Abstract
Strains of the Pasteurellaceae bacteria Pasteurella multocida and Mannheimia haemolytica are major etiological agents of bovine respiratory disease (BRD). Treatment of BRD with antimicrobials is becoming more challenging due to the increasing occurrence of resistance in infecting strains. In Pasteurellaceae strains exhibiting resistance to multiple antimicrobials including aminoglycosides, beta-lactams, macrolides and sulfonamides, the resistance determinants are often chromosomally encoded within integrative and conjugative elements (ICEs). To gain a more comprehensive picture of ICE structures, we sequenced the genomes of six strains of P. multocida and four strains of M. haemolytica; all strains were independent isolates and eight of them were multiple-resistant. ICE sequences varied in size from 49 to 79 kb, and were comprised of an array of conserved genes within a core region and varieties of resistance genes within accessory regions. These latter regions mainly account for the variation in the overall ICE sizes. From the sequence data, we developed a multiplex PCR assay targeting four conserved core genes required for integration and maintenance of ICE structures. Application of this assay on 75 isolates of P. multocida and M. haemolytica reveals how the presence and structures of ICEs are related to their antibiotic resistance phenotypes. The assay is also applicable to other members of the Pasteurellaceae family including Histophilus somni and indicates how clustering and dissemination of the resistance genes came about.
Keywords: antibiotic resistance, veterinary macrolides, Pasteurella, Mannheimia, genomics
Introduction
Bovine respiratory disease (BRD) is a common and complex form of pneumonia that affects beef cattle around the world. The disease can be caused by infection with several different viral and bacterial pathogens and is often exacerbated by environmental factors including stress (Taylor et al., 2010). In addition to the distress suffered by infected animals, BRD causes considerable economic losses to the beef cattle industry due to reduced meat yields and outlay associated with preventive measurements and treatment (Griffin, 1997). Numerous studies have therefore focused on identifying the major pathogens implicated in BRD in efforts to reduce animal morbidity and concomitant financial losses. Two of the most common bacterial species associated with this disease have been shown by microbiological and serological surveys to be Mannheimia haemolytica and Pasteurella multocida (Lillie, 1974; Welsh et al., 2004; Portis et al., 2012), both of which are members of the Pasteurellaceae family.
As with many other bacterial infections, antimicrobial regimens used to combat BRD have been compromised by resistance to numerous drugs including macrolides, aminoglycosides, beta-lactams, and sulfonamides (Kehrenberg et al., 2001, 2005; San Millan et al., 2009; Desmolaize et al., 2011b; Olsen et al., 2015). Macrolide resistance is of particular interest in P. multocida and M. haemolytica due to the varied mechanisms by which resistance can be conferred via point mutations in ribosomal operons (Poehlsgaard et al., 2012; Olsen et al., 2015) and by acquisition of a range of macrolide resistance genes (Desmolaize et al., 2011a,b; Kadlec et al., 2011). Collectively, resistance genes tend to be clustered within integrative and conjugative elements (ICEs) in the chromosomes of Pasteurellaceae. Macrolide resistance genes include erm(42) and msr(E)-mph(E) and can be acquired in various combinations within an ICE where they are generally interspersed amongst genes conferring resistance to other classes of drugs (Desmolaize et al., 2011a,b; Michael et al., 2012b). Each ICE encodes the machinery required to regulate is own excision, integration, and conjugative transfer (Burrus and Waldor, 2004; Carraro and Burrus, 2015), potentially to other Pasteurellaceae pathogens including Haemophilus somni (DeDonder et al., 2016), Haemophilus parasuis (Lei et al., 2017), and Haemophilus influenzae (Mohd-Zain et al., 2004; Dimopoulou et al., 2007; Juhas et al., 2007, 2013). Previous studies emphasize that Pasteurellaceae species can possess a broad repertoire of antimicrobial resistance genes together with the means of disseminating them (Desmolaize et al., 2011a,b; Michael et al., 2012b), and that the use of appropriate therapeutic agents is therefore of great importance in treating livestock illnesses including BRD.
Detection and characterization of ICE sequences would serve as an initial step in ascertaining whether Pasteurellaceae strains are likely to be equipped with multiple resistance determinants. Due to their large size and diverse composition, the identification of ICEs has up to now required whole genome sequencing coupled with bioinformatics analysis. Sequence characterization of ICEPmu1 in P. multocida (Michael et al., 2012a,b) and ICEMh1 in M. haemolytica (Eidam et al., 2013, 2015) revealed structures of 82 and 92 kb, respectively containing core- and accessory gene regions. The core regions encode genes for ICE maintenance and proliferation ensuring the essential functions of transfer, replication, regulation and integration. The accessory regions encode a range of additional traits including antibiotic resistance, metal-fixation and novel metabolic capacities (Wozniak and Waldor, 2010; Bi et al., 2012). The findings from the two P. multocida and M. haemolytica isolates gave the first clear indication that certain core region features could be common amongst Pasteurellaceae ICEs, whereas the antimicrobial resistance genes and their distribution within ICEs could differ significantly.
Here we report a more comprehensive picture of Pasteurellaceae ICE structures. Initially, we sequenced the genomes of 10 independent P. multocida and M. haemolytica isolates eight of which were selected due to their resistance to macrolides and in most cases to an array of other veterinary antimicrobials. For strains that had acquired exogenous resistance genes, these were located in nearly all cases within chromosomally-encoded ICEs. Alignment and comparative analysis of the chromosomes revealed sets of core genes common to ICEs and with highly conserved sequences. Conserved genes from different locations within the core regions were chosen to design PCR primer pairs for a multiplex assay to detect the presence of ICEs. Additional primer sets that target rRNA genes located outside the ICE sequences were included in the assay as a means of identifying false negatives and to differentiate between P. multocida and other gammaproteobacterial species. Here we describe the application of this assay on over 70 P. multocida and M. haemolytica isolates, many of which are resistant to multiple drugs. The composition of ICE antimicrobial resistance genes is shown to be highly variable, and while most regions connected with maintenance and transfer are conserved, a significant proportion of the M. haemolytica ICEs have lost genes essential for intercellular transfer. In silico interrogation of genomes presently available in databases using the multiplex primer sequences shows that the assay is applicable to other Pasteurellaceae members including Histophilus somni and confirms that ICE sequences are to be found throughout within this family of bacteria.
Materials and methods
Bacterial strains, growth, and macrolide resistance phenotypes
The P. multocida and M. haemolytica strains are field isolates obtained from nasal swabs of cattle suffering from BRD in Europe and USA, and were procured from the MSD Animal Health culture collection. Strains were plated onto agar containing brain-heart infusion broth (Oxoid, England) and grown at 37°C overnight; cell colonies were purified by restreaking on agar and were then grown again to form individual colonies for direct PCR testing. Standard CLSI procedures (Clinical and Laboratory Standards Institute, 2008) were applied to determine the minimal inhibitory concentrations (MICs) of antibiotics. The macrolides used were tilmicosin (TIL, Sigma-Aldrich, Germany), tildipirosin (TIP, MSD Animal Health, Germany), and gamithromycin (GAM) and tulathromycin (TUL), respectively extracted and purified from Zactran® (Merial, Germany), and Draxxin® (Pfizer, USA). Tildipirosin, gamithromycin and tulathromycin were purified as colorless powders; their structures were verified by liquid chromatography/mass spectrometry and nuclear magnetic resonance.
