Abstract
Human mitochondrial transcription factor A (TFAM) distorts DNA into a U-turn, as shown by crystallographic studies. The relevance of this U-turn is associated with transcription initiation at the mitochondrial light strand promoter (LSP). However, it has not been yet discerned whether a tight U-turn or an alternative conformation, such as a V-shape, is formed in solution. Here, single-molecule FRET experiments on freely diffusing TFAM/LSP complexes containing different DNA lengths show that a DNA U-turn is induced by progressive and cooperative binding of the two TFAM HMG-box domains and the linker between them. SAXS studies further show compaction of the protein upon complex formation. Finally, molecular dynamics simulations reveal that TFAM/LSP complexes are dynamic entities, and the HMG boxes induce the U-turn against the tendency of the DNA to adopt a straighter conformation. This tension is resolved by reversible unfolding of the linker, which is a singular mechanism that allows a flexible protein to stabilize a tight bending of DNA.
Introduction
Mitochondrial transcription factor A (TFAM) regulates the transcription and transcription-dependent replication of human mitochondrial DNA (mtDNA) (1). In this genome, both the light and the heavy DNA strands code for proteins. In addition, there is a noncoding region where most of the cis-regulatory elements of mtDNA are found, including the heavy and light strand promoters (HSP1 and light strand promoter (LSP), respectively). By binding to specific sequences at LSP and HSP1, TFAM recruits the RNA polymerase (2). Another equally important function of this abundant protein is the compaction of mtDNA into nucleoprotein structures named nucleoids (1). TFAM activities are essential for mtDNA maintenance, mitochondrial biogenesis, and organism viability (1). TFAM belongs to the High Mobility Group B protein family. It contains two HMG box domains (HMG1 and HMG2) separated by a linker and followed by a C-terminal tail. In general terms, the HMG box domains consist of three α-helices arranged in an L-shape. The concave surface of this L-shape binds to the DNA minor groove, inducing pronounced bending of the DNA double helix (3).
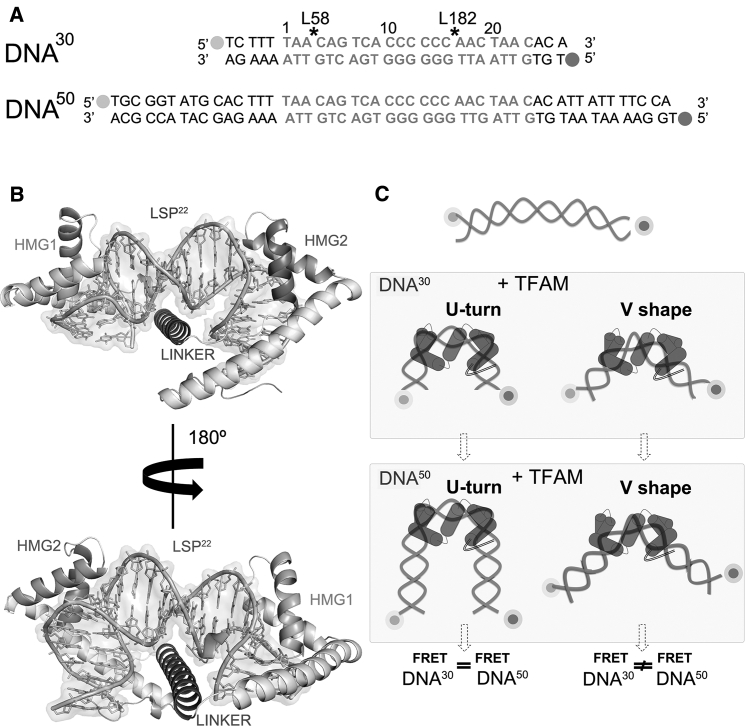
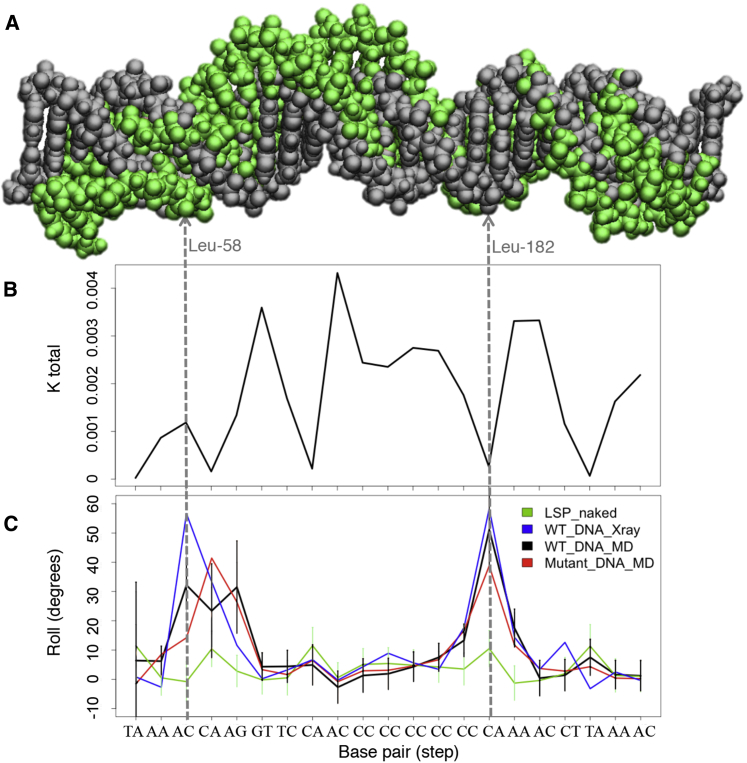
The structure of TFAM in complex with its cognate binding site at LSP was determined by crystallographic studies, using either 22-bp (4) or 28-bp (5) DNA fragments (PDB: 3TQ6 and 3TMM, respectively). Additional structures include TFAM in complex with 22-bp fragments that contain either mitochondrial heavy strand promoter HSP1 (PDB: 4NNU) or nonspecific DNA (nsDNA; PDB: 4NOD) (6). In all these structures, each HMG-box domain of TFAM induces 90° bending to the DNA minor groove and the insertion of Leu58 (from HMG1) and Leu182 (from HMG2) into two 10 bp-separated sites, thus inducing an overall U-turn to the double helix (Fig. 1, A and B; Fig. S1 A). Additional residues from each HMG box domain contact the DNA and further stabilize the interaction (Fig. S1 B). Both HMG domains are joined by a positively charged 30-residue linker. In the complex, the protein and the DNA are intertwined. The HMG-boxes contact the minor groove on the same face of the DNA, whereas the linker, in an α-helix conformation, nonspecifically interacts with the minor groove at the opposite side of the double helix. The C-terminal tail of TFAM contacts the mitochondrial RNA polymerase at the promoters (2). Both linker and C-terminal tail are unstructured in unbound TFAM, whereas, in a complex with DNA, the linker folds into the α-helix structure (4). These crystallographic studies have provided enormous insight into the atomic details of complex formation, but they present a static structure with potential lattice artifacts. This raises two questions: whether the x-ray structure represents the conformational ensemble of the TFAM/LSP complex in solution, and what underlying mechanism is used by the protein to bend the DNA.
Figure 1.
