Abstract
The isozymes of photosynthetic phosphoenolpyruvate carboxylase from C4 plants (PEPC-C4) play a critical role in their atmospheric CO2 assimilation and productivity. They are allosterically activated by phosphorylated trioses or hexoses, such as d-glucose 6-phosphate, and inhibited by l-malate or l-aspartate. Additionally, PEPC-C4 isozymes from grasses are activated by glycine, serine, or alanine, but the allosteric site for these compounds remains unknown. Here, we report a new crystal structure of the isozyme from Zea mays (ZmPEPC-C4) with glycine bound at the monomer–monomer interfaces of the two dimers of the tetramer, making interactions with residues of both monomers. This binding site is close to, but different from, the one proposed to bind glucose 6-phosphate. Docking experiments indicated that d/l-serine or d/l-alanine could also bind to this site, which does not exist in the PEPC-C4 isozyme from the eudicot plant Flaveria, mainly because of a lysyl residue at the equivalent position of Ser-100 in ZmPEPC-C4. Accordingly, the ZmPEPC-C4 S100K mutant is not activated by glycine, serine, or alanine. Amino acid sequence alignments showed that PEPC-C4 isozymes from the monocot family Poaceae have either serine or glycine at this position, whereas those from Cyperaceae and eudicot families have lysine. The size and charge of the residue equivalent to Ser-100 are not only crucial for the activation of PEPC-C4 isozymes by neutral amino acids but also affect their affinity for the substrate phosphoenolpyruvate and their allosteric regulation by glucose 6-phosphate and malate, accounting for the reported kinetic differences between PEPC-C4 isozymes from monocot and eudicot plants.
Keywords: allosteric regulation, protein structure, enzyme kinetics, crystal structure, site-directed mutagenesis, amino acid residue conservation, monocot and eudicot PEPC-C4 isozymes, Zea mays L.
Introduction
During photosynthesis, to fix and reduce carbon and to synthesize simple sugars, the ATP and NADPH generated in the light reactions are used in the carbon-fixation reactions. In many plant species, known as C3 type, these reactions occur exclusively in the chloroplast by means of the C3 pathway, also known as the Calvin-Benson cycle, in which the starting and ending compound is ribulose 1,5-bisphosphate. In this pathway, the carboxylation step is catalyzed by ribulose 1,5-bisphosphate carboxylase/oxygenase (Rubisco).3 The majority of angiosperms are of the C3 type, but there is a group of plants, called the C4 type, that include agronomically important crops, such as maize (Zea mays), sorghum (Sorghum bicolor), sugarcane (Saccharum officinarum), and common millet (Panicum miliaceum), as well as most of the weeds in which the Calvin-Benson cycle is not the only pathway used in the carbon-fixation reactions. In the C4 plants, two metabolic pathways are involved in CO2 assimilation: the C4 and the C3 pathways (1). The C4 pathway provides a CO2-concentrating mechanism that ensures high CO2 levels in the environment of the C3 pathway enzymes, greatly favoring the carboxylating activity of Rubisco over its oxygenating activity. This results in an important reduction in photorespiration, a process that in C3 plants causes significant losses in photosynthetic efficiency, as well as in a better use of water and nitrogen. As a consequence, C4 plants exhibit an increased productivity, especially in sunny, dry, and hot environments.
The initial and highly regulated step in the C4 pathway takes place in the cytosol of mesophyll cells and consists in the irreversible carboxylation of 2-phosphoenolpyruvate (PEP) by the C4 photosynthetic isozyme of phosphoenolpyruvate carboxylase (orthophosphate:oxaloacetate carboxylyase (phosphorylating), EC 4.1.1.31), yielding oxaloacetate and Pi. The carboxylating reactions of PEPC-C4 and Rubisco take place in different cell types in the leaves of those plants that have the two-cell C4 pathway (also called Kranz type because of the anatomy of their leaves), which is by far the most widespread, whereas these reactions take place in different intracellular compartments in those plants that have the single-cell C4 pathway (2, 3). In addition to transport of some of the pathway intermediates between cells or intracellular compartments, in the two kinds of C4 pathways, the three essential steps are the initial fixation of CO2 by a PEPC-C4 isozyme to form a C4 acid, the decarboxylation of the C4 acid to release CO2 in the Rubisco environment, and the regeneration of the primary CO2 acceptor PEP.
Because of the increased need of food to satisfy the requirements of the growing human population, much effort has been, and still is, devoted to the improvement of the photosynthetic performance and therefore the yield of C3 plants, particularly those of agronomically and economically important crops, such as wheat (Triticum aestivum), rice (Oryza sativa), barley (Hordeum vulgare), soybean (Glycine max), and potato (Solanum tuberosum). This goal has been attempted by introducing C4 pathway genes, particularly that coding for the PEPC-C4 isozyme from maize, using conventional transgenic techniques so they could have the CO2-concentrating mechanism of C4 plants.
The importance of the reaction catalyzed by PEPC-C4 isozymes in the photosynthetic metabolism of C4 plants is underscored by their allosteric regulation (for reviews, see Refs. 4–6), which is particularly complex in the case of isozymes from some monocot C4 plants, such as that from maize (ZmPEPC-C4) (Fig. 1). It has long been known that, at physiological pH, ZmPEPC-C4 is activated by phosphorylated trioses and hexoses, particularly d-glucose 6-phosphate (G6P) (7–13), by its substrate PEP in its free form, i.e. not complexed with Mg2+ ions (13, 14), as well as by the neutral amino acids glycine, serine, and alanine (8–11, 15–19). In addition, it is allosterically inhibited by l-malate (20). Whereas the activation by G6P and the inhibition by l-malate or l-aspartate are common to most PEPC enzymes biochemically characterized to date, the activation by neutral amino acids has only been found in PEPC-C4 isozymes from monocot grasses (Poaceae family) (8, 9, 11, 15, 16, 19). As exemplified by ZmPEPC-C4, the activation by neutral amino acids of the PEPC-C4 isozymes from grasses appears to be crucial for their activity under conditions close to those assumed to be physiological given that malate inhibition is significantly reverted by these compounds but not by G6P (18). The structural basis of these kinetic differences between monocot and eudicot PEPC-C4 isozymes is not known at present.
Figure 1.
Schematic representation of the allosteric regulation of ZmPEPC-C4. In C4 plants, the first carboxylation reaction that incorporates the atmospheric CO2 in an organic compound is catalyzed by PEPC-C4 isozymes located in the mesophyll cells. The product of this reaction, oxaloacetate, is then reduced to malate, as in maize shown here, or converted to aspartate in other plants; both compounds are important allosteric inhibitors of most PEPC enzymes. During the illumination period, l-malate accumulates to high concentrations in the mesophyll cells from where it passively diffuses to the bundle-sheath cells where it is decarboxylated to yield CO2, which is then incorporated in the Calvin-Benson cycle by Rubisco, and pyruvate, which moves back to the mesophyll cells, regenerating PEP. The allosteric activators trioses-P and hexoses-P are mainly produced in the bundle-sheath cells from the 3-phosphoglycerate (PGA) formed in the Rubisco-catalyzed reaction but also may be produced in mesophyll cells given that 3-phosphoglycerate may diffuse from the bundle-sheath cells. Neutral amino acids (glycine and serine) are intermediates of the photorespiration pathway; they may also be transported to the mesophyll cell where they activate PEPC-C4 isozymes from monocot grasses, thus counteracting malate inhibition.
The location in the three-dimensional structure of different PEPC enzymes of the allosteric sites for carboxylic acids and for phosphorylated sugars was predicted by site-directed mutagenesis and confirmed by X-ray crystallography (for a review, see Ref. 6). To understand the mechanism underlying allosteric regulation, it is of pivotal importance to identify the protein residues that interact with the allosteric effector. The allosteric site for malate was unequivocally identified in the three-dimensional structure of the PEPC-C4 isoenzyme from Flaveria trinervia (FtPEPC-C4), which was crystallized bound to an aspartate molecule (Ref. 21; Protein Data Bank code 3ZGE). This allosteric site appears to be highly conserved in PEPC enzymes because that of FtPEPC-C4 is very similar to the allosteric site previously found in the PEPC enzyme from Escherichia coli (Refs. 22–24; Protein Data Bank codes 1QB4, 1FIY, and 1JQN). The ZmPEPC-C4 allosteric site for G6P was proposed to be located between the two monomers of a dimeric unit of the native tetrameric PEPC structure because a sulfate anion was found bound at this site (24). This suggested location was in agreement with previous site-directed mutagenesis studies on residues assumed to be involved in binding G6P (25, 26). Moreover, this is a conserved region in other plant PEPCs as shown in the crystal structures of the PEPC-C3 isozymes from Flaveria pringlei (Ref. 21; Protein Data Bank code 3ZGB) and Arabidopsis thaliana (Protein Data Bank code 5FDN). However, Schlieper et al. (27) have recently questioned the relevance of this site for activation. With regard to the allosteric site for neutral amino acids, its existence has been predicted on the basis of biochemical data, which indicate that it is distinct from the allosteric site for G6P (11, 13, 17, 18, 28), but its position remains unknown. Site-directed mutagenesis experiments on ZmPEPC-C4 suggested that it might be formed by residues of the monomer–monomer interface of the dimeric units of the native tetramer, contiguous to the proposed G6P allosteric site (26), and by residues in a loop at the C-terminal region of the protein (29).
