Abstract
[URE3] is an amyloid-based prion of Ure2p, a regulator of nitrogen catabolism. While most “variants” of the [URE3] prion are toxic, mild variants that only slightly slow growth are more widely studied. The existence of several antiprion systems suggests that some components may be protecting cells from potential detrimental effects of mild [URE3] variants. Our extensive Hermes transposon mutagenesis showed that disruption of YLR352W dramatically slows the growth of [URE3-1] strains. Ylr352wp is an F-box protein, directing selection of substrates for ubiquitination by a “cullin”-containing E3 ligase. For efficient ubiquitylation, cullin-dependent E3 ubiquitin ligases must be NEDDylated, modified by a ubiquitin-related peptide called NEDD8 (Rub1p in yeast). Indeed, we find that disruption of NEDDylation-related genes RUB1, ULA1, UBA3, and UBC12 is also counterselected in our screen. We find that like ylr352wΔ [URE3] strains, ylr352wΔ ure2Δ strains do not grow on nonfermentable carbon sources. Overexpression of Hap4p, a transcription factor stimulating expression of mitochondrial proteins, or mutation of GLN1, encoding glutamine synthetase, allows growth of ylr352w∆ [URE3] strains on glycerol media. Supplying proline as a nitrogen source shuts off the nitrogen catabolite repression (NCR) function of Ure2p, but does not slow growth of ylr352wΔ strains, suggesting a distinct function of Ure2p in carbon catabolism. Also, gln1 mutations impair NCR, but actually relieve the growth defect of ylr352wΔ [URE3] and ylr352wΔ ure2Δ strains, again showing that loss of NCR is not producing the growth defect and suggesting that Ure2p has another function. YLR352W largely protects cells from the deleterious effects of otherwise mild [URE3] variants or of a ure2 mutation (the latter a rarer event), and we name it LUG1 (lets [URE3]/ure2 grow).
Keywords: F-box protein, Gln1p, Hap4p, LUG1/YLR352W, nitrogen catabolite repression, prion, Ure2p, [URE3]
THE prions (infectious proteins) [URE3] and [PSI+] are amyloidoses of Ure2p and Sup35p, respectively, in Saccharomyces cerevisiae [reviewed in Liebman and Chernoff (2012), Wickner et al. (2015), and Saupe et al. (2016)], and are important models for the human prion and amyloid diseases (Kraus et al. 2013; Prusiner 2017). Ure2p is necessary for nitrogen catabolite repression (NCR), the shut-off of transcription of genes for the utilization of poor nitrogen sources when a good nitrogen source is available (Cooper 2002). In a [URE3] strain, most of the Ure2p is sequestered in amyloid filaments and so genes for assimilation of poor nitrogen sources (such as DAL5, encoding the allantoin transporter) are inappropriately derepressed, a result detected as an Ade+ phenotype in a strain with a DAL5 promoter driving transcription of the ADE2 gene. Sup35p is a subunit of the translation termination factor that is essential for growth (Frolova et al. 1994; Stansfield and Tuite 1994). [PSI+] cells have most (but not all) of their Sup35p tied up in the amyloid filaments, and therefore frequently read through termination codons.
A single prion protein with a single amino acid sequence can form any of a large number of prion variants or strains, with different biological properties due to different conformations of the protein in the different variants (Derkatch et al. 1996; Collinge and Clarke 2007). Each prion variant is rather stably propagated, implying that a mechanism exists for templating of protein conformation. The parallel in-register folded β-sheet architecture known for several infectious yeast prion amyloids (Shewmaker et al. 2006; Baxa et al. 2007; Wickner et al. 2008; Gorkovskiy et al. 2014) naturally suggests a mechanism of conformational templating based on the favorable interactions of aligned identical polar or hydrophobic amino acid side chains (Wickner et al. 2007, 2015).
While a majority of variants of [URE3] or [PSI+] are highly toxic, or even lethal (McGlinchey et al. 2011), there are mild variants of each prion that are also in fact detrimental (Nakayashiki et al. 2005), but not severely so. Several antiprion systems have now been described in yeast, either preventing prions from arising (Chernoff et al. 1999) or curing most prions as they arise (Wickner et al. 2014, 2017; Gorkovskiy et al. 2017; Son and Wickner 2018). In a study of the role of the essential Hsp40 chaperone Sis1p in [PSI+] propagation, Kirkland, Reidy and Masison found that deletion of the C-terminal domain did not impair cell growth in the absence of [PSI+], but resulted in a severe growth defect on the introduction of an otherwise mild [PSI+] (Kirkland et al. 2011). Sis1p is necessary for [PSI+] propagation (Higurashi et al. 2008) and the C-terminal deletion mutants were not losing [PSI+], but were no longer protecting the cells from [PSI+] toxicity (Kirkland et al. 2011). We carried out a general screen for such genes that normally protect the cell from adverse effects of a prion. We used a transposon mutagenesis method based on the Hermes transposon originally from the house fly (Gangadharan et al. 2010; Guo et al. 2013).
Materials and Methods
The supplemental material has a detailed description of the culture conditions, induction of Hermes transposition, selection of colonies carrying a transposition, extraction of cellular DNA, PCR amplification and isolation of the junction points between transposon and chromosomal insertion site, next-generation sequencing of these sites, and analysis of the data by visual display and by counting insertions per open reading frame (ORF). The supplemental material also includes: “Exon Intron Counts.xlsx,” giving the insertions in every yeast ORF, distinguishing exons from introns where appropriate; “Sorted Hits.xlsx,” giving prominent hits sorted by functional group; “TY gag-pol Counts.xlsx,” comparing insertions in the Ty retrotransposons at different locations in the genome; “Count Insertions in ORFs and Introns.txt,” the Python program used for counting insertions; and “LUGsIGV-InsertDistributions.pptx,” a slide show of insert distributions in each of 500 genes for which insertions were recovered more frequently in [ure-o] than in [URE3] cultures.
Data availability
The authors affirm that all data necessary for confirming the conclusions of this article are represented fully within the article and its tables and figures, and in the supplemental material. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6205691.
Results
Transposon mutagenesis using Hermes
Hermes is a 2749-bp DNA transposon from the house fly related to the Drosophila hobo element and, distantly, to the Ac transposon of maize (Warren et al. 1994). Hermes transposes by excising itself from one site and inserting into another site in the host genome (Atkinson et al. 1993). Recently, Hermes has been adapted for use as a transposon in S. cerevisiae (Gangadharan et al. 2010) and in Schizosaccharomyces pombe (Guo et al. 2013) for genome-wide mutagenesis and insertion site analysis. We used Hermes transposon mutagenesis of strains with (YHE1609) and without (YHE1608) the [URE3-1] prion (Supplemental Material, Table S1), followed by a period of cell growth, PCR amplification of insertion sites from extracted DNA, and next-generation sequencing, seeking genes whose interruption by the transposon would impair growth or survival in the presence of the prion but not without it. The detailed protocol is given in Materials and Methods and in the Supplemental Material.
The transposon launch plasmid, pSG36, carries a GAL-promoted hyperactive form of the Hermes transposase (G366W, M286T) and a Hermes transposon with the NatMX gene (nourseothricin-resistance; NAT) on a URA3 CEN backbone (Gangadharan et al. 2010). Because Hermes excises from the plasmid to transpose into the chromosome, the plasmid is damaged and lost after transposition. Even when grown without uracil and without transposase synthesis (glucose), ∼10% of cells lack pSG36 because the CEN6 of pSG36 was intentionally destabilized by mutation (Gangadharan et al. 2010). To maintain the plasmid, the transposase induction is done without uracil. As a result, cells in which Hermes has transposed soon stop growing. Thus, each transposition is represented by one or a few cells (at this stage), and early transpositions are not unduly amplified. Cells lacking the plasmid were then amplified by growth with uracil and 5-FOA (5-fluoro-orotic acid) to kill Ura3+ cells, and glucose to shut off transposase synthesis and further transpositions. Finally, cells with an integrated transposon were further amplified by growth in media containing glucose, uracil, 5-FOA, and NAT. During this amplification, cells with transposon insertions in genes needed to protect cells from the prion are selectively lost in [URE3] cultures compared to [ure-o] cultures. Cellular DNA was extracted and used to PCR amplify the junctions of the integrated transposon and chromosomal DNA. An Illumina platform was used to obtain 50–130 × 106 reads from each sample. Methods of data analysis are detailed in the Methods section in the supplemental material. The numbers of total reads and unique insertions identified in each ORF are given in Excel file, “Exon Intron Counts.xlsx.”
