Falzone et al. interpret the mechanisms underlying the activity of TMEM16 family members from recent structural and functional work.
Abstract
The TMEM16 family of membrane proteins is composed of both Ca2+-gated Cl− channels and Ca2+-dependent phospholipid scramblases. The functional diversity of TMEM16s underlies their involvement in numerous signal transduction pathways that connect changes in cytosolic Ca2+ levels to cellular signaling networks. Indeed, defects in the function of several TMEM16s cause a variety of genetic disorders, highlighting their fundamental pathophysiological importance. Here, we review how our mechanistic understanding of TMEM16 function has been shaped by recent functional and structural work. Remarkably, the recent determination of near-atomic-resolution structures of TMEM16 proteins of both functional persuasions has revealed how relatively minimal rearrangements in the substrate translocation pathway are sufficient to precipitate the dramatic functional differences that characterize the family. These structures, when interpreted in the light of extensive functional analysis, point to an unusual mechanism for Ca2+-dependent activation of TMEM16 proteins in which substrate permeation is regulated by a combination of conformational rearrangements and electrostatics. These breakthroughs pave the way to elucidate the mechanistic bases of ion and lipid transport by the TMEM16 proteins and unravel the molecular links between these transport activities and their function in human pathophysiology.
Introduction
Membrane bilayers delimit and compartmentalize all living cells. In eukaryotes, lipid biogenesis occurs in the ER where lipids are randomly distributed between the two leaflets of the membrane (Balasubramanian and Schroit, 2003; Pomorski and Menon, 2006; Bevers and Williamson, 2010). As lipids travel from the ER to the plasma membrane through the Golgi apparatus, the lipid composition of the membranes becomes progressively asymmetric via the action of two classes of ATP-driven enzymes, flippases and floppases, which catalyze the slow transbilayer movement of lipids between leaflets and establish their chemical gradients (Balasubramanian and Schroit, 2003; Pomorski and Menon, 2006; Bevers and Williamson, 2010). The final result of their activity is the generation and maintenance of strict lipid asymmetry in the plasma membrane: choline-containing phospholipids, phosphatidylcholine (PC), and sphingomyelin, are more abundant in the outer leaflet whereas amino phospholipids, phosphatidylethanolamine (PE), phosphatidylserine (PS), and phosphoinositides (such as PI and PIP2), are confined to the inner leaflet (Holthuis and Levine, 2005; Bevers and Williamson, 2010, 2011). Flippases and floppases are lipid-specific, with the former mostly belonging to the P4 subfamily of ATPases tasked with the internalization of PE and PS, whereas the latter are ATP-binding cassette transporters and move PC and sphingomyelin from the inner to the outer leaflet (Seigneuret and Devaux, 1984; Pomorski and Menon, 2006; Quazi and Molday, 2013).
Maintenance of the lipid asymmetry of the plasma membrane is crucial for cellular homeostasis. However, a variety of physiological processes are triggered by externalization of PS and, to a more limited degree, of PE (Castoldi et al., 2011; Segawa et al., 2011; Whitlock and Hartzell, 2017). In contrast, constitutive or uncontrolled exposure of PS disrupts cellular homeostasis, rendering the maintenance of this asymmetry crucial for living cells. Exposure of PS is essential to activate platelets and promote blood coagulation as well as for the suppression of inflammatory responses (Nagata et al., 2016) and is a necessary, but not sufficient, marker of apoptotic cells for recognition and clearance by macrophages (Fadok et al., 1992; Balasubramanian and Schroit, 2003). The rapid collapse of lipid asymmetry on the plasma membrane, resulting in the exposure of PS, is known as phospholipid scrambling. Phospholipid scramblases, the class of protein responsible for scrambling, translocate lipids rapidly and bidirectionally down their concentration gradients. Unlike flippases and floppases, phospholipid scramblases are mostly not selective for lipids and can be thought of as “poorly selective” lipid channels.
To date, two major families of regulated scramblases have been identified: the TMEM16 family and the Xk-related (Xkr) family. Some TMEM16 proteins are Ca2+-activated scramblases, which control blood coagulation and membrane repair (Nagata et al., 2016). Members of the Xkr family are caspase-activated scramblases involved in PS exposure during apoptosis (Suzuki et al., 2014; Nagata et al., 2016). Other proteins, such as G protein–coupled receptors (GPCRs) and membrane-active peptides, moonlight as nonregulated scramblases (Kol et al., 2003a; Menon et al., 2011; Goren et al., 2014).
Interestingly, some TMEM16 family members function as ion channels rather than scramblases. Indeed, the first two functionally characterized TMEM16 homologues were TMEM16A and B, which are calcium-activated chloride channels (CaCCs; Caputo et al., 2008; Schroeder et al., 2008; Yang et al., 2008). The finding that most of the TMEM16 family members encode for scramblases rather than channels was therefore unexpected (Suzuki et al., 2010, 2013b; Malvezzi et al., 2013; Brunner et al., 2014). Furthermore, several of the TMEM16 scramblases also mediate nonselective ion transport, in addition to their lipid-scrambling activity (Malvezzi et al., 2013; Scudieri et al., 2015; Yu et al., 2015; Lee et al., 2016), suggesting that these two functions are closely linked in the TMEM16s. In the present review, we will discuss the current understanding of the mechanistic bases of phospholipid scrambling and ion transport by the TMEM16 proteins. We will place special emphasis on how the structures of the nhTMEM16 channel/scramblase from Nectria haematococca and of the TMEM16A CaCC influence our understanding of how lipid scrambling and ion permeation are inextricably intertwined in these proteins.
Unregulated scramblases
The first Ca2+-independent scramblase identified was rhodopsin, the light-sensitive GPCR found in rod photoreceptor discs whose primary function is to mediate visual phototransduction (Menon et al., 2011). Purified rhodopsin constitutively scrambles lipids when reconstituted in proteoliposomes (Menon et al., 2011) independent of its conformational state, because opsin, rhodopsin, and metarhodopsin II all have similar activities (Goren et al., 2014; Ernst and Menon, 2015), suggesting that the conformational changes underlying its light-driven cycle are not involved in lipid scrambling. Scrambling is likely to be performed by the monomeric form of the protein, because oligomerization-deficient rhodopsin mutants mediate constitutive lipid scrambling (Ploier et al., 2016). Lipid scrambling by opsin has been recently proposed to occur through a transient hydrophilic groove that is opened by a conformational rearrangement of TM6 and TM7 (Morra et al., 2018). Remarkably, several reconstituted GPCRs, the class A adenosine A2A receptor, the β1 and β2 adrenergic receptors, and the bacterial light-driven H+-pump bacteriorhodopsin (Menon et al., 2011; Goren et al., 2014; Verchère et al., 2017) also moonlight as scramblases. These findings suggest that the opening of a transient hydrophilic groove is an evolutionarily conserved feature of seven TM receptors. The physiological role of the moonlighting scrambling activity of GPCRs with full “daytime” jobs remains poorly understood. It has been speculated that opsin could be the scramblase that contributes to the lipid homeostasis of photoreceptor disc membranes by counteracting the action of the ATP-binding cassette A4 pump (Ernst and Menon, 2015). It is possible that other GPCR scramblases could contribute to the maintenance of symmetry in the ER membranes en route to their eventual plasma membrane targeting (Ernst and Menon, 2015). The question of how the constitutive scrambling activity of these GPCRs is silenced in the plasma membrane, a biological necessity to maintain lipid asymmetry, remains open.
In addition to GPCRs, single-helix peptides have been shown to mediate constitutive, unregulated phospholipid scrambling when reconstituted in artificial membranes. Indeed, several antimicrobial peptides combat bacteria by permeabilizing their membranes via the formation of toroidal pores, or half-pores (Gilbert et al., 2014). Oligomerization of these peptides promotes fusion of the two leaflets in the membrane, thereby enabling lipid scrambling. A different class of single-helix peptides scrambles lipids without higher-order oligomerization, suggesting that the simple structure of these peptides is sufficient to distort the membrane sufficiently to lower the energy barrier that prevents flipping of the lipid headgroups between the membrane leaflets (Kol et al., 2003a,b; Langer et al., 2013). These peptides share a common general design, where the hydrophobic core of the helix is composed of a repeated sequence and the N and C termini are decorated by positively charged residues.
The Xkr family of caspase-activated scramblases
Recent work has shown that members of the Xkr family of membrane proteins are the long-sought scramblases that mediate Ca2+-independent PS exposure during apoptosis (Suzuki et al., 2014, 2016). The Xkr family is composed of nine human members, each of which contains 6–10 predicted transmembrane domains. Deletion of the Xkr8 gene in cells results in lack of PS exposure in response to apoptotic stimuli, and transient expression of Xkr4, 8, or 9 rescues this response, promoting macrophage engulfment (Suzuki et al., 2014, 2016). These proteins are characterized by a caspase recognition signal at the C-terminal domain, accounting for their caspase-dependent activity. Similarly, the Caenorhabditis elegans Xkr orthologue Cell Death abnormal-8 gene is involved in caspase-dependent PS externalization of apoptotic cells (Chen et al., 2013; Li et al., 2015), suggesting an evolutionarily conserved scramblase function for these proteins.
The TMEM16 family of Ca2+-regulated channels and scramblases
The discovery that most TMEM16 family members are phospholipid scramblases was unexpected. The first characterized homologues, TMEM16A and B, were CaCCs (Caputo et al., 2008; Schroeder et al., 2008; Yang et al., 2008), leading to the proposal that the TMEM16s were a family of CaCCs. It was therefore a surprise that TMEM16F plays a role in PS exposure in mouse Ba/F3 cells and mouse lymphocytes (Suzuki et al., 2010, 2013b). Furthermore, mutations in this gene cause Scott syndrome, a bleeding disorder characterized by defective PS exposure in platelets (Suzuki et al., 2010; Castoldi et al., 2011; Lhermusier et al., 2011), and its ablation leads to impaired coagulation caused by reduced PS exposure in platelets from animal models (Yang et al., 2012; Brooks et al., 2015). These findings indicated that TMEM16F, and other family members, could be scramblases. The finding that some TMEM16s directly mediate lipid scrambling in reconstituted systems (Malvezzi et al., 2013; Brunner et al., 2014; Lee et al., 2016; Watanabe et al., 2018) together with the identification of a minimal region necessary and sufficient for scrambling activity (Yu et al., 2015; Gyobu et al., 2017) led to the conclusion that the TMEM16 family is composed of both ion channels and phospholipid scramblases.
The TMEM16s form a widespread protein family, with homologues found in all eukaryotic phyla but absent in prokaryotes, indicating that the family appeared relatively late during evolution (Hartzell et al., 2009; Pedemonte and Galietta, 2014). The TMEM16s were initially named anoctamins (Ano1-10), because they were thought to be a family of chloride channels with eight transmembrane domains (Yang et al., 2008). However, subsequent work showed that neither was the case: TMEM16 proteins have 10 membrane-spanning helices (Brunner et al., 2014; Dang et al., 2017; Paulino et al., 2017a,b), and the family is characterized by a high degree of functional divergence, including chloride channels, phospholipid scramblases, and dual-function nonselective ion channel/phospholipid scramblases (Huang et al., 2013; Malvezzi et al., 2013; Brunner et al., 2014; Gyobu et al., 2015; Yu et al., 2015). We therefore favor the TMEM16 nomenclature.
There are 10 human TMEM16 proteins, TMEM16A–K, excluding I (Fig. 1). The known CaCCs, TMEM16A, B, and the Drosophila melanogaster homologue Subdued (Wong et al., 2013; Jang et al., 2015), cluster to one branch of the phylogenetic tree (Fig. 1, blue), suggesting that this clade is likely composed exclusively of CaCCs. Two distal clades (Fig. 1, red) contain dual-function channels/scramblases, the mammalian TMEM16E, F, and the fungal nh- and afTMEM16 (from Aspergillus fumigatus; Malvezzi et al., 2013; Brunner et al., 2014; Gyobu et al., 2015; Scudieri et al., 2015; Yu et al., 2015; Lee et al., 2016). Three clades (Fig. 1, green) contain homologues that have been associated with scramblase activity, the mammalian TMEM16C, D, G, J, and ANOH-1 from C. elegans (Suzuki et al., 2013a; Li et al., 2015), but it remains to be seen whether they also have channel function. A recent study suggests that TMEM16J might be a cation channel (Kim et al., 2018). Finally, the function of the mammalian homologues TMEM16H and K and of the D. melanogaster homologue aberrant X segregation (AXS) is unknown (Fig. 1, black), but their sequence similarity to the fungal channel/scramblases suggests that they belong to this subgroup. It is worth noting that despite a low sequence similarity between distant family members, such as TMEM16F and nhTMEM16, there is a remarkable conservation of function, indicating that scramblase activity is robust and tolerates sequence divergence.
Figure 1.
