Abstract
The RNase MRP and RNase P ribonucleoprotein particles both function as endoribonucleases, have a similar RNA component, and share several protein subunits. RNase MRP has been implicated in pre-rRNA processing and mitochondrial DNA replication, whereas RNase P functions in pre-tRNA processing. Both RNase MRP and RNase P accumulate in the nucleolus of eukaryotic cells. In this report we show that for three protein subunits of the RNase MRP complex (hPop1, hPop4, and Rpp38) basic domains are responsible for their nucleolar accumulation and that they are able to accumulate in the nucleolus independently of their association with the RNase MRP and RNase P complexes. We also show that certain mutants of hPop4 accumulate in the Cajal bodies, suggesting that hPop4 traverses through these bodies to the nucleolus. Furthermore, we characterized a deletion mutant of Rpp38 that preferentially associates with the RNase MRP complex, giving a first clue about the difference in protein composition of the human RNase MRP and RNase P complexes. On the basis of all available data on nucleolar localization sequences, we hypothesize that nucleolar accumulation of proteins containing basic domains proceeds by diffusion and retention rather than by an active transport process. The existence of nucleolar localization sequences is discussed.
INTRODUCTION
The endoribonuclease MRP (mitochondrial RNA processing) complex is a member of the large family of small nucleolar ribonucleoprotein particles shown to be involved in precursor-rRNA processing. Three classes of snoRNPs can be distinguished based on structural elements: Box H/ACA snoRNPs, Box C/D snoRNPs, and RNase MRP/RNase P (Tollervey and Kiss, 1997).
RNase MRP and RNase P ribonucleoprotein particles are related in many aspects. Both complexes function as site-specific endonucleases: RNase P is involved in the generation of the mature 5′ end of tRNAs, whereas RNase MRP has been shown to be involved in the processing of pre-rRNA. The RNA components of both enzymes adopt a similar cage-shaped secondary structure. Furthermore, it became clear that most of the currently known human protein subunits of RNase MRP and RNase P are shared by both enzymes (see for review van Eenennaam et al., 2000).
Besides the reported function of RNase MRP in the processing of pre-rRNA, in vitro experiments have shown that RNase MRP is also involved in the processing of mitochondrial RNA that functions as a primer for mitochondrial DNA replication (Chang and Clayton, 1987). Only a relatively small amount of RNase MRP has been reported to reside in the mitochondria, whereas the majority (99%) of RNase MRP is located in the nucleolus (Li et al., 1994). By genetic depletion in yeast it has been shown that the nucleolar RNase MRP is involved in the formation of the short form of the 5.8S rRNA [5.8S(S)], by catalyzing the cleavage at site A3 in the first internal transcribed spacer (ITS1) of pre-rRNA (Schmitt and Clayton, 1993; Chu et al., 1994; Lygerou et al., 1994, 1996a). Although none of these functions have been demonstrated to apply to the human system as yet, they are thought to be evolutionarily conserved. Recently, it was demonstrated that mutations in the human gene coding for the RNase MRP RNA cause an autosomal recessive disease called cartilage-hair hypoplasia, suggesting that the RNase MRP complex and/or its substrates play an important role in embryogenesis (Ridanpaa et al., 2001).
The first identified element important for the targeting of RNase MRP to the nucleolus is the 5′-terminal region of the RNase MRP RNA (referred to as P3 domain or Th/To binding site; Jacobson et al., 1995). The RNase P complex has been detected in both nucleolus and nucleoplasm (Jacobson et al., 1997; Bertrand et al., 1998), and it has been determined that the evolutionarily conserved P3 domain of RNase P RNA functions in nucleolar localization as well (Jacobson et al., 1997). Recently, a domain in two protein subunits, Rpp29 (identical to hPop4) and Rpp38, of RNase MRP and RNase P has been reported to be involved in the nucleolar localization of these protein subunits (Jarrous et al., 1999a, 1999b). In both cases the highly basic nature of these domains rather than a distinct amino acid sequence appeared to be important.
In this study we analyzed several deletion mutants of three protein subunits associated with both RNase MRP and RNase P: hPop1 (Lygerou et al., 1996b), hPop4 (van Eenennaam et al., 1999), and Rpp38 (Eder et al., 1997; Pluk et al., 1999) and examined the accumulation of these mutants in the nucleolus, complex association, and RNase P enzymatic activity.
MATERIALS AND METHODS
hPop1 Deletion Mutant Constructs
hPop1 cDNA (Lygerou et al., 1996b) was mutated by PCR to introduce a XhoI site before the translational start codon. The PCR product was digested with XhoI/EcoRI and ligated into an XhoI/EcoRI-digested pCI-neo vector (Promega, Madison, WI) that contains a 5′ VSV-G tag encoding sequence inserted between the original NheI-XhoI sites of pCI-neo (Pluk et al., 1998), resulting in a construct referred to as VSV-hPop1.
Partial digestion of VSV-hPop1 with HpaI and XbaI results in the release of a DNA fragment of 900 bp. The vector with the remaining hPop1 cDNA sequence was treated with Klenow polymerase and religated, generating a construct encoding the VSV-hPop1 Δ(711–1024) deletion mutant. Constructs encoding VSV-hPop1 Δ(558–1024) and VSV-hPop1 Δ(339–1024) were obtained by partial EcoRV digestion of VSV-hPop1, followed by religation to the filled-in SalI site of pCI-neo. The later constructs contain a vector-encoded translational stop codon, thus encoding eight additional amino acids at the C terminus.
Digestion with SmaI of VSV-hPop1 releases a DNA fragment with a length of 2100 bp, which was ligated in filled-in XhoI/SmaI-digested VSV-pCI-neo, resulting in a construct designated VSV-hPop1 Δ(1–318). VSV-hPop1 Δ(129–1024) was obtained by cleavage of VSV-hPop1 with EcoRI/SmaI and subsequent religation of the vector-containing fragment. Synthesis of the mutant encoded by this construct is terminated by a vector-encoded translational stop codon, leading to six additional C-terminal amino acids.
A construct encoding hPop1 (128–319) was obtained by ligation of the EcoRI/SmaI DNA fragment of VSV-hPop1 in EcoRI/SmaI-digested pEGFP-C1 (Clontech, Palo Alto, CA). Ligation of the EcoRI/RsaI fragment of VSV-hPop1 in EcoRI/SmaI-digested pEGFP-C1 results in a construct designated hPop1 (128–167). Digestion of VSV-hPop1 with RsaI/SmaI and subsequent ligation of the resulting fragment in SmaI-digested pEGFP-C1 generates the hPop1 (168–319) construct. The EcoRI/PstI-digested fragment of VSV-hPop1 was ligated in EcoRI/PstI-digested pEGFP-C1, resulting in hPop1 (128–245).
The integrity of all DNA constructs described above was confirmed by DNA sequencing.
hPop4 Deletion Mutant Constructs
Wild-type hPop4 cDNA was subcloned in pCI-neo containing a DNA sequence coding for either an N-terminal or a C-terminal VSV-G tag as described (van Eenennaam et al., 1999); the resulting constructs were designated VSV-hPop4 and hPop4-VSV, respectively.