With the aim of unequivocally locating and characterizing the allosteric site for neutral amino acids and to determine the structural differences between monocot and eudicot PEPC-C4 isozymes accounting for their kinetic differences, especially in regard to their sensitivity to neutral amino acid activation, we obtained a new crystal structure of the ZmPEPC-C4 isozyme which showed, for the first time, a glycine molecule bound to its allosteric site at the predicted monomer–monomer interface, close to the proposed G6P allosteric site. In addition, we found that a serine residue (Ser-100), which was not previously identified by site-directed mutagenesis and does not directly interact with the bound glycine, is critical for allowing binding of the neutral amino acids, whereas a lysine at this position, as found in the FtPEPC-C4 isozyme would impede their binding. Accordingly, the ZmPEPC-C4 S100K mutant was insensitive to activation by neutral amino acids. Multiple alignment studies of the known PEPC-C4 amino acid sequences indicated that a serine or glycine is present at this position in PEPC-C4 isozymes from the Poaceae family, but a lysine is present at this position in the isozymes from the monocot sedges (Cyperaceae family) and eudicots families, thus explaining the reported lack of activation by neutral amino acids of eudicot PEPC-C4 isozymes. In addition, Ser-100 appears to play an important role in determining other kinetic properties of ZmPEPC-C4 and presumably of every PEPC-C4 from grasses, different from those of the eudicot PEPC-C4 isozymes.
Results
Crystallographic evidence of the allosteric site for neutral amino acids in ZmPEPC-C4
The structure of the recombinant ZmPEPC-C4 in complex with glycine was determined at 3.3 Å resolution. The atomic coordinates and structure factors have been deposited in the Protein Data Bank under accession code 5VYJ. All data collection and refinement statistics of the structure are summarized in Table 1.
Table 1.
Crystallographic data collection and refinement statistics of ZmPEPC-C4 in complex with Gly
APS, Advanced Photon Source; r.m.s., root mean square.
| Data collectiona | |
| X-ray source | 23-ID B, APS |
| Wavelength (Å) | 0.9793 |
| Space group | P212121 |
| Cell dimensions | |
| a, b, c (Å) | 156.95, 167.24, 242.90 |
| α, β, γ (°) | 90, 90, 90 |
| Rmerge (%) | 12.2 (43.7) |
| I/σ (I) | 11.3 (2.8) |
| Completeness (%) | 93.28 (62.19) |
| Unique reflections | 90,116 (5,966) |
| Redundancy | 3.9 (2.4) |
| Refinement | |
| Resolution range (Å) | 57.1–3.3b (3.4–3.3) |
| Rwork/Rfree (%) | 21.0/25.3 |
| No. of atoms | |
| Protein | 29,265 |
| Glycine | 25 |
| Acetate | 52 |
| Average B-factors (Å2) | |
| Protein | 27.50 |
| Glycine | 12.75 |
| Acetate | 22.77 |
| r.m.s. deviations from ideal stereochemistry | |
| Bond lengths (Å) | 0.007 |
| Bond angles (o) | 0.98 |
| Ramachandran plot | |
| Most favored regions (%) | 94.30 |
| Allowed regions (%) | 5.02 |
| Protein Data Bank code | 5VYJ |
a Values in parentheses correspond to the highest resolution shell.
b Diffracted anisotropically to a resolution of 3.7, 3.3, and 2.7 Å along a*, b*, and c*, respectively. Data were anisotropically truncated and scaled with the Diffraction Anisotropy Server (http://services.mbi.ucla.edu/anisoscale) (Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party-hosted site.).
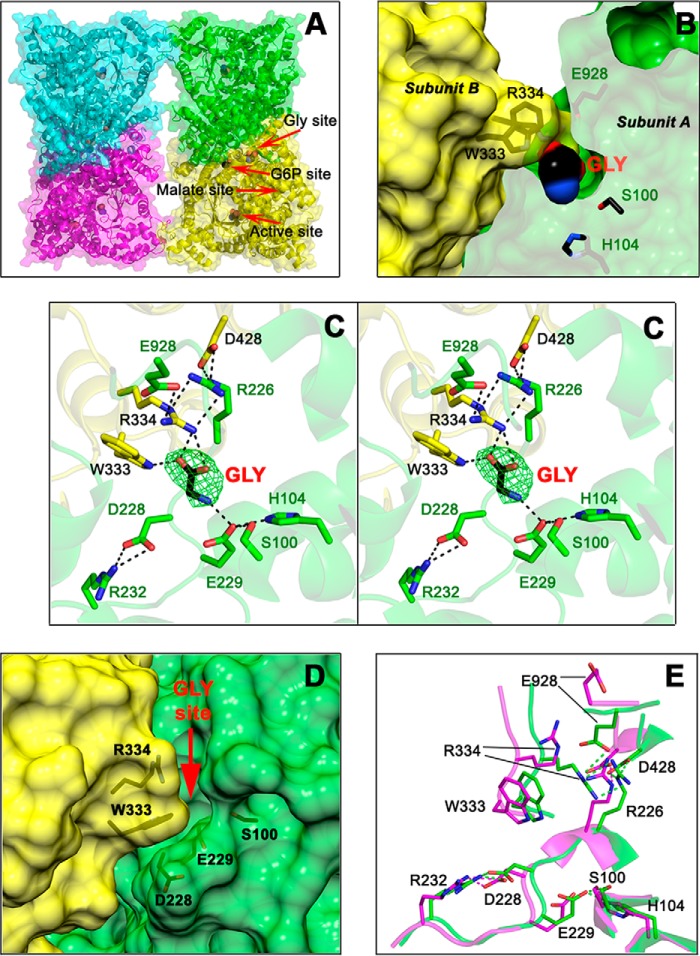
This new crystal structure consists of a αβ-barrel with eight central β-strands surrounded by at least 44 α-helices (Fig. 2A), very similar to the other known PEPC three-dimensional structures, including those of the enzyme from E. coli (22–24; Protein Data Bank codes 1QB4, 1FIY, and 1JQN), the previously reported ZmPEPC-C4 crystal structure (Ref. 24; Protein Data Bank code 1JQO), the FtPEPC-C4 isozyme (Refs. 21 and 27; Protein Data Bank codes 4BXH, 4BXC, and 3ZGE), and the C3 isozymes from F. pringlei (FpPEPC-C3; Ref. 21; Protein Data Bank code 3ZGB) and Arabidopsis thaliana (Protein Data Bank code 5FDN). The asymmetric unit of the crystal reported here contained the four subunits that constitute the biological unit of the enzyme, which can be considered a dimer of dimers, thus allowing the observation of the dimer–dimer interface for the first time. The asymmetric units of the previously reported PEPC crystals were either a dimer (1JQO) or two monomers belonging to different dimeric units of the tetramer (1QB4, 1JQN, 4BXH, 4BXC, 3ZGE, 3ZGB, and 5FDN). Also, for the first time, in each monomer of the 5VYJ crystal, a clear electron density was observed in crevices that open to the solvent at the monomer–monomer interface of the dimeric units of the tetramer, adjacent to the G6P allosteric site. A glycine molecule with occupation of 1 accounted for the extra electronic density as indicated by a simulated annealing omit map Fo − Fc at 3.0 σ (Fig. 2, B and C). Modeling other low-molecular-weight compounds present in the crystallization medium, glycerol, acetate, or ethylene glycol, gave no satisfactory results (Fig. S1).
Figure 2.
The allosteric-site for neutral amino acids of ZmPEPC-C4. A, cartoon and surface representations of the fold and secondary structure elements of the tetrameric ZmPEPC-C4 in complex with glycine. Red arrows indicate the position of the allosteric site for neutral amino acids with a glycine bound, of the active site with an acetate bound, of the putative allosteric site for G6P also with an acetate bound, and of the empty allosteric site for l-malate. B, transverse section of a dimeric unit showing the glycine molecule bound inside its allosteric site. Residues are colored green or yellow depending on the monomer. C, stereoview showing the residues and interactions of the allosteric site for neutral amino acids. The simulated annealing omit map (Fo − Fc) of the bound glycine molecule is shown contoured at 3.0 σ level (green mesh). D, surface representation of the monomer–monomer interface showing the crevice of the neutral amino acid allosteric site as seen in the previously reported 1JQO ZmPEPC-C4 crystal structure where this site is empty and open. The red arrow marks the position of the glycine activator in the 5VYJ ZmPEPC-C4 crystal structure reported here. E, differences in the conformation of critical residues of the allosteric site for amino acids between the 5VYJ ZmPEPC-C4 in complex with glycine (green carbon atoms) and the 1JQO ZmPEPC-C4 structure (magenta carbon atoms). Hydrogen bonds are depicted as black dashed lines (C) or green and magenta (E); cutoff is 3.0 Å. In B, C, and E, the side chains of relevant protein residues are shown as sticks with oxygen atoms in red, nitrogen in blue, and carbons in green, yellow, or magenta. Glycine and acetate molecules are depicted as spheres colored similarly but with black carbons. A, C, and E were generated using PyMOL. B and D were generated using the UCSF Chimera package (52).
The glycine molecule makes hydrogen bonds through its α-carboxyl group with the side chains of Arg-334 and Trp-333 of one subunit and through its α-amino group with the side-chain carboxyl group of Glu-229 of the neighboring subunit. Glu-229 in turn is hydrogen-bonded to the side chains of Ser-100 and His-104 of its own subunit. In addition to its binding to the α-carboxyl of the bound glycine, Arg-334 participates in a web of interactions with side chains of residues of the opposing subunit that contribute to the stabilization of the monomer–monomer interface. Of particular relevance appears to be interactions of Arg-334 with Glu-928 and Arg-226, which is in turn bound to the side-chain carboxyl of Asp-428 of the same subunit as Arg-334. In the ZmPEPC-C4 5VYJ crystal structure, Arg-334 and Glu-928 close the allosteric site, acting as a lid that should be open to allow the entrance of the amino acid activator. The “open” conformation of this allosteric site can actually be observed in the previously reported 1JQO ZmPEPC-C4 structure as shown in Fig. 2D in which no activator molecule is bound. In the empty allosteric site for neutral amino acids of the 1JQO crystal there is free access of the amino acid activator due to the “outside” conformations of both Arg-334 and Glu-928. The high B-factor values of the side chains of these two residues in the 1JQO structure (96 for Arg-334 and 132 for Glu-928, much higher than the average B-factors values) compared with those in the 5VYJ crystal (22 and 20, respectively, similar to the average B-factors value) indicate their mobility in the absence of the activator glycine and the flexibility of this region needed to allow the entry and binding of the activator in the allosteric site. Indeed, the conformation of the residues surrounding the α-amino group of glycine is similar in both structures except that of Arg-334, whose side chain moves about 5 Å from a position exposed to the solvent in the 1JQO structure toward the α-carboxyl group of glycine in the 5VYJ structure reported here (Fig. 2E). This movement, most likely induced by the binding of glycine, makes room for the side chain of Glu-928, whose carboxyl group also moves about 11 Å from its position in the protein surface toward the guanidinium group of Arg-334 of the neighboring subunit, as shown in Fig. 2E. Also, in the 5VYJ structure, the imidazole ring of His-53 rotates about 25° from its position in the 1JQO structure, interacting with Glu-928 of its own subunit (not shown in the figure for clarity). Therefore, the binding of glycine significantly contributes to the stabilization of the monomer–monomer interface of the ZmPEPC-C4 isozyme, not only by bridging the two monomers through the hydrogen bonds made by its α-amino and α-carboxyl groups but also by promoting the formation of several hydrogen bonds not present when the activator is not bound. This may account, at least in part, for the activation effect of glycine.
