act in several major tissues of L1 larvae to inhibit postembryonic developmental programs in the absence of food. Loss of mir-235 causes increased expression of its target nhr-91, a nuclear hormone receptor gene.
mir-71 was identified as being critical for L1 diapause animals to properly develop after feeding (Zhang et al. 2011). Interestingly, mir-71 mutants that did recover from L1 starvation often displayed retarded VPC divisions, similar to mutants that are defective in the regulation of lin-14 and other heterochronic genes. These results indicate that mir-71 contributes to the regulation of heterochronic pathway genes, perhaps in a fashion that is coupled to the stress of starvation and L1 diapause. In this context, it is noteworthy that several of the key heterochronic gene mRNAs, including lin-42 and hbl-1, contain mir-71 complementary sites in their 3′-UTRs.
mir-58 family microRNAs are redundantly required for dauer larva formation (Alvarez-Saavedra and Horvitz 2010). Other microRNAs were placed in dauer larva formation genetic pathways using a combination of genetic and biochemical strategies. Sensitized genetic backgrounds designed to compromise microRNA activity in the nervous system yielded phenotypic evidence for multiple dauer-regulating microRNAs (Than et al. 2013). Tissue-specific immunoprecipitation (IP) of miRISC identified the neuronally expressed microRNAs mir-80/81, mir-124, and mir-234, whose mutant phenotypes were subsequently determined to include effects on dauer formation, likely through multiple targets in the dauer regulatory pathways (Than et al. 2013). Similarly, mir-58 family microRNAs regulate specific target genes in the TGF-β dauer as well as TGF-β body size regulatory cascades, including dbl-1, daf-1, daf-4, sma-6 (de Lucas et al. 2015), and sta-1 (Lozano et al. 2016).
Heterochronic microRNAs also regulate dauer formation. lin-4 mutants are completely unable to form dauer larva due to the overexpression of lin-14, which is a potent regulator of the timing of dauer formation (Liu and Ambros 1989). lin-14 activity in the L1 stage prevents early dauer formation and, accordingly, the downregulation of lin-14 by lin-4 is critical for dauer formation to be permitted at the normal time, at the end of the L2 stage. let-7 family microRNAs also affect the decision to undergo dauer formation by modulating the levels of DAF-12 and HBL-1 proteins, suggesting that the upregulation of let-7 family microRNAs during the L2 stage may modulate the temporal response of the dauer entry program according to environmental signals (Hammell et al. 2009a; Karp and Ambros 2011).
In animals that develop through the dauer larval stage, microRNA pathways are reprogrammed in interesting ways. The temporal profile of expression of certain microRNAs is altered in L2 animals entering the dauer stage (“L2D” larvae), and in L3 and L4 animals developing after dauer arrest (“postdauer” larvae), compared to continuously developing larvae (Karp et al. 2011). Moreover, the relative functional contributions of lin-4 and let-7 family microRNAs to developmental cell fate specification are altered for postdauer development compared to continuous development (Karp and Ambros 2012).
Embryonic development
The mir-35-42 family of microRNAs are maternally contributed to the early embryo, expressed in the zygote shortly after fertilization (Wu et al. 2010), and contribute redundantly to embryonic development and viability. The precise nature of the essential functions of mir-35-42 are unknown and appear to be complex (Alvarez-Saavedra and Horvitz 2010). mir-35 family mutants exhibit diverse pleiotropic phenotypes at embryonic and postembryonic stages, suggesting functions for these microRNAs in multiple pathways. Among the characterized early embryonic functions of mir-35-42 is a role in sex determination, wherein these microRNAs act by regulating a set of RNA-binding protein targets to prevent the premature expression of the male developmental program in XX embryos (McJunkin and Ambros 2017). In this capacity, mir-35-42 serves as a sort of “timer” to delay sex determination until after the proper reading of the zygotic X/A ratio. It is possible that some of the essential functions of mir-35-42 in the early embryo could include analogous roles in preventing premature expression of other, “late” developmental programs. The fact that the mir-35-42 family microRNAs are downregulated during mid embryogenesis is consistent with the model that they may broadly control early-to-late developmental transitions in the embryo.
The mir-35 family microRNAs also act, together with mir-58/bantam microRNAs, to prevent inappropriate expression of the EGL-1 proapoptotic protein in certain embryonic cell lineages. In particular, these two microRNA families cooperate to target the egl-1 mRNA in the mothers of cells programmed to die, thereby preventing precocious apoptosis (Sherrard et al. 2017).
Another abundant microRNA family expressed in the worm embryo is the mir-51 family, the worm homolog of the deeply conserved miR-100. The mir-51 family functions redundantly with the mir-35 family to regulate embryonic viability (Alvarez-Saavedra and Horvitz 2010) and pharyngeal morphogenesis (Shaw et al. 2010). The pleiotropic phenotypes of mutants of the mir-35 family or the mir-51 family indicate that these abundant early embryo microRNAs are engaged with multiple essential developmental pathways. Interestingly, the mir-35 family microRNAs are relatively specific for the early embryo and are relatively nematode-specific compared to the mir-51 family, which are abundant in C. elegans larvae as well as in embryos, and are broadly conserved evolutionarily. mir-51/mir-100 may function in diverse and conserved genetic regulatory contexts, while mir-35-42 may be adapted for coping with gene regulatory challenges that are more particular to nematodes.
Germline development
C. elegans germline development and gametogenesis appear to be impacted by microRNA genes, although there is clearly much more to be learned about germline functions of microRNAs in the worm. Characterization of the phenotypes resulting from the depletion of ALG-1 and ALG-2 from the somatic distal tip cells (DTCs) suggests cell nonautonomous roles for microRNAs in processes where signals from the DTCs regulate the germline cell cycle and proliferation (Bukhari et al. 2012). Whether or not microRNAs expressed within the germline function cell-autonomously is less clear. Small RNA cDNA sequencing has identified over a dozen microRNAs that are enriched in the germline, including prominently the mir-35-42 family (McEwen et al. 2016). Although it is clear that the maternal contribution of mir-35-42 can affect embryonic viability (Alvarez-Saavedra and Horvitz 2010), it is not yet established whether these roles for maternally expressed mir-35-42 include the repression of targets within the maternal germline itself or only after deposition in the embryo. Among the postembryonic phenotypes of mir-35-42 mutants are defects in hermaphrodite fecundity, owing at least in part to impaired spermatogenesis (McJunkin and Ambros 2014). This function of mir-35-42 could be the result of a combination of germline and/or somatic gonad activity of these microRNAs.
An apparent direct function for microRNAs within the germline is suggested from the phenotype of loss-of-function mutants for ALG-5, a microRNA-associated Argonaute that is expressed primarily in the germline of hermaphrodites. alg-5(lf) mutants exhibit reduced fertility and a precocious developmental switch from spermatogenesis to oogenesis (Brown et al. 2017).
Neural development and behavior
One of the first C. elegans microRNAs that was found to affect behavior is mir-786, deletion mutations of which display abnormally long intestinal defecation cycles (Miska et al. 2007). Detailed genetic analysis showed that mir-786 regulates the expression of the fatty acid elongase, elo-2, in intestinal cells, and thereby ensures the proper rhythmic behavior of those cells in their role as pacemakers for the defecation cycle (Kemp et al. 2012).
mir-1 is an evolutionarily conserved muscle-expressed microRNA whose function in C. elegans was not apparent at first, as mir-1 was among those microRNAs for which mutants had no apparent defects. mir-1 mutant phenotypes identified from genetically sensitized screens (Brenner et al. 2010) have not yet been investigated in depth, but a window into mir-1 function in C. elegans was opened by challenging mir-1 mutants pharmacologically (Simon et al. 2008). mir-1 mutants show altered acetylcholine sensitivity and, based on that phenotype, roles for mir-1 were uncovered in controlling muscle–neuronal signaling at the neuromuscular junction. Another microRNA implicated in the regulation of neuromuscular signaling is mir-238; upregulation of the nicotinic acetylcholine receptor acr-19 during chronic exposure of C. elegans to nicotine was traced to a downregulation of mir-238, which was found to directly target acr-19 (Rauthan et al. 2017).
The formation of specific neurons and neuronal connections is coordinated with positional and temporal information in the developing worm. Certain microRNAs, including mir-54 and mir-56, have been implicated in the regulation of the Hox gene egl-5 in the context of specifying the posterior pattern of male sensory rays (Zhang and Emmons 2009). Neuronal development in response to temporal cues is exemplified by the hermaphrodite-specific neuron (HSN), which extends its axon in the L4 stage. The developmental timing microRNA lin-4 is critical for specifying the timing of HSN axon outgrowth through the developmental downregulation of two targets, lin-14 and lin-28, which inhibit HSN differentiation (Olsson-Carter and Slack 2010).
Another role for lin-4 in controlling the timing of steps in the outgrowth and migratory behavior of axons occurs for the anterior ventral microtubule (AVM) neurons, where lin-4 acts cell autonomously in AVM neurons to promote the proper formation of AVM connections, apparently by repressing its target LIN-14 and thereby terminating AVM axon migration (Zou et al. 2012). Another microRNA affecting neuronal migration is mir-79, which functions in the epidermis to control the properties of the extracellular matrix (Pedersen et al. 2013). MicroRNAs can also regulate the capacity of neurons to regenerate after injury, as exemplified by a role for let-7 in the developmental decline of AVM axon regeneration (Zou et al. 2013).
Mutants of the let-7 family microRNA mir-84 display defects in the stage-specific rewiring of the dorsal D (DD) motor neuron in the L1 larval stage, due to dysregulation of heterochronic genes including hbl-1, an apparent mir-84 target (Thompson-Peer et al. 2012). The heterochronic gene lin-14 also controls the timing of DD rewiring (Hallam and Jin 1998), although curiously it is not clear whether microRNAs that could target lin-14 (which include the let-7 family as well as lin-4) may act via lin-14 to participate in regulating the stage-specificity of DD rewiring.
Developmental decisions between alternative neuronal subtype fates often occur in response to the activity of developmental signals. In many cases, these decisions involve precise, yet subtle, distinctions in gene activity. One such situation is the stochastic left/right specialization of the two AWC neurons. nsy-4 and nsy-5 signals act stochastically to inhibit calcium signaling asymmetrically in the pair of AWC precursor cells to produce asymmetric alternative fates, AWC(OFF) and AWC(ON). However, the mechanism of coupling nsy-4 and nsy-5 to asymmetric calcium signaling, and hence cell fate, is not understood. mir-71 was identified genetically as an integral post-transcriptional switch for specifying distinct left vs. right AWC fates (Hsieh et al. 2012). mir-71 acts as a repressor of TIR-1/Sarm1, a critical calcium signaling component, to promote the AWC(ON) identity. Tests of epistasis and cell autonomy indicate that nsy-4 and nsy-5 promote mir-71 activity in one AWC, possibly by stabilizing mature mir-71, to promote the AWC(ON) fate (Hsieh et al. 2012). Similarly, the lsy-6 and mir-273 microRNAs are deployed asymmetrically in the left ASE (ASEL) vs. the right ASE (ASER) neurons (Johnston and Hobert 2005; Cochella and Hobert 2012), and control a bimodal developmental switch that specifies the distinct chemosensory properties of ASEL and ASER (Johnston and Hobert 2003; Chang et al. 2004).
A conceptually novel perspective on microRNAs in neuronal specialization emerges from studies of the functions of microRNAs expressed at high levels in a very limited set of neurons in C. elegans. mir-791 was found to be expressed exclusively in certain CO2-sensing neurons, and was shown to confer the CO2-sensing functionality of these neurons by repressing two otherwise broadly expressed genes (Drexel et al. 2016). This mode of action, where a microRNA expressed specifically in a particular cell modulates the level of otherwise broadly expressed (even essential) genes, could underlie the elaboration of neuronal diversity in more complex nervous systems.
MicroRNAs can also function to sharpen neuronal vs. nonneuronal gene expression patterns. For example, the mir-58 microRNA family functions to restrict the expression of pmk-2/p38 to the nervous system, where it is coexpressed with its ortholog pmk-1. Consequently, pmk-1 and pmk-2 function together and redundantly in the nervous system to control pathogen avoidance behavior, while pmk-1 functions on its own in the intestine to guard against pathogen infection (Pagano et al. 2015).
Regulation of the Biogenesis, Stability, and Activity of MicroRNAs
C. elegans research has led to many of the advances in our understanding of the expression and regulation of microRNA genes, how mature microRNAs are generated from primary transcripts of microRNA genes, and how the activity of a microRNA is regulated after biogenesis (Figure 1). Forward genetic screens (for example, Ding et al. 2005) and RNA interference (RNAi) screens [for example, Parry et al. (2007) and Rausch et al. (2015)] have enabled the identification of scores of genes encoding protein factors that positively or negatively contribute to microRNA activity.
