This work shows that Centrobin (CNB) mutant males assemble aberrant basal bodies and do not produce functional sperm. It also shows that CNB can act as a positive or negative regulator of ciliogenesis in a cell type–dependent manner.
Abstract
Centrobin homologues identified in different species localize on daughter centrioles. In Drosophila melanogaster sensory neurons, Centrobin (referred to as CNB in Drosophila) inhibits basal body function. These data open the question of CNB’s role in spermatocytes, where daughter and mother centrioles become basal bodies. In this study, we report that in these cells, CNB localizes equally to mother and daughter centrioles and is essential for C-tubules to attain the right position and remain attached to B-tubules as well as for centrioles to grow in length. CNB appears to be dispensable for meiosis, but flagellum development is severely compromised in Cnb mutant males. Remarkably, three N-terminal POLO phosphorylation sites that are critical for CNB function in neuroblasts are dispensable for spermatogenesis. Our results underpin the multifunctional nature of CNB that plays different roles in different cell types in Drosophila, and they identify CNB as an essential component for C-tubule assembly and flagellum development in Drosophila spermatogenesis.
Introduction
Centrobin was initially identified in humans through a yeast two-hybrid screen for proteins that interact with the tumor suppressor BRCA2 (Zou et al., 2005). Like human Centrobin (referred to as CNTROB in humans), its homologues in other species are components of daughter centrioles in different cell types (Zou et al., 2005; Januschke et al., 2011, 2013; Gottardo et al., 2015). In mammals, CNTROB has been reported to be required for centriole duplication and elongation as well as for microtubule nucleation and stability through its binding to tubulin and its effect in stabilizing centrosomal P4.1-associated protein (CPAP; Jeong et al., 2007; Jeffery et al., 2010, 2013; Gudi et al., 2011, 2014, 2015; Shin et al., 2015).
Drosophila melanogaster Centrobin (referred to as CNB in Drosophila) is a key determinant of mother/daughter centriole functional asymmetry in larval neuroblasts and type I sensory neurons. In neuroblasts, CNB functions as a positive regulator of microtubule organizing center (MTOC) activity during interphase: its depletion impedes daughter centrioles to assemble an MTOC, whereas mother centrioles carrying ectopic CNB become active MTOCs (Januschke et al., 2013). This activity is dependent on CNB phosphorylation by PLK1/POLO on three conserved sites (Januschke et al., 2013) and involves the function of pericentrin-like protein (PLP; Lerit and Rusan, 2013) and BLD10/CEP135 (Singh et al., 2014). In Drosophila type I sensory neurons, CNB functions as a negative regulator of ciliogenesis: CNB depletion enables daughter centrioles as functional basal bodies that template ectopic cilia, and mother centrioles modified to carry CNB cannot function as basal bodies (Gottardo et al., 2015).
The function of a daughter centriole protein like CNB as a negative regulator of ciliogenesis opens the question of CNB’s role in Drosophila primary spermatocytes where after centrosome duplication all four centrioles, mothers and daughters alike become basal bodies that assemble axoneme-based cilium-like structures, which are the precursors of sperm flagella (Tates, 1971; Riparbelli et al., 2012, 2013; Gottardo et al., 2013). To investigate this issue, we have studied CNB localization and function in spermatogenesis.
Results and discussion
CNB localizes equally to mother and daughter centrioles in Drosophila primary spermatocytes
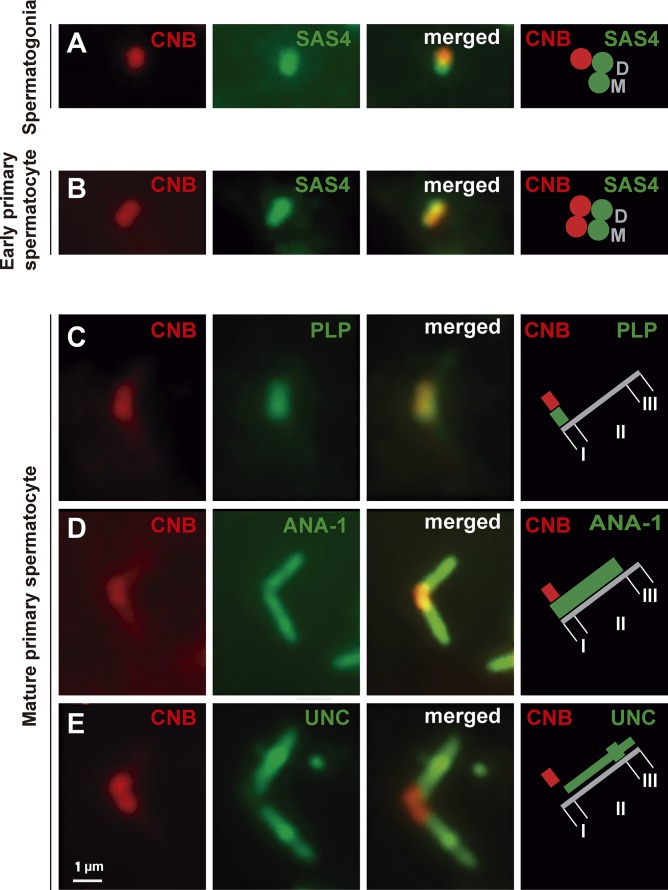
As in other cell types in Drosophila and other species, CNB localization in spermatogonial cells was asymmetric, restricted to only one of the two centrioles that were labeled with anti-SAS4 antibodies (Fig. 1 A, red and green, respectively; n = 86 cells from 25 spermatogonial cysts). However, after the last (fourth) round of spermatogonial mitosis, the resulting early primary spermatocytes presented CNB equally distributed on the two centrioles of each pair (Fig. 1 B; n = 163 cells from 15 cysts).
Figure 1.
CNB localizes equally in mother and daughter centrioles in primary spermatocytes. (A and B) CNB (red) localized to only one of the of the two SAS4-positive (green) centrioles, presumably the daughter (D in the cartoon), in spermatogonia (A; n = 86 cells from 25 cysts), but it colocalized with the two centrioles—mother (M in the cartoon) and daughter—in early primary spermatocytes (B; n = 163 cells from 15 cysts). (C–E) CNB signal (red) was equal in the two basal bodies of a pair. The region of highest concentration of CNB (region I) overlapped with PLP (C) on the proximal end of the ANA1 domain (D), where UNC concentration was lowest (E). Weak CNB and PLP signal could also be detected all along the basal body (C–E; region II; n = 247 cells from 98 cysts).
