The balance between inflammatory IFN-γ and antiinflammatory IL-10 plays a critical role in modulating type 1 immune responses. Yu et al. show that the transcription factor Bhlhe40 serves as a molecular switch between inflammatory and antiinflammatory Th1 cells.
Abstract
Type 1 T helper (Th1) cells play a critical role in host defense against intracellular pathogens and in autoimmune diseases by producing a key inflammatory cytokine interferon (IFN)–γ; some Th1 cells can also be antiinflammatory through producing IL-10. However, the molecular switch for regulating the differentiation of inflammatory and antiinflammatory Th1 cells is still elusive. Here, we show that Bhlhe40-deficient CD4 Th1 cells produced less IFN-γ but substantially more IL-10 than wild-type Th1 cells both in vitro and in vivo. Bhlhe40-mediated IFN-γ production was independent of transcription factor T-bet regulation. Mice with conditional deletion of Bhlhe40 in T cells succumbed to Toxoplasma gondii infection, and blockade of IL-10 signaling during infection rescued these mice from death. Thus, our results demonstrate that transcription factor Bhlhe40 is a molecular switch for determining the fate of inflammatory and antiinflammatory Th1 cells.
Introduction
Naive CD4 T cells differentiate into distinct subsets of T helper (Th) cells during immune responses (Zhu et al., 2010). Th subsets play a critical role in protective immunity against a variety of infections and are involved in different forms of inflammatory diseases. Type 1 Th (Th1) cells are indispensable for fighting against infections with intracellular pathogens. Th1 cells are also responsible for the pathogenesis of many autoimmune diseases. Transcription factor T-bet is the master transcriptional regulator for the development and functions of Th1 cells (Szabo et al., 2000; Lazarevic et al., 2013). T-bet directly regulates the expression of Th1 effector cytokine IFN-γ (Yagi et al., 2010; Zhu et al., 2012). Besides T-bet, other Th1 lineage–specific transcription factors, such as Runx3 and Hlx, either directly or indirectly regulate IFN-γ expression (Mullen et al., 2002; Djuretic et al., 2007; Yagi et al., 2010). It is possible that other lineage-specific transcription factors are also involved in this process (Hu et al., 2013).
IL-10 is an antiinflammatory cytokine. IL-10–producing CD4 T cells that possess regulatory functions are designated as TR1 cells (Roncarolo et al., 2006). However, Foxp3-expressing regulatory T (T reg) cells and GATA3-expressing Th2 cells also express IL-10 (Maynard et al., 2007; Wei et al., 2011). Furthermore, some Th1 cells are capable of expressing IL-10 during Leishmania major or Toxoplasma gondii infection, which elicits a very robust Th1 response (Anderson et al., 2007; Jankovic et al., 2007). The balance between the expression of inflammatory IFN-γ and antiinflammatory IL-10 by Th1 cells is critical for host mounting an appropriate immune response in controlling parasites. IFN-γ– or IL-10–deficient mice succumb to T. gondii infection as a result of either ineffective or excessive immune response, respectively (Hunter et al., 1994; Gazzinelli et al., 1996; Neyer et al., 1997). However, the molecular mechanism of regulating the balance between IFN-γ and IL-10 production in T cells is still elusive.
The transcription factor Bhlhe40, also known as Bhlhb2, Dec1, and Stra13, is up-regulated during T cell activation (Sun et al., 2001). In fact, IRF4 and Bhlhe40 are the top two transcription factors whose expression is highly induced within 4 h of T cell activation (Hu et al., 2013). It has been reported that Bhlhe40 is critically important for inducing autoimmune diseases, such as experimental autoimmune encephalomyelitis (EAE), an animal model of multiple sclerosis (Martínez-Llordella et al., 2013; Lin et al., 2014, 2016). However, the function of Bhlhe40 in type 1 immune response, particularly in vivo, has not been investigated.
Here, we report that transcription factor Bhlhe40 is required for optimal production of IFN-γ by Th1 cells both in vitro and in vivo, and this effect is independent of T-bet induction. However, Bhlhe40 suppresses IL-10 production by Th1 cells. Bhlhe40-deficient CD4 T cells, producing less IFN-γ but more IL-10, failed to induce colitis in mice in a transfer model. In addition, Bhlhe40 conditional knockout (cKO) mice are susceptible to T. gondii infection. Blockade of IL-10 signaling in Bhlhe40 cKO mice during T. gondii infection prevented these mice from death. Therefore, Bhlhe40 serves as an important molecular switch for the development of inflammatory and antiinflammatory Th1 cells.
Results and discussion
Characterization of Bhlhe40 cKO mice in the context of previous studies
Bhlhe40 is a transcription factor regulating circadian rhythms (Honma et al., 2002). Within the immune system, Bhlhe40 is not only expressed in activated T cells, but also expressed in eosinophils, macrophages, and dendritic cell subsets (Lin et al., 2016). To investigate the role of Bhlhe40 in T cells, we generated a cKO mouse strain, Bhlhe40fl/fl-CD4-Cre, in which the Bhlhe40 gene is deleted only in T cells (Fig. S1 A). Bhlhe40 cKO mice were born at the expected Mendelian ratio and appeared to be as healthy as their Bhlhe40fl/fl WT littermates and C57BL/6 WT mice.
Because it has been reported that Bhlhe40 germline KO (Bhlhe40−/−) mice are resistant to EAE induction, we first examined our Bhlhe40 cKO mice using this model. Consistent with the previous studies (Miyazaki et al., 2010; Martínez-Llordella et al., 2013; Lin et al., 2014, 2016), Bhlhe40 cKO mice immunized with MOG35–55 peptide in CFA were also markedly resistant to EAE compared with WT mice (Fig. S1 B). Paradoxically, it has also been shown that Bhlhe40−/− mice develop late-onset of lymphoproliferative disease associated with autoantibodies, which may be a result of a requirement for Bhlhe40 in T reg cell maintenance during aging (Sun et al., 2001; Miyazaki et al., 2010). Consistent with these studies, our Bhlhe40−/− mice also developed splenomegaly and lymphadenopathy starting at 4–5 mo of age (unpublished data). However, our Bhlhe40fl/fl-Foxp3-Cre mice kept for >6 mo did not develop any obvious abnormality before they were euthanized (unpublished data). These results suggest that T cells in Bhlhe40−/− mice may have a developmental defect, and/or Bhlhe40 expression in non-T cells may indirectly affect T cell activation in these mice. Therefore, our Bhlhe40 cKO mice offer a unique tool to study Bhlhe40 function in T cells.
To further assess whether the absence of Bhlhe40 could have functional consequences on the ability of T reg cells to control tissue inflammation, we used a well-described inflammatory bowel disease model induced by T cell transfer into Rag1−/− recipient (Powrie et al., 1994). As expected, transferring naive CD4 T cells alone led to severe inflammation associated with significant weight loss (Fig. S1 C). Cotransferring T reg cells from either Bhlhe40fl/+-Foxp3-Cre or Bhlhe40fl/fl-Foxp3-Cre mice with naive CD4 T cells effectively prevented disease. These results indicate that Bhlhe40 deficiency in T reg cells does not result in an obvious loss of T reg cell functions. Therefore, our subsequent studies focused on the effect of Bhlhe40 in effector T cells.
