Abstract
Regulated gene expression is necessary for developmental and homeostatic processes. The KDM5 family of transcriptional regulators are histone H3 lysine 4 demethylases that can function through both demethylase-dependent and -independent mechanisms. While loss and overexpression of KDM5 proteins are linked to intellectual disability and cancer, respectively, their normal developmental functions remain less characterized. Drosophila melanogaster provides an ideal system to investigate KDM5 function, as it encodes a single ortholog in contrast to the four paralogs found in mammalian cells. To examine the consequences of complete loss of KDM5, we generated a null allele of Drosophila kdm5, also known as little imaginal discs (lid), and show that it is essential for viability. Animals lacking KDM5 show a dramatically delayed larval development that coincides with decreased proliferation and increased cell death in wing imaginal discs. Interestingly, this developmental delay is independent of the well-characterized Jumonji C (JmjC) domain-encoded histone demethylase activity of KDM5, suggesting key functions for less characterized domains. Consistent with the phenotypes observed, transcriptome analyses of kdm5 null mutant wing imaginal discs revealed the dysregulation of genes involved in several cellular processes, including cell cycle progression and DNA repair. Together, our analyses reveal KDM5 as a key regulator of larval growth and offer an invaluable tool for defining the biological activities of KDM5 family proteins.
Keywords: KDM5, Lid, H3K4me3, chromatin, larval growth, imaginal disc
REGULATED gene expression is essential for growth and cell fate decisions that are critical to development and to the maintenance of tissues and organs during adulthood. Changes to chromatin, the structure that includes DNA and its associated histone proteins, is one key mechanism by which transcription is regulated (Swygert and Peterson 2014). Histones are extensively decorated by covalent modifications that can affect chromatin compaction to influence transcription factor binding, or affect the recruitment of proteins that recognize specific histone modifications to activate or repress promoter activity (Rothbart and Strahl 2014). One family of transcriptional regulators that both recognizes and enzymatically modifies chromatin is the lysine demethylase 5 (KDM5) family of evolutionarily conserved histone demethylases. Mammalian cells encode four KDM5 paralogs, KDM5A, KDM5B, KDM5C, and KDM5D, whereas organisms with smaller genomes, such as Drosophila and Caenorhabditis elegans, have a single KDM5 protein.
Dysregulation of KDM5 family genes is linked to human disorders, underscoring the importance of understanding the in vivo functions of KDM5 proteins. Specifically, loss-of-function mutations in KDM5A, KDM5B, and KDM5C are found in patients with intellectual disability, establishing KDM5 as a key regulator of neuronal development or function (Vallianatos and Iwase 2015). Altered expression of KDM5 family genes is also implicated in cancer (Blair et al. 2011). This is best illustrated by KDM5A and KDM5B, which are overexpressed in breast, ovarian, and lung cancers, and whose expression correlates with proliferation rate and propensity for metastatic invasion (Hayami et al. 2010; Hou et al. 2012; Teng et al. 2013; Yamamoto et al. 2014; Wang et al. 2015; Feng et al. 2017; Huang et al. 2018). Although the molecular mechanisms by which dysregulation of KDM5 proteins cause disease remain unknown, it is likely to involve their activities as transcriptional regulators. The Jumonji C (JmjC) catalytic domain that demethylates histone H3 that is trimethylated at lysine 4 (H3K4me3) is the most characterized domain of KDM5 proteins (Klose and Zhang 2007). High levels of H3K4me3 are found predominantly surrounding the transcriptional start site of actively expressed genes (Santos-Rosa et al. 2002). Thus, the demethylase activity of KDM5 primarily results in transcriptional repression. However, the importance of this enzymatic activity to disease initiation and progression remains enigmatic. For example, while a number of intellectual disability-associated missense mutations in KDM5C impair in vitro demethylase activity, this is not universally true (Tahiliani et al. 2007; Vallianatos and Iwase 2015; Vallianatos et al. 2018). Likewise, the proliferation and survival of some KDM5-overexpressing cancers rely on enzymatic activity (Teng et al. 2013), while others do not (Cao et al. 2014). Other domains of KDM5 are therefore likely to play key roles in KDM5-regulated expression. One such candidate is the C-terminal PHD domain (PHD3) of KDM5 that binds to H3K4me2/3 to regulate genes necessary for metabolic and mitochondrial functions (Wang et al. 2009; Li et al. 2010; Klein et al. 2014; Liu and Secombe 2015). The importance of this PHD motif to gene expression regulation is highlighted by the observation that this domain causes leukemia when fused to the nuclear pore protein NUP98 (van Zutven et al. 2006; Wang et al. 2009).
Despite a growing body of evidence linking altered expression of KDM5 proteins to human disorders, the normal developmental functions of KDM5 proteins remain unclear. To begin defining the physiological functions of KDM5, the genetic advantages of model organisms such as mice, flies, and worms have been utilized. Significantly, some phenotypes observed are similar to the clinical features found in patients with altered levels of KDM5 proteins, suggesting that further studies will aid in uncovering the molecular mechanisms underlying these disorders. For example, consistent with mutations in KDM5 genes causing intellectual disability in humans, mice and worms harboring mutations in kdm5 genes display cognitive and axonal guidance defects, respectively (Iwase et al. 2016; Lussi et al. 2016; Mariani et al. 2016; Scandaglia et al. 2017). Mouse knockout studies also link KDM5 to proliferation, highlighting a potential means by which dysregulation could contribute to tumorigenesis. For instance, while Kdm5A knockout mice are viable, cultured mouse embryonic fibroblasts from these animals show reduced proliferation, implicating KDM5A as a positive regulator of cell cycle progression (Lin et al. 2011). Kdm5B and Kdm5C knockout mice also survive, but they are smaller than their littermates, suggesting roles for these KDM5 family proteins in growth regulation (Zou et al. 2014; Iwase et al. 2016). Unfortunately, mouse studies carried out to-date have been complicated by phenotypes that are dependent on strain background, raising questions regarding whether phenotypes observed are specifically due to the loss of the Kdm5 gene being examined (Catchpole et al. 2011; Albert et al. 2013). Moreover, there is evidence that loss of one KDM5 family gene can cause compensatory upregulation of another family member, making it difficult to define the processes that rely on KDM5 function (Jensen et al. 2010; Zou et al. 2014).
Drosophila encodes a single kdm5 gene, also known as little imaginal discs (lid), that is broadly expressed in all tissues examined (Secombe et al. 2007; Liu et al. 2014; Zamurrad et al. 2018). Decreasing KDM5 levels using hypomorphic alleles or by RNA interference-mediated knockdown reduces viability, suggesting that in the absence of paralogs, kdm5 may be necessary for development (Gildea et al. 2000; Li et al. 2010; Lloret-Llinares et al. 2012). However, analyses of the phenotypes caused by a complete absence of KDM5 have been hampered by the absence of a kdm5 null allele. Here, we describe the generation of the amorphic allele kdm5140 and show that KDM5 is essential for viability. Specifically, we demonstrate that loss of KDM5 slows larval development in a manner that correlates with decreased rates of proliferation and increased cell death in larval wing imaginal discs. Transcriptome analyses of kdm5140 wing imaginal discs show that loss of KDM5 alters the expression of genes involved in a number of cellular processes, including cell cycle progression and DNA damage repair. Consistent with this, kdm5140 mutants are sensitive to DNA-damaging agents. Together, our results demonstrate that KDM5 is a key regulator of larval growth and provides an invaluable tool to further dissect the biological roles of KDM5 proteins.
Materials and Methods
Fly strains and husbandry
All flies were kept at 25° on standard food with 60% humidity and a 12-hr light/dark cycle. The kdm5:HAWT and kdm5:HAJmjC* transgenes are published elsewhere (Navarro-Costa et al. 2016; Zamurrad et al. 2018). Briefly, they are 11 kb constructs that encompass the kdm5 genomic region with the addition of three HA tags at the 3′ end of the open reading frame. Transgenes were inserted into the attP site at 86Fb and the white and RFP cassette were removed using germline expression of the Cre recombinase (Bischof et al. 2007). The kdm5:HAW1771A transgene is identical to kdm5:HAWT except for a codon change that alters tryptophan 1771 to alanine. The kdm5:HAW1771A transgene and corresponding control wild-type transgene were inserted into the attP site at 68A4 on chromosome III (generated at the “Rainbow transgenic flies”) (Groth et al. 2004). This kdm5:HAW1771A construct is similar to a previously published transgene that was not HA tagged, did not contain kdm5 introns, and was randomly integrated into the Drosophila genome (Liu and Secombe 2015). The homozygous viable kdm5NP4707 strain was obtained from the Kyoto Stock Center (stock #104754; Kyoto Institute of Technology) and kdm5140 was generated by imprecise excision of this P element using Δ2–3 transposase. The breakpoints of kdm5140 were molecularly mapped by genomic PCR and subsequent sequencing (Supplemental Material, Figure S1). All other strains were obtained from the Bloomington Drosophila Stock Center.
Developmental timing analyses
Ten females and males were crossed and allowed to lay eggs for 24 hr in food vials, with parental flies being subsequently removed. The number of animals that had pupariated was scored twice per day. Experiments were carried out in biological triplicate.
γ-Irradiation
Third-instar larvae were exposed to a 40 Gy dose of γ-radiation using a 137Cs irradiator (Shepherd Mark I Irradiator). After 4 hr at 25°, wings discs were dissected and used for Dcp-1 immunostaining. Fluorescence intensity of Dcp1 staining was quantified using ImageJ Software and divided by the area of that wing disc to provide a ratio of intensity/area.
Larval food ingestion analyses
To monitor food intake, adult flies were allowed to lay eggs on fly food containing 0.05% bromophenol blue. Larvae were removed from the food 36 hr later and examined by light microscopy.
