Ribosome biogenesis involves numerous pre-rRNA processing events to remove internal and external transcribed spacer sequences, ultimately yielding three mature rRNAs. Biedka et al. show that ribosomal proteins and assembly factors remodel several neighborhoods, including two 60S ribosomal subunit functional centers, during removal of the ITS2 spacer RNA.
Abstract
Ribosome biogenesis involves numerous preribosomal RNA (pre-rRNA) processing events to remove internal and external transcribed spacer sequences, ultimately yielding three mature rRNAs. Removal of the internal transcribed spacer 2 spacer RNA is the final step in large subunit pre-rRNA processing and begins with endonucleolytic cleavage at the C2 site of 27SB pre-rRNA. C2 cleavage requires the hierarchical recruitment of 11 ribosomal proteins and 14 ribosome assembly factors. However, the function of these proteins in C2 cleavage remained unclear. In this study, we have performed a detailed analysis of the effects of depleting proteins required for C2 cleavage and interpreted these results using cryo–electron microscopy structures of assembling 60S subunits. This work revealed that these proteins are required for remodeling of several neighborhoods, including two major functional centers of the 60S subunit, suggesting that these remodeling events form a checkpoint leading to C2 cleavage. Interestingly, when C2 cleavage is directly blocked by depleting or inactivating the C2 endonuclease, assembly progresses through all other subsequent steps.
Introduction
Ribosome biogenesis is a complex, dynamic process involving the coordinated transcription, processing, modification, and structural remodeling of immature ribosomal RNA (rRNA) and binding of ribosomal proteins (r-proteins; Woolford and Baserga, 2013; de la Cruz et al., 2015; Nerurkar et al., 2015; Kressler et al., 2017; Peña et al., 2017). In eukaryotes, ribosome assembly spans three cellular compartments, beginning in the nucleolus and continuing in the nucleoplasm, with final stages of maturation occurring in the cytoplasm. To ensure efficient and accurate construction of ribosomes, eukaryotic ribosome assembly is facilitated by several hundred protein assembly factors (AFs), which include nucleases, RNA helicases, nucleoside triphosphatases, and scaffolding proteins, among others.
Pioneering in vitro bacterial ribosomal subunit reconstitution experiments were the first to establish that ribosome assembly is hierarchical, with primary binding r-proteins participating in the formation of binding sites for later-entering r-proteins (Held et al., 1973; Nierhaus and Dohme, 1974). This model is supported by recent in vivo work with Escherichia coli that suggests that large ribosomal subunit assembly occurs in blockwise parallel pathways (Davis et al., 2016). Assembly of both the large and small ribosomal subunits in eukaryotes was shown to follow a similar hierarchical model (Ferreira-Cerca et al., 2007; O’Donohue et al., 2010; Gamalinda et al., 2014). In Saccharomyces cerevisiae, assembly of the large (60S) ribosomal subunit begins with stable association of early r-proteins bound to the solvent-exposed side of the subunit, followed by maturation of the region surrounding the polypeptide exit tunnel (PET). Construction of the subunit interface, containing the peptidyl transferase center (PTC), and the central protuberance is completed last (Gamalinda et al., 2014; Kater et al., 2017).
As assembling pre-60S subunits progress from the nucleolus to the nucleoplasm, major compositional changes occur as many early-acting AFs dissociate from the preribosome and additional AFs bind (Fig. 1). During this period of assembly, removal of the internal transcribed spacer 2 (ITS2) RNA is initiated by cleavage of 27SB pre-rRNA at the C2 site by the endonuclease Las1 (Fig. S1 A; Gasse et al., 2015). Las1 functions in a complex with Grc3, Rat1, and Rai1, and both Las1 and Grc3 are required for cleavage at the C2 site (Castle et al., 2010, 2013; Schillewaert et al., 2012; Gasse et al., 2015; Pillon et al., 2017). C2 cleavage is coordinated with transit of the pre-60S subunit from the nucleolus to the nucleoplasm by an unknown mechanism.
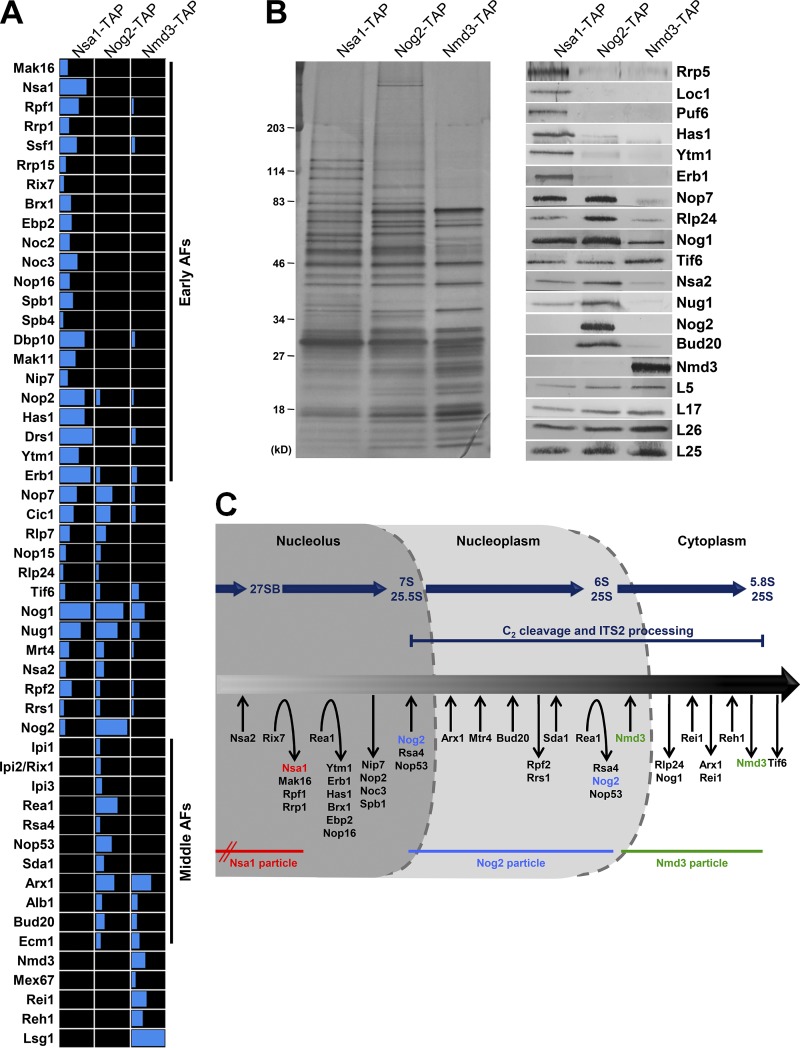
Figure 1.
Pre-60S subunit protein composition changes dramatically leading up to and during ITS2 processing. (A and B) Sequential pre-60S subunit assembly intermediates were purified from yeast via TAP-tagged AFs Nsa1, Nog2, or Nmd3. The protein composition of the purified pre-60S subunits was analyzed via mass spectrometry (A) and by SDS-PAGE followed by silver staining and Western blotting (B). (C) A working model for the pathway of 60S subunit assembly. The association and dissociation of AFs with pre-60S subunits are indicated as is the timing of pre-rRNA processing events. The lifetimes of the Nsa1, Nog2, and Nmd3 particles are shown; Nsa1 enters preribosomes during early stages of 60S subunit assembly.
The hierarchical recruitment and assembly of a group of 11 r-proteins (L9 [uL6], L17 [uL22], L19 [eL19], L23 [uL14], L25 [uL23], L26 [uL24], L27 [eL27], L31 [eL31], L34 [eL34], L35 [uL29], and L37 [eL37]) and 14 AFs (Nip7, Nop2, Rpf2, Rrs1, Spb4, Mak11, Dbp10, Rlp24, Tif6, Nog1, Nsa2, Nog2, and the dual A3/B-factors Has1 and Drs1) known as the B-factors are necessary for C2 cleavage. This B-factor hierarchy begins with function of the heterodimer Nip7-Nop2 and continues along two parallel pathways that ultimately lead to the recruitment of Nog2 (Fig. S1 B). Although most B-factors associate with the pre-60S subunit early, before processing of 27SA3 pre-rRNA, Nog2 assembles onto the pre-60S subunit just before C2 cleavage and transit into the nucleoplasm.
Recently determined near-atomic resolution cryo-EM structures of pre-60S subunit assembly intermediates identified the binding sites of 11 of the 14 B-factor AFs (Nip7, Nop2, Mak11, Has1, Rpf2, Rrs1, Rlp24, Tif6, Nog1, Nsa2, and Nog2; Fig. S1 C; Talkish et al., 2012; Wu et al., 2016; Kater et al., 2017; Sanghai et al., 2018). Intriguingly, 10 of these AFs are located in an arc along the subunit interface; this arc is on the opposite side of the pre-60S subunit from ITS2. This finding revealed a major gap in our understanding of late nucleolar stages of 60S subunit assembly because it was unclear how C2 cleavage could be affected or regulated by a group of AFs not bound near ITS2. Although it is known that B-factors are required for hierarchical recruitment of other B-factors, it remained unclear why exactly they are necessary for C2 cleavage and transit of preribosomes into the nucleoplasm.
In this study, we have expanded on previous research (Talkish et al., 2012) of the B-factor hierarchy by determining why, beyond their role in recruitment of other B-factors, the B-factor r-proteins and AFs are required for C2 cleavage. We have taken advantage of cryo-EM structures of pre-60S subunit assembly intermediates to gain a better understanding of the interdependence of AFs and r-proteins during 60S subunit biogenesis. We have established that the AFs and r-proteins required for C2 cleavage are necessary for the same remodeling events, where remodeling events are considered any change in pre-60S subunit protein composition or structure. Specifically, we show that, in most mutants that cannot carry out C2 cleavage, there is a block in nucleolar stages of assembly during the transition from the Nsa1 state C particle to the state D particle (Kater et al., 2017). This block may initially result from failure of the AFs Noc3 and Spb1 to bind to or stably interact with the pre-60S subunit. Subsequent defects in 60S subunit assembly include impaired formation of the PET exit platform, a block in the release of two groups of AFs by AAA-ATPases, and failure of a set of AFs that normally bind to 25S rRNA domain V to stably associate with the pre-60S subunit, likely preventing proper maturation of the PTC. In contrast, and consistent with its more direct role in C2 cleavage, we found that depletion of the C2 endonuclease Las1 or inactivation of its endonucleolytic activity does not result in any of these defects. Surprisingly, we found that when Las1 is depleted or inactivated, assembly progresses into the cytoplasm and results in the production of translation-competent 60S subunits containing the ITS2 spacer RNA and AFs bound to ITS2. These results suggest that, although these remodeling events are necessary for C2 cleavage, association of Las1 and C2 cleavage are not required for most, if any, subsequent steps of 60S subunit assembly.
Results
Dramatic changes in pre-60S ribosomal subunit protein composition occur before and after C2 cleavage
To better understand the period of 60S subunit assembly leading up to and after C2 cleavage and ITS2 processing, we analyzed the protein composition of three sets of assembly intermediates spanning early, middle, and late stages of 60S subunit assembly. These were purified via the tandem affinity purification (TAP)-tagged AFs Nsa1, Nog2, and Nmd3 (Fig. 1). Nsa1 associates with pre-60S subunits early in the nucleolus and is removed by the AAA-ATPase Rix7 before entry of Nog2 (Fig. 1 C; Kressler et al., 2008). Nog2 is the last protein to enter the pre-60S subunit before C2 cleavage (Talkish et al., 2012). The removal of Nog2 near the end of nucleoplasmic stages of assembly is coupled with entry of the nuclear export factor Nmd3 and subsequent nuclear export of pre-60S ribosomes (Matsuo et al., 2014). Nmd3 is one of the last AFs released from the pre-60S subunit in the cytoplasm (Ma et al., 2017; Malyutin et al., 2017).
