Abstract
Processing of the eukaryotic transcriptome is a dynamic regulatory mechanism that confers genetic diversity, and splicing and adenosine to inosine (A-to-I) RNA editing are well-characterized examples of such processing. Growing evidence reveals the cross-talk between the splicing and RNA editing, but there is a paucity of substantial evidence for its mechanistic details and contribution in a physiological context. Here, our findings demonstrate that tumor-associated differential RNA editing, in conjunction with splicing machinery, regulates the expression of variants of HNRPLL, a gene encoding splicing factor. We discovered an HNRPLL transcript variant containing an additional exon 12A (E12A), which is a substrate of ADAR1 and ADAR2. Adenosine deaminases acting on RNA (ADAR) direct deaminase-dependent expression of the E12A transcript, and ADAR-mediated regulation of E12A is largely splicing-based, and does not affect the stability or nucleocytoplasmic distribution of the transcript. Furthermore, ADAR-mediated modification of exon 12A generates an enhancer for the oncogenic splicing factor SRSF1 and consequently promotes the frequency of alternative splicing. Gene expression profiling by RNA-seq revealed that E12A acts distinctly from HNRPLL and regulates a set of growth-related genes, such as cyclin CCND1 and growth factor receptor TGFBR1. Accordingly, silencing E12A expression leads to impaired clonogenic ability and enhanced sensitivity to doxorubicin, thus highlighting the significance of this alternative isoform in tumor cell survival. In summary, we present the interplay of RNA editing and splicing as a regulatory mechanism of gene expression and also its physiological relevance. These findings extend our understanding of transcriptional dynamics and provide a mechanistic explanation to the link of RNA editors to tumorigenesis.
Keywords: RNA editing, alternative splicing, exonic splicing enhancer, cell proliferation, tumor cell biology
Introduction
Most eukaryotic genes are transcribed to precursor mRNA (pre-mRNA),3 which undergoes multiple rounds of RNA processing before becoming mature mRNA that can either be translated into proteins or perform biological functions directly. Thus, mRNA processing of pre-mRNA is a gene regulatory mechanism that confers genome dynamics and diversity. Examples of mRNA processing include capping, polyadenylation, adenosine to inosine (A-to-I) RNA editing, and splicing; the latter two occur during transcription and are organized in a spatiotemporal manner (1–3). There is growing evidence for interplay between these two molecular processes in the regulation of gene expression: RNA editing affects splicing patterns by disrupting exon definition or mimicking the splice acceptor via conversion of the intronic dinucleotides AA to AI (4), whereas splicing can direct editing frequency via control of the editable substrates (5). This cross-talk between RNA editing and splicing thus appears to serve as a mechanism for fine-tuning gene expression.
A-to-I RNA editing is mediated by adenosine deaminases acting on RNA (ADARs), which recognize repetitive elements in transcripts forming double-stranded structures, and catalyze the hydrolytic deamination of adenosine residues. ADAR proteins are highly conserved across species (6), and expression of all three family members: ADAR1, ADAR2, and ADAR3, is found in vertebrates. ADAR1 and ADAR2 are expressed in most tissues, and the ubiquitously expressed ADAR1 has two isoforms: a nuclear constitutive protein (ADAR1–p110) and a cytoplasmic interferon-inducible protein (ADAR1–p150). ADAR3 expression is restricted to brain tissues, and it appears to be enzymatically inactive (7). Resulting inosine by ADARs acts as guanosine in genetic translation machinery and forms bp with cytidine; thus, A-to-I editing is functionally equivalent to A-to-G editing. A-to-I modification is known to alter transcript characteristics such as structural stability, nucleocytoplasmic distribution, and protein coding (8).
Dysregulation of ADAR expression and ADAR-mediated editing is observed in physiological disorders and cancers (9, 10). The oncogenic and tumor-suppressive roles of ADARs have been reported and are dependent upon cell type. In this regard, ADAR-mediated regulation relies on the nonsynonymous substitution of oncogenic proteins and modulation of the microRNA interference pathway. For example, via changes in their protein codes and properties, enhanced ADAR-mediated RNA editing of AZIN1 and RHOQ confer augmented invasiveness and aggressiveness to liver and colorectal cancer cells, respectively (11, 12). In contrast, the ADAR-edited form of GABRA3 down-regulates AKT activation and suppresses breast cancer metastasis (13). In parallel with protein recoding, ADARs could also modulate the substrate recognition of microRNA and RNAi machinery (14). ADAR-edited miR-376a and unedited miR-376a individually function as tumor-suppressive and oncogenic molecules, respectively, and attenuated editing promotes the expression of unedited miR-376a and the metastasis of glioblastomas (15). However, there is a paucity of concrete evidence for the elucidation of the interplay between RNA editing and splicing and its relevance in a physiological context.
Splicing removes introns from pre-mRNA and ligates adjacent exons to form continuous mRNA. Splicing of the same pre-mRNA transcript to produce different exon arrangements is termed alternative splicing, and this process generates transcript diversity. Core splicing operators are composed of small nuclear ribonucleoprotein molecules, which cooperate with auxiliary components to recognize splice sites and catalyze intron excision via transesterification reactions. The nature of spliceosome assembly is dynamic and complex, and the composition of and interaction within the assembly architecture determines the splicing outcome (16, 17). Cancer-associated mutations and dysregulated splicing factors can drive oncogenic splicing patterns that produce loss-of-function proteins and increase susceptibility to disease and malignancy (18). Aberrant splicing can also promote tumor progression by producing noncanonical transcripts implicated in anti-apoptotic signaling, angiogenesis, and drug resistance (19).
The current study provides substantial evidence of the interplay between RNA editing and splicing, with an implication in cancer biology. We uncovered in different tumor types the splicing factor gene HNRPLL as a differentially edited target, which was further identified as a substrate of both ADAR1 and ADAR2. ADARs-mediated RNA editing of the HNRPLL pre-mRNA determined its splicing pattern, an inclusion of a novel exon 12A, and consequently the generation of a transcript variant we named HNRPLL–E12A (E12A). E12A up-regulation is coordinated with alternative splicing; RNA editing created an additional exonic enhancer for the oncogenic splicing factor SRSF1, which promotes exon 12A inclusion and E12A transcript abundance in an ADAR-dependent manner. Although no functional protein product was detected for the E12A transcript, this variant was found by RNA-seq-based transcriptome profiling to regulate the expression of several genes, revealing a cellular role distinct from HNRPLL and a functional link to proliferation. Further in line with this role in cell growth and survival, knockdown of E12A expression altered the expression of several cell growth regulators, impaired colony formation of tumor cells, and enhanced their apoptotic state. Taken together, these findings demonstrate that tumor-associated A-to-I modification of the E12A transcript, which occurs via altered splicing preferences, has a fundamental role in tumorigenesis. This study also sheds light on the functional relevance of the coordinated action of two co-transcriptional processes, RNA editing and alternative splicing.
Results
Identification of tumor-associated differential RNA editing events in the HNRPLL transcript
To provide further insights into the physiological relevance of RNA editing in tumor malignancy, we first sought to identify from a systems perspective A-to-I(G) events with differential incidence in tumors. Toward this end, we used public RNA-seq data on kidney and bladder tumors to obtain corresponding RNA editome profiles (20, 21). We subsequently discovered three editing events in close proximity on the HNRPLL transcript, with a distinct difference in the degree of editing between normal and tumor specimens (Fig. 1, A and B). Sequence alignment mapped this cluster of editing sites to intron 12 of the HNRPLL gene. Intriguingly, according to the EST sequences archived in the UCSC genome browser, these editing positions could be expressed in the exonic region of a particular HNRPLL transcript variant (Fig. S1A). Because this alternative exon is 3′ to the annotated exon 12, we hereafter termed this exon 12A and the transcript variant as HNRPLL–E12A (or E12A). The expression of the E12A transcript was confirmed by variant-specific amplification in both normal and cancer cell lines (Fig. 1C), whereas its RNA editing at the three nucleotide locations (annotated from 5′ to 3′ as A, B, and C) in various cancer cells was verified by Sanger sequencing (Fig. 1C, Fig. S1B). Several lines of evidence provided additional support to the notion that the E12A transcript is a substrate of ADARs-mediated RNA editing: 1) HNRPLL pre-mRNA was predicted by the RNAfold web server (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi)4 to form double-stranded structures via complementarity between two inverted Alu repetitive elements bordering on the 12A exon, which presumably favors ADAR catalysis (Fig. S1C). 2) RNAi-mediated repression of ADAR1 was found to reduce A-to-G transition at the first two sites (A and B) (Fig. 1D, Fig. S1, D and E), whereas ADAR2 repression decreased editing degrees of all three sites, with site C exhibiting the most notable change (Fig. 1E). These results further illustrated that E12A indeed undergoes ADARs-dependent RNA editing. 3) Next, RNA immunoprecipitation (RNA-IP) assays were conducted to document the physical interaction between ADARs and the E12A transcript. Anti-ADAR1 RNA-IP revealed specific and abundant levels of E12A RNA in the precipitates relative to the control IgG (Fig. 1F, Fig. S1F). RNA-IP with ectopic ADAR2 expression also showed similar results, illustrating the binding of ADAR2 to the pre-mRNA, but not the mature transcript, of HNRPLL (Fig. 1G, Fig. S1G). Our RNA-IP assays therefore substantiated the possibility that ADARs bind and enzymatically modify the E12A RNA. 4) Finally, as a further verification of the A-to-I(G) editing of E12A transcript, Sanger sequencing of the genomic region corresponding to the editing sites excluded the interference of SNP (Fig. 1H). Taken together, these findings demonstrated that the E12A transcript, a novel HNRPLL splicing variant, is targeted by ADARs-mediated RNA editing.