Experimental design for TFAM-LSP FRET experiments. (A) Sequences of DNA30 and DNA50 used for FRET assays were designed based on the TFAM/LSP22 crystal structure (4). DNA bases in light gray correspond to the crystallized fragment LSP22 (bent by 180°; see B), the additional bases in black correspond to the mtDNA sequence. Alexa 488 and Alexa 594 fluorophores are shown as light and dark gray dots, respectively. Black asterisks indicate the insertion sites of Leu58 and Leu182. (B) 180°-rotated views are given of the TFAM/LSP22 crystal structure (4). The DNA (LSP22, 22 bp) is bent in a U-turn by the protein domains HMG1 and HMG2 (labeled in light and dark gray, respectively), stabilized by the linker domain. (C) Shown here is a scheme of the FRET strategy based on DNA30 and DNA50. A U-turn (a V-shape) results in similar (different) FRET efficiencies for DNA30 and DNA50. In a V-shape, the distance between dyes increases dramatically with DNA length (thus energy transfer decreases), whereas in the U-turn the distance is similar (and so is the FRET efficiency).
Previous FRET studies on TFAM/LSP complexes showed consistency between the distance of the DNA ends in solution (∼60 Å) (5, 6, 7) and the distance found in the 28-bp DNA crystal structure (∼55 Å) (5). However, these and other studies (8, 9) showed weak or no binding of isolated HMG2 constructs to the DNA, and suggested that HMG1 and the linker direct DNA binding and bending. Furthermore, these FRET studies failed to confirm the double kink shown by the crystal structure, and did not provide any information on the mechanism of TFAM-induced bending. Single-molecule FRET (smFRET) reports on the mutual distance between dyes and can resolve subpopulations within a heterogeneous sample to distinguish molecules in different dynamic states. Pioneered in the late 1990s on immobilized DNA (10) and freely diffusing molecules in solution (11), the power of this technique has been shown by many subsequent studies. smFRET has proven particularly suitable for the analysis of DNA distortion on protein binding in, for example, the yeast Nhp6 protein (12) or the eukaryotic transcription factor TATA-box binding protein (13).
In this study, we combined data from smFRET on freely diffusing molecules with results from SAXS and atomistic MD simulations to obtain structural information on the TFAM/LSP complex in solution, and to understand the mechanism of DNA binding and bending by TFAM. Our first aim was to discern whether TFAM bends DNA into a U-shape, which would have two kinks, or in a V-form with a single kink. We then studied the role of each protein domain in DNA binding and bending and derived a stepwise model for the TFAM/LSP interaction. SAXS studies provided a further biophysical characterization of the complex, revealing intrinsic structural dynamics of the complex. Finally, the structural properties underlying these dynamics were investigated by MD simulations. These properties were compared for a wild-type TFAM/LSP complex and an in silico nine-residue mutant, to analyze the contribution of protein and DNA components to complex dynamics and flexibility. Our study reveals the molecular mechanism underlying LSP bending in which TFAM bends the DNA into a U-turn by synergic cooperation between its HMG box domains that kink the DNA, and the flexible linker that maintains the U-turn in a dynamic complex.
Materials and Methods
The Supporting Material provides further details, adding to the succinct methods below.
Design of DNA constructs for smFRET experiments
Two DNA constructs of 30-bp (DNA30) and 50-bp (DNA50) length containing the cognate binding sequence at LSP were labeled with donor (Alexa 488) and acceptor (Alexa 594) dyes attached to the 5′ ends of complementary strands. After purification, single-stranded DNAs were annealed and the duplex was separated from nonannealed DNA by HPLC. Control samples were prepared with the same DNA sequences carrying only the donor (donor-only) or acceptor (acceptor-only) fluorophore.
Protein preparation
TFAM (residues 43–246; UniProt Q00059) and TFAM domains HMG1 (aa 43–125) and HMG2-Cter (aa 149–246) were cloned into pET28b(+) (Novagen (EMD) Biosciences, Madison, WI). The HMG1-L domain (aa 43–152) was cloned in pOPINF vector. All proteins and domain constructs were prepared as previously reported (4). Their folding was assessed by circular dichroism at the Centre for Genomic Regulation (Barcelona, Spain).
Protein and DNA complex formation
DNA and protein (or protein domains) were mixed at different molar ratios (1:1, 2:1) and dialyzed stepwise to a final buffer containing low salt (100 mM NaCl). smFRET measurements were made at a 10-fold excess of unlabeled DNA (from 500 pM total DNA, 50 pM were labeled). Complex formation was assessed by electrophoretic mobility shift assay (EMSA).
Single-molecule FRET experiments
smFRET experiments were performed on a home-built confocal microscope as described in (14, 15). smFRET was measured on DNA and complexes in solution, in buffers containing 1 mM ascorbic acid and 0.01% surfactant Nonidet P40 to avoid photodamage and adhesion, by using multiplates (Greiner Bio-One, Kremsmünster, Austria) passivated with Sigmacote (Sigma-Aldrich, St. Louis, MO). For each fluorescence burst, the proximity ratio was analyzed by the sensitized emission of the acceptor upon selective donor excitation (14).
SAXS measurements
SAXS data of TFAM/LSP22 from 1.7 to 10.5 mg mL−1 were measured at the BioSAXS beamline from European Synchrotron Radiation Facility (Grenoble, France). Parameters were calculated from merged curves using appropriate programs. Models for comparison were generated with specific methods as detailed in the Supporting Material.
Molecular dynamics simulations
MD simulations of free DNA30 and DNA30 in complex with wild-type or mutant TFAM were performed using the AMBER 12 package. The starting coordinates of the TFAM/LSP complex were taken from the LSP22 crystal structure (PDB: 3TQ6). We created the mutant variant by mutating all residues of the HMG-boxes that directly interact with the DNA bases to alanine (L58A, Y57A, T77A, T78A, and I81A from HMG1, and Y162A, N163A, V166A, and L182A from HMG2-Cter). Details of preparation, equilibration, production, and analysis of the simulations can be found in the Supporting Material.
Results
Design of the DNA constructs for smFRET experiments
Until now, not much is known about the structure and the stability of the TFAM/LSP complex in solution. Thus, we first determined whether TFAM bends LSP into a U-shape—similar to the crystal structure (Fig. 1 B) (4, 5)—or into a V-shape, as previously suggested (7). The rationale behind our approach is shown in Fig. 1. We designed two DNA constructs of 30 bp (DNA30) and 50 bp (DNA50) length, whose central part includes the cognate binding site of TFAM at LSP (LSP22) (Fig. 1 A). The DNA ends were labeled with a FRET donor (Alexa 488) and acceptor (Alexa 594). Assuming a canonical B-DNA double helix in the absence of TFAM, the distance between dyes would exceed 10 nm for both constructs (102 Å for DNA30, 170 Å for DNA50), hence we expected very little or no FRET in unbound DNA. Upon binding to the central cognate sequence (hereafter denoted as TFAM/DNA30 and TFAM/DNA50 complexes), TFAM is expected to bend the DNA, reducing the distance between both dyes and generating a FRET signal. If TFAM bends the DNA at a single site, the DNA would assume a V-form and the FRET signal for TFAM/DNA50 would be much smaller than for TFAM/DNA30, because the distance between both fluorophores would increase with DNA length (Fig. 1 C, right column). However, if TFAM kinks LSP at two sites, a U-turn would be induced, and the distance between both fluorophores would be similar in TFAM/DNA30 and TFAM/DNA50 (Fig. 1 C, left column). Therefore, to probe the mechanism of TFAM-induced DNA bending, we performed smFRET experiments for DNA30 and DNA50 incubated with either full-length TFAM or individual TFAM domains.
