Abstract
Cytokinesis, the terminal step in cell division, in the protist human pathogen Trypanosoma brucei occurs along the longitudinal axis from the anterior tip of the new flagellum attachment zone (FAZ) toward the posterior cell tip. This process is regulated by a signaling cascade composed of the Polo-like kinase homolog TbPLK, the Aurora B kinase homolog TbAUK1, and the trypanosome-specific CIF1–CIF2 protein complex. However, the regulatory mechanism and the signaling pathway for this unusual mode of cytokinesis remain poorly understood. Here, we report another trypanosome-specific protein assembly, the CIF1–CIF3 complex, and its essential role in cytokinesis initiation. Through biochemical and genetic approaches, we demonstrate that CIF3 interacts with CIF1 in a TbPLK-dependent manner and maintains CIF1 localization at the new FAZ tip. Conversely, CIF1 maintains CIF3 stability at the new FAZ tip. We further show that TbPLK is required for CIF3 localization and that CIF3 is necessary for targeting TbAUK1 to the new FAZ tip during anaphase. These results suggest that two trypanosome-specific CIF1-containing protein complexes cooperate with the evolutionarily conserved Polo-like kinase and Aurora B kinase to promote cytokinesis in T. brucei.
Keywords: cell cycle, cell division, cytokinesis, parasite, Trypanosoma brucei, Aurora B kinase, kinetoplastid, polo-like kinase, sleeping sickness, trypanosomiasis, CIF1, CIF2, CIF3
Introduction
Cytokinesis occurs along the short axis in organisms across all three domains of life, and in many organisms it requires a constriction machinery assembled at the cell division plane. The constriction machinery is best known as the FtsZ ring in bacteria and some archaea species and as the actomyosin ring in animals, fungi, and amebae, and it ingresses toward the middle of the cell to divide the cell (1, 2). Higher plants do not assemble a constriction machinery at the cell division plane but construct, in the middle of the cell, a cell plate composed of the phragmoplast, an array of remnant spindle microtubules, and build a new cell wall outward to the cell cortex (3). Despite the drastic morphological differences among eukaryotic organisms, however, the regulators implicated in cytokinesis appear to be considerably conserved across the eukaryotic organisms from yeast to humans.
Many protists, including Giardia, trypanosomes, and Leishmania spp., undergo cell division along the longitudinal axis through binary cell fission. The mechanism underlying this unusual mode of cytokinesis remains poorly understood. Trypanosomes lack the type II myosin, a key component of the actomyosin contractile ring apparatus found in animals, fungi, and amebae, suggesting that cytokinesis cleavage furrow ingression in trypanosomes employs a distinct, yet unknown, mechanism. The cell division plane in trypanosomes is determined by the length of the newly assembled flagellum and its associated structure, termed flagellum attachment zone (FAZ)2 (4, 5). Cytokinesis is initiated from the anterior tip of the new FAZ and proceeds toward the posterior end of the cell along the division fold, which is formed by membrane invagination between the new and old flagella prior to cytokinesis initiation (6–8). However, it remains unclear how the formation of the division fold is controlled and what determines the unidirectionality of cleavage furrow ingression.
We recently delineated a novel signaling pathway that acts at the anterior tip of the new FAZ to promote cytokinesis unidirectionally along the longitudinal axis of a trypanosome cell (7, 8). This signaling cascade is composed of two evolutionarily conserved protein kinases, the Polo-like kinase TbPLK (9, 10) and the Aurora B kinase TbAUK1 (11, 12), and two trypanosome-specific proteins, the TbPLK-interacting protein CIF1 (7, 13), which is also known as TOEFAZ1 (14), and the CIF1-interacting protein CIF2 (8). CIF1 is a potential substrate of TbPLK and forms a complex with CIF2 during the S phase of the cell cycle (8), whereas TbAUK1 acts downstream of CIF1 in the cytokinesis regulatory pathway during late anaphase (7). CIF2 is required for maintaining CIF1 stability, and its level appears to be tightly regulated (8), although the mechanism for this regulation is still unknown. Given that CIF2 disappears from the new FAZ tip after S phase, it raises the questions of how CIF1 is maintained at the new FAZ tip after S phase and whether other CIF1 partners replace CIF2 to stabilize CIF1 at the new FAZ tip.
In this report, we identify another CIF1-interacting protein, named CIF3, which regulates cytokinesis initiation in the procyclic form of Trypanosoma brucei. CIF3 maintains CIF1 at the new FAZ tip and cooperates with CIF1 to recruit TbAUK1 to the new FAZ tip during late anaphase for TbAUK1 to drive cytokinesis initiation. These findings suggest the requirement of two CIF1-containing protein assemblies, the CIF1–CIF2 complex and the CIF1–CIF3 complex, for promoting the longitudinal binary cell fission through cooperation with the evolutionarily conserved Polo-like kinase and Aurora B kinase, and highlight the unusual mechanism of cytokinesis in T. brucei.
Results
Identification of CIF3 as a new binding partner of CIF1
To identify new cytokinesis regulators that function in the TbPLK–CIF1–CIF2–TbAUK1 pathway, we searched for new FAZ tip–localizing proteins by epitope tagging and fluorescence microscopic analysis of genes whose transcripts are enriched in S phase (15), based on the fact that both CIF1 and CIF2 first emerge at the S phase of the cell cycle (7, 8). Among the ∼30 proteins tagged, 1 protein, encoded by Tb927.10.13100, was found to localize to the new FAZ tip (Fig. 1) and was further confirmed to play an essential role in cytokinesis initiation (see below). We thus named this protein CIF3, after CIF1 and CIF2 (7, 8). CIF3 contains three coiled-coil motifs and is conserved in Trypanosoma cruzi (TcCLB.506359.90), but it appears to lack a homolog in Leishmania spp.
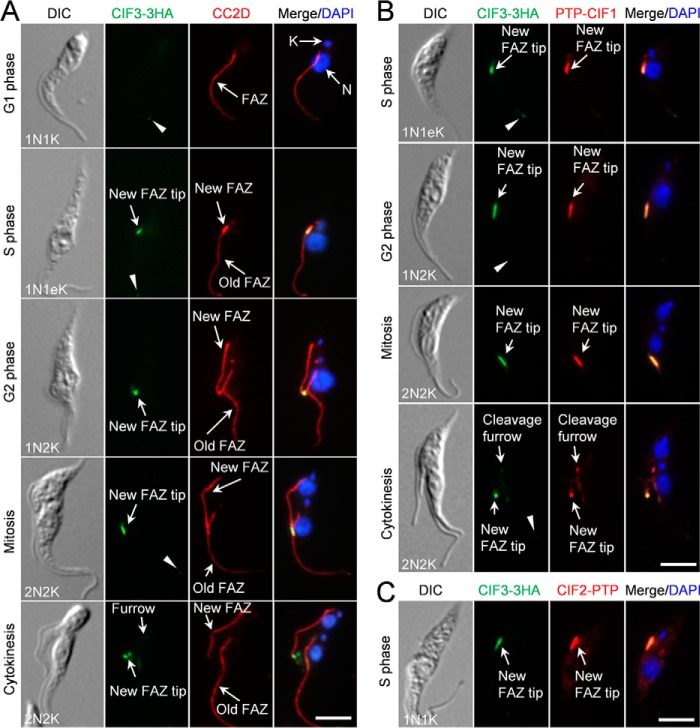
Figure 1.
CIF3 co-localizes with CIF1 and CIF2 at the new FAZ tip. A, subcellular localization of CIF3 during the cell cycle. Cells were co-immunostained with FITC-conjugated anti-HA mAb and anti-CC2D pAb to detect CIF3–3HA and the FAZ, respectively. The white arrowheads indicate the weak CIF3 fluorescence signal at the old FAZ tip. N, nucleus; K, kinetoplast. Scale bar: 5 μm. B, CIF3 co-localizes with CIF1 at the new FAZ tip from S phase to early cytokinesis. Cells expressing endogenously 3HA-tagged CIF3 and PTP-tagged CIF1 were co-immunostained with FITC-conjugated anti-HA mAb and anti-protein A pAb. The white arrowheads indicate the weak CIF3 fluorescence signal at the old FAZ tip. Scale bar: 5 μm. C, CIF3 co-localizes with CIF2 at the new FAZ tip during S phase. Cells co-expressing CIF3–3HA and CIF2–PTP from their respective endogenous locus were co-immunostained with FITC-conjugated anti-HA mAb and anti-protein A pAb. Scale bar: 5 μm.
CIF3 protein was not detectable at the new FAZ tip in G1 cells, which contain one nucleus and one kinetoplast (1N1K), by immunofluorescence microscopy, but weak CIF3 fluorescence signal was detected at the old FAZ tip (Fig. 1A, arrowhead). In S-phase cells, which contain one nucleus and one elongated kDNA (1N1eK) and a short new FAZ, CIF3 emerged at the new FAZ tip and remained at the new FAZ tip until early cytokinesis (Fig. 1A), similar to CIF1 (7). In these cells, weak CIF3 signal was detectable at the old FAZ tip (Fig. 1A, arrowheads). However, unlike CIF1 that is enriched at the cleavage furrow during cytokinesis (7) (Fig. 1B), a much lower level of CIF3 was detected at the cleavage furrow (Fig. 1, A and B). At the new FAZ tip, CIF3 co-localized with CIF1 from S phase to early cytokinesis (Fig. 1B) and with CIF2 during S phase (Fig. 1C).
