Trim33, a modulator of TGF-β signaling, mediates the proinflammatory function of Th17 cells by the promotion of IL-17 and repression of IL-10 production. These cytokine expressions are regulated through an epigenetic mechanism in cooperation with Smad2 and ROR-γ.
Abstract
Transforming growth factor–β (TGF-β) regulates reciprocal regulatory T cell (T reg) and T helper 17 (Th17) differentiation, the underlying mechanism of which is still not understood. Here, we report that tripartite motif-containing 33 (Trim33), a modulator of TGF-β signaling that associates with Smad2, regulates the proinflammatory function of Th17 cells. Trim33 deficiency in T cells ameliorated an autoimmune disease in vivo. Trim33 was required for induction in vitro of Th17, but not T reg cells. Moreover, Smad4 and Trim33 play contrasting roles in the regulation of IL-10 expression; loss of Trim33 enhanced IL-10 production. Furthermore, Trim33 was recruited to the Il17a and Il10 gene loci, dependent on Smad2, and mediated their chromatin remodeling during Th17 differentiation. Trim33 thus promotes the proinflammatory function of Th17 cells by inducing IL-17 and suppressing IL-10 expression.
Introduction
After activation, naive CD4+ T cells differentiate into several T helper (Th) cell subsets that have distinct effector functions. Th1 cells express IFN-γ and promote immunity against intracellular pathogens. Th2 cells secrete IL-4, IL-5, and IL-13 and contribute to immunity against extracellular pathogens. T follicular helper cells, localized in the germinal center, promote antibody production by B cells and germinal center reactions. Th17 cells, which express IL-17 and IL-17F, are crucial regulators of host defense against various infections (Dong, 2008). Moreover, Th17 cells have been increasingly associated with many human autoimmune disorders, such as psoriasis, inflammatory bowel disease, and multiple sclerosis, and are critical in animal models of autoimmune diseases including experimental autoimmune encephalomyelitis (EAE; Dong, 2008).
Although Th17 cells mediate autoimmunity, accumulating results have suggested that Th17 cells could be modulated in their pathogenic function by the microenvironment. Th17 cells cultured in the presence of IL-23 were more potent in order to induce EAE with decreased IL-10 expression (McGeachy et al., 2007; Ichiyama et al., 2016). TGF-β3, which is induced by IL-23 in T cells, has been reported to promote the pathogenic function of Th17 cells (Lee et al., 2012). In contrast, in a model of tolerance, a regulatory type of Th17 cells were induced that produce IL-10 (Esplugues et al., 2011). Thus, IL-10 expression by Th17 cells may balance out their proinflammatory function. However, molecular mechanisms that program the proinflammatory and regulatory phenotypes of Th17 cells remain unknown.
TGF-β is an important pleiotropic cytokine in the immune system, with both pro- and anti-inflammatory functions. TGF-β, in the presence of IL-6, plays a crucial role in driving Th17 cell differentiation (Bettelli et al., 2006; Mangan et al., 2006; Veldhoen et al., 2006). However, downstream signaling mechanisms underlying the TGF-β–mediated Th17 cell function are not well understood. Although Smad2, but not Smad4, has been genetically demonstrated to regulate Th17 cell differentiation (Yang et al., 2008; Martinez et al., 2009, 2010; Malhotra et al., 2010; Takimoto et al., 2010), how molecules associating TGF-β signaling regulate the function and differentiation of Th17 cells has not been well understood.
Tripartite motif-containing 33 (Trim33), also known as transcriptional intermediary factor 1-γ (TIF1-γ), was previously reported to act as a noncanonical branch of TGF-β/Smad signaling (He et al., 2006). During hematopoiesis, Trim33/Smad2/3 complex regulates a set of genes different from those governed by Smad4/2/3 complex (He et al., 2006; Xi et al., 2011). Interestingly, Trim33, with an E3 ubiquitin ligase domain, was reported to inhibit Smad4 function (Dupont et al., 2005, 2009; Agricola et al., 2011). However, a role of Trim33 in T cell differentiation is unknown.
In this study, we found that Trim33 regulates the proinflammatory function of Th17 cells. Deficiency of Trim33 in T cells resulted in decreased IL-17 but enhanced IL-10 production in CD4+ T cells, leading to amelioration of EAE diseases. Although Smad4 promoted IL-10 production in Th17 cells, Trim33 negatively regulated IL-10 by direct suppression of Il10 transcription. The chromatin immunoprecipitation sequencing (ChIP-seq) analysis showed that the genomic regions bound by Trim33 were largely co-occupied by retinoic acid orphan receptor γ (ROR-γ). Consistently, Trim33 physically associated with ROR-γ and Smad2 in Th17 cells. Loss of Trim33 impaired chromatin remodeling during Th17 cell differentiation. Our data thus indicate that Trim33 mediates proinflammatory T cell function by differential regulation of IL-17 and IL-10.
Results
Trim33 plays a crucial role in Th17 cell development in vivo
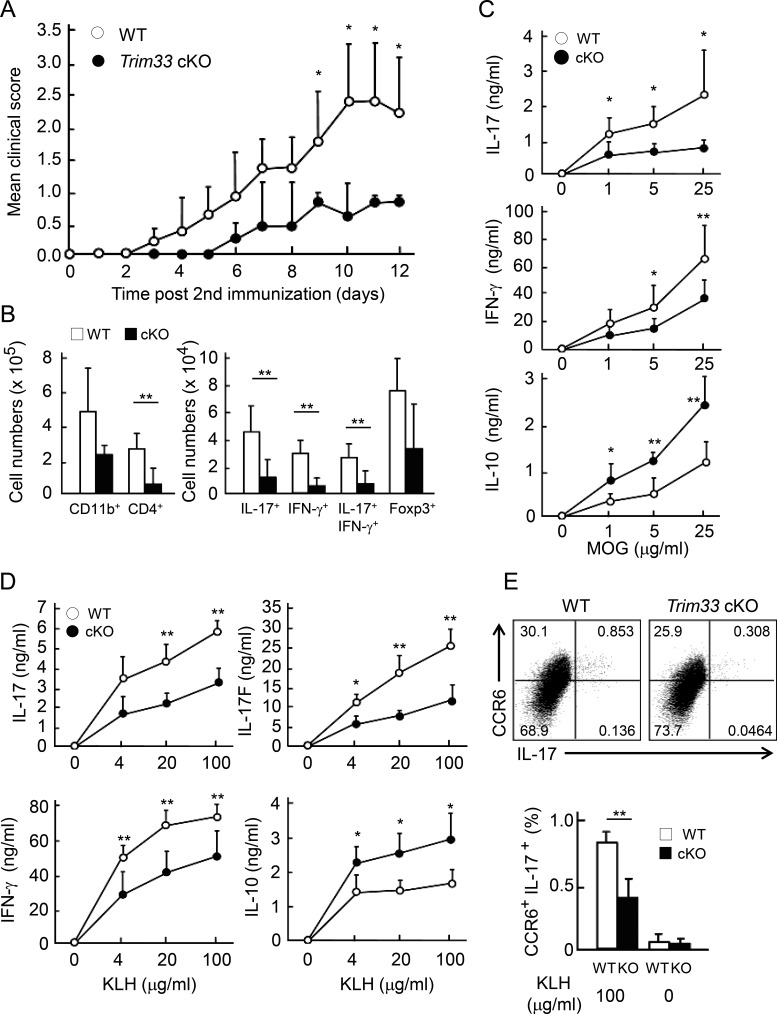
To analyze the role of Trim33 in T cells, Trim33 flox mice (Kim and Kaartinen, 2008) were crossed with CD4-Cre transgenic mice (Makar et al., 2003) to specifically disrupt the Trim33 gene in T cells (conditional KO [cKO]). Trim33 was efficiently deleted in CD4+ T cells isolated from Trim33 cKO mice at the protein level (Fig. S1 A). There was no obvious defect in T cell development in the cKO mice (unpublished data). To analyze the role of Trim33 in T cell differentiation and autoimmunity, we immunized Trim33 flox/flox mice with or without CD4-Cre to induce EAE. On day 3 after the second immunization with myelin oligodendrocyte glycoprotein (MOG) peptide in CFA, control mice started to develop EAE disease and reached a score of 2.5–3.0 by day 10 (Fig. 1 A). In contrast, Trim33 cKO mice first showed EAE symptoms on day 6. On day 10, a much milder disease (score 0.5–1.0) was observed in Trim33 cKO mice compared with WT control mice, indicating that deficiency of Trim33 in T cells led to an amelioration of EAE. To understand the underlying mechanism, we analyzed cell infiltration in the central nervous system and cytokine expression by CD4+ T cells in the spleen. Absolute numbers of central nervous system–infiltrating total and cytokine-expressing CD4+ cells (IL-17+, IFN-γ+, IL-17/IFN-γ+, IL-10+, IL-17/IL-10+ GM-CSF+, IL-17/GM-CSF+) were greatly reduced in Trim33 cKO mice compared with WT animals, although frequencies of these cytokine-expressing cells were largely unchanged (Figs. 1 B and S1, B and C). However, Foxp3+ cells were not significantly reduced (Fig. 1 B, right). In periphery, MOG-specific IL-17 and IFN-γ expression was significantly reduced by Trim33 KO T cells (Fig. 1 C). IL-10 has been reported to negatively regulate EAE development (Bettelli et al., 1998). Interestingly, antigen-specific IL-10 expression was enhanced in Trim33 cKO mice (Fig. 1 C).
Figure 1.
Trim33 is indispensable for Th17 response in vivo. (A) Trim33 f/f (WT n = 5; open circle) and Trim33 f/f CD4-Cre (Trim33 cKO n = 5; closed circle) mice were immunized with MOG peptides in CFA to induce EAE. Disease scores are shown. (B) Absolute numbers of central nervous system–infiltrating cells were analyzed. CD11b+or CD4+ cells are presented in left panel. IL-17+, IFN-γ+, IL-17+IFN-γ+, and Foxp3+ cells in CD4+ cells are shown in right panel. (C) Splenocytes of immunized mice shown in A were restimulated with MOG peptide for 72 h. IL-17, IL-10, and IFN-γ were measured by ELISA. (D) WT (n = 4; open circle) and Trim33 cKO (n = 4; closed circle) mice were immunized with KLH in CFA. After 7 d, dLN cells of immunized mice were restimulated with KLH for 72 h, and cytokine expression was assessed by ELISA. (E) dLN cells were restimulated for 24 h, as performed in D, followed by costaining of CCR6 and IL-17. Frequency of CCR6+ and IL-17+ double-positive cells in WT and the KO mice is shown. Data are representative of three independent experiments. Error bars show means ± SD. *, P < 0.05; **, P < 0.01.