We mainly focused on the transition between Nsa1-TAP particles and Nog2-TAP particles, where we observed dramatic changes in pre-60S subunit protein composition (Fig. 1, A and B). Specifically, a group of 22 early AFs was present in Nsa1-TAP particles but not in intermediates purified via Nog2-TAP, suggesting that they are removed before Nog2 enters (Fig. 1 A). There were also several middle AFs in Nog2-TAP particles that were absent in Nsa1-TAP particles, indicating that they enter pre-60S subunits during the transition from Nsa1 to Nog2 particles or during the lifetime of the Nog2 particle.
Depletion of various B-factors results in similar changes in pre-60S subunit protein composition
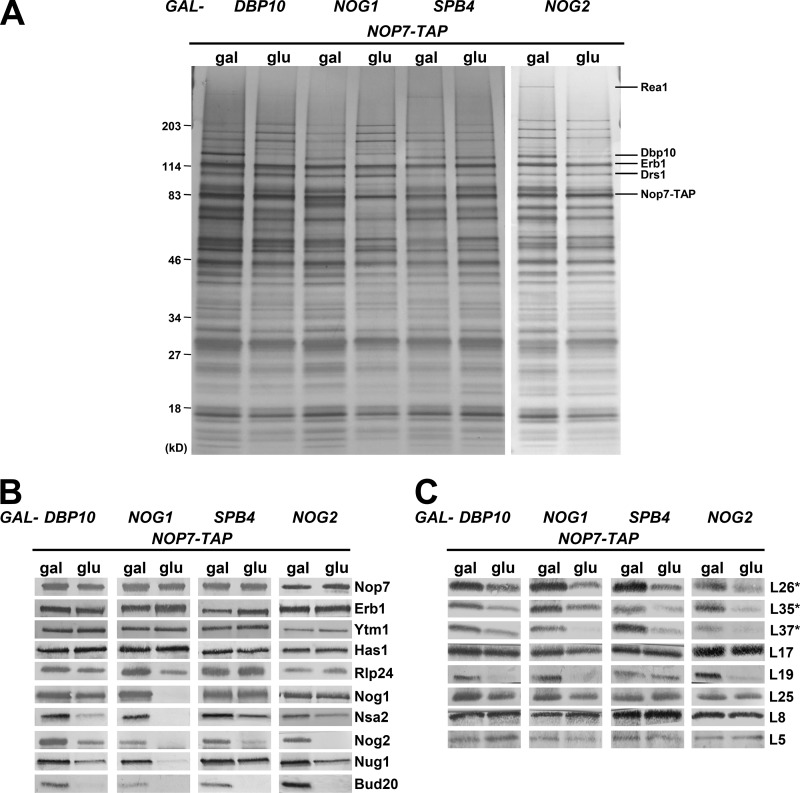
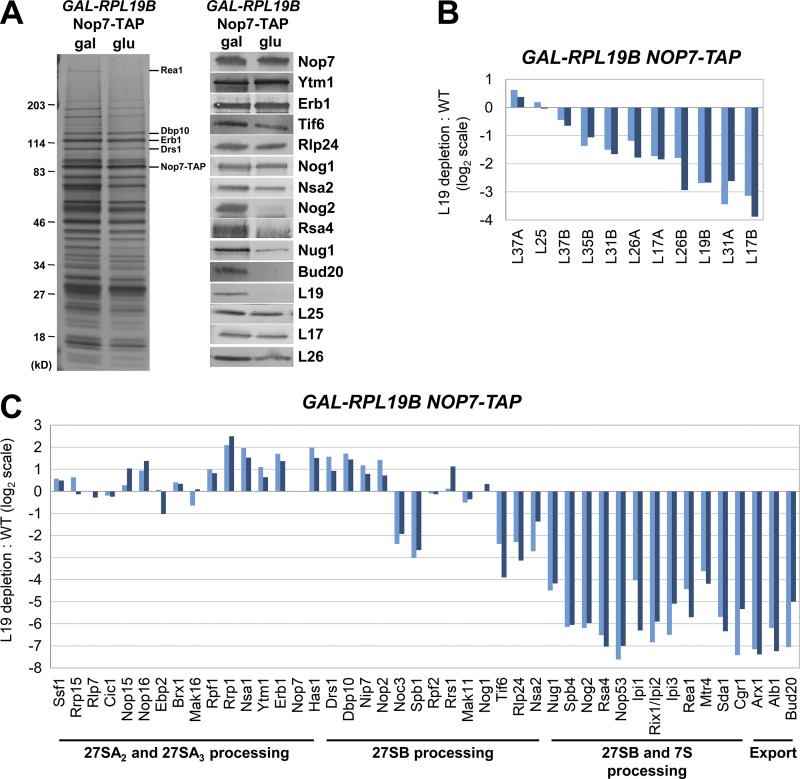
We previously showed that recruitment of Nog2 to the pre-60S subunit and cleavage of 27SB pre-rRNA at the C2 site require the hierarchical assembly of a group of 14 AFs termed B-factors (Fig. S1 B; Talkish et al., 2012). However, it remained unclear why exactly these B-factor AFs are necessary for C2 cleavage, especially after cryo-EM structures of pre-60S subunits revealed that only one of the resolved B-factor AFs (Has1) is located proximal to ITS2 (Fig. S1 C). To begin to determine the defects in 60S subunit assembly in mutants blocked in C2 cleavage, we examined the effects of depleting four B-factor AFs, Dbp10, Nog1, Spb4, and Nog2. Although the effects of depleting these AFs have been previously studied (Saveanu et al., 2003; Lebreton et al., 2008; Talkish et al., 2012), our goal was to carry out a more in-depth survey of the defects in 60S subunit assembly and, more specifically, changes in pre-60S subunit protein composition, induced by their absence. We began by assaying the protein constituents of pre-60S subunits depleted of Dbp10, Nog1, Spb4, or Nog2 by SDS-PAGE followed by silver staining and Western blotting (Fig. 2). We observed changes in the levels of both AFs and r-proteins after depletion of these four AFs, many of which are common among all of these mutants.
Figure 2.
Similar changes in pre-60S subunit protein composition occur upon depletion of the B-factors Dbp10, Nog1, Spb4, or Nog2. The TAP-tagged AF Nop7 was used as bait to purify pre-60S subunits from yeast expressing (gal) or depleted of (glu) the B-factors Dbp10, Nog1, Spb4, and Nog2. The preribosome protein composition was analyzed by SDS-PAGE followed by silver staining (A) and Western blotting for AFs (B) and r-proteins (C). Asterisks indicate that the protein being blotted for was tagged and detected with anti-HA antibody.
In each depletion mutant, levels of the A3 factors Erb1 and Ytm1 and the dual A3/B-factors Has1 and Drs1 were either increased or unaffected, consistent with the B-factors acting downstream of the A3 factors (Fig. 2, A and B). Interestingly, upon depletion of Spb4 and Nog2, we observed decreased levels of the AAA-ATPase Rea1 (Fig. 2 A). These results suggest that Rea1 fails to associate with preribosomes in these mutants, and thus, release of Erb1 and Ytm1 by Rea1 is impaired (Bassler et al., 2010).
The B-factors Rlp24 and Nog1 were decreased by depletion of Dbp10 and Nog1 but unaffected by the absence of Spb4 or Nog2, consistent with the previously determined B hierarchy (Fig. S1 B; Talkish et al., 2012). The levels of Nog2 were decreased in each mutant as expected, as were the levels of the AFs Nug1 and Bud20. Unexpectedly, Nsa2 was present in decreased levels after depletion of Dbp10, Nog1, Spb4, or Nog2; based on the B hierarchy (Fig. S1 B), we expected Nsa2 to be affected only by depletion of Dbp10 or Nog1. A possible explanation is that Nog2, which binds to the pre-60S subunit on top of Nsa2 (Fig. 4 A; Wu et al., 2016), may be necessary for stable association of Nsa2 and, in its absence, Nsa2 may dissociate from the pre-60S subunit during preribosome purifications.
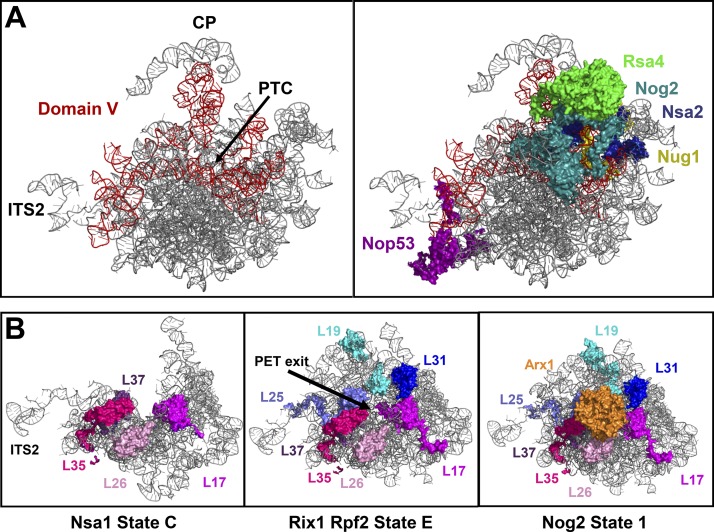
Figure 4.
AFs binding to 25S rRNA domain V and r-proteins surrounding the PET exit are present in decreased levels in B mutants. (A) The B-factors Nsa2 and Nog2 and the AFs Rsa4, Nug1, and Nop53 bind to 25S rRNA domain V. (B) As assembly proceeds, the globular bodies of the r-proteins L19, L25, L31, L17, L26, L35, and L37 form a platform surrounding the exit of the PET to which Arx1 can then bind. Structures shown are PDB IDs 6EM1, 6ELZ, and 3JCT.
Depletion of Dbp10, Nog1, Spb4, or Nog2 also affected the levels of certain r-proteins in preribosomes. Although L5 (uL18) and L8 (eL8) were consistently unaffected by depletion of the B-factors, one or more of the middle-acting r-proteins L17, L26, L35, L37, L19, and L25, which form a platform around the exit of the PET (Fig. 4 B), were present in decreased levels in all of these mutants (Fig. 2 C; Gamalinda et al., 2014). These PET exit r-proteins bind to the pre-60S subunit during early stages of biogenesis but are thought to become more strongly and stably associated with the pre-60S subunit as assembly progresses. As such, the decreased levels of these r-proteins indicate that they are not yet stably bound to the pre-60S subunit in the B-factor mutants. This suggests that the construction of the PET exit is impaired in mutants blocked at C2 cleavage.
Defects caused by depletion of the AF Nsa2 are similar to those observed in other B mutants
We also performed a more thorough investigation into the effects of depleting the B-factor Nsa2, which assembles onto preribosomes after 27SB pre-rRNA has been formed and is necessary for association of Nog2 (Figs. 1 C and S1 B; Lebreton et al., 2006; Talkish et al., 2012). In addition to SDS-PAGE followed by silver staining and Western blotting, we also analyzed the effects of Nsa2 depletion by isobaric tag for relative and absolute quantitation (iTRAQ) semiquantitative mass spectrometry (Figs. 3 and S2 A).