To examine whether CIF3 interacts with CIF1 and/or CIF2, we carried out co-immunoprecipitation experiments. The results showed that CIF1, but not CIF2, was able to pull down CIF3 from trypanosome lysate (Fig. 2, A and B), indicating that CIF3 forms a complex with CIF1. Given that CIF1 also forms a complex with CIF2 (8), these results suggest that the three proteins exist in two separate protein complexes, the CIF1–CIF2 complex and the CIF1–CIF3 complex.
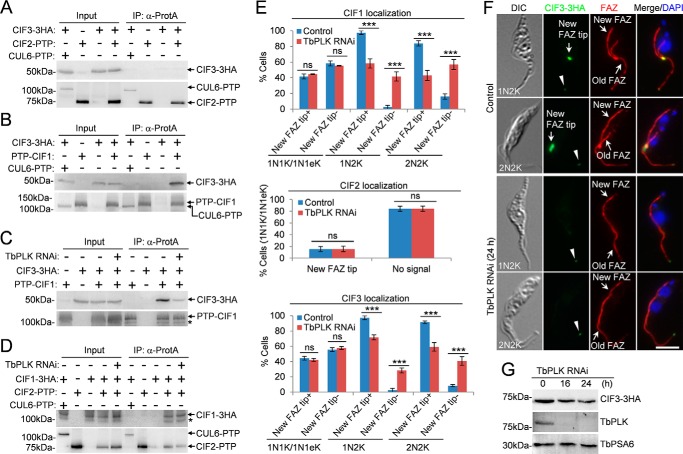
Figure 2.
CIF3 interacts with CIF1 in a TbPLK-dependent manner. A, immunoprecipitation of CIF2–PTP was unable to pull down CIF3–3HA from trypanosome cell lysate. Cells expressing CIF2–PTP alone, CIF3–3HA alone, or both CUL6–PTP and CIF3–3HA served as the controls. B, immunoprecipitation of PTP–CIF1 was able to pull down CIF3–3HA from trypanosome cell lysate. Co-immunoprecipitation was carried out as above. CUL6–PTP was co-expressed with CIF3–3HA, and co-immunoprecipitation was carried out to rule out the possibility that the PTP tag can pull down CIF3–3HA. C, CIF3–CIF1 interaction depends on TbPLK. TbPLK RNAi was induced for 24 h. The asterisk indicates a degradation fragment of PTP–CIF1. D, CIF2–CIF1 interaction is independent of TbPLK. TbPLK RNAi was induced for 24 h. Cells co-expressing CUL6–PTP and CIF1–3HA were used as a control to show that the PTP tag was not able to pull down CIF1–3HA. The asterisk indicates a degradation fragment of CIF1–3HA. E, effect of TbPLK RNAi on the localization of CIF1, CIF2, and CIF3 during different cell cycle stages. Total numbers of cells counted are as follows. For CIF1 localization: 1N1K/1N1eK, 344 (control) and 334 (TbPLK RNAi); 1N2K, 167 (control) and 154 (TbPLK RNAi); 2N2K, 211 (control) and 256 (TbPLK RNAi). For CIF2 localization: 1N1K/1N1eK, 619 (control) and 310 (TbPLK RNAi). For CIF3 localization: 1N1K/1N1eK, 325 (control) and 308 (TbPLK RNAi); 1N2K, 154 (control) and 152 (TbPLK RNAi); 2N2K, 216 (control) and 220 (TbPLK RNAi). Error bars represented S.D. from three independent experiments. ***, p < 0.001; ns, no statistical significance. The label New FAZ tip+ indicates that CIF proteins are detected at the new FAZ tip, whereas the label New FAZ tip− indicates that CIF proteins are not detected at the new FAZ tip. F, localization of CIF3–3HA in noninduced control and TbPLK RNAi cells. Cells were co-immunostained with FITC-conjugated anti-HA mAb and anti-CC2D pAb to label CIF3–3HA and the FAZ, respectively. The white arrowheads indicate the weak CIF3 fluorescence signal at the old FAZ tip. Scale bar: 5 μm. G, CIF3 protein levels in control and TbPLK RNAi cells. TbPSA6 served as the loading control.
TbPLK is required for CIF1–CIF3 complex formation and localization to the new FAZ tip
Because TbPLK RNAi disrupted CIF1 phosphorylation (7), we investigated the potential requirement of TbPLK for the formation of the CIF1–CIF3 complex and the CIF1–CIF2 complex. Depletion of TbPLK by RNAi significantly reduced the amount of CIF3 protein precipitated by CIF1 (Fig. 2C), indicating that TbPLK is important for CIF1–CIF3 complex formation. However, depletion of TbPLK did not affect CIF1–CIF2 complex formation (Fig. 2D). We next examined whether TbPLK is required for the localization of the two protein complexes to the new FAZ tip at different cell cycle stages by immunofluorescence microscopy. Knockdown of TbPLK did not affect the localization of CIF1, CIF2, and CIF3 in S-phase (1N1eK) cells, but significantly impaired CIF1 and CIF3 localization at the new FAZ tip in G2-phase cells, which contain one nucleus and two kinetoplasts (1N2K), and mitotic cells, which contain two nuclei and two kinetoplasts (2N2K) (Fig. 2, E and F). The weak CIF3 signal at the old FAZ tip was not affected by TbPLK RNAi (Fig. 2F, arrowheads). In TbPLK RNAi cells induced for no longer than 24 h, the level of CIF3 protein was not significantly changed (Fig. 2G). Given that CIF2 is only detectable at the new FAZ tip during S phase (8) and that formation of the CIF1–CIF2 complex is not dependent on TbPLK (Fig. 2D), these results suggest that the CIF1–CIF2 complex is targeted to the new FAZ tip at the S phase of the cell cycle in a TbPLK-independent manner. These results also suggest that localization of the CIF1–CIF3 complex at the new FAZ tip during G2 and mitotic phases depends on TbPLK.
CIF3 is required for cytokinesis initiation
To understand the function of CIF3, RNAi was carried out in the procyclic form of T. brucei. Four clonal CIF3 RNAi cell lines were characterized, which showed almost identical phenotypes. Thus, only the results from one clonal cell line were presented. The efficiency of RNAi was monitored by tagging CIF3 protein with an N-terminal PTP epitope at the endogenous locus in the CIF3 RNAi cell line and subsequent Western blotting with anti-protein A antibody. The results showed that CIF3 was gradually depleted upon tetracycline induction (Fig. 3A). This depletion of CIF3 protein caused a growth defect, resulting in the reduction of the doubling time from ∼10 h to ∼15 h (Fig. 3B). To characterize the potential defects in cell cycle progression, we quantified the cells at different cell cycle stages by counting the numbers of nuclei and kinetoplasts in control and CIF3 RNAi-induced cells. Cells with one nucleus and one kinetoplast (1N1K), which include G1 and S-phase cells, gradually decreased from ∼75% to ∼15% after 2 days of RNAi induction, whereas cells with two nuclei and two kinetoplasts (2N2K), which include mitotic cells, postmitotic cells, and cells undergoing cytokinesis, increased from ∼13% to ∼37% after RNAi induction for 1 day and then gradually decreased (Fig. 3C). Additionally, cells with two nuclei and four kinetoplasts (2N4K) also emerged, and cells with multiple (>2) nuclei and multiple (>2) kinetoplasts (XNXK, X>2) accumulated to ∼35% after RNAi induction for 2 days (Fig. 3C). These results suggest that CIF3 depletion caused a cytokinesis defect. Notably, cells without a nucleus (0N1K and 0N2K) accumulated to ∼30% of the total population after RNAi induction for 4 days (Fig. 3C), indicating that aberrant cytokinesis occurred after prolonged RNAi induction.
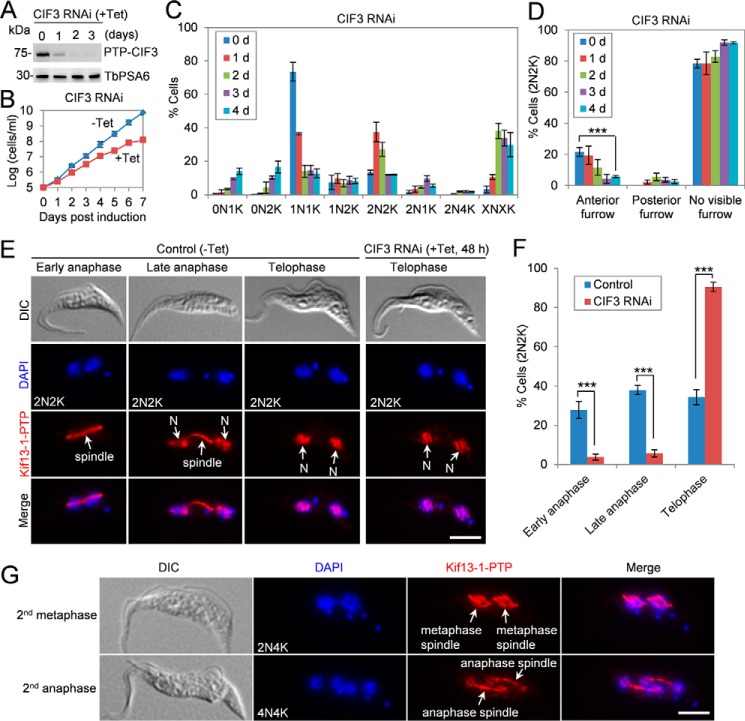
Figure 3.
