Abstract
Background: Emerging evidences have shown that the high-mobility group protein A2 (HMGA2) can aberrantly express in human cancers, and it could be an unfavorable prognostic factor in cancer patients. However, the prognostic value of HMGA2 was still unclear. Therefore, in this study, we explored the potential prognostic value of HMGA2 in human cancers by using meta-analysis based on published literatures and The Cancer Genome Atlas (TCGA) datasets.
Methods: Through searching PubMed, Embase, Web of Science and Cochrane Library databases, we were able to identify the studies evaluating the prognostic value of HMGA2 in cancers. Then, UALCAN and TCGA datasets were used to validate the results of our meta-analysis.
Results: In all, 15 types of cancers were included in this meta-analysis. Pooled results showed that high level of HMGA2 was significantly correlated with poor OS (HR = 1.88, 95% confidence interval (CI) = 1.68-2.11, P < 0.001) and poor DFS (HR = 2.49, 95% CI = 1.44-4.28, P = 0.001) in cancer patients. However, subgroup analyses revealed that the high expressed HMGA2 was associated with poor OS in head and neck cancer, gastric cancer and colorectal cancer, but not esophageal cancer and ovarian cancer. Based on TCGA datasets, we analyzed 9944 patients with 33 types of cancers. Significant association between HMGA2 overexpression and poor OS was found in 14 types of cancers. Taken together, consistent results were observed in clear cell renal cell carcinoma, esophageal adenocarcinoma, head and neck cancer, hepatocellular carcinoma, ovarian carcinoma, and pancreatic ductal adenocarcinoma.
Conclusion: Our meta-analysis showed the significance of HMGA2 and its prognostic value in various cancers. High level of HMGA2 could be associated with poor OS in patients with clear cell renal cell carcinoma, head and neck cancer, hepatocellular carcinoma and pancreatic ductal adenocarcinoma, but not esophageal adenocarcinoma and ovarian carcinoma.
Keywords: HMGA2, cancer, prognosis, TCGA, meta-analysis
Introduction
Cancer has been one of the major causes of death and threats to global health due to its high morbidity and mortality (Wu S. et al., 2016). Meanwhile, the global incidence of cancer is rapidly increasing (Torre et al., 2015). Study shows that in 2017 there are 1,688,780 new cancer cases and 600,920 cancer deaths in the United States (Siegel et al., 2017). Many researchers have been focusing on identifying new tumor biomarkers which can be associated with cancer screening, diagnosis, prognosis, and evaluation of treatment efficacy (Ribeiro et al., 2016; Gorin et al., 2017; Kojima et al., 2017). Currently, even though various therapeutic methods have made significant achievements in cancer therapy, the 5-year-survival rate still remains unsatisfactory in cancer patients (Gonzalez and Agudo, 2012). Therefore, it is necessary to identify new prognostic biomarkers for accurate prognosis prediction.
High mobility group protein A2 (HMGA2) is a small non-histone chromosomal protein. It has no intrinsic transcriptional activity, but can modulate transcription by altering chromatin architecture (Hock et al., 2007; Pallante et al., 2015). Normally, HMGA2 protein is highly expressed in embryogenesis, while its expression is almost undetectable in most adult and differentiated tissues (Chiappetta et al., 1995; Rogalla et al., 1996; Hirning-Folz et al., 1998). An increasing number of recent studies have suggested that HMGA2 could highly express in many human malignant cells, and it can participate in different cellular activities including cell cycle regulation, differentiation and senescence (Wood et al., 2000; Hristov et al., 2009). HMGA2 overexpression has been reported to be associated with tumorigenesis and progression in many human cancers such as esophageal squamous cell cancer (Zhang et al., 2016), lung cancer (Kumar et al., 2014), pancreatic cancer (Piscuoglio et al., 2012), breast cancer (Wu J. et al., 2016) and colorectal cancer (Wang et al., 2011). Meanwhile, plenty of studies showed that elevated HMGA2 expression in cancer tissues was associated with poor survival in patients such as lung cancer (Sarhadi et al., 2006), oral squamous cell carcinoma (Miyazawa et al., 2004), ovarian cancer (Shell et al., 2007) and metastatic breast cancer (Langelotz et al., 2003). However, due to the limitations of sample size and research programs, single studies are inadequate to obtain a reliable assessment of the potential prognostic value of HMGA2. Therefore, we conducted a meta-analysis of a large sample size to gain better insight into this problem. In addition, The Cancer Genome Atlas (TCGA) databases were utilized to validate the result of our meta-analysis.
Materials and Methods
Study Strategy
Method of the analysis was based on the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement (Moher et al., 2009). We searched PubMed, Embase, Web of Scienceand Cochrane Library databases for relevant articles from January 1, 2000 to August 17, 2017. The research subject and article language were limited to human and English. Both MeSH terms and free-text words were used as strategy to increase the sensitivity of the searching. The search terms included: (“cancer” OR “tumor” OR “neoplasm” OR “carcinoma”) AND (“High mobility group protein A2” OR “HMGA2”) AND (“prognosis” OR “prognostic” OR “outcome”). We also manually screened the references of retrieved articles to identify more eligible studies that may have been missed by the key word search.
Inclusion and Exclusion Criteria
Two authors (Ben Huang and Jiayi Yang) independently selected all eligible articles according to the criteria: (1) studies that showed the association between HMGA2 expression and prognosis of cancers patients as well as reporting survival data; (2) studies that provided related clinicopathological parameters; (3) studies in which number of patients were more than 40.
Exclusion criteria were as followed: (1) articles that lack data; (2) reviews, letters, case reports or duplicates.
Data Extraction and Quality Assessment
Two investigators independently extracted the relevant data from the included articles using a predefined standardized form. We extracted the following data from each study: (1) publication year; (2) countries; (3) first author’s name; (4) types of cancers; (5) detection methods; (6) number of patients; (7) cut off values; (8) clinicopathological parameters; (9) relevant data for OS/PFS/DFS. If the articles did not offer HRs and its 95% CIs directly, we then used the Engauge Digitizer 4.1 software (Tierney et al., 2007) to extract the data from the survival curves. Disagreements in the literature assessment were resolved through consensus with a third reviewer (June Wang). Furthermore, we used the Newcastle-Ottawa Scale (NOS) to assess the quality of these included studies (Zeng et al., 2015). According to the NOS criteria, studies with a score of ≥7 were considered to be high quality articles.
Extraction and Analysis of TCGA Datasets
Datas for HMGA2 expression and clinical information in TCGA were extracted from the UALCAN1 (Chandrashekar et al., 2017). In all, 33 types of cancers were analyzed. There were 9944 subjects that have both HMGA2 expression data and survival data. Accorrding to the transcripts per million (TPM) expression value, the HMGA2 expression levels were divided into high expression group (with TPM values above upper quartile) and low/median expression group (with TPM values below upper quartile). Then, according to the survival data of patients in TCGA datasets, Kaplan–Meier survival analyses were performed and overall survival plots were generated. The difference between high gene expression and low/medium gene expression was compared by Log-rank test.
Outcomes Analysis
Pooled HRs and corresponding 95% CIs were calculated to evaluate the impact of HMGA2 expression on overall survival and disease-free survival. ORs with corresponding 95% CIs were calculated to assess the correlation between HMGA2 expression and clinicopathological features. Chi square-based Q test and I2 test were used to evaluate the heterogeneity across the studies. A fixed-effect model was adopted if the heterogeneity was not significant (I2 < 50% or P-value > 0.05). Otherwise, a random-effect model was selected. Besides, subgroup analysis was performed to explore the sources of heterogeneity. Meanwhile, we used Stata12.0 software (Stata Corporation, College Station, TX, United States) to analyze the sensitivity and publication bias in this study. All the statistical tests were two-sided, and P < 0.05 was considered statistically significant.
