Abstract
Menstrual toxic shock syndrome (mTSS) is a severe disease that occurs in healthy women vaginally colonized by Staphylococcus aureus producing toxic shock toxin 1 and who use tampons. The aim of the present study was to determine the impact of the composition of vaginal microbial communities on tampon colonisation by S. aureus during menses. We analysed the microbiota in menstrual fluids extracted from tampons from 108 healthy women and 7 mTSS cases. Using culture, S. aureus was detected in menstrual fluids of 40% of healthy volunteers and 100% of mTSS patients. Between class analysis of culturomic and 16S rRNA gene metabarcoding data indicated that the composition of the tampons’ microbiota differs according to the presence or absence of S. aureus and identify discriminating genera. However, the bacterial communities of tampon fluid positive for S. aureus did not cluster together. No difference in tampon microbiome richness, diversity, and ecological distance was observed between tampon vaginal fluids with or without S. aureus, and between healthy donors carrying S. aureus and mTSS patients. Our results show that the vagina is a major niche of. S. aureus in tampon users and the composition of the tampon microbiota control its virulence though more complex interactions than simple inhibition by lactic acid-producing bacterial species.
Subject terms: Microbiome, Translational research
Introduction
The continuous interactions between an individual host and its microbial community (or microbiota) impact the development of the holobiont1–3. The host commensal microbiota is clearly important to host health. In healthy individuals, Bacteroidetes, Proteobacteria, and Firmicutes, such as Lactobacillales, are the major phyla among human microbiota4,5. These commensals develop complex networks with other microbes and the host, constituting the first line of defence against pathogen invasion by inhibiting colonization4,6,7. The contribution of human indigenous microbiota to the control of disease severity and the modulation of pathogen virulence is an emerging issue in the domain of clinical infectiology8,9.
Staphylococcus aureus, an opportunistic human pathogen, is a commensal species retrieved from the microbiota of almost 30% of humans10,11. S. aureus colonization is due to host factors and other members of the microbiota that may act as facilitators or antagonists. For example, both Corynebacterium spp. and Staphylococcus epidermidis have been shown to antagonize nasal colonization by S. aureus12,13.
A subpopulation of S. aureus is able to provoke toxic shock syndrome (TSS), an acute life-threatening illness characterized by sudden onset of fever, cutaneous signs (rash followed by desquamation), hypotension, and multiple organ failure, through the production of TSS toxin-1 (TSST-1). Menstrual TSS (mTSS), a rare but severe disease, is associated with menstruation and tampon use in previously healthy women vaginally colonized by S. aureus clones producing TSST-1 (TSST-1+)14–16. Colonizing S. aureus strain use the catamenial products collected in the tampon as a growth medium and proliferate17.
Although vaginal colonization by TSST-1+ S. aureus has been reported in 1% to 4% of the populations studied18–20, mTSS afflicts only 1 to 3 in 100,000 women18,19. The current assumption of this low prevalence relies on different factors that include the vaginal environment and its resident microbiota. For example, lactobacilli have been shown to be keystone species in vaginal health by strongly affecting the ecology of pathogenic bacteria, controlling their growth and/or virulence5,21.
However, most studies have examined variations in the vaginal microbiota during different microbial infections, such as bacterial vaginosis, or even HIV infection22–25, but none have examined the influence of vaginal microbial communities on the colonization of S. aureus in the vagina and whether an aberrant vaginal microbiota would favour vaginal colonization by S. aureus, especially in women using tampons during menses. The oldest studies essentially relied on prokaryote culture, whereas new sequencing techniques are enabling the detection of uncultivable parts of the microbiota. The current challenge is to reconcile both metataxonomic approaches with culturomic analysis in order to assess bacterial interaction theories developed from community analysis (i.e., metabarcoding) by performing experimental bacterial competitions (cultivable microbiome)26.
The aim of the present study was to determine the impact of the composition of vaginal microbial communities on tampon colonisation by S. aureus during menses. For this purpose, we examined used tampons from healthy women and mTSS cases utilizing culturomic and 16S rRNA gene metabarcoding analyses.
Methods
Subjects
Women recruitment, tampons collection and experimental protocols were approved by the ethical committee (Comités de Protection des Personnes (CPP), Interregion Sud-Est, n° L13–159). Ninety-one menstruating healthy volunteers were recruited between March 2014 and August 2016 through the gynaecology departments of Hospices Civil de Lyon. The only inclusion criterion was the use of tampon during menses, regardless of brand. Each volunteer signed informed consent forms. During the same period, 7 mTSS cases in France were spontaneously referred to the National Reference Center for Staphylococci and included in the study. The TSS diagnosis was based on the Centers for Disease Control and Prevention criteria (CDC) diagnostic criteria27. In brief, a woman was considered as mTSS if she met all CDC criteria as confirmed or probable TSS: The TSS occurs during menstruation with the use of tampon, the presence of S. aureus producing TSST-1 in vaginal specimen, and the absence of focal suppurative S. aureus infection and positive blood culture27. The clinical and biological characteristics of the cases of mTSS are described in Table S1 in supplementary data.
Sample collection
A total of 213 tampons were collected from the 91 healthy menstruating women. Forty-eight women provided three tampons (one tampon per menstrual cycle, not necessarily consecutive), 26 provided two tampons, and 17 provided only one tampon. Tampons were placed in a sterile flask and stored at 4 °C until analysis. The conservation at 4 °C never exceed 4 days for analyses in culturomic, beyond this limit the cultivable population decreased dramatically (data not shown). Each tampon was accompanied by an information form about the woman (i.e., age, history of salpingitis or mTSS, antibiotics within the last 15 days, methods of contraception, sexual intercourse within the last 5 days) and tampon (i.e., date of removal, tampon brand, carrying time). For mTSS cases, the incriminated tampon was withdrawn by the doctor or nurse in charge of the patient, placed in a sterile flask, and stored at 4 °C until analysis.
Menstrual fluid extraction
Menstrual fluid was extracted by suspending the tampon in 15 mL of sterile distilled water and then pressing it. The fluid was aliquoted and maintained as frozen stock (−20 °C) until further analysis. Stocks for the culturomic approach contained 30% glycerol.
S. aureus vaginal screening
S. aureus screening was performed by culturing, which has been shown to be more sensitive than PCR28, and isolates could be subsequently screened for TSST-1 status. Fifty microliters of menstrual fluid were spread on a SAID chromogenic plate selective for S. aureus (chromID™ S. aureus, Biomérieux, Marcy l’Étoile, France). Briefly, on this chromogenic medium, S. aureus forms distinctive green colonies due to production of α-glucosidase. Other Staphylococcus species are developing white colonies or occasionally produce pink colonies due to the hydrolysis of a second chromogenic substrate for β-glucosidase. Real-time SYBR®Green biplex PCR nuc/tst was performed for each green colony suspected of S. aureus for species identification and the co-detection of S. aureus nuclease gene (nuc) and TSST-1 gene (tst). The primers were as follows: tst1, 5′-GTA AAA GTG TCA GAC CCA CT-3′; tst2, 5′-ATT TTT TTA TCG TAA GCC CTT-3′29; nucF, 5′-CAT CCT AAA AAA GGT GTA GAG A-3′; and nucR, 5′-TTC AAT TTT MTT TGC ATT TTC TAC CA-3′30. PCR was performed using the LightCycler® Fast Start DNA Master SYBR Green kit (Roche, Meylan, FRANCE) and performed in the LightCycler® thermocycler (Version 4.1) with the following program: denaturation for 30 s at 95 °C, amplification in 35 cycles of 15 s at 95 °C, 20 s at 62 °C, and 30 s at 72 °C, and annealing at 95 °C, 15 s at 65 °C, and 90 °C. The tst + S. aureus HT20030749 strain belonging to the clone Geraldine was used as a positive control31.
