Dear Editor,
Non-Hodgkin’s lymphoma (NHL) is a type of cancer that mainly develops from B-cell malignancies, causing 231,400 deaths in 2015 globally. According to the American Cancer Society, around 66,000 new cases of NHL are diagnosed each year in the United States.1 Bruton’s tyrosine kinase (BTK) is an enzyme encoded by the BTK gene in human. BTK is expressed in all cell lineages of the hematopoietic system except for T cells.2 As a cytoplasmic tyrosine kinase of the TEC family, BTK plays a crucial role in B cell development, differentiation, and signaling. BTK is closely associated with chronic B-cell receptor (BCR) activation, and is critical for the survival of B-cell neoplasms.3
Inhibition of BTK kinase activity has been proven to be an important and practical way of treating NHL. Ibrutinib is a first-in-class, covalent BTK inhibitor that is able to bind C481 (Cysteine481) of BTK with an ideal IC50 of 0.5 nM.4,5 Ibrutinib was approved by the FDA in 2013 to 2015 for the treatment of several types of NHL, specifically relapsed/refractory mantle cell lymphoma (MCL), chronic lymphocytic leukemia (CLL), and Waldenström macroglobulinaemia (WM). Currently, there is an ongoing clinical trial on ibrutinib for its efficacy in DLBCL treatment. However, resistance to ibrutinib has been reported in various lymphomas, including CLL and MCL, due to a C481S (cysteine to serine mutation at position 481) BTK mutation.6 Because ibrutinib cannot form a covalent bond with the hydroxyl group of serine, C481S mutation increases the IC50 against BTK-C481S phosphorylation from 2.2 nM to 1 μM.7 Thus, there is an urgent need to develop a new strategy against C481S mutation-induced resistance. In addition, ibrutinib has been reported to induce a variety of side effects, including arthralgias, myalgias, atrial fibrillation, ecchymosis, and major hemorrhage.8 As ibrutinib shows inhibition of EGFR, ITK and TEC family kinases, with IC50s of around 10–100 nM, the pathogeny could be correlated to these known off-target effects of ibrutinib.
Proteolysis-targeting chimera (PROTAC) has emerged as a novel chemical approach for the selective degradation of cellular proteins, known as chemical knockdown of a protein of interest.9 PROTAC molecules (PROTACs) are small molecules capable of bringing a target protein into the proximity of an E3 ligase of interest, causing consequent degradation of the target protein (Fig. 1a). These heterobifunctional molecules consist of three components: a target protein-binding moiety (targeting arm, TA), a degradation machinery-recruiting unit (degradation arm, DA), and a linker that couples these two functionalities. Typically, the utilized degradation machinery is the ubiquitin–proteasome system (UPS) that recruits an E3 ubiquitin ligase followed by ubiquitination of the target protein and its subsequent degradation by the proteasome. In 2015, Crews10 and Bradner11 independently reported that BRD4, which has been implicated in several different cancers, could be efficiently degraded through the PROTAC technique. This presented the PROTAC technique as a promising alternative approach against cancer. Unlike traditional drugs, PROTACs aim to eliminate proteins with aberrant functions, rather than inhibiting their activity. Therefore, the acquired resistance caused by the C481S BTK mutant could, in principle, be overcome by using PROTAC molecules.