Results
The Description of the Included Studies
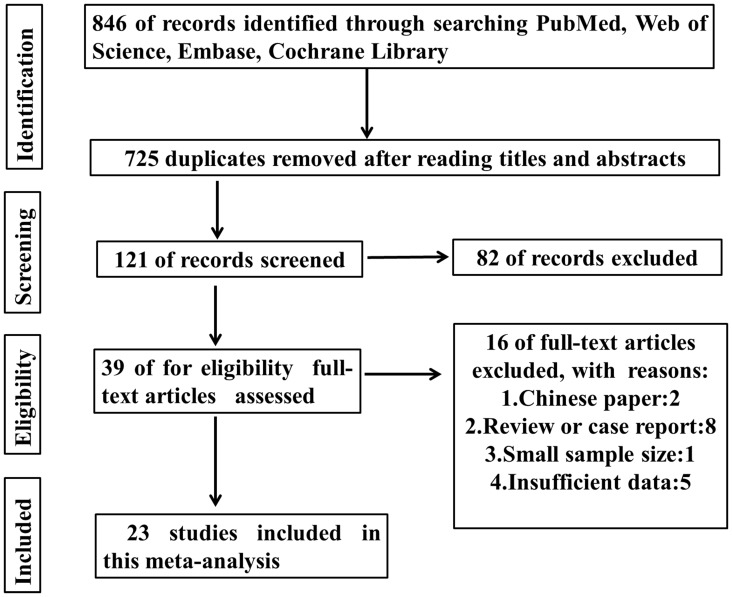
As shown in the Figure 1, 846 records were obtained by searching the databases. After screening the titles and abstracts, 807 articles were excluded. Then 16 papers were excluded because of no available data or non English papers. Eventually, 23 articles were enrolled (Motoyama et al., 2008; Wang et al., 2011; Yang et al., 2011; Wu et al., 2012; Rizzi et al., 2013; Yamazaki et al., 2013; Califano et al., 2014; Kong et al., 2014; Lee et al., 2014, 2015; Jun et al., 2015; Kim et al., 2015; Liu et al., 2015; Xia et al., 2015b; Na et al., 2016; Wei et al., 2016; Wu J. et al., 2016; Zhang et al., 2016; Zhao et al., 2016; Fang et al., 2017; Gunther et al., 2017; Mito et al., 2017; Strell et al., 2017). In total, 15 types of cancers were included in this meta-analysis including ampullary adenocarcinoma, breast cancer, colorectal cancer, clear cell renal cell carcinoma (ccRCC), esophageal adenocarcinoma, esophageal squamous cell carcinoma, gastric cancer, head and neck squamous cell carcinoma, hepatocellular carcinoma, intrahepatic cholangiocarcinomas, nasopharyngeal carcinoma, ovarian carcinoma, oral squamous cell carcinoma, pancreatic ductal adenocarcinoma, and tongue squamous cell carcinoma. In these studies, the level of HMGA2 expression was all detected in collected tumor tissues. Almost all the studies performed immunohistochemistry (IHC) to evaluate the expression of HMGA2 while only one of them used the relative quantitative reverse transcription-polymerase chain reaction (qT-PCR). The main features of the eligible studies were listed in Table 1.
FIGURE 1.
Flow diagram of study selection.
Table 1.
Characteristics of eligible studies in this meta-analysis.
| First author | Year | Country | No. of patients | Tumor type | Method | Cut-off | Outcome | Analysis | Nos |
|---|---|---|---|---|---|---|---|---|---|
| Strell | 2017 | Sweden | 253 | Pancreatic ductal denocarcinoma | ICH | IHC score ≥ 1 | OS | Multivariate | 7 |
| Strell | 2017 | Sweden | 155 | Ampullary adenocarcinoma | ICH | IHC score ≥ 1 | OS | Multivariate | 7 |
| Mito | 2017 | USA | 91 | Esophageal Adenocarcinoma | ICH | NR | OS | Multivariate | 8 |
| Fang | 2017 | China | 148 | Oral Squamous Cell Carcinoma | ICH | IHC ≥ 30% | OS/DFS | Multivariate | 9 |
| Wei | 2017 | China | 96 | Esophageal squamous cell carcinoma | ICH | IHC score ≥ 4 | OS | Multivariate | 8 |
| Gunther | 2016 | Germany | 202 | Head and neck squamous cell carcinoma | RT-PCR | upper quartile | OS/PFS | Multivariate | 8 |
| Wu | 2016 | China | 273 | Breast cancer | ICH | NR | OS | Multivariate | 7 |
| Zhao | 2016 | China | 60 | Tongue squamous cell carcinoma | ICH | NR | OS/DFS | Multivariate | 9 |
| Na | 2016 | China | 162 | Clear cell renal cell carcinoma | ICH | IHC score > 106 | OS | Multivariate | 9 |
| Zhang | 2016 | China | 127 | Esophageal squamous cell carcinoma | ICH | IHC score ≥ 4 | OS | Multivariate | 8 |
| Jun | 2015 | Korea | 110 | Gastric cancer | ICH | IHC score ≥ 5 | RFS | Multivariate | 9 |
| Kim | 2015 | Korea | 74 | Ovarian carcinoma | ICH | NR | OS | Multivariate | 9 |
| Liu | 2015 | China | 116 | Nasopharyngeal carcinoma | ICH | IHC score ≥ 4 | OS | Multivariate | 8 |
| Xia | 2015 | China | 124 | Nasopharyngeal carcinoma | ICH | IHC score ≥ 6 | OS | Multivariate | 8 |
| Lee | 2015 | Korea | 170 | Gastric cancer | ICH | IHC score ≥ 9 | OS | Multivariate | 9 |
| Kong | 2014 | China | 158 | Gastric cancer | ICH | IHC score ≥ 1 | OS | Multivariate | 8 |
| Lee | 2014 | China | 55 | Intrahepatic cholangiocarcinomas | ICH | IHC > 5% | OS | Multivariate | 7 |
| Rizzi | 2013 | Italy | 103 | Colorectal cancer | ICH | IHC ≥ 5% | OS | Kaplan-Meier curves | 7 |
| Califano | 2013 | Italy | 113 | Ovarian cancer | ICH | IHC ≥ 10% | OS/DFS | Multivariate | 7 |
| Yamazaki | 2013 | Japan | 91 | Head and neck squamous cell carcinoma | ICH | IHC score ≥ 1 | OS | Kaplan-Meier curves | 8 |
| Wu | 2012 | China | 107 | Hepatocellular Carcinoma | ICH | IHC ≥ 10% | OS | Multivariate | 8 |
| Yang | 2011 | China | 148 | Bladder cancer | ICH | IHC ≥ 50% | PFS/RFS | Multivariate | 9 |
| Wang | 2011 | China | 89 | Colorectal Cancer | ICH | IHC ≥ 5% | OS | Multivariate | 8 |
| Wang | 2011 | China | 191 | Colorectal Cancer | ICH | IHC ≥ 5% | OS | Multivariate | 8 |
| Motoyama | 2008 | Japan | 110 | Gastric cancer | ICH | NR | OS | Multivariate | 8 |
ICH, Immunohistochemistry; NR, Not Reported; NOS, Newcastle-Ottawa Scale; OS, overall survival; DFS, disease-free survival.