For contradictory data on the presence of S. aureus in a sample obtained between metabarcoding and culturomic analysis, real presence of S. aureus was checked by real-time PCR on metagenomic DNA.
For further analysis, only one tampon per women was included unless S. aureus detection differed between tampons (including also differences between S. aureus tst + and S. aureus tst−carriage). Thus, 108 tampons from healthy women and 7 tampons from mTSS cases were selected to analyse the diversity of the vaginal microbiota during menstruation.
Culturomic analysis
Culturomic methods consisted of plating menstrual fluids extracted from the 108 tampons on selective media in different atmospheres, followed by colony identification by matrix-associated laser desorption ionization - time of flight (MALDI-TOF) mass spectrometry and 16S rRNA gene sequencing when required. In brief, thawed menstrual fluids, that were less than 4 days old, were plated in diverse media with sterile 10 µL-calibrated spreaders. The media and growth conditions used in this approach are listed in Table 1. Briefly, culture media used are normative chromogenic media that are used in medical diagnosis. Thus, it is known that only a selected range of bacterial species will grow on these specific culture media by forming phenotypically different colony forming units (CFU). These CFU develop morphologies and colours specific to each species or genus on a given selective medium. For each phenotypically different CFU, semi-logarithmic quantification was performed32. The logarithmic quantification corresponds to the calibrated loop (10 µL) method that is commonly used to quantify the bacteriuria. The plate reading is in log.mL−1 with a limit of detection at 102 CFU.mL−1. Identification was performed by MALDI-TOF with the Vitek MS analyser (Biomérieux) and Vitek version 2.3.3 database. When identification failed, 16S rRNA gene (rrs) V5/V6/V7 variable region sequencing was performed using the following primers as described by33: Neck1, 5′-TCA AAK GAA TTG ACG GGG GC-3′ and Neck2, 5′-GCC CGG GAA CGT ATT CAC-3′. Sequences were analysed by leBIBIQBPP software (Bioinformatics Bacterial Identification - Quick BioInformatic Phylogeny of Prokaryotes, V1.0.1). When identical species was detected in multiple culture mediums, the highest determination was selected to approximate the relative ratio of genera by culturomic analysis. Among the 108 tampons, only 82 could be cultivated, with the other 26 resulting in no growth. The seven mTSS tampons were collected from different French hospitals and processed at the collection site for S. aureus isolation and DNA extraction. The culturomic approach was not carried out with these tampons because of the extended delay in transportation to the Centre National de Reference des Staphylocoques.
Table 1.
Media and growth conditions.
| Media | Isolation | Atmosphere | Incubation period (hours) |
|---|---|---|---|
| Candida ID 2 agar (CAN2, Biomérieux®) | Selective isolation of yeasts and the direct identification of Candida albicans | Aerobic | 48 |
| Chocolate Agar PolyViteX (PVX, Biomérieux®) | Isolation of fastidious bacteria | 5% CO2 | 48 |
| Columbia Blood Agar with 5% Sheep Blood (COS, Biomérieux®) | Selective isolation of fastidious bacteria, detection of haemolysis | Anaerobic | 72 |
| Columbia Blood Agar with 5% Sheep Blood, Colistin, and Nalidixic Acid (CNA, Biomérieux®) | Selective isolation of Gram-positive bacteria | 5% CO2 | 48 |
| Gardnerella Selective Agar with 5% Human Blood (Gadnerella, BD®) | Partially selective isolation of Gardnerella vaginalis | 5% CO2 | 48 |
| Lactobacillus Selection Agar (LBS, BD®) | Partially selective isolation of lactobacilli | Anaerobic | 72 |
| Schaedler Kanamycin-Vancomycin Agar with 5% Sheep Blood (Schaedler-KV, BD®) | Selective isolation of fastidious Gram-negative anaerobic microorganisms, especially Bacteroides and Prevotella species | Anaerobic | 72 |
| UriSelect 4 Agar (URI4, BioRad®) | Isolation of urinary pathogens and direct identification of Escherichia coli, Enterococcus spp., and Proteus mirabilis | Aerobic | 48 |
Metabarcoding analysis
Microbial DNA were efficiently extracted from 107 of the 108 tampons from healthy menstruating women and the 7 tampons from mTSS cases, which were analysed for the diversity of vaginal microbiota by 16S rRNA gene metabarcoding analysis. Briefly, to extract the total microbial DNA from menstrual fluid, 1 mL of the fluid sample was centrifuged at 11,000 × g at 4 °C for 10 min. The supernatant was discarded and the pellet suspended in 200 µL of high purity water. Total DNA was extracted using the NucleoSpin Tissue® DNA Isolation Kit (Macherey-Nagel, Hoerdt, France) according to the manufacturer’s instructions. The DNA quality and quantity were assessed by agarose gel electrophoresis and using the Quant-iT™ PicoGreen® dsDNA Assay Kit (Invitrogen, Carlsbad, CA, USA) in 96-well microplates and a TECAN infinite M200 Pro spectrofluorometer (Tecan Trading AG, Männedorf, Switzerland). DNA was adjusted to a concentration of 10 ng/µL and sent to the MR DNA laboratory (Shallowater, TX, USA) for sequencing. The 16S rRNA gene V3/V4 variable region PCR primers 341 F/785 R34,35 with a barcode on the forward primer were used in a 28-cycle PCR (5 cycles implemented on PCR products) using the HotStarTaq Plus Master Mix Kit (Qiagen, Valencia, USA) under the following conditions: 94 °C for 3 min, followed by 28 cycles of 94 °C for 30 s, 53 °C for 40 s, and 72 °C for 1 min, with a final elongation step at 72 °C for 5 min. PCR products were checked on a 2% agarose gel to determine amplification success and relative band intensity. Multiple samples were pooled together in equal proportions based on their molecular weight and DNA concentrations. Pooled samples were purified using calibrated Ampure XP beads and used to prepare an Illumina DNA library. Sequencing was performed on a MiSeq following the manufacturer’s guidelines. Sequence data were processed using the MR DNA analysis pipeline. Briefly, sequences were joined, depleted of barcodes, and sequences <150 bp or with ambiguous base calls removed. Sequences were denoised. Operational taxonomic units (OTUs) were defined by clustering at 3% divergence (97% similarity) and chimeras removed. Final OTUs were taxonomically classified using BLASTn against a curated database derived from Greengenes36, RDPII (http://rdp.cme.msu.edu), and NCBI (www.ncbi.nlm.nih.gov).
When S. aureus was detected in sample only by 16S rRNA gene sequencing, its presence was verified by qPCR targeting nuc gene.
Nucleotide sequence accession numbers
Reads have been deposited in the European Bioinformatics Institute (EBI) database under accession number PRJEB24013.
