Abstract
It remains challenging to boost statistical power of genome-wide association studies (GWASs) to identify more risk variants or loci that can account for “missing heritability.” Furthermore, since most identified variants are not in gene-coding regions, a biological interpretation of their function is largely lacking. On the other hand, recent biotechnological advances have made it feasible to experimentally measure the three-dimensional organization of the genome, including enhancer–promoter interactions in high resolutions. Due to the well-known critical roles of enhancer–promoter interactions in regulating gene expression programs, such data have been applied to link GWAS risk variants to their putative target genes, gaining insights into underlying biological mechanisms. However, their direct use in GWAS association testing is yet to be exploited. Here we propose integrating enhancer–promoter interactions into GWAS association analysis to both boost statistical power and enhance interpretability. We demonstrate that through an application to two large-scale schizophrenia (SCZ) GWAS summary data sets, the proposed method could identify some novel SCZ-associated genes and pathways (containing no significant SNPs). For example, after the Bonferroni correction, for the larger SCZ data set with 36,989 cases and 113,075 controls, our method applied to the gene body and enhancer regions identified 27 novel genes and 11 novel KEGG pathways to be significant, all missed by the transcriptome-wide association study (TWAS) approach. We conclude that our proposed method is potentially useful and is complementary to TWAS and other standard gene- and pathway-based methods.
Keywords: ChIA-PET, eQTL, gene-based testing, Hi-C, TWAS
SCHIZOPHRENIA (SCZ) is a chronic and severe mental disorder, affecting of the general population worldwide, characterized by cognitive impairment and increased mortality (Sullivan et al. 2012). Previous studies have demonstrated the high heritability of SCZ (Sullivan et al. 2012). Although >100 loci have been identified from some recent large genome-wide association studies (GWASs), the identified genetic variants can explain only a small proportion of the heritability (Sullivan et al. 2012; Ripke et al. 2013, 2014; Li et al. 2017). This phenomenon is common for other GWASs on other complex traits and diseases (Manolio et al. 2009; Welter et al. 2013). Furthermore, the majority of the identified risk variants are located outside gene-coding regions (Maurano et al. 2012), making it difficult to interpret the underlying biological mechanisms such as their target genes. Presumably, many risk variants are in regulatory regions, influencing the function of their target genes that are either nearby or distal (Corradin et al. 2014; Smemo et al. 2014). An alternative to the most popular single SNP-based analysis is gene-based testing (Pan 2009; Wu et al. 2011; Pan et al. 2014; Wang et al. 2017), in which a gene-coding region is extended up to several kilobase pairs to hopefully cover some regulatory elements, e.g., promoter regions. However, the distance between a target gene and its regulatory elements can be as far as 2 or 3 Mb (Krivega and Dean 2012), while a too large extension of a gene region to be tested may include too many nonassociated SNPs, leading to not only low statistical power but also difficulties in result interpretation. For example, an identified gene–trait association may be due to a far away causal SNP, which may not have any biological function linked to the identified significant gene.
A new approach is to use expression quantitative trait locus (eQTL) data to select and then weight gene expression-associated SNPs (i.e., eSNPs) in a largely expanded gene region (e.g., up to 1 Mb) in transcriptome-wide association studies (TWASs) (Gamazon et al. 2015; Gusev et al. 2016). However, there are still some shortcomings in TWASs. For example, due to linkage disequilibrium (LD) or reverse-causal effects, an eSNP of a gene may not necessarily have a direct biological function on the gene. In addition, due to low power in detecting trans-effects, TWAS cannot include far away regulatory regions of a gene (Wainberg et al. 2017). Furthermore, the effects of eSNPs or eQTL on target transcript levels could be too modest to be detected or estimated accurately (Corradin et al. 2014). On the other hand, it is known that GWAS risk loci are enriched in enhancers (Hawkins et al. 2013; Glodzik et al. 2017), implicating their regulatory roles in disease etiology. Through the overall three-dimensional structure of chromatin, distal enhancers can be brought into close proximity of promoters, leading to transcriptional regulation of the linked genes (Ong and Corces 2014). Recent biotechnological advances based on Chromatin Conformation Capture (3C), such as Hi-C (Van Berkum et al. 2010), ChIA-PET (Li et al. 2012), and promoter capture Hi-C (Javierre et al. 2016), have made it feasible to experimentally measure (Dryden et al. 2014; Burren et al. 2017) or computationally predict (Cao et al. 2017) nearby or distal enhancer–promoter interactions. Such data have been used to link GWAS risk loci to their target genes, thus gaining insights into the genetic basis of complex diseases (Dryden et al. 2014; Martin et al. 2015; Mishra and Hawkins 2017). In particular, it has been discovered (Mumbach et al. 2017) that for 684 autoimmune disease-associated variants studied and their 2597 target genes, only 14% of the target genes were the nearest gene to the disease-associated variant, which has been often incorrectly taken as the putative one in GWASs. Importantly, such data also offer a new opportunity to be directly used in gene-based association testing for GWASs: when testing on a gene, in addition to its coding and promoter regions, we can also include its enhancer regions. For simplicity, throughout this paper, we refer to a DNA fragment interacting with a promoter as an enhancer. Finally, since an enhancer may be associated with multiple target genes while the target genes are often functionally related (Corradin et al. 2014), a pathway analysis of a set of some functionally related genes may be more powerful than gene-based testing if the individual gene–trait associations are weak, as widely applied in practice without enhancer–promoter interaction information (Jia et al. 2010; Wang et al. 2010, 2011; Schaid et al. 2012; Huang et al. 2016). Our method is applicable to pathway analysis, albeit different from existing approaches by including the enhancers, in addition to the gene bodies and possibly promoters of the genes in a pathway.
In this paper, we propose a simple but powerful analysis strategy to integrate enhancer–promoter interactions with GWAS summary results to identify novel trait-associated genes and pathways; it can not only boost statistical power for new discoveries by focusing on enhancer regions enriched with risk variants, but also enhance interpretability of new discoveries by linking risk variants to their putative target genes. To further explain the missing heritability and better understand the mechanism of SCZ, we applied our proposed methods to perform gene- and pathway-based analyses to identify SCZ-associated genes and pathways.
Methods
Data
Although >100 loci have been identified from some recent large GWASs, the identified genetic variants can explain only a small proportion of the heritability (Sullivan et al. 2012; Ripke et al. 2013, 2014; Li et al. 2017), and most of these loci reside in relatively uncharacterized noncoding regions of the genome (Ripke et al. 2014). To further explain the missing heritability and better understand the underlying mechanism of SCZ, we performed gene- and pathway-based analyses to identify SCZ-associated genes and pathways by reanalyzing two SCZ GWAS summary data sets: a meta-analyzed SCZ GWAS data set with 8832 cases and 12,067 controls, denoted as SCZ1 (Ripke et al. 2013); and a more recent and larger one with 36,989 cases and 113,075 controls, denoted as SCZ2 (Ripke et al. 2014).