Phylogenetic organization of the TMEM16 family. The unrooted phylogenetic tree was derived from the original alignment from the Pfam database (PF04547) and constructed by using FastTree with the Jones-Taylor-Thornton model and CAT approximation for branch lengths (Price et al., 2010). Red, dual-function scramblases/channels; blue, CaCCs; green, scramblases with no reported channel activity; black, homologues with unknown function; *, homologues whose function has been demonstrated in vitro (afTMEM16, nhTMEM16, mTMEM16F, and hTMEM16A). For clarity, only the 10 human TMEM16 homologues are shown (TMEM16A–K), Subdued is a CaCC from D. melanogaster, AXS is a functionally uncharacterized homologue from D. melanogaster, and ANOH-1 is a scramblase from C. elegans.
CaCCs encoded by TMEM16 homologues
Native CaCCs play crucial roles in a variety of physiological processes, which include salt secretion in kidneys, lungs, and airway epithelia (Arreola et al., 1995; Nilius et al., 1997; Hartzell et al., 2005), modulation of the excitability of cardiac and smooth muscle cells (Hartzell et al., 2005; Thomas-Gatewood et al., 2011; Huang et al., 2012b), nociception in sensory neurons (Liu et al., 2010; Cho et al., 2012), cell proliferation (Stanich et al., 2011; Duvvuri et al., 2012; Liu et al., 2012), olfactory transduction (Kleene, 1993; Boccaccio and Menini, 2007), and photoreception (Barnes and Deschênes, 1992; MacLeish and Nurse, 2007; Pedemonte and Galietta, 2014). Despite their physiological importance, the molecular identity of the CaCCs remained unknown until 2008, when three groups used complementary cloning strategies to show that two members of the TMEM16 family, TMEM16A and B, mediate outwardly rectifying, Cl−-selective currents that are activated by physiological increases in intracellular Ca2+ (Caputo et al., 2008; Schroeder et al., 2008; Yang et al., 2008). These currents recapitulate the key functional features of native CaCCs, supporting the notion that TMEM16A and B encode for these channels. Further work showed that Subdued, a TMEM16A homologue from D. melanogaster, functions as a CaCC that is important for thermal nociception in the fly (Wong et al., 2013; Jang et al., 2015). Subsequent purification of mammalian TMEM16A confirmed that these proteins indeed are the pore-forming subunits of CaCCs (Terashima et al., 2013). Electrophysiological, biochemical, and fluorescent measurements suggested that these channels are dimers (Fallah et al., 2011; Sheridan et al., 2011; Tien et al., 2013), where each monomer is independently gated by Ca2+ and forms a physically distinct Cl− permeation pathway (Jeng et al., 2016; Lim et al., 2016). Indeed, recent structural work confirmed that TMEM16A is a double-barreled ion channel (Dang et al., 2017; Paulino et al., 2017a,b), like the CLC Cl− channels and transporters (Miller, 1982).
The fundamental properties of the TMEM16A and B channels are well characterized: their opening is synergistically driven by depolarizing voltages and binding of Ca2+ to two conserved sites located within the transmembrane region (Yu et al., 2012; Terashima et al., 2013; Brunner et al., 2014; Tien et al., 2014; Jeng et al., 2016; Lim et al., 2016), their anion selectivity sequence is I−>Br−>Cl−>F− (Caputo et al., 2008; Schroeder et al., 2008; Yang et al., 2008; Pifferi et al., 2009), and they are inhibited by nonspecific Cl− channel blockers such as niflumic acid, 4,4'-Diisothiocyano-2,2'-stilbenedisulfonic acid, and N-phenylanthranilic acid (Yang et al., 2008; Yu et al., 2012; Scudieri et al., 2015). Prolonged exposures to Ca2+ induce rundown or desensitization in both channels (Pifferi et al., 2009; Stephan et al., 2009; Ni et al., 2014; Dang et al., 2017; Pifferi, 2017). The mechanisms underlying this process are not clear at the moment, although it has been recently proposed that it might depend on PIP2 depletion (Tembo and Carlson, 2018). Further studies are needed to clarify this point.
It is less clear whether calmodulin also plays a role in modulating their Ca2+ dependence and whether these channels can also allow passage of cations in addition to anions. Several studies initially suggested that TMEM16A and B can form a stable complex with calmodulin in the presence of Ca2+ and that this association modulates their activation and/or inactivation properties (Tian et al., 2011; Vocke et al., 2013; Jung and Lee, 2015; Yang and Colecraft, 2016). However, each study identified a different calmodulin-binding site and mode of action complicating their interpretation. Furthermore, other groups could not recapitulate the finding that TMEM16A interacts with calmodulin in cells and in vitro (Terashima et al., 2013; Tien et al., 2014; Yu et al., 2014a,b). Therefore, the question of whether a TMEM16A-calmodulin complex exists in vivo remains open, and its possible role in regulating CaCC function remains unclear.
A similar uncertainty surrounds the question of whether the TMEM16 Cl− channels are ideally anion selective or if they are also permeable to cations. Some studies indicate that in whole-cell patch-clamp recordings, TMEM16A and B have anion-to-cation permeability ratios as high as 0.23, indicating that in physiological conditions, ∼20% of the total current mediated by these channels is carried by small cations such as Na+ or K+ (Schroeder et al., 2008; Pifferi et al., 2009; Yang et al., 2012; Peters et al., 2015, 2018). In contrast, others reported that TMEM16A-mediated currents are nearly ideally selective for Cl− over Na+ and K+ (Caputo et al., 2008; Yang et al., 2008; Terashima et al., 2013; Jeng et al., 2016; Lim et al., 2016; Paulino et al., 2017a). The origin of this discrepancy is not clear. Potential factors contributing to these divergent results are the presence of contaminating currents, different patch-clamp and recording solutions used to measure reversal potentials, incomplete junction potential subtractions, and the rectifying nature of the currents at low Ca2+ concentrations. We think that the preponderance of the evidence supports the hypothesis that TMEM16A is nearly ideally selective for anions over cations. The findings that the purified channel maintains a KCl gradient in liposomes (Terashima et al., 2013; Paulino et al., 2017a), which would be dissipated by a channel conductive to cations, and that the reversal potential of TMEM16A does not depend on the rectification of the currents (Lim et al., 2016) are strong indications in this regard. However, anion selectivity in TMEM16A is not as robust as that of other anion channels, such as the CLCs or CFTR (Chen and Hwang, 2008), as single-point mutations in the TMEM16A pore degrade its anion to cation selectivity (Yang et al., 2008, 2012; Peters et al., 2015) or convert it into a phospholipid scramblase that also mediates nonselective ion transport (Jiang et al., 2017). Thus, we cannot rule out the possibility that the TMEM16A pore could adopt a conformation that is permissive to cation permeation.
Some TMEM16 family members are Ca2+-dependent phospholipid scramblases
Soon after the initial cloning and identification of TMEM16A and B as CaCCs, the notion that the TMEM16s were a family of CaCCs was challenged by the unexpected discovery that TMEM16F is involved in phospholipid scrambling (Suzuki et al., 2010, 2013b; Segawa et al., 2011; Yang et al., 2012; Yu et al., 2015). Overexpression of wild-type TMEM16F enhances the Ca2+-dependent externalization of PS in cells and a constitutively active TMEM16F point mutant was identified (Suzuki et al., 2010; Segawa et al., 2011). Loss-of-function mutations in TMEM16F were found in patients with Scott syndrome, a rare bleeding disorder characterized by decreased PS exposure in platelets resulting in impaired blood clot formation (Suzuki et al., 2010; Castoldi et al., 2011; Boisseau et al., 2016). These mutations result in protein that is either partially deleted or truncated. Finally, platelets from mice or dogs lacking the TMEM16F gene have reduced Ca2+-dependent PS exposure and blood clot formation (Yang et al., 2012; Brooks et al., 2015).
Despite these initial lines of evidence, the role of TMEM16F in phospholipid scrambling became hotly debated as several groups reported that TMEM16F was an ion channel but without reaching a consensus as to the type of ion channel or its activating stimulus (Almaça et al., 2009; Martins et al., 2011; Yang et al., 2012; Grubb et al., 2013; Kmit et al., 2013; Sirianant et al., 2016; Ye et al., 2018). Among these, at least two studies did suggest that TMEM16F regulates the function of an unknown scramblase (Yang et al., 2012; Kmit et al., 2013), consistent with the idea that this homologue is involved in phospholipid scrambling. Further elements of controversy emerged when several other TMEM16 family members, TMEM16C, D, G, and J, were reported to have scrambling activity but no ion channel function (Suzuki et al., 2013b), again contrasting with previous indications that suggested that they function as CaCCs (Hartzell et al., 2009; Tian et al., 2012). Three complementary lines of evidence helped resolve this controversy and more generally clarify whether members of the TMEM16 family are channels, scramblases, or both. First, two fungal TMEM16 homologues, afTMEM16 from Aspergillus fumigatus and nhTMEM16 from Nectria hematococca, were successfully purified and functionally reconstituted, demonstrating that these proteins have intrinsic Ca2+-dependent phospholipid scramblase activity (Malvezzi et al., 2013; Brunner et al., 2016). More recently, a single-molecule fluorescence study showed that purified TMEM16F directly mediates phospholipid scrambling (Watanabe et al., 2018). Interestingly, both fungal and mammalian TMEM16 scramblases also mediate nonselective ion transport, suggesting that the two functions might be linked (Malvezzi et al., 2013; Scudieri et al., 2015; Yu et al., 2015; Lee et al., 2016; Di Zanni et al., 2017; Jiang et al., 2017). Second, combined phylogenetic, electrophysiological, and mutagenesis studies led to the identification of a short stretch of amino acids, called the “scramblase domain,” that is necessary and sufficient for lipid scrambling in TMEM16 proteins (Gyobu et al., 2015, 2017; Yu et al., 2015). Strikingly, grafting this domain from the TMEM16E and F scramblase homologues onto the TMEM16A Cl− channel transforms the latter into a dual-function Ca2+-activated scramblase and nonselective ion channel (Gyobu et al., 2015; Yu et al., 2015). Finally, ANOH-1, a TMEM16 homologue from C. elegans (Fig. 1), was shown to be important in PS exposure in necrotic touch neurons in the nematode (Li et al., 2015). Collectively, these lines of evidence indicate that the TMEM16s are a functionally divergent family, composed of CaCCs and of Ca2+-dependent scramblases, some of which also mediate nonselective ion transport.
Most TMEM16-type scramblases have been shown to transport equally well chemically diverse lipids, such as the negatively charged PS and PG, the positively charged 1,2-dioleoyl-3-trimethylammonium-propane, as well as polar (PE) and nonpolar lipids (PC and glucosylceramide) in cellular systems and in vitro reconstitutions (Malvezzi et al., 2013; Suzuki et al., 2013b; Brunner et al., 2014; Watanabe et al., 2018). Furthermore, afTMEM16 can scramble lipids conjugated to large polyethylene glycol molecules, of molecular weight up to 5,000 D, suggesting that the lipid pathway does not impose a strict size cut-off on lipid transport (Malvezzi et al., 2018). Finally, fungal and mammalian TMEM16 scramblases transport lipids very rapidly, with estimated transport rates ≥104 lipid/s (Malvezzi et al., 2013, 2018; Watanabe et al., 2018). The fast transport rate and poor selectivity suggest that during scrambling, the lipids do not interact tightly with the protein permeation pathway. It is important to note, however, that one family member, TMEM16C, has been reported to select against PS while transporting PC and glucosylceramide (Suzuki et al., 2013b), indicating that the TMEM16 lipid permeation pathway can be tuned to differentiate between lipids. The basis of this selectivity, or lack thereof, remains to be determined.
The physiological stimulus activating the TMEM16 scramblases is an increase in the cytosolic Ca2+ concentration (Suzuki et al., 2010; Yu et al., 2015). However, both fungal homologues retain measurable scramblase activity even in the absence of Ca2+ (Malvezzi et al., 2013; Brunner et al., 2014; Lee et al., 2016). It is unclear what the origin of this residual activity is. It could reflect incomplete removal of Ca2+, because nhTMEM16 maintains at least one bound Ca2+ ion in a putatively Ca2+-free structure (Brunner et al., 2014), or the lipid pathway could open in the absence of ligand. However, the presence of scramblases in the plasma membrane of mammalian cells does not lead to constitutive externalization of PS, suggesting that in cells, secondary regulatory mechanisms exist to prevent such a leak from occurring. Further, neither the mammalian homologues TMEM16E and F nor the fungal nhTMEM16 displays basal activity when overexpressed in cells (Yu et al., 2015; Di Zanni et al., 2017; Jiang et al., 2017), suggesting that additional regulatory components are present in cellular membranes. Although plasma membrane flippases and floppases could counteract the basal activity of native scramblases, it is unlikely that their expression levels and activity are sufficient to compensate the action of overexpressed TMEM16 scramblases. Indeed, purified and reconstituted TMEM16 scramblases respond to submicromolar changes in Ca2+ nearly instantaneously, whereas in cells, prolonged exposure to hundreds of micromoles are required for their activation, lending credibility to the idea that additional regulatory mechanisms might exist.
