Norris et al. show that microglia are the key phagocytes in removal of synaptic debris in the dorsal lateral geniculate nucleus after optic nerve injury. This microglial function is dependent on recognition of neurodegeneration and is mediated by the complement system.
Abstract
Phagocytosis of synaptic material by microglia is critical for central nervous system development. Less well understood is this microglial function in the injured adult brain. Assay of microglial phagocytosis is challenging, because peripheral myeloid cells engraft the site of injury, which could obscure interpretation of microglial roles. The model used here, optic nerve crush injury, results in degeneration of synapses in the dorsal lateral geniculate nucleus (dLGN), which stimulates rapid activation and engulfment of synaptic material by resident microglia without myeloid cell engraftment. Pharmacological depletion of microglia causes postinjury accumulation of synaptic debris, suggesting that microglia are the dominant postinjury phagocytes. Genetic or pharmacological manipulations revealed that neuronal activity does not trigger microglia phagocytosis after injury. RNA sequencing reveals C1q and CD11b/CR3 involvement in clearance of debris by dLGN-resident microglia. Indeed, C1qa−/− and Itgam−/− mice exhibit impaired postinjury debris clearance. Our results show how neurodegenerative debris is cleared by microglia and offers a model for studying its mechanisms and physiological roles.
Introduction
Professional phagocytes clear apoptotic cells and cellular debris both at baseline and during pathology, maintaining tissue homeostasis and preventing inflammation (Steinman, 2006; Arandjelovic and Ravichandran, 2015). The professional phagocytes in the central nervous system (CNS) are microglia, which are capable of responding rapidly to tissue damage by extending cell processes, engulfing debris, and undergoing proliferation (Davalos et al., 2005; Nimmerjahn et al., 2005; Aguzzi et al., 2013; Roth et al., 2014; Ulland et al., 2015; Colonna and Butovsky, 2017). Microglia were also shown to play a major role in CNS development through their engulfment of superfluous neuronal (presynaptic) material (Stevens et al., 2007; Schafer et al., 2012; Bialas and Stevens, 2013; Chung et al., 2013). Specifically, neuronal activity and the complement system were shown to regulate microglial engulfment of presynaptic terminals, thus ensuring optimal architecture of the visual system. Despite those important findings, our understanding of microglial phagocytic activity in adulthood is still limited. Up to now, researchers have studied microglial phagocytosis in adulthood mainly by using exogenous phagocytic targets, such as cells killed ex vivo or various types of beads. Such targets are usually delivered by their direct injection into the CNS, disrupting the blood–brain barrier, which results in parenchymal injury, peripheral monocyte engraftment (Ling, 1979), complement and IgG deposition, and activation of microglia (Obermeier et al., 2013). Documentation of microglial clearance of neuronal and myelin debris has been well characterized previously, with a heavy emphasis placed on cuprizone models of demyelination and spinal cord injury (Smith, 1999; Greenhalgh and David, 2014; Gudi et al., 2014). Further, the phagocytic receptor TREM2 was shown to be critical for efficient removal of myelin debris after cuprizone-mediated demyelination (Poliani et al., 2015). Although these models show robust roles for microglial clearance of debris, they are limited as a result of the potential contribution of peripheral myeloid cells in these models (Greenhalgh and David, 2014; Lampron et al., 2015).
Our laboratory and others have used crush injury of the optic nerve as a model to study neurodegeneration and neuroprotection, in both of which the immune system is a key player (Moalem et al., 1999; Schwartz and Cohen, 2000; Schwartz, 2004; London et al., 2011; Walsh et al., 2014, 2015; Gadani et al., 2015; Benhar et al., 2016). Over several weeks after the injury, there is a gradual decline in the population of retinal ganglion cells (RGCs) that innervate the superior colliculus, the suprachiasmatic nucleus of the hypothalamus, and the lateral geniculate nucleus (LGN) of the thalamus. Concurrently, the distal stump of the nerve and projections from the RGCs succumb to Wallerian degeneration (Levkovitch-Verbin et al., 2000; Vargas and Barres, 2007). Within the LGN, experiments using transmission electron microscopy have shown that optic nerve injury is followed by rapid degeneration of retinogeniculate terminals, where presynaptic debris is observed within activated glial cells (Vaccarezza et al., 1970; Ghetti et al., 1972; Wong-Riley, 1972). Accordingly, we postulated that after an optic nerve crush injury in the adult mouse, LGN microglia become activated and engulf synaptic material, a process similar to that seen in the developing animal during refinement of the binocular architecture of the dorsal lateral geniculate nucleus (dLGN; Schafer et al., 2012). With this system established, we could then proceed to assess microglial engulfment of endogenous debris, without introduction of exogenous material or breach of the blood–brain barrier.
Here, we show that after optic nerve crush injury, during Wallerian degeneration of retinogeniculate inputs, microglia serve as key players in clearance of the resulting presynaptic debris. We found that resident dLGN microglia exhibit a dynamic transcriptional signature after injury (up-regulating C1q and CD11b, both shown to be necessary for debris clearance in development; Stevens et al., 2007; Schafer et al., 2012) and that microglial depletion causes neuronal debris to accumulate. We also observed that postinjury activation of microglia and phagocytosis are regulated by synaptic integrity rather than by neuronal activity, a finding that distinguishes microglial function in neurodegeneration from that seen in development.
Results and discussion
Microglia are activated after optic nerve crush injury and phagocytose neuronal debris
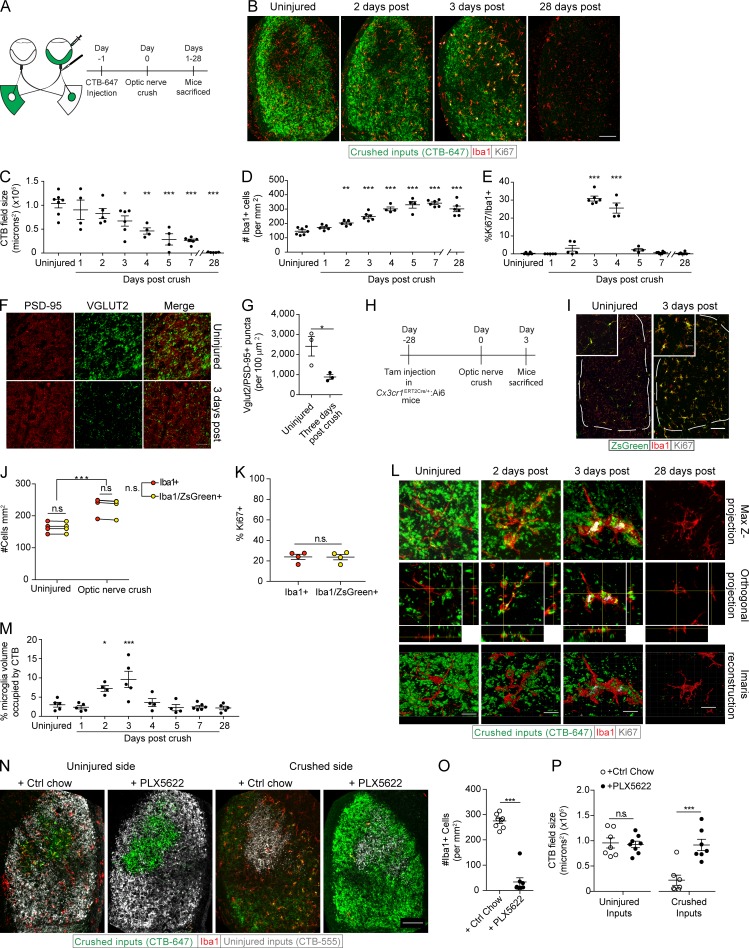
We first assessed the function and kinetics of dLGN microglia in response to neurodegeneration. To this end, we injected the right eyes of WT mice with cholera toxin subunit B (CTB) conjugated to Alexa Fluor 647, 24 h before subjecting the mice to crush injury (Schwartz, 2004; Fig. 1 A). After the injury, mice were killed at various time points (days 1–5, 7, and 28 after crush). Measurement of CTB-tagged retinogeniculate inputs into the dLGN, contralateral to the injected eye, revealed their progressive loss with substantial loss observed on day 3 and an even greater loss (∼70%) on day 7 after the crush injury (Fig. 1, B and C). By day 28, no CTB-647 signal could be detected (Fig. 1, B and C). We observed an increase in the numbers of Iba1+ cells in the dLGN, starting on day 2 after the crush injury and peaking on day 7 (Fig. 1, B and D), similar to what has been previously seen in nerve injury models (Bartel, 2012; Tay et al., 2017). Ki67+ (proliferating) microglia were detectable by postinjury day 2, with a significant increase in their numbers on days 3 and 4, followed by a marked reduction on day 5 (Fig. 1, B and E).
Figure 1.
