Abstract
Histone chaperones, chromatin remodelers, and histone modifying complexes play a critical role in alleviating the nucleosomal barrier for DNA-dependent processes. Here, we have examined the role of two highly conserved yeast (Saccharomyces cerevisiae) histone chaperones, facilitates chromatin transcription (FACT) and Spt6, in regulating transcription. We show that the H3 tail contributes to the recruitment of FACT to coding sequences in a manner dependent on acetylation. We found that deleting a H3 histone acetyltransferase Gcn5 or mutating lysines on the H3 tail impairs FACT recruitment at ADH1 and ARG1 genes. However, deleting the H4 tail or mutating the H4 lysines failed to dampen FACT occupancy in coding regions. Additionally, we show that FACT depletion reduces RNA polymerase II (Pol II) occupancy genome-wide. Spt6 depletion leads to a reduction in Pol II occupancy toward the 3′-end, in a manner dependent on the gene length. Severe transcription and histone-eviction defects were also observed in a strain that was impaired for Spt6 recruitment (spt6Δ202) and depleted of FACT. Importantly, the severity of the defect strongly correlated with wild-type Pol II occupancies at these genes, indicating critical roles for Spt6 and Spt16 in promoting high-level transcription. Collectively, our results show that both FACT and Spt6 are important for transcription globally and may participate during different stages of transcription.
Keywords: Spt6, Spt16, FACT, transcription elongation, histone chaperones, Gcn4, Saccharomyces cerevisiae, Gcn5, Esa1, histone acetyltransferases, genome-wide, ChIP-chip
THE nucleosome is the fundamental unit of chromatin and is composed of ∼147 bp of DNA wrapped around a histone octamer consisting of two copies of histones H2A, H2B, H3, and H4. Nucleosomes pose a significant impediment to all steps of transcription, including the steps of initiation and elongation of RNA polymerase II (Pol II) through the coding sequences (CDS) (Li et al. 2007). There are two principle mechanisms that are suggested to alleviate the nucleosomal barrier: (1) removal of the H2A/H2B dimer to generate hexamers that can be readily overcome by elongating polymerases (Kireeva et al. 2002) and (2) complete removal of histone octamers leading to reduced nucleosomal density across the transcribing genes (Lee et al. 2004; Dion et al. 2007).
Various factors and enzyme complexes have been implicated in removing the nucleosomal barriers in vivo (Li et al. 2007). For example, acetylation of histone tails facilitates histone eviction by weakening histone–DNA interactions (Govind et al. 2007, 2010) and by promoting recruitment of ATP-dependent remodelers such as RSC and SWI/SNF (Hassan et al. 2001; Dechassa et al. 2010; Spain et al. 2014). While nucleosome disassembly is important for transcription, it is also important to restore chromatin structure in the wake of transcription. Histone chaperones, many of which are cotranscriptionally recruited to transcribing genes, are implicated in performing this function (Gurard-Levin et al. 2014).
Two such chaperones, the facilitates chromatin transcription (FACT) complex (FACT; Spt16/SSRP1 in humans and Spt16/Pob3 in yeast) and Spt6, are enriched in transcribed coding regions (Andrulis et al. 2000; Kaplan et al. 2000; Krogan et al. 2002; Mason and Struhl 2003; Mayer et al. 2010; Formosa 2012; Burugula et al. 2014), consistent with a role for these factors in regulating transcription and maintaining chromatin integrity. Human FACT recognizes and displaces one of the H2A/H2B dimers from the nucleosome, and promotes transcription on a chromatin template in vitro. In addition, it can assemble all four histones on the DNA (Belotserkovskaya et al. 2003). Yeast (Saccharomyces cerevisiae) FACT also interacts with all four histones and displays a strong affinity toward intact nucleosomes (Formosa et al. 2001; VanDemark et al. 2008). Multiple domains within the FACT subunits Spt16 and Pob3 are implicated in binding and chaperoning histones (Hondele and Ladurner 2011; Formosa 2012). The N-terminal domain (peptidase-like) of Schizosaccharomyces pombe Spt16, for example, binds to core histones and histone N-terminal tails (Stuwe et al. 2008), and the M-domain of Chaetomium thermophilum Spt16 recognizes H2A/H2B with affinity similar to that observed with full-length Spt16 (Hondele et al. 2013). However, in S. cerevisiae, the C-terminal regions of both Spt16 and Pob3 were defined as H2A/H2B-binding domains (VanDemark et al. 2008; Kemble et al. 2015). Whether these different domains are required for interacting with nucleosomes in a context-dependent manner remains to be seen.
FACT has been shown to promote reassembly of the displaced H3/H4 in the ADH1, ADH2, and STE3 coding regions (Jamai et al. 2009), implicating FACT in restoring chromatin behind elongating Pol II. The role of FACT (and Spt6) in regulating histone occupancy has been examined at a genome-wide scale (van Bakel et al. 2013; Jeronimo et al. 2015). Impairing FACT function leads to reduced histone occupancy and also results in aberrant incorporation of histone variant H2AZ (Jeronimo et al. 2015). Additionally, recruitment of FACT to the HO promoter is shown to promote histone eviction (Takahata et al. 2009). Likewise, FACT helps in evicting H2A/H2B from the promoter of the PHO5 gene upon induction (Ransom et al. 2009).
Gene-specific studies have shown a role for FACT in regulating transcription. For example, FACT mutants impaired Pol II and TBP recruitment at the GAL1 promoter, implicating a role for FACT in regulating transcription at the initiation step (Biswas et al. 2006; Fleming et al. 2008). However, moderate reductions in Pol II occupancy were observed in the 3′ ORFs of GAL1 and PHO5, but not in LacZ or YAT1 genes expressed on plasmids under the control of the GAL1 promoter (Jimeno-Gonzalez et al. 2006). Likewise, transcription defects were observed only at ADH1 and not at ADH2 or STE3, despite all three genes showing a histone reassembly defect (Jamai et al. 2009), making it unclear if FACT is generally required for transcription. In addition to transcription initiation, FACT is shown to participate in promoting Pol II processivity and elongation rate at the GAL1 gene, in conjunction with H2B ubiquitination (Fleming et al. 2008).
FACT shares many functional similarities with another histone chaperone, known as Spt6, which interacts with H3/H4 (Bortvin and Winston 1996) and also with H2A/H2B (McCullough et al. 2015). Loss of Spt6 function leads to reduced histone occupancy over transcribed regions, suggesting a role for Spt6 in cotranscriptional histone reassembly (Ivanovska et al. 2011; Perales et al. 2013; van Bakel et al. 2013; Jeronimo et al. 2015). Spt6, along with the FACT complex, is also implicated in preventing spurious incorporation of H2AZ in coding regions (Jeronimo et al. 2015) and thereby helps to restrict H2AZ to promoter nucleosomes (Billon and Cote 2013). Therefore, Spt6 and FACT play important roles in maintaining chromatin integrity. Consistent with this, widespread aberrant transcription was observed in cells deficient of Spt6 or FACT (Cheung et al. 2008; van Bakel et al. 2013). Altered histone modifications and reduced transcription genome-wide has been observed in Spt6 mutants (DeGennaro et al. 2013; Kato et al. 2013; Perales et al. 2013). Spt6 possesses a tandem SH2 (tSH2) domain at its C-terminus that was shown to interact with the phosphorylated Pol II C-terminal domain in vitro (Dengl et al. 2009; Close et al. 2011; Liu et al. 2011). Recently, it was demonstrated that the tSH2 domain actually binds to the phosphorylated residues present in the linker region of Pol II rather than the C-terminal domain heptad repeats (Sdano et al. 2017). Consistent with this, deleting this domain significantly dampens occupancy of Spt6 in the transcribed CDS in vivo (Mayer et al. 2010, 2012; Burugula et al. 2014).
In this study, we show that the acetylated histone H3 tail contributes to efficient recruitment of FACT to transcribed CDS in S. cerevisiae. Depleting the Spt16 subunit of FACT elicits a global reduction in Pol II occupancies, consistent with its previously described role in transcription initiation. In contrast, Pol II occupancies were reduced toward the 3′-end in Spt6-depleted cells, suggesting processivity defects. Significant reductions in Pol II occupancies were also observed in a strain defective for Spt6 recruitment. Our study reveals that both FACT and Spt6 promote transcription genome-wide and, perhaps, participate in different stages of transcription.
Materials and Methods
Yeast strains and growth conditions
The yeast strains used in this study are listed in Supplemental Material, Table S1. The cells were grown to an absorbance A600 of 0.5–0.6 in synthetic complete (SC) media lacking isoleucine/valine and treated with sulfometuron methyl (SM; 0.6 µg/ml) for 30 min to induce the Gcn4 targets. Esa1 in the gcn5∆/esa1ts strain was inactivated by shifting the cultures grown at 25–37° for ∼1.5 hr prior to induction by SM. Spt16 and Spt6 were depleted in SPT16-TET and SPT6-TET (Hughes et al. 2000; Mnaimneh et al. 2004) cells by growing these strains in the presence of 10 μg/ml doxycycline (dox) overnight, and subculturing the overnight cultures in 100 ml of SC media with 10 μg/ml dox to an absorbance A600 of 0.6.