The 5VYJ crystal reported here also shows how the allosteric site for neutral amino acids and the proposed allosteric site for G6P are connected in the ZmPEPC-C4 isozyme, thus explaining the synergistic effects of both kinds of allosteric activators observed before (11, 12, 17, 18). This connection is established by Asp-228, whose side-chain carboxyl group makes hydrogen bonds with the side chain of Arg-232 (Fig. 2C), a conserved residue in PEPC-C4 isozymes (see below). This is in agreement with the previous observations that the affinity for glycine decreased when Asp-228 was changed to an asparagine and that the affinity for both glycine and G6P decreased when Arg-232 was changed to a glutamine (26). Therefore, the interactions of Asp-228 with Arg-232 appear to play an important role in the activation by both kinds of activators, neutral amino acids and G6P.
To explore whether l-serine or l-alanine, the other two neutral amino acids that have been reported as activators of ZmPEPC-C4 (11, 15, 18), could also be bound in the same site as Gly as well as to learn the possible mode of their binding, we constructed in silico energy-minimized models with l-alanine or l-serine docked in this region. We also docked d-serine and d-alanine inside the Gly-binding site given that they were also found to be activators of the ZmPEPC-C4 isozyme (11). In the models, shown in Fig. S2, the side chains of l/d-alanine or l/d-serine fit well into this site without producing any steric clash, and their α-carboxyl and α-amino groups interact with protein residues in the same manner as do the glycine groups in the crystal structure reported here. The results of these simulations support that the site where glycine was observed bound in the 5VYJ crystal is indeed the allosteric site for neutral amino acids.
Comparison of ZmPEPC-C4–Gly and FtPEPC-C4 crystal structures
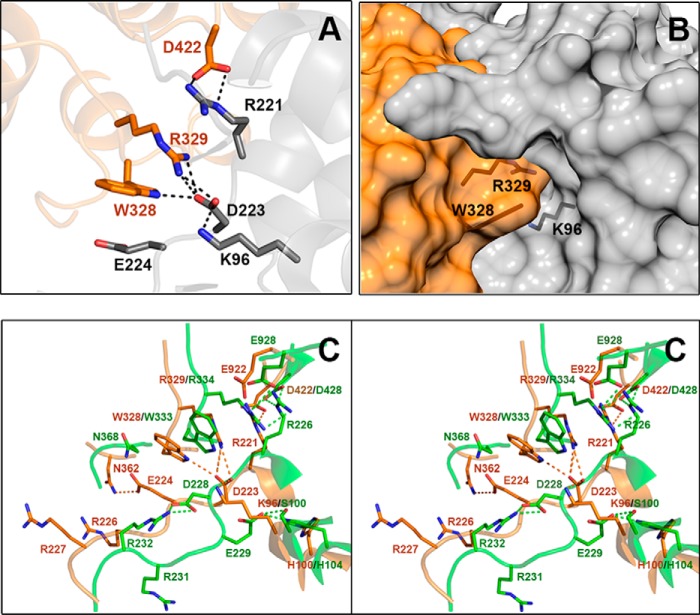
To determine the structural differences between the PEPC-C4 isozymes that are allosterically activated or nonactivated by neutral amino acids, we compared the region of the allosteric site for neutral amino acids of the 5VYJ crystal structure with the equivalent region in the reported crystal structures of FtPEPC-C4. Despite that every residue that interacts with glycine in the 5VYJ ZmPEPC-C4 crystal structure is conserved in the FtPEPC-C4 isozyme (although some of them have a different conformation), we observed a critical change in a nearby residue that does not make a direct interaction with the bound glycine: Ser-100 in the maize enzyme is changed to a lysine (Lys-96) in the Flaveria enzyme. Interestingly, the side-chain amino group of Lys-96 occupies a position similar to that of the α-amino group of the bound glycine in the ZmPEPC-C4 5VYJ crystal (Fig. 3A), but it is hydrogen-bonded to the carboxyl group of Asp-223 (equivalent to Asp-228 in the maize enzyme) instead of being bonded to the carboxyl group of Glu-224 (equivalent to Glu-229 in the maize enzyme). Moreover, the side-chain carboxyl of Asp-223 has a position comparable with that of the α-carboxyl of the glycine bound in the maize enzyme and forms hydrogen bonds with the same tryptophanyl (Trp-328 in FtPEPC-C4 equivalent to Trp-333 in ZmPEPC-C4) and arginyl (Arg-329 in FtPEPC-C4 equivalent to Arg-334 in ZmPEPC-C4) residues as the activator molecule in the maize enzyme. Consequently, in the Flaveria PEPC-C4 crystals, the neutral amino acid allosteric site does not exist because the side chains of Lys-96 and Asp-223 totally fill up the cavity where glycine binds in the ZmPEPC-C4 5VYJ crystal (Fig. 3B). The different position of Asp-223 in the FtPEPC-C4 crystals with respect to that of the equivalent Asp-228 in the two known ZmPEPC-C4 structures is due to a −95° change in the ψ angle of this aspartate, resulting in the movement of its side chain toward the region equivalent to the neutral amino acid allosteric site. Also, in the FtPEPC-C4 crystals, Glu-224 (equivalent to Glu-229 in the maize enzyme) has a different conformation than in the ZmPEPC-C4 crystals. Its side chain is outside the Gly-binding site, interacting with Asn-362 of the opposing subunit (Fig. 3C), whereas in the maize crystals the asparagine at the equivalent position (Asn-368) is exposed to the solvent. Interestingly, the interactions between the activator molecule and protein residues that stabilize the dimer–dimer interface in the region of the neutral amino acid allosteric site of the maize enzyme are made between protein residues in the Flaveria enzyme. Finally, the side chain of Glu-922 of FtPEPC-C4 (equivalent to ZmPEPC-C4 Glu-928, a residue that interacts with Arg-334 in the 5VYJ crystal) is only observed in two of the FtPEPC-C4 crystals where it is exposed to the solvent (Fig. S3).
Figure 3.
Region of FtPEPC-C4 equivalent to the allosteric-site for neutral amino acids in ZmPEPC-C4. A, cartoon representation showing that Lys-96 (equivalent to Ser-100 of ZmPEPC-C4) interacts with Asp-223 (equivalent to Asp-228 of ZmPEPC-C4), which in turn is hydrogen-bonded to Arg-329 (equivalent to Arg-334 of ZmPEPC-C4) and Trp-328 (equivalent to Trp-333 of ZmPEPC-C4). Amino acid side chains are depicted as sticks with carbon atoms in orange or gray (depending on the monomer of the dimeric unit), oxygen atoms in red, and nitrogen atoms in blue. B, surface representation of the monomer–monomer interface of the FtPEPC-C4 dimer, showing that the allosteric site for neutral amino acids does not exist because the position of the above mentioned residues does not leave a cavity between the two monomers. The side chains of relevant protein residues are shown as sticks with oxygen atoms in red, nitrogen in blue, and carbons in black. Of the three reported crystal structures of FtPEPC-C4, the one shown here is Protein Data Bank code 3ZGE. C, differences in the conformation of critical residues of the allosteric site for amino acids between the 5VYJ ZmPEPC-C4 (green carbon atoms) and FtPEPC-C4 3ZGE (orange carbon atoms) crystal structures. Hydrogen bonds are depicted as dashed lines, black in A and green (maize isozyme) or orange (Flaveria isozyme) in C; cutoff is 3.0 Å. A and C were generated using PyMOL, and B was generated using the UCSF Chimera package (52).
Biochemical evidence supporting the identification of the allosteric site for neutral amino acids in the ZmPEPC-C4–Gly crystal structure
To prove the critical role of the residue at position 100 of ZmPEPC-C4 in its allosteric activation by neutral amino acids, we substituted Ser-100 with a lysine by site-directed mutagenesis and compared the kinetics of activation by neutral amino acids of the S100K mutant with those of the WT ZmPEPC-C4 enzyme (Fig. 4). The assays were conducted at pH and concentrations of substrates believed to be close to those prevailing in vivo under illumination conditions (pH 7.4, 0.1 mm bicarbonate, 3 mm PEP, 0.4 mm free Mg2+, 20 mm l-malate) as discussed previously by Tovar-Méndez et al. (18). As shown in Fig. 4, A and B, glycine greatly increases the WT recombinant enzyme activity even in the presence of the high l-malate concentrations characteristic of the day period in a manner similar to that observed in the enzyme purified from maize leaves (18). In contrast, glycine was not able to activate the S100K mutant either in the absence or presence of the inhibitor, thus confirming the importance of the size and charge of the residue at position 100 for the amino acid activator to bind to its allosteric site. Neither was the S100K mutant enzyme activated by l-serine and l-alanine (Fig. 4, C and D). The kinetic parameters estimated from the neutral amino acid saturation data are given in Table 2.