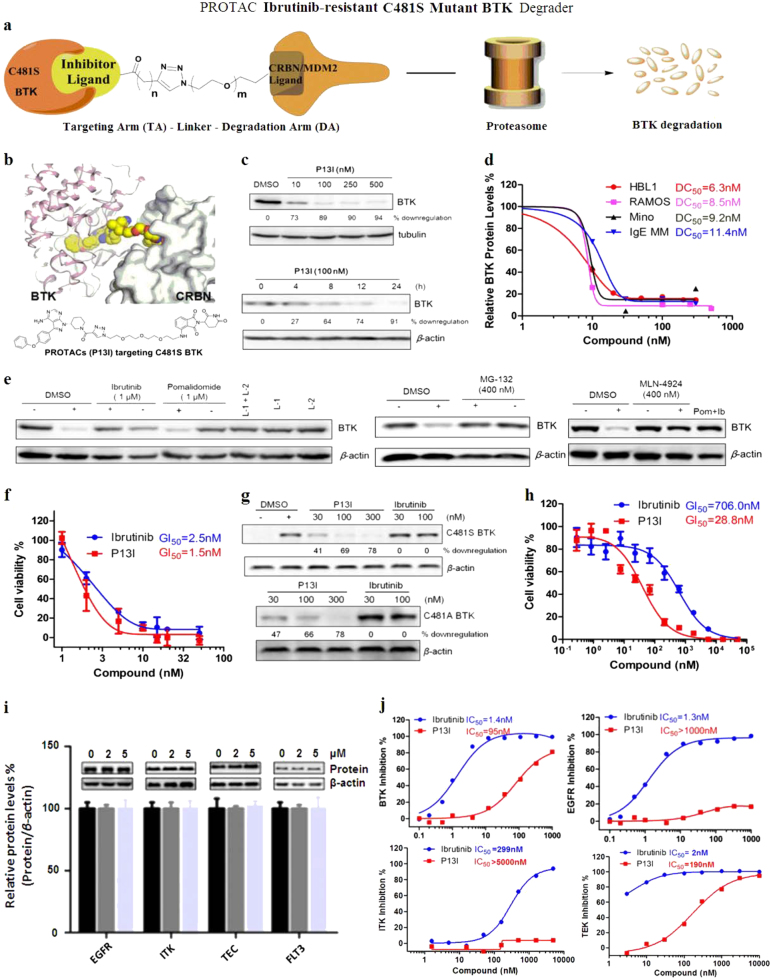
Fig. 1.
Schematic representation and characterization of the C481S mutant ibrutinib-resistant BTK degrader P13I. a Brief introduction of the newly developed C481S BTK degraders. b Schematic representation of BTK degrader design. c Immunoblot for BTK and β-actin after 48 h of treatment of RAMOS cells with the indicated concentrations of P13I. Immunoblot for BTK and β-actin after treatment of HBL-1 cells with 100 nM P13I for the indicated exposures. d DC50 (50% protein degradation concentration) of P13I for BTK in RAMOS, HBL-1, IgE MM, or Mino cells. e Immunoblot for BTK and β-actin after a 2-h pretreatment with DMSO, ibrutinib (1 μM), or pomalidomide (1 μM), followed by a 48-h P13I treatment (100 nM) in RAMOS cells. Concentration of L-1 and L-2 is 100 nM; Immunoblot for BTK and β-actin after a 4-h pretreatment with DMSO or MLN-4924 (400 nM), followed by a 48-h P13I treatment (100 nM) in RAMOS cells. Pom = Pomalidomide, Ib = Ibrutinib. Concentration of pomalidomide and ibrutinib is 1 μM; Immunoblot for BTK and β-actin after a 4-h pretreatment with DMSO or MG-132 (400 nM), followed by a 12-h P13I treatment (100 nM) in RAMOS cells. “+” refers to with treatment of P13I, “−” refers to without treatment of P13I. f In vitro study of inhibition effect on wild type HBL-1 cell lines by the BTK degrader P13I and ibrutinib. In 96-well plates, 3000 cells were incubated in each well at 37 °C for 72–96 h. The final GI50 was calculated by CCK-8. g Plasmids expressing full-length constructs of C481S or C481A BTK were transfected into HeLa cells by PEI, “+” refers to transfection, “−” refers to no transfection. Immunoblot for C481S or C481A BTK and β-actin after 4 days of treatment of HeLa cells with the indicated concentrations of P13I. h In vitro study of the inhibition effect on HBL-1 (C481S mutant BTK over-expression) cell lines by the BTK degrader P13I and ibrutinib. In 96-well plates, 3000 cells were incubated in each well at 37 °C for 72–96 h. The final EC50 was calculated by MTT. i Immunoblot for EGFR, ITK, TEC, FLT-3 and β-actin after 48 h of treatment of HeLa, Jurkat, or HBL-1 cells with the indicated concentrations of P13I. j IC50 of ibrutinib or P13I for the inhibition of WT BTK, EGFR, ITK and TEC
In this study, for the first time, we report the development of BTK-targeting degraders using the PROTAC strategy. These PROTACs could efficiently degrade ibrutinib-sensitive BTK-WT (wild type). More importantly, our newly designed PROTACs also significantly induced the degradation of ibrutinib-resistant BTK-C481S (50% degradation efficiency at 30 nM). Furthermore, our PROTAC molecules efficiently inhibited cell proliferation and colony formation, while exhibited no obvious inhibition (>1000 nM) of ITK, EGFR, and TEC, which are major off-targets of ibrutinib. These data demonstrate the strong potential for developing PROTAC-based therapeutic molecules.