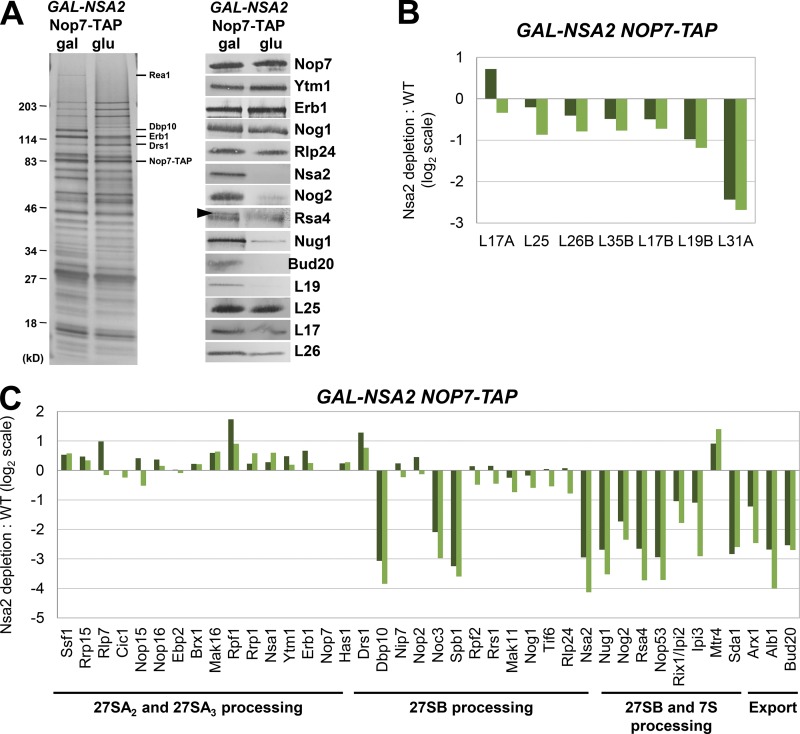
Figure 3.
Dramatic changes in pre-60S subunit protein composition are caused by depletion of AF Nsa2. (A–C) Preribosomes containing (gal) or lacking (glu) Nsa2 were purified using TAP-tagged Nop7 as bait. (A) The protein composition of the pre-60S subunits was analyzed by SDS-PAGE followed by silver staining and Western blotting. Rsa4 is indicated by a black arrowhead; the contaminating band is IgG. (B and C) Semiquantitative mass spectrometry (iTRAQ) was used to quantify the relative changes in levels of 60S subunit r-proteins (B) and AFs (C) in the presence and absence of Nsa2. The ratios were normalized to levels of Nop7, and the fold change in log2 scale is shown for two biological replicates. A complete dataset for AFs is shown in Fig. S2.
The overall pattern of preribosome protein composition changes observed upon depletion of Nsa2 was similar but not identical to that of depleting Dbp10, Nog1, Spb4, or Nog2. Similar to depletion of these four AFs, depletion of Nsa2 resulted in unchanged or slightly increased levels of most early-acting AFs, including the A3 factors (Nop7, Ytm1, Erb1, Pwp1, Brx1, Ebp2, Cic1, Rlp7, Nop15, Nop12, Drs1, and Has1). In addition, levels of the Mak16 group (Mak16, Nsa1, Rpf1, and Rrs1) and Ssf1 heterodimer (Ssf1 and Rrp15) were generally unaltered or increased (Figs. 3, A and C; and Fig. S2 A; Kater et al., 2017; Sanghai et al., 2018; Zhou et al., 2018).
Furthermore, the levels of most of the B-factors that enter preribosomes upstream of Nsa2 (Nip7, Nop2, Mak11, Rlp24, Tif6, and Nog1) were unaffected. One exception is Dbp10, which was originally placed upstream of Nsa2 in the B-factor hierarchy (Talkish et al., 2012) but is decreased upon depletion of Nsa2 (Fig. 3 A). As Dbp10 cross-links to rRNA in domain V that overlaps the binding site for Nsa2 (Manikas et al., 2016), Dbp10 may become more stably associated with the pre-60S subunit upon binding of Nsa2 and, therefore, may dissociate during preribosome purifications in the absence of Nsa2.
Depletion of Nsa2 also caused decreased levels of the PET platform r-proteins (L19, L31, L25, L17, L26, and L35) as was observed after depletion of Dbp10, Nog1, Spb4, and Nog2. Interestingly, levels of the AF Arx1 were also decreased upon depletion of Nsa2 (Fig. 3 C). Arx1 binds to the PET platform r-proteins L26, L35, L19, and L25 (Fig. 4 B), and probably cannot assemble onto the pre-60S subunit when these r-proteins are not stably associated.
The most obvious defect caused by depletion of Nsa2 is strongly decreased levels of AFs that bind to 25S rRNA domain V (Nsa2, Nog2, Rsa4, Nug1, and Nop53; Figs. 3 C and 4 A). This is consistent with changes observed upon depletion of Dbp10, Nog1, Spb4, or Nog2, and suggests that 25S rRNA domain V, which contains the PTC, may be improperly structured in the absence of Dbp10, Nog1, Nsa2, Spb4, or Nog2 (Fig. 4 A).
Changes in pre-60S subunit protein composition upon depletion of r-protein L19 are similar to those in other B mutants
Having studied several AFs required for C2 cleavage, we wanted to thoroughly examine the effects of depleting an r-protein necessary for this step of 60S subunit biogenesis. We noticed that the r-protein L19, which is part of the PET platform and is necessary for C2 cleavage (Gamalinda et al., 2014; Kisly et al., 2016), is strongly decreased upon depletion of Dbp10, Nog1, Nsa2, and Nog2 (Figs. 2 C and 3, A and B). Furthermore, it is the most commonly affected PET exit r-protein in all of the studied mutants blocked at C2 cleavage (Fig. 10 A). Therefore, we chose L19 as our representative B-factor r-protein.
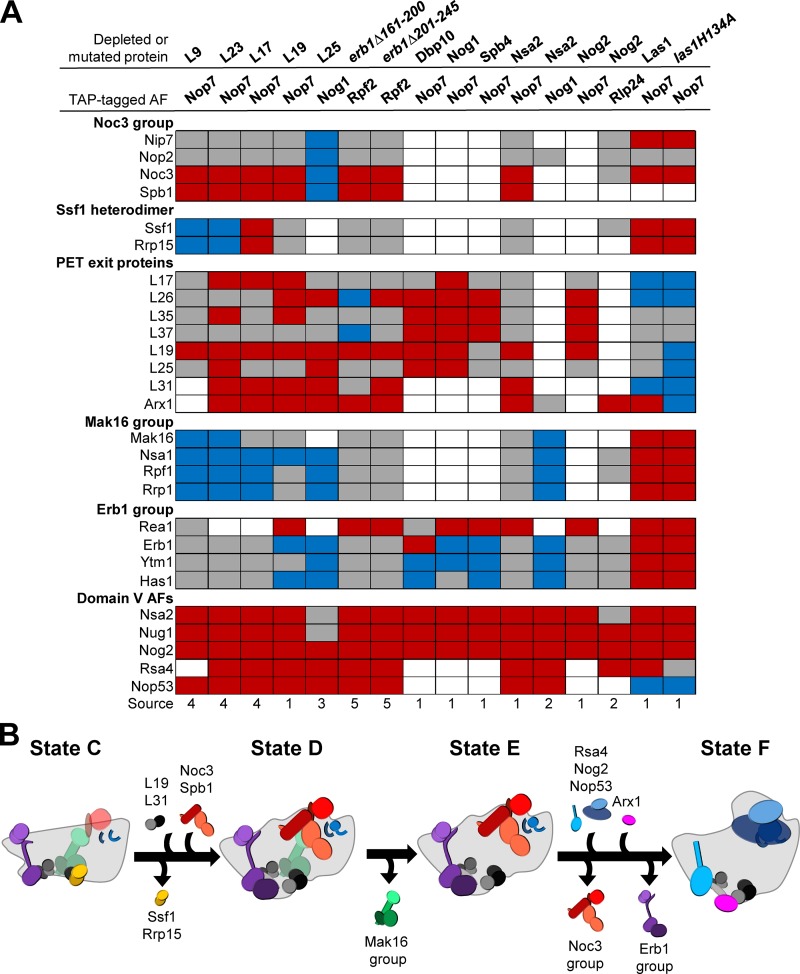
Figure 10.
Six common defects occur in most mutants blocked at C2 cleavage. (A) Comparisons of the data presented in this study with previously published data for mutants blocked at C2 cleavage revealed that there are six groups of proteins commonly affected in these mutants. Increased, decreased, or unchanged levels of proteins are indicated by blue, red, and gray boxes, respectively. White boxes indicate that data were not available. Sources: 1, this study; 2, Lebreton et al., 2008; 3, Ohmayer et al., 2013; 4, Gamalinda et al., 2014; 5, Konikkat et al., 2017. (B) A model of the nucleolar remodeling events affected in B mutants.
We found that the changes in pre-60S subunit protein composition induced by the absence of L19 were similar to those we observed upon depletion of Dbp10, Nog1, Spb4, Nog2, or Nsa2 (Figs. 2, 3, and 5). Levels of the Mak16 and Erb1 groups of AFs were increased or unchanged, with the exception of Rea1, which was present in decreased levels (Fig. 5, A and C). Furthermore, depleting L19 resulted in decreased levels of most of the PET platform r-proteins (L17, L26, L35, L19, and L31) as well as Arx1, and of the domain V AFs (Nsa2, Nog2, Rsa4, Nug1, Nop53; Fig. 5).
Figure 5.
Dramatic changes in pre-60S subunit protein composition are caused by depletion of r-protein L19. (A–C) Preribosomes containing (gal) or lacking (glu) L19 were purified using TAP-tagged Nop7 as bait. (A) The protein composition of pre-60S subunits was analyzed by SDS-PAGE followed by silver staining and Western blotting. (B and C) Semiquantitative mass spectrometry (iTRAQ) was used to quantify the relative changes in levels of 60S subunit r-proteins (B) and AFs (C) in the presence and absence of L19. The ratios were normalized to levels of Nop7, and the fold change in log2 scale is shown for two biological replicates. Both samples of yeast grown in glucose were compared with a single sample grown in galactose. A complete dataset for AFs is shown in Fig. S2.
The structure of the ITS2 spacer RNA is largely intact in the absence of L19
One possible explanation for blocked C2 cleavage in B mutants is that the B-factors may be necessary for allosteric structuring of the ITS2 spacer RNA to allow binding or recognition of the C2 site by the Las1 complex. To test this possibility, we assayed the structure of ITS2 in vivo in the presence and absence of L19 via chemical modification of RNA with 2-methylnicotinic acid imidazolide (NAI) followed by selective 2′-hydroxyl acylation analyzed primer extension (SHAPE; Wilkinson et al., 2006).
The structure of ITS2 was largely intact in the absence of L19 (Fig. S3). Several nucleotides in the binding sites of Nop15 and Cic1 displayed altered susceptibility to modification by NAI, indicating that the flexibility of these nucleotides was altered by the absence of L19. However, the levels of Nop15 and Cic1 were unaffected by depletion of L19, and thus, the significance of modification to these nucleotides is unclear. Modest effects on ITS2 structure were also observed after depletion of the PET platform r-proteins L17, L35, or L37 and in the erb1Δ161–200 and erb1Δ201–245 B mutants (Gamalinda et al., 2013; Konikkat et al., 2017). The modified nucleotides in ITS2 are not generally consistent across these B mutants. These results suggest that the block in C2 cleavage in B mutants is not caused by improper structuring of the ITS2 RNA.