Although enhancer–promoter interactions are generally believed to be tissue-specific (Andersson et al. 2014), due to the lack of data and shared enhancer–promoter interactions across multiple tissues and cell types, we expect and thus demonstrate that enhancer–promoter interaction data from other tissues might still be useful. For simplicity, we call any DNA fragment interacting with a promoter as an enhancer. Here we mainly used two publicly available data sets to determine the enhancers for each target gene based on its enhancer–promoter interactions: (i) experimentally measured from the MCF-7 cell line by genome-wide Chromatin Interaction Analysis with Paired-End-Tag sequencing (ChIA-PET) (Li et al. 2012), denoted as MCF7 in the following; and (ii) computationally predicted for the brain hippocampus region based on the ENCODE and Roadmap Epigenomic data (Cao et al. 2017), denoted as Hippo. Given our example application to SCZ and the relatedness of hippocampus to the neuropathology and pathophysiology of SCZ (Harrison 2004), we chose the predicted enhancer–promoter interactions for the hippocampus (Cao et al. 2017). In addition, we considered two publicly available Hi-C libraries from midgestation developing human cerebral cortex from two zones to determine the enhancers for each target gene (Won et al. 2016): (i) the cortical and subcortical plate, consisting primarily of postmitotic neurons, denoted as CP in the following; and (ii) the germinal zone, containing primarily mitotically active neural progenitors, denoted as GZ in the following.
We defined multiple SNP sets for each gene to be tested as the following. First, we obtained the genomic coordinates of the SNPs and genes based on the human reference genome hg19. Second, we defined two promoter regions of a gene by extending 500 bp (Andersson et al. 2014) upstream (from its TSS) or downstream (from its TES) of the gene. Note that, although a promoter region is generally located upstream of a gene, a gene might have several proximal promoter regions scattered around introns and TES (Goñi et al. 2007). Hence, we extended 500 bp both upstream TSS and downstream TES of each gene to include some possible cis-acting regulatory regions. Third, an enhancer region of a (target) gene was defined as one interacting with its promoter region (based on a data source of enhancer–promoter interactions). Note that, depending on the source of the data sets, such as MCF7 or Hippo, the defined enhancer regions for each target gene might be different. Fourth, a gene body region was defined as that flanking its TSS and TES, including both introns and exons, plus its two promoter regions (upstream its TSS and downstream its TES). Finally, to minimize the effect of collinearity and to reduce the computational burden, the SNPs were further pruned such that no pairs of SNPs were highly correlated (with ) within a set of the SNPs being tested. For simplicity, we denote a set of the SNPs inside a gene’s body and enhancer regions as “E + G,” while that inside a gene’s enhancer regions as “E only” or “E.” We further denote standard gene-based analysis, which tests a set of the SNPs inside a gene’s body, as “STD.”
Statistical tests
For a given set of SNPs for a target gene or pathway, to determine whether it is associated with a GWAS trait, for illustration we applied two popular SNP set-based tests, a burden test called the Sum or SPU(1) test and a variance-component score test called the SSU or SPU(2) test, which is equivalent to kernel machine regression or SKAT with a linear kernel (Pan 2009; Wu et al. 2011; Pan et al. 2014). Briefly, based on a GWAS summary data set, for each target gene (or target pathway) we have its Z-score vector for k SNPs in a defined SNP set; for each SNP j, we have the Z-score with being the estimated (marginal) effect size and its standard deviation. The burden test SPU(1) and the variance-component score test SPU(2) are defined as:
Under the null hypothesis that the SNP set (for a gene or a pathway) is not associated with the trait, SPU(1) and SPU(2) follow an asymptotically (or approximately) normal distribution and a mixture of chi-squared distributions, respectively. To calculate the P-values, we need the correlation matrix for Z, which can be estimated by LD among the SNPs based on a reference panel (e.g., the 1000 Genomes Project data) (Kwak and Pan 2015; Gusev et al. 2016).
To better illuminate the effects of enhancers, we applied both SPU(1) and SPU(2) to enhancer regions only (called “E only” or “E”), in addition to “E + G” regions and the standard gene body regions (called “STD”), respectively. For comparison, we also applied the TWAS method (Gusev et al. 2016) and its extension based on the (weighted) SPU(2) (Xu et al. 2017). Note that, since TWAS is equivalent to the weighted SPU(1) test with cis-eQTL-derived weights [with 500-kb extension; Xu et al. (2017)], we applied the weighted SPU(1) test to represent TWAS. Specifically, the weighted SPU(1) test uses a weighted sum of the z-scores of the SNPs with eQTL-derived weights to construct its test statistic, while, as an extension of TWAS, the weighted SPU(2) test is based on a weighted sum of the squared z-scores of the SNPs. We downloaded four sets of eQTL-derived weights from the TWAS website: microarray gene expression data measured in blood from 1245 unrelated subjects from the Netherlands Twin Registry (NTR), microarray expression array data measured in blood from 1264 individuals from the Young Finns Study (YFS), RNA-seq measured in adipose tissue from 563 individuals from the Metabolic Syndrome in Men study (METSIM), and RNA-seq measured in the dorsolateral prefrontal cortex from 621 individuals from CommonMind Consortium (CMC) (Gusev et al. 2016).
To control multiple testing, we used the Bonferroni correction. For the SCZ1 data, we analyzed 9127 and 4600 genes for MCF7- and Hippo-defined gene regions, respectively; we used a slightly more stringent Bonferroni cutoff (). For STD, we tested on ∼22,000 genes with a corresponding Bonferroni-adjusted cutoff. For TWAS, we applied the Bonferroni correction to each set of the eQTL-derived weights (around a few thousands), for which we ignored the fact that the four sets of the eQTL-derived weights were used in TWAS; unless specified otherwise, we took the union of the identified gene sets of TWAS across the four sets of the weights.
Following Gusev et al. (2016), we evaluated the performance of the methods by first identifying the significant and novel genes that did not overlap with any genome-wide significant SNP, both based on the SCZ1 data, then examining the replication rate of the identified genes that also contained one or more genome-wide significant SNPs in the larger SCZ2 data. To test for the statistical significance of such a replication rate or an enrichment, we applied a hyper-geometric test with the background probability estimated from the set of genes being tested. Note that, for a given GWAS data set, a novel gene is defined as a significant gene (extended 500 kb) that does not include any significant SNP.
