Abstract
Finding the keys to understanding the infectious process of Yersinia ruckeri was not a priority for many years due to the prompt development of an effective biotype 1 vaccine which was used mainly in Europe and USA. However, the gradual emergence of outbreaks in vaccinated fish, which have been reported since 2003, has awakened interest in the mechanism of virulence in this pathogen. Thus, during the last two decades, a large number of studies have considerably enriched our knowledge of many aspects of the pathogen and its interaction with the host. By means of both conventional and a variety of novel strategies, such as cell GFP tagging, bioluminescence imaging and optical projection tomography, it has been possible to determine three putative Y. ruckeri infection routes, the main point of entry for the bacterium being the gill lamellae. Moreover, a wide range of potential virulence factors have been highlighted by specific gene mutagenesis strategies or genome-wide transposon/plasmid insertion-based screening approaches, such us in vivo expression technology (IVET) and signature tagged mutagenesis (STM). Finally, recent proteomic and whole genomic analyses have allowed many of the genes and systems that are potentially implicated in the organism's pathogenicity and its adaptation to the host environmental conditions to be elucidated. Altogether, these studies contribute to a better understanding of the infectious process of Y. ruckeri in fish, which is crucial for the development of more effective strategies for preventing or treating enteric redmouth disease (ERM).
Keywords: Yersinia ruckeri, infection route, virulence genes, comparative genome, proteome analysis
Introduction
Yersinia ruckeri is able to infect different fish species such as carp, catfish, sturgeon, perch and burbot (Tobback et al., 2007; Kumar et al., 2015), although it mainly affects salmonids. Interestingly, Y. ruckeri has recently been isolated as an unusual microorganism in a human wound infection (De Keukeleire et al., 2014) as well as in milk, cheese, chicken and minced meat (Özdemir and Arslan, 2015). In salmonids it causes enteric red mouth disease (ERM), which is important owing to the economic losses it causes in the aquaculture industry. ERM is a systemic disease affecting fish in all stages of development, causing a high degree of mortality. The disease occurs acutely in juveniles and has a tendency to become chronic in adult fish (Tobback et al., 2007). Some infected fish are asymptomatic, becoming carriers of the pathogen and acting as reservoirs of the bacterium. It is important to highlight that Y. ruckeri is a facultative intracellular bacterium, capable of surviving inside macrophages (Ryckaert et al., 2010).
Y. ruckeri strains have been classified into four serotypes (Romalde et al., 1993), the main outbreaks in fish farms being caused by serotype O1 biotype I, although other serotypes, particularly serotype O2, can also be involved (Romalde et al., 2003). Vaccination is the most effective way to control the disease. ERM biotype 1 vaccine, which was the first to be available for fish (Amend et al., 1983), has been used since the early 80s and is very effective in the prevention of the disease. However, during recent years worldwide outbreaks have been produced mainly by novel non-motile and lipase-negative strains, belonging to biotype II (Austin et al., 2003; Fouz et al., 2006; Arias et al., 2007; Calvez et al., 2014). These phenotypes have no appreciable effect on the virulence of the pathogen (Evenhuis et al., 2009; Welch et al., 2011), but the biotype 2 strains are able to elude the protection provided by the vaccine, probably due to the antigenic differences existing in their O-antigen with respect to biotype 1 strains (Tinsley et al., 2011). This has promoted the development and subsequent commercialization of a new vaccine which simultaneously confers protection against the two biotypes (Tinsley et al., 2011).
Owing to the prompt development of the vaccine, the study of the virulence mechanisms of this bacterium has been side-lined for a long time. Nevertheless, during the last few years a considerable number of reports related to the route of Y. ruckeri infection or its virulence, as well as several comparative genomic and proteomic analyses have been published. All of this knowledge about the infectious process of this bacterium is condensed and critically reviewed in the present article.
Yersinia ruckeri infection route
Studies aimed at determining the infection path and invasion mechanism of fish by Y. ruckeri have been carried out with different strains, modes of infection and experimental conditions, which sometimes makes it difficult to draw a conclusion. In addition, various tools have been developed and used for elucidating the interaction between Y. ruckeri and rainbow trout, its natural host.
Bacterial counting in fish tissues at different times after experimental infection (Tobback et al., 2009, 2010); GFP tagging of Y. ruckeri and further visualization by epifluorescence or flow cytometry in tissues (Welch and Wiens, 2005); in situ hybridization (Khimmakthong et al., 2013); bioluminescence imaging using strains harboring the luxCDABE operon from Photorhabdus luminescens (Méndez and Guijarro, 2013) and optical projection tomography (OPT) (Ohtani et al., 2014), a recently developed three-dimensional bioimaging technique combined with immunohistochemical assay (IHC) (Tobback et al., 2009; Ohtani et al., 2014), have all been used. Each of these technologies provides a different approach to determining the route of entry and tissue distribution of Y. ruckeri. Although all of them have limitations, if we analyse the results obtained as a whole, it seems that the gills are the portal of entry for the bacterium into the fish. In effect, most studies have established this organ as the first to be infected (Tobback et al., 2009; Khimmakthong et al., 2013; Ohtani et al., 2014, 2015), and more specifically the pavement cells of the gill lamellae, from which the bacterium would spread to blood (Ohtani et al., 2014). Thus, Y. ruckeri was detected only 1 min post infection in the blood, where the number of bacteria increased for 40 min, reaching 2.3x105 CFU/ml (Ohtani et al., 2014). Although gills seem to be the main point of entry of Y. ruckeri, other sites in the fish cannot be excluded. In effect, most studies establish the presence of Y. ruckeri in the intestine shortly after infection (Khimmakthong et al., 2013; Ohtani et al., 2014, 2015). It is known that the mucus of the gills is twice as efficient as that of the intestine in enabling the adherence of the bacterium (Tobback et al., 2010). Moreover, after infection, twice as many bacteria were detected in the gills as in the intestine (Tobback et al., 2009; Khimmakthong et al., 2013). However, bacteria have been detected in the intestine from 1 min (Khimmakthong et al., 2013) to 30 min post-infection (Ohtani et al., 2014). Additionally, intestinal dissemination is the main sign of the pathology (Figure 1) (Méndez and Guijarro, 2013; Ohtani et al., 2014). This, together with the fact that it is unusual for an intestinal pathogen to enter the host by any other route than the oral one means that the digestive tract cannot be excluded as another portal of entry. In addition, Ohtani et al. (2014) used OPT analysis to establish a third route of infection for Y. ruckeri, through the skin surface on the lateral line, from which it would penetrate to internal cell layers, a similar result to that found by Khimmakthong et al. (2013). A recent study reported the ability of two isolates of Y. ruckeri to invade different types of salmon epithelial cells (Menanteau-Ledouble et al., 2018), the biotype 1 isolate (ATCC 29473) being more infectious than the non-motile biotype 2 strain (A7959-11). The authors suggested that the bacterium could enter host cells by means of several mechanisms that take advantage of cytoskeletal systems. The capacity to invade the epithelium would undoubtedly facilitate entry into the host.
Figure 1.