A DNA fragment of hPop4/pCR-II-Topo obtained by digestion with DdeI/SmaI (van Eenennaam et al., 1999) was treated with Klenow polymerase to fill in the overhangs and was ligated in VSV-hPop4 digested with XhoI/SmaI and treated with Klenow polymerase, resulting in the VSV-hPop4 Δ(1–10) construct. Ligation of a Klenow polymerase–treated DNA fragment resulting from digestion of hPop4/pCR-II-Topo with NcoI/XbaI in hPop4-VSV treated with Klenow polymerase after XhoI/XbaI digestion generated a construct encoding hPop4-VSV Δ(1–47).
Two DNA fragments generated by XhoI/RsaI and RsaI/XbaI digestion of hPop4/PCR-II-Topo were ligated in XhoI/XbaI-digested VSV-hPop4, resulting in the VSV-hPop4 Δ(61–109) construct.
Three C-terminal deletion mutants were generated with the use of PCR-based approaches. Besides the mutations introduced by PCR, a XbaI-site was introduced after the most 3′ codon, followed by an in-frame stop-codon and a SmaI site in the oligonucleotides used. With the use of this strategy DNA fragments were produced and digested with XhoI/SmaI and ligated in XhoI/SmaI-digested VSV-hPop4, generating VSV-hPop4 Δ(162–220), Δ(181–220), and Δ(210–220), respectively.
The integrity of these constructs was confirmed by DNA sequencing.
Rpp38 Deletion Mutant Constructs
Wild-type Rpp38 cDNA was subcloned in the pECFP-C3 vector (Clontech) containing a DNA sequence encoding an N-terminal ECFP tag.
Constructs encoding deletion mutants of Rpp38 were generated by PCR-based approaches. Besides the deletions introduced by the PCR strategy, an XhoI site was introduced in-frame at the 5′-end of the translational start codon, whereas an XbaI site was introduced at the 3′-end of the coding sequence. Digestion with XhoI/XbaI of the PCR products and ligation in XhoI/XbaI-digested pECFP-C3, generated the after constructs; Rpp38 Δ(1–40), Δ(1–98), Δ(1–141), Δ(181–283), and Δ(246–283).
The integrity of the constructs described above was confirmed by DNA sequencing.
Transient Transfection of HEp-2 Cells
HEp-2 monolayer cells were grown to 70% confluency by standard tissue culture techniques in either culture flasks or on coverslips. pCI-neo–derived constructs (3–4 μg) were transfected in HEp-2 cells with the use of LipofectAmine 2000 reagent (Life Technologies-BRL, Rockville, MD) according to the manufacturer's instructions. pEGFP/pECFP derived constructs (10 μg) were transfected into 3 × 106 HEp-2 cells by electroporation in a total volume of 400 μl of DMEM containing 10% fetal calf serum. Electroporation was performed at 276 V and a capacity of 950 μFa with a Gene Pulser II (Bio-Rad, Hercules, CA).
Transfected cells were grown overnight in DMEM supplemented with 10% fetal calf serum.
Cells grown on coverslips were washed twice with phosphate-buffered saline (PBS), fixed with either methanol (5 min at −20°C) or with 4% paraformaldehyde (20 min at room temperature), and permeabilized with the use of 0.2% Triton X-100 (5 min at room temperature).
Cells grown in flasks were harvested, washed once with PBS, and used to prepare extracts for immunoprecipitation assays (see below).
Immunofluorescence
Indirect immunofluorescence assays were performed on transfected HEp-2 cells. Fixed cells were incubated with affinity-purified mouse anti–VSV-tag antibodies (diluted 1:50 in PBS; Roche, Indianapolis, IN) for 1 h at room temperature, washed with PBS, and subsequently incubated with rabbit anti-mouse immunoglobulins coupled to FITC for 1 h at room temperature. Cells were mounted with PBS/glycerol, and bound antibodies were visualized by fluorescence microscopy.
Preparation of HEp-2 Cell Extracts
Extracts of HEp-2 cells were prepared by resuspending cell pellets in buffer A (25 mM Tris-HCl [pH 7.5], 100 mM KCl, 1 mM dithioerythritol, 2 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride, 0.05% NP-40) and lysis by sonication with the use of a Branson microtip (three times for 20 s). Insoluble material was removed by centrifugation (12,000 × g, 15 min), and supernatants were used directly for immunoprecipitation.
Immunoprecipitation and pre-tRNA Processing Assay
Monoclonal anti–VSV-tag (Roche) or anti-EGFP antibodies were coupled to protein A-agarose beads (Biozyme Laboratories, San Diego, CA) in IPP500 (500 mM NaCl, 10 mM Tris-HCl [pH 8.0], 0.05% NP-40) by incubation for 1 h at room temperature. Beads were washed twice with IPP500 and once with IPP150 (150 mM NaCl, 10 mM Tris-HCl [pH 8.0], 0.05% NP-40). For each immunoprecipitation the cell extract was incubated with the antibody-coupled beads for 2 h at 4°C. Subsequently, beads were washed three times with IPP150.
To analyze coprecipitating RNAs, the RNA was isolated from the beads by phenol–chloroform extraction and ethanol precipitation. RNAs were resolved on a denaturing polyacrylamide gel and blotted to a Hybond-N membrane (Amersham, Arlington Heights, IL). Northern blot hybridizations with riboprobes specific for human RNase P and RNase MRP RNAs were performed as previously described (Verheijen et al., 1994).
To assay for RNase P enzymatic activity in the immunoprecipitates (i.e., the beads), an internally 32P-labeled pre-tRNA substrate (S. pombe tRNASer SupS1; Krupp et al., 1986) was transcribed in vitro and gel-purified. This 110 nt-long substrate contains a 5′-extension of 28 nts in comparison with the mature tRNA. The immunoprecipitates were incubated with substrate RNA in assay buffer (20 mM Tris-HCl [pH 8.0], 10 mM MgCl2, 1 mM DTE, 50 mM KCl, 50 μg/ml BSA, 60 U/ml RNasin) for 10 min at 37°C. The products were analyzed by denaturing PAGE and autoradiography.
RESULTS
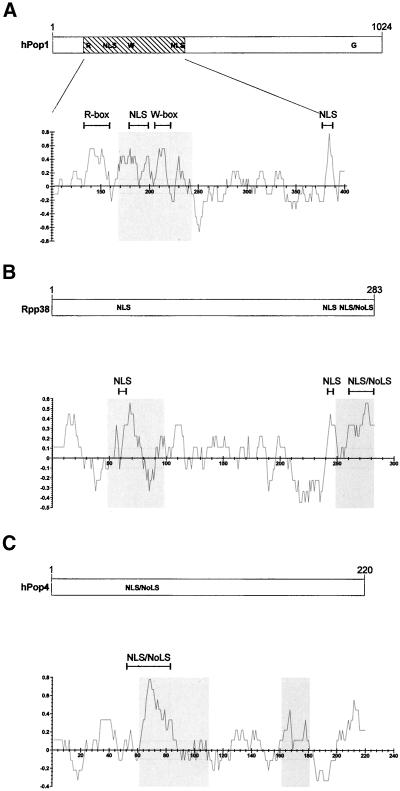
To learn more about the nucleolar targeting of the human RNase MRP and RNase P complexes, three of their protein subunits (hPop1, Rpp38, and hPop4) were mutated, and the effects of the mutations on their subcellular localization was analyzed by fluorescence microscopy. Figure 1 shows the main characteristics of the hPop1, Rpp38, and hPop4 protein subunits, including putative nuclear localization signals, identified nucleolar localization signals, and conserved sequence elements.