Depletion of CIF3 impairs cytokinesis initiation. A, Western blotting to monitor the level of CIF3 in CIF3 RNAi cells. TbPSA6 served as the loading control. B, depletion of CIF3 by RNAi caused a growth defect. C, depletion of CIF3 in procyclic trypanosome caused accumulation of binucleate (2N) cells initially and multinucleate cells (XN, X >2) subsequently. A total of 280 cells were counted for each time point, and the results were presented as mean percentage ± S.D. (n = 3). N, nucleus; K, kinetoplast. D, quantification of cells without or with a visible cleavage furrow in the binucleate cells from the control and CIF3 RNAi cells. A total of 100 cells were counted for each time point, and the results were presented as mean percentage ± S.D. (n = 3). ***, p < 0.001. E, determination of the cell cycle stages of the 2N2K cells by immunostaining the mitotic spindle using Kif13–1 as a spindle marker. CIF3 RNAi was induced for 48 h. Kif13–1-PTP was immunostained with the anti-protein A antibody. N, nucleus. Scale bar: 5 μm. F, quantitation of the 2N2K cells at different cell cycle stages from control and CIF3 RNAi (+Tet, 48 h) based on the results in panel E. The total numbers of 2N2K cells counted are 309 (control) and 316 (CIF3 RNAi). Error bars indicate S.D. from three independent experiments. ***, p < 0.001. G, CIF3-depleted cells entered next cell cycle in the absence of mitosis. Shown are the cells at the second metaphase and the second anaphase. Kif13–1 tagged with a C-terminal PTP epitope was immunostained with the anti-protein A antibody and served as a spindle marker. Scale bar: 5 μm.
The accumulation of 2N2K, 2N4K, and XNXK cells prompted us to examine whether cytokinesis cleavage furrow formation was defective in CIF3 RNAi cells. The 2N2K cells with a visible anterior cleavage furrow were significantly decreased after CIF3 RNAi for 4 days (Fig. 3D), indicating that cleavage furrow ingression was impaired by depletion of CIF3. Only ∼2% of the CIF3-depleted 2N2K cells contained a posterior cleavage furrow (Fig. 3D). This is in striking contrast to the CIF1-deficient 2N2K cells and the CIF2-deficient 2N2K cells, ∼30% of which contained a posterior cleavage furrow (7, 8). It suggests that CIF3 RNAi cells did not initiate the alternative cytokinesis that occurred in CIF1 RNAi cells and CIF2 RNAi cells (7, 8), although CIF3 forms a complex with CIF1 (Fig. 2, B and C).
To further confirm that CIF3 RNAi caused a cytokinesis defect but not a defect in late mitosis, we used Kif13–1 as a spindle marker (16, 17) to determine the cell cycle stages of the CIF3-depleted 2N2K cells. Kif13–1, which was endogenously tagged with a C-terminal PTP epitope, localized to the spindle during early anaphase, to the spindle and the two nuclei during late anaphase, and to the two nuclei during telophase in control cells (Fig. 3, E and F), as reported previously (16, 17). In CIF3 RNAi cells, however, the cells at the early anaphase stage (spindle-localized Kif13–1) and at the late anaphase stage (spindle- and nucleus-localized Kif13–1) were significantly decreased, but the cells at telophase (nucleus-localized Kif13–1) were significantly increased (Fig. 3, E and F). CIF3-deficient cells appeared to re-enter the next cell cycle, as shown by the emergence of 2N4K and 4N4K cells (Fig. 3, C and G). Using Kif13–1 as the spindle marker, two metaphase spindles of a “diamond” shape were detected in some of the 2N4K cells, and two anaphase spindles were readily detected in many of the 4N4K cells (Fig. 3G), indicating that these cells were undergoing the second round of mitosis in the absence of cytokinesis. Altogether, these results demonstrated that CIF3 depletion inhibited cytokinesis without affecting mitosis.
CIF3 maintains CIF1 localization at the new FAZ tip, whereas CIF1 maintains CIF3 stability
Given that CIF3 interacts with CIF1 in vivo in trypanosomes (Fig. 2, B and C), we investigated the effect of CIF3 depletion on the subcellular localization and stability of CIF1. To this end, CIF1 was endogenously tagged with a triple HA epitope in CIF3 RNAi cell line, and the level of CIF1 protein was examined by Western blotting and the localization of CIF1 was examined by immunofluorescence microscopy. Knockdown of CIF3 did not affect CIF1 protein level (Fig. 4A) and did not affect CIF1 localization in S-phase (1N1eK) cells, but depletion of CIF3 significantly disrupted CIF1 localization at the new FAZ tip in G2 (1N2K) and mitotic (2N2K) cells (Fig. 4, B and C). These results suggest that CIF3 is required for maintaining CIF1 localization at the new FAZ tip after the S phase of the cell cycle.
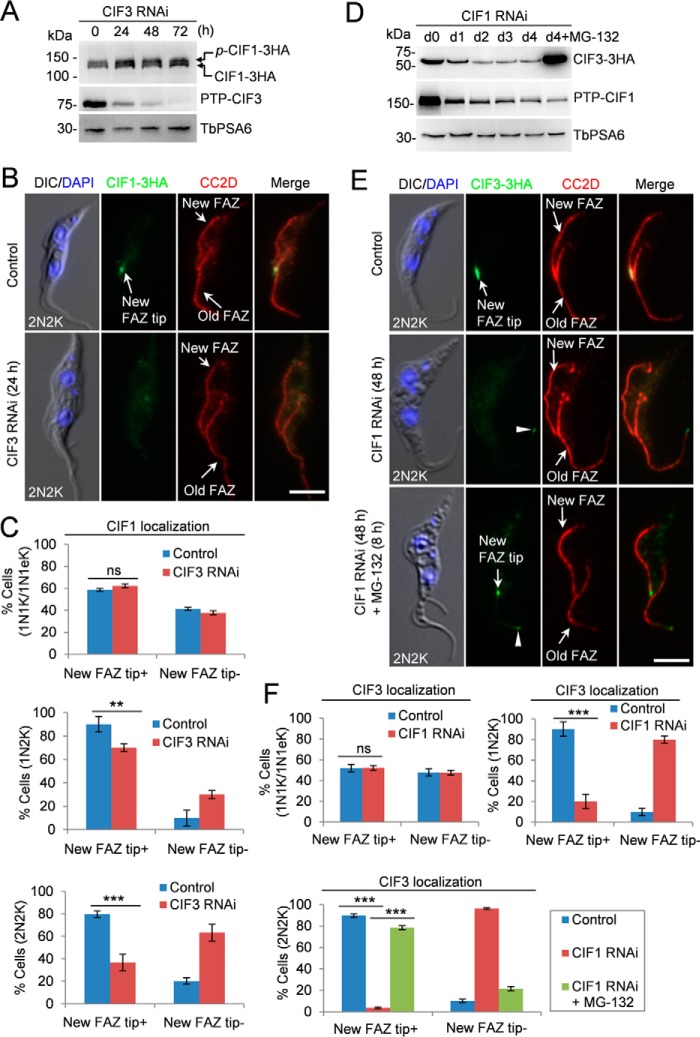
Figure 4.
CIF3 maintains CIF1 localization and CIF1 maintains CIF3 stability. A, effect of CIF3 depletion on CIF1 protein level. CIF3 RNAi was induced for 72 h. TbPSA6 served as the loading control. p-CIF1–3HA, phosphorylated CIF1–3HA. B, depletion of CIF3 disrupted CIF1 localization to the new FAZ tip. Cells were co-immunostained with FITC-conjugated anti-HA mAb and anti-CC2D antibody to label CIF1–3HA and the FAZ, respectively. Scale bar: 5 μm. C, quantification of cells with different CIF1 localization patterns. Total numbers of cell counted are as follows: 1N1K/1N1eK, 213 (control) and 212 (CIF3 RNAi); 1N2K, 91 (control) and 90 (CIF3 RNAi); 2N2K, 158 (control) and 161 (CIF3 RNAi). The results were presented as mean percentage ± S.D. (n = 3). **, p < 0.01; ***, p < 0.001; ns, no statistical significance. D, effect of CIF1 depletion on CIF3 protein level. CIF1 RNAi was induced for 96 h. The proteasome inhibitor MG-132 was added after CIF1 RNAi was induced for 88 h and incubated for an additional 8 h. TbPSA6 served as the loading control. E, effect of CIF1 depletion on CIF3 localization. CIF1 RNAi was induced for 48 h. For MG-132 treatment, cells were induced for RNAi for 40 h and then treated with MG-132 for an additional 8 h. Cells were co-immunostained with FITC-conjugated anti-HA mAb and anti-CC2D antibody to label CIF3–3HA and the FAZ, respectively. The white arrowheads indicate the weak CIF3 fluorescence signal at the old FAZ tip. Scale bar: 5 μm. F, quantification of the cells with different CIF3 localization patterns. Total numbers of cells counted are as follows: 1N1K/1N1eK, 285 (control) and 261 (CIF1 RNAi); 1N2K, 153 (control) and 164 (CIF1 RNAi); 2N2K, 206 (control), 205 (CIF1 RNAi) and 203 (CIF1 RNAi+MG-132). The results were presented as mean percentage ± S.D. (n = 3). ***, p < 0.001; ns, no statistical significance.