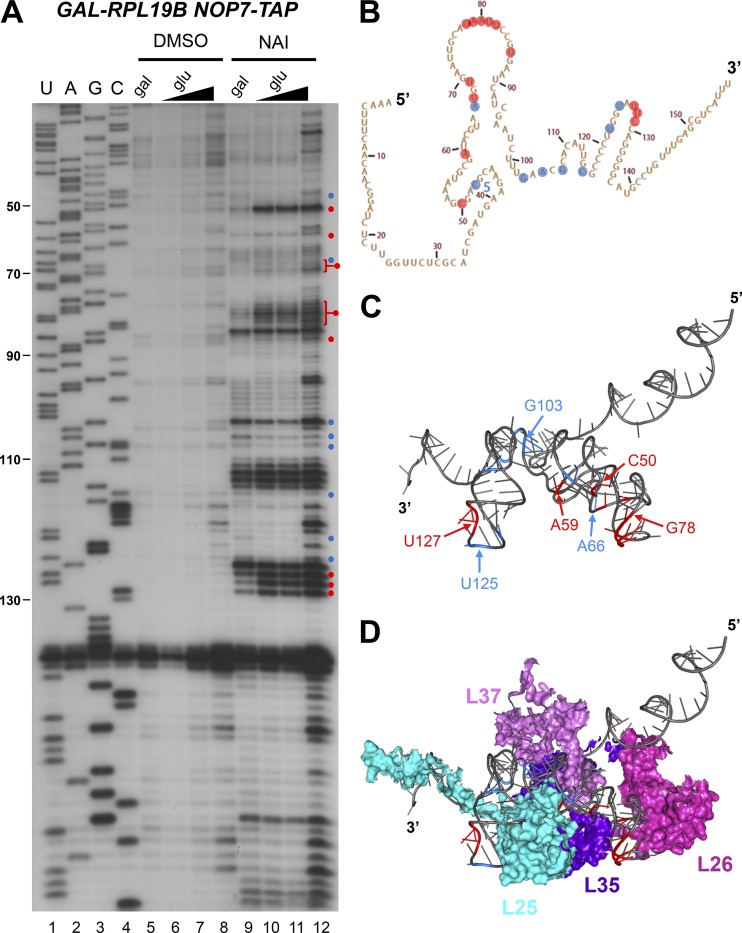
The structure of 5.8S rRNA is altered in the absence of L19
Although the structure of ITS2 was unaffected by the absence of L19, we reasoned that the structure of 5.8S rRNA, to which the PET platform r-proteins L17, L26, L35, L37, and L25 bind, might be altered when L19 is depleted because most of these r-proteins are affected in B mutants (Fig. 10 A). We assayed the effects of chemical modification on 5.8S rRNA using an oligonucleotide specific to the 5′ end of ITS2, ensuring that primer extension was specific to pre-rRNA and not mature rRNA. We found that several nucleotides in 5.8S rRNA displayed altered reactivity to NAI in the absence of L19 (Fig. 6, A and B). Interestingly, these modified nucleotides were clustered in the same stem and loop structures that were modified upon expression of erb1Δ161–200 or erb1Δ201–245 (Konikkat et al., 2017). The modified nucleotides were largely clustered around the binding sites of the PET platform r-proteins L25, L26, L35, and L37 (Fig. 6, C and D). Because depletion of L19 resulted in decreased levels of most of the PET platform r-proteins (Fig. 5, A and B), the alterations to the structure of 5.8S rRNA in the absence of L19 could be due to the absence of these proteins. Alternatively, the altered 5.8S rRNA structure could impair stable association of the PET exit r-proteins.
Figure 6.
The structure of 5.8S rRNA in preribosomes is altered in the absence of L19. (A) In vivo structure probing of 5.8S rRNA (SHAPE) was conducted by treating cells with NAI and assaying modified nucleotides via primer extension with an oligonucleotide complimentary to the 5′ half of ITS2. Cells were grown in galactose or grown in galactose and shifted to glucose for 16 h. Nucleotides with increased or decreased modification in the absence of L19 are indicated by red and blue circles, respectively. RNA extracted from cells treated with DMSO instead of NAI was used as a control. Increasing amounts of primer extension product from cells shifted to glucose were loaded in lanes 6–8 and 10–12 to achieve loading similar to the galactose lanes (5 and 9). (B and C) Nucleotides modified by NAI are indicated on the secondary (B) and tertiary (C) structures of 5.8S rRNA. (D) The PET exit r-proteins L25, L26, L35, and L37 are clustered on top of the modified nucleotides.
Depletion or inactivation of Las1 results in a phenotype that is dramatically different from other B mutants
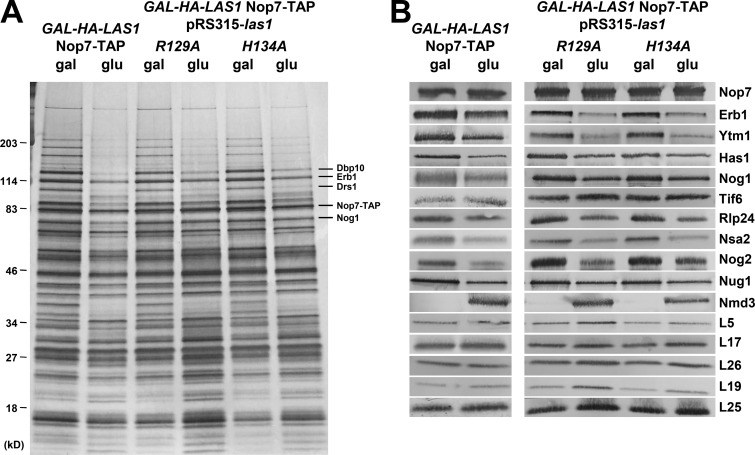
After indirectly blocking C2 cleavage by depleting several B-factors, we wanted to determine whether directly blocking cleavage from occurring would have a similar effect. To do this, we began by assaying the effects on pre-60S subunit protein composition of depletion of Las1, the endonuclease that cleaves at the C2 site, or upon expression of the las1R129A or las1H134A mutant proteins, which were previously shown to be unable to cleave 27SB pre-rRNA (Gasse et al., 2015). Surprisingly, we found that depletion of Las1 or expression of either mutant protein resulted in a unique phenotype unlike those we observed in any of the other B mutants (Fig. 7).
Figure 7.
Depletion or mutational inactivation of Las1 results in changes in pre-60S subunit protein composition unlike those observed in other B mutants. (A and B) Preribosomes were purified from yeast expressing (gal) or depleted of (glu) Las1 or from yeast expressing las1R129A or las1H134A using Nop7-TAP as bait. The protein composition of the pre-60S subunits was analyzed by SDS-PAGE followed by silver staining (A) and Western blotting (B). Yeast containing pRS315-las1R129A or pRS315-las1H134A were grown in C-leu minimal media.
Pre-60S subunits purified with TAP-tagged Nop7 from strains depleted of Las1 or expressing las1R129A or las1H134A displayed several key differences from those purified from other B mutants. For example, Ytm1 and Erb1 are decreased in the Las1 mutants, whereas they are unaffected or increased in other B mutants (Figs. 2, 3, 5, and 7). Also, the PET r-proteins are generally unaffected by depletion or mutation of Las1, unlike in other B mutants where these r-proteins tend to be present in decreased levels. Most strikingly, however, is the finding that levels of Nmd3 in Nop7 particles are strongly increased by depletion of Las1 or expression of the Las1 mutant proteins (Fig. 7 B). This contrasts with other B mutants and wild-type cells, where little or no Nmd3 is present in Nop7 particles (unpublished data). Furthermore, several cytoplasmic AFs, including Sqt1, Rei1, Reh1, and Lsg1, are present in Nop7-containing preribosomes after depletion of Las1 or expression of las1H134A, whereas these proteins are not present in wild-type Nop7 particles (Fig. S2, C and D). These results suggest that in the absence of Las1 protein or activity, 60S subunit maturation is blocked later than in the other B mutants. Because depletion of Las1 and expression of the Las1 mutants resulted in very similar phenotypes (Fig. 7), we decided to focus mainly on depletion of Las1 for further experiments.
AFs bound to ITS2 remain associated with cytoplasmic pre-60S subunits in the absence of Las1
We reasoned that the high levels of Nmd3 present in Nop7 particles upon depletion or mutational inactivation of Las1 could be caused either by Nmd3 entering pre-60S subunits earlier than it would under wild-type conditions or by Nop7 remaining associated with pre-60S subunits longer than normal. To distinguish between these possibilities, we purified preribosomes from Las1 depleted cells using Nmd3-TAP.
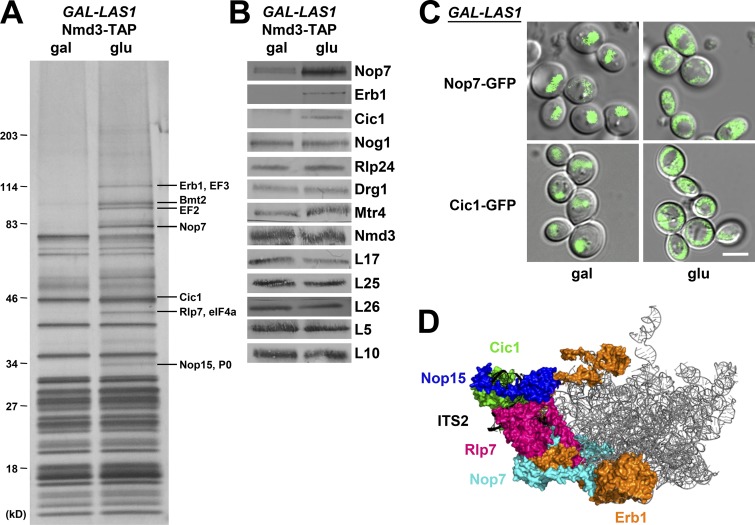
We found that Nmd3 particles purified from cells depleted of Las1 contain several additional proteins compared to wild-type. SDS-PAGE revealed that Nop15 and Rlp7, AFs that bind to ITS2 to form the “foot” structure, as well as Erb1 and Nop7, which bind proximal to ITS2, are present in Nmd3 particles when Las1 is depleted or when las1H134A is expressed but not in wild-type cells (Fig. 8, A and D). Western blotting and mass spectrometry showed that these additional proteins also include the ITS2 foot AF Cic1 (Fig. 8 B). We assayed the subcellular localization of Nop7 and Cic1 to determine whether these AFs are bound to cytoplasmic pre-60S subunits (Fig. 8 C). Both Nop7 and Cic1 are mislocalized to the cytoplasm upon depletion of Las1. Collectively, these results suggest that AFs bound to ITS2 (Nop15, Rlp7, Cic1) or proximal to ITS2 (Erb1, Nop7) are not properly removed from the pre-60S subunit in the absence of Las1 and that these aberrant preribosomes can be exported to the cytoplasm. While this manuscript was in preparation, similar results were published by the Hurt group (Sarkar et al., 2017).
Figure 8.
AFs bound to the ITS2 foot are present in cytoplasmic pre-60S subunits in the absence of Las1. (A and B) Pre-60S subunits containing (gal) or lacking (glu) Las1 were purified using TAP-tagged Nmd3 as bait. The protein composition of preribosomes was analyzed by SDS-PAGE followed by silver staining (A) and Western blotting (B). (C) The cellular localization of GFP-tagged Nop7 and Cic1 was monitored in live yeast cells expressing (gal) or depleted of (glu) Las1. Bar, 5 µm. (D) The locations of AFs bound on or proximal to ITS2 in the state E particle. The structure shown is PDB ID 6ELZ.