Immunostaining
For immunostaining, wing imaginal discs from third-instar larvae were dissected in 1× PBS and fixed in 4% paraformaldehyde for 30 min at room temperature. They were then washed three times with PBST (1× PBS, 0.2% Triton), blocked for 1 hr at 4° in 0.1% BSA, and incubated with the primary antibody overnight at 4°. Anti-phospho-histone H3 (#9701; Cell Signaling Technology) and anti-Dcp1 (#9578; Cell Signaling Technology) were used at 1:50 and 1:100, respectively. For pupal wing staining, white prepupae were picked and aged until 28–30 hr after puparium formation (APF) at 25° before fixing in 4% paraformaldehyde overnight at 4°. Samples were then washed three times with PBST, pupal wings were dissected, blocked for 1 hr at 4° with 0.1% BSA/PBST, and incubated with anti-Futsch (22C10, at 1:50; Developmental Studies Hybridoma Bank, University of Iowa) overnight at 4°. Wings were mounted in Vectashield and imaged on a Zeiss Axio Imager 2 microscope. For quantification, the number of phospho-histone H3- and Dcp1-positive cells in the wing pouch were counted using Image J software.
Western blotting
Western blotting was carried out as previously described, using LI-COR Odyssey (LI-COR Biosciences) (Liu and Secombe 2015). Antibodies used were anti-phospho-histone H3 (#9701, 1:1000; Cell Signaling Technology), anti-histone H3 (#39763 or #39163, 1:5000; Active Motif), anti-α-tubulin (1:5000; Developmental Studies Hybridoma Bank, University of Iowa). The rabbit polyclonal KDM5 antibody was raised to amino acids 1418–1760 and has been previously published (Secombe et al. 2007).
Translation quantification
Translation levels in wing imaginal discs was quantified as previously described (Deliu et al. 2017). Briefly, wing discs from third-instar larvae were dissected and incubated in 5 mg/ml puromycin (Sigma, St. Louis, MO) or puromycin plus 10 mg/ml cycloheximide (Sigma). Puromycin levels in wing discs were then assayed by Western blotting with anti-puromycin (3RH11, 1:1000; Kerafast). Anti-histone H3 (#39763, 1:1000; Active Motif) was used as a loading control. Quantification was carried out using LI-COR Odyssey software version 3.0.30 (LI-COR Biosciences) by determining the intensity of all puromycin-labeled proteins between 14 and 200 kDa and dividing this by the intensity of the histone H3 load control.
RNA sequencing
RNA sequencing (RNA-seq) was carried out at the New York Genome Center. RNA was prepared in biological triplicate from wild-type (kdm5140; kdm5:HAWT; referred to as kdm5WT) and kdm5140 third-instar mutant larval wing discs matched for developmental age using TRIzol and RNAeasy (Qiagen, Valencia, CA). An equal mix of male and female larvae were used. RNA-seq libraries were prepared using the TruSeq Stranded mRNA Library Preparation Kit in accordance with the manufacturer’s instructions. Briefly, 500 ng of total RNA was used for purification and fragmentation of messenger RNA (mRNA). Purified mRNA underwent first and second strand complementary DNA (cDNA) synthesis. cDNA was then adenylated, ligated to Illumina sequencing adapters, and amplified by PCR (eight cycles). Final libraries were evaluated using fluorescence-based assays, including PicoGreen (Life Technologies) or Qubit Fluorometer (Invitrogen, Carlsbad, CA) and Fragment Analyzer (Advanced Analytics) or BioAnalyzer (2100; Agilent), and were sequenced on an Illumina HiSeq2500 sequencer (v4 chemistry) using 2 × 50 bp cycles. Alignment of raw reads was carried out using STAR aligner, normalized, and differential expression determined with DESeq2. The accession number for the RNA-seq data described here is GSE109201. A volcano plot showing dysregulated genes was generated with ggplot2 package in R.
Real-time PCR
Real-time PCR was carried out as previously described (Liu and Secombe 2015), using cDNA from third-instar larval wing imaginal discs. Primer sequences are provided in Table S1.
Statistical analyses
All experiments were done in biological triplicate (minimum) and Ns are provided for each experiment. Fisher’s exact test was carried out with the program R (v3.3.2). Student’s t-test, chi-squared and Wilcoxon rank-sum tests were carried out using GraphPad Prism version 7.00 (GraphPad Software, La Jolla, CA).
Data availability
The fly strains described in this article are available upon request. RNA-seq data from kdm5140 mutant wing discs is available under the accession number GSE109201. A list of differential expressed genes (and log2 fold change) observed in kdm5140 compared to wild type is provided in Table S2 [5% false discovery rate (FDR)]. Table S2 also includes data from KDM5 chromatin immunoprecipitation sequencing (ChIP-seq) that is published and publicly available from adult flies (accession number GSE70591) (Liu and Secombe 2015) and wing imaginal discs (accession number GSE27081) (Lloret-Llinares et al. 2012). Table S3 shows genes dysregulated in kdm5140 RNA-seq and kdm510424 wing disc microarray data (accession number GSZ53881) (Liu et al. 2014). Table S4 shows genes dysregulated in kdm5140 RNA-seq and S2 cell KDM5 knockdown RNA-seq (accession number GSE68775) (Gajan et al. 2016). The overlap between genes affected in kdm5140 RNA-seq, kdm510424 microarray, S2 cell knockdown RNA-seq, and kdm5 knockdown wing disc microarray (accession number GSE27081) (Lloret-Llinares et al. 2012) is provided in Figure S2. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6239003.
Results
kdm5 is an essential gene required for developmental timing
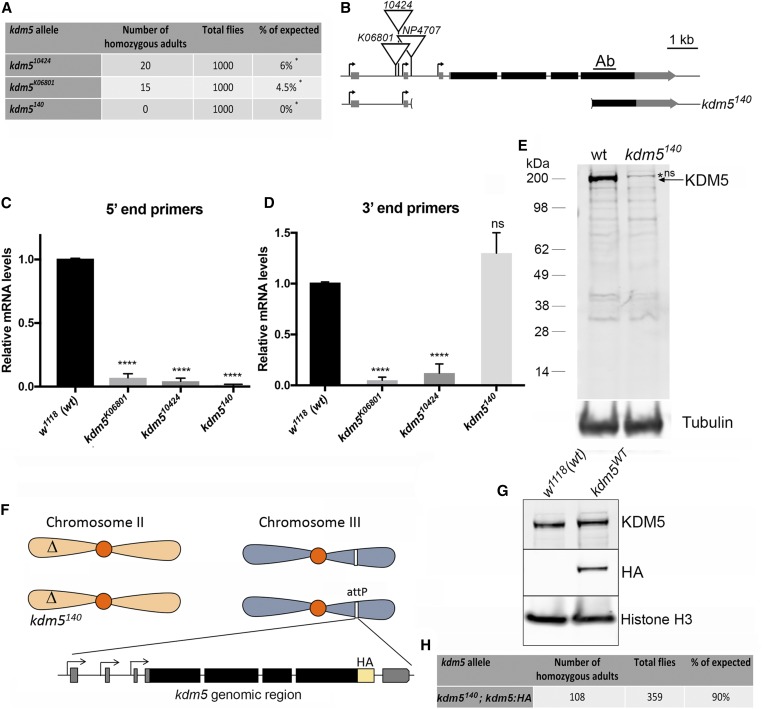
Existing hypomorphic P element alleles of kdm5 such as kdm510424 and kdm5K06801 are 95% lethal (Gildea et al. 2000; Secombe et al. 2007) (Figure 1, A and B). The small number of kdm5 mutant adult flies that do eclose are morphologically normal, but are short-lived (Liu and Secombe 2015). Because the effects of complete loss of KDM5 remain unknown, we generated a kdm5 null allele by imprecise excision of a P element inserted in the kdm5 promoter region (kdm5140; Figure 1B). Molecular mapping demonstrated that this allele deletes three of the four coding exons of the kdm5 gene, including the start codon (Figure 1B and Figure S1). Real-time PCR analyses of kdm5140 homozygous mutant larvae confirmed the loss of the full-length kdm5 transcript compared to the genetically similar w1118 wild-type strain (Figure 1C). Although a partial 3′ end transcript remains, with an open reading frame with the potential to encode a 51 kDa protein, no full-length or truncated KDM5 protein(s) were present in kdm5140 animals (Figure 1, D and E). kdm5140 null mutants are 100% lethal (Figure 1A). To confirm that this was due specifically to the loss of KDM5, we utilized a genomic rescue transgene containing a HA-tagged form of the kdm5 locus (Figure 1, F and G) (Navarro-Costa et al. 2016; Zamurrad et al. 2018). This transgene fully rescued the lethality of kdm5140 mutants, establishing kdm5 as an essential gene in Drosophila (Figure 1H).
Figure 1.
A kdm5140 null allele demonstrates that KDM5 is essential in Drosophila. (A) Lethality of kdm5K06801, kdm510424, and kdm5140 homozygous mutant animals generated from a cross between five female and five male heterozygous parents balanced using CyO-GFP. The column labeled total flies indicates the number of progeny (adult) flies scored from at least three independent crosses. Expected number of progeny is based on Mendelian frequencies and taking into account the lethality of CyO homozygotes, i.e., 33% of total adult flies. * P < 0.01 (chi-squared test). (B) Position of the NP4707, 10424, and K06801 P-element insertions and molecular mapping of the kdm5140 deletion. Ab indicates the region used to generate the rabbit polyclonal anti-KDM5 antibody (Secombe et al. 2007). (C) Real-time PCR using primers to the 5′ end of the gene using RNA from whole third-instar larvae. Animals homozygous for kdm5K06801 or kdm510424 show low levels of transcript while kdm5140 shows none. kdm5 mRNA normalized to wild type (w1118), using rp49. **** P < 0.0001. (D) Real-time PCR using primers to the 3′ end of the gene using RNA from whole third-instar larvae. kdm5140 has wild-type levels of the 3′ end of the transcript. **** P < 0.0001. (E) Western blot from wild-type (w1118) and kdm5140 homozygous mutant wing imaginal discs showing KDM5 and α-tubulin. kdm5140 animals have no detectable full-length or truncated KDM5 proteins. *ns indicates nonspecific band. (F) Schematic of strain genotype for rescue of kdm5140 with a genomic rescue transgene. Flies are homozygous for the kdm5140 mutation on the second chromosome and homozygous for an 11 kb genomic rescue transgene on the third chromosome. (G) Western blot showing KDM5 protein levels from third-instar larval wing imaginal discs from wild-type (w1118) and kdm5140 homozygotes that also have two copies of the kdm5:HA genomic rescue transgene. Anti-KDM5 (top), anti-HA (middle), and anti-histone H3 loading control (bottom). (H) kdm5140 lethality is rescued by a transgene encoding the kdm5 locus. These data were generated by crossing female and male flies heterozygous for kdm5140 and homozygous the wild-type genomic rescue transgene (intercross of kdm5140/CyO-GFP ; kdm5:HA/kdm5:HA males and females).