The above results suggest that Trim33 may regulate Th17 but not regulatory T cell (T reg cell) response in vivo. To better examine the role of Trim33 during the early differentiation of Th17 cells in vivo, we immunized WT and Trim33 cKO mice with KLH in CFA. After 1 wk, draining LN (dLN) cells isolated from immunized mice were restimulated with KLH. As seen in the EAE model, Trim33 KO CD4+ T cells showed significantly reduced expression of IL-17, IL-17F, and IFN-γ (Fig. 1 D). In contrast, KLH-specific IL-10 production by Trim33 KO T cells was enhanced compared with WT T cells (Fig. 1 D). CCR6-mediated cell migration to the central nervous system is a key in EAE induction (Yamazaki et al., 2008). In Trim33 cKO mice after KLH immunization, the frequency of antigen-specific CCR6+ IL-17-producing CD4+ T cells was significantly reduced (Fig. 1 E). These data suggest that Trim33 plays a crucial role in Th17-mediated autoimmunity by promoting IL-17 and CCR6 but dampening IL-10 expression. These phenotypes may result in reduced numbers of infiltrating lymphocytes into the central nervous system in the absence of Trim33 in T cells. To examine the significance of the enhanced production of IL-10 in EAE disease, we blocked IL-10 signal by administration of neutralizing antibody against IL-10 receptor (IL-10R). Trim33 cKO mice treated with isotype Ig showed milder symptoms and lower numbers of infiltrating cells in the central nervous system compared with isotype Ig-treated WT. Anti–IL-10R antibody treatment almost fully restored the disease scores and absolute numbers of total central nervous system–infiltrating and IL-17–expressing CD4+ cells in Trim33 cKO mice (Fig. S1, D and E). The results suggest that the enhanced IL-10 expression by Trim33 KO T cells protected against EAE disease in Trim33 cKO mice.
Trim33 is crucial for the differentiation of Th17 but not T reg cells in vitro
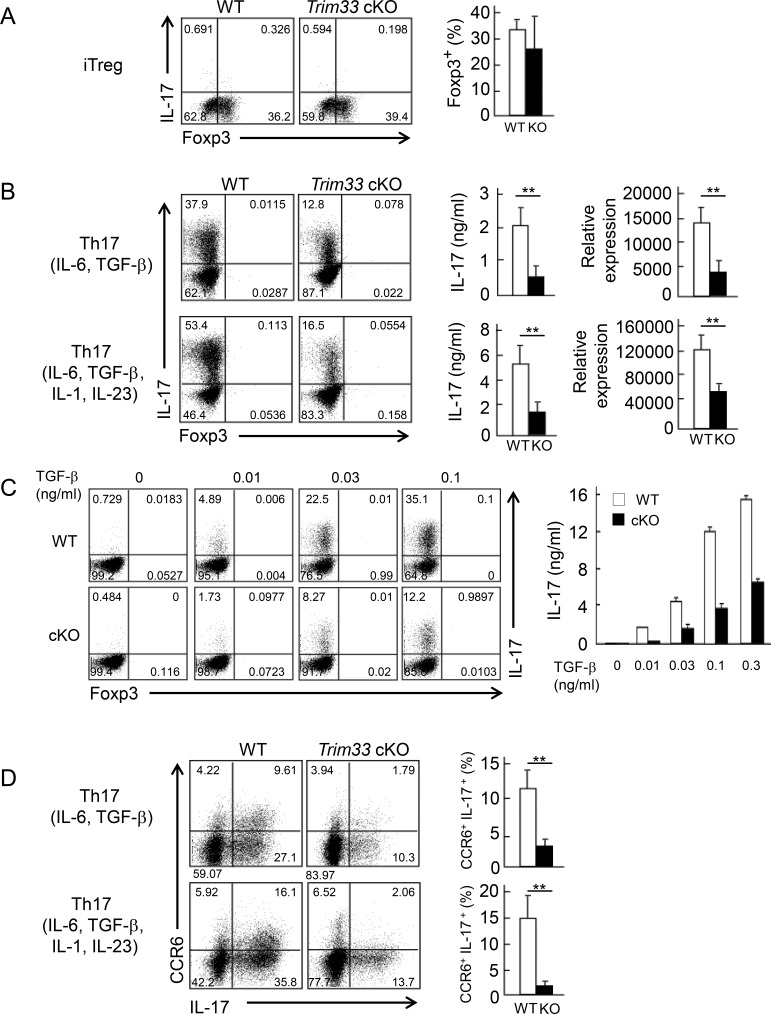
To further examine the regulation by Trim33 in T cells, we purified naive CD4+ T cells (CD44loCD62LhiCD25−) from cKO or WT mice by FACS and stimulated them with anti-CD3 and anti-CD28 antibodies under inducible T reg (iT reg) cell and Th17 skewing conditions. We found no difference in Foxp3 expression at any dose of TGF-β tested between WT and Trim33 KO T cells under iT reg conditions (Fig. 2 A and Fig. S2 A). IL-10 production by WT and Trim33 KO iT reg cells was quite low and, in both cells, comparable to naive CD4+ T cells at the mRNA level (unpublished data). Furthermore, we measured IL-10 expression in total and ROR-γ+ T reg cells (Kim et al., 2017) and found slightly increased IL-10 production in Trim33-deficient total and ROR-γ+ T reg cells compared with WT, but no difference in the frequency of these T reg cells (Fig. S2 B). In Th17 skewing conditions (IL-6, TGF-β, anti-IL-4, and anti–IFN-γ or IL-6, TGF-β, IL-1, IL-23, anti–IL-4, and anti–IFN-γ), we found that IL-17 production by Trim33 KO T cells was significantly reduced at both the protein and mRNA levels compared with WT cells; Foxp3 expression was low in both WT and Trim33 KO T cells (Fig. 2 B). Defects in IL-17 expression were observed at all concentrations of TGF-β tested (Fig. 2 C) or when TGF-β3 was used instead of TGF-β1 (Fig. S2, C and D). Moreover, CCR6+ IL-17–producing cells were almost completely absent in Trim33-deficient T cells (Fig. 2 D). The culture with IL-6, IL-23, and IL-1 showed a slight reduction of Th17 differentiation in the absence of Trim33, whereas addition of anti–TGF-β neutralizing antibody completely diminished Th17 differentiation (Fig. S2, E and F). These data indicate that Trim33 plays a crucial role in the differentiation of Th17 cells but not iT reg cells in vitro.
Figure 2.
Trim33 is required for Th17 but not iT reg cell differentiation in vitro. (A) Naive CD4+ T cells (CD25−CD44loCD62Lhi) isolated from WT (white) and Trim33 cKO (black) mice were stimulated by anti-CD3 and anti-CD28 mAbs under Th17 and iT reg skewing conditions. After 4 d, cultured cells were restimulated. Foxp3 and IL-17 expression in iT reg cell condition are shown (left). Frequency of Foxp3+ cells is presented (right). (B) Foxp3 and IL-17 expression in Th17 conditions are shown (left). IL-17 expression was measured by ELISA (middle) and quantitative PCR (right). Data were normalized to b-actin. (C) Naive CD4+ T cells isolated from WT and Trim33 cKO mice were cultured under Th17 skewing conditions with indicated concentrations of TGF-β. IL-17 expression was detected upon restimulation by intracellular cytokine staining (left) and ELISA (right). (D) Costaining of CCR6 and IL-17 in Th17 conditions is presented (left). Frequency of CCR6+ IL-17+ cells is shown (right). Data are representative of three independent experiments. Error bars show means ± SD. **, P < 0.01.
Microarray analysis identified IL-10 to be regulated by Trim33
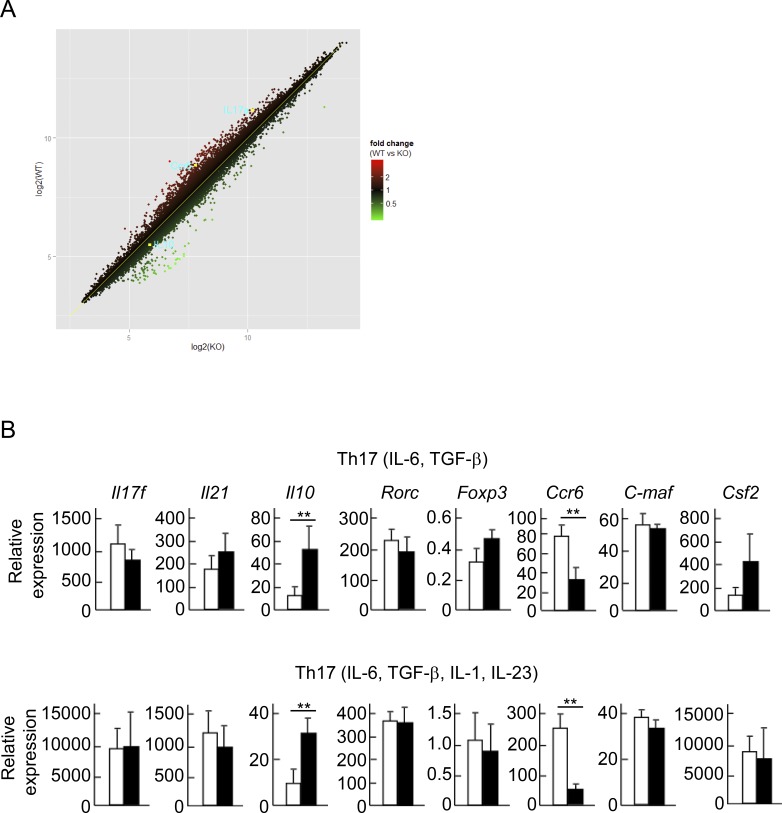
To further explore the functional targets of Trim33, we performed microarray analysis of WT and Trim33 KO T cells cultured under Th17 skewing conditions. We confirmed that mRNA expression of Il17 and Ccr6 was down-regulated. Il10 expression was increased in the absence of Trim33 (Fig. 3 A), consistent with the in vivo data shown in Fig. 1 (C and D). We further confirmed these results with real-time RT-PCR. Trim33 KO T cells showed reduced Il17a and Ccr6 and enhanced Il10 expression, although mRNA levels for Il21, retinoic acid orphan receptor c (Rorc), maf, and Il17f were comparable between WT and KO T cells (Fig. 3 B). GM-CSF produced by Th1 and Th17 cells was shown to play an essential role in EAE (Codarri et al., 2011; El-Behi et al., 2011). However, GM-CSF expression was not decreased in the absence of Trim33 in Th17 cells (Fig. 3 B).