After centrosome duplication, all four centrioles of each spermatocyte grow significantly in length, migrate to the cell surface, and become basal bodies that template a short axoneme that forms a protrusion covered by the cell membrane (Tates, 1971; Fritz-Niggli and Suda, 1972). Recent studies refer to this protrusion as the cilium-like region (CLR; Gottardo et al., 2013) or the ciliary cap (Vieillard et al., 2016).
To determine CNB localization in the basal body–CLR complex, we immunostained CNB together with PLP, ANA1, and UNC. PLP signal was strongest on the proximal end of the basal body (Fig. 1 C, region I; Fu and Glover, 2012; Galletta et al., 2014). ANA1 labelled the entire length of the basal body (Fig. 1 D, regions I and II; Blachon et al., 2009; Riparbelli et al., 2012, 2013; Basiri et al., 2014). UNC overlapped with ANA1 except on the PLP-positive proximal end, was strongest at the transition zone, and extended further distally into the axoneme (Fig. 1 E, regions II and III; Baker et al., 2004; Ma and Jarman, 2011; Riparbelli et al., 2012). We found that CNB signal was equal in all basal bodies (n = 247 cells from 98 cysts); most concentrated on the proximal end of the ANA1 domain, largely overlapping with PLP. Like PLP, a very weak CNB signal could also be detected all along the basal body (Fig. 1, C–E, region II). These observations reveal some remarkable differences regarding CNB localization, which in turn suggest important functional differences as far as ciliogenesis is concerned between early spermatocytes and sensory neurons in Drosophila.
CNB is required in primary spermatocytes for basal bodies to achieve normal length and for the proper assembly of the C-tubule
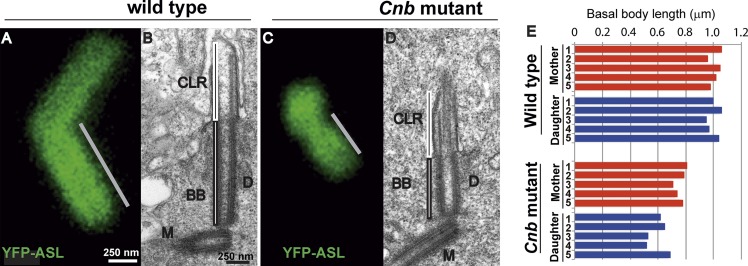
To determine whether CNB has a function in the basal body–CLR complex, we examined trans-heterozygous PBac(RB)Cnb[e00267]/Df(3L)ED4284 (henceforth referred to as Cnb mutant) spermatocytes. We found that the length of YFP-Asterless (ASL) signal (a proxy for centriole length) was significantly shorter (P < 0.001) in Cnb mutant (0.97 ± 0.28 µm) than in WT males (1.55 ± 0.11 µm; Fig. 2, A and C, gray bars). Longitudinal EM sections from centriole pairs in which both mother and daughter were cut along their entire length confirmed this observation (Fig. 2, B, D, and E) and revealed that daughter basal bodies were shorter than their mothers in Cnb mutant spermatocytes (0.62 ± 0.07 µm and 0.78 ± 0.04 µm, respectively; n = 5 centriole pairs; Fig. 2 E and Table S2). This was not the case in WT spermatocytes, where as previously reported (Riparbelli et al., 2012), we found that mother and daughter basal bodies were of equal length (1.02 ± 0.04 µm and 1.00 ± 0.05 µm, respectively; n = 5 centriole pairs; Fig. 2 E and Table S2). EM sections also revealed that 10 out of 16 mutant axonemes sectioned longitudinally were irregularly shaped and had abnormal axonemes with doublets that were aberrantly interrupted halfway through the CLR (Fig. 2 D).
Figure 2.
Basal bodies are shorter in Cnb mutants than in WT spermatocytes. (A–D) Basal bodies (BBs) revealed by YFP-ASL fluorescence (A and C, green) and basal body–CLR complexes observed in EM longitudinal sections (B and D) from WT (A and B) and Cnb mutant (C and D) mature spermatocytes. M and D indicate basal bodies derived from mother and daughter centrioles, respectively. Gray bars show the length of the YFP-ASL signal. (E) Basal body length measured from EM longitudinal sections of five mother (red)/daughter (blue) pairs (numbered) from WT and Cnb mutant mature spermatocytes.
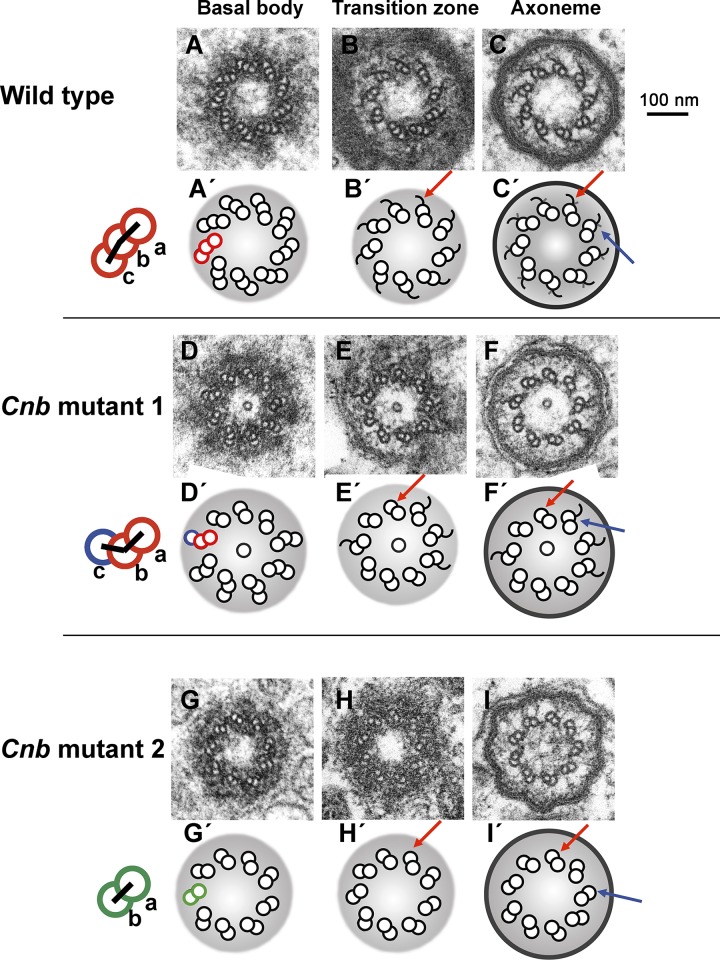
We then examined the ultrastructural details revealed by transversal EM sections. As reported previously (Gottardo et al., 2015), unlike basal bodies in sensory neurons that are made of doublets, basal bodies in WT spermatocytes are made of triplets (Fig. 3, A and A′). At the transition zone, the outermost tubule (C-tubule) progressively lost protofilaments (Fig. 3, B and B′), hence becoming an open longitudinal sheet that extended beyond the transition zone along the axoneme (Fig. 3, C and C′, red arrow). At the point where C-tubule remnants and B-tubules intersect, short radial projections could be observed (Fig. 3, C and C′, blue arrow; Riparbelli et al., 2012).