TFAM binds LSP in a U-turn
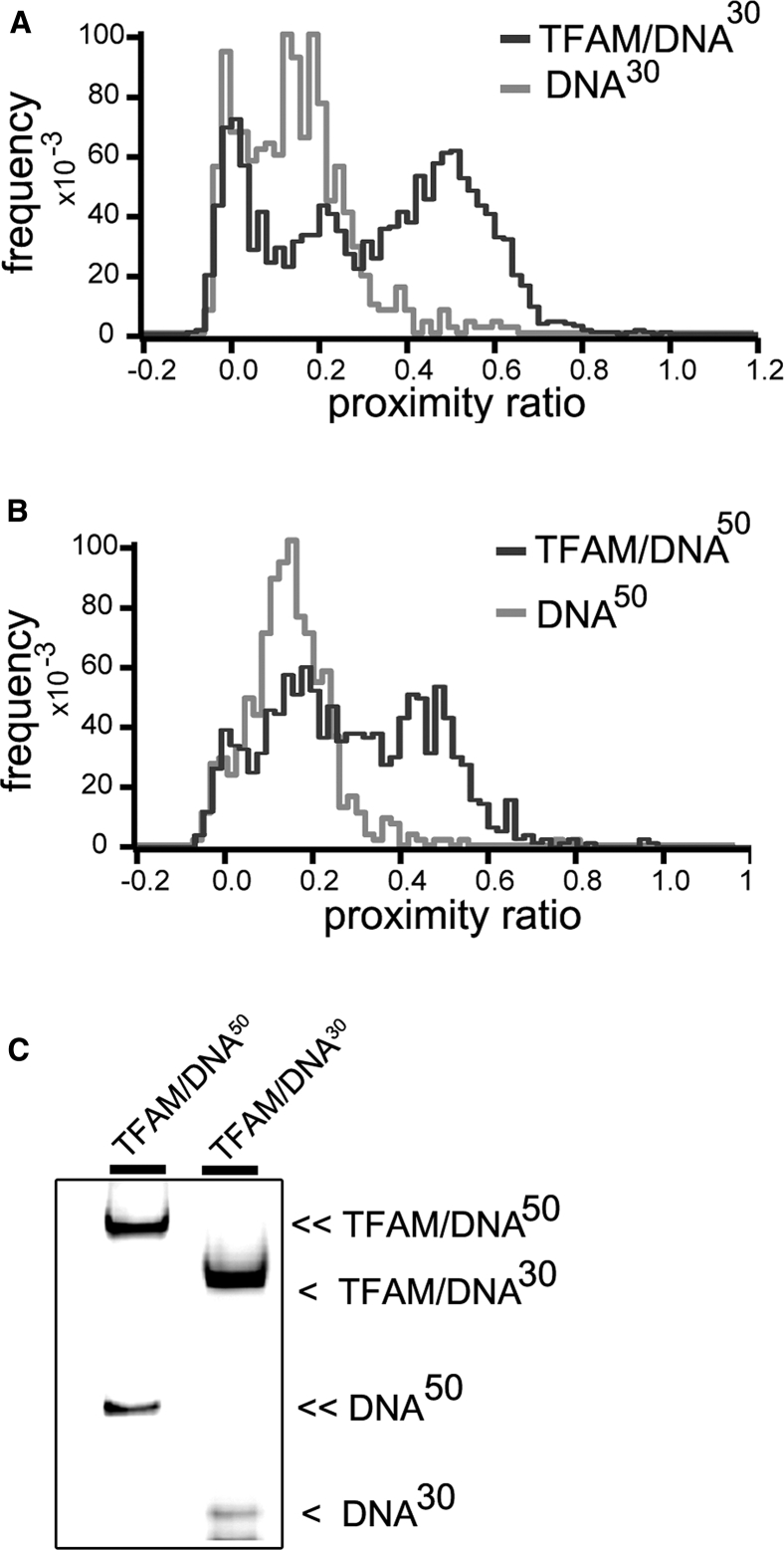
smFRET histograms of TFAM/DNA30 and TFAM/DNA50 complexes are shown in Fig. 2. The concentration of labeled DNA was set to 50 pM (corresponding to 500 pM DNA in total; see the Supporting Material). Data were acquired for 600 s, which is sufficient to achieve a good separation of subpopulations in the histogram. After fitting with multiple Gaussian distributions, the characteristic proximity ratio value (P) of each peak was determined, as well as their width (FWHM). Free DNA showed two peaks; a donor-only species, which represents molecules that lack an active acceptor dye and which is centered around P = 0, and a second population that corresponds to the donor-acceptor duplex DNA (Fig. 2 A, light gray line, Table 1). The nonzero proximity ratio of the second peak is a result of residual acceptor excitation and does not arise from energy transfer. In contrast, TFAM/DNA30 formed an additional species with higher proximity ratio at P ∼ 0.5 (Fig. 2 A, in dark gray). This population was broader (FWHM = 0.22 ± 0.08) than free DNA (FWHM = 0.13 ± 0.01). The excess width suggests either static heterogeneity, due to the presence of differently bent subconformations, or fast dynamics of the complex.
Figure 2.
Representative smFRET histograms for TFAM/DNA30 (A) and TFAM/DNA50 (B) complexes. The distributions of free DNA30 and DNA50 are shown in light gray, whereas complexes with TFAM (TFAM/DNA) are shown in dark gray. The donor-only peak at P = 0 represents molecules that lack an active acceptor. The p values of high FRET peaks are indicated in Table 1. (C) Shown here is nondenaturing polyacrylamide gel (5%) with TFAM/DNA30 and TFAM/DNA50 samples. The gel bands were visualized using the fluorescence signal from Alexa 594 (excitation at 532 nm, detection at 595–625 nm). The symbol (<) shows migration of DNA30 (free and in complex) and (<<) shows DNA50 (free and in complex). Note that the lower band below free DNA30 is due to a minor impurity of single-stranded DNA in the sample.
Table 1.
Characteristic Proximity Ratio Values of the High FRET Peak for TFAM and TFAM Domains in Complex with DNA30 and DNA50
| Sample | Proximity Ratio DNA30 | Proximity Ratio DNA50 |
|---|---|---|
| Free DNA | 0.160 ± 0.005 | 0.150 ± 0.003 |
| TFAM/DNA | 0.481 ± 0.011 | 0.437 ± 0.007 |
| HMG1-L/DNA | 0.277 ± 0.006 | 0.152 ± 0.004 |
| HMG1/DNA | 0.217 ± 0.007 | 0.156 ± 0.001 |
| HMG2-Cter/DNA | 0.175 ± 0.003 | 0.156 ± N/A |
For each peak, the proximity ratio (i.e., mean value over replicates) and the associated SE (σ/√n) are shown. Note that the free DNA sample is described by the peak of the donor-acceptor duplex DNA, as the donor-only peak at P = 0 does not convey any structural information.
Importantly, the smFRET distribution of TFAM/DNA50 was similar to that of TFAM/DNA30, with a slightly lower P for the TFAM/DNA50 complex (P ∼ 0.43), as shown in Fig. 2, A and B. This small difference could arise from electrostatic repulsion of the longer DNA arms of DNA50. For both DNA30 and DNA50, the complex formation was previously assessed by EMSA (Fig. 2 C). The fact that DNA30 and DNA50 yielded comparable P values upon TFAM binding strongly supports the U-turn model for TFAM-induced DNA bending.
