Abstract
Small noncoding RNAs (sncRNAs) regulate many genes in eukaryotic cells. Hua enhancer 1 (Hen1) is a 2′-O-methyltransferase that adds a methyl group to the 2′-OH of the 3′-terminal nucleotide of sncRNAs. The types and properties of sncRNAs may vary among different species, and the domain composition, structure, and function of Hen1 proteins differ accordingly. In mammals, Hen1 specifically methylates sncRNAs called P-element–induced wimpy testis-interacting RNAs (piRNAs). However, other types of sncRNAs that are methylated by Hen1 have not yet been reported, and the structures and the substrates of mammalian Hen1 remain unknown. Here, we report that mouse Hen1 (mHen1) performs 3′-end methylation of classical piRNAs, as well as those of most noncanonical piRNAs derived from rRNAs, small nuclear RNAs and tRNAs in murine spermatogonial stem cells. Moreover, we found that a distinct class of tRNA-derived sncRNAs are mHen1 substrates. We further determined the crystal structure of the putative methyltransferase domain of human Hen1 (HsHen1) in complex with its cofactor AdoMet at 2.0 Å resolution. We observed that HsHen1 has an active site similar to that of plant Hen1. We further found that the putative catalytic domain of HsHen1 alone exhibits no activity. However, an FXPP motif at its N terminus conferred full activity to this domain, and additional binding assays suggested that the FXPP motif is important for substrate binding. Our findings shed light on its methylation substrates in mouse spermatogonial stem cells and the substrate-recognition mechanism of mammalian Hen1.
Keywords: crystal structure, RNA methyltransferase, spermatogenesis, RNA, RNA modification, 3'-end methylation, Hen1, methyltransferase domain, piRNA, spermatogonial stem cell
Introduction
Small RNA-mediated RNA interference (RNAi) plays essential roles in regulating gene expression in eukaryotes (1, 2). Three major classes of small RNAs, microRNAs (miRNAs,4 ∼22 nucleotides (nt)), short-interfering RNAs (siRNAs, ∼21 nt), and P-element-induced wimpy testis (PIWI)-interacting RNAs (piRNAs, 24–31 nt), are defined according to their associated partners and functions (3–5). These small RNAs can associate with Argonaute (Ago) proteins to form RNA-induced silencing complexes (RISCs). Through base pairing between small RNAs and their RNA targets, RISCs can specifically recognize their targets and regulate gene expression transcriptionally or post-transcriptionally (1, 6).
Although biogenesis pathways are diverse, miRNAs and siRNAs in plants and piRNAs in animals, all contain a methylated 3′-terminal nucleotide (7–11). In Arabidopsis, the methylation of duplex miRNAs and siRNAs is mediated by the methyltransferase (MTase) protein Hen1 (HUA enhancer 1), which catalyzes the transfer of a methyl group from the cofactor S-adenosyl-l-methionine (AdoMet) to the 2′-hydroxyl of the 3′-terminal nucleotide of small RNAs (12). In hen1 mutant plants, miRNAs exhibit abnormal heterogeneous polyuridylation length or truncation at their 3′-unmethylated ends, and a considerably reduced abundance, resulting in pleiotropic phenotypes (13–17). The previously reported Arabidopsis thaliana Hen1 (AtHen1) crystal structure showed that AtHen1 consists of multiple domains: two dsRNA-binding domains (dsRBDs), La-motif–containing domain, protein peptidyl isomerase–like domain, and the C-terminal Rossmann fold methyltransferase (MTase) domain. The MTase domain can bind the product of the methyl donor S-adenylhomocysteine and exhibit MTase activity toward miRNA/miRNA* duplexes in a magnesium (Mg2+)-dependent manner. In addition, multiple domains in the N-terminal region can contribute to the recognition and binding of the target dsRNA (18).
In contrast to plants, Hen1 homologues in animals catalyze the methylation of piRNAs and single-stranded siRNAs but not small RNA duplexes (10, 11, 19–22). Mouse Hen1 (mHen1) is specifically expressed in testis and methylates the 3′-ends of piRNAs (9, 11, 21). Drosophila melanogaster Hen1 (DmHen1) uses Ago2-bound single-stranded siRNAs and PIWI-bound piRNAs as substrates. Loss of function of DmHen1 results in shortened length, decreased abundance, and perturbed functions of piRNAs (10). Similarly, the Hen1 homologue in zebrafish also methylates piRNAs in germline cells and is required for oocyte development (22). The substrates of mammalian Hen1 are quite different from the substrates in plants. The substrate of mammalian Hen1 is a single-stranded RNA; thus, the mammalian Hen1 lacks the dsRBD domains that exit in plant Hen1 to facilitate the binding of RNA duplex. The animal Hen1 is relatively small compared with plant Hen1, containing only an N-terminal MTase domain and a C-terminal domain (CTD). The CTD of Hen1 in zebrafish is responsible for the localization of Hen1 to nuage, a perinuclear area for piRNA biogenesis (22). In addition to animals and plants, Hen1 homologues have also been studied in bacteria, in which Hen1 is able to methylate single-stranded small regulatory RNAs and is involved in the bacterial RNA repair system (23, 24). The substrates and functions of Hen1 vary in different species, and the domain architectures of Hen1 may vary.
However, for mammalian Hen1 many questions remain unanswered, such as how many types of small noncoding RNAs (sncRNAs) bear 3′-end methylation, whether piRNAs are the sole substrate for mammalian Hen1, and the structure and molecular basis of the catalytic mechanism of mammalian Hen1. Here, we investigated the methylation pattern of sncRNAs in mouse spermatogonial stem cells (SSCs) by sodium periodate (NaIO4)–assisted deep sequencing. Knockout of hen1 in mouse SSCs resulted in loss of 3′-end methylation and shortening of ∼1 nucleotide of the 3′-end of piRNA. We identified a group of noncanonical piRNAs derived from rRNA (rRNAs), small nuclear RNAs (snRNAs), and transfer RNAs (tRNAs), most of which underwent mHen1-dependent 3′-methylation. We found a class of tRNA-derived sncRNAs that do not belong to the piRNAs and are ∼31–33 nt in length, and which are also methylated by mouse Hen1 (mHen1). Furthermore, we determined the crystal structure of the putative MTase domain of HsHen1 in complex with the cofactor AdoMet at a resolution of 2.0 Å. The structure revealed that this domain shares a similar allosteric arrangement as the MTase domain of AtHen1. Residues composing the active site as well as the FXPP motif at the N terminus of the putative MTase domain were required to methylate target RNAs. In addition, HsHen1 showed a preference of Mn2+ over Mg2+, similar to bacteria Hen1. Our structural and functional data led to the identification of a series of sites critical for the MTase activity of HsHen1 and also provide insight into the molecular basis for the role of Hen1 in methylation of piRNAs and other sncRNAs in mammals.
Results
Hen1-dependent methylation of piRNAs in SSCs
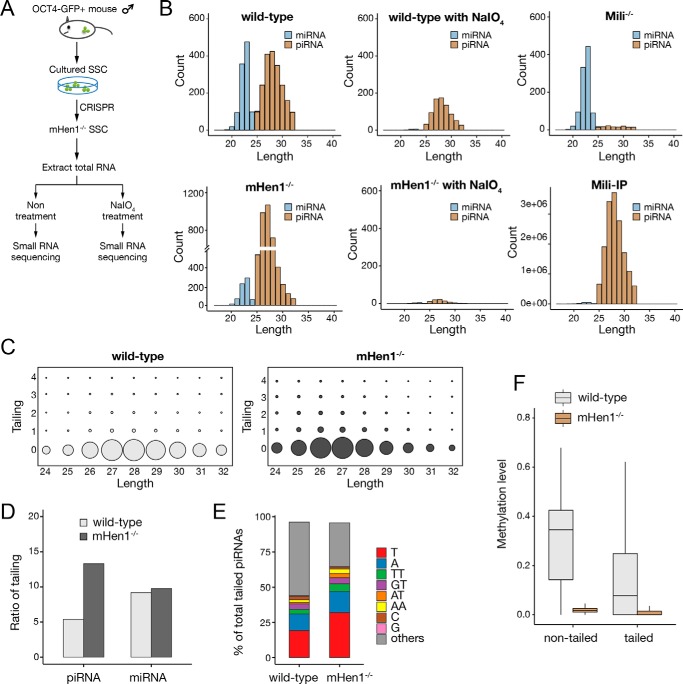
Spermatogonial stem cells have the capacity for self-renewal to maintain the stem-cell pool and differentiate into spermatocytes, spermatids, and finally spermatozoa. The isolated SSCs are able to grow in vitro and restore fertility after being transplanted into the seminiferous tubules of infertile recipient mice. Moreover, SSCs express high levels of piRNAs and mHen1, which make it an ideal cell model to study the Hen1 substrates. Previous studies in mouse testis showed that 3′-end of piRNA was methylated by mHen1 (21, 25). However, SSCs are rare in the adult testis (less than 0.1%), and the changes of sncRNAs in the SSCs upon mHen1 knockout were completely masked by the signal from other abundant cells in the testis. To explore the mHen1 substrates in mouse SSCs, we carried out sodium periodate (NaIO4)–assisted deep sequencing, a method previously used to investigate piRNA methylation (26, 27). We generated mHen1 knockout SSC lines by the CRISPR/Cas9 method as well as Mili (a piRNA-binding protein indispensable for piRNA biogenesis) knockout SSC lines (Fig. 1A). Total RNA from WT and mHen1−/− SSCs were treated with NaIO4 prior to constructing a cDNA library of the small RNAs. Lacking 2′-OH modifications, the hydroxyl groups on the 3′-terminal base of the sncRNAs were oxidized into a formyl group, which could not be ligated with a 3′ adaptor and were eliminated during reverse transcription with a 3′ adaptor–specific DNA primer during sncRNA cDNA library construction, whereas RNA molecules with a 2′-O-methyl modification at their 3′-end were unaffected (Figs. 1A and Fig. S1A). Equal amounts of synthetic single-stranded RNA (ssRNA) spike-ins with 2′-O-methylation at the 3′-end ribose, which were verified by a β-elimination reaction, were added into each sample before NaIO4 treatment for sncRNA quantification (Fig. S1A).