As a control for neutral genes, we examined the total reads and unique reads in both all ORFs and those in 42 nonessential (actually undesirable) Ty elements spread throughout the genome (Figure S2, Table 1, and supplemental file Ty gag-pol Counts.xlsx). The frequency of inserts in Ty elements varied substantially with considerably higher insertion frequencies in Ty2 elements than into Ty1, Ty3, Ty4, or Ty5 (Figure S2 and supplemental file Ty gag-pol Counts.xlsx). However, the ratios of insertions among the different strains and conditions used showed only modest variation among different Ty elements or (nearly) identical Ty elements at different locations (Figure S2). The distribution of insertions across each chromosome is quite uneven (chromosome I is shown in Figure S3), in part reflecting essential or desirable genes, and in part probably differences in chromatin access. However, the overall pattern is remarkably similar among the different cultures.
Table 1. Total sequence reads and unique insertions in ORFs of the entire genome and in Ty1 transposons.
| [URE3] − adenine | [ure-o] + adenine | [URE3] + adenine | [URE3] + adenine | |||||
|---|---|---|---|---|---|---|---|---|
| Total | Unique | Total | Unique | Total | Unique | Total | Unique | |
| Ty1 elements | ||||||||
| Reads | 4,843,313 | 12,005 | 3,893,719 | 21,962 | 4,215,067 | 21,170 | 2,167,896 | 13,446 |
| [URE3] − adenine /[ure-o] + adenine | 1.24 | 0.547 | ||||||
| Amplification: growth and PCR | 403.4 | 177.3 | 199.1 | 161.2 | ||||
| All ORFs | ||||||||
| Reads | 57,042,201 | 123,762 | 45,431,202 | 322,123 | 55,268,447 | 286,166 | 37,268,448 | 175,837 |
| [URE3] − adenine /[ure-o] + adenine | 1.26 | 0.384 | ||||||
| Amplification | 460.9 | 141.0 | 193.1 | 211.9 | ||||
In these experiments, [URE3] is scored using an integrated DAL5 promoter driving the ADE2 gene. Data were obtained from [ure-o] cells grown with adenine, [URE3] cells grown without adenine, and [URE3] cells grown with adenine. As expected, cells grown without adenine could not tolerate insertions in any gene in the ADE pathway, independent of [URE3] (Figure S4 and supplemental file Sorted Hits.xlsx). Because the [URE3]+Ade cultures eventually accumulated a substantial fraction of [ure-o] cells (Table S2), differences with the [ure-o]+Ade culture were reduced. Nonetheless, significant differences could still be observed and these data served to distinguish the effects of [URE3] from the effects of adenine deficiency.
Even if Ure2p was inactive because it was in prion form, cells growing without adenine need an active DAL5 promoter to drive ADE2. Insertions in the GAT1 and DAL81 genes, on which DAL5 expression depends (Turoscy and Cooper 1982; Georis et al. 2008), were rare and poorly expanded in [URE3] cells growing without adenine, as expected (Figure S5 and supplemental file Sorted Hits.xlsx). Both the number of distinct insertion sites (insertion events) and their representation among the sequences (reflecting relative abundance as a result of growth and PCR amplification) were decreased compared to genes nonessential under these conditions. GLN3 is also important for expression of DAL5 (Turoscy and Cooper 1982; Georis et al. 2008) and the distribution of Hermes insertions showed few sites of insertion in GLN3 in [URE3]−Ade compared to [ure-o]+Ade, as expected (Figure S5). However, an insertion at a single site in GLN3 was massively amplified in the [URE3]−Ade sample, exceeding total runs in the [ure-o]+Ade sample. This illustrates the importance of monitoring insertion distribution (supplemental file LUGsIGV-InsertDistribution.pptx) instead of simply relying on total ORF insertion numbers. Also, many insertions were found at the 3′ end of ORFs known to be essential for growth.
The strain used also carried CAN1, the arginine transporter, driven by the DAL5 promoter, so that loss of [URE3] could be selected as resistance to canavanine, a toxic arginine analog (Brachmann et al. 2005). As a result, arginine auxotrophs were selected against in [ure-o] strains (supplemental file Sorted Hits.xlsx).
Differences in total sequence reads in a gene can be due to differences in (1) availability for insertion (illustrated by the Ty data), (2) differences in cell growth after insertion, or (3) differences in amplification by PCR. Source 1 is assumed to be controlled by comparison of the same sites in [URE3] and [ure-o] cells, but a difference in an NCR-controlled gene might be a result of altered chromatin structure due to NCR. Source 3 is largely a problem of genes with few unique insertions in either strain. Source 2 is what we are trying to measure. All samples had roughly equal total read counts, so we have shown absolute read numbers in the tables.
Chaperone effects in [URE3] strains:
Table 2 shows that insertions in a wide array of chaperone genes were markedly disadvantageous to the [URE3] strain compared with the isogenic [ure-o] host. Of course, chaperone genes necessary for [URE3] propagation, such as Hsp104 (Moriyama et al. 2000), Ssa2p (Roberts et al. 2004; Sharma and Masison 2008), Sse1p (Kryndushkin and Wickner 2007), Fes1p (Kryndushkin and Wickner 2007), Cpr7p (Kumar et al. 2015), and Swa2p (Troisi et al. 2015), were expected to appear only rarely because we selected for retention of [URE3]. Insertions in SWA2 appeared rarely in any sample because mutants are known to grow slowly (Gall et al. 2000; Pishvaee et al. 2000). Unique insertions in HSP104 were not unusual in the [URE3] strain, but they were not amplified compared to the [ure-o] host because they eventually lost [URE3] and became Ade−. In cultures with added adenine, cells with insertions in HSP104 became abundant because they lacked the growth-slowing [URE3] prion (Wickner 1994; Schwimmer and Masison 2002).
Table 2. Transposon insertions in chaperone-encoding genes.
| Gene | Total reads | Unique inserts | |||||
|---|---|---|---|---|---|---|---|
| [URE3] − adenine | [ure-o] + adenine | [URE3] + adenine | [URE3] − adenine | [ure-o] + adenine | [URE3] + adenine | ||
| Genome total in ORFs | 57,042,201 | 45,431,202 | 55,692,869 | 303,010 | 710,420 | 627,085 | |
| HSP82 | YPL240c | 1,665 | 18,663 | 12,922 | 26 | 160 | 110 |
| HSC82 | YMR186W | 335 | 7,899 | 24,898 | 14 | 121 | 117 |
| HSP104 | YLL026W | 2,340 | 122,764 | 8,098,849 | 91 | 310 | 321 |
| YDJ1 | YNL064c | 0 | 2,175 | 1,554 | 0 | 20 | 7 |
| SWA2 | YDR320c | 0 | 2 | 8 | 0 | 2 | 50 |
| CAJ1 | YER048c | 112 | 957 | 343 | 3 | 32 | 20 |
| CPR7 | YJR032W | 13 | 224 | 605 | 4 | 19 | 21 |
| STI1 | YOR027W | 0 | 1,588 | 3,645 | 0 | 80 | 74 |
| FES1 | YBR101c | 0 | 344 | 581 | 0 | 22 | 17 |
| SSE1 | YPL106c | 2,674 | 5,498 | 30,377 | 20 | 115 | 130 |
| HSP42 | YDR171W | 806 | 14,004 | 24,364 | 21 | 104 | 95 |
| HSP26 | YBR072W | 3,630 | 9,741 | 5,921 | 10 | 59 | 28 |
| SSB1 | YDL229W | 2,007 | 14,463 | 29,150 | 38 | 182 | 177 |
| SSB2 | YNL209W | 1,659 | 11,839 | 23,213 | 37 | 166 | 162 |
| SSA1 | YAL005c | 176 | 8,562 | 46,571 | 11 | 219 | 203 |
| SSA2 | YLL024c | 287 | 6,775 | 36,604 | 12 | 147 | 126 |
| SBA1 | YKL117W | 559 | 1,800 | 1,846 | 7 | 35 | 24 |
However, many other chaperone genes, including those known not to be necessary for [URE3] propagation, were underrepresented in the collection of insertions. The duplicated Hsp90-encoding genes HSP82 and HSC82, and the duplicated SSB1 and SSB2, were all underrepresented in the [URE3] compared to the [ure-o] condition (Table 2). It is possible that the requirement for chaperones to deal with the stress of [URE3] prion infection, combined with a chaperone deficiency, gives a synthetic slow growth resulting in underrepresentation of this group in our competitive growth environment.
Particularly striking is the fact that no insertions in STI1 were recovered in the presence of [URE3] (Table 2). Sti1p is a cochaperone for Hsp90s (Hsp82 and Hsc82 in yeast), Hsp104, and Hsp70s (Scheufler et al. 2000; Abbas-Terki et al. 2001), which has already shown several antiprion activities. Overproduction of Sti1p cures some [PSI+] variants (Kryndushkin et al. 2002) and promotes loss of [PSI+] in an ssa1 mutant (Jones et al. 2004), although [URE3] is not cured (Lian et al. 2007). Sti1p is also necessary for the curing of [PSI+] by overproduction of Hsp104 (Moosavi et al. 2010; Reidy and Masison 2010) and for the curing of many [PSI+] variants by normal levels of Hsp104 (Gorkovskiy et al. 2017). Sti1p also protects cells from the toxicity produced in [PIN+] cells by the overproduction of Rnq1p or of Htt103Q, an expanded toxic version of Huntingtin (Wolfe et al. 2013).