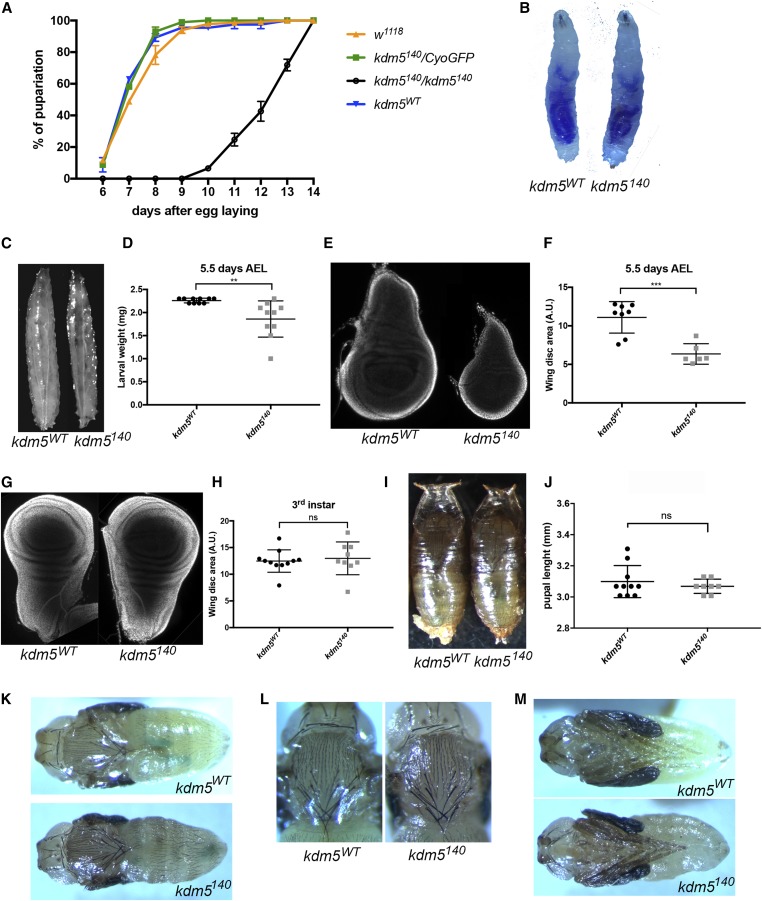
To understand the basis for the essential functions of KDM5, we quantified the developmental timing of kdm5140 homozygous mutants. To ensure that we were examining defects caused by the loss of KDM5, these and all subsequent experiments utilized a wild-type control strain in which the kdm5140 mutation was rescued by two copies of the kdm5 genomic rescue transgene (kdm5WT). Whereas w1118, kdm5WT, and kdm5140 heterozygous animals have indistinguishable developmental timings and took an average of 6.8 days for 50% of animals to pupariate, kdm5140 homozygous mutant larvae took an average of 12 days (Figure 2A). To rule out the possibility that kdm5140 mutants were delayed due to decreased food consumption, we fed first-instar larvae with food containing bromophenol blue. As shown in Figure 2B, KDM5 is not required for feeding, as kdm5WT and kdm5140 larvae show similar levels of ingested dye. Comparing age-matched larvae at 5.5 days after egg laying revealed that kdm5140 mutants weighed less and had significantly smaller imaginal discs than control larvae (Figure 2, C–F). Despite taking 5 days longer, the final larval, imaginal disc, and pupal size of kdm5140 mutants were normal (Figure 2, G–J). Indeed, kdm5140 mutant pharate adults have morphologically normal heads, thoraces, legs, and abdominal segmentation, but fail to eclose (Figure 2, K–M).
Figure 2.
kdm5140 null mutants show a developmental delay and pupal lethality. (A) Time taken for animals to pupariate for w1118 (N = 221), kdm5WT (N = 82), kdm5140 heterozygous (kdm5140/CyO-GFP; N = 184), or kdm5140 homozygous mutant (N = 49). (B) kdm5WT and kdm5140 mutant larvae fed food containing the dye bromophenol blue. (C) kdm5WT and kdm5140 mutant larvae at 5.5 days after egg laying (AEL), 12.5× magnification. (D) Weight in milligrams of kdm5WT (N = 10) and kdm5140 mutant (N = 10) larvae at 5.5 days. ** P = 0.005. (E) Wing imaginal discs from kdm5WT or kdm5140 mutants at 5.5 days AEL, 100× magnification. (F) Quantification of the size of kdm5WT (N = 8) and kdm5140 mutant (N = 6) wing discs at 5.5 days AEL. *** P = 0.0003. (G) Wing imaginal discs from kdm5WT or kdm5140 mutants at wandering third-instar larval stage (5.5 days for kdm5WT, 10 days for kdm5140), 100× magnification. (H) Quantification of wing disc size (area) of wing imaginal discs from kdm5WT (N = 11) or kdm5140 mutants (N = 9) at wandering third-instar larval stage. (I) Nine-day-old kdm5WT and 14-day-old kdm5140 mutant pupae, 12.5× magnification. (J) Quantification of pupal final size for kdm5WT (N = 10) and kdm5140 mutants (N = 7). (K) Dorsal view of 9-day-old kdm5WT (top) and 14-day-old kdm5140 mutant (bottom) pupae dissected from their pupal case, 12.5× magnification. (L) Thorax and head of kdm5WT (left) and kdm5140 mutant (right) pupae, 16× magnification. (M) Ventral view of kdm5WT (top) and kdm5140 mutant (bottom) pupae dissected from their pupal case showing normal morphology, 12.5× magnification.
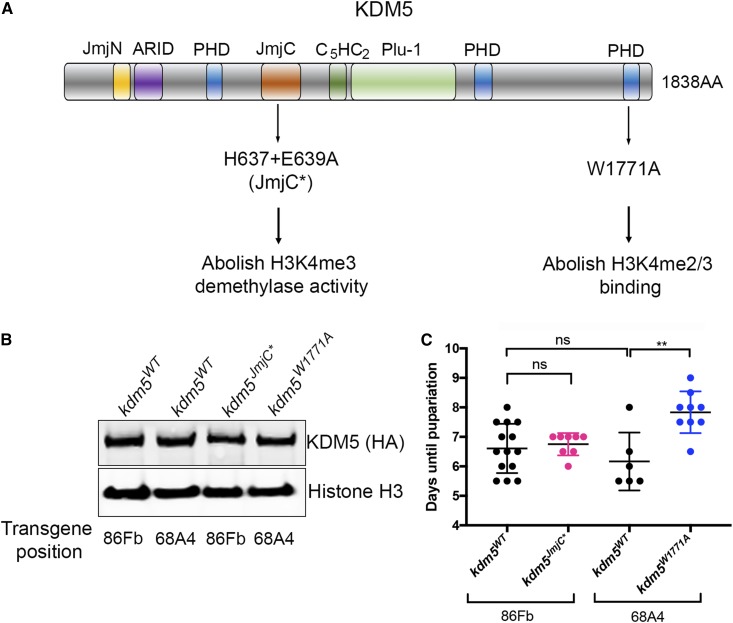
Two of the best defined functions of KDM5 are its JmjC domain-encoded demethylase activity that removes H3K4me3, and its C-terminal PHD motif that binds to H3K4me2/3 (PHD3) (Eissenberg et al. 2007; Lee et al. 2007; Secombe and Eisenman 2007; Liu et al. 2014). While both domains involve the same chromatin modification, H3K4me3, they regulate transcription independently of each other (Liu and Secombe 2015). We therefore tested whether the developmental delay phenotype observed in kdm5140 larvae was dependent on either of these domains. To do this, we utilized transgenes with point mutation forms of the kdm5 locus that abolish the function of the JmjC or PHD3 domains (kdm5JmjC* and kdm5W1771A; Figure 3A) (Liu and Secombe 2015; Navarro-Costa et al. 2016). By crossing these transgenes into the kdm5140 null allele background, the mutant form was the sole source of KDM5 (Figure 3B). kdm5JmjC* mutant animals showed a developmental timing profile that was indistinguishable from wild type, demonstrating that the slowed growth of kdm5140 mutant animals is independent of its demethylase activity (Figure 3C). Interestingly, while the PHD3 mutant strain kdm5W1771A took 1.5 days longer to pupariate than kdm5WT animals (P = 0.002), this was still significantly faster than the 5-day delay observed in kdm5140 mutant animals (P < 0.0001). Thus, while chromatin binding by PHD3 may contribute to the larval growth functions of KDM5, other activities are also critical.
Figure 3.
KDM5-mediated developmental delay is independent of its H3K4me3 removal or binding activities. (A) Schematic of the KDM5 protein showing the domain structure and the location of the JmjC* point mutations that abolish demethylase activity and the W1771A point mutation in the C-terminal PHD motif that prevents binding to H3K4me2/3 (Li et al. 2010). The kdm5JmjC* genomic transgene is inserted at the attP site at 86Fb while the kdm5W1771A transgene is located at the attP site at 68A4. Each mutant strain therefore has a separate control kdm5WT strain with a matching insertion of the wild-type kdm5 genomic region. (B) Western blot showing wild-type expression of KDM5 (using anti-HA; top) in kdm5JmjC* and kdm5W1771A wing imaginal discs. The kdm5WT strain at 86Fb is a control for kdm5JmjC* while the wild-type insertion at 68A4 is the control for kdm5W1771A. (C) Time for 50% of kdm5WT (86Fb; N = 415; 6.9 days), kdm5JmjC* (86Fb; N = 66; 6.75 days), kdm5WT (68A4; N = 379; 6.2 days), and kdm5W1771A (68A4; N = 126; 8.25 days) to pupariate (T1/2). Each data point represents animals counted from an independent cross. N values represent the total number of animals scored. ** P = 0.002.