Figure 1.
Schematic representation of the hPop1, Rpp38, and hPop4 proteins. The length, putative nuclear localization signals (NLS) and nucleolar localization signals (NoLSs) as established by Jarrous and coworkers (Jarrous et al., 1999b) for the hPop1 (A), Rpp38 (B), and hPop4 (C) protein subunits are depicted in the upper parts. The lower parts represent charge plots of these proteins. Values on the vertical axis represent positively charged and negatively charged regions, respectively. These values were calculated with the use of a window of nine amino acids. Gray regions in the charge plots represent the regions of the proteins important for their nucleolar localization. For the hPop1 protein (A) the charge plot of only amino acids 100–400 is depicted (hatched area in upper part). The R-, W- and G-box refer to conserved sequence elements observed in the hPop1 protein (Lygerou et al., 1996b).
Because the nucleolar accumulation of these protein subunits might be dependent on their association with the particles, the association of the mutants with the RNase MRP and RNase P RNAs was analyzed by Northern blot hybridization, and their assembly into functional particles was analyzed in an in vitro pre-tRNA processing assay.
Basic Domains of the hPop1 Protein Are Required for Its Nucleolar Targeting
DNA molecules encoding wild-type hPop1, and deletion mutants of hPop1 were cloned into the mammalian expression vector pCI-neo, in combination with a sequence encoding a VSV-G tag (Kreis, 1986) fused to the 5′-end of the coding sequences, and transiently expressed in HEp-2 cells. After overnight culturing, the cells were fixed and the subcellular localization of the VSV-tagged hPop1 (mutants) was determined by indirect immunofluorescence with the use of anti–VSV-tag antibodies.
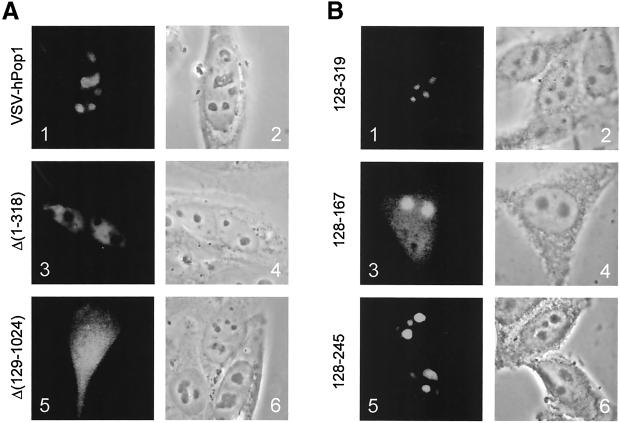
The results in Figure 2A, panels 1 and 2, show that the VSV-hPop1 construct expressed a protein that accumulated in the nucleoli, in accordance with previous observations (Lygerou et al., 1996b). A similar nucleolar staining pattern was found for VSV-tagged hPop1 deletion mutants Δ(711–1024), Δ(558–1024), and Δ(339–1024; see Table 1), indicating that the N-terminal region plays an important role in the nucleolar accumulation of the hPop1 protein. This was confirmed by the finding that a mutant in which most of these amino acids had been deleted, VSV-hPop1 Δ(1–318), was targeted to the nucleoplasm and did not accumulate in the nucleoli (Figure 2A, panels 3 and 4).
Figure 2.
Subcellular localization of deletion mutants of hPop1. (A) VSV-hPop1 constructs were transiently transfected into HEp-2 cells. Cells were fixed with paraformaldehyde, and the expressed proteins were visualized with the use of anti-VSV antibodies (panels 1, 3, and 5). A phase-contrast image of the same cells is shown in panels 2, 4, and 6. Panels 1 and 2: VSV-hPop1 construct; panels 3 and 4: VSV-hPop1 Δ(1–318); panels 5 and 6: VSV-hPop1 Δ(129–1024). (B) EGFP-hPop1 constructs were transiently transfected into HEp-2 cells. Cells were fixed with methanol/acetone, and the fluorescent proteins were visualized by direct fluorescence microscopy (panels 1, 3, and 5). Phase-contrast images of the same cells are shown in panels 2, 4, and 6. Panels 1 and 2: EGFP-hPop1 128–319; panels 3 and 4: EGFP-hPop1 128–167; panels 5 and 6: EGFP-hPop1 168–245.
Table 1.
Summary of subcellular localization, complex association with RNase MRP and RNase P, and RNase P activity of deletion mutants of hPop1, Rpp38, and hPop4 protein subunits
| Construct | Subcellular localization | Complex association with RNase MRP/P | Association with RNase P activity |
|---|---|---|---|
| hPop1 protein | |||
| Wild type | No | — | — |
| Δ(711–1024) | No | nd | nd |
| Δ(558–1024) | No | nd | nd |
| Δ(339–1024) | No | nd | nd |
| Δ(1–318) | Np | nd | nd |
| Δ(129–1024) | No + Np + Cy | nd | nd |
| 128–319 | No | nd | nd |
| 128–167 | No + Np + Cy | nd | nd |
| 168–319 | No | nd | nd |
| 128–245 | No | nd | nd |
| Rpp38 protein | |||
| Wild type | No + Np | + | + |
| Δ(1–40) | No + Np | Only MRP | − |
| Δ(1–98) | Np | − | − |
| Δ(1–141) | No + Np + Cy | − | − |
| Δ(181–283) | Np + Cy | − | − |
| Δ(246–283) | Np | + | + |
| hPop4 protein | |||
| Wild type | No | + | + |
| Δ(1–10) | No | + | + |
| Δ(1–47) | No | + | + |
| Δ(61–109) | No + CB | − | − |
| Δ(162–220) | No + CB | − | − |
| Δ(181–220) | No | − | − |
| Δ(210–220) | No | + | + |
No, Nucleolus; Np, Nucleoplasm; Cy, Cytoplasm; −, no association with RNase MRP and RNase P/no association with RNase P activity; +, association with RNase MRP and RNase P/association with active RNase P; nd, not determined; CB, Cajal bodies.
These results were further detailed by the finding that a deletion mutant containing only the N-terminal 128 amino acids, Δ(129–1024) was found uniformly distributed throughout the cell (Figure 2A, panels 5 and 6). Taken together, these results suggest that amino acids 129–318 are important for the nucleolar accumulation of the hPop1 protein.