Insertion in URE2 is rare in [ure-o] and common in the [URE3] host:
It may seem paradoxical that insertions in URE2 were far more common in the [URE3] strain (with or without adenine) than in the [ure-o] host, because in both cases, Ure2p is largely or completely inactivated (supplemental file Sorted Hits.xlsx). However, most [URE3] cells grow slower because of the prion than they would simply by losing Ure2p because of ure2Δ (McGlinchey et al. 2011). Deletion (ure2Δ) relieves the prion-induced growth inhibition and cells grow better than all the other ([URE3]) cells. In wild-type cells, ure2Δ makes cells grow slightly slower than normal, and so insertions in URE2 are counterselected. Essentially, the results follow from the fact (McGlinchey et al. 2011) that [URE3] is usually worse for cells than is ure2Δ.
YLR352W/LUG1 (lets [URE3] grow):
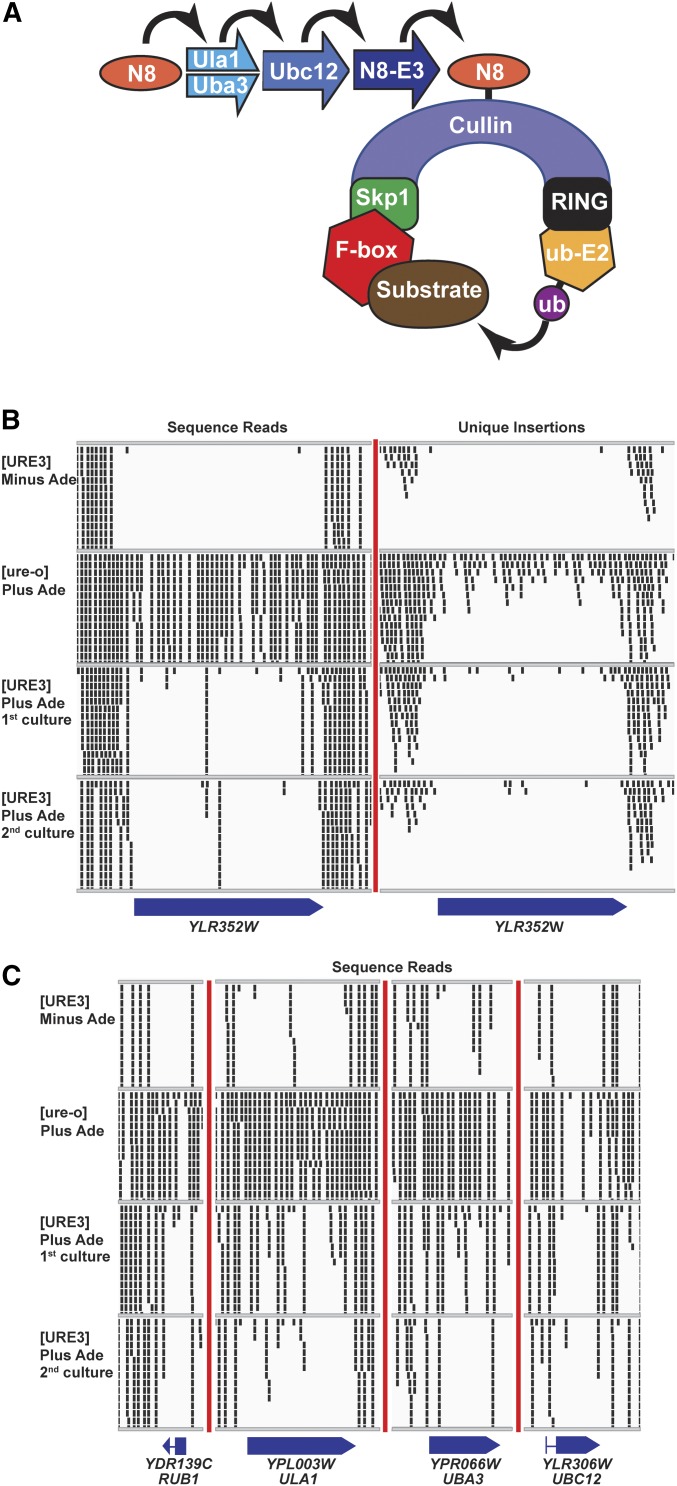
The strongest signal for a gene that improves fitness dramatically in a [URE3] strain was YLR352W (Figure 1B and Table 3). In a [ure-o] host, 127 distinct insertions (on average one per 19 bp) were found in YLR352W, which amplified to nearly 10,000 reads by cell growth and PCR. While PCR inevitably amplifies different fragments somewhat differently, it is likely that such differences even out over an entire ORF. In a [URE3] host grown without adenine, only one insertion, unamplified, was observed, suggesting a difference in amplification by cell growth after the transpositions into YLR352W and the selective loss of many insertions in this gene. When the transposition was induced in cells carrying [URE3] in the presence of adenine, there was still a substantial reduction of insertions and little amplification of those that did occur (Figure 1 and Table 3). However, the reduction was less dramatic in this case, perhaps because, in the absence of selection for the prion, a substantial fraction of [ure-o] cells accumulated (Table S2, C and D).
Figure 1.
(A) The SCF complex ubiquitination system. Ylr352wp/Lug1p is one of 20 yeast F-box proteins specifying substrates for ubiquitination by SCF complexes (see text). Yeast cullins include Rtt101p, Cdc53p, Cul3p, and Apc2p. The RING protein is Hrt1p. The E2-ubiquitin ligase is Cdc34p. NEDD8 (Rub1p in yeast; N8 in the figure) is a ubiquitin-like peptide whose attachment and removal from cullin is required for cycling of the SCF complex. (B) Insertions in YLR352W/LUG1 are recovered less frequently in [URE3] cells compared to [ure-o] cells. Although there were more total sequence reads for the [URE3]−Ade or [URE3]+Ade first cultures than for the [ure-o] culture, few were recovered in YLR352W/LUG1. The small rectangles represent the 50 bp to the right of the Hermes integration site. Only a maximum of 14 reads at one site, including overlapping sites, are shown. The totals in the tables include all reads/insertions. This figure is produced using IGV (http://software.broadinstitute.org/software/igv/). (C) NEDDylation genes are important for the ubiquitinylation process by SCF complexes and their mutations are also selectively underrepresented in [URE3] cultures. Total sequence reads are shown. Unique insertions are shown in Figure S5.
Table 3. Transposon insertions rare in YLR352W/LUG1 and NEDDylation-related genes.
| Gene | Total reads | Unique inserts | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| [URE3] − adenine | [ure-o] + adenine | [URE3] + adenine | [URE3] − adenine | [ure-o] + adenine | [URE3] + adenine | |||||
| Genome total in ORFs | 57,042,201 | 45,431,202 | 55,692,869 | 303,010 | 710,420 | 627,085 | ||||
| F-box protein | ||||||||||
| LUG1 | YLR352W | 1 | 9,882 | 344 | 1 | 127 | 17 | |||
| NEDDylation | ||||||||||
| RUB1 | YDR139C | 0 | 574 | 24 | 0 | 13 | 4 | |||
| ULA1 | YPL003W | 43 | 8,200 | 1,513 | 7 | 112 | 31 | |||
| UBA3 | YPR066W | 30 | 9,874 | 307 | 5 | 69 | 24 | |||
| UBC12 | YLR306W | 21 | 8,458 | 9,266 | 2 | 31 | 12 | |||
| DCN1 | YLR128W | 392 | 1,301 | 618 | 4 | 23 | 14 | |||
| Cullins | ||||||||||
| RTT101 | YJL047C | 488 | 3,358 | 1,511 | 14 | 61 | 71 | |||
| CDC53 | YDL132W | 8 | 571 | 242 | 1 | 6 | 7 | |||
| APC2 | YLR127C | 417 | 3,360 | 1,601 | 5 | 20 | 11 | |||
| deNEDDylation | ||||||||||
| LAG2 | YOL025W | 1143 | 3,622 | 1,934 | 14 | 59 | 53 | |||
| YUH1 | YJR099W | 520 | 5,145 | 838 | 9 | 31 | 22 | |||
Among the 20 yeast F-box protein genes, only YLR352W/LUG1 shows a dramatic deficit of insertions in the [URE3] host. The cullin gene RTT101 also showed only rare mutation in the [URE3] host without adenine. DeNEDDylation genes show a modest decrease in the [URE3] host. Frequent insertions in the [URE3] cells grown with adenine may be due to accumulation of [ure-o] cells in these cultures (Table S2, C and D).
