Abstract
Cis -regulatory elements (CREs) play a pivotal role in spatiotemporal control of tissue-specific gene expression, yet the molecular composition of the vast majority of CREs in native chromatin remains unknown. In this unit, we describe the CRISPR affinity purification in situ of regulatory elements (CAPTURE) approach to simultaneously identify locus-specific chromatin-regulating protein complexes and long-range DNA interactions. Using an in vivo biotinylated nuclease-deficient Cas9 (dCas9) protein and programmable single guide RNAs (sgRNAs), this approach allows for high-resolution and locus-specific isolation of protein complexes and long-range chromatin looping associated with single copy CREs in mammalian cells. Unbiased analysis of the compositional structure of developmentally regulated or disease-associated CREs identifies new features of transcriptional regulation. Hence, CAPTURE provides a versatile platform to study genomic locus-regulating chromatin composition in a mammalian genome.
Keywords: chromatin, cis-regulatory elements, enhancer, DNA looping, CRISPR
INTRODUCTION
CREs such as transcriptional enhancers regulate tissue-specific gene expression by recruiting transcription factors and chromatin complexes, and interact with target genes by long-range DNA looping. Dysregulation of CREs underlies the development of a variety of human disorders including cancer. Identifying the molecular composition of a specific CRE in situ can provide unprecedented insight into the mechanisms regulating its activity in physiological or pathological conditions. However, purifying a small chromatin segment from nucleus remains a significant challenge. Various affinity purification strategies have been developed to study the localization of trans-regulatory factors, including chromatin immunoprecipitation (ChIP) (Dedon et al., 1991; Moqtaderi and Struhl, 2004; Ren et al., 2000), affinity purification using integrated epitope tags (Agelopoulos et al., 2012; Fujita and Fujii, 2011; Griesenbeck et al., 2003), or sequence-specific molecules (Dejardin and Kingston, 2009; Fujita et al., 2013; Fujita and Fujii, 2013, 2014; Waldrip et al., 2014), but major technical difficulties limit the application of existing approaches in purifying macromolecules at a single copy genomic locus.
Clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 is a prokaryotic immune system that confers resistance to viruses by Cas9 nuclease-mediated cleavage of foreign genetic elements (Barrangou et al., 2007; Bhaya et al., 2011). Cas9 can assemble with a chimeric sgRNA to recognize and cleave DNA in a mammalian genome. The first 20 nucleotides of sgRNA guide Cas9 to recognize complementary genomic sequence and introduce double-strand breaks (Cong et al., 2013; Jinek et al., 2012; Mali et al., 2013). Sequence-specific sgRNAs can be easily constructed and multiplexed, thus Cas9 has been widely adapted as a tool for genome editing in mammalian cells (Doudna and Charpentier, 2014; Hsu et al., 2014). Mutations in the nuclease domains of Cas9 deactivate its exonuclease activity without affecting the sequence-specific DNA binding ability of the nuclease-deficient Cas9 (dCas9) protein (Gilbert et al., 2013; Qi et al., 2013). dCas9 has soon been adapted as a DNA-binding protein for a wide range of applications including transcriptional modulation, epigenome editing, and genome imaging (Chen et al., 2013; Dominguez et al., 2016; Gilbert et al., 2013; Qi et al., 2013; Thakore et al., 2016).
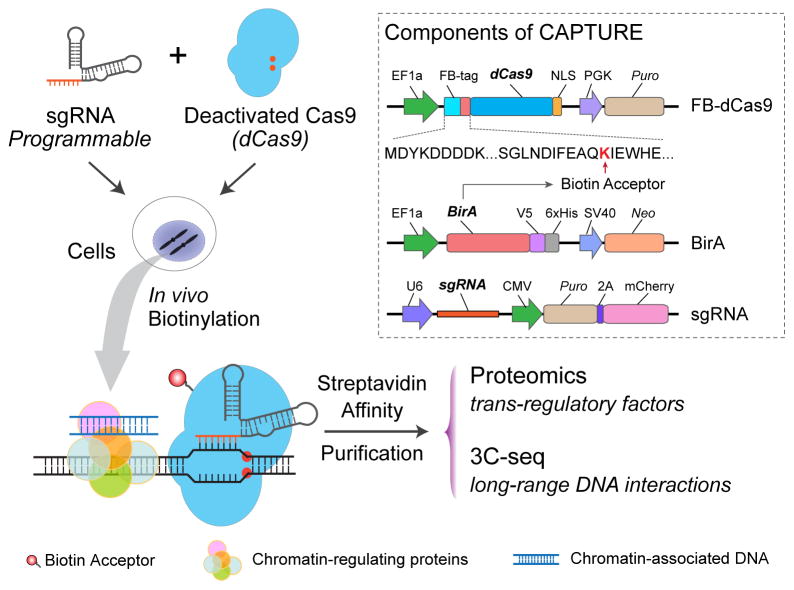
In this unit, we describe the CAPTURE approach utilizing sequence-specific binding of in vivo biotinylated dCas9 to simultaneously identify genomic locus-associated protein complexes and long-range DNA interactions in mammalian cells (Fig. 1) (Liu et al., 2017). The CAPTURE system consists of three core components: a dCas9 fused with an N-terminal FLAG and biotin acceptor tandem peptide (FB-dCas9), an E. coli. biotin holoenzyme synthetase (BirA) (Schatz, 1993), and a structure-optimized sgRNA (Fig. 1). The biotin acceptor peptide serves as an in vivo substrate mimic for BirA biotin ligase. Stably expressed FB-dCas9 proteins are biotinylated in vivo by BirA and guided to the genomic locus-of-interest by sequence-specific sgRNAs. By high affinity streptavidin-biotin-mediated purification, the dCas9-tethered chromatin segments together with associated protein and DNA complexes are isolated. The purified protein and DNA molecules are identified and analyzed by mass spectrometry (MS)-based quantitative proteomics and high-throughput sequencing for study of native genomic locus-regulating proteins and long-range DNA interactions, respectively.
Figure 1. In Situ Capture of Locus-Specific Chromatin Interactions by Biotinylated dCas9.
Schematic of dCas9-mediated CAPTURE of cis-element-associated chromatin interactions. The three components of the CAPTURE system: a FLAG-biotin acceptor (FB)-tagged dCas9, a biotin ligase BirA, and a target-specific sgRNA. This figure was reproduced from (Liu et al., 2017) with permission from Elsevier.
CAPTURE offers several advantages in isolating chromatin associated complexes compared to conventional antibody-based affinity purification approaches. First, due to extremely high affinity between biotin and streptavidin with Kd = 10−14 mol/L (Schatz, 1993), CAPTURE allows for more efficient and stable capture of protein-DNA complexes. Second, the high affinity between biotin and streptavidin also allows for stringent purification procedures to reduce co-purified protein contamination. Third, few naturally biotinylated proteins are localized in nucleus, thus streptavidin affinity purification helps reduce non-specific binding associated with antibody-based approaches. Fourth, the dCas9/sgRNA system can be easily modified by altering sgRNA sequences or using sgRNA combinations, thus allowing for high-throughput multiplexed analysis of chromatin-associated interactions. Moreover, an optimized sgRNA design (Chen et al., 2013) with increased stability and enhanced dCas9/sgRNA assembly is adapted to maximize on-target enrichment and eliminate non-specific binding.
The following protocols outline an easy-to-follow and scalable procedure for establishing the CAPTURE system in mammalian cell lines and applying the system to study chromatin composition associated with single copy genomic loci. The procedure can be divided into three parts: CAPTURE setup and sgRNA validation (see Basic Protocol 1), identification of chromatin-regulating protein complexes by CAPTURE-Proteomics (see Basic Protocol 2), and analysis of locus-specific long-range DNA interactions by CAPTURE-3C-seq (see Basic Protocol 3).
BASIC PROTOCOL 1. Establishment of FB-dCas9/BirA Stable Cell Lines and sgRNA Validation
The protocol is used to generate cell lines that stably express both BirA and FB-dCas9, and to validate the specificity of sgRNAs (Fig. 2) (Liu et al., 2017). Stable cells are established by transfecting cells with pEF1a-FB-dCas9-puro and pEF1a-BirA-V5-neo constructs, followed by selection with G418 and puromycin. After establishing single cell-derived stable clones expressing high level of BirA and in vivo biotinylated dCas9, sequence-specific or non-targeting control sgRNAs are introduced by lentiviral infection to direct biotinylated dCas9 proteins to sgRNA-targeted genomic loci. Using a common reverse primer and unique forward primers containing the protospacer sequences, any sgRNA sequence can be incorporated into PCR amplicons and subcloned into the lentiviral U6 promoter-driven expression vector. To evaluate the specificity and on-target enrichment of sgRNAs, FB-dCas9/BirA stable cells transduced with sequence-specific or control sgRNAs are cross-linked by formaldehyde, followed by nuclei extraction. Nuclei are then sonicated to solubilize chromatin and biotinylated dCas9-tethered chromatin is purified by streptavidin immunoprecipitation. dCas9 affinity purified DNA are recovered and analyzed by qPCR and high-throughput sequencing.