Figure 4.
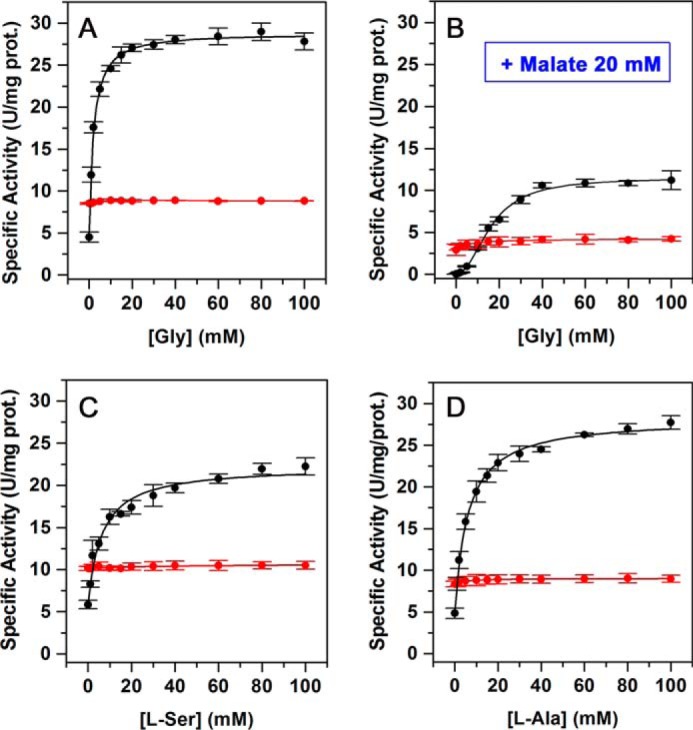
Loss of response to neutral amino acids of the ZmPEPC-C4 S100K mutant. A and B, saturation by Gly in the absence (A) or presence of 20 mm l-malate (B) of the WT and S100K mutant enzymes. C and D, saturation by l-Ala and l-Ser, respectively, of the WT and S100K mutant enzymes. Initial velocity data are depicted as black circles (WT) or red circles (S100K mutant). Activity assays were carried out at pH 7.4 and 30 °C, 3 mm total PEP, 0.4 mm free Mg2+, and 0.1 mm bicarbonate; 20 mm l-malate was included in the assay medium where indicated. The results are mean values from two independent experiments. The points in the figures are the experimentally determined values, whereas the curves are calculated from fits of these data to Equation 1 given under “Experimental procedures.” Error bars represent S.E.; prot., protein.
Table 2.
Apparent kinetic parameters of ZmPEPC-C4 wildtype and mutant S100K enzymes
| Parametera | Wildtype | S100K |
|---|---|---|
| Saturation by Gly | ||
| v0 | 4.5 | 8.5 |
| vamax | 24.3 ± 0.3 | NE |
| A0.5 | 2.0 ± 0.1 | NE |
| Saturation by Gly + 20 mm malate | ||
| v0 | 0.02 | 2.9 |
| vamax | 11.5 ± 0.2 | NE |
| A0.5 | 16.3 ± 0.6 | NE |
| Saturation by l-Ala | ||
| v0 | 4.8 | 8.3 |
| vamax | 23.4 ± 0.3 | NE |
| A0.5 | 5.9 ± 0.3 | NE |
| Saturation by l-Ser | ||
| v0 | 5.8 | 10.1 |
| vamax | 16.4 ± 0.6 | NE |
| A0.5 | 6.2 ± 0.9 | NE |
a Values ±S.D. were estimated by the best fit to Equation 1, fixing the h value to 1 and v0 to the experimentally determined values. Velocities (v0 and va max) are given as units/mg of protein, and A0.5 is given as mm. Assays were carried out at pH 7.3, 0.4 mm free Mg2+, 0.1 mm bicarbonate, and 3 mm total PEP. NE, values not estimated due to the absence of activation.
The ZmPEPC-C4 S100K mutant also showed other significant kinetic differences from the ZmPEPC-C4 WT (Fig. 5 and Table 3). These differences mainly consist in a higher affinity for the substrate and activator (Fig. 5, A and B) and a lower affinity for the inhibitor (Fig. 5C). Interestingly, G6P counteracted the inhibition by l-malate of the S100K mutant (Fig. 5D), whereas it did not in the WT enzyme as reported previously (18). Thus, the residue at position 100 of ZmPEPC-C4 appears to be determinant not only of its activation by neutral amino acids but also of its general kinetic behavior. Interestingly, the global kinetic properties of the S100K mutant resemble more those of the PEPC-C4 isozymes from eudicot plants (19) than of the WT ZmPEPC-C4. These kinetic differences could not be attributed to changes in the association state, folding, or stability of the mutant enzyme. Both the mutant and WT recombinant enzymes were obtained in the soluble fraction of the E. coli cells extracts with a similar yield, indicating that both recombinant proteins were properly folded. Also, the native structure of both proteins is tetrameric (Fig. S4A), and both exhibit a similar thermal stability in thermal-shift experiments with apparent transition temperatures (Tm) of 48.8 °C for the WT and 47.75 °C for the S100K mutant (Fig. S4B).
Figure 5.
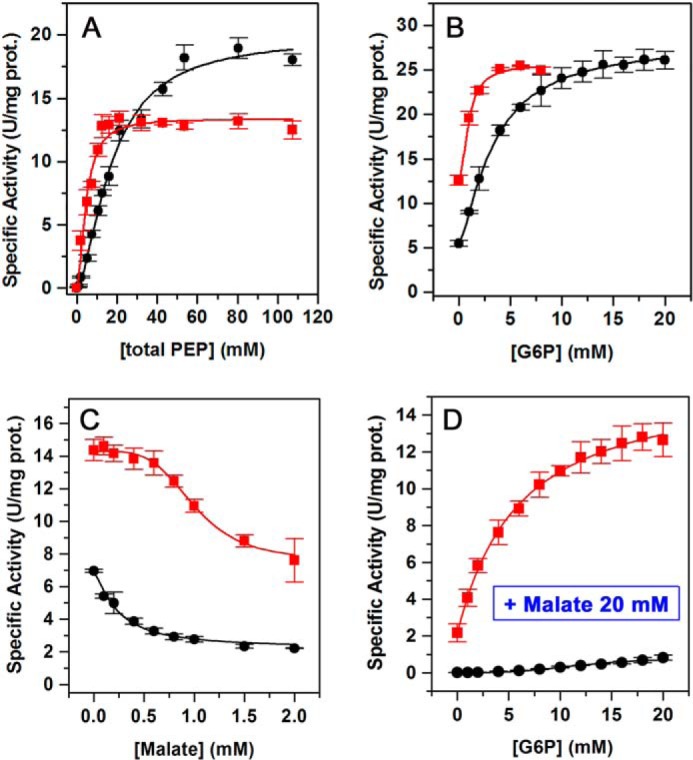
Kinetics of the ZmPEPC-C4 S100K mutant. A, saturation by total PEP. B, saturation by G6P. C, saturation by malate. D, saturation by G6P in the presence of 20 mm l-malate. Initial velocity data are depicted as black circles (WT) or red squares (S100K mutant). Experimental conditions were the same as those given in the legend of Fig. 4. The initial velocity data were fitted to Equation 1, 2, or 3 as appropriate. Error bars represent S.E.; prot., protein.
Table 3.
Apparent kinetic parameters of ZmPEPC-C4 wildtype and mutant S100K enzymes
| Parametera | Wildtype | S100K |
|---|---|---|
| Saturation by total PEP | ||
| Vmax | 19.9 ± 0.8 | 13.4 ± 0.4 |
| S0.5 | 17.5 ± 1.3 | 4.6 ± 0.5 |
| h | 1.6 ± 0.1 | 1.8 ± 0.3 |
| Saturation by G6Pb | ||
| v0 | 5.4 | 12.6 |
| vamax | 22.7 ± 0.2 | 13.1 ± 0.8 |
| A0.5 | 3.4 ± 0.1 | 0.9 ± 0.1 |
| h | 1.4 ± 0.0 | 1.8 ± 0.5 |
| Saturation by G6P + 20 mm malateb | ||
| v0 | 0.01 | 2.2 |
| va max | NE | 13.9 ± 0.7 |
| A0.5 | NE | 5.9 ± 0.7 |
| h | NE | 1.0 ± 0.0 |
| Saturation by malateb | ||
| v0 | 6.9 | 14.4 |
| vi min | 2.21 | 7.6 |
| I50 | 0.23 ± 0.0 | 1.0 ± 0.01 |
| h | 1.3 ± 0.1 | 4 |
a Values ±S.D. were estimated by the best fit to Equation 2 for saturation by total PEP, to Equation 1 for saturation by G6P in the absence and presence of malate, or to Equation 3 for saturation by malate; v0 was fixed to the experimentally determined values. Velocities (v0, Vmax, va max, and vi min) are given as units/mg of protein and S0.5, A0.5, and I0.5 are given as mm. Assays were carried out at pH 7.3, 0.4 mm free Mg2+, and 0.1 mm bicarbonate.
b The apparent kinetic parameters for the allosteric effectors were determined at fixed 3 mm total PEP. NE, values not estimated due to the low enzyme activity under these conditions.