Figure 3.
C-tubules are mispositioned or lacking in the basal bodies in Cnb mutant spermatocytes. (A–C′) Serial sections through a basal body (A and A′), transition zone (B and B′), and axoneme (C and C′) from a WT primary spermatocyte. Basal bodies presented triplets composed of A-, B-, and C-tubules arranged along a nearly straight line (cartoon, red). At the transition zone, the outermost tubule (C-tubule) progressively lost protofilaments and became an open sheet (B and B′, red arrow). C-tubule remnants extended along the axoneme (C and C′, red arrow). Short radial projections could be observed in the axoneme at the point where C-tubule remnants and B-tubules intersected (C and C′, blue arrow). (D–I′) Serial sections through basal bodies, transition zones, and axonemes from two Cnb mutant primary spermatocytes (Cnb mutants 1 and 2). In all 11 serially sectioned samples, C-tubules in Cnb mutant basal bodies were either mispositioned away (blue tubule) from the straight line defined by the A/B doublet (red tubules) or lacking leaving doublets behind (green). Cnb mutant transition zones often lacked C-tubule–derived longitudinal sheets (E, E′, H, and H′, red arrow) and so did mutant axonemes (F, F′, I, and I′, red arrows), which also lacked the short radial projections observed in WT axonemes (F, F′, I, and I′, blue arrows). Neither the WT cell shown in this figure (A–C′) nor Cnb mutant 2 (G–I′) presented a microtubule in the center of the lumen, but Cnb mutant 1 did (D–F′). The presence of this luminal singlet was as erratic in Cnb mutant as it was in WT males and does not represent a Cnb mutant phenotype.
In addition to this basic layout of triplets, doublets, and open sheaths, about half of basal body–CLR complexes in Drosophila WT spermatocytes present a luminal singlet microtubule independently of the Z position of the cross-section (Tates, 1971; Carvalho-Santos et al., 2012; Riparbelli et al., 2012). The presence of this luminal singlet is as erratic in Cnb mutant as it is in WT males and does not represent a Cnb mutant phenotype.
We serially sectioned 11 basal body–CLR complexes from two Cnb mutant males. In all 11 samples, C-tubules were either misplaced outwards, away from the nearly straight line defined by the A/B doublet (Fig. 3, D and D′, blue), or lost, leaving only duplets behind (Fig. 3, D, D′, G, and G′, green; Table S1). These abnormalities extended distally along the axoneme, where C-tubule–derived longitudinal sheets were often lacking in Cnb mutant spermatocytes (Fig. 3, E–F′ and H–I′, red arrow). The short radial projections observed in WT axonemes (Fig. 3, C and C′, blue arrow) also appeared to be lacking in Cnb mutant CLRs (Fig. 3, F, F′, I, and I′, blue arrow), but this observation must be interpreted with caution because of the technical difficulties of detecting such small structures that are located in regions of electron-dense material.
These observations reveal that CNB is required in Drosophila spermatocytes for centrioles to properly position and stabilize C-tubules, grow in length up to full-sized basal bodies, and template normal axonemes. Our results identify CNB as one of the very few proteins known so far to be required for C-tubule assembly, including Δ-tubulin in Paramecium tetraurelia (Garreau de Loubresse et al., 2001) and Chlamydomonas reinhardtii (Dutcher and Trabuco, 1998) along with UNC and SAS4 in Drosophila (Gottardo et al., 2013; Zheng et al., 2016). Notably, as in Cnb mutant spermatocytes, basal bodies are also shorter than normal in Sas4F112A and Unc mutant spermatocytes (Gottardo et al., 2013; Zheng et al., 2016), hence suggesting that short centrioles and lacking or mispositioned C-tubules may be causally linked.
Drosophila SAS4 and its human homologue CPAP are closely functionally related to the corresponding Centrobin homologues CNB/CNTRB. Mutants in the N-terminal helical motif of the PN2-3 domain of CPAP and SAS4 cause overelongation of newly formed centriolar/ciliary microtubules, and so does CNTRB overexpression by stabilizing and driving the centriolar localization of CPAP (Gudi et al., 2011, 2014, 2015). Moreover, the mutants in PN2-3 domain of CPAP referred to above result in biciliated cells, and so does Cnb loss of function in Drosophila sensory neurons. In Drosophila, SAS4 and CNB coimmunoprecipitate from embryo extracts (Januschke et al., 2013). These data strongly suggest that the role of CNB on basal body length may be SAS4 dependent.
Elongating axonemes are abnormal in Cnb mutant males
The ultrastructural phenotypes that we have observed in basal body–CLR complexes in Cnb mutant spermatocytes persist through meiosis and are present in early postmeiotic cells (Fig. S1 A). However, we did not detect any observable defects in meiosis. Cnb mutant males assembled fairly normal meiotic spindles and generated normal cysts of postmeiotic onion-stage spermatids that presented a uniform nuclear size (Fig. S1 B), which is a very sensitive readout of faithful meiotic chromosome segregation (González et al., 1989). These results strongly suggest that shortened centrioles and lacking C-tubules have no major observable consequences on meiosis progression.