Figure 1.
The HNRPLL pre-mRNA transcript is differentially edited in tumors and a substrate of ADAR1 and ADAR2. A and B, editing profiles of HNRPLL in clinical tumor specimens. Distribution of the A-to-G editing rates of three editing sites identified in the HNRPLL transcript is shown for paired kidney (a; n = 34) and bladder (b; n = 16) tumor patient samples. C, an exon E12A-containing HNRPLL variant was detected by end point PCR, using primers as shown by the schematic representation in the upper panel. PCR amplicons from the indicated cell lines were sequenced by the Sanger method shown in the lower panel, and the marked nucleotides denote the A-to-I(G) editing positions. D and E, vectors expressing control (pSUPER) or ADAR1/2-specific (shADAR1/2) shRNAs were transfected into HeLa cells, and protein knockdown in transfected cells was assessed by Western blotting, with GAPDH and β-actin as loading control. E12A editing in the transfected cells was determined by Sanger sequencing. F, for RNA-IP assay, HeLa cell lysates were immunoprecipitated with control IgG or ADAR1 antibody, and the precipitated RNA was measured by qPCR assay with specific primers to GAPDH and pre-mRNA of HNRPLL (n = 3). G, HeLa cells were transfected with the indicated plasmids and harvested for immunoprecipitation with anti-FLAG M2 beads. The RNA abundance in precipitated complexes was determined by a qPCR assay with specific primers to HNRPLL pre-mRNA, reference HNRPLL mRNA (WT), and the E12A variant (n = 3). H, Sanger sequencing of the genomic DNA region corresponding to the editing positions. The highlighted nucleotides indicate positions undergoing editing in the transcripts.
The editing status of the HNRPLL–E12A transcript is correlated with its expression
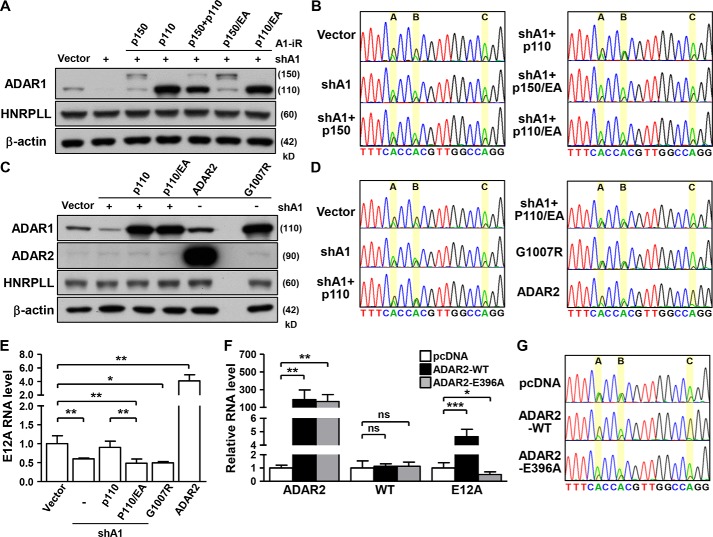
Given the possible involvement of both ADAR1 and ADAR2 in E12A RNA editing, we set out to delineate the mode and consequence of this enzymatic regulation. Because both the p110 and p150 isoforms of ADAR1 are known to possess deaminase activity, we first aimed to distinguish the isoform responsible for E12A editing. For this purpose, we generated isoform-specific expression constructs with synonymous mutations that render their ectopic expression resistant to RNAi-mediated silencing. Although ADAR1 knockdown cells exhibited reduced editing at positions A and B, as shown in Fig. 1D, restoration of ADAR1 p110 in these cells via the RNAi-resistant expression construct rescued RNA editing at both sites (Fig. 2, A and B). By contrast, replenishing ADAR1 with an enzyme-deficient mutant (E912A) had no effect on E12A editing, confirming enzyme catalysis as a requirement of this regulation. Re-expression of ADAR1 p150 did not restore editing, thus excluding a connection to E12A editing.
Figure 2.
ADARs control HNRPLL–E12A expression in a deaminase-dependent manner. A and B, HeLa cells were transfected with shADAR1 (shA1) and different ADAR1 expression vectors harboring resistance to shA1 (A1-iR). Specific protein expression and E12A editing status in transfected cells was analyzed, respectively, by Western blotting and Sanger sequencing. β-Actin serves as internal control. C–E, ADAR1 mutants, shADAR1 (shA1), and ADAR2 expression vectors were, respectively, transfected into HeLa cells, which were subsequently harvested for protein and RNA expression analyses. Specific protein expression was monitored by Western blotting (β-actin is the loading control). Expression and editing status of the E12A transcript was analyzed by qPCR and Sanger sequencing, respectively (n = 3). F and G, WT and mutant ADAR2 expression vectors were transfected into HeLa cells, and the E12A transcript was assessed for expression and editing status in the transfected cells, as above (n = 5).
Because of the possible role of ADARs in expression regulation of edited genes (8, 22, 23), we then analyzed E12A RNA abundance in ADAR1-altered cells. ADAR1 depletion decreased the expression levels of the E12A transcript, whereas ectopic re-expression of enzymatically inactive ADAR1 failed to discernably alter target abundance (Fig. 2, C and E, Fig. S2, A and B). To further highlight the importance of deaminase activity in this functional context, a dominant-negative ADAR1 mutant construct (G1007R) (24) was delivered into cells, and consequently shown to diminish both the editing and expression of the E12A transcript (Fig. 2, C–E).
In terms of ADAR2 and its editing activity, we found that overexpression of ADAR2 in HeLa cells markedly up-regulated the expression and editing of the E12A transcript (Fig. 2, C–G). This phenotype was similarly observed in HEK293 and MCF7 cells (Fig. S2, C and D). Incidentally, WT ADAR2 enhanced E12A expression and editing in a dose-dependent manner (Fig. S2, E and F). Overexpression of enzymatically inactive ADAR2 (E396A) in these cell lines led to reduced expression and editing of the E12A transcript, underscoring the requirement for deamination in ADAR2-mediated regulation. Conversely, ADAR-altered cells showed negligible changes in the expression of WT HNRPLL RNA (Fig. 2, E and F, Fig. S2B) and protein (Fig. 2, A and C), suggesting that the reference form of HNRPLL is not a target of ADARs. Taken together, our results implicated ADAR1 p110 and ADAR2 deaminase activities in RNA editing and expression of the HNRPLL–E12A transcript.
RNA editing mediates HNRPLL–E12A expression by triggering alternative splicing of HNRPLL
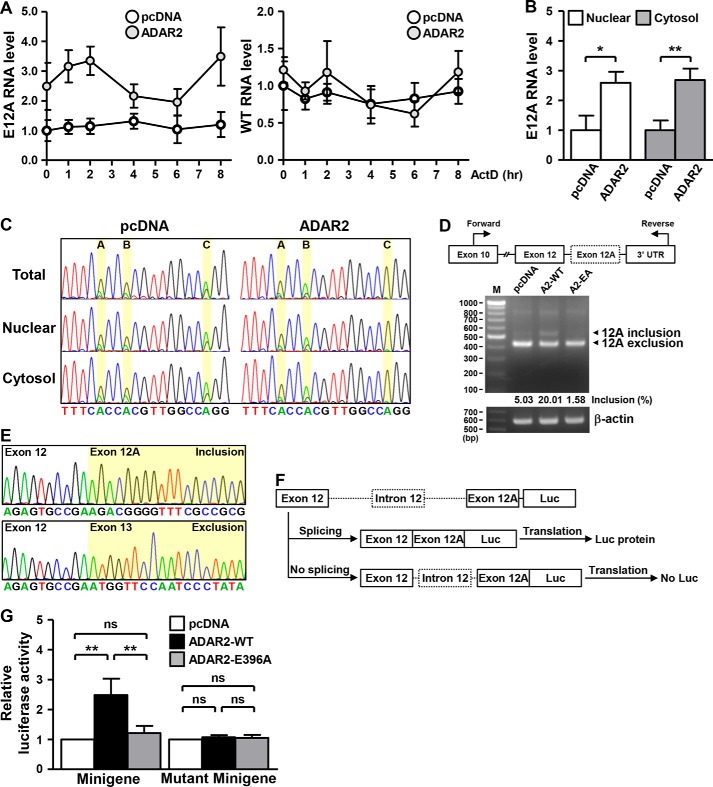
We next investigated the mechanistic basis underlying the RNA editing-dependent E12A expression changes. Because of the reported functional link of ADAR2 to mRNA decay machinery (22), we first assessed target transcript stability. To this end, we monitored temporal changes in RNA abundance in the presence of transcription inhibitor actinomycin D. EGR1, a known short-lived gene, served as an experimental control and displayed rapid decline in response to actinomycin D treatment (Fig. S3A). However, in ADAR2-overexpressing cells, time course detection of RNA abundance showed no difference in the turnover rates of both WT and E12A transcripts (Fig. 3A), therefore ruling out the involvement of the mRNA-decay mechanism in E12A regulation. Next, given that certain edited transcripts reportedly display distinct subcellular localizations via association with RNA shuttling factors such as P54 and STAU1 (25, 26), we analyzed the spatial distribution of E12A RNA under ADAR2 overexpression. We first performed subcellular fractionation, the extent of which was measured by expression of compartment-specific marker RNA (Fig. S3B). Subsequent qRT-PCR analysis first revealed that the E12A transcripts are expressed in both the nuclear and cytosolic fractions (Fig. 3B). Moreover, whereas ectopic ADAR2 elevated the overall abundance and site C editing of E12A (Fig. 3, B and C), it did not have any effect on the subcellular distribution of E12A (Fig. 3B). As a control, the relative levels of the HNRPLL reference RNA transcript between compartments remained unchanged in ADAR2-overexpressing cells (Fig. S3C). The lack of spatial changes thus indicated that RNA editing mediates E12A expression regulation irrespectively of subcellular localization of the transcript.