Genome sequencing
Eight P. multocida and M. haemolytica isolates Pmu3358, Pmu3361, Pmu12591, Pmu12601, Pmu14424, Mh6055, Mh12540, and Mh12565 that exhibited intermediate and high resistance to macrolides, plus two macrolide susceptible strains Pmu4407 and Mh11935, were selected for genome sequencing. Genomic DNA was prepared as previously described (Desmolaize et al., 2011a) and sequenced by a paired-end, shotgun approach using Illumina HiSeq equipment (BaseClear, Leiden, Netherlands). Sequence reads were assembled using the CLC genomics workbench (www.CLCbio.com) and the SSPACE premium scaffolder (Boetzer et al., 2011). Assembled contigs were aligned to the reference genomes P. multocida 36950 (Genbank: CP003022.1) and M. haemolytica 42548 (Genbank: CP005383.1) using the Multiple Genome alignment software, Mauve (Darling et al., 2010) and the genomes were annotated using RAST—Rapid annotation using subsystem technology (Overbeek et al., 2014). Integrative and conjugative elements were identified and their structural similarities were investigated using the bioinformatics tools Island viewer (Langille and Brinkman, 2009), Artemis sequence visualization (Rutherford et al., 2000), Artemis Comparison Tool ACT(Carver et al., 2005) and CMG—Comparative Microbial Genomic software (Vesth et al., 2013).
Multiplex PCR analyses
Four oligodeoxynucleotide primer pairs were designed to detect genes specific for ICE core regions (Table 1). These primers comprised: pInt1F/pInt1R that bind to int1 gene encoding a 35.6 kDa integrase and result in a PCR product of 301 bp; pInt2F/pInt2R that give a product of 215 bp and detect a second 29.9 kDa integrase encoded by int2; pRelF/pRelR that bind to an ICE-specific relaxase gene (ICE-rel1, but not to the homologous ICE-rel2) to give a 437 bp product; and pParBF/pParBR that detect parB with a product of 503 bp.
Table 1.
Oligonucleotide primers used for detection of Pasteurellaceae ICE sequences.
| Primer | Sequence (5′-3′) | Direction | Screening function | PCR fragment size (bp) |
|---|---|---|---|---|
| p84 | GACGGAAAGACCCCGTGAACCT | Forward | rrl sequence G2053 to T2074 in Gammaproteobacteria | |
| p85 | GGCAAGTTTCGTGCTTAGAT | Reverse | rrl sequence A2753 to C2772 in Gammaproteobacteria | 720 |
| p86 | GGAGCAGCCCCAATCAATCA | Reverse | rrl sequence T2633 to C2652 specific to P. multocida | 600 |
| pParBF | GCTTGGCTCTTCATTGCTCG | Forward | parB | |
| pParBR | TTTCTCCTCCTTGTTGGCGA | Reverse | parB | 503 |
| pRelF | GGCTCACGTTGGTTTGCTTG | Forward | ICE-rel1 | |
| pRelR | TCAGCGGCAGTTTTGCTAAC | Reverse | ICE-rel1 | 437 |
| pInt1F | TAGAACGGAATCATAGACCTGCC | Forward | int1 (35.6 kDa integrase) | |
| pInt1R | TGGATTTGCCTTTCTGTTAGTAGT | Reverse | int1 | 301 |
| pInt2F | TCAACATTTCCACATCGTGCTC | Forward | int2 (29.9 kDa integrase) | |
| pInt2R | AAGAGGACAGCCAATGAGCC | Reverse | int2 | 215 |
The ParB-primers screen for the parB gene; the Rel-primers amplify a region within an ICE-specific relaxase gene, ICE-rel1; and the pairs of Int-primers target two distinct integrase genes int1 and int2. The p85 and p86 primers in combination with the p84 primer are included to verify whether the PCR reaction has functioned (they give a product that is independent of the presence of ICE sequences) and additionally serve to differentiate between P. multocida from other species in the Gammaproteobacteria class. The PCR reactions produce easily distinguishable gel bands ranging from 215 to 720 bp.
In addition to the ICE-specific primers, three 23S rRNA gene-specific primers, p84, p85 and p86, were included in the multiplex assay (Table 1), and served as internal controls to check whether each PCR assay had functioned and to distinguish between P. multocida and other gammaproteobacterial species (Rose et al., 2012). Primers p84 and p85 are complementary to sequences that are conserved in the 23S rRNAs of all Gammaproteobacteria and give a PCR product of 720 bp for bacterial species including P. multocida, M. haemolytica, and enterics such as E. coli, with no product being formed for species outside the Gammaproteobacteria class. The p86 oligo is specific for P. multocida 23S rRNA genes and acts as a nested primer in combination with p84 to produce a 600 bp PCR fragment (Rose et al., 2012). Each of the 75 strains in the study was additionally tested with an independent multiplex assay to detect macrolide resistance genes erm(42), msr(E), and mph(E) (Rose et al., 2012).
Cell colonies were transferred from agar plates and resuspended in 100 μl water, boiled for 5 min, and 1 μl was taken for PCR analyses. Each ICE-specific multiplex PCR was carried out with 200 μM dATP, dCTP, dGTP, and dTTP, 1.0 U Taq polymerase (VWR International), 0.4 μM of the primers in 25 μl total volume of 10 mM Tris-HCl pH 8.3, 50 mM KCl, 3.5 mM MgCl2, 0.1% Triton X-100. A Mastercycler Personal apparatus (Eppendorf) was used with a denaturation step for 2 min at 95°C followed by 30 cycles of 30 s at 95°C, 30 s at 62°C, 1 min at 72°C, with the final cycle concluding after 5 min at 72°C. PCR fragments were analyzed on 2% agarose gels and the sizes were estimated from a GeneRuler 100 bp DNA Ladder (Fermentas). The multiplex PCR assays for erm(42), msr(E), and mph(E) (Rose et al., 2012) were carried out under the same conditions with primer hybridization at 60°C.
Results
Genome analyses
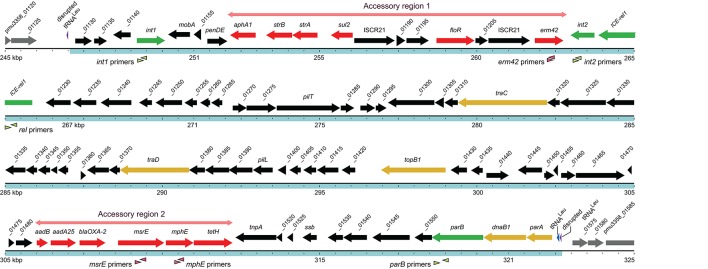
The genome sizes of the six P multocida strains fell within the range 2.24–2.33 Mb, with the smallest genomes belonging to Pmu4407 and Pmu14424 (2,243,572 and 2,245,345 bp, respectively) and the largest to Pmu3361 (2,321,102 bp). The genomes of the four M. haemolytica strains were between 2.56 and 2.69 Mb, where Mh11935 possessed the smallest (2,562,294 bp), and Mh6055 the largest (2,689,542 bp). These sizes are within the span of genome lengths considered typical for the two species; for instance, the P. multocida strains collectively possessed a core genome of 1,896 genes and these constituted between 88 and 91% of the total genes. The differences in about half of the remaining P. multocida genes reflected the presence or absence of an ICE, and this was also the case for the larger M. haemolytica genomes. The sizes of the ICEs ranged from 49 to 79 kb, and the most extensive of these were found in P. multocida strain 3358 (Figure 1) and M. haemolytica strain 6055. Alignment of the sequences showed that they contain regions of similarity with ICEMh1 and ICEPmu1 (Figure 2). The ICEs in strains Pmu3358 and Mh6055 showed the highest identity with ICEPmu1. Despite the general similarity between the strains, several gaps were evident in the alignments and corresponded mainly to the absence of certain resistance genes in the accessory regions 1 and 2 (Table 2).
Figure 1.
Structure of the 75.6 kb ICE within the chromosome of P. multocida strain 3358 is depicted by the blue region, with the extent of the two accessory regions indicated in pale red. The size marker dots are spaced at 500 bp. The int1, int2, ICE-rel1, and parB genes in the core regions are shown in green, other essential core genes in yellow, the remaining core genes in black, and antibiotic resistance genes in the accessory regions in red. Genes are annotated according to their known functions or by using the NCBI notation (Supplementary Table S1). All numbered genes are prefixed with pmu3358_ as shown for the first and last genes in this figure. The hybridization sites for the primers used in the multiplex PCR assays are indicated by the arrow heads below the respective genes.