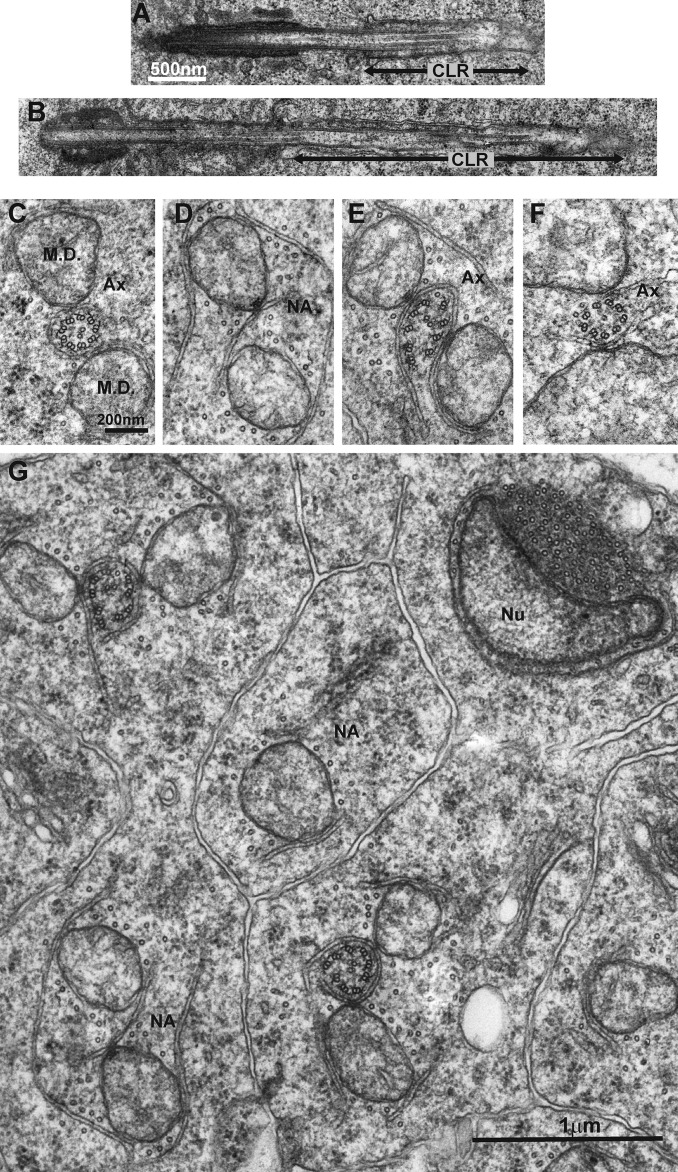
However, soon after meiosis, when the basal body attaches to the nucleus and the axoneme starts to grow, pushing the old CLR caudally, Cnb mutant spermatids presented CLRs that were much longer than those from WT cells: 5.6 ± 1.4 µm (n = 7) and 1.5 ± 0.8 µm (n = 6), respectively (Fig. 4, A and B). Abnormally long CLRs have also been reported in Unc mutant early spermatids (Gottardo et al., 2013). In WT cells at this stage, CNB signal was strongest on the basal body proximal to UNC and was also detectable over the perinuclear plasm (Tates, 1971), a cap over the nuclear hemisphere where the basal body is engaged (Fig. S1 C).
Figure 4.
Axonemes are often malformed or lost in Cnb mutant spermatids. (A and B) Longitudinal sections through the basal body and axoneme (Ax) in WT (A) and Cnb mutant (B) early spermatids. The CLR (black line) was longer than normal in Cnb mutant spermatids (5.6 ± 1.4 µm, n = 7, and 1.5 ± 0.8 µm, n = 6, respectively). (C) Section through the tail of a WT elongating spermatid showing the axoneme that presented the typical 9 + 2 configuration flanked by two mitochondrial derivatives (M.D.s). (D–F) Sections through Cnb mutant elongating spermatid showing one without axoneme (no axoneme [N.A.]; D) and axonemes that presented duplets with and without C remnants (E and F). (G) General view revealing phenotypic variation among Cnb mutant spermatids of the same cyst including a cell that had no axoneme and presented a single mitochondrial derivative and the nucleus (Nu) of an ectopic spermatid head.
Cnb mutant phenotypes were also conspicuous at later stages of elongation. Transversal sections through the tails of WT elongating spermatids showed two mitochondrial derivatives flanking the axoneme, which was almost completely surrounded by a double membrane (Fig. 4 C). Among the sample of 32 Cnb mutant elongating spermatids that we analyzed in detail (belonging to three cysts from two different males), we found a wide range of phenotypic variability (Fig. 4, D–G). Five presented a fairly WT 9 + 2 configuration, whereas the other 27 included cells that had no axoneme (n = 5; Fig. 4 D, NA) or axonemes with missing or highly disarrayed duplets and in which the surrounding membrane was widely open (n = 22; Fig. 4, E and F). Among the latter, six had some duplets that carried C remnants, and 16 did not. Interestingly, not all duplet-bound open sheaths observed in Cnb mutant cells were C remnants, such as for instance the one shown in Fig. 4 F that originated from an A-tubule and was oriented in the wrong direction. We also found four axonemes that presented one to three triplets with an attached open sheath (Fig. S2 A, arrows). No such structures have been reported in WT cells. The presence of a fraction of Cnb mutant spermatids with axonemes that presented C remnants suggests that a certain fraction of Cnb mutant primary spermatocytes retain C-tubules.
In addition to lacking and disarranged axonemes, Cnb mutant elongating spermatids also presented ectopic nuclei at sections where only tails were present in WT cysts (Fig. 4 G, Nu, and Fig. S2 B). Scattered nuclei have also been reported in other Drosophila mutants like Unc and Sas4 (Baker et al., 2004; Riparbelli and Callaini, 2011). Head-to-tail attachment defects have been documented in the sperm of a rat CNTROB mutant strain (hd rats) that was male sterile (Liška et al., 2013).
Our results reveal that CNB depletion brings about a spectrum of mutant phenotypes at different stages of spermatogenesis that includes shorter centrioles, mispositioned or absent C-tubules, lack of C-tubule derivatives, abnormal or absent axonemes, and scattered nuclei along elongating spermatid bundles. We cannot discard that CNB may have independent functions accounting for each of these phenotypes. However, the apparent pleiotropic effect of CNB loss may simply reflect the downstream consequences of centrioles that are shorter than normal and present C-tubule defects.