Conversely, the effect of CIF1 depletion on CIF3 stability and localization was investigated. CIF3 was endogenously tagged with a triple HA epitope in CIF1 RNAi cell line expressing endogenously PTP-tagged CIF1. Western blotting showed that upon knockdown of CIF1, the level of CIF3 protein gradually decreased, but it was stabilized in the presence of the proteasome inhibitor MG-132 (Fig. 4D), indicating that CIF1 depletion destabilized CIF3. Immunofluorescence microscopy showed that CIF3 was no longer detectable at the new FAZ tip in G2 (1N2K) and mitotic (2N2K) cells, but it was still detectable at the new FAZ tip in S-phase (1N1eK) cells (Fig. 4, E and F). The weak CIF3 signal at the old FAZ tip was not affected by CIF1 RNAi (Fig. 4E, arrowhead). Treatment of CIF1 RNAi cells with MG-132 restored CIF3 localization at the new FAZ tip (Fig. 4, E and F), demonstrating that the lack of CIF3 signal at the new FAZ tip was because of CIF3 degradation. Altogether, these results suggest that CIF1 is required for maintaining CIF3 protein stability after S phase.
The zinc finger motif 1 in CIF1 is required for CIF1–CIF3 interaction
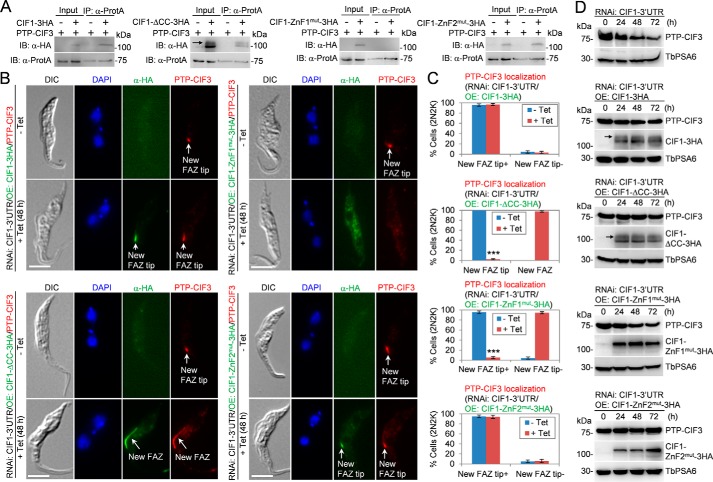
CIF1 contains a coiled-coil motif at the N-terminal portion and two CCHC-type zinc finger (ZnF) motifs, ZnF1 and ZnF2, at the C terminus (7). Our recent study demonstrated that ZnF1 is required for CIF1 localization to the new FAZ tip and that both zinc finger motifs are required for interaction with CIF2 (18). To investigate whether any of these motifs in CIF1 is required for interaction with CIF3, we co-expressed CIF1 mutants and CIF3 in T. brucei and carried out co-immunoprecipitation experiments. To this end, we first generated a CIF1 RNAi cell line by targeting the 3′UTR of CIF1 and then ectopically expressed triple HA–tagged CIF1, CIF1–ΔCC, CIF1–ZnF1mut, or CIF1–ZnF2mut, each of which contains the actin 3′UTR sequence (19), in the RNAi cell line. These cell lines have been reported in our recent work, which demonstrated that the coiled-coil motif and both zinc finger motifs are required for CIF1 function in cytokinesis (18). We then tagged CIF3 with an N-terminal PTP tag from one of its endogenous loci in these cell lines. RNAi by targeting CIF1–3′UTR and overexpression of WT and mutant CIF1 proteins were switched on by adding tetracycline to the cell culture and incubated for 24 h. Immunoprecipitation of CIF3 was able to pull down WT CIF1, CIF1–ΔCC, and CIF1–ZnF2mut, but not CIF1–ZnF1mut (Fig. 5A), demonstrating that ZnF1 in CIF1 is required for CIF1–CIF3 interaction. We previously showed that a phosphorylated form of CIF1 was detectable by Western blotting when cells were lysed by direct boiling in the SDS sampling buffer (7). However, when cells were lysed in the immunoprecipitation buffer and used for co-immunoprecipitation, the phosphorylated form was not detected by Western blotting in WT CIF1 and the two zinc finger mutants (Fig. 5A), likely because they were dephosphorylated during cell lysis. Intriguingly, under the same conditions, a slower migrating band was detected in CIF1–ΔCC mutant (Fig. 5A), suggesting that it may be a phosphorylated form of the CIF1–ΔCC mutant. It is unclear how the phosphorylated form of CIF1–ΔCC mutant, but not the phosphorylated form of WT CIF1, was preserved during cell lysis.
Figure 5.
CIF3 interaction with CIF1 requires zinc-finger motif 1 in CIF1. A, co-immunoprecipitation to investigate the interaction of CIF3 with various CIF1 mutants in T. brucei. CIF1 and its various mutants were either not expressed (−) or expressed (+) in the absence or presence of tetracycline. PTP-CIF3 was expressed from its endogenous locus. The black arrow in the CIF1–ΔCC–3HA panel indicates the phosphorylated form of CIF1–ΔCC–3HA protein. B, immunofluorescence microscopic analysis of PTP-tagged CIF3 in CIF1–3′UTR RNAi cells expressing 3HA-tagged CIF1, CIF1–ΔCC, CIF1–ZnF1mut, or CIF1–ZnF2mut. Noninduced control cells and tetracycline-induced cells were co-immunostained with FITC-conjugated anti-HA mAb and anti-protein A pAb. Scale bars: 5 μm. C, quantitation of 2N2K cells with different CIF3 localization in noninduced control cells and tetracycline-induced cells. Total numbers of cells counted are as follows: CIF1–3HA cell line, 596 (−Tet) and 613 (+Tet); CIF1–ΔCC cell line, 602 (−Tet) and 618 (+Tet); CIF1–ZnF1mut cell line, 588 (−Tet) and 593 (+Tet); CIF1–ZnF2mut cell line, 626 (−Tet) and 604 (+Tet). Error bars indicated S.D. from three independent experiments. ***, p < 0.001. D, Western blotting to examine the level of PTP-tagged CIF3 in CIF1–3′UTR RNAi cells expressing 3HA-tagged CIF1, CIF1–ΔCC, CIF1–ZnF1mut, or CIF1–ZnF2mut. TbPSA6 served as the loading control. The black arrows in the CIF1–3HA panel and the CIF1–ΔCC–3HA panel indicate the phosphorylated form of CIF1–3HA and CIF1–ΔCC–3HA, respectively.
Interaction with CIF1 is required for maintaining CIF3 stability
We next investigated the requirement of the structural motifs in CIF1 for CIF3 localization by immunofluorescence microscopy. In CIF1–3′UTR RNAi cells expressing WT CIF1, CIF3 localized to the new FAZ tip, similar to that in the noninduced control cells (Fig. 5, B and C). However, in CIF1–3′UTR RNAi cells expressing the CIF1–ΔCC mutant, CIF3 was not restricted to the distal tip of the new FAZ as in the noninduced controls cells, but instead it was spread over to either the anterior one-third length or the full length of the new FAZ in almost all of the cells examined (Fig. 5, B and C). In these cells, CIF1–ΔCC overlapped with CIF3 (Fig. 5B). In CIF1–3′UTR RNAi cells expressing CIF1–ZnF1mut, CIF3 was not detectable at the new FAZ tip, similar to CIF1–ZnF1mut (Fig. 5, B and C). In CIF1–3′UTR RNAi cells expressing CIF1–ZnF2mut, however, CIF3 still localized to the new FAZ tip, where it co-localized with CIF1–ZnF2mut (Fig. 5, B and C).