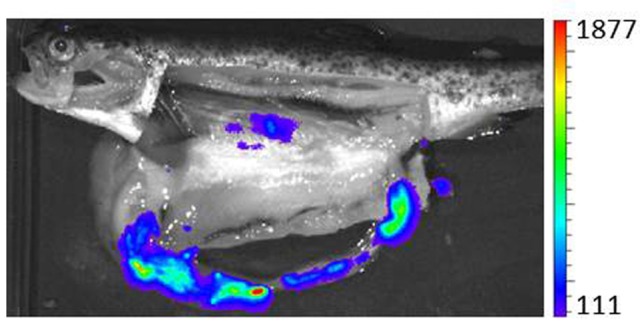
Bioluminescent tracking of Y. ruckeri harboring pCS26-Pac in a fish infected by bath immersion with 107 cfu ml−1 for 1 h. Bioluminescence emitted by the bacterium was captured three days postinfection by Ivis-Lumina equipment. Color standards represent “RLU max.” This measure expresses the highest number of counts in a pixel inside the region analyzed (Figure taken from Méndez and Guijarro, 2013).
In conclusion, all the results indicate that Y. ruckeri can access the interior of rainbow trout by at least three different paths: the gills, lateral line and digestive tract. From these locations bacteria would spread to the blood circulation system to further colonize and infect other internal organs, causing in many cases the death of the fish.
Defined genes related to virulence
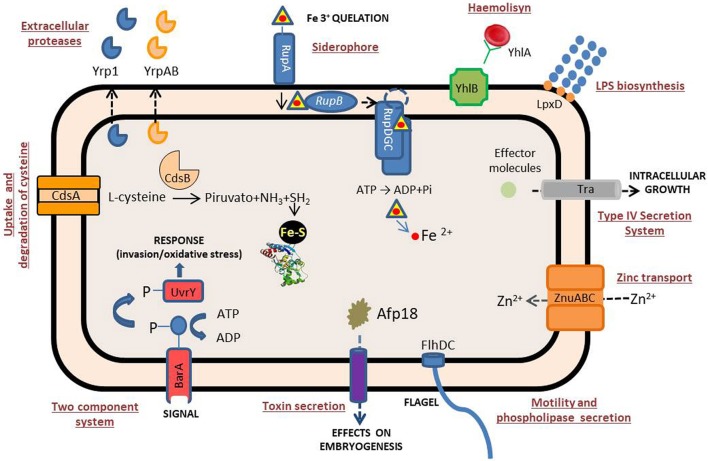
Despite its being a microorganism responsible for a high proportion of losses in continental aquaculture, there are few studies on Y. ruckeri virulence factors, perhaps as a consequence of the early development of a vaccine, which made their study unattractive. Many of the virulence factors described so far refer to extracellular factors (ECF) that are common to a wide range of gram-negative pathogenic bacteria, particularly the enterobacteria group (Table 1, Figure 2). Indeed, extracellular proteases, haemolysins and siderophores are among the main extracellular molecules involved in the Y. ruckeri infection process.
Table 1.
Virulence related genes of Y. ruckeri.
| Gene | Function | References |
|---|---|---|
| yrp1 | Extracellular protease | Secades and Guijarro, 1999 |
| yrpAB | Peptidases | Navais et al., 2014b |
| rucC | Iron captation. Ruckerbactin Siderophore synthesis | Fernandez et al., 2004 |
| yhlA | Hemolysine | Fernández et al., 2007b |
| traI | Type IV secretion system | Méndez et al., 2009 |
| cdsBA | Cysteine transport and degradation | Méndez et al., 2011 |
| znuA | Zinc transport | Dahiya and Stevenson, 2010b |
| uvrY | Response regulator. Two component system | Dahiya and Stevenson, 2010c |
| lpxD | Lipid A biosynthesis | Altinok et al., 2016 |
| afp18 | Toxin. Glycosyltransferase | Jank et al., 2014 |
| flhD | Motility regulation | Jozwick et al., 2016 |
Figure 2.
Schematic representation of the Y. ruckeri systems related to virulence. See text for further details.
Yrp1 is a serralysin family extracellular metalloprotease, widely characterized from an enzymatic point of view (Secades and Guijarro, 1999) and secreted through an ABC transport system (Fernández et al., 2002). It is able to degrade different matrix and muscle proteins such as laminin, fibrinogen, gelatin, actin, and myosin (Fernandez et al., 2003). Although it was shown not to be present in all virulent Y. ruckeri strains (Secades and Guijarro, 1999), its presence makes the bacteria more virulent. Transcriptional fusion between the luxCDABE operon and the yrp1 promoter demonstrated that yrp1 is induced more in vivo than in vitro (Méndez and Guijarro, 2013). This is consistent with the expression of yrp1 in fish tissues and the observation that an yrp1 mutant strain is significantly attenuated in virulence (Fernández et al., 2002). Interestingly, a heat-inactivated Yrp1 toxoid was able to elicit a strong protection against the ERM disease (Fernandez et al., 2003). All of this established a clear and important role for this protease in the pathogenic process.
Additional degrading enzymes were identified in Y. ruckeri, such as two putative peptidases belonging to the U32 family which are encoded by the yrpAB operon (Navais et al., 2014b). Both genes, yrpA and yrpB, are present in a similar genetic organization in different pathogens of the enterobacteria group, including human pathogenic Yersinia species (Navais et al., 2014b). A synergistic effect occurred in the induction of the yrpAB operon under low oxygen conditions when peptones were also present in the culture medium (Navais et al., 2014b). Infection studies with a yrpA mutant strain suggest that the YrpA peptidase contributes to virulence in a similar way to the U32 peptidases of Proteus mirabilis (Zhao et al., 1999) and Helicobacter pylori (Kavermann et al., 2003). A more detailed analysis of the role of these peptidases in the virulence of pathogenic Yersiniae could be interesting from a public health point of view.
Two different genetic approaches have been applied to allow the selection and further identification of Y. ruckeri genes related to the infection process: in vivo expression technology (IVET) (Fernandez et al., 2004) and signature-tagged mutagenesis (STM) (Dahiya and Stevenson, 2010a). Each of these has advantages and disadvantages, but undoubtedly, their application has led to the identification of a number of genes related to Y. ruckeri pathogenesis.
IVET allowed the identification of up to 14 genes specifically induced in vivo, during the infection process of Y. ruckeri. Among them, those related to two-component and type IV secretion systems, adherence, haemolytic activity and iron acquisition were further analyzed. It has been found that the genes involved in iron acquisition through the catechol siderophore ruckerbactin have a genetic organization similar to those of the E. coli enterobactin gene cluster (Fernandez et al., 2004). Regulation of this cluster depends on iron availability and temperature in such a way that low iron conditions and low temperature (18°C in relation to 28°C) induced its expression (Fernandez et al., 2004). It was shown that production of the siderophore is upregulated during the infection of fish (Fernandez et al., 2004; Fernández et al., 2007a) and it is also involved in virulence since 100-fold attenuation, in relation to the parental strain, was obtained when an isogenic mutant in rucC, a gene from the ruckerbactin cluster, was used in LD50 experiments (Fernandez et al., 2004).
A putative additional system of Y. ruckeri for obtaining iron is the yhlBA cluster, also identified by IVET. The yhlA gene encodes a Serratia-type haemolysin, whereas the yhlB product is related to the secretion/activation of YhlA (Fernández et al., 2007b) (Figure 2). Analysis of yhlB::lacZY transcriptional fusion indicated that higher levels of expression of yhlB were obtained under iron-starvation conditions, and also at 18°C than at 28°C (Fernández et al., 2007b). Interestingly, the involvement of the two genes in bacterial virulence is indicated not only by the virulence attenuation obtained when yhlA and yhlB insertional mutants were used, but also by the cytolytic properties of YhlA against the BF-2 fish cell line (Fernández et al., 2007b). In vivo monitoring of yhlBA promoter showed that it was highly expressed (Méndez and Guijarro, 2013), as was expected, since the system had been previously isolated as an IVET clone. In contrast to yrp1, the yhlAB operon is present and the haemolytic activity is detectable in different Y. ruckeri strains from different origins and geographical locations. This indicates that this operon is of importance for virulence in this species (Fernández et al., 2007b).