SCF ubiquitin ligases are protein complexes consisting of Skp1p, a cullin [a structural framework for the complex; in yeast Cdc53, RTT101, or CUL3 (Sarikas et al. 2011)], Cdc34p (the catalytic subunit), Hrt1 (RING protein, connects Cdc34p to Cdc53), and one of 20 F-box proteins (substrate-specifying subunit bound by the F-box sequence to Skp1p) (Jonkers and Rep 2009; Hua and Vierstra 2011) (Figure 1A). Lug1p/Ylr352wp is one such F-box protein, which has been shown to bind to Skp1p (by two-hybrid experiments) and to Cdc53p (by affinity purification), but whose target proteins are not known (Seol et al. 2001).
NEDD8 is a ubiquitin-like (60% identity) peptide that modifies a limited range of proteins, primarily the cullin subunits of SCF ubiquitin ligases (Enchev et al. 2015). This NEDDylation of cullins enhances the activity of SCF ubiquitin ligases by allowing the exchange of F-box proteins (Figure 1A). If Lug1p is needed for growth of [URE3] strains because of its role in SCF complex ubiquitin ligation, then the NEDDylation genes should also be necessary. The yeast NEDD8 is encoded by RUB1, and Ula1p and Uba3p form a complex that acts for NEDDylation like an E1 enzyme for ubiquitin. UBC12 encodes the E2 analog (Lammer et al. 1998; Liakopoulos et al. 1998). Indeed, a clear deficiency of reads was seen in RUB1, ULA1, UBA3, and UBC12 in [URE3] cells compared to [ure-o] cells (Figure 1C, Figure S6, and Table 3). Lag2p binds to cullins and inhibits NEDDylation (Liu et al. 2009), and Yuh1p is responsible for removal of NEDD (Rub1p) (Linghu et al. 2002), and both were significantly less often mutated by Hermes in the [URE3] strain, consistent with their roles in the NEDDylation cycle (Table 3).
Growth defect of lug1Δ [URE3-1] and NEDDylation defective [URE3] strains:
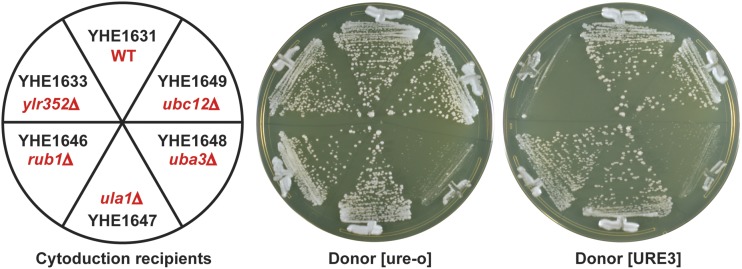
To confirm the apparent poor fitness of the normally mild prion variant [URE3-1] in strains lacking either the F-box protein Lug1p or one of the NEDDylation components Rub1p, Ula1p, Uba3p, or Ubc12p, we transferred cytoplasm (cytoduced) from [URE3-1] or [ure-o] donors into recipients deleted for one of each of the genes encoding these factors (Figure 2). Recipient cells were made ρo by growth on ethidium [so that growth on glycerol medium (YPG) could be used as an indication of receipt of donor cytoplasm] and carried a recessive cyh2 allele, making them cycloheximide-resistant. Cytoductants were selected on YPG containing cycloheximide. Note that diploids would be cycloheximide-sensitive because the cyh2 allele in the recipient is recessive. Unmated recipients will not grow on YPG because they are ρo. When [URE3] was introduced into the lug1Δ strain, no growth was seen, although cytoplasm from the same donor without the prion did not substantially affect growth (Figure 2). This result explains and confirms the outcome of the Hermes transposition experiment. Each deletion mutant in the NEDDylation genes could grow on the YPG + cycloheximide medium after receiving [URE3], except for uba3Δ (Figure 2), which is known to be respiratory incompetent independent of [URE3] (Merz and Westermann 2009). The receipt of [URE3] by these cytoductants, including the very slow-growing uba3Δ [URE3] and uba3Δ [ure-o] cytoductants, was confirmed by their ability to grow on –Ade plates and their white color on adenine-limiting (1/2 YPD) medium (Figure S7).
Figure 2.
Cytoduction (cytoplasmic transfer) from the [URE3] strain YHE1627 or the [ure-o] strain YHE1635 into wild-type and deletion strains was carried out. Cytoductants, carrying the nucleus of the recipients and a mixture of donor and recipient cytoplasm, were selected on rich glycerol medium (YPG) containing 3 μg/ml cycloheximide, and photographed after 7 days at 30°.
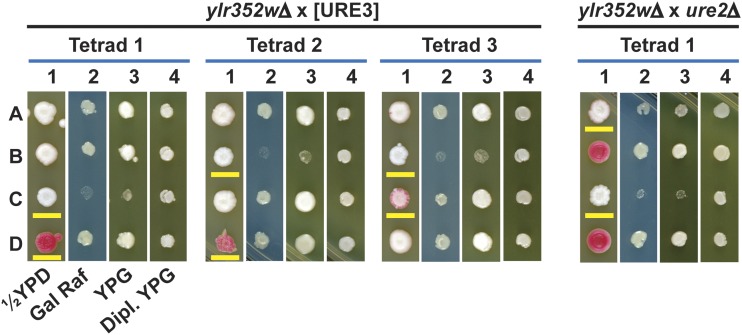
Meiotic analysis confirmed that lug1Δ strains grew poorly on glycerol medium, specifically if [URE3] was present (Figure 3). The lug1Δ [ure-o] strain YHE1633 was crossed with the LUG1 [URE3-1] strain YHE1627, the diploids were sporulated, and tetrads dissected and germinated on YPAD (rich dextrose medium = yeast extract peptone adenine dextrose). Spore clones with the lug1Δ disruption and [URE3] were viable, but were slow growing on rich dextrose medium and did not grow on rich glycerol medium (YPG) (Figure 3). In contrast, lug1Δ [ure-o] segregants (red on 1/2 YPD) grew well on YPAD or YPG [note that [URE3-1] × [ure-o] meiotic crosses are known to produce some [ure-o] segregants (Lacroute 1971)]. To test whether mitochondrial DNA was lost or damaged in the lug1Δ [URE3] segregants, all segregants were mated with LUG1 ρo strains and the diploids tested for growth on YPG. The diploids in each case could grow on YPG (Figure 3), showing that the mitochondrial DNA was intact. Also, mitochondria had a normal appearance when visualized with mitoDsRed (Figure S8).
Figure 3.
Meiotic crosses of lug1Δ strains with [URE3] and ure2Δ strains. Left: lug1Δ strain YHE1633 crossed with [URE3] strain YHE1627 showing three typical tetrads. Medium 1 = 1/2 YPD (limiting adenine), medium 2 = yeast nitrogen base medium with galactose (Gal) and raffinose (Raf) as carbon sources (2% each), medium 3 = YPG, and medium 4 = YPG growth of diploids (Dipl.) formed with the meiotic segregant and a ρo strain of opposite mating type as a check on loss of mitochondrial DNA. Spore clones with the lug1Δ allele are underlined in yellow. The occasional spore clone that spontaneously lost [URE3] appears red on 1/2 YPD. In general, lug1Δ [URE3] segregants could not grow on YPG and grew poorly on the minimal Gal/Raf medium. Right: A typical tetrad from a cross of lug1Δ strain YHE1630 and ure2Δ strain YHE1636. The media are as for the left cross. Segregants with lug1Δ are underlined in yellow and ure2Δ segregants appear white on 1/2 YPD (medium 1). In general, lug1Δ ure2Δ segregants did not grow on YPG and poorly on minimal Gal/Raf medium, and had not lost their mitochondrial DNA.