The individual protein domains orchestrate a stepwise DNA bending
Next, we studied the role of the individual TFAM domains in LSP bending. To do so, we generated a construct containing HMG1 domain alone (residues 43–125, called HMG1), HMG1 with the linker region (residues 43–152, HMG1-L), and HMG2 with the C-terminal tail (residues 149–246, HMG2-Cter) (Fig. S1 A). We first tested the ability of these protein fragments to bind fluorescently labeled DNA30 and DNA50 at nanomolar concentrations by EMSA. Increasing concentrations of full-length TFAM (>200 nM) resulted in a two-band shift (Fig. S2), which might be related to oligomerization of TFAM in the presence of DNA, as previously reported (8). Note that in subsequent smFRET studies, sample concentrations were in the subnanomolar range, so protein oligomerization should be negligible. Regarding the EMSAs, full-length TFAM showed higher DNA binding ability than the individual domains. HMG1 showed blurred band shifts, which suggests that it is a less stable complex. In contrast, the presence of the linker in HMG1-L resulted in better DNA binding, whereas HMG2-Cter showed very weak (but still detectable) binding, in agreement with previous reports (8, 9). The fact that bands shifted by full-length TFAM were better defined than those of HMG1-L indicates that HMG2 helps to stabilize the position of TFAM on DNA30, even though it has a low affinity for DNA on its own.
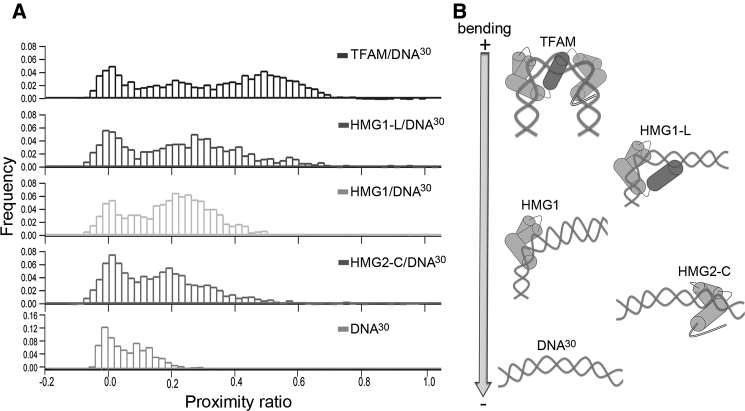
Whereas EMSA data provide information on protein binding, FRET serves as a local probe for DNA bending. smFRET histograms of the domain constructs in complex with DNA were asymmetric at proximity ratios higher than those of free DNA30 (Fig. 3 A; Table 1). Binding of HMG1 led to a broader distribution with a slightly higher proximity ratio (P ∼ 0.22) compared to the free DNA peak (P ∼ 0.16). For HMG1-L, the proximity ratio further increased (P ∼ 0.28) and showed a somewhat sharper distribution, which suggests that the linker stabilizes binding of HMG1 to DNA and enhances DNA bending. Still, the proximity ratio was lower than that observed for full-length TFAM, which indicates that the second HMG box is required to induce the final U-turn. Binding of HMG2-Cter to DNA30 showed a different picture. Although the center of the proximity ratio for the complex was similar to that of HMG1/DNA30, its relative area was much lower, which suggests weaker binding. Such a weaker binding is consistent with the marginal DNA shift in EMSA (Fig. S2 A). A hypothetical representation of DNA bending within each complex is shown beside the corresponding FRET histogram in Fig. 3 B. Notably, the complex of any of these TFAM fragments with longer DNA50 resulted in histograms similar to that of unbound DNA50 (see EMSA and FRET data in Fig. S2 and FRET values in Table 1), which further indicates that each subdomain induces only a single kink and bends DNA into a V-shape.
Figure 3.
The individual protein domains orchestrate stepwise DNA bending. (A) Representative smFRET histograms for TFAM and TFAM domains in complex with DNA30 are shown. The complex with full-length TFAM is shown at the top, followed by HMG1-L, HMG1, HMG2-Cter, and free DNA (bottom). (B) A model of bending is represented aligned with each histogram and ordered from top (sharpest) to bottom (lowest) according to the extent of DNA bending. The TFAM/DNA model is based on the U-turn structure, whereas the complexes with domains are hypothetical representations based on the proximity ratio distributions. The DNA is represented as prebent, in accordance with our MD results. From bottom to top, the free DNA and HMG2 complex show similar bending due to the marginal effect of HMG2. Above, HMG1 introduces the first kink, which is enhanced by the linker. At the top, full-length TFAM induces a U-turn, suggesting cooperativity, because HMG2 kinks the DNA only in the presence of the N-terminal protein fragment.
TFAM forms a compact and heterogeneous complex with DNA
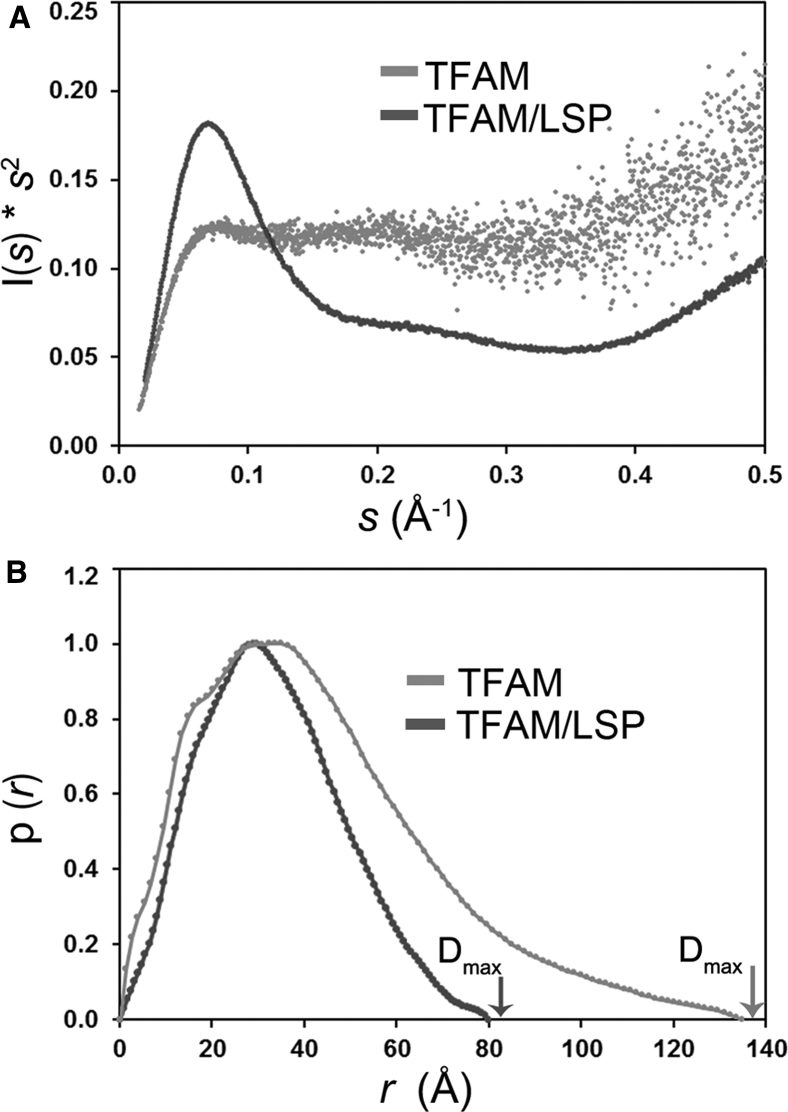
To further characterize the TFAM/LSP complex in solution, we performed SAXS measurements. From the TFAM/LSP scattering curve we could estimate a molecular mass of ∼30 kDa (16), which agrees with one protein bound to one DNA molecule (25.6 KDa TFAM + 13.5 kDa DNA = 39.1 kDa). The estimated radius of gyration (Rg) of the complex (Rg = 26.0 ± 0.1 Å) is smaller than that of the free protein (Rg = 32.0 ± 0.3 Å) (4). This is consistent with previous single-molecule (17) and multiangle light scattering (5) studies, which indicated that the protein/DNA particle is more compact than the free protein. The Kratky plot derived from the scattering curve has a bell-shaped profile consistent with an overall globular shape in solution (Fig. 4 A). This is in contrast to the Kratky plot for the free protein, which does not show a maximum and indicates a high degree of conformational flexibility that we attributed to the structural variability, due to both the linker and C-terminal tail (Fig. 4 A; (4)). The pairwise distance distribution p(r), which provides the overall shape of the particle in solution, displays a slightly asymmetric profile with a maximum intramolecular distance (Dmax) of 82 Å. Notably, this value is smaller than Dmax of unbound TFAM (Dmax = 135 ± 5 Å) (Fig. 4 B), which further demonstrates the compaction of the protein upon DNA binding.