Diversity analysis
Sequencing datasets were used to generate rarefaction curves and Shannon, Simpson, and Chao diversity indices. The indices are presented as the average of the individual sample sets (calculated at the OTU level). Extracted from tampons’ fluid, vaginal cultivable bacterial diversity and 16S rRNA gene diversity from the different groups of women (healthy women carrying or not carrying S. aureus that produced or did not produce TSST-1) were compared in between-class analysis (BCA)37 with the ADE4 and ggplot2 packages for R38,39. BCA is a robust alternative to linear discriminant analysis40 when the number of samples is too small compared to the number of predictors. When the number of samples is low, and particularly when it is lower than the number of predictors, discriminant analysis cannot be used. In this case, BCA can be very useful and provides illustrative graphical displays of between-group differences41. The significance of BCA was assessed using a Monte-Carlo test with 10,000 permutations (null hypothesis: absence of difference between groups). The genera contributing most to differentiating the group of women were identified. Unless otherwise stated, statistical analyses were performed using R version 3.3.2 at p < 0.05.
Comparison of the culturomic and metabarcoding approaches
A co-inertia analysis42,43 was performed with the ADE4 software on the 81 samples that were analysed by both culturomic and metabarcoding approaches. Co-inertia analysis comes from Ecological data analysis, and comprises two steps. In the first step, two separate PCA (Principal Component Analysis) are used to find inertia axes and sample scores in each of the two data sets (culturomic analysis and 16S rRNA gene metabarcoding). In a second step, the co-inertia analysis computes new axes that maximize the squared covariance (called “co-inertia”) between PCA sample scores. Co-inertia is a global measure of the relationship between samples of the two data sets: it is high when the two sample scores vary simultaneously (with either a positive or negative relationship), and low when they vary independently. The statistical significance of co-inertia analysis was assessed with a Monte-Carlo test using 10,000 permutations. The null hypothesis of this test was the absence of a relationship between culturomic analysis and 16S rRNA gene metabarcoding scores.
Ethics approval and consent to participate
Written informed consent was obtained from the volunteers and patients in accordance with the guidelines of the ethical committee (Comités de Protection des Personnes (CPP), Interrégion Sud-Est, France, approval L13–156).
Results
Cohort description
The median age of healthy volunteers was 25 years (range 18–52 years). Only one volunteer had a history of salpingitis and two were treated with antibiotics within the 15 days prior to sampling (Table 2). Sixty-nine percent used contraception: 56% (i.e. 60 volunteers) the oestroprogestative pill and 13% (i.e. 14 volunteers) intrauterine device. Tampax® was the most used tampon brand, followed by Nett® (Table 2). The majority of volunteers (93%; i.e. 100 volunteers) did not exceed the recommended maximum use duration of 8 hours. The median delay between tampon collection and analysis was 1 day (range 0–9 days; Table 2).
Table 2.
Characteristics of the 108 samples used for the diversity analysis of the healthy vaginal microbiota.
| Sample | n = 108 | |
|---|---|---|
| Age | ||
| ≤25 years | 46 | 42.59% |
| 26–35 years | 40 | 37.04% |
| >35 years | 22 | 20.37% |
| History of salpingitis | 2 | 1.85% |
| History of mTSS | 1 | 0.93% |
| Tampon brand | ||
| Tampax ® | 43 | 39.81% |
| Nett® | 29 | 26.85% |
| Carrefour® | 8 | 7.41% |
| OB® | 5 | 4.63% |
| Leader Price® | 5 | 4.63% |
| Others | 18 | 16.67% |
| Tampon use duration | ||
| median 4 h 30 [range 27 min-24 h] | ||
| 0–4 h | 49 | 45.37% |
| 4–8 h | 51 | 47.22% |
| >8 h | 7 | 6.48% |
| NA | 1 | 0.93% |
| Antibiotic use in the previous 15 days | ||
| Total | 2 | 1.85% |
| Doxycycline | 1 | 0.93% |
| Tetralysal | 1 | 0.93% |
| Contraceptive method | ||
| None | 33 | 30.56% |
| Oestroprogestative pill | 60 | 55.56% |
| Intra-uterine device | 14 | 12.96% |
| NA | 1 | 0.93% |
| Sexual intercourse in the previous 5 days | ||
| Yes | 40 | 37.04% |
| No | 66 | 61.11% |
| NA | 2 | 1.85% |
mTSS, menstrual toxic shock syndrome; NA, not available.
Collected information came from the survey accompanying each sample.
Four of the mTSS cases were confirmed and three probable (Table S1). The median age of mTSS cases was 17 years (range 14–30 years). None of them were treated with antibiotics within 15 days prior to sampling. The mode of contraception was not recorded. Tampax® was the most often used tampon brand.
S. aureus vaginal carriage during menses in women using tampon
The S. aureus vaginal carriage status of the 91 volunteers was determined by culturomic analysis from tampons. Sixty-five of the 213 tampons (30.5%) were colonized by S. aureus at a given time. Among these, 10 tampons (4.7%) were colonized by a S. aureus tst + isolate. But when we relate these values to the 91 volunteers the numbers become 44 volunteers were found to carry S. aureus (consistently or transiently; 48% of 91 volunteers), and 6 volunteers (6,6% of 91 volunteers) are carrying an S. aureus tst + isolate (consistently or transiently colonized).
Forty-eight volunteers performed follow-up over three menstrual cycles, most generally consecutive. Interestingly, 25 (52% of the 48 volunteers) were colonized by S. aureus on at least one of the cycles. Among these 25 carriers, 6 volunteers (24%) had colonization by S. aureus over 3 cycles and remaining 19 volunteers (76%) had transient colonization (1 or 2 of the 3 cycles, all combinations were retrieved). When detected, S. aureus reached a concentration of 300 to >3 × 107 CFU per tampon (outnumbering the detection technique).
The age of the participants, the tampon brand and duration of use, the delay between tampon collection and analysis, the contraceptive method, and sexual intercourse in the 5 days prior to sampling had no significant effect on S. aureus detection (Additional file 1: Table S2). For further analysis, only one tampon per woman was kept, unless a difference in S. aureus carriage was found between 2 months. Thus, 108 tampons were selected to analyse the diversity of the vaginal microbiota (Table 2). S. aureus strains carrying tst were detected in all tampons recovered from mTSS cases and exceeded 3 × 107 CFUs per tampon.
Culturomic versus metabarcoding to detect S. aureus carriage
Sequence analysis of the 16S rRNA gene V3/V4 variable region suggested that S. aureus could be retrieved in each samples (Additional file 1: Table S3). However, culturomic analysis determined presence of S. aureus only in 44 out of the 91 analysed samples. nuc gene (a thermonuclease gene specific to S. aureus) real-time PCR performed on the 91 samples confirmed the culturomic data, suggesting that V3/V4 variable region cannot specifically target S. aureus sensu stricto. False positive obtained through 16SrRNA gene diversity sequencing data analysis showed from 8 to 78 reads per samples, whereas S. aureus positive sample exhibit from 9 reads to 9577 reads. Cultivable analysis also showed to be globally more sensitive than 16S rRNA gene metabarcoding as example the 3 mTSS cases (patient 94, 95 and 96; Additional file 1: Table S3) that showed less than 60 reads (reads similar to false positives) affiliated to S. aureus whereas culturomic showed that the bacteria outnumbered the detection limit (>107 CFU/mL).
Analysis of tampon microbiota diversity during menses by culturomic analysis
Colonies were detected in 82 of the 108 menstrual fluid samples. A total of 51 genera and 96 different species were identified. The number of cultivable bacterial genera colonizing tampons varied from 2 to 11 (median 6), and the number of cultivable bacterial species colonizing tampons varied from 2 to 15 (median 8). The cultivable microbiota composition of 82 tampons is shown in the Additional file 1: Fig. S1B.