Amino acids T4, T9, and S82, which are critical for centriole asymmetry in Drosophila neuroblasts, are dispensable for CNB function in spermatogenesis
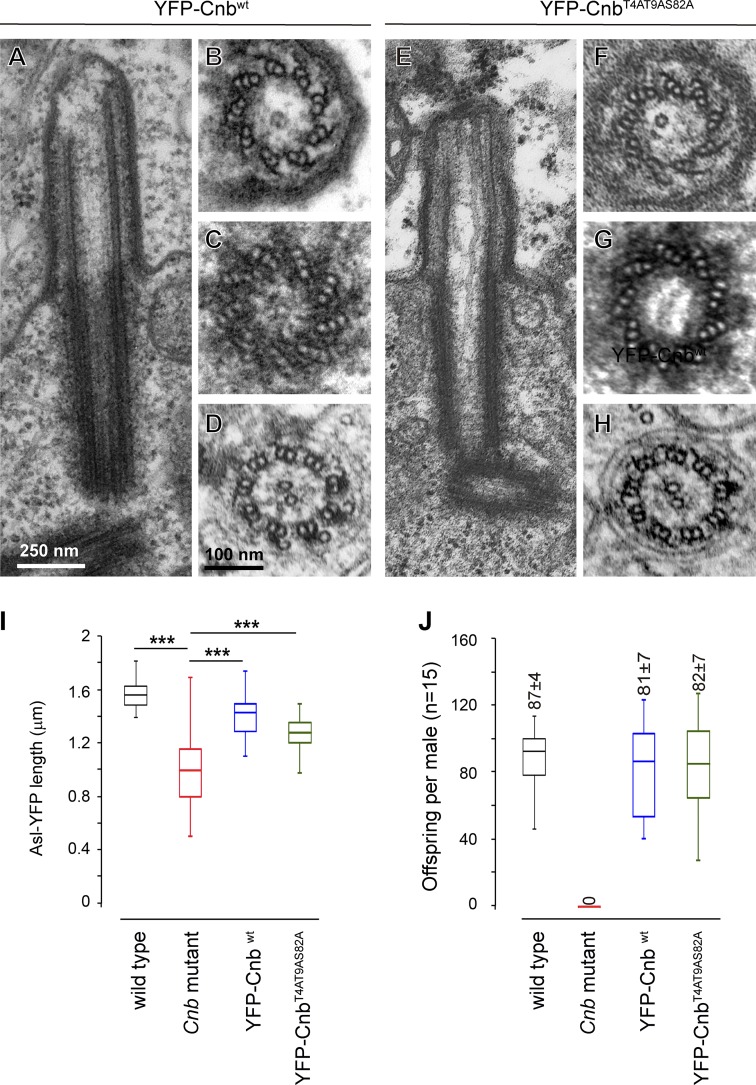
Expression of YFP-CNBWT, a fusion protein between YFP and WT CNB, rescued all the phenotypes that we observed in Cnb mutant males. These included (A) the ultrastructural abnormalities, which affected premeiotic basal body–CLR complexes and postmeiotic axonemes (Fig. 5, A–D), (B) centriole (YFP-ASL) length (Fig. 5 I), which was rescued from 0.97 ± 0.28 µm (n = 27) in Cnb mutant males to 1.47 ± 0.16 µm (n = 20), very close to WT length (1.55 ± 0.11 µm, n = 21), and (C) fertility (Fig. 5 J), which was rescued to 81 ± 7, which is indistinguishable from WT levels (87 ± 4; offspring per male in single-pair mating tests; n = 15). These results confirm that all these phenotypes are indeed brought about by Cnb loss of function and are not caused by other unrelated mutations.
Figure 5.
CNBWT and CNBT4A-T9A-S82A are equally capable of rescuing CNB mutant traits in spermatogenesis. (A–H) EM longitudinal sections through basal body–CLR complexes (A and E) and transversal sections through premeiotic axonemes (B and F) and basal bodies (C and G) as well as postmeiotic axonemes (D and H) from Cnb mutant males expressing the YFP-CNBWT (A–D; nine longitudinal sections through CLRs, five and 11 cross sections through basal bodies and CLRs, respectively, and 67 cross sections through elongating spermatids from six cysts) or the YFP-CNBT4A-T9A-S82A construct (E–H; seven longitudinal sections through CLRs, six and 13 cross sections through basal bodies and CLRs, respectively, and 38 cross sections through elongating spermatids from four cysts). (I) Basal body length (YFP-ASL fluorescence) in WT (n = 21), Cnb mutant (n = 27), Cnb mutant expressing YFP-CNBWT (n = 20), and Cnb mutant expressing YFP-CNBT4A-T9A-S82A males (n = 17). ***, P < 0.001 (ANOVA). (J) Fertility of WT, Cnb mutant, Cnb mutant expressing YFP-CNBWT, and Cnb mutant expressing YFP-CNBT4A-T9A-S82A single males (n = 15). Error bars indicate SEM.
Remarkably, all these Cnb mutant traits were also rescued by the YFP-CNBT4A-T9A-S82A transgene (Januschke et al., 2013). Basal bodies, CLRs, and spermatids recovered WT ultrastructure including properly positioned C-tubules and C-tubule remnants (Fig. 5, E–H); centriole (YFP-ASL) length was recovered to 1.26 ± 0.14 µm (n = 17); and, notably, the amount of offspring per male in single-pair mating experiments was also fully recovered to WT levels (82 ± 7; n = 15; Fig. 5 J).
YFP-CNBT4A-T9A-S82A carries a mutant version of CNB in which three consensus POLO phosphorylation sites have been substituted by alanine. In larval neuroblasts, YFP-CNBT4A-T9A-S82A retains specific daughter centriole binding, but unlike YFP-CNBWT, it does not rescue the loss of PCM and microtubule aster during interphase caused by Cnb loss of function (Januschke et al., 2013). The efficient rescue of Cnb mutant traits in spermatogenesis by YFP-CNBT4A-T9A-S82A strongly suggests that CNB phosphorylation by POLO in these three critical sites is not necessary for CNB function during spermatogenesis, hence underpinning the diversity of cell type–specific molecular functions of CNB.