Bhlhe40 is required for optimal IFN-γ production by Th1 cells
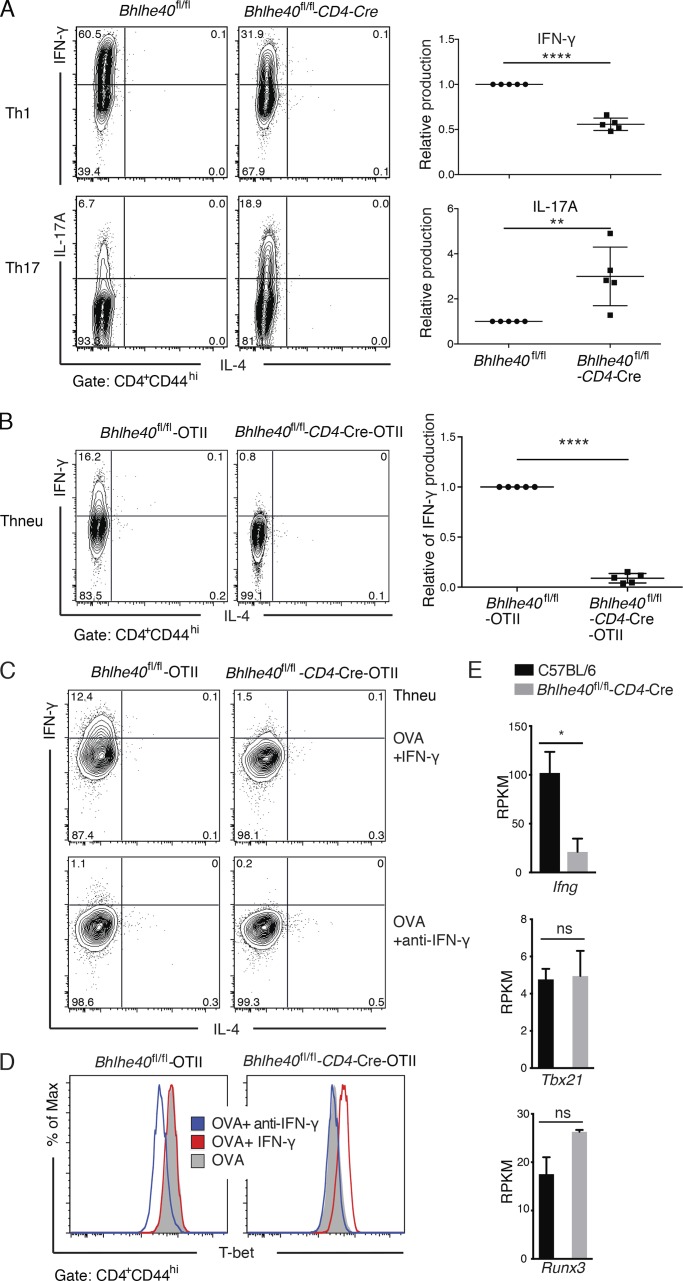
We first examined cytokine production by in vitro polarized Th cells. Although polarized Th cells of the Bhlhe40 cKO origin were able to produce signature cytokines of each lineage (Fig. S1 D), Bhlhe40 cKO cells showed an ∼50% reduction in IFN-γ production (for Th1; Fig. 1 A), but an approximately threefold-increased in IL-17A production (for Th17; Fig. 1 A). However, inducible T reg cell differentiation indicated by Foxp3 induction was similar in the presence or absence of Bhlhe40 (Fig. S1 D).
Figure 1.
Bhlhe40 regulates IFN-γ production by in vitro differentiated Th1 cells without affecting T-bet induction. (A) In vitro differentiation of Th1 and Th17 cells from sorted naive CD4 T cells with plate-bound anti-CD3/CD28 for 4 d. Flow cytometric analysis of IFN-γ and IL-17A production by CD4+CD44hi Th1 and Th17 cells, respectively, from WT and cKO mice (left). Graphical representation of relative IFN-γ and IL-17A production of cKO mice is compared with WT set as 1 (right). A representative of five independent experiments is shown. In each experiment at least two mice were used in each group (mean ± SEM, n = 5). Statistical significance was determined by a two-tailed unpaired Student’s t test. (B–D) Sorted naive OTII-CD4 T cells were stimulated with 10 µm OVA323–339 peptide under Thneu conditions with CD11c+ dendritic cells for 4 d in the presence or absence of IFN-γ or anti–IFN-γ antibody as indicated and restimulated with PMA-ionomycin in the presence of monensin for 4 h. (B) Flow cytometric analysis of IFN-γ production by CD4+CD44hi cells from Bhlhe40fl/fl-OTII and Bhlhe40fl/fl-CD4-Cre-OTII transgenic mice. Right: Graphical representation of relative IFN-γ production of cKO is compared with WT set as 1 (mean ± SEM, n = 5; ≥2 mice per group in each experiment). (C) Flow cytometric analysis of IFN-γ production by CD4+CD44hi cells from Bhlhe40fl/fl-OTII and Bhlhe40fl/fl-CD4-Cre-OTII transgenic mice in the presence of OVA323–339 peptide with either IFN-γ or anti–IFN-γ antibody as indicated. (D) Histogram plots of T-bet expression from same cells treated with OVA323–339 peptide alone, together with IFN-γ, or anti–IFN-γ antibody as indicated. Gray solid, OVA peptide alone; red line, OVA+ IFN-γ; blue line, OVA+ anti–IFN-γ antibody. Data are representative of three independent experiments, and in each experiment at least three mice in each group were used (C and D). (E) Gene expression values (RPKM) of Ifng, Tbx21, and Runx3 from RNA-Seq analysis of C57BL/6 WT and Bhlhe40 cKO Th1 cells (n = 2). ns, not significant; *, P < 0.05; **, P < 0.01; ****, P < 0.0001; Student’s t test.
It has been reported that Bhlhe40 promotes cell survival and proliferation induced by CD28-mediated signaling by using the Bhlhe40 germline KO (Martínez-Llordella et al., 2013). To test whether Bhlhe40 regulates cell proliferation, naive CD4 T cells from WT and Bhlhe40 cKO mice were labeled with CFSE before they were cultured under Th1, Th17, and neutral (Thneu) conditions. A similar cell division profile of WT and cKO cells, indicated by CFSE dilution, was observed, consistent with identical cell recovery by the end of the culture (Fig. S1, E and F). These results suggest that down-regulation of IFN-γ production in cKO Th1 cells is not a result of a defect in cell proliferation. In addition, we examined the expression of T-bet, the key transcription factor of Th1 cells, and found that T-bet expression in cKO Th1 cells was comparable to that in WT Th1 cells (Fig. S1 G).
To further test the function of Bhlhe40 during T cell differentiation in an antigen-specific model, we generated Bhlhe40fl/fl-OTII (WT-OTII) and Bhlhe40fl/fl-CD4-Cre-OTII (cKO-OTII) mice in which CD4 T cells express T cell receptor specific for OVA323–339 peptide. We found that cKO-OTII CD4 T cells stimulated with OVA323–339 peptide–loaded WT dendritic cells had a dramatic reduction in IFN-γ production under Thneu culture conditions (Fig. 1 B). Adding anti–IFN-γ antibody into the culture blocked the IFN-γ production in the WT-OTII group (Fig. 1 C), accompanied by a significant reduction in T-bet expression (Fig. 1 D), suggesting that the IFN-γ–/T-bet–/IFN-γ–positive feedback loop plays a major role in this culture system. Accordingly, cKO-OTII CD4 T cells cultured under Thneu conditions failed to up-regulate T-bet, possibly as a result of a defect in IFN-γ production (Fig. 1 D). Indeed, addition of exogenous IFN-γ restored T-bet expression in cKO-OTII cells to a level found in WT-OTII CD4 T cells (Fig. 1 D). However, despite of T-bet induction, IFN-γ production by CKO-OTII CD4 T cells was still defective (Fig. 1 C), indicating that Bhlhe40 is required for IFN-γ production independent of T-bet regulation.