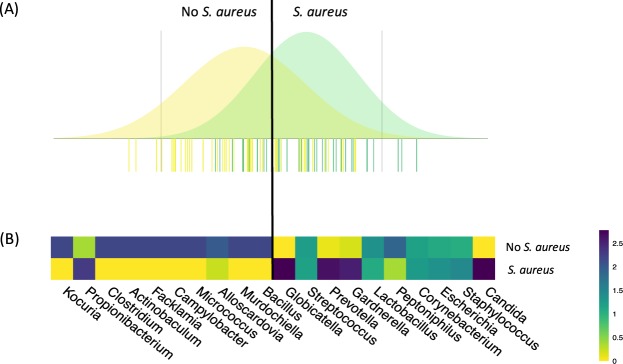
We examined the effect of age, tampon use duration, contraceptive method, and sexual intercourse in the 5 days prior to sampling on the composition of the vaginal cultivable microbiota by BCA. Only tampon use duration exhibited a significant effect on the composition of the vaginal cultivable microbiota (Additional file 1: Fig. S2). Next, the impact of S. aureus vaginal carriage on microbiota composition was assessed. BCA of culturomics data indicated that the composition of the vaginal microbiota differed according to the presence of S. aureus (Fig. 1A). Candida, non-aureus Staphylococcus, Escherichia, Corynebacterium, Peptoniphilus, Lactobacillus, Gardnerella, Prevotella, Streptococcus, and Globicatella were the bacterial genera most commonly associated with vaginal carriage of S. aureus, whereas Kocuria, Propionibacterium, Clostridium, Actinobaculum, Facklamia, Campylobacter, Micrococcus, Alloscardovia, Murdochiella, and Bacillus were the bacterial genera most often associated with the absence of S. aureus (Fig. 1B).
Figure 1.
Composition of the vaginal microbiota during menses as determined by culturomic analysis as a function of vaginal colonization by S. aureus. (A) Between-class analysis was performed at the bacterial genus level. Thirty-six samples were from women vaginally colonized by S. aureus (green) and 46 samples from women not colonized by S. aureus (yellow). The analysis was supported by a Monte Carlo test (p = 0.026). The data include the distribution of bacterial genera according to their contribution to differentiation of the vaginal microbiota of women carrying or not carrying S. aureus. (B) Heatmap showing the average relative proportion of the 10 microbial genera most commonly associated with vaginal carriage of S. aureus (left) and 10 microbial genera most often associated with the absence of S. aureus at the vaginal stage (right) in samples containing (lower line) and not containing S. aureus (upper line). Heatmap unit is a percentage of the average relative proportion of taxa.
Analysis of tampon microbiota diversity during menses by metabarcoding analysis
The bacterial diversity of the microbiota was determined by MiSeq Illumina sequencing of the V4 variable region of rrs. A total of 261 different bacterial genera were identified from the microbial DNA efficiently extracted from 107 of the 108 tampons from healthy donors and all 7 from mTSS cases. For the healthy donors, an average of 96,251 sequence reads was obtained from each sample (range 42,247–201,056; median 90,622). The number of genera per tampon varied from 29 to 96 (median 59). The proportions of bacterial genera/species found in vaginal microbiota isolated from the different tampons are shown in the Additional file 1: Figs S1A and S3. Clustering of the 107 samples revealed nine different clusters dominated by Lactobacillus iners (two clusters), Shigella sonnei/Escherichia coli, Lactobacillus gallinarum and Lactobacillus crispatus, Gardnerella vaginalis, Streptococcus agalactiae or Staphylococcus epidermidis, and a last cluster with vaginal microbiota showing no dominant species (Additional file 1: Fig. S3).
For the seven mTSS donors, an average of 103,808 sequence reads was obtained from each sample (range 45,417–151,011; median 101,049). The number of genera per tampon varied from 50 to 128 (median 72). The proportions of bacterial species found in vaginal microbiota isolated from the different tampons are shown in the Additional file 1: Fig. S3. The bacterial communities of the seven mTSS cases did not cluster together. Three patients had vaginal microbiota clustering with a known cluster. For example, in patient 98 (vaginal mTSS case), the microbiota was in the cluster dominated by L. iners, patient 94 in a cluster dominated by G. vaginalis, and patient 78 in the cluster dominated by S. sonnei/E. coli. The four other cases (96, 92, 93, and 97) formed unique branches with multiple bacterial species dominating their vaginal community (Additional file 1: Fig. S3).
Microbiota that were characterized from the tampons of women vaginally colonized with S. aureus that did or did not produce TSST-1 and mTSS cases were distributed in many of these groups and did not appear to share the same microbiota composition. Only patient 96, an mTSS case, exhibited a specific community dominated by Fusobacterium gonidiaformans and Prevotella bivia. The 10 women for whom two tampons were analysed, one colonized by S. aureus and the other not, presented vaginal microbiota profiles belonging to different clusters. In three cases, the community changed very little (women 43, 49, and 88), the dominant species remained the same, and the two tampons belonged to the same group. In others, different communities were observed between the two tampons, and the two communities were no longer dominated by the same species. This was the case for woman 28, whose tampon microbiota changed to a community dominated by L. gallinarum and L. crispatus in the absence of S. aureus to a community dominated by G. vaginalis when carrying S. aureus (Additional file 1: Fig. S3).
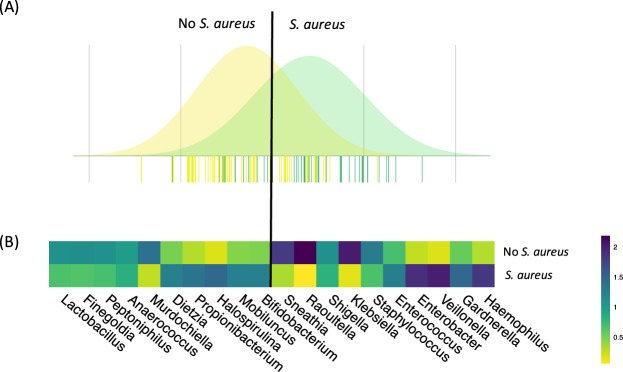
As was done for the culturomic analysis, we examined the effect of age, tampon use duration, contraceptive method, and sexual intercourse in the 5 days prior to sampling on the composition of the vaginal microbiota by BCA (Additional file 1: Fig. S2). In the case of the metabarcoding analysis, BCA of 16S rRNA gene sequences indicated that the composition of the vaginal microbiota during menses differed with age (Additional file 1: Fig. S2) and vaginal colonization by S. aureus (Fig. 2A). Haemophilus, Gardnerella, Veillonella, Enterobacter, Enterococcus, non-aureus Staphylococcus, Klebsellia, Shigella/Escherichia, Raoultella, and Sneathia were the bacterial genera most commonly associated with vaginal carriage of S. aureus, whereas Lactobacillus, Finegoldia, Peptoniphilus, Anaerococcus, Mudochiella, Dietzia, Propionibacterium, Halospirulina, Mobiluncus, and Bifidobacterium were the bacterial genera most often associated with the absence of S. aureus at the vaginal stage (Fig. 2B).
Figure 2.