Co-immunoprecipitation assay
The co-immunoprecipitation experiments were performed as described previously (Govind et al. 2010). The HA-tagged H2B or Spt16-Myc-tagged wild-type (WT) and H3∆1-28 cells were resuspended in 500 µl of lysis buffer [50 mM Tris-HCl (pH 7.5), 50 mM HEPES-KOH (pH 7.9), 10 mM MgSO4, 100 mM (NH4)2SO4, 12.5 mM KOAc, 0.01% NP-40, 20% glycerol, 1 µg/ml Pepstatin A, 100 mM PMSF, and 1 µg/ml Leupeptin] and 500 µl of glass beads, and disrupted by vortexing (18 sec × 8 times, and 150 sec on ice between each agitation cycle). Whole-cell extracts (WCEs) were incubated overnight with magnetic beads that were preconjugated to anti-Myc or anti-HA antibodies in 100 µl 4× MTB buffer [200 mM HEPES-KOH (pH 7.9), 800 mM KOAc, 54 mM MgOAc, 40% glycerol, 0.04% NP-40, 400 mM PMSF, 4 µg/ml Pepstatin, and 4 µg/ml Leupeptin], and washed five times with the wash buffer [50 mM Tris-HCl (pH 8.0), 0.3% NP-40, 500 mM NaCl, 10% glycerol, 1 mM PMSF, 1 µg/ml Leupeptin, and 1 µg/ml Pepstatin]. Immunoprecipitates were analyzed by western blot using the following antibodies: anti-Myc (Roche), anti-HA (Roche), and anti-Spt16 and anti-Spt6 (kindly provided by Tim Formosa). The signal intensities were quantified using Image Studio lite version 5.2 (LI-COR Biosciences).
Chromatin immunoprecipitation (ChIP) and ChIP-chip
The cultures were cross-linked with formaldehyde and processed for ChIP, as described previously (Govind et al. 2012). ChIPs were performed using antibodies anti-Myc (Roche), anti-Rpb3 (Neoclone), and anti-H3 (Abcam). ChIP DNA and the related input DNA was amplified using the primers against specific regions. First, 5 µl of ChIP dye (15% Ficol and 0.25% bromophenol blue in 1× TBE) and SYBR green dye were added to the PCR products, which were resolved on 8% TBE gels, visualized on a phosphorimager, and quantified using ImageQuant 5.1 software. Fold enrichments were determined by taking the ratios of the ChIP signal for the gene of interest and the signal obtained for POL1 (used as an internal control), and then dividing by the ratios obtained for the related input samples. Fold enrichment: (ChIP intensities of the gene of interest / POL1) / (input intensities of gene of interest / POL1). The ChIP experiments were performed using at least three independent cultures and PCR reactions were conducted at least in duplicate. The error bars represent SEM. The P-values were determined using two-tailed distribution with unequal variance. The oligos used for ChIP PCRs are provided in Table S2.
For ChIP-chip experiments, ChIP and related input DNA samples were amplified from two biological replicates using the GenomePlex complete whole-genome amplification kit (catalog number WGA2; Sigma [Sigma Chemical], St. Louis, MO), according to the manufacturer’s instructions. The amplified ChIP DNA and input DNA were purified using a PCR cleanup kit (catalog number 28104; QIAGEN, Valencia, CA), and the DNA was quantified by NanoDrop. The samples were hybridized on Agilent 4 × 44 arrays (G4493A) after labeling the ChIP and input DNA with Alexa555 and Alexa647 fluorescent dyes, respectively, as per the manufacturer’s instructions at the genomics core facility at Michigan State University. The arrays were scanned using an Agilent scanner (G2600D), and data were extracted with the Feature Extraction software (Agilent) as described previously (Spain et al. 2014).
Bioinformatics analysis
The data extracted with the Feature Extraction software (Agilent) was normalized using the Limma package from Bioconductor, as described previously (Venkatesh et al. 2012). The genes were divided into 10 equal sized bins, with two bins assigned to the region 500-bp upstream of the transcription start site (TSS) and two bins to the region 500-bp downstream of the transcription end site (TES). The average probe enrichment values were assigned to the closest bin according to the probe location, and a 10 bin matrix was generated using a PERL script. Genes corresponding to the majority of dubious ORFs, tRNA genes, and small nuclear RNA genes, as well as autonomously replicating sequences, were removed from the data set. The enrichments in the six bins between TSS and TES were averaged to obtain an average ORF occupancy. The genes for analysis were selected on the basis of ORF enrichment. The genes < 500 bp in length were removed from the analyses. The versatile aggregate profiler (Brunelle et al. 2015) was used to generate gene-average profiles. The genes were split in the middle, and the probe intensities were aligned to the TSS for the first-half and to the TES for the second-half of the genes.
Box plots
Center lines show the medians, and the box limits indicate the 25th and 75th percentiles, as determined by R software. Whiskers extend 1.5× the interquartile range from the 25th and 75th percentiles, and outliers are represented by dots.
Data availability
Strains generated in this study are available upon request. The Gene Expression Omnibus accession number for the ChIP-chip data reported in this paper is GSE69642. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6133802.
Results
Histone H3 N-terminal tails facilitate FACT interaction with chromatin in vivo
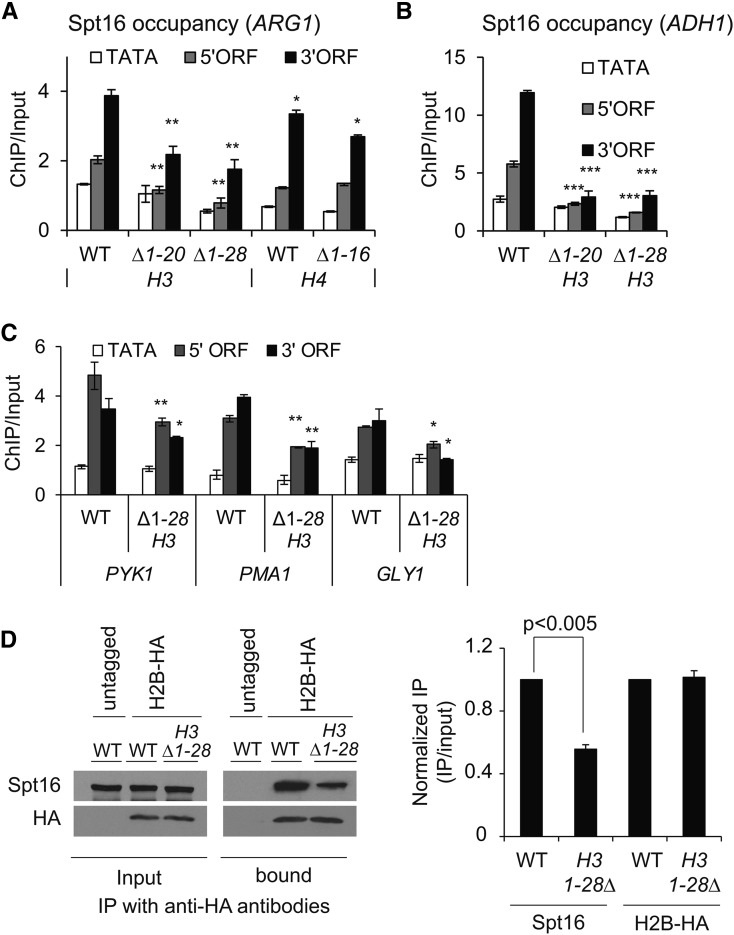
While Spt16 is enriched in coding regions of actively transcribing genes (Mason and Struhl 2003; Mayer et al. 2010), the mechanisms by which it is recruited remain to be established. Given that the FACT complex interacts with nucleosomes in vitro (Belotserkovskaya et al. 2003), it can be recruited through nucleosome interactions. Moreover, Spt16 interaction was greatly reduced with the nucleosomes lacking the histone N-terminal tails, implicating histone tails in promoting Spt16–nucleosome interactions (Stuwe et al. 2008; VanDemark et al. 2008). Significantly, removal of the H3 and H4 tails, but not of the H2A/H2B tails, abolished Spt16–histone interactions in vitro (Winkler et al. 2011). The H3 or H4 tails could therefore facilitate recruitment/retention of the FACT complex. To test this possibility, we examined yeast Spt16 occupancy by ChIP in histone mutants lacking the H3 or H4 N-terminal tail. Spt16 occupancy in the ARG1 5′ and 3′ ORFs was reduced by ∼50% in the H3 mutants lacking 1–20 (H3∆1-20) or 1–28 (H3∆1-28) N-terminal residues (Figure 1A). In comparison, in the H4 tail mutant (H4∆1-16), only a small (∼20%) reduction was observed in the ARG1 3′ ORF (Figure 1A). A substantial reduction in Spt16 occupancy (∼80%) was also observed at a constitutively expressed ADH1 gene in the H3 mutants (Figure 1B), but not in the H4 mutant (Figure S1A). Although FACT/Spt16 interacts with both H3 and H4 tail peptides in vitro, (Stuwe et al. 2008; VanDemark et al. 2008) it appears that the H3 tail may help in recruiting or retaining FACT to its target genes in vivo. Consistent with this idea, reduced occupancy of Spt16 was also observed in the coding regions of PYK1, PMA1 and GLY1 genes (Figure 1C). In contrast to the impaired Spt16 occupancy in the H3 mutant, for most genes the Pol II occupancies were comparable in the WT and H3 mutants, except for the GLY1 gene, which showed a small reduction in Pol II occupancy (Figure S1B).
Figure 1.
The H3 N-terminal tail promotes FACT recruitment. (A and B) Chromatin immunoprecipitation (ChIP) occupancy of Myc-tagged Spt16 in wild-type (WT) cells and mutants lacking the H3 N-terminal tail residues 1–20 (H3∆1-20) or 1–28 (H3∆1-28) and 1–16 residues of H4 (H4∆1-16) at ARG1 (A), and in H3 mutants at ADH1 (B). Graphs show mean and SEM. * P-value < 0.01, ** P-value < 0.001, and *** P-value < 0.0001. (C) ChIP occupancies of Myc-tagged Spt16 at the indicated genes in the WT and the H3 tail mutant, H3Δ1-28. * P-value < 0.01 and ** P-value < 0.001. (D) Whole-cell extracts prepared from HA-tagged H2B WT and H3∆1-28 strains were pulled down with the anti-HA beads via immunoprecipitation (IP), and the immunoprecipitates were subjected to western blot analysis using antibodies against Spt16 and HA. Untagged WT cells were used as a control. The representative blot is shown on the left and the quantified data on the right.