The analysis of a chromosomally located traHIJKCLMN operon of Y. ruckeri that was identified by IVET showed that it was structurally related to the DNA transfer system present in the pADAP virulence-related plasmid of Serratia entomophila (Hurst et al., 2003). The cluster was present in different Y. ruckeri strains but it was not found in the genomic analysis of human pathogenic Yersinia (Méndez et al., 2009). This operon is composed of at least eight genes displaying homology with type IV secretion systems (T4SS). Indeed, traH, traI, traJ, and traK genes encoded for proteins similar to those encoded by the dot/icm genes of the T4SS of Legionella pneumophila (Komano et al., 2000). Thus, it is likely that the gene products of the tra operon constitute parts of the T4SS involved in the transfer of virulence factors. In fact, insertional mutagenesis of the traI gene resulted in attenuation of the virulence (Méndez et al., 2009). Like other virulence factors, the tra operon was more highly expressed at 18°C than at 28°C.
A novel two-gene operon cdsBA involved in the transport and further degradation of L-cysteine was also selected by IVET. Attenuation of a cdsA mutant supported a role in virulence (Méndez et al., 2011). The cdsB gene encoded an L-cysteine desulfidase involved in cysteine degradation, previously characterized in the archaeal Methanocaldococcus jannaschii (Tchong et al., 2005), whereas cdsA is a cysteine permease (Méndez et al., 2011). Interestingly, the operon was shown to be present in several anaerobic and facultative bacterial groups, particularly in some species of enterobacteria, including Y. enterocolitica, although it was absent in Y. pseudotuberculosis and Y. pestis (Méndez et al., 2011). This operon was specifically induced by L-cysteine under low oxygen conditions and the uptake of this amino acid was an energy-dependent process (Méndez et al., 2011). Transcriptional fusion of the cdsBA promoter with the luxCDABE operon showed that the cdsA and cdsB genes were barely expressed under routine laboratory conditions but were found to have high activity inside the fish. Therefore, virulence attenuation of the cdsA mutant underlines the importance of this operon for the bacterium during the infection process. Although the precise role of the CdsAB system remains unknown, it was suggested that it could be involved in the generation of iron-sulfur-centers for FE-S proteins or in the accumulation of glutathione, an important detoxification and redox buffer molecule in the cell (Méndez et al., 2011).
Interestingly, more of these IVET-selected genes are induced at 18°C, the temperature around which outbreaks of the disease take place, than at 28°C, the optimal temperature for growth of the bacterium (Fernández et al., 2007b; Méndez et al., 2009). This finding points to the existence in Y. ruckeri of specific mechanisms (thermo-induced changes of DNA supercoiling, RNA and protein thermometer, etc.) for the temperature regulation of virulence at temperatures below the optimal for its growth, as seems to occur in a good number of microorganisms that infect ectothermic animals (Guijarro et al., 2015).
STM was also able to select Y. ruckeri mutants that can survive in vitro but not in the fish host (Dahiya and Stevenson, 2010a). Further identification of the inactivated genes in these mutants revealed up to 25 different ORFs (Dahiya and Stevenson, 2010a), among which the znuA gene, coding a zinc binding protein, and the uvrY gene, coding a response regulator, were chosen for deeper study. The znuA gene forms part of the znuABC operon encoding a high-affinity zinc transporter, which has a genetic organization similar to that found in E. coli (Patzer and Hantke, 2000) and S. Typhimurium (Campoy et al., 2002). In fact, the introduction of the Y. ruckeri znuABC locus was able to restore the growth of a znuABC mutant of E. coli in zinc-deficient media (Dahiya and Stevenson, 2010b). The znuABC system is widely distributed in bacteria such as Brucella abortus (Kim et al., 2004), Campylobacter jejuni (Davis et al., 2009) and Neisseria gonorrhoeae (Lim et al., 2008), contributing to their virulence. Interestingly, mutations in the znuABC system of Y. pestis did not affect virulence (Bobrov et al., 2014, 2017). The reason is that this bacterium has an additional zinc transport system mediated by some components of the yersiniabactin siderophore (Ybt) which is able to sequestrate zinc in addition to iron (Bobrov et al., 2014, 2017). In mouse models of bubonic and pneumonic plague this system compensates for the loss of znuABC. However, the simultaneous absence of both systems leads to a decrease in Y. pestis virulence (Bobrov et al., 2014, 2017). However, in Y. ruckeri it seems that the znuABC system is the main Zn acquisition system, since the znuA mutant had nearly 150- to 350-fold lower infection loads in the kidney of rainbow trout than the parental strain, which demonstrates that this Zn transport system has an important role in the infection process (Dahiya and Stevenson, 2010b).
In another mutant selected by STM, the BarA-UvrY two-component system was inactivated. This mutant was recovered from fish in 5- to 15-fold lower numbers compared to the parental strain at the beginning of the infection (Dahiya and Stevenson, 2010c), suggesting that this system is involved in the invasion of gills or gut tissues, as occurred in the uvrY mutants of Y. pseudotuberculosis (Heroven et al., 2008), E. coli (Herren et al., 2006) and S. Typhimurium (Johnston et al., 1996). In addition, the Y. ruckeri uvrY mutant was more sensitive to H2O2, which suggests an increasing susceptibility to oxidative killing by phagocytic cells. All of this resulted in a lower recovery (103- to 105-fold) of the uvrY mutant than the parental strain at the end of co-infection experiments which were carried out by immersion of rainbow trout in water containing both strains (Dahiya and Stevenson, 2010c).
As expected, LPS is important for the pathogenesis of Y. ruckeri (Altinok et al., 2016). Specific deletion of the lpxD gene, involved in the lipid A biosynthesis, resulted in an LPS-deficient strain. This lpxD mutant showed a significant attenuation of virulence (103-fold) in relation to the parental strain when LD50 determination was carried out by injection (Altinok et al., 2016). It is important to highlight that lpxD mutation reduced the overall number of invasive bacteria, which in turn, may influence the process of invasion and colonization, leading to a decrease in virulence. Special attention must be paid to a virulence factor related to legionaminic acid, a component of the Y. ruckeri LPS that will be addressed in the next section.
Another interesting virulence factor that has been described in Y. ruckeri is the Afp18 toxin, although it should be noted that its effect has hitherto only been tested in zebrafish embryos (Jank et al., 2014). This is encoded by a gene that is part of the antifeeding prophage gene cluster (afp), consisting in a 25 Kb DNA segment involved in the production of a prophage contractile tail. This cluster is similar to the one found in Serratia entomophila (Rybakova et al., 2013; Jank et al., 2014), which has common characteristics with the R-pyocins type VI secretion system delivery apparatus of Pseudomonas aeruginosa (Heymann et al., 2013). This contractile tail is thought to capture the Afp18 toxin and inject it into the cell cytoplasm of zebrafish blastoderm cells (Jank et al., 2014). The Afp18 protein has a glycosyltransferase domain. Its activity alters the early phase of zebrafish embryogenesis, whereas a non-functional Afp18 lacking the glycosyltransferase domain has no effect on development (Jank et al., 2014). The effect of the Afp18 glycosyltransferase is through a process of specific tyrosine mono-O-GlcNAcylation of RhoA, a regulatory GTPase associated with cytoskeleton development, resulting in actin depolymerization and the abrogation of early zebrafish development (Jank et al., 2014). Interestingly, genome analysis indicates that this antifeeding prophage system is widely distributed among prokaryotes and archaea (Sarris et al., 2014). In S. entomophila the Afp18 toxin acts as a virulence factor during the infection of insects (Rybakova et al., 2013), and could also be important as a virulence factor in human pathogenic bacteria. Since all this work was carried out on zebrafish embryos, it could be interesting to study the role of Afp18 on Y. ruckeri infection of rainbow trout by generating a mutant strain and following the progression of the disease.