Optic nerve crush induces proliferation of microglia and phagocytic clearance of neuronal debris. (A) Scheme of CTB-labeling and optic nerve crush paradigm. (B–E) Representative images (B) and quantification (C) of the degenerating dLGN presynaptic field, Iba1+ (D), and Iba1/Ki67+ cells (E; n = 4–7 mice per group; one-way ANOVA, Dunnett’s post hoc, representative of two independently performed experiments). (F and G) Representative images (F) and quantification (G) of VGlut2/PSD95+ retinogeniculate synapse loss 3 d after optic nerve crush (n = 3 mice per group; Student’s t test, representative of two independently performed experiments). (H–K) Scheme (H), representative images (I), and quantification of the number (J) and proliferation (K) of Iba1+ and Iba1/ZsGreen+ cells in Cx3cr1ERT2Cre/+:Ai6 mice prior and 3 d after optic nerve crush (n = 4 mice per group; two-way ANOVA with Holm-Sidak post hoc test [J], Student’s t test [K], representative of three independently performed experiments). (L and M) Z projections, orthogonal slices, Imaris three-dimensional reconstructions (L) and quantification (M) of microglial phagocytosis of crushed retinogeniculate inputs (n = 4–6 mice per group; one-way ANOVA, Dunnett’s post hoc test, representative of two independently performed experiments). (N–P) Representative images (N) and quantification of Iba1+ cells (O) and CTB+ fields (P) in uninjured and 3 d postinjury PLX5622 and control chow-fed mice (n = 7–8 mice per group; Student’s t test, representative of three independently performed experiments). Error bars represent mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; n.s., not significant. Bars: 100 µm (B, I, and N); 25 µm (F); 10 µm (L).
To demonstrate that the gradual disappearance of CTB labeling actually corresponded to the loss of presynaptic terminals, we quantified retinogeniculate synapses in areas of CTB loss within the dLGN contralateral to the crushed eye on day 3 after optic nerve crush injury. Using a method previously used to count synapses of the mouse visual system (Ippolito and Eroglu, 2010), we observed a significant decrease in retinogeniculate (VGlut2/PSD95 double-positive) synapses (Fig. 1, F and G), suggesting that the disappearance of CTB observed after the injury was indeed related to the loss of retinogeniculate terminals.
The optic nerve crush injury model is unique in that it allows for the study of cellular responses to neurodegeneration that occur far from the actual site of injury. This is particularly important when attempting to isolate the role of microglia, as the injury site is heavily populated by peripheral cells (Walsh et al., 2014) that may compete and even interfere with microglial function and complicate microglial identification. We first needed to confirm that, in the context of neurodegeneration, the microglia in our system could indeed be studied in isolation from contamination by peripheral macrophages. We did this by fate labeling microglia, using Cx3cr1ERT2Cre/+:Ai6 mice. Using a previously published protocol (Goldmann et al., 2013), mice were fed tamoxifen-containing chow for 1 wk and then waited 1 mo for peripheral myeloid cell turnover before we analyzed the brain and peripheral organs for macrophage expression of ZsGreen (Fig. S1 A). We observed that nearly 100% of microglia retained ZsGreen labeling in the uninjured cerebral cortex and cerebellum (Fig. S1, B–D). This contrasted with peripheral macrophage populations, which exhibited decreasing percentage of ZsGreen+ myeloid populations in the spleen, liver, and blood, respectively (Fig. S1, B–D). We then performed unilateral optic nerve crush and sacrificed the injured mice 3 d later (Fig. 1 H). Both before and after crush injury, the vast majority of myeloid cells (Iba1+) in the dLGN were ZsGreen+ (Fig. 1, I and J). Unlike in the dLGN, the injury site in the optic nerve had numerous Iba1+/ZsGreen– cells, indicative of peripheral myeloid engraftment there (Fig. S1 E), as has been previously reported (Walsh et al., 2014). Furthermore, there was no difference in the percentages of Iba1+ or Iba1+/ZsGreen+ cells expressing Ki67 in dLGN after injury (Fig. 1, I and K), implying that the Iba1+ cells were derived from endogenous microglial populations. Despite retention of ZsGreen labeling after nerve crush injury, a marked reduction in the microglia-specific marker TMEM119 (Bennett et al., 2016) was seen on microglia 3 d postinjury (Fig. S2, A and B). This shows that activation leads to loss of TMEM119 expression by endogenous microglia as has been previously shown in a mouse model of Alzheimer’s disease (Keren-Shaul et al., 2017). Therefore, caution should be used when interpreting TMEM119 labeling of microglia, as loss of this marker may be indicative of microglial activation in general.
The unique model we describe here incorporates crush injury of the optic nerve and the consequent neurodegeneration in the dLGN, distal to the injury site itself. Thus, this allows a “closed system” to study phagocytic uptake of neuronal material with no prior priming of microglia or peripheral myeloid “contamination.”
Because microglia have been shown to be prolific phagocytes of synaptic material (Schafer et al., 2012), we examined whether the observed phagocytosis and clearance of presynaptic terminals were attributable to dLGN microglia. Using a previously described method (Schafer et al., 2014), we observed that neuronal debris (presented as the percentage of microglial volume) was indeed present within the microglial soma (Fig. 1, L and M). In Cx3cr1GFP/+ mice, costaining for engulfed neuronal debris alongside the phagolysosomal marker CD68 revealed a high degree of internalized CTB colocalized with the microglial phagolysosome (Fig. S2 C), indicative of microglial phagocytosis.
Although microglia are known to play a role in the pruning of synapses during the developmental period of eye-specific segregation at postnatal day 5, it was suggested that, given the amount of consumed neuronal material, the dominant phagocytes at that stage were astrocytes (Chung et al., 2013). To assess the possibility that astrocytes could be phagocytic of neuronal debris after optic nerve crush injury, we quantified the number of cells positive for the astrocyte marker, glial fibrillary acidic protein (GFAP), after injury (Fig. S2, D and E). Neither a change in GFAP+ cell number nor CTB+ debris within GFAP+ cells was observed (Fig. S2, F and G).
As a result of the limitations of GFAP as a gray matter astrocyte marker (Lundgaard et al., 2014), we further assessed the phagocytic capabilities of LGN astrocytes and microglia during acute Wallerian degeneration using flow cytometry. Bilateral CTB-A647 injection and crush injury were performed, followed by LGN microdissection 3 d later (Fig. S2, H and I). Although a significant increase in the amount of CTB was only seen in the microglial population, we did see a small, yet not significant, increase in CTB+ astrocyte populations with CTB injected eyes in both uninjured and postcrush conditions (Fig. S2, J and K). To distinguish whether this positivity was a result of internalization of CTB or extracellular attachment, we used a CTB conjugated to the pH-sensitive dye CypHer 5E. This compound is maximally fluorescent at low pH with minimal fluorescence at neutral pH (Cooper et al., 2002). Thus, only cells with engulfed CTB should exhibit CypHer positivity. After bilateral eye injection with CTB-CypHer followed by optic crush, only CypHer+ microglia, and not astrocytes, were observed (Fig. S2, L and M).
To assess the necessity of microglia in the clearance of neuronal debris after injury in adult mice, we eliminated microglia in our system by using the CSF 1 receptor (CSF1R) inhibitor PLX5622, a compound previously shown to efficiently deplete microglia in the CNS (Elmore et al., 2014). After 3 wk of treatment with PLX5622 chow, mice received bilateral injections of CTB intravitreally and were subjected 24 h later to unilateral optic nerve crush injury. As previously shown (Hilla et al., 2017), PLX5622 treatment also had no effect on RGC survival 3 (Fig. S2, N and O) and 7 d after optic nerve crush injury (Fig. S2, P and Q). As expected, PLX5622 also resulted in a marked reduction in microglial counts (Fig. 1, N and O). Subsequently, CTB+ dLGN fields contralateral to crushed optic nerves were significantly larger 3 d after optic nerve crush injury in PLX5622-fed mice than in mice fed control chow, suggesting that after optic nerve crush injury, microglia are necessary for the optimal clearance of neuronal debris (Fig. 1, N and P). In fact, microglial depletion and debris presence persisted in PLX5622-fed mice up to 7 d after optic nerve crush injury (Fig. S2, R–T). No changes were seen in CTB-labeled presynaptic inputs into the dLGN contralateral to uninjured optic nerves (Fig. 1, N and P; and Fig. S2, R and T). These results suggest that microglia are the key phagocytes of dLGN presynaptic debris during Wallerian degeneration in our system. This is in contrast to the situation during developmental pruning, where astrocytes are the dominant phagocyte (Chung et al., 2013). A recent study has also shown that microglia are capable of modulating astrocyte function through soluble factors, allowing for their reactivity and enhanced function (Liddelow et al., 2017). Although microglial depletion may alter astrocyte function and thus removal of neuronal debris, it appears that microglia are the key cell responsible for debris engulfment after Wallerian degeneration of the optic nerve.