Residue conservation at the allosteric site for neutral amino acid in plant PEPC-C4 isozymes
Because PEPC-C4 isozymes from monocots are activated by neutral amino acids, whereas isozymes from eudicots are not, we considered it of interest to investigate the degree of conservation at the amino acid positions of the allosteric site for neutral amino acids found in the ZmPEPC-C4 crystal structure (Protein Data Bank 5VYJ). Using the proposed criterion to identify PEPC-C4 isozymes, that is the presence of serine at a position equivalent to 774 of the FtPEPC-C4 isozyme (30) or equivalent to position 780 of the ZmPEPC-C4 isozyme, a total of 172 nonredundant protein sequences (allelic forms excluded) were initially identified as PEPC-C4 in the RefSeq protein database of the National Center for Biotechnology Information (NCBI), and two were identified in the Phytozome version 12.1.5 database. Of these, only 138 sequences are from C4 plants (118 from monocots and 20 from eudicots), and thus we consider them to be true PEPC-C4 isozymes (Table S1); the rest were from eudicot C3 plants (three sequences) or CAM plants (31 sequences) (Table S2). The latter finding is consistent with the proposed shared origin of some C4 and CAM PEPC isozymes (31). Another PEPC sequence that possesses a Ser at position 780 is that encoded by the gene EEF27881 (GenBankTM accession number) from Ricinus communis; it is a bacterial-like PEPC, similar to those previously found in Arabidopsis and rice (32), and therefore was not included in our analysis. So far, the only known sequence for a PEPC-C4 isozyme from a C4 plant from the Hydrocharitaceae family (that from Hydrilla verticillata) does not have a serine at this position. Hence, the presence of Ser-780 is not a sufficient criterion to distinguish PEPC-C4 from other PEPC isozymes as previously pointed out by others (33).
Although our sequence analysis was restricted because most of the reported PEPC-C4 sequences were partial, lacking the N- and/or C-terminal regions (only 13 sequences from monocots and five sequences from eudicots were complete), and in the case of eudicots only sequences from four of the 14 families known to contain at least one C4 member have been reported to date, after multiple amino acid sequences alignments, we prepared sequence logos of selected residues of the monocot and eudicot sequences showing their degree of conservation (Fig. S5). Of the 16 PEPC-C4 sequences from grasses that show the residue at position 100, in 11 of them this residue is serine, as in ZmPEPC-C4, and in the other 5 the residue at this position is glycine, which is a conservative change from the point of view of size and charge. The H. verticillata PEPC-C4 sequence also has a serine at the position equivalent to Ser-100 of ZmPEPC-C4, but in the Hydrilla enzyme the possible activation by neutral amino acids has not been tested. The two PEPC-C4 sequences reported from sedges that include this position have lysine, similar to the only eight PEPC-C4 sequences from different eudicot families that include the residue at this position (Fig. S5A). The PEPC-C4 isozymes from sedges have not been biochemically characterized yet, so we do not know whether they are activated by neutral amino acids, although on the basis of the presence of lysine at position 100 we would predict that they are not. In regard to the PEPC-C4 isozymes from eudicots, those studied in this respect (Amaranthus retroflexus, Amaranthus tricolor, Amaranthus hypochondriacus, and Portulaca oleracea) are insensitive to neutral amino acids (15, 19). The evolutionary conservation of these two different kinds of residues, small and neutral or large and positively charged, at this critical position strongly supports the importance of this trait. We also found that the residues directly involved in the binding of the activator glycine (Glu-229, Trp-333, and Arg-334; ZmPEPC-C4 numbering) are absolutely conserved in both monocot and eudicot PEPC-C4 sequences (Fig. S5B). In regard to the residues involved in the web of hydrogen bonds that stabilize the conformation of the neutral amino acid allosteric site, His-104, Arg-226, Asp-428, Glu-928 (residues that interact with those that bind the activator molecule), Asp-228, and Arg-232 (the residue that links the neutral amino acid and the proposed G6P allosteric sites as mentioned above) also are totally conserved or conservatively changed in the case of His-104 (changed to asparagine in some monocots or to glutamine in some eudicots sequences) and of Glu-928 (which is aspartate in some of the eudicot sequences) (Fig. S5C).
The conservation of other ZmPEPC-C4 residues that have been related to the allosteric site for neutral amino acids by previous site-directed mutagenesis studies (Lys-927, Glu-932, Lys-934, Gly-937, and Lys-940) (29) as well as the two Pro residues (Pro-915 and Pro-949) that flank the highly flexible loop in which these residues are included was also investigated (Fig. S5D). The polar nature of the charged residues is highly conserved, and the two prolines are absolutely conserved, suggesting that this exposed loop exists in every PEPC-C4 isozyme and that the flexibility of the loop flanked by these prolines is important for their activity and/or allosteric regulation. The most conspicuous change is that of Gly-937, which is the most frequent residue in the PEPC-C4 monocot sequences, although some have aspartate or glutamate, and is glutamate and less often aspartate in the eudicot PEPC-C4 sequences.
Discussion
Identification of the allosteric site for neutral amino acids
To understand the mechanism underlying allosteric regulation, it is of pivotal importance to identify the protein residues that interact with the allosteric effectors. The crystal structure of ZmPEPC-C4 in complex with glycine reported here reveals the position in the three-dimensional structure and the structural details of the allosteric site for neutral amino acids, which remained unknown. The comparison of ZmPEPC-C4 5VYJ crystal structure with that of the FtPEPC-C4 isozyme already known indicated that the main structural reason for the lack of activation by amino acids in PEPC-C4 isozymes from eudicot plants is the presence of a lysine at position 100 of the maize enzyme instead of the serine or alanine residue that the isozymes from monocots have. The crystallographic data show that there is no room for the binding of the neutral amino acids in the Flaveria enzyme because of the large and positively charged side chain of this lysine and the interactions it makes with a nearby aspartate residue conserved in both monocot and eudicot PEPC-C4 isozymes. In fact, the cavity between dimers that forms the allosteric site for neutral amino acids in the ZmPEPC-C4 does not exist in the FtPEPC-C4 enzyme. Moreover, despite the scarce number of complete sequences reported to date, the conservation of serine at position 100 in the PEPC-C4 sequences from grasses and the presence of lysine at the equivalent position in those from sedges and eudicots strongly support the critical role played by this residue in determining the different susceptibility to activation by neutral amino acids of these two kinds of PEPC-C4 isozymes, which was confirmed by the lack of activation by these compounds of the ZmPEPC-C4 S100K mutant. Because other kinetic properties were also significantly affected in this mutant, we propose that in this respect there are two different kinds of PEPC-C4 isozymes: the Ser/Gly-100 type or the Lys-100 type. The available PEPC-C4 sequences indicate that of the three monocot families that include species with C4 metabolism (Cyperaceae, Hydrocharitaceae, and Poaceae) the isozymes from plants of the Poaceae family (grasses), which are economically the most important, and probably the only PEPC-C4 from the Hydrocharitaceae family are of the Ser/Gly-100 type, whereas those from plants of the Cyperaceae family (sedges) are of the Lys-100 type. Likewise, PEPC-C4 sequences from eudicot plants appears to be of the Lys-100 type according to the absolute conservation of this residue in the known sequences that show this position.
Ser-100 was not identified as an important residue for glycine activation in the previously reported site-directed mutagenesis studies, which concluded that Arg-226, Asp-228, Glu-229, and Arg-232 are the important residues (26). The ZmPEPC-C4 5VYJ crystal structure reported here with a glycine molecule bound shows that Glu-229 actually participates in the binding of the activator molecule and that the other three residues are involved in maintaining the proper conformation of the allosteric site by means of a web of hydrogen bonds (Fig. 2C). Later, it was suggested that Lys-927 and Gly-937 form the glycine-binding site of the maize enzyme and that other charged residues in the loop to which they belong are also important for binding of the activator (29). However, the ZmPEPC-C4 5VYJ crystal structure reported here shows that neither Lys-927 nor Gly-937 participate in the binding of the activator. The residues mentioned in the referenced work belong to an exposed loop that extends from Pro-915 to Pro-949 in ZmPEPC-C4 and from Pro-909 to Pro-945 in FtPEPC-C4. This loop is so flexible that a great part of it could not be observed in the reported crystal structures (Fig. S3A). Lys-927 is exposed to the solvent in every plant PEPC structure so far reported, and Lys-940 is also exposed to the solvent in the maize crystal structures but is not observed in the Flaveria crystal structures. The residues equivalent to Glu-932 and Lys-934 are not observed in any of the reported PEPC-C4 crystal structures, whereas the residue at the position equivalent to Gly-937 is not observed in the FtPEPC-C4 crystal structures. Gly-937 was proposed to be critical for the ability of monocot PEPC-C4 isozymes to be activated by neutral amino acids in contrast to the eudicot isozymes because it was assumed that the presence of glycine at this position is a distinct feature of monocot PEPC-C4 isozymes that distinguishes them from the eudicot PEPC-C4 isozymes where there is an aspartate or glutamate at this position (29). However, our sequence analysis indicated that not all monocot sequences have Gly-937; some of them, even of the Poaceae family, have aspartate or glutamate (Table S1). This finding suggests that the residue at position 937 is not as critical for neutral amino acid activation as is the residue at position 100. Nevertheless, to determine the possible structural reason for the reported effect that the change of Gly-937 of the maize enzyme to an aspartate had on the response to glycine (29), we performed the in silico mutations G937D and G937E (Fig. S6). Using the ZmPEPC-C4 5VYJ crystal structure with glycine bound, we found that either an aspartate or glutamate at the position equivalent to Gly-937 would be exposed to the solvent, not interacting with other protein residues in any of their possible rotamers (Fig. S6, A and B). But when we simulated the possible exposed conformations of Arg-334 and Glu-928 that could occur when the allosteric site for neutral amino acid is empty as indicated by the ZmPEPC-C4 1QJO structure, we found rotamers of Arg-334 and of Asp/Glu-937 that can interact (Fig. S6, C and D). This interaction would stabilize the exposed conformation of Arg-334, opposing its movement toward the bound activator molecule to form the critical hydrogen bond with the α-carboxyl group as well as the other hydrogen bonds with other protein residues that stabilize the activator-bound conformation of the allosteric site. Therefore, it appears that the presence of aspartate/glutamate at position 937 in PEPC-C4 isozymes from monocot plants could decrease the affinity to neutral amino acids by somehow limiting the movement of Arg-334 into the allosteric site where this residue importantly contributes to the binding of the carboxyl group of the activator. Additionally, the substitution of charged residues for neutral residues in the Pro-915 to Pro-949 loop could affect the dynamics of this loop, which in turn may negatively affect the allosteric transition induced by the binding of neutral amino acids. This dynamic effect could explain the decreased sensitivity to glycine caused by the amino acid substitutions in this loop.