Figure 3.
RNA editing determines HNRPLL alternative splicing. A, for RNA stability analysis, actinomycin D was added to HeLa cells transfected with ADAR2-encoding or pcDNA empty vectors. At the indicated time points post-treatment, the abundance of WT HNRPLL (WT) and E12A RNA in cells was detected by qPCR assay (representative sample, n = 2). B and C, HeLa cells were transfected with control vector or ADAR2 expression plasmid, and subsequently separated into nuclear and cytoplasmic fractions. Expression and editing status of the E12A transcript in these subcellular compartments was analyzed by the qPCR assay and Sanger method, respectively (n = 3). D, HeLa cells transfected with plasmids encoding the indicated ADAR2 variant proteins were collected, and analyzed for the extent of alternative splicing by PCR analysis of the E12A region (β-actin expression as internal control). Schematic depiction of the relative locations of primers is shown in upper panel, and the inclusion frequency (%) of E12A-specific PCR products was measured by GelQuantNet software and corresponds to the relative representation of the variant signals. E, PCR products shown in D were cloned and sequenced by Sanger method. F, schematic representation of the E12A splicing reporter construct. G, HeLa cells were transfected with different versions of reporter constructs, ADAR2 overexpression vectors, and β-gal expression plasmid, as indicated. Transfected cells were harvested and subjected to splicing efficiency analysis via measurement of luciferase activity. Relative luciferase intensity was obtained by normalizing to the β-gal levels in each sample, and shown compared with the control group (represented as 1). Four independent experiments were conducted and statistical significance was measured by paired Student's t test.
PCR amplification of cDNA using primers specific to exon 10 and 3′ UTR of HNRPLL resulted in a product that is longer than the reference form (Fig. 3D) and later confirmed to be the exon 12A-inclusion variant (Fig. 3E). Isoform-specific amplification of the adjoining sequences at 5′ and 3′ sides of exon 12A illustrated that, aside from the included exon 12A, the E12A variant sequences are identical to reference HNRPLL transcripts (Fig. S3, D and E). Additionally, whereas HNRPLL-targeting shRNAs triggered down-regulation of both the reference and E12A variant transcripts, shRNAs specifically targeting the E12A sequence resulted in limited repression of the reference transcript (Fig. S4, A and B), further pinpointing the novel exon 12A as the structural distinction between the two HNRPLL transcript forms. Based on these observations, together with the results that ADARs regulate E12A RNA levels without affecting the reference RNA level, we hypothesized that ADARs might modulate the splicing of this region to give rise to the alternative E12A transcript. In support of this notion, ectopic expression of WT ADAR2 promoted the incorporation of exon 12A in HNRPLL transcripts, but had no effect on the levels of the reference transcript (Fig. 3D). In line with the requirement for deamination in this regulation, expression of enzymatically-inactive ADAR2 conversely only marginally influenced the extent of exon 12A inclusion.
To further strengthen this ADAR-mediated regulation of E12A splicing, we next employed a splicing reporter assay. For this purpose, we established a minigene reporter construct (27), which encompasses the splice donor/acceptor sequences, the luciferase reporter, and the intronic fragment with the embedded Alu elements (Fig. 3F). Repetitive sequences of Alu elements that readily form double-stranded structures facilitate ADAR binding and editing (28), and the editing complementary Alu repeat for E12A was identified in intron 12 and thus part of the reporter construct (Fig. S4C). Additionally, the translation initiation codons in the acceptor and luciferase sequences were removed by mutagenesis to reduce interference from leaky reporter protein expression. Based on our design, expression of the luciferase reporter thus signify splicing of the ectopic HNRPLL exons into a coding sequence in-frame with the luciferase gene, therefore serving as a readout for our assay (Fig. 3F). The feasibility of our minigene reporter construct was confirmed by its editability, we were able to detect specific occurrence of A-to-G changes at the three editing locations in the ectopic expressed sequences (Fig. S4D).
Using this reporter construct, we found that overexpression of WT ADAR2 led to elevated luciferase activity (Fig. 3G), illustrating the role of RNA editing in HNRPLL splicing. The ADAR2 mutant had a negligible effect on the levels of the reporter as compared with the control vector group, thus affirming that deamination is requisite for HNRPLL alternative splicing (Fig. 3G). We also repeated the reporter assay using a noneditable version of the minigene reporter (termed Mutant Minigene), which was altered to express guanosines instead of adenosines at the editing positions. Intriguingly, ADAR2 overexpression did not induce an up-regulation of the noneditable reporter (Fig. 3G), indicating that ADAR-directed A-to-G transition on the transcript determines the splicing event. Furthermore, we did not observe any changes in the HNRPLL transcription rate in the ADAR2-overexpressing cells, thus excluding the possibility that the HNRPLL gene is transcriptionally regulated by ADAR2 (Fig. S4E). In conclusion, our results in this part demonstrated that ADAR-catalyzed deamination on the E12A transcript contributes to the splicing outcome and consequently variant expression.
Exon 12A inclusion is regulated coordinately by RNA editing and SRSF1
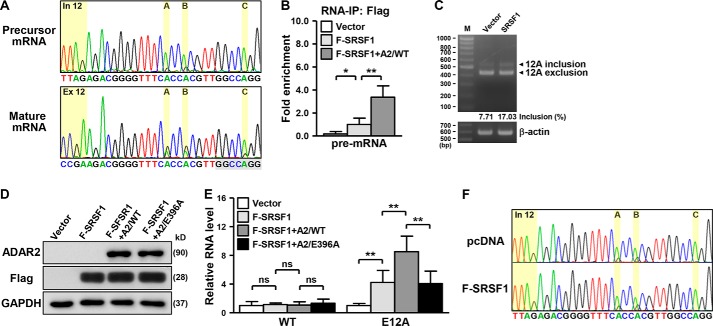
Upon establishing the positive role of RNA editing in alternative splicing of HNRPLL, we next examined whether the mature and the unspliced forms of E12A are differentially edited. Our Sanger sequencing results showed a greater extent of editing in the mature E12A than in the precursor counterpart (Fig. 4A, Fig. S5, A and B). Distinct differences in the degree of editing in precursor versus spliced E12A transcripts implied that the edited transcript might be prioritized in the recognition of spliceosome assembly and splicing outcome. Thus, to explore the possibility of a functional interaction between RNA editing and splicing, we used Human Splicing Finder (http://www.umd.be/HSF3/),4 an online bioinformatics web server, to search for cis-acting elements underlying the HNRPLL splicing event. As expected of a canonical exon, the exon 12A sequence was found to possess a competent donor splice site as well as a characteristic splice acceptor (Fig. S5C). Intriguingly, we detected an exonic splicing enhancer in the edited but not the reference version of the E12A sequence that also corresponds to the binding sequence of serine- and arginine-rich splicing factor 1 (SRSF1), a member of the SR protein family that modulates constitutive and alternative splicing (29, 30).
Figure 4.
Exon 12A inclusion is regulated coordinately by RNA editing and SRSF1. A, editing status of the E12A pre-mRNA and mature mRNA in HeLa cells was detected by the Sanger method. The marked nucleotides indicate the editing positions as well as the intronic/exonic regions, and the highlighted sequences at the bottom denote the binding motif of SRSF1. B, for RNA-IP experiments, the indicated transfected HeLa cells were collected and subsequently immunoprecipitated with anti-FLAG M2 beads, and the precipitated RNA was quantified by qPCR assay using specific primers to the pre-mRNA sequence of HNRPLL (n = 4). C, extent of exon 12A inclusion in control and SRSF1-overexpressing cells was analyzed by PCR analysis (β-actin expression as loading control). Schematic depiction of relative primer locations is shown in Fig. 3D, and the inclusion frequency (%) of E12A-specific PCR products was measured by GelQuantNet software and corresponds to the relative representation of the variant signals. D and E, FLAG-tagged SRSF1 was co-expressed with WT or mutant ADAR2 in HeLa cells, which were examined for ectopic proteins and RNA expression by Western blotting assay and qPCR experiment, respectively (n = 4). GAPDH was used as internal control. F, E12A editing status in transfected HeLa cells was analyzed by Sanger sequencing.