Meta-Analysis of HMGA2 Expression on OS/DFS
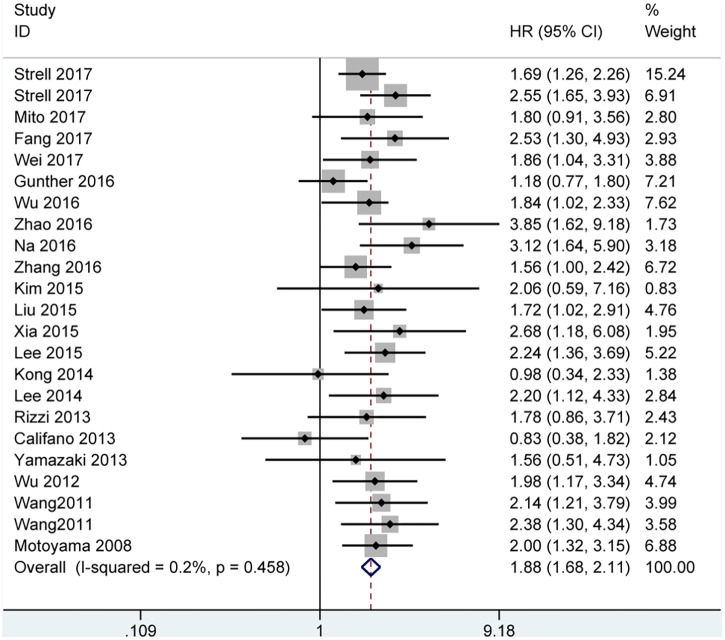
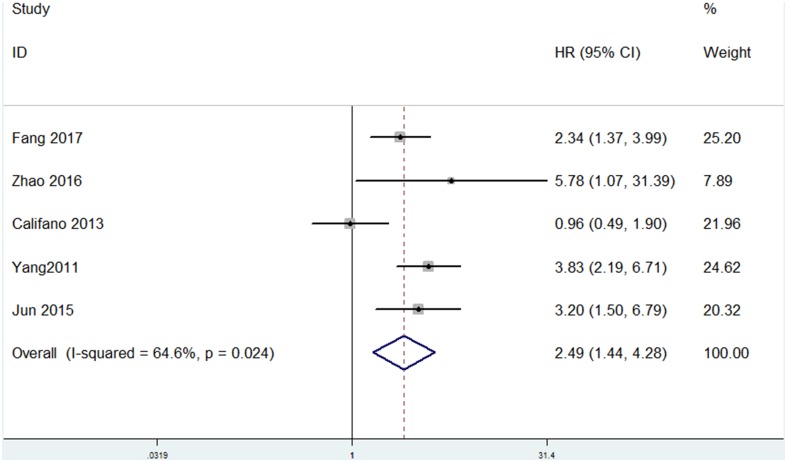
Among the included papers, 23 studies involving 3068 patients showed the data of both HMGA2 expression and OS of the patients. As displayed in Figure 2, there was no obvious heterogeneity across these studies (I2 = 0.2%, P = 0.458). Thus, we used the fixed-effect model to evaluate the pooled HRs and 95% CIs. As a result, the pooled data indicated that elevated HMGA2 was significantly associated with poor OS in patients with cancers (HR = 1.88, 95% CI = 1.68-2.11, P < 0.001). Meanwhile, there were five studies showed the association between HMGA2 expression level and DFS in the included studies. Heterogeneity test indicated there was an obvious heterogeneity (I2 = 64.6%), then the random-effect model was used. Pooled results also demonstrated that high HMGA2 expression was associated with shorter DFS in cancer patients (HR = 2.49, 95% CI = 1.44-4.28) (Figure 3).
FIGURE 2.
Forest plot of studies evaluating hazard ratios of high expressed HMGA2 and the overall survival of cancer patients.
FIGURE 3.
Forest plot of HR for the correlation between HMGA2 expression and Disease-free survival (DFS) in cancers.
Subgroup Analysis for OS
We then made a subgroup analysis for OS, in which the patients were stratified based on cancer type, analysis type and sample size (Table 2). There was only one study for each that evaluated the association between HMGA2 expression and OS in pancreatic ductal adenocarcinoma, ampullary adenocarcinoma, breast cancer, ccRCC, intrahepatic cholangiocarcinoma, and hepatocellular carcinoma. Therefore, we defined these cancers as “other cancers” in this subgroup. Results show that high expression level of HMGA2 was associated with poor OS in gastric cancer (HR = 1.94, 95% CI = 1.42-2.65, P < 0.001), head and neck cancer (HR = 1.77, 95% CI = 1.37-2.29, P < 0.001), colorectal cancer (HR = 2.13, 95% CI = 1.48-3.05, P < 0.001) and other cancers (HR = 2, 95% CI = 1.68-2.40, P < 0.001), but not esophageal cancer (HR = 1.15, 95% CI = 0.55-2.37, P = 0.712) and ovarian cancer (HR = 1.07, 95% CI = 0.55-2.37, P = 0.712). As a result, we found that high level of HMGA2 was related with poor OS in 13 types of cancers. Meanwhile, in the subgroups based on sample size and analysis type, we also found the association between HMGA2 and poor OS except for multivariate analysis.
Table 2.
Subgroup analyses of pooled HR for OS.
| Categories | No. of studies | No. of patients | Pooled HR (95% CI) |
Heterogeneity |
||
|---|---|---|---|---|---|---|
| Fix/Random | p-value | I2(%) | P-value | |||
| [1] OS | 23 | 3068 | 1.88 (1.68-2.11) | 0 | 0.2 | 0.458 |
| [2] Cancer type | ||||||
| 1) Colorectal cancer | 3 | 383 | 2.13(1.48-3.05) | 0 | 0 | 0.834 |
| 2) Esophageal cancer | 3 | 314 | 1.15 (0.55-2.37) | 0.712 | 80 | 0.007 |
| 3) Gastric cancer | 3 | 438 | 1.94 (1.42-2.65) | 0 | 11.4 | 0.324 |
| 5) Head and neck cancer | 6 | 741 | 1.77 (1.37-2.29) | 0 | 42.8 | 0.12 |
| 6) Ovarian cancer | 2 | 187 | 1.07 (0.55-2.08) | 0.835 | 31.6 | 0.227 |
| 7) Others | 8 | 1213 | 2.09 (1.76-2.47) | 0 | 0 | 0.43 |
| [3] Analysis | ||||||
| Multivatiate | 21 | 2874 | 1.80 (1.60-2.02) | 0 | 48.9 | 0.006 |
| Survival curves | 2 | 194 | 1.71 (0.93-3.15) | 0.085 | 0 | 0.864 |
| [4] Sample size | ||||||
| ≥115 | 12 | 2079 | 1.85 (1.61-2.13) | 0 | 22 | 0.227 |
| <115 | 11 | 989 | 1.68 (1.21-2.33) | 0.002 | 59.1 | 0.006 |
OS, overall survival; HR, hazard ratio.
HMGA2 Overexpression and Clinical Pathological Features
In order to gain further insight into the value of HMGA2, we investigated the association between HMGA2 level and certain clinicopathological parameters in cancers (Table 3). The expression level of HMGA2 was significantly associated with TNM stage (OR = 1.65, 95% CI = 1.12-2.44, P = 0.011, random-effect model), tumor differentiation (OR = 1.94, 95% CI = 1.51-2.51, P < 0.001, fix-effect model), tumor invasion depth (OR = 1.71, 95% CI = 1.35-3.16, P < 0.001, fix-effect model), lymph node metastasis (OR = 1.86, 95% CI = 1.27-2.72, P = 0.001, random-effect model), lymphovascular invasion (OR = 2.18, 95% CI = 1.49-3.18, P < 0.001, fix-effect model), vascular invasion (OR = 2.1, 95% CI = 1.42-3.10, P < 0.001, fix-effect model) and distant metastasis (OR = 3.45, 95% CI = 2.06-5.75, P < 0.001, fix-effect model). No significant correlations were found with age (OR = 0.99, 95% CI = 0.74-1.32, P = 0.931, fixed-effect model), gender (OR = 1, 95% CI = 0.84-1.18, P = 0.974, fixed-effect model) and tumor size (OR = 0.77, 95% CI = 0.53-1.14, P = 0.19, fixed-effect model). The results indicated that high HMGA2 expression in human cancers was linked to aggressive biological behavior.