Finally, two elements that should be taken into account are the flhDC operon, whose inactivation results in a more virulent strain and the heat sensitive factor (HSF), which has been considered as a virulence factor for a good number of years. The flhDC is a flagellar master operon of Y. ruckeri involved in the regulation of motility and phospholipase secretion, among other processes (Jozwick et al., 2016). In fact, mutation of the flhD gene resulted in significant changes in the bacterial transcriptome (Jozwick et al., 2016). Interestingly, no differences were found between the flhD mutant and the wild type strain when a standard experimental challenge model with mortalities as an endpoint was used. However, there was a significant increase in the flhD mutant density within the spleen in relation to the wild type strain in competition experiments, a characteristic which was reverted in the complemented strain (Jozwick et al., 2016). The result indicated that mutation of the flhDC operon gives a competitive advantage over the parental strain during the infection process. Although it is not known how the mutation in this operon affects bacterial virulence, the result is similar to that found in the S. Typhimurium flhD mutant in the sense that it was more virulent than the parental strain (Schmitt et al., 2001).
It is also worth mentioning that for many years it was thought HSF was required for virulence in Y. ruckeri (Furones et al., 1993). However, this assumption was recently discarded, since HSF was found to be an alkylsulphatase enzyme encoded by the yraS gene, whose mutation did not produce any modification in the virulence of the strain in relation to the wild type (Navais et al., 2014a).
Predicted virulence factors deduced from proteomic and genomic analysis
During recent years the development of alternative technologies based on proteomic, transcriptomic and genomic analyses has provided new tools to identify potential virulence factors in bacteria. It is important to consider that this type of analysis is based on comparison and only predicts the potential role of some of the genes or proteins identified as virulence factors. In Y. ruckeri, proteomic studies have been carried out under very specific culture conditions. Thus, differentially regulated proteins were identified and quantified in normal and iron-limited media using a label-free, gel-free shotgun proteomic approach (Kumar et al., 2016). Iron is an essential nutrient for bacteria that is difficult to obtain during the infection process, so a limited-iron environment is expected to induce a set of genes, some of which are related to pathogenesis. Under reduced iron availability, the majority of up-regulated proteins in biotype 1 and biotype 2 Y. ruckeri strains were related to iron-binding proteins (YfuA, YiuA and YfeA), iron-binding transporters (HemS and Feo), hemin transport (YhlBA) and TonB-dependent iron binding receptor, the latter probably being involved in the transport of ruckerbactin into the periplasm of cells and siderophore biosynthesis proteins. YfeA and YhlBA systems have previously been described as important virulence factors in Y. pestis (Bearden and Perry, 1999) and Y. ruckeri (Fernández et al., 2007b), respectively. Another up-regulated protein under iron-limited conditions was a superoxide dismutase, involved in protective mechanisms against oxidative stress, and particularly important in intracellular pathogens such as Y. ruckeri. Interestingly, this study revealed no significant differences in protein profile, using liquid chromatography-mass spectrometry analyses, between motile and non-motile Y. ruckeri strains, corresponding with biotype 1 and biotype 2, when they were cultured under iron-limited conditions, indicating that iron availability it is not involved in the differential virulence behavior of the two biotypes (Kumar et al., 2016).
A similar study was conducted in order to establish the differential protein profile between virulent and avirulent strains of Y. ruckeri grown under standard conditions (Kumar et al., 2017). Interestingly, in virulent strains, a total of 16 proteins were shown to be upregulated, including the haemolysis expression regulator HtrA, DegQ proteases, an anti-sigma regulator factor, different transcriptional regulators belonging to the LuxR, AsnC, and PhoP families, RNA-binding protein Hfq, invasion protein Inv, Cu-Zn superoxide dismutase, among others. All of these have been previously described as virulence-related proteins in different bacterial pathogens, which strongly suggests a role for them in the infection process of Y. ruckeri.
Invasins play an essential role in the initial colonization and attachment of Y. enterocolitica and Y. pseudotuberculosis (Chauhan et al., 2016). In Y. ruckeri, invasin (yrInv) and invasin-like molecule (yrllm) are two putative inverse autotransporter genes that, according to qRT-PCR analysis, were upregulated in a medium that mimics the environmental conditions present in the fish host (Wrobel et al., 2018), so they are likely to be involved in the infection process of Y. ruckeri although there are no conclusive data.
Whole genome analysis of Y. ruckeri strains provides a new approach to identifying putative virulence factors. Indeed, genome analysis of the Y. ruckeri SC09 strain isolated from catfish (Ictalurus punctatus) showed that it harbors a ysa locus containing all the genetic components of a type III secretion system (T3SS) quite similar in gene sequence and genetic structure to the Salmonella enterica pathogenicity island 1 and chromosomally encoded T3SS of Y. enterocolitica 1B (Liu et al., 2016). It should be remembered that T3SS are present only in virulent strains of human pathogenic Yersinia (Chen et al., 2010). For this reason, it is possible that the T3SS of Y. ruckeri contributes to the survival of the bacterium in fish macrophages and helps in the invasion of gill and gut epithelia. Other putative systems in Y. ruckeri SC09 that, according to the genomic analysis carried out by Liu et al. (2016), could be involved in the development of the infection in fish are: a type II secretion system, different two-component signal transduction systems and a phage shock protein system (Psp), which is distributed among enterobacteria and is implicated in the virulence of Salmonella, Shigella and Yersinia species (Huvet et al., 2011). Interestingly, a type IV secretion system described as a putative virulence factor by Liu et al. (2016) is unique to Y. ruckeri SC09 strain and it is not present in the other four Y. ruckeri strains analyzed by Cascales et al. (2017).
The comparative analysis of the genome sequences of five Y. ruckeri strains showed that, although they share approximately 75% of their genes, significant genetic differences were detected depending on the serotype or virulence of these strains (Cascales et al., 2017). Thus, a cluster of 18 genes related to the biosynthesis of legionaminic acid, the major component of the LPS, was exclusively present in serotype O1 strains. This cluster was absent in serotype O2 strains as well as in other Yersinia species, but present in different aquatic pathogenic bacteria such as Vibrio vulnificus, Aeromonas salmonicida, Vibrio fischeri (Cascales et al., 2017) and also in Campylobacter jejuni, where it is involved in virulence (Zebian et al., 2016). In addition to this cluster, serotype O1 strains share a total of 268 genes, among which are the invasin and the type IV secretion system corresponding with the traHIJKCLMN operon (Méndez et al., 2009).