Neuronal activity is not the trigger for microglia activation after optic nerve crush injury
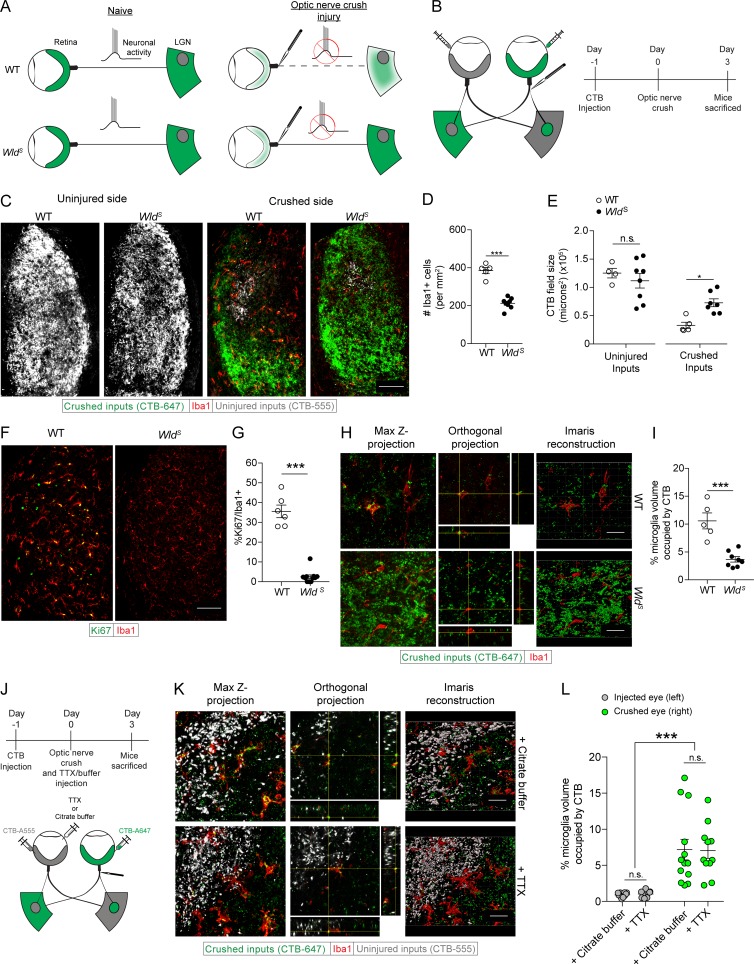
The rapid response of dLGN microglia observed after optic nerve crush prompted us to investigate whether neuronal activity from the retina, shown to be crucial for engulfment of neuronal material in development (Schafer et al., 2012), might regulate microglial activation in response to this injury, especially because all neuronal activity from the retina is known to be rapidly disrupted after optic nerve crush in the mouse. To address this question, we used WldS mice (Coleman et al., 1998), which demonstrate delayed axonal and synaptic degeneration after acute injury to the peripheral and central nervous systems (Gillingwater et al., 2006; Vargas and Barres, 2007; Wright et al., 2010). Thus, by studying the microglia in the acutely injured WldS dLGN, we were able to examine a system in which neuronal activity had ceased (as in the injured WT), but presynaptic degeneration was markedly delayed (Fig. 2, A and B). Before examining the dLGN we counted the RGCs in WldS mice 3 d after optic nerve crush and observed no changes in their numbers (Fig. S3, A and B). We then examined the dLGN and found significantly less microglia in the dLGN contralateral to the crushed eye of WldS mice than in their WT counterparts, indicating that little or no dLGN microglial proliferation had occurred in the WldS mice by day 3 after the optic nerve crush (Fig. 2, C and D). The CTB+ field of the dLGN contralateral to the crushed optic nerve was also significantly larger in the WldS mice, indicating a reduced microglial clearance of the CTB+ presynaptic field in WldS mice (Fig. 2, C and E). CTB+ field size in the dLGN of WldS mice contralateral to the uninjured optic nerve were unchanged (Fig. 2, C and E), indicating that, as shown previously (Hoopfer et al., 2006), there were no developmental changes in retinogeniculate projections in WldS mice. Coinciding with the reduction in microglia number was a decrease in Ki67+ microglia, implying that microglial proliferation is greatly diminished when neurodegeneration is delayed (Fig. 2, F and G). Our analysis of microglial activation and phagocytosis of crushed neuronal inputs in the WldS mice also revealed a less activated morphology and significantly less engulfed neuronal debris than in WT microglia at 3 d after injury (Fig. 2, H and I).
Figure 2.
Microglial activation and phagocytosis after optic nerve crush injury is triggered by synaptic degeneration and not cessation of neuronal activity. (A and B) Scheme of conceptual (A) and experimental (B) setup of optic nerve injury in WT and WldS mice. (C–E) Representative images (C) and quantification of Iba1+ cells (D) and dLGN CTB fields (E) 3 d after optic nerve crush in WT and WldS mice (n = 5–8 mice per group; Student’s t test, representative of three independent experiments). (F and G) Representative images (F) and quantification (G) of Ki67+ dLGN microglia 3 d after optic nerve crush in WT and WldS mice (n = 5–8 mice per group; Student’s t test, representative of three independent experiments). (H and I) Z projections, orthogonal slices, Imaris three-dimensional reconstructions (H), and quantification (I) of microglial phagocytosis of crushed retinogeniculate inputs in WT and WldS mice 3 d after optic nerve crush (n = 5–8 mice per group; Student’s t test, representative of three independent experiments). (J) Scheme of TTX application and crush injury. (K and L) Z projections, orthogonal slices, Imaris three-dimensional reconstructions (K), and quantification (L) of microglial phagocytosis after TTX application and crush injury (n = 7–13 mice per group, pooled from two independent experiments; two-way ANOVA for effect of optic nerve crush, Holm-Sidak post hoc test). Error bars represent mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001. n.s., not significant. Bars: 100 µm (C and F); 20 µm (H and K).
To further determine whether presynaptic terminals lacking neuronal activity from the retina could be engulfed by injury-activated microglia, we took advantage of the dLGN architecture (consisting of a small ipsilateral patch within the large contralateral projection area). Using pharmacological attenuation of neuronal activity in the ipsilateral eye concurrently with optic nerve crush in the contralateral eye, we tested whether injury-activated microglia would engulf weakened inputs (from the ipsilateral patch, Fig. 2 J). To this end, we labeled retinogeniculate inputs with CTB conjugated to Alexa Fluor dyes, and 24 h later applied tetrodotoxin (TTX) or its carrier (citrate buffer) to one eye, effectively blocking action potential–mediated activity from the eye, while crush injuring the contralateral optic nerve. To confirm that TTX had an effect on dampening neuronal activity from the eye, we examined the pupillary constriction response (Su et al., 2011; Shanks et al., 2016) in our mice 24 and 48 h after the above procedures. Significant effects of both the optic nerve crush and the TTX application on pupillary constriction reflexes were observed both at 24 and at 48 h after the injury, with no detectable effect on naive eyes or on citrate buffer–treated controls (Fig. S3, C–F). The mice were killed 3 d after crush, during peak phagocytosis of crushed neuronal inputs. Examination at that time (3 d after crush injury) showed that there were no effects of TTX on RGC survival (Fig. S3, G and H). dLGN microglia counts were also unchanged contralateral to eyes treated with TTX, with only dLGN microglia contralateral to the crushed eye showing an increase in number (Fig. S3, I and J). We also found that the CTB+ presynaptic field sizes contralateral to TTX-treated eyes were unchanged compared with either naive or citrate buffer–treated eyes, with only dLGN CTB fields contralateral to crushed eyes decreasing in size (Fig. S3, I and K). Next, we examined whether injury-activated microglia would phagocytose TTX-treated uninjured presynaptic terminals. We found that microglia engulfed neuronal material from the crushed eye but did not engulf TTX-treated inputs (Fig. 2, K and L), which were deficient in action potential–mediated activity from the eye.
Although TTX application did serve to block activity emanating from the eye, spontaneous release of neurotransmitter from presynaptic terminals would not be affected by TTX treatment (Chesselet, 1984). Still, this lack of microglia dependence on retinal activity distinguishes the dLGN microglial phagocytosis seen in development (Schafer et al., 2012) from that observed during CNS Wallerian degeneration. Reasons for this may include, for example, differences between the effect of neuronal activity on the membrane properties of presynaptic terminals of retinogeniculate inputs during adulthood and during a critical period in development. Another possibility is that there are physiological differences in postnatal day 5 dLGN microglia versus those seen in adult microglia during Wallerian degeneration. Finally, an actively degenerating presynaptic terminal seen after nerve crush might be more salient to activated microglia than a presynaptic terminal treated with TTX, allowing microglia to interact more robustly with degenerating terminals than with those having less neuronal activity. These results demonstrate that in the adult mouse, microglia become activated in response to synaptic degeneration, whereas cessation of neuronal activity has no effect on microglial phagocytic activity.