Table 3.
Clinicopathological features of the enrolled studies with high expressed HMGA2 in patients with cancer.
| Clinicopathological parameters | Studies | No. of patients | Risk of high HMGA2 OR (95% CI) | Significant Z | p-value | Heterogeneity I2 (%) | p-value | Model |
|---|---|---|---|---|---|---|---|---|
| Age (<50 vs ≥50) | 6 | 919 | 0.99 (0.74-1.32) | 0.09 | 0.931 | 0 | 0.836 | Fixed effects |
| Gender (male vs female) | 21 | 2803 | 1.00 (0.84-1.18) | 0.03 | 0.974 | 27.3 | 0.122 | Fixed effects |
| Tumor size (<3 cm vs ≥3 cm) | 4 | 543 | 0.77 (0.53-1.14) | 1.31 | 0.19 | 0 | 0.756 | Fixed effects |
| TNM stage (III-IV vs I-II) | 16 | 2292 | 1.65 (1.12-2.44) | 2.53 | 0.011 | 68.4 | 0 | Random effects |
| Tumor differentiation (moderate/well vs poor) | 10 | 1317 | 1.94 (1.51-2.51) | 5.11 | 0 | 46.9 | 0.049 | Fixed effects |
| Tumor invasion depth (T3–T4 vs T1–T2) | 13 | 1767 | 1.71 (1.35-3.16) | 4.52 | 0 | 40.7 | 0.063 | Fixed effects |
| Distant metastasis (Positive vs negative) | 6 | 721 | 3.45 (2.06-5.75) | 4.73 | 0 | 41.7 | 0.127 | Fixed effects |
| Lymph node metastasis (Positive vs negative) | 17 | 2289 | 1.86 (1.27-2.72) | 3.19 | 0.001 | 74.9 | 0 | Random effects |
| Lymphovascular invasion (Positive vs negative) | 5 | 629 | 2.18 (1.49-3.18) | 4.02 | 0 | 31.4 | 0.212 | Fixed effects |
| Vascular invasion (Positive vs negative) | 5 | 655 | 2.1 (1.42-3.10) | 3.73 | 0 | 0 | 0.464 | Fixed Effects |
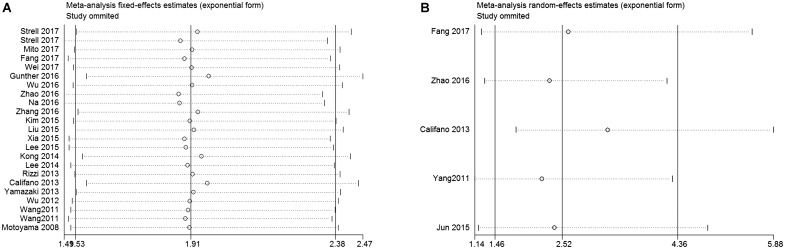
Sensitivity Analyses and Publication Bias
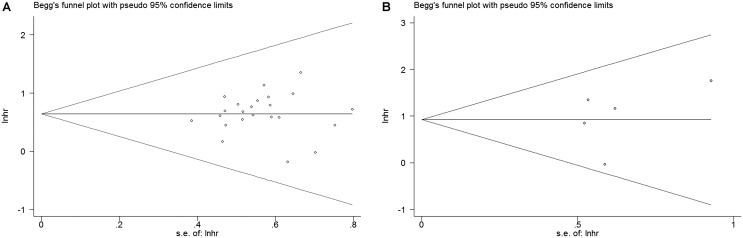
In order to assess whether a single study could significantly affect the overall result, we performed sensitivity analyses. Results demonstrated the individual study had no influence on our meta-analysis (Figure 4), which supported credibility of our analysis. In addition, Funnel plots and Begg’s test were used to evaluate the publication bias of this meta-analysis. Results showed that there was no publication bias existed in studies on HMGA2 overexpression in association with OS (P = 0.597. Figure 5A) and DFS (P = 0.462. Figure 5B).
FIGURE 4.
Sensitivity analysis of this meta-analysis. (A) Overall survival (OS). (B) Disease-free survival (DFS).
FIGURE 5.
Begg’s funnel plots for the studies involved in the meta-analysis of HMGA2 expression and the prognosis of patients with cancers. (A) Overall survival. (B) Disease-free survival. loghr, logarithm of hazard ratios; s.e., standard error.
The Expression of HMGA2 in Cancers Based on TCGA Datasets
We then analyzed the expression of HMGA2 in different cancers in TCGA datasets through UALCAN, which was an interactive web-portal to perform in-depth analyses of TCGA gene expression data (Chandrashekar et al., 2017). HMGA2 was detected in 21 types of cancers and corresponding normal tissues. According to P value obtained from log-rank test, we found that the HMGA2 expression was significantly higher than corresponding normal tissues in 17 types of cancers except glioblastoma multiforme, kidney chromophobe, kidney renal clear cell carcinoma and prostate adenocarcinoma (Table 4).
Table 4.
The difference of HMGA2 expression in cancers and corresponding normal tissues in TCGA datasets.
| Types of cancer | TCGA dataset | No. of cancer tissues | No. of normal tissues | P value |
|---|---|---|---|---|
| Bladder urothelial carcinoma | TCGA-BLCA | 408 | 19 | <0.0001 |
| Breast invasive carcinoma | TCGA-BRCA | 1097 | 114 | <0.0001 |
| Cervical squamous cell carcinoma | TCGA-CESC | 305 | 3 | <0.0001 |
| Cholangiocarcinoma | TCGA-CHOL | 36 | 9 | 0.032 |
| Colon adenocarcinoma | TCGA-COAD | 286 | 41 | <0.0001 |
| Esophageal carcinoma | TCGA-ESCA | 184 | 11 | <0.0001 |
| Glioblastoma multiforme | TCGA-GBM | 156 | 5 | 0.052 |
| Head and Neck squamous cell carcinoma | TCGA-HNSC | 520 | 44 | <0.0001 |
| Kidney chromophobe | TCGA-KICH | 67 | 25 | 0.316 |
| Kidney renal clear cell carcinoma | TCGA-KIRC | 533 | 72 | 0.071 |
| Kidney renal papillary cell carcinoma | TCGA-KIRP | 290 | 32 | <0.0001 |
| Liver hepatocellular carcinoma | TCGA-LIHC | 371 | 50 | <0.0001 |
| Lung adenocarcinoma | TCGA-LUAD | 515 | 59 | <0.0001 |
| Lung squamous cell carcinoma | TCGA-LUSC | 503 | 52 | <0.0001 |
| Ovarian serous cystadenocarcinoma | TCGA-OV | 178 | 4 | <0.0001 |
| Pheochromocytoma and Paraganglioma | TCGA-PCPG | 179 | 3 | 0.011 |
| Prostate adenocarcinoma | TCGA-PRAD | 497 | 52 | 0.201 |
| Rectum adenocarcinoma | TCGA-READ | 166 | 10 | <0.0001 |
| Stomach adenocarcinoma | TCGA-STAD | 415 | 34 | <0.0001 |
| Thyroid carcinoma | TCGA-THCA | 505 | 59 | <0.0001 |
| Uterine corpus endometrial carcinoma | TCGA-UCEC | 546 | 35 | <0.0001 |
Validation by TCGA Datasets
In order to validate the result of our meta-analysis, TCGA datasets were analyzed to explore whether HMGA2 could be involved in human cancers and affect patients’ survival. In total, 9944 patients with 33 types of cancer were obtained (Table 5). Significant association between high expressed HMGA2 and poor overall survival in patients was found in 14 types of cancers. They were adrenocortical carcinoma, bladder urothelial carcinoma, brain lower grade glioma, head and neck squamous cell carcinoma, kidney renal clear cell carcinoma, kidney renal papillary cell carcinoma, acute myeloid leukemia, liver hepatocellular carcinoma, lung adenocarcinoma, pancreatic adenocarcinoma, prostate adenocarcinoma, sarcoma, uterine corpus endometrial carcinoma and uveal melanoma (Figure 6).
