Abstract
Nucleosome-depleted regions around which nucleosomes order following the “statistical” positioning scenario were recently shown to be encoded in the DNA sequence in human. This intrinsic nucleosomal ordering strongly correlates with oscillations in the local GC content as well as with the interspecies and intraspecies mutation profiles, revealing the existence of both positive and negative selection. In this letter, we show that these predicted nucleosome inhibitory energy barriers (NIEBs) with compacted neighboring nucleosomes are indeed ubiquitous to all vertebrates tested. These 1 kb-sized chromatin patterns are widely distributed along vertebrate chromosomes, overall covering more than a third of the genome. We have previously observed in human deviations from neutral evolution at these genome-wide distributed regions, which we interpreted as a possible indication of the selection of an open, accessible, and dynamic nucleosomal array to constitutively facilitate the epigenetic regulation of nuclear functions in a cell-type-specific manner. As a first, very appealing observation supporting this hypothesis, we report evidence of a strong association between NIEB borders and the poly(A) tails of Alu sequences in human. These results suggest that NIEBs provide adequate chromatin patterns favorable to the integration of Alu retrotransposons and, more generally to various transposable elements in the genomes of primates and other vertebrates.
Main Text
During the past decade, in vivo and in vitro high-resolution mapping of nucleosomes along various genomes ranging from yeast to human and for different cell types have been made available and have progressively led scientists to deeply revise the original dogma concerning DNA-sequence-driven nucleosome positioning (1, 2, 3, 4, 5, 6, 7, 8). Indeed, an alternative to the tight histone binding obtained with favorable positioning sequences is the statistical positioning of nucleosomes near nucleosome inhibitory energy barriers (NIEBs) (7, 9, 10, 11). These excluding barriers can be encoded via either unfavorable sequences that potentially resist the structural distortions required by nucleosome formation or particular sequences that may recruit transcription factors and/or other protein complexes such as chromatin regulators that may compete with the nucleosomes (1, 3, 6, 7). In that context, a possible clue to the understanding of chromatin-mediated regulation of nuclear functions is the relative positioning of regulatory sites with respect to the NIEBs encoded in the DNA sequence. In Saccharomyces cerevisiae and related yeast species, most of the nucleosome-depleted regions (NDRs) observed in vivo at gene transcription start sites and transcription termination sites (10, 12, 13, 14, 15) and at active DNA replication origins (16, 17) indeed correspond to NIEBs, up to some local shape remodeling and phasing of the nucleosome occupancy profile (2, 3). Besides this intrinsic regulation of transcription and replication initiation, the remarkable nucleosome organization observed in yeast genes because of the collective confinement of nucleosomes between the bordering NIEBs was shown to play an important role in the regulation of gene expression (18, 19). This functional location of NIEBs is indeed quite consistent with the fact that they correspond to sequences that display the lowest level of evolutionary divergence along yeast chromosomes (20, 21, 22, 23, 24). The situation is totally different in mammals and higher eukaryotes, in whom gene promoters and replication origins are known to be GC-rich, strongly suggesting a nucleosome positioning preference at these regulatory sequences (4, 15, 25, 26, 27, 28). This is exactly what has been recently observed in human, for whom a high nucleosome affinity is directly programmed at regulatory sequences to intrinsically restrict access to regulatory information that will be mostly used in vivo in an epigenetically controlled cell-type-dependent manner (15, 29, 30, 31, 32, 33, 34). Interestingly, a higher-density ∼0.65 NIEB/kb of NIEBs has been observed along human chromosomes, as compared to ∼0.39 NIEB/kb in S. cerevisiae, with highly compacted flanking nucleosomes not only in vitro but also in vivo (33, 34). The analysis of intra- and interspecies divergence rates confirms that these ∼1 kb chromatin motifs have been imprinted in the DNA sequence during evolution and that they have not evolved neutrally (34). The fact that these chromatin motifs are equally found in GC-rich and GC-poor isochores, in early and late replicating regions, in euchromatin and heterochromatin regions, and in intergenic and genic regions, but not at gene promoters, raises the question of which chromatin structure has been selected during evolution, and if so, to favor or facilitate which function. An attractive scenario is the possible existence in the germline of an open and accessible basal nucleosomal array that would have been selected in human to intrinsically facilitate the epigenetic regulation of nuclear functions in a cell-type-specific manner (34, 35). To our knowledge, except for the reported experimental results (36) arguing against the existence of a highly ordered secondary structure as the 30 nm chromatin fiber in pluripotent as well as in differentiated cell types in human and mouse, there is no evidence of the existence of such a noncondensed highly accessible nucleosomal array in human, mammalian, or other vertebrate genomes.
Repeated sequences constitute a ubiquitous component of eukaryotic genomes (37). The prevalence of these sequences is highly variable in terms of copy number and type of sequences. It was recently estimated that repetitive or repeat-derived sequences compose more than two-thirds of the human genome (38). Repeated sequences are mainly grouped in two classes. Tandem repeats correspond to multiple adjacent repetitions of a DNA motif; they are often found at centromeres and telomeres. Interspersed repeats correspond to the dispersion of a DNA sequence throughout the genome. Interspersed repeats mainly come from transposable elements (TEs), typically in the 100–10,000 bp size range. In vertebrates, they constitute from ∼6% of the genome in Tetraodon (39) (a pufferfish with a compact genome) to nearly half of the genome in human (40) and more than half of the genome in, for example, zebrafish and opossum (39). TEs are considered to be major drivers of gene and genome evolution. But despite their central role in biological diversity and speciation (41, 42, 43, 44, 45, 46, 47), the interactions between TEs and their genomic ecological niche and the interplay between transposition targeting and chromatin structure remain poorly understood. Although chromatin structure is accepted as playing an important role in the regulation of transposon activity, almost nothing is known at the genome-wide level, and a fortiori in a multispecies context, concerning how TEs access places to transpose into chromatin and/or how genomes target the landing of TEs to specific zones of peculiar chromatin structure to restrict their deleterious effects on genes and other important genomic structures. Indeed, when not controlled, TE insertions can lead to a number of human diseases, including cancer (48, 49, 50).
