Plant heterotrimeric G-proteins function in important signaling pathways mediated by plant receptor kinases (RKs), however, the unique biochemical properties of Gα subunits have complicated our understanding of their regulation in plants. In their new paper in Cell Research, Liang et al. reveal that phosphorylation of the Gα regulator, RGS1, is critical for triggering G-protein signaling downstream of RK activation.
Plant growth and survival necessitates appropriate establishment of the plant body and timely response to environmental stressors. Many signaling pathways controlling plant development and environmental response are initiated by receptor kinases (RKs) at the plasma membrane. Downstream of RKs, signaling is propagated by different means including protein kinases/phosphatases, ion fluxes, reactive oxygen species (ROS) production, and heterotrimeric G-proteins1 (Fig. 1).
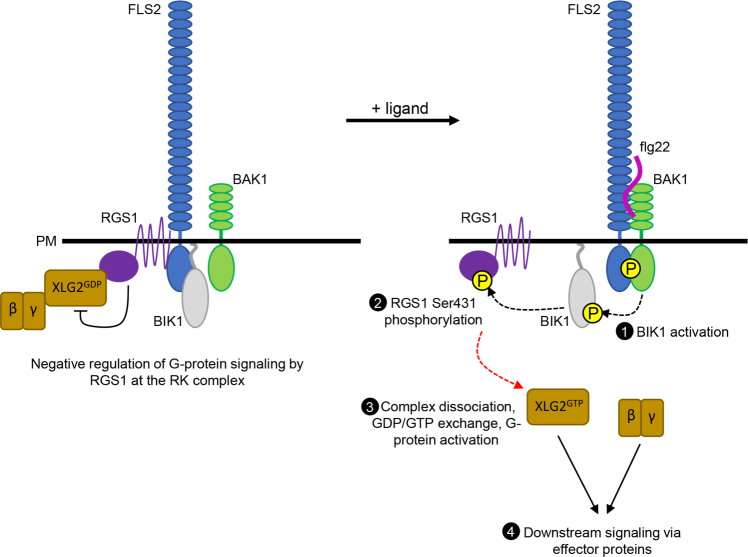
Fig. 1.
Regulatory phosphorylation of RGS1 initiates G-protein-dependent signaling downstream of flg22 perception. Under naïve conditions, RGS1 associated with FLS2 maintains the Gα protein XLG2 in the inactive, GDP-bound state. Upon PAMP perception, BAK1 phosphorylates and activates BIK1, which subsequently phosphorylates RGS1 at Ser431. RGS1 phosphorylation leads to dissociation of the heterotrimeric G-protein from RGS1, GDP-to-GTP exchange by XLG2, G-protein activation, and subsequent activation of downstream signaling via regulation of effector proteins
Heterotrimeric G-proteins are universal signaling components in eukaryotes, and their regulatory properties are well known in animals. Owing to the spontaneous guanine nucleotide exchange activity of plant Gα subunits,2 and the absence of G-protein-coupled receptors (GPCRs) in plants,3 the details of how heterotrimeric G-proteins are regulated in plants remain elusive. Furthermore, controversy over the genetic role of REGULATOR OF G-PROTEIN SIGNALING 1 (RGS1) in Arabidopsis has complicated our understanding of how these proteins function in plants.4 Recently, it has become apparent that plant G-proteins interact with multiple RKs, and that G-proteins are genetically required for RK-mediated responses.5–7 How RKs regulate G-protein signaling downstream of ligand perception remains unclear. Using the immune RK FLAGELLIN SENSING 2 (FLS2) as a model, Liang and colleagues address the ambiguity over RGS1 function, and identify RGS1 phosphorylation sites critical for de-repression of G-protein signaling.8 Overall, their study details an exciting new mechanism by which plant G-proteins are activated in response to extracellular ligand perception that may have broad implications for signaling via plant RKs.