To further examine the role of the H3 tails in promoting FACT association with chromatin, we performed co-immunoprecipitation experiments. The HA-tagged histone H2B (H2B-HA) efficiently pulled-down Spt16 from the WCEs prepared from HA-tagged WT cells, but not from untagged cells (Figure 1D, left). We also observed a reduced Spt16 pull-down from the H3∆1-28 WCEs (∼50%; Figure 1D, right). No such reduction in Spt16 occupancy was seen in the H4 tail deletion mutant (H4Δ1-16; Figure S1C), supporting the idea that the H3 tail promotes Spt16 association with chromatin in vivo. However, the extent to which the H3 tail contributes in this process may be variable, as observed by the differences in Spt16 occupancies at different genes in the H3 mutant (Figure 1, A–C).
Acetylation of the H3 tail promotes FACT occupancy at ADH1 and ARG1 genes
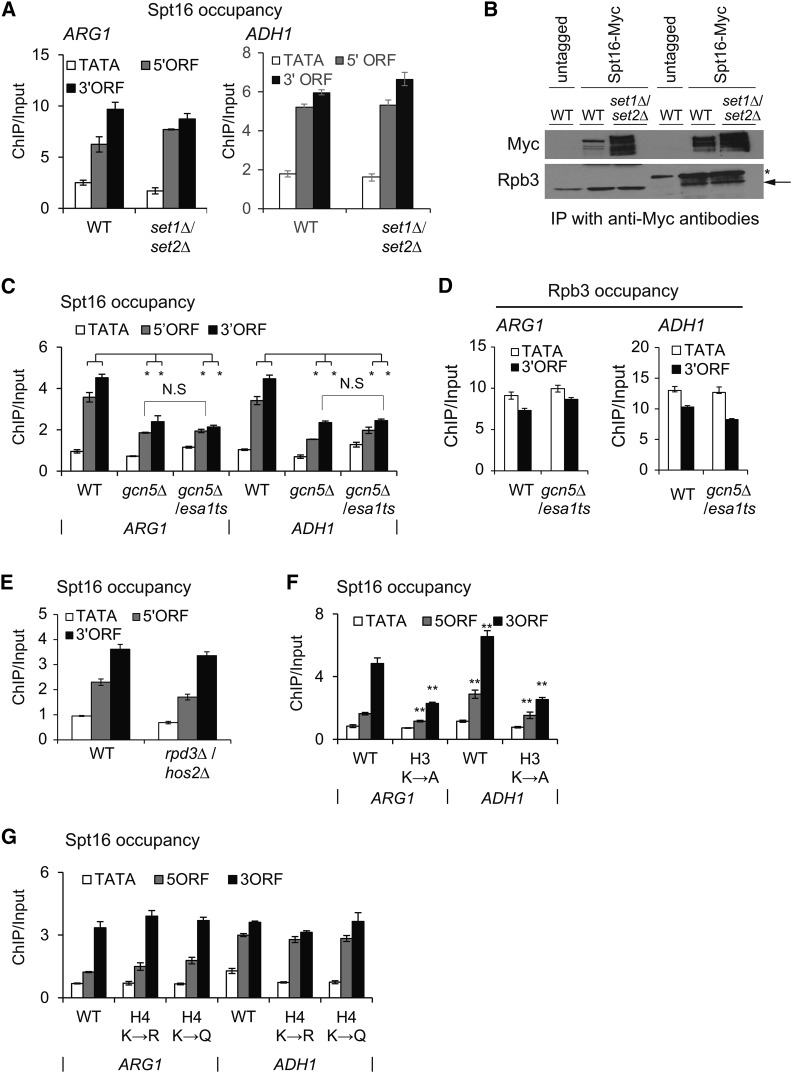
After observing that the H3 tail contributes to Spt16, we examined the role for posttranslational modifications on the H3 tail in regulating FACT localization to transcribed genes. The H3 tail lysines, K4 and K36, are methylated by Set1 and Set2, respectively (Briggs et al. 2001; Strahl et al. 2002). To examine whether the reduction in Spt16 occupancy in the H3 tail deletion mutant is due to the loss of H3 methylation, we measured Spt16 enrichment in the set1Δ/set2Δ double mutant. Comparable occupancies of Spt16 were observed in the ORFs of ARG1 and ADH1 genes in the WT and set1∆/set2∆ mutant (Figure 2A). Furthermore, we found a very similar signal for co-immunoprecipitation of Pol II with Spt16 in WT and set1∆/set2∆ WCEs (Figure 2B), suggesting that H3 methylation is likely dispensable for maintaining WT levels of FACT occupancy, at least at these two genes. The H3 and H4 tails are also acetylated by the Gcn5-containing SAGA and Esa1-containing NuA4 histone acetyltransferase (HAT) complexes, respectively. Accordingly, a gcn5∆/esa1ts double mutant elicits strong reductions in H3 and H4 acetylation (Ginsburg et al. 2009). The gcn5∆/esa1ts mutant, grown at the permissive temperature (25°) (represented as gcn5∆) or at the nonpermissive temperature (37°) to inactivate Esa1 (gcn5∆/esa1ts), produced comparable reductions in Spt16 occupancy in the ARG1 and ADH1 ORFs (Figure 2C and Figure S2, right). In contrast, both the WT and HAT mutant displayed comparable Pol II occupancies at ARG1 and ADH1 (Figure 2D and Figure S2, left). To examine whether FACT occupancy correlates to H3 acetylation levels, we determined Spt16 enrichment at ARG1 in a histone deacetylase mutant, rpd3Δ/hos2Δ. Interestingly, Spt16 occupancy was not increased in the histone deacetylase mutant rpd3∆/hos2∆ (Figure 2E). This is surprising given that H3 acetylation levels were shown to be elevated in this mutant (Govind et al. 2010), and that our current results reveal diminished FACT occupancy in the HAT mutant. It is possible that the WT level of histone acetylation is sufficient for normal FACT occupancy, suggesting that histone acetylation, but not deacetylation, plays a role in FACT recruitment/retention in coding regions. As such, any further increase does not necessarily increase FACT occupancy. Taken together, these results suggest a role for the acetylated H3 tail in promoting FACT occupancy in transcribed coding regions.
Figure 2.
Role of the acetylated H3 tail in modulating Spt16 occupancy. (A) Chromatin immunoprecipitation (ChIP) enrichment of Spt16-Myc at ARG1 in wild-type (WT) cells and a histone methyltransferase mutant set1∆/set2∆. (B) Whole-cell extracts prepared from Spt16-Myc-tagged WT and set1∆/set2∆ strains were subjected to immunoprecipitation (IP) with anti-Myc antibodies, and the immunoprecipitates were analyzed by western blot to detect signals for Myc and Rpb3. Untagged WT cells were used as a control. Rpb3 band in immunoprecipitated samples is shown by an arrow and the IgG heavy chain by an asterisk (*). (C) Spt16-Myc ChIP occupancy in WT cells and a gcn5∆/esa1 histone acetyltransferase (HAT) mutant. Spt16-Myc occupancies were measured by ChIP at ARG1 and ADH1 in a gcn5∆/esa1ts strain grown either at 25° (represented as gcn5∆) or at 37° to inactivate Esa1 (gcn5∆/esa1). Graphs show mean and SEM. * P-value < 0.001. Spt16 occupancy differences between gcn5Δ and gcn5∆/esa1ts were not significant (N.S). (D) Rpb3 occupancies in WT and gcn5Δ/esa1ts cells at ARG1 and ADH1. (E) Spt16 occupancies in WT and histone deacetylase mutant hos2Δ/rpd3Δ cells at ARG1. (F and G) Spt16-Myc enrichments at ARG1 and at ADH1 in histone H3 (F) and H4 tail mutants (G). H3 K4, K9, K14, and K18 substituted to alanine, H3K→A; H4 K5, K8, K12, and K16 substituted to arginine; H4K→R, or to glutamine H4K→Q. Graphs show mean and SEM. ** P-value < 0.0001.
To provide additional proof for the role of histone acetylation, we examined Spt16 occupancy in H3 and H4 tail point-mutants. The H3 mutant (K4, K9, K14, and K18 substituted to alanine; H3K→A) displayed reduced Spt16 occupancy in the ORFs of both ARG1 and ADH1 genes (Figure 2F). However, only minimal changes in Spt16 occupancy were observed in the H4 mutant (K5, K8, K12, and K16 substituted to arginine; H4K→R, or to glutamine; H4K→Q) (Figure 2G; H4K→A mutant exhibits a lethal phenotype). While Spt16 interacts with both H3 and H4 N-terminal tails and the histone tail mutants impair FACT function (Biswas et al. 2006; VanDemark et al. 2008), our results suggest that acetylation of the H3 tail makes a greater contribution to FACT occupancy in coding regions.
FACT and Spt6 are required for transcription genome-wide
Microarray analyses examining transcription defects have revealed that the loss of Spt16 function leads to aberrant transcription at many genomic locations, including within coding regions (Cheung et al. 2008; van Bakel et al. 2013). While it is evident that Spt16 functions to suppress widespread cryptic and antisense transcription, the role of FACT in regulating Pol II occupancy in coding regions is not well understood at a genome-wide scale. Gene-specific studies have suggested that FACT regulates transcription at the initiation step (Biswas et al. 2006; Jimeno-Gonzalez et al. 2006; Duina et al. 2007). Additionally, FACT, in cooperation with H2B ubiquitination, is important for the restoration of chromatin structure in the wake of Pol II elongation (Fleming et al. 2008). As mentioned earlier, similar to the FACT complex, Spt6 (a H3/H4 chaperone) is localized to the coding regions of strongly transcribed genes (Mayer et al. 2010; Ivanovska et al. 2011; Perales et al. 2013; Burugula et al. 2014) and is also important for suppressing aberrant transcription (Kaplan et al. 2003; Cheung et al. 2008; van Bakel et al. 2013). To compare the impact of FACT and Spt6 on transcription, we utilized strains in which the expression of SPT16 or SPT6 was under the control of a tetracycline (TET)-repressible promoter (SPT16-TET and SPT6-TET). These promoters can be repressed by growing cells in the presence of dox.