Although splicing mechanisms could antagonize the binding of ADARs to pre-mRNA and thus interfere with RNA editing frequency (5), our in silico analysis suggested that A-to-G changes on the E12A RNA creates a binding site for a splicing regulator and conceivably engenders an alternative regulation. To further clarify this functional coordination as well as the involvement of SRSF1 in alternative splicing of HNRPLL, we provided the following lines of experimental evidence. First, a direct regulation was illustrated by RNA-IP assay, which showed the occupancy of the HNRPLL pre-mRNA by SRSF1 (Fig. 4B). ADAR2 overexpression enhanced the interaction of SRSF1 with the transcript, further supporting our predicted gain of SRSF1-binding site upon RNA editing. Second, to demonstrate the involvement of SRSF1 in E12A splicing, we next characterized the effect of ectopic SRSF1 in cells. Increased exon 12A inclusion was evident as a result of SRSF1 expression (Fig. 4C). Concordantly, SRSF1 overexpression also elevated E12A transcript abundance but had no effect on the reference form, as shown by qRT-PCR (Fig. 4, D and E, Fig. S5D). This SRSF1-mediated up-regulation was further stimulated by overexpression of WT ADAR2. On the contrary, the splicing enhancing effect of ADAR2 was lost when the enzymatically inactive form was simultaneously introduced with SRSF1, again indicative of the importance of RNA editing in conferring SRSF1-dependent alternative splicing. Finally, we assessed the effect of ectopic SRSF1 on E12A editing and did not observe a notable alteration in RNA editing (Fig. 4F), suggesting that the formation of the E12A editing-competent duplex was independent, and likely upstream, of SRSF1 activity. Viewed together, these results substantiated the scenario that the occurrence of RNA editing on the E12A transcript provided preferential recognition to splicing factors that contribute to an alternative splicing outcome.
The E12A-encoded protein product is intrinsically unstable
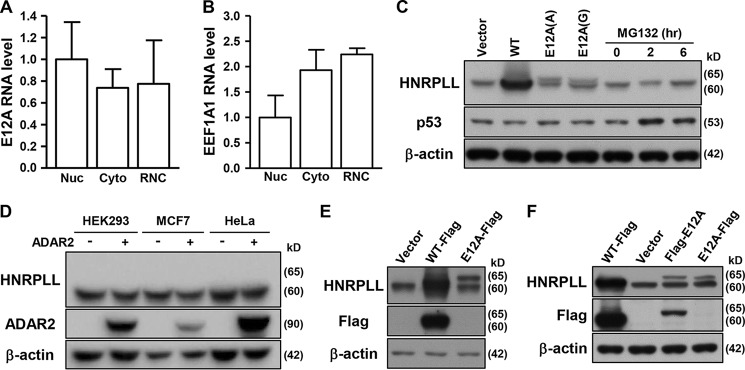
The additional exon 12A in the HNRPLL variant transcript creates an extended coding sequence that could hypothetically be translated into a 65-kDa polypeptide (595 amino acids). The association of this transcript with the isolated ribosome nascent chain mRNA complex (RNC-mRNA) fraction (Fig. 5A), which typically comprises translating mRNAs (Fig. 5B, Fig. S6A), served as another evidence for the protein coding potential of the E12A transcript (31). To verify the protein output of this variant, we then constructed unedited and edited versions of the E12A protein-coding region for ectopic expression analysis by Western blotting. To this end, ectopic E12A expression was detected using an antibody raised against a conserved region in the N terminus of HNRPLL (Fig. 5C, lanes 3 and 4); however, no signals of the size corresponding to E12A (∼65 kDa) or similar to the ectopic protein were present in the nontransfected samples (Fig. 5C, lane 1). Absence of these signals suggested the putative protein product is either unstable and/or minimally expressed endogenously. We then tested this possibility by treating cells with proteasome inhibitors, and subsequently found that, at doses that rescued expression of unstable protein (i.e. p53), no protein signals corresponding to E12A was distinctly observed (Fig. 5C, lanes 5–7). Moreover, whereas ectopic ADAR2 expression significantly increased the abundance of E12A RNA, no protein signals were correspondingly detected in ADAR2-overexpressing cells (Figs. 2C and 5D).
Figure 5.
The intrinsic instability of the E12A-encoded protein product. A and B, E12A and EEF1A1 RNA abundance in different subcellular fractions of HeLa cells was determined by qPCR experiment (n = 4). Nuc, nucleus; Cyto, cytosol. C, ectopic expression vectors encoding WT HNRPLL (WT), unedited (A) and edited (G) versions of E12A were transfected into cells. Cells were treated with MG132 at the indicated time point. Immunoblotting analysis was performed to assess expression levels of the indicated proteins in the transfected and treated cells. D, expression levels of HNRPLL in the control and ADAR2-overexpressing cells were detected by immunoblotting. E and F, HeLa cells were transfected with empty vector, or expression vectors for FLAG-tagged WT HNRPLL, and N terminus- or C terminus-tagged E12A. Protein expression was monitored by Western blotting assay using the indicated antibodies. For immunoblotting data shown in this figure, β-actin expression was used as loading control.
To ascertain expression of the E12A protein product, we generated a specific antibody against the putatively encoded peptide sequence of the E12A exon (Fig. S6B). Antigen ELISA and immunoblotting analysis of ectopic E12A overexpression verified the specificity of the E12A antibody (Fig. S6, B and C). Ectopic overexpression of the E12A protein was detected by the E12A-specific antibody, confirming the feasibility of this antibody (Fig. S6, D, left panel, and E). However, use of E12A-specific antibody did not yield distinct signals in the lysates of either the control or ADAR2-overexpressing cells (Fig. S6D, right panel), indicating a lack of endogenous expression of full-length protein product for this variant. Interestingly, in Western blot analysis of the overexpression of FLAG-tagged E12A constructs, anti-FLAG signals were detected in cells overexpressing N terminus-tagged E12A, but not the C terminus-tagged E12A (Fig. 5, E and F), implying an intrinsic instability of the protein product. Collectively, these results suggested that E12A-encoded polypeptides were targeted for proteolytic processing (Fig. S6F).
E12A contributes to tumor cell survival via expression regulation of CCND1
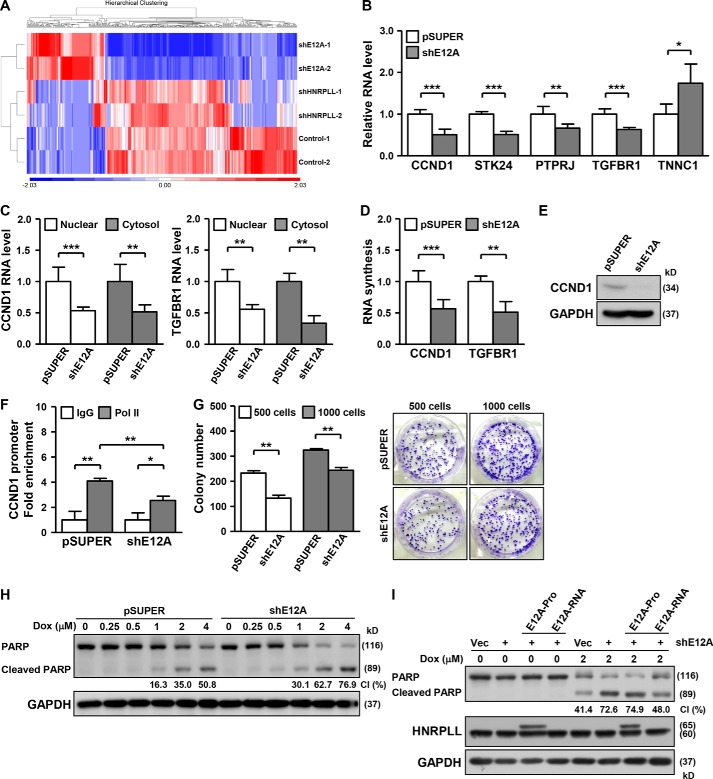
Given that SRSF1 amplification, which promotes the aberrant splicing patterns that confer oncogenic activity, is observed in most cancers (32), the regulation of HNRPLL alternative splicing by SRSF1 may hint at a function of E12A in tumorigenesis. To further dissect the functional relevance of the E12A variant, we conducted RNA-seq experiments to profile transcriptome-wide alterations in response to E12A knockdown (Fig. 6A). Differential expression analysis revealed that about 114 genes were up-regulated (p < 0.05, ≥1.5-fold change; Table S1) and 280 genes down-regulated (p < 0.05, ≥1.5-fold change; Table S1) significantly in these cells relative to the control. The overall distribution of the differentially altered genes, as depicted by heat map representation, showed distinct changes in the transcriptome landscape associated with E12A knockdown (Fig. 6A), indicative of a unique mode of action that is different from that of the HNRPLL reference gene product. By analyzing the functional and cellular attributes of the down-regulated gene, we found a subset of targets implicated in cell growth. We further performed qRT-PCR validation on a subset of the differentially expressed targets and confirmed the effect of E12A down-regulation on their expression (Fig. 6B, Fig. S7A). Reduction in the overall abundance of these targets in the E12A-knockdown cells was not mediated by changes in subcellular localization (Fig. 6C, Fig. S7B), but could be attributed to an attenuated transcription rate, as illustrated by a lower extent of nascent RNA synthesis (Fig. 6D, Fig. S7C).
Figure 6.