Because depletion of CIF1 caused CIF3 degradation (Fig. 4D), we asked whether disruption of CIF1–CIF3 interaction also caused CIF3 degradation. Knockdown of CIF1 by targeting CIF1–3′UTR destabilized CIF3 (Fig. 5D), similar to the effect through knockdown of CIF1 by targeting the CIF1 coding region (Fig. 4D). Ectopic expression of CIF1, CIF1–ΔCC, or CIF1–ZnF2mut in CIF1–3′UTR RNAi cells all stabilized CIF3 (Fig. 5D), but ectopic expression of CIF1–ZnF1mut in CIF1–3′UTR RNAi cells still caused CIF3 degradation, albeit the degree of CIF3 degradation was less severe than that in CIF1–3′UTR RNAi cells alone (Fig. 5D). When cells were lysed by boiling in the SDS sampling buffer, a slower migrating band or the phosphorylated form of WT CIF1 and CIF1–ΔCC mutant was detected (Fig. 5D, black arrows), but no slower migrating band of CIF1–ZnF1mut and CIF1–ZnF2mut was detected (Fig. 5D), suggesting that the two zinc finger mutants were not phosphorylated in vivo. The underlying mechanism remains to be investigated. Nevertheless, these results suggest that interaction with CIF1 is required for maintaining CIF3 protein stability.
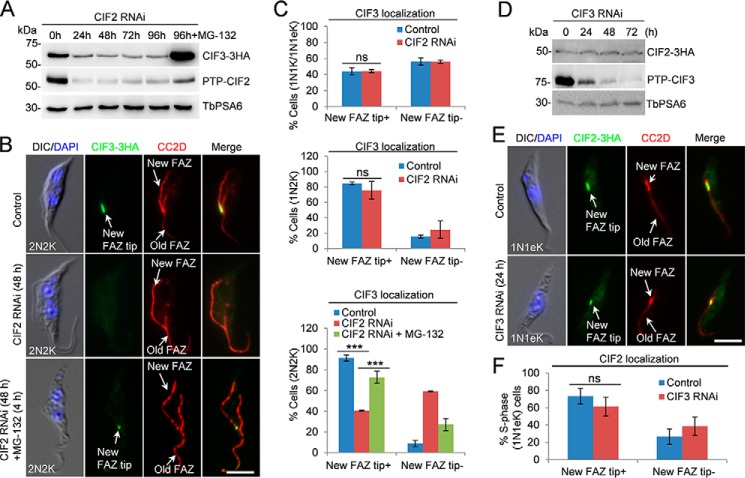
CIF2 is required for maintaining CIF3 stability
CIF2 is only detectable at the new FAZ tip during the S phase of the cell cycle (8) and interacts with CIF1 but not CIF3 (Fig. 2, A and B), suggesting that CIF2 may function upstream of CIF3 in the cytokinesis regulatory pathway. To test this hypothesis, we investigated whether CIF2 depletion affected CIF3 localization and stability. CIF3 was endogenously tagged with a triple HA epitope in CIF2 RNAi cell line, and Western blotting showed that upon CIF2 depletion, CIF3 protein was degraded (Fig. 6A), similar to that in CIF1 RNAi cells (Fig. 4D). Treatment of the CIF2 RNAi cells with the proteasome inhibitor MG-132 stabilized CIF3 (Fig. 6A), indicating that CIF3 was destabilized in CIF2 RNAi cells. Immunofluorescence microscopy showed that knockdown of CIF2 disrupted the localization of CIF3 in 2N2K cells, but not in 1N1K and 1N2K cells (Fig. 6, B and C). In the presence of MG-132, CIF3 localization to the new FAZ tip in 2N2K cells was restored (Fig. 6, B and C). These results suggest that the lack of CIF3 signal at the new FAZ tip in 2N2K cells was because of degradation of CIF3. Given that CIF2 depletion caused CIF1 degradation (8) and CIF1 depletion caused CIF3 degradation (Fig. 4D), the destabilization of CIF3 in CIF2 RNAi cells likely was attributed to the degradation of CIF1, but not because of the direct effect of CIF2 depletion, as CIF2 and CIF3 do not form a complex (Fig. 2A).
Figure 6.
CIF2 is required for maintaining CIF3 stability. A, effect of CIF2 depletion on CIF3 protein level. CIF2 RNAi was induced for 96 h. The proteasome inhibitor MG-132 was added after CIF2 RNAi was induced for 88 h and incubated for an additional 8 h. TbPSA6 served as the loading control. B, effect of CIF2 depletion on CIF3 localization. Cells were co-immunostained with FITC-conjugated anti-HA mAb and anti-CC2D antibody to label CIF3–3HA and the FAZ, respectively. Scale bar: 5 μm. C, quantification of the cells with different CIF3 localization patterns in control and CIF2 RNAi cells (48 h). Total numbers of cells counted are as follows: 1N1K/1N1eK, 256 (control) and 245 (CIF2 RNAi); 1N2K, 90 (control) and 112 (CIF2 RNAi); 2N2K, 215 (control), 226 (CIF2 RNAi) and 216 (CIF2 RNAi+MG-132). The results were presented as mean percentage ± S.D. (n = 3). ***, p < 0.01; ns, no statistical significance. D, effect of CIF3 depletion on CIF2 protein level. CIF3 RNAi was induced for 72 h. TbPSA6 served as the loading control. E, depletion of CIF3 did not affect CIF2 localization to the new FAZ tip. Cells were co-immunostained with FITC-conjugated anti-HA mAb and anti-CC2D antibody to label CIF2–3HA and the FAZ, respectively. CIF3 RNAi was induced for 24 h. Scale bar: 5 μm. F, quantification of cells with different CIF2 localization patterns in S-phase cells (1N1K cells with a short new FAZ). Total number of cells counted are as follows: 223 (control) and 219 (CIF3 RNAi). The results were presented as mean percentage ± S.D. (n = 3). ns, no statistical significance.
We next investigated the effect of CIF3 depletion on CIF2 localization and stability. CIF2 was endogenously tagged with a triple HA epitope in CIF3 RNAi cell line, and Western blotting showed that CIF3 depletion did not affect CIF2 protein stability (Fig. 6D). Immunofluorescence microscopy showed that CIF3 depletion did not affect CIF2 localization to the new FAZ tip in S-phase cells (Fig. 6, E and F). These results suggest that CIF2 functions upstream of CIF3 in the cytokinesis regulatory pathway by maintaining CIF1 at the new FAZ tip such that CIF1 maintains CIF3 stability.
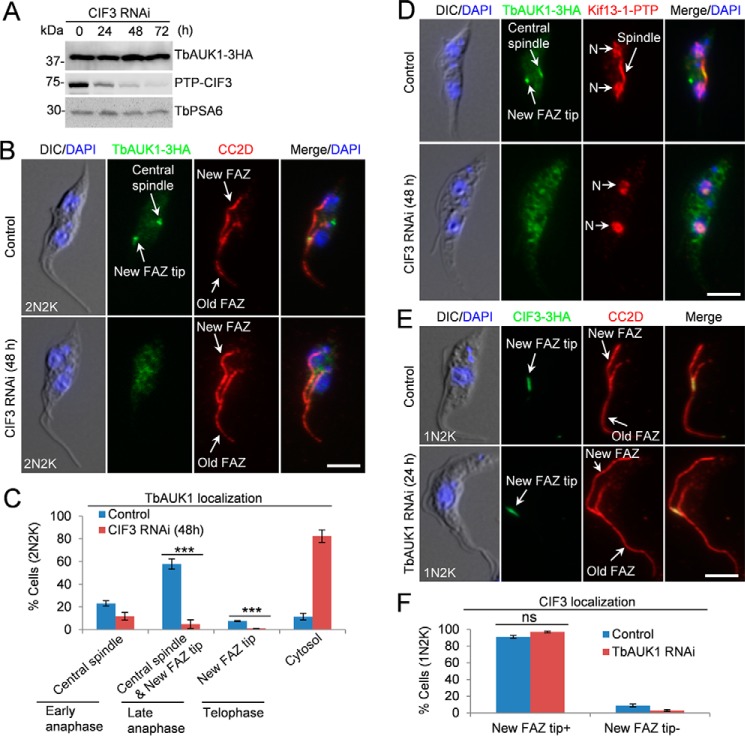
CIF3 is required for targeting TbAUK1 to the new FAZ tip at late anaphase
TbAUK1 is targeted to the new FAZ tip during late anaphase (20, 21), which depends on CIF1 and TbPLK (7). Given that CIF3 forms a complex with CIF1 (Fig. 2, B and C), we tested whether TbAUK1 localization to the new FAZ tip also depends on CIF3. To this end, TbAUK1 was endogenously tagged with a triple HA epitope in cells harboring the CIF3 RNAi construct. Western blotting demonstrated that TbAUK1 protein level was not affected by CIF3 RNAi (Fig. 7A). Immunofluorescence showed that CIF3 depletion impaired TbAUK1 localization to the new FAZ tip (Fig. 7, B and C). Among the 2N2K cells that represent different cell cycle stages from early anaphase to telophase, TbAUK1 was detected at the central spindle in early anaphase cells, at the spindle and the new FAZ tip in late anaphase cells, and at the new FAZ tip in telophase cells (20) (Fig. 7C). It was noted that TbAUK1 localization at the central spindle was also affected by CIF3 RNAi (Fig. 7, B and C). However, this was not a direct effect of CIF3 depletion, but rather because the 2N2K cells that accumulated in the CIF3 RNAi population had already passed through anaphase (disassembled the mitotic spindle) and most of them were telophase cells (Fig. 3, E and F). In control cells, Kif13–1 was detected in the nuclei and the spindle and TbAUK1 was detected at the central spindle and the new FAZ tip during late anaphase (Fig. 7D). In CIF3 RNAi cells, Kif13–1 was detected in the two nuclei, but not the spindle, in the majority (>90%) of the 2N2K cells (Fig. 7D and Fig. 3, E and F), and TbAUK1 was detected in the cytosol (Fig. 7D). These results confirmed that the lack of localization of TbAUK1 to the spindle in CIF3 RNAi cells is not a direct effect of CIF3 depletion.
