Abstract
Chronic inflammation is associated with multiple human disorders, such as rheumatoid arthritis, metabolic diseases, and neurodegenerative diseases. Therefore, alleviation of inflammation induced by environmental stimuli is important for disease prevention or treatment. Cereblon (CRBN) functions as a substrate receptor of the cullin-4 RING E3 ligase to mediate protein ubiquitination and degradation. Although it has been reported that CRBN reduces the inflammatory response through its nonenzymatic function, its role as a substrate receptor of the E3 ligase is not explored in mediating this process. Here we used a quantitative proteomics approach to find that the major component of the activator protein 1 (AP-1) complex, c-Jun, is significantly down-regulated upon CRBN expression. Biochemical approaches further discover that CRBN interacts and partially colocalizes with c-Jun and promotes the formation of Lys48-linked polyubiquitin chains on c-Jun, enhancing c-Jun degradation. We further reveal that CRBN attenuates the transcriptional activity of the AP-1 complex and reduces the mRNA expression and protein level of several pro-inflammatory cytokines. Moreover, flow cytometry analyses show that CRBN attenuates lipopolysaccharide-induced apoptosis in differentiated THP-1 cells. Through genetic manipulation and pharmacological inhibition, we uncover a new molecular mechanism by which CRBN regulates the inflammatory response and apoptosis induced by lipopolysaccharide. Our work and previous studies demonstrate that CRBN suppresses the inflammatory response by promoting or inhibiting the ubiquitination of two key molecules at different levels of the inflammatory cascade through its enzymatic function as a substrate receptor and its nonenzymatic function as a protein binding partner.
Keywords: inflammation, AP1 transcription factor (AP-1), ubiquitylation (ubiquitination), c-Jun transcription factor, protein degradation, apoptosis, cereblon, inflammatory response
Introduction
Inflammation is an adaptive response triggered by exogenous stimuli, such as UV light, environmental toxins, lipopolysaccharide (LPS),3 reactive oxygen species, and endogenous inducers resulting from stressed, damaged, and malfunctioning tissues (1). It has been reported that chronic inflammation is associated with many health problems, such as rheumatoid arthritis (2), inflammatory bowel disease (3), atherosclerosis (4), neurodegenerative diseases (5, 6), metabolic diseases (7), and acute pancreatitis (8). Two critical inflammatory signaling pathways frequently activated in the inflammatory response are the NF-κB and activator protein 1 (AP-1) pathways (9–11). Some triggers, such as ligands of Toll-like receptors (12), tumor necrosis factor (TNF) receptors (13), and interleukin-1 receptors (14), can activate both the NF-κB and AP-1 signaling pathways through regulation of their common upstream mediators (15). Alleviation of the inflammatory response by interfering with these mediators is a potential therapeutic strategy for the treatment of inflammation-associated diseases, including cancer (16). New regulators for the signaling pathways involved in the inflammatory response are still being uncovered, which may lead to the discovery of novel molecular targets for intervention with the inflammatory response.
NF-κB and AP-1 are the two most extensively studied transcription factor complexes that respond to inflammatory stimuli. Both the NF-κB and the AP-1 signaling pathways can be activated by LPS and TNFα through TNF receptor–associated factor 6 (TRAF6) and transforming growth factor β–activated kinase 1 (TAK1) (17, 18). In the NF-κB pathway, inflammatory stimuli trigger phosphorylation and promote the subsequent ubiquitination of inhibitor of NF-κB (IκB) or the NF-κB p100 subunit for their degradation, thus liberating and translocating the NF-κB complex to the nucleus (19). In the AP-1 pathway, the basic leucine zipper family protein c-Jun is first phosphorylated by c-Jun N-terminal kinase (JNK) or mitogen-activated protein kinase/extracellular signal–regulated kinase and then forms a transcriptionally active dimer with itself or other proteins, such as c-Fos (20–22). Activation of these two pathways enhances the expression of inflammatory cytokines, including inducible nitric oxide synthase (iNOS), cyclooxygenase-2 (COX-2 or prostaglandin G/H synthase 2), interleukin-1β (IL-1β), and IL-6 (23). AP-1 transcriptional activity participates in the regulation of cell proliferation, survival, and apoptosis (24, 25). Apoptosis is a programmed cell death process in which caspases are activated to cleave their downstream targets, leading to the DNA fragmentation and the formation of apoptotic bodies for further lysis. Both pro-apoptotic genes, such as caspases, and anti-apoptotic genes, such as BCL2 and survivin, are mediated during this process, and their overall effect determines the cell fate (25). Although LPS is an inflammatory stimulus, it can induce apoptosis of macrophages (26), and this process can be mediated by autocrine production of TNFα (26) and activation of the JNK/AP-1 signaling pathway (27, 28). Therefore, modulating the AP-1 signaling pathway is a promising therapeutic strategy for the treatment of inflammation-associated diseases (29).
Many studies have demonstrated that CRBN is a substrate receptor of a cullin-4 RING E3 ligase (CRL4), which consists of damage-specific DNA-binding protein 1 (DDB1), cullin-4A/B, and the RING-box protein ROC1 (30–34). This E3 ligase, CRL4-CRBN, can mediate the ubiquitination and degradation of endogenous substrates, such as the homeobox protein MEIS2 (32), calcium-activated potassium channel subunit α-1 (34), glutamine synthetase (35, 36), adenosine monophosphate–activated protein kinase subunit α1 (37), chloride channel protein 1 (38), argonaute 2 (39), and amyloid precursor protein (40). Immunomodulatory drugs (IMiDs), including thalidomide and its structural analogs lenalidomide and pomalidomide, are a class of compounds that can be used to alleviate the inflammatory response. However, how CRBN regulates inflammation has only begun to unfold (41). It has been reported that CRBN binds to the ubiquitination domain of TRAF6 and attenuates the ubiquitination of TRAF6 and TAK1-binding protein 2 (TAB2), leading to suppression of NF-κB activation by negatively regulating the Toll-like receptor 4 (TLR4) signaling pathway (41). This discovery revealed the important nonenzymatic function of CRBN in the regulation of inflammation. Indeed, it has also been found that the nonenzymatic function of CRBN prevents the formation of protein aggregates caused by pathological mutations of huntingtin (Htt) and TAR DNA-binding protein 43 (TDP43) in cell lines and animal tissues (42).
Although it has been demonstrated that CRBN regulates LPS-induced inflammation through its nonenzymatic function, whether the enzymatic activity of the CRBN-associated E3 ligase is also involved in mediating inflammation has not been explored. Because the CRL4 E3 ligases mainly target proteins for ubiquitination and proteasomal degradation, we hypothesize that CRBN may also exhibit its function as the substrate receptor of the CRL4-CRBN E3 ligase in response to inflammatory stimuli. In this work, we identify c-Jun as a significantly down-regulated protein upon CRBN expression using a quantitative proteomics approach. Biochemical experiments reveal that CRBN interacts with c-Jun, enhances its Lys48-linked polyubiquitination, promotes its degradation, and thus reduces its protein level. We also find that CRBN expression attenuates the transcriptional activity of the AP-1 complex and reduces the gene expression and protein level of four pro-inflammatory cytokines upon LPS stimulation. We further explore the molecular mechanism by which CRBN regulates the inflammatory response using siRNA and pharmacological inhibition. Flow cytometry analyses further discover that CRBN attenuates LPS-induced apoptosis. Taken together, this work reveals a new molecular mechanism by which CRBN regulates the inflammatory response and apoptosis upon LPS stimulation.
Results
CRBN down-regulates the c-Jun protein level
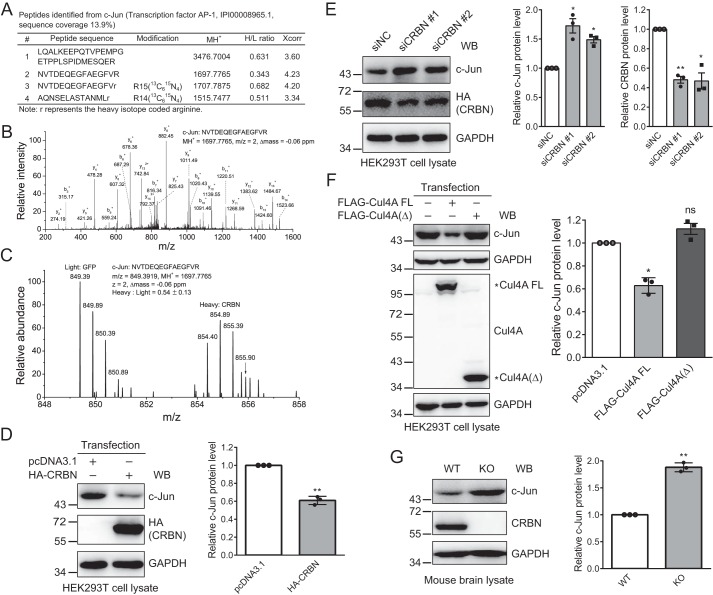
To explore the potential functions of CRBN and its associated E3 ligase CRL4-CRBN, we used stable isotope labeling of amino acids by cell culture to identify proteins that are down-regulated by expressing CRBN in HEK293T cells. Among the identified peptides, we detected three tryptic peptides derived from c-Jun (Fig. 1A). The MS/MS spectra of one peptide were detected in both the light and heavy arginine-labeled samples. The average relative ratio of the MS signals of these peptides from the CRBN-expressed sample to the mock-expressed sample was about 0.54. A representative MS/MS spectrum with the annotated b and y ions indicated confident identification of the peptide and protein (Fig. 1B). A representative MS spectrum provided the relative intensity of the light and heavy arginine-labeled peptides derived from two samples (Fig. 1C). Two biological replicates resulted in similar relative ratios of the MS signals for the c-Jun tryptic peptides derived from two samples.
Figure 1.
Quantitative proteomics and biochemical analysis reveal c-Jun as a protein down-regulated by CRBN. A, c-Jun tryptic peptides identified by the quantitative proteomics analysis. The peptide sequence, modification, MH+, heavy to light (H/L) ratio, and Sequest cross correlation score Xcorr were included in the peptide list. B, MS/MS spectrum of a representative tryptic peptide identified from c-Jun. The peptide sequence, MH+, m/z, and Δmass were depicted in the annotated MS/MS spectrum. C, representative MS spectrum of a tryptic peptide used for quantification of the c-Jun protein level. The peptide sequence, MH+, m/z, Δmass, and heavy to light ratio were depicted. The m/z value for the isotope peaks from the light and heavy samples were labeled. D, CRBN overexpression reduces the c-Jun protein level. HEK293T cells were transfected with the pcDNA3.1 or HA-CRBN plasmid using PEI transfection reagent for 48 h. Cell lysates were analyzed by Western blotting (WB) with the indicated antibodies. The means and standard deviations were depicted, and the p value was calculated using Student's t test with data from three biological replicates. **, p < 0.01. E, siRNA knockdown CRBN increases the c-Jun protein level. HEK293T cells were transfected with siNC and siCRBN using Lipofectamine 2000 for 48 h. Samples and data were analyzed as described in D. *, p < 0.05; **, p < 0.01. F, Cul4A FL, but not the deletion mutant Cul4A(Δ), decreases the c-Jun protein level. HEK293T cells were transfected with Cul4A FL or the Cul4A(Δ) plasmid using PEI transfection reagent for 48 h. Samples and data were analyzed as described in D. *, p < 0.05; ns, not significant. G, the c-Jun protein level is increased in brain tissue of Crbn knockout mice. Brain lysates obtained from WT and Crbn knockout mice were immunoblotted with the indicated antibodies. Experiments were carried out with three biological replicates, and quantification was performed as described in D. **, p < 0.01.