Physiological roles of the TMEM16 channels and scramblases
The involvement of the TMEM16 channels and scramblases in numerous genetic defects has provided hints to their physiological roles, and numerous insights have come from studies using animal models. Several excellent reviews cover in detail the physiological roles of TMEM16 family members (Huang et al., 2012a; Pedemonte and Galietta, 2014; Whitlock and Hartzell, 2016, 2017); therefore, we will only briefly summarize some key results here. The family-founding member TMEM16A is broadly expressed in airway epithelia, in the interstitial cells of Cajal in the gastrointestinal tract, in the trachea, and in smooth muscle cells of airways and reproductive tracts (Rock et al., 2008, 2009; Huang et al., 2009; Romanenko et al., 2010; Dixon et al., 2012; Cobine et al., 2017; He et al., 2017). Deletion of TMEM16A in mice impairs development of the trachea and is lethal (Rock et al., 2008). Conditional knockout studies have been used to probe the tissue-specific role of TMEM16A, indicating that Cl− transport mediated by this channel is essential in controlling processes that range from smooth muscle contractility to epithelial secretion (Huang et al., 2009; Rock et al., 2009; Romanenko et al., 2010; Dixon et al., 2012; Cobine et al., 2017; He et al., 2017).
The TMEM16 channel/scramblase homologue whose physiological role is best understood is TMEM16F, which plays a key role in controlling blood coagulation. In humans, loss-of-function mutations in the TMEM16F gene cause Scott syndrome, a mild bleeding disorder caused by defective Ca2+-dependent exposure of PS in activated platelets (Zwaal et al., 2004; Suzuki et al., 2010; Castoldi et al., 2011). Studies show that deletion of the TMEM16F gene in mice recapitulates well the Scott syndrome phenotype (Yang et al., 2012; Baig et al., 2016; Mattheij et al., 2016). Remarkably, other groups did not report a bleeding phenotype and identified additional effects associated with deletion of TMEM16F, such as defects in bone mineralization (Ehlen et al., 2013), in the immune response of macrophages (Ousingsawat et al., 2015), microglial response to injury (Batti et al., 2016), and defects in T-cell signaling and activation (Hu et al., 2016). It is unclear whether a bleeding phenotype was present but not described in these studies or if the effects of TMEM16F deletions depend on other factors.
A similarly complex panoply of phenotypes has been associated with the deletion of the TMEM16E channel/scramblase. This homologue prevalently localizes to intracellular compartments (Duran et al., 2012), rendering its study difficult. In humans, loss-of-function mutations in TMEM16E cause limb-girdle muscular dystrophy and Miyoshi myopathy type 3 (Bolduc et al., 2010; Hicks et al., 2011; Griffin et al., 2016), suggesting that this protein plays a role in membrane fusion and repair. Interestingly, a different cohort of dominant mutations was identified in TMEM16E as causative for gnathodiaphyseal dysplasia 1 (Marconi et al., 2013; Tran et al., 2014; Di Zanni et al., 2017), a bone malformation disorder. One of these mutations (T513I) has been characterized in vivo and was shown to be a gain of function for both channel and scramblase activities (Di Zanni et al., 2017). Three deletion studies of TMEM16E in mice found different phenotypes: one recapitulates the human dystrophic phenotype (Griffin et al., 2016), another leads to deficient sperm motility and male infertility (Gyobu et al., 2015), and the third described altered lipid metabolism and inflammation signaling (Xu et al., 2015). Thus, TMEM16E also plays a critical role in a variety of physiological processes, ranging from membrane repair and bone formation to sperm motility and lipid metabolism. However, a clear unifying picture of its functional properties is yet to be reached.
Although the diverse effects caused by the deletion of TMEM16E and 16F reveal the physiological roles played by these proteins, they also raise the question of what underlies the observed differences. It is possible that the genetic background of the mice strains, as well as the precise deletion strategy used in these studies matters. Whereas most studies were performed by using a C57B/6 background (Yang et al., 2012; Gyobu et al., 2015; Baig et al., 2016; Griffin et al., 2016; Mattheij et al., 2016), one used the similar but not identical strain C57BL/6J (Xu et al., 2015), and the rest were performed on SV129/J mice (Ousingsawat et al., 2015; Batti et al., 2016; Hu et al., 2016). Regardless of the origin of these discrepancies, all animal models display defective phospholipid scrambling, supporting the idea that TMEM16E and 16F are scramblases and identify a wide range of novel physiological processes where phospholipid scrambling plays a key role.
Several TMEM16 proteins, both CaCCs and scramblases, play key roles in controlling neuronal activity. For example, the mammalian TMEM16A and the D. melanogaster CaCC Subdued are required for heat sensing in nociception (Cho et al., 2012; Jang et al., 2015), whereas the TMEM16B CaCC is required for correct action-potential firing and axonal targeting in hippocampal, inferior olivary, and olfactory sensory neurons (Stephan et al., 2009; Huang et al., 2012b; Pietra et al., 2016; Zhang et al., 2017). Remarkably, the TMEM16 scramblases also play key roles in neuronal activity, because TMEM16F is required for injury response in microglia (Batti et al., 2016), whereas TMEM16C regulates pain processing in rat sensory neurons by facilitating the activation of the Na+-activated K+ channel SLACK (Huang et al., 2013). Whether this regulation is caused by its reported scrambling activity or if this homologue functions as a proper β subunit of SLACK remains to be established. Finally, mutations in the functionally uncharacterized TMEM16K cause autosomal recessive ataxia, attentional disorders, epilepsy, and porencephalic cysts (Chamova et al., 2012; Renaud et al., 2014), which have been linked to defects in Ca2+ signaling (Wanitchakool et al., 2017). Interestingly, this phenotype is similar to that associated with the deletion of the D. melanogaster TMEM16 homologue AXS, which is required for meiotic/mitotic spindle formation (Kramer and Hawley, 2003). Collectively, these results highlight the important roles played by the TMEM16 channels and scramblases in both peripheral and central nervous systems.
At least two TMEM16 homologues have been involved in different types of cancers. Specifically, TMEM16G (also known as NGEP), a prostate-specific homologue, was reported to be involved in the formation of cell–cell contacts in healthy and cancerous prostate tissues (Bera et al., 2004; Das et al., 2007), and its expression levels were correlated to prostate cancer progression (Cereda et al., 2010; Mohsenzadegan et al., 2013). Further, TMEM16A is highly expressed in multiple different cancer types, and its knockdown or inhibition leads to reduced malignancy of the tumors, consistent with its established role in cell proliferation (Stanich et al., 2011; Duvvuri et al., 2012; Liu et al., 2012; Wanitchakool et al., 2014). Collectively, these studies highlight the broad importance of the TMEM16 family members in human physiology and disease. Further work is needed to clarify their involvement in this multitude of physiological processes and clarify how their ion channel and/or scramblase activities fit in these diverse cellular functions.
Overall, the identification of the physiological roles of TMEM16 homologues remains a work in progress as novel functions are identified for known homologues and new roles are discovered for poorly characterized ones. For example, it has been proposed that the TMEM16J homologue might function as a cation channel that is activated by the cAMP/PKA signaling pathway (Kim et al., 2018). The scrambling activity of this homologue was not tested in this initial study.
Architecture of TMEM16 channels and scramblases
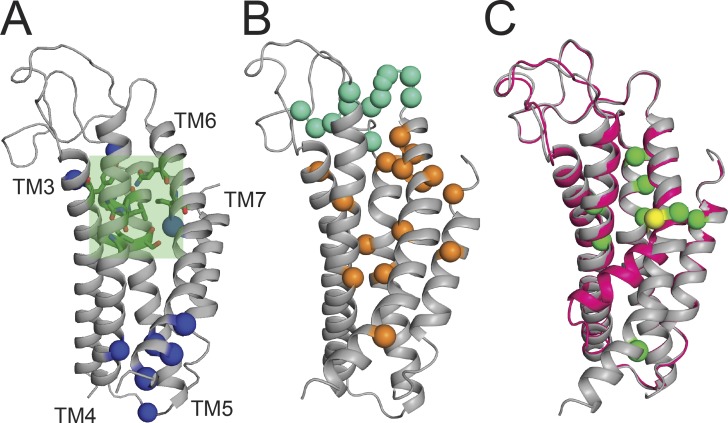
The breakthrough determination of the structures of the nhTMEM16 scramblase in a Ca2+-bound state (Brunner et al., 2014) and of the TMEM16A channel with zero, one, and two Ca2+ ions bound (Dang et al., 2017; Paulino et al., 2017a,b) provided an atomic-resolution framework to understand how these proteins mediate Ca2+-dependent ion and/or lipid transport. The TMEM16 channels and scramblases share an overall similar architecture: they are homodimers, where the transmembrane region of each monomer has 10 helices, labeled TM1–10, and an extensive cytosolic domain that adopts a ferredoxin-like fold (Fig. 2 B, orange). The TMEM16A channel has a structured extracellular domain that is absent in the nhTMEM16 scramblase (Fig. 2 A, red). This domain is stabilized by four pairs of cysteine residues engaged in constitutive disulfide bonds, explaining why mutation of these residues results in nonfunctional channels (Yu et al., 2012). Each TMEM16 monomer forms an independent substrate translocation pathway, called the subunit cavity, whose opening is regulated by two Ca2+-binding sites (Fig. 2, A and B, Ca2+ ions pink; Fig. 2, C and D). This pathway is formed by TM3–7 of each monomer and is hydrophilic, even though it is at least in part exposed to the hydrophobic core of the membrane (Fig. 2 A and B, green). These pathways have a conserved hourglass shape, where a wide intracellular vestibule and a smaller extracellular accessway are separated by a narrow constriction located just above the Ca2+-binding sites (Figs. 3 A and 4 A, green box). Multiple converging lines of evidence support the notion that this groove-like structure serves as the nonselective translocation pathway for phospholipids and ions in the TMEM16 scramblases and forms the anion-selective pore through which Cl− ions diffuse in the CaCCs (Brunner et al., 2014; Dang et al., 2017; Jiang et al., 2017; Paulino et al., 2017a,b).
Figure 2.
TMEM16 architecture. (A and B) The structures the mTMEM16A Cl− channel (A) and the nhTMEM16 scramblase (B) with two bound Ca2+ ions are viewed from the plane of the membrane. For clarity, one monomer is shown in gray. The cytosolic domain is in orange, the permeation pathway is in green, the extracellular domain in TMEM16A is in red, Ca2+ ions are in pink, and the remainder of the protein is in blue. (C and D) Close-up view of the Ca2+-binding site of TMEM16A (C) and nhTMEM16 (D). Conserved acidic residues are shown as yellow sticks, and polar residues are shown as green sticks. (E) Sequence conservation of the Ca2+-binding sites. Alignment of the regions shown in C and D of select TMEM16 family members. Coordinating acidic residues are highlighted in yellow, and polar residues in green. PDB accession nos.: 5OYB (mTMEM16A) and 4WIS (nhTMEM16).
Figure 3.
The mTMEM16A pore. (A) Close-up view of the mTMEM16A pore. Basic residues lining the pore are shown as blue spheres, the neck constriction region is shown as a green shaded area, and key residues are shown as green sticks (Paulino et al., 2017a). (B) Residues important for ion permeation and selectivity: positions within the pore are shown as orange spheres; positions outside the pore that play a role in permeation are shown in cyan (Yu et al., 2012; Dang et al., 2017; Paulino et al., 2017a,b). (C) Structural alignment of the Ca2+-bound structure of mTMEM16A (gray) with the zero-Ca2+ structure (pink). TM6 bends into the pore around G644, shown as a yellow sphere. Green spheres are residues important for Ca2+ gating (Dang et al., 2017; Paulino et al., 2017a).
Figure 4.
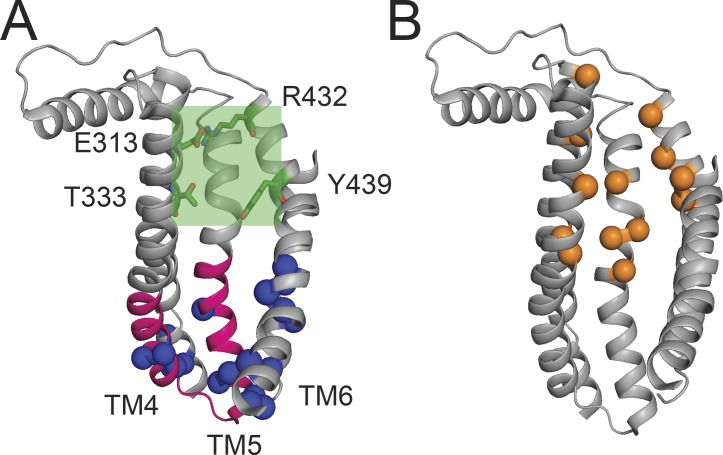
The lipid pathway of nhTMEM16. (A) Ribbon representation of the lipid pathway of nhTMEM16. Charged residues forming the intracellular lipid binding site are shown as blue spheres, the “scramblase domain” is shown in pink, and the narrowest region of the pathway is shown as shaded green box. Residues important in controlling this narrow region are shown as green sticks. (B) Residues important for lipid permeation (orange spheres; Bethel and Grabe, 2016; Jiang et al., 2017; Lee et al., 2018).