Figure 4.
SAXS analysis of TFAM/LSP complexes. (A) Kratky representation of experimental SAXS curves. Values have been normalized by I0. The TFAM/LSP complex shows the profile of a globular particle (dark gray), whereas free TFAM (in light gray) clearly shows the profile of a flexible protein. (B) Shown here is the pairwise distance distribution function p(r). Maximal particle dimensions are arrowed for both free and DNA-bound TFAM.
We used the crystallographic structure of the complex as a model to describe the SAXS curve. Despite the overall similarity of both curves, the fit is relatively poor (χ2 = 2.93) and shows systematic deviations throughout the complete momentum transfer range (Fig. S3 A). This suggests that TFAM/LSP is not a static complex. The aforementioned broadening of the TFAM/DNA peak in the smFRET distribution also hints at the conformational flexibility of the TFAM/LSP complex. A possible source of flexibility could be the intrinsic disorder of the C-terminal tail, as suggested by the weak electron density between Lys237 and Cys246 in both TFAM/LSP crystal structures. To check this, we applied the ensemble optimization method (18) using a large pool of TFAM/LSP structures with different C-terminal conformations added to the crystallographic structure (Fig. S3 B). However, the ensemble optimization method did not yield a better description of the experimental curve, which suggests that additional sources of flexibility apart from the C-terminal tail are present in TFAM/LSP.
Computational analysis shows the intrinsic curvature of the TFAM binding site at LSP
To better understand the flexibility of TFAM/LSP complexes at the molecular level, we characterized the structural and dynamic properties of free LSP-DNA and the protein/DNA complex by MD simulations. First, we assessed the sequence-dependent intrinsic curvature and flexibility of each base pair step of free DNA30 in water. During the simulation, the initially straight DNA30 spontaneously assumed a more curved conformation, with an average total bend of 72 ± 18° (Figs. 5 A and S4 A). In particular, the region specifically contacted by TFAM in the bound structure (represented by LSP22 in the crystal structure; see Fig. 1 A), showed an average curvature of 46 ± 12°, with an occasional high curvature conformation (113°) closer to that of the protein-bound DNA (169°) (Fig. S4 B). An analysis of the average minor groove width along the TFAM binding site showed sequence-dependent variability. In particular, the minor groove around the Leu58 insertion site is narrower than that around Leu182 in the free DNA (Fig. S4 C).
Figure 5.
Structural features of the TFAM binding site at LSP. (A) Given here is a DNA30 LSP sequence in the ideal B-DNA form (in gray) and a frame from the MD simulation (in green) that shows considerable distortions. (B) Values of the stiffness parameter Ktotal are for the naked LSP sequence, calculated from the MD simulation for each base pair step. (C) Given here is the base pair parameter roll (in degrees) for the DNA30 LSP sequence in the x-ray crystal structure (in blue) and averaged during the MD simulations for the naked DNA (in green), for the DNA in complex with the wild-type protein (in black), and in complex with the MD-mutant protein (in red).
An analysis of the basepair parameters shows that step A3C4/G19T20, into which HMG1 Leu58 inserts, is moderately flexible (left vertical line in stiffness plot in Fig. 5 B) and has roll values close to those of an ideal B-DNA (−0.84° on average; green line in Fig. 5 C). In contrast, the neighboring step C4A5/T18G19 and, importantly, the insertion site of HMG2 Leu182, step C15A16/T7G8, have a higher, positive roll (10.4 and 10.5°, respectively) and are more flexible (Fig. 5, B and C). In summary, DNA30 deviates moderately from the canonical B-DNA at the Leu58 insertion site, whereas at the following step and Leu182 site the deviations are much more pronounced.
LSP forms a nonstatic complex with TFAM
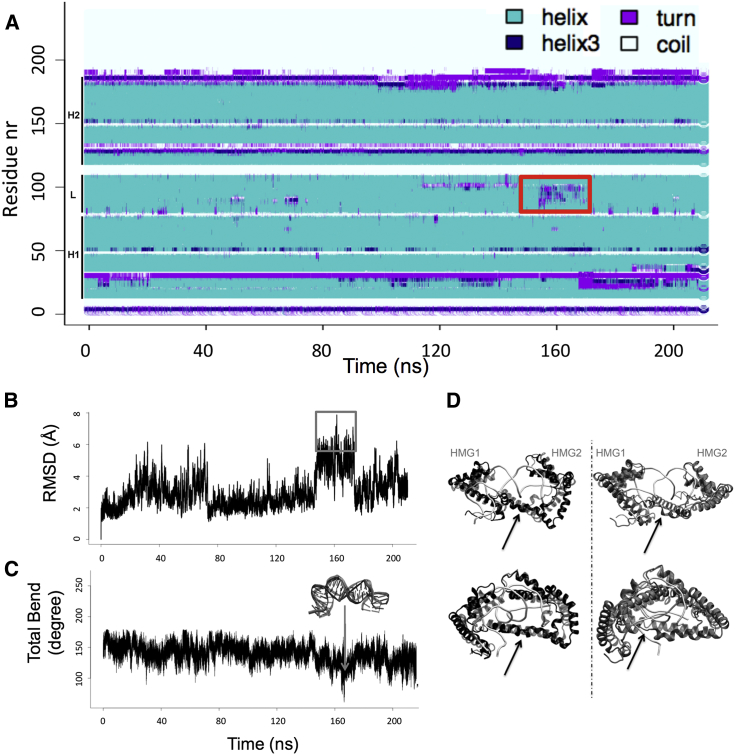
The influence of TFAM on DNA bending was analyzed by MD simulations of LSP in complex with wild-type protein. We monitored structural changes during the MD by secondary structure analysis (Fig. 6 A) and time-dependent root mean square deviation (RMSD) relative to the equilibrated initial structure (Fig. 6 B). During the MD simulation, the complex maintained the DNA-protein contacts, and converged to a structure similar to the initial one at the end of the simulation.
Figure 6.
Structural variability of the TFAM/LSP complex. (A) Shown here is a secondary structure plot of WT protein in complex with LSP during the simulation (every 100th snapshot). The secondary structure of the WT protein, i.e., the HMG1, HMG2, and linker domains (marked as H1, H2 and L, respectively), are maintained during the simulation, except for an unfolding event at the linker region at ∼160 ns (red box). Helical conformation is labeled in cyan (helix 310 in dark blue), turns in violet, and coils in white. (B) Shown here is the time-dependent RMSD of Cα atoms in the WT protein in complex with DNA. The reference structure is the x-ray crystal structure after MD relaxation. The box highlights the highest values, which coincide with unfolding of the linker (compare with (A)). (C) Given here is the total bend (in degrees) of the DNA in complex with the WT protein during the MD simulation. The arrow points at the event during which the DNA significantly reduces its bending, as shown in the overlay between the starting DNA structure (light gray) and the DNA at this time step (dark gray). (D) Shown here are plots showing the unfolding of the linker compared to the initial structure (in light gray) for the WT protein (in black, left panel) and the MD-mutant protein (in dark gray, right panel) throughout the simulation. In the images (front and side views of the complexes), the black arrows point to the linker region that unfolds during the MD. To see this figure in color, go online.