We further used biochemical approaches to validate the effect of CRBN on c-Jun protein level in cells and animal tissue. Immunoblotting of cell lysates obtained from HEK293T cells transfected with the pcDNA3.1 or HA-CRBN plasmid showed that CRBN expression significantly reduced the c-Jun protein level (Fig. 1D) and that this reduction was dose-dependent (Fig. S1). Similarly, siRNA knockdown of CRBN in HEK293T cells increased the c-Jun protein level (Fig. 1E). To further examine whether this effect was regulated by the CRL4 E3 ligase, we transfected full-length (FL) Cul4A and a deletion mutant Cul4A(Δ) (43), which did not form CRL4 E3 ligase, into HEK293T cells. Immunoblotting of cell lysates showed that Cul4A FL, but not Cul4A(Δ), attenuated the c-Jun protein level (Fig. 1F). Furthermore, the c-Jun protein level was significantly increased (Fig. 1G) in brain lysates of Crbn knockout mice, further supporting regulation of the c-Jun protein level by CRBN under physiological conditions. Taken together, these data indicate that CRBN reduces c-Jun protein level, suggesting that c-Jun might be a substrate of the CRL4-CRBN E3 ligase.
CRBN interacts and colocalizes with c-Jun
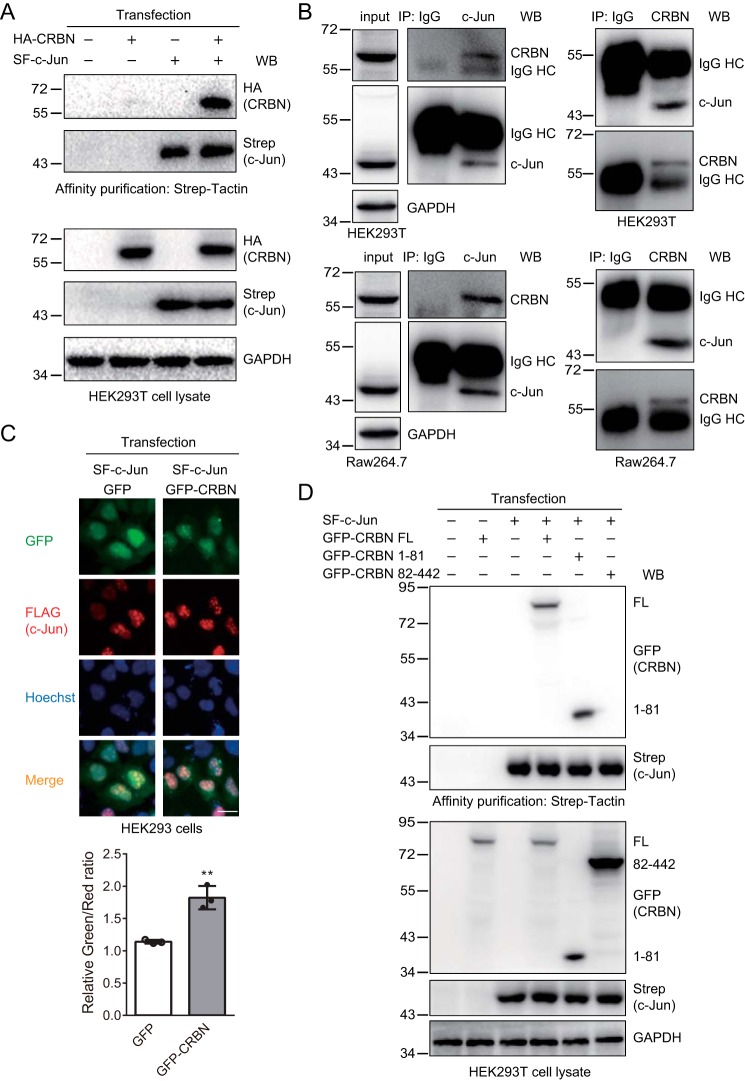
Because CRBN reduced the c-Jun protein level, we attempted to determine whether c-Jun was a substrate of the CRL4-CRBN E3 ligase. To do so, we first tested whether CRBN interacted with c-Jun. We transfected the pcDNA3.1, HA-CRBN, Strep-FLAG (SF)–c-Jun, or SF–c-Jun and HA-CRBN plasmids into HEK293T cells and purified SF–c-Jun with Strep-Tactin–agarose beads. Immunoblotting of cell lysates and affinity-purified samples indicated that c-Jun interacted with CRBN (Fig. 2A). To examine whether the endogenous proteins also interacted with each other, we immunoprecipitated either CRBN or c-Jun from cell lysates obtained from HEK293T cells or LPS-stimulated murine macrophage Raw264.7 cells. Here, Raw264.7 cells were treated with LPS to enhance c-Jun expression. The immunoblotting results clearly demonstrated that endogenous CRBN and c-Jun interacted with each other (Fig. 2B). To further support this result, we expressed SF–c-Jun along with either GFP or GFP-CRBN in HEK293 cells and immunostained the cells for FLAG (c-Jun). The resulting fluorescence images demonstrated that c-Jun colocalized with CRBN (Fig. 2C), which is consistent with their interaction obtained from affinity purification and immunoblotting analyses. The immunostaining also revealed that c-Jun was mainly localized in the nucleus, which is in agreement with a previous study (44) and the fact that c-Jun is a major component of the AP-1 transcription factor. To determine which region in CRBN interacts with c-Jun, we further constructed a series of C-terminal truncation mutants and transfected them into HEK293T cells. The affinity purification and immunoblotting experiments showed that the CRBN N-terminal region (1–81 amino acids) interacted with c-Jun (Fig. S2), whereas the mutant lacking the 1–81 amino acids, the 82–442 mutant, could not interact with c-Jun (Fig. 2D). Taken together, these results clearly demonstrate that CRBN interacts with c-Jun.
Figure 2.
c-Jun interacts and colocalizes with CRBN. A, c-Jun interacts with CRBN. HEK293T cells were transiently transfected with the pcDNA3.1, HA-CRBN, Strep-SF–c-Jun, or SF–c-Jun and HA-CRBN plasmids using PEI transfection reagent. 48 h post-transfection, cells were treated with MG132 (10 μm) for 12 h and lysed. SF–c-Jun was purified with Strep-Tactin–agarose beads. Cell lysates and purified samples were immunoblotted for Strep (c-Jun), HA (CRBN), and/or GAPDH. MG132 was added to prevent degradation of c-Jun upon CRBN expression, which resulted in approximately equal c-Jun protein levels in the cell lysates. The second lane served as a control for possible nonspecific binding of proteins to Strep-Tactin–agarose beads. WB, Western blot. B, endogenous CRBN and c-Jun interact with each other. HEK293T cells or Raw264.7 cells (treated with 200 ng/ml LPS for 12 h) were lysed and immunoprecipitated (IP) with IgG, CRBN, or c-Jun antibodies. The cell lysates and immunoprecipitates were blotted with the indicated antibodies. HC, heavy chain. C, c-Jun colocalizes with CRBN. HEK293 cells were transiently transfected with SF–c-Jun and GFP or the GFP-CRBN plasmid using Lipofectamine 3000 transfection reagent for 24 h. Cells were immunostained with FLAG (c-Jun) and Alexa Fluor 594 donkey anti-mouse IgG (red fluorescence) for immunofluorescence measurements. Hoechst was used to stain the nuclei. Scale bar = 20 μm. The quantification is presented with the relative ratio of green fluorescence (CRBN) signal colocalized with the red fluorescence (c-Jun) signal. The data were normalized with the green fluorescence (CRBN) signal that was not colocalized with the red fluorescence (c-Jun) signal. Three fields were counted for each experimental condition, and each field contained at least 10 cells. Quantitative data from three independent experiments were provided. Student's t test; **, p < 0.01. D, c-Jun interacts with the N-terminal region (1–81 amino acids) of CRBN. HEK293T cells were transiently transfected with SF–c-Jun and the pcDNA3.1, GFP-CRBN FL, 1–81, and 82–442 plasmids for 48 h. SF–c-Jun was purified with Strep-Tactin–agarose beads from cell lysates. Cell lysates and purified samples were immunoblotted for Strep (c-Jun), GFP (CRBN), and/or GAPDH.
Proteasomal inhibition attenuates CRBN-mediated c-Jun degradation
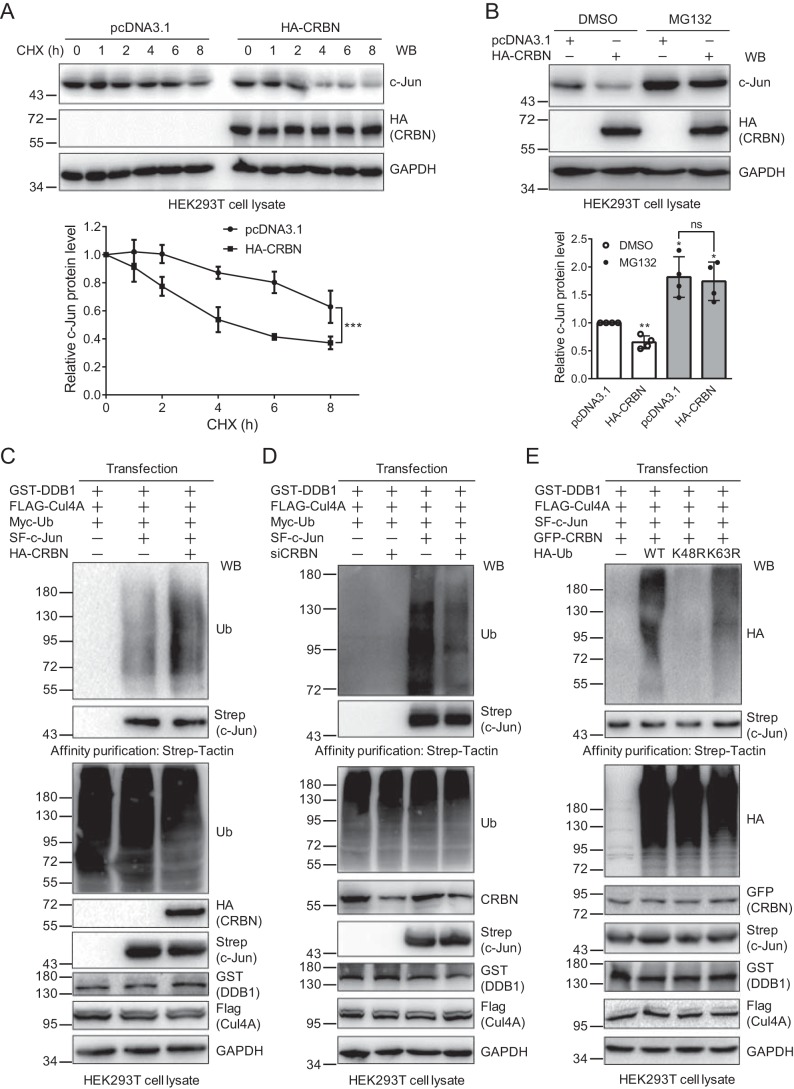
The protein level can be regulated both at the transcriptional level and at the posttranslational level. To determine which process affects the c-Jun protein level, we first performed a qPCR experiment for c-Jun upon expressing the control or CRBN plasmid in HEK293T cells. The result indicated that CRBN did not affect the c-Jun transcription level (Fig. S3), suggesting that this regulation might occur at the posttranslational level. We then used cycloheximide (CHX) to inhibit protein synthesis and immunoblotted the cell lysates obtained at different time points after CHX treatment to examine the regulation of c-Jun degradation by CRBN. The immunoblotting images showed that CRBN could accelerate c-Jun degradation, resulting in a reduction of its half-life from more than 8 h to about 5 h (Fig. 3A).