Implications for the understanding of the kinetics and allosteric regulation of the PEPC-C4 isozymes
In the absence of the inhibitor l-malate, neutral amino acids activate ZmPEPC-C4 roughly to the same extent as G6P (8, 10, 11, 18), but when this enzyme is assayed in the presence of malate, glycine or serine counteract the inhibition of the ZmPEPC-C4 enzyme by l-malate to a much higher degree than G6P (17, 34). This is particularly true when the assays are carried out at conditions close to those prevailing during the day, i.e. high l-malate concentrations (35, 36) and low CO2 concentration (37), when G6P is unable to significantly revert the inhibition caused by l-malate, whereas glycine or l-serine offset this inhibition, producing an enzyme almost as active as that in the absence of the inhibitor (Refs. 18 and 19 and results in the present work shown in Fig. 4B). At first sight, this result contradicts previous reports of G6P offsetting malate inhibition of ZmPEPC-C4 isozymes (10, 17) and other PEPC-C4 isozymes such as that from Hydrilla (38) and those from eudicot plants reported by Rosnow et al. (39). In the case of the eudicot isozymes, the reports are consistent with the results obtained with the ZmPEPC-C4 S100K mutant presented here. In the case of the monocot PEPC-C4 isozymes, it has to be taken into account that these previous results were obtained at nonphysiologically high Mg2+ ion and bicarbonate concentrations, and under those experimental conditions the concentration of the preferred substrate, the complex Mg-PEP (13), when the total PEP concentration is kept at 3 mm is almost saturating, which greatly increases the affinity for the activator G6P and greatly diminishes that for the inhibitor l-malate. But under conditions near those estimated to prevail during the day, when the concentration of Mg-PEP is well below saturation, the activation by glycine and/or l-serine, not by G6P, appears to be crucial for achieving appreciable levels of ZmPEPC-C4 activity and, therefore, a significant rate of CO2 assimilation (18). Given this, we believe that it is of pivotal importance that PEPC-C4 isozymes are in vitro tested at near physiological concentrations of their substrates to correctly evaluate the role played by their allosteric effectors.
If the activation by neutral amino acids is so critical, the question arises of how do those C4 plants whose PEPC-C4 isozyme is not activated by neutral amino acids contend with l-malate inhibition. Noteworthy, the kinetic properties of the ZmPEPC-C4 S100K mutant suggest that it would be more active than the WT enzyme at low PEP concentrations in the absence of the activator regardless of the absence or presence of l-malate. Also, of particular relevance in this respect, is that G6P is able to revert to a significant extent the inhibition by malate of the S100K mutant, contrary to the WT enzyme. These kinetic properties of the S100K mutant are, at least qualitatively, more similar to those of the PEPC-C4 isozyme from the eudicot plant A. hypochondriacus (AhPEPC-C4) than to those of the WT ZmPEPC-C4. Our findings suggest that these kinetic differences between the Ser/Gly-100–type and the Lys-100–type PEPC-C4 isozymes could explain why the PEPC-C4 isozymes from eudicot plants and likely also those from sedges, although they have not been kinetically characterized to date, do not require the activation by neutral amino acids to efficiently work under physiological conditions.
Conclusion
The crystallographic and site-directed mutagenesis data reported here unequivocally identify the location of the allosteric site for neutral amino acids in the ZmPEPC-C4 three-dimensional structure and point out the importance of the residue at position 100 of ZmPEPC-C4 for neutral amino acid activation and other kinetic properties, which was confirmed by site-directed mutagenesis experiments. Residue conservation analysis supports our proposal that the nature of the residue at this position plays a similar critical role on the catalysis and allosteric properties of every PEPC-C4 isozyme. We believe that our findings are of great biotechnological interest given the current efforts for the molecular engineering of grass C3 plants, such as rice, wheat, and barley, to improve their productivity.
Experimental procedures
DNA constructs and site-directed mutagenesis
The ppc-C4 gene coding for ZmPEPC-C4 with GenBank accession number CAD60555 cloned into plasmid pTM94 was a kind gift from Prof. Izui (Kyoto University). The protein encoded by this gene differs from that encoded by the gene with GenBank accession number CAA33317 in only two amino acid residues: Pro-482 and Asp-509 in CAD60555 are Ser-482 and Glu-509, respectively, in CAA33317. These are polymorphic changes that seem not to affect either the structure or function of the PEPC-C4 isozyme. The gene was transferred from plasmid pTM94 to the pET32a(+) (Novagen) expression vector by ligating the NcoI-HindIII–digested insert into the previously linearized plasmid with the same restriction enzymes. Because this gene contains an internal NcoI restriction site, a partial NcoI digestion was used to isolate the full-length gene prior to its insertion into pET32a(+). The plasmid was purified from transformed TOP-10 E. coli cells selected by growing the cells at 100 μg/ml ampicillin and analyzed by restriction analysis, PCR, and sequencing of the insert. The ZmPEPC-C4 sequence within pET32a(+) did not contain any mutations and was in-frame to be expressed as a fusion protein with a thrombin-cleavable double tag of thioredoxin and His6 at the N terminus of the ZmPEPC-C4 protein. Once the construct was verified, it was used to transform BL21-CodonPlus(DE3)-RIL E. coli cells (Novagen) to overexpress the gene.
To generate the ZmPEPC-C4 S100K mutant, the forward and reverse primers 5′-GCCATCCTCGTGGCGAAGTCCATCCTGCAC-3′ and 5′-GTGCAGGATGGACTTCGCCACGAGGATGGC-3′, respectively, were used to substitute serine at position 100 by lysine. The ZmPEPC-C4 gene cloned into the pET32a(+) plasmid was used as the template for site-directed mutagenesis by PCR using the QuikChange II site-directed mutagenesis kit (Agilent Technologies) following manufacturer's instructions. The DNA was sequenced to confirm that the desired mutation was present and that no other nucleotide changes occurred. Finally, the mutant protein was overexpressed in BL21-CodonPlus(DE3)-RIL E. coli cells.
ZmPEPC-C4 expression, purification, and assay
Overexpression of both the recombinant WT and S100K ZmPEPC-C4 proteins was achieved by inducing cells grown in Luria-Bertani (LB) broth with 100 μg ml−1 ampicillin at an A600 of 0.6 by the addition of 0.1 mm isopropyl β-d-thiogalactoside followed by incubation for 7 h at 25 °C. Cells were harvested by centrifugation at 6,000 × g for 10 min. The pellet was suspended in 10 ml of 50 mm HEPES-KOH buffer, pH 7.5, containing 2 mm 2-mercaptoethanol, 50 mm KCl, 1 mm phenylmethanesulfonyl fluoride (PMSF), and 10% (v/v) glycerol (Buffer A) and sonicated for 20 min. Cell debris was removed by centrifugation at 15,000 rpm for 20 min. The ZmPEPC-C4 protein was purified using Protino nickel-tris(carboxymethyl)ethylene diamine resin (Macherey-Nagel, Düren, Germany) from which it was eluted with 150 mm imidazole in Buffer A. Imidazole was then removed by centrifugal concentration using Amicon Ultra 30 (Millipore). The concentrated enzyme was digested for 2 h at 25 °C with enterokinase (New England Biolabs), then applied to a Mono Q chromatography column, and eluted with a gradient from 0 to 400 mm potassium phosphate, pH 7.4. Protein concentrations were determined spectrophotometrically by A280 using a extinction coefficient of 111,730 m−1 cm−1 predicted from the amino acid sequence using ExPASy ProtParam (40).
ZmPEPC-C4 activity was assayed spectrophotometrically at 30 °C in a coupled enzymatic assay with malate dehydrogenase by monitoring NADH oxidation at 340 nm (ϵ340 = 6,220 m−1 cm−1) as described (28). The standard assay medium consisted of 100 mm HEPES-KOH buffer, pH 7.3, containing 0.1 mm NaHCO3, 0.2 mm NADH, 3 mm total PEP, 0.4 mm free Mg2+, and 5 units of malate dehydrogenase. Enzyme activity in the presence of allosteric effectors was assayed using the standard assay with the indicated effector concentrations. Each assay was performed in duplicate and with at least two different enzyme preparations. One unit is defined as the amount of enzyme needed to catalyze the formation of 1 μmol of oxaloacetate/min under our experimental conditions.
Crystallization, X-ray data collection, structure solution, and refinement
Crystals of ZmPEPC-C4 with glycine bound were grown at 18 °C by the hanging-drop vapor-diffusion method, mixing equal volumes of protein and reservoir solution, which consisted of 100 mm Tris-HCl, pH 8.5, 200 mm sodium acetate trihydrate, 100 mm potassium/sodium tartrate 4-hydrate, and 15% (w/v) PEG 4000. Protein concentration was 10 mg ml−1 in 10 mm HEPES-KOH buffer, pH 7.5, 1 mm DTT, 10 mm MgCl2, and 100 mm glycine. The crystals were cryoprotected in reservoir solution plus 20% (v/v) glycerol and cryocooled in N2. X-ray diffraction data of the ZmPEPC-C4–Gly were collected at 100 K at the Advanced Photo Source, beamline 23 ID-B (GM/CA), at the Argonne National Laboratory, Chicago, IL.