The most significant motion as captured by principal component analysis of the MD trajectory was a reversible dynamic separation between the two HMG domains, which moved apart back and forth in a butterfly or “breathing” movement (Fig. S5 A). At the center of this movement, the linker is predicted to locate the hinge point of the entire complex (arrow in Fig. S5 A). The breathing of the protein was coupled to a bending-relaxation movement of DNA, but no changes in protein/DNA contacts were detected. The largest fluctuations of atomic positions took place at HMG1 and, remarkably, in the linker region (the average atomic mobility during MD is represented by the root mean square fluctuations in Fig. S5 B, black curve). Interestingly, during the simulation, an event with the highest RMSD occurred (indicated with a gray frame in Fig. 6 B). This corresponded to DNA straightening toward its ideal naked conformation (DNA bending ranges from 169 to 61°; Fig. 6 C), together with partial unfolding of the linker region (black square in Fig. 6, A and D, left column). Thus, the linker is the protein region that adapts most to DNA movements, and it unfolded precisely at the region that contacts the DNA. This conformational change suggests that the protein accommodates the DNA motions by virtue of its linker flexibility, without disrupting the contacts of the HMG boxes with the DNA. To relate these conformational changes to the smFRET broadening, we calculated interdye distances for those MD models that diverged most from the x-ray structure after relaxation (Fig. S3 C). This led to theoretical, MD-based FRET efficiency values, from 0.31 to 0.65 (Table 2). This broad range is in agreement with the increased peak width in the smFRET distributions, which indicates that the flexibility of the TFAM-LSP complex is reflected in the peak broadening.
Table 2.
Donor-Acceptor Distance Calculation for the Initially Relaxed Crystal Structure and Four Representative Conformations from the MD Simulations
| Sample | RDA | σDA | EFRET |
|---|---|---|---|
| 0_pdb | 48.1 | 10.3 | 0.31 |
| 121_pdb | 52.3 | 11.8 | 0.43 |
| 31_pdb | 56.6 | 7.2 | 0.54 |
| 26_pdb | 53.3 | 10.5 | 0.45 |
| 00_pdb | 61.2 | 8.9 | 0.65 |
The DNA from the MD structures was extended with the DNA30 sequence. All parameters were computed using the FPS toolkit. 0_pdb indicates the initially relaxed structure, RDA indicates the dye-to-dye distance in Å, σDA indicates the standard deviation, and EFRET values are FRET efficiencies calculated from the weighted RDA−s assuming a Förster radius of R0 = 55.6 Å.
We next took a closer look at the base pair parameters of the DNA. In the crystal structure, the protein-bound DNA displays the highest roll angle at the base pair steps contacted by Leu58 (at HMG1) or Leu182 (at HMG2) (4) (Fig. 5 C, blue line). During the MD simulation of the wild-type (WT) complex, the high roll at the Leu182 step was maintained, whereas the roll angle at the Leu58-contacted step A3C4/G19T20 diminished (Fig. 5 C, black line). The angle at the neighboring steps C4A5/T18G19 and A5G6/C17T18 was maintained or increased. Thus, in the HMG1 region, the stress at one point with high roll angle redistributes to the neighboring steps, keeping the bending of the DNA. These changes in roll at the HMG1 binding site may correspond to release of crystal packing constraints, which results in a less kinked and less stressed DNA than that found in the x-ray structure. Interestingly, the comparison of DNA in bound and unbound states shows that the major structural distortions found in the complex (such as the roll angle) are already present in the unbound form, yet less pronounced. In addition, the roll redistribution in the complex during the MD leads to a pattern that is more similar to that of naked DNA than the x-ray one. These similarities suggest that the free 22 bps in LSP that contact TFAM have a structural propensity toward the protein-bound conformation.
The dynamic tension between TFAM and LSP causes reversible unfolding of the linker
To further understand the influence of the DNA-contacting residues in HMG1 and HMG2 on DNA bending, we ran an MD simulation of an in silico TFAM mutant (MD-mutant) in complex with DNA. In the mutant, nine residues were substituted by alanines: L58A, Y57A, T77A, T78A, and I81A from HMG1; and Y162A, N163A, V166A, and L182A from HMG2 (Fig. S1 B). Comparison of the mutant and the WT trajectory should reveal the constraints imposed by the WT side chains to the bent DNA.
The MD trajectory of the MD-mutant/DNA complex showed higher deviation from the initial conformation (see the overall higher values of mutant RMSD, Fig. S5 C). Principal component analysis revealed a faster breathing motion of the HMG boxes around the linker hinge point than in the WT complex (Fig. S5 A). Compared to the WT complex, the root mean square fluctuation of DNA atoms increased significantly in regions that are contacted by the HMG boxes and, notably, by the linker (Fig. S5 B). During the simulation, the strain on the DNA diminished because the overall roll angles decreased, as did the global curvature (exemplified by lower roll values, Fig. 5 C). Most importantly, the linker irreversibly lost its helical structure due to unfolding and stretching at the region between residues 123–153 (Fig. 6 D right column, and Fig. S6), which on average increased its distance from the DNA (Fig. S5 D). Although these deformations were considerable, the contacts between the HMG boxes and DNA were preserved. Only a slight increase in the distance between HMG2 and the DNA was observed along the MD simulation, which did not result in disassembly of the complex at this timescale (Fig. S5 D).
The HMG2 DNA region has a kink characterized by the highest roll (red graph in Fig. 5 C), which only slightly diminished during the simulation, even though Leu182 was substituted by the much shorter alanine. On the other hand, the kink at the HMG1 region shifted from A3C4/G19T20 in the x-ray structure to the immediate neighbor C4A5/T18G19. Strikingly, the latter is also the step with the highest flexibility and roll in the naked DNA structure (compare green and red lines in Fig. 5 C). Thus, upon removal of the side chains that constrain the DNA in a kinked conformation, the DNA relocates the distortions to the steps that are intrinsically easier to distort, without affecting the binding or the bending. Therefore, in complex with the MD-mutant, the DNA tries to recover, even more closely, the intrinsic sequence-dependent structure of the naked double helix. This results in stronger distortions at the protein region that is most sensitive to structural variations in DNA, which is the linker.
Discussion
TFAM is a transcription factor that induces strong bending to LSP and HSP1 promoters (19), enabling mtRNAP recruitment (2, 5, 7). In addition, it is an architectural protein that compacts human mtDNA by introducing strong bends to the double helix (5, 6, 20, 21, 22). In this study, we investigated the mechanism by which the protein binds and bends DNA in solution and whether it does so at a single point, leading to a V-kink deformation, or at two points, leading to the U-turn conformation found in the crystal. By combining single-molecule FRET and SAXS data, we unambiguously showed that TFAM compacts LSP by inducing a U-turn, and that each individual HMG box imposes a single kink to the DNA. Our data revealed that the linker between the HMG boxes enhances the bending exerted by HMG1 and, as unveiled by MD, confers considerable flexibility to the complex. In the TFAM/LSP crystal structure, an electropositive surface of the linker C-terminal region binds the concave surface of the U-turn, thus stabilizing the bending by electrostatic interactions (4, 5). Similar contact might occur in the HMG1-L/DNA complex.