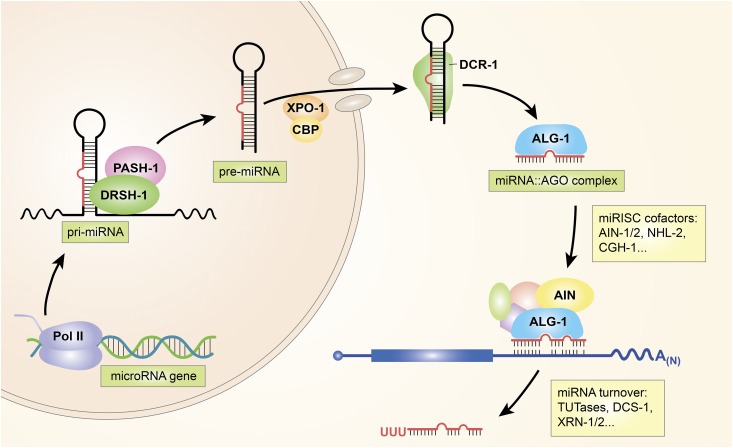
Figure 1.
MicroRNA metabolism and function in C. elegans. Current understanding of major factors involved with various steps in the transcription and processing of microRNA primary transcripts (pri-miRNA) in the nucleus (left), export of the hairpin RNA microRNA precursor (pre-miRNA) through the nuclear pore to the cytoplasm (top), processing of the pre-miRNA by Dicer/DCR-1, and loading of the mature miRNA into a core microRNA-Induced Silencing Complex (miRISC) Argonaute protein (ALG-1). Additional factors assemble with miRISC, including the general miRISC effector protein AIN-1/2. The miRISC complex binds to target mRNAs via complementary sites in their 3′-UTRs and represses protein production from the target by various mechanisms, as discussed in the text. MicroRNAs eventually undergo downregulation through processes involving 3′ terminal uridyl modifications and degradation by the cellular RNA turnover machinery (figure courtesy of Gloria Ha). Pol II, RNA polymerase II: TUTase, terminal uridyl transferase.
Genetic identification of Dicer, Argonautes ALG-1/2, and microRNA effectors AIN-1/2
The Argonaute RDE-1 emerged from a genetic screen for RNAi-defective mutants and provided the first evidence that the Argonaute class of proteins are intimately associated small RNAs (Tabara et al. 1999). Using an RNAi screen for heterochronic phenotypes similar to those caused by mutations in the microRNAs lin-4 or let-7, the Argonautes ALG-1 and ALG-2 (paralogs of RDE-1) were shown to be required for proper microRNA biogenesis and function (Grishok et al. 2001). The seminal discovery of the roles for specialized Argonautes in RNAi and microRNAs, together with the identification of microRNA-related phenotypes associated with a loss-of-function of dcr-1, the C. elegans gene encoding Dicer, cemented our understanding of the fundamental linkage between RNAi and microRNAs (Grishok et al. 2001).
There is intriguing evidence that ALG-1 and ALG-2 may not be the only C. elegans Argonautes that associate with microRNAs. Immunoprecipitation of epitope-tagged ALG-5 resulted in enrichment for a specific subset of germline microRNAs, indicating that the reduced fertility of alg-5(lf) hermaphrodites (see Germline development, above) may reflect a role for the ALG-5-associated miRISC in the germline (Brown et al. 2017). Similarly, HA-tagged RDE-1 was found to co-immunoprecipitate with a subset of microRNAs (Steiner et al. 2007; Corrêa et al. 2010), suggesting possible crossover between microRNA and RNAi pathways (see mRNA translational repression and/or mRNA turnover).
The AIN-1 and AIN-2 proteins were initially identified as suppressors of the lin-31 multivulva (Muv) phenotype and found to have more general heterochronic phenotypes (Ding et al. 2005). AIN-1 and AIN-2 function redundantly, and depletion of both proteins causes pleiotropic phenotypes consistent with general impaired microRNA activity. AIN-1 and AIN-2 are degenerate orthologs of the conserved miRISC component and microRNA activity effector GW182 (Ding et al. 2005). Tagged AIN-1 or AIN-2 can be used to immunoprecipitate miRISC from worms, and this approach has been a powerful means of identifying other miRISC-associated proteins, for profiling microRNAs associated with AIN-1 or AIN-2 in particular cell types at particular developmental stages, and for profiling mRNA targets engaged by miRISC (Zhang et al. 2007, 2009).
Transcriptional regulation of microRNA gene expression
As is the case for animals in general, some microRNAs in C. elegans are produced from a dedicated noncoding primary transcript (and therefore likely from a dedicated promoter), and other microRNAs are processed from pre-mRNAs of coding genes, so that a microRNA gene can share transcriptional regulatory sequences with one or more protein-coding genes. It is also not uncommon for several microRNAs to be expressed from the same primary transcript. For many of the C. elegans microRNAs, transcriptional regulatory sequences have been characterized using fluorescence reporter transgenes (Martinez et al. 2008b), but for several other microRNAs, primary transcript configurations and expression parameters have not been characterized.
Interestingly, in many cases, C. elegans microRNAs are located in an intron and in the sense direction relative to the host protein-coding gene, but nevertheless appear to be expressed from a dedicated intronic promoter (Lee et al. 1993; Martinez et al. 2008b). Obvious exceptions include the mirtrons, which are microRNAs whose precursor hairpins are processed out of pre-mRNA transcripts by the spliceosomal machinery, bypassing the requirement for Drosha processing (Ruby et al. 2007; Chung et al. 2011). There are at least 13 mirtrons encoded in the C. elegans genome whose expression patterns have been confirmed (Chung et al. 2011). These C. elegans mirtrons have not been well-studied genetically, and no functions have been yet ascribed to them.
In some cases, the transcriptional regulation of microRNA gene expression is coupled to developmental signals. let-7 expression is subject to complex transcriptional control (Kai et al. 2013), including temporal modulation by another heterochronic gene, hbl-1 (Roush and Slack 2009). Similarly, the expression of the let-7 family microRNAs mir-48, mir-84, and mir-241 appears to be restrained by the heterochronic TF LIN-14, such that the midlarval events triggered by those particular microRNAs, particularly the downregulation of LIN-28, are restricted to stages after the downregulation of LIN-14 (Tsialikas et al. 2017).
The heterochronic gene lin-42 encodes a Period homology protein (Jeon et al. 1999), and lin-42(lf) mutants exhibit precocious developmental timing phenotypes that appear to reflect the hyperactive transcription of certain microRNA genes, identifying LIN-42 as a transcription repressor of microRNAs that likely modulates their developmental expression (McCulloch and Rougvie 2014; Perales et al. 2014; Van Wynsberghe et al. 2014).
C. elegans is a good model for how microRNAs and TFs are organized into gene regulatory network motifs that provide feedback and/or feed forward functionality. A specific example is a motif consisting of the microRNA mir-57 and the Hox gene nob-1 (Zhao et al. 2010). nob-1 activates mir-57 expression in the posterior of the embryo, and nob-1 mRNA is also a direct target of mir-57, producing a negative feedback loop between the microRNA and the Hox gene, perhaps to sharpen positional cues in the embryo. A broader, genome-wide analysis of predicted interactions of TFs with microRNA regulatory sequences, combined with microRNA target prediction, led to the construction of a genome-scale model of TF→microRNA interactions, as well as predicted microRNA→TF interactions. More than 20 microRNA←→TF-predicted composite feedback loops were identified in C. elegans (Martinez et al. 2008a). Such mutually direct regulatory motifs containing microRNAs and TFs could help to coordinate the regulation of microRNA and TF target repertoires.
An example of a rather complex microRNA←→TF feedback motif, which acts during early larval development to integrate environmental and developmental signals, consists of a set of let-7-family microRNAs and the DAF-12 nuclear hormone receptor (Bethke et al. 2009; Hammell et al. 2009a). These microRNAs directly regulate DAF-12 levels and, in turn, their levels are transcriptionally regulated by DAF-12, which directly activates (in the presence of ligand) or represses (in the absence of ligand) transcription of the microRNA genes.
Post-transcriptional regulation of microRNA biogenesis and turnover
Much remains to be learned about how the C. elegans core microRNA biogenesis machinery (Figure 1) can be regulated to control microRNA levels in response to signals. There is evidence that microRNA biogenesis can be regulated at the level of the microprocessor complex, which consists of DRSH-1/Drosha and PASH-1/DGCR8 (Denli et al. 2004; Lehrbach et al. 2012). For example, trans-splicing of the let-7 primary transcript (pri-let-7) seems to modulate the processing of pri-let-7 by microprocessor (Mondol et al. 2015).
After release of the microRNA precursor hairpin (by microprocessor activity, in the case of conventional microRNAs, or by the spliceosome, in the case of mirtrons), subsequent steps include nuclear export of the pre-microRNA, followed by further processing by Dicer to produce the mature microRNA (Figure 1). In C. elegans, nuclear export of the majority of pre-microRNAs appears to depend on the nuclear export receptor XPO-1 and components of the cap-binding complex (CBC) (Büssing et al. 2010). Interestingly, the export of mirtrons seems to occur independently of XPO-1/CBC (Büssing et al. 2010). Little is known about potential modes of regulation of microRNA nuclear–cytoplasmic trafficking. There are indications that regulation of the Dicer (DCR-1) processing step can occur; for example, DCR-1 appears to be developmentally regulated by phosphorylation in oocytes, suggesting that some maternally deposited microRNAs may not be processed until fertilization (Drake et al. 2014).
Upon Dicer processing of the pre-microRNA, the mature microRNA is loaded into one of the principle miRISC Argonautes, ALG-1 and ALG-2 (Figure 1), or in rarer cases, into an alternative Argonaute such as ALG-5 (Brown et al. 2017) or RDE-1 (Steiner et al. 2007). Evidence that the Argonaute loading step can be regulated includes the observation that the developmental profiles of microRNAs associated with ALG-1 vs. ALG-2 differ (Vasquez-Rifo et al. 2012; Brown et al. 2017). Also, there is evidence that pre-microRNA hairpin structure can influence Argonaute loading specificity, such that certain microRNAs with precursors that have relatively few mismatches can be preferentially loaded into the (otherwise RNAi-specific) Argonaute RDE-1 (Steiner et al. 2007; Corrêa et al. 2010).
Evidence that Argonaute may actively participate in miRISC loading comes from studies of antimorphic alleles of ALG-1 that broadly impair the function of many microRNAs. ALG-1(anti) proteins show an increased association with Dicer and a decreased association with AIN-1/GW182, suggesting that these antimorphic mutations cause ALG-1 to stall in a microRNA loading state, prior to advancing to effector status. Tellingly, the alg-1(anti) mutants dramatically overaccumulate microRNA* (“star,” i.e., passenger) strands, suggesting that wild-type ALG-1 complexes recognize structural features of microRNAs in the context of the guide strand selection and passenger strand ejection steps of miRISC maturation (Zinovyeva et al. 2014, 2015).
Mechanisms involved in regulating the stability and degradation of microRNAs in C. elegans have been identified. The enzymes involved in these mechanisms include terminal uridyl transferase (Lehrbach et al. 2009), the decapping scavenger enzyme DCS-1 (Bossé et al. 2013), and the exonucleases XRN-1 and XRN-2 (Chatterjee and Grosshans 2009; Chatterjee et al. 2011; Miki et al. 2014). Interestingly, the degradation of microRNAs in worm lysates or in vivo can be modulated depending on the presence of target mRNA, consistent with the finding that microRNA homeostasis may be coupled to target recognition (Chatterjee and Grosshans 2009; Chatterjee et al. 2011).
There is evidence that the turnover of microRNAs in C. elegans could also be coupled to the turnover of miRISC protein components. The finding that microRNA-mediated gene regulation in C. elegans can be modulated by autophagy (Zhang and Zhang 2013) suggests that the degradation of miRISC components, including miRISC-bound microRNAs, could be a potent mechanism of controlling microRNA activity in response to signals that regulate autophagy. Similarly, genetic and biochemical evidence suggests that TEG-1, a conserved protein that can associate with miRISC (C. Wang et al. 2017), regulates the levels of miRISC proteins (particularly ALG-1 and VIG-1), and also regulates the levels of several microRNAs (C. Wang et al. 2017). teg-1(lf) mutants exhibit developmental defects consistent with reduced microRNA function, reinforcing the model that TEG-1 functions to stabilize miRISC complexes. Going forward, an interesting aspect of better understanding microRNA/miRISC turnover mechanisms will be to determine how signaling pathways may be coupled to the selective inactivation of miRISC complexes containing specific microRNAs.