Figure 1.
mHen1-dependent 3′-methylation of canonical piRNAs in mouse SSCs. A, strategies to profile the methylated pattern of sncRNAs in vivo and to determine whether methylation is Hen1-dependent in mouse SSCs. B, composition of miRNAs and piRNAs according to their length distribution in WT SSCs with or without NaIO4 treatment, mHen1−/− SSCs with or without NaIO4 treatment, and Mili−/− SSC and Mili-IP in WT SSCs. The abundance (count) of miRNAs and piRNAs was corrected by exogenous spike-in normalization, except Mili-IP. The count in Mili-IP represents raw sequencing reads. Mili-IP, immunoprecipitated sncRNAs with Mili-specific antibody. C, trimming and tailing matrix in WT and mHen1−/− SSC; the x axis represents length of the 5′ genome-matched component (head) of overall piRNAs related sequences; the y axis represents the length of the tail added to the head. The piRNA sequence that could be perfectly matched to genome is considered to have no tail and therefore is positioned at y axis = 0. The area within the circle for each position indicates the relative abundance of overall tailed piRNAs and nontailed piRNAs matching to that particular position. D, percentage of piRNAs and miRNAs with tailing in WT and mHen1−/− SSC. E, percentage of different nucleotides tailed at piRNAs 3′-end. F, methylation level of tailed and nontailed piRNAs in WT and mHen1−/− SSC.
The sequencing results revealed that miRNAs and piRNAs were two major populations of sncRNAs in WT mouse SSCs (Fig. 1B and Table S1). The piRNAs in Mili−/− SSCs were almost diminished, consistent with previous studies that Mili is indispensable for pre-pachytene piRNAs production in mouse testis, whereas the miRNAs remained unchanged (Figs. 1B and Fig. S1B) (28, 29). In mHen1−/− SSCs, the abundance of total miRNA was slightly reduced, whereas the relative expression level of individual miRNA after normalization by total miRNAs was not changed, indicating mHen1 knockout may have some secondary effect on miRNA processing or stability. The miRNAs completely disappeared after treatment with NaIO4 in WT SSC, indicating that the elimination of sncRNAs without 3′-end modification by NaIO4 treatment was complete. By contrast, approximately half of the piRNAs in WT SSCs remained after NaIO4 treatment, indicating that a significant proportion of piRNAs was unmethylated. These methylated piRNAs disappeared in mHen1−/− SSCs upon NaIO4 treatment, supporting the conclusion that piRNA 3′-methylation is Hen1-dependent in mammals (22, 25). In contrast to previous studies that mHen1 knockout reduced piRNAs levels in the fly, zebrafish ovary and mouse testis (10, 22, 30, 31), as detected by 5′-end labeling of sncRNAs with γ-32P followed by PAGE or Northern blotting, the relative abundance of piRNAs increased more than 2-fold in mHen1−/− SSCs as detected by deep sequencing with methylated ssRNA oligos as spike-ins (Fig. 1B). To investigate the cause of this discrepancy, we compared the ligation efficiency of 3′-end 2′-O-methylated and non-2′-O-methylated synthetic ssRNA with the 3′ adaptor by T4 RNA ligase II. The result showed that 2′-O-methylation at the 3′-end reduced ligation efficiency of ssRNA more than 4-fold (Fig. S1C), indicating that the appearance of a piRNA abundance increase upon mHen1 knockout was likely due to the higher adaptor ligation efficiency of piRNAs without 3′-methylation during the cDNA library construction procedure. However, the relative methylation level of different sncRNAs still can be accurately calculated by comparing the sncRNAs read counts with or without NaIO4 treatment.
Notably, in contrast to a previous study in which the piRNA length has almost no change in the testis of mHen1 knockout mice (25), upon mHen1 knockout in the SSCs, the length of piRNAs became ∼1 nucleotide shorter than that in WT SSCs (Fig. 1, B and C). This discrepancy indicates that mHen1-dependent 3′-methylation may have different functions on the pre-pachytene piRNAs predominantly expressed in the SSCs and the pachytene piRNAs predominantly expressed in the adult mouse testis. Our result in the mouse SSCs is consistent with the previous report in zebrafish upon Hen1 knockout (22) and supports the hypothesis that 2′-O-methylation at the 3′-end of the piRNAs by Hen1 may contribute to the resistance of piRNAs to 3′-exonuclease trimming in germ cells. Moreover, 3′-end trimming of piRNAs appears to occur prior to 3′ tailing (Fig. 1C), consistent with the observations of hen1 mutant in plants (17), indicating that this phenomenon is conserved in plants and mammals. The nontemplate tailing ratio of piRNAs was about half that of the miRNAs in SSCs, and most of the tailing was more than 1 nucleotide (Fig. 1, D and E). The tailing ratio of piRNAs increased more than 2-fold in mHen1−/− SSCs, to ∼15% of total piRNAs. Interestingly, this increase was mostly contributed by single-nucleotide uridylation and adenylation, which accounted for about 50% of total tailing in mHen1−/− SSCs (Fig. 1E), in agreement with the observation that RNAs without 2′-O-methylation at the 3′-end are favorable substrates for uridylation and adenylation (32). Compared with the increased tailing of piRNAs by the loss of mHen1, the tailing of miRNAs was not affected, consistent with the lack of 2′-O-methylation at the 3′-end of miRNAs in mammals (Fig. 1E). Interestingly, those tailed piRNAs could still be methylated by mHen1, even though the methylation level was less than that of the nontailed piRNAs (Fig. 1F). This observation indicates that the 3′-methylation of piRNAs may normally take place in a processing step that occurs earlier than tailing in the cells.
Novel sncRNA substrates of mHen1 in mouse SSCs
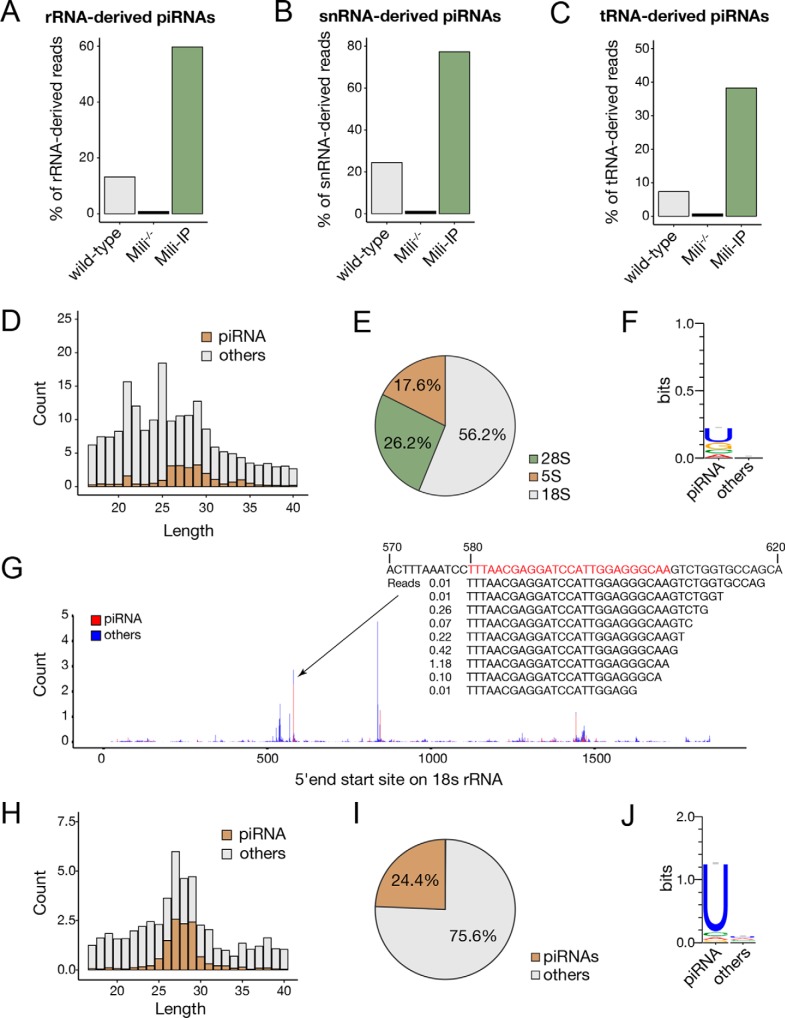
Beside the canonical piRNAs that generated from piRNA clusters, we found that a portion of piRNAs in SSCs was derived from rRNAs, snRNAs, and tRNAs, which were significantly enriched by anti-Mili immunoprecipitation (Mili-IP) and diminished under Mili knockout (Fig. 2, A–C; Table S1). Notably, ∼15% of rRNA-derived small RNAs in SSCs were piRNAs with a peak length of 25–32 nt (Fig. 2, A and D), whereas more than half were from 18S rRNA (Fig. 2E). Compared with other rRNA-derived sncRNAs, these rRNA-derived piRNAs had an obvious U bias at the 5′-end (Fig. 2F). The highly expressed rRNA-derived piRNAs were located in a few hotspots and had a defined 5′-end and a heterogeneous 3′-end, which were similar to the canonical piRNAs (Fig. 2G). Approximately one-quarter of snRNA-derived sncRNAs were piRNAs (Fig. 2, B, H, and I), with an even higher U bias at the 5′-end than the rRNA-derived piRNAs (Fig. 2J).