Composition of the vaginal microbiota during menses as determined by 16S rRNA gene metabarcoding analysis as a function of vaginal colonization by S. aureus. (A) Between-class analysis was performed at the bacterial genus level. Forty-four samples are from women vaginally colonized by S. aureus (green) and 63 from women not colonized by S. aureus (yellow). The analysis is supported by a Monte Carlo test (p = 0.028). Data include the distribution of bacterial OTU according to their contribution to differentiation of the vaginal microbiota of women carrying or not carrying S. aureus. (B) Heatmap showing the average relative proportion of the 10 bacterial genera most commonly associated with vaginal carriage of S. aureus (left) and 10 bacterial genera most often associated with the absence of S. aureus at the vaginal stage (right) in samples containing (lower line) and not containing S. aureus (upper line). Heatmap unit is a percentage of the average relative proportion of taxa.
The diversity indices calculated at the OTU level are presented in the Additional file 1: Table S4. No significant differences were found based on the presence of S. aureus TSST-1 status.
Correlation of the two methods
Culturomic and 16S rRNA gene metabarcoding analyses were also evaluated by co-inertia analysis (Fig. 3). The results showed a strong co-structure between the PCA resulting from the culturomic data and the PCA resulting from the metabarcoding data, supported by a significant Monte Carlo test (p < 0.05). This result confirms that the bacterial microbiota assessed by culturomic and metabarcoding analyses are closely linked.
Figure 3.
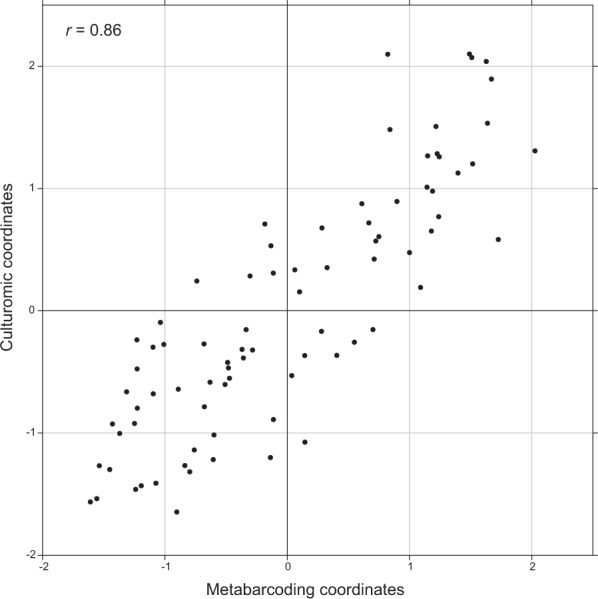
Coinertia analysis between bacterial community data obtained by 16S rRNA metabarcoding (abscissa) and culturomic (ordinate) analyses. Each point on the figure corresponds to one sample (tampon). The value of the correlation coefficient is 0.86. The samples in the upper right-hand corner have high values for the same bacterial species in both metabarcoding and culturomic analyses. Conversely, the samples at the bottom left have low values for the same bacterial species in both metabarcoding and culturomic analyses.
As the data from the BCAs analysis showed some common taxa between culturomic and metabarcoding, the microbiota fractions from the 81 samples tested by both culturomic and 16S rRNA gene metabarcoding analysis were compared (Additional file 1: Fig. S1). The relative proportion of each common genus between the two methods is represented for each sample and the ratio calculated for each genus, excluding other. When considering the common genera between both techniques, the mean ratios were not significantly different from 1 (Student’s test), indicating a correspondence between the two methods (Additional file 1: Fig. S1C).
Discussion
The vaginal bacterial community in apparently healthy reproductive age women has been shown to present a strong barrier against various infections5,44–46, but its role in preventing S. aureus colonization or virulence is poorly understood. mTSS is a rare but specific disease caused by TSST-1-producing S. aureus in women who use tampons or a menstrual cup as periodic protection15,47,48. Colonizing S. aureus strain use the catamenial products collected into the intra vaginal device as a growth medium and proliferate17,49,50.
The aim of the present study was to determine whether the composition of microbial communities that colonised tampon during menstruation is associated to the vaginal carriage of S. aureus and TSST-1-producing S. aureus.
mTSS is more prevalent in young women. We initially though that age, though hormonal changes, has an impact on vaginal microbiological communities that interfere with S. aureus carriage. Our results do not support this hypothesis and that the high prevalence of mTSS in young women population is linked to higher carriage of S. aureus in this population15.
S. aureus carriage was assessed by cultivable methods, which have been described to be more sensitive than PCR techniques28 and allowed subsequent screening of the presence of tst. Thus, we detected tampon colonization by S. aureus in 40.7% of tampons from healthy women, which is higher than the previous determination of 9–28% in menstruating women based on vaginal sampling by swab instead of tampons51,52. None of the clinical data recorded in our study appeared to affect vaginal S. aureus carriage in healthy women (i.e., age, tampon brand, duration of use, contraceptive method, sexual activity in the 5 days prior to sampling). Among the carriers of S. aureus, 24% were positive during three consecutive menstruations. This result is startlingly similar to that observed for S. aureus nasal colonization with approximately 30% of permanent S. aureus carriers and 70% intermittent carriers10,11.
We determined that TSST-1+ S. aureus colonized 5.7% of the tested tampons, which is a little higher but comparable to previous studies reporting vaginal colonization by TSST-1+ S. aureus in 1% to 4% of the woman studied18–20. Interestingly, although S. aureus was found by culture (from 300 CFU/mL to >105 CFU/mL), it could not always be correctly retrieved by the 16S RNA gene sequencing approach, suggesting that the bacteria did not reach the detection level. Even in tampons from mTSS cases, in which culture approaches estimated colonization of >107 CFU/mL, the bacterium was detected below 60 sequences by meta-analysis for patients 94, 95, and 96 (Additional file 1: Fig. S3). These data were consistent with previous studies showing that, in complex ecosystems such as the gut microbiota, culture-independent techniques are unable to detect bacteria <106 cells/mL28. The present data highlight that TSST-1+ S. aureus is not necessarily the most abundant taxa in the vaginal of mTSS cases.
Recent community profiling by metataxonomic sequencing techniques have shown that healthy vaginal microbial communities are usually dominated by Lactobacillus (L. crispatus, L. iners, L. gasseri, L. jensenii) or mixed profiles depleted of lactobacilli and consisting of bacteria such as Prevotella, Gardnerella, Atopobium, Megasphaera, and Streptococcus24,53–57. The results of our 16S RNA gene study demonstrated that besides the currently published community states type (CST; vaginal microbiota which are dominated by one or more species 50% of all sequences in a sample24,55,56), new state dominated by different populations exist on healthy women’s tampons. L. gasseri (CST II) and L. jensenii (CST V), which are prominent in other studies24,35, were not dominant organisms in any microbiota profile in the current study. Only L. crispatus (CST I), L. iners (CST III) and G. vaginalis (CST IV) dominating communities were already described in previous studies24,55,56. But new CST dominated by Escherichia/Shigella, Streptococcus agalactiae CST or S. epidermis were characterized (Fig. S3). These divergences from previous studies may rely on different factors, such as socio-demographics, sexual activity, health behaviour, and hygiene, but the most important factor that distinguishes the current study from previous studies was the sampling technique. To the best of our knowledge, previous studies all relied on swab sampling of the vaginal microbiota, whereas we directly studied the tampon colonization. Tampons were also in contact with skin hand microbiota when they were inserted and removed without any other protection than hand washing. Besides, tampons constitute a complex environment for bacteria. They are not uniformly impregnated by menstrual fluid, creating micro-niches enabling the development of specific metabolism and interactions, such as the production of TSST-114.