In animals, G-proteins are controlled through interaction with seven-transmembrane domain GPCRs that activate intracellular signaling upon ligand perception. The heterotrimeric G complex consists of α, β and γ subunits, which in animals are encoded by multi-gene families. For example, the human genome encodes 23 Gα, 5 Gβ and 12 Gγ subunits, in addition to greater than 800 GPCRs.3 The diverse subunits allow for myriad combinations and thus exquisite signaling specificity. By comparison, plant genomes encode no GPCRs and a limited number of G-protein subunits. Arabidopsis encodes four Gα (GPA1, XLG1, XLG2 and XLG3 where XLG is EXTRA LARGE G PROTEIN), one Gβ (AGB1) and three Gγ (AGG1, AGG2 and AGG3) proteins.3 Nevertheless, genetic analysis in Arabidopsis reveals that G-proteins function in many aspects of plant physiology.5
Plant Gα proteins are distinct from their animal counterparts in that they undergo spontaneous guanine nucleotide exchange,2 suggesting that in the absence of negative regulatory factors, plant Gα proteins would be constitutively active. Animal G-proteins are activated in response to ligand binding by an associated GPCR that promotes exchange of GDP for GTP by the Gα subunit.3 GTP binding induces both the dissociation of the G-protein complex from the GPCR and dissociation of Gα from the Gβγ subunits. Gα and Gβγ are then freed to initiate downstream signaling via interaction with diverse effector proteins. Attenuation of signaling occurs following GTP hydrolysis by Gα, leading to reconstitution of the inactive Gαβγ heterotrimer. GTPase accelerating RGS proteins promote return to the resting state by enhancing the GTPase activity of Gα.3 Because they undergo spontaneous guanine nucleotide exchange, plant Gα proteins must be held in the inactive state to limit downstream signaling. Maintenance of inactive G-protein complexes is achieved by interaction with plasma membrane-localized RGS proteins (RGS1 in Arabidopsis) that promote the GDP-bound state of Gα through enhancing GTP hydrolysis.3 How this repression is relieved in response to different stimuli is unknown.
Several recent studies have linked G-proteins to plant RKs, including FLS2.4,6,7,9 Upon flagellin (or its derived peptide flg22) perception, FLS2 forms a complex with its co-receptor BRASSINOSTEROID INSENSITIVE 1 ASSOCIATED KINASE 1 (BAK1), leading to activation of the receptor-like cytoplasmic kinase BOTRYTIS INDUCED KINASE 1 (BIK1) and subsequent downstream signaling.1 Notably, all the components of an inactive G-protein complex, i.e., RGS1 and a GDP-bound Gαβγ trimer, are associated with FLS2 in the resting state in vivo, suggesting the involvement of G-proteins in plant immune signaling. Indeed, genetic analysis implicates RGS1 and Gα proteins in immune signaling downstream of flg22 perception.4,7–10 While it is clear that the Gα protein XLG2 positively regulates flg22-induced ROS production and bacterial immunity, the role of RGS1 in immune signaling has been the subject of some controversy. In Arabidopsis, RGS1 has previously been shown to positively regulate the flg22-induced ROS burst.4 Now, detailed genetic studies of the rgs1-2 allele, including gene expression and bacterial infection assays, demonstrate that RGS1 negatively regulates signaling downstream of flg22 perception.8 An inhibitory role for RGS1 in immunity is supported by its function as a negative regulator of Gα proteins in other signaling pathways and the observation that XLG2 and GPA1 positively regulate different branches of immune responses.7,8,10 Positive and negative regulation of immunity by Gα proteins and RGS1, respectively, raises the question of how Gα repression by RGS1 is released to activate G-signaling in response to flg22 perception.