To study whether a hPop1 fragment comprising amino acids 128–319 was able to accumulate in the nucleolus, we fused this part of the hPop1 protein to EGFP and analyzed its localization. This fragment of hPop1 indeed proved to be sufficient for nucleolar targeting of EGFP (Figure 2B, panels 1 and 2). In this region 1 of the two putative nuclear localization sequences, a highly conserved arginine-rich motif (R-box) and a tryptophan-rich domain (W-box) have been identified (Lygerou et al., 1996b). In the same way, three subfragments of the amino acid 128–319 region were analyzed. Constructs were made that encode only the R-box (amino acids 128–167); the NLS and W-box together (amino acids 168–319); and the R-box, NLS, and W-box (amino acids 128–245), each fused to EGFP. Surprisingly, these fusion proteins all localized to the nucleolus (see Figure 2B, panels 3–6 for 128–167 and 128–245 and Table 1 for 168–319), although the efficiency of nucleolar accumulation for the 168–319 and 128–245 fusion proteins was much higher than that for the 128–167 fragment. The latter was also detected in the cytoplasm and nucleoplasm (panel 3).
Rpp38 Harbors Two Regions Important for Nucleolar Accumulation
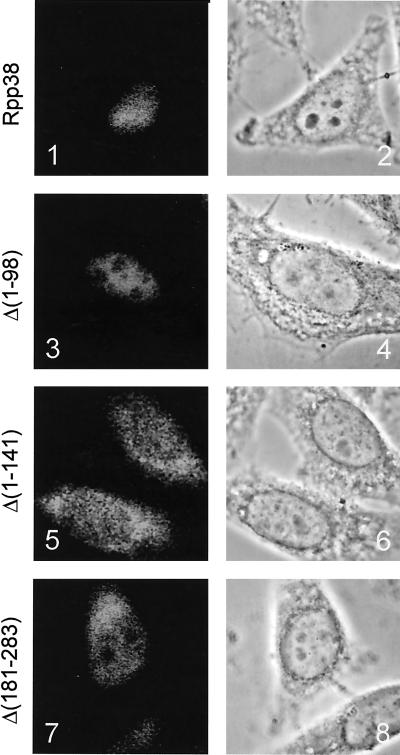
Recently, Jarrous and collaborators have identified a so-called nucleolar localization sequence (referred to as NoLS) near the C terminus of the Rpp38 protein subunit of the RNase MRP and RNase P complexes (Jarrous et al., 1999b). To investigate whether this NoLS was sufficient for the nucleolar localization of the Rpp38 protein, we made several deletion mutants of the Rpp38, fused them to ECFP, and expressed them transiently in HEp-2 cells. As is depicted in Figure 3, panel 1, the wild-type Rpp38-ECFP fusion protein was localized in both nucleoli and nucleoplasm. This distribution is very similar to that reported before for a GFP-Rpp38 fusion protein (Jarrous et al., 1999b). Although this distribution deviates from what has been described for the endogenous Rpp38 protein (localization to the nucleolus and Cajal bodies; Jarrous et al., 1999b), cells with a lower level of expression of this construct showed more prominent nucleolar staining, suggesting that the strong nucleoplasmic staining is caused by overexpression.
Figure 3.
Subcellular localization of deletion mutants of Rpp38. ECFP-Rpp38 constructs were transiently transfected into HEp-2 cells. Cells were fixed with methanol/acetone, and the fluorescent proteins were visualized by direct fluorescence microscopy (panels 1, 3, 5, and 7). Phase-contrast images of the same cells are shown in panels 2, 4, 6, and 8. Panels 1 and 2: ECFP-Rpp38; panels 3 and 4: ECFP-Rpp38 Δ(1–98); panels 5 and 6: ECFP-Rpp38 Δ(1–141); and panels 7 and 8: ECFP-Rpp38 Δ(181–283).
A construct that encodes Rpp38 lacking the N-terminal 40 amino acids showed an identical subcellular localization (Table 1). Surprisingly, deletion of the N-terminal 98 amino acids resulted in staining of only the nucleoplasm (Figure 3, panel 3), which suggests that in spite of the presence of the complete NoLS (amino acids 246–283), this mutant is not able to enter the nucleoli. Further truncation of the N-terminus of the Rpp38 protein (Rpp38 Δ (1–141)) resulted in a cytoplasmic accumulation and a weak nuclear staining (see Figure 3, panel 5). Finally, we showed that deletion of the C terminus (Rpp38 Δ (246–283) and Rpp38 Δ (181–283)) results in nucleoplasmic staining, loss of nucleolar accumulation, and staining of the cytoplasm (Figure 3, panel 7, and Table 1).
Taken together, these results indicate that two domains appear to be necessary for the nucleolar accumulation of the Rpp38 protein subunit and that each of these domains (amino acids 40–98 and 246–283) alone is not sufficient for nucleolar targeting.
The Amino-terminus of Rpp38 Differentiates between RNase MRP and RNase P Complexes
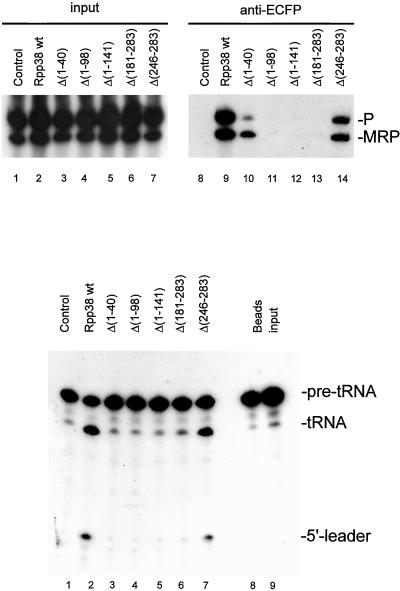
Because differences in the subcellular localization of various Rpp38 deletion mutants might be dependent on its ability to associate with the RNase MRP and/or RNase P particle, complex association was studied by immunoprecipitation with the use of anti-ECFP antibodies and extracts of transiently transfected cells. Complex association was monitored by the isolation of RNA from the immunoprecipitates and visualization of RNase MRP and RNase P RNA by Northern blot hybridization. As indicated in Figure 4A, lane 9, the wild-type Rpp38-ECFP fusion protein effectively associated with both RNase P and RNase MRP, in accordance with previously published data (Pluk et al., 1999). Surprisingly, the Rpp38 Δ (246–283) mutant also associated with both the RNase MRP and the RNase P complex (Figure 4A, lane 14), despite the loss of its ability to enter the nucleolus. Moreover, an N-terminal deletion mutant, which displayed a wild-type subcellular distribution, Rpp38 Δ (1–40), appeared to associate preferentially with the RNase MRP particle (Figure 4A, lane 10). With this mutant only a very inefficient coprecipitation of RNase P RNA was reproducibly observed, which suggests that the N-terminus of the Rpp38 is involved in its stable association with the RNase P complex but not with the RNase MRP complex.
Figure 4.