The role of the E12A variant in gene regulation and tumor cell survival. A, for global profiling of target genes, RNA-seq experiments were performed on HeLa cells with HNRPLL or E12A knockdown (see “Experimental procedures”). The heat map represents the distribution of genes differentially expressed compared with the control culture. The expression values were displayed in shades of red or blue relative to the means of all corresponding values within individual experimental groups. Hierarchical clustering was performed on the samples by Euclidian distance and average linkage method. B, vectors expressing control (pSUPER) or E12A-specific (shE12A) shRNAs were transfected into HeLa cells, and RNA expression of the indicated genes was analyzed by qPCR assay (n = 5). C, subcellular distribution of the CCND1 and TGFBR1 RNA transcripts in the indicated transfected HeLa cells was examined by qPCR experiment (n = 3). D, nascent RNA synthesis of the indicated genes in HeLa transfectants was determined by qPCR assay (n = 4). E, expression of the CCND1 protein in E12A-knockdown HeLa cells was analyzed by Western blotting assay, with GAPDH as loading control. F, HeLa cells with E12A-knockdown were harvested and subjected to ChIP assay, using control IgG or Pol II antibody. The abundance of the precipitated chromatin complexes was quantified by qPCR with a specific primer to the CCND1 promoter sequence (n = 3). G, colony formation assay was performed on E12A-knockdown HeLa cells, and images of colony formation assay shown in the right panel (representative sample, n = 2). Starting cell number in each experiment was noted. H, vectors expressing control (pSUPER) or E12A-specific (shE12A) shRNAs were transfected into HeLa cells, which were subsequently treated with different doses of doxorubicin, as indicated. The treated cells were collected for PARP protein expression analysis via Western blotting, and the cleaved PARP ratio (CI) was quantitatively determined by GelQuantNet software. GAPDH was used as loading control. I, to assess the apoptotic consequence of E12A mis-expression, HeLa cells were transfected with the control or E12A-targeting shRNA-expressing vector, together with E12A-specific RNA-only (E12A-RNA) and protein (E12A-protein) expression vectors, as indicated. The transfected cells were treated with the indicated dose of doxorubicin, and subsequently collected for PARP protein expression analysis as shown in H. GAPDH expression serves as internal control.
We then selected one of the targets, CCND1, for further characterization because of its close link to cancer cell growth and survival (33). Correspondingly to the effect on mRNA expression, E12A-knockdown markedly decreased the CCND1 protein level (Fig. 6E). Additionally, the ChIP assay showed that E12A-knockdown reduced the RNA polymerase II binding of the CCND1 promoter, in line with a possible transcriptional mechanism of E12A's action (Fig. 6F, Fig. S7, D and E). On the basis of the pro-growth function of CCND1, we carried out a colony formation assay and subsequently observed that E12A-suppressed cells exhibited reduced colony numbers but unaltered colony sizes (Fig. 6G). However, results from the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay did not reveal any difference in the metabolic state of E12A-knockdown versus control cells (Fig. S8A). Impaired clonogenic ability thus suggested that E12A was required for cell proliferation under selective pressure. In parallel to cell proliferation, we also examined the possible link of E12A to the cellular apoptotic response. We used in our study a clinical anti-cancer drug doxorubicin as an inducer of apoptosis, initiation of which entails the systematic activation of caspases, with cleavage of the PARP protein as a hallmark of the caspase cascade. In the course of doxorubicin treatment, the levels of cleaved PARP were augmented in E12A-repressed cells, indicating that loss of E12A could sensitize cells to doxorubicin-mediated apoptosis (Fig. 6H).
Due to the limited expression of the E12A protein product (Fig. 5), we next set out to determine whether E12A exerts its function via either the RNA transcript or the protein form. In this regard, we first generated a nonprotein-coding construct of E12A by removing the start codon and multiple Met codons from the E12A full-length sequence (termed E12A-RNA; see “Experimental procedures”). Further distinct from the protein-coding construct (E12A-Pro), the E12A-RNA construct also expresses intact 5′ UTR and 3′ UTR sequences. We then aimed to compare the effect of this “RNA-only” form with the protein-coding construct in a E12A-knockdown plus rescuing experiment (Fig. 6I). Upon treating transfected cells with doxorubicin, we analyzed the response of cells to apoptosis stimuli and subsequently found that replenishing ectopic E12A “protein” with the protein-coding construct in E12A-depleted cells does not influence enhanced apoptosis by E12A knockdown; however, rescuing E12A “RNA” expression protected cells from apoptotic signaling induced by E12A depletion (Fig. 6I). Moreover, we assessed the target gene expression in similar experimental conditions, and further found that the down-regulated expression of targets in E12A-knockdown cells was restored by over-expressing the E12A RNA-only but not the protein-coding form (Fig. S8B). These findings thus suggest that the RNA transcript of E12A acts as the predominant functional form of E12A.
Interestingly, we observed via Sanger sequencing analysis a more prominent E12A editing in response to a doxorubicin challenge (Fig. S8C). Expression of E12A also underwent an increase in the presence of doxorubicin, suggesting a distinct regulation of and requirement for this variant expression when cells encounter apoptotic stress (Fig. S8C). Furthermore, in support of the functional relevance of the SRSF1–E2A axis, we found that ectopic SRSF1 expression promoted CCND1 expression, which was reversed by simultaneous silencing of E12A expression (Fig. S8D). These observations hence evidenced that E12A expression may contribute in part to the SRSF1-mediated oncogenic process. In summary, our results uncovered an indispensable role of HNRPLL–E12A in proper expression of key cell growth promoters (i.e. CCND1) and the consequent preservation of a pro-growth and anti-apoptosis response.
Discussion
Advances in next-generation sequencing have improved the resolution and accuracy of the RNA editome and extended our understanding of RNA editing (34, 35). A-to-I transition by ADARs is the predominant RNA editing mechanism, and tens of thousands to millions of editing positions have been identified (36). However, only a limited number of editing targets have been functionally defined, and the vast majority of edits remain uncharacterized. Here, our findings have uncovered tumor-associated RNA editing of the HNRPLL transcript and addressed the role of ADARs in mediating the editing and consequent alternative splicing of HNRPLL. Mechanistically, A-to-I modification on the E12A transcript leads to up-regulation of E12A RNA expression by enhancing recognition of the splicing factor SRSF1. Functional relevance of the E12A variant was revealed by RNA-seq, which showed that E12A targets a subset of cell growth-related genes. Consistently, E12A repression altered expression of the cyclin CCND1, and led to compromised clonogenic ability and enhanced sensitivity to the doxorubicin challenge. RNA editing of E12A was observed in various cell lines and tumor tissues, demonstrating the requirement of A-to-I modification of the HNRPLL transcript for cell growth maintenance (Fig. 1, Fig. S1).
Biased splicing patterns in cancers, caused by the mutation and mis-expression of splicing factors, produce abnormal proteins that could generate detrimental effects (17). For example, cancer-associated SF3B1 mutations misdirect branch point usage, which leads to inappropriate splicing and malignancy (37–39). In line with the occurrence of splicing factor mutations, oncogene-driven regulation of splicing factors emerges as another cause of dysregulated splicing. For example, up-regulation of SRSF1 in tumors disrupts the function of the tumor suppressors BIM and BIN1, consequently triggering accelerated cell growth and delayed apoptosis (32, 40). The known oncogenic transcription factor, MYC, can also directly activate SRSF1 expression, and controls the splicing pattern of downstream genes (41). Thus, our observations of the SRSF1-directed production of the E12A transcript, as well as the impaired cell growth and survival in response to E12A knockdown, extend the substrate spectrum of SRSF1 and further strengthen the fundamental role for SRSF1 in E12A regulation and cancer biology.
The reference HNRPLL-encoded protein functions as a splicing regulator by recognizing negative cis-regulatory elements and promoting exon exclusion. The reference HNRPLL protein governs global splicing in T-cell activation, including the alternative splicing of CD45, a gene implicated in T-cell transition from naive to activated state (42). Patients with a higher proportion of CD45RO-positive cells have reduced pathological stages and prolonged survival (43), thus linking HNRPLL to cancer development with a tumor-suppressive role. Interestingly, several lines of evidence provided by this study indicate that the novel E12A variant seemingly exerts contrasting functions in the cells. First, global transcriptome profiling revealed alterations in gene expression in E12A-depleted cells distinct from the HNRPLL-knockdown cells (Fig. 6A), suggesting diverse modes of gene regulatory action between these two molecules of the same origin. Furthermore, the putative regulatory role of E12A in nascent RNA synthesis implies that it may function as a transcription regulator and to a lesser extent, if at all, as a splicing regulator. Second, the requirement for E12A in cell growth and survival (Fig. 6) contradicts the tumor-suppressive function of reference HNRPLL, and implies that these two molecules might act antagonistically in the context of cancer biology. Finally, whereas noncoding RNA could function through the modulation of the expression and regulatory pathways of the parental gene (44), our results showed that the expression levels of reference HNRPLL mRNA and protein remained unchanged under E12A mis-expression (Fig. 2). Taken together, these inconsistent findings strongly suggest that, despite their shared origin, the E12A variant may acquire a unique cellular role that is unrelated to the HNRPLL splicing function.