Transcriptional profiling of dLGN microglia after optic nerve crush
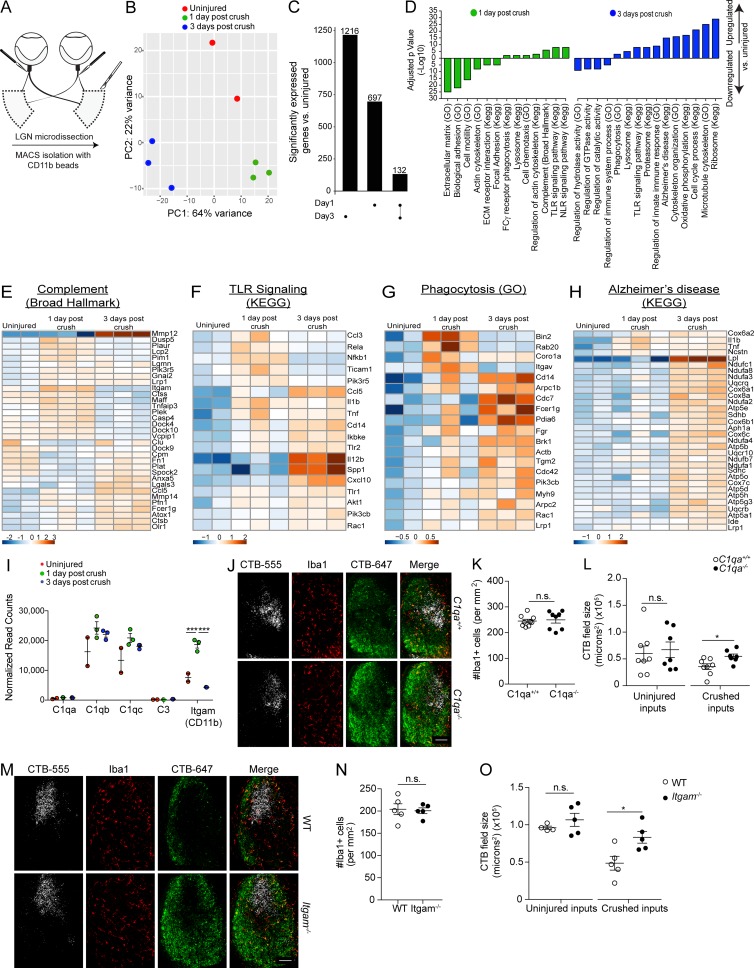
Because microglia are the main phagocytes in response to neurodegeneration in the dLGN, these findings provide an amenable system for studying molecular mechanisms governing microglial engulfment of endogenous material. Thus, to identify the genetic programs necessary for removal of neuronal debris by dLGN microglia, we isolated CD11b+ cells (using AutoMACS) from an uninjured state and at 1 and 3 d after bilateral optic nerve crush (Fig. 3 A). RNA sequencing (RNA-seq) was performed on our samples after flow cytometry confirmed that each sample was at least 90% CD11b+CD45+ (Fig. S3, L–N). Principal component analysis of all statistically significant transcripts revealed three discretely clustered populations, indicating a unique transcriptional signature for postinjury microglia at both time points tested (Fig. 3 B). Notably, day 1 and 3 microglial subsets did not cluster together more closely than in the uninjured condition, indicating a dynamic transcriptional profile of microglia in the acute phase of Wallerian degeneration. To assess the differences between genes expressed on days 1 and 3 after crush injury, we compared the uniquely enriched genes of each condition to uninjured controls (Fig. 3 C). We found 132 common genes enriched in microglia on both days 1 and 3 after crush injury (adjusted p-value < 0.05), with the 3-d postinjury microglia expressing the higher amount of uniquely enriched genes after the injury (1,216 unique genes on day 3 compared with 697 unique genes on day 1 after crush). To illustrate the functional differences between dLGN microglia at 1 and 3 d after crush injury, we identified gene sets enriched on days 1 and 3 by performing a gene set enrichment analysis. This revealed that several gene sets were down-regulated in microglia on day 1 after crush injury, including those associated with the extracellular matrix and biological adhesion (Fig. 3 D). Up-regulated gene sets on day 1 after injury included those associated with sensing molecular “danger,” such as complement (Fig. 3 E) and NOD-like receptor and Toll-like receptor signaling pathways (Fig. 3 F), indicating that microglia at 1 d after crush injury indeed sense and respond to the ongoing accumulation of neuronal debris. Gene sets down-regulated on day 3 after the injury included regulation of hydrolase, GTPase, and catalytic activity, as well as regulation of immune system processes. Up-regulated gene sets included genes necessary for ribosome and proteasome production, cell cycle processes, oxidative phosphorylation, phagocytosis (Fig. 3 G), and Alzheimer’s disease (Fig. 3 H). This diverse array of gene sets would be expected for microglial cells that are surrounded by neuronal debris, actively engaged in phagocytosis, and undergoing cell division. Interestingly, the transcriptional signature 3 d after injury is distinct from that of 1 d after injury, with just 132 genes enriched on both postinjury days compared with noninjured condition. That such a unique transcriptional signature of microglia could be displayed only 2 d apart is unique, as most transcriptional studies to date have examined microglial activation on a longer time scale (Chiu et al., 2013; Hickman et al., 2013; Butovsky et al., 2015; Keren-Shaul et al., 2017). This speed of change is likely indicative of the responsive and plastic nature of microglia. Further studies examining microglial transcriptomes on a more acute basis after insult or during chronic disease may yield even more insights into microglial biology in health and disease. Recently, microglial activation genes associated with neurodegeneration have been characterized (Keren-Shaul et al., 2017; Krasemann et al., 2017). After crush, several of these genes were also up-regulated in LGN microglia 3 d after injury (Fig. S3 O), indicating that there may be a core program of microglial activation central to both acute and chronic neurodegenerative processes. Despite this finding, key genes shown previously to be necessary for the chronic neurodegenerative signature (Trem2 and Apoe) were not up-regulated, indicating that there may still be differences in the activation profiles of microglia in acute and chronic neurodegenerative settings.
Figure 3.
dLGN microglia after optic nerve crush exhibit a dynamic transcriptional profile using C1q and CD11b for optimal clearance of neuronal debris. (A) Scheme of dLGN microglia isolation. (B) PCA analysis of microglia after injury. (C) UpSet plot detailing enriched genes after crush. (D) Gene sets enriched in dLGN microglia after crush. (E–H) Heat maps of complement (E), TLR signaling (F), phagocytosis (G), and Alzheimer’s disease (H) gene sets up-regulated in dLGN microglia after crush. (I) Normalized read counts of selected complement genes in microglia after crush (n = 2 samples for uninjured microglia and n = 3 samples for one and 3 d after crush, Fisher’s test, performed once). (J–L) Representative images (J) and quantification of microglia (K) and dLGN CTB fields (L) 3 d after optic nerve crush in C1qa+/+ and C1qa−/− mice (n = 8–9 mice per group; Student’s t test, representative of two independent experiments). (M–O) Representative images (M) and quantification of microglia (N) and dLGN CTB fields (O) 3 d after optic nerve crush in WT and Itgam−/− mice (n = 5 mice per group; Student’s t test, representative of two independent experiments). Error bars represent mean ± SEM. *, P < 0.05; ***, P < 0.001; n.s., not significant. Bars, 100 µm.
Because the complement gene set was up-regulated on day 1 after crush injury, we examined specific complement genes associated with the microglial engulfment of neuronal material during development (Fig. 3 I). The results showed that Itgam (CD11b or C3R) was highly up-regulated in microglia on day 1 after optic nerve crush. We also observed a slight (not statistically significant) increase in transcription of C1qb and C1qc. To determine whether the complement system plays a role in the microglial clearance of neuronal debris after optic nerve crush, we repeated optic nerve crush injury experiments using C1qa−/− and Itgam−/− mice. On day 3 after optic nerve crush, we observed no change in the number of microglia in C1qa−/− mice (Fig. 3, J and K), whereas significantly more residual neuronal debris was seen compared with WT mice (Fig. 3, J and L). Similarly, Itgam−/− mice showed no change in the number of microglia after injury (Fig. 3, M and N), and exhibited more neuronal debris in the dLGN contralateral to the crushed eye as compared with their WT counterparts (Fig. 3, M and O). Importantly, RGC survival was not affected by deficiency of C1q (Fig. S3, P and Q) or CD11b (Fig. S3, R and S). Altogether, these experiments indicated an active role for complement in the microglial clearance of neuronal debris pursuant to Wallerian degeneration, implicating complement as a key driver of neuronal material clearance by microglia in Wallerian degeneration, shown previously in development (Stephan et al., 2012), Alzheimer’s disease (Hong et al., 2016), schizophrenia (Sekar et al., 2016), and viral infection (Vasek et al., 2016).
In this study, we provide a novel platform for the study of physiological microglial phagocytosis pursuant to Wallerian degeneration. We further show that microglia are the key players in the clearance of neuronal debris, that their depletion results in failed debris clearance, and that neuronal activity is irrelevant for the activation and initiation of phagocytosis by microglia.
Notable in our transcriptional findings were microglial profiles characterized by up-regulation of TLR, NLR, and Alzheimer’s disease gene sets. An up-regulation of such programs could very well be a result of the release or exposure of damage-associated molecular patterns (DAMPs) by degenerating nerve terminals. Future studies attempting to identify microglial-activating DAMPs in the context of acute injury may yield important insights into microglial responses to neurodegeneration in a larger context. The importance of neurodegeneration and exposure of DAMPs was seen with crush in WldS mice, where the lack of neurodegeneration was accompanied by no induction of microglial proliferation. A possible mediator could be phosphatidyl serine exposure on presynaptic terminals, which could be diminished in the WldS LGN after crush. Phosphatidyl serine exposure has been shown previously to be essential to induce microglial proliferation in response to neurodegeneration in the context of CNS viral injection (Tufail et al., 2017).
Future research may yield further insights into the sensors that allow for microglial activation in response to neurodegeneration, as well as into the array of receptors needed to facilitate debris removal. Knowledge of these tools might aid in the tuning of microglial function by limiting the spread of secondary neurodegeneration or by boosting the microglial clearance of potentially toxic debris. Consequences of failed debris clearance in the periphery have been well characterized, and lead to inflammation and subsequent autoimmune disease (Elliott and Ravichandran, 2010). We suggest that the same may hold true in the CNS, where absence or dysfunction of microglial phagocytosis may ultimately lead to neuronal dysfunction and disruption of CNS homeostasis.