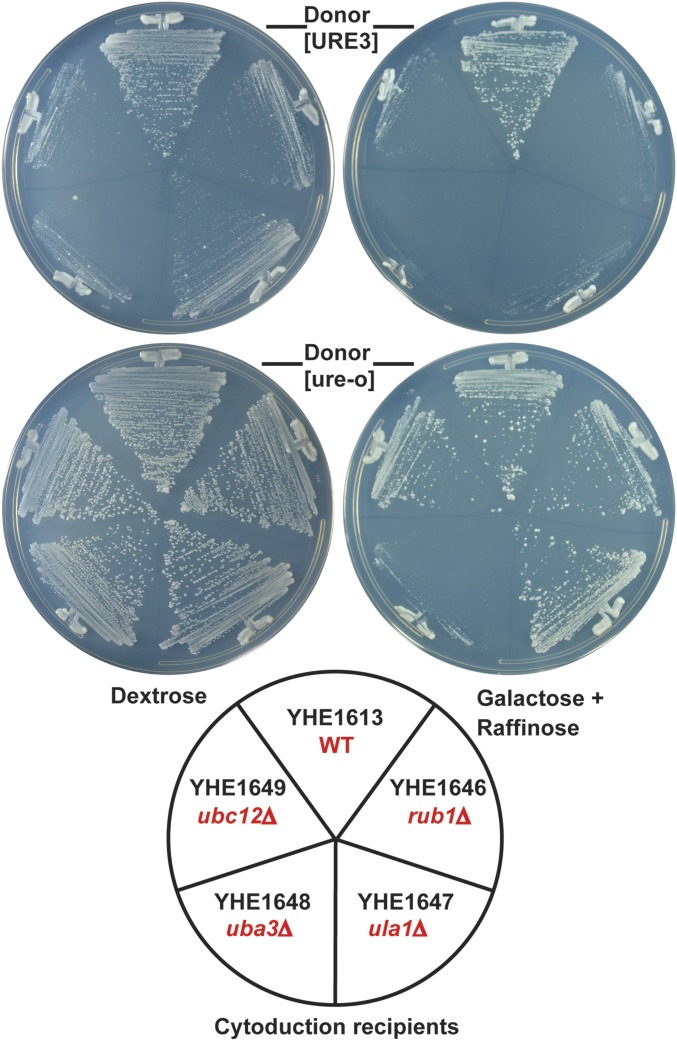
Cytoductants for the NEDDylation mutants and lug1Δ [URE3] meiotic segregants (see above) were also tested for growth on dextrose or galactose–raffinose, the two carbon sources used in the transposon experiment (Figure 4 and Figure 5). The lug1Δ and each of the NEDDylation-related deletion strains grew well on dextrose medium if [ure-o], but poorly if [URE3]. The same was true for galactose–raffinose medium, except that again the uba3Δ strain grew poorly even without [URE3] (Figure 4).
Figure 4.
Test of incompatibility of [URE3] and NEDDylation gene mutants rub1, uba3, ula1, and ubc12. Cytoductants of [URE3] (or the [ure-o] control) into knockouts of rub1, uba3, ula1, and ubc12 from Figure 2 were tested for growth on media with either dextrose or galactose/raffinose as a carbon source (2% each). Plates were minimal medium supplemented with Ura, Leu, Trp, and Ade, and incubation was at 30° for 2 days for dextrose plates and 3 days for galactose plates. WT, wild-type.
Figure 5.
Inactivation of Ure2p by using proline as a nitrogen source does not make lug1Δ strains slow growing. Although lug1Δ [URE3] and lug1Δ ure2Δ cells grow poorly on minimal proline glucose plates (or minimal ammonium glucose plates), lug1Δ URE2+ cells do not show slow growth on proline, although this condition is well known to inactivate the nitrogen catabolite repression activity of Ure2p (Cooper 2002).
Growth defect of lug1Δ ure2Δ strains:
The growth problem of lug1Δ [URE3] could be due to the presence of the amyloid filaments of Ure2p or due to the absence of Ure2p activity. To differentiate between these two possibilities, ure2Δ cells were mated with lug1Δ cells lacking [URE3] and, after sporulation, tetrads were analyzed. Spore clones that contained both ure2Δ and lug1Δ were viable, but were slow growing on dextrose-rich medium (YPAD) and did not grow on YPG (glycerol-rich medium) (Figure 3). Analysis of 34 tetrads showed that nearly all lug1Δ ure2Δ spore clones grow poorly on YPG and that nearly all subclones growing on YPG had a gln1 suppressor mutation (see below). Thus, it is the absence of Ure2p rather than the presence of Ure2p amyloid that causes the observed phenotypes. Expression in the lug1Δ ure2Δ cells of full-length Ure2p or the C-terminal portion responsible for NCR corrects the growth defect, but expression of the N-terminal prion domain does not (Figure S9).
Inactivation of Ure2p on a proline nitrogen source does not slow growth of lug1Δ strains:
When yeast has a good nitrogen source such as ammonia (present in Yeast Nitrogen Base), it shuts off the transcription of genes for the utilization of poor nitrogen sources, a process called NCR and requiring an active Ure2p. On a poor nitrogen source, such as proline, this repressing activity of Ure2p is inactivated. If it is this NCR-mediating activity of Ure2p that is needed for growth in a lug1Δ strain, then URE2lug1Δ cells should grow poorly on medium whose nitrogen source is proline. We find that lug1Δ cells grow as well as wild-type cells on either proline–glucose medium or on ammonia–glucose medium (Figure 5). Thus, the functional inactivation of Ure2p that occurs when NCR is shut off does not produce the toxicity seen for a ure2Δ strain or a [URE3] strain.
High copy HAP4 suppresses lug1 [URE3] growth defect:
To obtain some insight into the growth defect in lug1Δ [URE3] cells, we introduced a high-copy library (Jones et al. 2008) into the lug1Δ cyh2(Q38K) [ure-o] strain YHE1633 (MATa ura3leu2trp1kar1 [ure-o] lug1::kanMX cyhR rho0). We looked for cells, deleted for LUG1, that would tolerate [URE3] in the presence of a high-copy plasmid containing parts of the yeast genome. Plates with ∼200 transformants each were replica plated onto a lawn of [URE3] donor strain YHE1627 (MATα ura3leu2his3::TRP1albtrp1kar1 [URE3-1] rho+). This allows cells to mate and, due to the kar1 mutation, cytoductants to form. Cytoductants will contain the nuclear information of the lug1parent but have the cytoplasmic contents of both parents. After overnight incubation at 30°, the mating plate was replica plated onto YPG medium containing 3 μg/ml cycloheximide. This selects for lug1 deletion cells that have obtained mitochondria, and thus also [URE3], and are capable of growing on glycerol medium due to the presence of a plasmid from the library. In total, we screened 11,800 colonies covering the library of 1588 plasmids around seven times.
In addition to a plasmid carrying LUG1 and plasmids curing [URE3] because they overexpress Sse1p, Ydj1p, or Btn2p [see Moriyama et al. (2000), Kryndushkin and Wickner (2007), Kryndushkin et al. (2008)], the screen yielded the overlapping plasmids YGPM17p16 (four times) and YGPM3b23 (once). The overlap region of these plasmids contains two genes: HAP4, encoding a transcriptional activator of many mitochondrial genes on respiratory substrates (Lascaris et al. 2002), and SLD2, encoding a single-stranded DNA origin-binding and annealing protein. Overexpression of just the HAP4 gene was then found to be sufficient to suppress the growth defect of lug1::kanMX [URE3] strains (Figure S10). The suppression of the growth defect on glycerol by Hap4p overproduction indicates that the growth defect is in some aspect of mitochondrial carbon utilization.
Spontaneous gln1 mutations suppress the growth defect of lug1∆ [URE3] strains:
Strain YHE1674 (lug1::kanMX [URE3-1]) does not grow on YPG, but 36 rare YPG+ clones were isolated. Of these, eight clones had lost [URE3] (Ade−, red on YES medium), explaining their restored growth on YPG. Thirteen of the remaining clones were tested for the presence of [URE3] by cytoduction into a wild-type [ure-o] (YHE1714) recipient, and each transmitted [URE3] efficiently. As before, transmission to a lug1∆ recipient again produced failure of recipient growth on YPG, indicating that the [URE3] in the donors had not changed in this respect. After growth of each of these strains on rich medium with 3 or 5 mM guanidine to cure [URE3] [by inhibition of Hsp104 (Jung et al. 2002)], they were cytoduced into the same wild-type recipient (YHE1714) and none transmitted [URE3]. However, although [URE3] had evidently been cured, the strains that had been grown in the presence of guanidine remained Ade+ and white on adenine-limiting plates (Figure S11). A ure2 mutation could explain this phenomenon, but sequences of six of these clones showed normal sequence of URE2, including 280 bp upstream and 368 bp downstream of the ORF.
All 13 lug1Δ [URE3] YPG+ clones tested, when crossed with the lug1Δ strain YHE1721, produced diploids that grew poorly on YPG and, once cured of [URE3], became red on limiting adenine, implying that the mutation conferring growth on YPG and white color in spite of being [ure-o] is recessive. To find the affected gene, we transformed one of the YPG+ clones (YHE1760) cured of [URE3] (but still white, see above) with the same yeast genomic library (Jones et al. 2008) used above, looking for clones that made the cells turn red on limiting adenine. Plasmid-containing clones were selected on minimal dextrose medium containing 0.0006% adenine sulfate. This amount of adenine is sufficient to allow growth, but also allows red pigment development in Ade2-deficient cells. As explained above, in our strains, ADE2 is controlled by the NCR system. Among almost 40,000 colonies, we isolated the URE2-containing plasmid YGPM26i16 five times, and plasmids with the ammonia permease genes MEP1 (17 times) and MEP3 (23 times). Seven isolates carried YGPM23i14, which includes GLN1 encoding glutamine synthetase. We sequenced the GLN1 ORF from the same six strains whose URE2 gene is normal and found that all six had mutations in GLN1. YHE1760, as well as four others of these strains, had C896A, resulting in a threonine to asparagine change in the conserved amino acid residue 299. The sixth strain had G304T (D102Y) and A517G (I13V) mutations. The importance of GLN1 in this phenomenon was confirmed by isolation of YPG+ derivatives of five ure2Δ lug1Δ strains, four of which had mutations in GLN1, including three different mutant alleles (G263Δ, H250Q, and double mutant T345K M348I). Based on the fact that nearly all YPG+ clones were mutant in GLN1 and that the YPG+ and Ade+ phenotypes (even after curing [URE3]) coincided in all mutants, the complementation of both phenotypes by GLN1 implies that the gln1 mutations were the cause of both phenotypes.