In this letter, we investigated the genome-wide distribution of TEs along human autosomes relative to the spatial positioning of NIEBs encoded in the DNA sequence. In particular, we reveal a remarkable association between NIEB borders and Alu retroelements (40, 46) that strongly suggests that NIEBs preexist Alu insertions and constitute a favorable substrate to Alu integration. We further elaborate on the perspective that NIEBs and flanking nucleosomes constitute a chromatin platform for other TEs in other primates and possibly in most vertebrates.
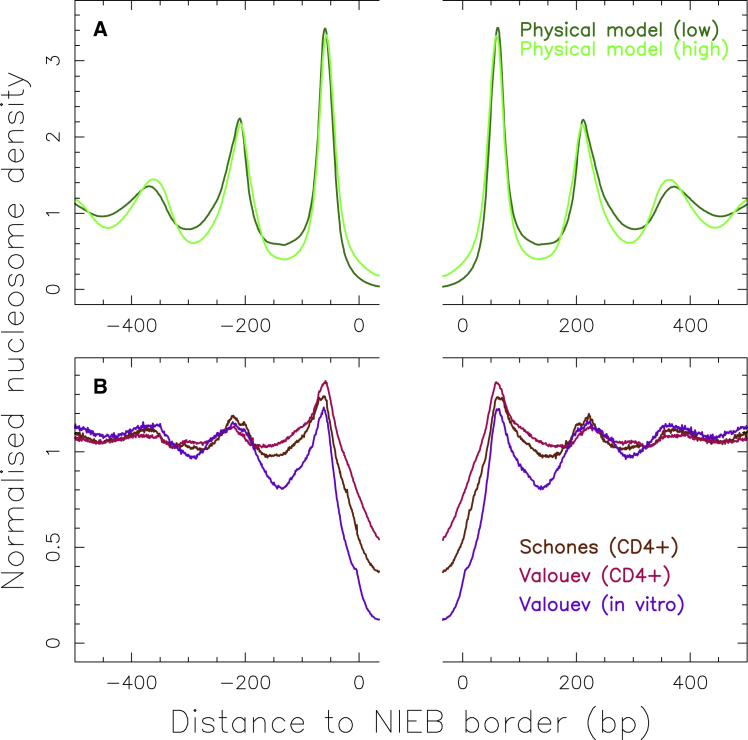
Most of the models proposed so far to mimic genome-wide nucleosome occupancy profiles were based on statistical learning (14, 25, 26, 27, 51). Recently, a simple physical model of nucleosome assembly, based on the computation of the free energy cost of bending a DNA fragment of a given sequence from its natural curvature to the final superhelical structure around the histone core, was shown to mimic in vitro nucleosome occupancy data remarkably well (3, 18, 19, 52, 53, 54). When compared to in vivo data in S. cerevisiae and Caenorhabditis elegans, this sequence-dependent thermodynamic model performs as well as models based on statistical learning, suggesting that in these organisms, the in vivo nucleosome array organization is to a large extent controlled by the underlying genomic sequence, although it is also subject to the finite-range remodeling action of external factors (3). This physical model was further used as a guide to identify NIEBs in the human genome (33, 34, 35). When combining the nucleosome occupancy probability profile obtained by fixing the chemical potential to reproduce the average nucleosome coverage observed experimentally and the original energy profile, NIEBs are defined as the genomic energy barriers that are high enough to induce an NDR in the nucleosome occupancy profile (defined by an occupancy cutoff) (3, 33). As reported in a previous work (34), this method allowed us to delineate an impressive ∼1.6 million NIEBs, demonstrating that NIEBs are an important feature of the human genome (Fig. 1). Importantly, we also observed that the model predictions around NIEBs (Fig. 1 A) at low genome coverage are in very good agreement with Valouev et al. (30) in vitro nucleosome occupancy data. Not only is a very low nucleosome occupancy observed within the NIEBs, but the compact positioning of ∼2–3 nucleosomes with a nucleosome repeat length (NRL) bp at each NIEB border predicted by the physical model (Fig. 1 A) is also observed in the experimental data (Fig. 1 B). This clearly demonstrates that this physical model also captures intrinsic sequence-dependent nucleosome positioning signals in human (33, 34), as previously reported for in vitro data for the yeast genome (3, 19, 53). But what makes a drastic difference with what has been observed for yeast is that as predicted by the physical model at high genome coverage (Fig. 1 A), this nucleosome ordering near NIEBs is also observed in the Valouev et al. (30) and Schones et al. (29) in vivo data (Fig. 1 B). The concordance between in vitro and in vivo nucleosome positioning suggests that chromatin remodeling is not necessary to establish nucleosome ordering at NIEBs borders (34), in contrast to yeast genes for which remodeler action is required to maintain nucleosome alignment with respect to transcriptional start sites (55). Note that the intrinsic nucleosome spacing predicted by the physical model and consistently observed in vitro and in vivo, namely, NRL ∼150–160 bp, corresponds to a highly compacted nucleosome arrangement as compared to the in vivo average NRL ∼203 bp, the average heterochromatin NRL bp, and also the average NRL observed in euchromatin around active promoters and enhancers in CD4+ cells (30).
Figure 1.
Normalized (with respect to genome average) mean nucleosome density on both sides of the 1,581,256 NIEBs previously predicted by the sequence-dependent physical model in humans (HG18) (33, 34). (A) This panel shows the numerical mean profiles predicted by the physical model at low (dark green) and high (light green) genomic nucleosome coverages (34). (B) This panel gives “Schones” in vivo (29) (brown), “Valouev” in vivo (30) (pink), and “Valouev” in vitro (30) (purple) data. All profiles at 1 bp resolution are from (34). To see this figure in color, go online.