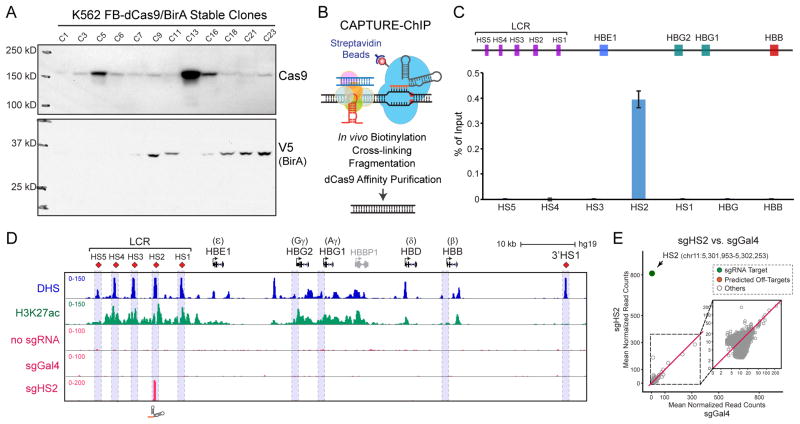
Figure 2. CAPTURE System Setup and Validation.
(A) Representative western blot analysis of dCas9 and BirA expression in G418 and puromycin selected single cell-derived K562 stable clones.
(B) Schematic of CAPTURE-ChIP-seq to identify dCas9-captured DNA sequences.
(C) CAPTURE-ChIP-qPCR analysis of sgHS2-captured DNA shows significant enrichment of HS2 compared to other LCR-HS regions and promoters of HBG and HBB. Results are mean ± SD and analyzed by a two-tailed t-test: **P < 0.01. The positions of LCR (HS1 to HS5), HBG1, HBG2 and HBB are indicated on the top.
(D) ChIP-seq density maps are shown for CAPTURE-ChIP-seq analysis in K562 cells with sgRNA targeting HS2. Cells expressing dCas9 only (no sgRNA) or dCas9 with non-targeting sgGal4 were analyzed as controls. DNase I hypersensitivity (DHS) and H3K27ac ChIP-seq profiles are shown for comparison.
(E) Genome-wide differential analysis of dCas9 binding in cells expressing sgHS2 versus the non-targeting sgGal4. Data points for sgRNA target regions and predicted off-targets are shown as green and red, respectively. Several panels of this figure were reproduced from (Liu et al., 2017) with permission from Elsevier.
Materials
pEF1α-BirA-V5-neo (Addgene plasmid 100548)
pEF1α-FB-Cas9-puro (Addgene plasmid 100547)
pSLQ1651-sgRNA(F+E)-sgGal4 (non-targeting control sgRNA, Addgene plasmid 100549)
VSV-G (Addgene plasmid 8454)
psPAX2 (Addgene plasmid 12260)
LB liquid medium and agar plates with 100 μg/ml ampicillin
QIAprep Spin Miniprep Kit (Qiagen)
QIAquick Gel Extraction Kit (Qiagen)
MinElute PCR Purification Kit (Qiagen)
10 U/μl BstxI restriction enzyme (New England Biolabs, Cat # R0113L)
10 U/μl BamHI restriction enzyme (New England Biolabs, Cat # R0136L)
10× NEBuffer 3.1 (New England Biolabs, Cat # B7203S)
Phusion® High-Fidelity DNA Polymerase (New England Biolabs, Cat # M0530L)
400 U/μl T4 ligase with 10× T4 ligase buffer (New England Biolabs, Cat # M0202L)
NEBNext ChIP-seq library prep kit (New England Biolabs, Cat # E6240L)
Chemically competent DH5a cells (Home-made)
SOC medium (Thermo-Fisher, Cat # 15544034)
37°C incubator (Thermo-Fisher)
SORVALL ST16R centrifuge (Thermo-Fisher)
Thermomixer (Eppendorf)
Thermocycler (Bio-Rad)
PEI (Sigma-Aldrich, Polyethylenimine, Branched, Cat # 408727)
16,16-dimethyl Prostaglandin E2 (PGE2, 10 mM in DMSO) (Cayman Chemical Company, Cat # 14750)
PEG-it Virus Precipitation Solution (System Bioscience, Cat # LV825A-1)
IMDM (Thermo-Fisher, Cat # 12440053)
Opti-MEM (Thermo-Fisher, Cat # 31985062)
Puromycin (Thermo-Fisher, Cat # A1003802)
G418 (Sigma-Aldrich, Cat # G1720)
37% formaldehyde solution (Calbiochem, Cat # 344198)
Protease inhibitor cocktail (Sigma-Aldrich, Cat # P8340)
Dynabeads® MyOne™ Streptavidin T1 (Invitrogen, Cat # 656-01)
RNase A, DNase-free (0.5 mg/ml, Roche, Cat # 11119915001)
Proteinase K (20 mg/ml, Ambion, Cat # AM2546)
Cas9 antibody (Abcam, ab191468, RRID:AB_2692325)
V5-HRP antibody (Thermo-Fisher, Cat # R961-25, RRID:AB_2556565)
Goat anti-mouse IgG-HRP (Santa Cruz, Cat # sc-2005, and RRID:AB_631736)
ECM 830 Square Wave Electroporation System (BTX)
2 mm electroporation cuvette (BTX, Cat # 45-0141)
Digital Sonifier (BRANSON, Models S-450)
sgGal4_sgRNA.fwd (non-targeting control sgRNA):
ggagaaCCACCTTGTTGGAACGACTAGTTAGGCGTGTAGTTTAAGAGCTATGCTGGAAACAGCA
(GNX is the spacer sequence which can be changed for targeting any locus)
Universal.rev: ctagtaCTCGAGAAAAAAAGCACCGACTCGGTGCCAC
Primer sequences used to validate sgRNA sequence by sanger DNA sequencing:
U6-Vector_seq.fwd: GAGATCCAGTTTGGTTAGTACCGGG
U6-Vector_seq.rev: ATGCATGGCGGTAATACGGTTAT
Electroporation and Screening for FB-dCas9 and BirA Stable Cell Clones
-
1
Culture human K562 cells in a T-25 flask in IMDM medium containing 10% FBS and 1% penicillin/streptomycin at 37°C and 5% CO2 until they reach ~0.5 × 106 cells/ml.
If other cell types are to be used in this protocol, growth and electroporation conditions will need to be adjusted for the specific cell type. -
2
Harvest 2 × 106 cells per electroporation by centrifuging at 400 × g for 5 min at 4°C. Wash cells once with PBS.
-
3
Resuspend cell pellet in 100 μl of IMDM and transfer cells to a 2 mm electroporation cuvette.
-
4
Add 2 μg of pEF1α-FB-dCas9 and pEF1α-BirA vectors (prepare 1 μg/μl of vectors in ddH2O) to the cell suspension and mix well by pipetting.
-
5
Perform electroporation at 0.25 kV, 15 mSec with the BTX 630 electroporator.
Other electroporation systems can be used; optimize for efficiency with the cell line of interest. -
6
Add 1 ml of warm IMDM containing 10% FBS to the cuvette and transfer electroporated cells to 15 ml tube.
-
7
Add 30 ml of IMDM medium and transfer cells into 96-well plates (100 μl/well) immediately.
-
8
48 h after transfection, treat cells with 1 μg/ml of puromycin and 600 μg/ml of G418.
-
9
Feed the cells with complete medium containing fresh G418 and puromycin every two days for 2 weeks until colonies become apparent in wells.
-
10
Look thoroughly in each well of 96-well plates to identify wells containing the single cell-derived colony and transfer the colonies to 12-well plates.
-
11
Harvest 1 × 106 cells from each picked clone and confirm dCas9 and BirA expression by western blot (WB) (Gallagher et al., 2008) using Cas9 and V5 antibody, respectively (Fig. 2A).
To establish FB-dCas9/BirA stable cell clones, we usually screen 24 clones selected by G418 and puromycin. Over half of the clones should show varying amounts of FB-dCas9 and BirA expression (Fig. 2A). We have found that high level BirA expression and medium to low level dCas9 expression clones can generate good signal to noise ratio in subsequent validation of dCas9 binding by CAPTURE-ChIP-qPCR and CAPTURE-ChIP-seq.
sgRNA Cloning and Transduction
-
12
Dilute forward primers containing non-targeting sgRNA sequence (sgGal4_sgRNA.fwd) or the specific protospacer sequence targeting the region of interest to 10 μM.
-
13
Set up a 50 μL PCR reaction in a 0.2 ml PCR tube on ice as the following:
10μl 5 × Phusion HF reaction buffer 2 μl 10 μM forward primer 2 μl 10 μM universal reverse primer 1 μl 10 mM dNTP mix 1 μl pSLQ1651-sgRNA(F+E)-sgGal4 (1ng, Addgene: 100549) 0.5 μl Phusion Polymerase (2 U/ μl) water to 50 μl -
14
Mix components thoroughly by vortexing, then run on a thermocycler with the following program:
1 cycle: 2 min 98°C (initial denaturation) 30 cycles: 10 sec 95°C (denaturation) 15 sec 60°C (annealing) 30 sec 72°C (extension) Final cycle: 2 min 72°C (final extension) Indefinite 4°C (hold) -
15
Run 5 μl of PCR product on a 2% TAE-agarose gel to ensure PCR specificity. The expected product size is about 120 bp.