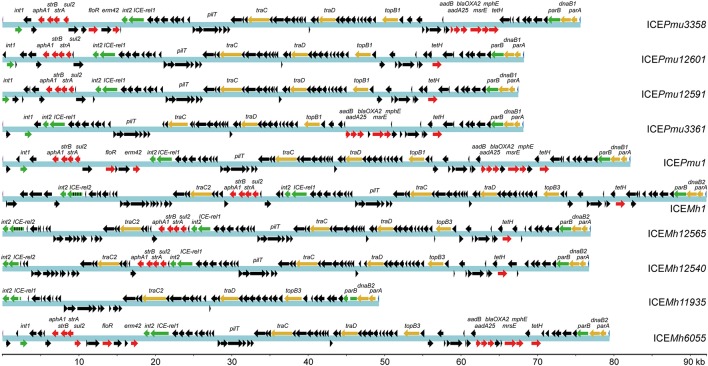
Figure 2.
Content of ICEs from the Pasteurellaceae strains determined by whole genome sequencing. The previously reported ICEPmu1 of P. multocida strain 36950 (Michael et al., 2012b) and ICEMh1 from M. haemolytica strain 42548 (Eidam et al., 2015) are included for comparison. The ICEs are annotated here according to the strains containing them, and their sizes can be read from the kilobase pair (kb) scale at the bottom. Color coding of the genes as in Figure 1 with the core genes targeted in the multiplex assay (green), other essential core genes (yellow), antibiotic resistance genes (red) plus additional open reading frames (black). The chromosomal site of ICE insertion left a disrupted tRNALeu gene at both ends of the sequence, which is replaced by in all cases by an intact copy of the tRNALeu gene located after parA (far right, in this sequence orientation). The multiplex assay was design to give a positive signal for ICE-rel1 but not for the ICE-rel2 gene (green/black stripes) that is evident in some M. haemolytica ICEs. The sensitive strain Pmu4407 contains no ICE genes. Strain Mh11935 also completely lacks resistance genes but contains numerous ICE core region genes, although key genes for ICE transfer and maintenance are missing. The ICE-rel1 gene in ICEMh11935 is truncated and presumably inactive, despite giving a positive multiplex PCR signal. The parB gene of ICEMh11935 is also truncated, and lacks one of the priming regions needed for a PCR signal. All of the four targeted genes int1, int2, ICE-rel1, and parB were 100% conserved in ICEPmu3358, ICEPmu3361, ICEPmu12591, and ICEPmu12601 of the P. multocida strains. Accession codes for the whole genome sequences are listed in Supplementary Table S1.
Table 2.
Antibiotic resistance genes and their respective ICE locations, determined by whole genome sequencing in this study.
| Accessory region 1 | Accessory region 2 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ICE | aphA1 | strB | strA | sul2 | floR | erm(42) | aadB | aadA25 | blaOXA−2 | msr(E) | mph(E) | tetH |
| ICEPmu3358 | + | + | + | + | + | + | + | + | + | + | + | + |
| ICEPmu3361 | – | – | – | – | – | – | + | + | + | + | + | + |
| ICEPmu12591 | + | + | + | + | – | – | – | – | – | – | – | + |
| ICEPmu12601 | + | + | + | + | – | – | – | – | – | – | – | + |
| ICEMh6055 | + | + | + | + | + | + | + | + | + | + | + | + |
| ICEMh12540 | + | + | + | + | – | – | – | – | – | – | – | + |
| ICEMh12565 | + | + | + | + | – | – | – | – | – | – | – | + |
The distribution of genes in accessory region 1 indicates a clustering of resistance determinants for aminoglycosides, florfenicols, macrolides, and sulfonamides. Accessory region 2 contained predominately genes conferring resistance against aminoglycosides, beta-lactams, macrolides, and tetracyclines. Strain Mh11935 harbored an ICE without any these resistance genes.
Genes associated with ICE core functionality
The most conserved genes are concerned with replication, conjugative transfer, and chromosomal integration of an ICE, and thereby ensuring its maintenance and dissemination. The ICEs examined here (Table 2) each carry the int2 gene, which encodes a 30 kDa integrase, while the isolates Pmu3358, Pmu3361, Pmu12591, Pmu12601, and Mh6055 have an additional integrase gene, int1, which encodes a paralogous integrase of 36 kDa. Both enzymes belong to the XerD-family of tyrosine integrases (Argos, 1986; Esposito and Scocca, 1997; Nunes-Düby et al., 1998). The int1 gene is located at the outer edge of the ICE flanking accessory region 1, while int2 is in the opposite orientation on the other side of accessory region 1 (as shown for ICEPmu3358, Figure 1). The genome sequences showed that the larger integrase is absent in Mh11935, Mh12540, Mh12565.
Between the two ICE accessory regions are the conserved relaxase ICE-rel1 and the traC-, traD-, and traG-like genes that encode a type IV secretion system associated with conjugative transfer. The genes topB, parA, parB, and dnaB are also highly conserved and linked with DNA replication, and the latter three genes are grouped outside accessory region 2 at the far end of the ICE (Figure 2). The M. haemolytica strains Mh12540 and Mh12565 possesses second copies the traC- and traG-like genes that are 80% identical to the homologous sequences present in all the ICEs. These traC- and traG-like paralogs were also evident in the M. haemolytica ICEMh1 sequence (Eidam et al., 2015), but absent in the P. multocida strains sequenced here. A paralogous relaxase gene ICE-rel2 was seen in M. haemolytica strains and sometimes duplicated, but was absent from our P. multocida strains. The ICE-rel2 enzyme has 90% sequence identity with ICE-rel1.
All the M. haemolytica and P. multocida ICE sequences were integrated within a chromosomal tRNALeu gene, and the disrupted gene is replaced by an intact tRNALeu copy at the end of the ICE (Figures 1, 2).
Antimicrobial resistance genes
The greatest diversity in the ICE structures was observed in the accessory regions 1 and 2, which contain genes conferring resistance to antimicrobials including aminoglycosides, beta-lactams, macrolides, phenicols, sulfonamides, and tetracyclines. The number of antibiotic resistance genes in the strains tested here range from zero to twelve, with the greatest abundance of these genes in strain Pmu3358 and Mh6055. The identities and locations of the resistance genes within the respective ICEs are listed in Table 2.
In accessory region 1, the combination of aphA1-strB-strA-sul2 genes were observed in several different ICEs, and these cluster with floR and erm(42) in Pmu3358 and Mh6055. In Pmu3361, region 1 is completely absent. In accessory region 2, the compositions of resistance genes also varied, with strains Pmu3358, Pmu3361, and Mh6055 possessing the combination of aadB-aadA-blaOXA−2-msr(E)-mph(E)-tetH genes, while the remaining strains have only tetH.
Primer design for the multiplex PCR system
Genome sequence comparisons showed that the highly conserved regions associated with ICE-functionality would provide a basis for rapid ICE detection using multiplex PCR, while the more variable ICE regions could be used to differentiate between strains. Primers were designed to target three conserved sequences in parB, ICE-rel1, and int2. A fourth primer pair detected the other integrase gene int1 that was present in a subset of ICEs. The locations of these target genes are spread through the length of the ICEs (Figure 2). In addition, three 23S rRNA gene-specific oligonucleotide primers, p84, p85, and p86, were included in the assay to serve as internal controls for the PCR assay and to distinguish between P. multocida and other gammaproteobacterial species (Rose et al., 2012).