The work by Liang and colleagues8 reveals that phosphorylation of RGS1 is the likely trigger for activation of G-protein signaling downstream of RK complexes. Early studies of RGS1 show that it undergoes endocytosis in response to glucose stimulation driven by with no lysine 8 (WNK8)-mediated phosphorylation.11 WNK8 phosphorylation of RGS1 on Ser428 and Ser435/436 is essential for RGS1 endocytosis. Following the discovery of RGS1/Gαβγ association with RKs, the immunity-related protein kinases BAK19 and BIK17 were shown to phosphorylate different components of the G-protein complex in response to flg22 perception, including phosphorylation of RGS1 by BAK1. BAK1 targets the same RGS1 residues as those phosphorylated by WNK8, suggesting that G-protein activation downstream of FLS2/BAK1 could be activated by phosphorylation-dependent endocytosis of RGS1, which would relieve Gα subunit inhibition and activate G-signaling. Importantly, Gα proteins dissociate from rather than follow RGS1 into the endocytic pathway,9 but how this occurs prior to RGS1 internalization has remained unclear. In their latest work, Liang et al.8 show that an additional phosphorylation site promotes dissociation of RGS1 from both FLS2 and XLG2. BIK1 phosphorylates RGS1 at Ser431, in the same vicinity as the previously known WNK8/BAK1 sites.8 Phosphorylation of RGS1 at Ser431 promotes dissociation of the FLS2/RGS1/XLG2/Gβγ complex, de-repressing RGS1-mediated inhibition of XLG2 and activating G-protein-dependent immune signaling. Simultaneous phosphorylation of RGS1 Ser428, Ser431 and Ser435/436 is required for complete de-repression of G-protein signaling, but phospho-mimetic mutation of all four sites does not cause constitutive signaling via XLG2.8 Notably, both BIK1 and BAK1 phosphorylate additional residues on XLG2 and GPA1 that are either required for function in immunity,7 or modulate Gα function.12 It will be interesting in future to understand at the mechanistic level how these additional phosphorylation events regulate Gα function following release from the receptor complex. Furthermore, it will be important to determine whether RGS1 phosphorylation operates to release G-proteins from repression in the context of other RK complexes.
References
- 1.Couto D, Zipfel C. Nat. Rev. Immunol. 2016;16:537–552. doi: 10.1038/nri.2016.77. [DOI] [PubMed] [Google Scholar]
- 2.Jones JC, et al. Sci. Signal. 2011;4:ra8. doi: 10.1126/scisignal.2001446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trusov Y, Botella JR. Front. Chem. 2016;4:24. doi: 10.3389/fchem.2016.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tunc-Ozdemir M, Jones AM. PLoS ONE. 2017;12:e0171854. doi: 10.1371/journal.pone.0171854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stateczny D, Oppenheimer J, Bommert P. Curr. Opin. Plant Biol. 2016;34:127–135. doi: 10.1016/j.pbi.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 6.Bommert P, Je BI, Goldshmidt A, Jackson D. Nature. 2013;502:555–558. doi: 10.1038/nature12583. [DOI] [PubMed] [Google Scholar]
- 7.Liang X, et al. eLife. 2016;5:e13568. doi: 10.7554/eLife.13568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liang Xiangxiu, Ma Miaomiao, Zhou Zhaoyang, Wang Jinlong, Yang Xinru, Rao Shaofei, Bi Guozhi, Li Lin, Zhang Xiaojuan, Chai Jijie, Chen She, Zhou Jian-Min. Ligand-triggered de-repression of Arabidopsis heterotrimeric G proteins coupled to immune receptor kinases. Cell Research. 2018;28(5):529–543. doi: 10.1038/s41422-018-0027-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tunc-Ozdemir M, Urano D, Jaiswal DK, Clouse SD, Jones AM. J. Biol. Chem. 2016;291:13918–13925. doi: 10.1074/jbc.C116.736702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maruta N, Trusov Y, Brenya E, Parekh U, Botella JR. Plant Physiol. 2015;167:1004–1016. doi: 10.1104/pp.114.255703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Urano D, et al. Nat. Cell Biol. 2012;14:1079–1088. doi: 10.1038/ncb2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li B, et al. J. Biol. Chem. 2018;293:4752–4766. doi: 10.1074/jbc.RA117.000163. [DOI] [PMC free article] [PubMed] [Google Scholar]