For pathway-based analysis, we extracted the candidate pathways from the KEGG pathway database (Kanehisa and Goto 2000) and restricted our analyses to the 191 KEGG pathways containing between 10 and 200 genes, which is widely adopted in practice for pathway-based analysis (O’Dushlaine et al. 2015). We used a stringent Bonferroni cutoff () for pathway-based analysis. For comparison, we applied a new method (Wu and Pan 2018), which extends TWAS from gene-based to pathway-based analysis. Briefly, we applied the weighted SPU(1) and SPU(2) tests, in which each of the SNPs in the genes (or their extended regions) belonging to a pathway is weighted by its estimated cis-effect size on the gene expression based on an eQTL data set.
Data availability
The original SCZ1 and SCZ2 GWAS summary data can be downloaded at the PGC site https://www.med.unc.edu/pgc/results-and-downloads. The LD reference data can be obtained from http://www.internationalgenome.org/data; TWAS and eQTL-based weights can be downloaded at http://gusevlab.org/projects/fusion/. The enhancer–promoter interaction data can be obtained from Li et al. (2012), Won et al. (2016), and Cao et al. (2017). The related computer scripts, examples, and processed enhancer information can be downloaded at https://figshare.com/articles/Enhancer_information_and_related_codes_for_a_new_gene-based_analysis/5995381. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6193055.
Results
Data summary
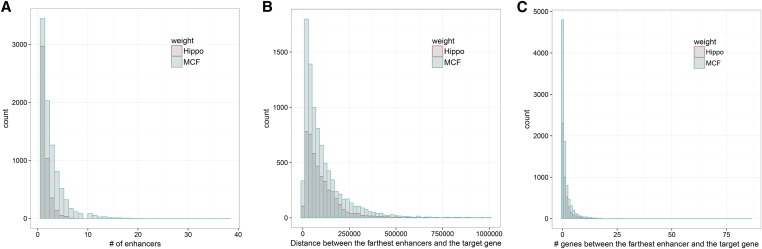
Figure 1 shows the distributions of some statistics for the two enhancer–promoter interaction data sets. The MCF7 and Hippo data contained 25,310 and 7245 pairs of enhancer–promoter interactions, respectively. On average, for each target gene there were 2.8 and 1.6 enhancer–promoter interactions in the two data sets, respectively. Some enhancers (e.g., 168 in the MCF7 data) located on chromosomes different from that of their target genes, confirming the potential usefulness of enhancer–promoter interaction data. For the MCF7 and Hippo data, the average distances between a target gene and its farthest enhancer were ∼246 and 99 kb, respectively, indicating that the usual practice of extending a gene body by several kilobase pairs (as in STD) might fail to cover some important regulatory elements. Furthermore, there were on average ∼1.5 (with the MCF7) and 1.3 genes (with the Hippo) between a target gene and its farthest enhancer, suggesting the pitfall of the usual practice of assigning an associated SNP to the nearest gene in GWASs. This phenomenon has been confirmed by other researchers as well (Won et al. 2016; Mumbach et al. 2017).
Figure 1.
Histograms of enhancer–promoter interaction data. In the middle panel, for better visualization, 12 pairs with distance >1 Mb are omitted.
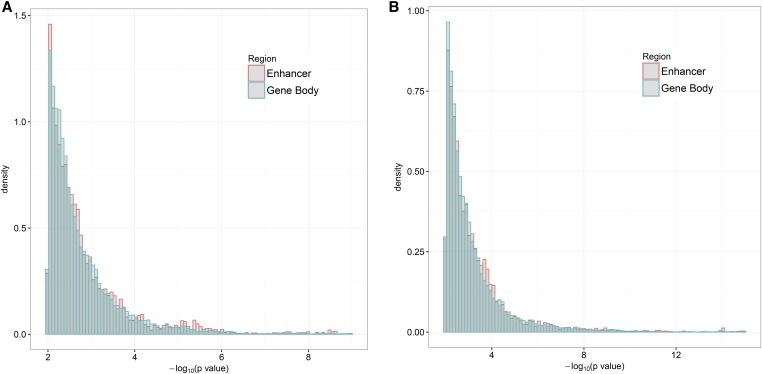
The Kolmogorov–Smirnov test showed that the empirical distribution of the P-values for SNPs in enhancers was significantly different from that for gene body regions (P-value ). Figure 2 depicts the distribution of P-values for SNPs in enhancers and in gene body regions, respectively, illustrating that there was an enrichment of small P-values for SCZ GWASs in enhancers. This phenomenon was more evident for the larger SCZ2 data.
Figure 2.
Histograms of P-values for SNPs in enhancers and gene body regions, respectively. The left and right panels are based on the SCZ1 and SCZ2 data, respectively.
Gene-based testing
We first applied the various methods to the SCZ1 data while using the larger (but overlapping) SCZ2 data to partially validate the results. First, the numbers of the significant genes are shown in Table 1. For fair comparisons, we applied the Bonferroni correction for each method (with possibly different numbers of the genes/SNP sets available) separately. It appears that our methods and TWAS identified fewer significant genes than that of the standard gene-based testing, which was likely due to differing numbers of the genes tested: the former applied to only ∼10,000 genes while the latter (STD) to ∼22,000 genes. If we focused on the common set of 5203 genes that could be analyzed by all methods, using a common and more stringent cutoff “E + G,” “E only,” STD, and TWAS identified 29, 20, 26, and 38 significant genes, respectively (Supplemental Material, Figure S1).
Table 1. Numbers of significant genes identified by analyzing the SCZ1 data.
| Enhancer | Enhancer + gene body | Gene body (STD) | TWAS | ||||||
|---|---|---|---|---|---|---|---|---|---|
| MCF7 | Hippo | MCF7 | Hippo | YFS | NTR | METSIM | CMC | ||
| # genes | 8589 | 3363 | 9127 | 4600 | 22842 | 4697 | 2452 | 4665 | 5412 |
| SPU(1) | 14/12/11a | 8/6/6 | 20/19/18 | 15/13/14 | 36/32/34 | 14/11/14 | 10/6/10 | 8/5/7 | 16/10/13 |
| SPU(2) | 35/25/29 | 9/9/9 | 39/29/33 | 46/34/40 | 89/77/84 | 31/25/26 | 27/19/26 | 23/14/23 | 39/25/34 |
The numbers a/b/c in each cell indicate the numbers of (a) significant genes; (b) significant genes that covered one or more genome-wide significant SNPs within an extended gene region kb in the SCZ1 data; (c) significant genes that covered one or more genome-wide significant SNPs within an extended gene region kb in the SCZ2 data.
Some genome-wide significant loci in the SCZ1 data were no longer significant in the SCZ2 data. For example, gene CUL9 contained some significant SNPs in the SCZ1 data (with the most significant SNP ) but did not contain any significant SNPs in the SCZ2 data (with the smallest ).