Direct screening of bacterial colonies by multiplex PCR
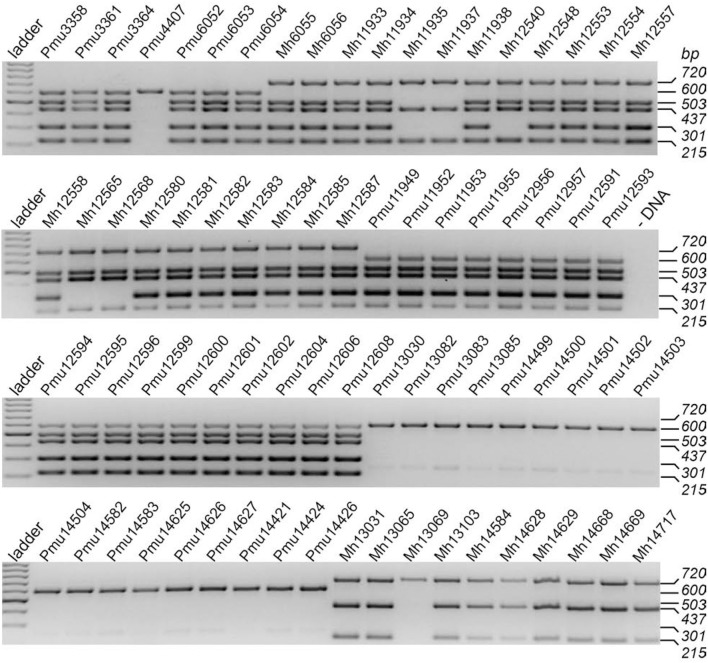
A total of 43 P. multocida and 32 M. haemolytica isolates were screened for ICE sequences using the multiplex assay (Table 3). Twenty-four of the P. multocida isolates showed a positive signal for int1, int2, ICE-rel1, and parB, while the remaining 19 strains produced no signal for any these genes indicating that they lacked an ICE (Figure 3). Several macrolide resistant P. multocida isolates were shown to lack all the ICE-specific genes.
Table 3.
Overview of the P. multocida (Pmu) and M. haemolytica (Mh) strains investigated in this study and their macrolide resistance profiles.
| Strains and species | Macrolide resistance genes | ICE-specific genes | MIC | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| erm(42) | msr(E) | mph(E) | int1 | int2 | ICE-rel1 | parB | TIP | TUL | TIL | GAM | ||
| 3358 | Pmu* | + | + | + | + | + | + | + | 128 | >128 | 128 | 64 |
| 3361 | Pmu* | – | + | + | + | + | + | + | 2 | 64 | 32 | 32 |
| 3364 | Pmu | – | – | – | + | + | + | + | 1 | 32 | 16 | 0.5 |
| 4407 | Pmu* | – | – | – | – | – | – | – | 1 | 0.5 | 4 | 0.5 |
| 6052 | Pmu | + | – | – | + | + | + | + | >128 | 8 | >128 | 8 |
| 6053 | Pmu | + | – | – | + | + | + | + | 128 | 8 | 128 | 16 |
| 6054 | Pmu | + | – | – | + | + | + | + | >128 | 8 | >128 | 8 |
| 6055 | Mh* | + | + | + | + | + | + | + | 128 | >128 | 128 | 128 |
| 6056 | Mh | + | + | + | + | + | + | + | >128 | >128 | >128 | 128 |
| 11933 | Mh | + | – | – | + | + | + | + | 128 | 8 | 64 | 4 |
| 11934 | Mh | + | + | + | + | + | + | + | 128 | 64 | 128 | 64 |
| 11935 | Mh* | – | – | – | – | + | + | – | 0.5 | 2 | 4 | 0.25 |
| 11937 | Mh | – | – | – | – | + | + | – | 0.5 | 2 | 8 | 0.5 |
| 11938 | Mh | + | + | + | + | + | + | + | 128 | 128 | 64 | 128 |
| 11949 | Pmu | + | + | + | + | + | + | + | >128 | >128 | >128 | 64 |
| 11952 | Pmu | + | – | – | + | + | + | + | >128 | 8 | >128 | 8 |
| 11953 | Pmu | + | – | – | + | + | + | + | >128 | 4 | 128 | 4 |
| 11955 | Pmu | + | – | – | + | + | + | + | >128 | 4 | 128 | 4 |
| 11956 | Pmu | + | – | – | + | + | + | + | >128 | 4 | 128 | 4 |
| 11957 | Pmu | + | + | + | + | + | + | + | >128 | >128 | 128 | 128 |
| 12540 | Mh* | – | – | – | – | + | + | + | 1 | 2 | 32 | 0.5 |
| 12548 | Mh | – | + | + | + | + | + | + | 0.5 | 128 | 32 | 64 |
| 12553 | Mh | – | + | + | + | + | + | + | 1 | 128 | 32 | 128 |
| 12554 | Mh | – | + | + | + | + | + | + | 1 | 128 | 32 | 128 |
| 12557 | Mh | – | + | + | + | + | + | + | 1 | 128 | 32 | 128 |
| 12558 | Mh | – | + | + | + | + | + | + | 2 | >128 | 32 | 128 |
| 12565 | Mh* | – | – | – | – | + | + | + | 2 | 2 | 32 | 1 |
| 12568 | Mh | – | – | – | – | + | + | + | 2 | 2 | 32 | 1 |
| 12580 | Mh | + | – | – | + | + | + | + | >128 | 16 | 128 | 8 |
| 12581 | Mh | + | – | – | + | + | + | + | >128 | 16 | 128 | 8 |
| 12582 | Mh | + | – | – | + | + | + | + | >128 | 16 | 128 | 8 |
| 12583 | Mh | + | – | – | + | + | + | + | >128 | 16 | 128 | 8 |
| 12584 | Mh | + | + | + | + | + | + | + | >128 | 128 | 128 | 128 |
| 12585 | Mh | + | – | – | + | + | + | + | >128 | 16 | 128 | 8 |
| 12587 | Mh | + | + | + | + | + | + | + | >128 | >128 | >128 | 128 |
| 12591 | Pmu* | – | – | – | + | + | + | + | 1 | 0.5 | 32 | 0.5 |
| 12593 | Pmu | – | + | + | + | + | + | + | 2 | >128 | 32 | 64 |
| 12594 | Pmu | – | + | + | + | + | + | + | 2 | >128 | 32 | 64 |
| 12595 | Pmu | – | + | + | + | + | + | + | 4 | >128 | 32 | 64 |
| 12596 | Pmu | – | + | + | + | + | + | + | 4 | >128 | 32 | 64 |
| 12599 | Pmu | – | – | – | + | + | + | + | 1 | 1 | 32 | 0.5 |
| 12600 | Pmu | – | – | – | + | + | + | + | 1 | 1 | 32 | 0.5 |
| 12601 | Pmu* | – | – | – | + | + | + | + | 1 | 0.5 | 32 | 0.5 |
| 12602 | Pmu | – | + | + | + | + | + | + | 4 | >128 | 32 | 32 |
| 12604 | Pmu | – | – | – | + | + | + | + | 1 | 0.5 | 32 | 0.5 |
| 12606 | Pmu | + | + | + | + | + | + | + | >128 | >128 | >128 | 128 |
| 12608 | Pmu | + | – | – | + | + | + | + | >128 | 8 | >128 | 16 |
| 13030 | Pmu | – | – | – | – | – | – | – | 0.5 | 0.5 | 1 | 0.25 |
| 13031 | Mh | – | – | – | – | + | + | – | 0.25 | 1 | 2 | nd |
| 13065 | Mh | – | – | – | – | + | + | – | 0.25 | 1 | 2 | nd |
| 13069 | Mh | – | – | – | – | – | – | – | 0.5 | 1 | 4 | nd |
| 13082 | Pmu | – | – | – | – | – | – | – | 0.5 | nd | nd | nd |
| 13083 | Pmu | – | – | – | – | – | – | – | 0.25 | nd | nd | nd |
| 13085 | Pmu | – | – | – | – | – | – | – | 0.5 | nd | nd | nd |
| 13103 | Mh | – | – | – | – | + | + | – | 0.25 | 0.5 | 1 | 0.25 |
| 14499 | Pmu | – | – | – | – | – | – | – | 4 | 4 | 16 | 2 |
| 14500 | Pmu | – | – | – | – | – | – | – | 4 | 4 | 16 | 2 |
| 14501 | Pmu | – | – | – | – | – | – | – | 8 | 4 | 32 | 2 |
| 14502 | Pmu | – | – | – | – | – | – | – | 8 | 4 | 32 | 2 |
| 14503 | Pmu | – | – | – | – | – | – | – | 8 | 4 | 32 | 2 |
| 14504 | Pmu | – | – | – | – | – | – | – | 8 | 4 | 32 | 2 |
| 14582 | Pmu | – | – | – | – | – | – | – | 8 | 2 | 16 | 1 |
| 14583 | Pmu | – | – | – | – | – | – | – | 16 | 4 | 16 | 2 |
| 14625 | Pmu | – | – | – | – | – | – | – | 8 | 0.5 | 16 | 0.5 |
| 14626 | Pmu | – | – | – | – | – | – | – | 32 | 16 | 32 | 4 |
| 14627 | Pmu | – | – | – | – | – | – | – | 32 | 16 | 32 | 4 |
| 14421 | Pmu | – | – | – | – | – | – | – | >64 | >64 | >64 | >64 |
| 14424 | Pmu* | – | – | – | – | – | – | – | >64 | >64 | >64 | >64 |
| 14426 | Pmu | – | – | – | – | – | – | – | >64 | >64 | >64 | >64 |
| 14584 | Mh | – | – | – | – | + | + | – | 16 | 4 | 32 | 4 |
| 14628 | Mh | – | – | – | – | + | + | – | >64 | 32 | 64 | 16 |
| 14629 | Mh | – | – | – | – | + | + | – | >64 | 32 | 64 | 32 |
| 14668 | Mh | – | – | – | – | + | + | – | 16 | 64 | 8 | 32 |
| 14669 | Mh | – | – | – | – | + | + | – | 8 | 32 | 8 | 8 |