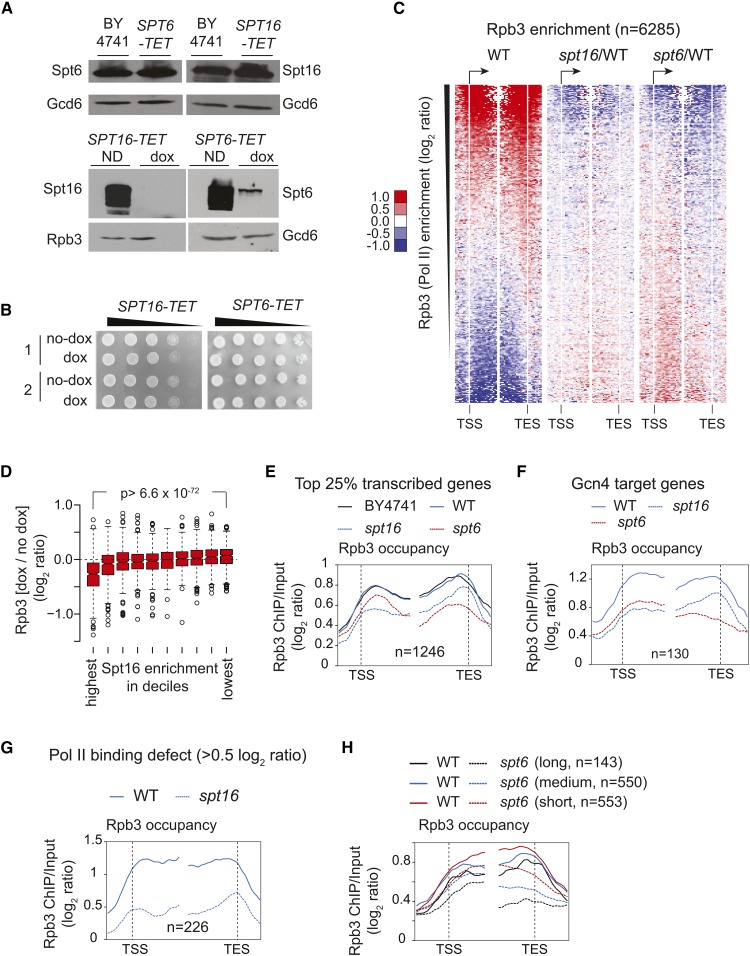
To rule out unexpected consequences of replacing the endogenous promoter with the TET promoter, we first compared Spt16 and Spt6 protein levels in TET strains and BY4741 (S. cerevisiae WT strain). Spt16 and Spt6 protein levels in the untreated (no dox) SPT16-TET and SPT6-TET, respectively, were very similar to those detected in the BY4741 cells (Figure 3A, top panel). As expected, treating SPT16-TET and SPT6-TET cells with dox led to reduced expression of Spt16 and Spt6, respectively (Figure 3A, bottom panel). We noted that Spt16 was depleted to a greater extent than Spt6 upon dox treatment. Since Spt16 mutants have been shown to cause cell cycle defects (Prendergast et al. 1990), we also measured the level of budded and unbudded cells in BY4741, SPT16-TET, and SPT6-TET dox-treated cells. We did not find any significant increase in the number of budded or unbudded cells under Spt6- or Spt16-depleted conditions (Figure S3A), suggesting that depleting Spt16 or Spt6 under the experimental conditions employed elicits minimal cell cycle defects. Moreover, the TET strains grown in the presence or absence of dox exhibited similar viability (Figure 3B). Altogether, these results indicate that the untreated SPT16-TET and SPT6-TET cells behave in a similar way to BY4741.
Figure 3.
Effect of depleting Spt16 and Spt6 on RNA polymerase II (Pol II) occupancy genome-wide. (A) BY4741, SPT16-TET, and SPT6-TET cells were grown in synthetic complete (SC) media and induced by sulfometuron methyl . Western blots show Spt16 and Spt6 protein levels in BY4741, SPT16-TET, and SPT6-TET strains without dox (doxycycline) treatment (top panel). The Spt16 and Spt6 levels in untreated (ND) and dox-treated TET strains are shown (bottom panel). (B) SPT16-TET and SPT6-TET cells grown and subcultured in SC media, with and without dox, were collected, serially 10-fold diluted, and spotted on SC plates. Growth for the two cultures of SPT16-TET (left) and SPT6-TET (right) with and without dox (no-dox) treatment are shown. (C) Heat maps depicting genome-wide Rpb3 (Pol II) enrichment in SPT16-TET [without dox; wild-type (WT)] (left), and changes in Rpb3 occupancies in dox-treated SPT16-TET (spt16/WT) (middle) and SPT6-TET (spt6/WT) (right) cells. Genes were sorted from highest to lowest ORF Rpb3 enrichment in WT cells. TES, transcription end site; TSS, transcription start site. (D) Box plot showing the changes in Rpb3 occupancy according to the Spt16 enrichment in deciles. The average ORF occupancy of Rpb3 in SPT16-TET dox-treated and untreated cells was determined, and the change in Rpb3 occupancy on depleting Spt16 (dox/no dox) was calculated for each gene. Genes were then grouped into deciles according to the Spt16 occupancy in WT cells. The first decile shows the highest Spt16 occupancy and the 10th shows the lowest. (E) Rpb3 occupancy profiles for the top 25% Pol II-occupied genes (n = 1246) in untreated BY4741 and SPT16-TET (WT), and in dox-treated SPT16-TET (spt16) and SPT6-TET (spt6), cells are shown. (F) Rpb3 occupancy profiles in untreated SPT16-TET (WT) and dox-treated SPT16-TET (spt16) and SPT6-TET (spt6) cells at the Gcn4 target genes enriched in the top 25% Pol II-occupied genes. (G) Rpb3 occupancy profile at the genes eliciting reduction in Rpb3 occupancy ≥ 0.5 log2 ratio [chromatin immunoprecipitation (ChIP)/input] on depleting Spt16 in WT and spt16. (H) The top 25% Pol II-occupied genes were grouped on the basis of their gene length, and Rpb3 occupancy profiles for the long (>2 kb), medium (1–2 kb), and short (0.5–1 kb) genes are shown in the WT and Spt6-depleted cells.
To examine the effects of depleting Spt16 and Spt6 on transcription, we determined Rpb3 occupancy in untreated cells (SPT16-TET; referred to as WT hereafter) and dox-treated SPT16-TET (spt16) and SPT6-TET (spt6) cells by ChIP-chip. We observed a strong correlation (Pearson correlation, r = 0.93) between the Pol II occupancies in the dox-untreated SPT16-TET and BY4741 strains genome-wide, which further indicated that replacing the endogenous SPT16 promoter with the TET promoter does not adversely affect transcription. We also determined Spt16 occupancy genome-wide by ChIP-chip and found that Spt16 occupancy in coding regions strongly correlated with Pol II occupancy (Pearson correlation, r = 0.85), in agreement with previous studies (Mayer et al. 2010).
The heat maps depicting changes in Rpb3 enrichment (spt16/WT) showed diminished ratios in coding regions of the genes displaying the greatest Rpb3 enrichments in WT cells (Figure 3C). Consistent with a strong correlation between Spt16 and Rpb3 occupancies, the genes with the highest Spt16 enrichments showed the greatest Rpb3 reductions (Figure 3D). We also noticed that depletion of Spt16 (and Spt6, described later) also revealed an increase in Pol II occupancies at those genes, which otherwise show very poor enrichment ratios in WT cells. Given that our ChIP-chip normalization was performed without spike-in control, it is difficult to ascertain the apparent increase in Rpb3 occupancy as biologically relevant.
To further analyze the impact of depleting Spt16 on Pol II occupancy, we selected the top 25% of genes showing greatest Rpb3 occupancy in WT cells (n = 1246). Nearly identical profiles for Rpb3 occupancy were observed in WT and BY4741 at the metagene comprised of these transcribed genes (Figure 3E). In contrast, Spt16 depletion evoked a reduction in Pol II occupancy in the coding regions of these sets of genes. Given that Gcn4 target genes are activated under the growth conditions used (see Materials and Methods), we additionally analyzed the effect of depleting Spt16 on the transcription of Gcn4 targets. Among the top 1246 transcribed genes, 130 Gcn4-regulated genes were enriched. Reduced Pol II occupancies were evident in the coding regions of these genes, as well as in Spt16-depleted cells (Figure 3F). Further, we identified 226 genes showing reduction in Pol II occupancy ≥ 0.5 log2 ratio (ChIP/input) in Spt16-depleted cells (Figure 3G). Interestingly, these genes were enriched among the top 10% transcribed genes (P-value = 10−117), suggesting that depletion of Spt16 imparts a significant effect on Pol II occupancy at highly expressed genes.
We noticed a small increase in Pol II occupancy in the Spt16-depleted cells at the 3′-ends of genes, analyzed above (Figure 3E). This increase may reflect increased cryptic transcription events that accumulate Pol II from the 5′- to 3′-end. Such an explanation would be consistent with the established role of histone chaperones in suppressing cryptic transcription (Kaplan et al. 2003; Cheung et al. 2008; van Bakel et al. 2013). A previous study utilizing a microarray predicted 960 and 1130 genes to have cryptic transcription in spt6-1004 and spt16-197 mutants, respectively (Cheung et al. 2008). We found that only 154 genes, predicted to express cryptic transcripts in the previous study, were among the 1246 genes exhibiting high levels of Pol II occupancy (Figure S3B). Replotting Pol II occupancy data after excluding these 154 genes (Figure S3C) displayed profiles similar to that observed in Figure 3E. Pol II pausing and queuing in the 3′-end superimposed on elongation defects at the very 5′-end could potentially explain an apparent increase in Pol II occupancy toward the 3′-end on depleting Spt16. Regardless, our result showing a reduction in Pol II occupancy across the coding regions is consistent with previous studies indicating a role for FACT in transcription initiation or the early stages of Pol II elongation.