Depletion of Las1 did not affect levels of the AFs Nog1 or Rlp24 or of the AAA-ATPase Drg1 that promotes release of Nog1 and Rlp24 in the cytoplasm (Pertschy et al., 2007), suggesting that removal of these AFs is not impaired by depletion of Las1 (Fig. 8 B). Levels of the r-protein L10 (uL16), which associates with pre-60S subunits during late stages of cytoplasmic maturation (West et al., 2005; Lo et al., 2010), were not affected by Las1 depletion. Thus, in the absence of Las1 and without removal of the ITS2 foot structure, assembly is likely to progress normally at least until association of L10.
Aberrant 60S subunits containing the ITS2 foot structure produced in the absence of Las1 engage in translation
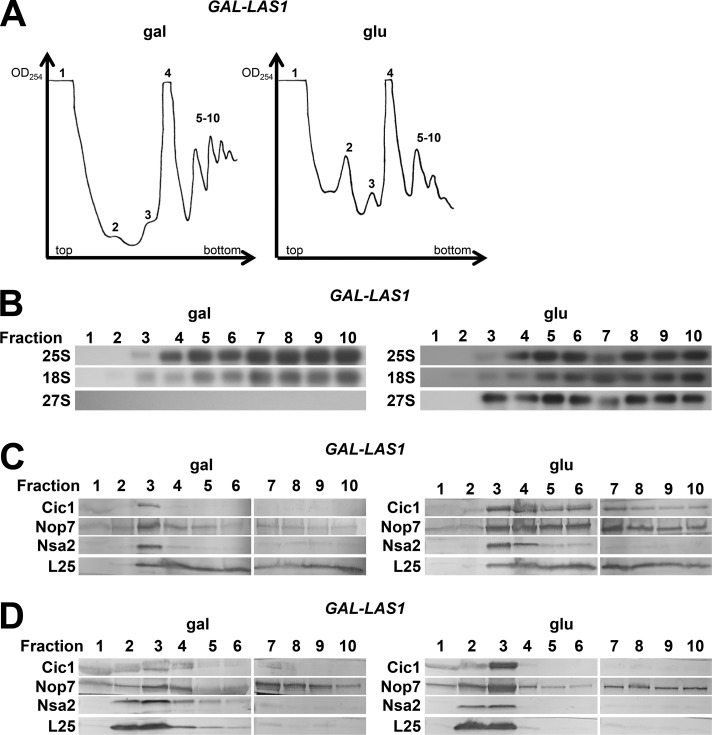
Next, we wanted to determine whether the cytoplasmic pre-60S particles containing the ITS2 foot structure produced in the absence of Las1 can be incorporated into translating ribosomes. To do this, we assayed for 27S pre-rRNA as well as the ITS2 AFs in polyribosomes when Las1 is present or depleted by sucrose gradient fractionation followed by Northern and Western blotting (Fig. 9).
Figure 9.
27S pre-rRNA and ITS2 AFs are present in polyribosomes in the absence of Las1. (A) Whole-cell lysates from cells expressing (gal) or depleted of (glu) Las1 were fractionated on a 7–47% sucrose gradient, and 1-ml fractions were collected from the indicated peaks. Fractions 1–4 were collected at the maximum of the indicated peak, and fractions 5–10 were collected continuously beginning with the first polyribosome peak. (B and C) Fractions were analyzed by Northern blotting (B) and Western blotting (C). (D) Whole-cell lysates prepared under polyribosome run-off conditions (omission of cycloheximide and MgCl2) from cells expressing or lacking Las1 were fractionated and analyzed by Western blotting.
After depletion of Las1, 27S pre-rRNA appears in both 80S ribosomes and polyribosomes (Fig. 9 B). In contrast, 27S pre-rRNA is only present in 66S preribosomes in wild-type cells (unpublished data). Similarly, Cic1 and Nop7 cosediment with 80S ribosomes and polyribosomes in the absence of Las1 (Fig. 9 C). Identical results were obtained upon expression of las1H134A (unpublished data). To confirm that the enrichment of Cic1 and Nop7 in polyribosome fractions after depletion of Las1 is not a result of their presence in structures other than polyribosomes, we prepared cell extracts under polyribosome run-off conditions (omission of cycloheximide and MgCl2). Under these conditions, Nop7 and Cic1 were shifted from high-molecular weight fractions to 60S and 80S fractions (Fig. 9 D). These results suggest that pre-60S subunits containing intact ITS2 and AFs bound to, or proximal to, ITS2 are able to engage in translation.
Discussion
Here we performed a thorough examination of the changes in pre-60S subunit protein composition in seven mutants blocked at cleavage of the C2 site in the ITS2 spacer. We compared the results described in this study with those previously published for depletion or mutation of a B-factor (Fig. 10 A; Lebreton et al., 2008; Gamalinda et al., 2013, 2014; Ohmayer et al., 2013; Konikkat et al., 2017). In almost all of these B mutants, six groups of proteins are consistently present in altered levels, suggesting that the B mutants cannot carry out C2 cleavage because several upstream remodeling events, including maturation of two functional centers of the 60S subunit, the PTC and PET, are impaired. Inspection of cryo-EM structures of nucleolar 60S subunit assembly intermediates suggests that the most parsimonious explanation for these effects is that each of the B mutants is blocked in transition from the Nsa1 state C particle to state D in the nucleolus (Fig. 10 B; Kater et al., 2017).
In contrast with the canonical B mutants, blocking ITS2 removal by depleting or inactivating Las1, the endonuclease that cleaves at the C2 site, did not affect these remodeling events. Unexpectedly, we found that removal of the ITS2 spacer RNA and the AFs bound to it or near it is not required for downstream steps of 60S subunit assembly, although it is necessary to allow ribosomes to function properly (Sarkar et al., 2017). When Las1 was depleted or inactivated, pre-60S subunits containing ITS2 and AFs bound to or proximal to ITS2 escaped into the cytoplasm, where they could be incorporated into translating ribosomes.
Stable association of Noc3 and Spb1 is impaired in B mutants
Cryo-EM analysis of Nsa1-containing pre-60S particles suggests that Nip7 and Nop2, along with Noc3 and Spb1, undergo stable incorporation into the pre-60S subunit at the transition from state C to state D particles (Kater et al., 2017). However, previous experiments suggest that Nip7 and Nop2 bind to 25S rRNA domain V cotranscriptionally, whereas Spb1 and Noc3 are predicted to bind posttranscriptionally (Chen et al., 2017). Although Nip7 and Nop2 were able to bind to a 27S pre-rRNA fragment containing 25S rRNAs I through V, Noc3 and Spb1 were unable to assemble onto any of the tested 27S pre-rRNA fragments.
Consistent with these AFs entering preribosomes at different points in assembly, we found that levels of Nip7 and Nop2 were generally unaffected in B mutants whereas levels of Noc3 and Spb1 were greatly decreased in most of the mutants (Fig. 10 A). Nip7 and Nop2 are likely unaffected in B mutants because they assemble onto pre-60S subunit assembly intermediates preceding Nsa1 state D but are flexibly bound and cannot be resolved by cryo-EM until they become stably associated at the transition from state C to state D. Noc3 and Spb1, however, likely do not bind preribosomes until the state C to D transition.
R-proteins surrounding the PET are not stably associated in B mutants
The decreased levels of the r-proteins surrounding the exit of the PET suggests that in B mutants these r-proteins have not yet become stably associated with the pre-60S subunit and that the rim around the PET exit is not properly matured (Figs. 4 B and 10). Of the 7 PET platform r-proteins, L19 and L31 were most commonly affected in B mutants. Although L17, L26, L35, and L37 can be visualized in Nsa1 state C particles, L19, L31, and L25 do not become stably associated until the transition from state C to state D particles (Kater et al., 2017). During this transition, 25S rRNA domain III becomes stabilized. Of the PET platform r-proteins, only L19, L31, and L25 exhibit extensive interactions with domain III (Gamalinda et al., 2014). However, L25 also interacts with 5.8S rRNA and domain I, both of which are stabilized early, which could account for L25 being less commonly affected than L19 and L31.
The entry of r-proteins L19 and L31 may be linked to the release of the AFs Ssf1 and Rrp15. The binding site of L31 was shown to overlap with that of Ssf1 in early nucleolar particles (Sanghai et al., 2018). Levels of the Ssf1-Rrp15 heterodimer were generally increased or unchanged in B mutants (Fig. 10 A). Thus, the strongly decreased levels of L31 as well as of L19, which is positioned proximal to L31, may be a result of Ssf1 and Rrp15 failing to exit the pre-60S subunit. Interestingly, levels of Ssf1 and Rrp15 were strongly decreased upon depletion of L17, which is positioned in close proximity to the Ssf1-Rrp15 heterodimer. Perhaps, then, in the absence of L17, Ssf1 and Rrp15 fail to tightly incorporate with the preribosome.
Furthermore, we observed a bilobal structure at the bottom of the pre-60S subunit present in the difference density of EMD-3891 (state E single particle reconstruction) and the state E fitted atomic model (PDB ID 6ELZ). We believe that this density belongs to the DEAD-box protein Spb4 based on the bilobal structure and the abundance of Spb4 in state E particles (Kater et al., 2017). Spb4, then, appears to dock on top of L19, suggesting that Spb4 may be unable to associate until the PET exit platform is constructed and that a failure to recruit Spb4 could be partially responsible for the phenotypes observed in the B mutants.
Similarly, we found that levels of the nuclear export factor Arx1 are decreased in most B mutants (Fig. 10 A). Arx1 binds to L26, L35, L19, and L25 (Fig. 4 B; Bradatsch et al., 2012; Wu et al., 2016). It is likely that, when these r-proteins cannot become tightly bound to the pre-60S subunit, Arx1 lacks the majority of its binding site and is unable to associate. Arx1 association with the properly matured PET platform could serve as a structural quality-control checkpoint to assess assembly of the PET exit. In this manner, an immature PET platform to which the surrounding r-proteins are not stably bound could impair export of the pre-60S subunit to the cytoplasm.
The Mak16 and Erb1 groups are not released from preribosomes in B mutants
The Mak16 (Mak16, Nsa1, Rpf1, and Rrp1) and Erb1 (Erb1, Ytm1, and Has1) groups exit preribosomes during the transitions from Nsa1 state D to E and state E to F, respectively (Kater et al., 2017). Levels of these seven AFs were generally increased or unaffected in mutants blocked at C2 cleavage, suggesting that their removal from preribosomes is impaired, consistent with most B mutants being blocked upstream of the state D particle (Fig. 10 A).
One member of the Mak16 group, Rpf1, extends into what will become the PET, suggesting that this set of AFs may play a role in initial structuring of the PET. Removal of the Mak16 group is likely performed by the AAA-ATPase Rix7, which has been shown to power release of Nsa1 (Kressler et al., 2008; Pratte et al., 2013). Thus, the failure to remove the Mak16 group in B mutants could be due to a failure of Rix7 to associate with the pre-60S subunit or become activated. Alternatively, other remodeling events may be necessary to correctly position Nsa1 such that Rix7 can properly interact with it.
Erb1 and Ytm1 are removed from the pre-60S subunit by Rea1, which interacts directly with the N-terminal ubiquitin-like domain of Ytm1 (Bassler et al., 2010). The generally decreased levels of Rea1 in B mutants along with increased or unchanged levels of Erb1 and Ytm1 suggest that removal of Erb1 and Ytm1 by Rea1 is impaired. Levels of Has1, a DEAD-box helicase that interacts with the N-terminal extension of Erb1 (Kater et al., 2017), were also unchanged or increased, suggesting that removal of Ytm1 and Erb1 may be linked to that of Has1. For example, removal of Erb1 may stimulate Has1 ATP hydrolysis and/or dissociation from the preribosome.