The TMEM16 transmembrane dimer interface is small, formed only by the interaction of the extracellular portion of TM10 from each monomer via two salt bridges and hydrophobic contacts (Fig. 2, A and B). In the nhTMEM16 scramblase, contact between the monomers mostly occurs in the cytosolic region, via extensive interactions between the protein’s N and C termini (Fig. 2 B). In contrast, in the TMEM16A channel this interaction is nearly abolished (Fig. 2 A), giving rise to a minimalistic dimer interface, mediated only by TM10, which buries only 1.3% of the protein (Paulino et al., 2017a). This arrangement gives rise to two large hydrophobic cavities along the major axis of the protein that could play a role in lipid binding and regulation of TMEM16 activity (Brunner et al., 2014; Dang et al., 2017; De Jesús-Pérez et al., 2018).
Structural basis of Ca2+ binding
Each TMEM16 monomer contains two evolutionarily conserved Ca2+ binding sites (Fig. 2, C and D, S1 and S2) situated in close proximity to the substrate permeation pathway, with which they share some structural constituents (Fig. 2, A and B). In remarkable agreement with early electrophysiological predictions (Arreola et al., 1995; Hartzell et al., 2005; Cruz-Rangel et al., 2015), the Ca2+-binding sites are located approximately one third of the way through the membrane, rationalizing the synergistic effects of voltage and Ca2+ binding on channel opening. These sites are formed by five highly conserved residues on TM6–8 (Fig. 2 E), consistent with electrophysiological data (Yu et al., 2012; Jeng et al., 2016) as well one to three variable ones, which might contribute to the modulation of the apparent affinities for Ca2+ in different family members (Caputo et al., 2008; Schroeder et al., 2008; Yang et al., 2008, 2012; Scudieri et al., 2013; Yu et al., 2015; Di Zanni et al., 2017; Jiang et al., 2017). The S1 binding site is closest to the intracellular solution, and a Ca2+ ion there is directly coordinated by three conserved acidic residues, E654, E702, and D738 in mouse TMEM16A (Fig. 2 C). A fourth acidic residue, E734, is shared between S1 and S2, which is also lined by the fifth conserved acidic residue, E705, as well as by three asparagine residues, N650, 651, and 730 (Fig. 2 C). The overall arrangement of the Ca2+-binding sites in the nhTMEM16 scramblase is very similar, with the main difference that one of the three asparagines coordinating S2 is replaced by a phenylalanine (Fig. 2 D). Indeed, mutation of any of these residues dramatically reduces the apparent affinity of the TMEM16s for Ca2+, supporting their role in Ca2+ binding (Yang et al., 2012; Yu et al., 2012; Malvezzi et al., 2013; Terashima et al., 2013; Brunner et al., 2014; Lim et al., 2016; Paulino et al., 2017a).
In the structural models of Ca2+-bound nhTMEM16 and TMEM16A, the pore-lining helix TM6 is distorted to accommodate the favorable interactions of N650 in TMEM16A (N448 in nhTMEM16) with the Ca2+ ion in the S2 site (Fig. 2 C). This favors the formation of a π-helix bulge one (two in nhTMEM16) helical turn above S2. Notably, in the TMEM16A structure in nanodiscs, TM6 is built as a slightly bent regular α-helix (Dang et al., 2017), resulting in the structural misalignment of the extracellular portions of TM6, which are off register by one-half helical turn. This causes a critical glycine residue (G644) to face the core of the protein or the permeation pathway in the two structures. It remains to be seen whether these discrepancies reflect different conformational states of TM6 and if they play a role in the gating rearrangements of the pathway.
Ion permeation and Ca2+ gating of the TMEM16A ion channel
The structure of the Ca2+-bound TMEM16A shows that the subunit cavity forms a pore directly lined by TM4–7, with minimal participation of TM3 (Fig. 3 A). Multiple basic residues in the intra- and extracellular vestibules make the pore electropositive, suggesting that this channel’s anion selectivity is at least in part caused by an electrostatic discrimination against cations (Fig. 3 A, blue spheres). When at least one Ca2+-binding site is occupied, the intracellular vestibule is wide, ∼30 Å across, possibly exposing part of the ion permeation pathway to the membrane. The extracellular portions of TM4 and TM6 come in close contact, isolating the ion permeation pore from the hydrocarbon core of the membrane and forming a proteinaceous pore that spans approximately half of the membrane (Fig. 3 A, shaded green). The central constriction defines the narrowest portion of the pore, which is only ∼2.5 Å across and thus smaller than the permeating ions (Fig. 3 A, green area), suggesting that the pore is closed. No chloride-binding sites have been identified so far, suggesting that the selectivity filter of TMEM16A is formed by multiple residues scattered throughout the length of the pore, rather than by a short stretch of conserved amino acids, as seen in most other channels (Doyle et al., 1998; Morais Cabral et al., 1998; Dutzler et al., 2002; Payandeh et al., 2011; Gao et al., 2016). Indeed, extensive mutagenesis of the amphipathic side chains lining the pore identified several positions that modulate ion permeation and selectivity, but none that are strictly essential for either process (Fig. 3 B, orange spheres). Ion permeation through TMEM16A is well described by a permeation model with three barriers but no deep binding sites (Paulino et al., 2017a), further supporting this idea. Interestingly, MTS modification of substituted cysteines within the TM5–6 loop, outside of the pore, suggested that this region is important for TMEM16A function and that it lines the extracellular vestibule and ion pathway (Yu et al., 2012). Some of the MTS-induced changes in the currents could reflect long-range electrostatic interactions of the added charges with the permeating ions. However, it is also possible that this region adopts a different conformation when the pore opens. Indeed, in all available TMEM16A structures, the pore is in a nonconductive state, as the central constriction is too narrow to allow passage of Cl− ions, and TMEM16A currents desensitize/“run down” in the presence of Ca2+ (Dang et al., 2017; Paulino et al., 2017a). This suggests that the central constriction and extracellular vestibule might undergo additional limited rearrangements to open a pathway for Cl− permeation.
The structures of TMEM16A in the presence of zero, one, and two Ca2+ ions revealed that the main consequence of Ca2+ unbinding from S1 and S2 is a pronounced rearrangement of TM6, although the rest of the protein remains largely unchanged (Dang et al., 2017; Paulino et al., 2017a). In the absence of Ca2+, the intracellular portion of TM6 bends around a glycine hinge residue, G644, moving away from TM7 and resulting in a pronounced narrowing of the intracellular vestibule and the pore constriction adjacent to the Ca2+-binding sites (Fig. 3 C). Alanine mutation of several residues near the glycine hinge (Fig. 3 C, green spheres) shifts the EC50 for Ca2+, further cementing the role of this region in Ca2+-dependent gating (Dang et al., 2017; Paulino et al., 2017a; Peters et al., 2018). Indeed, the TM6 helix appears to serve as a key allosteric nexus through which Ca2+, voltage, and extracellular Cl− act to regulate opening of the TMEM16A pore (Peters et al., 2018). Upon Ca2+ release, the N-terminal end of TM6 becomes unstructured, suggesting that this region might undergo a ligand-dependent order-to-disorder transition (Dang et al., 2017; Paulino et al., 2017a). The movement of TM6 disrupts the S1 binding site and opens a water-filled pathway directly connecting S2 to the intracellular solution, allowing the exchange of Ca2+. In addition to these structural rearrangements, unbinding of Ca2+ alters the electrostatic environment of the pathway, making it significantly more electronegative. This likely impedes permeation of the negatively charged Cl− ions (Paulino et al., 2017a). Thus, closing of the TMEM16A pore is mediated by a combination of steric and electrostatic effects. It is not known whether the lipid pathway of the scramblases undergoes a similar rearrangement of TM6 upon Ca2+ release. Notably, in the scramblases the glycine hinge is missing (Paulino et al., 2017a), raising the possibility that the lipid permeation might be regulated by a different conformational rearrangement. However, the remarkable structural and sequence conservation of the residues coordinating S1 and S2 strongly supports the hypothesis that the bases for Ca2+-sensing are evolutionarily conserved in these functionally diverse proteins.
The lipid pathway of the TMEM16 scramblases
In the nhTMEM16 scramblase, the subunit cavity is formed by TM3–7, is hydrophilic, and is open to the membrane core throughout its length. These unusual characteristics suggested that it might serve as the lipid permeation pathway (Brunner et al., 2014), a hypothesis that has found vast experimental and computational support (Yu et al., 2015; Bethel and Grabe, 2016; Gyobu et al., 2017; Jiang et al., 2017; Lee et al., 2018). The intracellular vestibule of the groove is wide, ∼35 Å across, and decorated with numerous charges (Fig. 4 A, blue spheres), which were proposed to help attract lipid headgroups from the intracellular leaflet and possibly form an intracellular lipid-binding site (Bethel and Grabe, 2016; Jiang et al., 2017). The portions of TM4 and TM5 lining the intracellular vestibule form the “scrambling domain” (Fig. 4 A, pink; Gyobu et al., 2015; Yu et al., 2015), supporting the role of this region in lipid scrambling. The extracellular vestibule (Fig. 4 A, green box, green sticks) is lined by charged residues whose interactions tether TM3 to TM6 (Fig. 4 A, E313/R432) and form the extracellular lipid-binding site (Bethel and Grabe, 2016; Jiang et al., 2017). These vestibules are separated by a constriction induced by bends in TM4 and TM6, which bring the side chains of T333 and Y439 within ∼7 Å of each other above the S2 Ca2+-binding site (Fig. 4 A). This narrowing is too small to allow the unhindered passage of lipid headgroups and extends vertically for ∼8 Å (Fig. 4 A). Multiple side chains within the vicinity of this constriction have been shown to be functionally important for scrambling (Fig. 4 B, orange spheres; Bethel and Grabe, 2016; Jiang et al., 2017; Lee et al., 2018), highlighting its critical role in lipid permeation.
The generally accepted mechanism for lipid scrambling by the TMEM16s closely resembles the so-called “credit card” model (Pomorski and Menon, 2006): the lipid headgroups enter and diffuse through the hydrophilic groove while their acyl tails remain embedded in the hydrocarbon core of the membrane (Fig. 5 B; Brunner et al., 2014; Bethel and Grabe, 2016; Jiang et al., 2017; Lee et al., 2018). Although the intra- and extracellular vestibules are sufficiently wide to allow lipids to partition into the permeation pathway, the central region is too narrow to allow passage of the headgroups. This could suggest that the constriction of the nhTMEM16 groove, like the TMEM16A pore, is in a nonconductive, desensitized state. However, no decrease of activity in the presence of ligand (i.e., desensitization) has been described for the TMEM16 scramblases (Suzuki et al., 2010; Malvezzi et al., 2013; Brunner et al., 2014; Yu et al., 2015). Alternatively, the groove could be in a nonconductive conformation because of the absence of a membrane-like environment in the crystallization conditions, like what is seen in the pore of K+ channels in the absence of the permeating ions (Zhou et al., 2001).
Figure 5.
Structural differences between the TMME16 ion and lipid pores. (A) Structural alignment of nhTMEM16 (gray) and TMEM16A (pink) pathways. (B) Credit card model of lipid scrambling. (C) Alignment of the permeation pathways of the high-resolution structures of nhTMEM16 and TMEM16A highlighting the different tilts of TM4. The TM3 and TM5 from each structure are shown as gray ribbons, TM6 are shown as black ribbons, and TM4 are shown in color. Colors of TM4 are as follows: nhTMEM16 is red, mTMEM16A +Ca2+ is orange, mTMEM16A −Ca2+ is yellow, mTMEM16A in nanodiscs is green, and mTMEM16A in Lauryl Maltose Neopentyl Glycol is blue. (D) Cartoon depicting the ‘dynamic switch’ model for the dual-function activity of the TMEM16 scramblases. See text for details.
Two studies have indeed proposed that this central constriction undergoes some conformational rearrangements that control lipid scrambling (Jiang et al., 2017; Lee et al., 2018) but differ in the stimuli triggering these rearrangements. Originally, it was proposed that this constriction could form the Ca2+-dependent gate of the scramblases, because MD simulations suggested it might narrow in the absence of Ca2+ (Jiang et al., 2017). However, this would predict different Ca2+-dependent rearrangements than those undergone by TMEM16A (Dang et al., 2017; Paulino et al., 2017a; Fig. 3 C). More recently, we proposed that the widening of the central constriction is part of a concerted rearrangement undergone by the pathway (Lee et al., 2018), which is triggered by the interactions of lipids with the E318/R313/R432 cluster of residues in the extracellular vestibule (Lee et al., 2018). We proposed that interactions of lipids with R432 promote its disengagement from the E313/E318 pair and unlatching TM6 from TM3. This allows TM6 to rotate, moving the Y439 side chain out of the pathway. This opens the central constriction so that water molecules and lipids can penetrate into and freely diffuse through it (Lee et al., 2018). Further work will be required to elucidate the conformational rearrangements undergone by the groove during lipid scrambling.
Does ion permeation through nhTMEM16 occur through a lipid-lined pathway?