KDM5 is required for optimal wing disc cell proliferation and survival
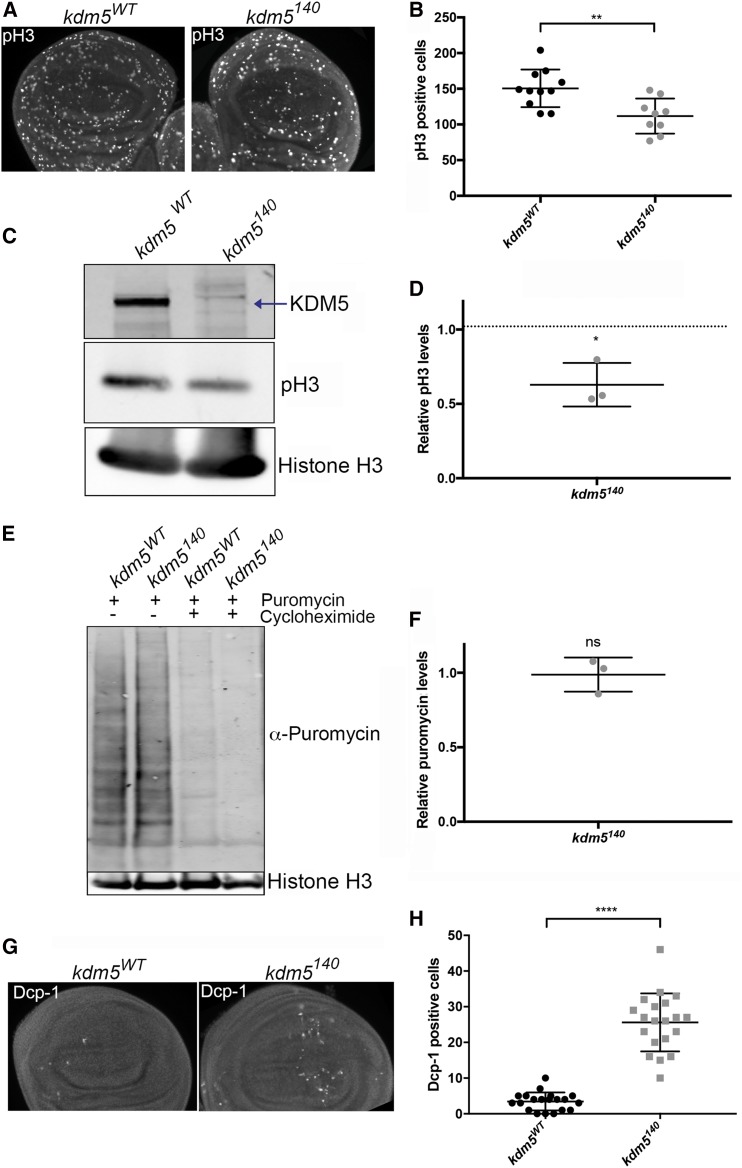
The prolonged larval growth period observed in kdm5140 mutants could, at least in part, be due to reduced proliferation of imaginal disc cells that are the precursors to adult structures. Quantifying the number of mitotic cells in kdm5140 mutant third-instar larval wing imaginal discs using an anti-phospho-histone H3 antibody revealed ∼25% fewer proliferating cells compared to control discs at the same developmental stage (Figure 4, A and B). This decreased wing disc proliferation is supported by Western blotting analyses showing significantly reduced levels of phospho-histone H3 (Figure 4, C and D). To test whether the proliferative defect was an indirect consequence of a general cell growth defect, we quantified nascent translation by incubating kdm5WT and kdm5140 wing discs with the transfer RNA analog puromycin (Deliu et al. 2017). Western blotting analyses to detect puromycin-labeled proteins confirmed that wing discs lacking KDM5 did not have altered translation rates (Figure 4, E and F).
Figure 4.
kdm5 mutant wing discs show reduced proliferation and increased cell death. (A) Anti-phospho-histone H3 (pH3) labeling of kdm5WT (left) and kdm5140 (right) mutant wing discs matched for developmental stage (third instar) (200× magnification). (B) Quantification of the number of pH3) cells in the wing pouch of kdm5WT (N = 11) and kdm5140 mutant (N = 9) wing discs. ** P = 0.003. (C) Western blot of kdm5WT and kdm5140 mutant wing discs (10 discs per lane) showing anti-KDM5 (top; arrow indicates KDM5 band), pH3 (middle), and histone H3 load control (bottom). (D) Quantification of three Western blots showing increased phosphorylated histone H3 in kdm5140. * P = 0.01. Dotted line indicates position expected if no change in pH3 levels. (E) Anti-puromycin Western blot after puromycin incorporation into kdm5WT or kdm5140 wing imaginal discs (developmental age-matched). Incorporation of puromycin is blocked by coincubation with cycloheximide, which serves as a specificity control. Histone H3 levels serve as a loading control. (F) Quantification of three anti-puromycin Western blots shown as a ratio, with anti-histone H3 loading control. (G) Dcp-1 staining to show cell death in kdm5WT and kdm5140 third-instar wing imaginal discs (200× magnification). (H) Quantification of the number of Dcp-1-positive cells in the pouch region of wild-type (kdm5WT; N = 18) and kdm5140 (N = 20) wing discs. **** P < 0.0001.
Because elevated cell death could also impede wing disc growth and lead to altered larval development, we examined levels of apoptosis using an antibody to the effector caspase Dcp-1 (Sarkissian et al. 2014). kdm5140 mutant wing discs showed an increased number of Dcp-1 positive cells, suggesting elevated rates of cell death (Figure 4, G and H). Combined, decreased proliferation and increased cell death of imaginal disc cells may contribute to the developmental delay of kdm5140 larvae.
Wing imaginal discs from kdm5 null mutants have gene expression defects
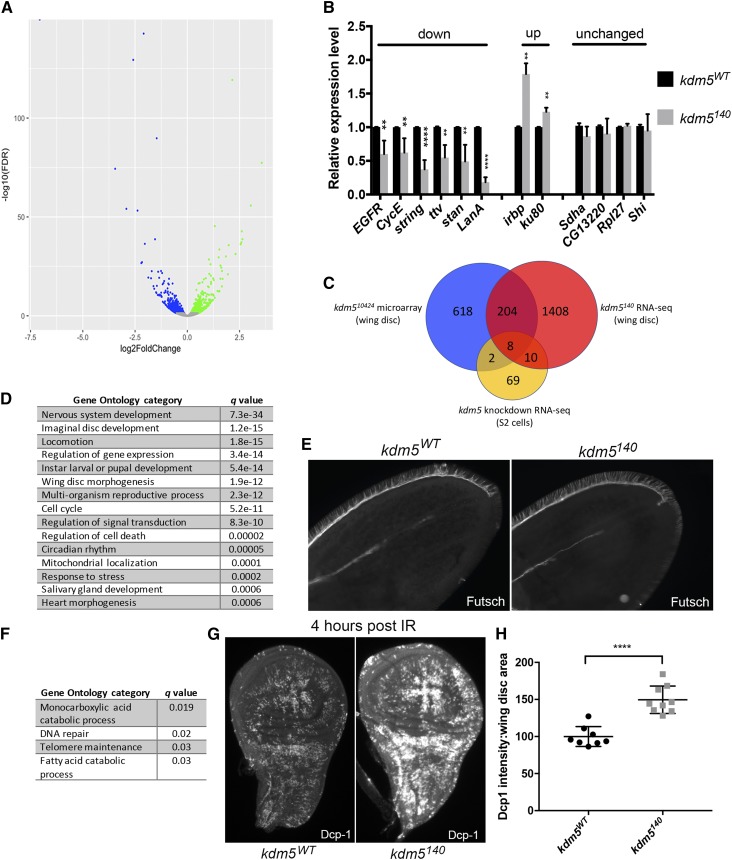
To gain insight into the gene expression defects caused by loss of KDM5, we carried out mRNA sequencing from kdm5WT and kdm5140 mutant larval wing discs that were matched for developmental age based on imaginal disc size rather than days after egg laying. These analyses identified 1630 dysregulated genes in kdm5140 wing discs (5% FDR), 883 of which were downregulated and 747 of which were upregulated (Figure 5A and Table S2). As with other gene expression defects caused by mutations in kdm5 across a diverse range of organisms, the changes to mRNA levels were predominantly mild, averaging a 1.5-fold change for both upregulated and downregulated genes (Lopez-Bigas et al. 2008; Lloret-Llinares et al. 2012; Liu and Secombe 2015; Iwase et al. 2016; Lussi et al. 2016).
Figure 5.
KDM5 is required for the normal transcriptional pattern of wing imaginal discs. (A) Volcano plot of genes significantly upregulated (green) and downregulated (blue) in kdm5140 RNA-seq data (FDR < 0.05). (B) Real-time PCR validation of genes that were downregulated, upregulated, or unaffected in RNA-seq data. **** P < 0.0001, *** P < 0.001, ** P < 0.01. (C) Venn diagram showing overlap between current RNA-seq data and previously published microarray data from kdm510424 (P = 3.7e−48) and S2 cell KDM5 knockdown RNA-seq (P = 0.0008). (D) Gene ontology categories enriched using FlyMine (Lyne et al. 2007) using genes significantly downregulated in kdm5140. (E) kdm5WT (left) kdm5140 (right) pupal wings stained 30 hr after puparium formation with an anti-Futsch antibody (also known as 22C10) (200× magnification). (F) Gene ontology categories enriched using FlyMine (Lyne et al. 2007) using genes significantly upregulated in kdm5140. (G) Third-instar larval wing imaginal disc 4 hr postirradiation, showing cell death using anti-Dcp1 in kdm5WT (left) and kdm5140 (right) (100× magnification). (H) Quantitation of Dcp-1 intensity in kdm5WT (N = 8) and kdm5140 (N = 9) wing imaginal discs. **** P < 0.0001.