Figure 7.
CIF3 is required for TbAUK1 localization to the new FAZ tip. A, TbAUK1 protein level was not changed in CIF3 RNAi cells. TbPSA6 served as the loading control. B, CIF3 depletion disrupted TbAUK1 localization to the new FAZ tip. CIF3 RNAi was induced for 48. Cells were co-immunostained with FITC-conjugated anti-HA mAb and anti-CC2D antibody to label TbAUK1–3HA and the FAZ, respectively. Scale bar: 5 μm. C, quantification of TbAUK1 localization in control and CIF3-depleted cells. Total number of cells counted are as follows: 307 (control) and 286 (CIF3 RNAi). The results were presented as mean percentage ± S.D. (n = 3). ***, p < 0.001. D, RNAi of CIF3 arrested cells at the post-mitotic phase prior to cytokinesis initiation. Kif13–1 was used as the spindle marker. N, nucleus. Scale bar: 5 μm. E, depletion of TbAUK1 did not affect CIF3 localization to the new FAZ tip. Cells were co-immunostained with FITC-conjugated anti-HA mAb and anti-CC2D antibody to label CIF3–3HA and the FAZ, respectively. Scale bar: 5 μm. F, quantification of CIF3 localization in control cells and TbAUK1 RNAi cells. Total number of cells counted are as follows: 301 (control) and 301 (TbAUK1 RNAi). Results were presented as mean percentage ± S.D. (n = 3). ns, no statistical significance.
Conversely, the effect of depletion and inhibition of TbAUK1 on CIF3 localization was also investigated. CIF3 was tagged with a triple HA epitope in TbAUK1 RNAi cells, and immunofluorescence microscopy showed that CIF3 remained at the new FAZ tip in TbAUK1 RNAi cells (Fig. 7, E and F). Altogether, these results suggest that CIF3 functions upstream of TbAUK1 in the cytokinesis regulatory pathway by cooperating with CIF1 to target TbAUK1 to the new FAZ tip during late anaphase. During early cell cycle stages, TbAUK1 plays additional roles in spindle assembly and chromosome segregation (20, 21); this mitotic function of TbAUK1 is independent of CIF3 and CIF1, which function at the new FAZ tip to regulate cytokinesis initiation. In this regard, it is only in the cytokinesis regulatory pathway that TbAUK1 acts downstream of the CIF1–CIF3 complex.
Discussion
Cytokinesis in T. brucei is known to be initiated from the anterior tip of the new FAZ. Therefore, the factors that play a direct role in cytokinesis initiation are anticipated to localize to the new FAZ tip prior to cytokinesis initiation. CIF3 is a new FAZ tip-localizing protein that plays an essential role in cytokinesis initiation in the procyclic form. Several lines of evidence support the role of CIF3 in cytokinesis initiation. First, RNAi-mediated ablation of CIF3 in the procyclic form of T. brucei inhibited cleavage furrow ingression (Fig. 3, C, D, and G). Secondly, depletion of CIF3 disrupted the localization of the cytokinesis regulator CIF1 to the new FAZ tip during G2 and mitotic phases (Fig. 4, B and C). Finally, depletion of CIF3 disrupted the localization of TbAUK1 to the new FAZ tip during late anaphase (Fig. 7, B and C). The effect of CIF3 depletion on TbAUK1 localization likely is through disrupting the localization of CIF1, which is known to be required for TbAUK1 localization to the new FAZ tip (7). Nevertheless, these results uncovered the mechanistic role of CIF3 in cytokinesis initiation through maintaining CIF1 and TbAUK1 at the new FAZ tip for TbAUK1 to initiate cleavage furrow ingression.
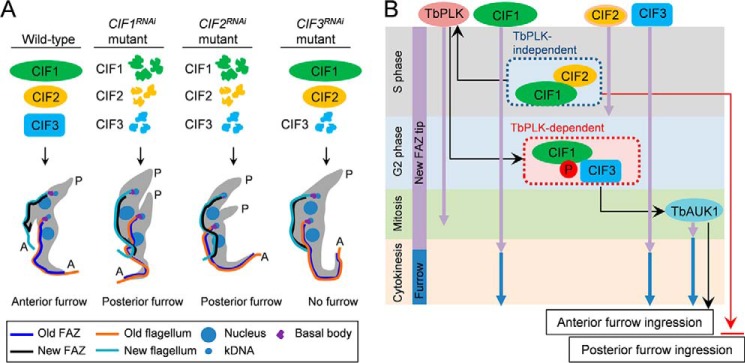
The finding that CIF3 RNAi cells did not initiate a posterior cleavage furrow (Fig. 3D) is surprising, as RNAi of CIF1 and RNAi of CIF2 both caused cleavage furrow ingression from the cell posterior (7, 8). A closer comparison of these three RNAi cell lines (Fig. 8A) revealed that CIF3 RNAi did not destabilize CIF1 and CIF2 proteins (Figs. 4A and 6D), whereas CIF1 RNAi destabilized CIF2 (8) and CIF3 (Fig. 4D) and CIF2 RNAi destabilized CIF1 (8) and CIF3 (Fig. 6A). Therefore, the difference between CIF3 RNAi cell line and CIF1 RNAi and CIF2 RNAi cell lines lies in the presence or absence of CIF1 and CIF2 proteins (Fig. 8A). It is thus likely that CIF1 and/or CIF2 may play an additional role in inhibiting cleavage furrow ingression from the cell posterior (Fig. 8B). We recently ectopically overexpressed CIF1 zinc finger 1 mutant (CIF1–ZnF1mut) in CIF1–3′UTR RNAi cells, and we found that despite the mislocalization of CIF1–ZnF1mut to the cytosol, the CIF1–3′UTR RNAi cells expressing CIF1–ZnF1mut failed to initiate cleavage furrow from the posterior end of the cell (18). Moreover, ectopic overexpression of CIF1 in CIF2 RNAi cells also repressed cleavage furrow ingression from the cell posterior.3 It appears that as long as CIF1 protein is absent or its level is decreased, either through RNAi of CIF1 (7) or because of destabilization by CIF2 RNAi (8), cleavage furrow ingression from the cell posterior will occur. Future work will be directed to understand the underlying mechanisms.
Figure 8.
Summary of the phenotypes of CIF1, CIF2, and CIF3 RNAi and a mode of action of cytokinesis regulators at the new FAZ tip in T. brucei. A, summary of the phenotypes caused by CIF1 RNAi, CIF2 RNAi, and CIF3 RNAi and the effect on the stability of the three proteins in the RNAi cell lines. B, a mode of action of cytokinesis regulators at the new FAZ tip and the cleavage furrow. Purple arrows indicate the duration of the time of the cytokinesis regulators at the new FAZ tip during the cell cycle, whereas blue arrows indicate the duration of the cytokinesis regulators at the cleavage furrow. Cell cycle stages are indicated at the left. Black arrows indicate the positive effect by TbPLK, the CIF1–CIF2 complex, the CIF1–CIF3 complex, and TbAUK1, whereas the red arrow indicates the inhibitory effect by CIF1.
Our immunoprecipitation results suggest that CIF1, CIF2, and CIF3 form two separate protein assemblies, the CIF1–CIF2 complex and the CIF1–CIF3 complex (Fig. 2). Immunoprecipitation of CIF2 did not pull down CIF3 (Fig. 2A) and vice versa (data not shown), suggesting that the two proteins either do not interact directly or interact in such a weak manner that the complex did not tolerate detergent treatment during cell lysis. It is possible that a tri-protein complex formed by CIF1, CIF2, and CIF3 is not present in T. brucei, or if the tri-protein complex does exist, it either is unstable or only is formed transiently during the S phase such that it was not readily precipitated by co-IP. CIF2 is only detectable at the new FAZ tip during the S phase of the cell cycle (8); therefore, the CIF1–CIF2 complex is likely only formed at the new FAZ tip during S phase. CIF1 and CIF3 both emerge at the new FAZ tip from early S phase and remain associated with the new FAZ tip until early cytokinesis (Fig. 1B). It is unclear whether CIF1 and CIF3 form a complex from early S phase or after S phase when CIF2 disappears from the new FAZ tip (8). Through its N-terminal EF-hand motifs, CIF2 binds to the two zinc finger motifs of CIF1 (18). CIF3 interaction with CIF1 also requires the zinc finger motif 1 of CIF1 (Fig. 5), but it remains to be determined whether CIF3 binds to the zinc finger motif directly. If CIF3 does bind to the zinc finger of CIF1 directly, it will be interesting to test whether CIF2 and CIF3 compete for binding to the zinc finger of CIF1.