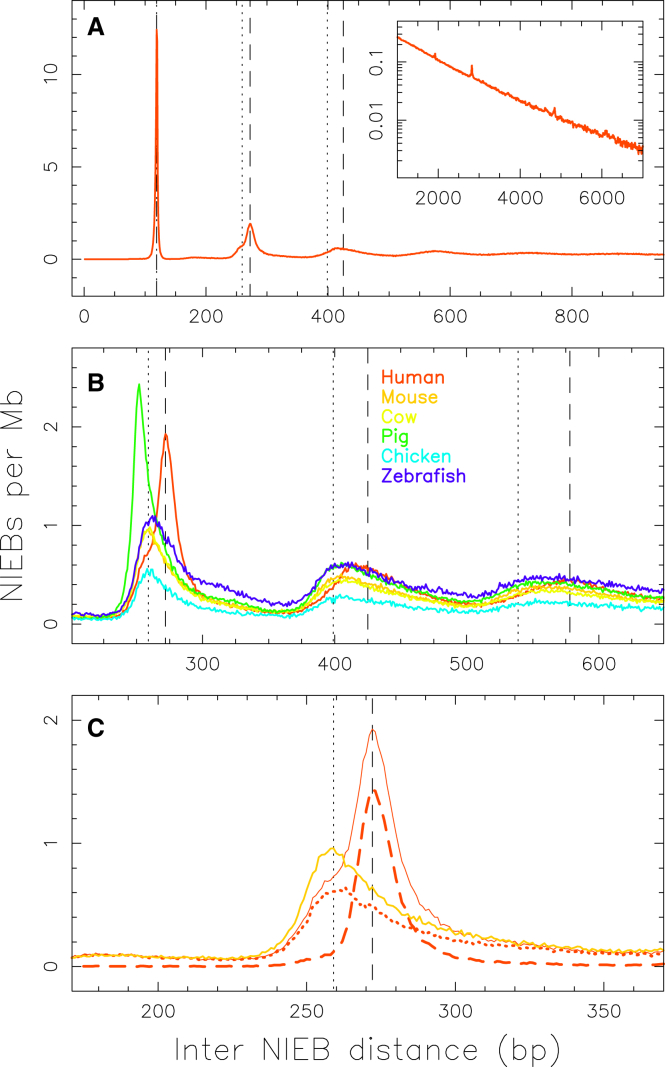
To test the relevance of NIEBs across eukaryotes, we ran the sequence-dependent physical model over various genomes, resulting in the prediction of 997,374 NIEBs in zebrafish, 426,500 NIEBs in chicken, 149,058 NIEBs in pig, 1,514,184 NIEBs in cow, 168,593 NIEBs in mouse, and 1,745,801 NIEBs in human (Table 1). The density of NIEBs (∼0.6–0.7 NIEB/kb) is thus higher in vertebrates than in the budding yeast with only 0.39 NIEB/kb (compared to a density of <0.01 NIEB/kb for a random sequence with equal proportions of A, G, C, and T). However, if a similar density of NIEBs (∼0.6–0.65 NIEB/kb) is consistently observed in mammalian genomes (primates, rodents, cow, pig), some signature of inhomogeneous evolution is obtained in other vertebrates. For fish, a higher density of NIEBs, ∼0.75 NIEB/kb, is observed in zebrafish, and on the opposite side, for birds, a lower density of NIEBs, ∼0.49 NIEB/kb, is observed in chicken. Interestingly, similar analysis of the most primitive jawed vertebrate to have its DNA analyzed—namely, the elephant shark (Callorhinchus milii), which is made of cartilage, not bone—reveals a quite low density of NIEBs, ∼0.45 NIEB/kb, only slightly larger than the one predicted in budding yeast. To characterize the spatial distribution of these NIEBs along vertebrate chromosomes, we performed a statistical analysis of the border-to-border interdistance between successive NIEBs (Fig. 2). The histogram obtained for human (Fig. 2 A) displays an exponential tail with a characteristic interdistance compatible with the mean distance kb as the signature of a Poisson-like distribution (34). Strikingly, for interdistances kb, the histogram switches to a quantized distribution with peaks equally separated by a remarkable and robust distance bp (Fig. 2, A and B), quite similar to the characteristic DNA length 147 bp involved in the nucleosome complex. Similar NIEB interdistance histograms are obtained for the other vertebrate genomes (Fig. 2 B) with again a remarkable quantification for interdistance kb, but with a significantly smaller interpeak distance of bp. As discussed in the following, this singularity of the human genome will be of first importance when investigating the correlation between NIEB borders and Alu retroelements. This robust quantization is an indication that, in vertebrates, NIEB positioning is constrained by nucleosome ordering. The somehow less-marked quantization in chicken (Fig. 2 B) is a direct consequence of the large mean NIEB interdistance kb (low NIEB density), with only a small percentage of successive NIEBs separated by interdistances kb small enough to promote statistical nucleosomal ordering.
Table 1.
Database of Six Vertebrate Genomes Analyzed for NIEB Presence
| Species | Assembly | GC Content (%) | DNA Length (Mb) | NIEB Number | Mean Distance (kb) | Mean Density (kb−1) |
|---|---|---|---|---|---|---|
| Human (Homo sapiens) | hsap_hg38 | 41.0 | 2756 | 1745,801 | 1.579 | 0.63 |
| Mouse (Mus musculus) | mmus_mm10 | 41.9 | 2396 | 1465,549 | 1.635 | 0.61 |
| Cow (Bos taurus) | bosTau8 | 41.9 | 2493 | 1514,184 | 1.647 | 0.61 |
| Pig (Sus scrofa) | susScr3 | 41.6 | 2322 | 1573,764 | 1.476 | 0.68 |
| Chicken (Gallus gallus) | galGal5 | 41.2 | 869 | 426,500 | 2.038 | 0.49 |
| Zebrafish (Danio rerio) | danRer10 | 36.6 | 1339 | 997,374 | 1.342 | 0.75 |
GC content and the total sequenced length are for the autosomes of size ≥10 Mb that were considered for the identification of NIEBs and the computation of the mean distance between successive NIEB centers and of the mean NIEB density. Note that for human, we used the latest assembly (HG38) of the human genome instead of the HG18 assembly as in previous work (34). All NIEB coordinates described in Table 1 can be downloaded from http://perso.ens-lyon.fr/benjamin.audit/Vertebrate_NIEBs.
Figure 2.
Distribution of border-to-border interdistances between successive NIEBs in the autosomes of size >10 Mb of human (red), mouse (orange), cow (yellow), pig (green), chicken (blue), and zebrafish (purple) (Table 1). To remove dependency on genome size, histograms were normalized by the total length of sequenced DNA. (A) This panel gives human data: the inset corresponds to a log-linear representation of the tail of this histogram, putting into light a Poisson-like exponential decay with a mean interdistance of kb. (B) This panel shows the comparative analysis in vertebrate genomes. (C) This panel zooms in on the second peak of the data for human (red solid line) and mouse (orange solid line); also shown for comparison are the normalized histograms in human for inter-NIEBs containing at least one Alu retroelement (red dashed line, 728,678) or no Alu retroelement (red dotted line, 1,016,864). The vertical dashed lines mark interdistances (bp) and the vertical dotted lines mark interdistances (bp), for k = 0, 1, 2, and 3. To see this figure in color, go online.