| 14717 | Mh | – | – | – | – | + | + | – | >64 | >64 | >64 | >64 |
Strains up to Pmu12608 were isolated in the USA from cattle suffering from respiratory infections (with the exception of Pmu4407, France); strains Pmu13030 to Mh14717 are from cases of bovine respiratory infection in Europe. The genomes of the strains marked with an asterisk were sequenced. All strains were screened using the multiplex PCR assay described here for ICE genes, and additionally with an independent multiplex assay to detect macrolide resistance genes (Rose et al., 2012). The presence (+) or absence (–) of the ICE-specific int1, int2, ICE-rel1, parB genes and the macrolide resistance genes erm(42), msr(E), and mph(E) is indicated. Some of the macrolide resistant strains, such as Pmu14421, Pmu14424, and Pmu14426 that lack ICE structures, have attained resistance via A2059G mutations in all six of their rrn operons; strain Mh14717 has the A2058G mutation in all six of its rrn operons (Olsen et al., 2015). The minimal inhibitory concentrations (MICs in μg/ml) of the macrolides tildipirosin (TIP), tulathromycin (TUL), tilmicosin (TIL), and gamithromycin (GAM), some of which have been reported previously (Rose et al., 2012; Olsen et al., 2015), are color coded to indicate whether the strains show susceptibility (green), intermediate resistance (orange), or resistance (red) according to CSLI breakpoints for these antibiotics: http://vet01s.edaptivedocs.info/GetDoc.aspx?doc=CLSI%20VET01S%20ED3:2015&scope=user. nd, not determined.
Figure 3.
Identification of ICE-related Pasteurellaceae genes using multiplex PCR. The gels show the analyses of 75 field isolates, 43 of which were P. multocida (identified by the 600 bp band that is specific for Pasteurella spp.) and 32 were M. haemolytica (band at 720 bp and lack of 600 bp band). The four ICE-related PCR products correspond to parB (503 bp), the ICE-specific relaxase gene (437 bp), and the longer and shorter versions of the integrase genes int1 (301 bp) and int2 (215 bp). In strains that lacked all these ICE genes (e.g., Pmu13083) a faint artifact band of 250 bp was sometimes apparent. This band was no longer evident after raising the primer annealing temperature 3°C above the optimal hybridization temperature for the canonical sites. The control lane (-DNA) shows a reaction without template DNA. The bands in the GeneRuler ladder on the left are in 100-bp steps.
In the M. haemolytica isolates, 17 gave positive signals for int1, int2, ICE-rel1, and parB, and a further three isolates contained int2, ICE-rel1 and parB (but not int1). Eleven of the strains gave clear signals for int2 and ICE-rel1, but parB was missing (Figure 3), suggesting that these strains possess remnants of ICEs which lack the ability to be disseminated. The multiplex assay was designed to give a positive signal for ICE-rel1, but not for the homologous ICE-rel2 gene present in M. haemolytica strains.
The occurrence of the macrolide resistance genes erm(42), msr(E), and mph(E) (or subsets thereof) in the M. haemolytica isolates correlates with the presence of ICE sequences containing both int1 and int2. Several M. haemolytica isolates with ICE remnants were also macrolide resistant without having any of the erm(42), msr(E), or mph(E) genes (Table 3). Only one M. haemolytica isolate tested here (Mh13069) possessed none of the four ICE-specific genes.
Discussion
The P. multocida and M. haemolytica strains included in this study were isolated in the USA and Europe from cattle with respiratory tract infections. Strains were initially selected on the basis of macrolide antibiotic resistance. However, the majority of strains associated with BRD did not exceed breakpoint values for macrolides, and some susceptible strains from the same locations were included in this study. Genome sequence analyses of a subset of the strains revealed a series of highly conserved genes that could serve as markers for the presence of an ICE and from these the int1, int2, ICE-rel1, and parB genes, which are distributed along the length of the ICE structures (Figure 1), were chosen for the multiplex PCR assay.
All the P. multocida and M. haemolytica strains that possessed one or more of the macrolide resistance genes erm(42), msr(E), and mph(E) also had a full complement of the int1, int2, ICE-rel1, and parB marker genes. The converse was not true, however, and five P. multocida strains possessed all four ICE marker genes but none of the macrolide resistance genes (Table 3). As previously reported, the different combinations of the erm(42), msr(E), and mph(E) genes conferred distinct resistance phenotypes to the 15-membered ring macrolides GAM and TUL and the 16-membered macrolides TIL and TIP (Rose et al., 2012), and a full complement of all three genes was required to attain high level resistance to all these drugs (Table 3).
Some strains possessed neither ICE nor resistance genes but were nevertheless highly resistant to macrolides. In these cases, resistance was achieved via point mutations in the drug binding site on the ribosome, as we have seen previously in the P. multocida strains Pmu14421, Pmu14424, and Pmu14426 with A2059G substitutions in the 23S rRNA genes in all six of their rrn operons, and the M. haemolytica strain Mh14717 which had A2058G mutations in all six of its rrn operons (Olsen et al., 2015). This same mechanism could explain the macrolide resistance phenotypes of Canadian M. haemolytica isolates that lack all the erm(42), msr(E), and mph(E) genes (Alexander et al., 2013). In the related Pasteurellaceae species Haemophilus parasuis, the A2059G mutation has been reported in an Australia isolate from swine respiratory infection (Dayao et al., 2016).