Figure 3.
CRBN mediates c-Jun degradation through the ubiquitin–proteasome system. A, CRBN promotes c-Jun degradation. HEK293T cells were transfected with the pcDNA3.1 or HA-CRBN plasmid and split into 6-well plates. 48 h post-transfection, cells were treated with CHX (200 μg/ml) for the indicated times. Cell lysates were immunoblotted with the indicated antibodies. Quantification (mean ± S.D.) was obtained from three biological replicates, and p values were calculated using two-way analysis of variance. ***, p < 0.001. WB, Western blot. B, proteasomal inhibition prevents CRBN-mediated c-Jun degradation. HEK293T cells were transfected with the pcDNA3.1 or HA-CRBN plasmid for 48 h and then treated with DMSO or MG132 (20 μm) for 8 h. Cell lysates were immunoblotted with the indicated antibodies. Experiments were repeated four times for quantification, and Student's t test was used to calculate the p values. *, p < 0.05; **, p < 0.01 compared with the first lane; ns, not significant. C, CRBN expression promotes c-Jun ubiquitination. HEK293T cells were transfected with the indicated plasmids for 48 h and then treated with MG132 (20 μm) for 8 h. c-Jun was purified with Strep-Tactin–agarose beads. The cell lysates and purified samples were immunoblotted with the indicated antibodies. D, CRBN knockdown reduces c-Jun ubiquitination. HEK293T cells were transfected with the indicated siRNAs and plasmids for 48 h using Lipofectamine 2000 transfection reagent and then treated with MG132 (20 μm) for 8 h. Samples were prepared as described in C, and the cell lysates and purified samples were immunoblotted with the indicated antibodies. E, CRBN promotes the formation of Lys48-linked polyubiquitin chains on c-Jun. HEK293T cells were transfected with the indicated plasmids for 48 h. Cells and samples were prepared and immunoblotted as described in C.
Because CRBN is a substrate receptor of the CRL4 E3 ligase, which recognizes specific targets for ubiquitination and subsequent degradation by the ubiquitin–proteasome system (UPS), we speculated that the regulation of c-Jun protein by CRBN might be caused by the same mechanism. We further transfected the pcDNA3.1 or HA-CRBN plasmid into HEK293T cells and detected the c-Jun protein level in the presence or absence of a proteasome inhibitor, MG132. The immunoblotting and statistical results demonstrate that MG132 prevents c-Jun from CRBN-mediated degradation (Fig. 3B), suggesting that the reduction of c-Jun protein by CRBN is most probably subjected to the UPS.
CRBN promotes the formation of Lys48-linked polyubiquitin chains on c-Jun
To further determine whether CRBN affects the ubiquitination of c-Jun, we cotransfected two plasmids expressing the CRL4 E3 ligase components glutathione S-transferase–DDB1 and FLAG-Cul4A along with the Myc-Ub, SF–c-Jun, and/or HA-CRBN plasmids into HEK293T cells. At 48 h post-transfection, cells were treated with MG132, and SF–c-Jun was affinity-purified. Immunoblotting of cell lysates and purified samples with an anti-ubiquitin antibody revealed that CRBN could markedly enhance c-Jun ubiquitination (Fig. 3C). However, the 82–442 mutant did not increase c-Jun ubiquitination (Fig. S4), which is consistent with the evidence showing that it cannot interact with c-Jun. To further validate this result, we knocked down CRBN using siRNA along with the expression of CRL4 E3 ligase components and ubiquitin. CRBN knockdown significantly decreased c-Jun ubiquitination (Fig. 3D), further supporting the fact that CRBN mediates c-Jun ubiquitination. Given that CRBN promotes c-Jun ubiquitination and degradation and that Lys48-linked polyubiquitin chains are the major type of ubiquitination targeted for proteasomal degradation, we thought that CRBN might promote the formation of the Lys48-linked polyubiquitin chains on c-Jun. To justify this hypothesis, we transfected WT ubiquitin and the K48R or K63R mutant, which cannot form Lys48- or Lys63-linked polyubiquitin chains, respectively, into HEK293T cells in the presence of CRBN, DDB1, and Cul4A. The immunoblotting of affinity-purified c-Jun showed that c-Jun ubiquitination almost completely disappeared in cells expressing the K48R mutant (Fig. 3E), whereas the K63R mutant did not affect c-Jun ubiquitination, suggesting that the ubiquitination formed on c-Jun by CRBN was mainly Lys48-linked polyubiquitin chains. These data further demonstrate that CRBN-mediated c-Jun ubiquitination leads to its degradation.
CRBN attenuates AP-1 transcriptional activity and reduces iNOS and COX-2 protein levels in LPS-stimulated Raw264.7 cells
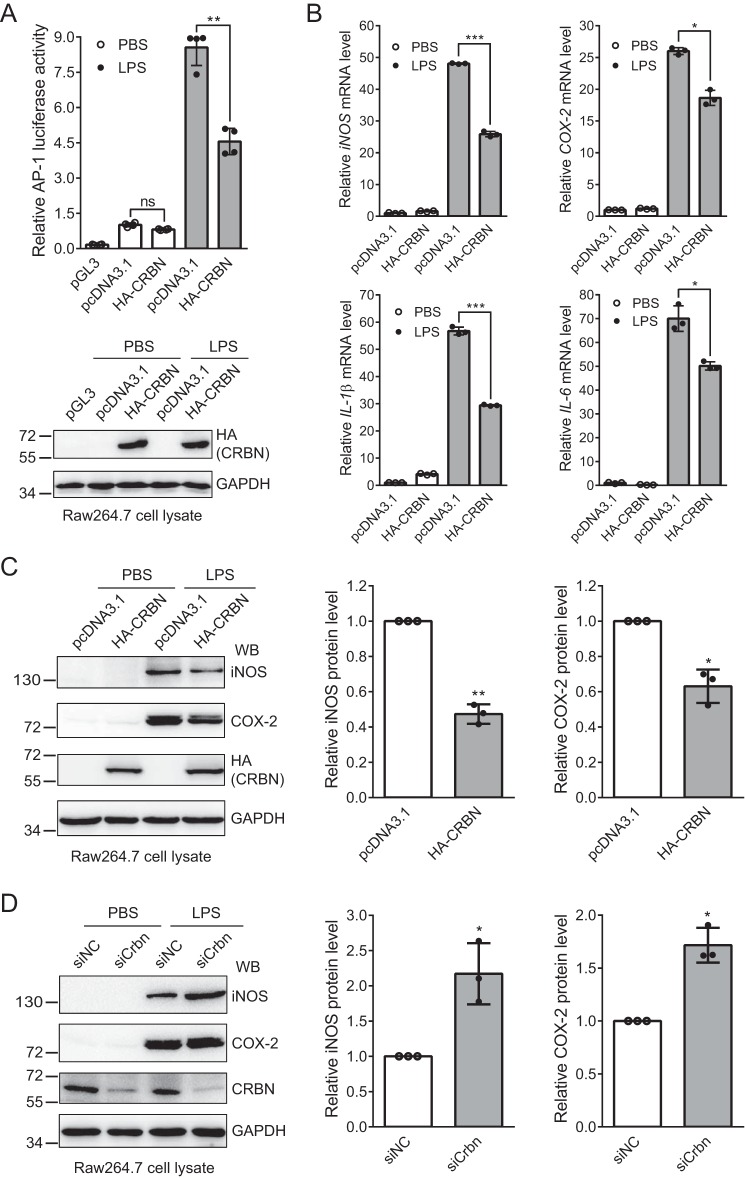
c-Jun forms the transcription factor AP-1 complex with its interacting partner, such as c-Jun itself or c-Fos (20–22). Activation of the AP-1 complex can be modulated by regulating the expression or activation of c-Jun because c-Jun is the major component of this complex (45). Our above experiments demonstrated that CRBN interacted with c-Jun and promoted its ubiquitination and proteasomal degradation. Therefore, we asked whether CRBN could regulate the inflammatory response. First, we carried out a luciferase reporter assay to test whether CRBN regulates AP-1 transcriptional activity. We transfected the pcDNA3.1 or HA-CRBN plasmid along with the AP-1 firefly luciferase reporter plasmid and the Renilla luciferase control reporter vector into Raw264.7 cells. 36 h post-transfection, cells were treated with or without LPS for 12 h, and AP-1 luciferase activity was measured with the Dual-Luciferase reporter assay system. During data processing, the firefly luciferase signal was first divided by the Renilla luciferase signal and further normalized to the ratio from the pcDNA3.1 control sample. The result indicates that CRBN significantly reduces AP-1 luciferase activity in LPS-stimulated cells (Fig. 4A).
Figure 4.
CRBN regulates the LPS-induced inflammatory response. A, CRBN reduces AP-1 luciferase activity in Raw264.7 cells upon LPS stimulation. The pcDNA3.1 or HA-CRBN plasmid, along with the AP-1 firefly luciferase reporter plasmid and Renilla luciferase control reporter vector, was transfected into Raw264.7 cells using Lipofectamine 3000 transfection reagent for 36 h. Cells were then treated with PBS or LPS (200 ng/ml) for 12 h. AP-1 luciferase activity was measured with the Dual-Luciferase reporter assay system. Mean ± S.D. values were obtained from four technical replicates, and p values were calculated using Student's t test. Two biological replicates were performed, and similar results were obtained. **, p < 0.01; ns, not significant. Western blotting was used to confirm protein expression. B, CRBN attenuates the mRNA level of four inflammatory cytokines in LPS-induced Raw264.7 cells. Raw264.7 cells were transfected with the pcDNA3.1 or HA-CRBN plasmid using Lipofectamine 3000 transfection reagent for 36 h and then treated with PBS or LPS (200 ng/ml) for 12 h. Total RNAs were extracted 48 h after transfection, and the first-strand cDNA was synthesized. The relative mRNA level of four inflammatory cytokines was measured by qPCR. Means ± S.D. were obtained from three technical replicates, and p values were calculated using Student's t test. The experiments were performed with three biological replicates, and similar results were obtained. *, p < 0.05; ***, p < 0.001. C, CRBN expression reduces iNOS and COX-2 protein levels upon LPS stimulation. The pcDNA3.1 or HA-CRBN plasmid was transiently transfected into Raw264.7 cells using Lipofectamine 3000 transfection reagent for 36 h, and cells were then treated with PBS (control) or LPS (200 ng/ml) for 12 h. Cell lysates were immunoblotted with the indicated antibodies. Means ± SD were obtained from three biological replicates, and p values were calculated using Student's t test for samples treated with LPS. *, p < 0.05; **, p < 0.01. Only LPS-treated samples were used for quantification because the levels of iNOS and COX-2 proteins in the PBS-treated samples were too low to be accurately quantified. WB, Western blot. D, Crbn knockdown increases iNOS and COX-2 protein levels upon LPS stimulation. Raw264.7 cells were transiently transfected with siNC (negative control) or siCRBN using Lipofectamine 3000 transfection reagent. Cells and samples were treated and analyzed as described in C. *, p < 0.05.