To confirm the robustness of the kdm5140 RNA-seq data, we verified the expression of downregulated, upregulated and unchanged genes by real-time PCR (Figure 5B). In addition, we compared our new data to our previously published microarray data generated using kdm510424 hypomorphic mutant wing imaginal discs (Liu et al. 2014). Despite differences in platform and allele severity, 26% of genes identified as significantly dysregulated in the wing disc microarray data were similarly affected in kdm5140 (212 out of 824; P = 3.7e−48; Figure 5C and Table S3). In contrast, we did not observe significant overlap between the current RNA-seq or previous microarray data and published microarray data from kdm5 knockdown wing discs (Figure S2) (Lloret-Llinares et al. 2012). It should be noted, however, that these comparisons were limited by the small number of genes that were dysregulated in kdm5 knockdown discs. RNA-seq data are also available from KDM5 knockdown cultured S2 cells, which are a macrophage-like lineage (Gajan et al. 2016). We therefore also determined the extent to which KDM5-regulated genes in S2 cells overlapped with the changes observed in mutant wing discs in vivo. Perhaps unsurprisingly given the difference in cell type and context, the overlap between kdm5140 and S2 cell knockdown data were modest (P = 0.0008; Figure 5C and Table S4). Similar to previous observations, these data are consistent with KDM5 regulating distinct targets in different cell types and at different stages of development (Liu and Secombe 2015; Zamurrad et al. 2018).
To define pathways affected in kdm5140 mutants, we carried out gene ontology (GO) analyses of genes dysregulated in kdm5140, using GOrilla (Eden et al. 2009) and FlyMine (Lyne et al. 2007). This revealed a large number of significantly enriched terms for downregulated genes, which are summarized in Figure 5D and provided in full in Table S5 (q ≤ 0.01). The reduced proliferation observed in kdm5140 mutant wing discs is reflected in our GO analyses, with cell cycle regulation being significantly enriched. This included genes such as cyclin E and cdc25 that mediate cell cycle progression (Bertoli et al. 2013) in addition to components of key growth signaling pathways such as the epidermal growth factor receptor (Lusk et al. 2017). Consistent with previous studies of kdm5 mutants, genes involved in circadian rhythm, mitochondrial function, and stress response were also enriched (DiTacchio et al. 2011; Liu et al. 2014; Gajan et al. 2016). Similarly, in keeping with the observation that mutations in KDM5 family genes are found in human patients with intellectual disability, genes involved in nervous system development were altered (van Bokhoven 2011). These encompassed a wide range of functions, including transcriptional regulators such as groucho (Agarwal et al. 2015), actin cytoskeletal organizers such as the Abl tyrosine kinase (Kannan et al. 2017), and cell-cell adhesion and communication regulators such as fascilin1, fascilin3, tout-velu, and starry night (Elkins et al. 1990; Kraut et al. 2001; Chanana et al. 2009). Because the neuronal-related genes that were downregulated did not affect a single pathway or process, the importance of their dysregulation to epithelial wing disc development was not clear. Indeed, it is notable that the wing disc includes neuronal lineages that are not affected by the loss of KDM5, such as sensory microchaete and macrochaete of the adult thorax (Figure 2L). To further examine the connection between KDM5 and neuronal development, we stained pupal wing discs with an antibody to the microtubule-binding protein Futsch (22C10) (Fujita et al. 1982). At 30 hr APF, kdm5WT and kdm5140 pupal wings show similar growth of the axon along the L3 wing vein, in addition to the correct number and position of the sensory bristles along the anterior margin of the wing (Figure 5E). While we have not identified obvious neuronal defects in the wing epithelium, it is possible that the observed gene expression defects also occur in cells of the nervous system where they may be functionally important.
Fewer GO categories were identified among the genes that were upregulated in kdm5140 using a less stringent q ≤ 0.05 cutoff (Figure 5F and Table S6). Interestingly, the inclusion of DNA repair as an enriched category suggested that kdm5140 mutants have increased levels of DNA damage in the absence of any exogenous mutagens. These included genes encoding Irbp/Ku70 and Ku80 that form a complex and are required for double-stranded break repair and telomere maintenance (Min et al. 2004; Melnikova et al. 2005). Loss of KDM5 may therefore be expected to increase sensitivity to mutagens such as γ-irradiation. Indeed, irradiating kdm5140 mutant larvae elevated levels of cell death in wing imaginal discs (Figure 5, G and H). Notably, we did not find any significantly enriched GO terms by analyzing genes found to be overlapping between the current RNA-seq analyses and previously generated microarray data. This emphasizes the power of our new analyses that utilizes a kdm5 null allele, as this enables us to better detect the relatively small changes to gene expression that are caused by loss of KDM5.
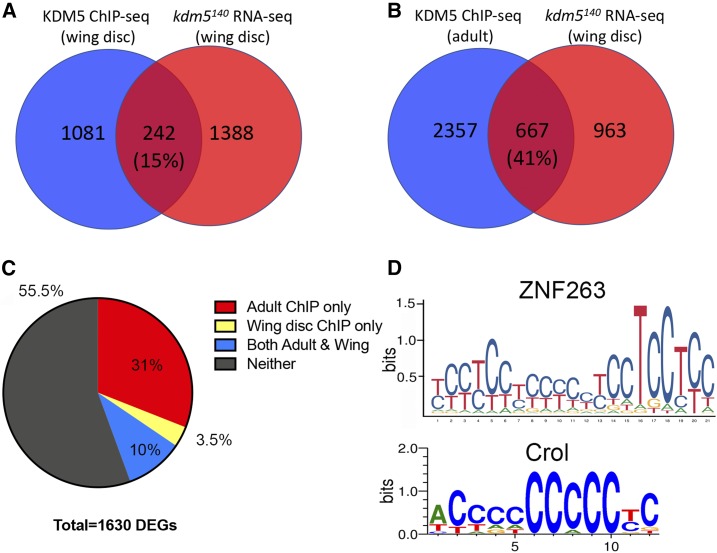
KDM5 directly regulates many genes dysregulated in kdm5 mutant wing discs
The 1630 genes identified as dysregulated in kdm5140 mutant wing discs include direct transcriptional targets of KDM5 in addition to indirect changes. Utilizing published anti-KDM5 ChIP-seq data from third-instar wing imaginal discs, we found that 15% of dysregulated genes had an associated ChIP peak and were therefore likely to be direct KDM5 targets (P = 8.8e−16; Figure 6A and Table S2) (Lloret-Llinares et al. 2012). In line with other studies showing that KDM5 proteins can activate or repress gene expression in a context-dependent manner, 16% of KDM5-bound genes were downregulated in kdm5140 (P = 8.5e−24) and 13% were upregulated (P = 4.53e−12). Dysregulated genes with a corresponding KDM5 ChIP signal included genes within key GO categories such as cell cycle (e.g., cyclin E and string), neurogenesis (e.g., groucho), transcription (e.g., Enhancer of polycomb), and circadian rhythm (e.g., no circadian temperature entrainment, nocte). These genes were all downregulated in kdm5140 mutant wing discs, suggesting the importance of KDM5-mediated gene activation. Upregulated genes, such as the DNA repair proteins ku70 and ku80, were not identified as direct KDM5 targets (Table S2).
Figure 6.
A subset of genes dysregulated in kdm5140 mutants are directly bound by KDM5. (A) Venn diagram showing overlap between dysregulated genes and those bound by KDM5 in ChIP-seq data from third-instar larval imaginal discs (Lloret-Llinares et al. 2012). P = 8.8e−16. (B) Venn diagram showing overlap between dysregulated genes and those bound by KDM5 in ChIP-seq data from adults (Liu and Secombe 2015). P = 3e−144. (C) Pie chart showing proportion of 1630 dysregulated genes in kdm5140 wing discs and those genes bound by KDM5 in wing disc ChIP (Lloret-Llinares et al. 2012), adult ChIP (Liu and Secombe 2015), both ChIP datasets, and those that did not have an associated ChIP peak (see also Table S2). (D) MEME analyses of 162 directly regulated genes in wing disc and adult KDM5 ChIP-seq data showed enrichment for the zinc finger transcription factor ZNF263, which binds to the consensus sequence shown based on ChIP-seq data (Frietze et al. 2010). Crol is the most similar Drosophila gene to ZNF263 and has a similar consensus DNA-binding sequence as defined by a bacterial 1-hybrid assay (Enuameh et al. 2013). DEGs, differentially expressed genes.
The availability of ChIP data allowed us to begin addressing the key question of how KDM5 is recruited to its target genes. No significant enrichment (q < 0.05) was found when using MEME software (Machanick and Bailey 2011) to analyze KDM5-bound regions for the DNA sequence bound by KDM5′s AT-rich interaction domain (ARID) motif in vitro (CCGCCC) (Tu et al. 2008; Yao et al. 2010), or binding sites for transcription factors that could mediate KDM5 recruitment. Because the peaks defined in the available KDM5 wing disc ChIP-seq data were short (Lloret-Llinares et al. 2012), we also utilized KDM5 ChIP-seq data generated using whole adults to identify candidate direct targets (Liu and Secombe 2015). Despite being from a later developmental stage, 41% of genes dysregulated in the wing imaginal disc had significant KDM5 binding at their promoters (Figure 6B; P = 3e−144; Table S2). Combining the single wing disc ChIP-seq and the triplicate ChIP-seq studies carried out using adults identified 162 high-confidence, directly regulated KDM5 target genes (Figure 6C and Table S2). Interrogating these KDM5-bound sequences for the known ARID DNA-binding motif did not reveal any significant enrichment (P = 0.6), suggesting that recognition of this sequence is not a key means by which KDM5 recognizes its target promoters. We did, however, identify significant enrichment for the DNA sequence bound by the C2H2 zinc finger protein ZNF263, using a general eukaryotic transcription factor binding site database (P = 3.1e−3) (Frietze et al. 2010) (Figure 6D). ZNF263 is most similar to the Drosophila Crooked legs (Crol) protein, which is expressed in the wing imaginal disc and has a similar in vitro DNA binding preference to ZNF263 (Enuameh et al. 2013) (Figure 6D). Interestingly, clones of crol mutant cells in the wing imaginal disc show reduced proliferation and increased apoptosis (Mitchell et al. 2008), reminiscent of the phenotypes we observe in kdm5 mutants. Crol is therefore a candidate transcription factor that mediates KDM5 recruitment to a subset of its target genes. Testing this hypothesis awaits the identification of Crol target genes.