-
16
Purify PCR product using Qiagen PCR purification kit and elute DNA in 15 μl of EB.
-
17
Set up a 15 μl restriction enzyme digestion reaction in a 0.2 ml PCR tube as the following:
12.5 μl purified PCR amplicon from step 16 or pSLQ1651-sgRNA(F+E)-sgGal4 vector (1 μg) 1.5 μl 10x NEB buffer 3.1 0.5 μl NEB BstXI 0.5 μl NEB XhoI water to 15 μl Incubate reactions at 37°C overnight. -
18
Purify the digested PCR amplicon by MinElute PCR purification kit.
-
19
Run digested pSLQ1651-sgRNA(F+E)-sgGal4 vector on a 1.2% TAE-agarose gel and purify the backbone by Qiagen Gel purification kit.
-
20
Set up a 10 μl ligation reaction in a 0.2 ml PCR tube as the following:
5 μl digested PCR amplicon (100 ng) 1 ul digested pSLQ1651 backbone (200 ng) 1 μl 10x NEB T4 ligase buffer 1 μl NEB T4 ligase water to 10 μl Incubate at room temperature for 1 h. -
21
Transform 100 μl of DH5α competent cells with 10 μl of ligation product (Seidman et al., 2001).
-
22
Pick 4 clones of each sgRNA and validate sgRNA sequence by Sanger DNA sequencing.
Lentiviruses Packaging and Transduction of K562 FB-dCas9/BirA Stable Cell Lines
-
23
Seed 3 × 106 293T cells in a 6 cm petri-dish in high glucose DMEM medium containing 10% FBS, and incubate at 37°C overnight.
-
24
Change medium (3 ml) 4 h before transfection.
-
25
Prepare transfection cocktail as the following:
1 μg VSV-G plasmid 2 μg psPAX2 plasmid 3 μg pSLQ1651-sgRNA plasmid 18 μg PEI (Plasmid:PEI ratio is 1:3) 300 μl Opti-MEM Vortex the tubes to mix, incubate at RT for 20 minus. -
26
Add cocktail to 293T cells dropwise and swirl medium to mix.
-
27
Change media 10 h after transfection.
-
28
Collect virus supernatant at 48 and 72 h post transfection.
-
29
Filter lentivirus supernatant through a disposable 0.45 um filter and concentrate virus using PEG-It Virus Precipitation Solution following manufacturer’s instruction.
-
30
Resuspend 0.5 × 106 K562 FB-dCas9/BirA expressing stable cells in 800 μl of fresh IMDM medium and plate cells in 12-well plates.
-
31
Add 200 μl of concentrated virus and 5 μM of PGE2 to cell suspension.
-
32
Seal the plate with parafilm and spin at 2,500 rpm for 90 min at 30°C.
-
33
Gently shake plates to resuspend cells, and incubate cells with virus overnight in 37°C incubator.
-
34
Wash infected cells 3 times with pre-warmed PBS 24 h post-infection and resuspend cells in fresh medium.
-
35
48 h post-infection, collect top 5% mCherry-positive (mCherryhi) cells by flow cytometry and expand for CAPTURE-ChIP (see Basic Protocol 1, Step 36 – 59), CAPTURE-Proteomics (see Basic Protocol 2), and CAPTURE-3C-seq (see Basic Protocol 3).
Quality Control of CAPTURE Specificity and Efficiency
-
36
Harvest 1 × 107 mCherryhi K562 FB-dCas9/BirA cells from step 35 and resuspend cells in 5 ml of ice-cold PBS.
-
37
Add 135 μl of 37% formaldehyde to the cell suspension to a final concentration of 1% and incubate for 10 min at RT with rotation.
-
38
Quench cross-linking by adding 250 μl of 2M glycine and incubate for 5 min at RT with rotation.
-
39
Wash cells twice in 10 ml of ice-cold PBS.
-
40
Resuspend cells in 1 ml of Nuclear Extraction Buffer and incubate for 30 min at 4°C with rotation.
-
41
Centrifuge at 2,500 × g for 5 min at 4°C to collect nuclei.
Nuclei can be snap frozen in liquid nitrogen and stored at −80°C for several months -
42
Resuspend nuclear pellets in 400 μl of 0.5% Nuclear Lysis Buffer.
-
43
Sonicate nuclei in an ice water bath using a Branson Sonifier S-450 at 20% amplitude for 60 pulses (0.5 sec ON and 1 sec OFF). Shear DNA to a size of 300–500bp.
Sonication condition needs to be optimized to achieve the desired size range.Take 5 μl of sheared chromatin, perform a quick reverse-crosslinking by diluting to 25 μl in TE Buffer with 0.1% SDS, and boil for 10 min. Purify DNA by phenol-chloroform extraction and run aqueous phase on a 2% TAE-agarose gel to check the size. There is no need for ethanol precipitation.The samples can be frozen at −80°C. -
44
Add 44 μl of 10% Triton X-100 to sonicated nuclei lysate and centrifuge for 10 min at 16,100 × g at 4°C.
-
45
Carefully transfer 400 μl of the supernatant to a new Eppendorf tube and add 24 μl of 5 M NaCl to the supernatant. This is the soluble chromatin.
Save 20 μl of residual supernatant as the input control. -
46
Thoroughly resuspend MyOne™ Streptavidin T1 Dynabeads and transfer 20 μl of beads to a new Eppendorf tube.
-
47
Collect beads on a magnet stand and discard supernatant by pipetting. Wash beads twice with 1 ml of RIPA 0.3 Buffer.
-
48
Separate beads on a magnet and discard supernatant.
-
49
Add soluble chromatin from step 45 directly to beads and incubate tubes on rotator at 4°C for at least 3 h to overnight.
-
50
Collect beads on a magnet stand and discard supernatant.
-
51
Wash the beads twice by adding 1 ml of 2% SDS, vortex at max speed for 15 sec and collect beads on a magnet. Transfer beads to a new Eppendorf LoBind microcentrifuge tube.
-
52
Repeat step 51 six times as the following:
Two washes with 1ml of High Salt Wash Buffer
Two washes with 1ml of LiCl Wash Buffer
Two washes with 1ml of TE Buffer
-
53
Resuspend beads in 100 μl of SDS Elution Buffer. Incubate tubes at 65°C for at least 6 h to overnight with shaking.
-
54
Briefly spin to collect beads. Separate beads on magnet for 3 min and transfer the supernatant to a new Eppendorf LoBind microcentrifuge tube.
-
55
Rinse beads with 50 μl of SDS Elution Buffer and pool supernatant with eluted chromatin from step 54.
-
56
Add 1 μl of 0.5 μg/μl RNase A and incubate at 37°C for 30 min.
-
57
Add 1 μl of 20 mg/ml Protease K. Incubate at 37°C for 2 h.
-
58
Recover ChIP DNA using Qiagen PCR Purification Kit and elute DNA in 50 μl of ddH2O.
-
59
To validate on-target enrichment of CAPTURE-ChIP, perform qPCR using DNA from step 58 as template or proceed for library prep using NEBNext ChIP-seq library prep kit for next-generation sequencing (NGS).
BASIC PROTOCOL 2. CAPTURE-Proteomics
This protocol is used to isolate protein complexes associated with biotinylated dCas9 captured CREs (Fig. 3A) (Liu et al., 2017). The method consists of chemical cross-linking by formaldehyde, followed by RNase digestion and extensive chromatin washes to remove molecules that do not bind to DNA. Chromatin preparation steps were performed as previously described with modifications (Kustatscher et al., 2014). The resulting material is subjected to sonication to solubilize cross-linked chromatin. Streptavidin-mediated affinity purification is then performed and dCas9-tethered chromatin segments are isolated. Chromatin-associated protein complexes from dCas9-captured genomic loci are recovered under denaturing conditions, and analyzed by western blot (WB) or digested to peptides for identification by liquid chromatography-mass spectrometry (LC-MS/MS). CAPTURE-Proteomics enable selective isolation of chromatin complexes associated with dCas9-targeted genomic loci in mammalian cells.