The different, and in some aspects opposite, roles of CNB in basal body structure and ciliogenesis in type I sensory neurons and spermiogenesis are remarkable. In neurons, CNB inhibits basal body function on the centriole to which it binds that in WT cells is only the daughter centriole (Gottardo et al., 2015). In primary spermatocytes, however, CNB localizes to both mother and daughter centrioles, which in these cells are equally able basal bodies, and acts as a positive regulator of ciliogenesis (Tates, 1971; Fritz-Niggli and Suda, 1972; Gottardo et al., 2013). One conspicuous difference between these two cell types is that basal bodies in spermatocytes contain C-tubules, whose assembly, positioning, and stability requires CNB. In vertebrates, where centrioles also have C-tubules, a recent study has found that in addition to its primary localization to daughter centrioles, CNTROB associates with mother centrioles at the onset of ciliogenesis, and CNTROB loss causes defective axonemal extension (Ogungbenro et al., 2018). These results suggest that the different effect that CNB depletion has on ciliogenesis in Drosophila neurons and spermatocytes may reflect the doublet versus triplet structure of the corresponding basal bodies.
Materials and methods
Fly stocks and husbandry
The following fly stocks were used in this study: PBac{RB}Cnbe00267 (Exelexis); Df(3L)ED4284 (Bloomington Drosophila Stock Center); pUbqYFP-ASL (Rebollo et al., 2007); ANA1-GFP (from T. Avidor-Reiss; Blachon et al., 2008); pUbq YFP-CNB and pUbq YFP-CNBT4A-T9A-S82A (Januschke et al., 2013); and UNC-GFP (Baker et al., 2004). Throughout the study, Cnb mutant refers to PBac{RB}Cnbe00267/Df(3L)ED4284. This allelic combination causes the loss of interphase asters in larval neuroblasts as effectively as Cnb-RNAi driven from ubiquitous promoters. However, unlike RNAi-driven CNB depletion, this allelic combination does not have any significant effect on adult viability (Januschke et al., 2013). All crosses were performed at 25°C.
Fertility tests
Fertility tests were performed by scoring the offspring from each of 15 males individually mated to three w1118 females.
Immunocytochemistry
Testes from pupae were dissected in PBS and placed in a small drop of 5% glycerol in PBS on a glass slide, squashed under a small coverslip, and frozen in liquid nitrogen. After removal of the coverslip, the samples were immersed in methanol for 10 min at −20°C followed by 15 min in PBS and 1 h in PBS containing 0.1% BSA (PBS-BSA). Samples were then incubated with primary antibodies in a humid chamber either for 1 h at room temperature or overnight at 4°C, washed in PBS-BSA three times for 10 min, incubated for 1 h at room temperature with Alexa Fluor–conjugated secondary antibodies (Invitrogen), and then washed again as before. Immunostained preparations were mounted in small drops of Vectashield (Vector Laboratories). Images were taken with either an AxioImager Z1 microscope equipped with an AxioCam HR cooled charge-coupled camera using a 100× objective (ZEISS) or an SP8 confocal lens mounted on a DMIRBE2 microscope using an 63× 1.47 NA objective (Leica Microsystems). Acquired images were processed using ImageJ (1.48s; National Institutes of Health). Grayscale digital images were collected separately and then pseudocolored and merged using Photoshop (7.0; Adobe). Final figures were prepared using Illustrator (Adobe). First antibodies: rabbit anti–DSas-4 (from J. Gopalakrishnan; Gopalakrishnan et al., 2011), DM1a anti–α-TUB (1:500; T9026; Sigma-Aldrich), rabbit anti-Asl (1:500; Januschke et al., 2013), mouse antiacetylated Tub (1:500; T6793; Sigma-Aldrich), chicken anti-PLP (from A. Rodrigues-Martins; Rodrigues-Martins et al., 2007), and rabbit anti-CNB (1:500). Alexa Fluor conjugates were used at 1:1,000 dilution. DNA was stained with DAPI.
Production of anti-CNB antibody
A PCR fragment encoding amino acids 235–549 of CNB was subcloned into the pHAT vector (European Molecular Biology Laboratory). The resulting 6×His protein fusion protein was purified using TALON resin (Takara Bio Inc.) according to the manufacturer’s instructions and was used to generate rabbit antibodies at Harlan (ENVIGO).
Fluorescence-based estimation of centriole length
As a proxy for centriole length, we measured the distance between the distal end of the YFP-ASL signal and the point at which both centrioles intersect on the internal part of the V shape. Because emitted light scatters over an area that is larger than the actual fluorescent particle, measurements based on fluorescent labels overestimate the actual particle’s size. EM-based measurements provided much more precise data.
Phase-contrast microscopy
Phase-contrast microscopy of nonfixed testes was performed as previously described (Glover and González, 1993). In brief, testes were dissected and placed in a small drop of PBS on a siliconized slide, cut at about one third from its apical end with tungsten needles, covered with a coverslip, and gently squashed by removing the PBS with a small piece of blotting paper. We used a 40× dry phase-contrast objective. Images were acquired with an AxioCam MRm camera (ZEISS).
Transmission EM
Testes isolated from larvae and pupae were prefixed in 2.5% glutaraldehyde buffered in PBS overnight at 4°C. After prefixation, the material was carefully rinsed in PBS and postfixed in 1% osmium tetroxide in PBS for 2 h at 4°C. Samples were then washed in the same buffer, dehydrated in a graded series of ethanol, embedded in a mixture of epon-Araldite, and polymerized at 60°C for 48 h. 50–60-nm-thick sections were obtained with an Ultracut E ultramicrotome (Reichert) equipped with a diamond knife, mounted on copper grids, and stained with uranyl acetate and lead citrate. Images were taken with either a CM 10 transmission electron microscope (Philips) operating at an accelerating voltage of 80 kV or a Tecnai Spirit transmission electron microscope (FEI) operating at 100 kV equipped with a Morada charge-coupled device camera (Olympus).
Statistical analysis
We performed one-way ANOVA tests using Prism (GraphPad Software). Error bars represent SEM.
Online supplemental material
Fig. S1 shows EM sections showing that basal body–CLR complexes in Cnb mutant early spermatids retain the ultrastructural defects observed in early spermatocytes, immunofluorescence and phase-contrast microscopy documenting normal meiosis in Cnb mutant males, and immunofluorescence staining of CNB in WT spermatids. Fig. S2 shows ultrastructural details of a Cnb mutant axoneme that presents triplets with open sheaths as well as immunofluorescence showing scattered nuclei over the tails in Cnb mutant spermatids. Table S1 shows the number of C-tubules observed in 11 Cnb mutant basal bodies. Table S2 shows the mother and daughter basal body length in WT and Cnb mutant males.