Regulators of miRISC activity
RNAi screens for enhancers of microRNA-related phenotypes in C. elegans have contributed to the identifications of proteins that could link developmental or physiological signals to the regulation of microRNA activity, without necessarily affecting microRNA abundance (Parry et al. 2007; Rausch et al. 2015). Similarly, candidate microRNA regulatory cofactors have been identified among proteins found to be associated with miRISC in C. elegans and verified functionally by genetics or RNAi knockdown. In this manner, the miRISC-associated proteins NHL-2 (a TRIM-NHL protein) and CGH-1 (an RNA helicase domain protein) were found to function as positive cofactors for microRNAs (Hammell et al. 2009b). These results are consistent with NHL-2 and CGH-1 having evolutionarily conserved roles in modulating the efficacy of microRNA–target interactions in vivo.
C. elegans casein kinase II (CK2) promotes miRISC function. kin-3 and kin-10 encode subunits of CK2. kin-10 is required for RNAi (Kim et al. 2005) and casein kinase subunits can be obtained via co-IP with AIN-1 (Alessi et al. 2015). Casein kinase inactivation causes developmental defects that phenocopy a loss of miRISC cofactors and enhance the loss of microRNA function in diverse cellular contexts. CK2 is dispensable for microRNA biogenesis and the stability of miRISC cofactors, but is required for miRISC target mRNA binding and silencing. The conserved DEAD-box RNA helicase, CGH-1/DDX6, is a key CK2 substrate within miRISC; CGH-1 phosphorylation is required for CGH-1 function in the microRNA pathway (Alessi et al. 2015).
Other candidate miRISC cofactors in C. elegans that were confirmed functionally by using sensitized genetic backgrounds include PUF-9 (Nolde et al. 2007) and poly(A)-binding protein (Hurschler et al. 2011). The latter finding corroborates the idea that miRISC can regulate mRNA translation and/or stability by affecting polyadenylation (Flamand et al. 2016). Also implicating microRNA function in translational control, a yeast two-hybrid screen for proteins that can interact with ALG-1 identified RACK1 (receptor for activated C-kinase), a protein known to interact with ribosomes. rack-1 knockdown resulted in developmental phenotypes attributable to defects in microRNA activity, suggesting that RACK-1 may mediate interactions between miRISC and ribosomes, possibly in the context of microRNA repression of translation (Jannot et al. 2011). The ribosome connection is further supported by reports that knockdown of ribosomal protein RPS-14 can modify let-7 phenotypes (Chan and Slack 2009).
Staufen (STAU-1) is a double-stranded RNA-binding protein with known functions in the regulation of mRNA activity, including translation (Micklem et al. 2000). STAU-1 binds to multiple mRNAs in C. elegans (LeGendre et al. 2013), suggesting that, in principle, STAU-1 could functionally interact with microRNAs for cotargeted mRNAs. Indeed, stau-1(lf) can suppress phenotypes associated with the depletion of certain microRNAs in C. elegans without discernably affecting microRNA levels, indicating that STAU-1 may function as a negative regulator of microRNA activity (Ren et al. 2016).
Certain microRNA cofactors that have emerged from genetic enhancer screens point to an intimate relationship between microRNAs and vesicular sorting pathways. Components of the Golgi-Associated Retrograde Protein (GARP) complex have been functionally implicated with miRISC activity in C. elegans, suggesting a miRISC connection with membranes that may affect the abundance of GW182/AIN proteins and/or microRNAs (Vasquez-Rifo et al. 2013). ER pathways such as HMG-CoA reductase have also emerged as genetic enhancers of weak let-7 mutations (Parry et al. 2007). These C. elegans findings are endorsed by genetic analysis of microRNA-defective mutations in Arabidopsis (Li et al. 2016). For example, the ER-associated mevalonate pathway of sterol and dolichol synthesis in protein glycosylation strongly regulates let-7 activity in C. elegans as well as microRNA function in plants (Shi and Ruvkun 2012), and Arabidopsis microRNAs are strongly associated with ER-associated polysomes (Li et al. 2016). It is tantalizing to think that the target of one of the world’s most prescribed class of drugs, the statins, may affect microRNA function in the regulation of secreted protein translation (Shi and Ruvkun 2012).
Reciprocal regulation between let-7 and LIN-28
lin-28 negatively regulates the accumulation of let-7 mature microRNA in C. elegans. Mature let-7 accumulates to dramatically elevated levels at abnormally early larval stages in lin-28(lf) mutants (Van Wynsberghe et al. 2011). Furthermore, biochemical evidence points to direct in vivo binding of LIN-28 to the let-7 primary transcript in the nucleus at early larval stages, suggesting that LIN-28 inhibits processing of the pri-let-7 transcript into the let-7 precursor (Van Wynsberghe et al. 2011; Stefani et al. 2015). This situation reflects an apparently evolutionarily conserved, mutually antagonistic and direct relationship between let-7 and lin-28, where LIN-28 binds to the let-7 transcript, and let-7 binds to lin-28 mRNA. Interestingly, in C. elegans, LIN-28 binding seems to be downstream of the let-7 hairpin (Stefani et al. 2015), indicating that the regulation of let-7 biogenesis by LIN-28 in C. elegans may occur exclusively in the nucleus.
Feedback autoregulation of let-7 and lin-4
There is evidence that lin-4 and let-7 in C. elegans may offer fascinating opportunities to study the ways that a microRNA may feedback and regulate its own expression, perhaps even by interacting with its own primary transcript in the nucleus. lin-4 complementary sites were identified upstream of the lin-4 hairpin in the lin-4 primary transcript, and tests using mutated transgenic reporters suggest that these sites (and, by implication, the base pairing of lin-4) could affect the developmental expression of lin-4 in vivo (Turner et al. 2014).
Similarly, a region downstream of the let-7 hairpin, within sequences expressed as part of the let-7 primary transcript, contains let-7 complementary sites that were shown to bind to miRISC (ALG-1) in vivo (Zisoulis et al. 2012). Moreover, functional tests using let-7 transgenes have shown that the presence of the downstream sequences containing the let-7 sites could positively impact let-7 microRNA expression, suggesting that mature let-7 microRNA could act in the nucleus to promote its own biogenesis (Zisoulis et al. 2012).
Identification and Validation of MicroRNA Targets
Studies using C. elegans have contributed substantially to our understanding of the underlying principles of target recognition by microRNAs. The primacy of the 5′ part of the microRNA (eventually termed the seed) in target binding was evident from the predicted base pairing between lin-4 and let-7 microRNAs and their first genetically identified targets (Lee et al. 1993; Wightman et al. 1993; Reinhart et al. 2000). Some of the first in vivo structure–function analyses of microRNA–target interactions were conducted in C. elegans (Ha et al. 1996), and computational target prediction algorithms were developed using data from C. elegans (Hammell et al. 2008).
Genetic epistasis of predicted microRNA–target mRNA pairs
As base pairing of a microRNA to a target mRNA causes a decrease in either the translation of the target mRNA and/or the abundance of that mRNA, the phenotypes caused by a microRNA mutation are expected to be due to an increase in the expression of the target’s protein product. Tests of epistasis—whether knockdown of a putative mRNA target can suppress the phenotype of microRNA loss-of-function—is a powerful approach for validating that a gene containing predicted microRNA target sites can function downstream of the microRNA. In some cases, major microRNA targets have been identified directly in screens for suppressors of microRNA mutants, as was the case for lin-14 (Ambros and Horvitz 1984; Wightman et al. 1993) for the lin-4 microRNA and hbl-1(Abrahante et al. 2003; Lin et al. 2003) for the let-7 microRNA. Similarly, lin-41(lf) mutations were identified by epistasis to let-7(lf) in screens for suppressors of let-7(lf) (Slack et al. 2000), and targeting of nhl-2 and sup-26 by the mir-35 family was discovered in an RNAi screen for suppressors of the subliminal masculinization of the mir-35-family(lf) animals (McJunkin and Ambros 2017). Genome-wide RNAi screens can also, in principle, identify targets of a microRNA from knockdowns that suppress the microRNA mutant phenotypes. For example, in RNAi screens for suppressors and enhancers of let-7 phenotypes (Hunter et al. 2013), suppressors can include targets of let-7 as well as negative modulators of let-7 activity (see Regulators of miRISC activity, above).
Epistasis, in itself, does not unequivocally establish a gene as a direct downstream target of a microRNA, and so the phylogenetic conservation of orthologous complementary target sites (in one or more Caenorhabditis species) is an additional criterion for directness. Another shortcoming of epistasis is that for redundant targets, of the sort where the overexpression of any one target can cause the phenotype, simultaneous knockdown of multiple targets would be required. Conversely, it can be possible to suppress a microRNA loss-of-function phenotype by knockdown of any one of a set of predicted targets (Grosshans et al. 2005), indicating that the phenotype of a microRNA loss-of-function could depend on the simultaneous hyperactivity of multiple genes of a coherent downstream gene network, where the levels of each gene are coupled to the levels of others in the network. Even in cases where such a set of hypothetically coupled genes all contain microRNA complementary sites (Grosshans et al. 2005), it is possible that only a subset of the network may be directly regulated by the microRNA.
In summary, while epistasis is a powerful means to support the supposition of a direct microRNA–target interaction (by indicating that the putative target functions downstream of the microRNA), and phylogenetic conservation of targeting can also endorse target validity (by indicating selection on the putative target sequence), further supporting evidence can come from the mutation of the microRNA complementary sites using clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 and from assaying for upregulation of the putative target protein, along with associated phenotypes.
Computational prediction of microRNA complementary target sites
The primary involvement of “seed pairing” (base pairing between target nucleotides and positions 2–8 of the microRNA) was apparent from the initial identification of targets for lin-4 (Lee et al. 1993; Wightman et al. 1993) and let-7 (Slack et al. 2000). When additional conserved microRNAs were identified (Lau et al. 2001; Lee and Ambros 2001), the primacy of the seed in target recognition was confirmed by the almost universal conservation of nucleotides 2–8 among evolutionarily related microRNAs. Therefore, target prediction algorithms rely heavily on the base pairing of nucleotides 2–8 or 2–7 of the microRNA, with additional provisions for filtering out false positive predictions by employing evolutionary conservation of UTR sequence alignment (Lall et al. 2006), and the conservation of targeting and/or other parameters derived from in vivo confirmatory data (Hammell et al. 2008; Agarwal et al. 2015).
A number of different microRNA target prediction tools are available, and generally all of them are convenient and powerful. The chief differences among them are how underlying assumptions are weighted, and different tools can yield nonidentical sets of putative targets. So, in general, it is advisable to employ the combined predictions of multiple computational tools. Another consideration is that some tools may be found to be more stringent than others, being tuned to yield fewer false positives (at the expense of perhaps missing many bona fide targets). Less-stringent prediction tools can be more comprehensive and sweep up most bona fide targets, but at the expense of more false positive predictions. Predicted targets must be validated by in vivo experiments, and so the choice of target prediction tool is in part governed by the logistics of target validation in a given situation. A stringent tool may be advisable when high-throughput validation tests are not available, while a more comprehensive tool, such as RNAhybrid (Rehmsmeier et al. 2004), could be the choice in situations where avoiding false negatives is a priority and where false positives can easily be screened out.
Direct identification of in vivo microRNA–target complexes
The caveats associated with the computational identification of microRNA targets are derived from two issues. First, even the more stringent prediction tools can yield a list of scores, or even hundreds of predicted targets for a single microRNA, yet in cases where genetic epistasis has been applied to identify functional targets of a microRNA, it is generally found that very few or only a single target is actually involved in a given context. Therefore, it appears that we do not yet understand what contextual factors govern which specific microRNA–target interactions, among all the computationally predicted potential interactions, are efficacious. Second, it is not clear that we have a comprehensive understanding of the various configurations of microRNA–target interactions (other than seed pairing) that can be functional in vivo. Therefore, significant advances in identifying bona fide microRNA target complexes in vivo will not only enable focused attention on functional targets for genetic evaluation, but will also permit the continued refinement of computational target prediction algorithms.
IP of miRISC using antisera against miRISC components, followed by the identification of bound mRNAs using microarray or RNA sequencing (RNAseq), has provided data sets of mRNAs stably associated with miRISC (Zhang et al. 2007). These data sets have been used to shape target prediction algorithms based on experimental evidence (Hammell et al. 2008). The shortcoming of miRISC IP followed by RNAseq is that the precise location of miRISC binding is not known, so the specific microRNAs responsible for miRISC binding to the mRNA sequences obtained by co-IP must be inferred from sequence complementarity.