Based on our earlier findings (7, 8) and the current work, the cytokinesis regulatory pathway in the procyclic form of T. brucei, which consists of two evolutionarily conserved protein kinases and three trypanosome-specific proteins, is further elaborated (Fig. 8B). This novel cytokinesis regulatory pathway operates at the anterior tip of the newly assembled FAZ from early S phase to cytokinesis initiation. Four of the five proteins, TbPLK, CIF1, CIF2, and CIF3, are targeted to the new FAZ tip from early S phase (Fig. 8B, black arrows), but they disappear from the new FAZ tip at different times of the cell cycle. CIF2 disappears from the new FAZ tip after S phase, TbPLK disappears from the new FAZ tip after early anaphase, and CIF1 and CIF3 disappear from the new FAZ tip after early cytokinesis (Fig. 8B, black arrows). CIF1 and a small amount of CIF3 protein additionally localize to the cleavage furrow during cytokinesis (Fig. 1B). TbAUK1, however, is only targeted to the new FAZ tip during late anaphase (20), and it co-localizes with CIF1 at the new FAZ tip from late anaphase to telophase and then at the cleavage furrow during cytokinesis (7). The dynamic localizations of these cytokinesis regulators suggest that they may act sequentially to control cytokinesis. Among these cytokinesis regulators, CIF1 appears to play a pivotal role in the cytokinesis regulatory pathway by targeting and/or maintaining multiple cytokinesis regulators, including TbPLK, CIF2, CIF3, and TbAUK1, to the new FAZ tip. Future work will focus on identifying new cytokinesis regulators and exploiting their interplay with the known regulators for a better understanding of the mechanism of the unusual mode of cytokinesis in T. brucei.
Experimental procedures
Trypanosome cell culture
The T. brucei strain 29–13 (19) was cultured in SDM-79 medium supplemented with 10% heat-inactivated fetal bovine serum (Atlanta Biologicals), 15 μg/ml G418, and 50 μg/ml hygromycin at 27 °C. The T. brucei strain 427 was grown in SDM-79 medium containing 10% heat-inactivated fetal bovine serum at 27 °C.
RNAi
To generate CIF3 RNAi cell line, a 500-bp DNA fragment (nucleotides 401–900) corresponding to the middle portion of the coding sequence of CIF3 was cloned into the pZJM vector (22). The pZJM–CIF3 plasmid was linearized with SacII and transfected into the 29–13 strain by electroporation according to our published procedures (23). Transfectants were selected with 2.5 μg/ml phleomycin and cloned by limiting dilution in a 96-well plate. Four clonal cell lines were selected and characterized. The CIF1 RNAi cell line, CIF2 RNAi cell line, TbPLK RNAi cell line, and TbAUK1 RNAi cell line have been reported previously (7, 8, 20, 24). To induce RNAi, cells were cultured in SDM-79 medium containing 1.0 μg/ml tetracycline, and cell growth was monitored daily by counting the cells with a hemacytometer.
In situ epitope tagging of proteins
For all the epitope-tagging cell lines described below, cells were transfected by electroporation, selected with appropriate antibiotics (10 μg/ml blasticidin for plasmids containing BSD gene, 1.0 μg/ml puromycin for plasmids containing PAC gene, 40 μg/ml G418 for plasmids containing NEO gene, or 50 μg/ml hygromycin for plasmids containing HYG gene), and cloned by limiting dilution in a 96-well plate.
For endogenous tagging of CIF3 with an N-terminal PTP epitope in CIF3 RNAi cell line, CIF1 with an N-terminal PTP epitope in CIF1 RNAi cell line, and CIF2 with an N-terminal PTP epitope in CIF2 RNAi cell line, the PCR-based epitope tagging method (25) was carried out. Transfectants were selected with blasticidin.
For C-terminal epitope tagging of CIF3 at one of its endogenous loci, a 653-bp fragment corresponding to the C-terminal coding region of CIF3 was cloned into pC–3HA–BSD and pC–3HA–PAC vectors. The pC–CIF3–3HA–BSD plasmid was linearized with BmgBI and transfected into the 427 cell line. The pC–CIF3–3HA–PAC plasmid was linearized with BmgBI and transfected into the cell lines harboring pZJM–CIF1, pZJM–CIF2, pZJM–TbPLK, or pZJM–TbAUK1 plasmid.
For co-localization of CIF1–PTP with CIF3–3HA, the cell line harboring pC–CIF3–3HA–BSD was transfected with pN–PTP–CIF1–PAC, which was linearized with NcoI. For co-localization of CIF2–PTP with CIF3–3HA, pC–CIF3–3HA–PAC was transfected into the cell line containing pC–CIF2–PTP–NEO.
For tagging of CIF1 in CIF3 RNAi cell line, pC–CIF1–3HA–PAC was linearized with XcmI and transfected into CIF3 RNAi cell line expressing endogenously PTP-tagged CIF3. For tagging of CIF2 in CIF3 RNAi cell line, the PCR-based tagging method (25) was used to transfect CIF3 RNAi cell line expressing endogenously PTP-tagged CIF3. Transfectants were selected with puromycin.
For endogenous epitope tagging of TbAUK1 in CIF3 RNAi cell line, the pC–TbAUK1–3HA–PAC vector (7) was linearized with SphI and transfected into CIF3 RNAi cell line expressing endogenously PTP-tagged CIF3. For endogenous PTP tagging of Kif13–1 in CIF3 RNAi cell line expressing 3HA-tagged TbAUK1, the PCR-based tagging method (25) was used. Transfectants were selected with blasticidin in addition to G418, hygromycin, phleomycin, and puromycin. For endogenous PTP tagging of CUL6 in cells expressing CIF1–3HA or CIF3–3HA, the pC–CUL6–PTP–NEO vector (26) was linearized with MfeI, and transfected into the appropriate cell line.
RNAi of CIF1 by targeting CIF1–3′UTR and complementation of CIF1–3′UTR RNAi
To generate a CIF1 RNAi cell line for complementation studies, a 500-bp DNA fragment from the 3′UTR of CIF1 was cloned into the pZJM–PAC vector. The 3′UTR of CIF1 is 861 bp, so the fragment used in our RNAi experiment does not overlap with the downstream gene. The resulting plasmid was electroporated into the 29–13 cell line. Transfectants were selected with 1.0 μg/ml puromycin in addition to 15 μg/ml G418, and 50 μg/ml hygromycin and cloned by limiting dilution in a 96-well plate. Subsequently, CIF1, CIF1–ΔCC, CIF1–ZnF1mut, and CIF1–ZnF2mut were each cloned into pLew100–3HA–BLE vector, which contains the 3′UTR from actin (19), and the resulting plasmids were electroporated into the pZJM–CIF1–3′UTR cell line. Transfectants were selected with 2.5 μg/ml phleomycin in addition to 1.0 μg/ml puromycin, 15 μg/ml G418, and 50 μg/ml hygromycin, and cloned by limiting dilution.
To tag CIF3 with a PTP epitope in CIF1–3′UTR RNAi and CIF1–3′UTR RNAi complementation cell lines, the PCR-based tagging method (25) was carried out by integrating a C-terminal PTP epitope and a blasticidin S deaminase (BSD) gene immediately downstream of CIF3. Transfectants were selected with 10 μg/ml blasticidin in addition to 2.5 μg/ml phleomycin, 1.0 μg/ml puromycin, 15 μg/ml G418, and 50 μg/ml hygromycin and cloned by limiting dilution.
Immunoprecipitation
Immunoprecipitation was carried out essentially as described previously (8). Briefly, cells (5 × 107) were lysed in in 0.5 ml immunoprecipitation buffer (25 mm Tris-HCl, pH 7.6, 500 mm NaCl, 1 mm DTT, 1% Nonidet P-40, and protease inhibitor mixture). Cell lysate was cleared by centrifugation, and the supernatant (0.4 ml) was incubated with 50 μl settled IgG Sepharose beads (GE Healthcare) for 1 h at 4 °C. The remaining 0.1 ml supernatant was used as the input. Beads were then washed six times with the immunoprecipitation buffer. Immunoprecipitated proteins were eluted with 40 μl SDS (10%), mixed with 10 μl 5× SDS sampling buffer, and 25 μl of the samples was loaded onto SDS-PAGE. Proteins were transferred onto a PVDF membrane, and immunoblotted with anti-HA mAb (1:2500 dilution) and anti-protein A polyclonal antibody (1:2000 dilution).