These genes shared by serotype O1 strains could adapt the bacterium to a particular host, since the three serotype O1 strains analyzed (150, CSF007-82, and ATCC29473) were isolated from rainbow trout. In the same way, a serotype O2 strain (Big Creek 74) together with Y. ruckeri SC09, a putative O2 strain, which were isolated from chinook salmon and catfish respectively, share 122 genes which are absent in serotype O1 strains. These include a cluster involved in fimbriae biosynthesis similar to the Stf cluster of S. Typhimurium, associated with virulence (Emmerth et al., 1999) and an insecticidal toxin complex similar to the one found in Vibrio parahaemolyticus, involved in hepatopancreatic necrosis disease in penaeid shrimps (Tang and Lightner, 2014).
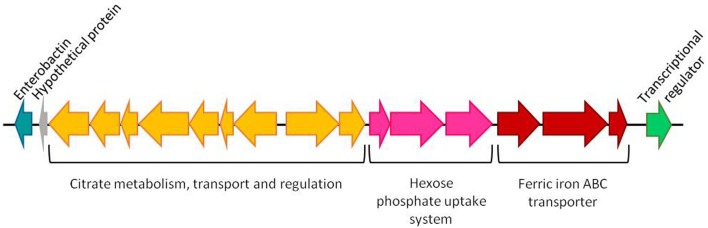
In the study of Cascales et al. (2017) it is proposed that the characteristic attenuation in virulence of the ATCC29473 type strain is a consequence of the lack of 21 genes that were present in the other virulent strains. It should be noted that 17 out of these 21 genes are clustered in all four genomes analyzed (Figure 3). This region includes genes encoding a Crp-Fnr family transcriptional regulator, enzymes related to enterobactin-like siderophore and gene clusters involved in iron transport, hexose phosphate uptake and citrate metabolism. Most of these genes are related to virulence in different bacteria (Gray et al., 2006; Urbany and Neuhaus, 2008; Moisi et al., 2013) so the absence of this region could be responsible for the attenuation of the type strain.
Figure 3.
Cluster of genes absent from Y. ruckeri ATCC29473 type strain and present in the strains Y. ruckeri 150, CSF007-82, Big Creek and SC09. The region contains genes encoding for an enterobactin-like siderophore (blue), nine genes involved in the uptake and metabolism of citrate (yellow), a group of three genes related to hexose phosphate uptake (pink), three genes involved in iron transport (red) and a Crp-Fnr family transcriptional regulator (green). (1) Citrate succinate antiporter, (2) 2-(5′-triphosphoribosyl)-3′-dephosphocoenzyme-A synthase, (3) Apo-citrate lyase phosphoribosyldephospho-CoA transferase, (4) Citrate lyase alpha chain, (5) Citrate lyase beta chain, (6) Citrate lyase gamma chain acyl carrier protein, (7) [Citrate[pro-3S]-lyase] ligase, (8) Sensor kinase, (9) Transcriptional regulatory protein, (10) Transcriptional regulatory protein, (11) Sensor histidine protein kinase glucose-6-phosphate specific, (12) Hexose phosphate uptake regulatory protein, (13) Ferric iron ABC transporter iron-binding protein, (14) Ferric iron ABC transporter permease protein, (15) Ferric iron ABC transporter binding subunit (Figure taken from Cascales et al., 2017).
Conclusions and further studies
The results lead to the conclusion that there exist at least three different routes of infection by Y. ruckeri. The most important pathway for the development of ERM has still to be elucidated, but it seems clear that, although the primary bacteremia is generated by the entry of the bacterium into the fish through the gills, the very significant presence of the bacterium in the intestine and also the signs of the disease, strongly suggest that the digestive route must play a relevant role. The virulence of Y. ruckeri is multifactorial and it depends not only on the presence of certain genes in each strain but also on certain environmental factors, particularly, temperature. In fact, a temperature below the optimal for bacterial growth and close to the one at which outbreaks occur seems to be important in the expression of some of the genes encoding virulence factors. This appears to be one of the areas for future research, since presently unknown regulation systems could be involved in this temperature-dependent response. Although a lot of genes involved in the pathogenesis of the bacterium have recently been identified, there are still many points that need to be addressed, such us the temporal and spatial expression pattern of each virulence factor and their specific function or the identification of the regulatory mechanisms which modulate the virulence gene expression. The new techniques and methodologies developed in recent years, including CLIQ-BID, a method allowing quantification of damage in eukaryotic cells infected by bacteria (Wallez et al., 2018), the new RNA-seq techniques such as Term-seq (Dar et al., 2016) and Dual RNA-seq (Westermann et al., 2016), useful for the analysis of pathogens under complex environmental conditions and Single Cell RNA-seq (Avraham et al., 2015; Saliba et al., 2017) can help to develop different approaches. Moreover, analysis of the data provided by the genetic and proteomic studies may be a good starting point for elucidating the role of new genes in the infectious process of Y. ruckeri. In fact, two specific gene clusters should be considered for their potential role in virulence. This is the case of a cluster involved in legionaminic acid biosynthesis that could be important in the pathogenesis of serotype O1 strains, and the 21 genes absent from the virulence-attenuated Y. ruckeri type strain, some of them encoding putative virulence factors.
Author contributions
JG and JM drafted the first version of the manuscript. DC, JM, AG-T, JG contributed to bibliography analysis and manuscript improvement. All authors have reviewed the final version of the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
DC and AG-T were the recipients of FPU and FPI fellowships, respectively.
Footnotes
Funding. This research was supported by the AGL2015-66018-R grant from the Ministerio de Economia y Competitividad of Spain.