To further illustrate the added value of using enhancer information, we generated random enhancer regions based on the Hippo data. Specifically, for each gene, we generated the same number of “enhancer regions” with the same lengths but different start and end positions as compared to the original enhancers. Both the SPU(1) and SPU(2) tests with the randomly generated “enhancer regions” plus the gene body identified fewer significant genes [eight for SPU(1) and 33 for SPU(2)] than those of using the original “E + G” regions [15 for SPU(1) and 46 for SPU(2)], showcasing that enhancer information indeed added the value. Note that, since gene body regions may contain some associated SNPs, with random “enhancer regions” both SPU(1) and SPU(2) could still identify some significant genes.
Next, we checked the novel genes among the significant genes as shown in Table 2; a novel gene is defined as one that does not cover any genome-wide significant SNP in an extended gene region kb upstream its TSS and downstream TES. We summarize the replication rates and their statistical significance by a hyper-geometric test in Table S1. SPU(2) applied to “E + G” based on MCF7 identified 10 novel genes in the SCZ1 data, of which 6 (60%) contained genome-wide significant SNPs in the SCZ2 data (P-value by the hyper-geometric test), offering a highly significant partial validation on the identified genes. Even though two significant and novel genes identified by applying SPU(1) to “E only” with MCF7 (or Hippo) data were not replicated in the SCZ2 data, SPU(1) is a widely used gene-based test with its well-controlled type 1 error rates established by many previous studies (Li and Leal 2008; Pan 2009; Kwak and Pan 2015; Gusev et al. 2016). In comparison, TWAS and its extension gave a similar replication rate. For example, the standard TWAS [i.e., SPU(1)] based on CMC identified six novel genes in the SCZ1 data, of which 4 (67%) contained genome-wide significant SNPs in the SCZ2 data (P-value by the hyper-geometric test). Importantly, Table S2 lists the significant and novel genes identified by analyzing the SCZ1 data, showing that most of the significant and novel genes (31 out of 37, ∼84%) identified by “E only” or “E + G” have been reported by other studies. Similarly, TWAS and its extension identified 41 significant and novel genes, of which 34 (∼83%) have been reported by other studies. In addition, applying SPU(1) and SPU(2) to “E + G” regions identified similar numbers of significant and novel genes to those of TWAS [i.e., SPU(1) and its extension SPU(2)] with each of the four sets of eQTL-derived weights. For a fair comparison, we also examined a common set of 2226 genes that could be analyzed by our methods with MCF7 data, TWAS with CMC-based weights, and STD. We applied the Bonferoni correction (). Figure S2 shows that using “E + G” and “E,” TWAS, and STD identified nine, six, seven, and eight significant and novel genes, respectively. Using “E + G” and “E only” identified two (CNOT7 and ACTR5) and three (SMG6, ANKRD44, and SH3RF1) significant and novel genes that were missed by the other two methods, respectively.
Table 2. Numbers of significant and novel genes identified by analyzing the SCZ1 data.
| Enhancer | Enhancer + gene body | STD | TWAS | ||||||
|---|---|---|---|---|---|---|---|---|---|
| MCF7 | Hippo | MCF7 | Hippo | YFS | NTR | METSIM | CMC | ||
| # genes | 8589 | 3363 | 9127 | 4600 | 22842 | 4697 | 2452 | 4665 | 5412 |
| SPU(1) | 2/0 | 2/0 | 1/1 | 2/2 | 4/4 | 3/3 | 4/4 | 3/2 | 6/4 |
| SPU(2) | 10/6 | 0/0 | 10/6 | 12/8 | 12/10 | 6/3 | 8/8 | 9/9 | 14/11 |
The numbers a/b in each cell indicate the numbers of (a) significant and novel genes with no genome-wide significant SNPs within an extended gene region kb in the SCZ1 data; (b) significant and novel genes that covered one or more genome-wide significant SNPs within an extended gene region kb in the SCZ2 data.
In summary, compared to TWAS and STD, our new methods (“E + G” and “E only”) identified similar numbers of the significant and novel genes with similar replication rates for the SCZ1 data. Importantly, our new methods could identify some significant and novel genes that were missed by both TWAS and STD. Equally, TWAS and its extension could also identify some significant and novel genes missed by our new methods. When a gene includes one or several far away enhancer regions with GWAS trait-associated SNPs, we expect that our new methods will be most useful. On the other hand, if one gene contains several cis-eQTLs that are not in annotated enhancer regions, we expect that TWAS will be more powerful than our new methods. In short, our new methods can be useful in using enhancer information to boost statistical power to identify novel trait-associated genes that could be missed by other methods.
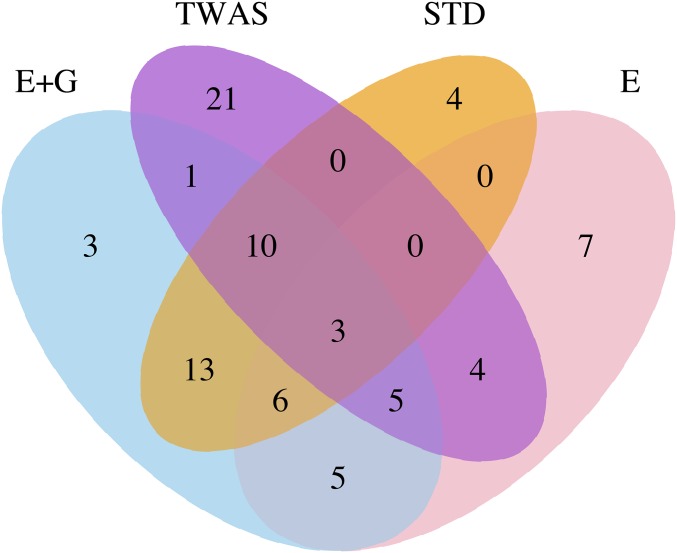
Having established the potential usefulness of our new method based on the smaller SCZ1 data, we applied the methods to the larger SCZ2 data to identify significant and novel genes. For a fair comparison, we mainly focused on the 5212 genes that could be analyzed by both our new methods and TWAS, using the same and more stringent cutoff (). Figure 3 shows the Venn diagram of the identified significant and novel genes by different methods. Our methods applied to “E + G” and “E only,” TWAS and STD identified 46, 30, 44, and 36 significant novel genes, respectively. Six novel genes have been identified by both TWAS and our new method, but missed by STD. For example, MRPL33 was identified by our methods; it contained eight SNPs in the gene body plus seven SNPs in three enhancers, of which the most distant enhancer was ∼618 kb away from the gene body. MRPL33 was reported to be associated with SCZ by Goes et al. (2015). However, a standard gene-based test with an extension of up to several kilobase pairs would fail to include some of its enhancers and thus miss its significant association. In addition, SCZ is associated with impairments in working memory that reflect dysfunction of dorsolateral prefrontal cortex (DLPFC) circuitry (Kahn and Keefe 2013; Arion et al. 2015); it has been shown that MRPL33 for cells dissected from the DLPFC of monkeys displayed significantly lower expression in SCZ subjects (Arion et al. 2015). Although TWAS/SPU(1) could not identify gene MRPL33 (P-value ), its extension SPU(2) could (P-value ). Table 3 highlights 27 significant and novel genes identified by “E + G”; none of the genes contained any genome-wide significant SNPs in its extended regions by kb in the SCZ2 data; they were also missed by TWAS and its extension with any of the four eQTL data sets. Twelve genes, such as MED19 and MAN2A1, have been reported by other independent studies (Goes et al. 2015; Li et al. 2017) as shown in the GWAS Catalog v1.0 (Welter et al. 2013). For example, gene FAM214A, reported to be associated with SCZ (Goes et al. 2015), contained 119 SNPs in the gene body plus 106 SNPs in 10 enhancer regions; its most distant enhancer region was ∼152 kb away. The most significant SNP (P-value ) within its E + G region was located in an enhancer region, explaining why our new method (when applied to either “E + G” or “E only”) could identify this gene while STD [P-value of SPU(1) ; P-value of SPU(2) ] failed, confirming GWAS signals in enhancer regions. Table 4 shows 18 significant and novel genes identified using “E only” regions; all of them were missed by TWAS, though 11 were also identified by our method applied to “E + G.” Again most of the genes have been reported to be SCZ-associated by other independent studies (Goes et al. 2015; Li et al. 2017). Because a gene body may contain many nonassociated SNPs, leading to nonsignificant gene-based testing, using enhancer regions only identified some genes that could have been missed by the standard gene-based or “E + G”-based testing. Tables S3–S6 list the significant and novel genes identified by “E + G”- and “E only”-based testing, TWAS, and STD (with 96, 60, 84, and 92 unique genes, respectively) when we focused on all available genes for each method.