The transcription factor AP-1 complex activates gene expression of inflammatory cytokines. Because CRBN regulated the c-Jun protein level and AP-1 luciferase activity, we next measured the mRNA level of four inflammatory cytokines: COX-2, iNOS, IL-1β, and IL-6. Again, we transfected the pcDNA3.1 or HA-CRBN plasmid into Raw264.7 cells and treated the cells with PBS or LPS for 12 h, followed by qPCR analysis. The result clearly demonstrated that CRBN could reduce the mRNA level of all four tested pro-inflammatory cytokines (Fig. 4B). Immunoblotting of cell lysates showed that CRBN expression markedly reduced COX-2 and iNOS protein levels in LPS-induced cells (Fig. 4C), although these two proteins were not detected in the PBS-treated samples. Consistently, knockdown of CRBN clearly increased these two proteins in LPS-stimulated cells (Fig. 4D). Taken together, these results demonstrate that CRBN could regulate the downstream targets of the AP-1 signaling pathway.
CRBN attenuates the inflammatory response by down-regulating c-Jun
Our above experiments revealed that CRBN reduced AP-1 transcriptional activity under LPS stimulation. c-Jun is the major component of the AP-1 complex, and CRBN down-regulates c-Jun. Therefore, we thought that this reduction might be regulated by down-regulation of c-Jun. It has also been reported that CRBN can inhibit the inflammatory response by attenuating the activity of the transcription factor NF-κB (41). Because both NF-κB and AP-1 play key roles in the inflammatory response, and LPS can stimulate both NF-κB and AP-1 transcriptional activity (17), we sought to test whether CRBN also regulates the inflammatory response through c-Jun.
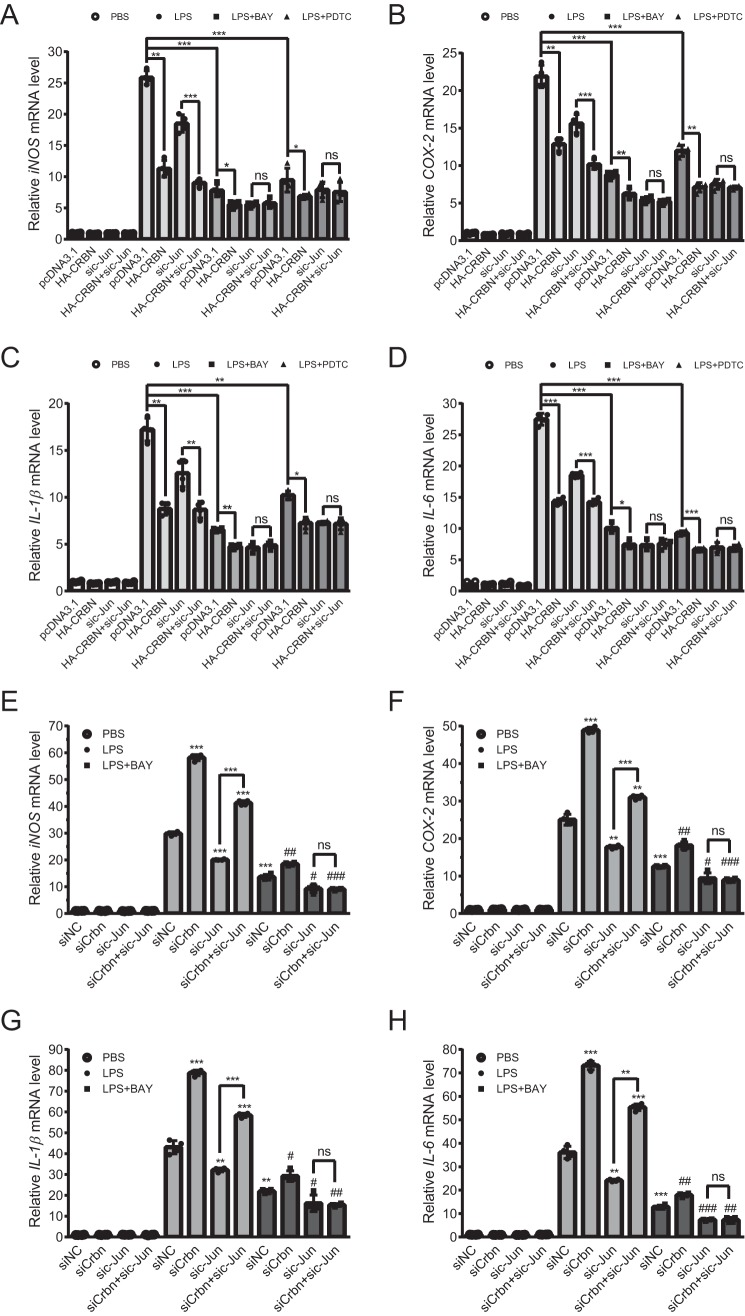
To explore this possibility, we transfected the pcDNA3.1 or HA-CRBN plasmid with either control siRNAs or c-Jun–specific siRNAs into Raw264.7 cells. Cells were then treated with DMSO or the NF-κB inhibitors BAY11-7082 (BAY) or ammonium pyrrolidine dithiocarbamate (PDTC) 1 h prior to PBS or LPS treatment. 12 h post-stimulation, half of the cells were used to verify transfection efficiency (Fig. S5A) and the other half was used for qPCR experiments (Fig. 5, A–D). Western blot images exhibited a reduction of the c-Jun protein level upon CRBN expression in Raw264.7 cells (Fig. S5A), in agreement with the data obtained from HEK293T cells (Fig. 1D).
Figure 5.
c-Jun knockdown eliminates the effect of CRBN expression and Crbn knockdown on the mRNA level of four inflammatory cytokines in the presence of NF-κB inhibitors. A–D, Raw264.7 cells were transfected with the pcDNA3.1 or HA-CRBN plasmid and siNC or sic-Jun using Lipofectamine 3000 transfection reagent for 36 h. Cells were then pretreated with DMSO or NF-κB inhibitors (15 μm BAY or 30 μm PDTC) 1 h prior to treatment with PBS or LPS (200 ng/ml, 12 h). Relative mRNA levels of cytokines were measured by qPCR. Western blotting was carried out to confirm the transfection (Fig. S5A). Means ± SD were obtained from four technical replicates, and p values were calculated using Student's t test. Two biological replicates were carried out, and similar results were obtained. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ns, not significant. A, iNOS. B, COX-2. C, IL-1β. D, IL-6. E–H, Raw264.7 cells were transfected with siNC or siCrbn with siNC or sic-Jun using Lipofectamine 3000 transfection reagent for 36 h. Cells were then pretreated with DMSO or the NF-κB inhibitor BAY (15 μm) for 1 h prior to treatment with PBS or LPS (200 ng/ml, 12 h). mRNA and data were analyzed as described in A–D. Knockdown efficiency was evaluated by Western blotting (Fig. S5B). Means ± SD were obtained from four technical replicates, and p values were calculated using Student's t test. Two biological replicates were carried out, and similar results were obtained. **, p < 0.01; ***, p < 0.001 compared with the fifth lane (transfected with siNC and treated with LPS). #, p < 0.05; ##, p < 0.01; ###, p < 0.001 compared with the ninth lane (transfected with siNC and treated with BAY and LPS). E, iNOS. F, COX-2. G, IL-1β. H, IL-6.
The qPCR results displayed several features. In the absence of LPS stimulation, the mRNA level for the tested pro-inflammatory cytokines (COX-2, iNOS, IL-1β, and IL-6) was almost not altered by CRBN expression or c-Jun knockdown. Upon LPS stimulation, the mRNA of all tested cytokines was significantly elevated. When CRBN was expressed, their mRNAs were markedly reduced, similar as observed in Fig. 4B. However, when c-Jun was knocked down, their mRNA level was significantly reduced, and CRBN expression could still down-regulate their mRNA level, albeit to a slightly lesser degree (Fig. 5, A–D, LPS-treated samples). This result suggested that CRBN could regulate cytokine expression and that part of this regulation was modulated in a c-Jun–independent manner.
Next we tested whether CRBN regulated cytokine expression via the c-Jun/AP-1 pathway. We used two NF-κB inhibitors to block the effect caused by activation of NF-κB because LPS can also activate inflammation through the NF-κB pathway (17, 18). Indeed, upon addition of these inhibitors, the mRNA level of the cytokines after LPS stimulation was remarkably reduced but still significantly higher than that from PBS-treated samples. CRBN expression further attenuated their mRNA level. Most importantly, c-Jun knockdown completely eliminated the effect of CRBN on the regulation of cytokine expression (Fig. 5, A–D). In addition, upon NF-κB inhibition, the mRNA level of cytokines in the CRBN-expressing sample was similar to that in the c-Jun knockdown sample, which was in agreement with the similar reduction of c-Jun protein level upon CRBN expression and c-Jun knockdown (Fig. S5A). c-Jun knockdown by siRNA completely abolished the effect of CRBN expression in the presence of NF-κB inhibitors. In addition, in the absence of these inhibitors, Crbn knockdown significantly increased the mRNA level of COX-2, iNOS, IL-1β, and IL-6 in Raw264.7 cells upon LPS stimulation both for the mock or c-Jun knockdown samples (Fig. 5, E–H, and Fig. S5B), indicating the presence of pathways other than the c-Jun/AP-1 pathway in CRBN-regulated inflammation. Pretreatment of cells with the NF-κB inhibitor BAY11-7082 reduced the inflammatory response. Further knockdown of c-Jun completely eliminated the effect of CRBN on the inflammatory response (Fig. 5, E–H). These results demonstrate that the modulation of cytokine expression by CRBN is, at least partially, regulated through c-Jun.
c-Jun reduces CRBN-mediated apoptosis induced by LPS
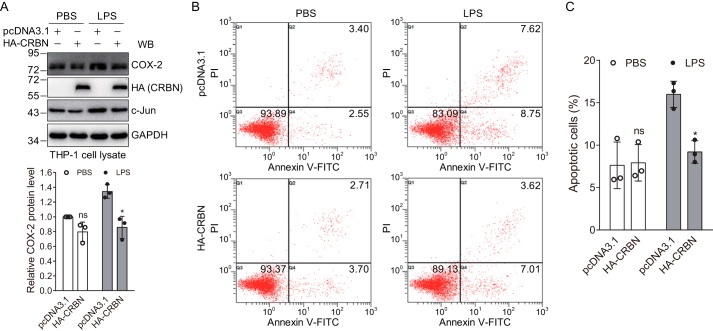
Because CRBN regulated the expression of cytokines induced by LPS, and LPS could modulate apoptosis through the JNK signaling pathway, we tested whether CRBN could regulate LPS-induced apoptosis. To do this, we first induced differentiation of human monocytic THP-1 cells using phorbol 12-myristate 13-acetate (PMA) and transfected the cells with the control or CRBN plasmid, respectively. 48 h post-transfection, cells were treated with PBS or LPS for 24 h, and cell lysates were used for immunoblot analysis. The results clearly indicated that LPS stimulation significantly enhanced the protein level of c-Jun and its downstream target COX-2 and that this enhancement was attenuated by CRBN expression (Fig. 6A), suggesting the existence of the same regulatory mechanism in THP-1 cells as discovered in HEK293T and Raw264.7 cells. The cells were also stained with annexin V–FITC/propidium iodide (PI) and analyzed by flow cytometry. The data showed that the percentage of apoptotic cells (including early and late apoptotic cells) was not affected by CRBN transfection in the PBS-treated sample. When the cells were stimulated with LPS, the percentage of apoptotic cells for the mock-transfected samples was increased as expected. However, CRBN expression significantly reduced the apoptotic cells induced by LPS (Fig. 6, B and C). Together, these data demonstrated that CRBN could protect cells from LPS-induced apoptosis, which was in concert with the observation that the expression of cytokines was attenuated (or increased) in CRBN-expressing (or Crbn knockdown) Raw264.7 cells under LPS stimulation (Fig. 5).