Table 5.
The difference of overall survival in cancer patients with high HMGA2 expression vs low/median expression.
| Cancer type | No. of cancer tissues |
P value | ||
|---|---|---|---|---|
| Total | High | Low/Median | ||
| ACC | 79 | 20 | 59 | <0.0001 |
| BLCA | 406 | 102 | 304 | 0.023 |
| BLGG | 511 | 128 | 383 | <0.0001 |
| BRCA | 1081 | 273 | 808 | 0.72 |
| CESC | 291 | 77 | 214 | 0.3 |
| CHOL | 36 | 9 | 27 | 0.14 |
| COAD | 279 | 67 | 212 | 0.38 |
| ESCA | 184 | 46 | 138 | 0.76 |
| GBM | 152 | 39 | 113 | 0.13 |
| HNSCC | 519 | 130 | 389 | 0.00016 |
| KICH | 65 | 17 | 48 | 0.39 |
| KIRC | 531 | 134 | 397 | <0.0001 |
| KIRP | 287 | 72 | 215 | 0.043 |
| LAML | 163 | 41 | 122 | 0.049 |
| LIHC | 365 | 89 | 276 | 0.0092 |
| LUAD | 502 | 123 | 379 | 0.015 |
| LUSC | 494 | 123 | 371 | 0.28 |
| DLBC | 47 | 11 | 36 | 0.21 |
| MESO | 85 | 22 | 63 | 0.56 |
| OVSC | 303 | 76 | 227 | 0.83 |
| PAAD | 177 | 45 | 132 | 0.019 |
| PCPG | 179 | 45 | 134 | 0.23 |
| PRAD | 497 | 125 | 372 | 0.0068 |
| READ | 165 | 41 | 124 | 0.52 |
| SARC | 259 | 65 | 194 | 0.019 |
| SKCM | 459 | 115 | 344 | 0.61 |
| STAD | 392 | 98 | 294 | 0.49 |
| TGCT | 134 | 30 | 104 | 0.26 |
| THYM | 119 | 30 | 89 | 0.052 |
| THCA | 504 | 127 | 377 | 0.64 |
| UCS | 56 | 15 | 41 | 0.57 |
| UCEC | 543 | 136 | 407 | 0.023 |
| UVM | 80 | 20 | 60 | 0.035 |
ACC, adrenocortical carcinoma; BLCA, bladder urothelial carcinoma; BLGG, brain lower grade glioma; BRCA, breast invasive carcinoma; CESC, cervical squamous cell carcinoma; CHOL, cholangiocarcinoma; COAD, colon adenocarcinoma; ESCA, esophageal carcinoma; GBM, glioblastoma multiforme; HNSCC, head and neck squamous cell carcinoma; KICH, kidney chromophobe; KIRC, kidney renal clear cell carcinoma; LAML, acute myeloid leukemia; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; DLBC, lymphoid neoplasm diffuse large B-cell lymphoma; MESO, mesothelioma; OVSC, ovarian serous cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; PCPG, pheochromocytoma and paraganglioma; PRAD, prostate adenocarcinoma; READ, rectum adenocarcinoma; SARC, sarcoma; SKCM, skin cutaneous melanoma; STAD, stomach adenocarcinoma; TGCT, testicular germ cell tumors; THYM, thymoma; THCA, thyroid carcinoma; UCS, uterine carcinosarcoma; UCEC, uterine corpus endometrial carcinoma; UVM, uveal melanoma.
FIGURE 6.
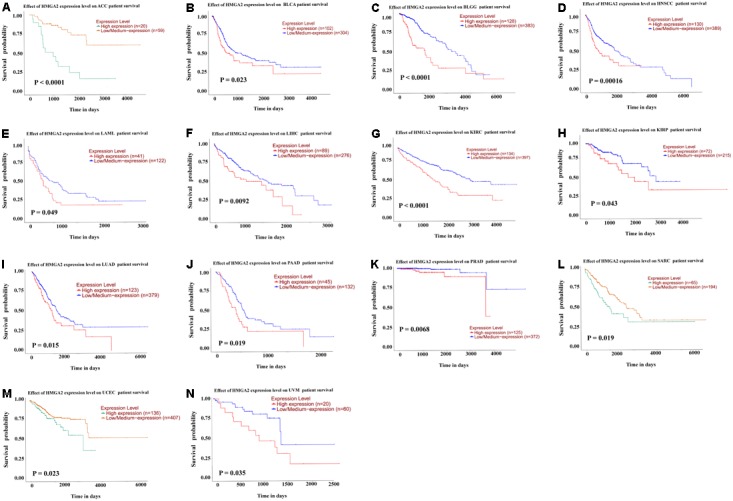
Kaplan–Meier survival curves for cancer patients based on TCGA datasets. (A) adrenocortical carcinoma. (B) bladder urothelial carcinoma. (C) brain lower grade glioma. (D) head and neck squamous cell carcinoma. (E) kidney renal clear cell carcinoma. (F) kidney renal papillary cell carcinoma. (G) acute myeloid leukemia. (H) liver hepatocellular carcinoma. (I) lung adenocarcinoma. (J) pancreatic adenocarcinoma. (K) prostate adenocarcinoma. (L) sarcoma. (M) uterine corpus endometrial carcinoma. (N) uveal melanoma.
Considering the TCGA datasets and our meta-analysis, we identified correlation between high HMGA2 expression and head and neck cancer, pancreatic ductal adenocarcinoma, ccRCC, hepatocellular carcinoma, esophageal adenocarcinoma and ovarian carcinoma, except breast cancer and gastric cancer.
Discussion
Recently, increasing evidences have suggested that HMGA2 protein could participate in aggressive tumor growth (Peng et al., 2008; Hristov et al., 2009), stem cell self-renewal (Nishino et al., 2008; Tzatsos and Bardeesy, 2008), DNA damage response (Li et al., 2009), and tumor cell differentiation (Shell et al., 2007). However, the precise role of HMGA2 in malignant transformation and the gene regulation of tumorigenesis were still unclear (Venkatesan et al., 2012). Data have collectively indicated that the high level of HMGA2 could serve as a novel biomarker to evaluate the prognosis of patients with various cancers such as breast cancer (Wu J. et al., 2016), lung cancer (Di Cello et al., 2008), ovarian cancer (Mahajan et al., 2010), colorectal cancer (Wang et al., 2011), and gastric cancer (Lee et al., 2015). However, single studies may not be sufficient and accurate and whether HMGA2 could be used as a prognostic biomarker in human cancers was still unclear.