Figure 2.
rRNA-, snRNA, and tRNA-derived piRNAs in SSCs. A–C, proportion of piRNAs derived from rRNA (A), snRNA (B), and tRNA (C) in WT SSCs, Mili−/− SSCs, and Mili-IP in WT SSCs. D, length distribution of rRNA-derived sncRNAs. E, ratio of piRNAs derived from different rRNA species. F, 5′-end nucleotide bias of rRNA-derived piRNAs. G, distribution of 18S rRNA-derived sncRNAs. Count at each position is added up with all reads at the same 5′-end mapped site. The count was normalized by exogenous spike-ins. For the region where the most piRNAs are derived, the piRNA sequences and the count of sequences are also indicated. H, length distribution of snRNA-derived sncRNAs. I, ratio of piRNAs derived from snRNAs. J, 5′-end nucleotide bias of snRNA-derived piRNAs.
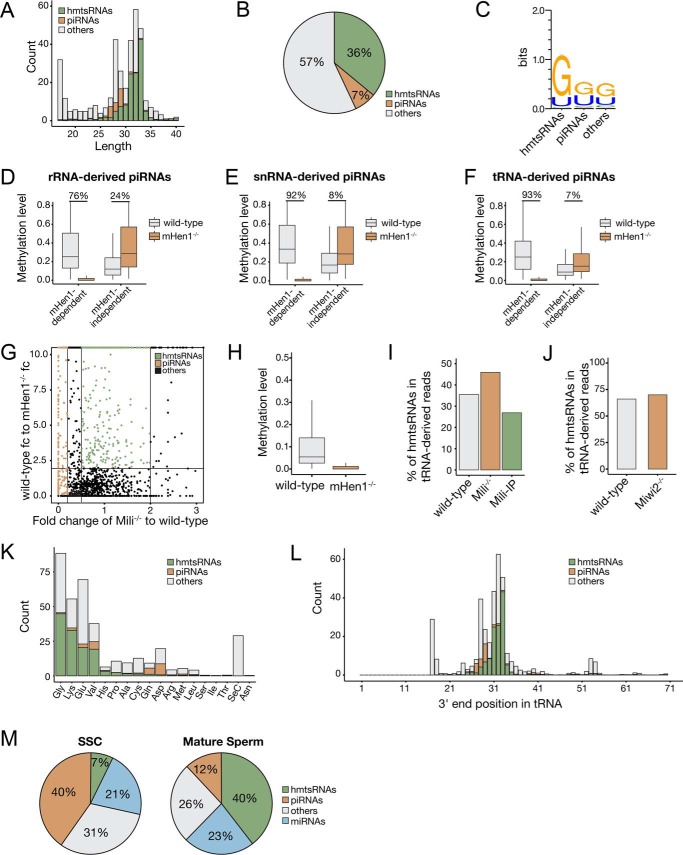
By contrast, we found that only 7% of tRNA-derived sncRNAs were piRNAs (Figs. 2C and 3, A and B). These tRNA-derived piRNAs had both of G and U biases at their 5′-end (Fig. 3C), which were different from the 5′ U bias of the Miwi homologue MARWI-bound tRNA-derived piRNAs in marmoset germ cells (33), but were similar to the Miwi2 homologue Hiwi2-bound piRNAs in human somatic cancer cells (34). As expected, the 3′-methylation of the majority of rRNA-, snRNA-, and tRNA-derived piRNAs were mHen1-dependent (Fig. 3, D–F). Interestingly, we found that some tRNA-derived sncRNAs also had mHen1-dependent methylation (Fig. 3, G and H), but their expression levels were not affected in Mili−/− SSCs (Fig. 3I) or Miwi2−/− SSCs (Fig. 3J) and were not enriched by Mili-IP (Fig. 3I), suggesting that in addition to piRNAs, other sncRNAs can act as mHen1 substrates in cells. We referred to this new type of small RNAs as mHen1 methylated tRNA-derived small RNAs (hmtsRNAs). These RNAs were ∼31–33 nt in length, longer than piRNAs in SSCs, and had a more obvious 5′ G bias than tRNA-derived piRNAs (Fig. 3, A and C). In addition, they were derived from different tRNAs compared with tRNA-derived piRNAs. Specifically, >95% of the hmtsRNAs were derived from four specific tRNA isotypes: Gly-, Lys-, Glu-, and Val-tRNA, whereas tRNA-derived piRNAs were mainly processed from Asp-, Gln-, and Val-tRNA (Fig. 3K). The majority of hmtsRNA sequences started from the 5′ first nucleotide and ended near the anti-codon loop of their corresponding tRNA (Fig. 3L). Notably, the abundance of hmtsRNA was greatly enriched in mature sperm, accounting for 40% of total small RNAs (Fig. 3M), which may be functional in the latter stages of spermatogenesis as well as in early embryonic development as reported (35, 36).
Figure 3.
mHen1-dependent 3′-methylation of noncanonical piRNAs and hmtsRNAs in SSCs. A, length distribution of tRNA-derived sncRNAs. B, ratio of piRNAs and hmtsRNAs derived from tRNAs. C, 5′-end nucleotide bias of tRNA-derived piRNAs and hmtsRNAs. D–F, boxplots represent the mHen1-dependent or mHen1-independent methylation level of rRNA-derived (D), snRNA–derived (E), and tRNA-derived (F) piRNAs at 3′-end in WT and mHen1−/− SSCs. G, dependences of the small RNAs derived from tRNAs. x axis represents the abundance change of sncRNAs in Mili−/− versus WT SSCs. y axis represents the methylation level change of sncRNAs in WT versus mHen1−/− SSC. Brown dots represent piRNAs, which were Mili-dependent. Green dots represent hmtsRNAs, which were mHen1-dependent but not Mili-dependent. H, methylation level of hmtsRNA in WT and mHen1−/− SSC. I, percentage of hmtsRNAs in tRNA-derived sncRNAs in the WT SSCs, Mili−/− SSCs, and Mili-IP of WT SSCs. J, percentage of hmtsRNAs in tRNA-derived sncRNAs in the WT and Miwi2−/− SSCs. K, abundance (count) of piRNAs and hmtsRNAs originating from each tRNA. L, 3′-end position of tRNA-derived piRNAs and hmtsRNA mapped on the intact tRNAs. The x axis represents the nucleotide position of intact tRNAs. K and L, abundance of sncRNAs was corrected by methylated spike-in normalization. M, relative abundance of different sncRNAs in WT SSC and mature sperm in mouse.
Human Hen1 exhibits higher activity in the presence of manganese ion over magnesium ion
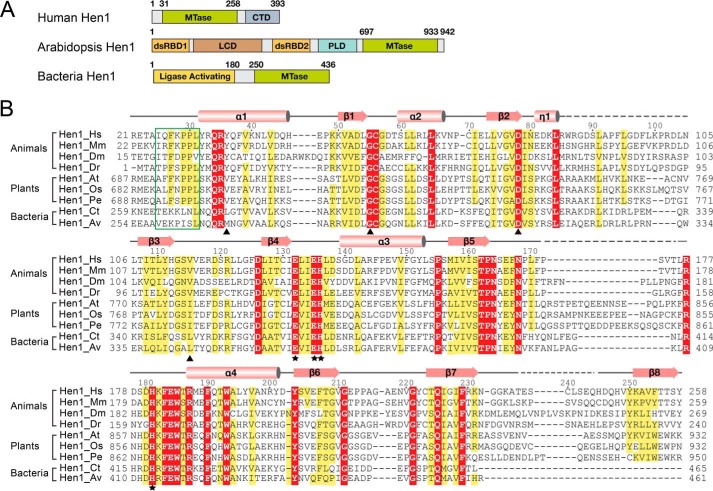
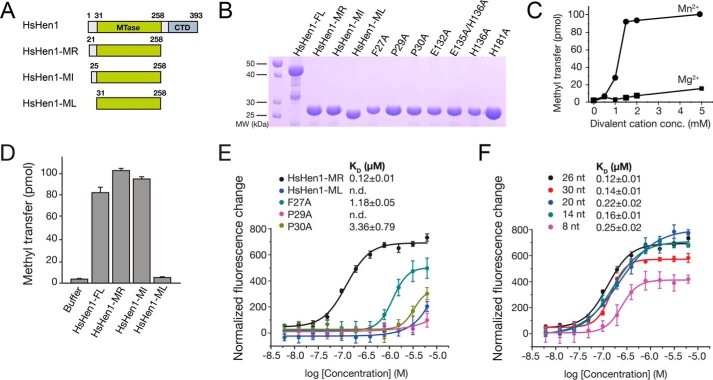
In addition to the canonical piRNAs, we showed that a portion of tRNA-, rRNA-, and snRNA-derived small RNAs associated with Mili in the SSCs were also methylated by Hen1, indicating the variety of substrates of mammalian Hen1. Hen1 shares a conversed MTase domain across species, although the domain architecture of Hen1 varies in plants, mammals, and bacteria (Fig. 4A). To investigate the molecular basis of the 3′-end methylation by animal Hen1, we attempted to crystallize human Hen1. Human Hen1 is 393 amino acids in length, which consists of an N-terminal MTase domain and a CTD. According to sequence alignment, the MTase domain is conserved across eukaryotic species. The catalytic domain of human Hen1 shares a sequence identity of 35% with that of AtHen1 (Fig. 4B). To understand the mechanism of the sncRNA methylation by Hen1, we first tried to assess its catalytic activity. Although the MTase activity of bacteria Hen1 was well characterized in previous studies, the enzymatic activities of mammalian Hen1 have not been studied yet (37). A 30-nt single-stranded RNA oligo was synthesized for the MTase assays. 3H-CH3–AdoMet was used as the cofactor to measure how many RNA substrates are methylated. Previous studies showed that bacterial Hen1 exhibited higher activity in the presence of Mn2+ over Mg2+ (37, 38). We purified the catalytic domain of Hen1 (HsHen1-MR; residues 21–258) (Fig. 5B). In a reaction containing 100 pmol of RNA oligo and 8 μm HsHen1-MR, only 15.6 pmol of RNA was methylated in the presence of 5 mm MgCl2 over 45 min, whereas under an equimolar concentration of MnCl2, ∼100% RNA (101 pmol as measured) was methylated. The catalytic activity has a huge leap between 1 and 1.5 mm MnCl2 and was basically saturated at 2 mm MnCl2. Also, the overall amount of transferred methyl group in the presence of different concentrations of MnCl2 is much higher than in the presence of MgCl2. Therefore, human Hen1 also showed a preference of Mn2+ similar to bacteria Hen1 (Fig. 5C). We performed the following methyltransferase experiments using 2 mm MnCl2.