The overall shape of the nhTMEM16 groove and the TMEM16A pore is similar, and the relatively small structural differences suggest a remarkably simple explanation for their functional diversity: the extracellular half of the TMEM16A pore is narrower and excludes the lipid headgroups from the pathway. The pore-lining TM3, 5, 6, and 7 from the two proteins overlay quite well (Fig. 5 A); however, in the TMEM16A channel the extracellular portion of TM4 slides in front of TM3 (Fig. 5, C and D), closing off the groove and generating the proteinaceous pore necessary for ion permeation. Interestingly, the TMEM16A structures suggest that the position of TM3 and TM4 is not fixed: three conformations of TM3 with different tilts were identified (Paulino et al., 2017a), and TM4 tilts at different angles depending on the number of Ca2+ ions bound (Dang et al., 2017; Paulino et al., 2017a; Fig. 5, C and D). These differences generate pathways with varying degrees of closure (Fig. 5 C). Furthermore, by comparing the TMEM16A and nhTMEM16 structures, it becomes apparent that TM4 is the only helix that moves significantly (Fig. 5 A) and that its positions suggest a transition pathway between the most open pore (Fig. 5, C and D, nhTMEM16 in red) and the narrowest one (Fig. 5, C and D, TMEM16A in blue).
Although the functional correlates of these structural rearrangements are not known, we are tempted to speculate that they might underlie one of the most surprising properties of the TMEM16 scramblases, their ability to mediate Ca2+-dependent ion transport. The current accepted model is that the ions move along the TMEM16 groove together with the lipids (Whitlock and Hartzell, 2016; Jiang et al., 2017), with the lipid headgroups providing a sufficiently hydrophilic environment for the ions. This idea is appealing in its provoking simplicity: a single pore, part-lipid and part-protein, underlies both transport functions of the TMEM16 scramblases. However, this model encounters some difficulties in reconciling three experimental observations. First, the pores of all known ion channel proteins shield the permeating ions from the hydrocarbon core of the membrane (Hille, 2001). On occasion, lipid-exposed fenestrations have been described but only to allow entry of phospholipid headgroups, not their tails (Payandeh et al., 2011; Brohawn et al., 2012, 2013; Miller and Long, 2012; Aryal et al., 2014). For the proposed lipid-lined channel to provide a similarly hydrophilic environment, the lipid headgroups would need to form a well-ordered single-file “shield” separating the ions from the membrane’s core. However, several converging lines of evidence suggest that scrambling is not an ordered, single-file process, but rather that it entails major rearrangements of the membrane around the groove so that lipid headgroups and tails interact with residues within and around the pathway (Bethel and Grabe, 2016; Jiang et al., 2017; Lee et al., 2018; Malvezzi et al., 2018). Second, the channel activity of purified TMEM16 scramblases is inhibited in some membrane compositions where lipid transport occurs (Malvezzi et al., 2013; Brunner et al., 2014; Lee et al., 2016). If a single water-filled pathway mediates transport of both substrates, then how can certain lipid compositions be permissive for one activity but not the other? Third, if the channel activity of the TMEM16 scramblases was enabled by the formation of a continuous file of lipid headgroups between the two leaflets, then we would expect that ion transport should be a general feature of all scramblases. However, not all TMEM16 scramblases have a coupled channel function, rather some like TMEM16C don’t mediate ion transport (Huang et al., 2013; Suzuki et al., 2013b). Further, other scramblases, such as the GPCRs, Xkr proteins, and scrambling peptides, do not function as ion channels. Thus, the proposed “lipid and ion pore” model does not easily explain the complex behavior of scramblases.
We would like to propose an alternative mechanism to explain how some TMEM16 scramblases also transport ions. Inspired by the structural diversity of the TMEM16 pores, we suggest that the dual-function activity of the TMEM16 channel/scramblases corresponds to a switch between an open, scramblase-like pathway and a more closed channel-like one via a rearrangement of TM4 (Fig. 5 E). This “dynamic switch” model accounts for several experimental observations that were otherwise difficult to explain. First, most mutations in the groove affect both functions, but some affect only one (Jiang et al., 2017; Lee et al., 2018). This is consistent with our proposal that ions and lipids go through similar—but not identical—pathways. Second, in our model ions would diffuse through a proteinaceous pore, not dissimilar to that of TMEM16A, accounting for the reported size cut-off on the permeating ions of both fungal and mammalian TMEM16 scramblases (Yang et al., 2012; Malvezzi et al., 2013; Scudieri et al., 2015; Yu et al., 2015; Lee et al., 2016). Conversely, passage of lipids through a hemi-channel should not incur in such size constraints, as has been reported (Malvezzi et al., 2018). Third, the movement of TM4 would require and depend on a rearrangement of the TM4–5 interface, accounting for the remarkable conversion of the TMEM16A channel into a scramblase via mutations in the “scramblase domain” and at the TM4–5 interface (Gyobu et al., 2015; Yu et al., 2015; Jiang et al., 2017). In the TMEM16 homologues that are only scramblases, such as TMEM16C or D (Huang et al., 2013; Suzuki et al., 2013b), these hypothetical rearrangements are inhibited, so that their lipid pathways cannot adopt a channel-like conformation. Fourth, the differential modulation of the channel and scramblase activities by lipids could be explained if the movement of TM4 was controlled by specific interactions with the lipid headgroups. Finally, within this framework, the TMEM16 channels would have evolved from the scramblases through mutations that stabilize the channel conformation of the pore. Although our idea has the merit of being consistent with several experimental observations, it is unburdened by the constraints of experimental tests and therefore should be taken as a conceptual model rather than a mechanistic proposal.
Conclusions and outlook
Phospholipid scrambling and ion transport have been regarded as mechanistically dissimilar, and the proteins that mediate them were thought to be structurally unrelated. However, the two processes are fundamentally similar: both entail the movement of charged or polar molecules—a lipid headgroup or an ion—across the hydrocarbon core of a lipid bilayer. To achieve this, scramblases and ion channels have developed similar solutions, because they create a transmembrane hydrophilic cavity through which their respective substrates diffuse down an electrochemical gradient. Thus, scramblases can be viewed as lipid channels. The only difference between a channel for ions and one for lipids is that in the former the pore is surrounded by protein, whereas in the latter an opening to the membrane is needed so that the lipid tails can access a hydrophobic environment. Considering these functional parallels, the existence of proteins straddling the two functions should not have been wholly unexpected.
The identification of the TMEM16s as CaCCs and scramblases and the determination of atomic-resolution structures of homologues from both subtypes allowed us to take the first steps toward unraveling the many questions surrounding their functions and mechanisms of action. These initial glimpses outline a fascinating puzzle, but the entire picture remains blurry. The TMEM16 scramblases regulate a wider-than-anticipated range of signaling pathways, raising the question of which cellular factors fine-tune their function to fill these diverse niches. Are these scramblases targeted to different cellular localization in different cells types? If so, is the local exposure of PS the main signal? Alternatively, does the ion transport mediated by the dual-function channel-scramblases also play a role? What molecules modulate their kinetics and apparent Ca2+ sensitivity in cells? The structures of TMEM16A and nhTMEM16 suggested conserved mechanisms for Ca2+-dependent activation, as well as of ion and lipid permeation. However, key conformations of both proteins remain unknown. What do the open pore of a TMEM16 channel and the closed pathway of a TMEM16 scramblase look like? Are minor movements sufficient, or do extensive rearrangements occur? Remarkably, none of the structures revealed the location of binding sites for the transported substrates. Do the TMEM16s really lack defined binding sites? Finally, the wide, membrane-exposed groove of nhTMEM16 suggested that scrambling occurs via a credit-card–like mechanism. Is this a general paradigm for all scramblases? Other scramblases lack such an obvious groove, suggesting other mechanisms might be at play. In conclusion, in the 10 yr since their initial discovery, the TMEM16s have fundamentally changed our way of thinking about the physiology of lipid scrambling, of ligand-gating of membrane proteins, and of ion and lipid permeation. We are excited to see the new surprises that the next 10 yr will bring.
Acknowledgments
This work was supported by National Institutes of Health grant GM106717 and an Irma T. Hirschl/Monique Weill-Caulier Scholar Award (to A. Accardi).
The authors declare no competing financial interests.
Author contributions: All authors conceived research and wrote and edited the manuscript.
Lesley C. Anson served as editor.
References
- Almaça J., Tian Y., Aldehni F., Ousingsawat J., Kongsuphol P., Rock J.R., Harfe B.D., Schreiber R., and Kunzelmann K.. 2009. TMEM16 proteins produce volume-regulated chloride currents that are reduced in mice lacking TMEM16A. J. Biol. Chem. 284:28571–28578. 10.1074/jbc.M109.010074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arreola J., Melvin J.E., and Begenisich T.. 1995. Inhibition of Ca(2+)-dependent Cl- channels from secretory epithelial cells by low internal pH. J. Membr. Biol. 147:95–104. 10.1007/BF00235400 [DOI] [PubMed] [Google Scholar]
- Aryal P., Abd-Wahab F., Bucci G., Sansom M.S.P., and Tucker S.J.. 2014. A hydrophobic barrier deep within the inner pore of the TWIK-1 K2P potassium channel. Nat. Commun. 5:4377 10.1038/ncomms5377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baig A.A., Haining E.J., Geuss E., Beck S., Swieringa F., Wanitchakool P., Schuhmann M.K., Stegner D., Kunzelmann K., Kleinschnitz C., et al. 2016. TMEM16F-mediated platelet membrane phospholipid scrambling is critical for hemostasis and thrombosis but not thromboinflammation in mice-brief report. Arterioscler. Thromb. Vasc. Biol. 36:2152–2157. 10.1161/ATVBAHA.116.307727 [DOI] [PubMed] [Google Scholar]
- Balasubramanian K., and Schroit A.J.. 2003. Aminophospholipid asymmetry: A matter of life and death. Annu. Rev. Physiol. 65:701–734. 10.1146/annurev.physiol.65.092101.142459 [DOI] [PubMed] [Google Scholar]
- Barnes S., and Deschênes M.C.. 1992. Contribution of Ca and Ca-activated Cl channels to regenerative depolarization and membrane bistability of cone photoreceptors. J. Neurophysiol. 68:745–755. 10.1152/jn.1992.68.3.745 [DOI] [PubMed] [Google Scholar]
- Batti L., Sundukova M., Murana E., Pimpinella S., De Castro Reis F., Pagani F., Wang H., Pellegrino E., Perlas E., Di Angelantonio S., et al. 2016. TMEM16F regulates spinal microglial function in neuropathic pain states. Cell Reports. 15:2608–2615. 10.1016/j.celrep.2016.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bera T.K., Das S., Maeda H., Beers R., Wolfgang C.D., Kumar V., Hahn Y., Lee B., and Pastan I.. 2004. NGEP, a gene encoding a membrane protein detected only in prostate cancer and normal prostate. Proc. Natl. Acad. Sci. USA. 101:3059–3064. 10.1073/pnas.0308746101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethel N.P., and Grabe M.. 2016. Atomistic insight into lipid translocation by a TMEM16 scramblase. Proc. Natl. Acad. Sci. USA. 113:14049–14054. 10.1073/pnas.1607574113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevers E.M., and Williamson P.L.. 2010. Phospholipid scramblase: An update. FEBS Lett. 584:2724–2730. 10.1016/j.febslet.2010.03.020 [DOI] [PubMed] [Google Scholar]