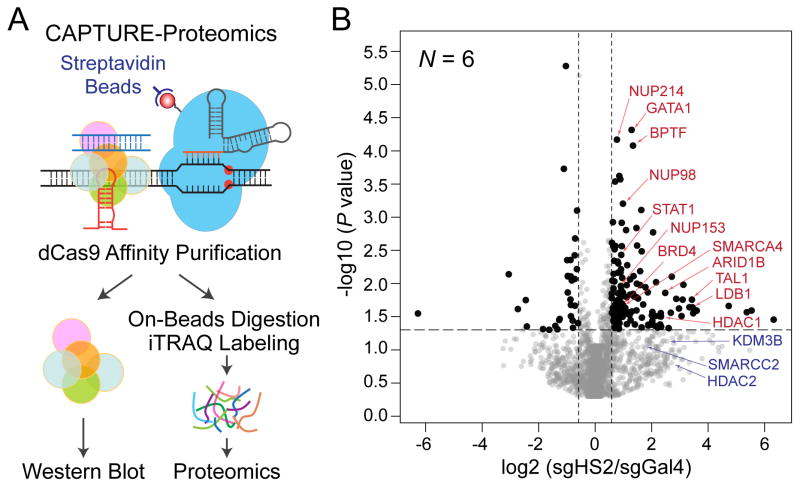
Figure 3. CAPTURE-Proteomics Analysis of β-Globin HS2 Enhancer-Associated Protein Complexes.
(A) Schematic of CAPTURE-Proteomics to identify CRE-associated protein complexes in situ.
(B) CAPTURE-Proteomics identified β-globin HS2-associated proteins. Volcano plots are shown for the iTRAQ-based quantitative proteomics of purifications in sgHS2 versus sgGal4-expressing cells. Relative protein levels in sgHS2 versus control (sgGal4) samples are plotted on the x-axis as mean log2 iTRAQ ratios across replicate experiments. Negative log10 transformed P values are plotted on the y-axis. Significantly enriched proteins (P & le; 0.05; iTRAQ ratio ≥ 1.5) are denoted by black dots, all others by grey dots. Dotted lines indicate 1.5-fold ratio (x-axis) and P value of 0.05 (y-axis). Representative locus-specific chromatin-regulating proteins are denoted by red arrowheads. Representative proteins with iTRAQ ratio ≥ 1.5 and P > 0.05 are denoted by blue arrowheads. This figure was reproduced from (Liu et al., 2017) with permission from Elsevier.
Materials
37% formaldehyde solution (Calbiochem, Cat # 344198)
Protease inhibitor cocktail (Sigma-Aldrich, Cat # P8340)
Urea (Sigma-Aldrich, Cat # U5128-500G)
TCEP (Sigma-Aldrich, Cat # 75259-1G)
MMTS (Sigma-Aldrich, Cat # 64306-1ML)
Trypsin (Promega, Cat # V511C)
Streptavidin agarose beads (Invitrogen, Cat # 15942-050)
Pierce cellulose acetate filter spin cup (Thermo-Fisher, Cat # 69702)
Screw Cap Micro Tube (SARSTEDT, Cat # 72.692.005)
Flat tip (Costar, Cat # 4884)
Thermomixer (Eppendorf)
Digital Sonifier (BRANSON, Models S-450)
Formaldehyde Crosslinking
-
1
Grow 1 × 108 cells from Basic Protocol 1 step 35 for CAPTURE-WB and 2.5 to 5 × 108 cells for CAPTURE-Proteomics analysis.
-
2
Aliquot cells to 15 ml tubes with 5 × 107 cells each tube. Centrifuge at 400 × g for 5 min at 4°C to collect cells.
-
3
Resuspend cell pellets in 5 ml of 2% formaldehyde in PBS, and incubate for 10 min at RT with rotation.
-
4
Add 0.5 ml of 2.5 M glycine and incubate at RT for 5 min with rotation to quench cross-linking.
-
5
Centrifuge at 400 × g for 5 min at 4 °C and carefully remove supernatant.
-
6
Wash cell pellets twice with 2 ml of ice-cold PBS.
Chromatin Preparation and Sonication
-
7
Resuspend cells with 2ml of ice-cold Cell Lysis Buffer and transfer cell suspension to a 2 ml tube. Incubate for 30 min at 4°C with rotation.
-
8
Spin at 2,500 × g for 5 min at 4°C to pellet nuclei.
-
9
Resuspend nuclei with 500 μl of Cell Lysis Buffer. Add 1 μl of 0.5 μg/μl RNase A and incubate at 37°C for 30 min with rotation.
-
10
Spin at 2,500 × g for 5 min at 4°C to collect nuclei.
-
11
Resuspend nuclear pellets in 0.4 ml of Nuclear Lysis Buffer by slowly pipetting up and down, and incubate for 10 min at RT.
To avoid damage to the chromatin, DO NOT vortex to mix. -
12
Add 1.2 ml of Urea Buffer and mix by inverting the tube.
-
13
Centrifuge the tube at 16,100 × g for 25 min at RT to pellet chromatin. Remove supernatant completely.
-
14
Repeat steps 11 to 13 three more time until the supernatant is clear.
-
15
Resuspend the pellets in 0.4 ml of Nuclear Lysis Buffer by slowly pipetting up and down, and add 1.2 ml of Nuclear Lysis Buffer to wash out urea.
-
16
Centrifuge the tube at 16,100 × g for 25 min at RT.
-
17
Repeat steps 15 and 16 once.
-
18
Resuspend the chromatin in 0.4 ml of Cell Lysis Buffer by slowly pipetting up and down, and add 1.2 ml of Cell Lysis Buffer to wash out SDS.
-
19
Centrifuge the tube at 16,100 × g for 25 min at RT to pellet chromatin.
-
20
Repeat steps 18 and 19 once.
-
21
Resuspend washed chromatin in 0.4 ml of IP Binding Buffer without NaCl by pipetting.
-
22
Sonicate nuclei in an ice water bath using a Branson Sonifier S-450 at 10% amplitude for 120 pulses (0.5 sec ON and 1 sec OFF). Shear chromatin to a size of 300–1000 bp.
Need to optimize the sonication condition for different cell types to achieve the desired size range. -
23
Centrifuge the tube at 16,100 × g for 5 min at 4°C. Check the size of sonicated chromatin fragments.
Take 5 μl of supernatant containing soluble chromatin, perform a quick reverse cross-linking by diluting to 25 μl in TE Buffer with 0.1% SDS, and boil for 10 min. Purify DNA by phenol-chloroform extraction and run aqueous phase on a 2% TAE-agarose gel to check the size.Desired size of chromatin is 300–1000 bp. If size of DNA fragment is within the range, transfer the supernatant to a new tube and keep sheared chromatin on ice. Resuspend residual chromatin pellet in 0.4 ml of IP Binding Buffer without NaCl and repeat steps 22 and 23 until desired size. -
24
Pool the supernatant from step 23 and centrifuge the tube at 16,100 × g for 25 min at 4°C. Carefully transfer supernatant to a new 1.5 ml microtube with screw cap for affinity purification.
Streptavidin Immunoprecipitation
-
25
Add 5 M NaCl to sheared chromatin to a final concentration of 150 mM and immediately invert tubes to mix.
-
26
Thoroughly mix streptavidin agarose beads slurry (50% beads slurry) by vortexing, and take 50 μl of slurry per IP.
-
27
Wash streptavidin agarose beads twice by 1 ml of IP Binding Buffer and centrifuge at 800 × g for 3 min at 4°C.
-
28
Resuspend beads in 25 μl of IP Binding Buffer and add to chromatin from step 24.
-
29
Incubate tubes at 4°C overnight with rotation.
-
30
Centrifuge at 800 × g for 3 min at 4 °C to collect beads.
-
31
Resuspend beads in 1 ml of IP binding Buffer and incubate for 10 min at 4°C with rotation. Spin at 800 × g for 3 min, and completely remove supernatant using a flat gel loading pipet tip.
-
32
Repeat steps 31 for 4 times.
After the last wash, resuspend beads in 100 μl of IP Binding Buffer. Transfer and pool all the beads from multiple tubes to a new Eppendorf LoBind tube. For WB, proceed to step 33, and for LC-MS, go to step 36.
Recover proteins for WB
-
33
Spin beads from step 32 at 800 × g for 3 min and remove the supernatant as much as possible using a flat gel loading pipet tip.
-
34
Add 30 μl of RIPA buffer and 10 ul of 4x XT loading buffer (Bio-Rad) supplied with 5% of β-mercaptoethanol to the dry beads. Incubate at 95°C for 20 min.
DO NOT incubate for more than 20 min. Extended heating may result in protein degradation. -
35
Spin at 12,000 × g for 3 min at RT to collect beads. The supernatant is ready to be loaded to SDS-PAGE gels (Gallagher et al., 2008).
Recover Peptides for LC-MS/MS
-
36
Resuspend beads from step 32 in 1 ml of IP Binding Buffer without NP-40. Spin at 800 × g for 3 min, and completely remove supernatant using a flat gel loading pipet tip.
-
37
Repeat step 36 twice to wash out detergent.
-
38
Resuspend beads in 500 μl of 0.5M Tris (pH 8.5). Add 0.5 M TCEP to suspension to a final concentration of 20 mM, and rotate tubes at RT for 1 h to reduce disulfide bonds.
-
39
Add 4 μl of MMTS and incubate tubes for 20 min with rotation at RT to mask reduced sulfhydryls.
-
40
Add 10 μl of 0.5 μg/μl sequencing grade Trypsin, and incubate at 37°C for 6 h with rotation. Add another 10 μl of Trypsin, and rotate at 37°C overnight.
-
41
Load all eluates from step 41 into the insert of Pierce cellulose membrane tube and centrifuge at 12,000 × g for 3 min at RT to remove beads.