Figure 3.
Venn diagram of the significant and novel genes identified by the different methods applied to the SCZ2 data. “E + G” and “E” combine the results (i.e., taking the union) of using MCF7 and Hippo data, while TWAS combines the results of using YFS-, NTR-, METSIM-, and CMC-based weights.
Table 3. Significant and novel genes identified by our new method applied to “enhancer + gene body” regions, but missed by TWAS, with the SCZ2 data.
| Gene | CHR | # SNPs | SPU(1) | SPU(2) | Sig SNP | Source | Ref | STD | E |
|---|---|---|---|---|---|---|---|---|---|
| ZBTB48 | 1 | 11 | Hippo | T | |||||
| RBBP5 | 1 | 64 | Hippo | T | T | ||||
| RBBP5 | 1 | 69 | MCF7 | T | T | ||||
| DSTYK | 1 | 147 | MCF7 | T | |||||
| HAT1 | 2 | 78 | MCF7 | ||||||
| MED19 | 3 | 15 | Hippo | [2] | T | T | |||
| UBE2D3 | 4 | 183 | MCF7 | T | |||||
| ZNF664 | 4 | 54 | Hippo | [1] | T | ||||
| NDFIP2 | 5 | 70 | Hippo | T | |||||
| MAN2A1 | 5 | 404 | MCF7 | [1,2] | T | ||||
| SRP54 | 6 | 144 | Hippo | T | |||||
| SLC16A10 | 6 | 163 | MCF7 | T | |||||
| TRAF3IP2-AS1 | 6 | 214 | MCF7 | [1] | T | T | |||
| DDX56 | 7 | 37 | MCF7 | [1] | T | T | |||
| LIPC | 7 | 309 | Hippo | [1] | T | ||||
| FAM63B | 7 | 123 | Hippo | [1] | T | ||||
| CNOT7 | 8 | 51 | MCF7 | T | |||||
| DYM | 10 | 759 | Hippo | T | |||||
| GSTO1 | 10 | 11 | MCF7 | T | |||||
| NDFIP2 | 13 | 74 | MCF7 | T | |||||
| DOPEY2 | 14 | 370 | Hippo | [1] | T | ||||
| FAM214A | 15 | 225 | MCF7 | [1] | T | ||||
| DNAJA3 | 16 | 41 | MCF7 | [1] | T | T | |||
| SPG7 | 16 | 237 | MCF7 | [1] | T | T | |||
| C16orf55 | 16 | 45 | MCF7 | ||||||
| SPATA2L | 16 | 50 | MCF7 | T | |||||
| VPS9D1 | 16 | 107 | MCF7 | T | |||||
| CDK5R1 | 17 | 12 | MCF7 | ||||||
| DIRAS1 | 19 | 23 | MCF7 | T | |||||
| DOPEY2 | 21 | 416 | MCF7 | [1] | T |
The P-value of the most significant SNP (“Sig SNP”) in the region and the source database used to construct enhancer–promoter interactions are also shown. The validated gene–trait associations appeared in the following references: [1] Goes et al. (2015); [2] Li et al. (2017). T stands for the gene has been identified by either STD or “E only.”
Table 4. Significant and novel genes identified by our new method applied to enhancer regions only (“E only”), but missed by TWAS, with the SCZ2 data.
| Gene | CHR | # SNPs | SPU(1) | SPU(2) | Sig SNP | Source | Ref | STD | E + G |
|---|---|---|---|---|---|---|---|---|---|
| NOL9 | 1 | 2 | Hippo | ||||||
| ZBTB48 | 1 | 5 | Hippo | T | |||||
| PSMB2 | 1 | 3 | MCF7 | ||||||
| RBBP5 | 1 | 11 | MCF7 | T | T | ||||
| MED19 | 3 | 5 | Hippo | [2] | T | T | |||
| SRP54 | 6 | 8 | Hippo | T | |||||
| REV3L | 6 | 48 | MCF7 | ||||||
| TRAF3IP2-AS1 | 6 | 48 | MCF7 | [1] | T | T | |||
| DDX56 | 7 | 21 | MCF7 | [1] | T | T | |||
| DEF8 | 8 | 9 | Hippo | ||||||
| ZNF623 | 8 | 46 | MCF7 | ||||||
| GNG7 | 11 | 3 | Hippo | [1] | |||||
| FAM214A | 15 | 106 | MCF7 | [1] | T | ||||
| DNAJA3 | 16 | 4 | MCF7 | [1] | T | T | |||
| SPG7 | 16 | 107 | MCF7 | [1] | T | T | |||
| SPATA2L | 16 | 40 | MCF7 | T | |||||
| VPS9D1 | 16 | 89 | MCF7 | T | |||||
| SLC35A4 | 18 | 4 | Hippo |
The P-value of the most significant SNP (“Sig SNP”) in the region and the source database used to construct enhancer–promoter interactions are also shown. The validated gene–trait associations appeared in the following references: [1] Goes et al. (2015); [2] Li et al. (2017). T stands for the gene has been identified by either STD or “E + G.”