Figure 4.
Domain architecture of human Hen1 that is different from plant Hen1. A, schematic representation of the domain structure of human Hen, Arabidopsis Hen1, and bacteria Hen1 (Trichormus variabilis). B, sequence alignment of MTase domain of Hen1 from different species. Hs, Homo sapiens; Mm, Mus musculus; Dm, Drosophila melanogaster; Dr, Danio rerio; At, Arabidopsis thaliana; Os, Oryza sativa Japonica Group; Pe, Populus euphratica; Ct, Clostridium thermocellum; Av, Anabaena variabilis. Numbers above sequences are labeled based on HsHen1. The secondary structure of the HsHen1 MTase domain is schematically represented above the sequences, with α-helices highlighted as cylinders, β-strands as arrows, loops as solid lines, and disordered regions as dotted lines. Strictly conserved residues are highlighted in red boxes and identical residues in yellow boxes. The FXPP motif (Phe-692–Leu-697) is marked in a green box. Residues binding AdoMet are marked with black triangles. Residues binding the divalent ion are marked with black asterisks.
Figure 5.
FXPP motif that is specific in eukaryotic Hen1 is important for RNA binding. A, schematic representation of human Hen1 and three truncated mutants. B, purified WT or mutant HsHen1 used in the methyltransferase assays or FP assays were detected by 15% SDS-PAGE and visualized by Coomassie Blue staining. Purified HsHen1-FL (residues 1–393), HsHen1-MR (residues 21–258), HsHen1-MI (residues 25–258), HsHen1-ML (residues 31–258), and HsHen1-MR mutants (F27A, P29A, P30A, E132A, E135A/E136A, H136A, and H181A) are indicated from left to right. C, in vitro methyltransferase assays showed that HsHen1-MR prefers manganese over magnesium. 0, 0.5, 1, 1.5, 2, or 5 mm either of MnCl2 or MgCl2 as specified were added and incubated in 37 °C for 45 min. D, in vitro methyltransferase assays of WT and mutant HsHen1. The averages of three independent experiments with S.D. values are shown. E and F, binding affinities between HsHen1 proteins and FAM-labeled RNA oligos. The fluorescence intensity changes under polarized illumination were plotted. Error bars indicate standard deviation of the mean (n = 3). E, fluorescence polarization assay showed affinities between HsHen1-ML, WT, or mutant HsHen1-MR and a 26-nt fluorescently-labeled RNA. Color scheme of different HsHen1 proteins is indicated. F, fluorescence polarization assay showed affinities between HsHen1-MR and different lengths of FAM-labeled RNA oligos. Color scheme of different length of FAM-labeled RNA oligos is indicated.
FXPP motif is essential for substrate recognition by HsHen1
To compare the catalytic activity of the full-length Hen1 (HsHen1-FL) with that of the catalytic domain, we purified the full-length protein and two truncated versions, HsHen1-ML (residues 31–258), spanning residues of the methyltransferase domain, and HsHen1-MI (residues 25–258) (Fig. 5, A and B). Both HsHen1-MR and HsHen1-MI showed a little higher activity than HsHen1-FL in the in vitro methyltransferase assay with triple repeated experiments. The measured activity for full-length Hen1 is 81%, compared with 100% for HsHen1-MR and 93% for HsHen1-ML, respectively. These data suggest that the CTD domain may have a negative impact on the methyltransferase activity. Surprisingly, HsHen1-ML showed no activity compared with the full-length construct (Fig. 5D). By comparing HsHen1-ML and HsHen1-MI, we found they are only 6 amino acids shorter at the N terminus of HsHen1-ML (Fig. 4B). Sequence alignment indicated that a conserved FXPP motif exists in the N terminus of the MTase domain of eukaryotic Hen1 but not in bacterial Hen1. In the crystal structure of AtHen1, the corresponding region (residues 692–697) specifically recognized and anchored the phosphate group connecting the second and the third nucleotides of the 3′-end guide RNA via two hydrogen bonds (Fig. S2). Therefore, we proposed that the FXPP motif may play an important role in substrate binding.
Next, we generated three mutants, namely F27A, P29A, and P30A, on the FXPP motif of HsHen1-MR (Fig. 5B). Fluorescence polarization (FP) assays were performed to measure the binding affinity between the RNA substrate and WT or mutant HsHen1-MR. A 26-nt RNA oligo with a fluorescein label at 5′-end was used. The measured dissociation constant (KD) for HsHen1-MR was 0.12 ± 0.01 μm. Mutation on the FXPP motif dramatically reduced the binding (Fig. 5E). P29A abolishes the binding. F27A and Pro-30 reduced the binding affinity about 10- and 30-fold, respectively. HsHen1-ML, which lacked the FXPP motif, showed no binding as expected, similar to the bacteria Hen1, although their sequences are not conserved (Fig. 4B) (37). In addition, different lengths of RNA oligos were also tried, including 30, 20, 14, and 8 nt in length. The FP results showed that variation in length has little impact on the binding affinity of HsHen1-MR to RNA oligos (Fig. 5F). Taken together, the FXPP motif was critical for the binding of the RNA substrate and thus indispensable for the enzymatic activity of HsHen1.
Overall structure of the putative MTase domain of human Hen1 in complex with AdoMet
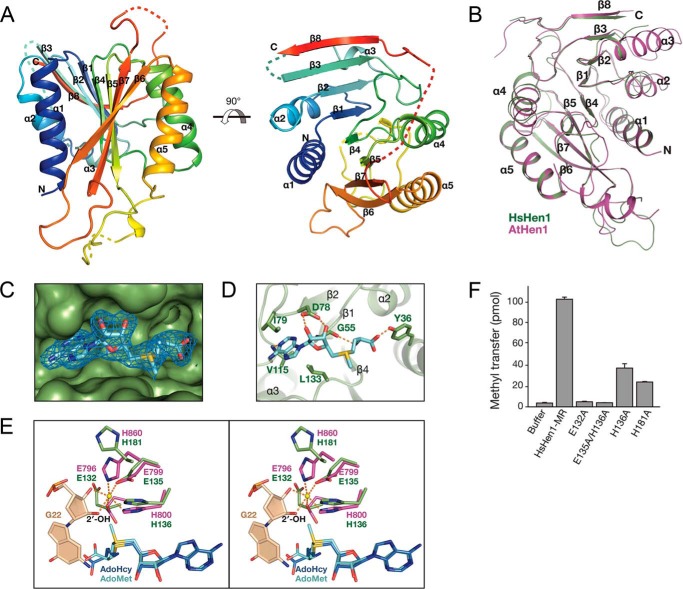
Next, we performed crystallization trials to explore the catalytic mechanism of human Hen1. We made many attempts to crystallize full-length Hen1 (HsHen1-FL) as well as three truncated versions, including HsHen1-MR, HsHen1-MI, and HsHen1-ML (Fig. 5A). However, only HsHen1-ML with its cofactor AdoMet was successfully crystallized. Because HsHen1-ML has no catalytic activity, we call this domain putative MTase domain. The structure was determined by molecular replacement using the structure of the AtHen1 MTase domain (PDB code 3HTX) as a search model. There is only one HsHen1-ML molecule in an asymmetric unit. All of the structural statistics were refined within a reasonable range (Table 1). Overall, the HsHen1 putative MTase domain adopts a classical Rossmann-fold, consisting of eight central β-strands (β1–β8) surrounded by five α-helices (α1–α4) (Fig. 6A). In detail, β1–β5 and β6 are parallel to each other, whereas β7, which is located between β5 and β6, is antiparallel to the other strands. β8 is followed by β7 connected by a long loop, which could not be modeled in this structure due to the poor electron density. Helices α1 and α2 are folded on one side of the β-sheet core, and the others are packed on the opposite side. Superimposition of the MTase domain of HsHen1 to AtHen1 (residues 696–933; PDB 3HTX) showed that these two domains could be aligned quite well with a root-mean-square-deviation (r.m.s.d.) of 1.4 Å, except that helix α3 in HsHen1 is much shorter than in the AtHen1 structure (Fig. 6B). The cofactor AdoMet is bound on the concave surface of HsHen1-ML formed by four loops connecting β1–α2, β2–β3, β3–β4, and β4–α3, respectively (Fig. 6, C and D).