Interestingly, in the free protein the linker is disordered but gains α-helix character in complex with DNA, as seen in the crystal structure, by SAXS (4) and by CD spectroscopy (7, 9). This transition is pivotal to the TFAM/LSP interaction. We found that the isolated HMG2-Cter domain has only low binding affinity to LSP. Previous studies showed that binding ability increases when HMG2 is fused to the linker C-terminal region (9), probably due to formation of a local hydrophobic core with the linker α-helix (4). This suggests that, for efficient DNA binding, HMG2 depends on the presence of the helical linker, whose fold in turn depends on HMG1 bound to the DNA. These findings support a unique stepwise binding mechanism, in which DNA binding is driven by HMG1, which initially bends the DNA into a V-shape. This reduces the accessible conformational space for the linker, which folds into a 29-residue α-helix that wraps around the DNA and reduces the distance between HMG boxes. This eventually facilitates HMG2 binding. The latter, in turn, introduces a second V-kink to the DNA, completing the U-shape conformation. The U-turn is thus a consequence of a double kink rather than a smooth DNA bending. To sum up, the system shows a sequential cooperative bending, in which the synergy between the individual domains and the folding of the linker mediate the correct alignment of protein and DNA.
An important question regarding the above model is whether binding follows a pure induced fit paradigm, in which the protein deforms a passive DNA, or if it has a certain degree of conformational selection. In the latter, the intrinsic deformability and conformation of the DNA help to achieve the distortion pattern required for binding. MD analysis shows that naked LSP has intrinsic overall curvature and conformation at the basepair level that may facilitate its bending into a U-turn. However, detailed analysis of trajectories shows the complexity of the contribution of the DNA to the protein binding. HMG1 inserts Leu58 into the DNA, but the strong distortion found in the crystal structure at the insertion site diminishes during the time course of the MD simulation and the neighboring steps, which have an intrinsic ability to curve, partially absorbed the distortion. Binding of HMG1 favors the formation of the linker α-helix that facilitates the insertion of HMG2 at a DNA step characterized by an extremely deformable and open base pair. Interestingly, the complex is not static due to the intrinsic flexibility of both LSP and the protein, as suggested by MD and distribution broadening in smFRET. This is backed up by the discrepancy between the experimental SAXS data and a model based on the static crystal structure. Additionally, an in silico mutant allowed us to gain information on such a flexible bending mechanism of TFAM. Mutations of key interacting residues in the two HMG domains lead to an increase in the tendency of DNA to recover a straighter conformation, which produces major corruption in the linker conformation and some departure of the HMG2 from the bound arrangement.
Taken together, all these observations strongly suggest that the TFAM/LSP complex is in a dynamic equilibrium governed by 1) protein-DNA contacts, especially those formed by the inserted leucines; 2) the equilibrium between disordered and α-helix conformation of the linker; and 3) the intrinsic tendency of DNA to fold back to a straighter structure. The global structure of the complex is subjected to opposing forces, leading to a flexible complex whose overall conformation varies considerably. The bending angles vary from 169° to 61°, a range that comprises the angles detected by previous studies using LSP (78° or 72°) or nsDNA (100° ± 20°) (7, 21). Because U-turn bending is required to recruit the mitochondrial RNA polymerase to the LSP transcription initiation site (2), it can be speculated that the degree of bending may condition transcription activation efficiency. The isoform TFAMΔexon5 lacks the C-terminal part of the linker and the first helix of HMG2 (residues 148–179) and is unable to activate transcription in vitro (23). After the linker, such a polypeptide chain may fold only into two consecutive α-helices and a disordered C-terminal tail. According to our data, this isoform might be unable to induce a U-turn on LSP, because only intact HMG2 might actively bend the DNA into the second V-shape. The C-terminal tail may contact the transcription machinery, but the partial bending may not be sufficient to bring it to the transcription initiation site. Such impaired transcription and transcription-dependent replication is expected to have a direct impact on mtDNA and mitochondria biogenesis.
TFAM is the main protein involved in mtDNA packaging, in which most of interactions are nonspecific. In this regard, it is highly plausible that variability in bending may depend on the properties of the sequence at the DNA binding sites. It was shown that HMG-box proteins increase DNA flexibility and decrease DNA persistence length (a parameter related to the stiffness of the DNA) by kinking the DNA. These HMG-box proteins include TFAM (17) and the Saccharomyces cerevisiae HMO1 (24) and Nhp6A (12) proteins. Two models have been proposed to explain the increase in flexibility of the kinked complexes ((12, 25, 26) and references therein): a static kink model, in which the protein binds to DNA creating fixed angle bends; and a flexible hinge model in which a highly dynamic complex has significant oscillations around a slightly preferred bending angle. Previous optical force experiments using long DNA molecules detected an increase in DNA flexibility upon TFAM binding (17). Based on the static crystal structure, the authors proposed that TFAM/DNA complexes were rigid entities and flexibility could arise from local denaturation of the DNA between protein/DNA kinks. However, our data clearly demonstrate that the TFAM/DNA complex is flexible and governed by dynamic tensions, as shown by our MD calculations, and suggests a rather flexible hinge model exhibiting variable bending. Thus, TFAM would introduce local flexible points at nsDNA during compaction, and such flexible assemblies should be compatible with formation of higher-order protein-DNA complexes.
In conclusion, our study provides important insights into the molecular mechanism underlying bending of LSP by TFAM. We show by FRET and SAXS that TFAM domains bind and bend LSP by a stepwise mechanism that induces a compact U-turn in solution. MD shows that the complex is dynamic and the DNA tends to recover its free state conformation, but this is counteracted by unfolding and refolding of the protein helical linker, which restores the U-turn. This tension results in variation of the bending angle. Our results point to a tight DNA bending mechanism based on the flexibility of TFAM. A similar mechanism may underlie DNA compaction in mitochondria, a process that is essential for cell life.
Author Contributions
A.R.-C. contributed to cloning. A.R.-C. and A.C. did protein production. A.R.-C. performed complex preparation and EMSA. A.R.-C., A.G., and J.L. performed the FRET studies. A.R.-C. and P.B. performed the SAXS studies. F.B. and M.O. performed molecular dynamics studies. M.S. and K.T. designed and supervised the project. All authors participated in writing the manuscript.
Acknowledgments
Jörg Langowski sadly passed away on May 6, 2017.
We thank Sébastien Lyonnais for guidance with the EMSA gels. M.O. is an ICREA Academia researcher.
This work was supported by the Ministry of Economy and Competitiveness (MINECO) (BFU2012-33516 and BFU2015-70645-R to M.S., and BIO2012-32868 and BFU2014-61670-EXP to M.O.); Generalitat de Catalunya (SGR2009-1366 and 2014-SGR-997 to M.S., and SGR2009-1348, 2014 SGR-134 to M.O.); the Instituto Nacional de Bioinformática; the European Union (FP7-HEALTH-2010-261460, FP7-PEOPLE-2011-290246, and FP7-HEALTH-2012-306029-2 to M.S., and H2020-EINFRA-2015-1-675728 and H2020-EINFRA-2015-676556 to M.O.); and the European Research Council (ERC-2011-ADG_20110209-291433 to M.O.). A.R.-C. was awarded with a “Junta para la Ampliación de Estudios” (Programa JAE) fellowship from Consejo Superior de Investigaciones Científicas (CSIC). The Structural Biology Unit at IBMB-CSIC is a “Maria de Maeztu” Unit of Excellence awarded by the Ministry of Economy and Competitiveness (MINECO) under MDM-2014-0435. IRB Barcelona is the recipient of a Severo Ochoa Award of Excellence from the Ministry of Economy and Competitiveness (MINECO). The CBS is a member of the French Infrastructure for Integrated Structural Biology (FRISBI), a national infrastructure supported by the French National Research Agency (ANR-10-INBS-05).