References
- Altinok I., Ozturk R. C., Kahraman U. C., Capkin E. (2016). Protection of rainbow trout against yersiniosis by lpxD mutant Yersinia ruckeri. Fish Shellfish Immunol. 55, 21–27. 10.1016/j.fsi.2016.04.018 [DOI] [PubMed] [Google Scholar]
- Amend D. F., Johnson K. A., Croy T. R., McCarthy D. H. (1983). Some factors affecting the potency of Yersinia ruckeri bacterins. J. Fish Dis. 6, 337–344. [Google Scholar]
- Arias C. R., Olivares-Fuster O., Hayden K., Shoemaker C. A., Grizzle J. M., Klesius P. H. (2007). First report of Yersinia ruckeri biotype 2 in the USA. J. Aquat. Anim. Health 19, 35–40. 10.1577/H06-011.1 [DOI] [PubMed] [Google Scholar]
- Austin D. A., Robertson P. A., Austin B. (2003). Recovery of a new biogroup of Yersinia ruckeri from diseases rainbow trout (Oncorhynchus mykiss, Walbaum). Syst. Appl. Microbiol. 26, 127–131. 10.1078/072320203322337416 [DOI] [PubMed] [Google Scholar]
- Avraham R., Haseley N., Brown D., Penaranda C., Jijon H. B., et al. (2015). Pathogen cell-to-cell variability drives heterogeneity in host immune response. Cell 162, 1309–1321. 10.1016/j.cell.2015.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearden S. W., Perry R. D. (1999). The Yfe system of Yersinia pestis transports iron and manganese and is required for full virulence of plague. Mol. Microbiol. 32, 403–414. [DOI] [PubMed] [Google Scholar]
- Bobrov A. G., Kirillina O., Feterston D., Miller M. C., Burlison J. A., Perry R. D. (2014). The Yersinia pestis siderophore, yersiniabactin, and the ZnuABC system both contribute to zinc acquisition and the development of lethal septicaemic plague in mice. Mol. Microbiol. 93, 759–775. 10.1111/mmi.12693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobrov A. G., Kirillina O., Fosso M. Y., Fetherston J. D., Miller M. C., VanCleave T. T., et al. (2017). Zinc transporters YbtX and ZnuABC are required for the virulence of Yersinia pestis in bubonic and pneumonic plague in mice. Metallomics 21, 757–772. 10.1039/c7mt00126f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvez S., Gantelet H., Blanc G., Douet D. G., Daniel P. (2014).Yersinia ruckeri biotypes 1 and 2 in France: presence and antibiotic susceptibility. Dis. Aquat. Organ. 109, 117–126. 10.3354/dao02725 [DOI] [PubMed] [Google Scholar]
- Campoy S., Jara M., Busquets N., de Rozas A. M. P., Badiola I., Barbe J. (2002). Role of the high-affinity zinc uptake znuACB system in Salmonella enterica serovar Typhimurium virulence. Infect. Immun. 70, 4721–4725. 10.1128/IAI.70.8.4721-4725.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascales D., Guijarro J. A., García-Torrico A. I., Méndez J. (2017). Comparative genome analysis reveals important genetic differences among serotype O1 and serotype O2 strains of Yersina ruckeri and provides insights into host adaptation and virulence. Microbiologyopen 6. 10.1002/mbo3.460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan N., Wrobel A., Skurnik M., Leo J. C. (2016). Yersinia adhesins: an arsenal for infection. Proteomics Clin. Appl. 10, 949–963. 10.1002/prca.201600012 [DOI] [PubMed] [Google Scholar]
- Chen P. E., Cook C., Stewart A. C., Nagarajan N., Sommer D. D., et al. (2010). Genomic characterization of the Yersinia genus. Genome Biol. 4:11 10.1186/gb-2010-11-1-r1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahiya I., Stevenson R. M. W. (2010a). Yersinia ruckeri genes that attenuate survival in rainbow trout (Oncorhynchus mykiss) are identified using signature-tagged mutants. Vet. Microbiol. 144, 399–404. 10.1016/j.vertmic.2010.02.003 [DOI] [PubMed] [Google Scholar]
- Dahiya I., Stevenson R. M. W. (2010b). The ZnuABC operon is important for Yersinia ruckeri infections of rainbow trout, Oncorhynchus mykiss (Walbaum). J. Fish Dis. 33, 331–340. 10.1111/j.1365-2761.2009.01125x [DOI] [PubMed] [Google Scholar]
- Dahiya I., Stevenson R. M. W. (2010c). The UvrY response regulator of the BarA–UvrY two-component system contributes to Yersinia ruckeri infection of rainbow trout (Oncorhynchus mykiss). Arch. Microbiol. 192, 541–547. 10.1007/s00203-010-0582-8 [DOI] [PubMed] [Google Scholar]
- Dar D., Shamir M., Mellin J. R., Koutero M., Stern-Ginossar N., Cossart P., et al. (2016). Term-seq reveals abundant ribo-regulation of antibiotics resistance in bacteria. Science 352:aad9822. 10.1126/science.aad9822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis L. M., Kakuda T., DiRita V. J. (2009). A Campylobacter jejuni znuA orthologue is essential for growth in low-zinc environments and chick colonization. J. Bacteriol. 191, 1631–1640. 10.1128/JB.01394-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Keukeleire S., De Bel A., Jansen Y., Wauters G., Pierard D. (2014). Yersinia ruckeri, an unusual microorganism isolated from a human wound infection. New Microbe. New Infect. 2, 134–135. 10.1002/nmi2.56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmerth M., Goebel W., Miller S. I., Hueck C. J. (1999). Genomic subtraction identifies Salmonella typhimurium prophages, F-related plasmid sequences, and a novel fimbrial operon, stf, which are absent in Salmonella typhi. J. Bacteriol. 181, 5652–5661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evenhuis J. P., La Patra S. E., Verner-Jeffreys D. W., Dalgaard I., Well T. J. (2009). Identification of flagellar motility genes in Yersinia ruckeri by transposon mutagenesis. Appl. Environm. Microbiol. 75, 6630–6633. 10.1128/AEM.01415-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez L., Lopez J. R., Secades P., Menendez A., Marquez I., Guijarro J. A. (2003). In vitro and in vivo studies of the Yrp1 protease from Yersinia ruckeri and its role in protective immunity against enteric red mouth disease of salmonids. Appl. Environ. Microbiol. 69, 7328–7335. 10.1128/AEM.69.12.7328-7335.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez L., Marquez I., Guijarro J. A. (2004). Identification of specific in vivo-induced (ivi) genes in Yersinia ruckeri and analysis of ruckerbactin, a catecholate siderophore iron acquisition system. Appl. Environ. Microbiol. 70, 5199–5207. 10.1128/AEM.70.9.5199-5207.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández L., Méndez J., Guijarro J. A. (2007a). Molecular virulence mechanisms of the fish pathogen Yersinia ruckeri. Vet. Microbiol. 125, 1–10. 10.1016/j.vetmic.2007.06.013 [DOI] [PubMed] [Google Scholar]
- Fernández L., Prieto M., Guijarro J. A. (2007b). The iron- and temperature-regulated haemolysin YhlA is a virulence factor of Yersinia ruckeri. Microbiology 153, 483–489. 10.1099/mic.0.29284-0 [DOI] [PubMed] [Google Scholar]
- Fernández L., Secades P., Lopez J. R., Márquez I., Guijarro J. A. (2002). Isolation and analysis of a protease gene with an ABC transport system in the fish pathogen Yersinia ruckeri: insertional mutagenesis and involvement in virulence. Microbiology 148, 2233–2243. 10.1099/00221287-148-7-2233 [DOI] [PubMed] [Google Scholar]
- Fouz B., Zarza C., Amaro C. (2006). First description of non-motile Yersinia ruckeri serovar I strains causing disease in rainbow trout, Oncorhynchus mykiis (Walbaum), cultured in Spain. J. Fis. Dis. 29, 339–346. 10.1111/j.1365-2761.2006.00723.x [DOI] [PubMed] [Google Scholar]
- Furones M. D., Gilpin M. L., Munn C. B. (1993). Culture media for the differentiation of isolated of Yersinia ruckeri, based on detection of a virulence factor. J. Appl. Microbiol. 74, 360–366. [DOI] [PubMed] [Google Scholar]