Several P. multocida and M. haemolytica strains (including Pmu12599, Pmu12600, Pmu12601, Pmu12604, Mh12540, Mh12558, Mh12565, and Mh12658, Table 3) were sensitive to both GAM and TIP but showed intermediate resistance to TIL, and in one case (Pmu3364) also to TUL. None of these strains possessed any of the erm(42), msr(E), and mph(E) genes, and they contained variable numbers (either none, two or all four) of the ICE gene markers (Table 3), indicating that their macrolide resistance phenotypes were unrelated to the presence of an ICE. The genomes of two of these strains, Pmu12591 and Pmu12601, were sequenced without revealing any changes in their rRNAs or ribosomal proteins, or other relevant genes that could account for elevated MICs to TIL (Vester and Douthwaite, 2001; Peric et al., 2003). Similar observations for M. haemolytica strains with no obvious molecular explanation have been made elsewhere for TUL resistance (Alexander et al., 2013) and also for GAM resistance (DeDonder et al., 2016). Up-regulation of one or more endogenous efflux system could possibly account for these anomalous macrolide phenotypes.
Similar to previous accounts (Michael et al., 2012b; Eidam et al., 2015; Clawson et al., 2016), the ICEs analyzed here contained a broad range of genes encoding resistance to other antibiotics that have been widely used in veterinary medicine. Genes conferring resistance to aminoglycoside, beta-lactam, florfenicol, sulfonamide, and tetracycline were distributed throughout accessory regions 1 and 2, with each specific gene maintaining its particular location within an accessory region. For instance, the aphA1-strA-strB-sul2 combination, previously identified in plasmids (Hirsh et al., 1989; Yamamoto et al., 1990) are found in the same order in accessory region 1, and in some cases followed by floR and erm(42) (Table 2). This latter region shares 96% sequence identity with plasmid pPDP9106b (Michael et al., 2012b), suggesting that floR and erm(42) also descended from plasmids. More common in the ICEs were resistance genes for the older aminoglycoside and sulfonamide drugs, consistent with sulfonamide resistance being one of the traits most often detected in Pasteurella and Mannheimia isolates (Kehrenberg et al., 2001). Strain Pmu14424 contains no ICE, but nevertheless encodes Sul2 (100% identical to the structure in Figure 1), StrA (94% identical), and TetR (65% identical) within a Tn10 transposon adjacent to an inactive mu-bacteriophage at a different chromosomal location (1842728–1846225) than the site of ICE integration (Figure 1).
In accessory region 2, the aadB-aadA-blaOXA−2 combination appears to have originated from the transposon Tn5706 and is generally flanked by the tetR and tetH genes (Michael et al., 2012b). Although the full-length tetH gene was evident at the same location in all the sequenced ICE structures (Table 2), we only observed remnants of tetR. The neighboring msr(E)-mph(E) pair was previously detected in plasmids (González-Zorn et al., 2005; Gołebiewski et al., 2007) and more recently in the Pasteurellaceae (Desmolaize et al., 2011b; Kadlec et al., 2011). In some strains, the msr(E)-mph(E) genes are flanked by IS26 elements, while in other strains the IS26 sequences are truncated (Pmu3361) or lost (Pmu3358), which could indicate that these macrolide resistance genes were inserted into accessory region 2 prior to ICE acquisition by these strains. The ICE locations of erm(42) and/or msr(E)-mph(E) (Figure 1) and their incidence in the different isolates (Table 3) indicates that they have been integrated into ICE structures from separate plasmids by independent recombination events.
Insertion of ICEs appears to be guided into the same chromosomal site by one or more phage-like integrases. All the ICE strains in this study possessed the int2 gene encoding a 30 kDa integrase, and in some strains an int1 gene encoding a larger 36 kDa enzyme was also present (Table 3). Alignment of these paralogs shows they have 49% amino acid identity, with particularly high conservation of residues within the active site that is common to members of the tyrosine recombinase XerD family (Esposito and Scocca, 1997; Nunes-Düby et al., 1998). These enzymes typically target tRNA genes for site-specific integration events (Reiter et al., 1989) and occur here at a tRNALeu gene, as initially observed for ICE integration in H. influenzae (Dimopoulou et al., 2002). A remnant tRNALeu anticodon loop sequence of 13 nucleotides (5′-GATTTTGAATCAA) remains at all the attL sites after ICE integration (Figure 1), while the attR site varies in size between 10 and 13 nucleotides with the sequence 5′-GATTTTGAAT(CAA). An intact tRNALeu copy is located at the end of the ICE immediately after parA (Figure 1), and replaces the disrupted gene.
The isolates studied here show that Pasteurellaceae ICE sequences can vary greatly in size and structure with some strains, exemplified by Pmu3358, possess what appears to be a fully functional ICE that encodes multiple resistance determinants. Despite the overall variations in the sizes and content of the ICEs, the core and resistance genes that remained generally showed >99% sequence conservation in the strains analyzed here. A similar picture emerges from the Pasteurellaceae genomes available at Genbank. Interrogating these Pasteurellaceae genomes in silico with the probes used in this study showed that a surprisingly high proportion possesses at least two of the int1, int2, ICE-rel1, and parB genes (Supplementary Table S2). Furthermore, the key proteins Int2 and ICE-Rel1 are generally 100% identical to those in our strains, and are evident in numerous Genbank sequences from H. somni isolates in addition to P. multocida and M. haemolytica (Supplementary Figure S1).
The data presented here on the conservation of key ICE genes suggest that propagation of these sequences is a relatively recent event within the Pasteurellaceae. The differences in ICE sizes found here reflect the plasticity and relatively rapid sequence losses subsequent to ICE acquisition. The compositions of several M. haemolytica ICEs (Figure 4), and also those in the database (Supplementary Figure 1), appear to be partially degenerate and incapable of being disseminated. This contrasts with the P. multocida ICE sequences (Table 3 and Figure 4) that have retained their function. In silico analysis of other P. multocida genomes in the database (Supplementary Table S2) present a similar but not identical picture where, although their ICE sequences appear functional, a proportion has lost int2 while retaining int1 (Supplementary Figure 1). This could reflect the wider range of hosts, which include swine and poultry, from which these latter strains were isolated. The overall picture suggests that some of the M. haemolytica ICE sequences are of older origin and were acquired before those found in P. multocida. In the case of the strains isolated from cattle, this is consistent with M. haemolytica being a primary etiologic agent associated with BRD (Klima et al., 2016; Snyder et al., 2017).
Figure 4.
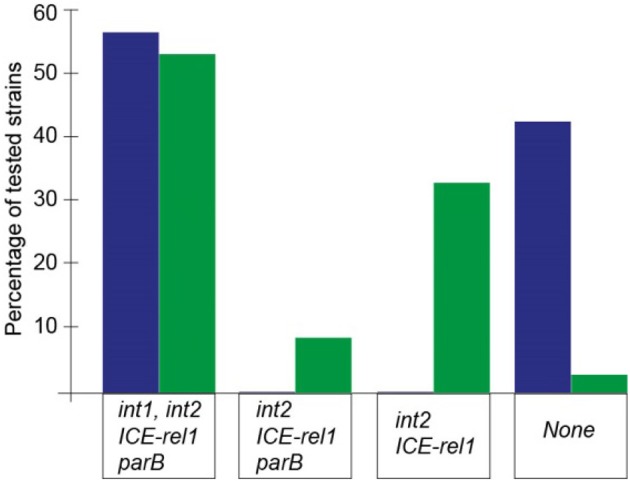
Relative numbers of the P. multocida (blue) and M. haemolytica strains (green) containing the ICE core genes screened in the multiplex PCR assay. Twenty-four of the 43 P. multocida isolates gave a positive signal with each of the int1, int2, ICE-rel1, and parB primer combinations; and a similar proportion of M. haemolytica strains (17 of 32) was also shown to contain these four ICE genes. The remainder of the P. multocida strains tested lacked all of the int1, int2, ICE-rel1, and parB genes, but no P. multocida strain was found to harbor a truncated, nonfunctional ICE structure. In contrast, 14 M. haemolytica strains possessed degenerate ICE sequences that would presumably not be capable of promoting intercellular transfer.