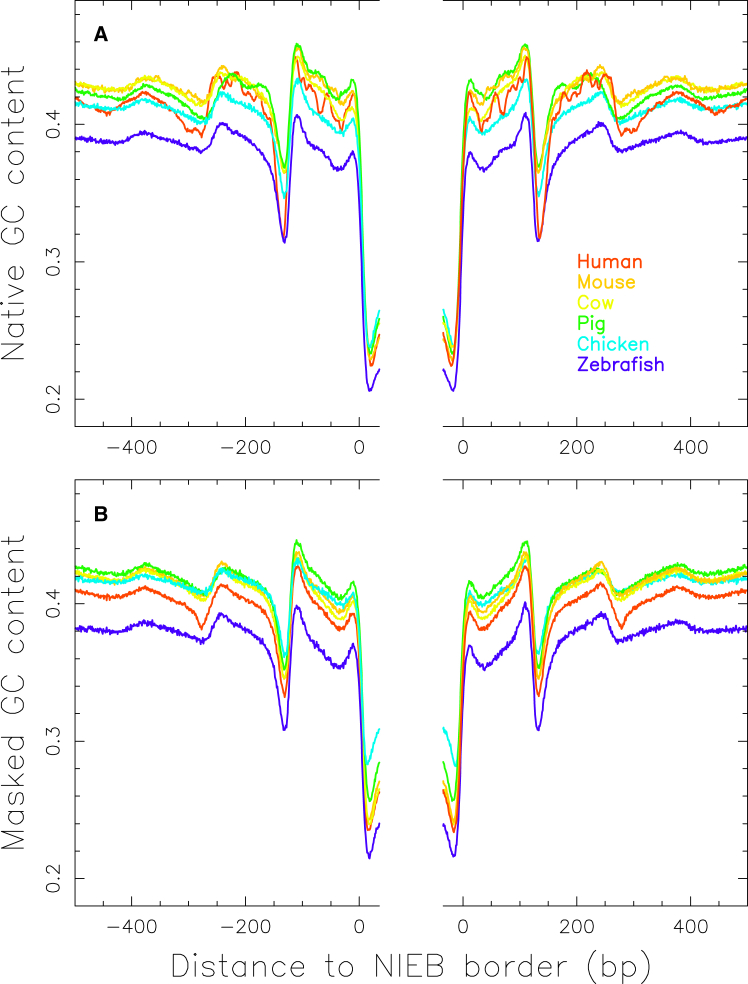
The fact that the NIEBs are ubiquitous to all vertebrates raises the issue of how robust and universal the neighboring compacted nucleosome arrangement is as predicted by our sequence-dependent physical model. As previously pointed out in various organisms including S. cerevisiae (3, 26, 27, 56), C. elegans (3, 26, 56, 57), and human (15, 30), the local GC content provides a good prediction of the mean nucleosome occupancy profiles observed in vitro. Importantly, consistent with the predictions of our physical model (Fig. 1 A), the mean GC content (Fig. 3 A) and repeat-masked GC content (Fig. 3 B) reproduce quite well the mean nucleosome occupancy profiles observed in vivo in human (33, 34) (Fig. 1 B), confirming that not only the NIEBs but also the flanking nucleosome positions are programmed in the DNA sequence. This study reveals that similar mean GC profiles are obtained in primates and more generally in all vertebrates with an NRL of ∼150 bp (distance between two successive GC minima) (Fig. 3). According to geometrical modeling of the constitutive 30 nm chromatin fiber (58, 59, 60, 61, 62, 63, 64, 65, 66), such a small nucleosome spacing with a rather short DNA linker size of ∼10–20 bp is likely to impair the condensation of the nucleosomal array into the chromatin fiber, leaving a well-organized accessible nucleosomal array. One possible scenario is that the nucleosomal array adopts a zig-zag configuration, in which interaction between nucleosomes at a distance of two nucleosomes is larger than between neighboring nucleosomes or between nucleosomes further apart, which has been recently shown to be a persistent pattern in interphase and metaphase nuclei of living cells (67). The fact that in human complex selection patterns involving positive and purifying selections were shown to maintain a high difference in GC composition between the lowest GC composition in the NIEBs and the highest composition in the closest flanking nucleosomes (34) (Figs. 1 and 3) suggests that an open and accessible basal nucleosomal array has been selected to intrinsically facilitate the epigenetic regulation of nuclear functions in a cell-type-specific manner (68, 69, 70). The remarkable stability of the GC profile near NIEBs across vertebrates (Fig. 3) is a strong indication that this accessible nucleosomal array likely has been encoded during evolution in all vertebrate genomes.
Figure 3.
Mean GC content at the borders between NIEBs of size >70 bp and inter-NIEBs of length >1000 bp in the autosomes of size >10 Mb of human (red), mouse (orange), cow (yellow), pig (green), chicken (blue), and zebrafish (purple) (Table 1). (A) This panel shows native GC content. (B) This panel shows repeat-masked GC content. To see this figure in color, go online.