We next performed differentially expressed gene analysis based on RNA-Seq data for WT and cKO Th1 cells to assess gene expression regulated by Bhlhe40. Consistent with intracellular staining results, RNA-Seq analysis showed a significant reduction of Ifng expression in cKO Th1 cells, whereas the expression of Tbx21 (encoding T-bet) and Runx3 was comparable between WT and cKO Th1 cells (Fig. 1 E). Collectively, these results indicate Bhlhe40 is required for optimal IFN-γ expression by Th1 cells in a T-bet regulation–independent manner. It has been reported that Bhlhe40 interacts with T-bet and thus serves as a cofactor to regulate IFN-γ expression in NKT cells (Kanda et al., 2016); however, we failed to detect protein–protein interaction between Bhlhe40 and T-bet (unpublished data), suggesting that Bhlhe40 may work on the Ifng gene in a T-bet–independent manner.
Bhlhe40 cKO Th1 cells overproduce IL-10
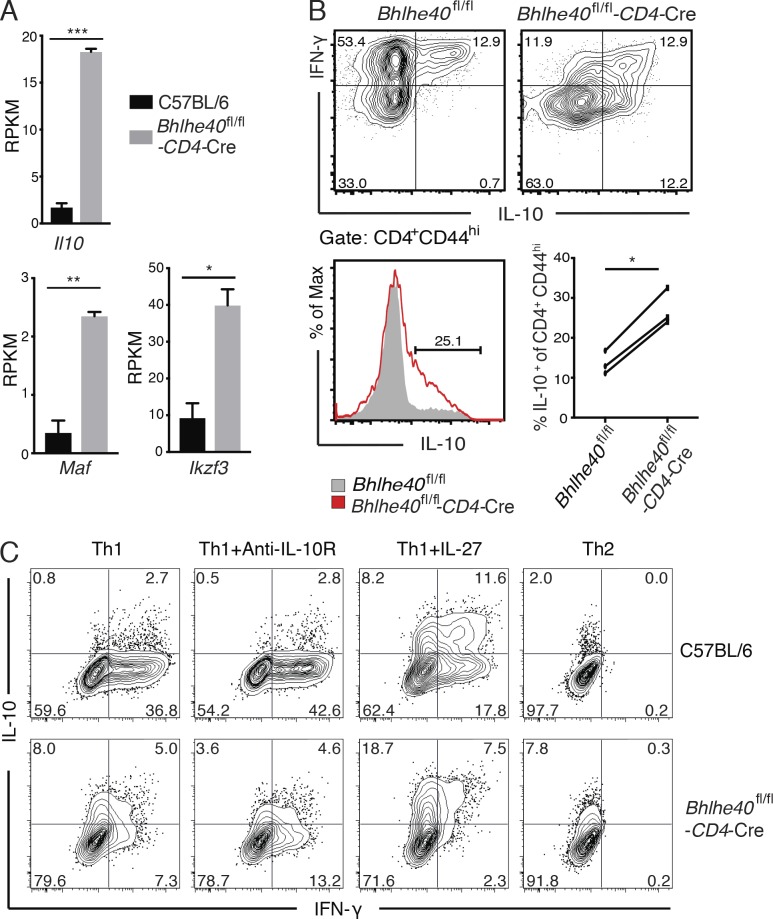
Our RNA-Seq analysis revealed 245 genes that are regulated by Bhlhe40 in Th1 cells (Table S1; fold change > 2; false discovery rate < 0.01; reads per kilobase of exon model per million reads [RPKM] > 2). Strikingly, Il10 is the most up-regulated genes in cKO Th1 cells compared with WT Th1 cells (Fig. 2 A and Table S1). Interestingly, two IL-10–inducing transcription factors, c-Maf (Pot et al., 2009) and IKZF3 (Evans et al., 2014), were increased in Bhlhe40-deficient Th1 cells (Fig. 2 A and Table S1), suggesting that Bhlhe40’s effect on IL-10 expression may be indirect. Whether up-regulation of c-Maf and/or IKZF3 is responsible for increased IL-10 expression in Bhlhe40-deficient Th1 cells requires further investigation. The increased IL-10 production in cKO Th1 cells was confirmed at the protein level by intracellular staining (Fig. 2 B). Interestingly, although IL-10 was produced by WT Th1 cells that also expressed IFN-γ as previously reported (Jankovic et al., 2007), both IFN-γ–expressing and nonexpressing cKO Th1 cells were capable of producing IL-10.
Figure 2.
Bhlhe40 suppresses IL-10 production by Th1 cells. (A) Gene expression values (RPKM) of Il10, Maf, and Ikzf3 from RNA-Seq analysis of C57BL/6 WT and Bhlhe40 cKO Th1 cells (n = 2). (B) Sorted naive CD4 T cells cultured under Th1-polarizing conditions with plate-bound anti-CD3/CD28 for 4 d and restimulated with PMA-ionomycin for 4 h. Flow cytometric analysis of IFN-γ/IL-10 production (top) and IL-10 expression (bottom left) by CD4+CD44hi WT and cKO Th1 cells. Gray solid, WT; red line, cKO. Bottom right: IL-10 production from three independent experiments (≥2 mice per group). *, P < 0.05; **, P < 0.01; ***, P < 0.001; Student’s t test. (C) Sorted naive CD4 T cells cultured under indicated conditions with plate-bound anti-CD3/CD28 for 4 d and restimulated with PMA-ionomycin for 4 h. Flow cytometric analysis of IFN-γ/IL-10 production by CD4+CD44hi WT and Bhlhe40 cKO cells. Data are representative of two independent experiments (≥2 mice per group).
To test whether increased IL-10 production is responsible for reduced IFN-γ by Bhlhe40-deficient Th1 cells, we used anti–IL-10R to block IL-10 signaling during the culture. Even in the presence of anti–IL-10R, Bhlhe40-deficient Th1 cells still produced less IFN-γ but more IL-10 compared with WT Th1 cells (Fig. 2 C), indicating that regulation of IFN-γ production by Bhlhe40 is largely independent of IL-10 up-regulation. The results from coculture of WT cells with Bhlhe40-deficient cells further confirm this conclusion (Fig. S2 A). Ifng mRNA was induced in both WT and Bhlhe40-deficient cells at 48 h of activation; however, Ifng transcription was transient in Bhlhe40-deficient cells (Fig. S2 B, upper panel). Il10 transcription kept rising over time in Bhlhe40-deficient Th1 cells, and WT Th1 cells expressed much less IL-10 (Fig. S2 B, lower panel). In addition, more IL-10 production was detected in Bhlhe40-deficient cells compared with WT cells when they were cultured in the presence of IL-27 or under Th2 conditions (Fig. 2 C). Overall, our data suggest that Bhlhe40 may serve as a molecular switch in determining Th1 cell fates for the expression of either inflammatory cytokine IFN-γ or antiinflammatory cytokine IL-10.