Table 1.
Summary of diffraction data and refinement statistics
| Data collection | |
| Space group | P3121 |
| Cell dimensions | |
| a, b, c (Å) | 38.5, 38.5, 249.4 |
| α, β, γ (°) | 90, 90, 120 |
| Wavelength (Å) | 0.9791 |
| Resolution (Å)a | 30.00–2.00 (2.07–2.00)a |
| Rmergea | 8.1 (25.1) |
| I/σ(I)a | 15.9 (5.5) |
| Completeness (%)a | 98.9 (100.0) |
| Redundancya | 6.6 (6.9) |
| Refinement | |
| Resolution (Å) | 30–2.0 |
| No. of reflections | 15,327 |
| Rwork/Rfree | 0.20/0.25 |
| No. of atoms | |
| No. of protein | 1599 |
| No. of AdoMet | 27 |
| No. of solvent atoms | 115 |
| Average B-factor (Å2) | |
| Protein | 27.8 |
| AdoMet | 29.0 |
| Solvent | 36.0 |
| r.m.s.d. | |
| Bonds length (Å) | 0.008 |
| Bond angles (°) | 1.138 |
| Ramachandran plot | |
| Favored (%) | 97.9 |
| Allowed (%) | 2.1 |
| Outlier (%) | 0.0 |
a Values in parentheses are for the highest resolution shell.
Figure 6.
Crystal structure of the putative MTase domain of human Hen1. A, cartoon representation of the crystal structure of HsHen1-MR in complex with AdoMet. HsHen1-MR is colored from blue to red (from N to C terminus). Loop regions that could not be modeled are shown as dotted lines. View of 90° rotation around the horizontal axis is shown on the right. B, structural comparison of HsHen1-MR with the MTase domain of Arabidopsis Hen1 (residues 696–933; PDB code 3HTX). HsHen1-MR is colored in green. AtHen1 is colored in purple. C, 2Fo − Fc electron density map (contoured at 1.0 σ cutoff, sky blue mesh) of the AdoMet bound on the concave surface of HsHen1-MR. D, details of AdoMet binding by the HsHen1 MTase domain. The hydrogen bonds are shown as orange-dotted lines. E, stereo view of Mg2+ coordination of AtHen1 and corresponding conserved residues in HsHen1. The Mg2+ is shown as a yellow sphere. The penultimate nucleotide A21 in the 3′-end of the guide RNA is colored in wheat. AdoMet and S-adenosylhomocysteine (AdoHcy) are colored in cyan and blue, respectively. F, in vitro methyltransferase assays of WT and mutant HsHen1. The averages of three independent experiments with S.D. values are shown.
Conserved active sites in human Hen1
Previously, studies have illustrated the catalytic sites in AtHen1 and bacteria Hen1 comprise the AdoMet-binding site, the divalent metal, and the 3′ adenine nucleotide of the RNA methyl acceptor (18, 23). In our crystal structure, the cofactor AdoMet was clearly observed (Fig. 6C). AdoMet was bound in a pocket formed by Tyr-36, Gly-55, Asp-78, Ile-79, Val-115, and Leu-133 (Fig. 6D). The adenine ring of AdoMet was stabilized by the main-chain amides of Gly-55 and Val-115 via two hydrogen bonds. The carboxyl group of Asp-78 forms two hydrogen bonds with the ribose of AdoMet. The carboxyl group of AdoMet was further stabilized by the phenolic hydroxyl group of Tyr-36. Moreover, Ile-79, Val-115, and Leu-133 contact the indole ring of AdoMet through hydrophobic interactions. In the previously reported AtHen1 structure, two glutamic acids and two histidine residues were involved in the chelation of the divalent metal ion (18). However, the divalent metal and the RNA substrate were not observed in our structure. Superimposition of the MTase domains of HsHen1 and AtHen1 showed that in the absence of the divalent ion, the side chains of Glu-132, Glu-135, His-136, and His-181 of HsHen1 flipped away to some degree, especially Glu-132 and His-181 (Fig. 6E). We also noticed that the methyl group of AdoMet in the HsHen1 structure was oriented toward the corresponding position of the 2′-OH of the 3′-terminal nucleotide in AtHen1 structure with a distance of 1.8 Å, making the transfer of the methyl group from AdoMet to RNA spatially possible (Fig. 6E).
To validate whether these residues are critical for HsHen1 MTase activity, site-specific mutagenesis was performed. Glu-132, Glu-135, His-136, and His-181 are four residues that are supposed to chelate the metal ion (Fig. 6E). Mutations of these residues decreased the methyl transfer activity considerably (Figs. 5B, 6F). H136A and H181A showed significant loss of MTase activity. The measured methylated RNA oligos for H136A and H181A are 28 and 24 pmol, respectively, which are 3.5- and 4.1-fold less effective than that of the WT HsHen1-ML. No activity was observed for the E132A mutant and the double mutant E135A/H136A. Therefore, the residues in the active site of human Hen1 are important for the activity just as in AtHen1 and bacteria Hen1.
Discussion
In this study, we systemically investigated the Hen1-dependent 2′-O-methylation of diverse types of sncRNAs in SSCs. In the absence of mHen1, piRNAs were trimmed from their 3′-ends by one nucleotide and had increased tailing ratio with one or more additional U and A residues at their 3′-end. We also identified a novel class of small RNA substrates of mHen1 in SSC, which are 31–33 nt long and derived from a few types of tRNAs with a significant 5′ G bias (named hmtsRNAs). We further determined the crystal structure of the putative catalytic domain of human Hen1 with its cofactor AdoMet. We found that similar to bacteria Hen1, HsHen1 prefers Mn2+ over Mg2+. The methyltransferase assay result indicated that with the N-terminal six additional amino acids containing the eukaryotic Hen1 conserved the FXPP motif, the putative MTase restored full activity as the full-length protein. Site-specific mutagenesis and the binding assays showed that the FXPP motif was critical for the substrate RNA binding. Moreover, our results provided structural and biochemical insights into the catalytic domain of mammalian Hen1 and may shed light on the Hen1 substrates in mouse SSCs.
The hmtsRNAs identified in this study accounted for 36% of the tRNA-derived sequences in the mouse SSCs. Compared with the canonical piRNAs, these hmtsRNAs had lower methylation levels, which may be due to their unfavorable interaction with Hen1 without the assistance of PIWI or its associated proteins. The majority of hmtsRNA sequences 3′-ended near the anti-codon loop. Because Hen1 protein can only catalyze methylation at the 3′-end of RNAs but not internally, the hmtsRNAs are likely to be initially cleaved from partially processed tRNA precursors in which nucleotides have not yet been modified and then been methylated at 3′-end by Hen1. Previous studies have shown that tRNA-derived small RNAs in mature sperm have important functions in epigenetic transgenerational regulation of gene expression, and the 3′-methylation by Hen1 may selectively stabilize the certain types of tRNA fragments or facilitate their recognition by some unknown proteins that are functional in spermatogenesis and embryonic development, which warrant future investigation. We also found that in addition to the previously reported tRNA-derived piRNAs (33, 34), a portion of rRNA- and snRNA-derived small RNAs was also associated with Mili in the SSCs, and possibly function as piRNAs. Moreover, we observed that most of the rRNA-, snRNA-, and tRNA-derived piRNAs were 3′-methylated by Hen1, although their methylation levels were slightly lower than those of canonical piRNAs. A portion of the 3′-modification of the rRNA-, snRNA-, and tRNA-derived piRNAs was not dependent on mHen1, suggesting that they may be processed from their precursors bearing modifications or methylated by a yet unknown MTase.
We also showed that the N-terminal FXPP motif of the MTase domain is important for the binding of substrate RNAs. Sequence alignment indicated that this motif is conserved in eukaryotic Hen1 proteins, including plant Hen1 and mammalian Hen1. However, the substrates of plant Hen1 and mammalian Hen1 are not the same. The substrate for plant Hen1 is duplex RNAs, whereas the substrate for Human Hen1 is single-stranded RNAs. Our FP results show that the FXPP motif is important for single-stranded RNA binding. By analyzing the structure of AtHen1, we found that in addition to the MTase domain, AtHen1 contains two dsRBD domains, which bind and stabilize the miRNA/miRNA* duplex. However, only the 3′-end of the target strand is inserted into the active site. The FXPP motif in AtHen1 interacts with the backbone of the 3′-terminal nucleotides of the target strand. Therefore, for AtHen1, the recognition of the substrate RNA can be divided into two parts. Two dsRBD domains bind to the major grooves of the duplex RNA, whereas the FXPP motif binds to the 3′-end of the target strand, suggesting the same role of the FXPP motif in AtHen1 and human Hen1.
Our results showed a similar binding affinity for the single-stranded RNA substrates from 30 to 8 nt, indicating that the binding affinity between Hen1 and its RNA substrate would not be affected by the length of the RNA oligo. Recent studies of piRNA biogenesis revealed a trimming mechanism of piRNA intermediates (40–43). In silkworms and mice, the production of mature piRNAs required a poly(A)-specific RNase family deadenylase PNLDC1 to trim the 3′-end of piRNA intermediates to an optimal length. Hen1 may be coupled with PNLDC1 to add the 2′-O-methyl group to the trimmed piRNAs. Therefore, we proposed that Hen1 itself has no capability to distinguish single-stranded RNAs of different size. Mammalian Hen1 is likely to distinguish its substrates in the assistance of other factors in piRNA pathway.