Association of ECFP-Rpp38 (mutants) with RNase MRP and RNase P ribonucleoprotein particles and RNase P activity associated with ECFP-Rpp38 deletion mutants. (A) Constructs encoding (deletion mutants of) ECFP-Rpp38 were transiently transfected in HEp-2 cells. Extracts from these cells used for immunoprecipitations with anti-ECFP antibodies. RNA isolated from total cell extracts (lanes 1–7) and immunoprecipitates (lanes 8–14) was analyzed by Northern blot hybridization with the use of riboprobes specific for RNase MRP and RNase P RNA. Lanes 1 and 8: material from control cells expressing ECFP alone; lanes 2 and 9: ECFP-Rpp38; lanes 3 and 10: ECFP-Rpp38 Δ(1–40); lanes 4 and 11: ECFP-Rpp38 Δ(1–98); lanes 5 and 12: ECFP-Rpp38 Δ(1–141); lanes 6 and 13: ECFP-Rpp38 Δ(181–283) and lanes 7 and 14, ECFP-Rpp38 Δ(246–283). The positions of RNase P and RNase MRP RNA are indicated. (B) RNase P activity assay associated with anti-ECFP immunoprecipitates from extracts of cells transiently transfected with ECFP-Rpp38 (deletion mutants). Lane 1: material from control cells expressing ECFP alone; lane 2: ECFP-Rpp38; lane 3: ECFP-Rpp38 Δ(1–40); lane 4: ECFP-Rpp38 Δ(1–98); lane 5: ECFP-Rpp38 Δ(1–141); lane 6: ECFP-Rpp38 Δ(181–283); lane 7: ECFP-Rpp38 Δ(246–283); lane 8: beads alone; and lane 9, substrate RNA. On the right, the positions of the pre-tRNA, the mature tRNA and the 5′-leader are indicated.
In contrast, no complex association was observed for the remaining deletion mutants of Rpp38, Rpp38 Δ (1–98), Δ (1–141), and Δ (181–283), which was not due to lack of expression as demonstrated by Western blotting (our unpublished results).
The Carboxy-terminus of Rpp38 Is Dispensable for RNase P Function
To investigate whether the complexes bound by the Rpp38 deletion mutants were functionally active, we analyzed the immunoprecipitated complexes in an in vitro precursor-tRNA processing assay. A 32P-labeled precursor-tRNA was incubated at 37°C with the anti-ECFP immunoprecipitates and the resulting products were analyzed by denaturing PAGE and autoradiography.
Particles associated with either the wild-type ECFP-Rpp38 fusion protein or the C-terminal deletion mutant Δ (246–283) displayed RNase P enzymatic activity (Figure 4B, lanes 2 and 7, respectively), whereas control material obtained from extracts of cells expressing ECFP alone did not (lane 1). As expected, no RNase P activity was detected with the remaining deletion mutants of Rpp38, including Δ (1–40), in agreement with the lack of stable association with the RNase P complex (Figure 4B, lanes 3–6).
hPop4 Contains Two Regions Important for Nucleolar Localization
Next, the subcellular localization and the association with the RNase MRP and (catalytically active) RNase P complexes were studied for six deletion mutants of the hPop4 protein subunit, following the procedures described above.
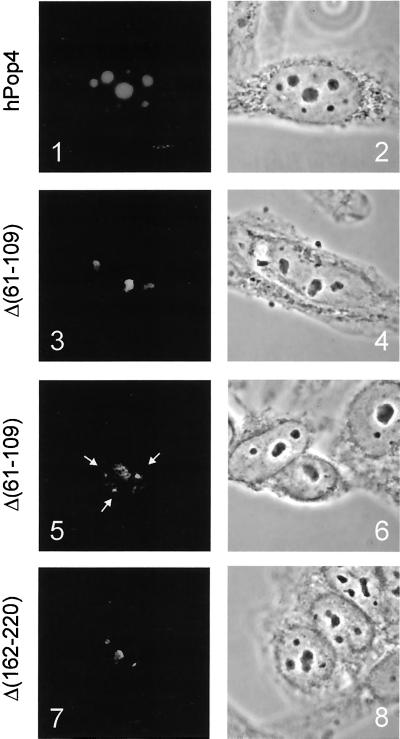
Indirect immunofluorescence showed that the VSV-tagged hPop4 protein subunit is localized in the nucleoli as has been established previously (Van Eenennaam et al., 1999; Figure 5, panel 1). The subcellular localization of the Δ (1–10), Δ (1–47), Δ (181–220), and Δ (210–220) deletion mutants (all VSV-tagged) showed that deletion of either the N- or the C-terminal regions of the hPop4 does not affect the localization of this protein as they all accumulated in the nucleoli (see Table 1).
Figure 5.
Subcellular localization of deletion mutants of hPop4. VSV-hPop4 constructs were transiently transfected into HEp-2 cells. Cells were fixed with methanol/acetone, and the expressed proteins were visualized by indirect fluorescence microscopy with the use of anti-VSV antibodies (panels 1, 3, 5, and 7). Phase-contrast images of the same cells are shown in panels 2, 4, 6, and 8. Panels 1 and 2: VSV-hPop4; panels 3 and 4: VSV-hPop4 Δ(61–109), nucleolar accumulation; panels 5 and 6: VSV-hPop4 Δ(61–109); accumulation in the Cajal bodies; and panels 7 and 8: VSV-hPop4 Δ(162–220), nucleolar pattern.
Two different subcellular localization patterns were observed for two other deletion mutants, one lacking an internal region (Δ (61–109)) and another in which a relatively large part of the C terminus had been deleted (Δ (162–220)). As shown in Figure 5, panels 3 and 7, both mutants do have the ability to accumulate in the nucleolus. However, in addition to strong nucleolar staining observed in a subset of cells, cells with weaker or no nucleolar staining were present as well. In many of these cells the fluorescent staining was concentrated in nucleoplasmic foci, as is depicted in Figure 5, panel 5, for Δ (61–109). These foci appeared to be Cajal bodies (formerly designated coiled bodies), which was established by colocalization experiments with the use of anti-p80 coilin antibodies (our unpublished results).