Supplementary Material
Acknowledgments
We thank T. Avidor-Reiss, J. Gopalakrishnan, A. Rodrigues-Martins, and the Bloomington Drosophila Stock Center for providing antibodies and fly lines, and G. Pollarolo for helpful discussions and suggestions.
Work in our laboratory is supported by Ministerio de Economía y Competitividad (MINECO; Government of Spain) grants BFU2015-66304-P and Redes de Excelencia BFU2014-52125-REDT-CellSYS as well as Generalitat de Catalunya grant SGR Agaur 2014-100. The Institute for Research in Biomedicine (IRB Barcelona) is the recipient of a Severo Ochoa Award of Excellence from MINECO. M. Gottardo is supported by a fellowship from the Alexander von Humboldt Foundation.
The authors declare no competing financial interests.
Author contributions: M. Gottardo, S. Llamazares, J. Reina, M.G. Riparbelli, G. Callaini, and C. Gonzalez conceived and designed the experiments and analyzed the data. J. Reina generated the antibodies. S. Llamazares and G. Callaini performed immunofluorescence microscopy. M. Gottardo, M.G. Riparbelli, and G. Callaini performed EM. C. Gonzalez wrote the manuscript.
References
- Baker J.D., Adhikarakunnathu S., and Kernan M.J.. 2004. Mechanosensory-defective, male-sterile unc mutants identify a novel basal body protein required for ciliogenesis in Drosophila. Development. 131:3411–3422. 10.1242/dev.01229 [DOI] [PubMed] [Google Scholar]
- Basiri M.L., Ha A., Chadha A., Clark N.M., Polyanovsky A., Cook B., and Avidor-Reiss T.. 2014. A migrating ciliary gate compartmentalizes the site of axoneme assembly in Drosophila spermatids. Curr. Biol. 24:2622–2631. 10.1016/j.cub.2014.09.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blachon S., Gopalakrishnan J., Omori Y., Polyanovsky A., Church A., Nicastro D., Malicki J., and Avidor-Reiss T.. 2008. Drosophila asterless and vertebrate Cep152 Are orthologs essential for centriole duplication. Genetics. 180:2081–2094. 10.1534/genetics.108.095141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blachon S., Cai X., Roberts K.A., Yang K., Polyanovsky A., Church A., and Avidor-Reiss T.. 2009. A proximal centriole-like structure is present in Drosophila spermatids and can serve as a model to study centriole duplication. Genetics. 182:133–144. 10.1534/genetics.109.101709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho-Santos Z., Machado P., Alvarez-Martins I., Gouveia S.M., Jana S.C., Duarte P., Amado T., Branco P., Freitas M.C., Silva S.T., et al. 2012. BLD10/CEP135 is a microtubule-associated protein that controls the formation of the flagellum central microtubule pair. Dev. Cell. 23:412–424. 10.1016/j.devcel.2012.06.001 [DOI] [PubMed] [Google Scholar]
- Dutcher S.K., and Trabuco E.C.. 1998. The UNI3 gene is required for assembly of basal bodies of Chlamydomonas and encodes delta-tubulin, a new member of the tubulin superfamily. Mol. Biol. Cell. 9:1293–1308. 10.1091/mbc.9.6.1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz-Niggli H., and Suda T.. 1972. Bildung und Bedeutung der Zentriolen: Eine Studie und Neuinterpretation der Meiose von Drosophila [Formation and significance of centrioles: A study and new interpretation of the meiosis of Drosophila]. Cytobiologie. 5:12–41. [Google Scholar]
- Fu J., and Glover D.M.. 2012. Structured illumination of the interface between centriole and peri-centriolar material. Open Biol. 2:120104 10.1098/rsob.120104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galletta B.J., Guillen R.X., Fagerstrom C.J., Brownlee C.W., Lerit D.A., Megraw T.L., Rogers G.C., and Rusan N.M.. 2014. Drosophila pericentrin requires interaction with calmodulin for its function at centrosomes and neuronal basal bodies but not at sperm basal bodies. Mol. Biol. Cell. 25:2682–2694. 10.1091/mbc.E13-10-0617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garreau de Loubresse N., Ruiz F., Beisson J., and Klotz C.. 2001. Role of delta-tubulin and the C-tubule in assembly of Paramecium basal bodies. BMC Cell Biol. 2:4 10.1186/1471-2121-2-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover D.M., and González C.. 1993. Techniques for studying mitosis in Drosophila. In The Cell Cycle: a Practical Approach. Brookes R., and Fantes P., editors. IRL Press, Oxford, England, UK: 163–168. [Google Scholar]
- González C., Casal J., and Ripoll P.. 1989. Relationship between chromosome content and nuclear diameter in early spermatids of Drosophila melanogaster. Genet. Res. 54:205–212. 10.1017/S0016672300028664 [DOI] [PubMed] [Google Scholar]
- Gopalakrishnan J., Mennella V., Blachon S., Zhai B., Smith A.H., Megraw T.L., Nicastro D., Gygi S.P., Agard D.A., and Avidor-Reiss T.. 2011. Sas-4 provides a scaffold for cytoplasmic complexes and tethers them in a centrosome. Nat. Commun. 2:359 10.1038/ncomms1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottardo M., Callaini G., and Riparbelli M.G.. 2013. The cilium-like region of the Drosophila spermatocyte: an emerging flagellum? J. Cell Sci. 126:5441–5452. 10.1242/jcs.136523 [DOI] [PubMed] [Google Scholar]
- Gottardo M., Pollarolo G., Llamazares S., Reina J., Riparbelli M.G., Callaini G., and Gonzalez C.. 2015. Loss of Centrobin Enables Daughter Centrioles to Form Sensory Cilia in Drosophila. Curr. Biol. 25:2319–2324. 10.1016/j.cub.2015.07.038 [DOI] [PubMed] [Google Scholar]