Discussion
Here we describe the phenotypes associated with a null allele of the transcriptional regulator kdm5. In contrast to individual mouse knockouts of Kdm5A, Kdm5B, and Kdm5C that are viable (Lin et al. 2011; Zou et al. 2014; Iwase et al. 2016), loss of the sole kdm5 gene in Drosophila results in lethality. In mice, the upregulation of other KDM5 paralogs could be a key confounding factor in the analysis of individual Kdm5 gene knockouts. For example, cells lacking KDM5B can upregulate KDM5A (Zou et al. 2014) and KDM5B is upregulated in cells harboring mutations in KDM5C (Jensen et al. 2010). It is also possible that the four mammalian paralogs have evolved specialized functions, and that the phenotype(s) of true loss of KDM5 function awaits combinatorial knockout strains. Interestingly, while C. elegans also has a single kdm5 ortholog (rbr-2), strong loss-of-function mutations are viable, suggesting that KDM5-regulated transcription during development may be more important in flies than in worms (Lussi et al. 2016). These data also point to Drosophila being an ideal model in which to define the essential functions of KDM5 proteins.
Drosophila kdm5 was originally named lid based on the size of tissues in kdm510424 homozygous mutant larvae (Gildea et al. 2000). However, our analyses of kdm5140 show that it would more aptly be described as a developmental delay phenotype affecting the whole animal rather than a defect in imaginal disc growth. Although they take significantly longer to grow, kdm5140 mutant larvae develop to the same size as wild type before they pupate. Metamorphosis is also largely unaffected, as kdm5140 mutant pharate adults are morphologically normal. Zygotic expression of kdm5 is therefore necessary for survival, but not for the cell fate decisions required to develop the structures that comprise the adult fly. Indeed, based on their appearance, it is unclear why kdm5140 mutants fail to eclose from their pupal case. Because kdm5 null mutants die late in development, it is possible that maternally contributed kdm5 transcript powers embryonic or early larval development. Determining the extent to which this is true requires examining the phenotypes of animals lacking both maternal and zygotic kdm5. While germline clones have been generated using hypomorphic kdm5 alleles, a requirement for KDM5 in female meiosis prevented the phenotypes of embryos lacking maternally deposited kdm5 from being characterized (Navarro-Costa et al. 2016; Zhaunova et al. 2016). Similar meiotic phenotypes are expected to be observed using the kdm5 null allele, leaving the question of KDM5′s embryonic functions unresolved.
Like several other described functions of KDM5, the developmental delay caused by loss of KDM5 is independent of its JmjC-encoded H3K4me3 demethylase function. In addition, while abolishing the H3K4me2/3 binding activity of KDM5′s PHD motif mildly slows development, this chromatin recognition function does not account for the pronounced larval growth extension seen in kdm5140 animals. Mutant strains lacking either JmjC or PHD domain activity are adult viable, suggesting that delayed development and pupal lethality are linked (Li et al. 2010; Liu et al. 2014). These data also point to the activity of one or more other domains of KDM5 being critical during larval development. Previous experiments based on rescue of a kdm5 hypomorphic allele by overexpressing wild-type or domain deletion versions of KDM5 showed that the JmjN and ARID motifs were essential for viability (Li et al. 2010). While these domains are good candidates for mediating key KDM5 functions, their in vivo activities are unclear. JmjN domains are found exclusively in proteins with a JmjC motif and the functions of these two motifs are assumed to be interdependent (Klose et al. 2006). This is based on crystal structure data showing that the JmjN and JmjC domains of KDM5A make extensive contacts, and that deletion of the JmjN motif abolishes the demethylase activity of KDM5 proteins (Chen et al. 2006; Xiang et al. 2007; Yamane et al. 2007; Li et al. 2010; Vinogradova et al. 2016). However, the JmjN domain may have additional functions based on the observation that this domain is essential for viability while the JmjC domain is not (Li et al. 2010). Similarly, while ARID motif is assumed to have physiological DNA binding functions based on in vitro assays (Tu et al. 2008), the extent to which this occurs in vivo remains untested. Indeed, structural modeling of this domain led to the suggestion that the ARID may also be a protein–protein interaction motif (Peng et al. 2015). The ARID or the JmjN motifs of KDM5 could, for example, mediate an interaction with the transcription factor as Crol to facilitate activation of genes required for wing disc proliferation and development (Mitchell et al. 2008). More detailed analyses of the in vivo functions of both of these KDM5 domains is needed to clarify their roles in gene expression and development.
While nutrient deprivation can delay development (Zinke et al. 1999), kdm5140 larvae ingest food normally and do not show transcriptional changes that would indicate starvation. kdm5140 mutant wing discs do, however, show decreased proliferation. This is consistent with previous studies showing that kdm5 genetically interacts with cyclin E in a manner that implicates KDM5 as a positive cell cycle regulator (Brumby et al. 2004). This proliferative function of KDM5 is likely to extend beyond larval epithelial cells, as cell cycle progression is affected by knocking down kdm5 in S2 cells, which are a macrophage-like cell culture line (Gajan et al. 2016). While the expression of cell cycle regulators was not significantly altered in S2 cells, we observe changes to numerous cell regulators in kdm5140 wing discs, including the G1-S phase regulator cyclin E and the G2-M regulator string (cdc25). Both cyclin E and string are direct KDM5 targets in wing imaginal disc cells (Lloret-Llinares et al. 2012) but not in the adult fly (Liu and Secombe 2015). These genes may therefore be bound and regulated by KDM5 only in tissues or cells that are actively growing and dividing. Significantly, human KDM5A activates the transcription of cyclin E1 in lung cancer cells, suggesting that the regulation of cell cycle genes is an evolutionarily conserved feature of KDM5 proteins (Teng et al. 2013).
Many factors are integrated to control the regulation of larval growth and to sense when the correct tissue and larval size has been reached for pupariation to occur. Imaginal disc size is one determinant of developmental timing (Stieper et al. 2008). For example, slowing imaginal disc growth by knocking down the expression of genes encoding ribosomal subunits extends larval development in a comparable manner to that seen for kdm5140. However, ribosomal protein genes were not enriched in our transcriptome analyses, nor was translation rate reduced in kdm5140 wing discs. Thus, while the larval phenotypes may be superficially similar, KDM5 likely affects larval development through a different mechanism. One possible mechanism is through the regulation of gene cycle genes, since a hypomorphic allele of cyclin E delays larval development (Secombe et al. 1998). It is also possible that the proliferative changes we observe in the wing imaginal disc are an indirect consequence of a signaling defect originating elsewhere in the larva. The larval prothoracic gland is one such signaling tissue that regulates developmental timing and imaginal disc proliferation through the regulated secretion of growth-regulatory hormones such as ecdysone (Colombani et al. 2005; Stieper et al. 2008).
We also observed increased cell death in kdm5140 wing discs, and this occurred coincidentally with the upregulation of genes required for DNA repair. These data suggest that loss of KDM5 results in higher than normal levels of DNA damage in the absence of any exogenous mutagen. Consistent with this, kdm5140 mutant animals were more sensitive to γ-irradiation than controls. Our previous analysis using a lacZ reporter transgene in kdm5 hypomorphic mutant larvae also revealed an increased mutation frequency (Liu et al. 2014). Whether this occurs in all tissues or affects some tissues more than others remains an open question. Similar to its role in regulating key cell cycle genes, the function of KDM5 in maintaining genome stability may also be conserved in mammalian cells. Knockdown of KDM5B in a range of transformed cell lines increases levels of spontaneous DNA damage in a manner consistent with defective DNA double-stranded break repair pathways (Li et al. 2014). Thus, while the molecular mechanism remains unclear, our data provide a clear in vivo link between KDM5 and its promotion of cell survival by restricting DNA damage.
Acknowledgments
The authors thank Kiera Brennan for help with initial mapping of the kdm5 mutant in addition to members of the Secombe laboratory for insights at all stages of this project. Stocks obtained from the Bloomington Drosophila Stock Center (National Institutes of Health grant P400D018537) were used in this study. We thank the Developmental Studies Hybridoma Bank for antibodies. Funding was received from the National Institutes of Health (grant R01 GM112783), the March of Dimes (grant 6-FY17-315), and the Einstein Cancer Center (support grant P30 CA013330).
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6239003.