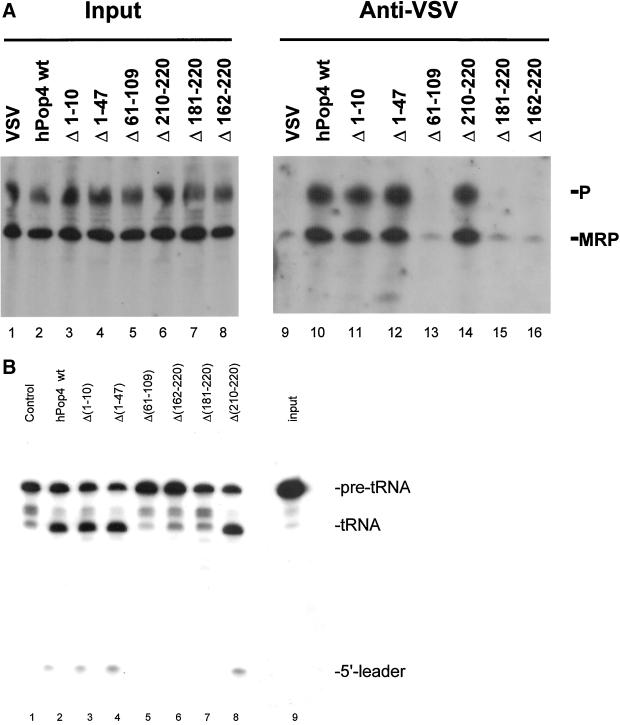
To investigate whether the subcellular distribution of these deletion mutants was somehow correlated with the ability to associate with RNase MRP and RNase P ribonucleoprotein particles, coimmunoprecipitation experiments were performed with anti–VSV-tag antibodies with the use of extracts from transiently transfected cells. As described previously (van Eenennaam et al., 1999), the wild-type hPop4 protein was found to be associated with both RNase MRP and RNase P complexes (Figure 6A, lane 10), whereas control precipitations did not display RNase MRP and RNase P RNA (lane 9). Deletion of the N-terminus of the hPop4 protein subunit did not abolish the association with both complexes, because deletion mutants Δ (1–10) and Δ (1–47) coprecipitated wild-type levels of both RNAs (lanes 11 and 12). Also deletion of 11 amino acids from the C terminus did not affect the association with both particles (lane 14). In contrast, the internal deletion mutant and the mutants having more than 10 amino acids deleted from the C terminus of the hPop4 protein did not detectably associate with RNase MRP and RNase P RNA, as demonstrated by the results in lanes 13 and 15–16 (Figure 6A). Control experiments (Western blotting) showed that this was not due to lack of expression (our unpublished results).
Figure 6.
Association of VSV-hPop4 (mutants) with RNase MRP and RNase P ribonucleoprotein particles and RNase P activity associated with VSV-hPop4 deletion mutants. (A) Constructs encoding (deletion mutants) of VSV-hPop4 were transiently transfected into HEp-2 cells. Extracts from these cells were used for immunoprecipitation with anti-ECFP antibodies. RNA isolated from total cell extracts (lanes 1–8) and immunoprecipitates (lanes 9–16) was analyzed by Northern blot hybridizations with the use of riboprobes specific for RNase MRP and RNase P RNA. Lanes 1 and 9: material from cells transfected with empty vector alone; lanes 2 and 10: VSV-hPop4; lanes 3 and 11: VSV-hPop4 Δ(1–10); lanes 4 and 12: VSV-hPop4 Δ(1–47); lanes 5 and 13: VSV-hPop4 Δ(61–109); lanes 6 and 14: VSV-hPop4 Δ(210–220); lanes 7 and 15: VSV-hPop4 Δ(181–220); and lanes 8 and 16, VSV-hPpop4 Δ(162–220). The positions of RNase MRP and RNase P RNA are indicated on the right. (B) RNase P activity assay associated with immunoprecipitates from extracts of cells transiently transfected with VSV-hPop4 (deletion mutants). Lane 1: material from cells transfected with empty vector alone; lane 2: material from cells expressing VSV-hPop4; lane 3: VSV-hPop4 Δ(1–10); lane 4: VSV-hPpop4 Δ(1–47); lane 5: VSV-hPop4 Δ(61–109); lane 6: VSV-hPop4 Δ(162–220); lane 7: VSV-hPop4 Δ(181–220); lane 8: VSV-hPop4 Δ(210–220); and lane 9: input. On the right, the positions of the pre-tRNA, the mature tRNA and the 5′-leader are indicated.
Finally, to investigate the association of the hPop4 deletion mutants with catalytically active RNase P complexes the immunoprecipitates were analyzed in the pre-tRNA processing assay. As demonstrated in Figure 6B, lanes 1 and 2, particles containing the wild-type VSV-tagged hPop4 protein displayed enzymatic activity. The analysis of the deletion mutants showed a complete correlation between the capacity to associate with the RNase P particle and the enzymatic activity (lanes 3–8). This result implies that neither the N-terminal 48 nor the C-terminal 11 amino acids of hPop4 are required for RNase P activity.
DISCUSSION
We have analyzed the effects of deletions in the hPop1, hPop4, and Rpp38 protein subunits of RNase MRP and RNase P on their subcellular localization, on their association with RNase MRP and RNase P and on the enzymatic activity of RNase P (see Table 1 for a summary). The results identified regions in each of these proteins important for their nucleolar accumulation. The nucleolar localization of hPop1 appeared to be dependent on a region near the N-terminus of this protein. The Rpp38 protein subunit needs two distinct regions to accumulate in the nucleolus. Although none of the hPop4 deletion mutants analyzed failed to enter the nucleolus, two mutants of this protein accumulated in the Cajal bodies in a subset of the cells. Importantly, complex association did not appear to be a requirement for nucleolar accumulation. All deletion mutants of Rpp38 and hPop4 that retained the capacity to associate with the RNase MRP and P complexes were bound to enzymatically active RNase P, suggesting that the deleted amino acids are dispensable for this activity. Finally, the N-terminus of Rpp38 appeared to be required for a stable association with RNase P but not with RNase MRP.
The N-terminus of Rpp38 Discriminates between RNase P and RNase MRP
The RNase MRP and RNase P ribonucleoprotein complexes show a high degree of similarity. Their RNA components can be folded in similar cage-shaped structures, and most of the currently known protein subunits have been shown to be associated with both complexes (for review see Van Eenennaam et al., 2000). In yeast only a single protein subunit has been reported to be specifically associated with either RNase MRP (Schmitt and Clayton, 1992) or RNase P (Chamberlain et al., 1998), whereas the other seven proteins are common to both particles. So far no human orthologues of the particle-specific yeast proteins have been found.
The N-terminal deletion mutant Δ (1–40) of Rpp38 appeared to associate preferentially with the RNase MRP complex. This implicates that the N-terminus of this protein plays an important role in the association with the RNase P complex. Whether the lack of binding of this mutant to RNase P is due to the loss of interactions with the RNA component (Pluk et al., 1999) or with another, most likely particle-specific protein subunit is presently not known.
The selective interaction of Rpp38 Δ (1–40) with the RNase MRP complex in combination with the lack of RNase P activity associated with this complex implies that the RNase MRP particle is unable to cleave the precursor-tRNA (substrate for RNase P), unless the N-terminus of Rpp38 is required for this activity.
Basic Domains in the Protein Subunits of RNase MRP and P Are Involved in Their Nucleolar Accumulation
A comparison of the regions in the three proteins that affect their nucleolar accumulation showed that all were highly enriched in basic amino acids (see Figure 1), suggesting that basic regions are important for nucleolar localization. Basic sequence elements previously have been implicated in nucleolar localization of the Rpp38 and Rpp29/hPop4 protein subunits (Jarrous et al., 1999b). Jarrous and coworkers proposed that nucleolar localization sequences (referred to as NoLS) are present at positions 260–283 in Rpp38 and 63–85 in hPop4. They showed that these elements are able to direct a reporter protein to the nucleolus. Our data confirm that these elements are required for nucleolar targeting of these proteins but also show that in the case of Rpp38 an additional element (amino acids 40–98) is required, whereas in the case of hPop4 a deletion of the previously identified NoLS (Δ6 1–109) does not abrogate its transport to the nucleolus. Apparently, the NoLS of Rpp38 identified previously is required but not sufficient for nucleolar accumulation, and hPop4 contains more than one functional NoLS.