The data were integrated using XDS (41) and scaled and truncated with programs from the CCP4 suite (42). Because of anisotropy, the data set was submitted to the Diffraction Anisotropy Server (http://services.mbi.ucla.edu/anisoscale)4 (43) to perform ellipsoidal truncation and anisotropic scaling. The ellipsoidal truncated data set, with the upper resolution limit cut at 3.7, 3.3, and 2.7 Å along a*, b*, and c*, respectively, was used in later stages of the model refinement. The initial phases were obtained by molecular replacement with the program Phaser (44) using the coordinates of the reported ZmPEPC-C4 structure (Protein Data Bank code 1JQO) as a starting model. Alternating cycles of automatic and manual refinement were carried out with the standard protocols of Phenix (45), monitoring the Rwork and Rfree split during the whole process. Phenix was also used for atomic positions; group atomic displacement parameters; and translation, libration, and screw-rotation displacement. The simulated annealing omit map was calculated with Phenix following simulated annealing refinement at 3,000 K. The program Coot (46) was used to analyze the electron density maps (2Fo − Fc and Fo − Fc). Structural alignments were performed with Coot and PyMOL.
Kinetic data analysis
Kinetic data were analyzed by nonlinear regression calculations. Initial velocity data obtained varying the concentration of activator at constant concentration of substrates were fitted to Equation 1.
| (1) |
where va and v0 are the experimentally determined initial velocities in the presence and absence of activator, respectively; va max is the estimated maximum activity at saturating activator concentrations; [A] is the activator concentration; A0.5 is the concentration of activator that gives half-maximum activation at fixed concentrations of substrate; and h is the Hill number indicative of the degree of cooperativity in the binding of the activator.
Kinetic data obtained at varied concentrations of substrate PEP were fitted to a Hill equation (Equation 2).
| (2) |
where v0 is the experimentally determined initial velocity, Vmax is the estimated Vmax, [S] is the concentration of the substrate, S0.5 is the concentration of substrate that gives half-Vmax, and h is the Hill number indicative of the degree of cooperativity in the binding of the substrate.
When the concentration of l-malate was varied at constant concentration of substrates, the experimental data were fitted to Equation 3.
| (3) |
where vi and v0 are the experimentally determined initial velocities in the presence and absence of inhibitor, respectively; vi min is the estimated residual velocity at saturation of the inhibitor; [I] is the inhibitor concentration; I50 is the concentration of inhibitor that gives half-maximum inhibition at fixed concentrations of substrates; and h the Hill number indicative of the degree of cooperativity in the binding of the inhibitor.
Retrieval and sequence analysis of PEPC-C4 orthologs
Plant genomes typically contain several sequences coding for members of the PEPC family. Some of them were considered alleles given their high similarity of the nucleotide and amino acid sequences or because they mapped to a single locus (when mapping information was available), but others were considered true isozymes because they are located in different loci. To clarify this, we searched all available PEPC orthologs from plants, performing a blastp search on the RefSeq collection of the NCBI (https://www.ncbi.nlm.nih.gov/protein) protein database using as query the amino acid sequence of ZmPEPC-C4 (GenBank accession number CAA33317) that corresponds to the PEPC isozyme involved in C4 photosynthesis in maize (47) coded by ppc-C4 gene (48). To not exclude any candidate plant PEPC-C4, the E-value threshold was set at 10; the scoring matrix used was BLOSUM62 with gap opening and gap extension costs of 11 and 1, respectively. All retrieved sequences belong to Viridiplantae. To include sequences for which we would have mapping, allelism, and paralogy information, we also performed blastp searches on version 12.1.5 of the Phytozome database (https://phytozome.jgi.doe.gov/),4 which contains data for 63 wholly sequenced Viridiplantae genomes. Only hits with an E-value higher than 10e−3 were excluded. The resulting sequences were aligned with ClustalX2 (49), and the alignment was refined by hand, in some cases with the help of structural alignments using homology models, and stripped of redundant sequences.
For the construction of the sequence logos that show the conservation of residues at positions related to the neutral amino acid allosteric site, plant PEPC-C4 sequences were chosen based on the criterion of having serine at the position equivalent to ZmPEPC-C4 780 (30). Only sequences belonging to a C4 plant were considered; PEPC sequences from C3 or CAM plants that have this residue were excluded. Sequence logos were constructed with WebLogo3 (50, 51).
Author contributions
L. G.-S. and R. A. M.-C. formal analysis; L. G.-S., C. M.-J., J. A. J.-D., R. G.-T., and L. P. M.-C. investigation; L. G.-S. visualization; J. A. J.-D. and R. A. M.-C. writing-review and editing; R. A. M.-C. conceptualization; R. A. M.-C. supervision; R. A. M.-C. funding acquisition; R. A. M.-C. writing-original draft; R. A. M.-C. project administration.
Supplementary Material
Acknowledgments
We are indebted to Prof. Izui (Kyoto University) for providing us the ZmPEPC-C4 cDNA. We are also very grateful to F. Barreda-Huerta, M.Sc., and Dr. I. Bustos-Jaimes for subcloning the ppc-C4 gene, to S. P. Rojas-Trejo, B.Sc., and Dr. H. Serrano-Posada for preliminary crystallization attempts of the ZmPEPC-C4 isozyme, and to Dr. E. Rudiño-Piñera and Dr. A. Zárate-Romero for helpful discussions. We thank the staff of beamline 23 ID-B (GM/CA) of the Advanced Photon Source at Argonne National Laboratory for the use of and assistance with the X-ray beamline and advice during collection. We also thank the lecturers of the “2013 CCP4/APS Crystallographic School” at Argonne National Laboratory for helpful discussions of data processing and refinement strategies and Dr. Dominika Borek for assistance with structure solution.
This work was supported in part by Dirección General de Asuntos del Personal Académico–Universidad Nacional Autónoma de México Programa de Apoyo a Proyectos de Investigación e Innovación Tecnológica PAPIIT, Grant IN216911 and Consejo Nacional de Ciencia y Tecnología (CONACYT) Grant 252123 (to R. A. M.-C.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S6 and Tables S1 and S2.
The atomic coordinates and structure factors (code 5VYJ) have been deposited in the Protein Data Bank (http://wwpdb.org/).
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party-hosted site.
- Rubisco
- ribulose-1,5-bisphosphate carboxylase/oxygenase
- PEP
- 2-phosphoenolpyruvate
- PEPC
- phosphoenolpyruvate carboxylase
- PEPC-C4
- photosynthetic phosphoenolpyruvate carboxylase isozyme from C4 plants
- Zm
- Zea mays
- G6P
- d-glucose 6-phosphate
- Ft
- Flaveria trinervia
- Fp
- Flaveria pringlei
- CAM
- crassulacean acid metabolism
- Ah
- Amaranthus hypochondriacus.
References
- 1. Hatch M. D. (1987) C4 photosynthesis: a unique blend of modified biochemistry, anatomy and ultrastructure. Biochim. Biophys. Acta 895, 81–106 10.1016/S0304-4173(87)80009-5 [DOI] [Google Scholar]
- 2. Bowes G., Rao S. K., Estavillo G. M., and Reiskind J. B. (2002) C4 mechanisms in aquatic angiosperms: comparisons with terrestrial C4 systems. Funct. Plant Biol. 29, 379–392 10.1071/PP01219 [DOI] [PubMed] [Google Scholar]
- 3. Edwards G. E., Franceschi V. R., and Voznesenskaya E. V. (2004) Single cell C4 photosynthesis versus the dual-cell (Kranz) paradigm. Annu. Rev. Plant Biol. 55, 173–196 10.1146/annurev.arplant.55.031903.141725 [DOI] [PubMed] [Google Scholar]
- 4. Andreo C. S., González D. H., and Iglesias A. A. (1987) Higher plant phosphoenolpyruvate carboxylase: Structure and regulation. FEBS Lett. 213, 1–8 10.1016/0014-5793(87)81454-0 [DOI] [Google Scholar]
- 5. Chollet R., Vidal J., and O'Leary M. H. (1996) Phosphoenolpyruvate carboxylase: A ubiquitous, highly regulated enzyme in plants. Annu. Rev. Plant Physiol. Plant. Mol. Biol. 47, 273–298 10.1146/annurev.arplant.47.1.273 [DOI] [PubMed] [Google Scholar]
- 6. Izui K., Matsumura H., Furumoto T., and Kai Y. (2004) Phosphoenolpyruvate carboxylase: A new era of structural biology. Annu. Rev. Plant Biol. 55, 69–84 10.1146/annurev.arplant.55.031903.141619 [DOI] [PubMed] [Google Scholar]
- 7. Wong K. F., and Davies D. D. (1973) Regulation of phosphoenolpyruvate carboxylase of Zea mays by metabolites. Biochem. J. 131, 451–458 10.1042/bj1310451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Uedan K., and Sugiyama T. (1976) Purification and characterization of phosphoenolpyruvate carboxylase from maize leaves. Plant Physiol. 57, 906–910 10.1104/pp.57.6.906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Stiborová M., and Leblová S. (1985) Activation of maize (Zea mays L.) phosphoenolpyruvate carboxylase by glucose-6-phosphate and glycine: effects of pH and Mg2. Photosynthetica 19, 177–182 [Google Scholar]