- Gudi R., Zou C., Li J., and Gao Q.. 2011. Centrobin-tubulin interaction is required for centriole elongation and stability. J. Cell Biol. 193:711–725. 10.1083/jcb.201006135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudi R., Zou C., Dhar J., Gao Q., and Vasu C.. 2014. Centrobin-centrosomal protein 4.1-associated protein (CPAP) interaction promotes CPAP localization to the centrioles during centriole duplication. J. Biol. Chem. 289:15166–15178. 10.1074/jbc.M113.531152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudi R., Haycraft C.J., Bell P.D., Li Z., and Vasu C.. 2015. Centrobin-mediated regulation of the centrosomal protein 4.1-associated protein (CPAP) level limits centriole length during elongation stage. J. Biol. Chem. 290:6890–6902. 10.1074/jbc.M114.603423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Januschke J., Llamazares S., Reina J., and Gonzalez C.. 2011. Drosophila neuroblasts retain the daughter centrosome. Nat. Commun. 2:243 10.1038/ncomms1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Januschke J., Reina J., Llamazares S., Bertran T., Rossi F., Roig J., and Gonzalez C.. 2013. Centrobin controls mother-daughter centriole asymmetry in Drosophila neuroblasts. Nat. Cell Biol. 15:241–248. 10.1038/ncb2671 [DOI] [PubMed] [Google Scholar]
- Jeffery J.M., Urquhart A.J., Subramaniam V.N., Parton R.G., and Khanna K.K.. 2010. Centrobin regulates the assembly of functional mitotic spindles. Oncogene. 29:2649–2658. 10.1038/onc.2010.37 [DOI] [PubMed] [Google Scholar]
- Jeffery J.M., Grigoriev I., Poser I., van der Horst A., Hamilton N., Waterhouse N., Bleier J., Subramaniam V.N., Maly I.V., Akhmanova A., and Khanna K.K.. 2013. Centrobin regulates centrosome function in interphase cells by limiting pericentriolar matrix recruitment. Cell Cycle. 12:899–906. 10.4161/cc.23879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong Y., Lee J., Kim K., Yoo J.C., and Rhee K.. 2007. Characterization of NIP2/centrobin, a novel substrate of Nek2, and its potential role in microtubule stabilization. J. Cell Sci. 120:2106–2116. 10.1242/jcs.03458 [DOI] [PubMed] [Google Scholar]
- Lerit D.A., and Rusan N.M.. 2013. PLP inhibits the activity of interphase centrosomes to ensure their proper segregation in stem cells. J. Cell Biol. 202:1013–1022. 10.1083/jcb.201303141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liška F., Gosele C., Popova E., Chylíková B., Křenová D., Křen V., Bader M., Tres L.L., Hubner N., and Kierszenbaum A.L.. 2013. Overexpression of full-length centrobin rescues limb malformation but not male fertility of the hypodactylous (hd) rats. PLoS One. 8:e60859 10.1371/journal.pone.0060859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., and Jarman A.P.. 2011. Dilatory is a Drosophila protein related to AZI1 (CEP131) that is located at the ciliary base and required for cilium formation. J. Cell Sci. 124:2622–2630. 10.1242/jcs.084798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogungbenro Y.A., Tena T.C., Gaboriau D., Lalor P., Dockery P., Philipp M., and Morrison C.G.. 2018. Centrobin controls primary ciliogenesis in vertebrates. J. Cell Biol. 217:1205–1215. 10.1083/jcb.201706095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebollo E., Sampaio P., Januschke J., Llamazares S., Varmark H., and González C.. 2007. Functionally unequal centrosomes drive spindle orientation in asymmetrically dividing Drosophila neural stem cells. Dev. Cell. 12:467–474. 10.1016/j.devcel.2007.01.021 [DOI] [PubMed] [Google Scholar]
- Riparbelli M.G., and Callaini G.. 2011. Male gametogenesis without centrioles. Dev. Biol. 349:427–439. 10.1016/j.ydbio.2010.10.021 [DOI] [PubMed] [Google Scholar]
- Riparbelli M.G., Callaini G., and Megraw T.L.. 2012. Assembly and persistence of primary cilia in dividing Drosophila spermatocytes. Dev. Cell. 23:425–432. 10.1016/j.devcel.2012.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riparbelli M.G., Cabrera O.A., Callaini G., and Megraw T.L.. 2013. Unique properties of Drosophila spermatocyte primary cilia. Biol. Open. 2:1137–1147. 10.1242/bio.20135355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues-Martins A., Bettencourt-Dias M., Riparbelli M., Ferreira C., Ferreira I., Callaini G., and Glover D.M.. 2007. DSAS-6 organizes a tube-like centriole precursor, and its absence suggests modularity in centriole assembly. Curr. Biol. 17:1465–1472. 10.1016/j.cub.2007.07.034 [DOI] [PubMed] [Google Scholar]
- Shin W., Yu N.K., Kaang B.K., and Rhee K.. 2015. The microtubule nucleation activity of centrobin in both the centrosome and cytoplasm. Cell Cycle. 14:1925–1931. 10.1080/15384101.2015.1041683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P., Ramdas Nair A., and Cabernard C.. 2014. The centriolar protein Bld10/Cep135 is required to establish centrosome asymmetry in Drosophila neuroblasts. Curr. Biol. 24:1548–1555. 10.1016/j.cub.2014.05.050 [DOI] [PubMed] [Google Scholar]
- Tates A.D. 1971. Cytodifferentiation during spermatogenesis in Drosophila melanogaster: an electron microscope study. Proefschrift, Rijksuniversiteit te Leiden, Leiden, The Netherlands: 162. [Google Scholar]
- Vieillard J., Paschaki M., Duteyrat J.L., Augière C., Cortier E., Lapart J.A., Thomas J., and Durand B.. 2016. Transition zone assembly and its contribution to axoneme formation in Drosophila male germ cells. J. Cell Biol. 214:875–889. 10.1083/jcb.201603086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Ramani A., Soni K., Gottardo M., Zheng S., Ming Gooi L., Li W., Feng S., Mariappan A., Wason A., et al. 2016. Molecular basis for CPAP-tubulin interaction in controlling centriolar and ciliary length. Nat. Commun. 7:11874 10.1038/ncomms11874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou C., Li J., Bai Y., Gunning W.T., Wazer D.E., Band V., and Gao Q.. 2005. Centrobin: a novel daughter centriole-associated protein that is required for centriole duplication. J. Cell Biol. 171:437–445. 10.1083/jcb.200506185 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.