However, consistent with previous studies assessing vaginal microbial diversity, we found that the tampon microbiota varied largely among healthy women and were, in a certain manner, specific to each woman24,35,54–56,58. Most women (11/16) with differential carriage of S. aureus between two menses presented changes in the microbiota composition. Tampons from two different cycles had a conserved community dominated by the same species in only five women. This corroborated previous studies showing that the vaginal microbiota may shifted to different community state through menstrual cycles35,55,58. In contrast to our expectations, clustering analysis did not distinguish a specific cluster grouping a microbial community carrying TSST-1+ S. aureus or the microbiota of mTSS cases. In addition, the richness, diversity, and ecological distance were similar between S. aureus carriers and non-carriers. These results suggest that neither variation in community composition nor constantly high levels of apparent diversity are indicative of S. aureus carriage status. Thus, the tampon microbiota composition that may control the ecology of TSST-1+ S. aureus and its virulence during toxic shock syndrome either directly by controlling (positively or negatively) its growth and/or TSST-1 production21, or indirectly by modifying the vaginal environmental conditions (i.e., pH, carbon, O2 levels)59, is not completely reshaped when S. aureus is present.
Carriage of S. aureus has a greater impact on the non-dominant population in the microbiota. BCA of the microbiota defined by the culturomic approach and rrs metabarcoding analysis showed that the microbiota of healthy menstruating women differed according to their S. aureus carriage status (Figs 1 and 2). Several bacterial genera, such as Murdochiella and Enterobacter, were preferentially found in the absence of S. aureus carriage, whereas non-aureus Staphylococcus and Bifidobacterium were more abundant in the presence of S. aureus carriage. Notably, Lactobacillus, a genus often described as an inhibitor of the growth of several pathogens, including S. aureus58,59,60,61, was not specifically associated with the carriage status. Here, we found that, even in the microbiota of tampons from mTSS cases, lactobacilli could be predominant. This observation contradicts some studies suggesting that the vaginal microbiota of healthy women is predominantly colonized by lactobacilli and protect against mTSS, whereas in the case of dysbiosis, such as vaginosis, populations of lactobacilli are overgrown by other bacterial populations21,62. Unlike lactobacilli, S. agalactiae significantly co-presents with S. aureus in pregnant women63 and has been shown to induce toxin production in S. aureus21. Although S. agalactiae was co-identified with S. aureus in the current study, it was not abundant in mTSS cases, suggesting that abundance of this bacterium does not correlate with an increased risk of developing mTSS.
These observations call into question whether tampon microbiota profiling studies may be reliably used to link vaginal bacterial community composition to health outcomes. Taxonomic composition is not sufficient to decipher the functional outcomes of a given bacterial community. A possible way of studying microbiota functioning and interactions is to return to in vitro studies combining larger sets of bacterial populations. To date, the vaginal microbiota has been shaped predominantly by culture-based or 16S rRNA gene diversity sequencing studies. Although both approaches have some limitations, they contribute to a wealth of information on microbiota composition. 16S rRNA gene-based studies are known to gain access to a large diversity of uncultivable bacteria, but have amplification biases and, for some, taxa-limited resolution63. Culturomic analysis associating bacteriological culture with characterization by MALDI-TOF enables the identification of large sets of bacteria populations28,64–67, but is heavy to handle since large sets of culture media and culture conditions have to be tested and give a approximation of target the cultivable fractions (approximation for enumeration). Here, we analysed the bacterial community by rrs gene sequencing and a culturomic approach for the menstrual fluids extracted from 81 tampons. We demonstrated that both techniques provided data that significantly correlated and their analyses reached the same conclusions. Thus, in future studies, it would be interesting to obtain a set of cultivable bacteria to pursue competition assays combining culture and sequencing approaches.
In conclusion, we showed that shifts in microbiota composition consist not only of changes in the dominance of lactic acid-producing bacterial species55, but also in metabarcoding-undetectable species, such as TSST-1+ S. aureus, which could greatly impact the health of the patient.
Electronic supplementary material
Acknowledgements
We are grateful to all of the volunteers, François Vandenesch and Frederic Laurent for fruitful comments and the Ibio (UMR Écologie Microbienne 5557) for bioinformatics facilities. This work was supported by the LABEX ECOFECT (ANR-11-LABX-0048) of Université de Lyon within the programme “Investissements d’Avenir” (ANR-11-IDEX-0007) operated by the French National Research Agency (ANR).
Author Contributions
All authors have contributed as followed: Isaline Jacquemond, Jean Thioulouse, Claire Prigent-Combaret, Daniel Muller, Gérard Lina, Anne Tristan, Yves Gillet, Pierre-Adrien Bolze, François Golfier, and Tristan Ferry conceived and designed the experiments; Gérard Lina, Yves Gillet, Pierre-Adrien Bolze, Gery Lamblin, Anaëlle Muggeo Tristan Ferry, Anne Tristan, Claude-Alexandre Gustave, Michèle Bes and Isaline Jacquemond recruited women; Isaline Jacquemond and Anaëlle Muggeo performed the experiments; Isaline Jacquemond, Jean Thioulouse, Audrey Dubost, Danis Abrouk, Samuel Barreto, Daniel Muller, Jean-Philippe Rasigade, Michèle Bes and Gérard Lina analysed the data; Isaline Jacquemond, Anaëlle Muggeo, Jean Thioulouse, Claire Prigent-Combaret, Daniel Muller, and Gérard Lina wrote the paper; Isaline Jacquemond, Samuel Barreto and Jean Thioulouse prepared the figures and/or tables; and all authors reviewed the drafts of the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
1/30/2020
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
Contributor Information
Gérard Lina, Email: gerard.lina@univ-lyon1.fr.
Daniel Muller, Email: daniel.muller@univ-lyon1.fr.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-28116-3.