Figure 6.
CRBN suppresses LPS-induced apoptosis in THP-1 cells. A, CRBN reduces COX-2 and c-Jun protein levels in THP-1 cells upon LPS stimulation. THP-1 cells were differentiated by PMA (50 ng/ml) for 24 h and transfected with the pcDNA3.1 or CRBN plasmid, respectively, using Lipofectamine 2000 transfection reagent. 48 h post-transfection, cells were treated with PBS or LPS (40 μg/ml) for 24 h, and cell lysates were immunoblotted. Densitometry quantification was performed for three biological replicates, and p values were calculated using Student's t test. *, p < 0.05; ns, not significant compared with samples transfected with the pcDNA3.1 vector. WB, Western blot. B, flow cytometry analyses of mock- and CRBN-transfected cells upon PBS or LPS treatment. THP-1 cells were treated as described in A, stained with annexin V–FITC/PI, and analyzed by flow cytometry. The percentage of cells in different states is depicted in the scatterplot. C, quantification of apoptotic cells from three biological replicates of flow cytometry analyses. The p value was calculated using Student's t test. *, p < 0.05.
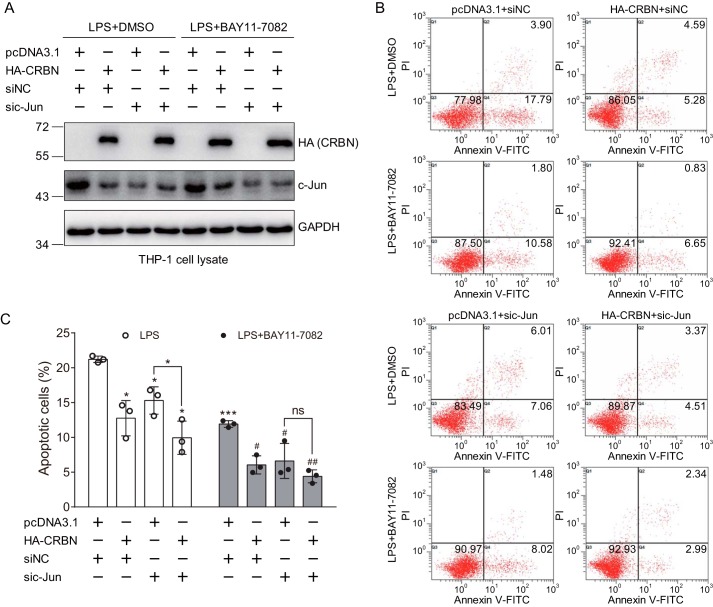
Because LPS can activate the JNK signaling pathway and lead to apoptosis, we explore whether CRBN regulates LPS-induced apoptosis through c-Jun. For this, we used siRNA to knock down c-Jun during transfection of the control or CRBN plasmid in THP-1 cells and then used BAY11-7082 to inhibit the NF-κB pathway prior to LPS stimulation. Western blot analyses were used to further confirm CRBN expression, c-Jun knockdown, and the regulation of c-Jun by CRBN (Fig. 7A). Without BAY11-7082 treatment, the cells exhibited a similar degree of apoptosis as shown in Fig. 6B when transfected with the control or CRBN plasmid and control siRNAs upon LPS stimulation (Fig. 7B, first row). In the presence of BAY11-7082, CRBN also significantly attenuated LPS-induced apoptosis, although the overall apoptosis level was lower than that in samples without BAY11-7082 treatment (Fig. 7, B, second row, and C). When c-Jun was knocked down, the percentage of apoptotic cells was also decreased for cells transfected with the pcDNA3.1 plasmid (Fig. 7C), indicating participation of the c-Jun signaling pathway in the regulation of LPS-induced apoptosis. However, when CRBN was expressed, the percentage of apoptotic cells was further reduced (Fig. 7, B, third row, and C). This may be explained by the fact that siRNA did not completely eliminate c-Jun or that LPS-induced apoptosis may also be influenced by other signaling pathways, such as the NF-κB signaling pathway. To eliminate the effect of the NF-κB signaling pathway on LPS-induced apoptosis, we treated cells with BAY11-7082 prior to LPS stimulation. Although CRBN alone still reduced apoptosis (Fig. 7, B, second row, and C), upon c-Jun knockdown, further expression of CRBN did not significantly alter the percentage of apoptotic cells (Fig. 7, B, fourth row, and C). Taken together, these results suggest that c-Jun mediates the effect of CRBN on LPS-induced apoptosis.
Figure 7.
c-Jun knockdown abolishes the effect of CRBN expression on LPS-induced apoptosis upon NF-κB inhibition. A, Western blot analysis for mock- and CRBN-transfected cells with or without pretreatment with the NF-κB inhibitor BAY11-7082. THP-1 cells were differentiated by PMA (50 ng/ml) for 24 h and transfected with the pcDNA3.1 or CRBN plasmid along with siNC or sic-Jun, respectively, using Lipofectamine 2000 transfection reagent. 48 h post-transfection, cells were treated with DMSO or BAY11-7082 for 1 h prior to treatment with PBS or LPS (40 μg/ml) for 24 h. Cell lysates were immunoblotted for the indicated antibodies. B, flow cytometry analyses of mock- and CRBN-transfected THP-1 cells treated with LPS in the absence or presence of the NF-κB inhibitor BAY11-7082. Cells were treated as described in A, stained, and analyzed as described in Fig. 6B. C, quantification of apoptotic cells obtained from three biological replicates of flow cytometry analyses. Student's t test was used to calculate the p value. *, p < 0.05; ***, p < 0.001 compared with the first column (mock-transfected cells with DMSO and LPS treatment) or as indicated. #, p < 0.05; ##, p < 0.01 compared with the fifth column (pcDNA3.1- and siNC-transfected cells with BAY11-7082 and LPS treatment). ns, not significant.
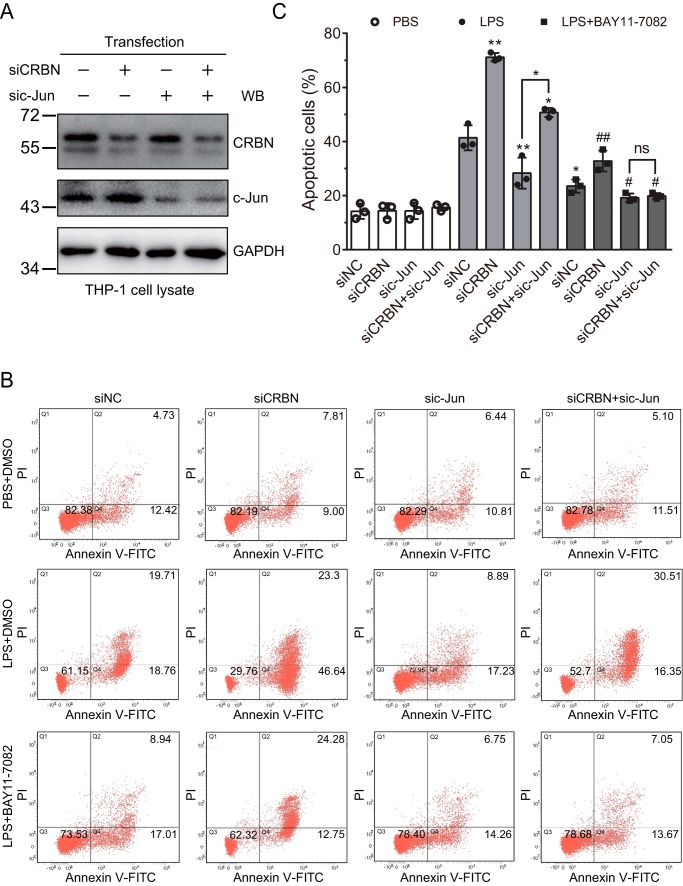
To further validate this mechanism, we also carried out the above experiment in THP-1 cells in which CRBN was knocked down by siRNA. Western blot analysis demonstrated efficient knockdown of CRBN and c-Jun in THP-1 cells (Fig. 8A). Similarly, without LPS stimulation, knockdown of CRBN and/or c-Jun did not affect cell apoptosis. Under mock knockdown, LPS stimulation significantly increased apoptotic cells, and pretreatment of cells with the NF-κB inhibitor BAY11-7082 slightly reduced apoptotic cells. When CRBN was knocked down, overall apoptotic cells were significantly increased upon LPS treatment in the absence or presence of BAY11-7082 compared with mock knockdown samples. Further knockdown of c-Jun significantly decreased the percentage of apoptotic cells under LPS stimulation. After inhibiting the NF-κB signaling pathway, although knockdown of CRBN alone increased apoptosis, upon c-Jun knockdown, further reduction of CRBN did not affect the percentage of apoptotic cells (Fig. 8, B and C). Again, these data support the idea that the CRBN-regulated inflammatory response is at least partially mediated through c-Jun.
Figure 8.
c-Jun knockdown eliminates the effect of CRBN knockdown on LPS-induced apoptosis upon NF-κB inhibition. A, Western blot (WB) analysis of c-Jun and CRBN knockdown efficiency. THP-1 cells were differentiated by PMA (50 ng/ml) for 24 h and transfected with siNC, sic-Jun, siCRBN, or sic-Jun and siCRBN using Lipofectamine 2000 transfection reagent for 48 h. Cells were treated with DMSO or BAY11-7082 for 1 h prior to treatment with PBS or LPS (40 μg/ml) for 24 h. Cell lysates were immunoblotted for the indicated antibodies. B, flow cytometry analyses of THP-1 cells transfected with siNC, sic-Jun, and siCRBN and treated with PBS or LPS in the absence or presence of the NF-κB inhibitor BAY11-7082. Cells were treated as described in A, stained, and analyzed as described in Fig. 6B. C, quantification of apoptotic cells obtained from three biological replicates of flow cytometry analyses. Student's t test was used to calculate the p values. *, p < 0.05; **, p < 0.001 compared with the fifth column (mock-transfected cells with DMSO and LPS treatment) or as indicated in the figure. #, p < 0.05; ##, p < 0.01 compared with the ninth column (mock-transfected cells with BAY11-7082 and LPS treatment). ns, not significant.