Materials and methods
Mice
C57/Bl6 and Cx3cr1-eGFP mice were obtained from Jackson Laboratory, stocks 000664 and 005582, respectively. B6.Cg-Gt(ROSA)26Sortm6(CAG-ZsGreen1)Hze/J (Ai6) mice were generously provided by the Harris Lab (University of Virginia) and originally obtained from Jackson Laboratories (Stock 007906). WldS mice were provided by the Deppmann Lab (University of Virginia, Charlottesville, VA) and originally obtained from Jackson Laboratories (008820) and bred with congenic FVB/N mice to obtain heterozygous WldS mice and WT controls. Itgam−/− mice and their congenic C57/Bl6J controls were purchased from Jackson Laboratories (stock 003991 and 000664, respectively). Heterozygous C1qa+/− mice were obtained from Jackson Laboratories (stock 022307) and bred to obtain C1qa−/− and C1qa+/+ control mice. All strains were kept in identical housing conditions. All animals were housed in temperature and humidity-controlled rooms, maintained on a 12-h light/dark cycle (lights on 7 a.m.); mice were used from 2 to 4 mo old with mice age-matched in each experiment described. For survival surgeries, mice were anesthetized with either 200 µl of Ketamine/Xylazine (1 ml Ketamine HCl [1 mg/ml], 1 ml of 2% Xylazine, and 8 ml saline), or inhaled isoflurane. All procedures complied with regulations of the Institutional Animal Care and Use Committee at the University of Virginia.
Fate mapping of microglia
B6.Cg-Gt(ROSA)26Sortm6(CAG-ZsGreen1)Hze/J mice (Ai6) were crossed to the Cx3cr1ERT2cre/ERT2cre mice to obtain Cx3cr1ERT2/+:Ai6 heterozygotes. Mice were placed on a tamoxifen-supplemented (250 mg/kg) diet for 1 wk (TD.130856 Envigo). After return to normal chow, a period of 1 mo was allowed to ensure turnover of the peripheral immune system, allowing only microglia to retain the ZsGreen label (Goldmann et al., 2013). Mice were then subjected to analysis or optic nerve crush injury and sacrificed 3 d after injury.
PLX5622 administration
PLX5622 was provided by Plexxikon Inc. and formulated in AIN-76A standard chow by Research Diets Inc. at 1,200 mg/kg. Mice were kept on PLX5622 or AIN-76A control diets for 3 wk before experimentation and maintained on their respective diets until sacrifice.
CTB-CypHer preparation
Cholera toxin β subunit (C9903; Sigma-Aldrich) was resuspended at 10 mg/ml in PBS and labeled with CypHer5 (10 µM; GE Healthcare) for 1 h at 20°C. Free dye was removed by gel filtration using Zeba desalting column (89890; Thermo Scientific) according to manufacturer’s instructions. Reagent preparations were performed by the Gaultier Lab (University of Virginia).
CTB injection and optic nerve injury
After anesthesia, mice received intravitreal injections of CTB conjugated to Alexa Fluor 555 dye into the left eye and CTB conjugated to Alexa Fluor 647 (Invitrogen) dye into the right eye (1 µl per eye at a concentration of 1 µg/µl in sterile saline). Optic nerve crush was performed 24 h after CTB injection. In brief, mice were anesthetized and the tissue around the nerve was carefully dissected and the optic nerve exposed. N5 self-closing forceps were closed around the optic nerve 5 mm behind the globe and held for 3 s. The mice were then allowed to recover at 37°C on a warming pad before returning to their cages and sacrificed at time points indicated in the text. (Mice that did not receive sufficient dLGN dye fills were omitted from analysis.)
TTX administration and measurement of pupil response
Once anesthetized, 1 µl TTX (5 µM) or its carrier solution (20 mM citrate buffer) was injected intravitreally with a Hamilton syringe contralateral to the eye used for optic nerve crush. Pupil response was measured as described previously (Shanks et al., 2016). In brief, at time points 24 and 48 h after optic nerve crush injury and TTX administration, mice were acclimated to a dark room for 30 min before testing. While under a red light (Black Diamond Cosmos), the pupil size was obtained using a Leica IC80HD camera fixed to a Leica M220 dissecting microscope. The mouse was then subjected to a 15-s exposure of bright white light and the pupil size was imaged instantly after return to red light conditions. The percentage pupil constriction was calculated as the difference in pupil size between red and bright white light conditions.
Tissue preparation and immunofluorescence
The brain and eyes were harvested from animals after perfusion with heparinized (5 U/ml) PBS and 4% paraformaldehyde (PFA). After 24 h postfixation in 4% PFA, the eye was dissected by cutting through the sclera just posterior to the ciliary body. The lens was removed, and four cuts were made toward the optic disc to allow the retina to lay flat. The retina was then separated from the sclera and pigmented epithelium and placed in to 96-well plates for staining. Brain tissue was postfixed for 48 h in 4% PFA, followed by cryoprotection in 30% sucrose for 48 h. Brains were then frozen in optimal cutting temperature compound (Tissue-Tek) on the freezing element of a Leica CM3050 S cryostat. 40-µm sections were sliced into 24-well plates containing PBS with 0.05% sodium azide. Slices were permeabilized with 0.5% Triton X-100 for 15 min then washed two times for 5 min in PBS and blocked for 1 h at room temperature in blocking buffer (5% serum of the secondary's species) followed by overnight incubation in 0.5% BSA with primary antibody at 4° C. The following antibodies were used for immunofluorescence staining: rabbit anti-Iba1 (1:300; CP-290; Biocare Medical), goat anti-BRN3a (1:300; sc-31984; Santa Cruz), rabbit anti-Ki67 (1:300; ab15580; Abcam), Goat anti-Iba1(1:300; Abcam; ab5076), rat anti-CD68 (FA-11; Biolegend), rabbit anti-TMEM119 (ab209064; Abcam) guinea pig anti-Vglut2 (AB2251; EMD Millipore) rabbit anti–PSD-95 (51-6900; Invitrogen). Slices were washed three times for 5 min, incubated for 2 h at room temperature with the appropriate secondary antibodies (all from Life Technologies; 1:1,000), washed again three times for 5 min and mounted with Aquamount (Thermo Scientific) and DAPI.
RGC image acquisition and quantification
For assessing RGC survival, images of Brn3a+ RGCs from four quadrants of the retina were taken at equal distances from the optic disc using an Olympus IX-71 microscope with 40× objective. The pictures were counted in a blinded fashion to determine the number of RGCs per field. RGC counts were established from each image using a modified method described previously (Danias et al., 2002). In brief, each image was inverted with the contrast enhanced (1%) and background subtracted (rolling ball 55). Each image was then set to a threshold to encompass all Brn3a+ cells. Once binary, the Watershed feature was applied and the Analyze Particle function used to count all RGCs. All images were acquired and quantified by a user blinded to the experiment.
dLGN image quantification and analysis
For each animal, three to four sections of medial dLGN were chosen for imaging and quantification of RGC inputs to the dLGN, Iba1+ cell counts and microglial engulfment analysis. Images were acquired on a Leica SP8 microscope at 20× magnification with 1-µm z steps for CTB field size measurements and Iba1+ cell counts and at 60× magnification using 0.2-µm z steps for engulfment analyses. For each animal, four to seven dLGN fields were imaged. Obtained images were processed and quantified using ImageJ (National Institutes of Health) for CTB field size and Iba1+ cell number and Imaris software (Bitplane) for engulfment analyses. For CTB field size and Iba1+ cell number, ImageJ was used to obtain regions of interest (ROIs) of contralateral dLGN fields for crushed and uninjured inputs. Each ROI was then thresholded to include all CTB+ inputs, and the area was measured. Iba1+ counts were made using the Cell Counter plugin (ImageJ). In vivo microglia phagocytosis assays and analyses were performed as previously described in detail (Schafer et al., 2012, 2014). Imaris software (Bitplane) was used to create three-dimensional volume surface renderings for microglia and engulf debris. To visualize and measure debris volume, any fluorescence that was not within the microglia volume was subtracted from the image using the mask function of Imaris. The remaining engulfed fluorescence was surface rendered and total volume of engulfed debris was calculated. To determine percentage engulfment the following calculation was used: volume of internalized debris /volume of microglial cell × 100. Image acquisition, quantification, and analyses were performed blind.
Retinogeniculate synapse quantification
Quantification of retinogeniculate synapses was performed as described previously (Ippolito and Eroglu, 2010). In brief, dLGN brain sections from uninjured and 3-d postcrush mice were stained with pre (VGlut2) and postsynaptic (PSD95) markers. 5-µm-thick confocal z stacks (optical section depth, 0.33 µm; 15 sections per z stack) within the outer shell were imaged at 60× magnification on a Leica SP8 confocal laser-scanning microscope. Maximum projections of three consecutive sections (corresponding to 1 µm total depth) were generated from the original z stack. The Puncta Analyzer plugin that was developed by Barry Wark for ImageJ (available at https://github.com/physion/puncta-analyzer) was used to count the number of colocalized puncta.