Editor: David Rueda.
Footnotes
Supporting Materials and Methods and seven figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)34975-5.
Contributor Information
Katalin Tóth, Email: kt@dkfz-heidelberg.de.
Maria Solà, Email: maria.sola@ibmb.csic.es.
Supporting Material
References
- 1.Gustafsson C.M., Falkenberg M., Larsson N.G. Maintenance and expression of mammalian mitochondrial DNA. Annu. Rev. Biochem. 2016;85:133–160. doi: 10.1146/annurev-biochem-060815-014402. [DOI] [PubMed] [Google Scholar]
- 2.Morozov Y.I., Parshin A.V., Temiakov D. A model for transcription initiation in human mitochondria. Nucleic Acids Res. 2015;43:3726–3735. doi: 10.1093/nar/gkv235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stros M., Launholt D., Grasser K.D. The HMG-box: a versatile protein domain occurring in a wide variety of DNA-binding proteins. Cell. Mol. Life Sci. 2007;64:2590–2606. doi: 10.1007/s00018-007-7162-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rubio-Cosials A., Sidow J.F., Solà M. Human mitochondrial transcription factor A induces a U-turn structure in the light strand promoter. Nat. Struct. Mol. Biol. 2011;18:1281–1289. doi: 10.1038/nsmb.2160. [DOI] [PubMed] [Google Scholar]
- 5.Ngo H.B., Kaiser J.T., Chan D.C. The mitochondrial transcription and packaging factor TFAM imposes a U-turn on mitochondrial DNA. Nat. Struct. Mol. Biol. 2011;18:1290–1296. doi: 10.1038/nsmb.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ngo H.B., Lovely G.A., Chan D.C. Distinct structural features of TFAM drive mitochondrial DNA packaging versus transcriptional activation. Nat. Commun. 2014;5:3077. doi: 10.1038/ncomms4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malarkey C.S., Bestwick M., Churchill M.E. Transcriptional activation by mitochondrial transcription factor A involves preferential distortion of promoter DNA. Nucleic Acids Res. 2012;40:614–624. doi: 10.1093/nar/gkr787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gangelhoff T.A., Mungalachetty P.S., Churchill M.E. Structural analysis and DNA binding of the HMG domains of the human mitochondrial transcription factor A. Nucleic Acids Res. 2009;37:3153–3164. doi: 10.1093/nar/gkp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wong T.S., Rajagopalan S., Fersht A.R. Biophysical characterizations of human mitochondrial transcription factor A and its binding to tumor suppressor p53. Nucleic Acids Res. 2009;37:6765–6783. doi: 10.1093/nar/gkp750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ha T., Enderle T., Weiss S. Probing the interaction between two single molecules: fluorescence resonance energy transfer between a single donor and a single acceptor. Proc. Natl. Acad. Sci. USA. 1996;93:6264–6268. doi: 10.1073/pnas.93.13.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deniz A.A., Dahan M., Schultz P.G. Single-pair fluorescence resonance energy transfer on freely diffusing molecules: observation of Förster distance dependence and subpopulations. Proc. Natl. Acad. Sci. USA. 1999;96:3670–3675. doi: 10.1073/pnas.96.7.3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coats J.E., Lin Y., Rasnik I. Single-molecule FRET analysis of DNA binding and bending by yeast HMGB protein Nhp6A. Nucleic Acids Res. 2013;41:1372–1381. doi: 10.1093/nar/gks1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hieb A.R., Gansen A., Langowski J. The conformational state of the nucleosome entry-exit site modulates TATA box-specific TBP binding. Nucleic Acids Res. 2014;42:7561–7576. doi: 10.1093/nar/gku423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clegg R.M. Fluorescence resonance energy transfer and nucleic acids. Methods Enzymol. 1992;211:353–388. doi: 10.1016/0076-6879(92)11020-j. [DOI] [PubMed] [Google Scholar]
- 15.Gansen A., Tóth K., Langowski J. Structural variability of nucleosomes detected by single-pair Förster resonance energy transfer: histone acetylation, sequence variation, and salt effects. J. Phys. Chem. B. 2009;113:2604–2613. doi: 10.1021/jp7114737. [DOI] [PubMed] [Google Scholar]
- 16.Rambo R.P., Tainer J.A. Accurate assessment of mass, models and resolution by small-angle scattering. Nature. 2013;496:477–481. doi: 10.1038/nature12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farge G., Laurens N., Wuite G.J. Protein sliding and DNA denaturation are essential for DNA organization by human mitochondrial transcription factor A. Nat. Commun. 2012;3:1013. doi: 10.1038/ncomms2001. [DOI] [PubMed] [Google Scholar]
- 18.Bernadó P., Mylonas E., Svergun D.I. Structural characterization of flexible proteins using small-angle x-ray scattering. J. Am. Chem. Soc. 2007;129:5656–5664. doi: 10.1021/ja069124n. [DOI] [PubMed] [Google Scholar]
- 19.Bonawitz N.D., Clayton D.A., Shadel G.S. Initiation and beyond: multiple functions of the human mitochondrial transcription machinery. Mol. Cell. 2006;24:813–825. doi: 10.1016/j.molcel.2006.11.024. [DOI] [PubMed] [Google Scholar]
- 20.Farge G., Mehmedovic M., Falkenberg M. In vitro-reconstituted nucleoids can block mitochondrial DNA replication and transcription. Cell Reports. 2014;8:66–74. doi: 10.1016/j.celrep.2014.05.046. [DOI] [PubMed] [Google Scholar]
- 21.Kaufman B.A., Durisic N., Shoubridge E.A. The mitochondrial transcription factor TFAM coordinates the assembly of multiple DNA molecules into nucleoid-like structures. Mol. Biol. Cell. 2007;18:3225–3236. doi: 10.1091/mbc.E07-05-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kukat C., Davies K.M., Larsson N.G. Cross-strand binding of TFAM to a single mtDNA molecule forms the mitochondrial nucleoid. Proc. Natl. Acad. Sci. USA. 2015;112:11288–11293. doi: 10.1073/pnas.1512131112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dairaghi D.J., Shadel G.S., Clayton D.A. Addition of a 29-residue carboxyl-terminal tail converts a simple HMG box-containing protein into a transcriptional activator. J. Mol. Biol. 1995;249:11–28. doi: 10.1006/jmbi.1995.9889. [DOI] [PubMed] [Google Scholar]
- 24.Murugesapillai D., McCauley M.J., Williams M.C. DNA bridging and looping by HMO1 provides a mechanism for stabilizing nucleosome-free chromatin. Nucleic Acids Res. 2014;42:8996–9004. doi: 10.1093/nar/gku635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCauley M.J., Zimmerman J., Williams M.C. HMGB binding to DNA: single and double box motifs. J. Mol. Biol. 2007;374:993–1004. doi: 10.1016/j.jmb.2007.09.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Noort J., Verbrugge S., Dame R.T. Dual architectural roles of HU: formation of flexible hinges and rigid filaments. Proc. Natl. Acad. Sci. USA. 2004;101:6969–6974. doi: 10.1073/pnas.0308230101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.