Experimental procedures
Cloning, expression, and purification of human Hen1
The MTase domain (residues 21–258, 25–258, and 31–258) and full-length domain (residues 1–393) of HsHen1 were amplified by PCR and cloned into the pET-SMT3 vector that contains an N-terminal Ulp1-cleavable 6×His–Sumo tag. Point mutations were performed by using the site-directed mutagenesis kit (New England Biolabs). All of the constructs were verified by sequencing and were expressed in Escherichia coli BL21 (DE3). After being induced with 0.2 mm isopropyl β-d-thiogalactopyranoside, the cells were incubated overnight at 18 °C. The cells were collected by centrifugation at 4000 rpm for 15 min and were lysed by a cell disruptor (JNBIO) at 4 °C. Proteins were purified by affinity chromatography using a HisTrap column (GE Healthcare) and then removed the 6×His-Sumo tag. The proteins were further purified by gel filtration with Superdex G75 Hiload 16/60 column (GE Healthcare) in 10 mm Tris buffer, pH 8.0, 100 mm NaCl, 1 mm DTT.
Crystallization, data collection, and structural determination
Purified HsHen1-ML was concentrated to 15 mg/ml and crystallized by vapor diffusion with the reservoir solution of 100 mm Tris-HCl, 200 mm MgCl2, 10% (w/v) PEG 3350, pH 7.5, at 16 °C. Diffraction data were collected at BL17U of Shanghai Synchrotron Radiation Facility (SSRF) and processed with HKL 2000 (44). The model was built using the MR method with the structure of the AtHen1 MTase domain (residues 697–933, PDB code 3HTX) as the search model. Model building and structural refinement were carried out using COOT (45) and PHENIX (46). All the structure figures were generated with PyMOL (The PyMOL Molecular Graphics System, Version 1.8 Schrödinger, LLC). The statistics of the diffraction data and the refinement data are summarized in Table 1. The coordinate has been deposited under PDB accession code 5WY0.
In vitro methyltransferase assays
The methyltransferase assays were carried out in a 10-μl reaction mixture containing 25 mm Tris-HCl, pH 8.5, 0.1 mm EDTA, 20 μm 3H-CH3-AdoMet, 2 mm MnCl2 with 10 μm 30 nt RNA (5′-UGUCUGACUGAAGGACCAGGUGCUGUCUGA-3′) and 8 μm purified protein. WT or mutant HsHen1 proteins were pretreated with 1 mm EDTA on ice for 30 min to remove any endogenous divalent metal ions and then dialyzed against reaction buffer for further methyltransferase assays. The reaction mixtures were incubated at 37 °C for 45 min. The reaction was stopped by adding 1 μl of 1 mm EDTA. Then the samples were spotted onto a Whatman 3 MM filter disk. The disk was washed twice with 5% TCA solution and three times with ethanol for 5 min per wash. After washing, the disk was completely dried. The radioactivity was measured by liquid scintillation counting (Beckman). For the methyltransferase assays showing that HsHen1-MR prefers manganese over magnesium, 0, 0.5, 1, 1.5, 2, or 5 mm either of MnCl2 or MgCl2 as specified were added.
Fluorescence polarization assays
The RNA oligos were synthesized with fluorescein amidite (FAM) labeled at 5′-end (30 nt, 5′-FAM-UGUCUGACUGAAGGACCAGGUGCUGUCUGA-3′; 26 nt, 5′-FAM-UAGACAAGUGCUUCUCACGAUCCAGA-3′; 20 nt, 5′-FAM-AGUGCUUCUCACGAUCCAGA-3′; 14 nt, 5′-FAM-UCUCACGAUCCAGA-3′; and 8 nt, 5′-FAM-GAUCCAGA-3′) (Bioneer).
FP experiments were performed in 96-well plates (Corning) with the fluorescence reader SynergyTM NEO (BioTek). Serial dilutions of purified HsHen1 proteins were prepared in FP assay buffer (100 mm NaCl, 10 mm Tris-HCl, pH 8.0, 1 mm MnCl2) with a final concentration ranging from 0.006 to 6.25 μm. FAM-labeled RNA oligos was then added to a final concentration of 4 nm for a final assay volume of 100 μl. The reaction mixture was incubated for 30 min at room temperature. Polarization was measured at an excitation wavelength of 485 nm and emission wavelength of 528 nm. Each plate was read three times, and the values were averaged prior to analysis. All dissociation constant (KD) values were determined by fitting the titration curve with Origin 8.0 software.
Isolation and manipulation of mouse SSCs
SSCs were isolated and cultured according to a previous protocol (47). In brief, testes were dissociated from a 15-day-old C57BL/6×DBA2 F1 male mouse carrying transgenic actin-enhanced GFP genes, and single-cell suspensions were obtained. Briefly, ∼2.0 × 105 cells were suspended in 1 ml of StemPro34 SFM medium containing 5 mg/ml BSA, 6 mg/ml glucose, 2 mm glutamine, 1× antibiotic/antimycotic, 1× minimal essential medium vitamins, 1× nonessential amino acids, 10 μg/ml biotin, 25 μg/ml insulin, 30 μg/ml sodium pyruvate, 0.06% lactic acid, 100 μm ascorbic acid, 30 nm sodium selenite, 60 μm putrescine, 100 μg/ml bovine apo-transferrin, 10 μm 2-mercaptoethanol, 1% fetal bovine serum, 20 ng/ml recombinant mouse EGF, 10 ng/ml recombinant human basic fibroblast growth factor, and 10 ng/ml recombinant rat glial cell line-derived neurotrophic factor. Cells were cultured in a 12-well plate in a humidified incubator containing 5% CO2 at 37 °C for 24 h. The cells were pipetted 10 times, and the floating cells were collected by centrifugation at 270 × g for 5 min. The pellets were suspended in SF medium and incubated in a humidified incubator for 10 days during which time the medium was refreshed every 3 days. The SSC colonies were trypsin-digested and plated on mouse embryo fibroblast feeder cells in a 24-well plate with SF medium. Plasmids encoding spCas9 and sgRNAs (20 μg) targeting mHen1, Mili, and Miwi2 were transiently transfected into SSCs with the Amaxa Cell Line Nucleofector Kit L, as described previously. Single SSC colonies were picked and verified by Sanger sequencing. WT and knockout SSC colonies were expanded and used for small RNA sequencing. The sgRNA sequences were shown as following: mHen1, GGAAATGGCAGAAAGCATACGTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGTGGCACCGAGTCGGTGCTTTTTT; Mili, GCCACCCATCTCAGTGTGTGGTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGTGGCACCGAGTCGGTGCTTTTTT; and Miwi2, GCCACCCATCTCAGTGTGTGGTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGTGGCACCGAGTCGGTGCTTTTTT.
NaIO4 treatment and small RNA library preparation
Total cellular RNA was extracted using TRIzol reagent (Takara). A total of 4 μg of total RNA with a 2 × 10−6 pmol spike-in mixture and 2 μl of freshly dissolved NaIO4 (200 mm) were kept on ice for 1 h in the dark. Then the RNAs were precipitated and dissolved in DEPC-treated water for a small RNA library construction according to the Illumina protocol. High-throughput RNA sequencing was performed using HiSeq 2500 with 50 running circles (GENEWIZ). The four spike-in RNA oligonucleotides containing 5′-phosphorylation with or without 3′-end 2′-O-methylation were chemically synthesized (Integrated DNA Technologies, IDT). The sequences were shown as follows: spike-in #1, 5Phos/rCrCrUrGrGrArCrUrArGrUrCrGrUrCrArGrCrArUrU, and 3′-methylated spike-in #1m, 5Phos/rCrCrUrGrGrArCrUrArGrUrCrGrUrCrArGrUrGrUmU; spike-in #2, 5Phos/rArArCrUrUrCrArGrGrGrUrCrArGrCrUrUrGrCrCrG, and 3′-methylated spike-in #2m, 5Phos/rUrUrGrUrArArGrArCrArUrGrArArArGrUrArCrUmG.
Immunoprecipitation
In vitro-cultured SSCs were digested and washed with PBS and harvested in lysis buffer containing 50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm EDTA, 0.5 mm DTT, 0.5% Nonidet P-40, 0.1 unit/μl RNase inhibitor (Fermentas), and 1:100 protease inhibitor mixture (Sigma). After rotation at 4 °C for 20 min, the lysate was clarified by centrifugation at 14,000 × g at 4 °C for 15 min. A total of 400 μl of supernatant was mixed with 5 μg of Mili antibody (MABE363, Millipore), coupled with protein G beads (Invitrogen), and incubated overnight with gentle rotation at 4 °C. The beads were washed four times with TBS before TRIzol reagent (Invitrogen) was added for RNA extraction.
β-Elimination treatment and Northern blotting
Synthetic RNAs were treated with 1× borate buffer (30 mm borax and 30 mm boric acid, pH 8.6) and 25 mm freshly dissolved NaIO4 on ice for 30 min in the dark. Then 2 μl of glycerol was added to the reaction to quench NaIO4 and incubated for 10 min. RNAs were precipitated and dissolved in 50 μl of 1× borax buffer (30 mm borax, 30 mm boric acid, and 50 mm NaOH, pH 9.5) and incubated at 45 °C for 90 min. Finally, the RNAs were precipitated and dissolved in DEPC-treated water. The RNA samples were denatured, fractionated by electrophoresis on a 20% polyacrylamide, 8 m urea gel at 500 V until the bromphenol blue reached the bottom of the gel, and then electroblotted and cross-linked to a nylon membrane (Roche Applied Science). The membranes were probed at 50 °C in DIG Easy Hyb buffer (Roche Applied Science) with terminally digoxigenin-labeled DNA oligonucleotides overnight, and then washed with 2× SSC and 0.1× SSC, 0.1% SDS buffer at 37 °C. The membranes were incubated with an anti-digoxigenin antibody (Roche Applied Science) (1:20,000 diluted) and then CDP-Star (ABI), after which the signal was detected on X-ray film.