Next, we analyzed Pol II occupancy in the Spt6-depleted cells. Interestingly, at the top 25% of Pol II-occupied genes, Spt6 depletion elicited a greater reduction in Rpb3 occupancy toward the 3′-end (Figure 3C, right and Figure 3E), consistent with previous studies showing the greatest Spt6 enrichment in the 3′-ends of transcribed genes (Perales et al. 2013; Burugula et al. 2014). A 5′ to 3′ bias in Pol II occupancy was also evident at the Gcn4 targets (Figure 3F) and at 238 genes that showed a reduction in Pol II occupancy ≥ 0.5 log2 ratio (ChIP/input) (Figure S3D). A progressive reduction in Pol II occupancy in the 5′ to 3′ direction in Spt6-depleted cells suggests that Spt6 may regulate Pol II processivity. Alternately, given the role of Spt6 activity in 3′-mRNA processing, diminished Spt6 levels could also result in reduced Pol II occupancy at the 3′-end (Kaplan et al. 2005). To distinguish between these two possibilities, we analyzed Rpb3 occupancy for the top 25% of transcribing genes based on their gene length. All three groups of genes—long (>2 kb), medium (1–2 kb) and short (0.5–1 kb)—showed a 5′ to 3′ bias in Pol II occupancies (Figure 3H). Interestingly, however, long and medium genes displayed a greater reduction at the 3′-end compared to the short genes (0.5–1 kb) (Figure 3H and Figure S3E). A simpler explanation for this observation is that Spt6 promotes Pol II processivity, in agreement with previous studies (Endoh et al. 2004; Ardehali et al. 2009; Perales et al. 2013). A 3′-mRNA processing defect (Kaplan et al. 2005) would be expected to produce similar reductions in Pol II occupancies at the 3′-end irrespective of gene length. Collectively, our data suggest that loss of FACT and Spt6 functions produces distinct effects on Pol II occupancy. However, the role of FACT and Spt6 in suppressing aberrant transcription could have an effect, to some extent, on Pol II occupancy under depletion conditions.
FACT and Spt6 differentially impact transcription and histone occupancy
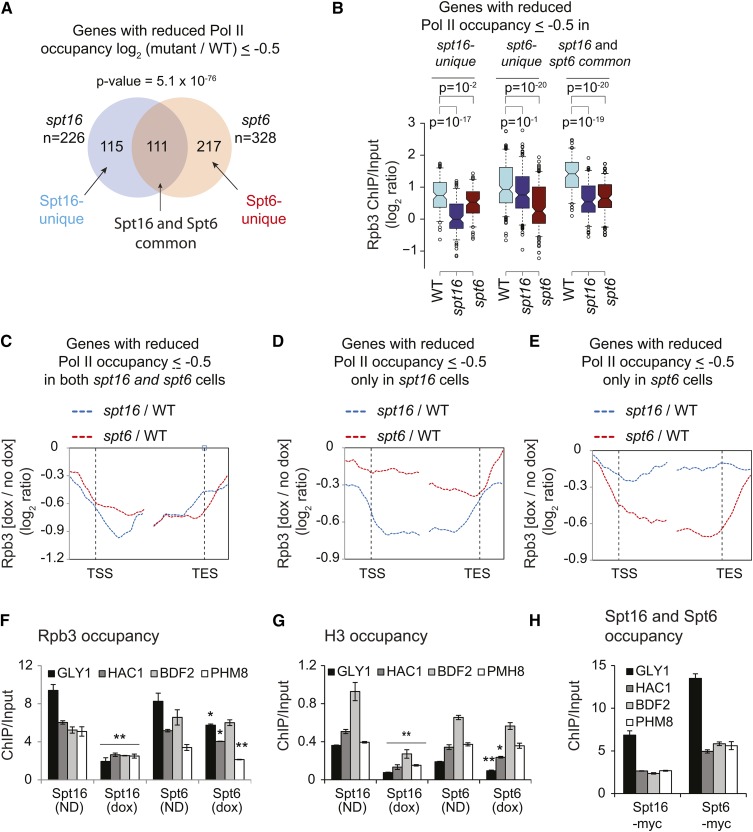
To further address the functional overlap between FACT and Spt6, we examined genes that showed a reduction in Pol II occupancy (≤ −0.5 log2 ratio) upon depleting these factors. We found a significant overlap between the genes exhibiting Pol II fold-change log2 ≥ 0.5 upon depleting either Spt16 or Spt6 (P-value = 5.1 × 10−76, n = 111) (Figure 4A). The genes showing Pol II occupancy defects upon depleting either Spt16 or Spt6 (common; n = 111) exhibited, on average, higher Pol II occupancy (in WT cells) than those genes that showed defects only after depleting either Spt16 or Spt6 (unique) (Figure 4B). This observation suggests that strongly transcribed genes may need full function of Spt6 and Spt16 for a high level of transcription. It is also interesting to note that while Spt6 depletion was less efficient compared to that of Spt16, it nonetheless evoked very similar Pol II occupancy defects on these genes (Figure 4C). Rpb3 profiles at the genes uniquely affected by either Spt6 or Spt16 depletion showed expected profiles (Figure 4, D and E). Considering that Spt6 exhibits higher occupancy toward the 3′-ends (Mayer et al. 2010; Perales et al. 2013; Burugula et al. 2014), we analyzed the distribution of gene length in the three classes of genes. The longer genes were enriched in the Spt6-unique and common genes, whereas Spt16-unique genes were shorter in comparison (Figure S4, A and B). The differential effect of Spt16 and Spt6 depletion nicely correlates with their respective localization patterns over genes, further suggesting their roles in different steps of transcription.
Figure 4.
FACT promotes nucleosome reassembly. (A) Venn diagram showing overlap among the genes that displayed Rpb3 occupancy defect > 0.5 log2 ratio chromatin immunoprecipitation [(ChIP)/Input] in Spt16- and Spt6-depleted cells. P-value for the overlap is shown. (B) Box plot showing Rpb3 average ChIP-chip enrichments at Spt16-unique, Spt6-unique, and Spt16 and Spt6 common genes in wild-type (WT), Spt16-depleted, and Spt6-depleted cells. (C–E) Metagene analysis for changes in RNA polymerase II (Pol II) enrichment under Spt16- and Spt6-depeleted cells, for genes showing reduction > 0.5 log2 ratio (ChIP/input) under Spt16- or Spt6-depleted conditions: (C) (common), only under Spt16 depletion (D), or only under Spt6 depletion (E). TES, transcription end site; TSS, transcription start site. (F–H) ChIP occupancies of Rpb3 (F), histone H3 (G), and of Spt6 and Spt16 (H) in 5′ ORFs of the indicated genes in SPT16-TET and SPT6-TET untreated (ND) or doxycycline (dox)-treated cells. * P-value < 0.05 and ** P-value < 0.001.
We further examined the differential effect of depletion by determining Rpb3 ChIP occupancy at four genes that showed comparable Spt6 and Spt16 occupancy in our ChIP-chip experiments. Pol II occupancies in the ORFs of GLY1, HAC1, BDF2, and PHM8 were substantially reduced (∼two- to fivefold) upon Spt16 depletion (Figure 4F). By contrast, only a moderate to negligible reduction was observed after depleting Spt6. For example, Rpb3 was reduced by <1.5-fold at GLY1, HAC1, and PHM8 upon Spt6 depletion. Similarly, we found that histone H3 occupancy was more severely reduced in the 5′ ORFs of these genes upon depleting Spt16 than Spt6 (Figure 4G). Reduced H3 occupancies upon depletion of Spt16 and Spt6 are consistent with their role in histone reassembly (Ivanovska et al. 2011; Perales et al. 2013; Jeronimo et al. 2015). Bigger reductions in Pol II and H3 occupancies were not due to higher Spt16 occupancies at these genes compared to that of Spt6 (Figure 4H). These analyses are consistent with the role of both FACT and Spt6 in regulating transcription globally.
FACT cooperates with Spt6 to promote transcription
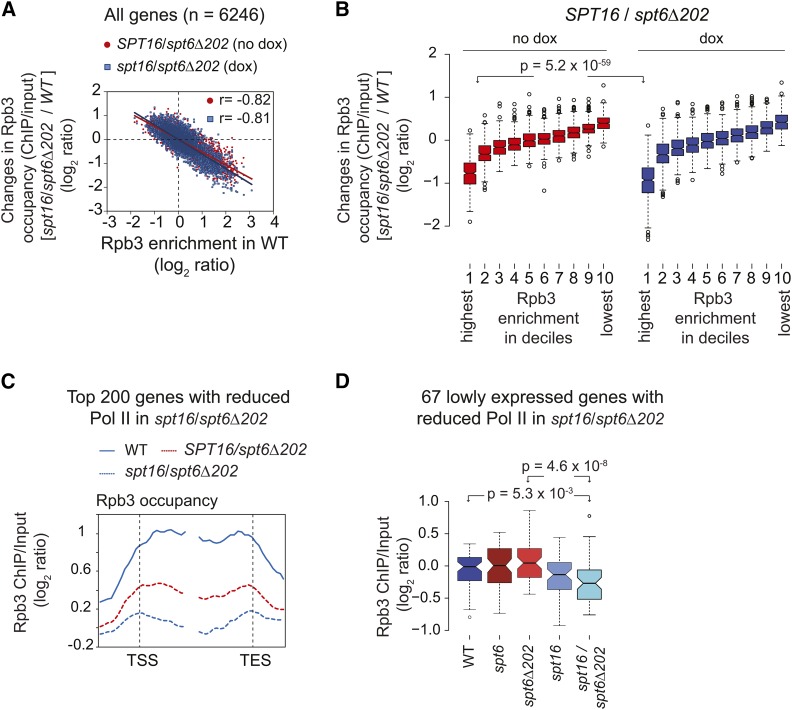
To further investigate whether Spt6 and FACT cooperate in promoting transcription, we deleted the Spt6 tSH2 domain (202 residues from the C-terminus), which mediates Spt6 recruitment genome-wide (Mayer et al. 2010; Burugula et al. 2014), in the SPT16-TET background (SPT16/spt6∆202). As expected, treating the SPT16/spt6∆202 mutant with dox resulted in reduced Spt16 protein levels (Figure S5A). We then determined Rpb3 occupancies by ChIP-chip in untreated (SPT16/spt6Δ202) and dox-treated (spt16/spt6Δ202) cells, and compared the occupancies with that of the untreated SPT16-TET cells (WT). The changes in Rpb3 occupancy in untreated SPT16/spt6∆202 cells were significantly anticorrelated with the Rpb3 occupancy in the WT cell (r = −0.81) (Figure 5A), indicating a strong requirement for Spt6 in transcription genome-wide. The reductions in Pol II occupancies in the dox-treated SPT16/spt6Δ202 cells were very similar to those in the untreated cells. However, a modest but statistically significant reduction in Rpb3 occupancy was observed in dox-treated (spt16/spt6∆202) cells when compared to the untreated cells only for the first decile (P = 5.2 × 10−59), representing the most highly transcribed genes (Figure 5B). This is consistent with the idea that both Spt6 and Spt16 are necessary to elicit high transcription levels.