Bhlhe40-deficient CD4 T cells fail to induce colitis
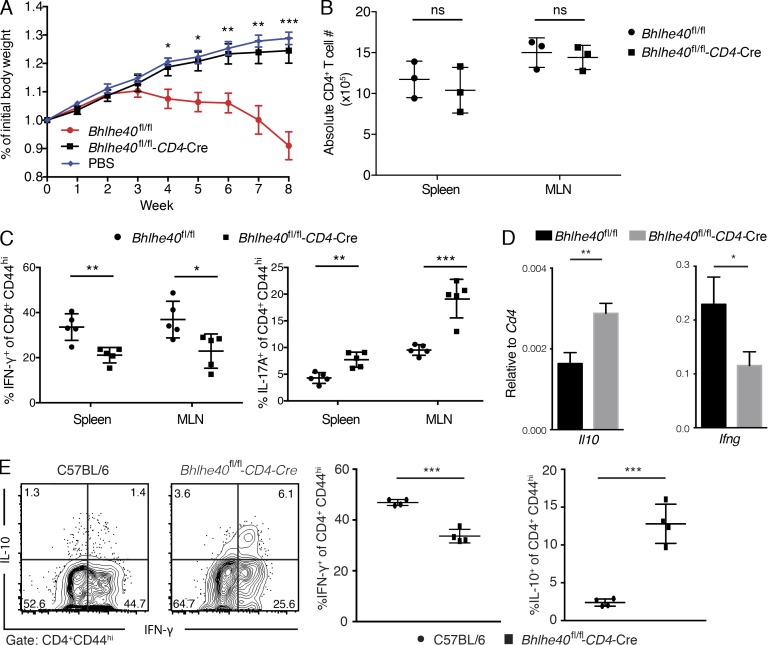
To assess Bhlhe40 function in Th1 cells in vivo, we used the inflammatory bowel disease model induced by transferring naive CD4 T cells into Rag1−/− recipients. As expected, transferring WT naive CD4 T cells led to severe inflammation associated with significant weight loss and colon thickening, whereas transferring cKO-naive CD4 T cells did not cause weight loss (Fig. 3 A). The failure of cKO CD4 T cells in inducing disease was not a result of a defect in cell expansion in vivo, because absolute cell number of CD4 T cells recovered from the recipients transferred with either WT or cKO naive cells was similar (Fig. 3 B).
Figure 3.
Bhlhe40 expression in T cells is required for colitis induction. (A) Rag1−/− mice received sorted naive CD4+CD25−CD45RBhi T cells from either WT (red dots) or Bhlhe40 cKO (black squares) or injected i.v. with PBS (blue diamonds) as the control group. Mice were weighed weekly. Statistical significance of body weight of Bhlhe40 cKO versus WT (mean ± SEM, n = 5) at different time points was determined by a two-tailed unpaired Student’s t test. Data are representative of three independent experiments. (B) Graphical representation of the absolute number of CD4 T cells harvested from spleen and mesenteric lymph node (MLN) of Rag1−/− mice, in which sorted naive CD4+CD25−CD45RBhi T cells from either WT (dots) or Bhlhe40 cKO mice (squares) were i.v. transferred for 4 wk (n = 3 per group) are shown. Data are representative of two independent experiments. (C) Percentages of IFN-γ+ and IL-17A+ cells among the CD4+CD44hi T cells in spleen and MLN from Rag1−/− mice received WT (dots) or Bhlhe40 cKO transfer (squares) cells for 8 wk (mean ± SEM, n = 5, right). Data are representative of three independent experiments. (D) Sorted naive CD4 T cells from WT or Bhlhe40 cKO were transferred into Rag1−/− mice. 2 wk after transfer, CD4 T cells were sorted from the spleens and RNAs were prepared from sorted cells. Real time PCR analysis of Ifng and Il10 mRNA was performed (mean ± SEM, n = 5). Data are representative of two independent experiments. (E) Sorted naive CD4 T cells from C57BL/6 or Bhlhe40 cKO were transferred into Rag1−/− mice. 2 wk after transfer, cells from MLN were restimulated with PMA plus ionomycin, and then intracellular staining for IFN-γ and IL-10 was performed. Flow plots were gated on CD4+CD44hi cells. ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001; Student’s t test. Data are representative of two independent experiments.
Very few regulatory T cells were generated in this transfer model (<5%), and there was no difference between the WT and cKO groups (unpublished data). We also measured cytokine production by CD4 T cells recovered from spleen and lymph nodes of recipients at the end of experiments. Consistent with the data from in vitro studies, cKO CD4 T cells showed decreased IFN-γ but increased IL-17A production compared with WT CD4 T cells (Fig. 3 C). We also assessed IFN-γ and IL-10 expression 2 wk after transfer. Consistent with in vitro findings, IL-10 expression by cKO Th1 cells generated in vivo was increased, whereas IFN-γ expression was reduced compared with WT Th1 cells both at the mRNA and protein level (Fig. 3, D and E).
Although more experiments are needed to determine the role of IL-10 overproduction by cKO cells in this model, our data strongly indicate that Bhlhe40 plays an important role in inducing the expression of proinflammatory cytokine IFN-γ, but suppressing the expression of antiinflammatory cytokine IL-10 in Th1 cells in vivo.
Altered balance between IFN-γ and IL-10 in Bhlhe40 cKO mice that are susceptible to T. gondii infection
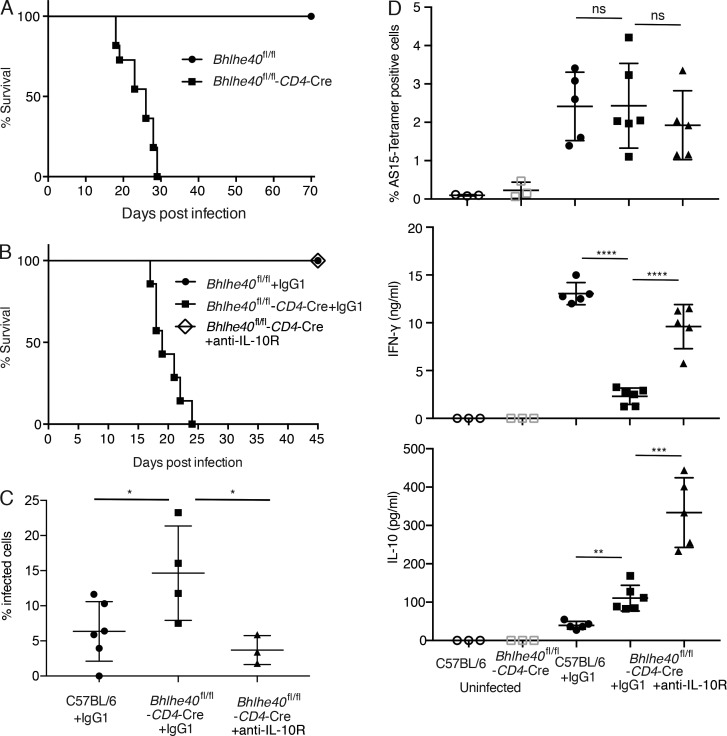
Previous studies have shown that balance between the production of inflammatory IFN-γ and antiinflammatory IL-10 is critical for mounting an appropriate immune response against T. gondii infection (Gazzinelli et al., 1996; Neyer et al., 1997; Jankovic et al., 2007). To test whether altered expression of IFN-γ and IL-10 in the absence of Bhlhe40 has any physiological consequence, we infected WT and Bhlhe40 cKO mice with T. gondii. As a consequence of T. gondii exposure, serum levels of IL-12p40 were increased dramatically in both infected WT and cKO mice (Fig. S2 C). In contrast to WT mice, which survived for the entire period of the experiments, all the cKO mice infected with an avirulent strain of T. gondii succumbed to infection (Fig. 4 A). The mortality was observed in between acute and chronic phase of infection (between 2–4 wk after infection). Interestingly, treating cKO mice with anti–IL-10R–blocking mAb before and after infection rescued these mice from death (Fig. 4 B). As expected, Bhlhe40 cKO mice were less efficient than WT mice in clearing parasites; however, anti–IL-10R treatment rescued this defect (Fig. 4 C). Furthermore, the levels of a liver enzyme aspartate transaminase (AST) in response to infection were identical between WT and Bhlhe40 cKO mice, indicating that Bhlhe40 cKO mice do not die of excessive immune responses (Fig. S2 D). We also found no defect in the generation of antigen-specific cells in the absence of Bhlhe40 (Fig. 4 D, upper panel). However, IFN-γ production by cKO cells upon antigen stimulation was substantially lower than that detected in the similarly treated WT cells (Fig. 4 D, middle panel), whereas IL-10 production by cKO cells were higher than that of WT cells (Fig.4 D, lower panel). IFN-γ from cKO mice injected with anti–IL-10R were dramatically increased compared with that in control mAb–treated cKO mice, indicating IL-10 blockade enhances IFN-γ production (Fig. 4 D, middle panel). However, such levels were still significantly lower than that found in the infected WT mice, indicating that Bhlhe40 is required for optimal IFN-γ production by Th1 cells in response to T. gondii infection. Whether Bhlhe40 also regulates IFN-γ production in antigen-specific CD8 T cells and, if so, how this defect may add to the defect of Bhlhe40-deficient CD4 T cells in host defense requires further investigation. Nevertheless, partial restoration of the balance between IFN-γ and IL-10 in cKO mice is sufficient to elicit protective immunity.