Several new questions can now be posed about the transfer of resistance genes in Pasteurellaceae. These include why the presence of the macrolide resistance genes erm(42), msr(E), and mph(E) correlates with that of int1 despite their relatively distant ICE locations. Possibly the acquisition of these macrolide resistance genes is a recent event and the presence of int1 is indicative of ICE functionality, which has not yet been lost in the sequences that have been transferred more recently. Finally, although lack of detection is not a proof of absence, we note that the erm(42), msr(E), and mph(E) genes have yet to be reported in European (Table 3) (Rose et al., 2012), Australian (Dayao et al., 2016), and Canadian animals (Alexander et al., 2013).
Author contributions
MB, SR, and CL: methodology and investigation. SD and MB: writing, review and editing. SD: resources, funding acquisition and supervision.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Nelson Enrique Arenas Suarez for advice with software for genome assembly. The research was funded by grants from the Danish Research Agency (FNU-rammebevilling 10-084554) to SD.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01329/full#supplementary-material
References
- Alexander T. W., Cook S., Klima C. L., Topp E., Mcallister T. A. (2013). Susceptibility to tulathromycin in Mannheimia haemolytica isolated from feedlot cattle over a 3-year period. Front. Microbiol. 4:297. 10.3389/fmicb.2013.00297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argos P. (1986). The integrase family of site-specific recombinases: regional similarities and global diversity. EMBO J. 5, 433–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi D., Xu Z., Harrison E. M., Tai C., Wei Y., He X., et al. (2012). ICEberg: a web-based resource for integrative and conjugative elements found in Bacteria. Nucleic Acids Res. 40, D621–D626. 10.1093/nar/gkr846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boetzer M., Henkel C. V., Jansen H. J., Butler D., Pirovano W. (2011). Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 27, 578–579. 10.1093/bioinformatics/btq683 [DOI] [PubMed] [Google Scholar]
- Burrus V., Waldor M. K. (2004). Shaping bacterial genomes with integrative and conjugative elements. Res. Microbiol. 155, 376–386. 10.1016/j.resmic.2004.01.012 [DOI] [PubMed] [Google Scholar]
- Carraro N., Burrus V. (2015). The dualistic nature of integrative and conjugative elements. Mob. Genet. Elements 5, 98–102. 10.1080/2159256X.2015.1102796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver T. J., Rutherford K. M., Berriman M., Rajandream M. A., Barrell B. G., Parkhill J. (2005). ACT: the artemis comparison tool. Bioinformatics 21, 3422–3423. 10.1093/bioinformatics/bti553 [DOI] [PubMed] [Google Scholar]
- Clawson M. L., Murray R. W., Sweeney M. T., Apley M. D., DeDonder K. D., Capik S. F., et al. (2016). Genomic signatures of Mannheimia haemolytica that associate with the lungs of cattle with respiratory disease, an integrative conjugative element, and antibiotic resistance genes. BMC Genomics 17:982. 10.1186/s12864-016-3316-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI) (2008). Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals; Approved Standard - 3rd Edn. Wayne, PA: CLSI. [Google Scholar]
- Darling A. E., Mau B., Perna N. T. (2010). progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 5:e11147. 10.1371/journal.pone.0011147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayao D. A., Seddon J. M., Gibson J. S., Blackall P. J., Turni C. (2016). Whole genome sequence analysis of pig respiratory bacterial pathogens with elevated minimum inhibitory concentrations for macrolides. Microb. Drug Resist. 22, 531–537. 10.1089/mdr.2015.0214 [DOI] [PubMed] [Google Scholar]
- DeDonder K. D., Harhay D. M., Apley M. D., Lubbers B. V., Clawson M. L., Schuller G., et al. (2016). Observations on macrolide resistance and susceptibility testing performance in field isolates collected from clinical bovine respiratory disease cases. Vet. Microbiol. 192, 186–193. 10.1016/j.vetmic.2016.07.021 [DOI] [PubMed] [Google Scholar]
- Desmolaize B., Rose S., Warrass R., Douthwaite S. (2011a). A novel Erm monomethyltransferase in antibiotic-resistant isolates of Mannheimia haemolytica and Pasteurella multocida. Mol. Microbiol. 80, 184–194. 10.1111/j.1365-2958.2011.07567.x [DOI] [PubMed] [Google Scholar]
- Desmolaize B., Rose S., Wilhelm C., Warrass R., Douthwaite S. (2011b). Combinations of macrolide resistance determinants in field isolates of Mannheimia haemolytica and Pasteurella multocida. Antimicrob. Agents Chemother. 55, 4128–4133. 10.1128/AAC.00450-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimopoulou I. D., Kartali S. I., Harding R. M., Peto T. E. A., Crook D. W. (2007). Diversity of antibiotic resistance integrative and conjugative elements among haemophili. J. Med. Microbiol. 56, 838–846. 10.1099/jmm.0.47125-0 [DOI] [PubMed] [Google Scholar]
- Dimopoulou I. D., Russell J. E., Mohd-Zain Z., Herbert R., Crook D. W. (2002). Site-specific recombination with the chromosomal tRNALeu gene by the large conjugative Haemophilus resistance plasmid. Antimicrob. Agents Chemother. 46, 1602–1603. 10.1128/AAC.46.5.1602-1603.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eidam C., Poehlein A., Brenner Michael G., Kadlec K., Liesegang H., Brzuszkiewicz E., et al. (2013). Complete genome sequence of Mannheimia haemolytica strain 42548 from a case of bovine respiratory disease. Genome Announc 1:e00318-13. 10.1128/genomeA.00318-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eidam C., Poehlein A., Leimbach A., Michael G. B., Kadlec K., Liesegang H., et al. (2015). Analysis and comparative genomics of ICEMh1, a novel integrative and conjugative element (ICE) of Mannheimia haemolytica. J. Antimicrob. Chemother. 70, 93–97. 10.1093/jac/dku361 [DOI] [PubMed] [Google Scholar]
- Esposito D., Scocca J. J. (1997). The integrase family of tyrosine recombinases: evolution of a conserved active site domain. Nucleic Acids Res. 25, 3605–3614. 10.1093/nar/25.18.3605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gołebiewski M., Kern-Zdanowicz I., Zienkiewicz M., Adamczyk M., Zylinska J., Baraniak A., et al. (2007). Complete nucleotide sequence of the pCTX-M3 plasmid and its involvement in spread of the extended-spectrum beta-lactamase gene blaCTX-M-3. Antimicrob. Agents Chemother. 51, 3789–3795. 10.1128/AAC.00457-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Zorn B., Catalan A., Escudero J. A., Dominguez L., Teshager T., Porrero C., et al. (2005). Genetic basis for dissemination of armA. J. Antimicrob. Chemother. 56, 583–585. 10.1093/jac/dki246 [DOI] [PubMed] [Google Scholar]
- Griffin D. (1997). Economic impact associated with respiratory disease in beef cattle. Vet. Clin. North Am. Food Anim. Pract. 13, 367–377. 10.1016/S0749-0720(15)30302-9 [DOI] [PubMed] [Google Scholar]