Figure 3.
Identification of Trim33-target genes during Th17 differentiation. (A) Naive CD4+ T cells isolated from WT and Trim33 cKO mice were cultured in Th17 skewing conditions (IL-6, TGF-β, anti–IL-4 antibody, and anti–IFN-γ antibody). The cells were restimulated with anti-CD3 antibody and subjected to microarray analysis. (B) Expression of Th17-related genes in WT and Trim33 KO T cells cultured under Th17 conditions, as cultured in Fig. 2, was assessed by quantitative PCR. Data shown combine results from three independent experiments. Error bars show means ± SD. **, P < 0.01.
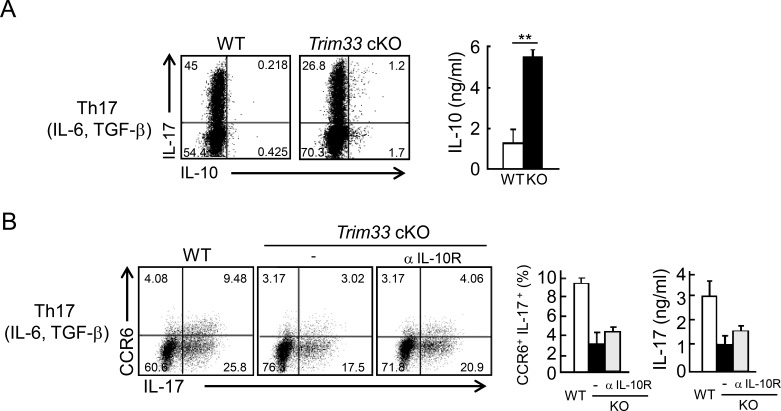
To confirm dysregulated IL-10 expression in Th17 cells, we cultured WT and Trim33 KO T cells under Th17 skewing conditions to stain for intracellular IL-17 and IL-10. In Trim33-deficient T cells, frequencies of IL-17/IL-10 double-positive T cells and IL-10 single-positive cells were increased compared with WT cells (Fig. 4 A). In agreement with the mRNA data shown in Fig. 3 B, IL-10 protein expression by Trim33 KO T cells was highly enhanced compared with WT T cells (Fig. 4 A), indicating IL-10 de-repression in the absence of Trim33 differentiation.
Figure 4.
Trim33 deficiency results in IL-10 up-regulation in Th17 cells. (A) Naive CD4+ T cells isolated from WT (white) and Trim33 cKO (black) mice were stimulated by anti-CD3 and anti-CD28 antibodies under Th17 skewing conditions. After restimulation, costaining of IL-17 and IL-10 was performed by intracellular staining (left), and IL-10 production was measured by ELISA (right). (B) Naive CD4+ T cells isolated from WT and Trim33 cKO mice were stimulated by anti-CD3 and anti-CD28 antibodies under Th17 skewing conditions with or without anti–IL-10R antibody (WT, white; KO, black; and KO+anti–IL-10R, gray). Costaining of CCR6 and IL-17 was performed after restimulation (left). The statistical analysis of CCR6+IL-17+ cells is shown (middle). IL-17 expression was measured by ELISA (right). Data are representative of three independent experiments. Error bars show means ± SD. **, P < 0.01.
Because IL-10 is a potent immunosuppressive cytokine, we examined the possibility that enhanced IL-10 expression by Trim33 KO CD4+ T cells inhibited their differentiation to Th17 cells in an autocrine and/or paracrine manner. We cultured control and Trim33 KO naive CD4+ T cells under Th17 conditions with or without anti–IL-10R antibody. IL-10 blockade could not restore IL-17 and CCR6 expression in Trim33 KO T cells (Fig. 4 B), excluding an indirect effect of de-repressed IL-10 in IL-17 and CCR6 regulation during Th17 development in vitro.
Genome-wide analysis on Trim33-bound genes
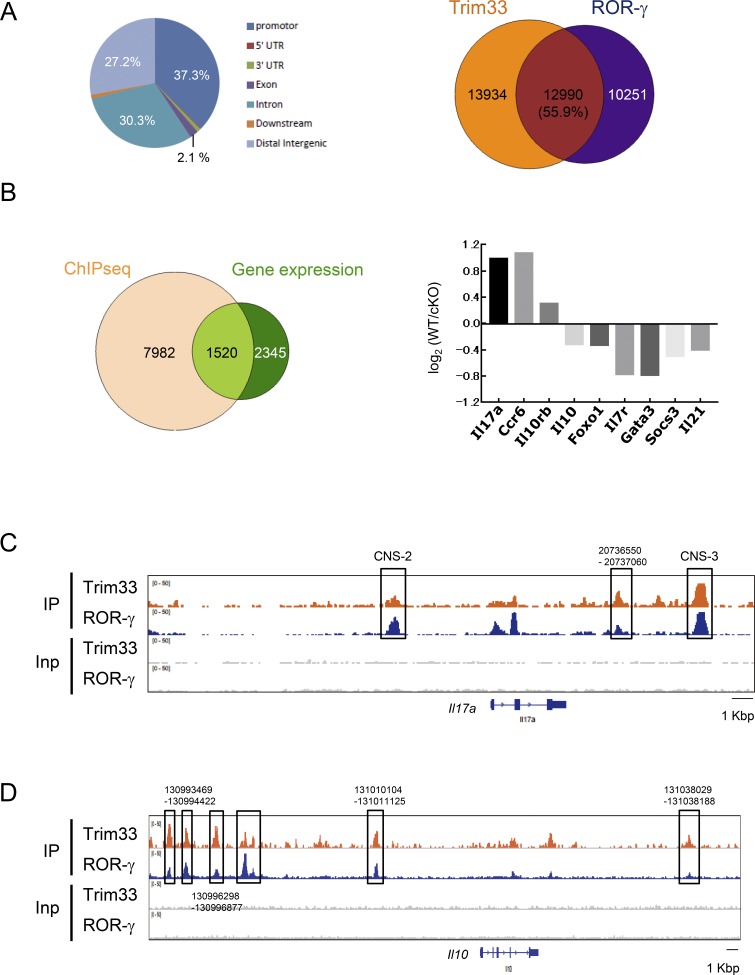
Our results demonstrated that expression of the molecules that regulate IL-17 and IL-10 expression, such as Rorc and maf, was not affected by the lack of Trim33 in Th17 cells, suggesting that Trim33 may directly regulate these genes in Th17 cells. To assess this, we performed ChIP-seq with anti-Trim33 antibody in developing Th17 cells. The computational analysis demonstrated that >94% of Trim33 peaks were located in noncoding genomic sequences such as promoter, intron, and intergenic regions, suggesting that Trim33 may regulate gene transcription by binding to regulatory cis-elements (Fig. 5 A, left). Trim33 has been shown to mediate the TGF-β signaling pathway, which is commonly used in both Th17 and T reg cell differentiations. To understand the specific function of Trim33 in Th17 but not T reg cell differentiation, we computationally analyzed colocalization of peaks obtained from our Trim33 ChIP-seq and ROR-γ ChIP-seq previously reported (Ciofani et al., 2012). Interestingly, approximately half of binding regions of Trim33 and ROR-γ was co-occupied with each other (Fig. 5 A, right). This result suggests collaborative regulation by Trim33 and ROR-γ.
Figure 5.
Trim33 ChIP-seq analysis. (A) Distribution of Trim33 ChIP-seq peaks in Th17 cells (left). Occupancy of ChIP-seq binding peaks of Trim33 and ROR-γ (right). (B) Integration of Trim33 binding peak data with gene expression analysis (left). Among 1,520 genes, genes related to the function and differentiation of Th cells are shown with their expression changes of WT versus KO (right). (C and D) ChIP-seq peaks of Trim33 and ROR-γ in Il17a (C) and Il10 (D) gene loci were shown. Data are representative of two independent experiments.
We then worked to identify direct targets of Trim33 by integration of the gene expression dataset with Trim33 binding data. With a default setting of parameters, 9,502 of 27,050 Trim33 peaks were annotated within genes by the PAVIS program. The integration analysis revealed that 1,520 genes were possible direct targets regulated by Trim33 (Fig. 5 B, left). We further selected genes that were suggested to regulate the function and/or differentiation of Th cells and showed them with differential expressions of WT versus KO (Fig. 5 B, right). The result suggests the direct transcriptional regulation of IL-17 and IL-10 by Trim33. We observed coaccumulation of Trim33 and ROR-γ on Il17a and Il10 but not Foxp3 loci (Fig. 5, C and D; and Fig. S3), further supporting our idea of cooperative regulation of the expression of IL-17 and IL-10 but not Foxp3 by Trim33 with ROR-γ. On the other hand, the analysis also raised a possible indirect mechanism: the expression of IL-17 and IL-10 may be regulated by GATA3, SOCS3, and FOXO1, suggesting dual regulatory pathways by Trim33.
Trim33 regulates IL-17 and IL-10 expression at the chromatin level
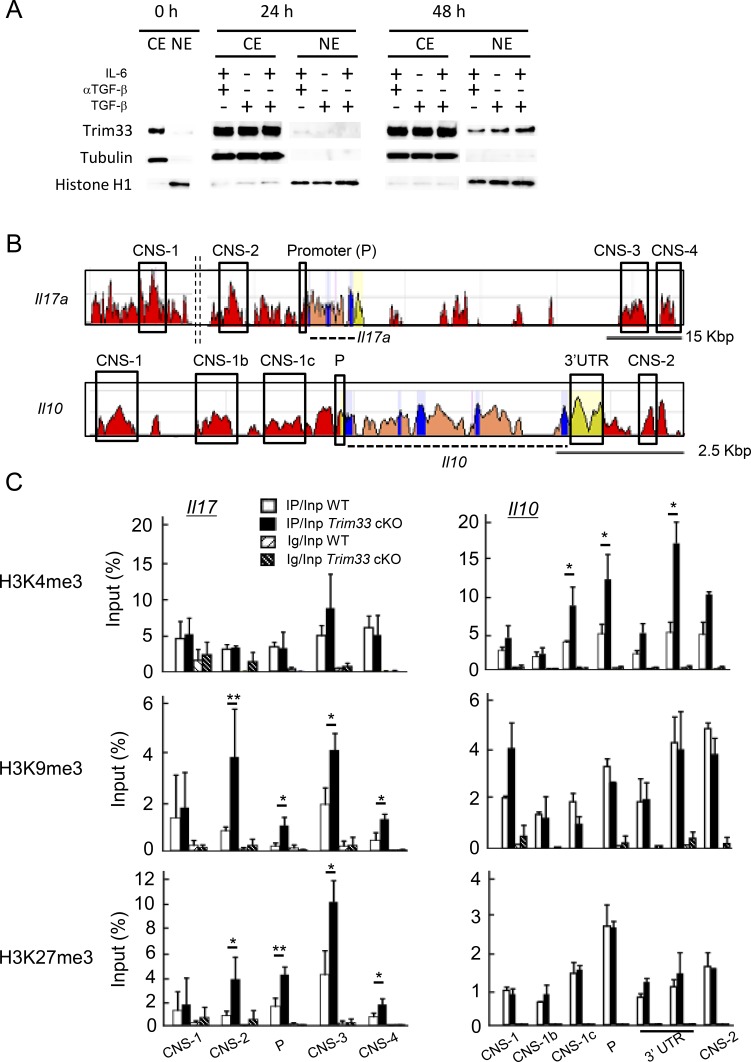
Our aforementioned results suggest that Trim33 controls the expression of IL-17 and IL-10 at the transcription level. To address this, we assessed localization of Trim33 before and after priming with the indicated cytokine combinations in the presence of antigen stimulation. We separated cytoplasmic and nuclear fractions 0, 24, and 48 h after stimulation and detected Trim33 protein by immunoblot. Trim33 was localized mainly in cytoplasm before priming, then it was found in the nuclear fraction after 48 h but not 24 h. In addition, its localization in nuclei was largely independent of the cytokine combinations but rather as a result of antigen stimulation (Fig. 6 A).