Flow cytometry of LGN astrocytes and microglia to assess engulfment of neuronal material
After anesthesia, mice received intravitreal injections of CTB conjugated to Alexa Fluor 647 dye or CypHer in to both eyes (1 µl per eye at a concentration of 1 µg/µl in sterile saline). Optic nerve crush was performed 24 h after CTB injection. 3 d after bilateral optic nerve crush, mice were perfused with PBS containing heparin (5 U/ml) until livers and tongues were exsanguinated to minimize peripheral immune cell contamination. The entire brain was then removed, and meninges and choroid plexus were carefully separated and discarded from the rest of the brain tissue. The brain was then placed in a coronal brain matrix (SA-2165; Roboz), and a razor blade was used to isolate a brain slice containing the dLGN. The dLGN was dissected from surrounding brain regions and placed in 1.5 ml Accutase (A1110501; Stempro) in a 1.5-ml Eppendorf tube. Samples were then incubated at 37°C for 15 min, followed by gentle trituration five times up and down with a 1,000-µl Eppendorf pipette. Samples were again incubated at 37°C for 10 min, followed by a second gentle trituration five times up and down with a 1,000-µl Eppendorf pipette. At this point, cells were well dissociated. The 1.5-ml suspension was then filtered gently through a 70-µm cell strainer and placed in 15-ml tubes containing 14 ml DMEM/F12/ with 10% FBS. Cells were then pelleted at 300 relative centrifugal force (RCF) for 10 min. Cells were then stained for 15 min at 4°C using antibodies against CD11b (PercpCy5.5), CD45 (FITC; both from R&D Systems), GLAST (PE, Miltenyi) as well as the viability dye ZombieNIR (Biolegend) and Hoechst dye (Molecular Probes) to label all nucleated cells. Cells were then washed and spun at 300 RCF for 10 min at 4°C and resuspended in FACS buffer (2 mM EDTA and 0.5% BSA in PBS) and ran on a Gallios flow cytometer (Becton Dickinson). Positive fraction cells gated on Hoechst positivity, singlet events, and live cells before their respective gates. CTB positivity was set on naive populations.
dLGN dissection, MACS isolation of microglia, and RNA extraction
3 d after bilateral optic nerve crush, mice were perfused with PBS containing heparin (5 U/ml) until livers and tongues were exsanguinated to minimize peripheral immune cell contamination. The entire brain was then removed and meninges and choroid plexus carefully separated and discarded from the rest of the brain tissue. The brain was then placed in a coronal brain matrix (Roboz SA-2165) and a razor blade used to isolate a brain slice containing the dLGN. The dLGN was dissected from surrounding brain regions and placed in 1 ml HBSS (containing Mg and Ca), 4 U/ml papain, and 50 U/ml DNASE-I (Sigma-Aldrich) in a 1.5-ml Eppendorf tube. Bilateral dLGNs were pooled from four mice to serve as one biological sample (three samples per condition). Samples were then incubated at 37°C for 15 m, followed by gentle trituration five times up and down with a 1,000-µl Eppendorf pipette. Samples were again incubated at 37°C for 15 min, followed by a second gentle trituration five times up and down with a 1,000-µl Eppendorf pipette. At this point, cells were well dissociated. The 1-ml suspension was then filtered gently through a 70-µm cell strainer and placed in 15-ml tubes containing 14 ml DMEM/F12 with 10% FBS. Cells were then pelleted at 300 RCF for 10 min. The top of the gradient was then removed. Cells were then labeled with microglia CD11b+ magnetic selection beads (Miltenyi). Cells were positively selected by AutoMACS twice using the Possel setting. All 15-ml tubes before use were coated with sterile BSA in diethyl pyrocarbonate–treated PBS to ensure optimal recovery of microglia. Flow cytometry was used to ensure purity of the positive and negative fraction of AutoMACS sorted cells. 5% of each fraction was isolated and stained for 15 min at 4°C using antibodies against CD11b (PE-Cy7) CD45 (BB515; both from R&D Systems), as well as the viability dye ZombieNIR (Biolegend) and Hoechst dye (Molecular Probes) to label all nucleated cells. Cells were then washed and spun at 300 RCF for 10 min at 4°C and resuspended in 1% PFA in PBS, and samples were run on a Gallios flow cytometer (Becton Dickinson). Positive fraction cells gated on Hoechst positivity, singlet events, and live cells were >90% pure CD45/CD11b+ microglia with the negative fraction <1%. RNA was collected using the RNAqueous micro kit (Ambion) according to the manufacturer’s protocol and stored at −80°C until use.
RNA-seq
RNA integrity was verified using the Qubit RNA assay kit (Life Technologies) and the Agilent RNA Pico kit (Agilent). Amplification and cDNA construction was performed using Nugen Ovation RNA-seq V2 kit with manufacturer's recommended protocol. Sample clean-up after amplification was performed using Qiagen PCR purification kit (Qiagen). Paired-end (50-bp reads) RNA-seq was performed using an Illumina HiSeq (Illumina) yielding 50–70 million reads per sample. All procedures were performed by HudsonAlpha.
RNA-seq analysis
The raw sequencing reads (FASTQ files) went through two stages of preprocessing to remove low-quality reads and bases. First, they were chastity filtered, which removes any clusters that have a higher than expected intensity of the called base compared with other bases. Then they were trimmed with Trimmomatic (Bolger et al., 2014) to remove low-quality bases (minimum read length after trimming = 36). After preprocessing, the quality of the reads was evaluated using FastQC (Andrews, 2010), and after passing quality control, were aligned to the UCSC mm9 genome (Harrow et al., 2012) using the splice-aware read aligner STAR (Dobin et al., 2013). The quality of the alignments was next assessed by SAMStat (Lassmann et al., 2011), and any low-quality alignments were removed with samtools (Li et al., 2009; mapping quality <10). Next, the number of reads aligning to each gene was quantified with HTSeq (Anders et al., 2015), and then the Bioconductor package DESeq2 (Love et al., 2014) was used to normalize the raw counts, perform exploratory analysis (e.g., principal component analysis), and differential expression analysis. The Benjamini–Hochberg false discovery rate procedure was used to correct the p-values for multiple testing. Heat maps of the differentially expressed genes generated with the R package pheatmap (Kolde, 2015). UpSet plots were created with the R package UpSetR (Conway et al., 2017), and the R implementation of Fisher’s exact test, fisher.test, was used to identify enriched gene sets in the differentially expressed genes using the gene set collections from MSigDB (Subramanian et al., 2005)and the gene families from the Hugo Gene Nomenclature Committee (Gray et al., 2015).
Statistical analyses
Statistical tests were performed using Prism (GraphPad) as described in the text and figure legends. In all figures, error bars represent mean ± SEM; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
Accession number
All sequencing data has been uploaded to the GEO repository under accession no. GSE107635.
Online supplemental material
Fig. S1 shows labeling of macrophage populations with ZsGreen label 1 mo after tamoxifen induction in Cx3cr1ERT2/+:Ai6 mice at baseline and at the optic nerve crush site 3 d after optic nerve crush injury. Fig. S2 details loss of TMEM119 positivity in dLGN microglia postoptic nerve crush injury and localization of ingested CTB to the CD68+ phagolysosome. Quantification of GFAP+ cell number and engulfment as well as astrocyte phagocytosis measure by flow cytometry is also provided. Finally, RGC counts 3 d after optic nerve crush injury and RGCs counts and CTB field measurements 7 d after injury are also provided. Fig. S3 shows RGC counts in WldS mice 3 d after optic nerve crush injury as well as pupil responses, RGC counts, and CTB field measurements in TTX-treated animals with optic nerve crush. Representative gating strategy, flow plots, and quantification of MACS efficiency for RNaseq of dLGN microglia are also shown along with changes in canonical microglial activation genes. Finally, RGC counts of C1qa−/− and Itgam−/− mice 3 d after injury are also provided.
Acknowledgments
We would like to thank Shirley Smith for editing the manuscript. We also thank all the members of the Kipnis laboratory and the members of the Center for Brain Immunology and Glia (BIG) for their valuable comments during multiple discussions of this work. Special thanks to Dr. Christopher Deppmann for providing us with WldS mice.
This work was supported by grants from the National Institutes of Health (MH096484 and NS096967) to J. Kipnis.
Authors declare no competing financial interests.
Author contributions: G.T. Norris, A.J. Filiano, and J. Kipnis designed the study. J.A. Thompson and T.H. Harris designed the experiments of microglia fate labeling using Cx3cr1ERT2Cre/+:Ai6 mice. G.T. Norris, I. Smirnov, H.M. Shadowen, and K.R. Cody performed the experiments. A. Gaultier provided the CTB-CypHer 5E reagent and designed the experiments associated with it. G.T. Norris, C.C. Overall, and J. Kipnis analyzed the data. G.T. Norris and J. Kipnis wrote the manuscript with input from the other authors. J. Kipnis oversaw the project.
References
- Aguzzi A., Barres B.A., and Bennett M.L.. 2013. Microglia: scapegoat, saboteur, or something else? Science. 339:156–161. 10.1126/science.1227901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P.T., and Huber W.. 2015. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 31:166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. (2010). FastQC A Quality Control tool for High Throughput Sequence Data http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ by S. Andrews.