- Hirsh D. C., Hansen L. M., Dorfman L. C., Snipes K. P., Carpenter T. E., Hird D. W., et al. (1989). Resistance to antimicrobial agents and prevalence of R plasmids in Pasteurella multocida from turkeys. Antimicrob. Agents Chemother. 33, 670–673. 10.1128/AAC.33.5.670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhas M., Dimopoulou I. D., Robinson E., Elamin A. R., Harding R. M., Hood D. W., et al. (2013). Identification of another module involved in the horizontal transfer of the Haemophilus genomic island ICEHin1056. Plasmid 70, 277–283. 10.1016/j.plasmid.2013.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhas M., Power P. M., Harding R. M., Ferguson D. J., Dimopoulou I. D., Elamin A. R., et al. (2007). Sequence and functional analyses of Haemophilus spp. genomic islands. Genome Biol. 8:R237. 10.1186/gb-2007-8-11-r237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadlec K., Brenner Michael G., Sweeney M. T., Brzuszkiewicz E., Liesegang H., Daniel R., et al. (2011). Molecular basis of macrolide, triamilide, and lincosamide resistance in Pasteurella multocida from bovine respiratory disease. Antimicrob. Agents Chemother. 55, 2475–2477. 10.1128/AAC.00092-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehrenberg C., Catry B., Haesebrouck F., De Kruif A., Schwarz S. (2005). tet(L)-mediated tetracycline resistance in bovine Mannheimia and Pasteurella isolates. J. Antimicrob. Chemother. 56, 403–406. 10.1093/jac/dki210 [DOI] [PubMed] [Google Scholar]
- Kehrenberg C., Schulze-Tanzil G., Martel J. L., Chaslus-Dancla E. S. S.. (2001). Antimicrobial resistance in Pasteurella and Mannheimia: epidemiology and genetic basis. Vet. Res. 32, 323–339. 10.1051/vetres:2001128 [DOI] [PubMed] [Google Scholar]
- Klima C. L., Cook S. R., Zaheer R., Laing C., Gannon V. P., Xu Y., et al. (2016). Comparative genomic analysis of Mannheimia haemolytica from bovine sources. PLoS ONE 11:e0149520. 10.1371/journal.pone.0149520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langille M. G., Brinkman F. S. (2009). IslandViewer: an integrated interface for computational identification and visualization of genomic islands. Bioinformatics 25, 664–665. 10.1093/bioinformatics/btp030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Z., Fu S., Yang B., Liu Q., Ahmed S., Xu L., et al. (2017). Comparative transcriptional profiling of tildipirosin-resistant and sensitive Haemophilus parasuis. Sci. Rep. 7:7517. 10.1038/s41598-017-07972-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillie L. E. (1974). The bovine respiratory disease complex. Canad. Vet. J. 15, 233–242. [PMC free article] [PubMed] [Google Scholar]
- Michael G. B., Eidam C., Kadlec K., Meyer K., Sweeney M. T., Murray R. W., et al. (2012a). Increased MICs of gamithromycin and tildipirosin in the presence of the genes erm(42) and msr(E)-mph(E) for bovine Pasteurella multocida and Mannheimia haemolytica. J. Antimicrob. Chemother. 67, 1555–1557. 10.1093/jac/dks076 [DOI] [PubMed] [Google Scholar]
- Michael G. B., Kadlec K., Sweeney M. T., Brzuszkiewicz E., Liesegang H., Daniel R., et al. (2012b). ICEPmu1, an integrative conjugative element (ICE) of Pasteurella multocida: analysis of the regions that comprise 12 antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 84–90. 10.1093/jac/dkr406 [DOI] [PubMed] [Google Scholar]
- Mohd-Zain Z., Turner S. L., Cerdeño-Tárraga A. M., Lilley A. K., Inzana T. J., Duncan A. J., et al. (2004). Transferable antibiotic resistance elements in Haemophilus influenzae share a common evolutionary origin with a diverse family of syntenic genomic islands. J. Bacteriol. 186, 8114–8122. 10.1128/JB.186.23.8114-8122.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes-Düby S. E., Kwon H. J., Tirumalai R. S., Ellenberger T., Landy A. (1998). Similarities and differences among 105 members of the Int family of site-specific recombinases. Nucleic Acids Res. 26, 391–406. 10.1093/nar/26.2.391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen A. S., Warrass R., Douthwaite S. (2015). Macrolide resistance conferred by rRNA mutations in field isolates of Mannheimia haemolytica and Pasteurella multocida. J. Antimicrob. Chemother. 70, 420–423. 10.1093/jac/dku385 [DOI] [PubMed] [Google Scholar]
- Overbeek R., Olson R., Pusch G. D., Olsen G. J., Davis J. J., Disz T., et al. (2014). The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res. 42, D206–D214. 10.1093/nar/gkt1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peric M., Bozdogan B., Jacobs M. R., Appelbaum P. C. (2003). Effects of an efflux mechanism and ribosomal mutations on macrolide susceptibility of Haemophilus influenzae clinical isolates. Antimicrob. Agents Chemother. 47, 1017–1022. 10.1128/AAC.47.3.1017-1022.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poehlsgaard J., Andersen N. M., Warrass R., Douthwaite S. (2012). Visualizing the 16-membered ring macrolides tildipirosin and tilmicosin bound to their ribosomal site. ACS Chem. Biol. 7, 1351–1355. 10.1021/cb300105p [DOI] [PubMed] [Google Scholar]
- Portis E., Lindeman C., Johansen L., Stoltman G. (2012). A ten-year (2000-2009) study of antimicrobial susceptibility of bacteria that cause bovine respiratory disease complex -Mannheimia haemolytica, Pasteurella multocida and Histophilus somni-in the United States and Canada. J. Vet. Diagn. Invest. 24, 932–944. 10.1177/1040638712457559 [DOI] [PubMed] [Google Scholar]
- Reiter W. D., Palm P., Yeats S. (1989). Transfer RNA genes frequently serve as integration sites for prokaryotic genetic elements. Nucleic Acids Res. 17, 1907–1914. 10.1093/nar/17.5.1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose S., Desmolaize B., Jaju P., Wilhelm C., Warrass R., Douthwaite S. (2012). Multiplex PCR to identify macrolide resistance determinants in Mannheimia haemolytica and Pasteurella multocida. Antimicrob. Agents Chemother. 56, 3664–3669. 10.1128/AAC.00266-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford K., Parkhill J., Crook J., Horsnell T., Rice P., Rajandream M. A., et al. (2000). Artemis: sequence visualization and annotation. Bioinformatics 16, 944–945. 10.1093/bioinformatics/16.10.944 [DOI] [PubMed] [Google Scholar]
- San Millan A., Escudero J. A., Gutierrez B., Hidalgo L., Garcia N., Llagostera M., et al. (2009). Multiresistance in Pasteurella multocida is mediated by coexistence of small plasmids. Antimicrob. Agents Chemother. 53, 3399–3404. 10.1128/AAC.01522-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder E., Credille B., Berghaus R., Giguere S. (2017). Prevalence of multi drug antimicrobial resistance in Mannheimia haemolytica isolated from high-risk stocker cattle at arrival and two weeks after processing. J. Anim. Sci. 95, 1124–1131. 10.2527/jas.2016.1110 [DOI] [PubMed] [Google Scholar]
- Taylor J. D., Fulton R. W., Lehenbauer T. W., Step D. L., Confer A. W. (2010). The epidemiology of bovine respiratory disease: what is the evidence for predisposing factors? Canad. Vet. J. 51, 1095–1102. [PMC free article] [PubMed] [Google Scholar]
- Vester B., Douthwaite S. (2001). Macrolide resistance conferred by base substitutions in 23S rRNA. Antimicrob. Agents Chemother. 45, 1–12. 10.1128/AAC.45.1.1-12.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesth T., Lagesen K., Acar O., Ussery D. (2013). CMG-biotools, a free workbench for basic comparative microbial genomics. PLoS ONE 8:e60120. 10.1371/journal.pone.0060120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh R. D., Dye L. B., Payton M. E., Confer A. W. (2004). Isolation and antimicrobial susceptibilities of bacterial pathogen from bovine pneumonia: 1994–2002. J. Vet. Diagnost. Investig. 16, 426–431. 10.1177/104063870401600510 [DOI] [PubMed] [Google Scholar]
- Wozniak R. A., Waldor M. K. (2010). Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat. Rev. Microbiol. 8, 552–563. 10.1038/nrmicro2382 [DOI] [PubMed] [Google Scholar]
- Yamamoto J., Sakano T., Shimizu M. (1990). Drug resistance and R plasmids in Pasteurella multocida isolates from swine. Microbiol. Immunol. 34, 715–721. 10.1111/j.1348-0421.1990.tb01049.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.