Discussion
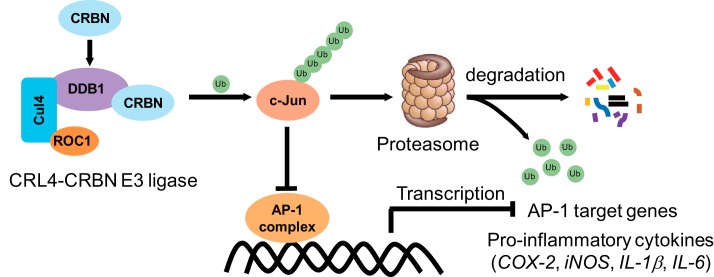
One of the most extensively studied functions for CRBN is its role as a substrate receptor for the CRL4 E3 ligase, which recognizes specific targets for their ubiquitination and degradation. CRBN not only targets constitutive endogenous substrates but also recruits neosubstrates for ubiquitination by interacting with IMiDs, such as thalidomide and its structural analogs lenalidomide and pomalidomide, and forming a new substrate binding pocket (32). These neosubstrates include two transcription factors, IKZF1 and IKZF3 (46, 47), and casein kinase 1α (48). Their ubiquitination and degradation induced by IMiDs is the molecular mechanism for the treatment of multiple diseases, such as myeloma and myelodysplastic syndrome with deletion of chromosome 5q (46–48). Although CRBN is the primary target of these IMiDs, whether the E3 activity of the CRL4-CRBN ligase regulates the inflammatory response has not yet been explored. Here, using biochemical approaches, we find that, together with DDB1, Cul4A, and ROC1, CRBN promotes the formation of the Lys48-linked polyubiquitin chains on c-Jun and enhances its proteasomal degradation. Therefore, CRBN reduces the c-Jun protein level and attenuates the transcriptional activity of the AP-1 transcription factor complex, in which c-Jun is the critical component. Using murine macrophage and human monocytic cell lines as model systems, we further discover that CRBN reduces the mRNA expression of pro-inflammatory cytokines downstream of the AP-1 signaling pathway and attenuates LPS-induced apoptosis. Together, our work reveals a new molecular mechanism by which CRBN suppresses inflammation and apoptosis under stress. In this mechanism, CRBN forms an E3 ligase with DDB1, Cul4A, and ROC1 to ubiquitinate c-Jun and regulate its proteasomal degradation, thus inhibiting the formation of the active AP-1 complex and reducing its transcriptional activity, leading to a reduced inflammatory response upon LPS stimulation (Fig. 9).
Figure 9.
A proposed model for regulation of the inflammatory response by CRBN. CRBN assembles to an active E3 ligase with DDB1, Cul4A, and ROC1; promotes c-Jun ubiquitination; and mediates its degradation by the 26S proteasome. The reduction of c-Jun protein diminishes AP-1 transcriptional activity and thus reduces LPS-stimulated expression of inflammatory cytokines such as iNOS, COX-2, IL-1β, IL-6, etc.
Several reports have revealed that CRBN also has functions that are not associated with CRL4-CRBN E3 ligase activity. These functions are achieved through the regulation of protein–DNA or protein–protein interaction, which further affects downstream signaling pathways. For example, CRBN epigenetically suppresses the expression of the Kv1.3 potassium channel by its binding to the conserved DNA elements adjacent to the Kcna3 gene, thus limiting T cell activation and IL-2 production upon T cell receptor stimulation (49). CRBN also exhibits a chaperone-like function that promotes maturation of CD147 and MCT1 and activation of the CD147–MCT1 transmembrane complex. Thalidomide and its structural analogs can compete their binding with CRBN and destabilize the CD147–MCT1 complex. Along with their function in the recruitment of neosubstrates for protein degradation, these IMiDs exhibit both teratogenic and pleiotropic antitumor effects (50). Under UPS dysfunction, CRBN is recruited to the aggresome and exhibits cytoprotective roles in a neuronal cell line (51). Further study reveals that CRBN interacts with sequestosome 1/p62 and prevents its interaction with polyubiquitinated proteins, thus reducing protein aggregates formed under proteasomal stress (42). In that study, we also find that CRBN expression protects neuronal cells against Htt60Q- and TDP43C-induced cell death.
It has also been reported that CRBN interacts with the ubiquitination domain of TRAF6 and, in turn, prevents formation of Lys63-linked polyubiquitin chains on TRAF6, inhibiting activation of NF-κB complex and reducing the LPS-induced inflammatory response (41). That and this work indicate that CRBN regulates the LPS-induced inflammatory response through both the nonenzymatic and the enzymatic functions of the CRL4-CRBN E3 ligases. These functions are involved in the regulation of two different types of ubiquitination in two opposite manners, inhibiting and promoting the ubiquitination of downstream signaling molecules. Based on the localization of these proteins, the reduction of TRAF6 and TAB2 ubiquitination by CRBN most probably occurs in the cytoplasm, whereas enhancement of CRBN-mediated c-Jun ubiquitination may happen in the nucleus. Therefore, CRBN can inhibit the inflammatory response in two different cellular compartments through two distinct molecular mechanisms. It should be noted that, although we used LPS as a stimulus to trigger the inflammatory response in this work, CRBN may also protect cells from inflammation induced by other triggers, such as TNFα and transforming growth factor β, that activate c-Jun/AP-1 and/or TRAF6/TAK1 signaling pathways (52, 53). Our experiments and several previous publications have demonstrated that CRBN exhibits protective roles against cell death upon activation of inflammation and under a variety of stresses but not under basal conditions, suggesting that it might be a more prevalent phenomenon that CRBN plays regulatory roles in response to stresses.
Activation of the AP-1 signaling pathway can regulate many cellular processes, such as cell survival and death (25). In this signaling pathway, c-Jun can modulate the expression of both pro-apoptotic and anti-apoptotic genes, such as FasL, Bim, Bcl3, and Fas, through the transcriptional activity of the AP-1 complex. The overall effect of these gene products determines whether cells undergo survival or apoptosis. These effects may be cell type-, developmental stage-, and stimulus-specific (25). Our work discovered that CRBN could negatively regulate the c-Jun protein level under LPS stimulation, suggesting that CRBN regulates cell survival and cell death under certain circumstances. It has been reported that LPS induces apoptosis of both undifferentiated and differentiated THP-1 cells (54). Several studies have demonstrated that the JNK signaling pathway is activated in LPS-induced apoptosis (27, 28). Indeed, our experiments also indicate that, under normal physiological conditions, c-Jun is not activated, and, therefore, apoptosis is maintained at a very low level, independent of CRBN expression. However, CRBN can significantly attenuate apoptosis of monocytic cells under LPS stimulation. Our data further revealed that c-Jun knockdown diminished the effect of CRBN on LPS-stimulated apoptosis. Indeed, in a neuronal cell line, CRBN also exhibited a cytoprotective effect against the UPS dysfunction–induced cell death, although the detailed molecular mechanism was not provided (51). Taken together, these results demonstrated that CRBN regulated apoptosis under different stresses through multiple signaling pathways, dependent and independent of the activity of the CRL4-CRBN E3 ligase.
Experimental procedures
Materials
Human embryonic kidney (HEK) 293 cells, HEK293T cells, murine macrophage Raw264.7 cells, and human monocytic THP-1 cells were cultured in Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% fetal bovine serum (PAN Biotech and Lonsera), 100 units/ml penicillin, and 100 μg/ml streptomycin (Gibco). Brain lysates from WT and Crbn knockout mice were obtained from a previous study (42).
Antibodies used in this work were from the following companies: anti-HA (sc-7392) and anti-ubiquitin (Ub, sc-8017) antibodies from Santa Cruz Biotechnology (USA); anti-Cul4A (2699S) antibody from Cell Signaling Technology (USA); anti-c-Jun (CPA1634), anti-COX-2 (CPA1969), and anti-Strep tag (CPA9045) antibodies from Cohesion Biosciences (China); anti-FLAG (0912–1) and anti-iNOS (RT1332) antibodies from HuaAn Biotechnology (China); anti-GAPDH (abs830030) antibody from Absin Bioscience (China). Mouse anti-CRBN antibody (39) was a gift from Dr. Xiu-Bao Chang (Mayo Clinic College of Medicine, USA) and rabbit anti-CRBN antibody (11435–1-AP) from ProteinTech Group (China). Secondary antibodies were from Beyotime Biotechnology (China).
MG132 (s8410) was from Selleck, LPS (S1732), BAY11-7082 (S1523, an NF-κB inhibitor), PDTC (S1808, an NF-κB inhibitor), and phorbol-12-myristate-13-acetate (PMA, S1819) were from Beyotime Biotechnology (China).
Plasmids
c-Jun was amplified from total RNA obtained from HEK293T cells and cloned to the pcDNA3.1 vector with an SF tag at the N terminus. HA-CRBN, GFP-CRBN, and CRBN mutants were subcloned from a CRBN plasmid obtained previously (34). Myc-Ub and HA-Ub (WT, K48R, and K63R) plasmids were from Dr. Xinliang Mao at Soochow University, the glutathione S-transferase–DDB1 plasmid was from Dr. Feng Rao at Southern University of Science and Technology, the FLAG-Cul4A and Cul4A(Δ) plasmids were from Dr. Pengbo Zhou at Weill Cornell Medical College, and the pGL3-AP-1 firefly luciferase reporter plasmid was from Dr. Guanghui Wang at Soochow University.
Mass spectrometry analysis
Protein quantification was carried out using the stable isotope labeling of amino acids by cell culture technique (55, 56). HEK293T cells expressing GFP (control) or CRBN/GFP (experimental) were prepared according to a method published previously (34). Control cells were grown in light medium (containing light Lys0: 12C6, 14N2 and light arginine Arg0: 12C6, 14N4), and experimental cells were grown in heavy medium (containing heavy lysine Lys8: 13C6, 15N2 and heavy arginine Arg10: 13C6, 15N4). The same numbers of cells from control and experimental samples were mixed, lysed, and digested with trypsin. The resulting peptide samples were analyzed by MS as described previously (57). Peptide quantification was obtained from the relative intensity of MS peaks from light medium– and heavy medium–cultured cells. Two biological replicates were performed to obtain confident quantification.
Immunofluorescence
Immunofluorescence measurements were carried out according to a method published previously (58). Briefly, SF–c-Jun was cotransfected with either the GFP or GFP-CRBN plasmid into HEK293 cells. Cells were washed, fixed, permeabilized, blocked, incubated with an anti-FLAG (mouse) antibody, and washed with TBST (TBS with 0.1% Tween 20) three times. Cells were further incubated with Alexa Fluor 594 donkey anti-mouse IgG (red fluorescence) and washed again. Cells were stained with Hoechst, and images were taken under an inverted microscope.
CHX treatment
The pcDNA3.1 (control) or HA-CRBN plasmid was transiently transfected into HEK293T cells. Cells were equally split to 6-well plates 6 h after transfection. 48 h after transfection, cells were treated with CHX (200 μg/ml, Sigma) for the indicated time, washed with ice-cold PBS, and lysed in modified RIPA buffer (150 mm Tris-HCl (pH 7.4), 150 mm NaCl, 1% Triton X-100, 0.1% SDS, and 1 mm EDTA) containing freshly added protease inhibitor mixture (Selleck, B14012) with brief sonication to obtain cell lysates for immunoblot analyses.