To develop BTK-targeting PROTAC degraders, a BTK-targeting arm was conjugated to a BTK-degradation arm by linkers with variable lengths (Fig. 1a, b, for details, please see Supplementary Information). As a result, the PROTAC molecules should bind both BTK and E3 ligase through the corresponding targeting arm and degradation arm, respectively. The drugs ibrutinib and spebrutinib were selected as the BTK-binding ligands, and pomalidomide (CRBN ligand) and RG-7112 (MDM2 ligand) were employed as corresponding E3 ligase binding partners. Based on our design principles (for details, please see Supplementary Information), a variety of BTK-targeting PROTAC molecules were prepared and evaluated. We found that CRBN-recruiting PROTACs were generally more effective than MDM2-recruiting ones. Among these CRBN-recruiting PROTACs, P13I with the conjugation of ibrutinib and pomalidomide demonstrated the best degrading ability (Fig. 1b). P13I induced 73% degradation of BTK at 10 nM and 89% at 100 nM in human Burkitt’s lymphoma, RAMOS cells. As a further validation, we examined the kinetics of P13I-induced BTK degradation in human ABC-DLBCL, HBL-1 cells. Western blot analysis showed that BTK degradation began at roughly 4 h, and was completed by around 24 h (Fig. 1c). With P13I treatment, half-lives of BTK and C481S BTK were 4 h and 3 h, respectively. The results demonstrated that P13I could accelerate BTK degradation (for details, please see SI). Moreover, P13I could also efficiently degrade BTK in other NHL cell lines including MCL (Mino cells) and MM cell lines with a DC50 (50% protein degradation concentration) of 9.2 nM and 11.4 nM, respectively (Fig. 1d).
Control experiments clearly demonstrated that ibrutinib, pomalidomide or unconjugated PROTAC arms (TA and DA) could not induce degradation of BTK (Fig. 1e). In contrast, ibrutinib or pomalidomide could competitively inhibit the degradation effect of P13I. Additionally, MG-132, a proteasome inhibitor, could completely disable the PROTAC effect. The same competitive inhibition was also observed with the neddylation inhibitor MLN-4924, which is in accordance with the requirement by CRBN for neddylation for processive E3 ligase activity (Fig. 1e). The above observations confirmed that degradation of BTK was mediated by the ubiquitin–proteasome system (UPS). In order to assess the effect of P13I in vitro, the DLBCL cell line HBL-1 was used since its viability and growth is dependent on BTK.3 HBL-1 cells were treated with increasing concentrations of P13I and ibrutinib for 72 h and the corresponding inhibition of cell proliferation was detected with CCK-8. Notably, P13I had a GI50 (50% growth inhibition concentration) of 1.5 nM, which is slightly better than that of ibrutinib (Fig. 1f). It is also noteworthy that the known IC50 of ibrutinib is 0.5 nM against pBTK, and is considerably less than P13I (95 nM) (Fig. 1j), suggesting that the observed effect of P13I is via degrading BTK rather than inhibiting it. In the RAMOS cell line, whose viability and growth is not dependent on BTK,3 P13I and ibrutinib did not show any inhibitory effect, indicating that P13I may not display any non-specific cytotoxic effects.