Figure 4.
Bhlhe40 cKO mice are susceptible to T. gondii infection. (A) Bhlhe40 WT and cKO mice were infected i.p. with a mean of 15–20 ME49 cysts of T. gondii. Survival of infected mice was monitored. The survival curves shown are one representative of two independent experiments preformed (n = 11 per group). (B) T. gondii–infected mice were treated i.p. with anti–IL-10R or control IgG1 at 2 d before and 2 d after infection. Survival of infected mice (WT with control IgG1, dots; cKO with control IgG1, squares; cKO with anti–IL-10R, diamonds) was monitored daily (n = 7–8). The data shown are the pooled results from two independent experiments. (C) T. gondii–infected mice were treated i.p. with anti–IL-10R or control IgG1 at 2 d before and 2 d after infection. Parasite burden was determined by counting infected cells in cytospin smears obtained from PECs on day 8 (mean ± SEM, n = 3–6). *, P < 0.05; Student’s t test.Data are representative of two independent experiments, and similar results were obtained in a third experiment analyzed on day 13. (D) PECs from 13-d infected mice were stained with tetramer to assess the frequency of T. gondii AS15 peptide-specific CD4 T cells. These PECs were also cultured with STAg for 3 d. IFN-γ and IL-10 production by T. gondii antigen-specific cells were measured by ELISA (mean ± SEM, n = 3–6). ns, not significant; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; Student’s t test. Data are representative of two independent experiments.
Collectively, our results indicate that Bhlhe40 serves as a molecular switch in determining the balance between the inflammatory cytokine IFN-γ and the antiinflammatory cytokine IL-10 in Th1 cells both in vitro and in vivo. The regulation of IFN-γ by Bhlhe40 is independent of T-bet regulation. Future understanding of pathways and/or molecules that regulate the expression of Bhlhe40 may offer new drug targets aiming to boost immune response or to limit inflammation.
Materials and methods
Mice
C57BL/6 mice and C57BL/6 Flpe (Taconic Line 7089) were purchased from Taconic Farms. C57BL/6-CD45.1 mice (Taconic Line 7), C57BL/6 Rag1−/− mice (Taconic Line 146), C57BL/6-OT-II (Taconic Line 187), and C57BL/6 CD4-Cre (Taconic Line 4196) were obtained from the National Institute of Allergy and Infectious Diseases (NIAID)–Taconic repository. To generate Bhlhe40fl/fl-CD4-Cre mice, mouse embryonic stem cells containing the Bhlhe40tm1a(KOMP)Wtsi allele (clone EPD0208_6_A02) were obtained from the University of California, Davis, KOMP Repository. This allele has a trapping cassette, SA-βneo-pA (splice acceptor-β-neo-polyA), flanked by flippase recombinase (Flp) target FRT sites upstream of exon4, resulting in truncation of the endogenous transcript and thus creating a constitutive null mutation. The cassette also tags the gene with a lacZ reporter. The FRT flanked region can be removed by Flp, but exon4, a critical exon, remains intact. Exon4 can be further removed by the Cre recombinase to achieve a cKO. The embryonic stem cells were injected into C57BL/6 blastocysts. Chimeric mice were born and mice were genotyped by Southern blotting as per instructions of the IKMC project 24480. After successful germline transmission, Bhlhe40+/flox-frt-neo mice were crossed with Flpe mice to facilitate in vivo FRT-neo deletion. The Bhlhe40flox/flox (fl/fl will be used herein to indicate Bhlhe40flox/flox for simplicity) mice were further crossed with CD4-Cre transgenic mice to generate Bhlhe40fl/fl-CD4-Cre cKO mice in C57BL/6 background. To generate mice capable of Bhlhe40 inactivation in T reg cells, Bhlhe40fl/fl mice were crossed with Foxp3-IRES-YFP-Cre mice (Foxp3-Cre mice, gift from A. Rudensky, Sloan-Kettering Institute, New York, NY; Rubtsov et al., 2008) to generate Bhlhe40fl/fl–Foxp3-Cre mice. All mice were genotyped as instruction of IKMC project 24480 by PCR using primer pairs: 5′-GGGGGAGATGCTCTTCTGAT-3′ and 5′-GGGGAAGTCTCCCATAACCT-3′; 5′-CACTTGCTGATGCGGTGCTGATTAC-3′ and 5′-GACACCAGACCAACTGGTAATGGTAG-3′; 5′-CTGACCGTACACCAAAATTTGC-3′ and 5′-GATCCTGGCAATTTCGGCTATACG-3′. Bhlhe40fl/fl and Bhlhe40fl/fl-CD4-Cre mice were maintained by genotyping using primer pairs: to check last exon4 deletion using 5′-GGGGGAGATGCTCTTCTGAT-3′ and 5′-GGGGAAGTCTCCCATAACCT-3′; to select CD4-Cre positive using 5′-CTGACCGTACACCAAAATTTGC-3′ and 5′-GATCCTGGCAATTTCGGCTATACG-3′. For Foxp3-Cre, primer pair-1: 5′-CCAGATGTTGTGGGTGAGTG-3′ and 5′-TGGACCGTAGATGAATTTGAGTT-3′; and primer pair-2: 5′-AGGATGTGAGGGACTACCTCCTGTA-3′ and 5′-TCCTTCACTCTGATTCTGGCAATTT-3′.
To generate Bhlhe40fl/fl-OT-II and Bhlhe40fl/fl-CD4-Cre-OT-II mice, Bhlhe40fl/fl and Bhlhe40fl/fl-CD4-Cre mice were crossed with C57BL/6-OT-II transgenic mice. OT-II transgene was genotyped by PCR using primer pairs: 5′-AAAGGGAGAAAAAGCTCTCC-3′ and 5′-ACACAGCAGGTTCTGGGTTC-3′; 5′-GCTGCTGCACAGACCTACT-3′ and 5′-CAGCTCACCTAACACGAGGA-3′.
All the mice were bred and/or maintained in the NIAID-specific pathogen free animal facility, and the experiments were done when mice were 8- to 14-wk old under protocols approved by the NIAID Animal Care and Use Committee.