The wild-type Rpp29/hPop4 has been reported to be present in the Cajal bodies previously (Jarrous et al., 1999b). Surprisingly, two deletion mutants of hPop4 appeared to be retained in the Cajal bodies in a subset of the transfected cells. Transport of the Rpp29/hPop4 protein subunit through the Cajal bodies on its way to the nucleolus fits very well in the model proposing that snoRNPs assemble into larger complexes in the Cajal bodies and are subsequently transported to the nucleolus (Gall, 2000). A defect in this assembly process caused by the deletions in our mutants might lead to their retention in the Cajal bodies. However, in addition to the retention in the Cajal bodies both mutants did enter the nucleoli in some cells. More importantly, all three proteins studied in this report have the capacity to enter the nucleus and nucleolus independent of an association with the cognate RNAs strongly suggesting that the association with the cognate particles is not an absolute requirement for transport of these proteins to the nucleoli.
Does a NoLS Exist?
The involvement of regions rich in basic amino acids in nucleolar localization has been found by others as well. For the poly(A)-binding protein II (PABP2) it has been shown that the basic C-terminal region of this nucleoplasmic protein directs a reporter protein to the nucleolus (Calado et al., 2000), and more recently it was shown that a basic element in p80-coilin, a marker protein for Cajal bodies, when uncovered by the deletion of an acidic region, also leads to a nucleolar localization (Hebert and Matera, 2000). In addition, two viral proteins, herpesvirus MEQ protein and PRRSV nucleocapsid protein (Liu et al., 1997; Rowland et al., 1999), and two cellular proteins, rat spermatidal protein TP2 and human I-mfa domain containing protein HICp40 (Meetei et al., 2000; Thebault et al., 2000) have been reported to contain NoLSs consisting of stretches of basic residues as well. Comparison of these various basic sequence elements does not reveal a consensus sequence. Taken together, our results and the work of others indicate that stretches of basic residues, even when these are derived from proteins that normally do not reside in the nucleoli, may target proteins to the nucleolus independently of functionality (e.g., complex association).
A prerequisite for nucleolar accumulation is translocation of a protein from the cytoplasm to the nucleoplasm. In the nucleoplasm proteins may bind to their functional entities. Nucleolar accumulation can subsequently proceed by two mechanisms: one in which the translocation to the nucleolus is an active process (either mediated by a signal in the protein that directs it to the nucleolus or mediated by its association with other molecules/complexes that are actively transported to the nucleolus) and another in which nucleolar entry is a passive process mediated by diffusion. In the latter case accumulation in the nucleolus may be due to retention by binding to nucleolar components.
The existence of a specific signal that directs proteins to the nucleolus is not supported by the data discussed above. The deletion of the previously described NoLS in hPop4 does not block nucleolar localization (hPop4 Δ (61–109)) and a mutant that does contain the NoLS identified in Rpp38 is not localized to the nucleolus (Rpp38 Δ (1–98)). Moreover, the NoLS identified in PABP2 is not able to direct this protein to the nucleolus (Calado et al., 2000). In conclusion, these data strongly suggest that the identified basic domains are not functioning in an active nucleolar transport pathway. Transport to the nucleolus via assembly into functional complexes is also not supported by our data. Mutants of all three proteins studied (hPop1, hPop4, and Rpp38) are able to localize to the nucleolus independently of complex association. This is substantiated by the finding that different proteins composed of 10–20 basic amino acids fused to EGFP are able to localize to the nucleolus as well (Jarrous et al., 1999b; Calado et al., 2000; Thebault et al., 2000). Nucleolar accumulation by diffusion and retention is consistent with all available data. Whether nucleolar retention is caused by the specific binding to nucleolar components is not clear at present. Most likely the RNase MRP complex is bound in the nucleolus because it is involved in the processing of the precursor-rRNA, and it associates with either pre-rRNA or the processing machinery. However, such a mechanism does not explain the nucleolar localization mediated by small stretches of basic amino acids. This type of nucleolar accumulation might be explained by the high concentration of negatively charged molecules in the nucleolar compartment (e.g., precursor-rRNA, rDNA) to which these basic elements may nonspecifically bind. Thus, in our opinion data obtained by these kind of experiments should be interpreted with great care.
In conclusion, we suggest that nucleolar localization signals that mediate active transport to the nucleolus may not exist and that the previously identified basic elements localize proteins to the nucleolus by virtue of their nucleolar retention capacity after diffusion into the nucleolus.
ACKNOWLEDGMENTS
We thank Dr. Bertrand Seraphin (EMBL, Heidelberg, Germany) for providing us with the pre-tRNA construct, Dr. Sidney Altman (Yale University, New Haven, CT), and Dr. C. Liew (University of Toronto, Canada) for their kind gift of Rpp38 cDNA and Dr. Wiljan Hendriks (Department of Cell Biology, University of Nijmegen, The Netherlands) for providing us with anti-EGFP/ECFP antibodies. This work was supported in part by the Netherlands Foundation of Chemical Research (NWO-CW) with financial aid from the Netherlands Organization for Scientific Research (NWO).