As mentioned above, the BTK C481S mutant is the main cause of ibrutinib resistance in reported clinical cases.6 Therefore, we transduced a recombinant BTK mutant plasmid into 293T and HeLa cell lines. The results showed that P13I could effectively degrade the C481S mutant BTK at a low concentration of 30 nM (Fig. 1g). However, ibrutinib could not inhibit the self-phosphorylation of C481S mutant BTK even at a 1 μM concentration.7 As reported, BTKCys481Ser DLBCL cell line was used as a model system to address ibrutinib resistance.12 Indeed, the proliferation of a DLBCL cell line, HBL-1 cells expressing BTK C481S mutant was significantly inhibited by P13I with a GI50 of around 28 nM. In contrast, ibrutinib lost its efficacy, with a GI50 of around 700 nM (Fig. 1h). These results strongly indicate that the BTK-targeting PROTAC degraders represent a new strategy for ibrutinib-resistant lymphoma treatment. Besides BTK, it has been reported that EGFR, ITK, and TEC family kinases can also be inhibited by low concentrations of ibrutinib, resulting in serious side effects in patients. In order to test the potential side effects of the BTK degraders, we analyzed the interaction between these other proteins and P13I in different cell lines. There was no degradation even at 5000 nM of PI3I (Fig. 1i). Furthermore, P13I demonstrated almost no inhibitory activity against ITK or EGFR (>1000 nM), which indicates that BTK degraders will unlikely lead to the aforementioned side effects of ibrutinib (Fig. 1j). These data collectively demonstrate that the BTK-targeting PROTAC molecule P13I could be of clinical significance in the treatment of B-cell malignancies.
In summary, a novel PROTAC strategy was developed with high efficacy and specificity for ibrutinib-resistant BTK degradation. Remarkably, the representative ibrutinib-mediated PROTAC P13I could efficiently degrade both wild type and the newly reported ibrutinib-resistant C481S BTK in lymphoma cell lines with high efficacy. Furthermore, P13I significantly inhibited the proliferation and colony formation potential of BTK C481S mutant HBL-1 cells (ibrutinib-resistant) in vitro. Moreover, the BTK degrader P13I had almost no effect on ITK, EGFR and TEC family kinases, which are known off-targets of ibrutinib and lead to serious side effects. These findings offer new opportunities to develop more efficient treatments for Non-Hodgkin lymphoma, including ibrutinib-resistant cases. More importantly, our results indicate that the PROTAC strategy (protein degradation instead of inhibition) could be adopted as a general and powerful therapeutic treatment for drug-resistant cancers in the future.
Electronic supplementary material
Acknowledgements
This work was supported by the National Natural Science Foundation of China (#81573277, 81622042, 81773567), National Major Scientific and Technological Special Project for “Significant New Drugs Development” (#SQ2017ZX095003) and Tsinghua University Initiative Scientific Research Program. This work was also financially supported by the NSFC (No. 81670187) and Beijing Natural Science Foundation (No. 7172047). We thank Prof. Haitao Li and Prof. Qinghua Tao of Tsinghua University for helpful discussions.
Competing interests
The authors declare no competing interests.
Contributor Information
Wanli Liu, Email: liuwanli@biomed.tsinghua.edu.cn.
Yu Rao, Email: yrao@tsinghua.edu.cn.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41422-018-0055-1.
References
- 1.Morton LM, et al. Blood. 2006;107:265–276. doi: 10.1182/blood-2005-06-2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mohamed AJ, et al. Immunol. Rev. 2009;228:58–73. doi: 10.1111/j.1600-065X.2008.00741.x. [DOI] [PubMed] [Google Scholar]
- 3.Davis RE, et al. Nature. 2010;463:88–92. doi: 10.1038/nature08638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pan Z, et al. ChemMedChem. 2007;2:58–61. doi: 10.1002/cmdc.200600221. [DOI] [PubMed] [Google Scholar]
- 5.Honigberg LA, et al. Proc Natl Acad Sci USA. 2010;107:13075–13080. doi: 10.1073/pnas.1004594107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woyach A, et al. N. Engl. J. Med. 2014;370:2286–2294. doi: 10.1056/NEJMoa1400029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Furman RR, et al. N. Engl. J. Med. 2014;370:2352–2354. doi: 10.1056/NEJMc1402716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Byrd JC, et al. N. Engl. J. Med. 2016;374:323–332. doi: 10.1056/NEJMoa1509981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sakamoto KM, et al. Proc Natl Acad Sci USA. 2001;98:8554–8559. doi: 10.1073/pnas.141230798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu J, et al. Chem. Biol. 2015;22:755–763. doi: 10.1016/j.chembiol.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winter GE, et al. Science. 2015;348:1376–1381. doi: 10.1126/science.aab1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen JG, et al. Blood. 2018;131:2047–2059. doi: 10.1182/blood-2017-10-811752. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.