- Bevers E.M., and Williamson P.L.. 2011. Phospholipid scramblase: When phospholipid asymmetry goes away. In Transmembrane Dynamics of Lipids. Devaux P., and Herrmann A., editors. Wiley, New York: 119–146. 10.1002/9781118120118.ch7 [DOI] [Google Scholar]
- Boccaccio A., and Menini A.. 2007. Temporal development of cyclic nucleotide-gated and Ca2+ -activated Cl- currents in isolated mouse olfactory sensory neurons. J. Neurophysiol. 98:153–160. 10.1152/jn.00270.2007 [DOI] [PubMed] [Google Scholar]
- Boisseau P., Bene M.C., Besnard T., Pachchek S., Giraud M., Talarmain P., Robillard N., Gourlaouen M.A., Bezieau S., and Fouassier M.. 2016. A new mutation of ANO6 in two familial cases of Scott syndrome. Br. J. Haematol. 180:750–752. [DOI] [PubMed] [Google Scholar]
- Bolduc V., Marlow G., Boycott K.M., Saleki K., Inoue H., Kroon J., Itakura M., Robitaille Y., Parent L., Baas F., et al. 2010. Recessive mutations in the putative calcium-activated chloride channel Anoctamin 5 cause proximal LGMD2L and distal MMD3 muscular dystrophies. Am. J. Hum. Genet. 86:213–221. 10.1016/j.ajhg.2009.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohawn S.G., del Mármol J., and MacKinnon R.. 2012. Crystal structure of the human K2P TRAAK, a lipid- and mechano-sensitive K+ ion channel. Science. 335:436–441. 10.1126/science.1213808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohawn S.G., Campbell E.B., and MacKinnon R.. 2013. Domain-swapped chain connectivity and gated membrane access in a Fab-mediated crystal of the human TRAAK K+ channel. Proc. Natl. Acad. Sci. USA. 110:2129–2134. 10.1073/pnas.1218950110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks M.B., Catalfamo J.L., MacNguyen R., Tim D., Fancher S., and McCardle J.A.. 2015. A TMEM16F point mutation causes an absence of canine platelet TMEM16F and ineffective activation and death-induced phospholipid scrambling. J. Thromb. Haemost. 13:2240–2252. 10.1111/jth.13157 [DOI] [PubMed] [Google Scholar]
- Brunner J.D., Lim N.K., Schenck S., Duerst A., and Dutzler R.. 2014. X-ray structure of a calcium-activated TMEM16 lipid scramblase. Nature. 516:207–212. 10.1038/nature13984 [DOI] [PubMed] [Google Scholar]
- Brunner J.D., Schenck S., and Dutzler R.. 2016. Structural basis for phospholipid scrambling in the TMEM16 family. Curr. Opin. Struct. Biol. 39:61–70. 10.1016/j.sbi.2016.05.020 [DOI] [PubMed] [Google Scholar]
- Caputo A., Caci E., Ferrera L., Pedemonte N., Barsanti C., Sondo E., Pfeffer U., Ravazzolo R., Zegarra-Moran O., and Galietta L.J.V.. 2008. TMEM16A, a membrane protein associated with calcium-dependent chloride channel activity. Science. 322:590–594. 10.1126/science.1163518 [DOI] [PubMed] [Google Scholar]
- Castoldi E., Collins P.W., Williamson P.L., and Bevers E.M.. 2011. Compound heterozygosity for 2 novel TMEM16F mutations in a patient with Scott syndrome. Blood. 117:4399–4400. 10.1182/blood-2011-01-332502 [DOI] [PubMed] [Google Scholar]
- Cereda V., Poole D.J., Palena C., Das S., Bera T.K., Remondo C., Gulley J.L., Arlen P.M., Yokokawa J., Pastan I., et al. 2010. New gene expressed in prostate: A potential target for T cell-mediated prostate cancer immunotherapy. Cancer Immunol. Immunother. 59:63–71. 10.1007/s00262-009-0723-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamova T., Florez L., Guergueltcheva V., Raycheva M., Kaneva R., Lochmüller H., Kalaydjieva L., and Tournev I.. 2012. ANO10 c.1150_1151del is a founder mutation causing autosomal recessive cerebellar ataxia in Roma/Gypsies. J. Neurol. 259:906–911. 10.1007/s00415-011-6276-6 [DOI] [PubMed] [Google Scholar]
- Chen T.-Y., and Hwang T.-C.. 2008. CLC-0 and CFTR: Chloride channels evolved from transporters. Physiol. Rev. 88:351–387. 10.1152/physrev.00058.2006 [DOI] [PubMed] [Google Scholar]
- Chen Y.Z., Mapes J., Lee E.S., Skeen-Gaar R.R., and Xue D.. 2013. Caspase-mediated activation of Caenorhabditis elegans CED-8 promotes apoptosis and phosphatidylserine externalization. Nat. Commun. 4:2726 10.1038/ncomms3726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H., Yang Y.D., Lee J., Lee B., Kim T., Jang Y., Back S.K., Na H.S., Harfe B.D., Wang F., et al. 2012. The calcium-activated chloride channel anoctamin 1 acts as a heat sensor in nociceptive neurons. Nat. Neurosci. 15:1015–1021. 10.1038/nn.3111 [DOI] [PubMed] [Google Scholar]
- Cobine C.A., Hannah E.E., Zhu M.H., Lyle H.E., Rock J.R., Sanders K.M., Ward S.M., and Keef K.D.. 2017. ANO1 in intramuscular interstitial cells of Cajal plays a key role in the generation of slow waves and tone in the internal anal sphincter. J. Physiol. 595:2021–2041. 10.1113/JP273618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Rangel S., De Jesús-Pérez J.J., Contreras-Vite J.A., Pérez-Cornejo P., Hartzell H.C., and Arreola J.. 2015. Gating modes of calcium-activated chloride channels TMEM16A and TMEM16B. J. Physiol. 593:5283–5298. 10.1113/JP271256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang S., Feng S., Tien J., Peters C.J., Bulkley D., Lolicato M., Zhao J., Zuberbühler K., Ye W., Qi L., et al. 2017. Cryo-EM structures of the TMEM16A calcium-activated chloride channel. Nature. 552:426–429. 10.1038/nature25024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S., Hahn Y., Nagata S., Willingham M.C., Bera T.K., Lee B., and Pastan I.. 2007. NGEP, a prostate-specific plasma membrane protein that promotes the association of LNCaP cells. Cancer Res. 67:1594–1601. 10.1158/0008-5472.CAN-06-2673 [DOI] [PubMed] [Google Scholar]
- De Jesús-Pérez J.J., Cruz-Rangel S., Espino-Saldaña Á.E., Martínez-Torres A., Qu Z., Hartzell H.C., Corral-Fernandez N.E., Pérez-Cornejo P., and Arreola J.. 2018. Phosphatidylinositol 4,5-bisphosphate, cholesterol, and fatty acids modulate the calcium-activated chloride channel TMEM16A (ANO1). Biochim. Biophys. Acta. 1863:299–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon R.E., Hennig G.W., Baker S.A., Britton F.C., Harfe B.D., Rock J.R., Sanders K.M., and Ward S.M.. 2012. Electrical slow waves in the mouse oviduct are dependent upon a calcium activated chloride conductance encoded by Tmem16a1. Biol. Reprod. 86:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Zanni E., Gradogna A., Scholz-Starke J., and Boccaccio A.. 2017. Gain of function of TMEM16E/ANO5 scrambling activity caused by a mutation associated with gnathodiaphyseal dysplasia. Cell. Mol. Life Sci. 75:1657–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle D.A., Morais Cabral J., Pfuetzner R.A., Kuo A., Gulbis J.M., Cohen S.L., Chait B.T., and MacKinnon R.. 1998. The structure of the potassium channel: Molecular basis of K+ conduction and selectivity. Science. 280:69–77. 10.1126/science.280.5360.69 [DOI] [PubMed] [Google Scholar]
- Duran C., Qu Z., Osunkoya A.O., Cui Y., and Hartzell H.C.. 2012. ANOs 3-7 in the anoctamin/Tmem16 Cl- channel family are intracellular proteins. Am. J. Physiol. Cell Physiol. 302:C482–C493. 10.1152/ajpcell.00140.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutzler R., Campbell E.B., Cadene M., Chait B.T., and MacKinnon R.. 2002. X-ray structure of a ClC chloride channel at 3.0 A reveals the molecular basis of anion selectivity. Nature. 415:287–294. 10.1038/415287a [DOI] [PubMed] [Google Scholar]
- Duvvuri U., Shiwarski D.J., Xiao D., Bertrand C., Huang X., Edinger R.S., Rock J.R., Harfe B.D., Henson B.J., Kunzelmann K., et al. 2012. TMEM16A induces MAPK and contributes directly to tumorigenesis and cancer progression. Cancer Res. 72:3270–3281. 10.1158/0008-5472.CAN-12-0475-T [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlen H.W., Chinenkova M., Moser M., Munter H.M., Krause Y., Gross S., Brachvogel B., Wuelling M., Kornak U., and Vortkamp A.. 2013. Inactivation of anoctamin-6/Tmem16f, a regulator of phosphatidylserine scrambling in osteoblasts, leads to decreased mineral deposition in skeletal tissues. J. Bone Miner. Res. 28:246–259. 10.1002/jbmr.1751 [DOI] [PubMed] [Google Scholar]
- Ernst O.P., and Menon A.K.. 2015. Phospholipid scrambling by rhodopsin. Photochem. Photobiol. Sci. 14:1922–1931. 10.1039/C5PP00195A [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadok V.A., Voelker D.R., Campbell P.A., Cohen J.J., Bratton D.L., and Henson P.M.. 1992. Exposure of phosphatidylserine on the surface of apoptotic lymphocytes triggers specific recognition and removal by macrophages. J. Immunol. 148:2207–2216. [PubMed] [Google Scholar]
- Fallah G., Römer T., Detro-Dassen S., Braam U., Markwardt F., and Schmalzing G.. 2011. TMEM16A(a)/anoctamin-1 shares a homodimeric architecture with CLC chloride channels. Mol. Cell. Proteomics. 10:M110.004697. [DOI] [PMC free article] [PubMed]
- Gao Y., Cao E., Julius D., and Cheng Y.. 2016. TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Nature. 534:347–351. 10.1038/nature17964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert R.J.C., Dalla Serra M., Froelich C.J., Wallace M.I., and Anderluh G.. 2014. Membrane pore formation at protein-lipid interfaces. Trends Biochem. Sci. 39:510–516. 10.1016/j.tibs.2014.09.002 [DOI] [PubMed] [Google Scholar]
- Goren M.A., Morizumi T., Menon I., Joseph J.S., Dittman J.S., Cherezov V., Stevens R.C., Ernst O.P., and Menon A.K.. 2014. Constitutive phospholipid scramblase activity of a G protein-coupled receptor. Nat. Commun. 5:5115 10.1038/ncomms6115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin D.A., Johnson R.W., Whitlock J.M., Pozsgai E.R., Heller K.N., Grose W.E., Arnold W.D., Sahenk Z., Hartzell H.C., and Rodino-Klapac L.R.. 2016. Defective membrane fusion and repair in Anoctamin5-deficient muscular dystrophy. Hum. Mol. Genet. 25:1900–1911. 10.1093/hmg/ddw063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubb S., Poulsen K.A., Juul C.A., Kyed T., Klausen T.K., Larsen E.H., and Hoffmann E.K.. 2013. TMEM16F (Anoctamin 6), an anion channel of delayed Ca(2+) activation. J. Gen. Physiol. 141:585–600. 10.1085/jgp.201210861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyobu S., Miyata H., Ikawa M., Yamazaki D., Takeshima H., Suzuki J., and Nagata S.. 2015. A role of TMEM16E carrying a scrambling domain in sperm motility. Mol. Cell. Biol. 36:645–659. 10.1128/MCB.00919-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyobu S., Ishihara K., Suzuki J., Segawa K., and Nagata S.. 2017. Characterization of the scrambling domain of the TMEM16 family. Proc. Natl. Acad. Sci. USA. 114:6274–6279. 10.1073/pnas.1703391114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartzell C., Putzier I., and Arreola J.. 2005. Calcium-activated chloride channels. Annu. Rev. Physiol. 67:719–758. 10.1146/annurev.physiol.67.032003.154341 [DOI] [PubMed] [Google Scholar]
- Hartzell H.C., Yu K., Xiao Q., Chien L.T., and Qu Z.. 2009. Anoctamin/TMEM16 family members are Ca2+-activated Cl- channels. J. Physiol. 587:2127–2139. 10.1113/jphysiol.2008.163709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M., Ye W., Wang W.-J., Sison E.S., Jan Y.N., and Jan L.Y.. 2017. Cytoplasmic Cl- couples membrane remodeling to epithelial morphogenesis. Proc. Natl. Acad. Sci. USA. 114:E11161–E11169. 10.1073/pnas.1714448115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks D., Sarkozy A., Muelas N., Köehler K., Huebner A., Hudson G., Chinnery P.F., Barresi R., Eagle M., Polvikoski T., et al. 2011. A founder mutation in Anoctamin 5 is a major cause of limb-girdle muscular dystrophy. Brain. 134:171–182. 10.1093/brain/awq294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille B. 2001. Ion channels of excitable membranes. 3rd edition. Sinauer, Sunderland, Mass. 814 pp. [Google Scholar]
- Holthuis J.C.M., and Levine T.P.. 2005. Lipid traffic: Floppy drives and a superhighway. Nat. Rev. Mol. Cell Biol. 6:209–220. 10.1038/nrm1591 [DOI] [PubMed] [Google Scholar]