-
42
Add 5M NaCl to flow-through to a final concentration of 3M, seal tubes tightly and boil samples at 95°C for 1 h to reverse cross-linking.
-
43
Dry samples on a Speedvac and follow manufacturer’s instructions for peptide purification using C18 column and iTRAQ labelling (SCIEX).
Salt crystal should become apparent in the tube after Speedvac if detergent is completely washed out.
BASIC PROTOCOL 3. CAPTURE-3C-seq
The protocol for CAPTURE-3C-seq is developed to identify long-range chromatin looping associated with dCas9-captured genomic loci (Fig. 4A) (Liu et al., 2017). In this protocol, cross-linked nuclei are first digested with a restriction enzyme (DpnII), followed by in situ proximity ligation to create chimeric DNA molecules. The digestion and ligation in fixed nucleus can generate chimeric molecules between DNA fragments that are far away in genome, but physically tethered by chromatin complexes. The ligated DNA are then sheared into smaller fragments by sonication, followed by streptavidin-based affinity purification of dCas9-targeted genomic regions. The purified chimeric DNA fragments are then paired-end sequenced to identify DNA interactions associated with dCas9-captured genomic loci. Biotinylated dCas9-mediated affinity purification can specifically enrich sgRNA-targeted DNA fragments, thus providing high resolution analysis of locus-specific chromatin structure. CAPTURE-3C-seq does not require PCR or oligonucleotide-based pre-selection steps and can be multiplexed using multiple sgRNAs in mammalian cells.
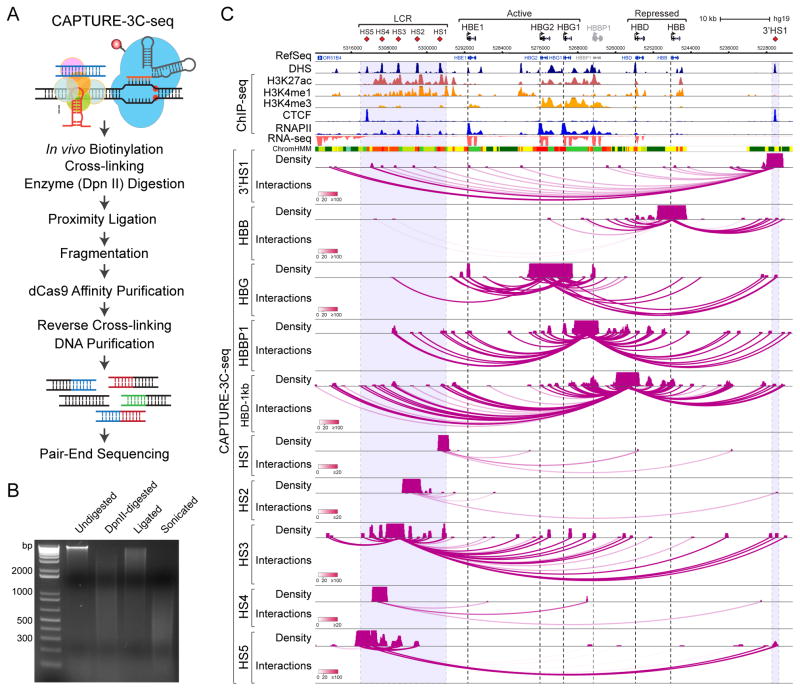
Figure 4. CAPTURE-3C-seq Analysis of Locus-specific DNA Interactions at Human β-Globin Gene Cluster.
(A) Schematic of CAPTURE-3C-seq to identify locus-specific long-range DNA interactions.
(B) Representative quality control analysis of CAPTURE-3C digestion and proximity ligation is shown.
(C) Browser view of the long-range DNA interaction profiles at β-Globin CREs (chr11:5,222,500–5,323,700; hg19) is shown. Contact profiles including the density map and interactions (or loops) are shown. The statistical significance of interactions between the captured region and other genomic regions was determined by the FDR-controlled Bayes factor (BF), and is indicated by the darkness of the interaction loops according to the color scale bars. Interactions with BF ≥ 20 were considered high-confidence long-range DNA interactions. DHS, ChIP-seq (H3K27ac, H3K4me1, H3K4me3, CTCF and RNAPII), RNA-seq, and chromatin state (ChromHMM) data are shown for comparison. Several panels of this figure were reproduced from (Liu et al., 2017) with permission from Elsevier.
Materials
37% formaldehyde solution (Calbiochem, Cat #344198)
Protease inhibitor cocktail (Sigma-Aldrich, Cat #P8340)
DpnII restriction enzyme with 10x DpnII Buffer (New England Biolabs, Cat #R0543M)
10× NEB T4 Ligase Buffer (New England Biolabs, Cat #B7203S)
400 U/μl T4 ligase with 10× T4 ligase buffer (New England Biolabs, Cat #M0202L)
Dynabeads® MyOne™ Streptavidin T1 (Invitrogen, Cat #656-01)
RNase A, DNase-free (0.5 mg/ml, Roche, Cat #11119915001)
Proteinase K (20 mg/ml, Ambion, Cat #AM2546)
37°C incubator (Thermo-Fisher)
16°C incubator (Thermo-Fisher)
Thermomixer (Eppendorf)
Digital Sonifier (BRANSON, Models S-450)
Formaldehyde Cross-linking and Nuclear Extraction
-
1
Expend 5 × 107 cells from Basic Protocol 1 step 35, and wash cells twice in ice-cold PBS.
-
2
Resuspend cell pellet in 10 ml PBS and add 37% formaldehyde to cell suspension to a final concentration of 1% formaldehyde. Incubate at RT for 10 min with rotation.
-
3
Transfer the reaction tubes to ice and add 2 M glycine to a final concentration of 0.125M to quench cross-linking. Incubate at RT for 10 min with rotation.
-
4
Centrifuge at 400 × g for 5 min at RT and carefully discard supernatant.
-
5
Wash cells twice with ice-cold PBS.
-
6
Resuspend cells in 1 ml ice-cold Cell Lysis Buffer and incubate 30 min at 4°C with rotation.
-
7
Centrifuge at 2,300 × g for 5 min at 4°C and discard supernatant.
The pelleted nuclei can be frozen in liquid nitrogen and store at −80°C for several months.
Digestion and In Situ Ligation
-
8
Rinse nuclei once in 500 μl of ice-cold 1x NEB DpnII Buffer and centrifuge at 2,300 × g for 5 min at 4°C to collect nuclei.
-
9
Gently resuspend nuclear pellet in 0.1 ml of 0.5% SDS by slowly pipetting, and incubate tubes at 62°C for 10 min. Immediately place the tube on ice and cool nuclei for 5 min.
-
10
To sequester SDS, add 60 μl of 10% Triton X-100 and ddH2O to a final volume of 534 μl. Mix carefully by slowly pipetting, and incubate at 37°C for 30 min.
Save a 2 μl aliquot of samples as undigested genomic DNA control. -
11
Add 60 μl of 10x NEB DpnII Buffer and 6 μl of DpnII (50 U/μl) to the sample and digest chromatin overnight at 37°C with rotation.
Take a 2 μl aliquot of samples, add 7 μl of TE buffer and 1 μl of 0.5 mg/ml RNase A, and incubate at 37°C for 30 min. Then add 20 μl of Proteinase K buffer and 20 mg/ml of Proteinase K and incubate at 55°C for 2 h. Run on a TAE-agarose gel to check the extent of digestion. Digested DNA should run as a smear with a size ranging from 400 bp to 3 kb (Fig. 4B). -
12
Incubate tubes at 62°C for 20 min to inactivate DpnII. Place tubes on ice immediately after incubation.
-
13
Transfer the digested nuclei to a 15 ml tube. Add the following ligation mix to digested nuclei:
300 μl 10x NEB T4 ligase buffer 240 μl 10% Triton X-100 1.845 ml ddH2O 15 μl NEB T4 DNA ligase (400 U/μl). -
14
Aliquot ligation mix into 3 × 1.5 ml microtubes and perform in situ ligation by incubating overnight at 16°C followed by 30 min at RT with rotation.
Save a 2 μl aliquot of samples. Purify DNA and run on a TAE-agarose gel to check ligation efficiency. After ligation, the DNA should shift to a higher molecular weight than the digested samples (Fig. 4B) -
15
Centrifuge at 2,300 × g for 5 min at 4°C to collect nuclei.
-
16
Resuspend nuclei in 500 μl of RIPA 0 buffer supplied with 0.25% Sarkosyl.
-
17
Sonicate nuclei in an ice water bath using Branson Sonifier S-450 at 10% amplitude for 120 pulses (0.5 sec ON and 1 sec OFF).
Need to optimize sonication conditions for different cell types. -
18
Centrifuge sonicated samples at 16,000 × g for 5 min at 4°C. Transfer supernatant to a new Eppendorf LoBind tube.
Streptavidin Affinity Purification and DNA Isolation
-
19
Add 5 M NaCl to sonicated chromatin from step 18 to a final concentration of 0.3 M for IP.