- 10. Doncaster H. D., and Leegood R. C. (1987) Regulation of phosphoenolpyruvate carboxylase activity in maize leaves. Plant Physiol. 84, 82–87 10.1104/pp.84.1.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bandarian V., Poehner W. J., and Grover S. D. (1992) Metabolite activation of crassulacean acid metabolism and C4 phosphoenolpyruvate carboxylase. Plant Physiol. 100, 1411–1416 10.1104/pp.100.3.1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mújica-Jiménez C., Castellanos-Martínez A., and Muñoz-Clares R. A. (1998) Studies of the allosteric properties of maize leaf phosphoenolpyruvate carboxylase with the phosphoenolpyruvate analog phosphomycin as activator. Biochim. Biophys. Acta 1386, 132–144 10.1016/S0167-4838(98)00093-4 [DOI] [PubMed] [Google Scholar]
- 13. Tovar-Méndez A., Rodríguez-Sotres R., López-Valentín D. M., and Muñoz-Clares R. A. (1998) Re-examination of the roles of PEP and Mg2+ in the reaction catalysed by the phosphorylated and non-phosphorylated forms of phosphoenolpyruvate carboxylase from leaves of Zea mays: Effects of the activators glucose 6-phosphate and glycine. Biochem. J. 332, 633–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rodríguez-Sotres R., and Muñoz-Clares R. A. (1990) Kinetic evidence of the existence of a regulatory phosphoenolpyruvate binding site in maize leaf phosphoenolpyruvate carboxylase. Arch. Biochem. Biophys. 276, 180–190 10.1016/0003-9861(90)90025-T [DOI] [PubMed] [Google Scholar]
- 15. Nishikido T., and Takanashi H. (1973) Glycine activation of PEP carboxylase from monocotyledoneous C4 plants. Biochem. Biophys. Res. Commun. 53, 126–133 10.1016/0006-291X(73)91410-1 [DOI] [PubMed] [Google Scholar]
- 16. Garson L., and Gray V. (1991) Regulation of phosphoenolpyruvate carboxylase from maize leaves by nitrate and alanine. Biochem. Int. 23, 299–305 [PubMed] [Google Scholar]
- 17. Gillinta J., and Grover S. D. (1995) Kinetic interactions of glycine with substrates and effectors of phosphoenolpyruvate carboxylase from maize leaves. Photosynth. Res. 45, 121–126 10.1007/BF00032583 [DOI] [PubMed] [Google Scholar]
- 18. Tovar-Méndez A., Mújica-Jiménez C., and Muñoz-Clares R. A. (2000) Physiological implications of the kinetics of maize leaf phosphoenolpyruvate carboxylase. Plant Physiol. 123, 149–160 10.1104/pp.123.1.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Güemez-Toro R., Mújica-Jiménez C., and Muñoz-Clares R. A. (2012) Allosteric regulation of the photosynthetic C4 isoenzyme of phosphoenolpyruvate carboxylase: A comparative study between enzymes from monocot and eudicots. J. Mex. Chem. Soc. 56, 58–66 [Google Scholar]
- 20. Gonzalez D. H., Iglesias A. A., and Andreo C. S. (1984) On the regulation of phosphoenolpyruvate carboxylase activity from maize leaves by L-malate. Effect of pH. J. Plant Physiol. 116, 425–434 10.1016/S0176-1617(84)80134-0 [DOI] [PubMed] [Google Scholar]
- 21. Paulus J. K., Schlieper D., and Groth G. (2013) Greater efficiency of photosynthetic carbon fixation due to single amino-acid substitution. Nat. Commun. 4, 1518 10.1038/ncomms2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Matsumura H., Terada M., Shirakata S., Inoue T., Yoshinaga T., Izui K., and Kai Y. (1999) Plausible phosphoenolpyruvate binding site revealed by 2.6 Å structure of Mn2+-bound phosphoenolpyruvate carboxylase from Escherichia coli. FEBS Lett. 458, 93–96 10.1016/S0014-5793(99)01103-5 [DOI] [PubMed] [Google Scholar]
- 23. Kai Y., Matsumura H., Inoue T., Terada K., Nagara Y., Yoshinaga T., Kihara A., Tsumura K., and Izui K. (1999) Three-dimensional structure of phosphoenolpyruvate carboxylase: A proposed mechanism for allosteric inhibition. Proc. Natl. Acad. Sci. U.S.A. 96, 823–828 10.1073/pnas.96.3.823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Matsumura H., Xie Y., Shirakata S., Inoue T., Yoshinaga T., Ueno Y., Izui K., and Kai Y. (2002) Crystal structures of C4 form maize and quaternary complex of E. coli phosphoenolpyruvate carboxylases. Structure 10, 1721–1730 10.1016/S0969-2126(02)00913-9 [DOI] [PubMed] [Google Scholar]
- 25. Takahashi-Terada A., Kotera M., Ohshima K., Furumoto T., Matsumura H., Kai Y., and Izui K. (2005) Maize phosphoenolpyruvate carboxylase. Mutations at the putative binding site for glucose 6-phosphate caused desensitization and abolished responsiveness to regulatory phosphorylation. J. Biol. Chem. 280, 11798–11806 10.1074/jbc.M408768200 [DOI] [PubMed] [Google Scholar]
- 26. Yuan J., Sayegh J., Mendez J., Sward L., Sanchez N., Sanchez S., Waldrop G., and Grover S. (2006) The regulatory role of residues 226–232 in phosphoenolpyruvate carboxylase from maize. Photosynth. Res. 88, 73–81 10.1007/s11120-005-9032-x [DOI] [PubMed] [Google Scholar]
- 27. Schlieper D., Förster K., Paulus J. K., and Groth G. (2014) Resolving the activation site of positive regulators in the plant phosphoenolpyruvate carboxylase. Mol. Plant 7, 437–440 10.1093/mp/sst130 [DOI] [PubMed] [Google Scholar]
- 28. Tovar-Méndez A., Mújica-Jiménez C., and Muñoz-Clares R. A. (1997) Desensitization to glucose 6-phosphate of phosphoenolpyruvate carboxylase from maize leaves by pyridoxal 5′phosphate. Biochim. Biophys. Acta 1337, 207–216 10.1016/S0167-4838(96)00166-5 [DOI] [PubMed] [Google Scholar]
- 29. Gonzalez L., Sánchez S., Home J., Kanzaki G., and Grover S. (2007) Mutational analysis of a proposed glycine binding site in PEP carboxylase. FASEB J. 21, 806.3 [Google Scholar]
- 30. Bläsing O. E., Westhoff P., and Svensson P. (2000) Evolution of C4 phosphoenolpyruvate carboxylase in Flaveria, a conserved serine in the carboxyterminal part of the enzyme is a major determinant for C4-specific characteristics. J. Biol. Chem. 275, 27917–27923 10.1074/jbc.M909832199 [DOI] [PubMed] [Google Scholar]
- 31. Christin P. A., Arakaki M., Osborne C. P., Bräutigam A., Sage R. F., Hibberd J. M., Kelly S., Covshoff S., Wong G. K., Hancock L., and Edwards E. J. (2014) Shared origins of a key enzyme during of C4 and CAM metabolism. J. Exp. Bot. 65, 3609–36021 10.1093/jxb/eru087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sánchez R., and Cejudo F. J. (2003) Identification and expression analysis of a gene encoding a bacterial-type phosphoenolpyruvate carboxylase from Arabidopsis and rice. Plant Physiol. 132, 949–957 10.1104/pp.102.019653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rosnow J. J., Edwards G. E., and Roalson E. H. (2014) Positive selection of Kranz and non-Kranz C4 phosphoenolpyruvate carboxylase amino acids in Suaedoideae (Chenopodiaceae). J. Exp. Bot. 65, 3595–3607 10.1093/jxb/eru053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gao Y., and Woo K. C. (1996) Regulation of phosphoenolpyruvate carboxylase in Zea mays by protein phosphorylation and metabolites and their roles in photosynthesis. Aust. J. Plant Physiol. 23, 25–32 10.1071/PP9960025 [DOI] [Google Scholar]
- 35. Leegood R. C. (1985) The intercellular compartmentation of metabolites in leaves of Zea mays L. Planta 164, 163–171 10.1007/BF00396078 [DOI] [PubMed] [Google Scholar]
- 36. Stitt M., and Heldt H. W. (1985) Generation and maintenance of concentration gradients between the mesophyll and bundle sheath in maize leaves. Biochim. Biophys. Acta 808, 400–414 [Google Scholar]
- 37. Heineke D., Riens B., Grosse H., Hoferichter P., Peter U., Flügge U. I., and Heldt H. W. (1991) Redox transfer across the inner chloroplast envelope membrane. Plant Physiol. 95, 1131–1137 10.1104/pp.95.4.1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rao S. K., Reiskind J. B., and Bowes G. (2008) Kinetic analyses of recombinant isoforms of phosphoenolpyruvate carboxylase from Hydrilla verticillata leaves and the impact of substituting a C4-signature serine. Plant Sci. 174, 475–483 10.1016/j.plantsci.2008.01.010 [DOI] [Google Scholar]
- 39. Rosnow J. J., Evans M. A., Kapralov M. V., Cousins A. B., Edwards G. E., and Roalson E. H. (2015) Kranz and single-cell forms of C4 plants in the subfamily Suaedoideae show kinetic C4 convergence for PEPC and Rubisco with divergent amino acid substitutions. J. Exp. Bot. 66, 7347–7358 10.1093/jxb/erv431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gasteiger E., Gattiker A., Hoogland C., Ivanyi I., Appel R. D., and Bairoch A. (2003) ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788 10.1093/nar/gkg563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 10.1107/S0907444994003112 [DOI] [PubMed] [Google Scholar]
- 43. Strong M., Sawaya M. R., Wang S., Phillips M., Cascio D., and Eisenberg D. (2006) Toward the structural genomics of complexes: Crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 103, 8060–8065 10.1073/pnas.0602606103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., and Terwilliger T. C. (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 10.1107/S0907444902016657 [DOI] [PubMed] [Google Scholar]
- 46. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 47. Hudspeth R. L., and Grula J. W. (1989) Structure and expression of the maize gene encoding the phosphoenolpyruvate carboxylase isoenzyme involved in C4 photosynthesis. Plant Mol. Biol. 12, 579–589 10.1007/BF00036971 [DOI] [PubMed] [Google Scholar]
- 48. Christin P. A., Salamin N., Savolainen V., Duvall M. R., and Besnard G. (2007) C4 photosynthesis evolved in grasses via parallel adaptative genetic changes. Curr. Biol. 17, 1241–1247 10.1016/j.cub.2007.06.036 [DOI] [PubMed] [Google Scholar]
- 49. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., and Higgins D. G. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- 50. Crooks G. E., Hon G., Chandonia J. M., and Brenner S. E. (2004) WebLogo: A sequence logo generator. Genome Res. 14, 1188–1190 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schneider T. D., and Stephens R. M. (1990) Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 18, 6097–6100 10.1093/nar/18.20.6097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera: A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.