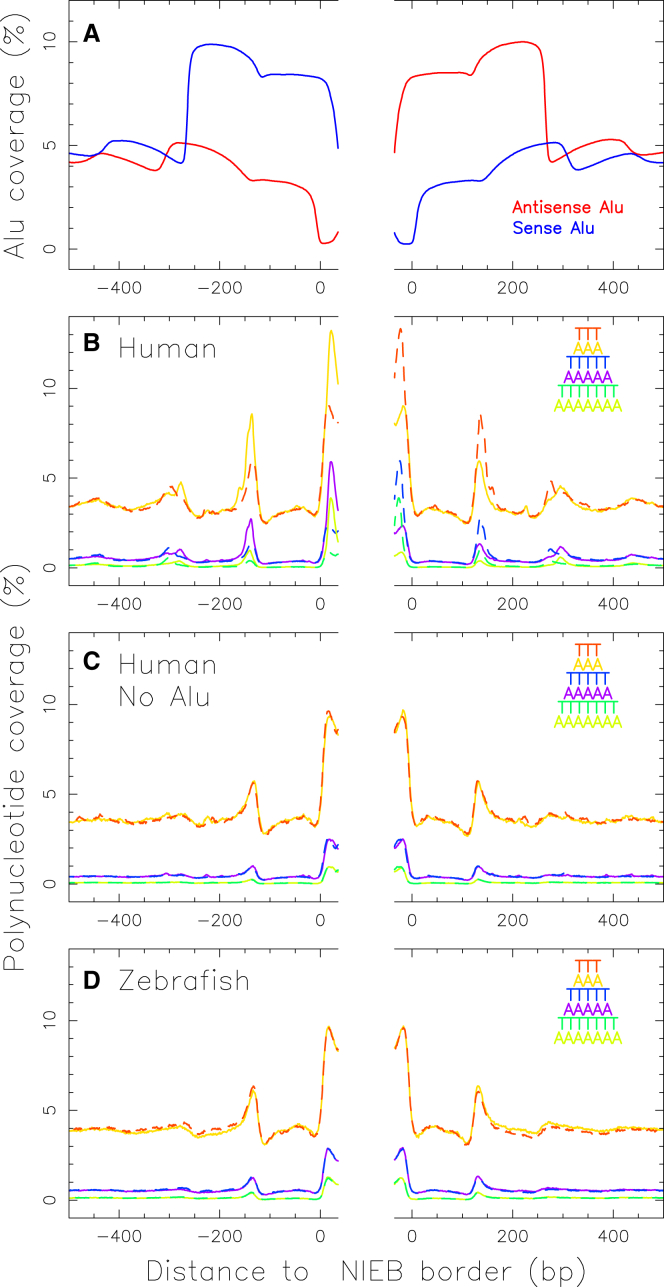
When looking carefully at the native GC content profiles around the NIEBs (Fig. 3 A), we can see slight differences from the ones obtained with the repeat masked sequences (Fig. 3 B). This is particularly true in human (34) in whom, as compared to the rather smooth two-bump-masked GC profile that remarkably matches in vitro and in vivo nucleosome occupancy data, the native GC profile displays some striking oscillatory internal patterns, suggesting the presence of some repeat sequences near a non-negligible subset of the predicted NIEBs. When systematically investigating the principal families of interspersed repeats, short interspersed nuclear elements (Alu, MIR), and long interspersed nuclear elements (LINE1, LINE2), we found that a lot of (∼52%) Alu retroelements were inserted flanking a NIEB (34). Alu retrotransposons constitute one of the best examples of the successful emergence of a lineage-specific TE family. Alu sequences are 7SL RNA-derived short interspersed nuclear elements specific to primates (71). They are nonautonomous and require the transposition machinery of LINE1 elements (72). The long interspersed nuclear elements have been dated back to the beginning of the eukaryotes (73). In terms of copy number, Alu is the most prolific family of elements in primates: these sequences have propagated extensively to reach more than one million copies over the past 65 million years (40, 74), accounting for more than 10% of the human genome (38, 75). A typical Alu retroelement is a dimer ∼300 bp long, composed of two distinct GC- and CpG-rich monomers separated by a short AT-rich region. Importantly, the 3′ end of an Alu retroelement has a longer poly(A) track that plays a critical role in its amplification mechanism (76, 77). Interestingly, we found that the orientation of the Alu retroelements is strongly dependent on which NIEB side they were inserted in (34). They are mainly sense at the NIEB 5′ end and antisense at the NIEB 3′ end (Fig. 4 A), so that the body of the Alu retroelement is external to the NIEB. The remarkable phasing of the Alu at the NIEB 5′ end (respectively NIEB 3′ end) results from the matching of the poly(A) (respectively poly(T)) tracks that were shown to define the edges of some of the predicted NIEBs (Fig. 4 B). This suggests that the Alu RNA brings the ORF2 protein to the region of the genome where there is a NIEB and where its endonuclease activity is going to cleave the poly(A) or poly(T) bordering sequence. Moreover, the external orientation of the Alu sequence with two associated, well-positioned nucleosomes will keep maintaining the NIEB without disturbing the flanking nucleosome ordering too much. Actually, these Alu retroelements were shown to have some affinity to core histones and to possess nucleosome-positioning signals (78). In vivo studies confirmed that two rotationally positioned nucleosomes are indeed formed on both sides of the central A-rich region with a rather small NRL (∼167 bp) (79, 80). This could explain the increase in the mean inter-NIEB distance observed for the subset of NIEBs with a flanking Alu retroelement (Fig. 2 C). However, a majority (∼61%) of NIEBs are free of Alu on either side, an indication that NIEBs do not result from the mechanisms underlying Alu insertions (34). Indeed, for these Alu-free NIEBs, we observed a symmetric enrichment of poly(A) and poly(T) tracks at the NIEB border (Fig. 4 C), including the ones that have a poly(A) and poly(T) track unfavorable to Alu insertion. Note that, as a control, a similar symmetric enrichment is observed for zebrafish (Fig. 4 D). A systematic analysis of the spatial distribution of the three major subfamilies of Alu that were active at different times during primate evolution (81, 82)—namely, AluJ (64–40 million years), AluS (45–25 million years), and AluY (30 million years–present)—in relation to NIEBs and related chromatin motifs should shed a new light on Alu integration and evolution as well as on their role in the chromatin-mediated regulation of gene expression and of the replication spatiotemporal program.
Figure 4.
(A) Mean coverage by sense (blue) and antisense (red) Alu retroelements at the borders between NIEBs of size >70 bp and inter-NIEBs of length >1000 bp in human (Table 1; 709,948 and 709,772 borders with the NIEBs on the 3′ and 5′ border sides, respectively). (B) This panel shows the mean polynucleotide coverage at the same NIEB borders as in (A): AAA (solid orange), TTT (dashed red), AAAAA (solid purple), TTTTT (dashed blue), AAAAAAA (solid lime green), TTTTTTT (dashed dark green). (C) This panel shows the same as in (B) but restricted to borders of inter-NIEBs not containing any Alu retroelements (as in Fig. 2C; 389,218 and 388,650 borders with the NIEBs on the 3′ and 5′ border sides, respectively). (D) This panel shows the same as in (B) for the zebrafish (Table 1; 385,640 and 385,554 borders with the NIEBs on the 3′ and 5′ border sides, respectively). To see this figure in color, go online.