Communicating editor: P. Geyer
Literature Cited
- Agarwal M., Kumar P., Mathew S. J., 2015. The Groucho/transducin-like enhancer of split protein family in animal development. IUBMB Life 67: 472–481. 10.1002/iub.1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert M., Schmitz S. U., Kooistra S. M., Malatesta M., Morales Torres C., et al. , 2013. The histone demethylase Jarid1b ensures faithful mouse development by protecting developmental genes from aberrant H3K4me3. PLoS Genet. 9: e1003461 10.1371/journal.pgen.1003461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertoli C., Skotheim J. M., de Bruin R. A., 2013. Control of cell cycle transcription during G1 and S phases. Nat. Rev. Mol. Cell Biol. 14: 518–528. 10.1038/nrm3629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J., Maeda R. K., Hediger M., Karch F., Basler K., 2007. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. USA 104: 3312–3317. 10.1073/pnas.0611511104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair L. P., Cao J., Zou M. R., Sayegh J., Yan Q., 2011. Epigenetic regulation by lysine demethylase 5 (KDM5) enzymes in cancer. Cancers (Basel) 3: 1383–1404. 10.3390/cancers3011383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumby A., Secombe J., Horsfield J., Coombe M., Amin N., et al. , 2004. A genetic screen for dominant modifiers of a cyclin E hypomorphic mutation identifies novel regulators of S-phase entry in Drosophila. Genetics 168: 227–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., Liu Z., Cheung W. K., Zhao M., Chen S. Y., et al. , 2014. Histone demethylase RBP2 is critical for breast cancer progression and metastasis. Cell Rep. 6: 868–877. 10.1016/j.celrep.2014.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catchpole S., Spencer-Dene B., Hall D., Santangelo S., Rosewell I., et al. , 2011. PLU-1/JARID1B/KDM5B is required for embryonic survival and contributes to cell proliferation in the mammary gland and in ER+ breast cancer cells. Int. J. Oncol. 38: 1267–1277. [DOI] [PubMed] [Google Scholar]
- Chanana B., Steigemann P., Jackle H., Vorbruggen G., 2009. Reception of slit requires only the chondroitin-sulphate-modified extracellular domain of Syndecan at the target cell surface. Proc. Natl. Acad. Sci. USA 106: 11984–11988. 10.1073/pnas.0901148106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. Z., Zang J. Y., Whetstine J., Hong X., Davrazou F., et al. , 2006. Structural insights into histone demethylation by JMJD2 family members. Cell 125: 691–702. 10.1016/j.cell.2006.04.024 [DOI] [PubMed] [Google Scholar]
- Colombani J., Bianchini L., Layalle S., Pondeville E., Dauphin-Villemant C., et al. , 2005. Antagonistic actions of ecdysone and insulins determine final size in Drosophila. Science 310: 667–670. 10.1126/science.1119432 [DOI] [PubMed] [Google Scholar]
- Deliu L. P., Ghosh A., Grewal S. S., 2017. Investigation of protein synthesis in Drosophila larvae using puromycin labelling. Biol. Open 6: 1229–1234. 10.1242/bio.026294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiTacchio L., Le H. D., Vollmers C., Hatori M., Witcher M., et al. , 2011. Histone lysine demethylase JARID1a activates CLOCK-BMAL1 and influences the circadian clock. Science 333: 1881–1885. 10.1126/science.1206022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden E., Navon R., Steinfeld I., Lipson D., Yakhini Z., 2009. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10: 48 10.1186/1471-2105-10-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg J. C., Lee M. G., Schneider J., Ilvarsonn A., Shiekhattar R., et al. , 2007. The trithorax-group gene in Drosophila little imaginal discs encodes a trimethylated histone H3 Lys4 demethylase. Nat. Struct. Mol. Biol. 14: 344–346. 10.1038/nsmb1217 [DOI] [PubMed] [Google Scholar]
- Elkins T., Zinn K., McAllister L., Hoffmann F. M., Goodman C. S., 1990. Genetic analysis of a Drosophila neural cell adhesion molecule: interaction of fasciclin I and Abelson tyrosine kinase mutations. Cell 60: 565–575. 10.1016/0092-8674(90)90660-7 [DOI] [PubMed] [Google Scholar]
- Enuameh M. S., Asriyan Y., Richards A., Christensen R. G., Hall V. L., et al. , 2013. Global analysis of Drosophila Cys(2)-His(2) zinc finger proteins reveals a multitude of novel recognition motifs and binding determinants. Genome Res. 23: 928–940. 10.1101/gr.151472.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J., Li L., Zhang N., Liu J., Zhang L., et al. , 2017. Androgen and AR contribute to breast cancer development and metastasis: an insight of mechanisms. Oncogene 36: 2775–2790. 10.1038/onc.2016.432 [DOI] [PubMed] [Google Scholar]
- Frietze S., Lan X., Jin V. X., Farnham P. J., 2010. Genomic targets of the KRAB and SCAN domain-containing zinc finger protein 263. J. Biol. Chem. 285: 1393–1403. 10.1074/jbc.M109.063032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita S. C., Zipursky S. L., Benzer S., Ferrus A., Shotwell S. L., 1982. Monoclonal antibodies against the Drosophila nervous system. Proc. Natl. Acad. Sci. USA 79: 7929–7933. 10.1073/pnas.79.24.7929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajan A., Barnes V. L., Liu M., Saha N., Pile L. A., 2016. The histone demethylase dKDM5/LID interacts with the SIN3 histone deacetylase complex and shares functional similarities with SIN3. Epigenetics Chromatin 9: 4 10.1186/s13072-016-0053-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gildea J. J., Lopez R., Shearn A., 2000. A screen for new trithorax group genes identified little imaginal discs, the Drosophila melanogaster homologue of human retinoblastoma binding protein 2. Genetics 156: 645–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth A. C., Fish M., Nusse R., Calos M. P., 2004. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics 166: 1775–1782. 10.1534/genetics.166.4.1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayami S., Yoshimatsu M., Veerakumarasivam A., Unoki M., Iwai Y., et al. , 2010. Overexpression of the JmjC histone demethylase KDM5B in human carcinogenesis: involvement in the proliferation of cancer cells through the E2F/RB pathway. Mol. Cancer 9: 59 10.1186/1476-4598-9-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J., Wu J., Dombkowski A., Zhang K., Holowatyj A., et al. , 2012. Genomic amplification and a role in drug-resistance for the KDM5A histone demethylase in breast cancer. Am. J. Transl. Res. 4: 247–256. [PMC free article] [PubMed] [Google Scholar]
- Huang D., Qiu Y., Li G., Liu C., She L., et al. , 2018. KDM5B overexpression predicts a poor prognosis in patients with squamous cell carcinoma of the head and neck. J. Cancer 9: 198–204. 10.7150/jca.22145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase S., Brookes E., Agarwal S., Badeaux A. I., Ito H., et al. , 2016. A mouse model of X-linked intellectual disability associated with impaired removal of histone methylation. Cell Rep. 14: 1000–1009. 10.1016/j.celrep.2015.12.091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen L. R., Bartenschlager H., Rujirabanjerd S., Tzschach A., Numann A., et al. , 2010. A distinctive gene expression fingerprint in mentally retarded male patients reflects disease-causing defects in the histone demethylase KDM5C. PathoGenetics 3: 2 10.1186/1755-8417-3-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan R., Song J. K., Karpova T., Clarke A., Shivalkar M., et al. , 2017. The Abl pathway bifurcates to balance enabled and Rac signaling in axon patterning in Drosophila. Development 144: 487–498. 10.1242/dev.143776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein B. J., Piao L., Xi Y., Rincon-Arano H., Rothbart S. B., et al. , 2014. The histone-H3K4-specific demethylase KDM5B binds to its substrate and product through distinct PHD fingers. Cell Rep. 6: 325–335. 10.1016/j.celrep.2013.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose R. J., Zhang Y., 2007. Regulation of histone methylation by demethylimination and demethylation. Nat. Rev. Mol. Cell Biol. 8: 307–318. 10.1038/nrm2143 [DOI] [PubMed] [Google Scholar]
- Klose R. J., Kallin E. M., Zhang Y., 2006. JmjC-domain-containing proteins and histone demethylation. Nat. Rev. Genet. 7: 715–727. 10.1038/nrg1945 [DOI] [PubMed] [Google Scholar]
- Kraut R., Menon K., Zinn K., 2001. A gain-of-function screen for genes controlling motor axon guidance and synaptogenesis in Drosophila. Curr. Biol. 11: 417–430. 10.1016/S0960-9822(01)00124-5 [DOI] [PubMed] [Google Scholar]
- Lee N., Zhang J. Y., Klose R. J., Erdjument-Bromage H., Tempst P., et al. , 2007. The trithorax-group protein Lid is a histone H3 trimethyl-Lys4 demethylase. Nat. Struct. Mol. Biol. 14: 341–343. 10.1038/nsmb1216 [DOI] [PubMed] [Google Scholar]
- Li L., Greer C., Eisenman R. N., Secombe J., 2010. Essential functions of the histone demethylase lid. PLoS Genet. 6: e1001221 10.1371/journal.pgen.1001221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Liu L., Yang S., Song N., Zhou X., et al. , 2014. Histone demethylase KDM5B is a key regulator of genome stability. Proc. Natl. Acad. Sci. USA 111: 7096–7101. 10.1073/pnas.1324036111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W., Cao J., Liu J., Beshiri M. L., Fujiwara Y., et al. , 2011. Loss of the retinoblastoma binding protein 2 (RBP2) histone demethylase suppresses tumorigenesis in mice lacking Rb1 or Men1. Proc. Natl. Acad. Sci. USA 108: 13379–13386. 10.1073/pnas.1110104108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Secombe J., 2015. The histone demethylase KDM5 activates gene expression by recognizing chromatin context through its PHD reader motif. Cell Rep. 13: 2219–2231. 10.1016/j.celrep.2015.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Greer C., Secombe J., 2014. KDM5 interacts with Foxo to modulate cellular levels of oxidative stress. PLoS Genet. 10: e1004676 10.1371/journal.pgen.1004676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloret-Llinares M., Perez-Lluch S., Rossell D., Moran T., Ponsa-Cobas J., et al. , 2012. dKDM5/LID regulates H3K4me3 dynamics at the transcription-start site (TSS) of actively transcribed developmental genes. Nucleic Acids Res. 40: 9493–9505. 10.1093/nar/gks773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Bigas N., Kisiel T. A., DeWaal D. C., Holmes K. B., Volkert T. L., et al. , 2008. Genome-wide analysis of the H3K4 histone demethylase RBP2 reveals a transcriptional program controlling differentiation. Mol. Cell 31: 520–530. 10.1016/j.molcel.2008.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusk J. B., Lam V. Y., Tolwinski N. S., 2017. Epidermal growth factor pathway signaling in Drosophila embryogenesis: tools for understanding cancer. Cancers (Basel) 9: 16 10.3390/cancers9020016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lussi Y. C., Mariani L., Friis C., Peltonen J., Myers T. R., et al. , 2016. Impaired removal of H3K4 methylation affects cell fate determination and gene transcription. Development 143: 3751–3762. 10.1242/dev.139139 [DOI] [PubMed] [Google Scholar]