- Arandjelovic S., and Ravichandran K.S.. 2015. Phagocytosis of apoptotic cells in homeostasis. Nat. Immunol. 16:907–917. 10.1038/ni.3253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D.L. 2012. Glial responses after chorda tympani nerve injury. J. Comp. Neurol. 520:2712–2729. 10.1002/cne.23069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhar I., Reemst K., Kalchenko V., and Schwartz M.. 2016. The retinal pigment epithelium as a gateway for monocyte trafficking into the eye. EMBO J. 35:1219–1235. 10.15252/embj.201694202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett M.L., Bennett F.C., Liddelow S.A., Ajami B., Zamanian J.L., Fernhoff N.B., Mulinyawe S.B., Bohlen C.J., Adil A., Tucker A., et al. 2016. New tools for studying microglia in the mouse and human CNS. Proc. Natl. Acad. Sci. USA. 113:E1738–E1746. 10.1073/pnas.1525528113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialas A.R., and Stevens B.. 2013. TGF-β signaling regulates neuronal C1q expression and developmental synaptic refinement. Nat. Neurosci. 16:1773–1782. 10.1038/nn.3560 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Bolger A.M., Lohse M., and Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butovsky O., Jedrychowski M.P., Cialic R., Krasemann S., Murugaiyan G., Fanek Z., Greco D.J., Wu P.M., Doykan C.E., Kiner O., et al. 2015. Targeting miR-155 restores abnormal microglia and attenuates disease in SOD1 mice. Ann. Neurol. 77:75–99. 10.1002/ana.24304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesselet M.F. 1984. Presynaptic regulation of neurotransmitter release in the brain: facts and hypothesis. Neuroscience. 12:347–375. 10.1016/0306-4522(84)90058-7 [DOI] [PubMed] [Google Scholar]
- Chiu I.M., Morimoto E.T.A., Goodarzi H., Liao J.T., O’Keeffe S., Phatnani H.P., Muratet M., Carroll M.C., Levy S., Tavazoie S., et al. 2013. A neurodegeneration-specific gene-expression signature of acutely isolated microglia from an amyotrophic lateral sclerosis mouse model. Cell Reports. 4:385–401. 10.1016/j.celrep.2013.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung W.-S., Clarke L.E., Wang G.X., Stafford B.K., Sher A., Chakraborty C., Joung J., Foo L.C., Thompson A., Chen C., et al. 2013. Astrocytes mediate synapse elimination through MEGF10 and MERTK pathways. Nature. 504:394–400. 10.1038/nature12776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman M.P., Conforti L., Buckmaster E.A., Tarlton A., Ewing R.M., Brown M.C., Lyon M.F., and Perry V.H.. 1998. An 85-kb tandem triplication in the slow Wallerian degeneration (Wlds) mouse. Proc. Natl. Acad. Sci. USA. 95:9985–9990. 10.1073/pnas.95.17.9985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colonna M., and Butovsky O.. 2017. Microglia Function in the Central Nervous System During Health and Neurodegeneration. Annu. Rev. Immunol. 35:441–468. 10.1146/annurev-immunol-051116-052358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway J.R., Lex A., and Gehlenborg N.. 2017. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics. 33:2938–2940. 10.1093/bioinformatics/btx364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper M.E., Gregory S., Adie E., and Kalinka S.. 2002. pH-Sensitive Cyanine Dyes for Biological Applications. J. Fluoresc. 12:425–429. 10.1023/A:1021366010681 [DOI] [Google Scholar]
- Danias J., Shen F., Goldblum D., Chen B., Ramos-Esteban J., Podos S.M., and Mittag T.. 2002. Cytoarchitecture of the retinal ganglion cells in the rat. Invest. Ophthalmol. Vis. Sci. 43:587–594. [PubMed] [Google Scholar]
- Davalos D., Grutzendler J., Yang G., Kim J.V., Zuo Y., Jung S., Littman D.R., Dustin M.L., and Gan W.-B.. 2005. ATP mediates rapid microglial response to local brain injury in vivo. Nat. Neurosci. 8:752–758. 10.1038/nn1472 [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T.R.. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott M.R., and Ravichandran K.S.. 2010. Clearance of apoptotic cells: implications in health and disease. J. Cell Biol. 189:1059–1070. 10.1083/jcb.201004096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore M.R.P., Najafi A.R., Koike M.A., Dagher N.N., Spangenberg E.E., Rice R.A., Kitazawa M., Matusow B., Nguyen H., West B.L., and Green K.N.. 2014. Colony-stimulating factor 1 receptor signaling is necessary for microglia viability, unmasking a microglia progenitor cell in the adult brain. Neuron. 82:380–397. 10.1016/j.neuron.2014.02.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadani S.P., Walsh J.T., Smirnov I., Zheng J., and Kipnis J.. 2015. The glia-derived alarmin IL-33 orchestrates the immune response and promotes recovery following CNS injury. Neuron. 85:703–709. 10.1016/j.neuron.2015.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghetti B., Horoupian D.S., and Wiśniewski H.M.. 1972. Transsynaptic response of the lateral geniculate nucleus and the pattern of degeneration of the nerve terminals in the rhesus monkey after eye enucleation. Brain Res. 45:31–48. 10.1016/0006-8993(72)90214-4 [DOI] [PubMed] [Google Scholar]
- Gillingwater T.H., Ingham C.A., Parry K.E., Wright A.K., Haley J.E., Wishart T.M., Arbuthnott G.W., and Ribchester R.R.. 2006. Delayed synaptic degeneration in the CNS of Wlds mice after cortical lesion. Brain. 129:1546–1556. 10.1093/brain/awl101 [DOI] [PubMed] [Google Scholar]
- Goldmann T., Wieghofer P., Müller P.F., Wolf Y., Varol D., Yona S., Brendecke S.M., Kierdorf K., Staszewski O., Datta M., et al. 2013. A new type of microglia gene targeting shows TAK1 to be pivotal in CNS autoimmune inflammation. Nat. Neurosci. 16:1618–1626. 10.1038/nn.3531 [DOI] [PubMed] [Google Scholar]
- Gray K.A., Yates B., Seal R.L., Wright M.W., and Bruford E.A.. 2015. Genenames.org: the HGNC resources in 2015. Nucleic Acids Res. 43(Database issue, D1):D1079–D1085. 10.1093/nar/gku1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenhalgh A.D., and David S.. 2014. Differences in the phagocytic response of microglia and peripheral macrophages after spinal cord injury and its effects on cell death. J. Neurosci. 34:6316–6322. 10.1523/JNEUROSCI.4912-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudi V., Gingele S., Skripuletz T., and Stangel M.. 2014. Glial response during cuprizone-induced de- and remyelination in the CNS: lessons learned. Front. Cell. Neurosci. 8:73 10.3389/fncel.2014.00073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J., Frankish A., Gonzalez J.M., Tapanari E., Diekhans M., Kokocinski F., Aken B.L., Barrell D., Zadissa A., Searle S., et al. 2012. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 22:1760–1774. 10.1101/gr.135350.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman S.E., Kingery N.D., Ohsumi T.K., Borowsky M.L., Wang L.C., Means T.K., and El Khoury J.. 2013. The microglial sensome revealed by direct RNA sequencing. Nat. Neurosci. 16:1896–1905. 10.1038/nn.3554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilla A.M., Diekmann H., and Fischer D.. 2017. Microglia Are Irrelevant for Neuronal Degeneration and Axon Regeneration after Acute Injury. J. Neurosci. 37:6113–6124. 10.1523/JNEUROSCI.0584-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S., Beja-Glasser V.F., Nfonoyim B.M., Frouin A., Li S., Ramakrishnan S., Merry K.M., Shi Q., Rosenthal A., Barres B.A., et al. 2016. Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science. 352:712–716. 10.1126/science.aad8373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoopfer E.D., McLaughlin T., Watts R.J., Schuldiner O., O’Leary D.D.M., and Luo L.. 2006. Wlds protection distinguishes axon degeneration following injury from naturally occurring developmental pruning. Neuron. 50:883–895. 10.1016/j.neuron.2006.05.013 [DOI] [PubMed] [Google Scholar]
- Ippolito D.M., and Eroglu C.. 2010. Quantifying synapses: an immunocytochemistry-based assay to quantify synapse number. J. Vis. Exp. 45:e2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren-Shaul H., Spinrad A., Weiner A., Matcovitch-Natan O., Dvir-Szternfeld R., Ulland T.K., David E., Baruch K., Lara-Astaiso D., Toth B., et al. 2017. A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell. 169:1276–1290.e17. 10.1016/j.cell.2017.05.018 [DOI] [PubMed] [Google Scholar]
- Kolde R. (2015). pheatmap: Pretty Heatmaps. Implementation of heatmaps that offers more control over dimensions and appearance. https://CRAN.R-project.org/package=pheatmap.