To summarize, we have reported in this letter very promising results on the universal sequence encoding of an accessible nucleosomal array in human, primate, nonprimate, mammalian, and non-mammalian vertebrate genomes. As a very plausible interpretation, this open and accessible chromatin structure would have been selected during evolution to intrinsically facilitate the epigenetic regulation of nuclear functions in a cell-type-specific manner. We have further shown that this accessible nucleosomal array with intrinsic NDRs constitutes an evolutionary stable substrate for Alu insertion in human. By systematically exploring the localization of mobile elements at NIEB borders across the vertebrate tree in species that do not possess Alu retroelements, we expect to confirm the fundamental role of this intrinsically open and accessible nucleosomal array on transposable element integration. This will likely provide important clues to our understanding of genome evolution and epigenetic regulation in both health and disease.
Author Contributions
All authors designed the study. F.G.B., B.A., and G.D. performed the analysis. B.A. and F.A. contributed to elaborate analysis tools. B.A., J.-N.V., and A.A. supervised the study. A.A. wrote the manuscript with contributions from all coauthors. All authors read and approved the final manuscript.
Acknowledgments
We are very grateful to C. Vaillant for helpful discussions and to BioSyl Federation and Ecofect LabEx (ANR-11-LABX-0048) for inspiring scientific events.
This work was supported by the Institut National du Cancer (PLBIO16-302), the Fondation pour la Recherche Médicale (DEI20151234404), the Agence Nationale de la Recherche (ANR-15-CE12-0011-01), and the ENS de Lyon “Projets Emergents” Program. B.A. acknowledges support from the Science and Technology Commission of Shanghai Municipality (15520711500) and the Joint Research Institute for Science and Society. The funding body had no role in the design of the study and collection, analysis, and interpretation of data, or in writing the manuscript.
Editor: Tamar Schlick.
References
- 1.Segal E., Widom J. What controls nucleosome positions? Trends Genet. 2009;25:335–343. doi: 10.1016/j.tig.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arneodo A., Vaillant C., Thermes C. Multi-scale coding of genomic information: from DNA sequence to genome structure and function. Phys. Rep. 2011;498:45–188. [Google Scholar]
- 3.Chevereau G., Arneodo A., Vaillant C. Influence of the genomic sequence on the primary structure of chromatin. Front. Life Sci. 2011;5:28–68. [Google Scholar]
- 4.Iyer V.R. Nucleosome positioning: bringing order to the eukaryotic genome. Trends Cell Biol. 2012;22:250–256. doi: 10.1016/j.tcb.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Travers A.A., Vaillant C., Muskhelishvili G. DNA structure, nucleosome placement and chromatin remodelling: a perspective. Biochem. Soc. Trans. 2012;40:335–340. doi: 10.1042/BST20110757. [DOI] [PubMed] [Google Scholar]
- 6.Struhl K., Segal E. Determinants of nucleosome positioning. Nat. Struct. Mol. Biol. 2013;20:267–273. doi: 10.1038/nsmb.2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hughes A.L., Rando O.J. Mechanisms underlying nucleosome positioning in vivo. Annu. Rev. Biophys. 2014;43:41–63. doi: 10.1146/annurev-biophys-051013-023114. [DOI] [PubMed] [Google Scholar]
- 8.Tompitak M., Vaillant C., Schiessel H. Genomes of multicellular organisms have evolved to attract nucleosomes to promoter regions. Biophys. J. 2017;112:505–511. doi: 10.1016/j.bpj.2016.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kornberg R.D., Stryer L. Statistical distributions of nucleosomes: nonrandom locations by a stochastic mechanism. Nucleic Acids Res. 1988;16:6677–6690. doi: 10.1093/nar/16.14.6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mavrich T.N., Ioshikhes I.P., Pugh B.F. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res. 2008;18:1073–1083. doi: 10.1101/gr.078261.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Möbius W., Gerland U. Quantitative test of the barrier nucleosome model for statistical positioning of nucleosomes up- and downstream of transcription start sites. PLoS Comput. Biol. 2010;6:e1000891. doi: 10.1371/journal.pcbi.1000891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuan G.C., Liu Y.J., Rando O.J. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- 13.Lee W., Tillo D., Nislow C. A high-resolution atlas of nucleosome occupancy in yeast. Nat. Genet. 2007;39:1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- 14.Field Y., Kaplan N., Segal E. Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLoS Comput. Biol. 2008;4:e1000216. doi: 10.1371/journal.pcbi.1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tillo D., Kaplan N., Hughes T.R. High nucleosome occupancy is encoded at human regulatory sequences. PLoS One. 2010;5:e9129. doi: 10.1371/journal.pone.0009129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berbenetz N.M., Nislow C., Brown G.W. Diversity of eukaryotic DNA replication origins revealed by genome-wide analysis of chromatin structure. PLoS Genet. 2010;6:e1001092. doi: 10.1371/journal.pgen.1001092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eaton M.L., Galani K., MacAlpine D.M. Conserved nucleosome positioning defines replication origins. Genes Dev. 2010;24:748–753. doi: 10.1101/gad.1913210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chevereau G., Palmeira L., Vaillant C. Thermodynamics of intragenic nucleosome ordering. Phys. Rev. Lett. 2009;103:188103. doi: 10.1103/PhysRevLett.103.188103. [DOI] [PubMed] [Google Scholar]
- 19.Vaillant C., Palmeira L., Arneodo A. A novel strategy of transcription regulation by intragenic nucleosome ordering. Genome Res. 2010;20:59–67. doi: 10.1101/gr.096644.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Warnecke T., Batada N.N., Hurst L.D. The impact of the nucleosome code on protein-coding sequence evolution in yeast. PLoS Genet. 2008;4:e1000250. doi: 10.1371/journal.pgen.1000250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Washietl S., Machné R., Goldman N. Evolutionary footprints of nucleosome positions in yeast. Trends Genet. 2008;24:583–587. doi: 10.1016/j.tig.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 22.Babbitt G.A., Kim Y. Inferring natural selection on fine-scale chromatin organization in yeast. Mol. Biol. Evol. 2008;25:1714–1727. doi: 10.1093/molbev/msn127. [DOI] [PubMed] [Google Scholar]