Strategies such as cross-linking immunoprecipitation with high-throughput sequencing (CLIP-seq) (Zisoulis et al. 2010), individual nucleotide-resolution cross-linking and immunoprecipitation (iCLIP) (Broughton and Pasquinelli 2013), and chimera PCR (ChimP) (Broughton et al. 2016), which employ UV-cross-linking of protein–RNA complexes in vivo, nuclease digestion of unprotected RNA, followed by IP of ALG-1 with its bound RNA and cDNA sequencing of the ALG-1-linked mRNA sequences, have provided genome-scale data sets of microRNA-binding sites. As these methods become more widely used, particularly for specific cell types and for specific microRNA mutants, it will be possible to more definitively match microRNA to specific targets in particular contexts. The iCLIP and CLIP-seq methods are powerful strategies for identifying miRISC-binding sites, but unambiguous assignment of the microRNA recognizing those sites is not always possible, especially for microRNAs of the same seed family. Such ambiguities in assigning specific microRNAs to specific mRNA sites are overcome by analyzing the rare sequence reads that result from the ligation of a microRNA to a fragment of mRNA target that is cocross-linked to miRISC, so that microRNA–target tandem sequences are obtained from single-cDNA sequencing reads. Such microRNA–target chimeric sequences have been found in C. elegans iCLIP data sets (Broughton et al. 2016), as well as in data sets from protocols designed to enrich for the intermolecular ligation events (Helwak et al. 2013; Grosswendt et al. 2014).
So far, microRNA-target chimera sequencing has been applied in a limited fashion for C. elegans, but going forward, these approaches that identify chimeric microRNA–target sequences, especially if applied with improved efficiency compared to current applications and in a tissue-specific fashion, should permit high-confidence analysis of microRNA–target regulatory networks.
While approaches that identify microRNA–target site chimeras can confirm whether or not a particular microRNA actually binds to particular targets in vivo, measurements of the ribosome occupancy of target mRNAs and quantitation of the levels of proteins by mass spectrometry can provide additional evidence for the efficacy of the microRNA interaction with specific mRNAs. For example, the combined application of ribosome profiling and targeted quantitative proteomics, combined with 3′-UTR reporter assays, has enabled the discovery and validation of numerous functionally relevant let-7 and miR-58 targets (Jovanovic et al. 2010).
It should be noted that nothing is perfect and there are caveats attached to every method: ribosome occupancy does not necessarily reflect translation rate (see below) and indirect effects on protein turnover can confound interpreting protein levels. Nevertheless, the experimental arsenal available to C. elegans researchers—including genetic epistasis, mRNA sequencing, proteomics, ribosome profiling, CLASH, and CRISPR/Cas9 for the tagging of target genes in loco and for surgical mutagenesis of mRNA and microRNA complementary sequences (see below)—offers a gold standard for microRNA target discovery and validation.
Mechanisms of MicroRNA Repression of Target mRNAs
How do microRNAs repress the production of proteins from target mRNAs? In addition to microRNA and Argonaute, the miRISC complex contains other effector proteins, including notably GW182 (AIN-1/2), which are understood to mediate the repression of translation and/or accelerate mRNA turnover. Studies using C. elegans have contributed fundamentally to our understanding of the range of mRNA regulatory mechanisms that can be elicited by microRNA and have highlighted areas for future study, namely, how it is that miRISC can be programmed for different outcomes, depending on the microRNA, the particular mRNA sequence that it recognizes, and interactions with miRISC cofactors and RNA-binding proteins (Regulators of miRISC activity).
mRNA translational repression and/or mRNA turnover
The current understanding of microRNA repression, from invertebrate and vertebrate experimental systems, is that the chief mode of microRNA action is via interactions in cis, between the miRISC complex and the polyadenylation/deadenylation machinery, resulting in shortening of the poly(A) tail, a decrease in translation, 5′ end decapping, and degradation of the mRNA.
Numerous investigators, using C. elegans and other systems, have sought to determine whether the primary activity of miRISC is to trigger mRNA turnover, and hence to indirectly inhibit protein output, or whether miRISC can inhibit translation independently of mRNA turnover. Many studies have focused on the effects of microRNAs on target mRNA levels, perhaps because mRNAs are so much more easily quantified than proteins, especially at the genomic scale, but also because in assays for protein production (e.g., measurement of luciferase activity produced from 3′-UTR reporters), microRNA regulation of translation activity and mRNA levels have often correlated.
Measuring the impact of microRNAs on the proteome, simultaneously with the quantitation of mRNA levels, can, in principle, resolve effects of microRNAs on mRNA abundance from translational repression and can be applied in high-throughput in C. elegans. For example, candidate targets of mir-58/bantam in C. elegans were identified by differential co-IP of mRNAs in wild-type vs. mir-58 family mutants and validated by targeted proteomics. In this study, the targets that were confidently validated displayed behaviors consistent with regulation primarily at the level of protein abundance rather than mRNA stability (Jovanovic et al. 2012). Interestingly, mRNA and proteomic quantitation of mir-58/bantam targets in response to a progressive depletion of mir-58 family members uncovered additional complexity, where translational inhibition was most evident in single mir-58 family mutants; however, with depletion of the whole family, mRNA degradation was predominant.
Because the heterochronic microRNAs are deployed in a timed sequence, the heterochronic gene pathway is well suited for exploring the dynamics of target gene protein and mRNA levels after the initiation of microRNA-mediated repression. By monitoring lin-14 protein and mRNA in finely staged larvae, a complex dynamic of mRNA and protein decline after developmental the induction of lin-4 microRNA could be resolved, revealing the repression of mRNA levels and a decline of protein that proceeds with distinct kinetics (Shi et al. 2013). For lin-14, a modest (two- to threefold decrease) in mRNA level was observed between the L1 (when lin-4 is absent) and the L2 (when lin-4 is present) stages, while at the same time, the level of lin-14 protein decreased much more (Shi et al. 2013). Similarly, the developmental dynamics of let-7 target levels support an acute regulation of protein synthesis, followed by subsequent changes in mRNA levels (Stadler et al. 2012). This dynamic—a relatively rapid translational repression of targets after appearance of the microRNA, accompanied by a longer time course of target mRNA decay—is similar to that observed for certain cases of microRNA repression in zebrafish embryos (Bazzini et al. 2012) and Drosophila S2 cells (Djuranovic et al. 2012).
Interestingly, the apparent impact of microRNA repression on target protein vs. mRNA can vary, depending on the experimental context. For example, studies comparing wild-type to lin-4 or let-7 microRNA mutants at corresponding developmental stages indicated a more potent contribution of mRNA turnover than did other studies that followed the mRNA and protein dynamics of those targets during wild-type development (Bagga et al. 2005; Ding and Grosshans 2009; Holtz and Pasquinelli 2009; Stadler et al. 2012; Shi et al. 2013). These contrasting findings from the same system, using different experimental approaches, highlight how examining the kinetics of stage-specific microRNA-mediated repression in wild-type worms can resolve time-dependent components of the mechanism (translational repression followed by mRNA turnover), whereas comparing mutant to wild-type worms can emphasize the final state of the system (the eventual loss of the mRNA).
There is also evidence that the physiological context can modulate the relative contribution of translational repression and mRNA destabilization. For example, in larvae developing under conditions of limited nutrients, repression of lin-14 by lin-4 primarily involves repression of LIN-14 protein levels, and less so repression of lin-14 mRNA levels (Holtz and Pasquinelli 2009), in contrast to nonstarved conditions, where mRNA degradation is more evident (Bagga et al. 2005).
What sorts of translational repression mechanisms can be invoked by miRISC? There are indications that microRNAs in C. elegans can impact translation at the initiation step (Ding et al. 2008; Ding and Grosshans 2009), and it is reasonable to suppose that the inhibition of translational initiation would be a predominant mechanism in C. elegans, as in other animals. However, there is also evidence that translational repression by C. elegans microRNAs, at least in some contexts, could occur postinitiation. lin-14 mRNA remains associated with polyribosomes, even at stages where lin-4 microRNA is abundant, suggesting a mode of translational repression in this case that occurs after translation initiation (Olsen and Ambros 1999). Polyribosome fractionation analysis of lin-28 mRNA supported a similar mechanism for lin-28 repression by lin-4 (Seggerson et al. 2002). Genome-wide ribosomal profiling across multiple developmental time points also indicates that many of the heterochronic gene microRNA targets are subject to postinitiation translational repression (Stadler et al. 2012).
This apparent polyribosome-associated mode of translational repression by microRNAs may not be specific to nematodes, as a similar phenomenon was observed for let-7 in mammalian cells (Nottrott et al. 2006). It seems relevant here to also mention that, at least in some systems, microRNA-mediated target mRNA destabilization can occur in association with ribosomes (Antic et al. 2015; Tat et al. 2016), which could provide one explanation for how postinitiation mechanisms for microRNA activity may not be incompatible with target degradation.
The association of a subset of microRNAs with RDE-1 (Steiner et al. 2007; Corrêa et al. 2010) suggests that RDE-1 may have roles in the activity of at least certain microRNAs, in addition to its major role as an siRNA effector in RNAi. Among the microRNAs found to complex with RDE-1 in vivo, mir-243 is particularly noteworthy, in that mir-243 is preferentially loaded into RDE-1 (compared to ALG-1/2) and is completely complementary to its primary mRNA target, Y47H10AA.5. Therefore, mir-243 is essentially an endogenous siRNA. As expected for an siRNA carried by RDE-1, mir-243 elicits a bloom of secondary endo-siRNAs that efficiently silence Y47H10AA.5 by RNAi (Corrêa et al. 2010). mir-243 is highly unusual among C. elegans microRNAs (and among animal microRNAs in general) in having perfect complementarity to a target; animals seem to generally stick to the partial-complementarity mode of microRNA–target base pairing, likely to avoid obligatory target destruction. The function of the silencing of Y47H10AA.5 by mir-243 is unknown, as mir-243 mutant animals appear superficially normal (Miska et al. 2007). Another microRNA that seems to cause the cleavage of a target is mir-249; degradome sequencing of RNA from wild-type vs. mir-249(lf) animals has revealed evidence of mir-249-dependent cleavage of the transcript ZK637.6 (Park et al. 2013). The functions of mir-249 and ZK637.6 are presently unknown.
MicroRNA–target base pairing
It is possible that distinct repressive outcomes could be mediated by structural properties of the base pairing between microRNAs and their targets. Whether the microRNA–Argonaute complex engages in seed-only base pairing or 3′ compensatory base pairing could in principle affect miRISC composition and therefore outcome. Distinct outcomes could be governed by accessory factors that associate with certain miRISC-containing microRNAs (for example by sequence-specific recognition of microRNAs) and/or the effects of miRISC–target conformation on the binding of specific cofactors. Conformational modeling of the C. elegans let-7 miRISC bound to its target (Gan and Gunsalus 2015) supports the concept that its pairing configuration could affect the conformation ALG-2, and perhaps thereby effect miRISC assembly and function. An influence of the configuration of 3′-UTR elements on microRNA repression has also been noted (Flamand et al. 2016). Structural analyses of microRNA–target complexes, exemplified by the NMR analysis of let-7 bound to synthetic lin-41 3′-UTR (Cevec et al. 2008, 2010) and modeling of let-7::lin-41 (Gan and Gunsalus 2013), promise to reveal principles of miRISC assembly and function that can be efficiently tested using C. elegans in vivo genetics.
Whether and how the members of microRNA seed families, which can differ in their nonseed nucleotides, might be deployed to regulate distinct target sets has been explored in C. elegans using the let-7::lin-41 interaction. The evidence suggests that the configuration of base pairing between the microRNA and its target site, particularly in the 3′ part of the microRNA, can govern the specificity with which a particular microRNA family member associates with the site (Broughton et al. 2016).
Although it is likely that the majority of microRNA–target interactions involve primarily perfect seed pairing, there is evidence that noncanonical base pairing interactions are not uncommon. Sites for lin-4 in the 3′-UTR of lin-14 are predicted to contain bulged seed nucleotides, the presence of which impacts target repression, suggesting that microRNA–target structure is important in certain contexts (Ha et al. 1996) and that perfect seed pairing is not a hard rule. Structure–function experiments on the lsy-6::cog-1 regulatory interaction support the idea that perfect seed pairing is not required (Didiano and Hobert 2006). Further, in the configuration of the two sites for let-7 in the lin-41 3′-UTR, extensive predicted 3′ pairing and imperfect seed pairing (one site has a bulge, while the other has a G:U base pair) indicate that perfect seed pairing is not required as long as there is sufficient 3′ compensatory pairing (Vella et al. 2004).