- Gray M. J., Freitag N. E., Boor K. J. (2006). How the bacterial pathogen Listeria monocytogenes mediates the switch from environmental Dr. Jekyll to pathogenic Mr. Hyde. Infect. Immun. 74, 2505–2512. 10.1128/IAI.74.5.2505-2512.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guijarro J. A., Cascales D., García-Torrico A. I., García-Domínguez M., Méndez J. (2015). Temperature-dependent expression of virulence genes in fish-pathogenic bacteria. Front. Microbiol. 6:700. 10.3389/fmicb.2015.00700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heroven K. A., Bohme K., Rhode M., Dersch P. A. (2008). Csr-type regulatory system, including small non-coding RNAs, regulates the global virulence regulator RovA of Yersinia pseudotuberculosis through RovM. Mol. Microbiol. 68, 1179–1195. 10.1111/j.1365-2958.2008.06218.x [DOI] [PubMed] [Google Scholar]
- Herren D., Mitra A., Palaniyandi S. K., Coleman A., Elankumaran S., Mukhopadhyay S. (2006). The Bar-UvrY two-component system regulates virulence in avian pathogenic Escherichia coli O78:K80:H9. Infect. Immun. 74, 4900–4909. 10.1128/IAI.00412-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heymann J. B., Bartho J. D., Rybakova D., Venugopal H. P., Winkler D. C., Sen A., et al. (2013). Three-dimensional structure of the toxin-delivery particle antifeeding prophage of Serratia entomophila. J. Biol. Chem. 288, 25276–25284. 10.1074/jbc.M113.456145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst M. R. H., O'Callaghan M., Glare T. R. (2003). Peripheral sequences of the Serratia entomophila pADAP virulence-associated region. Plasmid 50, 213–229. 10.1016/S0147-619X(03)00062-3 [DOI] [PubMed] [Google Scholar]
- Huvet M., Toni T., Sheng X., Thorne T., Jovanovic G., Engl C., et al. (2011). The evolution of the phage shock protein response system: interplay between protein function, genomic organization, and system function. Mol. Biol. Evol. 28, 1141–1155. 10.1093/molbev/msq301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jank T., Eckerle S., Steinemenn M., Trillhaase C., Schimpl M., et al. (2014). Tyrosine glycosylation of Rho by Yersinia toxin impairs blastomere cell behavior in zebrafish embryos. Nat. Comm. 6:7807 10.1038/ncomms8807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston C., Pegues D. A., Hueck C. J., Lee C. A., Miller S. I. (1996). Transcriptional activation of Salmonella typhimurium genes by a member of the phosphorylated response-regulator superfamily. Mol. Microbiol. 22, 715–727. [DOI] [PubMed] [Google Scholar]
- Jozwick A. K., Graf J., Welch T. J. (2016). The flagellar master operon flhDC is a pleiotropic regulator involved in motility and virulence of the fish pathogen Yersinia ruckeri. J. Appl. Microbiol. 122, 578–588. 10.1111/jam.13374. [DOI] [PubMed] [Google Scholar]
- Kavermann H., Burns B. P., Angermuller K., Odenbreir S., Fischer W., Melchers K., et al. (2003). Identification and characterization of Helicobacter pylori genes essential for gastric colonization. J. Exp. Med. 197, 813–822. 10.1084/jem.20021531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khimmakthong U., Deshmukh S., Chettri J. K., Bojesen A. M., Kania P. W., Dalsgaard I., et al. (2013). Tissue specific uptake of inactivated and live Yersina ruckeri in rainbow trout (Oncorhynchus mykiss): visualization by immunohistochemistry and in situ hybridization. Microbial. Pathog. 59–60. 10.1016/j.micpath.2013.03.001 [DOI] [PubMed] [Google Scholar]
- Kim S., Watanabe K., Shirahata T., Watarai M. (2004). Zinc uptake system (znuA locus) of Brucella abortus is essential for intracellular survival and virulence in mice. J. Vet. Med. Sci. 66, 1059–1063. 10.1292/jvms.66.1059 [DOI] [PubMed] [Google Scholar]
- Komano T., Yoshida T., Narahara K., Furuya N. (2000). The transfer region of Incl1 plasmid R64: similarities between R64 tra and Legionella icm/dot genes. Mol. Microbiol. 35, 1348–1359. 10.1046/j.1365-2958.2000.01769.x [DOI] [PubMed] [Google Scholar]
- Kumar G., Hummel K., Ahrens M., Mennateau-Ledoble S., Welch T. J., Eisenacher M., et al. (2016). Shotgun proteomic analysis of Yersinia ruckeri strains under normal and iron-limited conditions. Vet. Res. 47, 100–113. 10.1186/s13567-016-0384-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar G., Hummel K., Welch T. J., Razzazi-Fazeli W., El-Matbouli M. (2017). Global proteomic profiling of Yersinia ruckeri strains. Vet. Res. 48, 55–66. 10.1186/s13567-017-0460-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar G., Menanteau-Ledouble S., Saleh M., El-Matbouli M. (2015). Yersinia ruckeri, the causative agent of enteric redmouth disease in fish. Vet. Microbiol. 46:103. 10.1186/s13567-015-0238-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim K. H., Jones C. E., vanden Hoven R. N., Edwards J. L., Falsetta M. L., Apicella M. A., et al. (2008). Metal binding specificity of the MntABC permease of Neisseria gonorrhoeae and its influence on bacterial growth and interaction with cervical epithelial cells. Infect. Immun. 76, 3569–3576. 10.1128/IAI.01725-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Wang K.-Y., Wang J., Chen D.-F., Huang X.-L., Ouyang P., et al. (2016). Genome sequence of the fish pathogen Yersinia ruckeri SC09 provides insights into niche adaptation and pathogenic mechanism. Int. J. Mol. Sci. 17:557. 10.3390/ijms17040557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menanteau-Ledouble S., Lawrence M. L., El-Matbouli M. (2018). Invasion and replication of Yersinia ruckeri in fish cell cultures. BMC Vet. Res. 14:81. 10.1186/s12917-018-1408-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méndez J., Fernandez L., Menendez A., Reimundo P., Perez-Pascual D., Navais R., et al. (2009). A chromosomally located traHIJKCLMN operon encoding a putative Type IV secretion system is involved in the virulence of Yersinia ruckeri. Appl. Environ. Microbiol. 75, 937–945. 10.1128/AEM.01377-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méndez J., Guijarro J. A. (2013). In vivo monitoring of Yersinia ruckeri in fish tissues: progression and virulence gene expression. Environ. Microbiol. Rep. 5, 179–185. 10.1111/1758-2229.12030 [DOI] [PubMed] [Google Scholar]
- Méndez J., Reimundo O., Perez-Pascual D., Navais R., Gomez E., Guijarro J. A. (2011). A novel cdsAB operon is involved in the uptake of l-cysteine and participates in the pathogenesis of Yersinia ruckeri. J. Bacteriol. 193, 944–951. 10.1128/JB.01058-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moisi M., Lichtenegger S., Tutz S., Seper A., Schild S., Reidl J. (2013). Characterizing the hexose-6-phosphate transport system of Vibrio cholerae, a utilization system for carbon and phosphate sources. J. Bacteriol. 195, 1800–1808. 10.1128/JB.01952-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navais R., Méndez J., Cascales D., Reimundo P., Guijarro J. A. (2014a). The heat sensitive factor (HSF) of Yersinia ruckeri is produced by an alkyl sulphatase involved in sodium dodecyl sulphate (SDS) degradation but not in virulence. BMC Microbiol. 14:221 10.1186/s12866-014-0221-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navais R., Mendez J., Perez-Pascual D., Cascales D., Guijarro J. A. (2014b). The yrpAB operon of Yersinia ruckeri encoding two putative U32 peptidases is involved in virulence and induced under microaerobic conditions. Virulence 5, 619–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtani M., Villumsen R., Koppang E. O., Raida M. K. (2015). Global 3D imaging of Yersinia ruckeri bacterin uptake in rainbow trout fry. PLoS ONE 10:e0117263. 10.1371/journal.pone.0117263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtani M., Villumsen R., Kragelund H., Raida M. K. (2014). 3D visualization of the initial Yersinia ruckeri infection route in rainbow trout (Oncorhynchus mykiss) by optical projection tomography. PLoS ONE 9:e89672. 10.1371/journal.pone.0093845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özdemir F., Arslan S. (2015). Genotypic and phenotypic virulence characteristics and antimicrobial resistance of Yersinia spp. Isolated from meat and milk products. J. Food Sci. 80, 1306–1313. 10.1111/1750-3841.12911 [DOI] [PubMed] [Google Scholar]