Affinity purification
SF–c-Jun was purified with the Strep-Tactin–agarose beads. Cell lysate was incubated with prewashed Strep-Tactin–agarose beads (IBA) at 4 °C overnight. The beads were centrifuged at 900 × g for 1 min and washed with the modified RIPA buffer and RIPA buffer containing 300 mm NaCl, each three times. SF–c-Jun and its interacting proteins were eluted with 5 mm desthiobiotin twice, each time with a 30-min gentle incubation. The eluate was combined for subsequent analyses.
c-Jun and CRBN antibodies were used for immunoprecipitation of endogenous proteins according to a procedure described previously (59). Briefly, cell lysates were precleared with protein A/G–agarose beads and incubated with 1.6 μg of control IgG, c-Jun, or CRBN antibodies at 4 °C for 16 h. Prewashed protein A/G–agarose beads (40 μl) were mixed with the above cell lysate and incubated at 4 °C for 4 h. The protein A/G–agarose beads were washed three times with TBST and once with modified RIPA buffer, and proteins were eluted by heating the beads with 80 μl of 2× SDS sample loading buffer.
siRNA
The control siRNAs (ID 35164) and CRBN and c-Jun siRNAs were synthesized by GenePharma or RiboBio Co. The siRNA sequences were as follows: mouse siCRBN, 5′-CAGUGUCUCAUGUACAUAUCCAUGA-3′ (sense) and 5′-UCAUGGAUAUGUACAUGAGACACUG-3′ (antisense); human siCRBN 1, 5′-CCCAGACACUGAAGAUGAAAU-3′ (sense) and 5′-AUUUCAUCUUCAGUGUCUGGG-3′ (antisense); human siCRBN 2, 5′-CAGCUUAUGUGAAUCCUCAUGGAUA-3′ (sense) and 5′-UAUCCAUGAGGAUUCACAUAAGCUG-3′ (antisense); human sic-Jun, 5′-CCUUCUAUGACGAUGCCCUTT-3′ (sense) and 5′-AGGGCAUCGUCAUAGAAGGTT-3′ (antisense); mouse sic-Jun, 5′-GGCACAGCUUAAACAGAAATT-3′ (sense) and 5′-UUUCUGUUUAAGCUGUGCCTT-3′ (antisense). Raw264.7 cells cultured in 12-well plates were transfected with siRNA (200 pmol) using Lipofectamine 3000 transfection reagent (Life Technologies) according to the manufacturer's instructions. The culture medium was changed 12 h after transfection. Cells were treated further and collected for subsequent experiments.
Western blot analysis
Cell lysates or affinity-purified samples were mixed with appropriate amounts of 5× SDS sample loading buffer, boiled, centrifuged, and separated on SDS-PAGE. Proteins were transferred to a polyvinylidene difluoride membrane, which was further blocked with 5% nonfat milk in TBST, incubated with the indicated primary and secondary antibodies, and washed with TBST three times after each antibody incubation. Proteins on the membrane were visualized with Immobilon Western chemiluminescent HRP substrate (Millipore), and signals were recorded in a ChemiDoc (Bio-Rad) or Tanon 5200 imaging system.
qPCR
Total RNA was isolated from Raw264.7 cells using TRIzol reagent (Vazyme), and mRNA was reverse-transcribed into cDNA with a HiScript II Q RT SuperMix kit (Vazyme) according to the manufacturer's instructions. qPCR was performed on an Applied Biosystems 7500 real-time PCR system using the Biotool 2× SYBR Green qPCR Master Kit. The qPCR primers were synthesized by Sangon Biotech Co. The primer sequences were as follows: COX-2, 5′-CAGTTTATGTTGTCTGTCCAGAGTTTC-3′ (forward) and 5′-CCAGCACTTCACCCATCAGTT-3′ (reverse); iNOS, 5′-GAACTGTAGCACAGCACAGGAAAT-3′ (forward) and 5′-CGTACCGGATGAGCTGTGAAT-3′ (reverse); IL-1β, 5′-GTTCCCATTAGACAACTGCACTACAG-3′ (forward) and 5′-GTCGTTGCTTGGTTCTCCTTGTA-3′ (reverse); IL-6, 5′-CCAGAAACCGCTATGAAGTTCC-3′ (forward) and 5′-GTTGGGAGTGGTATCCTCTGTGA-3′ (reverse); β-actin, 5′-GTCAGGTCATCACTATCGGCAAT-3′ (forward) and 5′-AGAGGTCTTTACGGATGTCAACGT-3′ (reverse). β-Actin was used as the control. All qPCR experiments were repeated for the indicated times, and p values were calculated using Student's t test.
Luciferase reporter assay
The pcDNA3.1 or HA-CRBN plasmid along with the pGL3-AP-1 firefly luciferase reporter plasmid and the Renilla luciferase control reporter vector was transfected into Raw264.7 cells using Lipofectamine 3000 transfection reagent. pcDNA3.1 was used to balance the total amount of plasmids. 36 h post-transfection, cells were stimulated with PBS (control) or LPS (200 ng/ml) for 12 h and lysed in passive lysis buffer. The bioluminescence signals were measured with the Dual-Luciferase reporter assay system (Promega) after sequential addition of luciferase assay reagent II and Stop & Glo reagent. The signal from the firefly luciferase reporter gene was normalized to that of the Renilla luciferase control reporter vector. The data obtained from the pcDNA3.1 control samples were set to 1 to obtain the relative luciferase signal.
Flow cytometry analyses
Human monocytic THP-1 cells were first cultured with medium containing PMA (50 ng/ml) in 12-well plates for 24 h. Then some wells of the differentiated cells were transfected with the pcDNA3.1 or HA-CRBN plasmid using PEI transfection reagent. 48 h post-transfection, cells were treated with PBS or LPS (40 μg/ml) for 24 h. Other wells of the differentiated cells were transfected with the corresponding plasmids or siRNAs using Lipofectamine 2000 transfection reagent. Then cells were pretreated with DMSO or the NF-κB inhibitor BAY11-7082 (20 μm) for 1 h before LPS stimulation (40 μg/ml, 24 h). Cells were collected along with supernatant, washed with ice-cold PBS three times, and stained with an annexin V-FITC/PI apoptosis detection kit (MultiSciences) according to the manufacturer's instructions. Cells were further analyzed by flow cytometry (Beckman Coulter FC500 or BD FACSVerse).
Author contributions
J. Y., M. H., L. Z., X. H., X. J., and Y. Z. data curation; J. Y. and M. H. investigation; J. Y. and G. X. writing-original draft; J. Y., L. Z., and G. X. writing-review and editing; G. X. conceptualization; G. X. supervision; L. Z. and G. X. funding acquisition; G. X. project administration.
Supplementary Material
Acknowledgments
We thank Dr. Xiu-Bao Chang (Mayo Clinic College of Medicine) for generously providing the mouse CRBN antibody; Dr. Guanghui Wang, Dr. Xinliang Mao, Dr. Feng Rao, and Dr. Pengbo Zhou for kindly providing plasmids, and Dr. Guanghui Wang and Longtai Zheng (Soochow University) for fruitful discussions.
This work was supported by the National Natural Science Foundation of China (31670833 and 81703535), the China Postdoctoral Science Foundation (2017M611895), the Natural Science Foundation of Jiangsu Higher Education Institutions of China (17KJD180005), the Jiangsu Postdoctoral Science Foundation (1701130C), the Jiangsu Key Laboratory of Neuropsychiatric Diseases (BM2013003), and a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S5.
- LPS
- lipopolysaccharide
- TNF
- tumor necrosis factor
- JNK
- c-Jun N-terminal kinase
- iNOS
- inducible nitric oxide synthase
- IL
- interleukin
- CRBN
- cereblon
- IMiD
- immunomodulatory drug
- HA
- hemagglutinin
- FL
- full-length
- SF
- Strep-FLAG
- qPCR
- quantitative PCR
- CHX
- cycloheximide
- UPS
- ubiquitin–proteasome system
- Ub
- ubiquitin
- BAY
- BAY11-7082
- PDTC
- pyrrolidine dithiocarbamate
- PMA
- phorbol 12-myristate 13-acetate
- PI
- propidium iodide
- HEK
- human embryonic kidney
- RIPA
- radioimmune precipitation assay
- PEI
- polyethyleneimine
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- cDNA
- complementary DNA.
References
- 1. Medzhitov R. (2008) Origin and physiological roles of inflammation. Nature 454, 428–435 10.1038/nature07201 [DOI] [PubMed] [Google Scholar]
- 2. Demoruelle M. K., Deane K. D., and Holers V. M. (2014) When and where does inflammation begin in rheumatoid arthritis? Curr. Opin. Rheumatol. 26, 64–71 10.1097/BOR.0000000000000017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Geremia A., Biancheri P., Allan P., Corazza G. R., and Di Sabatino A. (2014) Innate and adaptive immunity in inflammatory bowel disease. Autoimmun. Rev. 13, 3–10 10.1016/j.autrev.2013.06.004 [DOI] [PubMed] [Google Scholar]
- 4. Wu M. Y., Li C. J., Hou M. F., and Chu P. Y. (2017) New insights into the role of inflammation in the pathogenesis of atherosclerosis. Int. J. Mol. Sci. 18, 2034 10.3390/ijms18102034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Block M. L., and Hong J. S. (2005) Microglia and inflammation-mediated neurodegeneration: multiple triggers with a common mechanism. Prog. Neurobiol. 76, 77–98 10.1016/j.pneurobio.2005.06.004 [DOI] [PubMed] [Google Scholar]
- 6. Akiyama H. (1994) Inflammatory response in Alzheimer's disease. Tohoku J. Exp. Med. 174, 295–303 10.1620/tjem.174.295 [DOI] [PubMed] [Google Scholar]
- 7. Monteiro R., and Azevedo I. (2010) Chronic inflammation in obesity and the metabolic syndrome. Mediators Inflamm. 2010, 289645 10.1155/2010/289645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Manohar M., Verma A. K., Venkateshaiah S. U., Sanders N. L., and Mishra A. (2017) Pathogenic mechanisms of pancreatitis. World J. Gastrointest. Pharmacol. Ther. 8, 10–25 10.4292/wjgpt.v8.i1.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Adcock I. M. (1997) Transcription factors as activators of gene transcription: AP-1 and NF-κB. Monaldi Arch. Chest Dis. 52, 178–186 [PubMed] [Google Scholar]
- 10. Hsu T. C., Young M. R., Cmarik J., and Colburn N. H. (2000) Activator protein 1 (AP-1)- and nuclear factor κB (NF-κΒ)-dependent transcriptional events in carcinogenesis. Free Radic. Biol. Med. 28, 1338–1348 10.1016/S0891-5849(00)00220-3 [DOI] [PubMed] [Google Scholar]
- 11. Karin M., and Greten F. R. (2005) NF-κΒ: linking inflammation and immunity to cancer development and progression. Nat. Rev. Immunol. 5, 749–759 10.1038/nri1703 [DOI] [PubMed] [Google Scholar]
- 12. Takeuchi O., and Akira S. (2001) Toll-like receptors; their physiological role and signal transduction system. Int. Immunopharmacol. 1, 625–635 10.1016/S1567-5769(01)00010-8 [DOI] [PubMed] [Google Scholar]
- 13. Brenner D. A., O'Hara M., Angel P., Chojkier M., and Karin M. (1989) Prolonged activation of jun and collagenase genes by tumour necrosis factor-α. Nature 337, 661–663 10.1038/337661a0 [DOI] [PubMed] [Google Scholar]