Structure–function studies of the interactions of lin-4 and let-7 with their chief targets, in the context of their in vivo developmental phenotypes, have revealed interesting differences in the constraints on these two microRNAs, suggesting that some microRNAs depend on seed sequences more than others. For lin-4, a strong requirement for seed sequences was apparent, while for let-7 there was a surprising tolerance for seed mutations, suggesting that let-7 has a greater capacity for engaging in functional noncanonical interactions than does lin-4 (Zhang et al. 2015).
In vitro analysis of microRNA mechanisms
MicroRNAs are stubbornly in vivo entities, with many connections, partners, and modulators that determine how they function in cells and animals. For this reason, C. elegans is a particularly appropriate system for the exploitation of in vivo genetics and cell biology to probe how microRNAs function in the context of an intact developing and behaving animal. However, biochemical approaches are ultimately required to fully understand how microRNAs work. An extraordinarily promising system was been devised using C. elegans embryo extracts programmed with synthetic mRNAs whose translation, stability, and poly(A) status can be monitored quantitatively (Wu et al. 2010). In this system, reporters containing 3′-UTR sequences from putative microRNA targets were regulated by endogenous embryonic microRNAs, enabling the characterization of microRNA mechanisms. The reporters containing natural 3′-UTRs exhibited rapid microRNA-dependent deadenylation and were translationally repressed. Interestingly, single sites did not work well, suggesting that cooperativity exists between miRISC complexes colocated in a UTR.
Surprisingly, many of the targets that were rapidly deadenylated in response to microRNA activity in vitro were stable, suggesting a novel mechanism of microRNA repression where targets appear to be converted (perhaps reversibly) to a translationally quiescent status. It is possible that the C. elegans embryo extract system is an example of a context where deadenylation can be uncoupled from decapping and degradation (Wu et al. 2010). It is also possible that microRNA-mediated gene regulation could differ substantially between the embryo and other stages. For example, targeted mutation of ALG-1 to eliminate the tryptophan-binding sites for AIN-1 and AIN-2 has been found to have little effect on embryonic development, but disrupts microRNA function in larval stages (Jannot et al. 2016).
The C. elegans embryo cell-free system (Wu et al. 2010) can be leveraged for the biochemical characterization of miRISC complexes and genetic tests of candidate microRNA cofactors and effectors (by employing extracts treated with RNAi or from mutants). In this fashion, roles have been uncovered for cytoplasmic poly(A)-binding proteins in microRNA-mediated deadenylation and in poly(A)-dependent and poly(A)-independent translational repression (Flamand et al. 2016).
Conclusions
C. elegans has remained in the thick of microRNA research and should continue to do so. The experimental toolkit continues to grow, and the use of CRISPR/Cas9 genome editing permits essentially any defined modification being made to microRNAs and target sequences in their natural genomic context. It should be possible to explore and uncover the principles of microRNA recognition in vivo using C. elegans genetics with greater efficiency than in any other organism.
Regulators of microRNA biogenesis, stability, and activity should continue to emerge from genetic screens, miRISC proteomics, and RNAi modifier screens. Whether and how specific miRISC complexes may be customized for specific outcomes remains an open question, and much remains to be learned from C. elegans about how specific microRNAs can be deployed to regulate particular targets, depending on the cellular or physiological context.
Further questions remain about the potential for cell nonautonomous activity of microRNAs in multicellular animals. In C. elegans, cell autonomy of microRNA function has been tested for only a handful of microRNAs: lin-4 (Zhang and Fire 2010), let-7 (Zhi et al. 2017), and mir-34 and mir-83 (Burke et al. 2015). The fact that RNAi can spread among cells in C. elegans indicates that the worm possesses mechanisms for the intracellular transport of RNA, and so it is conceivable that microRNAs could be similarly deployed extracellularly in some contexts.
Mechanisms of how microRNAs are regulated post-transcriptionally in response to developmental and physiological signals are ripe for investigation. For example, certain C. elegans microRNAs undergo rapid developmental downregulation, while others remain constant or increase. It is essentially unknown in any system how the rates of biogenesis and turnover of different microRNAs are programmed and regulated and, as usual, C. elegans is the ideal system to investigate such questions in vivo.
Acknowledgments
Research in VA’s laboratory is supported by NIH GM34028. Research in GR’s laboratory is supported by NIH GM44619. The authors thank Gloria Ha for help with Figure 1. The authors also thank John Kim and the WormBook editors and anonymous reviewers for advice and criticism.
Footnotes
Communicating editor: J. Kim
Literature Cited
- Abbott A. L., Alvarez-Saavedra E., Miska E. A., Lau N. C., Bartel D. P., et al. , 2005. The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev. Cell 9: 403–414. 10.1016/j.devcel.2005.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahante J. E., Daul A. L., Li M., Volk M. L., Tennessen J. M., et al. , 2003. The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev. Cell 4: 625–637. [DOI] [PubMed] [Google Scholar]
- Agarwal V., Bell G. W., Nam J. W., Bartel D. P., 2015. Predicting effective microRNA target sites in mammalian mRNAs. Elife 4: e05005 10.7554/eLife.05005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alessi A. F., Khivansara V., Han T., Freeberg M. A., Moresco J. J., et al. , 2015. Casein kinase II promotes target silencing by miRISC through direct phosphorylation of the DEAD-box RNA helicase CGH-1. Proc. Natl. Acad. Sci. USA 112: E7213–E7222. 10.1073/pnas.1509499112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Saavedra E., Horvitz H. R., 2010. Many families of C. elegans microRNAs are not essential for development or viability. Curr. Biol. 20: 367–373. 10.1016/j.cub.2009.12.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V., 1989. A hierarchy of regulatory genes controls a larva-to-adult developmental switch in C. elegans. Cell 57: 49–57. 10.1016/0092-8674(89)90171-2 [DOI] [PubMed] [Google Scholar]
- Ambros V., Horvitz H. R., 1984. Heterochronic mutants of the nematode Caenorhabditis elegans. Science 226: 409–416 [DOI] [PubMed] [Google Scholar]
- Ambros V., Horvitz H. R., 1987. The lin-14 locus of Caenorhabditis elegans controls the time of expression of specific postembryonic developmental events. Genes Dev. 1: 398–414. 10.1101/gad.1.4.398 [DOI] [PubMed] [Google Scholar]
- Antic S., Wolfinger M. T., Skucha A., Hosiner S., Dorner S., 2015. General and microRNA-mediated mRNA degradation occurs on ribosome complexes in Drosophila cells. Mol. Cell. Biol. 35: 2309–2320. 10.1128/MCB.01346-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagga S., Bracht J., Hunter S., Massirer K., Holtz J., et al. , 2005. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell 122: 553–563. [DOI] [PubMed] [Google Scholar]
- Bazzini A. A., Lee M. T., Giraldez A. J., 2012. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science 336: 233–237. 10.1126/science.1215704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethke A., Fielenbach N., Wang Z., Mangelsdorf D. J., Antebi A., 2009. Nuclear hormone receptor regulation of microRNAs controls developmental progression. Science 324: 95–98. 10.1126/science.1164899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm M., Slack F., 2005. A developmental timing microRNA and its target regulate life span in C. elegans. Science 310: 1954–1957. 10.1126/science.1115596 [DOI] [PubMed] [Google Scholar]
- Bossé G. D., Rüegger S., Ow M. C., Vasquez-Rifo A., Rondeau E. L., et al. , 2013. The decapping scavenger enzyme DCS-1 controls microRNA levels in Caenorhabditis elegans. Mol. Cell 50: 281–287. 10.1016/j.molcel.2013.02.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulias K., Horvitz H. R., 2012. The C. elegans microRNA mir-71 acts in neurons to promote germline-mediated longevity through regulation of DAF-16/FOXO. Cell Metab. 15: 439–450. 10.1016/j.cmet.2012.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner J. L., Jasiewicz K. L., Fahley A. F., Kemp B. J., Abbott A. L., 2010. Loss of individual microRNAs causes mutant phenotypes in sensitized genetic backgrounds in C. elegans. Curr. Biol. 20: 1321–1325. 10.1016/j.cub.2010.05.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner J. L., Kemp B. J., Abbott A. L., 2012. The mir-51 family of microRNAs functions in diverse regulatory pathways in Caenorhabditis elegans. PLoS One 7: e37185 10.1371/journal.pone.0037185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton J. P., Pasquinelli A. E., 2013. Identifying Argonaute binding sites in Caenorhabditis elegans using iCLIP. Methods 63: 119–125. 10.1016/j.ymeth.2013.03.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton J. P., Lovci M. T., Huang J. L., Yeo G. W., Pasquinelli A. E., 2016. Pairing beyond the seed supports microRNA targeting specificity. Mol. Cell 64: 320–333. 10.1016/j.molcel.2016.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K. C., Svendsen J. M., Tucci R. M., Montgomery B. E., Montgomery T. A., 2017. ALG-5 is a miRNA-associated Argonaute required for proper developmental timing in the Caenorhabditis elegans germline. Nucleic Acids Res. 45: 9093–9107. 10.1093/nar/gkx536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukhari S. I., Vasquez-Rifo A., Gagne D., Paquet E. R., Zetka M., et al. , 2012. The microRNA pathway controls germ cell proliferation and differentiation in C. elegans. Cell Res. 22: 1034–1045. 10.1038/cr.2012.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke S. L., Hammell M., Ambros V., 2015. Robust distal tip cell pathfinding in the face of temperature stress is ensured by two conserved microRNAs in Caenorhabditis elegans. Genetics 200: 1201–1218. 10.1534/genetics.115.179184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büssing I., Yang J. S., Lai E. C., Grosshans H., 2010. The nuclear export receptor XPO-1 supports primary miRNA processing in C. elegans and Drosophila. EMBO J. 29: 1830–1839. 10.1038/emboj.2010.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cevec M., Thibaudeau C., Plavec J., 2008. Solution structure of a let-7 miRNA:lin-41 mRNA complex from C. elegans. Nucleic Acids Res. 36: 2330–2337. 10.1093/nar/gkn088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cevec M., Thibaudeau C., Plavec J., 2010. NMR structure of the let-7 miRNA interacting with the site LCS1 of lin-41 mRNA from Caenorhabditis elegans. Nucleic Acids Res. 38: 7814–7821. 10.1093/nar/gkq640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M., Horvitz H. R., Sulston J. E., 1981. Mutations that lead to reiterations in the cell lineages of C. elegans. Cell 24: 59–69. [DOI] [PubMed] [Google Scholar]
- Chan S. P., Slack F. J., 2009. Ribosomal protein RPS-14 modulates let-7 microRNA function in Caenorhabditis elegans. Dev. Biol. 334: 152–160. 10.1016/j.ydbio.2009.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S., Johnston R. J., Jr., Frokjaer-Jensen C., Lockery S., Hobert O., 2004. MicroRNAs act sequentially and asymmetrically to control chemosensory laterality in the nematode. Nature 430: 785–789. 10.1038/nature02752 [DOI] [PubMed] [Google Scholar]
- Chatterjee S., Grosshans H., 2009. Active turnover modulates mature microRNA activity in Caenorhabditis elegans. Nature 461: 546–549. 10.1038/nature08349 [DOI] [PubMed] [Google Scholar]
- Chatterjee S., Fasler M., Bussing I., Grosshans H., 2011. Target-mediated protection of endogenous microRNAs in C. elegans. Dev. Cell 20: 388–396. 10.1016/j.devcel.2011.02.008 [DOI] [PubMed] [Google Scholar]