- Patzer S. I., Hantke K. (2000). The zinc-responsive regulator Zur and its control of the znu gene cluster encoding the ZnuABC zinc uptake system in Escherichia coli. J. Biol. Chem. 275, 24321–24332. 10.1074/jbc.M001775200 [DOI] [PubMed] [Google Scholar]
- Romalde J. L., Magariños B., Barja J. L., Toranzo A. E. (1993). Antigenic and molecular characterization of Yersinia ruckeri proposal for a new intraspecies classification. Syst. Appl. Microbiol. 16, 411–419. [Google Scholar]
- Romalde J. L., Planas E., Sotelo J. M., Toranzo A. E. (2003). First description of Yersinia ruckeri serotype O2 in Spain. Bull. Eur. Ass. Fish Pathol. 23, 135–138. [Google Scholar]
- Rybakova D., Radjainia M., Turner A., Sen A., Mitra A. K., Hurst M. R. (2013). Role of antifeeding prophage (Afp) protein Afp16 in terminating the length of the Afp tailocin and stabilizing its sheath. Mol. Microbiol. 89, 702–714. 10.1111/mmi.12305 [DOI] [PubMed] [Google Scholar]
- Ryckaert J., Bossier P., D'Herde K., Diez-Fraile A., Sorgeloos P., Hasebrouck F., et al. (2010). Persistence of Yersinia ruckeri in trout macrophages. Fish Shellfish Immunol. 29, 648–655. 10.1016/j.fsi.2010.06.009 [DOI] [PubMed] [Google Scholar]
- Saliba A.-E., Santos S. C., Vogel J. (2017). New RNA-seq approaches for the study of bacterial pathogens. Curr. Opin. Microbiol. 35, 78–87. 10.1016/j.mib.2017.01.001 [DOI] [PubMed] [Google Scholar]
- Sarris P. F., Ladoukakis E. D., Panopoulos N. J., Scoulica E. V. (2014). A phage tail-delivered element with wide distribution among both prokaryotic domains: a comparative genomic and phylogenetic study. Genome Biol. Evol. 6, 1739–1747. 10.1093/gbe/evu136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt C. K., Ikeda J. S., Darnell S. C., Watson P. R., Bispham J., Wallis T. S., Weinstein D. L., Metcalf E. S., et al. (2001). Absence of all components of the flagellar export and synthesis machinery differentially alters virulence of Salmonella enterica serovar Typhimurium in models of typhoid fever, survival in macrophages, tissue culture invasiveness, and calf enterocolitis. Infect. Immun. 9, 5619–5625. 10.1128/IAI.69.9.5619-5625.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secades P., Guijarro J. A. (1999). Purification and characterization of an extracellular protease from the fish pathogen Yersinia ruckeri and effect of culture conditions on production. Appl. Environ. Microbiol. 65, 3969–3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang K. F., Lightner D. V. (2014). Homologues of insecticidal toxin complex genes within a genomic island in the marine bacterium Vibrio parahaemolyticus. FEMS Microbiol. Lett. 361, 34–42. 10.1111/1574-6968.12609 [DOI] [PubMed] [Google Scholar]
- Tchong S.-I., Xu H., White R. H. (2005). L-cysteine desulfidase: and [4Fe-4S] enzyme isolated from Methanocaldococcus jannaschii that catalyzes the breakdown of L-cysteine into pyruvate, ammonia, and sulfide. Biochemistry 44, 1659–1670. 10.1021/bi0484769 [DOI] [PubMed] [Google Scholar]
- Tinsley J. W., Lyndon A. R., Austin B. (2011). Antigenic and cross-protection studies of biotype 1 and biotype 2 isolates of Yersinia ruckeri in rainbow trout, Oncorhynchus mykiss (Walbaum). J. Appl. Microbiol. 111, 8–16. 10.1111/j.1365-2672.2011.05020.x [DOI] [PubMed] [Google Scholar]
- Tobback E., Decostere A., Hermans K., Haesebrouck F., Chiers K. (2007). Yersinia ruckeri infections in salmonid fish. J. Fish Dis. 30, 257–268. [DOI] [PubMed] [Google Scholar]
- Tobback E., Decostere A., Hermans K., Ryckaert J., Duchateau L., Haesebrouck F., et al. (2009). Route of entry and tissue distribution of Yersinia ruckeri in experimentally infected rainbow trout Oncorhynchus mykiss. Dis. Aquat. Org. 84, 219–228. 10.3354/dao02057 [DOI] [PubMed] [Google Scholar]
- Tobback E., Hermans K., Decostere A., Broeck Van de Haesebrouck, F., Chiers K. (2010). Interactions of virulent and avirulent Yersinia ruckeri strains with isolated gill arches and intestinal explants of rainbow trout Oncorhynchus mykiss. Dis. Aquat. Org. 90, 175–179. 10.3354/dao02230 [DOI] [PubMed] [Google Scholar]
- Urbany C., Neuhaus H. E. (2008). Citrate uptake into Pectobacterium atrosepticum is critical for bacterial virulence. Mol. Plant. Microbe. Interact. 21, 547–554. 10.1094/MPMI-21-5-0547 [DOI] [PubMed] [Google Scholar]
- Wallez Y., Bouillot S., Soleihac E., Huber P., Atrée I., Faudry E. (2018). CLIQ-BID: A method to quantify bacteria-induced damage to eukaryotic cells by automated live-imaging of bright nuclei. Sci. Rep. 8:5. 10.1038/s41598-017-18501-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch T. J., Verner-Jeffreys D. W., Dalsgaard I., Wiklund T., Eenthuis J. P., Garcia Cabrera J. A., et al. (2011). Independent emergence of Yersinia ruckeri biotype 2 in the United States and Europe. Appl. Environm. Microbiol. 77, 3493–3499. 10.1128/AEM.02997-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch T. J., Wiens G. D. (2005). Construction of a virulent, green fluorescent protein-tagged Yersinia ruckeri and detection in trout tissues after intraperitoneal and immersion challenge. Dis. Aquat. Org. 67, 267–272. 10.3354/dao067267 [DOI] [PubMed] [Google Scholar]
- Westermann A. J., Forster K. U., Amaman F., Barquist L., Chao Y., et al. (2016). Dual RNA-seq unveils noncoding RNA functions in host-pathogen interactions. Nature 529, 496–501. 10.1038/nature16547 [DOI] [PubMed] [Google Scholar]
- Wrobel A., Ottoni C., Leo J. C., Gulla S., Linke D. (2018). The repeat structure of two paralogous genes, Yersinia ruckeri invasin (yrInv) and a “Y. ruckeri invasin-like molecule”, (yrIlm) sheds light on the evolution of adhesive capacities of a fish pathogen. J. Struct. Biol. 201, 171–183. 10.1016/j.jsb.2017.08.008 [DOI] [PubMed] [Google Scholar]
- Zebian N., Merkx-Jacques A., Pittock P. P., Houle S., Dozois C. M., Lajoie G. A., et al. (2016). Comprehensive analysis of flagellin glycosylation in Campylobacter jejuni NCTC 11168 reveals incorporation of legionaminic acid and its importance for host. Colonization 26, 386–397. 10.1093/glycob/cwv104 [DOI] [PubMed] [Google Scholar]
- Zhao H., Li X., Johnson D. E., Mobley H. L. (1999). Identification of protease and rpoN-associated genes of urophatogenic Proteus mirabilis by negative selection in a mouse model of ascending urinary tract infection. Microbiology 145, 185–195. 10.1999/13500872-145-1-185 [DOI] [PubMed] [Google Scholar]