-
20
Thoroughly resuspend MyOne™ Streptavidin T1 Dynabeads and transfer 20 μl of beads to a new Eppendorf tube.
-
21
Collect beads on a magnet stand and discard supernatant by pipetting. Wash beads twice with 1 ml of RIPA 0.3 Buffer.
-
22
Separate beads on a magnet and discard supernatant.
-
23
Add soluble chromatin from step 19 directly to beads and incubate tubes on rotator at 4°C for at least 3 h to overnight.
-
24
Collect beads on a magnet stand and discard supernatant.
-
25
Wash beads twice by adding 1 ml of 2% SDS, vortex at max speed for 15 sec and collect beads on a magnet. Transfer beads to a new Eppendorf tube.
-
26
Repeat step 18 six times as the following:
-
27
Two washes with 1ml of High Salt Wash Buffer
Two washes with 1ml of LiCl Wash Buffer
Two washes with 1ml of TE Buffer
-
27
Resuspend beads in 100 μl of SDS Elution Buffer. Incubate tubes at 65°C for at least 6 h to overnight with shaking.
-
28
Briefly spin to collect beads. Separate beads on magnet for 3 min and transfer supernatant to a new Eppendorf LoBind tube.
-
29
Rinse beads with 50 μl of SDS Elution Buffer and pool supernatant with previous eluted chromatin from step 28.
-
30
Add 1 μl of 0.5 μg/μl RNase A and incubate at 37°C for 30 min.
-
31
Add 1 μl of 20 mg/ml Proteinase K. Incubate at 37°C for 2 h.
-
32
Recover ChIP DNA using MinElute PCR Purification Kit and elute DNA in 50 μl of ddH2O.
-
33
Proceed for library prep using NEBNext ChIP-seq library prep kit and perform paired-end sequencing.
REAGENTS AND SOLUTIONS
RIPA Buffer
50 mM Tris-HCl (pH 7.4)
1% NP-40
0.25% Sodium deoxycholate (NaDOC)
150 mM NaCl
0.1% SDS
2 mM EDTA
Store at 4°C
Add 1 mM DTT, 1 mM PMSF and Protease Inhibitor Cocktail (1:1000) freshly.
Cell Lysis Buffer
25 mM Tris-HCl (pH 7.4)
85 mM KCl
0.1% Triton X-100
Store at 4°C
Add 1 mM DTT, 1 mM PMSF and Protease Inhibitor Cocktail (1:1000) freshly.
Nuclear Lysis Buffer
50 mM Tris-HCl (pH 8.1)
1 mM EDTA (pH 8.0)
0.5% SDS
Store at RT
Add 1 mM DTT, 1 mM PMSF and Protease Inhibitor Cocktail (1:1000) freshly.
RIPA 0 Buffer
10 mM Tris-HCl (pH 7.4)
1 mM EDTA (pH 8.0)
0.1% SDS
1% Triton X-100
0.1% NaDOC
Store at 4°C
Add 1 mM DTT, 1 mM PMSF and Protease Inhibitor Cocktail (1:1000) freshly.
RIPA 0.3 Buffer
10 mM Tris-HCl (pH 7.4)
1 mM EDTA (pH 8.0)
0.3 M NaCl
0.1% SDS
1% Triton X-100
0.1% NaDOC
Store at 4°C
Add 1 mM DTT, 1 mM PMSF and Protease Inhibitor Cocktail (1:1000) freshly.
2% SDS Buffer
2% SDS
Store at RT
High Salt Wash Buffer
50 mM HEPES (pH 7.5)
1 mM EDTA (pH 8.0)
0.5 M NaCl
1% Triton X-100
0.1% NaDOC
Store at 4°C
LiCl Wash Buffer
10 mM Tris-HCl (pH 8.1)
1 mM EDTA (pH 8.0)
250 mM LiCl
0.5% NP-40
0.5% NaDOC
Store at 4°C
TE Buffer
10 mM Tris-HCl (pH 7.5)
1 mM EDTA (pH 8.0)
Store at RT
SDS Elution Buffer
50 mM Tris-HCl (pH 8.1)
10 mM EDTA (pH 8.0)
1% SDS
Store at RT
4% SDS Nuclear Lysis Buffer
50 mM Tris-HCl (pH 7.4)
10 mM EDTA
4% SDS
Store at RT
8M Urea Buffer
10 mM Tris-HCl (pH 7.4)
1 mM EDTA
8M Urea
Store at RT
IP Binding Buffer
20 mM Tris-HCl (pH 7.5)
150 mM NaCl
1 mM EDTA
0.1% NP-40
10% Glycerol
Store at 4°C
IP Binding Buffer without NaCl
20 mM Tris-HCl (pH 7.5)
1 mM EDTA
0.1% NP-40
10% Glycerol
Store at 4°C
0.5M TCEP
0.287 g TCEP-HCl
Dissolve in 2 ml of 1M NaOH
Aliquot and store at −20°C
0.5 μg/μl Trypsin
100 μg Trypsin
Dissolve in 200 μl of 50mM Acetic Acid
Aliquot and store at −80°C
COMMENTARY
Background Information
CREs such as enhancers regulate temporal and tissue-specific gene expression by recruiting trans-acting factors and mediating long-range chromatin interactions. Despite recent advances in genome-scale annotation of CREs (Andersson et al., 2014; Heintzman et al., 2009; Heintzman et al., 2007; Visel et al., 2009), the biological importance and in situ molecular composition of the vast majority of CREs remain unknown. Over the years, various approaches have been developed to study CREs and associated trans-acting factors. ChIP assays can provide crucial insights into the global distribution of chromatin bound factors and the co-localization of multiple factors, but they rely on a priori identification of molecular targets. By coupling ChIP with MS, ChIP-MS approaches have been applied to identify chromatin-associated protein complexes (Ji et al., 2015); however, ChIP-MS pulls down protein complexes from many genomic regions simultaneously thus it does not provide information about proteins associated with specific genomic loci. To identify locus-specific chromatin complexes, targeted affinity purification of endogenous genomic loci has been developed by genetically engineered binding sites that enable purification of the targeted genomic regions (Agelopoulos et al., 2012; Byrum et al., 2012; Fujita and Fujii, 2011; Griesenbeck et al., 2003; Pourfarzad et al., 2013); however, these approaches require knock-in gene targeting which remains inefficient and low throughput. Recently, DNA sequence-specific binding molecules have been employed to enrich chromatin segments from specific genomic loci. For example, the proteomics of isolated chromatin segments (PICh) approach utilizes locked nucleic acids (LNA) to isolate telomere-associated proteins in human cells (Dejardin and Kingston, 2009). Transcription activator-like (TAL) effector proteins were also modified to target specific genomic loci for purification and proteomic analysis (Byrum et al., 2013; Fujita et al., 2013); however the engineering and validation of TAL proteins for each targeted locus remains inefficient and laborious. The development of the CRISPR system containing a nuclease-inactive dCas9 protein facilitated isolation of native genomic loci (Fujita and Fujii, 2013, 2014; Waldrip et al., 2014); however, previous studies relied on antibody-based affinity purification, and the genome-scale specificity and the utility in identifying both cis- and trans-acting factors were not fully evaluated.
Besides trans-acting factors, chromatin structure also plays an important role in regulating gene expression. Various methods have been developed to study long-range DNA interactions based on nuclear proximity ligation that allows for detection of distant interacting genomic regions tethered together by high order architectures. The chromatin interaction analysis by paired-end tag sequencing (ChIA-PET) can detect high resolution genome-wide chromatin interactions mediated by specific protein factors (Fullwood et al., 2009; Li et al., 2012), but it relies on a priori identification of the chromatin factors and existence of ChIP-quality antibodies. Hi-C was developed to unbiasedly capture genome-wide chromatin contacts such as the topologically associated domains (TADs) (Dixon et al., 2012); however, it is limited by the resolution required to analyze locus-specific interactions. By using additional PCR or oligonucleotide probes to enrich DNA contacts from individual genomic sites, 4C-based technologies were developed to obtain high resolution locus-specific chromatin interactions (Hughes et al., 2014; Simonis et al., 2006; Zhao et al., 2006). However, PCR or oligonucleotide-based pre-selection steps may introduce additional biases in quantitative analysis of interactions between CREs.
By directing in vivo biotinylated dCas9/sgRNA complex to specific CREs, CAPTURE provides an easier and more cost-efficient approach to isolate locus-specific chromatin interactions (Liu et al., 2017). CAPTURE does not rely on predefined protein factors, available reagents, or a priori knowledge of target loci. The biotin-streptavidin-mediated high affinity purification displays higher sensitivity and specificity than antibody-based strategies. Coupled with chromatin conformation assays, CAPTURE also allows for unbiased identification and quantitative analysis of long-range DNA interactions associated with targeted genomic loci. Therefore, CAPTURE provides an integrated platform for simultaneous analysis of a single copy genomic locus-associated proteins and long-range DNA interactions, and has the potential to significantly advance our understanding of the molecular composition of non-coding regulatory genome.