- Chung W. J., Agius P., Westholm J. O., Chen M., Okamura K., et al. , 2011. Computational and experimental identification of mirtrons in Drosophila melanogaster and Caenorhabditis elegans. Genome Res. 21: 286–300. 10.1101/gr.113050.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochella L., Hobert O., 2012. Embryonic priming of a miRNA locus predetermines postmitotic neuronal left/right asymmetry in C. elegans. Cell 151: 1229–1242. 10.1016/j.cell.2012.10.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrêa R. L., Steiner F. A., Berezikov E., Ketting R. F., 2010. MicroRNA-directed siRNA biogenesis in Caenorhabditis elegans. PLoS Genet. 6: e1000903 10.1371/journal.pgen.1000903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L. L., Gao J. X., Zou C. G., Ma Y. C., Zhang K. Q., 2015. mir-233 modulates the unfolded protein response in C. elegans during Pseudomonas aeruginosa infection. PLoS Pathog. 11: e1004606 10.1371/journal.ppat.1004606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lencastre A., Pincus Z., Zhou K., Kato M., Lee S. S., et al. , 2010. MicroRNAs both promote and antagonize longevity in C. elegans. Curr. Biol. 20: 2159–2168. 10.1016/j.cub.2010.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lucas M. P., Saez A. G., Lozano E., 2015. miR-58 family and TGF-beta pathways regulate each other in Caenorhabditis elegans. Nucleic Acids Res. 43: 9978–9993. 10.1093/nar/gkv923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli A. M., Tops B. B., Plasterk R. H., Ketting R. F., Hannon G. J., 2004. Processing of primary microRNAs by the microprocessor complex. Nature 432: 231–235. 10.1038/nature03049 [DOI] [PubMed] [Google Scholar]
- Didiano D., Hobert O., 2006. Perfect seed pairing is not a generally reliable predictor for miRNA-target interactions. Nat. Struct. Mol. Biol. 13: 849–851. 10.1038/nsmb1138 [DOI] [PubMed] [Google Scholar]
- Ding L., Spencer A., Morita K., Han M., 2005. The developmental timing regulator AIN-1 interacts with miRISCs and may target the argonaute protein ALG-1 to cytoplasmic P bodies in C. elegans. Mol. Cell 19: 437–447. 10.1016/j.molcel.2005.07.013 [DOI] [PubMed] [Google Scholar]
- Ding X. C., Grosshans H., 2009. Repression of C. elegans microRNA targets at the initiation level of translation requires GW182 proteins. EMBO J. 28: 213–222. 10.1038/emboj.2008.275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X. C., Slack F. J., Grosshans H., 2008. The let-7 microRNA interfaces extensively with the translation machinery to regulate cell differentiation. Cell Cycle 7: 3083–3090. 10.4161/cc.7.19.6778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djuranovic S., Nahvi A., Green R., 2012. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science 336: 237–240. 10.1126/science.1215691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowen R. H., Breen P. C., Tullius T., Conery A. L., Ruvkun G., 2016. A microRNA program in the C. elegans hypodermis couples to intestinal mTORC2/PQM-1 signaling to modulate fat transport. Genes Dev. 30: 1515–1528. 10.1101/gad.283895.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake M., Furuta T., Suen K. M., Gonzalez G., Liu B., et al. , 2014. A requirement for ERK-dependent Dicer phosphorylation in coordinating oocyte-to-embryo transition in C. elegans. Dev. Cell 31: 614–628. 10.1016/j.devcel.2014.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexel T., Mahofsky K., Latham R., Zimmer M., Cochella L., 2016. Neuron type-specific miRNA represses two broadly expressed genes to modulate an avoidance behavior in C. elegans. Genes Dev. 30: 2042–2047. 10.1101/gad.287904.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecsedi M., Rausch M., Großhans H., 2015. The let-7 microRNA directs vulval development through a single target. Dev. Cell 32: 335–344. 10.1016/j.devcel.2014.12.018 [DOI] [PubMed] [Google Scholar]
- Euling S., Ambros V., 1996. Heterochronic genes control cell cycle progress and developmental competence of C. elegans vulva precursor cells. Cell 84: 667–676. 10.1016/S0092-8674(00)81045-4 [DOI] [PubMed] [Google Scholar]
- Ferguson E. L., Horvitz H. R., 1985. Identification and characterization of 22 genes that affect the vulval cell lineages of the nematode Caenorhabditis elegans. Genetics 110: 17–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flamand M. N., Wu E., Vashisht A., Jannot G., Keiper B. D., et al. , 2016. Poly(A)-binding proteins are required for microRNA-mediated silencing and to promote target deadenylation in C. elegans. Nucleic Acids Res. 44: 5924–5935. 10.1093/nar/gkw276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan H. H., Gunsalus K. C., 2013. Tertiary structure-based analysis of microRNA-target interactions. RNA 19: 539–551. 10.1261/rna.035691.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan H. H., Gunsalus K. C., 2015. Assembly and analysis of eukaryotic Argonaute-RNA complexes in microRNA-target recognition. Nucleic Acids Res. 43: 9613–9625. 10.1093/nar/gkv990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A., Pasquinelli A. E., Conte D., Li N., Parrish S., et al. , 2001. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106: 23–34. 10.1016/S0092-8674(01)00431-7 [DOI] [PubMed] [Google Scholar]
- Grosshans H., Johnson T., Reinert K. L., Gerstein M., Slack F. J., 2005. The temporal patterning microRNA let-7 regulates several transcription factors at the larval to adult transition in C. elegans. Dev. Cell 8: 321–330. 10.1016/j.devcel.2004.12.019 [DOI] [PubMed] [Google Scholar]
- Grosswendt S., Filipchyk A., Manzano M., Klironomos F., Schilling M., et al. , 2014. Unambiguous identification of miRNA:target site interactions by different types of ligation reactions. Mol. Cell 54: 1042–1054. 10.1016/j.molcel.2014.03.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha I., Wightman B., Ruvkun G., 1996. A bulged lin-4/lin-14 RNA duplex is sufficient for Caenorhabditis elegans lin-14 temporal gradient formation. Genes Dev. 10: 3041–3050. 10.1101/gad.10.23.3041 [DOI] [PubMed] [Google Scholar]
- Hallam S. J., Jin Y., 1998. lin-14 regulates the timing of synaptic remodelling in Caenorhabditis elegans. Nature 395: 78–82. 10.1038/25757 [DOI] [PubMed] [Google Scholar]
- Hammell C. M., Karp X., Ambros V., 2009a A feedback circuit involving let-7-family miRNAs and DAF-12 integrates environmental signals and developmental timing in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 106: 18668–18673. 10.1073/pnas.0908131106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammell C. M., Lubin I., Boag P. R., Blackwell T. K., Ambros V., 2009b nhl-2 modulates microRNA activity in Caenorhabditis elegans. Cell 136: 926–938. 10.1016/j.cell.2009.01.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammell M., Long D., Zhang L., Lee A., Carmack C. S., et al. , 2008. mirWIP: microRNA target prediction based on microRNA-containing ribonucleoprotein-enriched transcripts. Nat. Methods 5: 813–819. 10.1038/nmeth.1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes G. D., Frand A. R., Ruvkun G., 2006. The mir-84 and let-7 paralogous microRNA genes of Caenorhabditis elegans direct the cessation of molting via the conserved nuclear hormone receptors NHR-23 and NHR-25. Development 133: 4631–4641. 10.1242/dev.02655 [DOI] [PubMed] [Google Scholar]
- Helwak A., Kudla G., Dudnakova T., Tollervey D., 2013. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell 153: 654–665. 10.1016/j.cell.2013.03.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtz J., Pasquinelli A. E., 2009. Uncoupling of lin-14 mRNA and protein repression by nutrient deprivation in Caenorhabditis elegans. RNA 15: 400–405. 10.1261/rna.1258309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh Y.-W., Chang C., Chuang C.-F., 2012. The microRNA mir-71 inhibits calcium signaling by targeting the TIR-1/Sarm1 adaptor protein to control stochastic L/R neuronal asymmetry in C. elegans. PLoS Genet. 8: e1002864 10.1371/journal.pgen.1002864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter S. E., Finnegan E. F., Zisoulis D. G., Lovci M. T., Melnik-Martinez K. V., et al. , 2013. Functional genomic analysis of the let-7 regulatory network in Caenorhabditis elegans. PLoS Genet. 9: e1003353 10.1371/journal.pgen.1003353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurschler B. A., Harris D. T., Grosshans H., 2011. The type II poly(A)-binding protein PABP-2 genetically interacts with the let-7 miRNA and elicits heterochronic phenotypes in Caenorhabditis elegans. Nucleic Acids Res. 39: 5647–5657. 10.1093/nar/gkr145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iatsenko I., Sinha A., Rodelsperger C., Sommer R. J., 2013. New role for DCR-1/dicer in Caenorhabditis elegans innate immunity against the highly virulent bacterium Bacillus thuringiensis DB27. Infect. Immun. 81: 3942–3957. 10.1128/IAI.00700-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibáñez-Ventoso C., Yang M., Guo S., Robins H., Padgett R. W., et al. , 2006. Modulated microRNA expression during adult lifespan in Caenorhabditis elegans. Aging Cell 5: 235–246. 10.1111/j.1474-9726.2006.00210.x [DOI] [PubMed] [Google Scholar]
- Isik M., Blackwell T. K., Berezikov E., 2016. MicroRNA mir-34 provides robustness to environmental stress response via the DAF-16 network in C. elegans. Sci. Rep. 6: 36766 10.1038/srep36766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jannot G., Bajan S., Giguère N. J., Bouasker S., Banville I. H., et al. , 2011. The ribosomal protein RACK1 is required for microRNA function in both C. elegans and humans. EMBO Rep. 12: 581–586. 10.1038/embor.2011.66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jannot G., Michaud P., Quevillon Huberdeau M., Morel-Berryman L., Brackbill J. A., et al. , 2016. GW182-free microRNA silencing complex controls post-transcriptional gene expression during Caenorhabditis elegans embryogenesis. PLoS Genet. 12: e1006484 (erratum: PLoS Genet. 12: e1006534) 10.1371/journal.pgen.1006484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon M., Gardner H. F., Miller E. A., Deshler J., Rougvie A. E., 1999. Similarity of the C. elegans developmental timing protein LIN-42 to circadian rhythm proteins. Science 286: 1141–1146. 10.1126/science.286.5442.1141 [DOI] [PubMed] [Google Scholar]
- Johnson S. M., Grosshans H., Shingara J., Byrom M., Jarvis R., et al. , 2005. RAS is regulated by the let-7 microRNA family. Cell 120: 635–647. 10.1016/j.cell.2005.01.014 [DOI] [PubMed] [Google Scholar]
- Johnston R. J., Hobert O., 2003. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature 426: 845–849. 10.1038/nature02255 [DOI] [PubMed] [Google Scholar]
- Johnston R. J., Hobert O., 2005. A novel C. elegans zinc finger transcription factor, lsy-2, required for the cell type-specific expression of the lsy-6 microRNA. Development 132: 5451–5460. 10.1242/dev.02163 [DOI] [PubMed] [Google Scholar]
- Jovanovic M., Reiter L., Picotti P., Lange V., Bogan E., et al. , 2010. A quantitative targeted proteomics approach to validate predicted microRNA targets in C. elegans. Nat. Methods 7: 837–842. 10.1038/nmeth.1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovanovic M., Reiter L., Clark A., Weiss M., Picotti P., et al. , 2012. RIP-chip-SRM–a new combinatorial large-scale approach identifies a set of translationally regulated bantam/miR-58 targets in C. elegans. Genome Res. 22: 1360–1371. 10.1101/gr.133330.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagias K., Pocock R., 2015. microRNA regulation of the embryonic hypoxic response in Caenorhabditis elegans. Sci. Rep. 5: 11284 10.1038/srep11284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai Z. S., Finnegan E. F., Huang S., Pasquinelli A. E., 2013. Multiple cis-elements and trans-acting factors regulate dynamic spatio-temporal transcription of let-7 in Caenorhabditis elegans. Dev. Biol. 374: 223–233. 10.1016/j.ydbio.2012.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp X., Ambros V., 2011. The developmental timing regulator HBL-1 modulates the dauer formation decision in Caenorhabditis elegans. Genetics 187: 345–353. 10.1534/genetics.110.123992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp X., Ambros V., 2012. Dauer larva quiescence alters the circuitry of microRNA pathways regulating cell fate progression in C. elegans. Development 139: 2177–2186. 10.1242/dev.075986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp X., Hammell M., Ow M. C., Ambros V., 2011. Effect of life history on microRNA expression during C. elegans development. RNA 17: 639–651. 10.1261/rna.2310111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga H., Fukuyama M., Kitazawa A., Kontani K., Katada T., 2013. The microRNA miR-235 couples blast-cell quiescence to the nutritional state. Nature 497: 503–506. 10.1038/nature12117 [DOI] [PubMed] [Google Scholar]