Domain V of 25S rRNA is not matured in B mutants
Levels of several AFs that bind 25S rRNA domain V (Nsa2, Nug1, Nog2, Rsa4, and Nop53) were strongly decreased in almost every B mutant, suggesting that there is a failure to carry out maturation of domain V in mutants blocked at C2 cleavage (Fig. 10 A). This is in agreement with the Nsa1 cryo-EM structures that suggest stabilization of domain V is performed predominantly after the transition from state C to D particles (Kater et al., 2017). Nog2, Nop53, and Rsa4 enter preribosomes after release of Nsa1 (Fig. 1 C). Thus, decreased levels of these AFs are simply because of assembly failing to progress to the point at which they would associate. Nsa2 and Nug1, however, are present in Nsa1 state C particles. Yet, these proteins are not fully resolved in state C, suggesting that they may not be stably associated in B mutants and may dissociate from pre-60S subunits during preribosome purifications.
Why is C2 cleavage blocked in B mutants?
A remaining question is why these B mutants are blocked in the transition from Nsa1 state C to state D, and how this prevents C2 cleavage. The most upstream defects in the B mutants are failure to assemble Noc3 and Spb1 as well as impaired formation of the PET exit platform. Although it is not yet entirely clear how depletion of each of the B-factors triggers these effects, we can build a model based on the available, but incomplete, collection of nucleolar pre-60S subunit assembly intermediate structures.
Depletion of the early-entering PET r-proteins (L17, L26, L35, and L37) may prevent early structuring of the PET exit and, therefore, could impair release of Ssf1 and Rrp15, thereby preventing entry or stabilization of L19 and L31. Because L19 and L31 exhibit significant interactions with 25S rRNA domain III, they may be necessary for the structuring of domain III observed during the transition from Nsa1 state C to state D particles. In state D particles, Noc3 interacts with domain III. Therefore, association of Noc3 with the pre-60S subunit may depend on construction of the PET exit platform.
Many of the B-factor AFs bind to 25S rRNA domain V (Nip7, Nop2, Nog1, Nsa2, and Nog2). Three others (Rlp24, Tif6, and Mak11) are mutually interdependent with Nog1 for entry into preribosomes. Depletion of any one of these B-factors, therefore, likely affects the structure of domain V, to which Spb1 binds in Nsa1 state D particles. Furthermore, L23 is located adjacent to the binding site of Rlp24 and is necessary for assembly of Nog1, Rlp24, and Tif6. The B-factor r-protein L9 binds adjacent to the binding site of Nsa2 and may be necessary for stable association of Nsa2. In each of these B mutants, a failure to properly structure domain V could prevent entry of Spb1.
Ultimately, C2 cleavage relies on entry and function of the Las1 complex; if one or more of the remodeling events preceding C2 cleavage fail to occur, the Las1 complex may be unable to enter the pre-60S subunit because of a failure to construct its binding site. Removal of the Erb1 group is an attractive candidate for a remodeling event necessary to facilitate entry of the Las1 complex. The N-terminal extension of Erb1 was found to extend across half of the early pre-60S subunit, interacting with at least eight AFs along the way (Kater et al., 2017). It seems likely that remodeling on a large scale would be initiated by removal of the Erb1 group, which binds proximal to ITS2. Indeed, it was found that removal of Erb1 initiates rotation of the N-terminal α helix of Rlp7, allowing the L1 stalk to assume its mature position; this is just one of many remodeling events that likely occur upon exit of Erb1 and may be necessary for entry of the Las1 complex.
C2 cleavage is not necessary for subsequent 60S subunit assembly events
The finding that depletion of Las1 results in polyribosomes containing the ITS2 foot structure suggests that C2 cleavage and subsequent removal of ITS2 are not necessary for late nuclear and cytoplasmic stages of 60S subunit assembly or for incorporation of the 60S subunit into an 80S ribosome. A similar phenomenon was observed with mutants that cannot carry out various stages of 7S pre-rRNA processing, including the dob1-1 mutant (Rodríguez-Galán et al., 2015).
In both the dob1-1 mutant and the Las1 depletion mutant, the aberrant 60S particles containing either 7S pre-rRNA or 27S pre-rRNA are able to assemble into polyribosomes, suggesting that they are able to carry out translation without stalling elongation. It was previously suggested that 7S pre-rRNA processing may not be necessary for export into the cytoplasm and that there is no quality-control mechanism to block pre-60S subunits containing 7S pre-rRNA from engaging in translation (Rodríguez-Galán et al., 2015). The results presented in this study suggest that the same can be true of immature 60S subunits containing 27S pre-rRNA. Consistent with this, we found that the translation factors EF2, EF3, and eIF4a were present in Nmd3-containing pre-60S subunits upon depletion of Las1 (Fig. 8 A).
However, both depletion and mutational inactivation of Las1 are lethal. There are several possible explanations for this lethality. Depletion of Las1 results in sequestration on cytoplasmic 60S subunits of Cic1, Nop7, Erb1, Rlp7, and Nop15 (Fig. 8, A–C), all of which are essential AFs required for processing of 27SA3 pre-rRNA (Sahasranaman et al., 2011). This could ultimately cause a block in production of additional pre-60S subunits and, therefore, result in lethality. Consistent with this notion, we found that depletion of Las1 results in accumulation of 27SA3 pre-rRNA as well as 27SB pre-rRNA, indicating that processing of 27SA3 pre-rRNA is slowed down or blocked upon depletion of Las1 (Fig. S4 C).
Additionally, it was recently determined that depletion of Las1 induces a translation defect, in which ribosomes containing aberrant 60S subunits from which the ITS2 foot has not been removed become trapped in a hybrid translation state (Sarkar et al., 2017). These ribosomes are subsequently targeted for turnover by the RQC and Ski–exosome complexes to alleviate translational stress.
Las1 depletion or inactivation versus other B mutants
An obvious question raised by our work is why depletion or mutational inactivation of Las1 results in a phenotype so dramatically different from those observed in every other B mutant discussed in this study. Depletion or mutation of Las1 likely causes a very specific block in C2 cleavage, without affecting assembly or maturation of other regions of the pre-60S subunit. The other B mutants, however, indirectly impair C2 cleavage by preventing one or more of the upstream remodeling events described in this study that are necessary for C2 cleavage to occur.
A remaining question posed by this work is why the failure to cleave at the C2 site in ITS2 in the absence of Las1 does not seem to impair later stages of 60S subunit assembly. We have long thought of C2 cleavage and subsequent removal of ITS2 as a checkpoint in 60S subunit assembly because it is a significant remodeling event that is initiated coincident with exit of the pre-60S subunit from the nucleolus. However, the results presented in this study suggest that it may simply be an easily assayed landmark. Perhaps C2 cleavage and subsequent ITS2 processing normally serve as a means to coordinate various remodeling events by acting as a timer. If this timer fails, there could be alternative assembly pathways that bypass the need for C2 cleavage. Alternatively, assembly events during later stages of 60S subunit biogenesis could in some way be separated from and uninfluenced by remodeling of the foot structure and ITS2 processing. Further work will be necessary to begin to understand what roles the ITS2 foot and ITS2 processing actually play in 60S subunit assembly.
The finding that C2 cleavage is not strictly required for subsequent steps of 60S subunit assembly was wholly unexpected. However, the six common defects observed in every B mutant with the exception of the Las1 mutants indicates that there are one or more checkpoints in assembly upstream of C2 cleavage by which construction of the PET platform and the PTC is monitored.
Materials and methods
Construction and growth of yeast strains
S. cerevisiae strains used in this study are listed in Table S1. Yeast strains conditional for expression of Dbp10, Nog1, Spb4, Nog2, Nsa2, and Las1 were constructed as described by Longtine et al. (1998) by replacing the endogenous promoter of each gene with the glucose-repressible GAL1 promoter and the selectable marker TRP1, with or without three copies of the 3HA epitope. Strains expressing C-terminal TAP-tagged Nop7 or Nmd3 were generated by the same method using URA3 as a selectable marker as described by Longtine et al. (1998) and Rigaut et al. (1999). Transformants were selected for by growth on C-trp + galactose or C-ura + galactose media and were screened by colony PCR or by Western blotting by using anti-HA (Sigma-Aldrich) or anti-TAP antisera (Promega) to confirm correct in-frame incorporation of the GAL1 or TAP cassettes.
Unless otherwise noted, yeast strains were grown at 30°C in either YEPGal (2% galactose, 2% peptone, and 1% yeast extract) or YEPGlu (2% dextrose, 2% peptone, and 1% yeast extract). To deplete Dbp10, Nog1, Spb4, Nog2, Nsa2, or L19, the conditional yeast strains were grown in media containing galactose and were then shifted to glucose-containing media for 16–17 h. Cells were harvested at mid-log phase.
Affinity purification of preribosomes
Pre-60S subunit assembly intermediates were affinity-purified from whole-cell extracts with magnetic Dynabeads (Invitrogen) using TAP-tagged Nop7. Cell pellets were resuspended in RNP buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10 mM MgCl2, 0.075% NP-40) and subjected to glass bead lysis. Lysates were incubated with IgG-coated Dynabeads for 1 h at 4°C with rocking, then the beads were washed three times with RNP buffer. Preribosomes were eluted from the Dynabeads by incubation at room temperature for 1 h with 10 U TEV protease (Invitrogen), which cleaves the TEV protease site within the TAP-tag. Preribosomes were precipitated with 10% TCA.
SDS-PAGE, silver staining, and Western blotting
Proteins in whole-cell extracts were prepared for PAGE as described by Ausubel et al. (1994). Proteins from affinity-purified preribosomes were precipitated with 10% TCA and resuspended in SDS sample buffer. Proteins were separated by SDS-PAGE on 4–20% Tris-Glycine Novex precast gels (Invitrogen) or 4–12% Bis-Tris gels (Invitrogen) and stained with silver by standard methods. Western blotting was performed with a protocol modified from Ausubel et al. (1994). To allow detection of several different proteins on a single blot, the nitrocellulose membranes were cut into smaller sections based on the previously established mobility of each protein. Each section of the membrane was then individually probed and developed. HA-tagged proteins were detected with mouse monoclonal antibody 12CA5 (Sigma-Aldrich), and TAP-tagged proteins were detected with anti-TAP antibody (Promega). All other proteins were detected with antibodies specific to the given AF or r-protein. Secondary antibodies used for detection were AP-conjugated anti–mouse or anti–rabbit (Promega), and colorimetric detection was performed by using nitro blue tetrazolium (NBT) and 5-bromo-4-chloro-3-indolyl-phosphate (BCIP; Promega).