Due to the limited space in which transcription occurs, the sequential order of RNA editing and splicing is still debated, despite accumulating evidence for their functional interaction (1, 3). Splicing reportedly can affect editing efficiency by regulating the presentation of pre-mRNA to the editing machinery (5). However, in this study, we found that A-to-I modification of the HNRPLL transcript facilitates recognition by the splicing factor SRSF1, leading to the inclusion of exon 12A. These findings thus imply that the interplay between the RNA editing and splicing mechanisms may be context-dependent and/or gene-specific. In this capacity, the dynamics of transcription may impinge on this cross-talk as an additional layer of regulation. Precise spatiotemporal regulation of transcription and post-transcriptional processing rely on the coordination between the RNA-binding protein (RBP) and the appropriate phosphorylation of the C-terminal domain of RNA polymerase II (1). These RBPs, notable examples of which include ELAVL1, are known to interact with pre-mRNA and impart splicing regulation, among other post-transcriptional mechanisms (45). Interestingly, ADAR enzyme-catalyzed A-to-I transition on transcripts is known to modify RBP functions via substrate accessibility (22). Thus, the kinetic mode of transcription may, directly or indirectly, act as part of the regulatory hierarchy that determines the outcome of the RNA editing and splicing processes. Additional global analyses using existing deep sequencing platforms and datasets may shed light on the interpretation of the dynamic interaction and cooperation between the RNA editing and splicing machineries.
Experimental procedures
Cell culture
HeLa and HEK293 cells were cultured in high-glucose Dulbecco's modified Eagle's medium, and 1× nonessential amino acid and 1 mm sodium pyruvate was added to medium for HEK293 cells. MCF7 cells were cultured in minimum essential media containing 1× nonessential amino acid and 1 mm sodium pyruvate. All culture media were supplemented with 10% heat-inactivated fetal bovine serum and 1 unit/ml of penicillin-streptomycin. All media and reagents were purchased from Thermo Fisher Scientific. The cells were incubated at 37 °C with 5% CO2 in a humidified incubator.
Plasmid construction for gene knockdown and overexpression
RNAi-mediated gene silencing was performed using the pSUPER RNAi system, with the target sequences annealed and ligated into the pSUPER vectors. The coding sequences of ADAR1, ADAR2, HNRPLL, and SRSF1 genes were amplified from cDNA with primers containing flanking restriction enzyme sequences (EcoRV-HindIII for ADAR1, XbaI-EcoRV for ADAR2, XbaI-EcoRV for HNRPLL, and XbaI-HindIII for SRSF1). The resulting PCR products were ligated into cloning vector using the HE Swift Cloning Kit (TOOLS Life Science) and subsequently subcloned into the expression vector pcDNA3.1(−). The ADAR mutants and RNAi-resistant variants were generated by PCR-directed mutagenesis, and constructed as described above. RNAi-resistant constructs were established by introducing synonymous nucleotide substitution within the shADAR1-targeting sequences, with the following primers: forward, 5′-GAACTAGTAAGGTATCTTAACACCAACCCTGTGGG; reverse, 5′-TAAGATACCTTACTAGTTCGCCAATCTTCCTGACC (altered nucleotides are underlined). For generating the nonprotein-coding, or RNA-only, construct of E12A, the start codon and multiple Met codons (at Met-106, Met-107, and Met-108) within the full-length E12A transcript sequence were mutated to stop codons, as a means to block protein translation of the E12A sequence. However, this construct still retains intact 5′ UTR and 3′ UTR sequences. To this end, PCR-directed mutagenesis was used to introduce nucleotide substitution with the designed primer sets, and the resulting PCR products were subcloned into the expression vector pcDNA3.1(−). All constructed plasmids were delivered to cells by Lipofectamine 2000 according to the manufacturer's instructions (Invitrogen). Primer sequences are listed in Table S2.
RNA extraction, RT-PCR, and quantitative PCR (qPCR)
Total RNA was isolated by TRIzol reagent (Invitrogen), and reverse transcribed into complementary DNA (cDNA) by Moloney murine leukemia virus reverse transcriptase (Invitrogen) with random hexamer. Editing status on target transcript was monitored by Sanger sequencing analysis of end point PCR products, which were amplified from cDNA with designed primers. The editing frequency (%) of sites shown in the figures was quantitatively determined based on Sanger sequencing chromatograms and summarized in Table S3. The transcript splicing ratio was analyzed by gel electrophoresis of the end point PCR products generated from cDNA with particular primers. Individual gene expression was detected by real-time qPCR (iQ5 Gradient Real Time SYBR Green PCR system) with specific primers, and quantitatively analyzed by CFX Manager Software (Bio-Rad). The relative gene expression was determined by normalization to the abundance of the indicated housekeeping gene expression, with the control group being represented as 1. All results were obtained from at least three independent experiments and presented as mean ± S.D., and statistical significance was measured by Student's t test and presented in p value. Sequences of primers used in end point PCR and real-time qPCR experiments are listed in Table S2, and the MIQE (46) (Minimum Information for Publication of Quantitative Real-Time PCR Experiments) checklist is shown in Table S4.
Chemicals and antibodies
Chemicals were mostly purchased from Sigma, except where indicated otherwise. Primary antibodies against ADAR1 (sc-73408), ADAR2 (sc-73409), GAPDH (sc-32233), β-actin (sc-47778), HNRPLL (sc-132712), P53 (sc-263), and PARP (sc-7150) were purchased from Santa Cruz Biotechnology. Anti-FLAG primary antibody (F1804) and anti-FLAG affinity agarose gel (A2220) were acquired from Sigma. Secondary antibodies for immunoblotting were obtained from Vector Laboratories.
RNA-IP experiment
RNA-IP was performed according to a previous report (47). Briefly, cells were washed once with PBS, and lysed by Polysomal Lysis Buffer (100 mm KCl, 5 mm MgCl2, 10 mm HEPES (pH 7.0), 0.5% Nonidet P-40, 1 mm DTT, 50 units/ml of RNaseOUT, and protease inhibitor (Roche)). Lysate was passed through a 27-gauge needle eight times for complete lysis, and subsequently centrifuged at 12,000 × g for 15 min at 4 °C. The supernatant was collected and incubated with Dynabeads Protein G (Invitrogen) for 1 h at 4 °C. The pre-cleared lysate was incubated with control IgG or antibody-coated Dynabeads for 3 h at 4 °C. The precipitated complex was washed with Polysomal Lysis Buffer, and treated with 10 units of DNase I (Fermentas) at 37 °C for 15 min. TRIzol reagent was added for RNA extraction. The immunoprecipitated RNA was reverse transcribed to cDNA, and subsequently analyzed by qPCR assay.
Nuclear and cytoplasmic fractionation
Cells were washed and collected with PBS, and lysate was centrifuged at 1,000 × g for 10 min. The supernatant was removed and the nuclear fractionation buffer was added to pellet. The nuclear fractionation buffer contained 10 mm Tris-HCl (pH 7.8), 140 mm NaCl, 1.5 mm MgCl2, 0.5% Nonidet P-40, and 3 units/ml of RNaseOUT (Invitrogen). The suspended pellet was incubated for 5 min at 4 °C, and subsequently centrifuged at 500 × g for 4 min. The supernatant was isolated and served as a cytoplasmic fraction. The pellet was washed with fractionation buffer twice, centrifuged at 3,000 × g for 10 min at 4 °C, and used as nuclear fraction. Total RNA of cytoplasmic and nuclear fractions were extracted by TRIzol reagent, and subsequently reverse transcribed to cDNA. Gene expression in cDNA sample was determined by qPCR, and U48 and 7SL were served as nuclear and cytoplasmic markers.
Splicing luciferase reporter assay
The chimeric reporter constructs were generated by fusing the E12A sequences with the luciferase gene. For reporter assay, the indicated plasmids were co-transfected with β-gal expression vector into HeLa cells for 2 days. Transfected cells were washed with PBS, and collected by Reporter Lysis 5X Buffer (Promega). Luciferase and β-gal activity in the indicated transfectant was measured by a Dual Luciferase Reporter Assay System (Promega), and the relative luciferase activity was obtained by normalized to β-gal intensity. All of the samples were analyzed in triplicate, and four independent experiments were performed.
Isolation of RNC-mRNA complex
Experimental procedures of the RNC-mRNA complex extraction were based on Wang's report and modified (31). Briefly, cells were pre-treated with 100 mg/ml of cycloheximide for 15 min, and subsequently separated into nuclear and cytosolic fractions. The cytosolic RNC-mRNA complex was further isolated by centrifuging at 185,000 × g for 5 h at 4 °C with sucrose solution. Sucrose solution contained 30% sucrose, ribosome buffer (20 mm HEPES/KOH (pH 7.4), 15 mm MgCl2, and 200 mm KCl), 2 mm DTT, and 100 mg/ml of cycloheximide. The sediment fraction was washed twice with ribosome buffer, and TRIzol reagent was used to isolate RNA. Collected RNA was reverse transcribed into the cDNA sample, and abundance of the target gene was quantified by a qPCR experiment. EEF1A1 expression was analyzed as the control.
RNA-Seq
Cells were transfected with control vector pSUPER and shRNA targeting to E12A variant, and total RNA of transfected cells was isolated by TRIzol reagent. cDNA libraries were prepared based on the TruSeq® Stranded Total RNA Sample Preparation Guide (Illumina, part number 15031048). Equal concentrations of each library were sequenced using a NextSeq 500 (Illumina) platform to create pair-end 75-bp reads.
Gene expression analysis
All RNA reads were mapped to hg19 with STAR 2.5.2b (48). The expression levels of genes in the three samples and the corresponding fold-changes were estimated by DESeq2 1.14.1 (49) with GENCODE V19 annotation (50).