Glutamine is a signal of nitrogen source sufficiency in yeast (Cooper 1982; Stanbrough et al. 1995; Crespo et al. 2002; Magasanik and Kaiser 2002; Stracka et al. 2014; Fayyad-Kazan et al. 2016). The fact that gln1 mutations overcome the YPG-negative phenotype of the lug1Δ ure2Δ strains suggests that from a point in the NCR pathway at or after glutamine, but before Ure2p, a signal is sent to the carbon-assimilation control systems.
Other genes whose mutation is rarely recovered in [URE3] cells:
No insertions in TKL1, encoding transketolase, were recovered in the [URE3] host, while a substantial number were found in the [ure-o] strain, and meiotic analysis confirmed a negative genetic interaction between ure2::URA3 and tkl1::kanMX. In addition, a large number of genes, representing many functional groups were significantly less often recovered in the [URE3] host (supplemental file Sorted Hits.xlsx). Further work will be required to determine the significance of these results.
Discussion
We have adopted the house fly transposon Hermes system, developed by Gangadharan et al. in S. cerevisiae (Gangadharan et al. 2010) and used in S. pombe (Guo et al. 2013), to search for genes that become more important when a cell becomes infected with a prion. This method is particularly sensitive because the competitive growth phase allows detection of all degrees of relative fitness of the many mutants generated, as well as, of course, addressing the entire genome at once. The utility of transposon mutagenesis and a method to use the maize Ac transposon for such work has recently appeared (Michel et al. 2017). As with transposon mutagenesis in general, different cultures accumulate insertions at a distinct distribution of sites. Thus, only those genes with a substantial number of independent insertions in at least one of the samples can be compared. Very small genes or genes in cold spots for Hermes integration are not well tested. In addition, there are many possible reasons why a gene may appear to be often mutated in [ure-o] strains and only rarely in [URE3] cells. The DAL5:ADE2 and DAL5:CAN1 constructs used to assay [URE3] differentially affect the recovery of insertions in ADE and ARG genes, as well as the NCR genes GAT1, GLN3, and DAL81. These results serve as controls showing that the method works. Since the insertions in one culture are not actually the same as those in another, differences in the efficiency of PCR amplification can affect the results. Even essentially identical sequences (Ty1) are differentially targeted by Hermes depending on their location in the genome. There are evidently differences in the accessibility of regions of the genome (Guo et al. 2013). However, when compared across cultures, the differences with location were consistent (Figure S2). Strains with differing chromatin structure (presumably NCR-sensitive genes in this case) could produce deceptive results, but no NCR-sensitive genes turned up in our screen.
Comparison of genes whose mutation is rarely recovered in [URE3] cells with known negative genetic interactors with ure2Δ
The Saccharomyces Genome Database lists 187 genes whose mutation shows negative genetic interaction with ure2Δ in mass screens (summarized at https://www.yeastgenome.org/locus/S000005173/interaction#annotations). Among these, 36 were detected in our screen as showing diminished frequency of mutant appearance in the [URE3] host, including lug1 and tkl1 (Costanzo et al. 2010) (Table S4). As expected, many of the known negative interactors with ure2Δ detected in our screen were transcription factors, or genes affecting histone modification or chromatin assembly. However, another substantial group involved the ubiquitin/proteasome system. Notably, none of the many chaperone genes detected in our screen with [URE3] had been identified in the ure2Δ negative interaction screens, suggesting that their role is protecting against [URE3] prion toxicity, rather than the deficiency of active Ure2p, but further work will be needed to establish this conclusion.
Our screen for genes protecting [URE3] cells from a growth defect revealed that lug1∆ [URE3] cells could not grow on glycerol medium and grew slowly on other carbon sources. The same growth defect is seen for lug1∆ ure2∆ cells, but [URE3] arises far more frequently than do ure2 mutations (79 of 93 spontaneous Ade+ mutants of YHE1608 were [URE3]), so we infer that Lug1p is mostly protecting cells from the adverse effects of [URE3]. Lug1p encodes an F-box protein, a substrate-specifying subunit of the Skp1-Cullin-Cdc34- Hrt1 E3 ubiquitin ligase.
Why do lug1 [URE3] and lug1 ure2 strains not grow on glycerol?
Lug1p is an F-box protein presumed to direct the degradation of some other protein. Ure2p binding to Gln3p (and Gat1p) mediates NCR when glutamine (made by Gln1p or glutamine in the medium) is in full supply. The lug1Δ [URE3] and lug1Δ ure2Δ strains can grow on glycerol if either Hap4p is overexpressed or Gln1p is defective. Utilization of glycerol requires oxidation by mitochondria and Hap4p is a major transcription factor promoting the expression of a myriad of mitochondrial proteins. It is possible that the lack of Lug1p and Ure2p results in an inadequate supply of some mitochondrial components needed for glycerol utilization. A gln1 mutant, presumably by not supplying the nitrogen sufficiency signal glutamine, allows the lug1 [URE3] and lug1Δ ure2Δ strains to grow on glycerol. Glutamine seems to inhibit glycerol utilization when Ure2p and Lug1p are absent.
Does Ure2p have a function independent of NCR?
The NCR pathway is: extracellular NH3 → intracellular NH3 → Gln1p makes glutamine → Ure2p binds Gln3p and Gat1p → reduced transcription of genes for using poor nitrogen sources. Like [URE3] or a ure2Δ mutation, a gln1 mutation or proline as the nitrogen source results in derepression of nitrogen catabolism genes. The following observations argue that Ure2p has a newly discovered role affecting carbon utilization, in addition to its well-known role in regulating nitrogen catabolism. (1) lug1Δ [URE3] or lug1Δ ure2Δ strains fail to grow on nonfermentable carbon sources. (2) The fact that gln1 mutations suppress the growth defect on YPG of lug1Δ [URE3] and lug1Δ ure2Δ strains implies that it is not the derepressed nitrogen catabolism of these strains that is producing the growth defect, and points to an alternate function of Ure2p. (3) We find that growth of lug1Δ strains on proline, which inactivates Ure2p for NCR, does not produce the growth-slowing effects that result from [URE3] or a ure2 mutation. This result again argues that there is a distinct function of Ure2p that is independent of the NCR pathway.
While our results suggest some cross talk between the NCR and carbon pathways, we lack a clear image of the mechanism. What is the target of Lug1p that mediates these effects? What mitochondrial factor(s) deficiency is (are) making these cells unable to use glycerol? What component does glutamine act on to inhibit glycerol utilization and would that target be degraded if Lug1p were present? We have succeeded in finding a cell component, the F-box protein Lug1p, that mitigates the adverse consequences of the [URE3] prion, but further studies will be needed to understand the mechanism of this effect.
Acknowledgments
We thank Nancy Craig (Johns Hopkins University) and Henry Levin (Eunice Kennedy Shriver National Institute of Child Health and Human Development) for plasmids and help with transposon mutagenesis, Harold Smith of the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) Genomics Core for help with this work, and Terry Cooper for helpful comments. This work was supported by the Intramural Program of the NIDDK of the National Insitutes of Health.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6205691.