To the best of our knowledge, our meta-analysis was the first study to evaluate the significance of HMGA2 and prognostic value in various cancers through drawing data from both the TCGA datasets and published studies. In this paper, 15 types of cancers involving 3068 patients were included. Then, based on TCGA datasets, we analyzed 9944 patients with 33 types of cancers. The meta-analysis results suggested that high expression of HMGA2 was associated with shorter OS and DFS in patients with cancers. However, in the subgroup analysis for OS, we only found the association in 13 types of cancers except for the ones in esophageal adenocarcinoma and ovarian carcinoma. In addition, through TCGA datasets, we observed the poor prognosis in 14 types of cancers. Consistent results were found in six types of cancers. Patients with high expressed HMGA2 in ccRCC, head and neck cancer, hepatocellular carcinoma and pancreatic ductal adenocarcinoma showed a significant shorter OS than patients with a low level of HMGA2 expression. However, in esophageal adenocarcinoma and ovarian carcinoma, we did not observe any significant correlation.
We then explored the relationship between clinicopathological features and high HMGA2 expression in our enrolled studies. We found the level of HMGA2 was positively associated with TNM stage, tumor differentiation, tumor invasion depth, lymph node metastasis, lymphovascular invasion, and vascular invasion, which indicated that HMGA2 might have a significant relationship with advanced features of cancer. Previous research performed by Di Cello et al. (2008) found that HMGA2 protein levels were increased in all metastatic lung cancer cell lines compared with benign tumors and normal cells. According to Piscuoglio et al. (2012), HMGA2 might play a significant role in the late stages of pancreatic carcinogenesis and in the progression towards a more aggressive tumor phenotype. In addition, HMGA2 was demonstrated to play a critical role in epithelial-to-mesenchymal transition (EMT) in various cancers such as gastric cancer (Zha et al., 2013), hepatocellular carcinoma (Luo et al., 2013) and nasopharyngeal cancer (Xia et al., 2015a), thus inducing epithelial cancer invasion and metastasis.
Since the high expression of HMGA2 could be potentially an indicator of poor prognosis in patients with certain cancers, HMGA2 was expected to be a new therapeutic biomarker in cancers. Recently, Zhao et al. demonstrated that by directly targeting HMGA2, miR-599 could serve tumor suppressive roles in ccRCC (Zhao et al., 2018). A study conducted by Malek et al. (2008) suggested silencing HMGA2 expression in ovarian cancer cells could have a therapeutic effect on ovarian cancer. Liu et al. (2014) reported the raf kinase inhibitor protein can inhibit the survival and invasion of gastric cancer cells and promote apoptosis through regulating the expression of HMGA2 and Osteopontin. The findings of Chau et al. (2003) suggested that HMGA2 could become a therapeutic target by blocking HMGA2 protein expression in retinoblastoma cells or through inhibiting expression of the HMGA2 gene by targeting its promoters. Wang et al. (2011) found radiotherapy significantly reduced the relative risk death in HMGA2-positive colorectal cancers (CRCs), but not in HMGA2-negative CRCs. All these studies showed HMGA2 could play a key role in cancers, which supported it potential role as a biomarker for cancer therapy.
However, some limitations of this study should be acknowledged. Firstly, heterogeneity in our study was substantial, which might be attributed to differences in types of cancers, study areas and cut-off values. Secondly, in the process of data extraction, we evaluated the HRs and 95% CIs from the Kaplan–Meier survival curves in five studies rather than directly obtained from the studies, which might be less accurate than extracting directly from published statistics. Thirdly, since negative results would have little chance to be published, there may be a bias in the published studies. Therefore, although no significant publication bias was detected in this meta-analysis, the results of our meta-analysis still need to be verified by a larger number of studies.
Despite the limitations described, our meta-analysis revealed the significance of HMGA2. Our meta-analysis showed that HMGA2 likely played an important role in human cancers and overexpression of HMGA2 could be associated with aggressive biological behavior although its prognostic values varied in different types of cancers. Specifically, overexpression of HMGA2 was significantly associated with poor prognosis in patients with ccRCC, head and neck cancer, hepatocellular carcinoma and pancreatic ductal adenocarcinoma, but not esophageal adenocarcinoma and ovarian carcinoma.
Author Contributions
BH, JY, and MY conceived the study. BH, JY, and JW searched the databases and extracted the data. WF, ZZ, and PX analyzed the data. BH wrote the draft of the paper. QC, PW, and MY reviewed the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. The study was supported by National Natural Science Foundation of China (Nos. 81472033 and 30901308), the National Science Foundation of Hubei Province (Nos. 2013CFB233 and 2013CFB235), the Scientific and Technological Project of Wuhan City (No. 2014060101010045), Hubei Province Health and Family Planning Scientific Research Project (WJ2015Q021), and Training Program of the Science and Technology Innovation from Zhongnan Hospital of Wuhan University (cxpy20160054).
References
- Califano D., Pignata S., Losito N. S., Ottaiano A., Greggi S., De Simone V., et al. (2014). High HMGA2 expression and high body mass index negatively affect the prognosis of patients with ovarian cancer. J. Cell. Physiol. 229 53–59. 10.1002/jcp.24416 [DOI] [PubMed] [Google Scholar]
- Chandrashekar D. S., Bashel B., Balasubramanya S. A. H., Creighton C. J., Ponce-Rodriguez I., Chakravarthi B., et al. (2017). UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 19 649–658. 10.1016/j.neo.2017.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau K. Y., Manfioletti G., Cheung-Chau K. W., Fusco A., Dhomen N., Sowden J. C., et al. (2003). Derepression of HMGA2 gene expression in retinoblastoma is associated with cell proliferation. Mol. Med. 9 154–165. 10.2119/2003-00020.Ono [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiappetta G., Bandiera A., Berlingieri M. T., Visconti R., Manfioletti G., Battista S., et al. (1995). The expression of the high mobility group HMGI (Y) proteins correlates with the malignant phenotype of human thyroid neoplasias. Oncogene 10 1307–1314. [PubMed] [Google Scholar]
- Di Cello F., Hillion J., Hristov A., Wood L. J., Mukherjee M., Schuldenfrei A., et al. (2008). HMGA2 participates in transformation in human lung cancer. Mol. Cancer Res. 6 743–750. 10.1158/1541-7786.mcr-07-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang C. Y., Liew P. L., Chen C. L., Lin Y. H., Fang C. L., Chen W. Y. (2017). High HMGA2 expression correlates with reduced recurrence-free survival and poor overall survival in oral squamous cell carcinoma. Anticancer. Res. 37 1891–1899. 10.21873/anticanres.11527 [DOI] [PubMed] [Google Scholar]
