Autophagy can promote the lysosomal degradation of specific cytoplasmic substrates recognized by cargo receptors. A recent study published in Science identifies a ribosome receptor required for the autophagic targeting of ribosomes to the lysosome.
Autophagy is the catabolic process that delivers cytoplasmic contents engulfed in a double-membrane vesicle, the autophagosome, to the lysosome for degradation and recycling. The targeting of cytoplasmic materials to the autophagosome was originally thought to promote non-selective bulk degradation, but it is now apparent that many forms of autophagy can target selective substrates dependent on specific cargo receptors.1 First described in yeast following nitrogen starvation, ribophagy refers to the autophagy-mediated selective engulfment of ribosome for delivery to the vacuole.2, 3 Recent studies have identified ribophagy in mammalian cells4 and the findings by Wyant et al.5 highlighted here report the identification of a putative ribosome receptor, nuclear fragile X mental retardation-interacting protein 1 (NUFIP1), required for the selective autophagy of ribosomes. (Fig. 1)
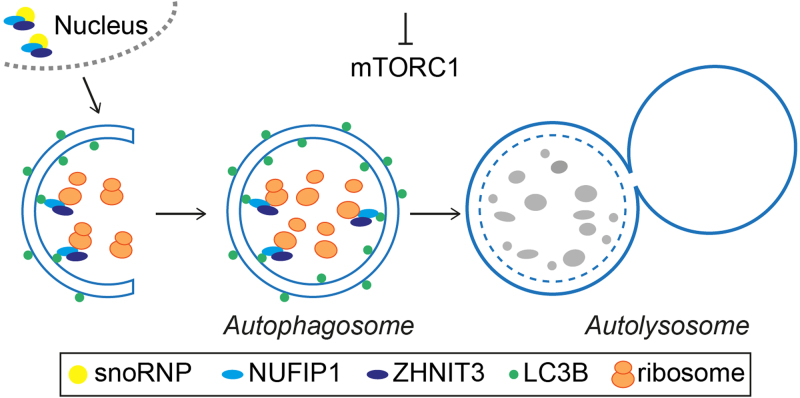
Fig. 1.
Schematic representation of ribophagy in mammalian cells. Following mTORC1 inhibition, the cytoplasmic cargo receptor NUFIP1 in association with ZNHIT3 binds to ribosomes and promotes their targeting to autophagosome by interacting with LC3B
The mTOR complex 1 (mTORC1) is a central mediator in regulating the response to nutrients and growth signaling, and is a key regulator of autophagy.6 The localization of mTORC1 to the lysosome promotes its activation and suppresses autophagy. To understand the relationship between mTORC1 activation and the lysosome, Wyant et al.5 compared the lysosomal proteome under conditions that inhibit mTOC1 signaling, nutrient starvation, and Torin1 inhibition. By isolating highly pure lysosomes (LysoIP) followed by mass spectrometry, they identified 828 proteins, including known lysosomal proteins, and over 40% were present under all conditions. Several identified proteins increased upon Torin1 treatment, including NUFIP1.
Previous studies have shown that NUFIP1 and its binding partner zinc finger HIT domain-containing protein 3 (ZNHIT3) are involved in ribonucleoprotein (RNP) complex formation (in the assembly of the box C/D small nucleolar RNPs).7–9 While ZNHIT3 was similarly increased in the lysosomal proteome upon Torin1 treatment, other components of the RNP complex were not identified, prompting the investigation of a lysosome-dependent function of NUFIP1-ZNHIT3. The increase of NUFIP1 and ZNHIT3 in the lysosome in response to mTORC1 inhibition occurred without a change in the total cellular level due to their redistribution from the nucleus. As mTOCR1 inhibition also strongly induces autophagy, the authors examined the contribution of autophagy to this localization. The Torin1-mediated lysosomal accumulation of NUFIP1 and ZNHIT3 was dependent on autophagy as it could be prevented by genetically blocking autophagy by using Atg7-null cells.
During autophagy, cargo receptors are important for the engulfment of selective substrates and contain an LC3-interacting region (LIR) motif to associate with the LC3 family members. Identification of putative LIR motifs in NUFIP1 prompted the investigation of the relationship between NUFIP1 and LC3 proteins. Following Torin1 treatment, NUFIP1 co-localized with LC3B-positive autophagosomes and NUFIP1 interacted with LC3B, but not with other LC3 family member GABARAP, dependent on the presence of ZNHIT3. Mutagenesis of an LIR motif of NUFIP1 abolished the binding to LC3B, and the mutant NUFIP1 or ZHNIT3 was no longer associated with lysosomes following Torin1 treatment.