Detection of nascent RNA synthesis rate
Metabolic labeling was used in the analysis of nascent RNA synthesis, and experimental procedures were performed as described previously (51), with modifications. Briefly, 500 μm 4-thiouridine was added to cells for 30 min, and labeled RNA was isolated by TRIzol reagent and dissolved in TE buffer (10 mm Tris-HCl, 1 mm EDTA (pH 7.5)). Isolated RNA was thiol-specific biotinylated (biotin-HPDP) and incubated for 1.5 h with rotation, and the RNA sample was extracted by Phase Lock Gel (QuantaBio) with chloroform/isoamyl alcohol (24:1), and re-suspended with TE buffer. Biotinylated RNA was immobilized by streptavidin-coated magnetic beads (Invitrogen) in a 30-min incubation with rotation, and subsequently washed and eluted with 100 mm DTT. Eluted RNA sample was purified with RNeasy Mini Kit (Qiagen), and converted into cDNA by reverse transcription. Nascent RNA synthesis rate was determined by qPCR experiment, with ACTIN expression as the internal control.
ChIP assay
Experimental procedures were performed as described before (52). Cells were transfected with the indicated plasmids, and transfected cells were subjected to formaldehyde cross-linking. Cross-linked lysate was sonicated and precleared before being incubated with the control IgG or the indicated antibodies with rotation at 4 °C overnight. The immunocomplexes were washed, treated with proteinase K, and decross-linked. Bound DNA as well as input DNA (1/10 fragmented chromatin) were purified and extracted. The isolated DNA was quantified by qPCR experiment using primers corresponding to the genomic loci.
Author contributions
Y.-T. C., L. K., and B. C.-M. T. conceptualization; Y.-T. C. and H. L. data curation; Y.-T. C., C.-P. M., Y.-P. K., C.-T. S., and Y.-H. S. validation; Y.-T. C., I. Y.-F .C., Y.-H. S., and B. C.-M. T. investigation; Y.-T. C. and B. C.-M. T. writing-original draft; H. L. methodology; B. C.-M. T. supervision; B. C.-M. T. writing-review and editing.
Supplementary Material
Acknowledgments
We are grateful to members of the BC-MT laboratory for critical reading of the article and important discussions.
This work was supported by Ministry of Science and Technology of Taiwan Grants MOST105-2320-B-182-017 (to H. L.), MOST104-2320-B-182-029-MY3, and MOST105-2314-B-182-061-MY4 (to B. C.-M. T.), MOST103-2632-B-182-001, MOST104-2632-B-182-001, and MOST105-2632-B-182-001, Chang Gung Memorial Hospital Grants CMRPD1F0571 (to H. L.), CMRPD1C0843, CMRPD3E0153, CMRPD1F0442, and BMRP960 (to B. C-M. T.), National Health Research Institute of Taiwan Grant NHRI-EX105-10321SI, and a grant from the Ministry of Education of Taiwan. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S8 and Tables S1–S4.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- pre-mRNA
- precursor mRNA
- E12A
- exon 12A
- ADAR
- adenosine deaminases acting on RNA
- qRT
- quantitative RT
- PARP
- poly(ADP-ribose) polymerase
- miRNA
- microRNA
- RBP
- RNA-binding protein
- β-gal
- β-galactosidase
- RNC
- ribosome nascent chain
- IP
- immunoprecipitation
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase.
References
- 1. Bentley D. L. (2014) Coupling mRNA processing with transcription in time and space. Nat. Rev. Genet. 15, 163–175 10.1038/nrm3753,10.1038/nrg3662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Raitskin O., Cho D. S., Sperling J., Nishikura K., and Sperling R. (2001) RNA editing activity is associated with splicing factors in lnRNP particles: the nuclear pre-mRNA processing machinery. Proc. Natl. Acad. Sci. U.S.A. 98, 6571–6576 10.1073/pnas.111153798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Laurencikiene J., Källman A. M., Fong N., Bentley D. L., and Ohman M. (2006) RNA editing and alternative splicing: the importance of co-transcriptional coordination. EMBO Rep. 7, 303–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rueter S. M., Dawson T. R., and Emeson R. B. (1999) Regulation of alternative splicing by RNA editing. Nature 399, 75–80 10.1038/19992 [DOI] [PubMed] [Google Scholar]
- 5. Licht K., Kapoor U., Mayrhofer E., and Jantsch M. F. (2016) Adenosine to inosine editing frequency controlled by splicing efficiency. Nucleic Acids Res. 44, 6398–6408 10.1093/nar/gkw325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Slavov D., Crnogorac-Jurcević T., Clark M., and Gardiner K. (2000) Comparative analysis of the DRADA A-to-I RNA editing gene from mammals, pufferfish and zebrafish. Gene 250, 53–60 10.1016/S0378-1119(00)00175-X [DOI] [PubMed] [Google Scholar]
- 7. Chen C. X., Cho D. S., Wang Q., Lai F., Carter K. C., and Nishikura K. (2000) A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single- and double-stranded RNA binding domains. Rna 6, 755–767 10.1017/S1355838200000170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang I. X., So E., Devlin J. L., Zhao Y., Wu M., and Cheung V. G. (2013) ADAR regulates RNA editing, transcript stability, and gene expression. Cell Rep. 5, 849–860 10.1016/j.celrep.2013.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Han L., Diao L., Yu S., Xu X., Li J., Zhang R., Yang Y., Werner H. M., Eterovic A. K., Yuan Y., Li J., Nair N., Minelli R., Tsang Y. H., Cheung L. W., et al. (2015) The genomic landscape and clinical relevance of A-to-I RNA editing in human cancers. Cancer Cell 28, 515–528 10.1016/j.ccell.2015.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zipeto M. A., Jiang Q., Melese E., and Jamieson C. H. (2015) RNA rewriting, recoding, and rewiring in human disease. Trends Mol. Med. 21, 549–559 10.1016/j.molmed.2015.07.001 [DOI] [PubMed] [Google Scholar]
- 11. Chen L., Li Y., Lin C. H., Chan T. H., Chow R. K., Song Y., Liu M., Yuan Y. F., Fu L., Kong K. L., Qi L., Li Y., Zhang N., Tong A. H., Kwong D. L., et al. (2013) Recoding RNA editing of AZIN1 predisposes to hepatocellular carcinoma. Nat. Med. 19, 209–216 10.1038/nm.3043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Han S. W., Kim H. P., Shin J. Y., Jeong E. G., Lee W. C., Kim K. Y., Park S. Y., Lee D. W., Won J. K., Jeong S. Y., Park K. J., Park J. G., Kang G. H., Seo J. S., Kim J. I., and Kim T. Y. (2014) RNA editing in RHOQ promotes invasion potential in colorectal cancer. J. Exp. Med. 211, 613–621 10.1084/jem.20132209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gumireddy K., Li A., Kossenkov A. V., Sakurai M., Yan J., Li Y., Xu H., Wang J., Zhang P. J., Zhang L., Showe L. C., Nishikura K., and Huang Q. (2016) The mRNA-edited form of GABRA3 suppresses GABRA3-mediated Akt activation and breast cancer metastasis. Nat. Commun. 7, 10715 10.1038/ncomms10715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ota H., Sakurai M., Gupta R., Valente L., Wulff B. E., Ariyoshi K., Iizasa H., Davuluri R. V., and Nishikura K. (2013) ADAR1 forms a complex with Dicer to promote microRNA processing and RNA-induced gene silencing. Cell 153, 575–589 10.1016/j.cell.2013.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Choudhury Y., Tay F. C., Lam D. H., Sandanaraj E., Tang C., Ang B. T., and Wang S. (2012) Attenuated adenosine-to-inosine editing of microRNA-376a* promotes invasiveness of glioblastoma cells. J. Clin. Investig. 122, 4059–4076 10.1172/JCI62925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen M., and Manley J. L. (2009) Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat. Rev. Mol. Cell Biol. 10, 741–754 10.1038/nrm2777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Salton M., and Misteli T. (2016) Small molecule modulators of pre-mRNA splicing in cancer therapy. Trends Mol. Med. 22, 28–37 10.1016/j.molmed.2015.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xu Q., and Lee C. (2003) Discovery of novel splice forms and functional analysis of cancer-specific alternative splicing in human expressed sequences. Nucleic Acids Res. 31, 5635–5643 10.1093/nar/gkg786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Salton M., Kasprzak W. K., Voss T., Shapiro B. A., Poulikakos P. I., and Misteli T. (2015) Inhibition of vemurafenib-resistant melanoma by interference with pre-mRNA splicing. Nat. Commun. 6, 7103 10.1038/ncomms8103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gui Y., Guo G., Huang Y., Hu X., Tang A., Gao S., Wu R., Chen C., Li X., Zhou L., He M., Li Z., Sun X., Jia W., Chen J., et al. (2011) Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat. Genet. 43, 875–878 10.1038/ng.907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Guo G., Gui Y., Gao S., Tang A., Hu X., Huang Y., Jia W., Li Z., He M., Sun L., Song P., Sun X., Zhao X., Yang S., Liang C., et al. (2011) Frequent mutations of genes encoding ubiquitin-mediated proteolysis pathway components in clear cell renal cell carcinoma. Nat. Genet. 44, 17–19 [DOI] [PubMed] [Google Scholar]