- Gonzalez C. A., Agudo A. (2012). Carcinogenesis, prevention and early detection of gastric cancer: where we are and where we should go. Int. J. Cancer 130 745–753. 10.1002/ijc.26430 [DOI] [PubMed] [Google Scholar]
- Gorin M. A., Verdone J. E., van der Toom E., Bivalacqua T. J., Allaf M. E., Pienta K. J. (2017). Circulating tumour cells as biomarkers of prostate, bladder, and kidney cancer. Nat. Rev. Urol 14 90–97. 10.1038/nrurol.2016.224 [DOI] [PubMed] [Google Scholar]
- Gunther K., Foraita R., Friemel J., Gunther F., Bullerdiek J., Nimzyk R., et al. (2017). The stem cell factor HMGA2 is expressed in Non-HPV-associated head and neck squamous cell carcinoma and predicts patient survival of distinct subsites. Cancer Epidemiol. Biomarkers. Prev. 26 197–205. 10.1158/1055-9965.EPI-16-0492 [DOI] [PubMed] [Google Scholar]
- Hirning-Folz U., Wilda M., Rippe V., Bullerdiek J., Hameister H. (1998). The expression pattern of the Hmgic gene during development. Genes Chromosomes Cancer 23 350–357. [DOI] [PubMed] [Google Scholar]
- Hock R., Furusawa T., Ueda T., Bustin M. (2007). HMG chromosomal proteins in development and disease. Trends Cell Biol. 17 72–79. 10.1016/j.tcb.2006.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hristov A. C., Cope L., Reyes M. D., Singh M., Iacobuzio-Donahue C., Maitra A., et al. (2009). HMGA2 protein expression correlates with lymph node metastasis and increased tumor grade in pancreatic ductal adenocarcinoma. Mod. Pathol. 22 43–49. 10.1038/modpathol.2008.140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun K. H., Jung J. H., Choi H. J., Shin E. Y., Chin H. M. (2015). HMGA1/HMGA2 protein expression and prognostic implications in gastric cancer. Int. J. Surg. 24(Pt A), 39–44. 10.1016/j.ijsu.2015.10.031 [DOI] [PubMed] [Google Scholar]
- Kim T. H., Song J. Y., Park H., Jeong J. Y., Kwon A. Y., Heo J. H., et al. (2015). miR-145, targeting high-mobility group A2, is a powerful predictor of patient outcome in ovarian carcinoma. Cancer Lett 356(2 Pt B), 937–945. 10.1016/j.canlet.2014.11.011 [DOI] [PubMed] [Google Scholar]
- Kojima T., Kawai K., Miyazaki J., Nishiyama H. (2017). Biomarkers for precision medicine in bladder cancer. Int. J. Clin. Oncol. 22 207–213. 10.1007/s10147-016-1068-8 [DOI] [PubMed] [Google Scholar]
- Kong D., Su G., Zha L., Zhang H., Xiang J., Xu W., et al. (2014). Coexpression of HMGA2 and Oct4 predicts an unfavorable prognosis in human gastric cancer. Med. Oncol. 31:130. 10.1007/s12032-014-0130-5 [DOI] [PubMed] [Google Scholar]
- Kumar M. S., Armenteros-Monterroso E., East P., Chakravorty P., Matthews N., Winslow M. M., et al. (2014). HMGA2 functions as a competing endogenous RNA to promote lung cancer progression. Nature 505 212–217. 10.1038/nature12785 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Langelotz C., Schmid P., Jakob C., Heider U., Wernecke K. D., Possinger K., et al. (2003). Expression of high-mobility-group-protein HMGI-C mRNA in the peripheral blood is an independent poor prognostic indicator for survival in metastatic breast cancer. Br. J. Cancer 88 1406–1410. 10.1038/sj.bjc.6600935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. T., Wu T. T., Lohse C. M., Zhang L. (2014). High-mobility group AT-hook 2: an independent marker of poor prognosis in intrahepatic cholangiocarcinoma. Hum. Pathol. 45 2334–2340. 10.1016/j.humpath.2014.04.026 [DOI] [PubMed] [Google Scholar]
- Lee J., Ha S., Jung C. K., Lee H. H. (2015). High-mobility-group A2 overexpression provokes a poor prognosis of gastric cancer through the epithelial-mesenchymal transition. Int. J. Oncol. 46 2431–2438. 10.3892/ijo.2015.2947 [DOI] [PubMed] [Google Scholar]
- Li A. Y., Boo L. M., Wang S. Y., Lin H. H., Wang C. C., Yen Y., et al. (2009). Suppression of nonhomologous end joining repair by overexpression of HMGA2. Cancer Res. 69 5699–5706. 10.1158/0008-5472.can-08-4833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Li P., Li B., Sun P., Zhang J., Wang B., et al. (2014). RKIP inhibits gastric cancer cell survival and invasion by regulating the expression of HMGA2 and OPN. Tumour Biol. 35 11949–11958. 10.1007/s13277-014-2486-8 [DOI] [PubMed] [Google Scholar]
- Liu Z., Wu K., Yang Z., Wu A. (2015). High-mobility group A2 overexpression is an unfavorable prognostic biomarker for nasopharyngeal carcinoma patients. Mol. Cell. Biochem. 409 155–162. 10.1007/s11010-015-2521-0 [DOI] [PubMed] [Google Scholar]
- Luo Y., Li W., Liao H. (2013). HMGA2 induces epithelial-to-mesenchymal transition in human hepatocellular carcinoma cells. Oncol. Lett. 5 1353–1356. 10.3892/ol.2013.1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan A., Liu Z., Gellert L., Zou X., Yang G., Lee P., et al. (2010). HMGA2: a biomarker significantly overexpressed in high-grade ovarian serous carcinoma. Mod. Pathol. 23 673–681. 10.1038/modpathol.2010.49 [DOI] [PubMed] [Google Scholar]
- Malek A., Bakhidze E., Noske A., Sers C., Aigner A., Schafer R., et al. (2008). HMGA2 gene is a promising target for ovarian cancer silencing therapy. Int. J. Cancer 123 348–356. 10.1002/ijc.23491 [DOI] [PubMed] [Google Scholar]
- Mito J. K., Agoston A. T., Dal Cin P., Srivastava A. (2017). Prevalence and significance of HMGA2 expression in oesophageal adenocarcinoma. Histopathology 71 909–917. 10.1111/his.13310 [DOI] [PubMed] [Google Scholar]
- Miyazawa J., Mitoro A., Kawashiri S., Chada K. K., Imai K. (2004). Expression of mesenchyme-specific gene HMGA2 in squamous cell carcinomas of the oral cavity. Cancer Res. 64 2024–2029. 10.1158/0008-5472.CAN-03-1855 [DOI] [PubMed] [Google Scholar]
- Moher D., Liberati A., Tetzlaff J., Altman D. G. (2009). Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann. Intern. Med. 151 264–269, W64. 10.7326/0003-4819-151-4-200908180-00135 [DOI] [PubMed] [Google Scholar]
- Motoyama K., Inoue H., Nakamura Y., Uetake H., Sugihara K., Mori M. (2008). Clinical significance of high mobility group A2 in human gastric cancer and its relationship to let-7 microRNA family. Clin. Cancer Res. 14 2334–2340. 10.1158/1078-0432.CCR-07-4667 [DOI] [PubMed] [Google Scholar]
- Na N., Si T., Huang Z., Miao B., Hong L., Li H., et al. (2016). High expression of HMGA2 predicts poor survival in patients with clear cell renal cell carcinoma. Onco Targets Ther. 9 7199–7205. 10.2147/OTT.S116953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino J., Kim I., Chada K., Morrison S. J. (2008). Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducing p16Ink4a and p19Arf Expression. Cell 135 227–239. 10.1016/j.cell.2008.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallante P., Sepe R., Puca F., Fusco A. (2015). High mobility group a proteins as tumor markers. Front. Med. 2:15. 10.3389/fmed.2015.00015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y., Laser J., Shi G., Mittal K., Melamed J., Lee P., et al. (2008). Antiproliferative effects by Let-7 repression of high-mobility group A2 in uterine leiomyoma. Mol. Cancer Res. 6 663–673. 10.1158/1541-7786.mcr-07-0370 [DOI] [PubMed] [Google Scholar]