- Krasemann S., Madore C., Cialic R., Baufeld C., Calcagno N., El Fatimy R., Beckers L., O’Loughlin E., Xu Y., Fanek Z., et al. 2017. The TREM2-APOE Pathway Drives the Transcriptional Phenotype of Dysfunctional Microglia in Neurodegenerative Diseases. Immunity. 47:566–581.e9. 10.1016/j.immuni.2017.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampron A., Larochelle A., Laflamme N., Préfontaine P., Plante M.-M., Sánchez M.G., Yong V.W., Stys P.K., Tremblay M.-È., and Rivest S.. 2015. Inefficient clearance of myelin debris by microglia impairs remyelinating processes. J. Exp. Med. 212:481–495. 10.1084/jem.20141656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassmann T., Hayashizaki Y., and Daub C.O.. 2011. SAMStat: monitoring biases in next generation sequencing data. Bioinformatics. 27:130–131. 10.1093/bioinformatics/btq614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levkovitch-Verbin H., Harris-Cerruti C., Groner Y., Wheeler L.A., Schwartz M., and Yoles E.. 2000. RGC death in mice after optic nerve crush injury: oxidative stress and neuroprotection. Invest. Ophthalmol. Vis. Sci. 41:4169–4174. [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., and Durbin R.. 1000 Genome Project Data Processing Subgroup . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 25:2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddelow S.A., Guttenplan K.A., Clarke L.E., Bennett F.C., Bohlen C.J., Schirmer L., Bennett M.L., Münch A.E., Chung W.-S., Peterson T.C., et al. 2017. Neurotoxic reactive astrocytes are induced by activated microglia. Nature. 541:481–487. 10.1038/nature21029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling E.A. 1979. Electron microscopic study of macrophages appearing in a stab wound of the brain of rats following intravenous injection of carbon particles. Arch. Histol. Jpn. 42:41–50. 10.1679/aohc1950.42.41 [DOI] [PubMed] [Google Scholar]
- London A., Itskovich E., Benhar I., Kalchenko V., Mack M., Jung S., and Schwartz M.. 2011. Neuroprotection and progenitor cell renewal in the injured adult murine retina requires healing monocyte-derived macrophages. J. Exp. Med. 208:23–39. 10.1084/jem.20101202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundgaard I., Osório M.J., Kress B.T., Sanggaard S., and Nedergaard M.. 2014. White matter astrocytes in health and disease. Neuroscience. 276:161–173. 10.1016/j.neuroscience.2013.10.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moalem G., Monsonego A., Shani Y., Cohen I.R., and Schwartz M.. 1999. Differential T cell response in central and peripheral nerve injury: connection with immune privilege. FASEB J. 13:1207–1217. 10.1096/fasebj.13.10.1207 [DOI] [PubMed] [Google Scholar]
- Nimmerjahn A., Kirchhoff F., and Helmchen F.. 2005. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science. 308:1314–1318. 10.1126/science.1110647 [DOI] [PubMed] [Google Scholar]
- Obermeier B., Daneman R., and Ransohoff R.M.. 2013. Development, maintenance and disruption of the blood-brain barrier. Nat. Med. 19:1584–1596. 10.1038/nm.3407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliani P.L., Wang Y., Fontana E., Robinette M.L., Yamanishi Y., Gilfillan S., and Colonna M.. 2015. TREM2 sustains microglial expansion during aging and response to demyelination. J. Clin. Invest. 125:2161–2170. 10.1172/JCI77983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth T.L., Nayak D., Atanasijevic T., Koretsky A.P., Latour L.L., and McGavern D.B.. 2014. Transcranial amelioration of inflammation and cell death after brain injury. Nature. 505:223–228. 10.1038/nature12808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer D.P., Lehrman E.K., Kautzman A.G., Koyama R., Mardinly A.R., Yamasaki R., Ransohoff R.M., Greenberg M.E., Barres B.A., and Stevens B.. 2012. Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron. 74:691–705. 10.1016/j.neuron.2012.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer D.P., Lehrman E.K., Heller C.T., and Stevens B.. 2014. An engulfment assay: a protocol to assess interactions between CNS phagocytes and neurons. J. Vis. Exp. 88:e51482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz M. 2004. Optic nerve crush: protection and regeneration. Brain Res. Bull. 62:467–471. 10.1016/S0361-9230(03)00076-5 [DOI] [PubMed] [Google Scholar]
- Schwartz M., and Cohen I.R.. 2000. Autoimmunity can benefit self-maintenance. Immunol. Today. 21:265–268. 10.1016/S0167-5699(00)01633-9 [DOI] [PubMed] [Google Scholar]
- Sekar A., Bialas A.R., de Rivera H., Davis A., Hammond T.R., Kamitaki N., Tooley K., Presumey J., Baum M., Van Doren V., et al. Schizophrenia Working Group of the Psychiatric Genomics Consortium . 2016. Schizophrenia risk from complex variation of complement component 4. Nature. 530:177–183. 10.1038/nature16549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanks J.A., Ito S., Schaevitz L., Yamada J., Chen B., Litke A., and Feldheim D.A.. 2016. Corticothalamic Axons Are Essential for Retinal Ganglion Cell Axon Targeting to the Mouse Dorsal Lateral Geniculate Nucleus. J. Neurosci. 36:5252–5263. 10.1523/JNEUROSCI.4599-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M.E. 1999. Phagocytosis of myelin in demyelinative disease: a review. Neurochem. Res. 24:261–268. 10.1023/A:1022566121967 [DOI] [PubMed] [Google Scholar]
- Steinman L. 2006. State of the art. Four easy pieces: interconnections between tissue injury, intermediary metabolism, autoimmunity, and chronic degeneration. Proc. Am. Thorac. Soc. 3:484–486. 10.1513/pats.200603-061MS [DOI] [PubMed] [Google Scholar]
- Stephan A.H., Barres B.A., and Stevens B.. 2012. The complement system: an unexpected role in synaptic pruning during development and disease. Annu. Rev. Neurosci. 35:369–389. 10.1146/annurev-neuro-061010-113810 [DOI] [PubMed] [Google Scholar]
- Stevens B., Allen N.J., Vazquez L.E., Howell G.R., Christopherson K.S., Nouri N., Micheva K.D., Mehalow A.K., Huberman A.D., Stafford B., et al. 2007. The classical complement cascade mediates CNS synapse elimination. Cell. 131:1164–1178. 10.1016/j.cell.2007.10.036 [DOI] [PubMed] [Google Scholar]
- Su J., Haner C.V., Imbery T.E., Brooks J.M., Morhardt D.R., Gorse K., Guido W., and Fox M.A.. 2011. Reelin is required for class-specific retinogeniculate targeting. J. Neurosci. 31:575–586. 10.1523/JNEUROSCI.4227-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., and Mesirov J.P.. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay T.L., Mai D., Dautzenberg J., Fernández-Klett F., Lin G., Sagar M., Datta M., Drougard A., Stempfl T., Ardura-Fabregat A., et al. 2017. A new fate mapping system reveals context-dependent random or clonal expansion of microglia. Nat. Neurosci. 20:793–803. 10.1038/nn.4547 [DOI] [PubMed] [Google Scholar]
- Tufail Y., Cook D., Fourgeaud L., Powers C.J., Merten K., Clark C.L., Hoffman E., Ngo A., Sekiguchi K.J., O’Shea C.C., et al. 2017. Phosphatidylserine Exposure Controls Viral Innate Immune Responses by Microglia. Neuron. 93:574–586.e8. 10.1016/j.neuron.2016.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulland T.K., Wang Y., and Colonna M.. 2015. Regulation of microglial survival and proliferation in health and diseases. Semin. Immunol. 27:410–415. 10.1016/j.smim.2016.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaccarezza O.L., Reader T.A., Pasqualini E., and Pecci-Saavedra J.. 1970. Temporal course of synaptic degeneration in the lateral geniculate nucleus. Its dependence on axonal stump length. Exp. Neurol. 28:277–285. 10.1016/0014-4886(70)90236-0 [DOI] [PubMed] [Google Scholar]
- Vargas M.E., and Barres B.A.. 2007. Why is Wallerian degeneration in the CNS so slow? Annu. Rev. Neurosci. 30:153–179. 10.1146/annurev.neuro.30.051606.094354 [DOI] [PubMed] [Google Scholar]
- Vasek M.J., Garber C., Dorsey D., Durrant D.M., Bollman B., Soung A., Yu J., Perez-Torres C., Frouin A., Wilton D.K., et al. 2016. A complement-microglial axis drives synapse loss during virus-induced memory impairment. Nature. 534:538–543. 10.1038/nature18283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh J.T., Zheng J., Smirnov I., Lorenz U., Tung K., and Kipnis J.. 2014. Regulatory T cells in central nervous system injury: a double-edged sword. J. Immunol. 193:5013–5022. 10.4049/jimmunol.1302401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh J.T., Hendrix S., Boato F., Smirnov I., Zheng J., Lukens J.R., Gadani S., Hechler D., Gölz G., Rosenberger K., et al. 2015. MHCII-independent CD4+ T cells protect injured CNS neurons via IL-4. J. Clin. Invest. 125:699–714. 10.1172/JCI76210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong-Riley M.T.T. 1972. Terminal degeneration and glial reactions in the lateral geniculate nucleus of the squirrel monkey after eye removal. J. Comp. Neurol. 144:61–91. 10.1002/cne.901440104 [DOI] [PubMed] [Google Scholar]
- Wright A.K., Wishart T.M., Ingham C.A., and Gillingwater T.H.. 2010. Synaptic protection in the brain of WldS mice occurs independently of age but is sensitive to gene-dose. PLoS One. 5:e15108 10.1371/journal.pone.0015108 [DOI] [PMC free article] [PubMed] [Google Scholar]