- 23.Babbitt G.A., Tolstorukov M.Y., Kim Y. The molecular evolution of nucleosome positioning through sequence-dependent deformation of the DNA polymer. J. Biomol. Struct. Dyn. 2010;27:765–780. doi: 10.1080/07391102.2010.10508584. [DOI] [PubMed] [Google Scholar]
- 24.Babbitt G.A., Cotter C.R. Functional conservation of nucleosome formation selectively biases presumably neutral molecular variation in yeast genomes. Genome Biol. Evol. 2011;3:15–22. doi: 10.1093/gbe/evq081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peckham H.E., Thurman R.E., Weng Z. Nucleosome positioning signals in genomic DNA. Genome Res. 2007;17:1170–1177. doi: 10.1101/gr.6101007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tillo D., Hughes T.R. G+C content dominates intrinsic nucleosome occupancy. BMC Bioinformatics. 2009;10:442. doi: 10.1186/1471-2105-10-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaplan N., Moore I.K., Segal E. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prendergast J.G., Semple C.A. Widespread signatures of recent selection linked to nucleosome positioning in the human lineage. Genome Res. 2011;21:1777–1787. doi: 10.1101/gr.122275.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schones D.E., Cui K., Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valouev A., Johnson S.M., Sidow A. Determinants of nucleosome organization in primary human cells. Nature. 2011;474:516–520. doi: 10.1038/nature10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cayrou C., Coulombe P., Méchali M. New insights into replication origin characteristics in metazoans. Cell Cycle. 2012;11:658–667. doi: 10.4161/cc.11.4.19097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lombraña R., Almeida R., Gómez M. High-resolution analysis of DNA synthesis start sites and nucleosome architecture at efficient mammalian replication origins. EMBO J. 2013;32:2631–2644. doi: 10.1038/emboj.2013.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Drillon G., Audit B., Arneodo A. Ubiquitous human ‘master’ origins of replication are encoded in the DNA sequence via a local enrichment in nucleosome excluding energy barriers. J. Phys. Condens. Matter. 2015;27:064102. doi: 10.1088/0953-8984/27/6/064102. [DOI] [PubMed] [Google Scholar]
- 34.Drillon G., Audit B., Arneodo A. Evidence of selection for an accessible nucleosomal array in human. BMC Genomics. 2016;17:526. doi: 10.1186/s12864-016-2880-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Audit B., Zaghloul L., Arneodo A. Open chromatin encoded in DNA sequence is the signature of ‘master’ replication origins in human cells. Nucleic Acids Res. 2009;37:6064–6075. doi: 10.1093/nar/gkp631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ricci M.A., Manzo C., Cosma M.P. Chromatin fibers are formed by heterogeneous groups of nucleosomes in vivo. Cell. 2015;160:1145–1158. doi: 10.1016/j.cell.2015.01.054. [DOI] [PubMed] [Google Scholar]
- 37.Richard G.F., Kerrest A., Dujon B. Comparative genomics and molecular dynamics of DNA repeats in eukaryotes. Microbiol. Mol. Biol. Rev. 2008;72:686–727. doi: 10.1128/MMBR.00011-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Koning A.P., Gu W., Pollock D.D. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 2011;7:e1002384. doi: 10.1371/journal.pgen.1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chalopin D., Naville M., Volff J.N. Comparative analysis of transposable elements highlights mobilome diversity and evolution in vertebrates. Genome Biol. Evol. 2015;7:567–580. doi: 10.1093/gbe/evv005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lander E.S., Linton L.M., Szustakowki J., International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 41.McClintock B. Controlling elements and the gene. Cold Spring Harb. Symp. Quant. Biol. 1956;21:197–216. doi: 10.1101/sqb.1956.021.01.017. [DOI] [PubMed] [Google Scholar]
- 42.Coyne J.A., Orr H.A. The evolutionary genetics of speciation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1998;353:287–305. doi: 10.1098/rstb.1998.0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xing J., Zhang Y., Jorde L.B. Mobile elements create structural variation: analysis of a complete human genome. Genome Res. 2009;19:1516–1526. doi: 10.1101/gr.091827.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kraaijeveld K. Genome size and species diversification. Evol. Biol. 2010;37:227–233. doi: 10.1007/s11692-010-9093-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beck C.R., Collier P., Moran J.V. LINE-1 retrotransposition activity in human genomes. Cell. 2010;141:1159–1170. doi: 10.1016/j.cell.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hormozdiari F., Alkan C., Eichler E.E. Alu repeat discovery and characterization within human genomes. Genome Res. 2011;21:840–849. doi: 10.1101/gr.115956.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stewart C., Kural D., Marth G.T., 1000 Genomes Project A comprehensive map of mobile element insertion polymorphisms in humans. PLoS Genet. 2011;7:e1002236. doi: 10.1371/journal.pgen.1002236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deininger P.L., Batzer M.A. Alu repeats and human disease. Mol. Genet. Metab. 1999;67:183–193. doi: 10.1006/mgme.1999.2864. [DOI] [PubMed] [Google Scholar]
- 49.Callinan P.A., Batzer M.A. Retrotransposable elements and human disease. Genome Dyn. 2006;1:104–115. doi: 10.1159/000092503. [DOI] [PubMed] [Google Scholar]
- 50.Belancio V.P., Roy-Engel A.M., Deininger P. The impact of multiple splice sites in human L1 elements. Gene. 2008;411:38–45. doi: 10.1016/j.gene.2007.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yuan G.C., Liu J.S. Genomic sequence is highly predictive of local nucleosome depletion. PLoS Comput. Biol. 2008;4:e13. doi: 10.1371/journal.pcbi.0040013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vaillant C., Audit B., Arneodo A. Experiments confirm the influence of genome long-range correlations on nucleosome positioning. Phys. Rev. Lett. 2007;99:218103. doi: 10.1103/PhysRevLett.99.218103. [DOI] [PubMed] [Google Scholar]