Communicating editor: A. Mitchell
Literature Cited
- Abbas-Terki T., Donze O., Briand P.-A., Picard D., 2001. Hsp104 interacts with Hsp90 cochaperones in respiring yeast. Mol. Cell. Biol. 21: 7569–7575. 10.1128/MCB.21.22.7569-7575.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson P. W., Warren W. D., O’Brochta D. A., 1993. The hobo transposable element of Drosophila can be cross-mobilized in houseflies and excises like the Ac element of maize. Proc. Natl. Acad. Sci. USA 90: 9693–9697. 10.1073/pnas.90.20.9693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxa U., Wickner R. B., Steven A. C., Anderson D., Marekov L., et al. , 2007. Characterization of β-sheet structure in Ure2p1–89 yeast prion fibrils by solid state nuclear magnetic resonance. Biochemistry 46: 13149–13162. 10.1021/bi700826b [DOI] [PubMed] [Google Scholar]
- Brachmann A., Baxa U., Wickner R. B., 2005. Prion generation in vitro: amyloid of Ure2p is infectious. EMBO J. 24: 3082–3092. 10.1038/sj.emboj.7600772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff Y. O., Newnam G. P., Kumar J., Allen K., Zink A. D., 1999. Evidence for a protein mutator in yeast: role of the Hsp70-related chaperone Ssb in formation, stability and toxicity of the [PSI+] prion. Mol. Cell. Biol. 19: 8103–8112. 10.1128/MCB.19.12.8103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinge J., Clarke A. R., 2007. A general model of prion strains and their pathogenicity. Science 318: 930–936. 10.1126/science.1138718 [DOI] [PubMed] [Google Scholar]
- Cooper T. G., 1982. Nitrogen metabolism in Saccharomyces cerevisiae, pp. 39–99 in The Molecular Biology of the Yeast Saccharomyces: Metabolism and Gene Expression, edited by Strathern J. N., Jones E. W., Broach J. R. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Cooper T. G., 2002. Transmitting the signal of excess nitrogen in Saccharomyces cerevisiae from the Tor proteins to th GATA factors: connecting the dots. FEMS Microbiol. Rev. 26: 223–238. 10.1111/j.1574-6976.2002.tb00612.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M., Baryshnikova A., Bellay J., Kim Y., Spear E. D., et al. , 2010. The genetic landscape of a cell. Science 327: 425–431. 10.1126/science.1180823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespo J. L., Powers T., Fowler B., Hall M. N., 2002. The TOR-controlled transcription activators GLN3, RTG1 and RTG3 are regulated in response to intracellular levels of glutamine. Proc. Natl. Acad. Sci. USA 99: 6784–6789. 10.1073/pnas.102687599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch I. L., Chernoff Y. O., Kushnirov V. V., Inge-Vechtomov S. G., Liebman S. W., 1996. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics 144: 1375–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enchev R. I., Schulman B. A., Peter M., 2015. Protein neddylation: beyond cullin-RING ligases. Nat. Rev. Mol. Cell Biol. 16: 30–44. 10.1038/nrm3919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayyad-Kazan M., Feller A., Bodo E., Boeckstaens M., Marini A. M., et al. , 2016. Yeast nitrogen catabolite repression is sustained by signals distinct from glutamine and glutamate reservoirs. Mol. Microbiol. 99: 360–379. 10.1111/mmi.13236 [DOI] [PubMed] [Google Scholar]
- Frolova L., Legoff X., Rasmussen H. H., Cheperegin S., Drugeon G., et al. , 1994. A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature 372: 701–703. 10.1038/372701a0 [DOI] [PubMed] [Google Scholar]
- Gall W. E., Higgenbotham M. A., Chen C.-Y., Ingram M. F., Cyr D. M., et al. , 2000. The auxilin-like phosphoprotein Swa2p is required for clathrin function in yeast. Curr. Biol. 10: 1349–1358. 10.1016/S0960-9822(00)00771-5 [DOI] [PubMed] [Google Scholar]
- Gangadharan S., Mularoni L., Fain-Thornton J., Wheelan S. J., Craig N. L., 2010. DNA transposon Hermes inserts into DNA in nucleosome-free regions in vivo. Proc. Natl. Acad. Sci. USA 107: 21966–21972. 10.1073/pnas.1016382107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georis I., Tate J. J., Cooper T. G., Dubois E., 2008. Tor pathway control of the nitrogen-responsive DAL5 gene bifurcates at the level of Gln3 and Gat1 regulation in Saccharomyces cerevisiae. J. Biol. Chem. 283: 8919–8929. 10.1074/jbc.M708811200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorkovskiy A., Thurber K. R., Tycko R., Wickner R. B., 2014. Locating folds of the in-register parallel β-sheet of the Sup35p prion domain infectious amyloid. Proc. Natl. Acad. Sci. USA 111: E4615–E4622. 10.1073/pnas.1417974111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorkovskiy A., Reidy M., Masison D. C., Wickner R. B., 2017. Hsp104 disaggregase at normal levels cures many [PSI+] prion variants in a process promoted by Sti1p, Hsp90, and Sis1p. Proc. Natl. Acad. Sci. USA 114: E4193–E4202. 10.1073/pnas.1704016114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Park J. M., Cui B., Humes E., Gangadharan S., et al. , 2013. Integration profiling of gene function with dense maps of transposon integration. Genetics 195: 599–609. 10.1534/genetics.113.152744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higurashi T., Hines J. K., Sahi C., Aron R., Craig E. A., 2008. Specificity of the J-protein Sis1 in the propagation of 3 yeast prions. Proc. Natl. Acad. Sci. USA 105: 16596–16601. 10.1073/pnas.0808934105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Z., Vierstra R. D., 2011. The cullin-ring ubiquitin-protein ligases. Ann. Rev. Plant Path. 62: 299–334. 10.1146/annurev-arplant-042809-112256 [DOI] [PubMed] [Google Scholar]
- Jones G., Song Y., Chung S., Masison D. C., 2004. Propagation of Saccharomyces cerevisiae [PSI+] prion is impaired by factors that regulate Hsp70 substrate binding. Mol. Cell. Biol. 24: 3928–3937. 10.1128/MCB.24.9.3928-3937.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones G. M., Stalker J., Humphray S., West A., Cox T., et al. , 2008. A systematic library for comprehensive overexpression screens in Saccharomyces cerevisiae. Nat. Methods 5: 239–241. 10.1038/nmeth.1181 [DOI] [PubMed] [Google Scholar]
- Jonkers W., Rep M., 2009. Lessons from fungal F-box proteins. Eukaryot. Cell 8: 677–695. 10.1128/EC.00386-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung G., Jones G., Masison D. C., 2002. Amino acid residue 184 of yeast Hsp104 chaperone is critical for prion-curing by guanidine, prion propagation, and thermotolerance. Proc. Natl. Acad. Sci. USA 99: 9936–9941. 10.1073/pnas.152333299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkland P. A., Reidy M., Masison D. C., 2011. Functions of yeast Hsp40 chaperone Sis1p dispensable for prion propagation but important for prion curing and protection from prion toxicity. Genetics 188: 565–577. 10.1534/genetics.111.129460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus A., Groveman B. R., Caughey B., 2013. Prions and the potential transmissibility of protein misfolding diseases. Annu. Rev. Microbiol. 67: 543–564. 10.1146/annurev-micro-092412-155735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryndushkin D., Wickner R. B., 2007. Nucleotide exchange factors for Hsp70s are required for [URE3] prion propagation in Saccharomyces cerevisiae. Mol. Biol. Cell 18: 2149–2154. 10.1091/mbc.e07-02-0128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryndushkin D., Smirnov V. N., Ter-Avanesyan M. D., Kushnirov V. V., 2002. Increased expression of Hsp40 chaperones, transcriptional factors, and ribosomal protein Rpp0 can cure yeast prions. J. Biol. Chem. 277: 23702–23708. 10.1074/jbc.M111547200 [DOI] [PubMed] [Google Scholar]
- Kryndushkin D., Shewmaker F., Wickner R. B., 2008. Curing of the [URE3] prion by Btn2p, a Batten disease-related protein. EMBO J. 27: 2725–2735. 10.1038/emboj.2008.198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N., Gaur D., Gupta A., Puri A., Sharma D., 2015. Hsp90-associated immunophilin homolog Cpr7 is required for the mitotic stability of [URE3] prion in Saccharomyces cerevisiae. PLoS Genet. 11: e1005567 10.1371/journal.pgen.1005567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroute F., 1971. Non-Mendelian mutation allowing ureidosuccinic acid uptake in yeast. J. Bacteriol. 106: 519–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammer D., Mathias N., Laplaza J. M., Jiang W., Liu Y., et al. , 1998. Modification of yeast Cdc53p by the ubiquitin-related protein Rub1p affects function of the SCFCdc4 complex. Genes Dev. 12: 914–926. 10.1101/gad.12.7.914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lascaris R., Bussemaker H. J., Boorsma A., Piper M., Van Der Spek H., et al. , 2002. Hap4p overexpression in glucose-grown Saccharomyces cerevisiae induces cells to enter a novel metabolic state. Genome Biol. 4: R3 10.1186/gb-2002-4-1-r3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liakopoulos D., Doenges G., Matuschewski K., Jentsch S., 1998. A novel protein modification pathway related to the ubiquitin system. EMBO J. 17: 2208–2214. 10.1093/emboj/17.8.2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian H. Y., Zhang H., Zhang Z. R., Loovers H. M., Jones G. W., et al. , 2007. Hsp40 interacts directly with the native state of the yeast prion protein Ure2 and inhibits formation of amyloid-like fibrils. J. Biol. Chem. 282: 11931–11940. 10.1074/jbc.M606856200 [DOI] [PubMed] [Google Scholar]