In addition to the nuclear role of NUFIP1 in ribosomal RNA modification, it has also been reported to localize in the cytoplasm associated with ribosomes.10 With the identification of an interaction with LC3B, the authors investigated the possibility that NUFIP1-ZNHIT3 may also associate with ribosomes. They detected NUFIP1-ZNHIT3 in ribosome-containing cell fractions that correlated with the large ribosomal subunit under conditions of mTORC1 inhibition. The interaction between NUFIP1 and ribosomes was confirmed by co-immunoprecipitation.
Having established that NUFIP1-ZNHIT3 binds both LC3 and ribosomes under conditions of mTORC1 inhibition, Wyant et al. examined the consequence of these interactions. They found a decrease in ribosomal proteins following mTORC1 inhibition that required Atg7 function. In the absence of NUFIP1, there was no longer a decrease in ribosomal proteins following mTORC1 inhibition, whereas autophagy induction still occurred. This could be rescued by re-expression of wild-type NUFIP1 but not the LIR motif mutant. Furthermore, while maintaining nuclear localization of NUFIP1 prevented ribosome degradation, the presence of NUFIP1 in the cytoplasm did not trigger ribosomal loss without mTORC1 inhibition, suggesting that mTORC1 inhibition is required to promote the association between NUFIP1 and ribosomes. Importantly, NUFIP1-deficient cells showed reduced survival following nutrient starvation that could not be rescued by expression of the LC3B-binding-deficient mutant. Thus, under nutrient limitation, NUFIP1 targeting of ribosomes to the lysosome provides metabolites that contribute to cell survival.
Based on the description by Galluzzi et al.,1 “Autophagy receptors are proteins that bind autophagy substrates, allow for their recognition by the autophagy machinery, and get degraded within lysosomes in the course of functional autophagic responses”, Wyant et al.5 have identified a receptor for ribophagy. Their findings reveal that ribosomes in mammalian cells can be degraded by autophagy requiring the specific cargo receptor NUFIP1. This can be triggered by a well-established signal of non-selective autophagy induction, mTOCR1 inhibition. This raises several questions about the broader role of ribophagy: whether it is simultaneously induced in parallel with other selective/non-selective autophagy pathways, and whether there are other unique components of the autophagy pathway downstream of mTORC1 inhibition that promote ribophagy? Also, does the induction of ribophagy by mTORC1 inhibition reflect a “basal” level of ribosomal turnover that occurs upon the induction of non-selective autophagy and can alternative stress signals trigger higher levels of ribophagy? Interestingly, ribophagy is also induced under conditions of proteotoxic stress and chromosome mis-segregation in mammalian cells.4 Therefore, it will be important to further explore the molecular mechanism of ribophagy and the potential for degradation of non-selective cargo during selective autophagy. These outcomes may be context-specific and further in vivo analysis of the function of NUFIP1-ZNHIT3 and ribophagy will be critical to establish the physiological context.
References
- 1.Galluzzi L, et al. EMBO J. 2017;36:1811–1836. doi: 10.15252/embj.201796697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kraft C, Deplazes A, Sohrmann M, Peter M. Nat. Cell Biol. 2008;10:602–610. doi: 10.1038/ncb1723. [DOI] [PubMed] [Google Scholar]
- 3.Ossareh-Nazari B, et al. EMBO Rep. 2010;11:548–554. doi: 10.1038/embor.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.An H, Harper JW. Nat. Cell Biol. 2018;20:135–143. doi: 10.1038/s41556-017-0007-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wyant GA, et al. Science. 2018;360:751–758. doi: 10.1126/science.aar2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saxton RA, Sabatini DM. Cell. 2017;168:960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boulon S, et al. J. Cell Biol. 2008;180:579–595. doi: 10.1083/jcb.200708110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McKeegan KS, Debieux CM, Boulon SE, Bertrand, Watkins NJ. Mol. Cell. Biol. 2007;27:6782–6793. doi: 10.1128/MCB.01097-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quinternet M, et al. J. Mol. Biol. 2015;427:2816–2839. doi: 10.1016/j.jmb.2015.07.012. [DOI] [PubMed] [Google Scholar]
- 10.Bardoni B, et al. Exp. Cell Res. 2003;289:95–107. doi: 10.1016/S0014-4827(03)00222-2. [DOI] [PubMed] [Google Scholar]