- 53.Milani P., Chevereau G., Arneodo A. Nucleosome positioning by genomic excluding-energy barriers. Proc. Natl. Acad. Sci. USA. 2009;106:22257–22262. doi: 10.1073/pnas.0909511106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Milani P., Marilley M., Arneodo A. Mechanics of the IL2RA gene activation revealed by modeling and atomic force microscopy. PLoS One. 2011;6:e18811. doi: 10.1371/journal.pone.0018811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gkikopoulos T., Schofield P., Owen-Hughes T. A role for Snf2-related nucleosome-spacing enzymes in genome-wide nucleosome organization. Science. 2011;333:1758–1760. doi: 10.1126/science.1206097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miele V., Vaillant C., Grange T. DNA physical properties determine nucleosome occupancy from yeast to fly. Nucleic Acids Res. 2008;36:3746–3756. doi: 10.1093/nar/gkn262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Valouev A., Ichikawa J., Johnson S.M. A high-resolution, nucleosome position map of C. elegans reveals a lack of universal sequence-dictated positioning. Genome Res. 2008;18:1051–1063. doi: 10.1101/gr.076463.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schiessel H. Topical review: the physics of chromatin. J. Phys. Condens. Matter. 2003;15:R699–R774. doi: 10.1088/0953-8984/27/6/060301. [DOI] [PubMed] [Google Scholar]
- 59.Lesne A., Victor J.M. Chromatin fiber functional organization: some plausible models. Eur Phys J E Soft Matter. 2006;19:279–290. doi: 10.1140/epje/i2005-10050-6. [DOI] [PubMed] [Google Scholar]
- 60.Langowski J. Polymer chain models of DNA and chromatin. Eur Phys J E Soft Matter. 2006;19:241–249. doi: 10.1140/epje/i2005-10067-9. [DOI] [PubMed] [Google Scholar]
- 61.Langowski J., Heermann D.W. Computational modeling of the chromatin fiber. Semin. Cell Dev. Biol. 2007;18:659–667. doi: 10.1016/j.semcdb.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 62.Kepper N., Foethke D., Rippe K. Nucleosome geometry and internucleosomal interactions control the chromatin fiber conformation. Biophys. J. 2008;95:3692–3705. doi: 10.1529/biophysj.107.121079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Diesinger P.M., Heermann D.W. Depletion effects massively change chromatin properties and influence genome folding. Biophys. J. 2009;97:2146–2153. doi: 10.1016/j.bpj.2009.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Beshnova D.A., Cherstvy A.G., Teif V.B. Regulation of the nucleosome repeat length in vivo by the DNA sequence, protein concentrations and long-range interactions. PLoS Comput. Biol. 2014;10:e1003698. doi: 10.1371/journal.pcbi.1003698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Todolli S., Perez P.J., Olson W.K. Contributions of sequence to the higher-order structures of DNA. Biophys. J. 2017;112:416–426. doi: 10.1016/j.bpj.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bascom G.D., Kim T., Schlick T. Kilobase pair chromatin fiber contacts promoted by living-system-like DNA linker length distributions and nucleosome depletion. J. Phys. Chem. B. 2017;121:3882–3894. doi: 10.1021/acs.jpcb.7b00998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grigoryev S.A., Bascom G., Schlick T. Hierarchical looping of zigzag nucleosome chains in metaphase chromosomes. Proc. Natl. Acad. Sci. USA. 2016;113:1238–1243. doi: 10.1073/pnas.1518280113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Julienne H., Zoufir A., Arneodo A. Human genome replication proceeds through four chromatin states. PLoS Comput. Biol. 2013;9:e1003233. doi: 10.1371/journal.pcbi.1003233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Picard F., Cadoret J.C., Prioleau M.N. The spatiotemporal program of DNA replication is associated with specific combinations of chromatin marks in human cells. PLoS Genet. 2014;10:e1004282. doi: 10.1371/journal.pgen.1004282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Julienne H., Audit B., Arneodo A. Embryonic stem cell specific “master” replication origins at the heart of the loss of pluripotency. PLoS Comput. Biol. 2015;11:e1003969. doi: 10.1371/journal.pcbi.1003969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ullu E., Tschudi C. Alu sequences are processed 7SL RNA genes. Nature. 1984;312:171–172. doi: 10.1038/312171a0. [DOI] [PubMed] [Google Scholar]
- 72.Dewannieux M., Esnault C., Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat. Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- 73.Ostertag E.M., Kazazian H.H., Jr. Biology of mammalian L1 retrotransposons. Annu. Rev. Genet. 2001;35:501–538. doi: 10.1146/annurev.genet.35.102401.091032. [DOI] [PubMed] [Google Scholar]
- 74.Batzer M.A., Deininger P.L. Alu repeats and human genomic diversity. Nat. Rev. Genet. 2002;3:370–379. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- 75.Cordaux R., Batzer M.A. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mighell A.J., Markham A.F., Robinson P.A. Alu sequences. FEBS Lett. 1997;417:1–5. doi: 10.1016/s0014-5793(97)01259-3. [DOI] [PubMed] [Google Scholar]
- 77.Deininger P. Alu elements: know the SINEs. Genome Biol. 2011;12:236. doi: 10.1186/gb-2011-12-12-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Englander E.W., Howard B.H. Nucleosome positioning by human Alu elements in chromatin. J. Biol. Chem. 1995;270:10091–10096. doi: 10.1074/jbc.270.17.10091. [DOI] [PubMed] [Google Scholar]
- 79.Englander E.W., Wolffe A.P., Howard B.H. Nucleosome interactions with a human Alu element. Transcriptional repression and effects of template methylation. J. Biol. Chem. 1993;268:19565–19573. [PubMed] [Google Scholar]
- 80.Tanaka Y., Yamashita R., Nakai K. Effects of Alu elements on global nucleosome positioning in the human genome. BMC Genomics. 2010;11:309. doi: 10.1186/1471-2164-11-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kapitonov V., Jurka J. The age of Alu subfamilies. J. Mol. Evol. 1996;42:59–65. doi: 10.1007/BF00163212. [DOI] [PubMed] [Google Scholar]
- 82.Batzer M.A., Deininger P.L., Zuckerkandl E. Standardized nomenclature for Alu repeats. J. Mol. Evol. 1996;42:3–6. doi: 10.1007/BF00163204. [DOI] [PubMed] [Google Scholar]