- Lyne R., Smith R., Rutherford K., Wakeling M., Varley A., et al. , 2007. FlyMine: an integrated database for Drosophila and Anopheles genomics. Genome Biol. 8: R129 10.1186/gb-2007-8-7-r129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machanick P., Bailey T. L., 2011. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics 27: 1696–1697. 10.1093/bioinformatics/btr189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani L., Lussi Y. C., Vandamme J., Riveiro A., Salcini A. E., 2016. The H3K4me3/2 histone demethylase RBR-2 controls axon guidance by repressing the actin-remodeling gene wsp-1. Development 143: 851–863. 10.1242/dev.132985 [DOI] [PubMed] [Google Scholar]
- Melnikova L., Biessmann H., Georgiev P., 2005. The Ku protein complex is involved in length regulation of Drosophila telomeres. Genetics 170: 221–235. 10.1534/genetics.104.034538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min B., Weinert B. T., Rio D. C., 2004. Interplay between Drosophila Bloom’s syndrome helicase and Ku autoantigen during nonhomologous end joining repair of P element-induced DNA breaks. Proc. Natl. Acad. Sci. USA 101: 8906–8911. 10.1073/pnas.0403000101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell N., Cranna N., Richardson H., Quinn L., 2008. The Ecdysone-inducible zinc-finger transcription factor Crol regulates Wg transcription and cell cycle progression in Drosophila. Development 135: 2707–2716. 10.1242/dev.021766 [DOI] [PubMed] [Google Scholar]
- Navarro-Costa P., McCarthy A., Prudencio P., Greer C., Guilgur L. G., et al. , 2016. Early programming of the oocyte epigenome temporally controls late prophase I transcription and chromatin remodelling. Nat. Commun. 7: 12331 10.1038/ncomms12331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y., Suryadi J., Yang Y., Kucukkal T. G., Cao W., et al. , 2015. Mutations in the KDM5C ARID domain and their plausible association with syndromic Claes-Jensen-type disease. Int. J. Mol. Sci. 16: 27270–27287. 10.3390/ijms161126022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbart S. B., Strahl B. D., 2014. Interpreting the language of histone and DNA modifications. Biochim. Biophys. Acta 1839: 627–643. 10.1016/j.bbagrm.2014.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H., Schneider R., Bannister A. J., Sherriff J., Bernstein B. E., et al. , 2002. Active genes are tri-methylated at K4 of histone H3. Nature 419: 407–411. 10.1038/nature01080 [DOI] [PubMed] [Google Scholar]
- Sarkissian T., Timmons A., Arya R., Abdelwahid E., White K., 2014. Detecting apoptosis in Drosophila tissues and cells. Methods 68: 89–96. 10.1016/j.ymeth.2014.02.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scandaglia M., Lopez-Atalaya J. P., Medrano-Fernandez A., Lopez-Cascales M. T., Del Blanco B., et al. , 2017. Loss of Kdm5c causes spurious transcription and prevents the fine-tuning of activity-regulated enhancers in neurons. Cell Rep. 21: 47–59. 10.1016/j.celrep.2017.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secombe J., Eisenman R. N., 2007. The function and regulation of the JARID1 family of histone H3 lysine 4 demethylases - the Myc connection. Cell Cycle 6: 1324–1328. 10.4161/cc.6.11.4269 [DOI] [PubMed] [Google Scholar]
- Secombe J., Pispa J., Saint R., Richardson H., 1998. Analysis of a Drosophila cyclin E hypomorphic mutation suggests a novel role for Cyclin E in cell proliferation control during eye imaginal disc development. Genetics 149: 1867–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secombe J., Li L., Carlos L. S., Eisenman R. N., 2007. The trithorax group protein Lid is a trimethyl histone H3K4 demethylase required for dMyc-induced cell growth. Genes Dev. 21: 537–551. 10.1101/gad.1523007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stieper B. C., Kupershtok M., Driscoll M. V., Shingleton A. W., 2008. Imaginal discs regulate developmental timing in Drosophila melanogaster. Dev. Biol. 321: 18–26. 10.1016/j.ydbio.2008.05.556 [DOI] [PubMed] [Google Scholar]
- Swygert S. G., Peterson C. L., 2014. Chromatin dynamics: interplay between remodeling enzymes and histone modifications. Biochim. Biophys. Acta 1839: 728–736. 10.1016/j.bbagrm.2014.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M., Mei P. C., Fang R., Leonor T., Rutenberg M., et al. , 2007. The histone H3K4 demethylase SMCX links REST target genes to X-linked mental retardation. Nature 447: 601–605. 10.1038/nature05823 [DOI] [PubMed] [Google Scholar]
- Teng Y. C., Lee C. F., Li Y. S., Chen Y. R., Hsiao P. W., et al. , 2013. Histone demethylase RBP2 promotes lung tumorigenesis and cancer metastasis. Cancer Res. 73: 4711–4721. 10.1158/0008-5472.CAN-12-3165 [DOI] [PubMed] [Google Scholar]
- Tu S. J., Teng Y. C., Yuan C. H., Wu Y. T., Chan M. Y., et al. , 2008. The ARID domain of the H3K4 demethylase RBP2 binds to a DNA CCGCCC motif. Nat. Struct. Mol. Biol. 15: 419–421. 10.1038/nsmb.1400 [DOI] [PubMed] [Google Scholar]
- Vallianatos C. N., Iwase S., 2015. Disrupted intricacy of histone H3K4 methylation in neurodevelopmental disorders. Epigenomics 7: 503–519. 10.2217/epi.15.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallianatos C. N., Farrehi C., Friez M. J., Burmeister M., Keegan C. E., et al. , 2018. Altered gene-regulatory function of KDM5C by a novel mutation associated with autism and intellectual disability. Front. Mol. Neurosci. 11: 104 10.3389/fnmol.2018.00104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bokhoven H., 2011. Genetic and epigenetic networks in intellectual disabilities. Annu. Rev. Genet. 45: 81–104. 10.1146/annurev-genet-110410-132512 [DOI] [PubMed] [Google Scholar]
- van Zutven L. J., Onen E., Velthuizen S. C., van Drunen E., von Bergh A. R., et al. , 2006. Identification of NUP98 abnormalities in acute leukemia: JARID1A (12p13) as a new partner gene. Genes Chromosomes Cancer 45: 437–446. 10.1002/gcc.20308 [DOI] [PubMed] [Google Scholar]
- Vinogradova M., Gehling V. S., Gustafson A., Arora S., Tindell C. A., et al. , 2016. An inhibitor of KDM5 demethylases reduces survival of drug-tolerant cancer cells. Nat. Chem. Biol. 12: 531–538. 10.1038/nchembio.2085 [DOI] [PubMed] [Google Scholar]
- Wang G. G., Song J., Wang Z., Dormann H. L., Casadio F., et al. , 2009. Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature 459: 847–851. 10.1038/nature08036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Wei J., Su P., Gao P., 2015. Histone demethylase JARID1C promotes breast cancer metastasis cells via down regulating BRMS1 expression. Biochem. Biophys. Res. Commun. 464: 659–666. 10.1016/j.bbrc.2015.07.049 [DOI] [PubMed] [Google Scholar]
- Xiang Y., Zhu Z., Han G., Ye X., Xu B., et al. , 2007. JARID1B is a histone H3 lysine 4 demethylase up-regulated in prostate cancer. Proc. Natl. Acad. Sci. USA 104: 19226–19231. 10.1073/pnas.0700735104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S., Wu Z., Russnes H. G., Takagi S., Peluffo G., et al. , 2014. JARID1B is a luminal lineage-driving oncogene in breast cancer. Cancer Cell 25: 762–777. 10.1016/j.ccr.2014.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane K., Tateishi K., Klose R. J., Fang J., Fabrizio L. A., et al. , 2007. PLU-1 is a H3K4 demethylase involved in transcriptional repression and breast cancer cell proliferation. Mol. Cell 25: 801–812. 10.1016/j.molcel.2007.03.001 [DOI] [PubMed] [Google Scholar]
- Yao W., Peng Y., Lin D., 2010. The flexible loop L1 of the H3K4 demethylase JARID1B ARID domain has a crucial role in DNA-binding activity. Biochem. Biophys. Res. Commun. 396: 323–328. 10.1016/j.bbrc.2010.04.091 [DOI] [PubMed] [Google Scholar]
- Zamurrad S., Hatch H. A. M., Drelon C., Belalcazar H. M., Secombe J., 2018. A Drosophila model of intellectual disability caused by mutations in the histone demethylase KDM5. Cell Rep. 22: 2359–2369. 10.1016/j.celrep.2018.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhaunova L., Ohkura H., Breuer M., 2016. Kdm5/Lid regulates chromosome architecture in meiotic prophase I independently of its histone demethylase activity. PLoS Genet. 12: e1006241 10.1371/journal.pgen.1006241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinke I., Kirchner C., Chao L. C., Tetzlaff M. T., Pankratz M. J., 1999. Suppression of food intake and growth by amino acids in Drosophila: the role of pumpless, a fat body expressed gene with homology to vertebrate glycine cleavage system. Development 126: 5275–5284. [DOI] [PubMed] [Google Scholar]
- Zou M. R., Cao J., Liu Z., Huh S. J., Polyak K., et al. , 2014. Histone demethylase jumonji AT-rich interactive domain 1B (JARID1B) controls mammary gland development by regulating key developmental and lineage specification genes. J. Biol. Chem. 289: 17620–17633. 10.1074/jbc.M114.570853 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The fly strains described in this article are available upon request. RNA-seq data from kdm5140 mutant wing discs is available under the accession number GSE109201. A list of differential expressed genes (and log2 fold change) observed in kdm5140 compared to wild type is provided in Table S2 [5% false discovery rate (FDR)]. Table S2 also includes data from KDM5 chromatin immunoprecipitation sequencing (ChIP-seq) that is published and publicly available from adult flies (accession number GSE70591) (Liu and Secombe 2015) and wing imaginal discs (accession number GSE27081) (Lloret-Llinares et al. 2012). Table S3 shows genes dysregulated in kdm5140 RNA-seq and kdm510424 wing disc microarray data (accession number GSZ53881) (Liu et al. 2014). Table S4 shows genes dysregulated in kdm5140 RNA-seq and S2 cell KDM5 knockdown RNA-seq (accession number GSE68775) (Gajan et al. 2016). The overlap between genes affected in kdm5140 RNA-seq, kdm510424 microarray, S2 cell knockdown RNA-seq, and kdm5 knockdown wing disc microarray (accession number GSE27081) (Lloret-Llinares et al. 2012) is provided in Figure S2. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6239003.