- Hu Y., Kim J.H., He K., Wan Q., Kim J., Flach M., Kirchhausen T., Vortkamp A., and Winau F.. 2016. Scramblase TMEM16F terminates T cell receptor signaling to restrict T cell exhaustion. J. Exp. Med. 213:2759–2772. 10.1084/jem.20160612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F., Rock J.R., Harfe B.D., Cheng T., Huang X., Jan Y.N., and Jan L.Y.. 2009. Studies on expression and function of the TMEM16A calcium-activated chloride channel. Proc. Natl. Acad. Sci. USA. 106:21413–21418. 10.1073/pnas.0911935106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F., Wong X., and Jan L.Y.. 2012a International Union of Basic and Clinical Pharmacology. LXXXV: Calcium-activated chloride channels. Pharmacol. Rev. 64:1–15. 10.1124/pr.111.005009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F., Wang X., Ostertag E.M., Nuwal T., Huang B., Jan Y.-N., Basbaum A.I., and Jan L.Y.. 2013. TMEM16C facilitates Na(+)-activated K+ currents in rat sensory neurons and regulates pain processing. Nat. Neurosci. 16:1284–1290. 10.1038/nn.3468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W.C., Xiao S., Huang F., Harfe B.D., Jan Y.N., and Jan L.Y.. 2012b Calcium-activated chloride channels (CaCCs) regulate action potential and synaptic response in hippocampal neurons. Neuron. 74:179–192. 10.1016/j.neuron.2012.01.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang W., Kim J.Y., Cui S., Jo J., Lee B.C., Lee Y., Kwon K.S., Park C.S., and Kim C.. 2015. The anoctamin family channel subdued mediates thermal nociception in Drosophila. J. Biol. Chem. 290:2521–2528. 10.1074/jbc.M114.592758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeng G., Aggarwal M., Yu W.-P., and Chen T.-Y.. 2016. Independent activation of distinct pores in dimeric TMEM16A channels. J. Gen. Physiol. 148:393–404. 10.1085/jgp.201611651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang T., Yu K., Hartzell H.C., and Tajkhorshid E.. 2017. Lipids and ions traverse the membrane by the same physical pathway in the nhTMEM16 scramblase. Elife. 6:1–26. 10.7554/eLife.28671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J., and Lee M.G.. 2015. Does calmodulin regulate the bicarbonate permeability of ANO1/TMEM16A or not? J. Gen. Physiol. 145:75–77. 10.1085/jgp.201411283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Kim H., Lee J., Lee B., Kim H.-R., Jung J., Lee M.-O., and Oh U.. 2018. Anoctamin 9/TMEM16J is a cation channel activated by cAMP/PKA signal. Cell Calcium. 71:75–85. 10.1016/j.ceca.2017.12.003 [DOI] [PubMed] [Google Scholar]
- Kleene S.J. 1993. Origin of the chloride current in olfactory transduction. Neuron. 11:123–132. 10.1016/0896-6273(93)90276-W [DOI] [PubMed] [Google Scholar]
- Kmit A., van Kruchten R., Ousingsawat J., Mattheij N.J., Senden-Gijsbers B., Heemskerk J.W., Schreiber R., Bevers E.M., and Kunzelmann K.. 2013. Calcium-activated and apoptotic phospholipid scrambling induced by Ano6 can occur independently of Ano6 ion currents. Cell Death Dis. 4:e611 10.1038/cddis.2013.135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kol M.A., van Dalen A., de Kroon A.I.P.M., and de Kruijff B.. 2003a Translocation of phospholipids is facilitated by a subset of membrane-spanning proteins of the bacterial cytoplasmic membrane. J. Biol. Chem. 278:24586–24593. 10.1074/jbc.M301875200 [DOI] [PubMed] [Google Scholar]
- Kol M.A., van Laak A.N.C., Rijkers D.T.S., Killian J.A., de Kroon A.I.P.M., and de Kruijff B.. 2003b Phospholipid flop induced by transmembrane peptides in model membranes is modulated by lipid composition. Biochemistry. 42:231–237. 10.1021/bi0268403 [DOI] [PubMed] [Google Scholar]
- Kramer J., and Hawley R.S.. 2003. The spindle-associated transmembrane protein Axs identifies a new family of transmembrane proteins in eukaryotes. Cell Cycle. 2:174–176. 10.4161/cc.2.3.368 [DOI] [PubMed] [Google Scholar]
- Langer M., Sah R., Veser A., Gütlich M., and Langosch D.. 2013. Structural properties of model phosphatidylcholine flippases. Chem. Biol. 20:63–72. 10.1016/j.chembiol.2012.11.006 [DOI] [PubMed] [Google Scholar]
- Lee B.-C., Menon A.K., and Accardi A.. 2016. The nhTMEM16 scramblase is also a nonselective ion channel. Biophys. J. 111:1919–1924. 10.1016/j.bpj.2016.09.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B.C., Kelashvili G., Falzone M., Menon A.K., Weinstein H., and Accardi A.. 2018. Gating mechanism of the lipid pathway in a TMEM16 scramblase. Nat. Comms. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lhermusier T., Chap H., and Payrastre B.. 2011. Platelet membrane phospholipid asymmetry: From the characterization of a scramblase activity to the identification of an essential protein mutated in Scott syndrome. J. Thromb. Haemost. 9:1883–1891. 10.1111/j.1538-7836.2011.04478.x [DOI] [PubMed] [Google Scholar]
- Li Z., Venegas V., Nagaoka Y., Morino E., Raghavan P., Audhya A., Nakanishi Y., and Zhou Z.. 2015. Necrotic cells actively attract phagocytes through the collaborative action of two distinct PS-exposure mechanisms. PLoS Genet. 11:e1005285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim N.K., Lam A.K.M., and Dutzler R.. 2016. Independent activation of ion conduction pores in the double-barreled calcium-activated chloride channel TMEM16A. J. Gen. Physiol. 148:375–392. 10.1085/jgp.201611650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Linley J.E., Du X., Zhang X., Ooi L., Zhang H., and Gamper N.. 2010. The acute nociceptive signals induced by bradykinin in rat sensory neurons are mediated by inhibition of M-type K+ channels and activation of Ca2+-activated Cl– channels. J. Clin. Invest. 120:1240–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Lu M., Liu B., Huang Y., and Wang K.. 2012. Inhibition of Ca(2+)-activated Cl(-) channel ANO1/TMEM16A expression suppresses tumor growth and invasiveness in human prostate carcinoma. Cancer Lett. 326:41–51. 10.1016/j.canlet.2012.07.015 [DOI] [PubMed] [Google Scholar]
- MacLeish P.R., and Nurse C.A.. 2007. Ion channel compartments in photoreceptors: evidence from salamander rods with intact and ablated terminals. J. Neurophysiol. 98:86–95. 10.1152/jn.00775.2006 [DOI] [PubMed] [Google Scholar]
- Malvezzi M., Chalat M., Janjusevic R., Picollo A., Terashima H., Menon A.K., and Accardi A.. 2013. Ca2+-dependent phospholipid scrambling by a reconstituted TMEM16 ion channel. Nat. Commun. 4:2367 10.1038/ncomms3367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malvezzi M., Andra K.K., Pandey K., Brown A., Iqbal R., Menon A.K., and Accardi A.. 2018. Out of the groove transport of lipids by TMEM16 and GPCR scramblases. PNAS. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marconi C., Brunamonti Binello P., Badiali G., Caci E., Cusano R., Garibaldi J., Pippucci T., Merlini A., Marchetti C., Rhoden K.J., et al. 2013. A novel missense mutation in ANO5/TMEM16E is causative for gnathodiaphyseal dyplasia in a large Italian pedigree. Eur. J. Hum. Genet. 21:613–619. 10.1038/ejhg.2012.224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins J.R., Faria D., Kongsuphol P., Reisch B., Schreiber R., and Kunzelmann K.. 2011. Anoctamin 6 is an essential component of the outwardly rectifying chloride channel. Proc. Natl. Acad. Sci. USA. 108:18168–18172. 10.1073/pnas.1108094108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattheij N.J., Braun A., van Kruchten R., Castoldi E., Pircher J., Baaten C.C., Wülling M., Kuijpers M.J., Köhler R., Poole A.W., et al. 2016. Survival protein anoctamin-6 controls multiple platelet responses including phospholipid scrambling, swelling, and protein cleavage. FASEB J. 30:727–737. 10.1096/fj.15-280446 [DOI] [PubMed] [Google Scholar]
- Menon I., Huber T., Sanyal S., Banerjee S., Barré P., Canis S., Warren J.D., Hwa J., Sakmar T.P., and Menon A.K.. 2011. Opsin is a phospholipid flippase. Curr. Biol. 21:149–153. 10.1016/j.cub.2010.12.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C. 1982. Open-state substructure of single chloride channels from Torpedo electroplax. Philos. Trans. R. Soc. Lond. B Biol. Sci. 299:401–411. 10.1098/rstb.1982.0140 [DOI] [PubMed] [Google Scholar]
- Miller A.N., and Long S.B.. 2012. Crystal structure of the human two-pore domain potassium channel K2P1. Science. 335:432–436. 10.1126/science.1213274 [DOI] [PubMed] [Google Scholar]
- Mohsenzadegan M., Madjd Z., Asgari M., Abolhasani M., Shekarabi M., Taeb J., and Shariftabrizi A.. 2013. Reduced expression of NGEP is associated with high-grade prostate cancers: A tissue microarray analysis. Cancer Immunol. Immunother. 62:1609–1618. 10.1007/s00262-013-1463-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morais Cabral J.H., Lee A., Cohen S.L., Chait B.T., Li M., and Mackinnon R.. 1998. Crystal structure and functional analysis of the HERG potassium channel N terminus: A eukaryotic PAS domain. Cell. 95:649–655. 10.1016/S0092-8674(00)81635-9 [DOI] [PubMed] [Google Scholar]
- Morra G., Razavi A.M., Pandey K., Weinstein H., Menon A.K., and Khelashvili G.. 2018. Mechanisms of lipid scrambling by the G protein-coupled receptor opsin. Structure. 26:356–367.e3. 10.1016/j.str.2017.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata S., Suzuki J., Segawa K., and Fujii T.. 2016. Exposure of phosphatidylserine on the cell surface. Cell Death Differ. 23:952–961. 10.1038/cdd.2016.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Y.-L., Kuan A.-S., and Chen T.-Y.. 2014. Activation and inhibition of TMEM16A calcium-activated chloride channels. PLoS One. 9:e86734 10.1371/journal.pone.0086734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilius B., Szücs G., Heinke S., Voets T., and Droogmans G.. 1997. Multiple types of chloride channels in bovine pulmonary artery endothelial cells. J. Vasc. Res. 34:220–228. 10.1159/000159226 [DOI] [PubMed] [Google Scholar]
- Ousingsawat J., Wanitchakool P., Kmit A., Romao A.M., Jantarajit W., Schreiber R., and Kunzelmann K.. 2015. Anoctamin 6 mediates effects essential for innate immunity downstream of P2X7 receptors in macrophages. Nat. Commun. 6:6245 10.1038/ncomms7245 [DOI] [PubMed] [Google Scholar]
- Paulino C., Kalienkova V., Lam A.K.M., Neldner Y., and Dutzler R.. 2017a Activation mechanism of the calcium-activated chloride channel TMEM16A revealed by cryo-EM. Nature. 552:421–425. 10.1038/nature24652 [DOI] [PubMed] [Google Scholar]
- Paulino C., Neldner Y., Lam A.K.M., Kalienkova V., Brunner J.D., Schenck S., and Dutzler R.. 2017b Structural basis for anion conduction in the calcium-activated chloride channel TMEM16A. Elife. 6:1–23. 10.7554/eLife.26232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payandeh J., Scheuer T., Zheng N., and Catterall W.A.. 2011. The crystal structure of a voltage-gated sodium channel. Nature. 475:353–358. 10.1038/nature10238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedemonte N., and Galietta L.J.V.. 2014. Structure and function of TMEM16 proteins (anoctamins). Physiol. Rev. 94:419–459. 10.1152/physrev.00039.2011 [DOI] [PubMed] [Google Scholar]
- Peters C.J., Yu H., Tien J., Jan Y.N., Li M., and Jan L.Y.. 2015. Four basic residues critical for the ion selectivity and pore blocker sensitivity of TMEM16A calcium-activated chloride channels. Proc. Natl. Acad. Sci. USA. 112:3547–3552. 10.1073/pnas.1502291112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters C.J., Gilchrist J.M., Tien J., Bethel N.P., Qi L., Chen T., Wang L., Jan Y.N., Grabe M., and Jan L.Y.. 2018. The sixth transmembrane segment is a major gating component of the TMEM16A calcium-activated chloride channel. Neuron. 97:1063–1077.e4. 10.1016/j.neuron.2018.01.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietra G., Dibattista M., Menini A., Reisert J., and Boccaccio A.. 2016. The Ca2+-activated Cl- channel TMEM16B regulates action potential firing and axonal targeting in olfactory sensory neurons. J. Gen. Physiol. 148:293–311. 10.1085/jgp.201611622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pifferi S. 2017. Permeation mechanisms in the TMEM16B calcium-activated chloride channels. PLoS One. 12:e0169572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pifferi S., Dibattista M., and Menini A.. 2009. TMEM16B induces chloride currents activated by calcium in mammalian cells. Pflugers Arch. 458:1023–1038. 10.1007/s00424-009-0684-9 [DOI] [PubMed] [Google Scholar]
- Ploier B., Caro L.N., Morizumi T., Pandey K., Pearring J.N., Goren M.A., Finnemann S.C., Graumann J., Arshavsky V.Y., Dittman J.S., et al. 2016. Dimerization deficiency of enigmatic retinitis pigmentosa-linked rhodopsin mutants. Nat. Commun. 7:12832 10.1038/ncomms12832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomorski T., and Menon A.K.. 2006. Lipid flippases and their biological functions. Cell. Mol. Life Sci. 63:2908–2921. 10.1007/s00018-006-6167-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price M.N., Dehal P.S., and Arkin A.P.. 2010. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One. 5:e9490 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quazi F., and Molday R.S.. 2013. Differential phospholipid substrates and directional transport by ATP-binding cassette proteins ABCA1, ABCA7, and ABCA4 and disease-causing mutants. J. Biol. Chem. 288:34414–34426. 10.1074/jbc.M113.508812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaud M., Anheim M., Kamsteeg E.-J., Martial M., Mochel F., Sascha V., Drouot N., Pouget J., Redin C., Salort-Campana E., et al. 2014. Autosomal recessive cerebellar ataxia type 3 due to ANO10 mutations: Delineation and genotype-phenotype correlation study. JAMA Neurol. 71:1305–1310. 10.1001/jamaneurol.2014.193 [DOI] [PubMed] [Google Scholar]