Computational categorization of small RNAs
The raw fastq data were pre-processed using a common procedure. After quality filtering, sequencing reads were clipped from the 3′ adaptor allowing a minimum match of 10 nt from the 5′-end. Reads unable to match the adaptor sequence or with lengths shorter than 17 bp after the 3′ adaptor was clipped were discarded. The useful reads were mapped to the mouse genome by bowtie (48). The mapped genome sequences were further aligned to known miRNAs, tRNAs, rRNAs, snoRNAs, and snRNAs by bowtie. The remaining 25–32-nt sequences were used to identify piRNAs following the method described previously (49) with slight modifications. The clustering parameters were determined as MinReads = 4 and Eps = 2500 bp by running a series of k-dist analysis with different Eps and MinReads of our own data. All of the candidate clusters that satisfied these parameters were considered as piRNA clusters, and the sequences located in these clusters were defined as piRNA without any further scoring and filtering. Only the reads that exactly matched the 5′ start site of annotated miRNAs and 3′-ends with ≤2-nt deletions or additional sequences derived from pri-miRNAs were counted as miRNAs. The count of each miRNA, piRNA, tRNA-, rRNA-, and snRNA-derived sequences was corrected by the count of methylated spike-ins. The small RNAs were mapped in the following order: miRNA, tRNA, rRNA, snoRNA, snRNA, and piRNA. The rRNA-, tRNA-, and snRNA-derived sncRNAs for which the expression levels decreased more than 10-fold upon Mili knockout in SSCs were identified as piRNAs. The identified piRNA clusters are listed in Table S2. The count of individual miRNAs normalized by methylated spike-ins in each sample is listed in Table S3. The information of noncanonical piRNAs derived from rRNAs, tRNAs, and snRNAs is listed in Table S4. The information of hmtsRNAs is listed in Table S5.
Nontemplate tailing
The sequences unable to be mapped to the mouse genome without any mismatch were used for further analyses of nontemplate tailing. Only those with a 5′-end identical to the annotated miRNAs and piRNAs and with consecutive mismatches at 3′-end were defined as tailed. The ratio of tailing might have been underestimated because the last residue of some small RNAs that aligned to the genome was in fact added through 3′-tailing but was indistinguishable by bioinformatics analysis alone.
Sources of sequences and genome assemblies
Mouse genome annotation mm10 were downloaded from the University of California Santa Cruz (UCSC) Genome Browser (http://genome.ucsc.edu/)5 (50, 51). Known RNA sequences were retrieved from the following databases: miRNA, miRBase (39); tRNAs, genomic tRNA database (52); rRNAs, 18S and 28S from NCBI; 5S and 5.8S from Ensembl (http://www.ensembl.org/index.html)5 (53); snoRNAs and snRNA from Ensembl (http://www.ensembl.org/index.html) (53).5
Data and material availability
All data used to obtain the conclusions in this paper are presented in the paper and the supporting Materials. The deep sequencing data have been deposited in the National Center for Biotechnology Information under accession number GSE97595. Atomic coordinates and structure factors for the reported crystal structures have been deposited with the Protein Data bank under accession number 5WY0. Other data may be requested from the authors.
Author contributions
L. P., F. Z., R. S., X. W., and J. C. data curation; L. P., F. Z., and R. S. formal analysis; J. J. C. resources; J. M., L. W., and Y. H. conceptualization; J. M. writing-review and editing; L. W. and Y. H. supervision; L. W. and Y. H. writing-original draft; Y. H. funding acquisition.
Supplementary Material
Acknowledgments
We thank Jinsong Li for providing Oct4-GFP mice. We thank Jun Yan and Minghan Tong for their advice and criticisms. We are extremely grateful to the Shanghai Scientific Research Center for their instrument support and technical assistance. We thank the staff from BL17U of the Shanghai Synchrotron Radiation Facility (SSRF) for assistance during data collection.
This work was supported by Strategic Priority Research Program Grants of the Chinese Academy of Sciences XDB08010202 (to Y. H.) and XDB19040102 (to L. W.), Ministry of Science and Technology of China Grant 2014CB943100 (to L. W.), National Natural Science Foundation of China Grants 91640102 (to Y. H.), 91440107 and 31470781 (to L. W.), and Grant 31230041 (to J. M.), Shanghai Municipal Commission for Science and Technology Grant 12JC1409400 (to L. W.), the Open Research Project on Major Infrastructure of Science and Technology of the Chinese Academy of Sciences, and the State Key Laboratory of Molecular Biology. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S2 and Tables S1–S5.
The atomic coordinates and structure factors (code 5WY0) have been deposited in the Protein Data Bank (http://wwpdb.org/).
Data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) under accession number GSE97595.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- miRNA
- microRNA
- nt
- nucleotide
- MTase
- methyltransferase
- FP
- fluorescent polarization
- AdoMet
- S-adenosyl-l-methionine
- r.m.s.d.
- root-mean-square-deviation
- piRNA
- PIWI-interacting RNA
- ssRNA
- single-strand RNA
- IP
- immunoprecipitation
- NaIO4
- sodium periodate
- sncRNA
- small noncoding RNA
- snRNA
- small nuclear RNA
- hmtsRNA
- Hen1 methylated tRNA-derived small RNA
- SSC
- mouse spermatogonial stem cells
- RISC
- RNA-induced silencing complex
- PIWI
- P-element-induced wimpy testis
- PDB
- Protein Data Bank
- dsRBD
- dsRNA-binding domain
- oligo
- oligonucleotide
- FAM
- fluorescein amidite
- CTD
- C-terminal domain
- MR
- molecular replacement
- DEPC
- diethyl pyrocarbonate.
References
- 1. Moazed D. (2009) Small RNAs in transcriptional gene silencing and genome defence. Nature 457, 413–420 10.1038/nature07756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Holoch D., and Moazed D. (2015) RNA-mediated epigenetic regulation of gene expression. Nat. Rev. Genet. 16, 71–84 10.1038/nrg3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bartel D. P. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 4. Carthew R. W., and Sontheimer E. J. (2009) Origins and mechanisms of miRNAs and siRNAs. Cell 136, 642–655 10.1016/j.cell.2009.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Iwasaki Y. W., Siomi M. C., and Siomi H. (2015) PIWI-interacting RNA: its biogenesis and functions. Annu. Rev. Biochem. 84, 405–433 10.1146/annurev-biochem-060614-034258 [DOI] [PubMed] [Google Scholar]
- 6. Luteijn M. J., and Ketting R. F. (2013) PIWI-interacting RNAs: from generation to transgenerational epigenetics. Nat. Rev. Genet. 14, 523–534 10.1038/nrg3495 [DOI] [PubMed] [Google Scholar]
- 7. Yu B., Yang Z., Li J., Minakhina S., Yang M., Padgett R. W., Steward R., and Chen X. (2005) Methylation as a crucial step in plant microRNA biogenesis. Science 307, 932–935 10.1126/science.1107130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vagin V. V., Sigova A., Li C., Seitz H., Gvozdev V., and Zamore P. D. (2006) A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313, 320–324 10.1126/science.1129333 [DOI] [PubMed] [Google Scholar]
- 9. Ohara T., Sakaguchi Y., Suzuki T., Ueda H., Miyauchi K., and Suzuki T. (2007) The 3′ termini of mouse Piwi-interacting RNAs are 2′-O-methylated. Nat. Struct. Mol. Biol. 14, 349–350 10.1038/nsmb1220 [DOI] [PubMed] [Google Scholar]
- 10. Horwich M. D., Li C., Matranga C., Vagin V., Farley G., Wang P., and Zamore P. D. (2007) The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr. Biol. 17, 1265–1272 10.1016/j.cub.2007.06.030 [DOI] [PubMed] [Google Scholar]
- 11. Kirino Y., and Mourelatos Z. (2007) Mouse Piwi-interacting RNAs are 2′-O-methylated at their 3′ termini. Nat. Struct. Mol. Biol. 14, 347–348 10.1038/nsmb1218 [DOI] [PubMed] [Google Scholar]
- 12. Yang Z., Ebright Y. W., Yu B., and Chen X. (2006) HEN1 recognizes 21–24 nt small RNA duplexes and deposits a methyl group onto the 2′ OH of the 3′ terminal nucleotide. Nucleic Acids Res. 34, 667–675 10.1093/nar/gkj474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen X., Liu J., Cheng Y., and Jia D. (2002) HEN1 functions pleiotropically in Arabidopsis development and acts in C function in the flower. Development 129, 1085–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yu B., Bi L., Zhai J., Agarwal M., Li S., Wu Q., Ding S.-W., Meyers B. C., Vaucheret H., and Chen X. (2010) siRNAs compete with miRNAs for methylation by HEN1 in Arabidopsis. Nucleic Acids Res. 38, 5844–5850 10.1093/nar/gkq348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li J., Yang Z., Yu B., Liu J., and Chen X. (2005) Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr. Biol. 15, 1501–1507 10.1016/j.cub.2005.07.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhai J., and Meyers B. C. (2012) Deep sequencing from hen1 mutants to identify small RNA 3′ modifications. Cold Spring Harb. Symp. Quant. Biol. 77, 213–219 10.1101/sqb.2013.77.014779 [DOI] [PubMed] [Google Scholar]