Using enhancer–promoter interaction data in developing human brain:
We applied CP- and GZ-based “E only” and “E + G” testing to both the SCZ1 and SCZ2 data. Tables S7 and S8 show the numbers of the significant genes identified by analyzing the SCZ1 and SCZ2 data, respectively. For fair comparisons, we used the Bonferroni correction for each method separately. Perhaps due to the numbers of genes tested being much smaller here (∼1000), testing with “E + G” identified fewer significant genes than that with the MCF7 data. This was also true for testing with “E only.” However, the CP and GZ data indeed provided some useful information. For the SCZ2 data, testing with CP- or GZ-based “E + G” could identify 52 significant and novel genes, among which 40 were missed by “E + G” with MCF7 or Hippo, “E only” with MCF7 or Hippo, TWAS, and STD (Table S9).
Pathway-based analysis
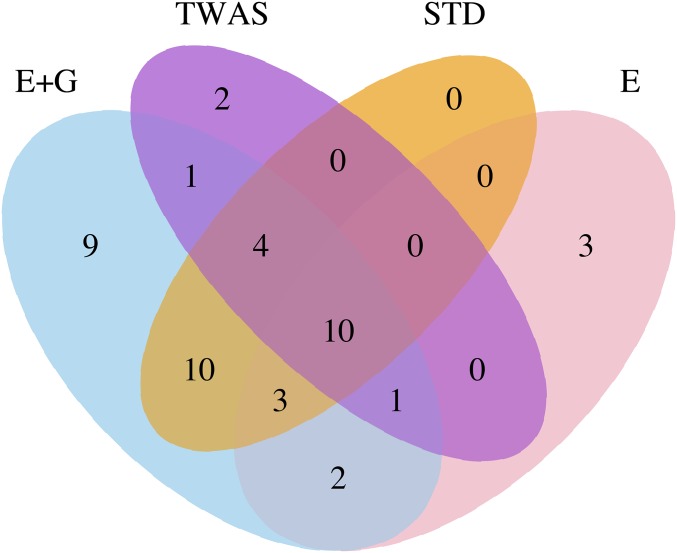
We applied the pathway-based methods to the SCZ2 data. We defined a significant gene as the one identified by applying the SPU(1) and SPU(2) tests to the SCZ2 data with the gene body regions (i.e., the STD method). For simplicity, we defined a novel pathway as the one with no known significant gene. Figure 4 shows the Venn diagram of the identified significant and novel pathways by the different methods. Our methods applied to “E + G” and “E only,” TWAS, and STD identified 40, 19, 18, and 27 significant and novel pathways, respectively. Table 5 highlights 11 novel pathways identified by our method with “E + G” regions but missed by both TWAS and STD. Pathways NOD-like receptor signaling (hsa04621) and Pathogenic Escherichia coli infection (hsa05130) have been reported by others to be associated with SCZ (Szatkiewicz et al. 2014; Wu et al. 2016). Table 6 shows five significant and novel pathways identified by using “E only” regions but missed by both TWAS and STD, of which three were also missed by using “E + G” regions. Again, because the gene bodies in a pathway may contain no or few associated SNPs, leading to nonsignificant pathway-based testing, using enhancer regions only identified some pathways that could be missed by the standard (STD) pathway-based or “E + G”-based testing. In summary, the pathways in Tables 5 and 6 represent some new discoveries gained by using enhancer–promoter interaction information.
Figure 4.
Venn diagram of the significant and novel pathways identified by the different methods applied to the SCZ2 data.
Table 5. Significant and novel pathways identified by our new method applied to “enhancer + gene body” regions, but missed by TWAS and STD, with the SCZ2 data.
| ID | Pathway name | # gen | SPU(1) | SPU(2) | Source |
|---|---|---|---|---|---|
| hsa00071 | Fatty acid degradation | 42 | Hippo | ||
| hsa00511 | Other glycan degradation | 15 | Hippo | ||
| hsa00534 | Glycosaminoglycan biosynthesis | 26 | Hippo | ||
| hsa03320 | PPAR signaling | 66 | Hippo | ||
| hsa04621 | NOD-like receptor signaling | 57 | Hippo | ||
| MCF7 | |||||
| hsa04960 | Aldosterone-regulated sodium reabsorption | 40 | Hippo | ||
| hsa04966 | Collecting duct acid secretion | 25 | Hippo | ||
| hsa00562 | Inositol phosphate metabolism | 53 | MCF7 | ||
| hsa03022 | Basal transcription factors | 33 | MCF7 | ||
| hsa03450 | Nonhomologous end-joining | 13 | MCF7 | ||
| hsa05130 | Pathogenic Escherichia coli infection | 52 | MCF7 |
Table 6. Significant and novel pathways identified by our new method applied to enhancer regions only (“E only”), but missed by TWAS and STD, with the SCZ2 data.
| ID | Pathway name | # gen | SPU(1) | SPU(2) | Source |
|---|---|---|---|---|---|
| hsa00340 | Histidine metabolism | 29 | Hippo | ||
| hsa00380 | Tryptophan metabolism | 37 | Hippo | ||
| hsa00740 | Riboflavin metabolism | 16 | Hippo | ||
| hsa03320 | PPAR signaling | 66 | Hippo | ||
| hsa03022 | Basal transcription factors | 33 | MCF7 |
Discussion
It has become increasingly important to measure enhancer–promoter interactions, or more generally the three-dimensional organization of the human genome, to understand gene expression regulation. In particular, such data have been used to link GWAS risk loci to their (putative) target genes, enhancing the interpretation of GWAS discoveries. Since the target genes may not be the ones nearest to GWAS risk variants, the usual practice of assigning the gene nearest to a risk variant as the (putative) target gene is generally problematic. Here we directly incorporate enhancer–promoter interactions into gene-based association testing for GWAS, which is expected to not only boost statistical power, but also enhance biological interpretation at the target gene level. In particular, complementary to the standard gene-based and TWAS approaches, testing with annotated enhancer regions could identify some significant and novel genes that would be missed by the other two approaches; these novel genes did not contain any significant SNPs inside or near the regions. Our proposed two variants of using gene body and enhancer regions (“E + G”) and using only enhancer regions (“E only”) are also complementary to each other: in general “E + G” is expected to be more powerful by taking advantage of information with gene body regions, while “E only” is more specific with a focus on enhancers, which might yield significant results that would be missed by “E + G.” Furthermore, the proposed method is applicable to pathway-based analysis. For its relative performance as compared to the standard or TWAS-based pathway analyses, we reach the same conclusions as that for gene-based testing.