Figure 6.
Trim33 regulates histone modifications at the Il17a and Il10 gene loci. (A) Extracts from both nuclei and cytosol were isolated from CD4+ T cells before stimulation (0 h) and stimulated with indicated conditions after 24 and 48 h. Trim33, tubulin (cytosolic protein) and histone H1 (nucleic protein) were detected by immunoblot. Data are representative of three independent experiments. (B) Schematic diagrams indicate comparison of genomic sequence of murine Il17 and Il10 gene loci with human. (C) Histone modifications were assessed by ChIP analysis. Naive CD4+ T cells isolated from WT and Trim33 cKO mice were stimulated under Th17 skewing conditions. Chromatin isolated from these cells was immunoprecipitated with antibodies against H3K4me3, H3K9me3, H3K27me3, and control Ig. Genomic regions modified with histone marks were detected by PCR with primer sets of each CNS. Data are representative of two independent experiments. Error bars show means ± SD. *, P < 0.05; **, P < 0.01.
Because Trim33 was implicated in epigenetic regulation, we examined several histone markers in conserved cis-elements in the Il17 and Il10 loci in WT and Trim33 KO CD4+ T cells cultured under Th17 conditions (Fig. 6 B). In the absence of Trim33, we found enhanced suppressive marks, H3K9me3 and H3K27me3, in multiple regions at the Il17 locus including its promoter, whereas H3K4me3 and H3Ac in the Il17 gene were not affected (Fig. 6 C, left; and not depicted). In contrast, H3K4me3 was enhanced in multiple conserved regions at the Il10 locus (Fig. 6 C, right). These data suggest that Trim33 controls the expression of the Il17 and Il10 genes through the regulation of histone modifications.
Trim33 recruitment is dependent on Smad2
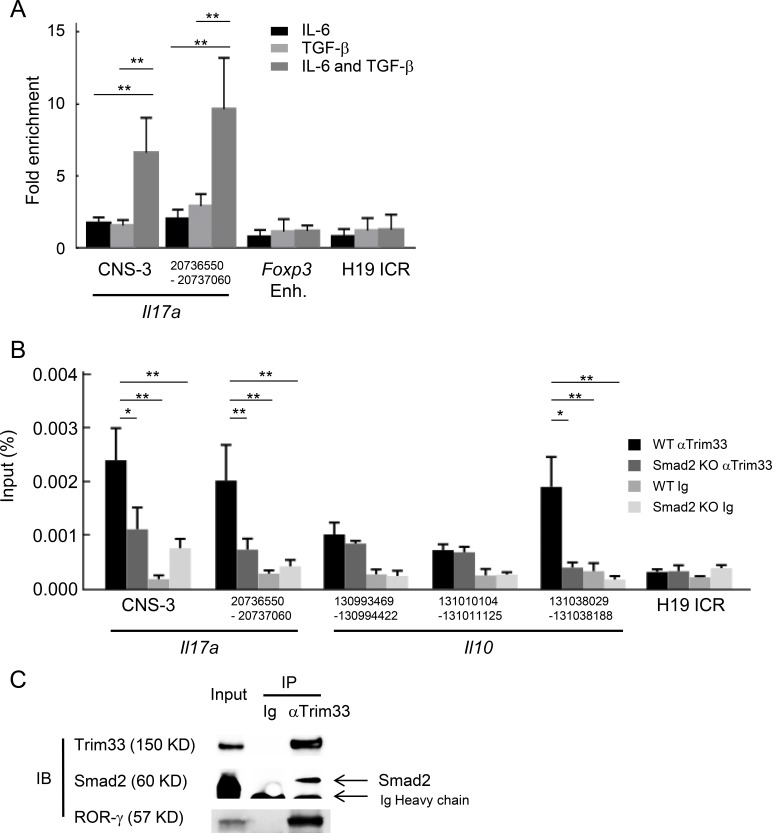
We then examined how Trim33, without DNA binding directly, was recruited to target genes. We first tested the binding of Trim33 at the Il17a locus in CD4+ T cells stimulated with different cytokine combinations by ChIP-PCR. We observed the binding at the Il17a locus only when T cells were stimulated with IL-6 and TGF-β in which ROR-γ is strongly induced, but not at Foxp3 enhancer or imprinting region, H19 ICR, as an unrelated genomic region (Fig. 7 A). Trim33 was reported to mediate Smad4-independent, but Smad2-dependent, signaling of TGF-β (Xi et al., 2011). We thus tested Trim33 recruitment to the Il17a and Il10 loci in T cells lacking Smad2 and found that the binding of Trim33 was attenuated (Fig. 7 B), supporting that Smad2 is crucial for Trim33 binding to the Il17 and Il10 gene loci.
Figure 7.
Cooperative regulation of IL-17 and IL-10 by Trim33, Smad2, and ROR-γ. (A) Trim33 binding was assessed by ChIP-PCR analysis. PCR amplification was conducted with input DNA and I.P. DNA prepared from chromatin precipitated with anti-Trim33 antibodies, with C57BL/6 CD4+ T cells stimulated by anti-CD3 and anti-CD28 antibodies under indicated conditions. Primer for the H19 ICR locus was used as a negative control. Data are representative of three independent experiments. (B) Trim33 ChIP was performed with WT and Smad2 KO CD4+ T cells cultured under Th17 skewing conditions. (C) Physical association of Trim33 with Smad2 and ROR-γ in Th17 cells. Lysates of Th17 cells were subjected to immunoprecipitation with anti-Trim33 antibody or isotype Ig, followed by immunoblot analysis with anti-Trim33, Smad2, and ROR-γ antibodies. Data are representative of four independent experiments. Error bars show means ± SD. *, P < 0.05; **, P < 0.01.
Our Trim33 ChIP-seq analysis revealed colocalization of Trim33 and ROR-γ in Th17 cells. We further examined the association of Trim33 with ROR-γ. Coimmunoprecipitation (coIP) assay with whole-cell lysate of Th17 cells revealed a physical association of Trim33 with Smad2 and ROR-γ, whereas the association of Trim33 with ROR-γ was lost in the absence of Smad2 (Figs. 7 C and S4 A). Furthermore, we confirmed that both Smad2 KO and ROR-γ KO T cells showed defects in the expression of IL-17, CCR6, and IL-10, resembling the phenotype caused by Trim33 deficiency to a greater or lesser extent (Fig. S4, B–F). In line with these results, we previously reported that Smad2 was associated with ROR-γ and these factors cooperatively promoted IL-17 expression (Martinez et al., 2010). Taken together, it is suggested that the complex of Trim33/Smad2/ROR-γ is necessary for an optimal expression of IL-17 and repression of IL-10 in Th17 cells.
Reciprocal regulation of IL-10 expression by Trim33 and Smad4
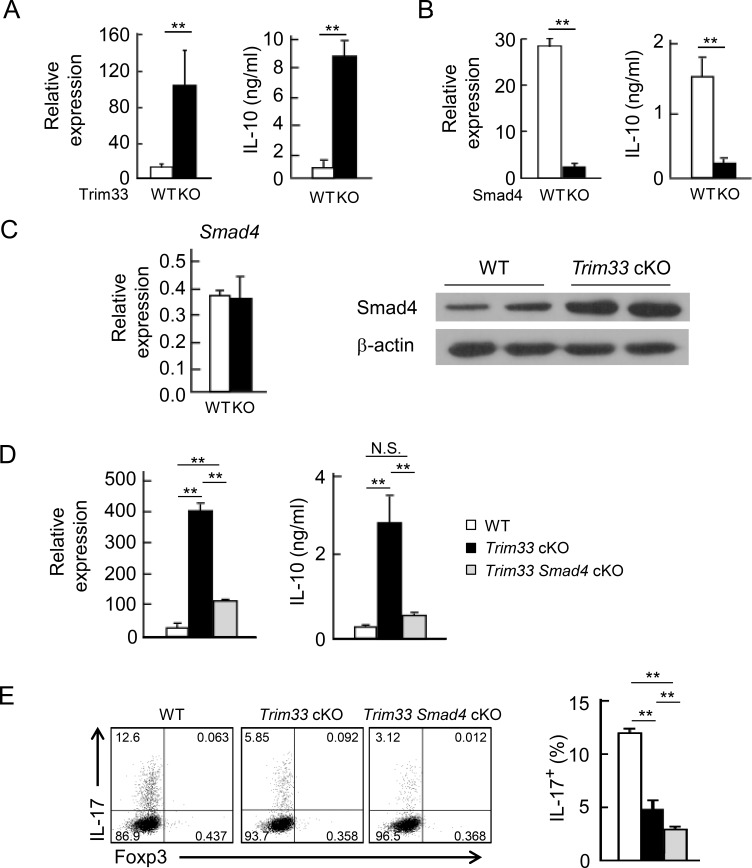
It has been reported that TGF-β regulated IL-10 expression during Th17 differentiation (Stumhofer et al., 2007). To further clarify the mechanism downstream of TGF-β signaling in the expression of IL-10, we isolated naive CD4+ T cells from Trim33 or Smad4 cKO mice and their appropriate control mice and cultured them under Th17 skewing conditions. Because IL-10 expression was observed after primary stimulation as well as during restimulation, we tested the IL-10 expression on day 3. Trim33-deficient T cells expressed increased amounts of IL-10 at both protein and mRNA levels (Fig. 8 A). In contrast, Smad4 deficiency led to reduced IL-10 production both in priming and after stimulation (Figs. 8 B and S5 A).