- 22. Anantharaman A., Tripathi V., Khan A., Yoon J. H., Singh D. K., Gholamalamdari O., Guang S., Ohlson J., Wahlstedt H., Öhman M., Jantsch M. F., Conrad N. K., Ma J., Gorospe M., Prasanth S. G., and Prasanth K. V. (2017) ADAR2 regulates RNA stability by modifying access of decay-promoting RNA-binding proteins. Nucleic Acids Res. 45, 4189–4201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nakano M., Fukami T., Gotoh S., and Nakajima M. (2017) A-to-I RNA editing up-regulates human dihydrofolate reductase in breast cancer. J. Biol. Chem. 292, 4873–4884 10.1074/jbc.M117.775684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rice G. I., Kasher P. R., Forte G. M., Mannion N. M., Greenwood S. M., Szynkiewicz M., Dickerson J. E., Bhaskar S. S., Zampini M., Briggs T. A., Jenkinson E. M., Bacino C. A., Battini R., Bertini E., Brogan P. A., et al. (2012) Mutations in ADAR1 cause Aicardi-Goutieres syndrome associated with a type I interferon signature. Nat. Genet. 44, 1243–1248 10.1038/ng.2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang Z., and Carmichael G. G. (2001) The fate of dsRNA in the nucleus: a p54(nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell 106, 465–475 10.1016/S0092-8674(01)00466-4 [DOI] [PubMed] [Google Scholar]
- 26. Yang C. C., Chen Y. T., Chang Y. F., Liu H., Kuo Y. P., Shih C. T., Liao W. C., Chen H. W., Tsai W. S., and Tan B. C. (2017) ADAR1-mediated 3′ UTR editing and expression control of antiapoptosis genes fine-tunes cellular apoptosis response. Cell Death Dis. 8, e2833 10.1038/cddis.2017.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nasim M. T., and Eperon I. C. (2006) A double-reporter splicing assay for determining splicing efficiency in mammalian cells. Nat. Protocols 1, 1022–1028 10.1038/nprot.2006.148 [DOI] [PubMed] [Google Scholar]
- 28. Kim D. D., Kim T. T., Walsh T., Kobayashi Y., Matise T. C., Buyske S., and Gabriel A. (2004) Widespread RNA editing of embedded Alu elements in the human transcriptome. Genome Res. 14, 1719–1725 10.1101/gr.2855504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Krainer A. R., Conway G. C., and Kozak D. (1990) Purification and characterization of pre-mRNA splicing factor SF2 from HeLa cells. Genes Dev. 4, 1158–1171 10.1101/gad.4.7.1158 [DOI] [PubMed] [Google Scholar]
- 30. Long J. C., and Caceres J. F. (2009) The SR protein family of splicing factors: master regulators of gene expression. Biochem. J. 417, 15–27 10.1042/BJ20081501 [DOI] [PubMed] [Google Scholar]
- 31. Wang T., Cui Y., Jin J., Guo J., Wang G., Yin X., He Q. Y., and Zhang G. (2013) Translating mRNAs strongly correlate to proteins in a multivariate manner and their translation ratios are phenotype specific. Nucleic Acids Res. 41, 4743–4754 10.1093/nar/gkt178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Anczukow O., Rosenberg A. Z., Akerman M., Das S., Zhan L., Karni R., Muthuswamy S. K., and Krainer A. R. (2012) The splicing factor SRSF1 regulates apoptosis and proliferation to promote mammary epithelial cell transformation. Nat. Struct. Mol. Biol. 19, 220–228 10.1038/nsmb.2207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Musgrove E. A., Caldon C. E., Barraclough J., Stone A., and Sutherland R. L. (2011) Cyclin D as a therapeutic target in cancer. Nat. Rev. Cancer 11, 558–572 10.1038/nrc3090 [DOI] [PubMed] [Google Scholar]
- 34. Peng Z., Cheng Y., Tan B. C., Kang L., Tian Z., Zhu Y., Zhang W., Liang Y., Hu X., Tan X., Guo J., Dong Z., Liang Y., Bao L., and Wang J. (2012) Comprehensive analysis of RNA-Seq data reveals extensive RNA editing in a human transcriptome. Nat. Biotechnol. 30, 253–260 10.1038/nbt.2122 [DOI] [PubMed] [Google Scholar]
- 35. Li J. B., Levanon E. Y., Yoon J. K., Aach J., Xie B., Leproust E., Zhang K., Gao Y., and Church G. M. (2009) Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science 324, 1210–1213 10.1126/science.1170995 [DOI] [PubMed] [Google Scholar]
- 36. Bazak L., Haviv A., Barak M., Jacob-Hirsch J., Deng P., Zhang R., Isaacs F. J., Rechavi G., Li J. B., Eisenberg E., and Levanon E. Y. (2014) A-to-I RNA editing occurs at over a hundred million genomic sites, located in a majority of human genes. Genome Res. 24, 365–376 10.1101/gr.164749.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Alsafadi S., Houy A., Battistella A., Popova T., Wassef M., Henry E., Tirode F., Constantinou A., Piperno-Neumann S., Roman-Roman S., Dutertre M., and Stern M. H. (2016) Cancer-associated SF3B1 mutations affect alternative splicing by promoting alternative branchpoint usage. Nat. Commun. 7, 10615 10.1038/ncomms10615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang L., Lawrence M. S., Wan Y., Stojanov P., Sougnez C., Stevenson K., Werner L., Sivachenko A., DeLuca D. S., Zhang L., Zhang W., Vartanov A. R., Fernandes S. M., Goldstein N. R., Folco E. G., et al. (2011) SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N. Engl. J. Med. 365, 2497–2506 10.1056/NEJMoa1109016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang L., Brooks A. N., Fan J., Wan Y., Gambe R., Li S., Hergert S., Yin S., Freeman S. S., Levin J. Z., Fan L., Seiler M., Buonamici S., Smith P. G., Chau K. F., et al. (2016) Transcriptomic characterization of SF3B1 mutation reveals its pleiotropic effects in chronic lymphocytic leukemia. Cancer Cell 30, 750–763 10.1016/j.ccell.2016.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Karni R., de Stanchina E., Lowe S. W., Sinha R., Mu D., and Krainer A. R. (2007) The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat. Struct. Mol. Biol. 14, 185–193 10.1038/nsmb1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Das S., Anczuków O., Akerman M., and Krainer A. R. (2012) Oncogenic splicing factor SRSF1 is a critical transcriptional target of MYC. Cell Rep. 1, 110–117 10.1016/j.celrep.2011.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Oberdoerffer S., Moita L. F., Neems D., Freitas R. P., Hacohen N., and Rao A. (2008) Regulation of CD45 alternative splicing by heterogeneous ribonucleoprotein, hnRNPLL. Science 321, 686–691 10.1126/science.1157610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pages F., Berger A., Camus M., Sanchez-Cabo F., Costes A., Molidor R., Mlecnik B., Kirilovsky A., Nilsson M., Damotte D., Meatchi T., Bruneval P., Cugnenc P. H., Trajanoski Z., Fridman W. H., and Galon J. (2005) Effector memory T cells, early metastasis, and survival in colorectal cancer. N. Engl. J. Med. 353, 2654–2666 10.1056/NEJMoa051424 [DOI] [PubMed] [Google Scholar]
- 44. Johnsson P., Ackley A., Vidarsdottir L., Lui W. O., Corcoran M., Grander D., and Morris K. V. (2013) A pseudogene long-noncoding-RNA network regulates PTEN transcription and translation in human cells. Nat. Struct. Mol. Biol. 20, 440–446 10.1038/nsmb.2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lebedeva S., Jens M., Theil K., Schwanhäusser B., Selbach M., Landthaler M., and Rajewsky N. (2011) Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol. Cell 43, 340–352 10.1016/j.molcel.2011.06.008 [DOI] [PubMed] [Google Scholar]
- 46. Bustin S. A., Benes V., Garson J. A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M. W., Shipley G. L., Vandesompele J., and Wittwer C. T. (2009) The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622 10.1373/clinchem.2008.112797 [DOI] [PubMed] [Google Scholar]
- 47. Hsieh C. L., Liu H., Huang Y., Kang L., Chen H. W., Chen Y. T., Wee Y. R., Chen S. J., and Tan B. C. (2014) ADAR1 deaminase contributes to scheduled skeletal myogenesis progression via stage-specific functions. Cell Death Differ. 21, 707–719 10.1038/cdd.2013.197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T. R. (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Love M. I., Huber W., and Anders S. (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Harrow J., Frankish A., Gonzalez J. M., Tapanari E., Diekhans M., Kokocinski F., Aken B. L., Barrell D., Zadissa A., Searle S., Barnes I., Bignell A., Boychenko V., Hunt T., Kay M., et al. (2012) GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 22, 1760–1774 10.1101/gr.135350.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dölken L., Ruzsics Z., Rädle B., Friedel C. C., Zimmer R., Mages J., Hoffmann R., Dickinson P., Forster T., Ghazal P., and Koszinowski U. H. (2008) High-resolution gene expression profiling for simultaneous kinetic parameter analysis of RNA synthesis and decay. Rna 14, 1959–1972 10.1261/rna.1136108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yang C. C., Liu H., Chen S. L., Wang T. H., Hsieh C. L., Huang Y., Chen S. J., Chen H. C., Yung B. Y., and Chin-Ming Tan B. (2012) Epigenetic silencing of myogenic gene program by Myb-binding protein 1a suppresses myogenesis. EMBO J. 31, 1739–1751 10.1038/emboj.2012.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.