- Piscuoglio S., Zlobec I., Pallante P., Sepe R., Esposito F., Zimmermann A., et al. (2012). HMGA1 and HMGA2 protein expression correlates with advanced tumour grade and lymph node metastasis in pancreatic adenocarcinoma. Histopathology 60 397–404. 10.1111/j.1365-2559.2011.04121.x [DOI] [PubMed] [Google Scholar]
- Ribeiro I. P., Barroso L., Marques F., Melo J. B., Carreira I. M. (2016). Early detection and personalized treatment in oral cancer: the impact of omics approaches. Mol. Cytogenet. 9:85. 10.1186/s13039-016-0293-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzi C., Cataldi P., Iop A., Isola M., Sgarra R., Manfioletti G., et al. (2013). The expression of the high-mobility group A2 protein in colorectal cancer and surrounding fibroblasts is linked to tumor invasiveness. Hum. Pathol. 44 122–132. 10.1016/j.humpath.2012.05.001 [DOI] [PubMed] [Google Scholar]
- Rogalla P., Drechsler K., Frey G., Hennig Y., Helmke B., Bonk U., et al. (1996). HMGI-C expression patterns in human tissues. Implications for the genesis of frequent mesenchymal tumors. Am. J. Pathol. 149 775–779. [PMC free article] [PubMed] [Google Scholar]
- Sarhadi V. K., Wikman H., Salmenkivi K., Kuosma E., Sioris T., Salo J., et al. (2006). Increased expression of high mobility group A proteins in lung cancer. J. Pathol. 209 206–212. 10.1002/path.1960 [DOI] [PubMed] [Google Scholar]
- Shell S., Park S. M., Radjabi A. R., Schickel R., Kistner E. O., Jewell D. A., et al. (2007). Let-7 expression defines two differentiation stages of cancer. Proc. Natl. Acad. Sci. U.S.A. 104 11400–11405. 10.1073/pnas.0704372104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R. L., Miller K. D., Jemal A. (2017). Cancer Statistics, 2017. CA Cancer J. Clin. 67 7–30. 10.3322/caac.21387 [DOI] [PubMed] [Google Scholar]
- Strell C., Norberg K. J., Mezheyeuski A., Schnittert J., Kuninty P. R., Moro C. F., et al. (2017). Stroma-regulated HMGA2 is an independent prognostic marker in PDAC and AAC. Br. J. Cancer 117 65–77. 10.1038/bjc.2017.140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tierney J. F., Stewart L. A., Ghersi D., Burdett S., Sydes M. R. (2007). Practical methods for incorporating summary time-to-event data into meta-analysis. Trials 8:16. 10.1186/1745-6215-8-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torre L. A., Bray F., Siegel R. L., Ferlay J., Lortet-Tieulent J., Jemal A. (2015). Global cancer statistics, 2012. CA Cancer J. Clin. 65 87–108. 10.3322/caac.21262 [DOI] [PubMed] [Google Scholar]
- Tzatsos A., Bardeesy N. (2008). Ink4a/Arf regulation by let-7b and Hmga2: a genetic pathway governing stem cell aging. Cell Stem Cell 3 469–470. 10.1016/j.stem.2008.10.008 [DOI] [PubMed] [Google Scholar]
- Venkatesan N., Krishnakumar S., Deepa P. R., Deepa M., Khetan V., Reddy M. A. (2012). Molecular deregulation induced by silencing of the high mobility group protein A2 gene in retinoblastoma cells. Mol. Vis. 18 2420–2437. [PMC free article] [PubMed] [Google Scholar]
- Wang X., Liu X., Li A. Y., Chen L., Lai L., Lin H. H., et al. (2011). Overexpression of HMGA2 promotes metastasis and impacts survival of colorectal cancers. Clin. Cancer Res. 17 2570–2580. 10.1158/1078-0432.CCR-10-2542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei R., Shang Z., Leng J., Cui L. (2016). Increased expression of high-mobility group A2: a novel independent indicator of poor prognosis in patients with esophageal squamous cell carcinoma. J. Cancer Res. Ther. 12 1291–1297. 10.4103/0973-1482.180616 [DOI] [PubMed] [Google Scholar]
- Wood L. J., Maher J. F., Bunton T. E., Resar L. M. (2000). The oncogenic properties of the HMG-I gene family. Cancer Res. 60 4256–4261. [PubMed] [Google Scholar]
- Wu J., Zhang S., Shan J., Hu Z., Liu X., Chen L., et al. (2016). Elevated HMGA2 expression is associated with cancer aggressiveness and predicts poor outcome in breast cancer. Cancer Lett. 376 284–292. 10.1016/j.canlet.2016.04.005 [DOI] [PubMed] [Google Scholar]
- Wu S., Powers S., Zhu W., Hannun Y. A. (2016). Substantial contribution of extrinsic risk factors to cancer development. Nature 529 43–47. 10.1038/nature16166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Wang Z., Lu R., Jiang W. (2012). Expression of high mobility group A2 is associated with poor survival in hepatocellular carcinoma. Pathol. Oncol. Res. 18 983–987. 10.1007/s12253-012-9514-z [DOI] [PubMed] [Google Scholar]
- Xia Y. Y., Yin L., Jiang N., Guo W. J., Tian H., Jiang X. S., et al. (2015a). Downregulating HMGA2 attenuates epithelial-mesenchymal transition-induced invasion and migration in nasopharyngeal cancer cells. Biochem. Biophys. Res. Commun. 463 357–363. 10.1016/j.bbrc.2015.05.068 [DOI] [PubMed] [Google Scholar]
- Xia Y. Y., Yin L., Tian H., Guo W. J., Jiang N., Jiang X. S., et al. (2015b). HMGA2 is associated with epithelial-mesenchymal transition and can predict poor prognosis in nasopharyngeal carcinoma. Onco Targets Ther. 8 169–176. 10.2147/OTT.S74397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki H., Mori T., Yazawa M., Maeshima A. M., Matsumoto F., Yoshimoto S., et al. (2013). Stem cell self-renewal factors Bmi1 and HMGA2 in head and neck squamous cell carcinoma: clues for diagnosis. Lab. Invest. 93 1331–1338. 10.1038/labinvest.2013.120 [DOI] [PubMed] [Google Scholar]
- Yang G. L., Zhang L. H., Bo J. J., Hou K. L., Cai X., Chen Y. Y., et al. (2011). Overexpression of HMGA2 in bladder cancer and its association with clinicopathologic features and prognosis HMGA2 as a prognostic marker of bladder cancer. Eur. J. Surg. Oncol. 37 265–271. 10.1016/j.ejso.2011.01.004 [DOI] [PubMed] [Google Scholar]
- Zeng X., Zhang Y., Kwong J. S., Zhang C., Li S., Sun F., et al. (2015). The methodological quality assessment tools for preclinical and clinical studies, systematic review and meta-analysis, and clinical practice guideline: a systematic review. J. Evid. Based Med. 8 2–10. 10.1111/jebm.12141 [DOI] [PubMed] [Google Scholar]
- Zha L., Zhang J., Tang W., Zhang N., He M., Guo Y., et al. (2013). HMGA2 elicits EMT by activating the Wnt/beta-catenin pathway in gastric cancer. Dig. Dis. Sci. 58 724–733. 10.1007/s10620-012-2399-6 [DOI] [PubMed] [Google Scholar]
- Zhang M., Hu D., Wang S., Qin C. (2016). Clinicopathologic significance of HMGA2 expression’s correlation with prognosis of esophageal squamous cell carcinoma after Ivor Lewis esophagectomy. Minerva Chir. 71 239–244. [PubMed] [Google Scholar]
- Zhao H., Zhao H., Xia X., Liu X. (2018). MicroRNA-599 targets high-mobility group AT-hook 2 to inhibit cell proliferation and invasion in clear cell renal carcinoma. Mol. Med. Rep. 17 7451–7459. 10.3892/mmr.2018.8755 [DOI] [PubMed] [Google Scholar]
- Zhao X. P., Zhang H., Jiao J. Y., Tang D. X., Wu Y. L., Pan C. B. (2016). Overexpression of HMGA2 promotes tongue cancer metastasis through EMT pathway. J. Transl. Med. 14:26. 10.1186/s12967-016-0777-0 [DOI] [PMC free article] [PubMed] [Google Scholar]