Although it would be ideal to use enhancer–promoter interaction data drawn from a disease- or trait-related tissue, we mainly used the data from the tissues not necessarily most relevant to schizophrenia but that still demonstrated their potential usefulness due to the lack of disease-related tissue data and expected commonalities of the DNA three-dimensional organizations across multiple tissue and cell types. Nevertheless, we also applied our method to an enhancer–promoter interaction data set based on the developing human brain, uncovering some significant genes that would be missed based on the other two data sets. Although the results confirmed the usefulness of using tissue-specific data, due to varying sensitivities and specificities of different biotechnologies (e.g., ChIA-PET vs. Hi-C, experimental vs. computational), we found that it was useful and complementary to use different tissue-based data sets. In addition, as in TWAS, we could apply our method to and then combine the results from multiple tissues, or apply other more powerful and adaptive tests (Gusev et al. 2016; Xu et al. 2017). The issue with the choice of the tissue or cell type is similar to that in TWAS: a recent study (Qi et al. 2018) has shown that, for brain-related traits, using blood cis-eQTL (with larger sample sizes) could gain power over using (smaller) brain eQTL data sets, while the genetic effects of cis-eQTL are highly correlated between independent brain and blood samples. Finally, although our application was focused on schizophrenia, the proposed method is quite general and applicable to other traits based on either individual-level or summary GWAS data.
Acknowledgments
We are grateful to the reviewers for constructive comments. We thank Hui Li for helping with the MCF7 data. This research was supported by National Institutes of Health (NIH) grants R21AG057038, R01HL116720, R01GM113250, R01HL105397, and R01GM126002, NSF grant DMS 1711226, and by the Minnesota Supercomputing Institute.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6193055.
Communicating editor: C. Kendziorski
Literature Cited
- Andersson R., Gebhard C., Miguel-Escalada I., Hoof I., Bornholdt J., et al. , 2014. An atlas of active enhancers across human cell types and tissues. Nature 507: 455–461. 10.1038/nature12787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arion D., Corradi J. P., Tang S., Datta D., Boothe F., et al. , 2015. Distinctive transcriptome alterations of prefrontal pyramidal neurons in schizophrenia and schizoaffective disorder. Mol. Psychiatry 20: 1397–1405. 10.1038/mp.2014.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burren O. S., García A. R., Javierre B.-M., Rainbow D. B., Cairns J., et al. , 2017. Chromosome contacts in activated T cells identify autoimmune disease candidate genes. Genome Biol. 18: 165 10.1186/s13059-017-1285-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Q., Anyansi C., Hu X., Xu L., Xiong L., et al. , 2017. Reconstruction of enhancer-target networks in 935 samples of human primary cells, tissues and cell lines. Nat. Genet. 49: 1428–1436. 10.1038/ng.3950 [DOI] [PubMed] [Google Scholar]
- Corradin O., Saiakhova A., Akhtar-Zaidi B., Myeroff L., Willis J., et al. , 2014. Combinatorial effects of multiple enhancer variants in linkage disequilibrium dictate levels of gene expression to confer susceptibility to common traits. Genome Res. 24: 1–13. 10.1101/gr.164079.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dryden N. H., Broome L. R., Dudbridge F., Johnson N., Orr N., et al. , 2014. Unbiased analysis of potential targets of breast cancer susceptibility loci by capture Hi-C. Genome Res. 24: 1854–1868. 10.1101/gr.175034.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamazon E. R., Wheeler H. E., Shah K. P., Mozaffari S. V., Aquino-Michaels K., et al. , 2015. A gene-based association method for mapping traits using reference transcriptome data. Nat. Genet. 47: 1091–1098. 10.1038/ng.3367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glodzik D., Morganella S., Davies H., Simpson P. T., Li Y., et al. , 2017. A somatic-mutational process recurrently duplicates germline susceptibility loci and tissue-specific super-enhancers in breast cancers. Nat. Genet. 49: 341–348. 10.1038/ng.3771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goes F. S., McGrath J., Avramopoulos D., Wolyniec P., Pirooznia M., et al. , 2015. Genome-wide association study of schizophrenia in Ashkenazi Jews. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 168: 649–659. 10.1002/ajmg.b.32349 [DOI] [PubMed] [Google Scholar]
- Goñi J. R., Pérez A., Torrents D., Orozco M., 2007. Determining promoter location based on DNA structure first-principles calculations. Genome Biol. 8: R263 10.1186/gb-2007-8-12-r263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusev A., Ko A., Shi H., Bhatia G., Chung W., et al. , 2016. Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet. 48: 245–252. 10.1038/ng.3506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison P. J., 2004. The hippocampus in schizophrenia: a review of the neuropathological evidence and its pathophysiological implications. Psychopharmacology (Berl.) 174: 151–162. 10.1007/s00213-003-1761-y [DOI] [PubMed] [Google Scholar]
- Hawkins R. D., Larjo A., Tripathi S. K., Wagner U., Luu Y., et al. , 2013. Global chromatin state analysis reveals lineage-specific enhancers during the initiation of human T helper 1 and T helper 2 cell polarization. Immunity 38: 1271–1284. 10.1016/j.immuni.2013.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Wang K., Wei P., Liu X., Liu X., et al. , 2016. FLAGS: a flexible and adaptive association test for gene sets using summary statistics. Genetics 202: 919–929. 10.1534/genetics.115.185009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javierre B. M., Burren O. S., Wilder S. P., Kreuzhuber R., Hill S. M., et al. , 2016. Lineage-specific genome architecture links enhancers and non-coding disease variants to target gene promoters. Cell 167: 1369–1384.e19. 10.1016/j.cell.2016.09.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia P., Wang L., Meltzer H. Y., Zhao Z., 2010. Common variants conferring risk of schizophrenia: a pathway analysis of GWAS data. Schizophr. Res. 122: 38–42. 10.1016/j.schres.2010.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn R. S., Keefe R. S., 2013. Schizophrenia is a cognitive illness: time for a change in focus. JAMA Psychiatry 70: 1107–1112. 10.1001/jamapsychiatry.2013.155 [DOI] [PubMed] [Google Scholar]
- Kanehisa M., Goto S., 2000. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28: 27–30. 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivega I., Dean A., 2012. Enhancer and promoter interactions-long distance calls. Curr. Opin. Genet. Dev. 22: 79–85. 10.1016/j.gde.2011.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak I.-Y., Pan W., 2015. Adaptive gene-and pathway-trait association testing with GWAS summary statistics. Bioinformatics 32: 1178–1184. 10.1093/bioinformatics/btv719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Leal S. M., 2008. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am. J. Hum. Genet. 83: 311–321. 10.1016/j.ajhg.2008.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Ruan X., Auerbach R. K., Sandhu K. S., Zheng M., et al. , 2012. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell 148: 84–98. 10.1016/j.cell.2011.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Chen J., Yu H., He L., Xu Y., et al. , 2017. Genome-wide association analysis identifies 30 new susceptibility loci for schizophrenia. Nat. Genet. 49: 1576–1583. 10.1038/ng.3973 [DOI] [PubMed] [Google Scholar]