- Kato M., Paranjape T., Müller R. U., Nallur S., Gillespie E., et al. , 2009. The mir-34 microRNA is required for the DNA damage response in vivo in C. elegans and in vitro in human breast cancer cells. Oncogene 28: 2419–2424 (erratum: Oncogene 28: 3008) 10.1038/onc.2009.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M., Chen X., Inukai S., Zhao H., Slack F. J., 2011. Age-associated changes in expression of small, noncoding RNAs, including microRNAs, in C. elegans. RNA 17: 1804–1820. 10.1261/rna.2714411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M., Kashem M. A., Cheng C., 2016. An intestinal microRNA modulates the homeostatic adaptation to chronic oxidative stress in C. elegans. Aging (Albany N.Y.) 8: 1979–2005. 10.18632/aging.101029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp B. J., Allman E., Immerman L., Mohnen M., Peters M. A., et al. , 2012. miR-786 regulation of a fatty-acid elongase contributes to rhythmic calcium-wave initiation in C. elegans. Curr. Biol. 22: 2213–2220. 10.1016/j.cub.2012.09.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. K., Gabel H. W., Kamath R. S., Tewari M., Pasquinelli A., et al. , 2005. Functional genomic analysis of RNA interference in C. elegans. Science 308: 1164–1167. 10.1126/science.1109267 [DOI] [PubMed] [Google Scholar]
- Kudlow B. A., Zhang L., Han M., 2012. Systematic analysis of tissue-restricted miRISCs reveals a broad role for microRNAs in suppressing basal activity of the C. elegans pathogen response. Mol. Cell 46: 530–541. 10.1016/j.molcel.2012.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M., Rauhut R., Lendeckel W., Tuschl T., 2001. Identification of novel genes coding for small expressed RNAs. Science 294: 853–858. 10.1126/science.1064921 [DOI] [PubMed] [Google Scholar]
- Lall S., Grun D., Krek A., Chen K., Wang Y. L., et al. , 2006. A genome-wide map of conserved microRNA targets in C. elegans. Curr. Biol. 16: 460–471. 10.1016/j.cub.2006.01.050 [DOI] [PubMed] [Google Scholar]
- Lau N. C., Lim L. P., Weinstein E. G., Bartel D. P., 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294: 858–862. 10.1126/science.1065062 [DOI] [PubMed] [Google Scholar]
- Lee R. C., Ambros V., 2001. An extensive class of small RNAs in Caenorhabditis elegans. Science 294: 862–864. 10.1126/science.1065329 [DOI] [PubMed] [Google Scholar]
- Lee R. C., Feinbaum R. L., Ambros V., 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75: 843–854. 10.1016/0092-8674(93)90529-Y [DOI] [PubMed] [Google Scholar]
- LeGendre J. B., Campbell Z. T., Kroll-Conner P., Anderson P., Kimble J., et al. , 2013. RNA targets and specificity of Staufen, a double-stranded RNA-binding protein in Caenorhabditis elegans. J. Biol. Chem. 288: 2532–2545. 10.1074/jbc.M112.397349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrbach N. J., Armisen J., Lightfoot H. L., Murfitt K. J., Bugaut A., et al. , 2009. LIN-28 and the poly(U) polymerase PUP-2 regulate let-7 microRNA processing in Caenorhabditis elegans. Nat. Struct. Mol. Biol. 16: 1016–1020. 10.1038/nsmb.1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrbach N. J., Castro C., Murfitt K. J., Abreu-Goodger C., Griffin J. L., et al. , 2012. Post-developmental microRNA expression is required for normal physiology, and regulates aging in parallel to insulin/IGF-1 signaling in C. elegans. RNA 18: 2220–2235. 10.1261/rna.035402.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis B. P., Burge C. B., Bartel D. P., 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20. 10.1016/j.cell.2004.12.035 [DOI] [PubMed] [Google Scholar]
- Li J., Greenwald I., 2010. LIN-14 inhibition of LIN-12 contributes to precision and timing of C. elegans vulval fate patterning. Curr. Biol. 20: 1875–1879. 10.1016/j.cub.2010.09.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Le B., Ma X., Li S., You C., et al. , 2016. Biogenesis of phased siRNAs on membrane-bound polysomes in Arabidopsis. Elife 5: e22750 10.7554/eLife.22750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S. Y., Johnson S. M., Abraham M., Vella M. C., Pasquinelli A., et al. , 2003. The C elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable microRNA target. Dev. Cell 4: 639–650. 10.1016/S1534-5807(03)00124-2 [DOI] [PubMed] [Google Scholar]
- Liu F., He C.-X., Luo L.-J., Zou Q.-L., Zhao Y.-X., et al. , 2013. Nuclear hormone receptor regulation of microRNAs controls innate immune responses in C. elegans. PLoS Pathog. 9: e1003545 10.1371/journal.ppat.1003545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Kirch S., Ambros V., 1995. The Caenorhabditis elegans heterochronic gene pathway controls stage-specific transcription of collagen genes. Development 121: 2471–2478. [DOI] [PubMed] [Google Scholar]
- Liu Z. C., Ambros V., 1989. Heterochronic genes control the stage-specific initiation and expression of the dauer larva developmental program in Caenorhabditis elegans. Genes Dev. 3: 2039–2049. 10.1101/gad.3.12b.2039 [DOI] [PubMed] [Google Scholar]
- Lozano E., de Lucas M. P., Saez A. G., 2016. sta-1 is repressed by mir-58 family in Caenorhabditis elegans. Worm 5: e1238560 10.1080/21624054.2016.1238560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y. C., Zhang L., Dai L. L., Khan R. U., Zou C. G., 2017. mir-67 regulates P. aeruginosa avoidance behavior in C. elegans. Biochem. Biophys. Res. Commun. 494: 120–125. 10.1016/j.bbrc.2017.10.069 [DOI] [PubMed] [Google Scholar]
- Martinez N. J., Ow M. C., Barrasa M. I., Hammell M., Sequerra R., et al. , 2008a A C. elegans genome-scale microRNA network contains composite feedback motifs with high flux capacity. Genes Dev. 22: 2535–2549. 10.1101/gad.1678608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez N. J., Ow M. C., Reece-Hoyes J. S., Barrasa M. I., Ambros V. R., et al. , 2008b Genome-scale spatiotemporal analysis of Caenorhabditis elegans microRNA promoter activity. Genome Res. 18: 2005–2015. 10.1101/gr.083055.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulloch K. A., Rougvie A. E., 2014. Caenorhabditis elegans period homolog lin-42 regulates the timing of heterochronic miRNA expression. Proc. Natl. Acad. Sci. USA 111: 15450–15455. 10.1073/pnas.1414856111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen T. J., Yao Q., Yun S., Lee C. Y., Bennett K. L., 2016. Small RNA in situ hybridization in Caenorhabditis elegans, combined with RNA-seq, identifies germline-enriched microRNAs. Dev. Biol. 418: 248–257. 10.1016/j.ydbio.2016.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McJunkin K., Ambros V., 2014. The embryonic mir-35 family of microRNAs promotes multiple aspects of fecundity in Caenorhabditis elegans. G3 (Bethesda) 4: 1747–1754. 10.1534/g3.114.011973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McJunkin K., Ambros V., 2017. A microRNA family exerts maternal control on sex determination in C. elegans. Genes Dev. 31: 422–437. 10.1101/gad.290155.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metheetrairut C., Adams B. D., Nallur S., Weidhaas J. B., Slack F. J., 2017. cel-mir-237 and its homologue, hsa-miR-125b, modulate the cellular response to ionizing radiation. Oncogene 36: 512–524. 10.1038/onc.2016.222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micklem D. R., Adams J., Grunert S., St Johnston D., 2000. Distinct roles of two conserved Staufen domains in oskar mRNA localization and translation. EMBO J. 19: 1366–1377. 10.1093/emboj/19.6.1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki T. S., Ruegger S., Gaidatzis D., Stadler M. B., Grosshans H., 2014. Engineering of a conditional allele reveals multiple roles of XRN2 in Caenorhabditis elegans development and substrate specificity in microRNA turnover. Nucleic Acids Res. 42: 4056–4067. 10.1093/nar/gkt1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miska E. A., Alvarez-Saavedra E., Abbott A. L., Lau N. C., Hellman A. B., et al. , 2007. Most Caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS Genet. 3: e215 10.1371/journal.pgen.0030215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondol V., Ahn B. C., Pasquinelli A. E., 2015. Splicing remodels the let-7 primary microRNA to facilitate Drosha processing in Caenorhabditis elegans. RNA 21: 1396–1403. 10.1261/rna.052118.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss E. G., Lee R. C., Ambros V., 1997. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell 88: 637–646. 10.1016/S0092-8674(00)81906-6 [DOI] [PubMed] [Google Scholar]
- Nehammer C., Podolska A., Mackowiak S. D., Kagias K., Pocock R., 2015. Specific microRNAs regulate heat stress responses in Caenorhabditis elegans. Sci. Rep. 5: 8866 10.1038/srep08866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolde M. J., Saka N., Reinert K. L., Slack F. J., 2007. The Caenorhabditis elegans pumilio homolog, puf-9, is required for the 3′UTR-mediated repression of the let-7 microRNA target gene, hbl-1. Dev. Biol. 305: 551–563. 10.1016/j.ydbio.2007.02.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nottrott S., Simard M. J., Richter J. D., 2006. Human let-7a miRNA blocks protein production on actively translating polyribosomes. Nat. Struct. Mol. Biol. 13: 1108–1114. 10.1038/nsmb1173 [DOI] [PubMed] [Google Scholar]
- Olsen P. H., Ambros V., 1999. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev. Biol. 216: 671–680. 10.1006/dbio.1999.9523 [DOI] [PubMed] [Google Scholar]
- Olsson-Carter K., Slack F. J., 2010. A developmental timing switch promotes axon outgrowth independent of known guidance receptors. PLoS Genet. 6: e1001054 10.1371/journal.pgen.1001054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano D. J., Kingston E. R., Kim D. H., 2015. Tissue expression pattern of PMK-2 p38 MAPK is established by the miR-58 family in C. elegans. PLoS Genet. 11: e1004997 (erratum: PLoS Genet. 11: e1005317) 10.1371/journal.pgen.1004997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit A., Jain V., Kumar N., Mukhopadhyay A., 2014. PHA-4/FOXA-regulated microRNA feed forward loops during Caenorhabditis elegans dietary restriction. Aging (Albany N.Y.) 6: 835–855. 10.18632/aging.100697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. H., Ahn S., Kim S., Lee J., Nam J.-W., et al. , 2013. Degradome sequencing reveals an endogenous microRNA target in C. elegans. FEBS Lett. 587: 964–969. 10.1016/j.febslet.2013.02.029 [DOI] [PubMed] [Google Scholar]
- Parry D. H., Xu J., Ruvkun G., 2007. A whole-genome RNAi Screen for C. elegans miRNA pathway genes. Curr. Biol. 17: 2013–2022. 10.1016/j.cub.2007.10.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli A. E., Reinhart B. J., Slack F., Martindale M. Q., Kuroda M. I., et al. , 2000. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 408: 86–89. 10.1038/35040556 [DOI] [PubMed] [Google Scholar]
- Pedersen M. E., Snieckute G., Kagias K., Nehammer C., Multhaupt H. A. B., et al. , 2013. An epidermal microRNA regulates neuronal migration through control of the cellular glycosylation state. Science 341: 1404–1408. 10.1126/science.1242528 [DOI] [PubMed] [Google Scholar]
- Perales R., King D. M., Aguirre-Chen C., Hammell C. M., 2014. LIN-42, the Caenorhabditis elegans PERIOD homolog, negatively regulates microRNA transcription. PLoS Genet. 10: e1004486 10.1371/journal.pgen.1004486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch M., Ecsedi M., Bartake H., Mullner A., Grosshans H., 2015. A genetic interactome of the let-7 microRNA in C. elegans. Dev. Biol. 401: 276–286. 10.1016/j.ydbio.2015.02.013 [DOI] [PubMed] [Google Scholar]
- Rauthan M., Gong J., Liu J., Li Z., Wescott S. A., et al. , 2017. MicroRNA regulation of nAChR expression and nicotine-dependent behavior in C. elegans. Cell Rep. 21: 1434–1441. 10.1016/j.celrep.2017.10.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehmsmeier M., Steffen P., Hochsmann M., Giegerich R., 2004. Fast and effective prediction of microRNA/target duplexes. RNA 10: 1507–1517. 10.1261/rna.5248604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart B. J., Slack F. J., Basson M., Pasquinelli A. E., Bettinger J. C., et al. , 2000. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403: 901–906. 10.1038/35002607 [DOI] [PubMed] [Google Scholar]
- Ren Z., Ambros V. R., 2015. Caenorhabditis elegans microRNAs of the let-7 family act in innate immune response circuits and confer robust developmental timing against pathogen stress. Proc. Natl. Acad. Sci. USA 112: E2366–E2375. 10.1073/pnas.1422858112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Z., Veksler-Lublinsky I., Morrissey D., Ambros V., 2016. Staufen negatively modulates microRNA activity in Caenorhabditis elegans. G3 (Bethesda) 6: 1227–1237. 10.1534/g3.116.027300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades M. W., Reinhart B. J., Lim L. P., Burge C. B., Bartel B., et al. , 2002. Prediction of plant microRNA targets. Cell 110: 513–520. [DOI] [PubMed] [Google Scholar]