Figure 5.
Spt6 promotes histone eviction and transcription. (A) The changes in Rpb3 occupancies [log2 ratio, chromatin immunoprecipitation (ChIP)/input] in the untreated [no dox (doxycycline)] and treated (dox) spt16/spt6∆202 cells relative to occupancies in the untreated SPT16-TET [wild-type (WT)] cells are plotted against the Rpb3 ORF occupancies in WT. Pearson correlations are shown. (B) Box plot showing changes in Rpb3 occupancy in SPT16/spt6∆202 (dox and no dox) relative to the SPT16-TET (no dox) WT cells, at the genes grouped in deciles on the basis of the average ORF Rpb3 occupancies observed in WT cells. The first decile shows the highest Rpb3 occupancy and the 10th shows the lowest. (C) Top 200 genes showing the greatest reduction in RNA polymerase II (Pol II) occupancy in dox-treated SPT16/spt6Δ202 relative to the untreated cells were selected based on the average Rpb3 enrichments in coding regions. A Pol II occupancy profile for these genes is plotted for WT, SPT16/spt6Δ202, and spt16/spt6Δ202. (D) Box plot showing average Rpb3 enrichment (log2 ratio, ChIP/input) for 67 genes of the 200 genes. These genes were not among the top 25% of genes showing greatest Rpb3 enrichment in WT cells. P-values are shown.
Rpb3 profiles in spt16/spt6∆202 mutant cells revealed greatly diminished occupancy across the coding regions of the top 25% of Pol II-occupied genes (Figure S5B). It appeared that untreated SPT16/spt6∆202 elicited a greater reduction in Pol II occupancy than observed upon depleting Spt6 (compare Figure 3E and Figure S5B). An incomplete depletion of Spt6 (Figure 3A) or different experimental conditions (no dox in spt6Δ202 vs. dox treatment for Spt6 depletion) could account for this observation. Nonetheless, these results indicate the importance of Spt6 in stimulating high-level transcription genome-wide. However, depleting Spt16 in SPT16/spt6∆202 cells produced only modest reductions in Pol II occupancy in the top 25% of Pol II-occupied genes (Figure S5B). This observation raises the possibility that FACT may require certain aspects of Spt6 function to stimulate transcription. Such an explanation is consistent with the observation that Spt16 depletion elicited reduced Pol II occupancy in an, otherwise, WT strain (Figure 3, D–G).
To further address functional cooperation between Spt16 and Spt6, we focused on the genes eliciting greater reduction in dox-treated spt16/spt6Δ202 cells than in untreated cells. The top 200 genes showing the greatest Pol II occupancy defect revealed that depleting Spt16 in an spt6Δ202 background significantly reduced Rpb3 occupancy across the coding region (Figure 5C). Of these 200 genes, 137 were among the top 25% expressed genes (n = 1246) in WT cells. The double mutant exhibited greater reduction in Pol II occupancy than observed in spt6Δ202 or spt16 cells at these 137 genes (Figure S5C). These observations support the idea that both FACT and Spt6 are needed to elicit high levels of transcription. At the 63 lowly expressed genes (among the 200 genes), a significant reduction in Pol II occupancy was seen only in the double mutant spt16/spt6Δ202 (Figure 5D). This could suggest that FACT acts redundantly with Spt6 in promoting the transcription of a subset of lowly expressed genes. More sensitive methods will be required to fully comprehend the extent to which Spt6 and FACT coordinate the transcription of lowly expressed genes, especially considering the technical challenges in accurately measuring Pol II occupancies at genes expressed at very low levels.
Discussion
In this study, we have examined the role of two highly conserved histone chaperones, Spt16 and Spt6, in regulating genome-wide transcription under amino acid starvation conditions. Spt6 recruitment to coding regions is stimulated by the phosphorylated Pol II and by the histone deacetylases Rpd3 and Hos2 (Mayer et al. 2012; Burugula et al. 2014; Sdano et al. 2017). However, the mechanism by which the FACT complex associates with chromatin is not well understood. Our results showing diminished occupancy of Spt16 at the ORFs of several genes and reduced interaction with nucleosomes in the H3 tail mutant (Figure 1) suggest a role for the H3 tail in promoting FACT association with transcribed regions. Our results additionally suggest that histone acetylation enhances the ability of the FACT complex to associate with transcribed ORFs, since deleting Gcn5 (an H3 HAT) or mutating the H3 tail lysine residues significantly dampened Spt16 enrichment in the CDS of ARG1 and ADH1 (Figure 2, C and F). These results provide in vivo evidence for the previous studies showing impaired FACT binding to the nucleosomes/histones lacking N-terminal tails (VanDemark et al. 2008; Winkler et al. 2011). However, which regions of Spt16 are required for histone tail interactions is currently unclear. While the Spt16 N-terminal domain of S. pombe was shown to be important for interacting with H3 and H4 tails (Stuwe et al. 2008), this domain was found to be dispensable in S. cerevisiae (VanDemark et al. 2008).
Considering that Spt16 exhibits similar affinity toward the acetylated and unacetylated histone peptides in vitro (Stuwe et al. 2008), it is plausible that acetylation-mediated changes to the nucleosome structure allow FACT to stably bind chromatin at transcribing loci. Consistent with this idea, Gcn5 promotes histone eviction, Pol II elongation, and stimulates recruitment of the bromodomain-containing chromatin remodelers RSC and SWI/SNF (Govind et al. 2007; Dutta et al. 2014; Spain et al. 2014). Additional contacts with the core domains of H3/H4, and with H2A/H2B through its C-terminal domain (Winkler et al. 2011; Kemble et al. 2015), could further stabilize FACT–chromatin interactions, thereby maintaining chromatin in an accessible conformation to promote transcription and concomitantly aiding the reassembly of evicted histones in the wake of transcription (Jamai et al. 2009). Additional factors may act cooperatively with other factors to enhance FACT enrichment. For instance, FACT interacts with the Paf1 complex and chromatin remodeler Chd1, both of which are enriched in transcribed regions (Krogan et al. 2002; Squazzo et al. 2002; Simic et al. 2003).
The enrichment of FACT in coding regions (Mayer et al. 2010) (data not shown), and its ability to promote Pol II transcription through the nucleosomal templates in vitro, (Belotserkovskaya et al. 2003; Hsieh et al. 2013), strongly suggests a role for FACT in the elongation step of transcription. FACT is linked to the reestablishment of the disrupted chromatin structure in the wake of transcription (Jamai et al. 2009) and, consequently, in suppressing aberrant transcription by preventing the utilization of cryptic promoters (Kaplan et al. 2003; Cheung et al. 2008; van Bakel et al. 2013). Our data indicate that FACT is globally required for the promotion of transcription in the coding regions. Spt16 deficiency reduced Pol II occupancy in coding regions of highly transcribed genes, including ribosomal protein genes and Gcn4 targets (Figure 3). It was previously reported that histone mutants that perturb the association of Spt16 (a shift in occupancy toward the 3′-end) also reduced Pol II occupancy at the 5′-end of PMA1 and FBA1 genes (Nguyen et al. 2013). Thus, it seems that impairing FACT activity through the depletion of Spt16 or by altering FACT association in the coding region (Nguyen et al. 2013) elicits greater defects in transcription in the early transcribed regions. These results raise a possibility that FACT may help polymerases negotiate the nucleosomal barrier downstream of the TSS. Such an idea is consistent with biochemical studies showing that FACT relieves Pol II pauses well within the nucleosomes and acts primarily to overcome tetramer–DNA contacts (Bondarenko et al. 2006). Considering that the rate of elongation in the early transcribed regions is slower than that observed in the middle or 3′-end regions (Danko et al. 2013), our findings suggest that FACT might help polymerases to overcome the nucleosomal barrier at the 5′-ends of the transcribed genes. Although nucleosomes are expected to pose a similar block to elongating Pol II irrespective of their position along the CDS, a nucleosomal impediment to transcription at the distal end may be additionally relieved by factors such as the chromatin remodelers RSC and SWI/SNF, which are also enriched in the ORFs of many genes (Dutta et al. 2014; Spain et al. 2014). Moreover, post-translational histone modifications, which are not uniform across coding regions, may have a differential effect on FACT activity to stimulate transcription. In support of such a possibility, it was shown that the FACT complex cooperates with H2B ubiquitination to regulate transcription both in vitro and in vivo (Pavri et al. 2006; Fleming et al. 2008).
In contrast to FACT, Spt6 depletion diminished Pol II occupancy at the 3′-end (Figure 3, E, F, and H). Lower Pol II occupancy toward the 3′-end could result from a processivity defect and/or from a specific defect at the 3′-end. Our analyses, based on gene length, favor the idea that Spt6 enhances Pol II processivity, possibly through stimulating nucleosome eviction or by preventing premature dissociation during Pol II traversal through coding regions. Such an interpretation is consistent with previous studies showing reduced Pol II occupancy at the 3′-ends of exceptionally long genes (Perales et al. 2013). Spt6 has also been shown to enhance the rate of Pol II elongation at heat-shock genes in Drosophila (Ardehali et al. 2009). Therefore, reduced Pol II occupancy near the 3′-ends of yeast genes suggests that Spt6 may function to help the elongating polymerase traverse coding regions through multiple mechanisms.