REFERENCES
- Bertrand E, Houser Scott F, Kendall A, Singer RH, Engelke DR. Nucleolar localization of early tRNA processing. Genes Dev. 1998;12:2463–2468. doi: 10.1101/gad.12.16.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calado A, Kutay U, Kuhn U, Wahle E, Carmo-Fonseca M. Deciphering the cellular pathway for transport of poly(A)-binding protein II. RNA. 2000;6:245–256. doi: 10.1017/s1355838200991908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain JR, Lee Y, Lane WS, Engelke DR. Purification and characterization of the nuclear RNase P holoenzyme complex reveals extensive subunit overlap with RNase MRP. Genes Dev. 1998;12:1678–1690. doi: 10.1101/gad.12.11.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang DD, Clayton DA. A novel endoribonuclease cleaves at a priming site of mouse mitochondrial DNA replication. EMBO J. 1987;6:409–417. doi: 10.1002/j.1460-2075.1987.tb04770.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S, Archer RH, Zengel JM, Lindahl L. The RNA of RNase MRP is required for normal processing of ribosomal RNA. Proc Natl Acad Sci USA. 1994;91:659–663. doi: 10.1073/pnas.91.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eder PS, Kekuda R, Stolc V, Altman S. Characterization of two scleroderma autoimmune antigens that copurify with human Ribonuclease P. Proc Natl Acad Sci USA. 1997;94:1101–1106. doi: 10.1073/pnas.94.4.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall JG. Cajal bodies: the first 100 years. Annu Rev Cell Dev Biol. 2000;16:273–300. doi: 10.1146/annurev.cellbio.16.1.273. [DOI] [PubMed] [Google Scholar]
- Hebert MD, Matera AG. Self-association of coilin reveals a common theme in nuclear body localization. Mol Biol Cell. 2000;11:4159–4171. doi: 10.1091/mbc.11.12.4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson MR, Cao LG, Taneja K, Singer RH, Wang YL, Pederson T. Nuclear domains of the RNA subunit of RNase P. J Cell Sci. 1997;110:829–837. doi: 10.1242/jcs.110.7.829. [DOI] [PubMed] [Google Scholar]
- Jacobson MR, Cao LG, Wang YL, Pederson T. Dynamic localization of RNase MRP RNA in the nucleolus observed by fluorescent RNA cytochemistry in living cells. J Cell Biol. 1995;131:1649–1658. doi: 10.1083/jcb.131.6.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrous N, Eder PS, Wesolowski D, Altman S. Rpp14 and Rpp29, two protein subunits of human ribonuclease P. RNA. 1999a;5:153–157. doi: 10.1017/s135583829800185x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrous N, Wolenski JS, Wesolowski D, Lee C, Altman S. Localization in the nucleolus and coiled bodies of protein subunits of the ribonucleoprotein ribonuclease P. J Cell Biol. 1999b;146:559–571. doi: 10.1083/jcb.146.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreis TE. Microinjected antibodies against the cytoplasmic domain of vesicular stomatitis virus glycoprotein block its transport to the cell surface. EMBO J. 1986;5:931–941. doi: 10.1002/j.1460-2075.1986.tb04306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupp G, Cherayil B, Frendewey D, Nishikawa S, Soll D. Two RNA species co-purify with RNase P from the fission yeast Schizosaccharomyces pombe. EMBO J. 1986;5:1697–1703. doi: 10.1002/j.1460-2075.1986.tb04413.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Smagula CS, Parsons WJ, Richardson JA, Gonzalez M, Hagler HK, Williams RS. Subcellular partitioning of MRP RNA assessed by ultrastructural and biochemical analysis. J Cell Biol. 1994;124:871–882. doi: 10.1083/jcb.124.6.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JL, Lee LF, Ye Y, Qian Z, Kung HJ. Nucleolar and nuclear localization properties of a herpesvirus bZIP oncoprotein, MEQ. J Virol. 1997;71:3188–3196. doi: 10.1128/jvi.71.4.3188-3196.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lygerou Z, Mitchell P, Petfalski E, Seraphin B, Tollervey D. The POP1 gene encodes a protein component common to the RNase MRP and RNase P ribonucleoproteins. Genes Dev. 1994;8:1423–1433. doi: 10.1101/gad.8.12.1423. [DOI] [PubMed] [Google Scholar]
- Lygerou Z, Allmang C, Tollervey D, Seraphin B. Accurate processing of a eukaryotic precursor ribosomal RNA by ribonuclease MRP in vitro. Science. 1996a;272:268–270. doi: 10.1126/science.272.5259.268. [DOI] [PubMed] [Google Scholar]
- Lygerou Z, Pluk H, van Venrooij WJ, Seraphin B. hPop1: an autoantigenic protein subunit shared by the human RNase P and RNase MRP ribonucleoproteins. EMBO J. 1996b;15:5936–5948. [PMC free article] [PubMed] [Google Scholar]
- Meetei AR, Ullas KS, Rao MR. Identification of two novel zinc finger modules and nuclear localization signal in rat spermatidal protein TP2 by site-directed mutagenesis. J Biol Chem. 2000;275:38500–38507. doi: 10.1074/jbc.M002734200. [DOI] [PubMed] [Google Scholar]
- Pluk H, Soffner J, Luhrmann R, van Venrooij WJ. cDNA cloning and characterization of the human U3 small nucleolar ribonucleoprotein complex-associated 55-kilodalton protein. Mol Cell Biol. 1998;18:488–498. doi: 10.1128/mcb.18.1.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluk H, Van Eenennaam H, Rutjes SA, Pruijn GJM, van Venrooij WJ. RNA-protein interactions in the human RNase MRP Ribonucleoprotein complex. RNA. 1999;5:512–524. doi: 10.1017/s1355838299982079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridanpaa M, van Eenennaam H, Pelin K, Chadwick R, Johnson C, Yuan B, van Venrooij W, Pruijn G, Salmela R, Rockas S, Makitie O, Kaitila I, de la Chapelle A. Mutations in the RNA component of RNase MRP cause a pleiotropic human disease, cartilage-hair hypoplasia. Cell. 2001;104:195–203. doi: 10.1016/s0092-8674(01)00205-7. [DOI] [PubMed] [Google Scholar]
- Rowland RR, Kervin R, Kuckleburg C, Sperlich A, Benfield DA. The localization of porcine reproductive and respiratory syndrome virus nucleocapsid protein to the nucleolus of infected cells and identification of a potential nucleolar localization signal sequence. Virus Res. 1999;64:1–12. doi: 10.1016/s0168-1702(99)00048-9. [DOI] [PubMed] [Google Scholar]
- Schmitt ME, Clayton DA. Yeast site-specific ribonucleoprotein endoribonuclease MRP contains an RNA component homologous to mammalian RNase MRP RNA and essential for cell viability. Genes Dev. 1992;6:1975–1985. doi: 10.1101/gad.6.10.1975. [DOI] [PubMed] [Google Scholar]
- Schmitt ME, Clayton DA. Nuclear RNase MRP is required for correct processing of pre-5.8S rRNA in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:7935–7941. doi: 10.1128/mcb.13.12.7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thebault S, Basbous J, Gay B, Devaux C, Mesnard JM. Sequence requirement for the nucleolar localization of human I-mfa domain-containing protein (HIC p40) Eur J Cell Biol, 2000;79:834–838. doi: 10.1078/0171-9335-00111. [DOI] [PubMed] [Google Scholar]
- Tollervey D, Kiss T. Function and synthesis of small nucleolar RNAs. Curr Opin Cell Biol. 1997;9:337–342. doi: 10.1016/s0955-0674(97)80005-1. [DOI] [PubMed] [Google Scholar]
- Van Eenennaam H, Jarrous N, van Venrooij WJ, Pruijn GJM. Architecture and function of the human endonucleases RNase P and RNase MRP. IUBMB LIFE. 2000;49:265–272. doi: 10.1080/15216540050033113. [DOI] [PubMed] [Google Scholar]
- Van Eenennaam H, Pruijn GJ, van Venrooij WJ. hPop4: a new protein subunit of the human RNase MRP and RNase P ribonucleoprotein complexes. Nucleic Acids Res, 1999;27:2465–2472. doi: 10.1093/nar/27.12.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheijen R, Wiik A, De Jong BA, Hoier Madsen M, Ullman S, Halberg P, van Venrooij WJ. Screening for autoantibodies to the nucleolar U3- and Th(7–2) ribonucleoproteins in patients' sera using antisense riboprobes. J Immunol Methods. 1994;169:173–182. doi: 10.1016/0022-1759(94)90261-5. [DOI] [PubMed] [Google Scholar]