Antibodies specific to AFs or r-proteins were provided by: J. de la Cruz (University of Seville, Seville, Spain), P. Linder (University of Geneva, Geneva, Switzerland), and J. Bravo (Spanish National Research Council, Madrid, Spain) provided Erb1, Ytm1, Has1, and Rsa4; C. Saveanu (Pasteur Institute, Paris, France) and M. Fromont-Racine (Pasteur Institute, Paris, France) provided Rlp24, Nsa2, Tif6, and Nog2; V. Panse (ETH Zurich, Zurich, Switzerland) provided Bud20 and Nug1; E. Tosta (University of Stuttgart, Stuttgart, Germany) provided Cic1/Nsa3; J. Maddock (University of Michigan, Ann Arbor, MI) provided Nog1; K.-Y. Lo (National Taiwan University, Taipei, Taiwan) provided Puf6 and Loc1; D. Tollervey (Wellcome Centre for Cell Biology, Edinburgh, Scotland, UK) provided Mtr4; K. Karbstein (The Scripps Research Institute, Jupiter, FL) provided Rrp5; H. Bergler (University of Graz, Graz, Austria) provided Drg1; A. Johnson (The University of Texas at Austin, Austin, TX) provided Nmd3, rpL8, and rpL10; F. Lacroute (Center for Molecular Genetics, Montpellier, France) provided rpL1; S. Rospert (University of Freiburg, Freiburg, Germany) provided rpL17, rpL19, and rpL26; and K. Siegers (Max Planck Institute of Biochemistry, Munich, Germany) provided rpL25.
Affinity purification of preribosomes and analysis by iTRAQ mass spectrometry
For semiquantitative iTRAQ mass spectrometry, preribosomes were purified as described above from 1-liter cultures with the following modifications. Cell pellets were resuspended in TNM150 buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1.5 mM MgCl2, 0.1% NP-40, and 5 mM 2-mercaptoethanol [Sigma-Aldrich]). NP-40 was excluded from the buffer for all steps after incubation of the lysates with IgG-coated Dynabeads for 1 h at 4°C.
Purified samples were sent to the Penn State Hershey Core Research Facilities for trypsin digestion and 4-plex labeling with iTRAQ reagents 114, 115, 116, and 117 (Applied Biosystems) or 8-plex labeling with iTRAQ reagents 113, 114, 115, 116, 117, 118, 119, and 121 (Applied Biosystems). Peptides were separated by 2D liquid chromatography, and parent ions were identified on a Sciex 5600 liquid chromatography mass spectrometry mass spectrometer. Protein Pilot 5.0 was used to obtain iTRAQ ratios as an average of all peptides for each protein. Proteins identified with >99.9% confidence were used for further data analysis. For each galactose-glucose pairwise comparison, data were normalized to the change in ratio of the TAP-tagged protein (Nop7), which serves as a loading control.
Growth assays
Yeast grown on galactose-containing solid media were suspended in galactose-containing liquid media to OD600 of 0.5–0.7. Cells were serially diluted and 20 µl of each dilution was spotted onto appropriate galactose- and glucose-containing solid media.
Sucrose gradient centrifugation
Preribosomes, 40S and 60S ribosomal subunits, 80S ribosomes, and polyribosomes were fractionated from yeast by using a protocol modified from Deshmukh et al. (1993). In brief, 100–200-ml cultures grown to log phase were treated with 5–10 mg cycloheximide for 30 min before harvesting. Approximately 20 OD254 units of whole-cell extracts for GAL-RPL19B or 50–60 OD254 units for GAL-LAS1 and GAL-LAS1 NOP7-TAP pRS315-las1H134A yeast were loaded on 7–47% (wt/vol) sucrose gradients and centrifuged at 27,000 rpm for 4 h by using a Sorvall AH-629 swinging bucket rotor. A Teledyne ISCO Foxy R1 density gradient fractionator was used to continuously monitor A254. For GAL-LAS1 and GAL-LAS1 NOP7-TAP pRS315-las1H134A yeast, 1-ml fractions were collected and used either for phenol-chloroform extraction of RNA or precipitation of proteins with 10% TCA. For polyribosome run-off experiments, cells were not treated with cycloheximide, cell lysates were prepared in buffer lacking cycloheximide and MgCl2, and the cell lysates were run on gradients lacking MgCl2.
Fluorescence microscopy
Pre-60S subunit localization in GAL-LAS1 was assayed by monitoring the localization of Nop7-GFP and Cic1-GFP. Live cells were imaged at room temperature in C-leu + galactose or C-leu + glucose fluid media by using a Zeiss LSM 880 laser scanning confocal microscope at 600× magnification. Images were acquired by using ZEN (blue edition) acquisition software (Zeiss). Images were processed using ImageJ (National Institutes of Health).
Assaying pre-rRNA processing
Steady-state levels of pre-rRNAs were assayed by primer extension and Northern blotting as described by Horsey et al. (2004) with the following modifications. RNA was quantified by using a Nano Drop 2000C spectrophotometer (Thermo Fisher Scientific) before aliquoting; 5 µg RNA was used per sample. The annealing step of the primer extension reactions was reduced to 10 min. Signal intensities of Northern blot bands were quantified by using ImageJ. Sequences of oligonucleotides used for primer extension and Northern blotting are available upon request.
In vivo RNA structure probing (SHAPE)
Cells were grown to an OD600 of 0.5–0.6, washed with PBS, and treated with 100 mM NAI or 100 mM DMSO for 20 min at 30°C with rocking. Total RNA was extracted, and primer extensions with Transcriptor Reverse Transcription (Roche) were performed with oligonucleotides that are complimentary to various regions in ITS2 (Fig. S3 B). For each reverse transcription reaction, 2.5 µl (5 µg) whole-cell RNA was incubated with 1.0 µl (0.2 µM) 32P-labeled oligonucleotide and 1.0 µl 4.5× hybridization buffer (225 mM Hepes, pH 7.0, and 450 mM KCl) at 80°C for 5 min. Reactions were cooled to an appropriate annealing temperature (49–55°C) and incubated for 20 min. Next, 15.5 µl prewarmed (49–55°C) extension mix containing 4.0 µl 5× hybridization buffer (Roche; 250 mM Tris-HCl, 150 mM KCl, 40 mM MgCl2), 2.0 µl (2.5 mM) deoxynucleoside triphosphate mixture, 1.0 µl (0.1 M) DTT, 0.5 µl (20 U) RNasin (Promega), 0.5 µl (10 U) Transcriptor Reverse Transcription (Roche), and 7.5 µl nuclease-free water was added to each reaction. For sequencing reactions only, 2.0 µl of the appropriate dideoxynucleotide (10 mM; Roche) was also added. Reaction mixtures were incubated at the annealing temperature (49–55°C) for 1 h. To degrade the RNA, 3.0 µl (1 M) NaOH was added to each reaction mixture, and the reaction mixtures were incubated at 50°C for 45 min. The reactions were then neutralized by adding 3.0 µl (1 M) HCl. The cDNAs were precipitated with 1.0 µl glycogen (10 mg/ml), 1.0 µl 0.5 M EDTA (pH 8.0), 2.8 µl (3 M) sodium acetate (pH 5.0), and 84 µl 100% ethanol. After ethanol precipitation, dried cDNA pellets were resuspended in 6.0 µl 1× loading dye (45% formamide and 0.01 M EDTA, pH 8.0), resolved on 6% polyacrylamide, 7 M urea gels, and visualized by autoradiography. Oligonucleotide sequences are available upon request.
PyMOL
PyMOL images of the structure of yeast mature 60S ribosomal subunits were generated by using PDB file 4V88 (Ben-Shem et al., 2011). PyMOL images of the structure of pre-60S subunit assembly intermediates were generated by using PDB files 6EM1, 6ELZ, and 3JCT (Wu et al., 2016; Kater et al., 2017).
Online supplemental material
Fig. S1 shows background material including the pre-rRNA processing pathway, the hierarchy of B-factor assembly, and the locations of 11 B-factors on pre-60S subunit intermediates determined by cryo-EM. Fig. S2 shows iTRAQ data for changes in amounts of 60S subunit AFs in preribosomes when Nsa2, L19, or Las1 are depleted or inactivated. Fig. S3 shows that there are modest changes in the structure of ITS2 after depletion of L19. Fig. S4 shows effects on cell growth and pre-rRNA processing after depletion or inactivation of Las1. Table S1 contains a list of yeast strains used in this study.
Supplementary Material
Acknowledgments
We thank Amber LaPeruta and Fiona Fitzgerald for critical reading of this manuscript and for helpful discussions. We thank the following people for their generous gifts of antibodies: Jesús de la Cruz, Patrick Linder, Jeronimo Bravo, Cosmin Saveanu, Micheline Fromont-Racine, Vikram Panse, Elizabeth Tosta, Janine Maddock, Kai-Yin Lo, David Tollervey, Katrin Karbstein, Helmut Bergler, Arlen Johnson, Francois Lacroute, Sabine Rospert, and Katja Siegers. We also thank Ed Hurt for providing the pRS315-las1R129A and pRS315-las1H134A plasmids.
This work was supported by National Institutes of Health grant R01GM028301 (to J.L. Woolford Jr.), the Richard King Mellon Foundation Presidential Fellowship in the Life Sciences (to S. Biedka), and the Roche/ARCS Foundation Scholar Award (to S. Biedka).
The authors declare no competing financial interests.
Author contributions: S. Biedka and J.L. Woolford Jr. conceived and designed experiments and wrote the manuscript. S. Biedka performed most of the experiments with help from J. Micic, D. Wilson (fluorescence microscopy), H. Brown (Nsa1-TAP, Nog2-TAP, and Nmd3-TAP mass spectrometry), and L. Diorio-Toth (initial Western blot assays).