Pol II occupancy in the Spt6 mutant lacking the C-terminal tandem SH2 domain was strongly correlated with Pol II occupancy in the WT cells, indicating a strong requirement for Spt6 in stimulating transcription genome-wide. Even though depleting Spt16 in otherwise WT cells evoked a reduction in Pol II occupancy from highly transcribed genes, this surprisingly had only a minor impact on Pol II occupancy at the majority of transcribed genes in spt6Δ202 cells. This suggests that both factors might have overlapping roles and that FACT may rely on certain aspects of Spt6 function to promote transcription. This makes sense, considering that FACT and Spt6 are recruited to transcribed regions (Mayer et al. 2010; Burugula et al. 2014), and that both factors can interact with H2A/H2B and H3/H4 (McCullough et al. 2015). Despite interacting with nucleosomes as well as with histones, FACT (unlike Spt6) can reorganize nucleosome structure in a manner that increases DNA accessibility. This distinct ability of FACT could explain the reduction in Pol II occupancy observed in a subset of transcribed genes in the spt16/spt6Δ202 double mutant, but not in the single mutant (Figure 5C). Overall, our study strongly implicates both chaperones in promoting transcription genome-wide.
Acknowledgments
We thank Tim Formosa for providing antibodies against Spt6 and Spt16, and Alan Hinnebusch, Randy Morse, and Jeena Kinney for useful discussions and providing valuable comments on the manuscript. We also acknowledge the genomic core facility at Michigan State University. C.K.G. is supported by grants from the National Institutes of Health (GM-095514) and the Center for Biomedical Research (Oakland University).
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6133802.
Communicating editor: C. Kaplan
Literature Cited
- Andrulis E. D., Guzman E., Doring P., Werner J., Lis J. T., 2000. High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: roles in promoter proximal pausing and transcription elongation. Genes Dev. 14: 2635–2649. 10.1101/gad.844200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardehali M. B., Yao J., Adelman K., Fuda N. J., Petesch S. J., et al. , 2009. Spt6 enhances the elongation rate of RNA polymerase II in vivo. EMBO J. 28: 1067–1077. 10.1038/emboj.2009.56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belotserkovskaya R., Oh S., Bondarenko V. A., Orphanides G., Studitsky V. M., et al. , 2003. FACT facilitates transcription-dependent nucleosome alteration. Science 301: 1090–1093. 10.1126/science.1085703 [DOI] [PubMed] [Google Scholar]
- Billon P., Cote J., 2013. Precise deposition of histone H2A.Z in chromatin for genome expression and maintenance. Biochim. Biophys. Acta 1819: 290–302. 10.1016/j.bbagrm.2011.10.004 [DOI] [PubMed] [Google Scholar]
- Biswas D., Dutta-Biswas R., Mitra D., Shibata Y., Strahl B. D., et al. , 2006. Opposing roles for Set2 and yFACT in regulating TBP binding at promoters. EMBO J. 25: 4479–4489. 10.1038/sj.emboj.7601333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondarenko V. A., Steele L. M., Ujvari A., Gaykalova D. A., Kulaeva O. I., et al. , 2006. Nucleosomes can form a polar barrier to transcript elongation by RNA polymerase II. Mol. Cell 24: 469–479. 10.1016/j.molcel.2006.09.009 [DOI] [PubMed] [Google Scholar]
- Bortvin A., Winston F., 1996. Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science 272: 1473–1476. 10.1126/science.272.5267.1473 [DOI] [PubMed] [Google Scholar]
- Briggs S. D., Bryk M., Strahl B. D., Cheung W. L., Davie J. K., et al. , 2001. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 15: 3286–3295. 10.1101/gad.940201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunelle M., Coulombe C., Poitras C., Robert M. A., Markovits A. N., et al. , 2015. Aggregate and heatmap representations of genome-wide localization data using VAP, a versatile aggregate profiler. Methods Mol. Biol. 1334: 273–298. 10.1007/978-1-4939-2877-4_18 [DOI] [PubMed] [Google Scholar]
- Burugula B. B., Jeronimo C., Pathak R., Jones J. W., Robert F., et al. , 2014. Histone deacetylases and phosphorylated polymerase II C-terminal domain recruit Spt6 for cotranscriptional histone reassembly. Mol. Cell. Biol. 34: 4115–4129. 10.1128/MCB.00695-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung V., Chua G., Batada N. N., Landry C. R., Michnick S. W., et al. , 2008. Chromatin- and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome. PLoS Biol. 6: e277 10.1371/journal.pbio.0060277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Close D., Johnson S. J., Sdano M. A., McDonald S. M., Robinson H., et al. , 2011. Crystal structures of the S. cerevisiae Spt6 core and C-terminal tandem SH2 domain. J. Mol. Biol. 408: 697–713. 10.1016/j.jmb.2011.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danko C. G., Hah N., Luo X., Martins A. L., Core L., et al. , 2013. Signaling pathways differentially affect RNA polymerase II initiation, pausing, and elongation rate in cells. Mol. Cell 50: 212–222 (erratum: Mol. Cell 50: 778) 10.1016/j.molcel.2013.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechassa M. L., Sabri A., Pondugula S., Kassabov S. R., Chatterjee N., et al. , 2010. SWI/SNF has intrinsic nucleosome disassembly activity that is dependent on adjacent nucleosomes. Mol. Cell 38: 590–602. 10.1016/j.molcel.2010.02.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGennaro C. M., Alver B. H., Marguerat S., Stepanova E., Davis C. P., et al. , 2013. Spt6 regulates intragenic and antisense transcription, nucleosome positioning, and histone modifications genome-wide in fission yeast. Mol. Cell. Biol. 33: 4779–4792. 10.1128/MCB.01068-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dengl S., Mayer A., Sun M., Cramer P., 2009. Structure and in vivo requirement of the yeast Spt6 SH2 domain. J. Mol. Biol. 389: 211–225. 10.1016/j.jmb.2009.04.016 [DOI] [PubMed] [Google Scholar]
- Dion M. F., Kaplan T., Kim M., Buratowski S., Friedman N., et al. , 2007. Dynamics of replication-independent histone turnover in budding yeast. Science 315: 1405–1408. 10.1126/science.1134053 [DOI] [PubMed] [Google Scholar]
- Duina A. A., Rufiange A., Bracey J., Hall J., Nourani A., et al. , 2007. Evidence that the localization of the elongation factor Spt16 across transcribed genes is dependent upon histone H3 integrity in Saccharomyces cerevisiae. Genetics 177: 101–112. 10.1534/genetics.106.067140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta A., Gogol M., Kim J.-H., Smolle M., Venkatesh S., et al. , 2014. Swi/Snf dynamics on stress-responsive genes is governed by competitive bromodomain interactions. Genes Dev. 28: 2314–2330. 10.1101/gad.243584.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endoh M., Zhu W., Hasegawa J., Watanabe H., Kim D. K., et al. , 2004. Human Spt6 stimulates transcription elongation by RNA polymerase II in vitro. Mol. Cell. Biol. 24: 3324–3336. 10.1128/MCB.24.8.3324-3336.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming A. B., Kao C.-F., Hillyer C., Pikaart M., Osley M. A., 2008. H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol. Cell 31: 57–66. 10.1016/j.molcel.2008.04.025 [DOI] [PubMed] [Google Scholar]
- Formosa T., 2012. The role of FACT in making and breaking nucleosomes. Biochim. Biophys. Acta 1819: 247–255. 10.1016/j.bbagrm.2011.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formosa T., Eriksson P., Wittmeyer J., Ginn J., Yu Y., et al. , 2001. Spt16-Pob3 and the HMG protein Nhp6 combine to form the nucleosome-binding factor SPN. EMBO J. 20: 3506–3517. 10.1093/emboj/20.13.3506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsburg D. S., Govind C. K., Hinnebusch A. G., 2009. NuA4 lysine acetyltransferase Esa1 is targeted to coding regions and stimulates transcription elongation with Gcn5. Mol. Cell. Biol. 29: 6473–6487. 10.1128/MCB.01033-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govind C. K., Zhang F., Qiu H., Hofmeyer K., Hinnebusch A. G., 2007. Gcn5 promotes acetylation, eviction, and methylation of nucleosomes in transcribed coding regions. Mol. Cell 25: 31–42. 10.1016/j.molcel.2006.11.020 [DOI] [PubMed] [Google Scholar]
- Govind C. K., Qiu H., Ginsburg D. S., Ruan C., Hofmeyer K., et al. , 2010. Phosphorylated Pol II CTD recruits multiple HDACs, including Rpd3C(S), for methylation-dependent deacetylation of ORF nucleosomes. Mol. Cell 39: 234–246. 10.1016/j.molcel.2010.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govind C. K., Ginsburg D., Hinnebusch A. G., 2012. Measuring dynamic changes in histone modifications and nucleosome density during activated transcription in budding yeast. Methods Mol. Biol. 833: 15–27. 10.1007/978-1-61779-477-3_2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurard-Levin Z. A., Quivy J. P., Almouzni G., 2014. Histone chaperones: assisting histone traffic and nucleosome dynamics. Annu. Rev. Biochem. 83: 487–517. 10.1146/annurev-biochem-060713-035536 [DOI] [PubMed] [Google Scholar]
- Hassan A. H., Neely K. E., Workman J. L., 2001. Histone acetyltransferase complexes stabilize swi/snf binding to promoter nucleosomes. Cell 104: 817–827. 10.1016/S0092-8674(01)00279-3 [DOI] [PubMed] [Google Scholar]
- Hondele M., Ladurner A. G., 2011. The chaperone-histone partnership: for the greater good of histone traffic and chromatin plasticity. Curr. Opin. Struct. Biol. 21: 698–708. 10.1016/j.sbi.2011.10.003 [DOI] [PubMed] [Google Scholar]
- Hondele M., Stuwe T., Hassler M., Halbach F., Bowman A., et al. , 2013. Structural basis of histone H2A–H2B recognition by the essential chaperone FACT. Nature 499: 111–114. 10.1038/nature12242 [DOI] [PubMed] [Google Scholar]