Figure 8.
Reciprocal IL-10 regulation by Trim33 and Smad4. (A) Naive CD4+ T cells isolated from WT and Trim33 cKO mice were stimulated by anti-CD3 and anti-CD28 antibodies under Th17 skewing conditions. On day 3, the cultured cells and supernatant were harvested, followed by measurement of IL-10 at the mRNA (left) and protein (right) levels. (B) IL-10 production was measured with WT and Smad4 KO Th17 cells, as done in A. (C) Smad4 expression was detected at the mRNA (left) and protein (right) levels in WT and Trim33 KO Th17 cells. β-Actin was used as loading control. Data are representative of three independent experiments. (D) IL-10 production was measured in WT, Trim33 KO, and Trim33 × Smad4 KO Th17 cells, as described in A. (E) IL-17 production by WT, Trim33 KO, and Trim33 × Smad4 KO Th17 cells, after restimulation. Data are representative of two independent experiments. Error bars show means ± SD. **, P < 0.01.
Trim33 contains an E3 ubiquitin ligase domain, which was suggested to target Smad4 for degradation (Dupont et al., 2005). In the absence of Trim33, the expression of Smad4 at the protein but not the mRNA level was increased in Th17 cells compared with WT. However, the increment was not observed in iT reg cells (Figs. 8 C and S5 B). These observations raised another possibility that Trim33 may suppress IL-10 expression by reduction of Smad4 protein in T cells. To address this possibility, we tested whether the enhanced production of IL-10 caused by the deletion of Trim33 could be reversed by Smad4 deficiency. We isolated naive CD4+ T cells from Trim33 cKO, Trim33 × Smad4 cKO, and WT mice and cultured them under Th17 skewing conditions. As shown above, Trim33 KO T cells expressed increased amounts of IL-10, whereas the enhanced IL-10 expression was almost completely suppressed by compound deletion of Smad4 (Fig. 8 D). These results were largely true even after restimulation (Fig. S5 C). The data support our idea of an interplay between Trim33 and Smad4 in IL-10 regulation during Th17 differentiation. Because Trim33 was localized in cytoplasm 24 h after primary activation, we compared cytokine expression between WT and the KO T cells 20 h after priming. IL-10 but not IL-17 expression was slightly enhanced in the absence of Trim33 (Fig. S5 D). Moreover, when RING domain was deleted, although not affecting its mRNA expression, the suppressive activity of Trim33 for IL-10 expression was partially attenuated, whereas IL-17 induction was comparable to that by the full-length one (Fig. S5 E). These results suggest a partial requirement of E3 activity for IL-10 suppression but not IL-17 induction.
Finally, we examined IL-17 in T cells in which Trim33 and Smad4 were doubly deleted during Th17 differentiation. Smad4 deficiency on its own did not significantly impact IL-17 expression, whereas Smad4/Trim33 double deficiency led to further reduction of IL-17 than Trim33 single-KO cells (Fig. 8 E), suggesting the redundant but minor role of Smad4 in IL-17 expression during Th17 differentiation in the absence of Trim33.
Discussion
The requirement of TGF-β signaling in the differentiation of Th17 cell differentiation has been shown in the literature (Bettelli et al., 2006; Veldhoen et al., 2006; Li et al., 2007; McGeachy et al., 2007; Ghoreschi et al., 2010; Lee et al., 2012). We previously showed that neutralization of TGF-β signaling with anti–TGF-β antibody (Chung et al., 2009) or with an inhibitor against the TGF-β receptor (unpublished data) blocked Th17 differentiation of mouse naive T cells cultured with IL-6, IL-1, and IL-23 in the presence of antigen stimulation. We confirmed that result in the current study, too. Furthermore, it has been reported that the major downstream signal transducer of TGF-β signaling, Smad2, but not Smad4, plays a crucial role in the differentiation of Th17 cells (Yang et al., 2008; Martinez et al., 2009, 2010; Malhotra et al., 2010; Takimoto et al., 2010). These results definitely support the requirement of TGF-β for Th17 differentiation. However, it is unclear how TGF-β signaling regulates the development of Th17 cells. In this study, we have shown that a modulator of TGF-β signaling, Trim33, regulates the proinflammatory function of Th17 cells by enhancing IL-17 and repressing IL-10 production.
Although Trim33 has been reported to associate with receptor-activated Smad (R-Smad) upon TGF-β stimulation (He et al., 2006; Xi et al., 2011), our current data indicate that Trim33 plays a crucial role in IL-17 production during Th17 differentiation but not Foxp3 induction in iT reg cells. In addition, Trim33 ChIP assay revealed that Trim33 did not bind to the Foxp3 gene locus in any conditions tested. These results support that Trim33 specifically regulates Th17 cells but not iT reg cells.
During Th17 cell differentiation, we found that Trim33 deficiency did not affect the expression level of ROR-γ, a Th17-specific master transcription factor. Also, the expression level of Trim33 mRNA in CD4+ T cells cultured in Th17 skewing conditions was similar to that of other Th subsets, including iT reg. Therefore, we sought the mechanisms underlying a selective requirement of Trim33 in Th17 differentiation. The computational analysis of global binding of Trim33 and ROR-γ demonstrated that Trim33 accumulated with close proximity to ROR-γ across the genome, including at the Il17a and Il10 gene loci. In addition, the binding of Trim33 on Il17a locus was observed only when CD4+ T cells received the signaling of both IL-6 and TGF-β. Furthermore, we found that Trim33 physically associated with ROR-γ as well as Smad2 by coIP assay. In the previous study, we found that the association of Smad2 and ROR-γ synergistically promoted IL-17 expression in Th17 cells (Martinez et al., 2010). In addition, our current study indicates that the binding of Trim33 at the Il17a and Il10 loci and physical association of Trim33 with ROR-γ was attenuated in the absence of Smad2, suggesting that Smad2 activated by TGF-β plays an important role in the recruitment of Trim33 to the target elements of ROR-γ. These observations indicate the regulation of appropriate IL-17 and IL-10 expression by a Trim33/Smad2/ROR-γ complex. In addition to Trim33, Smad2, and ROR-γ, other transcription factors essential for Th17 differentiation such as BATF, IRF4, STAT3, and c-Maf accumulate at the Il17a conserved noncoding sequence 3 (CNS-3) region (Ciofani et al., 2012), indicating that a functional supercomplex formed with these multiple factors may be required for proper expression of IL-17. In support of this idea, we observed reduced permissive chromatin markers at the Il17a locus in Trim33 KO T cells compared with WT cells, suggesting that Trim33 may play an important role in chromatin remodeling. It has been reported that a methyltransferase for H3K9me, G9a, negatively regulated IL-17 production in T cells and that a demethylase for H3K27me3, jumonji domain-containing protein 3 (JMJD3), was indispensable for Th17 differentiation (Lehnertz et al., 2010; Ciofani et al., 2012), indicating that these modifications play a suppressive role in IL-17 expression in T cells. Our data fit well with these observations. We observed that H3K9me3 and H3K27me3 at the Il17 locus were enhanced in the KO Th17 cells. Taken together, it is possible that Trim33 may be required for recruitment of demethylases for H3K9me3 and H3K27me3 to generate a permissive chromatin configuration at the Il17 locus. A detailed mechanism whereby Trim33 regulates histone modification during Th17 differentiation needs to be clarified in the future.
The integration of Trim33 ChIP-seq data with microarray analysis identified other possible factors, SOCS3, GATA3, and FOXO1, as Trim33 direct targets in Th17 cells. SOCS3 is a negative feedback regulator for IL-6/STAT3 signaling pathway (Croker et al., 2003; Yasukawa et al., 2003). STAT3 plays an essential role in Th17 differentiation by binding to the Il17 gene locus (Yang et al., 2007, 2011). GATA3 has been reported to be a potent IL-10 inducer (Shoemaker et al., 2006). Our group recently reported that FOXO1 negatively regulated the pathogenicity of Th17 cells via suppression of IL-1R1 (Ichiyama et al., 2016). Gene expression analysis revealed the enhanced expression of these genes in the absence of Trim33, suggesting a possible indirect regulation of IL-17 or IL-10 expression by Trim33. On the other hand, a previous study demonstrated that deficiency of ROR-γ did not greatly affect the expression of these molecules at the mRNA level (Ciofani et al., 2012). Therefore, the suppressive activity of Trim33 on the expression of these three factors could be exerted in an ROR-γ–independent way. A ROR-γ–independent role of Trim33 in Th17 cells should be examined in future work.
We demonstrated in this study that Smad4 is a pivotal positive regulator for IL-10 during Th17 differentiation. Trim33 proteins have been reported to attenuate Smad4 protein expression by degradation with their E3 ligase domain (Dupont et al., 2005). In line with that study, Smad4 expression was increased in the Trim33-deficient Th17 cells but not iT reg cells, compared with WT at the protein level but not at the mRNA level. As described, IL-10 expression was dramatically enhanced in Th17 skewing conditions in the absence of Trim33. Importantly, this enhancement was strongly suppressed by further deletion of Smad4 in T cells. These results suggest that Trim33 suppresses IL-10 expression, probably by dual mechanisms of targeting of Smad4 for degradation and direct transcriptional suppression in conjunction with ROR-γ and Smad2 during Th17 differentiation. In contrast, the enhanced Smad4 expression in Trim33-deficient Th17 cells did not result in up-regulation of Foxp3. This may be because STAT5 is inhibited by IL-6–activated STAT3 during Th17 cell differentiation. Early production of IL-10 20 h after stimulation, when Trim33 is localized mainly in cytoplasm, was slightly enhanced in the absence of Trim33 in Th17 cells. In addition, deletion of RING domain of Trim33 resulted in partial attenuation of IL-10 suppression. Taken together, these data suggest that Trim33 functions in the cytoplasmic compartment for IL-10 suppression probably by degradation of Smad4 with E3 activity in early phases. In addition, IL-10 expression is suppressed at chromatin level at later phases, supported by the data of Trim33 ChIP analysis. In contrast, IL-17 expression at 20 h was not defective by Trim33 deficiency, and RING domain was not required for IL-17 expression. These data suggest that IL-17 production is regulated mainly by transcriptional regulation after nuclear translocation but not E3 activity, and Trim33 may play a crucial role in the maintenance of IL-17 expression rather than its initiation. Overall, the transcriptional control for IL-17 and IL-10 in the late phase of Th17 cell differentiation is presumably more important than Trim33 function exerted in cytoplasm, because the function of Trim33 for IL-17 and IL-10 in cytoplasm is subtle. We compared IL-17 expression between Trim33 single-KO and Trim33/Smad4 double-KO cells. Consistent with our previous study (Yang et al., 2008), Smad4 had little effect on IL-17 expression during Th17 differentiation, in comparison to Trim33. Our ChIP-PCR assay of histone modification revealed increased permissive chromatin markers at the Il10 gene locus in Trim33 KO Th17 cells compared with WT cells, suggesting that the enhanced Smad4 expression may generate a permissive histone modification at the Il10 locus in the absence of Trim33 in Th17 cells. It has been reported that STAT3 activation induces IL-10 expression in T cells (Stumhofer et al., 2007). Interestingly, TGF-β without IL-6 induced quite low levels of IL-10 expression, comparable to unstimulated naive T cells in vitro (unpublished data), indicating that at least activation of both STAT3 and Smad4 is required for IL-10 expression. Although both signaling pathways of IL-6 and TGF-β are definitely required for Th17 differentiation, the activation of these signaling pathways results in strong induction of IL-10 by both STAT3 and Smad4. For proinflammatory function of Th17 cells, immune-suppressive IL-10 expression needs to be down-regulated by a certain mechanism. Our current results support the idea that Trim33 is in charge of the suppressive machinery for IL-10 expression for generation of proinflammatory Th17 cells. Taken together, these data show that Trim33 utilizes at least two distinct mechanisms to regulate IL-17 and IL-10 for the generation of Th17 cells with proinflammatory phenotype.