- 17. Zhai J., Zhao Y., Simon S. A., Huang S., Petsch K., Arikit S., Pillay M., Ji L., Xie M., Cao X., Yu B., Timmermans M., Yang B., Chen X., and Meyers B. C. (2013) Plant microRNAs display differential 3′ truncation and tailing modifications that are ARGONAUTE1 dependent and conserved across species. Plant Cell 25, 2417–2428 10.1105/tpc.113.114603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Huang Y., Ji L., Huang Q., Vassylyev D. G., Chen X., and Ma J.-B. (2009) Structural insights into mechanisms of the small RNA methyltransferase HEN1. Nature 461, 823–827 10.1038/nature08433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Montgomery T. A., Rim Y.-S., Zhang C., Dowen R. H., Phillips C. M., Fischer S. E., and Ruvkun G. (2012) PIWI associated siRNAs and piRNAs specifically require the Caenorhabditis elegans HEN1 ortholog henn-1. PLoS Genet. 8, e1002616 10.1371/journal.pgen.1002616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Billi A. C., Alessi A. F., Khivansara V., Han T., Freeberg M., Mitani S., and Kim J. K. (2012) The Caenorhabditis elegans HEN1 ortholog, HENN-1, methylates and stabilizes select subclasses of germline small RNAs. PLoS Genet. 8, e1002617 10.1371/journal.pgen.1002617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kirino Y., and Mourelatos Z. (2007) The mouse homolog of HEN1 is a potential methylase for Piwi-interacting RNAs. RNA 13, 1397–1401 10.1261/rna.659307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kamminga L. M., Luteijn M. J., den Broeder M. J., Redl S., Kaaij L. J., Roovers E. F., Ladurner P., Berezikov E., and Ketting R. F. (2010) Hen1 is required for oocyte development and piRNA stability in zebrafish. EMBO J. 29, 3688–3700 10.1038/emboj.2010.233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mui Chan C., Zhou C., Brunzelle J. S., and Huang R. H. (2009) Structural and biochemical insights into 2′-O-methylation at the 3′-terminal nucleotide of RNA by Hen1. Proc. Natl. Acad. Sci. U.S.A. 106, 17699–17704 10.1073/pnas.0907540106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang P., Selvadurai K., and Huang R. H. (2015) Reconstitution and structure of a bacterial Pnkp1-Rnl-Hen1 RNA repair complex. Nat. Commun. 6, 6876 10.1038/ncomms7876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lim S. L., Qu Z. P., Kortschak R. D., Lawrence D. M., Geoghegan J., Hempfling A.-L., Bergmann M., Goodnow C. C., Ormandy C. J., Wong L., Mann J., Scott H. S., Jamsai D., Adelson D. L., and O'Bryan M. K. (2015) HENMT1 and piRNA stability are required for adult male germ cell transposon repression and to define the spermatogenic program in the mouse. PLoS Genet. 11, e1005620 10.1371/journal.pgen.1005620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li X. Z., Roy C. K., Dong X., Bolcun-Filas E., Wang J., Han B. W., Xu J., Moore M. J., Schimenti J. C., Weng Z., and Zamore P. D. (2013) An ancient transcription factor initiates the burst of piRNA production during early meiosis in mouse testes. Mol. Cell 50, 67–81 10.1016/j.molcel.2013.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sun Y. H., Xie L. H., Zhuo X., Chen Q., Ghoneim D., Zhang B., Jagne J., Yang C., and Li X. Z. (2017) Domestic chickens activate a piRNA defense against avian leukosis virus. Elife 6, e24695 10.7554/eLife.24695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Aravin A. A., Sachidanandam R., Bourc'his D., Schaefer C., Pezic D., Toth K. F., Bestor T., and Hannon G. J. (2008) A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol. Cell 31, 785–799 10.1016/j.molcel.2008.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ishizu H., Siomi H., and Siomi M. C. (2012) Biology of PIWI-interacting RNAs: new insights into biogenesis and function inside and outside of germlines. Genes Dev. 26, 2361–2373 10.1101/gad.203786.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lim R. S. M., and Kai T. (2015) A piece of the pi(e): The diverse roles of animal piRNAs and their PIWI partners. Semin. Cell Dev. Biol. 47–48, 17–31 [DOI] [PubMed] [Google Scholar]
- 31. Saito K., Sakaguchi Y., Suzuki T., Suzuki T., Siomi H., and Siomi M. C. (2007) Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi-interacting RNAs at their 3′-ends. Genes Dev. 21, 1603–1608 10.1101/gad.1563607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ji L., and Chen X. (2012) Regulation of small RNA stability: methylation and beyond. Cell Res. 22, 624–636 10.1038/cr.2012.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hirano T., Iwasaki Y. W., Lin Z. Y., Imamura M., Seki N. M., Sasaki E., Saito K., Okano H., Siomi M. C., and Siomi H. (2014) Small RNA profiling and characterization of piRNA clusters in the adult testes of the common marmoset, a model primate. RNA 20, 1223–1237 10.1261/rna.045310.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Keam S. P., Young P. E., McCorkindale A. L., Dang T. H., Clancy J. L., Humphreys D. T., Preiss T., Hutvagner G., Martin D. I., Cropley J. E., and Suter C. M. (2014) The human Piwi protein Hiwi2 associates with tRNA-derived piRNAs in somatic cells. Nucleic Acids Res. 42, 8984–8995 10.1093/nar/gku620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sharma U., Conine C. C., Shea J. M., Boskovic A., Derr A. G., Bing X. Y., Belleannee C., Kucukural A., Serra R. W., Sun F., Song L., Carone B. R., Ricci E. P., Li X. Z., Fauquier L., et al. (2016) Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science 351, 391–396 10.1126/science.aad6780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen Q., Yan M., Cao Z., Li X., Zhang Y., Shi J., Feng G.-H., Peng H., Zhang X., Zhang Y., Qian J., Duan E., Zhai Q., and Zhou Q. (2016) Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science 351, 397–400 10.1126/science.aad7977 [DOI] [PubMed] [Google Scholar]
- 37. Jain R., and Shuman S. (2011) Active site mapping and substrate specificity of bacterial Hen1, a manganese-dependent 3′ terminal RNA ribose 2′-O-methyltransferase. RNA 17, 429–438 10.1261/rna.2500711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jain R., and Shuman S. (2010) Bacterial Hen1 is a 3′ terminal RNA ribose 2′-O-methyltransferase component of a bacterial RNA repair cassette. RNA 16, 316–323 10.1261/rna.1926510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kozomara A., and Griffiths-Jones S. (2011) miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 39, D152–D157 10.1093/nar/gkq1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ding D., Liu J., Dong K., Midic U., Hess R. A., Xie H., Demireva E. Y., and Chen C. (2017) PNLDC1 is essential for piRNA 3′-end trimming and transposon silencing during spermatogenesis in mice. Nat. Commun. 8, 819 10.1038/s41467-017-00854-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang Y., Guo R., Cui Y., Zhu Z., Zhang Y., Wu H., Zheng B., Yue Q., Bai S., Zeng W., Guo X., Zhou Z., Shen B., Zheng K., Liu M., Ye L., and Sha J. (2017) An essential role for PNLDC1 in piRNA 3′-end trimming and male fertility in mice. Cell Res. 27,1392–1396 10.1038/cr.2017.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Izumi N., Shoji K., Sakaguchi Y., Honda S., Kirino Y., Suzuki T., Katsuma S., and Tomari Y. (2016) Identification and functional analysis of the pre-piRNA 3′ trimmer in silkworms. Cell 164, 962–973 10.1016/j.cell.2016.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hayashi R., Schnabl J., Handler D., Mohn F., Ameres S. L., and Brennecke J. (2016) Genetic and mechanistic diversity of piRNA 3′-end formation. Nature 539, 588–592 10.1038/nature20162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Otwinowski Z., and Minor W. (1997) [20] Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 10.1016/S0076-6879(97)76066-X [DOI] [PubMed] [Google Scholar]
- 45. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 46. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L.-W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Falciatori I., Borsellino G., Haliassos N., Boitani C., Corallini S., Battistini L., Bernardi G., Stefanini M., and Vicini E. (2004) Identification and enrichment of spermatogonial stem cells displaying side-population phenotype in immature mouse testis. FASEB J. 18, 376–378 10.1096/fj.03-0744fje [DOI] [PubMed] [Google Scholar]
- 48. Langmead B., Trapnell C., Pop M., and Salzberg S. L. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jung I., Park J. C., and Kim S. (2014) piClust: a density based piRNA clustering algorithm. Comput. Biol. Chem. 50, 60–67 10.1016/j.compbiolchem.2014.01.008 [DOI] [PubMed] [Google Scholar]
- 50. Kent W. J., Sugnet C. W., Furey T. S., Roskin K. M., Pringle T. H., Zahler A. M., and Haussler D. (2002) The human genome browser at UCSC. Genome Res. 12, 996–1006 10.1101/gr.229102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kuhn R. M., Karolchik D., Zweig A. S., Wang T., Smith K. E., Rosenbloom K. R., Rhead B., Raney B. J., Pohl A., Pheasant M., Meyer L., Hsu F., Hinrichs A. S., Harte R. A., Giardine B., et al. (2009) The UCSC Genome Browser Database: update 2009. Nucleic Acids Res. 37, D755–D761 10.1093/nar/gkn875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chan P. P., and Lowe T. M. (2009) GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acid Res. 37, D93–D97 10.1093/nar/gkn787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cunningham F., Amode M. R., Barrell D., Beal K., Billis K., Brent S., Carvalho-Silva D., Clapham P., Coates G., Fitzgerald S., Gil L., Girón C. G., Gordon L., Hourlier T., Hunt S. E., et al. (2015) Ensembl 2015. Nucleic Acid Res. 43, D662–D669 10.1093/nar/gku1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.