Cell Preparation and cell sorting
Total CD4 T cells were purified by using mouse CD4 (L3T4) microbeads (Miltenyi Biotec Inc.) according to the manufacturer’s instructions. Naive CD4 T cells were isolated from lymph nodes. Lymph node cells were stained with FITC-anti-CD4, PE-anti-CD25, and APC-anti-CD45Rb and then sorted for CD4+CD25−CD45Rbhi population using FACSAria (BD Biosciences). For some in vitro culture experiments, lymph node cells were stained with APC-anti-CD4, APC-Cy7-anti-CD44, PE-anti-CD25, and PB-anti-CD62L and then sorted for CD4+CD44loCD25−CD62Lhi naive CD4 population.
Cell culture and proliferation assay
Cell culture medium and conditions were described previously (Yu et al., 2015). CFSE (Invitrogen Corporation) was used to detect the proliferation of naive CD4 T cells according to the manufacturer’s instructions. Cells were harvested on day 4, and the intensity of CFSE staining of CD4 T cells was determined by using a flow cytometer LSRII (BD Biosciences) and analysis was performed with FlowJo software (Tree Star). Equal numbers of sorted naive CD4 T cells were stimulated with plate-bound anti-CD3 (145-2C11) and anti-CD28 (37.51) in the absence or presence of IL-2 (50 U/ml) containing RPMI 1640 media. The number of live cells was counted on day 4 after trypan blue staining.
In vitro T cell differentiation and staining
Sorted naive CD4 T were stimulated with plate-bound anti-CD3 (145-2C11) and anti-CD28 (37.51) or with APC prepared from T cell–depleted splenocytes (Zhu et al., 2012). All cytokines used in cell culture were purchased from PeproTech. T cell–depleted splenocytes were prepared by incubation with anti-Thy1.2 mAb supernatant and rabbit complement (Cedarlane Laboratories Limited) at 37°C for 45 min followed by irradiation at 30 Gy (3,000 rad). Naive CD4 T cells were cultured with irradiated T cell–depleted splenocytes at a ratio of 1:5 in the presence of 1 µg/ml of anti-CD3 and 3 µg/ml of anti-CD28 for 3–4 d with various combinations of antibodies and cytokines: for Th1 cell conditions, 50 IU/ml of hIL-2, 10 ng/ml of IL-12, and 10 µg/ml anti-IL-4 (11B11; some Th1 cell cultures include 10 ng/ml IFN-γ or IL-27 10ng/ml as specified); for Th17 cell conditions, 5 ng/ml TGF-β, 10 ng/ml IL-6, 10 ng/ml IL-1β, 10 µg/ml anti-IL-4, 10 µg/ml anti-IL-12, and 10 µg/ml anti-IFN-γ; for Th2 cell conditions, 50 IU/ml of hIL-2, 10 ng/ml of IL-4, 10 µg/ml anti-IL-12, and 10 µg/ml anti–IFN-γ. For inducible T reg cell–polarizing condition, cells were cultured with plate-bound anti-CD3/CD28 in the presence of 100 IU/ml of hIL-2, 5 ng/ml TGF-β, 10 µg/ml anti–IL-4, 10 µg/ml anti–IL-12, and 10 µg/ml anti–IFN-γ for 4 d. For cells cultured in neutral conditions (Thneu), 50 IU/ml of hIL-2. In some Thneu cell cultures, naive CD4 T cells from OTII mice with dendritic cells (DCs) purified by CD11c microbeads (Miltenyi Biotec Inc.) were activated by OVA323–339 peptide (10 µM) in the absence or presence of either 10 ng/ml IFN-γ or 10 µg/ml anti–IFN-γ as indicated. All cytokines used in cell culture were purchased from PeproTech. All antibodies used in cell culture were purchased from Harlan. Cell surface molecules were stained in PBS with 2% FBS. Cells were stimulated with 10 ng/ml PMA and 500 nM ionomycin for 4 h or stimulated with plate-bound anti-CD3/anti-CD28 for 5 h in the presence of 2 mM monensin during the last 2 h. Cells were stained with a cocktail of fixable viability dye (eBioscience) and antibodies to cell surface markers and then fixed with 4% paraformaldehyde for 10 min at room temperature and permeabilized in PBS containing 0.5% Triton X-100 and 0.1% BSA. They were then stained intracellularly for cytokines. Staining for transcription factors was performed with the Foxp3 Staining Buffer Set (eBioscience) according to the manufacturer’s instructions. Flow cytometry data were collected with an LSR II (BD Biosciences) and the results were analyzed by using FlowJo software (Tree Star). Antibodies specific for mouse CD4 (RM4-5), CD25 (PC61.5), CD44 (IM7), CD45.1 (A20), CD45Rb (C363.16A), IL-4 (11B11), IFN-γ (XMG1.2), IL-17A (TC11-18H10), Foxp3 (FJK-16s), T-bet (4B10), and Fixable Viability Dye eFluor 506 were purchased from eBioscience; antibodies specific for mouse FCγII/III (2.4G2) were obtained from Harlan.
T cell transfer model of colitis
Sorted naive CD4+CD25−CD45RBhi T cells from either C57BL/6, Bhlhe40fl/fl, or Bhlhe40fl/fl-CD4-Cre mice were adoptively transfer to Rag1−/− recipient mice (Line 146). Each mouse received 2∼3 × 105 naive CD4 T cells intravenously. In some experiments, CD4+CD25+ YFP+ T reg cell population were sorted from lymph nodes of Bhlhe40fl/+-Foxp3-Cre, Bhlhe40fl/fl-Foxp3-Cre mice stained with APC-anti-CD4 and PE-anti-CD25. They were cotransferred with sorted naive CD4+CD25−CD45RBhi T cells from CD45.1 congenic mice into Rag1−/− recipients at a 1:4 ratio. The body weight of mice was monitored weekly for all experiments.
Induction of EAE
For induction of active EAE, 8- to 10-wk-old female Bhlhe40fl/fl WT or Bhlhe40fl/fl-CD4-Cre mice were immunized at two sites subcutaneously with 150 µg MOG35–55 in complete Freund’s adjuvant (Sigma) containing Mycobacterium tuberculosis strain H37Ra (5 mg/ml; Difco). Mice were injected i.p. on days 0 and 2 with 200 ng pertussis toxin (Difco) in PBS. EAE was clinically assessed by daily assignment of scores on a scale of 0–5 as follows: partially limp tail, 0.5; completely limp tail, 1; limp tail and waddling gait, 1.5; paralysis of one hind limb, 2; paralysis of one hind limb and partial paralysis of the other hind limb, 2.5; paralysis of both hind limbs, 3; ascending paralysis, 3.5; paralysis of trunk, 4; moribund, 4.5; and death, 5.
T. gondii infection
Type II avirulent strain ME49 cysts were obtained from the brains of chronically infected C57BL/6 mice. To eliminate host cell contamination, the cyst preparations were treated with pepsin. Mice were inoculated i.p. with a mean of 15–20 cysts per animal. WT and Bhlhe40 CKO mice were injected i.p. with 1 mg per animal of the anti–IL-10R (1B1.3a) or the IgG1 isotype control antibody (BioXCell) on days −2 and 2 of T. gondii infection. Parasite burden was determined by counting infected cells in cytospin smears obtained from peritoneal exudate cells (PECs) on day 8 and 13. PECs harvested 13 d after infection were stained at 37°C for 1 h with Tetramer I-Ab-AVEIHRPVPGTAPPS-APC or -BV421 provided by the National Institutes of Health Tetramer Core Facility to assess AS15 peptide-specific CD4 T cells (Grover et al., 2012). Serum levels of AST were measured using a commercial kit (Roche Diagnostic) in an automatic analyzer (Cobas C501; Roche).