- Rokavec M., Li H., Jiang L., Hermeking H., 2014. The p53/miR-34 axis in development and disease. J. Mol. Cell Biol. 6: 214–230. 10.1093/jmcb/mju003 [DOI] [PubMed] [Google Scholar]
- Roush S. F., Slack F. J., 2009. Transcription of the C. elegans let-7 microRNA is temporally regulated by one of its targets, hbl-1. Dev. Biol. 334: 523–534. 10.1016/j.ydbio.2009.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby J. G., Jan C. H., Bartel D. P., 2007. Intronic microRNA precursors that bypass Drosha processing. Nature 448: 83–86. 10.1038/nature05983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seggerson K., Tang L., Moss E. G., 2002. Two genetic circuits repress the Caenorhabditis elegans heterochronic gene lin-28 after translation initiation. Dev. Biol. 243: 215–225. 10.1006/dbio.2001.0563 [DOI] [PubMed] [Google Scholar]
- Shaw W. R., Armisen J., Lehrbach N. J., Miska E. A., 2010. The conserved miR-51 microRNA family is redundantly required for embryonic development and pharynx attachment in Caenorhabditis elegans. Genetics 185: 897–905. 10.1534/genetics.110.117515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherrard R., Luehr S., Holzkamp H., McJunkin K., Memar N., et al. , 2017. miRNAs cooperate in apoptosis regulation during C. elegans development. Genes Dev. 31: 209–222. 10.1101/gad.288555.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z., Ruvkun G., 2012. The mevalonate pathway regulates microRNA activity in Caenorhabditis elegans. Proc. Natl. Acad. Sci USA 109: 4568–4573. 10.1073/pnas.1202421109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z., Hayes G., Ruvkun G., 2013. Dual regulation of the lin-14 target mRNA by the lin-4 miRNA. PLoS One 8: e75475 10.1371/journal.pone.0075475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon D. J., Madison J. M., Conery A. L., Thompson-Peer K. L., Soskis M., et al. , 2008. The microRNA miR-1 regulates a MEF-2-dependent retrograde signal at neuromuscular junctions. Cell 133: 903–915. 10.1016/j.cell.2008.04.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack F. J., Basson M., Liu Z., Ambros V., Horvitz H. R., et al. , 2000. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol. Cell 5: 659–669. 10.1016/S1097-2765(00)80245-2 [DOI] [PubMed] [Google Scholar]
- Stadler M., Artiles K., Pak J., Fire A., 2012. Contributions of mRNA abundance, ribosome loading, and post- or peri-translational effects to temporal repression of C. elegans heterochronic miRNA targets. Genome Res. 22: 2418–2426. 10.1101/gr.136515.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefani G., Chen X., Zhao H., Slack F. J., 2015. A novel mechanism of LIN-28 regulation of let-7 microRNA expression revealed by in vivo HITS-CLIP in C. elegans. RNA 21: 985–996. 10.1261/rna.045542.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner F. A., Hoogstrate S. W., Okihara K. L., Thijssen K. L., Ketting R. F., et al. , 2007. Structural features of small RNA precursors determine Argonaute loading in Caenorhabditis elegans. Nat. Struct. Mol. Biol. 14: 927–933. 10.1038/nsmb1308 [DOI] [PubMed] [Google Scholar]
- Tabara H., Sarkissian M., Kelly W. G., Fleenor J., Grishok A., et al. , 1999. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99: 123–132. [DOI] [PubMed] [Google Scholar]
- Tat T. T., Maroney P. A., Chamnongpol S., Coller J., Nilsen T. W., 2016. Cotranslational microRNA mediated messenger RNA destabilization. Elife 5: e12880 10.7554/eLife.12880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Than M. T., Kudlow B. A., Han M., 2013. Functional analysis of neuronal microRNAs in Caenorhabditis elegans dauer formation by combinational genetics and Neuronal miRISC immunoprecipitation. PLoS Genet. 9: e1003592 10.1371/journal.pgen.1003592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson-Peer K. L., Bai J., Hu Z., Kaplan J. M., 2012. HBL-1 patterns synaptic remodeling in C. elegans. Neuron 73: 453–465. 10.1016/j.neuron.2011.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsialikas J., Romens M. A., Abbott A., Moss E. G., 2017. Stage-specific timing of the microRNA regulation of lin-28 by the heterochronic gene lin-14 in Caenorhabditis elegans. Genetics 205: 251–262. 10.1534/genetics.116.195040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner M. J., Jiao A. L., Slack F. J., 2014. Autoregulation of lin-4 microRNA transcription by RNA activation (RNAa) in C. elegans. Cell Cycle 13: 772–781. 10.4161/cc.27679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wynsberghe P. M., Kai Z. S., Massirer K. B., Burton V. H., Yeo G. W., et al. , 2011. LIN-28 co-transcriptionally binds primary let-7 to regulate miRNA maturation in Caenorhabditis elegans. Nat. Struct. Mol. Biol. 18: 302–308. 10.1038/nsmb.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wynsberghe P. M., Finnegan E. F., Stark T., Angelus E. P., Homan K. E., et al. , 2014. The Period protein homolog LIN-42 negatively regulates microRNA biogenesis in C. elegans. Dev. Biol. 390: 126–135. 10.1016/j.ydbio.2014.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasquez-Rifo A., Jannot G., Armisen J., Labouesse M., Bukhari S. I., et al. , 2012. Developmental characterization of the microRNA-specific C. elegans Argonautes alg-1 and alg-2. PLoS One 7: e33750 10.1371/journal.pone.0033750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasquez-Rifo A., Bosse G. D., Rondeau E. L., Jannot G., Dallaire A., et al. , 2013. A new role for the GARP complex in microRNA-mediated gene regulation. PLoS Genet. 9: e1003961 10.1371/journal.pgen.1003961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vella M. C., Choi E. Y., Lin S. Y., Reinert K., Slack F. J., 2004. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3′UTR. Genes Dev. 18: 132–137. 10.1101/gad.1165404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vora M., Shah M., Ostafi S., Onken B., Xue J., et al. , 2013. Deletion of microRNA-80 activates dietary restriction to extend C. elegans healthspan and lifespan. PLoS Genet. 9: e1003737 10.1371/journal.pgen.1003737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Gupta P., Fressigne L., Bossé G. D., Wang X., et al. , 2017. TEG-1 CD2BP2 controls miRNA levels by regulating miRISC stability in C. elegans and human cells. Nucleic Acids Res. 45: 1488–1500. 10.1093/nar/gkw836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Hou L., Nakamura S., Su M., Li F., et al. , 2017. LIN-28 balances longevity and germline stem cell number in Caenorhabditis elegans through let-7/AKT/DAF-16 axis. Aging Cell 16: 113–124. 10.1111/acel.12539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman B., Ha I., Ruvkun G., 1993. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75: 855–862. [DOI] [PubMed] [Google Scholar]
- Wu E., Thivierge C., Flamand M., Mathonnet G., Vashisht A. A., et al. , 2010. Pervasive and cooperative deadenylation of 3′UTRs by embryonic microRNA families. Mol. Cell 40: 558–570. 10.1016/j.molcel.2010.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Huang C., Taki F. A., Zhang Y., Dobbins D. L., et al. , 2015. Benzo-alpha-pyrene induced oxidative stress in Caenorhabditis elegans and the potential involvements of microRNA. Chemosphere 139: 496–503. 10.1016/j.chemosphere.2015.08.031 [DOI] [PubMed] [Google Scholar]
- Wu Q., Zhao Y., Zhao G., Wang D., 2014. microRNAs control of in vivo toxicity from graphene oxide in Caenorhabditis elegans. Nanomedicine 10: 1401–1410. 10.1016/j.nano.2014.04.005 [DOI] [PubMed] [Google Scholar]
- Yang J., Chen D., He Y., Melendez A., Feng Z., et al. , 2013. MiR-34 modulates Caenorhabditis elegans lifespan via repressing the autophagy gene atg9. Age (Dordr.) 35: 11–22. 10.1007/s11357-011-9324-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi Y. H., Ma T. H., Lee L. W., Chiou P. T., Chen P. H., et al. , 2015. A genetic cascade of let-7-ncl-1-fib-1 modulates nucleolar size and rRNA pool in Caenorhabditis elegans. PLoS Genet. 11: e1005580 10.1371/journal.pgen.1005580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo A. S., Greenwald I., 2005. LIN-12/Notch activation leads to microRNA-mediated down-regulation of Vav in C. elegans. Science 310: 1330–1333. 10.1126/science.1119481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Emmons S. W., 2009. Regulation of the Caenorhabditis elegans posterior Hox gene egl-5 by microRNA and the polycomb-like gene sop-2. Dev. Dyn. 238: 595–603. 10.1002/dvdy.21876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Fire A. Z., 2010. Cell autonomous specification of temporal identity by Caenorhabditis elegans microRNA lin-4. Dev. Biol. 344: 603–610. 10.1016/j.ydbio.2010.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Artiles K. L., Fire A. Z., 2015. Functional relevance of “seed” and “non-seed” sequences in microRNA-mediated promotion of C. elegans developmental progression. RNA 21: 1980–1992. 10.1261/rna.053793.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Ding L., Cheung T. H., Dong M.-Q., Chen J., et al. , 2007. Systematic identification of C. elegans miRISC proteins, miRNAs, and mRNA targets by their interactions with GW182 proteins AIN-1 and AIN-2. Mol. Cell 28: 598–613. 10.1016/j.molcel.2007.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Hammell M., Kudlow B. A., Ambros V., Han M., 2009. Systematic analysis of dynamic miRNA-target interactions during C. elegans development. Development 136: 3043–3055. 10.1242/dev.039008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Zhang H., 2013. Autophagy modulates miRNA-mediated gene silencing and selectively degrades AIN-1/GW182 in C. elegans. EMBO Rep. 14: 568–576. 10.1038/embor.2013.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Zabinsky R., Teng Y., Cui M., Han M., 2011. microRNAs play critical roles in the survival and recovery of Caenorhabditis elegans from starvation-induced L1 diapause. Proc. Natl. Acad. Sci. USA 108: 17997–18002. 10.1073/pnas.1105982108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Boyle T. J., Liu Z., Murray J. I., Wood W. B., et al. , 2010. A negative regulatory loop between microRNA and Hox gene controls posterior identities in Caenorhabditis elegans. PLoS Genet. 6: e1001089 10.1371/journal.pgen.1001089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhi L., Yu Y., Li X., Wang D., Wang D., 2017. Molecular control of innate immune response to Pseudomonas aeruginosa infection by intestinal let-7 in Caenorhabditis elegans. PLoS Pathog. 13: e1006152 10.1371/journal.ppat.1006152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinovyeva A. Y., Bouasker S., Simard M. J., Hammell C. M., Ambros V., 2014. Mutations in conserved residues of the C. elegans microRNA Argonaute ALG-1 identify separable functions in ALG-1 miRISC loading and target repression. PLoS Genet. 10: e1004286 10.1371/journal.pgen.1004286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinovyeva A. Y., Veksler-Lublinsky I., Vashisht A. A., Wohlschlegel J. A., Ambros V. R., 2015. Caenorhabditis elegans ALG-1 antimorphic mutations uncover functions for Argonaute in microRNA guide strand selection and passenger strand disposal. Proc. Natl. Acad. Sci. USA 112: E5271–E5280. 10.1073/pnas.1506576112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zisoulis D. G., Lovci M. T., Wilbert M. L., Hutt K. R., Liang T. Y., et al. , 2010. Comprehensive discovery of endogenous Argonaute binding sites in Caenorhabditis elegans. Nat. Struct. Mol. Biol. 17: 173–179. 10.1038/nsmb.1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zisoulis D. G., Kai Z. S., Chang R. K., Pasquinelli A. E., 2012. Autoregulation of microRNA biogenesis by let-7 and Argonaute. Nature 486: 541–544. 10.1038/nature11134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y., Chiu H., Domenger D., Chuang C.-F., Chang C., 2012. The lin-4 microRNA targets the LIN-14 transcription factor to inhibit netrin-mediated axon attraction. Sci. Signal. 5: ra43 10.1126/scisignal.2002437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y., Chiu H., Zinovyeva A., Ambros V., Chuang C.-F., et al. , 2013. Developmental decline in neuronal regeneration by the progressive change of two intrinsic timers. Science 340: 372–376. 10.1126/science.1231321 [DOI] [PMC free article] [PubMed] [Google Scholar]