- Liebman S. W., Chernoff Y. O., 2012. Prions in yeast. Genetics 191: 1041–1072. 10.1534/genetics.111.137760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linghu B., Callis J., Goebl M. G., 2002. Rub1p processing by Yuh1p is required for wild-type levels of Rub1p conjugation to Cdc53p. Eukaryot. Cell 1: 491–494. 10.1128/EC.1.3.491-494.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Mimura S., Kishi T., Kamura T., 2009. A longevity protein, Lag2, interacts with SCF complex and regulates SCF function. EMBO J. 28: 3366–3377. 10.1038/emboj.2009.268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magasanik B., Kaiser C. A., 2002. Nitrogen regulation in Saccharomyces cerevisiae. Gene 290: 1–18. 10.1016/S0378-1119(02)00558-9 [DOI] [PubMed] [Google Scholar]
- McGlinchey R., Kryndushkin D., Wickner R. B., 2011. Suicidal [PSI+] is a lethal yeast prion. Proc. Natl. Acad. Sci. USA 108: 5337–5341. 10.1073/pnas.1102762108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merz S., Westermann B., 2009. Genome-wide deletion mutant analysis reveals genes required for respiratory growth, mitochondrial genome maintenance and mitochondrial protein synthesis in Saccharomyces cerevisiae. Genome Biol. 10: R95 10.1186/gb-2009-10-9-r95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel A. H., Hatakeyama R., Kimmig P., Arter M., Peter M., et al. , 2017. Functional mapping of yeast genomes by saturated transposition. Elife 6: e23570 10.7554/eLife.23570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moosavi B., Wongwigkam J., Tuite M. F., 2010. Hsp70/Hsp90 co-chaperones are required for efficient Hsp104-mediated elimination of the yeast [PSI+] prion but not for prion propagation. Yeast 27: 167–179. [DOI] [PubMed] [Google Scholar]
- Moriyama H., Edskes H. K., Wickner R. B., 2000. [URE3] prion propagation in Saccharomyces cerevisiae: requirement for chaperone Hsp104 and curing by overexpressed chaperone Ydj1p. Mol. Cell. Biol. 20: 8916–8922. 10.1128/MCB.20.23.8916-8922.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayashiki T., Kurtzman C. P., Edskes H. K., Wickner R. B., 2005. Yeast prions [URE3] and [PSI+] are diseases. Proc. Natl. Acad. Sci. USA 102: 10575–10580. 10.1073/pnas.0504882102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pishvaee B., Costaguta G., Yeung B. G., Ryazantsev S., Greener T., et al. , 2000. A yeast DNA J protein required for uncoating of clathrin-coated vesicles in vivo. Nat. Cell Biol. 2: 958–963. 10.1038/35046619 [DOI] [PubMed] [Google Scholar]
- Prusiner S. B. (Editor), 2017. Prion Biology. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Reidy M., Masison D. C., 2010. Sti1 regulation of Hsp70 and Hsp90 is critical for curing of Saccharomyces cerevisiae [PSI+] prions by Hsp104. Mol. Cell. Biol. 30: 3542–3552. 10.1128/MCB.01292-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts B. T., Moriyama H., Wickner R. B., 2004. [URE3] prion propagation is abolished by a mutation of the primary cytosolic Hsp70 of budding yeast. Yeast 21: 107–117. 10.1002/yea.1062 [DOI] [PubMed] [Google Scholar]
- Sarikas A., Hartmann T., Pan Z. Q., 2011. The cullin protein family. Genome Biol. 12: 220 10.1186/gb-2011-12-4-220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saupe S. J., Jarosz D. F., True H. L., 2016. Amyloid prions in fungi. Microbiol. Spectr. 4 Available at: http://www.asmscience.org/content/journal/microbiolspec/10.1128/microbiolspec.FUNK-0029-2016 10.1128/microbiolspec.FUNK-0029-2016 [DOI] [PubMed] [Google Scholar]
- Scheufler C., Brinker A., Bourenkov G., Pegoraro S., Moroder L., et al. , 2000. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell 101: 199–210. 10.1016/S0092-8674(00)80830-2 [DOI] [PubMed] [Google Scholar]
- Schwimmer C., Masison D. C., 2002. Antagonistic interactions between yeast [PSI+] and [URE3] prions and curing of [URE3] by Hsp70 protein chaperone Ssa1p but not by Ssa2p. Mol. Cell. Biol. 22: 3590–3598. 10.1128/MCB.22.11.3590-3598.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seol J. H., Shevchenko A., Shevchenko A., Deshales R. J., 2001. Skip1 forms multiple protein complexes, including RAVE, a regulator of V-ATPase assembly. Nat. Cell Biol. 3: 384–391. 10.1038/35070067 [DOI] [PubMed] [Google Scholar]
- Sharma D., Masison D. C., 2008. Functionally redundant isoforms of a yeast Hsp70 chaperone subfamily have different antiprion effects. Genetics 179: 1301–1311. 10.1534/genetics.108.089458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewmaker F., Wickner R. B., Tycko R., 2006. Amyloid of the prion domain of Sup35p has an in-register parallel β-sheet structure. Proc. Natl. Acad. Sci. USA 103: 19754–19759. 10.1073/pnas.0609638103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son M., Wickner R. B., 2018. Nonsense-mediated mRNA decay factors cure most [PSI+] prion variants. Proc. Natl. Acad. Sci. USA 115: E1184–E1193. DOI: 201717410.201711073/pnas.1717495115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanbrough M., Rowen D. W., Magasanik B., 1995. Role of the GATA factors Gln3p and Nil1p of Saccharomyces cerevisiae in the expression of nitrogen-regulated genes. Proc. Natl. Acad. Sci. USA 92: 9450–9454. 10.1073/pnas.92.21.9450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stansfield I., Tuite M. F., 1994. Polypeptide chain termination in Saccharomyces cerevisiae. Curr. Genet. 25: 385–395. 10.1007/BF00351776 [DOI] [PubMed] [Google Scholar]
- Stracka D., Jozefczuk S., Rudroff F., Sauer U., Hall M. N., 2014. Nitrogen source activates TOR (target of rapamycin) complex I by glutamine and independently of Gtr/Rag proteins. J. Biol. Chem. 289: 25010–25020. 10.1074/jbc.M114.574335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troisi E. M., Rockman M. E., Nguyen P. P., Oliver E. E., Hines J. K., 2015. Swa2, the yeast homolog of mammalian auxilin, is specifically required for the propagation of the prion variant. Mol. Microbiol. 97: 926–941. 10.1111/mmi.13076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turoscy V., Cooper T. G., 1982. Pleiotropic control of five eucaryotic genes by multiple regulatory elements. J. Bacteriol. 151: 1237–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren W. D., Atkinson P. W., O’Brochta D. A., 1994. The Hermes transposable element from the house fly, Musca domestica, is a short inverted repeat-type element of the hobo, Ac, and Tam3 (hAT) element family. Genet. Res. 64: 87–97. 10.1017/S0016672300032699 [DOI] [PubMed] [Google Scholar]
- Wickner R. B., 1994. [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science 264: 566–569. 10.1126/science.7909170 [DOI] [PubMed] [Google Scholar]
- Wickner R. B., Edskes H. K., Shewmaker F., Nakayashiki T., 2007. Prions of fungi: inherited structures and biological roles. Nat. Rev. Microbiol. 5: 611–618. 10.1038/nrmicro1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner R. B., Dyda F., Tycko R., 2008. Amyloid of Rnq1p, the basis of the [PIN+] prion, has a parallel in-register β-sheet structure. Proc. Natl. Acad. Sci. USA 105: 2403–2408. 10.1073/pnas.0712032105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner R. B., Beszonov E., Bateman D. A., 2014. Normal levels of the antiprion proteins Btn2 and Cur1 cure most newly formed [URE3] prion variants. Proc. Natl. Acad. Sci. USA 111: E2711–E2720. 10.1073/pnas.1409582111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner R. B., Shewmaker F., Bateman D. A., Edskes H. E., Gorkovskiy A., et al. , 2015. Yeast prions: structure, biology and prion-handling systems. Microbiol. Mol. Biol. Rev. 79: 1–17. 10.1128/MMBR.00041-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner R. B., Kelly A. C., Bezsonov E. E., Edskes H. E., 2017. Prion propagation is controlled by inositol polyphosphates. Proc. Natl. Acad. Sci. USA 114: E8402–E8410. 10.1073/pnas.1714361114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe K. J., Ren H. Y., Trepte P., Cyr D. M., 2013. The Hsp70/90 cochaperone, Sti1, suppresses proteotoxicity by regulating spatial quality control of amyloid-like proteins. Mol. Biol. Cell 24: 3588–3602. 10.1091/mbc.e13-06-0315 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors affirm that all data necessary for confirming the conclusions of this article are represented fully within the article and its tables and figures, and in the supplemental material. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6205691.