- Hsieh F. K., Kulaeva O. I., Patel S. S., Dyer P. N., Luger K., et al. , 2013. Histone chaperone FACT action during transcription through chromatin by RNA polymerase II. Proc. Natl. Acad. Sci. USA 110: 7654–7659. 10.1073/pnas.1222198110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes T. R., Marton M. J., Jones A. R., Roberts C. J., Stoughton R., et al. , 2000. Functional discovery via a compendium of expression profiles. Cell 102: 109–126. 10.1016/S0092-8674(00)00015-5 [DOI] [PubMed] [Google Scholar]
- Ivanovska I., Jacques P. E., Rando O. J., Robert F., Winston F., 2011. Control of chromatin structure by spt6: different consequences in coding and regulatory regions. Mol. Cell. Biol. 31: 531–541. 10.1128/MCB.01068-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamai A., Puglisi A., Strubin M., 2009. Histone chaperone Spt16 promotes redeposition of the original H3–H4 histones evicted by elongating RNA polymerase. Mol. Cell 35: 377–383. 10.1016/j.molcel.2009.07.001 [DOI] [PubMed] [Google Scholar]
- Jeronimo C., Watanabe S., Kaplan C. D., Peterson C. L., Robert F., 2015. The histone chaperones FACT and Spt6 restrict H2A.Z from intragenic locations. Mol. Cell 58: 1113–1123. 10.1016/j.molcel.2015.03.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimeno-Gonzalez S., Gomez-Herreros F., Alepuz P. M., Chavez S., 2006. A gene-specific requirement for FACT during transcription is related to the chromatin organization of the transcribed region. Mol. Cell. Biol. 26: 8710–8721. 10.1128/MCB.01129-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan C. D., Morris J. R., Wu C., Winston F., 2000. Spt5 and spt6 are associated with active transcription and have characteristics of general elongation factors in D. melanogaster. Genes Dev. 14: 2623–2634. 10.1101/gad.831900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan C. D., Laprade L., Winston F., 2003. Transcription elongation factors repress transcription initiation from cryptic sites. Science 301: 1096–1099. 10.1126/science.1087374 [DOI] [PubMed] [Google Scholar]
- Kaplan C. D., Holland M. J., Winston F., 2005. Interaction between transcription elongation factors and mRNA 3′-end formation at the Saccharomyces cerevisiae GAL10–GAL7 locus. J. Biol. Chem. 280: 913–922. 10.1074/jbc.M411108200 [DOI] [PubMed] [Google Scholar]
- Kato H., Okazaki K., Iida T., Nakayama J., Murakami Y., et al. , 2013. Spt6 prevents transcription-coupled loss of posttranslationally modified histone H3. Sci. Rep. 3: 2186 10.1038/srep02186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemble D. J., McCullough L. L., Whitby F. G., Formosa T., Hill C. P., 2015. FACT disrupts nucleosome structure by binding H2A–H2B with conserved peptide motifs. Mol. Cell 60: 294–306. 10.1016/j.molcel.2015.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kireeva M. L., Walter W., Tchernajenko V., Bondarenko V., Kashlev M., et al. , 2002. Nucleosome remodeling induced by RNA polymerase II: loss of the H2A/H2B dimer during transcription. Mol. Cell 9: 541–552. 10.1016/S1097-2765(02)00472-0 [DOI] [PubMed] [Google Scholar]
- Krogan N. J., Kim M., Ahn S. H., Zhong G., Kobor M. S., et al. , 2002. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol. Cell. Biol. 22: 6979–6992. 10.1128/MCB.22.20.6979-6992.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.-K., Shibata Y., Rao B., Strahl B. D., Lieb J. D., 2004. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat. Genet. 36: 900–905. 10.1038/ng1400 [DOI] [PubMed] [Google Scholar]
- Li B., Carey M., Workman J. L., 2007. The role of chromatin during transcription. Cell 128: 707–719. 10.1016/j.cell.2007.01.015 [DOI] [PubMed] [Google Scholar]
- Liu J., Zhang J., Gong Q., Xiong P., Huang H., et al. , 2011. Solution structure of tandem SH2 domains from Spt6 protein and their binding to the phosphorylated RNA polymerase II C-terminal domain. J. Biol. Chem. 286: 29218–29226. 10.1074/jbc.M111.252130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason P. B., Struhl K., 2003. The FACT complex travels with elongating RNA polymerase II and is important for the fidelity of transcriptional initiation in vivo. Mol. Cell. Biol. 23: 8323–8333. 10.1128/MCB.23.22.8323-8333.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A., Lidschreiber M., Siebert M., Leike K., Soding J., et al. , 2010. Uniform transitions of the general RNA polymerase II transcription complex. Nat. Struct. Mol. Biol. 17: 1272–1278. 10.1038/nsmb.1903 [DOI] [PubMed] [Google Scholar]
- Mayer A., Heidemann M., Lidschreiber M., Schreieck A., Sun M., et al. , 2012. CTD tyrosine phosphorylation impairs termination factor recruitment to RNA polymerase II. Science 336: 1723–1725. 10.1126/science.1219651 [DOI] [PubMed] [Google Scholar]
- McCullough L., Connell Z., Petersen C., Formosa T., 2015. The abundant histone chaperones Spt6 and FACT collaborate to assemble, inspect, and maintain chromatin structure in Saccharomyces cerevisiae. Genetics 201: 1031–1045. 10.1534/genetics.115.180794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mnaimneh S., Davierwala A. P., Haynes J., Moffat J., Peng W. T., et al. , 2004. Exploration of essential gene functions via titratable promoter alleles. Cell 118: 31–44. 10.1016/j.cell.2004.06.013 [DOI] [PubMed] [Google Scholar]
- Nguyen H. T., Wharton W., II, Harper J. A., Dornhoffer J. R., Duina A. A., 2013. A nucleosomal region important for ensuring proper interactions between the transcription elongation factor Spt16 and transcribed genes in Saccharomyces cerevisiae. G3 (Bethesda) 3: 929–940. 10.1534/g3.113.005926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavri R., Zhu B., Li G., Trojer P., Mandal S., et al. , 2006. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell 125: 703–717. 10.1016/j.cell.2006.04.029 [DOI] [PubMed] [Google Scholar]
- Perales R., Erickson B., Zhang L., Kim H., Valiquett E., et al. , 2013. Gene promoters dictate histone occupancy within genes. EMBO J. 32: 2645–2656. 10.1038/emboj.2013.194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prendergast J. A., Murray L. E., Rowley A., Carruthers D. R., Singer R. A., et al. , 1990. Size selection identifies new genes that regulate Saccharomyces cerevisiae cell proliferation. Genetics 124: 81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransom M., Williams S. K., Dechassa M. L., Das C., Linger J., et al. , 2009. FACT and the proteasome promote promoter chromatin disassembly and transcriptional initiation. J. Biol. Chem. 284: 23461–23471. 10.1074/jbc.M109.019562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sdano M. A., Fulcher J. M., Palani S., Chandrasekharan M. B., Parnell T. J., et al. , 2017. A novel SH2 recognition mechanism recruits Spt6 to the doubly phosphorylated RNA polymerase II linker at sites of transcription. Elife 6: e28723 10.7554/eLife.28723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simic R., Lindstrom D. L., Tran H. G., Roinick K. L., Costa P. J., et al. , 2003. Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. EMBO J. 22: 1846–1856. 10.1093/emboj/cdg179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spain M. M., Ansari S. A., Pathak R., Palumbo M. J., Morse R. H., et al. , 2014. The RSC complex localizes to coding sequences to regulate Pol II and histone occupancy. Mol. Cell 56: 653–666. 10.1016/j.molcel.2014.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squazzo S. L., Costa P. J., Lindstrom D. L., Kumer K. E., Simic R., et al. , 2002. The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. EMBO J. 21: 1764–1774. 10.1093/emboj/21.7.1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl B. D., Grant P. A., Briggs S. D., Sun Z. W., Bone J. R., et al. , 2002. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol. Cell. Biol. 22: 1298–1306. 10.1128/MCB.22.5.1298-1306.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuwe T., Hothorn M., Lejeune E., Rybin V., Bortfeld M., et al. , 2008. The FACT Spt16 “peptidase” domain is a histone H3–H4 binding module. Proc. Natl. Acad. Sci. USA 105: 8884–8889. 10.1073/pnas.0712293105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata S., Yu Y., Stillman D. J., 2009. FACT and Asf1 regulate nucleosome dynamics and coactivator binding at the HO promoter. Mol. Cell 34: 405–415. 10.1016/j.molcel.2009.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bakel H., Tsui K., Gebbia M., Mnaimneh S., Hughes T. R., et al. , 2013. A compendium of nucleosome and transcript profiles reveals determinants of chromatin architecture and transcription. PLoS Genet. 9: e1003479 10.1371/journal.pgen.1003479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanDemark A. P., Xin H., McCullough L., Rawlins R., Bentley S., et al. , 2008. Structural and functional analysis of the Spt16p N-terminal domain reveals overlapping roles of yFACT subunits. J. Biol. Chem. 283: 5058–5068. 10.1074/jbc.M708682200 [DOI] [PubMed] [Google Scholar]
- Venkatesh S., Smolle M., Li H., Gogol M. M., Saint M., et al. , 2012. Set2 methylation of histone H3 lysine 36 suppresses histone exchange on transcribed genes. Nature 489: 452–455. 10.1038/nature11326 [DOI] [PubMed] [Google Scholar]
- Winkler D. D., Muthurajan U. M., Hieb A. R., Luger K., 2011. Histone chaperone FACT coordinates nucleosome interaction through multiple synergistic binding events. J. Biol. Chem. 286: 41883–41892. 10.1074/jbc.M111.301465 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains generated in this study are available upon request. The Gene Expression Omnibus accession number for the ChIP-chip data reported in this paper is GSE69642. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6133802.