ELISA
Total PEC samples were cultured with T. gondii–soluble tachyzoite antigen (STAg, 5 µg/ml) in complete RPMI for 3 d. The samples were centrifuged and the supernatant was used to measure the levels of IFN-γ and IL-10 using mouse Quantikine ELISA kits for IFN-γ and IL-10 (R&D systems). Experiments were performed following the manufacturer’s instructions.
RNA Purification and Quantitative PCR
Total RNAs were isolated using a combination of TRIzol (Invitrogen) and RNeasy kit (QIAGEN). cDNAs were prepared using SuperScript III Reverse transcription (Invitrogen). Quantitative PCR was performed on a 7900HT Sequence Detection System (Applied Biosystems) using the following predesigned primer/probe sets: Ifng (Mm99999071_m1), Il10 (Mm00439616_m1), and Cd4 (Mm00442754_m1; all purchased from Thermo Fisher).
RNA-Seq
Naive CD4 T cells sorted from WT C57BL/6 or Bhlhe40fl/fl-CD4-Cre mice were cultured with plate-bound anti-CD3 and anti-CD28 under Th1-polarizing conditions for 3 d and then restimulated with PMA plus ionomycin for 2 h. Samples were prepared in duplicates. Total RNAs were isolated using Qiagen’s miRNeasy micro kit (217084; Qiagen). PolyA-tailed RNAs were purified from pure total RNA using Dynabeads mRNA DIRECT kit (61012; Ambion Life Technologies). Library constructions and RNA-Seq follow the protocols described previously (Chepelev et al., 2009). 50-bp reads were generated by the National Heart, Lung, and Blood Institute DNA Sequencing and Computational Biology Core. Sequence reads were mapped to mouse genome (mm9) by using bowtie 2 with default settings (Langmead and Salzberg, 2012). Reads mapped to multiple positions (MAPQ < 10) were discarded. The mRNA expression level of a gene was quantified by RPKM (Mortazavi et al., 2008) with in-house script. Differentially expressed genes were identified by edgeR 3 (Robinson et al., 2010) with the following criteria: false discovery rate < 0.01, fold change log2 ≥ 1, and RPKM ≥ 2.
Statistics
Groups were compared using the Prism 6 software (GraphPad) using a two-tailed unpaired Student’s t test or an ordinary one-way ANOVA. Data were presented as mean ± SEM. A p-value <0.05 was considered statistically significant and indicated as *; P < 0.01 was indicated as **; P < 0.001 was indicated as ***; and P < 0.0001 was indicated as ****. Not statistically significant was indicated as ns.
Accession code
The RNA-Seq datasets are available in the Gene Expression Omnibus database under the accession no. GSE103173.
Online supplemental material
Fig. S1 shows schematic of Bhlhe40fl/fl-CD4-Cre (Bhlhe40 cKO) mouse construction; EAE induction in WT and Bhlhe40 cKO mice; functions of Bhlhe40-deficient T reg cells in suppressing colitis; in vitro differentiation of WT and Bhlhe40 cKO cells under Th1, Th2, Th17, and inducible T reg cell–polarizing conditions; cell proliferation and expansion of WT and Bhlhe40 cKO cells; and T-bet expression in WT and Bhlhe40 cKO Th1 cells. Fig. S2 shows WT and Bhlhe40 cKO cell coculture under Th1 conditions; RT-PCR analysis of Ifng and Il10 mRNAs in WT and Bhlhe40 cKO Th1 cells; serum IL-12 and AST levels in WT and Bhlhe40 cKO mice after T. gondii infection. Table S1 is an Excel file of differentially expressed genes between in vitro–generated WT and Bhlhe40 cKO Th1 cells with their RPKM values.
Acknowledgments
We thank Julie Edwards for her excellent assistance in cell sorting and the National Heart, Lung, and Blood Institute DNA Sequencing Core facility for sequencing the RNA-Seq libraries.
This work is supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health (grant 1ZIA-AI-001169) and the US-China Biomedical Collaborative Research Program (grant AI-129775). P. Su and B. Sun are supported by a grant from the National Natural Science Foundation of China (grant 81761128009).
The authors declare no competing financial interests.
Author contributions: F. Yu, B. Sun, and J. Zhu conceived the project. F. Yu and S. Sharma performed the majority of experiments. D. Jankovic contributed to T. gondii infection experiments. G. Hu and R. Li performed bioinformatics analysis. R.K. Gurram, P. Su, and S. Rieder helped in some experiments and data analysis. K. Zhao provided critical advice to the project and edited the manuscript. F. Yu and J. Zhu wrote the manuscript. S. Sharma, D. Jankovic, and B. Sun edited the manuscript. B. Sun and J. Zhu supervised the project.
References
- Anderson C.F., Oukka M., Kuchroo V.J., and Sacks D.. 2007. CD4+CD25-Foxp3- Th1 cells are the source of IL-10-mediated immune suppression in chronic cutaneous leishmaniasis. J. Exp. Med. 204:285–297. 10.1084/jem.20061886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chepelev I., Wei G., Tang Q., and Zhao K.. 2009. Detection of single nucleotide variations in expressed exons of the human genome using RNA-Seq. Nucleic Acids Res. 37:e106 10.1093/nar/gkp507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djuretic I.M., Levanon D., Negreanu V., Groner Y., Rao A., and Ansel K.M.. 2007. Transcription factors T-bet and Runx3 cooperate to activate Ifng and silence Il4 in T helper type 1 cells. Nat. Immunol. 8:145–153. 10.1038/ni1424 [DOI] [PubMed] [Google Scholar]
- Evans H.G., Roostalu U., Walter G.J., Gullick N.J., Frederiksen K.S., Roberts C.A., Sumner J., Baeten D.L., Gerwien J.G., Cope A.P., et al. 2014. TNF-α blockade induces IL-10 expression in human CD4+ T cells. Nat. Commun. 5:3199 10.1038/ncomms4199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzinelli R.T., Wysocka M., Hieny S., Scharton-Kersten T., Cheever A., Kühn R., Müller W., Trinchieri G., and Sher A.. 1996. In the absence of endogenous IL-10, mice acutely infected with Toxoplasma gondii succumb to a lethal immune response dependent on CD4+ T cells and accompanied by overproduction of IL-12, IFN-gamma and TNF-alpha. J. Immunol. 157:798–805. [PubMed] [Google Scholar]
- Grover H.S., Blanchard N., Gonzalez F., Chan S., Robey E.A., and Shastri N.. 2012. The Toxoplasma gondii peptide AS15 elicits CD4 T cells that can control parasite burden. Infect. Immun. 80:3279–3288. 10.1128/IAI.00425-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honma S., Kawamoto T., Takagi Y., Fujimoto K., Sato F., Noshiro M., Kato Y., and Honma K.. 2002. Dec1 and Dec2 are regulators of the mammalian molecular clock. Nature. 419:841–844. 10.1038/nature01123 [DOI] [PubMed] [Google Scholar]