- Manolio T. A., Collins F. S., Cox N. J., Goldstein D. B., Hindorff L. A., et al. , 2009. Finding the missing heritability of complex diseases. Nature 461: 747–753. 10.1038/nature08494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin P., McGovern A., Orozco G., Duffus K., Yarwood A., et al. , 2015. Capture Hi-C reveals novel candidate genes and complex long-range interactions with related autoimmune risk loci. Nat. Commun. 6: 10069 10.1038/ncomms10069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurano M. T., Humbert R., Rynes E., Thurman R. E., Haugen E., et al. , 2012. Systematic localization of common disease-associated variation in regulatory DNA. Science 337: 1190–1195. 10.1126/science.1222794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra A., Hawkins R. D., 2017. Three-dimensional genome architecture and emerging technologies: looping in disease. Genome Med. 9: 87 10.1186/s13073-017-0477-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumbach M. R., Satpathy A. T., Boyle E. A., Dai C., Gowen B. G., et al. , 2017. Enhancer connectome in primary human cells identifies target genes of disease-associated DNA elements. Nat. Genet. 49: 1602–1612. 10.1038/ng.3963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Dushlaine C., Rossin L., Lee P. H., Duncan L., Parikshak N. N., et al. , 2015. Psychiatric genome-wide association study analyses implicate neuronal, immune and histone pathways. Nat. Neurosci. 18: 199–209. 10.1038/nn.3922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong C.-T., Corces V. G., 2014. CTCF: an architectural protein bridging genome topology and function. Nat. Rev. Genet. 15: 234–246. 10.1038/nrg3663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan W., 2009. Asymptotic tests of association with multiple SNPs in linkage disequilibrium. Genet. Epidemiol. 33: 497–507. 10.1002/gepi.20402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan W., Kim J., Zhang Y., Shen X., Wei P., 2014. A powerful and adaptive association test for rare variants. Genetics 197: 1081–1095. 10.1534/genetics.114.165035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi T., Wu Y., Zeng J., Zhang F., Xue A., et al. , 2018. Identifying gene targets for brain-related traits using transcriptomic and methylomic data from blood. bioRxiv. Available at: https://www.biorxiv.org/content/early/2018/03/07/274472. [DOI] [PMC free article] [PubMed]
- Ripke S., O’Dushlaine C., Chambert K., Moran J. L., Kähler A. K., et al. , 2013. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat. Genet. 45: 1150–1159. 10.1038/ng.2742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke S., Neale B. M., Corvin A., Walters J. T., Farh K. H., et al. , 2014. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511: 421–427. 10.1038/nature13595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaid D. J., Sinnwell J. P., Jenkins G. D., McDonnell S. K., Ingle J. N., et al. , 2012. Using the gene ontology to scan multilevel gene sets for associations in genome wide association studies. Genet. Epidemiol. 36: 3–16. 10.1002/gepi.20632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smemo S., Tena J. J., Kim K.-H., Gamazon E. R., Sakabe N. J., et al. , 2014. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature 507: 371–375. 10.1038/nature13138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan P. F., Daly M. J., O’Donovan M., 2012. Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nat. Rev. Genet. 13: 537–551. 10.1038/nrg3240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szatkiewicz J. P., O’Dushlaine C., Chen G., Chambert K., Moran J. L., et al. , 2014. Copy number variation in schizophrenia in Sweden. Mol. Psychiatry 19: 762–773. 10.1038/mp.2014.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Berkum N. L., Lieberman-Aiden E., Williams L., Imakaev M., Gnirke A., et al. , 2010. Hi-C: a method to study the three-dimensional architecture of genomes. J. Vis. Exp. 6: 1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainberg M., Sinnott-Armstrong N., Knowles D., Golan D., Ermel R., et al. , 2017. Vulnerabilities of transcriptome-wide association studies. bioRxiv. Available at: https://www.biorxiv.org/content/early/2017/10/26/206961. [DOI] [PMC free article] [PubMed]
- Wang K., Li M., Hakonarson H., 2010. Analysing biological pathways in genome-wide association studies. Nat. Rev. Genet. 11: 843–854. 10.1038/nrg2884 [DOI] [PubMed] [Google Scholar]
- Wang L., Jia P., Wolfinger R. D., Chen X., Zhao Z., 2011. Gene set analysis of genome-wide association studies: methodological issues and perspectives. Genomics 98: 1–8. 10.1016/j.ygeno.2011.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Huang J., Liu Y., Ma L., Potash J. B., et al. , 2017. COMBAT: a combined association test for genes using summary statistics. Genetics 207: 883–891. 10.1534/genetics.117.300257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welter D., MacArthur J., Morales J., Burdett T., Hall P., et al. , 2013. The NHGRI GWAS catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 42: D1001–D1006. 10.1093/nar/gkt1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won H., de La Torre-Ubieta L., Stein J. L., Parikshak N. N., Huang J., et al. , 2016. Chromosome conformation elucidates regulatory relationships in developing human brain. Nature 538: 523–527. 10.1038/nature19847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Pan W., 2018. Integrating eQTL data with GWAS summary statistics in pathway-based analysis with application to schizophrenia. Genet. Epidemiol. 42: 303–316. 10.1002/gepi.22110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. Q., Green M. J., Gardiner E. J., Tooney P. A., Scott R. J., et al. , 2016. Altered neural signaling and immune pathways in peripheral blood mononuclear cells of schizophrenia patients with cognitive impairment: a transcriptome analysis. Brain Behav. Immun. 53: 194–206. 10.1016/j.bbi.2015.12.010 [DOI] [PubMed] [Google Scholar]
- Wu M. C., Lee S., Cai T., Li Y., Boehnke M., et al. , 2011. Rare variant association testing for sequencing data with the sequence kernel association test. Am. J. Hum. Genet. 89: 82–93. 10.1016/j.ajhg.2011.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Wu C., Wei P., Pan W., 2017. A powerful framework for integrating eQTL and GWAS summary data. Genetics 207: 893–902. 10.1534/genetics.117.300270 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original SCZ1 and SCZ2 GWAS summary data can be downloaded at the PGC site https://www.med.unc.edu/pgc/results-and-downloads. The LD reference data can be obtained from http://www.internationalgenome.org/data; TWAS and eQTL-based weights can be downloaded at http://gusevlab.org/projects/fusion/. The enhancer–promoter interaction data can be obtained from Li et al. (2012), Won et al. (2016), and Cao et al. (2017). The related computer scripts, examples, and processed enhancer information can be downloaded at https://figshare.com/articles/Enhancer_information_and_related_codes_for_a_new_gene-based_analysis/5995381. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6193055.