References
- 1.McFall-Ngai M, et al. Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl. Acad. Sci. USA. 2013;110:3229. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rosenberg E, Zilber-Rosenberg I. Symbiosis and development: The hologenome concept. Birth Defects Res. Part C Embryo Today Rev. 2011;93:56–66. doi: 10.1002/bdrc.20196. [DOI] [PubMed] [Google Scholar]
- 3.Theis KR, et al. Getting the Hologenome Concept Right: an Eco-Evolutionary Framework for Hosts and Their Microbiomes. mSystems. 2016;1:e00028–16. doi: 10.1128/mSystems.00028-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kamada N, Chen GY, Inohara N, Núñez G. Control of pathogens and pathobionts by the gut microbiota. Nat. Immunol. 2013;14:685–690. doi: 10.1038/ni.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma B, Forney LJ, Ravel J. The vaginal microbiome: rethinking health and diseases. Annu. Rev. Microbiol. 2012;66:371–389. doi: 10.1146/annurev-micro-092611-150157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wen L, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perry, S. et al. Infection with Helicobacter pylori Is Associated with Protection against Tuberculosis. PLoS ONE5 (2010). [DOI] [PMC free article] [PubMed]
- 8.Berrilli, F., Di Cave, D., Cavallero, S. & D’Amelio, S. Interactions between parasites and microbial communities in the human gut. Front. Cell. Infect. Microbiol. 2 (2012). [DOI] [PMC free article] [PubMed]
- 9.Clemente JC, Ursell LK, Parfrey LW, Knight R. The Impact of the Gut Microbiota on Human Health: An Integrative View. Cell. 2012;148:1258–1270. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Belkum A. Staphylococcal colonization and infection: homeostasis versus disbalance of human (innate) immunity and bacterial virulence. Curr. Opin. Infect. Dis. 2006;19:339–344. doi: 10.1097/01.qco.0000235159.40184.61. [DOI] [PubMed] [Google Scholar]
- 11.Wertheim HF, et al. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect. Dis. 2005;5:751–762. doi: 10.1016/S1473-3099(05)70295-4. [DOI] [PubMed] [Google Scholar]
- 12.Uehara Y, et al. Bacterial interference among nasal inhabitants: eradication of Staphylococcus aureus from nasal cavities by artificial implantation of Corynebacterium sp. J. Hosp. Infect. 2000;44:127–133. doi: 10.1053/jhin.1999.0680. [DOI] [PubMed] [Google Scholar]
- 13.Lina G, et al. Bacterial Competition for Human Nasal Cavity Colonization: Role of Staphylococcal agr Alleles. Appl. Environ. Microbiol. 2003;69:18–23. doi: 10.1128/AEM.69.1.18-23.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schlievert PM, Nemeth KA, Davis CC, Peterson ML, Jones BE. Staphylococcus aureus Exotoxins Are Present In Vivo in Tampons. Clin. Vaccine Immunol. CVI. 2010;17:722–727. doi: 10.1128/CVI.00483-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shands KN, et al. Toxic-Shock Syndrome in Menstruating Women. N. Engl. J. Med. 1980;303:1436–1442. doi: 10.1056/NEJM198012183032502. [DOI] [PubMed] [Google Scholar]
- 16.Todd J, Fishaut M, Kapral F, Welch T. Toxic-Shock Syndrome Associated with Phage-Group-I StaphylococcI. The Lancet. 1978;312:1116–1118. doi: 10.1016/s0140-6736(78)92274-2. [DOI] [PubMed] [Google Scholar]
- 17.Melish ME, Murata S, Fukunaga C, Frogner K, McKissick C. Vaginal Tampon Model for Toxic Shock Syndrome. Rev. Infect. Dis. 1989;11:S238–S247. doi: 10.1093/clinids/11.supplement_1.s238. [DOI] [PubMed] [Google Scholar]
- 18.DeVries AS, et al. Staphylococcal Toxic Shock Syndrome 2000–2006: Epidemiology, Clinical Features, and Molecular Characteristics. PLoS ONE. 2011;6:e22997. doi: 10.1371/journal.pone.0022997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hajjeh RA, et al. Toxic shock syndrome in the United States: surveillance update, 1979–1996. Emerg. Infect. Dis. 1999;5:807. doi: 10.3201/eid0506.990611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parsonnet J, et al. Prevalence of Toxic Shock Syndrome Toxin 1-Producing Staphylococcus aureus and the Presence of Antibodies to This Superantigen in Menstruating Women. J. Clin. Microbiol. 2005;43:4628–4634. doi: 10.1128/JCM.43.9.4628-4634.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.MacPhee RA, et al. Influence of the Vaginal Microbiota on Toxic Shock Syndrome Toxin 1 Production by Staphylococcus aureus. Appl. Environ. Microbiol. 2013;79:1835–1842. doi: 10.1128/AEM.02908-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nunn KL, Forney LJ. Focus: Microbiome: Unraveling the Dynamics of the Human Vaginal Microbiome. Yale J. Biol. Med. 2016;89:331. [PMC free article] [PubMed] [Google Scholar]
- 23.Onderdonk AB, et al. Normal vaginal microflora during use of various forms of catamenial protection. Rev. Infect. Dis. 1989;11:S61–S67. doi: 10.1093/clinids/11.supplement_1.s61. [DOI] [PubMed] [Google Scholar]
- 24.Ravel J, et al. Vaginal microbiome of reproductive-age women. Proc. Natl. Acad. Sci. USA. 2011;108:4680–4687. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou X, et al. Vaginal Microbiota of Women with Frequent Vulvovaginal Candidiasis. Infect. Immun. 2009;77:4130. doi: 10.1128/IAI.00436-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greub G. Culturomics: a new approach to study the human microbiome. Clin. Microbiol. Infect. 2012;18:1157–1159. doi: 10.1111/1469-0691.12032. [DOI] [PubMed] [Google Scholar]
- 27.Centers for Disease Control (CDC). Update: Toxic-shock syndrome–United States. MMWR. Morbidity and mortality weekly report, 32, 398-400 (1983). Available at: https://www.cdc.gov/mmwr/preview/mmwrhtml/00000119.htm. (Accessed: 20th October 2017). [PubMed]
- 28.Lagier J-C, et al. Microbial culturomics: paradigm shift in the human gut microbiome study. Clin. Microbiol. Infect. 2012;18:1185–1193. doi: 10.1111/1469-0691.12023. [DOI] [PubMed] [Google Scholar]
- 29.Jarraud S. Relationships between Staphylococcus aureus Genetic Background, Virulence Factors, agr Groups (Alleles), and Human Disease. Infect. Immun. 2002;70:631–641. doi: 10.1128/IAI.70.2.631-641.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brakstad OG, Mæland JA, Tveten Y. Multiplex polymerase chain reaction for detection of genes for Staphylococcus aureus thermonuclease and methicillin resistance and correlation with oxacillin resistance. APMIS. 1993;101:681–688. doi: 10.1111/j.1699-0463.1993.tb00165.x. [DOI] [PubMed] [Google Scholar]
- 31.Patot S, et al. The TIR homologue lies near resistance genes in Staphylococcus aureus, coupling modulation of virulence and antimicrobial susceptibility. PLoS Pathog. 2017;13:e1006092. doi: 10.1371/journal.ppat.1006092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cavallo. European Manuel of Clinical Microbiology. In European Manual ofClinical Microbiology (eds Cornaglia, G., Courcol, R., Herrmann, J. L. & Kahlmeter, G.) Ch. 14 Urinary tract infections, 133–144 (European Society for Clinical Microbiology and Infections Diseases, 2012).
- 33.Ferroni, A. et al. Prospective survey of acute osteoarticular infections in a French paediatric orthopedic surgery unit: Acute osteoarticular infections in children. Clin. Microbiol. Infect. no–no 10.1111/1469-0691.12031 (2012). [DOI] [PubMed]
- 34.Klindworth A, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41:e1–e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vodstrcil Lenka A., Twin Jimmy, Garland Suzanne M., Fairley Christopher K., Hocking Jane S., Law Matthew G., Plummer Erica L., Fethers Katherine A., Chow Eric P. F., Tabrizi Sepehr N., Bradshaw Catriona S. The influence of sexual activity on the vaginal microbiota and Gardnerella vaginalis clade diversity in young women. PLOS ONE. 2017;12(2):e0171856. doi: 10.1371/journal.pone.0171856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DeSantis TZ, et al. Greengenes, a Chimera-Checked 16S rRNA Gene Database and Workbench Compatible with ARB. Appl. Environ. Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dolédec S, Chessel D. Rythmes saisonniers et composantes stationnelles en milieu aquatique. I: Description d’un plan d’observation complet par projection de variables. Acta Oecologica Oecologia Gen. 1987;8:403–426. [Google Scholar]
- 38.Chessel D, Dufour A-B, Thioulouse J. The ade4 package - I: One-table methods. R News. 2004;4:5–10. [Google Scholar]
- 39.Dray S, Dufour AB, Chessel D. The ade4 Package–II: Two-table and K-tableMethods. R News. 2007;7:47–52. [Google Scholar]
- 40.Huberty, C. J. Applied discriminant analysis. (New York: Wiley, 1994).