Acquisition of a pathogenic phenotype in Th17 cells after IL-23 stimulation reported several years ago is the outcome of not only the induction of proinflammatory effector molecules but also the down-regulation of anti-inflammatory molecules. We hypothesized that Trim33 may mediate the pathogenic function of Th17 cells. However, comparative analysis of our microarray gene expression datasets with those reported for pathogenic Th17 cells (Ghoreschi et al., 2010; Lee et al., 2012) did not support this hypothesis (unpublished data), suggesting that Trim33 may regulate a distinct set of molecules that mediate proinflammatory function of Th17 cells.
As described above, Trim33 deficiency in T cells resulted in reduction of IL-17 and CCR6 but increased expression of IL-10 in Th17 cells in vitro. This is true with antigen-specific T cell response in vivo. On the other hand, we had different in vitro and in vivo results in the expression of IFN-γ (unpublished data) and IL-17F. This is probably due to secondary effect of IL-10. We assume that Trim33-deficient T cells were less activated, compared with WT cells, by APCs with attenuated T cell-priming activity, because APCs were exposed to an enhanced amount of IL-10 produced by Trim33 KO T cells in vivo. This is probably why IL-17F and IFN-γ expressions were decreased in the absence of Trim33 in vivo. The reduction of IL-17F was not observed in vitro, probably because we used APC-free culture. Consistent with reduced T cell activation and cytokine expression, EAE severity and the absolute numbers of cytokine-expressing, central nervous system–infiltrating T cells were greatly reduced in the absence of Trim33. The neutralization of IL-10 signaling almost fully restored EAE disease and absolute numbers of total central nervous system–infiltrating and IL-17-expressing CD4+ lymphocytes in Trim33 cKO mice, suggesting that enhanced expression of IL-10 by Trim33 KO T cells may be largely attributed to suppressed EAE disease, partly through inhibition of IL-17 expression in Trim33 cKO mice. By specific deletion of Trim33 in in vivo T reg cells including ROR-γ+ T reg, IL-10 mRNA expression was slightly enhanced. Although the effect in T reg cells was relatively minor, compared with Th17 cells, the de-repressed IL-10 by T reg cells might contribute to the milder EAE symptoms. The efficient and systemic blocking of IL-10 signaling possibly overcomes the IL-17 and CCR6 defect found in Trim33 KO T cells.
In summary, our current study has demonstrated a novel signaling mechanism in Th17 cells regulated by Trim33. Deficiency of Trim33 greatly reduced the proinflammatory function of Th17 cells, with attenuated IL-17 and CCR6 expression and enhanced IL-10 production. Therefore, our results suggest that Trim33 as a modulator of TGF-β–Smad signaling is crucial in the regulation of Th17 cell function. In other words, it serves as a pivotal switch regulator for proinflammatory versus regulatory phenotype of Th17 cells. Suggested by our data, inhibition of Trim33 function may help generate Th17 cells with regulatory phenotype. Trim33 thus would be a novel potential therapeutic target in inflammatory diseases.
Materials and methods
Mice
T cell– or T reg cell–specific Trim33 cKO mice were produced by breeding Trim33 f/f mice (Kim and Kaartinen, 2008) with CD4-Cre Tg mice (Makar et al., 2003) or Foxp3-Cre mice, respectively (Rubtsov et al., 2008). The flox mice were backcrossed onto C57BL/6 more than seven times. Smad2 f/f, Smad4 f/f, and Rorc f/f mice were previously described (Vincent et al., 2003; Chu et al., 2004; Ichiyama et al., 2015). They were crossed with CD4-Cre Tg mice. Mice at 6–10 wk of age were used in experiments following protocols approved by the institutional animal care and use committees of MD Anderson Cancer Center and Tsinghua University.
Th cell differentiation and antibodies
CD44loCD62LhiCD25− naive CD4+ T cells from lymph nodes and spleens of mice were purified by FACS sorting. For Th differentiation, naive CD4 T cells were stimulated with plate-bound anti-CD3 (0.5 µg/ml; 2C11; BioXcell) plus anti-CD28 (0.5 µg/ml; 37.51; BioXcell) in the presence of neutralizing antibodies (10 µg/ml anti–IL-4; 11B11; BE0045; BioXcell), 10 µg/ml anti–IFN-γ (XMG 1.2; BE0055; BioXcell), or polarizing cytokines (10 µg/ml anti–IL-4, 10 ng/ml IL-12 [210-12; Peprotech], and 50 U/ml human IL-2 for Th1; 10 µg/ml anti–IFN-γ, 10 ng/ml IL-4, and 50 U/ml human IL-2 for Th2; 10 ng/ml IL-6 [216-16; Peprotech], 0.01–1 ng/ml TGF-β [R&D Systems], 40 ng/ml IL-23 [culture sup], anti–IFN-γ, and anti–IL-4 for Th17; 50 U/ml human IL-2, 0.03–0.3 ng/ml TGF-β, anti–IFN-γ, and anti–IL-4 for iT reg cells). After 4 d of culture, cells were washed and restimulated with plate-bound anti-CD3 (0.5 µg/ml) for 4 h, and cells were then collected for RNA extraction. For cytokine measurement by ELISA, culture supernatants were collected either on day 3 (accumulating cytokine in culture sup) or after 24 h of restimulation on day 4. For intracellular cytokine analysis, cells were restimulated with 500 ng/ml ionomycin and 50 ng/ml PMA in the presence of Golgi Stop (BD PharMingen) for 5 h. Cells were then permeabilized with Cytofix/Cytoperm Kit (BD PharMingen) or Foxp3 staining buffer set (eBioscience) and analyzed for the expression of intracellular cytokines with anti–IFN-γ (XMG1.2), IL-4 (11B11), and IL-17 (TC11-18H10) antibodies (BD). Intracellular Foxp3 expression was detected with anti-Foxp3 (FJK-16s) antibody (BD). The reagents for ELISA, anti–IFN-γ (R4-6A2 and XMG1.2 biotin), anti–IL-2 (JES6-1A12 and JES6-5H4 biotin), anti–IL-4 (BVD4-1D11 and BVD6-24G2 biotin), anti–IL-10 (JES5-2A5 and JES5-16E3 biotin), anti–IL-17 (TC11-18H10 and TC11-8H4.1 biotin), and anti–IL-17F (AF2057 and BAF2057) were purchased from BD or R&D Systems. CCR6 expression was detected with anti-CCR6 antibody (140706) from BD. For blocking of IL-10 signaling, anti–IL-10R antibody (1B1.3a) from BioLegend was added in culture. Anit-Smad4 antibody (H-552) was purchased from Santa Cruz Biotechnology for Western blotting. For retroviral transduction of human Trim33 (aa 1–1,127) and ΔRING Trim33 (aa Δ2–188) were cloned into pMCs IRES GFP vector. The expression of these genes was measured by quantitative RT-PCR with primers forward (F) 5′-GAAGCTCCTAGCAGTTCTGATGA-3′ and reverse (R) 5′-GCCACTCTCCACATTCTACACA-3′.
Immunization
Mice were immunized with KLH emulsified in CFA at the base of the tail. For analysis of cytokine expression, dLN cells from KLH-immunized mice were stimulated as triplicates with 0, 4, 20, and 100 µg/ml KLH. After 4 d of restimulation, measurement of IL-10, IFN-γ, IL-17, and IL-17F was performed by ELISA. For intracellular cytokine staining, dLN cells from immunized mice were restimulated with 0 or 100 µg KLH for 24 h. CCR6 was stained with anti-CCR6 antibody (BD). For EAE induction, mice were immunized with MOG peptide emulsified in CFA on days 0 and 7. Pertussis toxin was intraperitoneally injected on days 2 and 8. Clinical scores are assigned as described before (Martinez et al., 2010). To examine infiltrating immune cells in the central nervous system, brain and spinal cord were collected from perfused mice, and mononuclear cells were isolated with Percoll gradient. Splenocytes were stimulated as triplicates with 0, 1, 5, and 25 µg/ml MOG peptides. EAE with IL-10R blocking was performed as described previously (Ichiyama et al., 2015).
Quantitative RT-PCR and microarray analysis
Total RNA extracted using Trizol reagent (Invitrogen) was used to generate cDNA using oligo(dT), random hexamers, and MMLV reverse transcription (Invitrogen). For quantitation of cytokines, cDNA samples were amplified in IQ SYBR Green Supermix (Bio-Rad Laboratories). The data were normalized to an Actb reference. The primer pairs for analysis were previously described (Yang et al., 2007). Primers for Trim33 were as follows: F 5′-CTTCTGCCTGCGCTGTCT-3′ and R 5′-TGCACTTGCATTATCTTCACAA-3′. The DNA microarray was performed with the Affymetrix Mouse Gene 1.0 ST Array (MoGene-1_0-st). RNA samples isolated from WT and Trim33 KO CD4+ T cells cultured under Th17 skewing conditions were labeled according to the manufacturer's instructions by the One-Cycle Target Labeling method, which consists of oligo(dT)-primed cDNA synthesis followed by in vitro transcription that incorporates biotinylated nucleotides. The microarray data were deposited in GEO under accession no. GSE113925.