References
- Ausubel F.M., Brent R., Kingston R.E., Moore D.D., Seidman J.G., Smith J.A., and Struhl K.. 1994. Current protocols in molecular biology. John Wiley & Sons Inc, New York. [Google Scholar]
- Bassler J., Kallas M., Pertschy B., Ulbrich C., Thoms M., and Hurt E.. 2010. The AAA-ATPase Rea1 drives removal of biogenesis factors during multiple stages of 60S ribosome assembly. Mol. Cell. 38:712–721. 10.1016/j.molcel.2010.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shem A., de Loubresse N.G., Melnikov S., Jenner L., Yusupova G., and Yusupov M.. 2011. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science. 334:1524–1529. 10.1126/science.1212642 [DOI] [PubMed] [Google Scholar]
- Bradatsch B., Leidig C., Granneman S., Gnädig M., Tollervey D., Böttcher B., Beckmann R., and Hurt E.. 2012. Structure of the pre-60S ribosomal subunit with nuclear export factor Arx1 bound at the exit tunnel. Nat. Struct. Mol. Biol. 19:1234–1241. 10.1038/nsmb.2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle C.D., Cassimere E.K., Lee J., and Denicourt C.. 2010. Las1L is a nucleolar protein required for cell proliferation and ribosome biogenesis. Mol. Cell. Biol. 30:4404–4414. 10.1128/MCB.00358-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle C.D., Sardana R., Dandekar V., Borgianini V., Johnson A.W., and Denicourt C.. 2013. Las1 interacts with Grc3 polynucleotide kinase and is required for ribosome synthesis in Saccharomyces cerevisiae. Nucleic Acids Res. 41:1135–1150. 10.1093/nar/gks1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Xie Z., Yang F., and Ye K.. 2017. Stepwise assembly of the earliest precursors of large ribosomal subunits in yeast. Nucleic Acids Res. 45:6837–6847. 10.1093/nar/gkx254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis J.H., Tan Y.Z., Carragher B., Potter C.S., Lyumkis D., and Williamson J.R.. 2016. Modular assembly of the bacterial large ribosomal subunit. Cell. 167:1610–1622.e15. 10.1016/j.cell.2016.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz J., Karbstein K., and Woolford J.L. Jr. 2015. Functions of ribosomal proteins in assembly of eukaryotic ribosomes in vivo. Annu. Rev. Biochem. 84:93–129. 10.1146/annurev-biochem-060614-033917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshmukh M., Tsay Y.F., Paulovich A.G., and Woolford J.L. Jr. 1993. Yeast ribosomal protein L1 is required for the stability of newly synthesized 5S rRNA and the assembly of 60S ribosomal subunits. Mol. Cell. Biol. 13:2835–2845. 10.1128/MCB.13.5.2835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira-Cerca S., Pöll G., Kühn H., Neueder A., Jakob S., Tschochner H., and Milkereit P.. 2007. Analysis of the in vivo assembly pathway of eukaryotic 40S ribosomal proteins. Mol. Cell. 28:446–457. 10.1016/j.molcel.2007.09.029 [DOI] [PubMed] [Google Scholar]
- Gamalinda M., Jakovljevic J., Babiano R., Talkish J., de la Cruz J., and Woolford J.L. Jr. 2013. Yeast polypeptide exit tunnel ribosomal proteins L17, L35 and L37 are necessary to recruit late-assembling factors required for 27SB pre-rRNA processing. Nucleic Acids Res. 41:1965–1983. 10.1093/nar/gks1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamalinda M., Ohmayer U., Jakovljevic J., Kumcuoglu B., Woolford J., Mbom B., Lin L., and Woolford J.L. Jr. 2014. A hierarchical model for assembly of eukaryotic 60S ribosomal subunit domains. Genes Dev. 28:198–210. 10.1101/gad.228825.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasse L., Flemming D., and Hurt E.. 2015. Coordinated ribosomal ITS2 RNA processing by the Las1 complex integrating endonuclease, polynucleotide kinase, and exonuclease activities. Mol. Cell. 60:808–815. 10.1016/j.molcel.2015.10.021 [DOI] [PubMed] [Google Scholar]
- Held W.A., Mizushima S., and Nomura M.. 1973. Reconstitution of Escherichia coli 30 S ribosomal subunits from purified molecular components. J. Biol. Chem. 248:5720–5730. [PubMed] [Google Scholar]
- Horsey E.W., Jakovljevic J., Miles T.D., Harnpicharnchai P., and Woolford J.L. Jr. 2004. Role of the yeast Rrp1 protein in the dynamics of pre-ribosome maturation. RNA. 10:813–827. 10.1261/rna.5255804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kater L., Thoms M., Barrio-Garcia C., Cheng J., Ismail S., Ahmed Y.L., Bange G., Kressler D., Berninghausen O., Sinning I., et al. 2017. Visualizing the assembly pathway of nucleolar pre-60s ribosomes. Cell. 171:1599–1610.e14. 10.1016/j.cell.2017.11.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisly I., Gulay S.P., Mäeorg U., Dinman J.D., Remme J., and Tamm T.. 2016. The functional role of eL19 and eB12 intersubunit bridge in the eukaryotic ribosome. J. Mol. Biol. 428(10, 10 Pt B):2203–2216. 10.1016/j.jmb.2016.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konikkat S., Biedka S., and Woolford J.L. Jr. 2017. The assembly factor Erb1 functions in multiple remodeling events during 60S ribosomal subunit assembly in S. cerevisiae. Nucleic Acids Res. 45:4853–4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kressler D., Roser D., Pertschy B., and Hurt E.. 2008. The AAA ATPase Rix7 powers progression of ribosome biogenesis by stripping Nsa1 from pre-60S particles. J. Cell Biol. 181:935–944. 10.1083/jcb.200801181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kressler D., Hurt E., and Baßler J.. 2017. A puzzle of life: Crafting ribosomal subunits. Trends Biochem. Sci. 42:640–654. 10.1016/j.tibs.2017.05.005 [DOI] [PubMed] [Google Scholar]
- Lebreton A., Saveanu C., Decourty L., Jacquier A., and Fromont-Racine M.. 2006. Nsa2 is an unstable, conserved factor required for the maturation of 27 SB pre-rRNAs. J. Biol. Chem. 281:27099–27108. 10.1074/jbc.M602199200 [DOI] [PubMed] [Google Scholar]
- Lebreton A., Rousselle J.C., Lenormand P., Namane A., Jacquier A., Fromont-Racine M., and Saveanu C.. 2008. 60S ribosomal subunit assembly dynamics defined by semi-quantitative mass spectrometry of purified complexes. Nucleic Acids Res. 36:4988–4999. 10.1093/nar/gkn469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo K.Y., Li Z., Bussiere C., Bresson S., Marcotte E.M., and Johnson A.W.. 2010. Defining the pathway of cytoplasmic maturation of the 60S ribosomal subunit. Mol. Cell. 39:196–208. 10.1016/j.molcel.2010.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A. III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., and Pringle J.R.. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961. [DOI] [PubMed] [Google Scholar]
- Ma C., Wu S., Li N., Chen Y., Yan K., Li Z., Zheng L., Lei J., Woolford J.L. Jr., and Gao N.. 2017. Structural snapshot of cytoplasmic pre-60S ribosomal particles bound by Nmd3, Lsg1, Tif6 and Reh1. Nat. Struct. Mol. Biol. 24:214–220. 10.1038/nsmb.3364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malyutin A.G., Musalgaonkar S., Patchett S., Frank J., and Johnson A.W.. 2017. Nmd3 is a structural mimic of eIF5A, and activates the cpGTPase Lsg1 during 60S ribosome biogenesis. EMBO J. 36:854–868. 10.15252/embj.201696012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manikas R.-G., Thomson E., Thoms M., and Hurt E.. 2016. The K+-dependent GTPase Nug1 is implicated in the association of the helicase Dbp10 to the immature peptidyl transferase centre during ribosome maturation. Nucleic Acids Res. 44:1800–1812. 10.1093/nar/gkw045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo Y., Granneman S., Thoms M., Manikas R.-G., Tollervey D., and Hurt E.. 2014. Coupled GTPase and remodelling ATPase activities form a checkpoint for ribosome export. Nature. 505:112–116. 10.1038/nature12731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nerurkar P., Altvater M., Gerhardy S., Schütz S., Fischer U., Weirich C., and Panse V.G.. 2015. Eukaryotic ribosome assembly and nuclear export. Int. Rev. Cell Mol. Biol. 319:107–140. 10.1016/bs.ircmb.2015.07.002 [DOI] [PubMed] [Google Scholar]
- Nierhaus K.H., and Dohme F.. 1974. Total reconstitution of functionally active 50S ribosomal subunits from Escherichia coli. Proc. Natl. Acad. Sci. USA. 71:4713–4717. 10.1073/pnas.71.12.4713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donohue M.F., Choesmel V., Faubladier M., Fichant G., and Gleizes P.E.. 2010. Functional dichotomy of ribosomal proteins during the synthesis of mammalian 40S ribosomal subunits. J. Cell Biol. 190:853–866. 10.1083/jcb.201005117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmayer U., Gamalinda M., Sauert M., Ossowski J., Pöll G., Linnemann J., Hierlmeier T., Perez-Fernandez J., Kumcuoglu B., Leger-Silvestre I., et al. 2013. Studies on the assembly characteristics of large subunit ribosomal proteins in S. cerevisae. PLoS One. 8:e68412 10.1371/journal.pone.0068412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña C., Hurt E., and Panse V.G.. 2017. Eukaryotic ribosome assembly, transport and quality control. Nat. Struct. Mol. Biol. 24:689–699. 10.1038/nsmb.3454 [DOI] [PubMed] [Google Scholar]
- Pertschy B., Saveanu C., Zisser G., Lebreton A., Tengg M., Jacquier A., Liebminger E., Nobis B., Kappel L., van der Klei I., et al. 2007. Cytoplasmic recycling of 60S preribosomal factors depends on the AAA protein Drg1. Mol. Cell. Biol. 27:6581–6592. 10.1128/MCB.00668-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillon M.C., Sobhany M., Borgnia M.J., Williams J.G., and Stanley R.E.. 2017. Grc3 programs the essential endoribonuclease Las1 for specific RNA cleavage. Proc. Natl. Acad. Sci. USA. 114:E5530–E5538. 10.1073/pnas.1703133114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratte D., Singh U., Murat G., and Kressler D.. 2013. Mak5 and Ebp2 act together on early pre-60S particles and their reduced functionality bypasses the requirement for the essential pre-60S factor Nsa1. PLoS One. 8:e82741 10.1371/journal.pone.0082741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., and Séraphin B.. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030–1032. 10.1038/13732 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Galán O., García-Gómez J.J., Kressler D., and de la Cruz J.. 2015. Immature large ribosomal subunits containing the 7S pre-rRNA can engage in translation in Saccharomyces cerevisiae. RNA Biol. 12:838–846. 10.1080/15476286.2015.1058477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahasranaman A., Dembowski J., Strahler J., Andrews P., Maddock J., and Woolford J.L. Jr. 2011. Assembly of Saccharomyces cerevisiae 60S ribosomal subunits: Role of factors required for 27S pre-rRNA processing. EMBO J. 30:4020–4032. 10.1038/emboj.2011.338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanghai Z.A., Miller L., Molloy K.R., Barandun J., Hunziker M., Chaker-Margot M., Wang J., Chait B.T., and Klinge S.. 2018. Modular assembly of the nucleolar large subunit processome. Nature. 556:126–129. 10.1038/nature26156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar A., Thoms M., Barrio-Garcia C., Thomson E., Flemming D., Beckmann R., and Hurt E.. 2017. Preribosomes escaping from the nucleus are caught during translation by cytoplasmic quality control. Nat. Struct. Mol. Biol. 24:1107–1115. 10.1038/nsmb.3495 [DOI] [PubMed] [Google Scholar]
- Saveanu C., Namane A., Gleizes P.-E., Lebreton A., Rousselle J.-C., Noaillac-Depeyre J., Gas N., Jacquier A., and Fromont-Racine M.. 2003. Sequential protein association with nascent 60S ribosomal particles. Mol. Cell. Biol. 23:4449–4460. 10.1128/MCB.23.13.4449-4460.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schillewaert S., Wacheul L., Lhomme F., and Lafontaine D.L.J.. 2012. The evolutionarily conserved protein Las1 is required for pre-rRNA processing at both ends of ITS2. Mol. Cell. Biol. 32:430–444. 10.1128/MCB.06019-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talkish J., Zhang J., Jakovljevic J., Horsey E.W., and Woolford J.L. Jr. 2012. Hierarchical recruitment into nascent ribosomes of assembly factors required for 27SB pre-rRNA processing in Saccharomyces cerevisiae. Nucleic Acids Res. 40:8646–8661. 10.1093/nar/gks609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West M., Hedges J.B., Chen A., and Johnson A.W.. 2005. Defining the order in which Nmd3p and Rpl10p load onto nascent 60S ribosomal subunits. Mol. Cell. Biol. 25:3802–3813. 10.1128/MCB.25.9.3802-3813.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson K.A., Merino E.J., and Weeks K.M.. 2006. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 1:1610–1616. 10.1038/nprot.2006.249 [DOI] [PubMed] [Google Scholar]
- Woolford J.L. Jr., and Baserga S.J.. 2013. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics. 195:643–681. 10.1534/genetics.113.153197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Tutuncuoglu B., Yan K., Brown H., Zhang Y., Tan D., Gamalinda M., Yuan Y., Li Z., Jakovljevic J., et al. 2016. Diverse roles of assembly factors revealed by structures of late nuclear pre-60S ribosomes. Nature. 534:133–137. 10.1038/nature17942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Zhu X., Zheng S., Tan D., Dong M.-Q., and Ye K.. 2018. Cryo-EM structure of an early precursor of large ribosomal subunit reveals a half-assembled intermediate. Protein Cell. 10.1007/s13238-018-0526-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.