Critical Parameters and Troubleshooting
sgRNA Design
sgRNA is one of the most important components of the CAPTURE system. The following parameters need to be taken into consideration for sgRNA design. First, the sgRNA should target the sequence located in close proximity to the captured CRE to maximize the capture efficiency, but not overlap with binding sites of trans-acting factors to avoid potential interference with protein binding. Second, capture of locus-specific chromatin looping requires restriction enzyme (DpnII) digestion followed by proximity ligation. Therefore, the sgRNA target sequence should localize in the same DpnII digested fragment as the CRE and in close proximity to DpnII sites. Third, the on-target enrichment and genome-wide specificity of individual sgRNAs should be carefully evaluated by CAPTURE-ChIP-qPCR and/or CAPTURE-ChIP-seq before proceeding to CAPTURE-Proteomics. Finally, the use of multiplexed sgRNAs targeting the same CRE may increase the on-target enrichment and help distinguish consistent interactions from rare interactions of individual sgRNAs. However, the on-target enrichment for each sgRNA should be comparable to minimize variation in capture efficiency by multiplexed sgRNAs.
In Vivo Biotinylation Efficiency
Affinity purification by CAPTURE relies on a biotin acceptor peptide fused to dCas9 that is biotinylated by a co-expressed BirA enzyme. Efficient in vivo biotinylation of FB-dCas9 requires an appropriate ratio of BirA and FB-dCas9 proteins. Since streptavidin only recognizes biotinylated FB-dCas9 protein, it is important to achieve high efficiency of in vivo biotinylation by adjusting the expression ratio of stably expressed BirA and FB-dCas9 proteins. Excessive FB-dCas9 protein expression with insufficient biotinylation may increase non-specific DNA binding and low capture efficiency. Therefore, it is critical to select single cell-derived stable clones expressing relatively high level of BirA and to evaluate the biotinylation efficiency of dCas9 by streptavidin IP-WB.
Proper Controls and Replicates
The inclusion of proper controls is critical for the identification and quantitative analysis of locus-specific chromatin-regulating proteins by CAPTURE-Proteomics. Cells expressing non-targeting sgGal4 and no sgRNA parental cells should always be processed in parallel with cells expressing sequence-specific sgRNAs. Side-by-side comparison of results in samples with sequence-specific sgRNAs and control sgRNAs allows for the identification of co-purified contaminating proteins, such as the endogenous biotinylated proteins and proteins associated with off-target dCas9 binding. The identification of high-confidence non-specific proteins in control samples is also required for quantitative and statistical analysis of candidate locus-specific proteins. To minimize sampling variation from LS-MS/MS, quantitative proteomic methods such as iTRAQ are preferred to analyze CAPTURE purified protein complexes. Control cells expressing non-targeting sgGal4 and sequence-specific sgRNAs should always be analyzed by LS-MS/MS in parallel. Multiple technical replicates of LC-MS/MS may help identify variability of mass spectrometry runs. Independent biological replicates are required to establish statistical models for downstream data analysis.
Quality Control of CAPTURE-3C-seq
In the CAPTURE-3C-seq protocol, small aliquots of chromatin were kept after four key steps: lysis, digestion, ligation, and sonication. DNA isolated from these aliquots need to be analyzed on an agarose gel to assess the intactness of DNA prior to digestion, the efficiency of digestion and subsequent ligation (Fig. 4B). Undigested genomic DNA should appear as a sharp band of over 12 kb. After DpnII digestion, DNA run as a smear with a size ranging from 400 bp to 3 kb. After ligation, the ligated chimera DNA should shift to a higher molecular weight than the digested samples. Then the large DNA molecules should be fragmented into smaller sizes ranging from 300 bp to 500 bp by sonication (Fig. 4B).
Data Processing
CAPTURE-ChIP-seq
Align ChIP-seq raw reads to human genome assembly (hg19) using Bowtie1 (Langmead et al., 2009) with default parameters. Trim the first 10 nucleotides and last 3 nucleotides from each read before alignment. Use MACS to perform peak calling using the “--nomodel” parameter (Zhang et al., 2008). Remove peaks that overlap with the blacklist regions annotated by the ENCODE project (Consortium, 2012). To compare ChIP-seq signal intensities in samples from cells expressing target-specific sgRNAs versus non-targeting sgGal4, apply MAnorm (Shao et al., 2012) to remove systematic bias between samples and calculate the normalized ChIP-seq read densities of each peak for all samples. Use 300 bp as the window size which matches the average width of the identified ChIP-seq peaks.
CAPTURE-3C-seq
Use a customized pipeline as previously described (Liu et al., 2017) to process pair-end sequencing data. The software package is publically available (https://github.com/YONGCHENUTD/C3S). Map pair-end sequencing reads individually to human (hg19) or mouse (mm9) genome assembly using Bowtie2 (Langmead and Salzberg, 2012) with the default parameters. Since DpnII digestion followed by proximity ligation generates chimeric molecules which cannot be mapped, trim the reads with DpnII digestion position, collect and remap the longer fragment with length ≥ 20 bp. Combine the mapped reads and perform quality trimming (MAPQ ≥ 30). Pair mapped reads from pair-end sequencing and removed PCR duplicates by discarding reads with the same positions at both paired ends. Use preprocessed read pairs to define the interactions at each sgRNA-targeted (or bait) region to other chromosomal regions. Define bait region by calling local peak surrounding the sgRNA target site using MACS2 with default parameters (Zhang et al., 2008). Filter self-ligated reads if the read pairs locate within the bait region. Only keep the pairs of reads that locate in two different regions to define interactions. Use Bayes statistical model to call significant interactions between the captured region and other genomic regions.
CAPTURE-Proteomics
Search raw mass spectrometry data against SwissProt database with ProteinPilot V4.5 (AB SCIEX, Framingham, MA) with parameter “iTRAQ 4-plex (peptides labeling) with 5600 TripleTOF” as previously described (Liu et al., 2017). Only keep peptides that can be assigned to unique genes. To determine the peptide spectra match (PSM) false discovery rate (FDR), use the statistical model based on the target-decoy search strategy to evaluate the confidence level of peptide identification (Elias and Gygi, 2007). Only keep the peptides with scores at or below a PSM FDR threshold of 1% for data analysis. Sum up the intensities of iTRAQ reporter ions for peptides to generate the ion intensity of the genes. Only retain genes with total signal intensity of iTRAQ reporter ions more than 50 which were considered confidently detected for further analysis. Normalize the ion intensity of iTRAQ mass spectrometry signal based on the cumulative intensity of the high-confidence non-specific proteins identified from control samples, including cell lines expressing BirA alone, BirA with FB-dCas9, BirA with FB-dCas9 and the non-targeting sgRNAs (sgGal4), and BirA with FB-dCas9 and sequence-specific sgRNAs with deletion of the sgRNA-targeted genomic region, by principal component analysis (PCA). After the global normalization of each sample, calculate the ratios of the iTRAQ reporter ion intensity for each protein in samples with target-specific sgRNA across replicate experiments relative to the non-targeting sgGal4, and use a paired t-test to calculate P values to measure the statistical significance of the log2 iTRAQ ratios of each identified protein. Define a protein as significantly enriched if the iTRAQ ratio ≥ 1.5 and P value ≤ 0.05 in samples prepared from cells expressing sequence-specific sgRNAs versus the non-targeting sgGal4 control.
Understanding Results
For CAPTURE-ChIP-seq, we usually purify less than 0.5 ng of ChIP DNA for a single locus. 10% of the ChIP DNA is used for qPCR analysis to test the quality of samples and on-target enrichment. Fig. 2B–E shows the results of representative CAPTURE-ChIP-qPCR and CAPTURE-ChIP-seq at the DNase I hypersensitive site 2 (HS2) region of the human β-globin locus control region (LCR). Enrichment of the HS2 genomic region was > 1000-fold compared to other neighboring HS sites (Fig. 2C). In most cases, we observed over 1000-fold enrichment of the targeted genomic loci compared to surrounding genomic regions. To evaluate genome-wide dCas9 binding, high-throughput sequencing by CAPTURE-ChIP-seq should be used. Co-expression of sgHS2 with dCas9 resulted in highly specific enrichment of HS2 with no or little dCas9 binding in the genome other than the sgRNA target site. None of the predicted off-target regions were significantly enriched (Fig. 2D,E).
For CAPTURE-Proteomics, we usually detect some background signals from some endogenous biotinylated proteins in samples with no sgRNA or non-targeting sgGal4 controls. However, samples with locus-specific sgRNAs are significantly enriched with nuclear proteins. Relative protein abundance levels associated with the captured region (HS2) versus the non-targeting sgGal4 control are determined by the ratio of the iTRAQ reporter ion intensity (Fig. 3B).
With a successful CAPTURE-3C-seq library, 20 million of 38 bp pair-end reads should generate enough information for analysis of locus-specific interactions. Using various CREs at human β-globin gene cluster as examples, CAPTURE-3C-seq can identify high resolution, locus-specific long-range DNA interactions at each captured β-globin CRE (Fig. 4C). All tracks were viewed using the WashU Epigenome Browser (https://epigenomegateway.wustl.edu/).