ChIP assay
ChIP assays were done as described (Akimzhanov et al., 2007) with anti-Trim33 antibody (A301-059A/060A; Bethyl Laboratories), anti-H3K9/14Ac (06–599; Upstate), anti-H3K4me antibody (07-473; Upstate), anti-H3K9me3 antibody (ab8898; Abcam), anti-H3K27me3 (07-449; Upstate) antibody, or control IgG. The immunoprecipitated DNA was analyzed by real-time PCR. Primers for Trim33 ChIP-PCR were as follows: Il17a CNS-3 F 5′-TCCAAGGGTGGCTGTTTATC-3′ and R 5′-ACCAGTCATGTCACCTGTGC-3′; Il17a 20736550–20737060 F 5′-AAGCGGCAAGAAAGAGTCAG-3′ and R 5′-GAAACTGACCTGTTGGCAAAT-3′; Il10 130993469–130994422 F 5′-TGAGTCAGATGACGCACACA-3′ and R 5′-TGTGGACGGAAGAAGGTAGG-3′; Il10 131010104–131011125 F 5′-CTTGAGGAAAAGCCAGCATC-3′ and R 5′-GAGCATCGGAAACCAGAGAG-3′; Il10 131038029–131038188 F 5′-CAGAACCAAGTCGGGATGTT-3′ and R 5′-GCCACCGTTCTGATTTAGGA-3′; and H19 ICR F 5′-GCATGGTCCTCAAATTCTGCA-3′ and R 5′-GCATCTGAACGCCCCAATTA-3′. Primers for ChIP for histone modification were as follows: Il10 CNS-1 F 5′-GCCACGATTCTCAGGACATT-3′ and R 5′-CTTCCTTGGTGGCTTCTCTG-3′; Il10 CNS-1b F 5′-CGCCAAACACTTCTCACAGA-3′ and R 5′-CCCATTGGCTTCAAGTTGTT-3′; Il10 CNS-1c F 5′-TCAAAAGGGAAGGAGAAAGTGA-3′ and R 5′-CAGGGCTGGTAGAACAGGAA-3′; Il10 promoter F 5′-AGGGAGGAGGAGCCTGAATA-3′ and R 5′-TGTGGCTTTGGTAGTGCAAG-3′; Il10 3′ UTR F 5′-ATAGGAGAAACAGGGGAAGG-3′ and R 5′-TGCAGTTGATGAAGATGTCAAA-3′; and Il10 CNS-2 F 5′-AGGTCACAGGCAGCAGAGTT-3′ and R 5′-CTACCCCAGGCAGAAGTCAG-3′. Primers for Il17 and Foxp3 locus were previously described (Akimzhanov et al., 2007; Tone et al., 2008).
Coimmunoprecipitation and immunoblot assay
Proteins were immunoprecipitated by incubation of cell lysates with anti-Trim33 antibody or control Ig overnight, followed by conjugation with protein A magnetic beads (10001D; Thermo Fisher Scientific). Immunoprecipitates were washed and boiled in 2× SDS loading buffer for elution. Physical association was assessed by Western blot with antibodies against Smad2 (5339; Cell Signaling) and ROR-γ (14-6988; eBioscience). Nuclear and cytoplasmic extract was isolated with NE-PER Nuclear and Cytoplasmic Extraction Regents (78833; Thermo Fisher Scientific). Tubulin and histone H1 in the extracts was detected with anti-tubulin antibody (DM1A; Santa Cruz Biotechnology) and histone H1 (ab134914; Abcam).
ChIP-seq and computational analysis
Input and Trim33 I.P. DNA obtained by the ChIP procedure above were subjected to library preparation using KAPA hyper prep kit (KAPA Biosystems). Size selection (50–400 bp) was performed for all samples with AMpure XP beads (Beckman Coulter) for library preparation. The DNA libraries were sequenced with single-end 50-bp reads on an Illumina Hiseq 2000. For data analysis, low-quality sequences and Illumina adaptor sequences were trimmed using TrimGalore (v.0.3.7) with default parameters. Reads were mapped to the Mus musculus genome (mm10) using Bowtie (v.1.2.1.1, option: –p 4 –n 3 –m 1 –a–best–strata). Peaks were identified using MACS2 (v.2.1.1.20160309) with q-value cutoff of 0.01 for narrow peaks. For analysis of ROR-γ global binding, we processed the fastq file published previously (Ciofani et al., 2012). The introns, exons, intergenic regions, and UTRs were defined by UCSC known gene (mm10) database. Genome-wide distribution analysis was performed with ChIPseeker with default settings except for a setting for promoter defined as genomic fragment between 5 Kbp upstream and 1 Kbp downstream of TSS. The overlapping peaks of Trim33 and ROR-γ were obtained by intersecting of bed files for ChIP-seq data. For integration of ChIP-seq data with gene expression data (threshold: log2 fold change ± 0.3), data of genomic positions of peaks were subjected to PAVIS program with default settings for annotation of peaks on gene loci. The ChIP-seq data were deposited in GEO under accession no. GSE114119.
Statistical analysis
P values were calculated using two-tailed Student’s t test or one-way ANOVA followed by Tukey’s multiple comparison test (GraphPad software). P values are denoted in figures by *, P < 0.05 and **, P < 0.01.
Online supplemental material
Fig. S1 shows deletion efficiency of Trim33 in T cells and cellular analysis of EAE. Fig. S2 shows roles of Trim33 in in vitro–cultured Th cell subsets and IL-10 expression in ex vivo T reg cells. Fig. S3 shows Trim33 binding on Foxp3 gene locus by ChIP-seq analysis. Fig. S4 shows in vitro analysis of Th17 cells with Smad2 and ROR-γ KO CD4+ T cells. Fig. S5 shows cytokine regulation by Trim33 and Smad4.
Acknowledgments
We thank Dr. Chris Wilson (University of Washington, Seattle, WA) for providing the CD4-Cre mice, Dr. Elizabeth J. Robertson (Weatherall Institute of Molecular Medicine, University of Oxford, John Radcliffe Hospital, Oxford, UK) for Smad2 flox and Smad4 flox mice, Dr. Alexander Rudensky (Sloan Kettering Institute and Ludwig Center at Memorial Sloan Kettering Cancer Center, New York, NY) for Foxp3-Cre mice, the flow cytometry core facility at the M.D. Anderson Cancer Center for cell sorting, K. Tanaka for assistance in computational analysis, and the entire Dong laboratory for their help and discussions.
This work was supported by grants from the National Natural Science Foundation of China (31630022 and 91642201 to C. Dong) and Ministry of Science and Technology of China (2016YFC0906200 to C. Dong). C. Dong is a Bayer Chair Professor at Tsinghua University.
The authors declare no competing financial interests.
Author contributions: S. Tanaka designed the study, performed experiments, analyzed data, prepared the figures, and wrote the manuscript. Y. Jiang, G.J. Martinez, and K. Tanaka designed and performed experiments. X Yan performed the microarray experiment and analyzed the data. T. Kurosaki supported experiments. V. Kaartinen and X.-H. Feng provided gene-modified mice. Q. Tian organized the microarray experiment. X. Wang supported the preparation and submission of the manuscript. C. Dong designed the study and wrote the manuscript.
References
- Agricola E., Randall R.A., Gaarenstroom T., Dupont S., and Hill C.S.. 2011. Recruitment of TIF1γ to chromatin via its PHD finger-bromodomain activates its ubiquitin ligase and transcriptional repressor activities. Mol. Cell. 43:85–96. 10.1016/j.molcel.2011.05.020 [DOI] [PubMed] [Google Scholar]
- Akimzhanov A.M., Yang X.O., and Dong C.. 2007. Chromatin remodeling of interleukin-17 (IL-17)-IL-17F cytokine gene locus during inflammatory helper T cell differentiation. J. Biol. Chem. 282:5969–5972. 10.1074/jbc.C600322200 [DOI] [PubMed] [Google Scholar]
- Bettelli E., Das M.P., Howard E.D., Weiner H.L., Sobel R.A., and Kuchroo V.K.. 1998. IL-10 is critical in the regulation of autoimmune encephalomyelitis as demonstrated by studies of IL-10- and IL-4-deficient and transgenic mice. J. Immunol. 161:3299–3306. [PubMed] [Google Scholar]
- Bettelli E., Carrier Y., Gao W., Korn T., Strom T.B., Oukka M., Weiner H.L., and Kuchroo V.K.. 2006. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 441:235–238. 10.1038/nature04753 [DOI] [PubMed] [Google Scholar]
- Chu G.C., Dunn N.R., Anderson D.C., Oxburgh L., and Robertson E.J.. 2004. Differential requirements for Smad4 in TGFbeta-dependent patterning of the early mouse embryo. Development. 131:3501–3512. 10.1242/dev.01248 [DOI] [PubMed] [Google Scholar]
- Chung Y., Chang S.H., Martinez G.J., Yang X.O., Nurieva R., Kang H.S., Ma L., Watowich S.S., Jetten A.M., Tian Q., and Dong C.. 2009. Critical regulation of early Th17 cell differentiation by interleukin-1 signaling. Immunity. 30:576–587. 10.1016/j.immuni.2009.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciofani M., Madar A., Galan C., Sellars M., Mace K., Pauli F., Agarwal A., Huang W., Parkhurst C.N., Muratet M., et al. 2012. A validated regulatory network for Th17 cell specification. Cell. 151:289–303. 10.1016/j.cell.2012.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codarri L., Gyülvészi G., Tosevski V., Hesske L., Fontana A., Magnenat L., Suter T., and Becher B.. 2011. RORγt drives production of the cytokine GM-CSF in helper T cells, which is essential for the effector phase of autoimmune neuroinflammation. Nat. Immunol. 12:560–567. 10.1038/ni.2027 [DOI] [PubMed] [Google Scholar]
- Croker B.A., Krebs D.L., Zhang J.G., Wormald S., Willson T.A., Stanley E.G., Robb L., Greenhalgh C.J., Förster I., Clausen B.E., et al. 2003. SOCS3 negatively regulates IL-6 signaling in vivo. Nat. Immunol. 4:540–545. 10.1038/ni931 [DOI] [PubMed] [Google Scholar]
- Dong C. 2008. TH17 cells in development: An updated view of their molecular identity and genetic programming. Nat. Rev. Immunol. 8:337–348. 10.1038/nri2295 [DOI] [PubMed] [Google Scholar]
- Dupont S., Zacchigna L., Cordenonsi M., Soligo S., Adorno M., Rugge M., and Piccolo S.. 2005. Germ-layer specification and control of cell growth by Ectodermin, a Smad4 ubiquitin ligase. Cell. 121:87–99. 10.1016/j.cell.2005.01.033 [DOI] [PubMed] [Google Scholar]