- Rock J.R., Futtner C.R., and Harfe B.D.. 2008. The transmembrane protein TMEM16A is required for normal development of the murine trachea. Dev. Biol. 321:141–149. 10.1016/j.ydbio.2008.06.009 [DOI] [PubMed] [Google Scholar]
- Rock J.R., O’Neal W.K., Gabriel S.E., Randell S.H., Harfe B.D., Boucher R.C., and Grubb B.R.. 2009. Transmembrane protein 16A (TMEM16A) is a Ca2+-regulated Cl- secretory channel in mouse airways. J. Biol. Chem. 284:14875–14880. 10.1074/jbc.C109.000869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanenko V.G., Catalán M.A., Brown D.A., Putzier I., Hartzell H.C., Marmorstein A.D., Gonzalez-Begne M., Rock J.R., Harfe B.D., and Melvin J.E.. 2010. Tmem16A encodes the Ca2+-activated Cl- channel in mouse submandibular salivary gland acinar cells. J. Biol. Chem. 285:12990–13001. 10.1074/jbc.M109.068544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder B.C., Cheng T., Jan Y.N., and Jan L.Y.. 2008. Expression cloning of TMEM16A as a calcium-activated chloride channel subunit. Cell. 134:1019–1029. 10.1016/j.cell.2008.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scudieri P., Sondo E., Caci E., Ravazzolo R., and Galietta L.J.. 2013. TMEM16A-TMEM16B chimaeras to investigate the structure-function relationship of calcium-activated chloride channels. Biochem. J. 452:443–455. 10.1042/BJ20130348 [DOI] [PubMed] [Google Scholar]
- Scudieri P., Caci E., Venturini A., Sondo E., Pianigiani G., Marchetti C., Ravazzolo R., Pagani F., and Galietta L.J.V.. 2015. Ion channel and lipid scramblase activity associated with expression of TMEM16F/ANO6 isoforms. J. Physiol. 593:3829–3848. 10.1113/JP270691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segawa K., Suzuki J., and Nagata S.. 2011. Constitutive exposure of phosphatidylserine on viable cells. Proc. Natl. Acad. Sci. USA. 108:19246–19251. 10.1073/pnas.1114799108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seigneuret M., and Devaux P.F.. 1984. ATP-dependent asymmetric distribution of spin-labeled phospholipids in the erythrocyte membrane: relation to shape changes. Proc. Natl. Acad. Sci. USA. 81:3751–3755. 10.1073/pnas.81.12.3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan J.T., Worthington E.N., Yu K., Gabriel S.E., Hartzell H.C., and Tarran R.. 2011. Characterization of the oligomeric structure of the Ca(2+)-activated Cl- channel Ano1/TMEM16A. J. Biol. Chem. 286:1381–1388. 10.1074/jbc.M110.174847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirianant L., Ousingsawat J., Wanitchakool P., Schreiber R., and Kunzelmann K.. 2016. Cellular volume regulation by anoctamin 6: Ca2+, phospholipase A2 and osmosensing. Pflugers Arch. 468:335–349. 10.1007/s00424-015-1739-8 [DOI] [PubMed] [Google Scholar]
- Stanich J.E., Gibbons S.J., Eisenman S.T., Bardsley M.R., Rock J.R., Harfe B.D., Ordog T., and Farrugia G.. 2011. Ano1 as a regulator of proliferation. Am. J. Physiol. Gastrointest. Liver Physiol. 301:G1044–G1051. 10.1152/ajpgi.00196.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan A.B., Shum E.Y., Hirsh S., Cygnar K.D., Reisert J., and Zhao H.. 2009. ANO2 is the cilial calcium-activated chloride channel that may mediate olfactory amplification. Proc. Natl. Acad. Sci. USA. 106:11776–11781. 10.1073/pnas.0903304106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki J., Umeda M., Sims P.J., and Nagata S.. 2010. Calcium-dependent phospholipid scrambling by TMEM16F. Nature. 468:834–838. 10.1038/nature09583 [DOI] [PubMed] [Google Scholar]
- Suzuki J., Denning D.P., Imanishi E., Horvitz H.R., and Nagata S.. 2013a Xk-related protein 8 and CED-8 promote phosphatidylserine exposure in apoptotic cells. Science. 341:403–406. 10.1126/science.1236758 [DOI] [PubMed] [Google Scholar]
- Suzuki J., Fujii T., Imao T., Ishihara K., Kuba H., and Nagata S.. 2013b Calcium-dependent phospholipid scramblase activity of TMEM16 protein family members. J. Biol. Chem. 288:13305–13316. 10.1074/jbc.M113.457937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki J., Imanishi E., and Nagata S.. 2014. Exposure of phosphatidylserine by Xk-related protein family members during apoptosis. J. Biol. Chem. 289:30257–30267. 10.1074/jbc.M114.583419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki J., Imanishi E., and Nagata S.. 2016. Xkr8 phospholipid scrambling complex in apoptotic phosphatidylserine exposure. Proc. Natl. Acad. Sci. USA. 113:9509–9514. 10.1073/pnas.1610403113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tembo M., and Carlson A.E.. 2018. PIP2 and Ca2+ are Both Required to Open TMEM16a Channels in Xenopus Laevis Oocytes. Biophys. J. 114:610a 10.1016/j.bpj.2017.11.3336 [DOI] [Google Scholar]
- Terashima H., Picollo A., and Accardi A.. 2013. Purified TMEM16A is sufficient to form Ca2+-activated Cl- channels. Proc. Natl. Acad. Sci. USA. 110:19354–19359. 10.1073/pnas.1312014110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas-Gatewood C., Neeb Z.P., Bulley S., Adebiyi A., Bannister J.P., Leo M.D., and Jaggar J.H.. 2011. TMEM16A channels generate Ca2+-activated Cl− currents in cerebral artery smooth muscle cells. Am. J. Physiol. Heart Circ. Physiol. 301:H1819–H1827. 10.1152/ajpheart.00404.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y., Kongsuphol P., Hug M., Ousingsawat J., Witzgall R., Schreiber R., and Kunzelmann K.. 2011. Calmodulin-dependent activation of the epithelial calcium-dependent chloride channel TMEM16A. FASEB J. 25:1058–1068. 10.1096/fj.10-166884 [DOI] [PubMed] [Google Scholar]
- Tian Y., Schreiber R., and Kunzelmann K.. 2012. Anoctamins are a family of Ca2+-activated Cl- channels. J. Cell Sci. 125:4991–4998. 10.1242/jcs.109553 [DOI] [PubMed] [Google Scholar]
- Tien J., Lee H.Y., Minor D.L. Jr., Jan Y.N., and Jan L.Y.. 2013. Identification of a dimerization domain in the TMEM16A calcium-activated chloride channel (CaCC). Proc. Natl. Acad. Sci. USA. 110:6352–6357. 10.1073/pnas.1303672110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tien J., Peters C.J., Wong X.M., Cheng T., Jan Y.N., Jan L.Y., and Yang H.. 2014. A comprehensive search for calcium binding sites critical for TMEM16A calcium-activated chloride channel activity. Elife. 3:02772 10.7554/eLife.02772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran T.T., Tobiume K., Hirono C., Fujimoto S., Mizuta K., Kubozono K., Inoue H., Itakura M., Sugita M., and Kamata N.. 2014. TMEM16E (GDD1) exhibits protein instability and distinct characteristics in chloride channel/pore forming ability. J. Cell. Physiol. 229:181–190. 10.1002/jcp.24431 [DOI] [PubMed] [Google Scholar]
- Verchère A., Ou W.-L., Ploier B., Morizumi T., Goren M.A., Bütikofer P., Ernst O.P., Khelashvili G., and Menon A.K.. 2017. Light-independent phospholipid scramblase activity of bacteriorhodopsin from Halobacterium salinarum. Sci. Rep. 7:9522 10.1038/s41598-017-09835-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vocke K., Dauner K., Hahn A., Ulbrich A., Broecker J., Keller S., Frings S., and Möhrlen F.. 2013. Calmodulin-dependent activation and inactivation of anoctamin calcium-gated chloride channels. J. Gen. Physiol. 142:381–404. 10.1085/jgp.201311015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanitchakool P., Wolf L., Koehl G.E., Sirianant L., Schreiber R., Kulkarni S., Duvvuri U., and Kunzelmann K.. 2014. Role of anoctamins in cancer and apoptosis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369:20130096 10.1098/rstb.2013.0096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanitchakool P., Ousingsawat J., Sirianant L., Cabrita I., Faria D., Schreiber R., and Kunzelmann K.. 2017. Cellular defects by deletion of ANO10 are due to deregulated local calcium signaling. Cell. Signal. 30:41–49. 10.1016/j.cellsig.2016.11.006 [DOI] [PubMed] [Google Scholar]
- Watanabe R., Sakuragi T., Noji H., and Nagata S.. 2018. Single-molecule analysis of phospholipid scrambling by TMEM16F. Proc. Natl. Acad. Sci. USA. 115:3066–3071. 10.1073/pnas.1717956115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock J.M., and Hartzell H.C.. 2016. A Pore Idea: the ion conduction pathway of TMEM16/ANO proteins is composed partly of lipid. Pflugers Arch. 468:455–473. 10.1007/s00424-015-1777-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock J.M., and Hartzell H.C.. 2017. Anoctamins/TMEM16 proteins: Chloride channels flirting with lipids and extracellular vesicles. Annu. Rev. Physiol. 79:119–143. 10.1146/annurev-physiol-022516-034031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong X.M., Younger S., Peters C.J., Jan Y.N., and Jan L.Y.. 2013. Subdued, a TMEM16 family Ca2+-activated Cl−channel in Drosophila melanogaster with an unexpected role in host defense. Elife. 2:e00862 10.7554/eLife.00862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., El Refaey M., Xu L., Zhao L., Gao Y., Floyd K., Karaze T., Janssen P.M.L., and Han R.. 2015. Genetic disruption of Ano5 in mice does not recapitulate human ANO5-deficient muscular dystrophy. Skelet. Muscle. 5:43 10.1186/s13395-015-0069-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T., and Colecraft H.M.. 2016. Calmodulin regulation of TMEM16A and 16B Ca(2+)-activated chloride channels. Channels (Austin). 10:38–44. 10.1080/19336950.2015.1058455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Kim A., David T., Palmer D., Jin T., Tien J., Huang F., Cheng T., Coughlin S.R., Jan Y.N., and Jan L.Y.. 2012. TMEM16F forms a Ca2+-activated cation channel required for lipid scrambling in platelets during blood coagulation. Cell. 151:111–122. 10.1016/j.cell.2012.07.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.D., Cho H., Koo J.Y., Tak M.H., Cho Y., Shim W.-S., Park S.P., Lee J., Lee B., Kim B.-M., et al. 2008. TMEM16A confers receptor-activated calcium-dependent chloride conductance. Nature. 455:1210–1215. 10.1038/nature07313 [DOI] [PubMed] [Google Scholar]
- Ye W., Han T.W., Nassar L.M., Zubia M., Jan Y.N., and Jan L.Y.. 2018. Phosphatidylinositol-(4, 5)-bisphosphate regulates calcium gating of small-conductance cation channel TMEM16F. Proc. Natl. Acad. Sci. USA. 115:E1667–E1674. 10.1073/pnas.1718728115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K., Duran C., Qu Z., Cui Y.-Y., and Hartzell H.C.. 2012. Explaining calcium-dependent gating of anoctamin-1 chloride channels requires a revised topology. Circ. Res. 110:990–999. 10.1161/CIRCRESAHA.112.264440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K., Zhu J., Qu Z., Cui Y.Y., and Hartzell H.C.. 2014a Activation of the Ano1 (TMEM16A) chloride channel by calcium is not mediated by calmodulin. J. Gen. Physiol. 143:253–267. 10.1085/jgp.201311047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K., Whitlock J.M., Lee K., Ortlund E.A., Cui Y.Y., and Hartzell H.C.. 2015. Identification of a lipid scrambling domain in ANO6/TMEM16F. Elife. 4:e06901 10.7554/eLife.06901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Kuan A.S., and Chen T.Y.. 2014b Calcium-calmodulin does not alter the anion permeability of the mouse TMEM16A calcium-activated chloride channel. J. Gen. Physiol. 144:115–124. 10.1085/jgp.201411179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zhang Z., Xiao S., Tien J., Le S., Le T., Jan L.Y., and Yang H.. 2017. Inferior Olivary TMEM16B Mediates Cerebellar Motor Learning. Neuron. 95:1103–1111.e4. 10.1016/j.neuron.2017.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Morais-Cabral J.H., Kaufman A., and MacKinnon R.. 2001. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 A resolution. Nature. 414:43–48. 10.1038/35102009 [DOI] [PubMed] [Google Scholar]
- Zwaal R.F., Comfurius P., and Bevers E.M.. 2004. Scott syndrome, a bleeding disorder caused by defective scrambling of membrane phospholipids. Biochim. Biophys. Acta. 1636:119–128. 10.1016/j.bbalip.2003.07.003 [DOI] [PubMed] [Google Scholar]