Immunofluorescence microscopy
Cells (5 × 104) were adhered to the glass coverslips for 30 min at room temperature, fixed with 2 ml cold methanol at −20 °C for 20 min, and then rehydrated with PBS for 5 min at room temperature. Cells were blocked with 3% BSA in PBS for 1 h at room temperature, and then incubated with the primary antibody for 1 h at room temperature. The following primary antibodies were used: FITC-conjugated anti-HA mAb (1:400 dilution, Sigma-Aldrich), anti-protein A polyclonal antibody (1:400 dilution, Sigma-Aldrich), anti-CC2D polyclonal antibody for the FAZ (1:2000 dilution) (5). Cells were washed three times with PBS and then incubated with FITC-conjugated anti-mouse IgG (1:400 dilution, Sigma-Aldrich) or Alexa Fluor®–conjugated anti-rabbit IgG (1:400 dilution, Molecular Probes) for 1 h at room temperature. Cells on the coverslips were washed three times with PBS, mounted with DAPI-containing VECTASHIELD mounting medium (Vector Labs), and imaged using an inverted fluorescence microscope (Olympus IX71) equipped with a cooled charge-coupled device camera (model Orca-ER, Hamamatsu) and a PlanApo N 60 × 1.42 numerical aperture lens. Images were acquired using the Slidebook 5 software.
Statistical analysis
Statistical analysis was performed using the Student's t test in the Microsoft Excel software. Detailed n values for each panel in the figures were stated in the corresponding legends. For immunofluorescence microscopy, images were randomly taken and all cells in each image were counted.
Author contributions
Y. K. and Z. L. conceptualization; Y. K., H. H., and Z. L. formal analysis; Y. K. and H. H. validation; Y. K., H. H., and Q. Z. visualization; Y. K., H. H., and Q. Z. methodology; Y. K., H. H., and Z. L. writing-review and editing; Z. L. supervision; Z. L. funding acquisition; Z. L. investigation; Z. L. writing-original draft; Z. L. project administration.
Acknowledgments
We are grateful to Drs. Arthur Günzl and Cynthia Y. He for providing the epitope tagging vector and anti-CC2D antibody, respectively.
This work was supported by National Institutes of Health Grants AI101437 and AI118736. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
H. Hu and Z. Li, unpublished data.
- FAZ
- flagellum attachment zone.
References
- 1. Haeusser D. P., and Margolin W. (2016) Splitsville: Structural and functional insights into the dynamic bacterial Z ring. Nat. Rev. Microbiol. 14, 305–319 10.1038/nrmicro.2016.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cheffings T. H., Burroughs N. J., and Balasubramanian M. K. (2016) Actomyosin ring formation and tension generation in eukaryotic cytokinesis. Curr. Biol. 26, R719–R737 10.1016/j.cub.2016.06.071 [DOI] [PubMed] [Google Scholar]
- 3. Müller S., and Jürgens G. (2016) Plant cytokinesis—no ring, no constriction but centrifugal construction of the partitioning membrane. Semin. Cell Dev. Biol. 53, 10–18 10.1016/j.semcdb.2015.10.037 [DOI] [PubMed] [Google Scholar]
- 4. Kohl L., Robinson D., and Bastin P. (2003) Novel roles for the flagellum in cell morphogenesis and cytokinesis of trypanosomes. EMBO J. 22, 5336–5346 10.1093/emboj/cdg518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhou Q., Liu B., Sun Y., and He C. Y. (2011) A coiled-coil- and C2-domain-containing protein is required for FAZ assembly and cell morphology in Trypanosoma brucei. J. Cell Sci. 124, 3848–3858 10.1242/jcs.087676 [DOI] [PubMed] [Google Scholar]
- 6. Wheeler R. J., Scheumann N., Wickstead B., Gull K., and Vaughan S. (2013) Cytokinesis in Trypanosoma brucei differs between bloodstream and tsetse trypomastigote forms: Implications for microtubule-based morphogenesis and mutant analysis. Mol. Microbiol. 90, 1339–1355 10.1111/mmi.12436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhou Q., Gu J., Lun Z. R., Ayala F. J., and Li Z. (2016) Two distinct cytokinesis pathways drive trypanosome cell division initiation from opposite cell ends. Proc. Natl. Acad. Sci. U.S.A. 113, 3287–3292 10.1073/pnas.1601596113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhou Q., Hu H., and Li Z. (2016) An EF-hand–containing protein in Trypanosoma brucei regulates cytokinesis initiation by maintaining the stability of the cytokinesis initiation factor CIF1. J. Biol. Chem. 291, 14395–14409 10.1074/jbc.M116.726133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kumar P., and Wang C. C. (2006) Dissociation of cytokinesis initiation from mitotic control in a eukaryote. Eukaryot. Cell 5, 92–102 10.1128/EC.5.1.92-102.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hammarton T. C., Kramer S., Tetley L., Boshart M., and Mottram J. C. (2007) Trypanosoma brucei Polo-like kinase is essential for basal body duplication, kDNA segregation and cytokinesis. Mol. Microbiol. 65, 1229–1248 10.1111/j.1365-2958.2007.05866.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tu X., Kumar P., Li Z., and Wang C. C. (2006) An Aurora kinase homologue is involved in regulating both mitosis and cytokinesis in Trypanosoma brucei. J. Biol. Chem. 281, 9677–9687 10.1074/jbc.M511504200 [DOI] [PubMed] [Google Scholar]
- 12. Li Z., and Wang C. C. (2006) Changing roles of Aurora-B kinase in two life cycle stages of Trypanosoma brucei. Eukaryot. Cell 5, 1026–1035 10.1128/EC.00129-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hu H., Zhou Q., and Li Z. (2015) A novel basal body protein that is a Polo-like kinase substrate is required for basal body segregation and flagellum adhesion in Trypanosoma brucei. J. Biol. Chem. 290, 25012–25022 10.1074/jbc.M115.674796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. McAllaster M. R., Ikeda K. N., Lozano-Núñez A., Anrather D., Unterwurzacher V., Gossenreiter T., Perry J. A., Crickley R., Mercadante C. J., Vaughan S., and de Graffenried C. L. (2015) Proteomic identification of novel cytoskeletal proteins associated with TbPLK, an essential regulator of cell morphogenesis in Trypanosoma brucei. Mol. Biol. Cell 26, 3013–3029 10.1091/mbc.e15-04-0219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Archer S. K., Inchaustegui D., Queiroz R., and Clayton C. (2011) The cell cycle regulated transcriptome of Trypanosoma brucei. PLoS One 6, e18425 10.1371/journal.pone.0018425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chan K. Y., Matthews K. R., and Ersfeld K. (2010) Functional characterisation and drug target validation of a mitotic kinesin-13 in Trypanosoma brucei. PLoS Pathog. 6, e1001050 10.1371/journal.ppat.1001050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wickstead B., Carrington J. T., Gluenz E., and Gull K. (2010) The expanded Kinesin-13 repertoire of trypanosomes contains only one mitotic Kinesin indicating multiple extra-nuclear roles. PLoS One 5, e15020 10.1371/journal.pone.0015020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hu H., Majneri P., Li D., Kurasawa Y., An T., Dong G., and Li Z. (2017) Functional analyses of the CIF1-CIF2 complex in trypanosomes identify the structural motifs required for cytokinesis. J. Cell Sci. 130, 4108–4119 10.1242/jcs.207134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wirtz E., Leal S., Ochatt C., and Cross G. A. (1999) A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99, 89–101 [DOI] [PubMed] [Google Scholar]
- 20. Li Z., Lee J. H., Chu F., Burlingame A. L., Günzl A., and Wang C. C. (2008) Identification of a novel chromosomal passenger complex and its unique localization during cytokinesis in Trypanosoma brucei. PLoS One 3, e2354 10.1371/journal.pone.0002354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li Z., Umeyama T., and Wang C. C. (2009) The Aurora kinase in Trypanosoma brucei plays distinctive roles in metaphase-anaphase transition and cytokinetic initiation. PLoS Pathog. 5, e1000575 10.1371/journal.ppat.1000575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang Z., Morris J. C., Drew M. E., and Englund P. T. (2000) Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J. Biol. Chem. 275, 40174–40179 10.1074/jbc.M008405200 [DOI] [PubMed] [Google Scholar]
- 23. Li Z., Zou C. B., Yao Y., Hoyt M. A., McDonough S., Mackey Z. B., Coffino P., and Wang C. C. (2002) An easily dissociated 26 S proteasome catalyzes an essential ubiquitin-mediated protein degradation pathway in Trypanosoma brucei. J. Biol. Chem. 277, 15486–15498 10.1074/jbc.M109029200 [DOI] [PubMed] [Google Scholar]
- 24. Li Z., Umeyama T., Li Z., and Wang C. C. (2010) Polo-like kinase guides cytokinesis in Trypanosoma brucei through an indirect means. Eukaryot. Cell 9, 705–716 10.1128/EC.00330-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shen S., Arhin G. K., Ullu E., and Tschudi C. (2001) In vivo epitope tagging of Trypanosoma brucei genes using a one step PCR-based strategy. Mol. Biochem. Parasitol. 113, 171–173 10.1016/S0166-6851(00)00383-2 [DOI] [PubMed] [Google Scholar]
- 26. Hu H., Zhou Q., Han X., and Li Z. (2017) CRL4WDR1 controls Polo-like kinase protein abundance to promote bilobe duplication, basal body segregation and flagellum attachment in Trypanosoma brucei. PLoS Pathog. 13, e1006146 10.1371/journal.ppat.1006146 [DOI] [PMC free article] [PubMed] [Google Scholar]