- 41.Thioulouse J, Prin Y, Duponnois R. Multivariate analyses in soil microbial ecology: a new paradigm. Environ. Ecol. Stat. 2012;19:499–520. [Google Scholar]
- 42.Dolédec S, Chessel D. Co-inertia analysis: an alternative method for studying species–environment relationships. Freshw. Biol. 1994;31:277–294. [Google Scholar]
- 43.Dray S, Chessel D, Thioulouse J. Co-inertia analysis and the linking of ecological data tables. Ecology. 2003;84:3078–3089. [Google Scholar]
- 44.Stumpf RM, et al. The primate vaginal microbiome: Comparative context and implications for human health and disease. Am. J. Phys. Anthropol. 2013;152:119–134. doi: 10.1002/ajpa.22395. [DOI] [PubMed] [Google Scholar]
- 45.Tibaldi C, et al. Vaginal and endocervical microorganisms in symptomatic and asymptomatic non-pregnant females: risk factors and rates of occurrence. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2009;15:670–679. doi: 10.1111/j.1469-0691.2009.02842.x. [DOI] [PubMed] [Google Scholar]
- 46.Srinivasan S, et al. Bacterial Communities in Women with Bacterial Vaginosis: High Resolution Phylogenetic Analyses Reveal Relationships of Microbiota to Clinical Criteria. PLoS ONE. 2012;7:e37818. doi: 10.1371/journal.pone.0037818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Todd JK, Todd BH, Franco-Buff A, Smith CM, Lawellin DW. Influence of focal growth conditions on the pathogenesis of toxic shock syndrome. J. Infect. Dis. 1987;155:673–681. doi: 10.1093/infdis/155.4.673. [DOI] [PubMed] [Google Scholar]
- 48.Mitchell MA, Bisch S, Arntfield S, Hosseini-Moghaddam SM. A Confirmed Case of Toxic Shock Syndrome Associated with the Use of a Menstrual Cup. Can. J. Infect. Dis. Med. Microbiol. 2015;26:218–220. doi: 10.1155/2015/560959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robbins RN, Reiser RF, Hehl GL, Bergdoll MS. Production of toxic shock syndrome toxin 1 by Staphylococcus aureus as determined by tampon disk-membrane-agar method. J. Clin. Microbiol. 1987;25:1446–1449. doi: 10.1128/jcm.25.8.1446-1449.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nonfoux Louis, Chiaruzzi Myriam, Badiou Cédric, Baude Jessica, Tristan Anne, Thioulouse Jean, Muller Daniel, Prigent-Combaret Claire, Lina Gérard. Impact of Currently Marketed Tampons and Menstrual Cups onStaphylococcus aureusGrowth and Toxic Shock Syndrome Toxin 1 ProductionIn Vitro. Applied and Environmental Microbiology. 2018;84(12):e00351–18. doi: 10.1128/AEM.00351-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guinan, M. et al. Vaginal colonization with Staphylococcus aureus in healthy women: a review of four studies (1982). [DOI] [PubMed]
- 52.Lansdell LW, Taplin D, Aldrich TE. Recovery of Staphylococcus aureus from multiple body sites in menstruating women. J. Clin. Microbiol. 1984;20:307. doi: 10.1128/jcm.20.3.307-310.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou X, et al. The vaginal bacterial communities of Japanese women resemble those of women in other racial groups. FEMS Immunol. Med. Microbiol. 2010;58:169–181. doi: 10.1111/j.1574-695X.2009.00618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hickey R, et al. Effects of tampons and menses on the composition and diversity of vaginal microbial communities over time. BJOG Int. J. Obstet. Gynaecol. 2013;120:695–706. doi: 10.1111/1471-0528.12151. [DOI] [PubMed] [Google Scholar]
- 55.Gajer P, et al. Temporal Dynamics of the Human Vaginal Microbiota. Sci. Transl. Med. 2012;4:132ra52. doi: 10.1126/scitranslmed.3003605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chaban B, et al. Characterization of the vaginal microbiota of healthy Canadian women through the menstrual cycle. Microbiome. 2014;2:23. doi: 10.1186/2049-2618-2-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fettweis JM, et al. Differences in vaginal microbiome in African American women versus women of European ancestry. Microbiology. 2014;160:2272–2282. doi: 10.1099/mic.0.081034-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Albert Arianne Y. K., Chaban Bonnie, Wagner Emily C., Schellenberg John J., Links Matthew G., van Schalkwyk Julie, Reid Gregor, Hemmingsen Sean M., Hill Janet E., Money Deborah. A Study of the Vaginal Microbiome in Healthy Canadian Women Utilizing cpn60-Based Molecular Profiling Reveals Distinct Gardnerella Subgroup Community State Types. PLOS ONE. 2015;10(8):e0135620. doi: 10.1371/journal.pone.0135620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sarafian SK, Morse SA. Environmental factors affecting toxic shock syndrome toxin-1 (TSST-1) synthesis. J. Med. Microbiol. 1987;24:75–81. doi: 10.1099/00222615-24-1-75. [DOI] [PubMed] [Google Scholar]
- 60.Liu H, et al. Inhibition of Staphylococcus aureus by Lysostaphin-Expressing Lactobacillus plantarum WCFS1 in a Modified Genital Tract Secretion Medium∇. Appl. Environ. Microbiol. 2011;77:8500–8508. doi: 10.1128/AEM.06755-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Malik, S. et al. High mannose-specific lectin Msl mediates key interactions of the vaginal Lactobacillus plantarum isolate CMPG5300. Sci. Rep. 6 (2016). [DOI] [PMC free article] [PubMed]
- 62.van de Wijgert Janneke H.H.M., Jespers Vicky. The global health impact of vaginal dysbiosis. Research in Microbiology. 2017;168(9-10):859–864. doi: 10.1016/j.resmic.2017.02.003. [DOI] [PubMed] [Google Scholar]
- 63.Chen KT, Huard RC, Della-Latta P, Saiman L. Prevalence of methicillin-sensitive and methicillin-resistant Staphylococcus aureus in pregnant women. Obstet. Gynecol. 2006;108:482–487. doi: 10.1097/01.AOG.0000227964.22439.e3. [DOI] [PubMed] [Google Scholar]
- 64.Lagier J-C, et al. The Rebirth of Culture in Microbiology through the Example of Culturomics To Study Human Gut Microbiota. Clin. Microbiol. Rev. 2015;28:237–264. doi: 10.1128/CMR.00014-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dubourg G, et al. The gut microbiota of a patient with resistant tuberculosis is more comprehensively studied by culturomics than by metagenomics. Eur. J. Clin. Microbiol. Infect. Dis. 2013;32:637–645. doi: 10.1007/s10096-012-1787-3. [DOI] [PubMed] [Google Scholar]
- 66.Dubourg G, et al. Culturomics and pyrosequencing evidence of the reduction in gut microbiota diversity in patients with broad-spectrum antibiotics. Int. J. Antimicrob. Agents. 2014;44:117–124. doi: 10.1016/j.ijantimicag.2014.04.020. [DOI] [PubMed] [Google Scholar]
- 67.Lagier J-C, et al. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat. Microbiol. 2016;1:16203. doi: 10.1038/nmicrobiol.2016.203. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.