- Hu G., Tang Q., Sharma S., Yu F., Escobar T.M., Muljo S.A., Zhu J., and Zhao K.. 2013. Expression and regulation of intergenic long noncoding RNAs during T cell development and differentiation. Nat. Immunol. 14:1190–1198. 10.1038/ni.2712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter C.A., Subauste C.S., Van Cleave V.H., and Remington J.S.. 1994. Production of gamma interferon by natural killer cells from Toxoplasma gondii-infected SCID mice: regulation by interleukin-10, interleukin-12, and tumor necrosis factor alpha. Infect. Immun. 62:2818–2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankovic D., Kullberg M.C., Feng C.G., Goldszmid R.S., Collazo C.M., Wilson M., Wynn T.A., Kamanaka M., Flavell R.A., and Sher A.. 2007. Conventional T-bet+Foxp3- Th1 cells are the major source of host-protective regulatory IL-10 during intracellular protozoan infection. J. Exp. Med. 204:273–283. 10.1084/jem.20062175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda M., Yamanaka H., Kojo S., Usui Y., Honda H., Sotomaru Y., Harada M., Taniguchi M., Suzuki N., Atsumi T., et al. 2016. Transcriptional regulator Bhlhe40 works as a cofactor of T-bet in the regulation of IFN-γ production in iNKT cells. Proc. Natl. Acad. Sci. USA. 113:E3394–E3402. 10.1073/pnas.1604178113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., and Salzberg S.L.. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 9:357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarevic V., Glimcher L.H., and Lord G.M.. 2013. T-bet: a bridge between innate and adaptive immunity. Nat. Rev. Immunol. 13:777–789. 10.1038/nri3536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.C., Bradstreet T.R., Schwarzkopf E.A., Sim J., Carrero J.A., Chou C., Cook L.E., Egawa T., Taneja R., Murphy T.L., et al. 2014. Bhlhe40 controls cytokine production by T cells and is essential for pathogenicity in autoimmune neuroinflammation. Nat. Commun. 5:3551 10.1038/ncomms4551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.C., Bradstreet T.R., Schwarzkopf E.A., Jarjour N.N., Chou C., Archambault A.S., Sim J., Zinselmeyer B.H., Carrero J.A., Wu G.F., et al. 2016. IL-1-induced Bhlhe40 identifies pathogenic T helper cells in a model of autoimmune neuroinflammation. J. Exp. Med. 213:251–271. 10.1084/jem.20150568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Llordella M., Esensten J.H., Bailey-Bucktrout S.L., Lipsky R.H., Marini A., Chen J., Mughal M., Mattson M.P., Taub D.D., and Bluestone J.A.. 2013. CD28-inducible transcription factor DEC1 is required for efficient autoreactive CD4+ T cell response. J. Exp. Med. 210:1603–1619. 10.1084/jem.20122387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard C.L., Harrington L.E., Janowski K.M., Oliver J.R., Zindl C.L., Rudensky A.Y., and Weaver C.T.. 2007. Regulatory T cells expressing interleukin 10 develop from Foxp3+ and Foxp3- precursor cells in the absence of interleukin 10. Nat. Immunol. 8:931–941. 10.1038/ni1504 [DOI] [PubMed] [Google Scholar]
- Miyazaki K., Miyazaki M., Guo Y., Yamasaki N., Kanno M., Honda Z., Oda H., Kawamoto H., and Honda H.. 2010. The role of the basic helix-loop-helix transcription factor Dec1 in the regulatory T cells. J. Immunol. 185:7330–7339. 10.4049/jimmunol.1001381 [DOI] [PubMed] [Google Scholar]
- Mortazavi A., Williams B.A., McCue K., Schaeffer L., and Wold B.. 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 5:621–628. 10.1038/nmeth.1226 [DOI] [PubMed] [Google Scholar]
- Mullen A.C., Hutchins A.S., High F.A., Lee H.W., Sykes K.J., Chodosh L.A., and Reiner S.L.. 2002. Hlx is induced by and genetically interacts with T-bet to promote heritable T(H)1 gene induction. Nat. Immunol. 3:652–658. 10.1038/ni807 [DOI] [PubMed] [Google Scholar]
- Neyer L.E., Grunig G., Fort M., Remington J.S., Rennick D., and Hunter C.A.. 1997. Role of interleukin-10 in regulation of T-cell-dependent and T-cell-independent mechanisms of resistance to Toxoplasma gondii. Infect. Immun. 65:1675–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pot C., Jin H., Awasthi A., Liu S.M., Lai C.Y., Madan R., Sharpe A.H., Karp C.L., Miaw S.C., Ho I.C., and Kuchroo V.K.. 2009. Cutting edge: IL-27 induces the transcription factor c-Maf, cytokine IL-21, and the costimulatory receptor ICOS that coordinately act together to promote differentiation of IL-10-producing Tr1 cells. J. Immunol. 183:797–801. 10.4049/jimmunol.0901233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powrie F., Leach M.W., Mauze S., Menon S., Caddle L.B., and Coffman R.L.. 1994. Inhibition of Th1 responses prevents inflammatory bowel disease in scid mice reconstituted with CD45RBhi CD4+ T cells. Immunity. 1:553–562. 10.1016/1074-7613(94)90045-0 [DOI] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., and Smyth G.K.. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26:139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roncarolo M.G., Gregori S., Battaglia M., Bacchetta R., Fleischhauer K., and Levings M.K.. 2006. Interleukin-10-secreting type 1 regulatory T cells in rodents and humans. Immunol. Rev. 212:28–50. 10.1111/j.0105-2896.2006.00420.x [DOI] [PubMed] [Google Scholar]
- Rubtsov Y.P., Rasmussen J.P., Chi E.Y., Fontenot J., Castelli L., Ye X., Treuting P., Siewe L., Roers A., Henderson W.R. Jr., et al. 2008. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity. 28:546–558. 10.1016/j.immuni.2008.02.017 [DOI] [PubMed] [Google Scholar]
- Sun H., Lu B., Li R.Q., Flavell R.A., and Taneja R.. 2001. Defective T cell activation and autoimmune disorder in Stra13-deficient mice. Nat. Immunol. 2:1040–1047. 10.1038/ni721 [DOI] [PubMed] [Google Scholar]
- Szabo S.J., Kim S.T., Costa G.L., Zhang X., Fathman C.G., and Glimcher L.H.. 2000. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 100:655–669. 10.1016/S0092-8674(00)80702-3 [DOI] [PubMed] [Google Scholar]
- Wei G., Abraham B.J., Yagi R., Jothi R., Cui K., Sharma S., Narlikar L., Northrup D.L., Tang Q., Paul W.E., et al. 2011. Genome-wide analyses of transcription factor GATA3-mediated gene regulation in distinct T cell types. Immunity. 35:299–311. 10.1016/j.immuni.2011.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi R., Junttila I.S., Wei G., Urban J.F. Jr., Zhao K., Paul W.E., and Zhu J.. 2010. The transcription factor GATA3 actively represses RUNX3 protein-regulated production of interferon-gamma. Immunity. 32:507–517. 10.1016/j.immuni.2010.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Sharma S., Edwards J., Feigenbaum L., and Zhu J.. 2015. Dynamic expression of transcription factors T-bet and GATA-3 by regulatory T cells maintains immunotolerance. Nat. Immunol. 16:197–206. 10.1038/ni.3053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Yamane H., and Paul W.E.. 2010. Differentiation of effector CD4 T cell populations (*). Annu. Rev. Immunol. 28:445–489. 10.1146/annurev-immunol-030409-101212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Jankovic D., Oler A.J., Wei G., Sharma S., Hu G., Guo L., Yagi R., Yamane H., Punkosdy G., et al. 2012. The transcription factor T-bet is induced by multiple pathways and prevents an endogenous Th2 cell program during Th1 cell responses. Immunity. 37:660–673. 10.1016/j.immuni.2012.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]