Time Considerations
CAPTURE approach takes about 4 to 6 weeks for the generation and validation of FB-dCas9 and BirA expressing stable cell lines. Lentiviral infection of sgRNAs, selection and expansion of transduced cells usually takes about 3 weeks. Once the proper number of cells is achieved, CAPTURE protocols usually take only 2 to 4 days. The first day consists of cross-linking and nuclear extraction. The nuclei can be stored at −80°C at this point. For CAPTURE-ChIP-seq, nuclei can be sonicated and processed through streptavidin affinity purification steps in 2 days. For CAPTURE-Proteomics, chromatin preparation takes one additional day prior to chromatin fragmentation. For CAPTURE-3C-seq, enzyme digestion and proximity ligation take 2 days before the collection of nuclei for IP. Reverse cross-linked peptide samples can be stored at −80°C for several weeks. DNA recovered from streptavidin purification can be stored at −20°C for several months before NGS library prep.
Acknowledgments
Due to space limit, we apologize for not being able to cite all the relevant references. We thank B. Chen and H. Chen (UCSF) for sharing reagents and protocols, and F. Zhou (Fudan University) for helpful advice regarding peptide preparation and mass spectrometry. This work was supported by the NIH grant R01MH102616, the Cecil H. and Ida Green Endowment, and the NSFC grants (91519326, 31671384) (to M.Q.Z.), by NIH/NIDDK grants K01DK093543, R03DK101665 and R01DK111430, by a Cancer Prevention and Research Institute of Texas (CPRIT) grants (RR140025 and RP180504), by the American Cancer Society (IRG-02-196) award and the Harold C. Simmons Comprehensive Cancer Center at UT Southwestern, by the Welch Foundation grant I-1942-20170325, and by an American Society of Hematology Scholar Award (to J.X.), and by a CSRM fellowship at UT Southwestern (to X. L.).
LITERATURE CITED
- Agelopoulos M, McKay DJ, Mann RS. Developmental regulation of chromatin conformation by Hox proteins in Drosophila. Cell reports. 2012;1:350–359. doi: 10.1016/j.celrep.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, Chen Y, Zhao X, Schmidl C, Suzuki T, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Bhaya D, Davison M, Barrangou R. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu Rev Genet. 2011;45:273–297. doi: 10.1146/annurev-genet-110410-132430. [DOI] [PubMed] [Google Scholar]
- Byrum SD, Raman A, Taverna SD, Tackett AJ. ChAP-MS: a method for identification of proteins and histone posttranslational modifications at a single genomic locus. Cell reports. 2012;2:198–205. doi: 10.1016/j.celrep.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrum SD, Taverna SD, Tackett AJ. Purification of a specific native genomic locus for proteomic analysis. Nucleic acids research. 2013;41:e195. doi: 10.1093/nar/gkt822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li GW, Park J, Blackburn EH, Weissman JS, Qi LS, et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex genome engineering using CRISPR/Cas systems. Science (New York, NY) 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium TEP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedon PC, Soults JA, Allis CD, Gorovsky MA. Formaldehyde cross-linking and immunoprecipitation demonstrate developmental changes in H1 association with transcriptionally active genes. Molecular and cellular biology. 1991;11:1729–1733. doi: 10.1128/mcb.11.3.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejardin J, Kingston RE. Purification of proteins associated with specific genomic Loci. Cell. 2009;136:175–186. doi: 10.1016/j.cell.2008.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez AA, Lim WA, Qi LS. Beyond editing: repurposing CRISPR-Cas9 for precision genome regulation and interrogation. Nature reviews Molecular cell biology. 2016;17:5–15. doi: 10.1038/nrm.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science (New York, NY) 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nature methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- Fujita T, Asano Y, Ohtsuka J, Takada Y, Saito K, Ohki R, Fujii H. Identification of telomere-associated molecules by engineered DNA-binding molecule-mediated chromatin immunoprecipitation (enChIP) Scientific reports. 2013;3:3171. doi: 10.1038/srep03171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita T, Fujii H. Direct identification of insulator components by insertional chromatin immunoprecipitation. PloS one. 2011;6:e26109. doi: 10.1371/journal.pone.0026109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita T, Fujii H. Efficient isolation of specific genomic regions and identification of associated proteins by engineered DNA-binding molecule-mediated chromatin immunoprecipitation (enChIP) using CRISPR. Biochemical and biophysical research communications. 2013;439:132–136. doi: 10.1016/j.bbrc.2013.08.013. [DOI] [PubMed] [Google Scholar]
- Fujita T, Fujii H. Identification of proteins associated with an IFNgamma-responsive promoter by a retroviral expression system for enChIP using CRISPR. PloS one. 2014;9:e103084. doi: 10.1371/journal.pone.0103084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, Orlov YL, Velkov S, Ho A, Mei PH, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher S, Winston SE, Fuller SA, Hurrell JG. Immunoblotting and immunodetection. Current protocols in molecular biology. 2008;Chapter 10(Unit 10.18) doi: 10.1002/0471142727.mb1008s83. [DOI] [PubMed] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griesenbeck J, Boeger H, Strattan JS, Kornberg RD. Affinity purification of specific chromatin segments from chromosomal loci in yeast. Molecular and cellular biology. 2003;23:9275–9282. doi: 10.1128/MCB.23.24.9275-9282.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nature genetics. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JR, Roberts N, McGowan S, Hay D, Giannoulatou E, Lynch M, De Gobbi M, Taylor S, Gibbons R, Higgs DR. Analysis of hundreds of cis-regulatory landscapes at high resolution in a single, high-throughput experiment. Nature genetics. 2014;46:205–212. doi: 10.1038/ng.2871. [DOI] [PubMed] [Google Scholar]
- Ji X, Dadon DB, Abraham BJ, Lee TI, Jaenisch R, Bradner JE, Young RA. Chromatin proteomic profiling reveals novel proteins associated with histone-marked genomic regions. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:3841–3846. doi: 10.1073/pnas.1502971112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kustatscher G, Wills KL, Furlan C, Rappsilber J. Chromatin enrichment for proteomics. Nat Protoc. 2014;9:2090–2099. doi: 10.1038/nprot.2014.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Ruan X, Auerbach RK, Sandhu KS, Zheng M, Wang P, Poh HM, Goh Y, Lim J, Zhang J, et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148:84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zhang Y, Chen Y, Li M, Zhou F, Li K, Cao H, Ni M, Liu Y, Gu Z, et al. In Situ Capture of Chromatin Interactions by Biotinylated dCas9. Cell. 2017;170:1028–1043. e1019. doi: 10.1016/j.cell.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science (New York, NY) 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moqtaderi Z, Struhl K. Defining in vivo targets of nuclear proteins by chromatin immunoprecipitation and microarray analysis. Current protocols in molecular biology. 2004;Chapter 21(Unit 21.29) doi: 10.1002/0471142727.mb2109s68. [DOI] [PubMed] [Google Scholar]
- Pourfarzad F, Aghajanirefah A, de Boer E, Ten Have S, Bryn van Dijk T, Kheradmandkia S, Stadhouders R, Thongjuea S, Soler E, Gillemans N, et al. Locus-specific proteomics by TChP: targeted chromatin purification. Cell reports. 2013;4:589–600. doi: 10.1016/j.celrep.2013.07.004. [DOI] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, et al. Genome-wide location and function of DNA binding proteins. Science (New York, NY) 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- Schatz PJ. Use of peptide libraries to map the substrate specificity of a peptide-modifying enzyme: a 13 residue consensus peptide specifies biotinylation in Escherichia coli. Bio/technology (Nature Publishing Company) 1993;11:1138–1143. doi: 10.1038/nbt1093-1138. [DOI] [PubMed] [Google Scholar]
- Seidman CE, Struhl K, Sheen J, Jessen T. Introduction of plasmid DNA into cells. Current protocols in molecular biology. 2001;Chapter 1(Unit1.8) doi: 10.1002/0471142727.mb0108s37. [DOI] [PubMed] [Google Scholar]
- Shao Z, Zhang Y, Yuan GC, Orkin SH, Waxman DJ. MAnorm: a robust model for quantitative comparison of ChIP-Seq data sets. Genome biology. 2012;13:R16. doi: 10.1186/gb-2012-13-3-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, de Laat W. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nature genetics. 2006;38:1348–1354. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- Thakore PI, Black JB, Hilton IB, Gersbach CA. Editing the epigenome: technologies for programmable transcription and epigenetic modulation. Nature methods. 2016;13:127–137. doi: 10.1038/nmeth.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldrip ZJ, Byrum SD, Storey AJ, Gao J, Byrd AK, Mackintosh SG, Wahls WP, Taverna SD, Raney KD, Tackett AJ. A CRISPR-based approach for proteomic analysis of a single genomic locus. Epigenetics. 2014;9:1207–1211. doi: 10.4161/epi.29919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome biology. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Tavoosidana G, Sjolinder M, Gondor A, Mariano P, Wang S, Kanduri C, Lezcano M, Sandhu KS, Singh U, et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nature genetics. 2006;38:1341–1347. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]