- Dupont S., Mamidi A., Cordenonsi M., Montagner M., Zacchigna L., Adorno M., Martello G., Stinchfield M.J., Soligo S., Morsut L., et al. 2009. FAM/USP9x, a deubiquitinating enzyme essential for TGFbeta signaling, controls Smad4 monoubiquitination. Cell. 136:123–135. 10.1016/j.cell.2008.10.051 [DOI] [PubMed] [Google Scholar]
- El-Behi M., Ciric B., Dai H., Yan Y., Cullimore M., Safavi F., Zhang G.X., Dittel B.N., and Rostami A.. 2011. The encephalitogenicity of T(H)17 cells is dependent on IL-1- and IL-23-induced production of the cytokine GM-CSF. Nat. Immunol. 12:568–575. 10.1038/ni.2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esplugues E., Huber S., Gagliani N., Hauser A.E., Town T., Wan Y.Y., O’Connor W. Jr., Rongvaux A., Van Rooijen N., Haberman A.M., et al. 2011. Control of TH17 cells occurs in the small intestine. Nature. 475:514–518. 10.1038/nature10228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoreschi K., Laurence A., Yang X.P., Tato C.M., McGeachy M.J., Konkel J.E., Ramos H.L., Wei L., Davidson T.S., Bouladoux N., et al. 2010. Generation of pathogenic T(H)17 cells in the absence of TGF-β signalling. Nature. 467:967–971. 10.1038/nature09447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W., Dorn D.C., Erdjument-Bromage H., Tempst P., Moore M.A., and Massagué J.. 2006. Hematopoiesis controlled by distinct TIF1gamma and Smad4 branches of the TGFbeta pathway. Cell. 125:929–941. 10.1016/j.cell.2006.03.045 [DOI] [PubMed] [Google Scholar]
- Ichiyama K., Chen T., Wang X., Yan X., Kim B.S., Tanaka S., Ndiaye-Lobry D., Deng Y., Zou Y., Zheng P., et al. 2015. The methylcytosine dioxygenase Tet2 promotes DNA demethylation and activation of cytokine gene expression in T cells. Immunity. 42:613–626. 10.1016/j.immuni.2015.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichiyama K., Gonzalez-Martin A., Kim B.S., Jin H.Y., Jin W., Xu W., Sabouri-Ghomi M., Xu S., Zheng P., Xiao C., and Dong C.. 2016. The microRNA-183-96-182 cluster promotes T helper 17 cell pathogenicity by negatively regulating transcription factor Foxo1 expression. Immunity. 44:1284–1298. 10.1016/j.immuni.2016.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., and Kaartinen V.. 2008. Generation of mice with a conditional allele for Trim33. Genesis. 46:329–333. 10.1002/dvg.20401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B.S., Lu H., Ichiyama K., Chen X., Zhang Y.B., Mistry N.A., Tanaka K., Lee Y.H., Nurieva R., Zhang L., et al. 2017. Generation of RORγt+ antigen-specific T regulatory 17 cells from Foxp3+ precursors in autoimmunity. Cell Reports. 21:195–207. 10.1016/j.celrep.2017.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Awasthi A., Yosef N., Quintana F.J., Xiao S., Peters A., Wu C., Kleinewietfeld M., Kunder S., Hafler D.A., et al. 2012. Induction and molecular signature of pathogenic TH17 cells. Nat. Immunol. 13:991–999. 10.1038/ni.2416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnertz B., Northrop J.P., Antignano F., Burrows K., Hadidi S., Mullaly S.C., Rossi F.M., and Zaph C.. 2010. Activating and inhibitory functions for the histone lysine methyltransferase G9a in T helper cell differentiation and function. J. Exp. Med. 207:915–922. 10.1084/jem.20100363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.O., Wan Y.Y., and Flavell R.A.. 2007. T cell-produced transforming growth factor-beta1 controls T cell tolerance and regulates Th1- and Th17-cell differentiation. Immunity. 26:579–591. 10.1016/j.immuni.2007.03.014 [DOI] [PubMed] [Google Scholar]
- Makar K.W., Pérez-Melgosa M., Shnyreva M., Weaver W.M., Fitzpatrick D.R., and Wilson C.B.. 2003. Active recruitment of DNA methyltransferases regulates interleukin 4 in thymocytes and T cells. Nat. Immunol. 4:1183–1190. 10.1038/ni1004 [DOI] [PubMed] [Google Scholar]
- Malhotra N., Robertson E., and Kang J.. 2010. SMAD2 is essential for TGF beta-mediated Th17 cell generation. J. Biol. Chem. 285:29044–29048. 10.1074/jbc.C110.156745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan P.R., Harrington L.E., O’Quinn D.B., Helms W.S., Bullard D.C., Elson C.O., Hatton R.D., Wahl S.M., Schoeb T.R., and Weaver C.T.. 2006. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature. 441:231–234. 10.1038/nature04754 [DOI] [PubMed] [Google Scholar]
- Martinez G.J., Zhang Z., Chung Y., Reynolds J.M., Lin X., Jetten A.M., Feng X.H., and Dong C.. 2009. Smad3 differentially regulates the induction of regulatory and inflammatory T cell differentiation. J. Biol. Chem. 284:35283–35286. 10.1074/jbc.C109.078238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez G.J., Zhang Z., Reynolds J.M., Tanaka S., Chung Y., Liu T., Robertson E., Lin X., Feng X.H., and Dong C.. 2010. Smad2 positively regulates the generation of Th17 cells. J. Biol. Chem. 285:29039–29043. 10.1074/jbc.C110.155820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeachy M.J., Bak-Jensen K.S., Chen Y., Tato C.M., Blumenschein W., McClanahan T., and Cua D.J.. 2007. TGF-beta and IL-6 drive the production of IL-17 and IL-10 by T cells and restrain T(H)-17 cell-mediated pathology. Nat. Immunol. 8:1390–1397. 10.1038/ni1539 [DOI] [PubMed] [Google Scholar]
- Rubtsov Y.P., Rasmussen J.P., Chi E.Y., Fontenot J., Castelli L., Ye X., Treuting P., Siewe L., Roers A., Henderson W.R. Jr., et al. 2008. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity. 28:546–558. 10.1016/j.immuni.2008.02.017 [DOI] [PubMed] [Google Scholar]
- Shoemaker J., Saraiva M., and O’Garra A.. 2006. GATA-3 directly remodels the IL-10 locus independently of IL-4 in CD4+ T cells. J. Immunol. 176:3470–3479. 10.4049/jimmunol.176.6.3470 [DOI] [PubMed] [Google Scholar]
- Stumhofer J.S., Silver J.S., Laurence A., Porrett P.M., Harris T.H., Turka L.A., Ernst M., Saris C.J., O’Shea J.J., and Hunter C.A.. 2007. Interleukins 27 and 6 induce STAT3-mediated T cell production of interleukin 10. Nat. Immunol. 8:1363–1371. 10.1038/ni1537 [DOI] [PubMed] [Google Scholar]
- Takimoto T., Wakabayashi Y., Sekiya T., Inoue N., Morita R., Ichiyama K., Takahashi R., Asakawa M., Muto G., Mori T., et al. 2010. Smad2 and Smad3 are redundantly essential for the TGF-beta-mediated regulation of regulatory T plasticity and Th1 development. J. Immunol. 185:842–855. 10.4049/jimmunol.0904100 [DOI] [PubMed] [Google Scholar]
- Tone Y., Furuuchi K., Kojima Y., Tykocinski M.L., Greene M.I., and Tone M.. 2008. Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer. Nat. Immunol. 9:194–202. 10.1038/ni1549 [DOI] [PubMed] [Google Scholar]
- Veldhoen M., Hocking R.J., Flavell R.A., and Stockinger B.. 2006. Signals mediated by transforming growth factor-beta initiate autoimmune encephalomyelitis, but chronic inflammation is needed to sustain disease. Nat. Immunol. 7:1151–1156. 10.1038/ni1391 [DOI] [PubMed] [Google Scholar]
- Vincent S.D., Dunn N.R., Hayashi S., Norris D.P., and Robertson E.J.. 2003. Cell fate decisions within the mouse organizer are governed by graded Nodal signals. Genes Dev. 17:1646–1662. 10.1101/gad.1100503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Q., Wang Z., Zaromytidou A.I., Zhang X.H., Chow-Tsang L.F., Liu J.X., Kim H., Barlas A., Manova-Todorova K., Kaartinen V., et al. 2011. A poised chromatin platform for TGF-β access to master regulators. Cell. 147:1511–1524. 10.1016/j.cell.2011.11.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki T., Yang X.O., Chung Y., Fukunaga A., Nurieva R., Pappu B., Martin-Orozco N., Kang H.S., Ma L., Panopoulos A.D., et al. 2008. CCR6 regulates the migration of inflammatory and regulatory T cells. J. Immunol. 181:8391–8401. 10.4049/jimmunol.181.12.8391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.O., Panopoulos A.D., Nurieva R., Chang S.H., Wang D., Watowich S.S., and Dong C.. 2007. STAT3 regulates cytokine-mediated generation of inflammatory helper T cells. J. Biol. Chem. 282:9358–9363. 10.1074/jbc.C600321200 [DOI] [PubMed] [Google Scholar]
- Yang X.O., Nurieva R., Martinez G.J., Kang H.S., Chung Y., Pappu B.P., Shah B., Chang S.H., Schluns K.S., Watowich S.S., et al. 2008. Molecular antagonism and plasticity of regulatory and inflammatory T cell programs. Immunity. 29:44–56. 10.1016/j.immuni.2008.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.P., Ghoreschi K., Steward-Tharp S.M., Rodriguez-Canales J., Zhu J., Grainger J.R., Hirahara K., Sun H.W., Wei L., Vahedi G., et al. 2011. Opposing regulation of the locus encoding IL-17 through direct, reciprocal actions of STAT3 and STAT5. Nat. Immunol. 12:247–254. 10.1038/ni.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasukawa H., Ohishi M., Mori H., Murakami M., Chinen T., Aki D., Hanada T., Takeda K., Akira S., Hoshijima M., et al. 2003. IL-6 induces an anti-inflammatory response in the absence of SOCS3 in macrophages. Nat. Immunol. 4:551–556. 10.1038/ni938 [DOI] [PubMed] [Google Scholar]