- 14. Yu Y., Ge N., Xie M., Sun W., Burlingame S., Pass A. K., Nuchtern J. G., Zhang D., Fu S., Schneider M. D., Fan J., and Yang J. (2008) Phosphorylation of Thr-178 and Thr-184 in the TAK1 T-loop is required for interleukin (IL)-1-mediated optimal NF-κB and AP-1 activation as well as IL-6 gene expression. J. Biol. Chem. 283, 24497–24505 10.1074/jbc.M802825200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shim J. H., Xiao C., Paschal A. E., Bailey S. T., Rao P., Hayden M. S., Lee K. Y., Bussey C., Steckel M., Tanaka N., Yamada G., Akira S., Matsumoto K., and Ghosh S. (2005) TAK1, but not TAB1 or TAB2, plays an essential role in multiple signaling pathways in vivo. Genes Dev. 19, 2668–2681 10.1101/gad.1360605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Afonina I. S., Zhong Z., Karin M., and Beyaert R. (2017) Limiting inflammation-the negative regulation of NF-κB and the NLRP3 inflammasome. Nat. Immunol. 18, 861–869 10.1038/ni.3772 [DOI] [PubMed] [Google Scholar]
- 17. Mackman N., Brand K., and Edgington T. S. (1991) Lipopolysaccharide-mediated transcriptional activation of the human tissue factor gene in THP-1 monocytic cells requires both activator protein 1 and nuclear factor κB binding sites. J. Exp. Med. 174, 1517–1526 10.1084/jem.174.6.1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mao R., Fan Y., Mou Y., Zhang H., Fu S., and Yang J. (2011) TAK1 lysine 158 is required for TGF-β-induced TRAF6-mediated Smad-independent IKK/NF-κB and JNK/AP-1 activation. Cell Signal. 23, 222–227 10.1016/j.cellsig.2010.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li Q., and Verma I. M. (2002) NF-κB regulation in the immune system. Nat. Rev. Immunol. 2, 725–734 10.1038/nri910 [DOI] [PubMed] [Google Scholar]
- 20. Angel P., and Karin M. (1991) The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim. Biophys. Acta 1072, 129–157 [DOI] [PubMed] [Google Scholar]
- 21. Halazonetis T. D., Georgopoulos K., Greenberg M. E., and Leder P. (1988) c-Jun dimerizes with itself and with c-Fos, forming complexes of different DNA binding affinities. Cell 55, 917–924 10.1016/0092-8674(88)90147-X [DOI] [PubMed] [Google Scholar]
- 22. Kouzarides T., and Ziff E. (1988) The role of the leucine zipper in the fos-jun interaction. Nature 336, 646–651 10.1038/336646a0 [DOI] [PubMed] [Google Scholar]
- 23. Guha M., and Mackman N. (2001) LPS induction of gene expression in human monocytes. Cell Signal. 13, 85–94 10.1016/S0898-6568(00)00149-2 [DOI] [PubMed] [Google Scholar]
- 24. Meng Q., and Xia Y. (2011) c-Jun, at the crossroad of the signaling network. Protein Cell 2, 889–898 10.1007/s13238-011-1113-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shaulian E., and Karin M. (2002) AP-1 as a regulator of cell life and death. Nat. Cell Biol. 4, E131–136 10.1038/ncb0502-e131 [DOI] [PubMed] [Google Scholar]
- 26. Xaus J., Comalada M., Valledor A. F., Lloberas J., López-Soriano F., Argilés J. M., Bogdan C., and Celada A. (2000) LPS induces apoptosis in macrophages mostly through the autocrine production of TNF-α. Blood 95, 3823–3831 [PubMed] [Google Scholar]
- 27. Hull C., McLean G., Wong F., Duriez P. J., and Karsan A. (2002) Lipopolysaccharide signals an endothelial apoptosis pathway through TNF receptor-associated factor 6-mediated activation of c-Jun NH2-terminal kinase. J. Immunol. 169, 2611–2618 10.4049/jimmunol.169.5.2611 [DOI] [PubMed] [Google Scholar]
- 28. Guo C., Yuan L., Wang J. G., Wang F., Yang X. K., Zhang F. H., Song J. L., Ma X. Y., Cheng Q., and Song G. H. (2014) Lipopolysaccharide (LPS) induces the apoptosis and inhibits osteoblast differentiation through JNK pathway in MC3T3-E1 cells. Inflammation 37, 621–631 10.1007/s10753-013-9778-9 [DOI] [PubMed] [Google Scholar]
- 29. Ye N., Ding Y., Wild C., Shen Q., and Zhou J. (2014) Small molecule inhibitors targeting activator protein 1 (AP-1). J. Med. Chem. 57, 6930–6948 10.1021/jm5004733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Angers S., Li T., Yi X., MacCoss M. J., Moon R. T., and Zheng N. (2006) Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature 443, 590–593 [DOI] [PubMed] [Google Scholar]
- 31. Chamberlain P. P., Lopez-Girona A., Miller K., Carmel G., Pagarigan B., Chie-Leon B., Rychak E., Corral L. G., Ren Y. J., Wang M., Riley M., Delker S. L., Ito T., Ando H., Mori T., et al. (2014) Structure of the human Cereblon-DDB1-lenalidomide complex reveals basis for responsiveness to thalidomide analogs. Nat. Struct. Mol. Biol. 21, 803–809 10.1038/nsmb.2874 [DOI] [PubMed] [Google Scholar]
- 32. Fischer E. S., Böhm K., Lydeard J. R., Yang H., Stadler M. B., Cavadini S., Nagel J., Serluca F., Acker V., Lingaraju G. M., Tichkule R. B., Schebesta M., Forrester W. C., Schirle M., Hassiepen U., et al. (2014) Structure of the DDB1-CRBN E3 ubiquitin ligase in complex with thalidomide. Nature 512, 49–53 10.1038/nature13527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ito T., Ando H., Suzuki T., Ogura T., Hotta K., Imamura Y., Yamaguchi Y., and Handa H. (2010) Identification of a primary target of thalidomide teratogenicity. Science 327, 1345–1350 10.1126/science.1177319 [DOI] [PubMed] [Google Scholar]
- 34. Xu G., Jiang X., and Jaffrey S. R. (2013) A mental retardation-linked nonsense mutation in cereblon is rescued by proteasome inhibition. J. Biol. Chem. 288, 29573–29585 10.1074/jbc.M113.472092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nguyen T. V., Lee J. E., Sweredoski M. J., Yang S. J., Jeon S. J., Harrison J. S., Yim J. H., Lee S. G., Handa H., Kuhlman B., Jeong J. S., Reitsma J. M., Park C. S., Hess S., and Deshaies R. J. (2016) Glutamine triggers acetylation-dependent degradation of glutamine synthetase via the thalidomide receptor cereblon. Mol. Cell 61, 809–820 10.1016/j.molcel.2016.02.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nguyen T. V., Li J., Lu C. J., Mamrosh J. L., Lu G., Cathers B. E., and Deshaies R. J. (2017) p97/VCP promotes degradation of CRBN substrate glutamine synthetase and neosubstrates. Proc. Natl. Acad. Sci. U.S.A. 114, 3565–3571 10.1073/pnas.1700949114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lee K. M., Yang S. J., Choi J. H., and Park C. S. (2014) Functional effects of a pathogenic mutation in Cereblon (CRBN) on the regulation of protein synthesis via the AMPK-mTOR cascade. J. Biol. Chem. 289, 23343–23352 10.1074/jbc.M113.523423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chen Y. A., Peng Y. J., Hu M. C., Huang J. J., Chien Y. C., Wu J. T., Chen T. Y., and Tang C. Y. (2015) The cullin 4A/B-DDB1-cereblon E3 ubiquitin ligase complex mediates the degradation of CLC-1 chloride channels. Sci. Rep. 5, 10667 10.1038/srep10667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xu Q., Hou Y. X., Langlais P., Erickson P., Zhu J., Shi C. X., Luo M., Zhu Y., Xu Y., Mandarino L. J., Stewart K., and Chang X. B. (2016) Expression of the cereblon binding protein argonaute 2 plays an important role for multiple myeloma cell growth and survival. BMC Cancer 16, 297 10.1186/s12885-016-2331-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Del Prete D., Rice R. C., Rajadhyaksha A. M., and D'Adamio L. (2016) Amyloid precursor protein (APP) may act as a substrate and a recognition unit for CRL4CRBN and Stub1 E3 ligases facilitating ubiquitination of proteins involved in presynaptic functions and neurodegeneration. J. Biol. Chem. 291, 17209–17227 10.1074/jbc.M116.733626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Min Y., Wi S. M., Kang J. A., Yang T., Park C. S., Park S. G., Chung S., Shim J. H., Chun E., and Lee K. Y. (2016) Cereblon negatively regulates TLR4 signaling through the attenuation of ubiquitination of TRAF6. Cell Death Dis. 7, e2313 10.1038/cddis.2016.226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhou L., Hao Z., Wang G., and Xu G. (2018) Cereblon suppresses the formation of pathogenic protein aggregates in a p62-dependent manner. Hum. Mol. Genet. 27, 667–678 10.1093/hmg/ddx433 [DOI] [PubMed] [Google Scholar]
- 43. Chen X., Zhang Y., Douglas L., and Zhou P. (2001) UV-damaged DNA-binding proteins are targets of CUL-4A-mediated ubiquitination and degradation. J. Biol. Chem. 276, 48175–48182 10.1074/jbc.M106808200 [DOI] [PubMed] [Google Scholar]
- 44. Ransone L. J., and Verma I. M. (1990) Nuclear proto-oncogenes fos and jun. Annu. Rev. Cell Biol. 6, 539–557 10.1146/annurev.cb.06.110190.002543 [DOI] [PubMed] [Google Scholar]
- 45. Karin M., Liu Z., and Zandi E. (1997) AP-1 function and regulation. Curr. Opin. Cell Biol. 9, 240–246 10.1016/S0955-0674(97)80068-3 [DOI] [PubMed] [Google Scholar]
- 46. Krönke J., Udeshi N. D., Narla A., Grauman P., Hurst S. N., McConkey M., Svinkina T., Heckl D., Comer E., Li X., Ciarlo C., Hartman E., Munshi N., Schenone M., Schreiber S. L., et al. (2014) Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science 343, 301–305 10.1126/science.1244851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lu G., Middleton R. E., Sun H., Naniong M., Ott C. J., Mitsiades C. S., Wong K. K., Bradner J. E., and Kaelin W. G. Jr. (2014) The myeloma drug lenalidomide promotes the cereblon-dependent destruction of Ikaros proteins. Science 343, 305–309 10.1126/science.1244917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Krönke J., Fink E. C., Hollenbach P. W., MacBeth K. J., Hurst S. N., Udeshi N. D., Chamberlain P. P., Mani D. R., Man H. W., Gandhi A. K., Svinkina T., Schneider R. K., McConkey M., Järås M., Griffiths E., et al. (2015) Lenalidomide induces ubiquitination and degradation of CK1α in del(5q) MDS. Nature 523, 183–188 10.1038/nature14610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kang J. A., Park S. H., Jeong S. P., Han M. H., Lee C. R., Lee K. M., Kim N., Song M. R., Choi M., Ye M., Jung G., Lee W. W., Eom S. H., Park C. S., and Park S. G. (2016) Epigenetic regulation of Kcna3-encoding Kv1.3 potassium channel by cereblon contributes to regulation of CD4+ T-cell activation. Proc. Natl. Acad. Sci. U.S.A. 113, 8771–8776 10.1073/pnas.1502166113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Eichner R., Heider M., Fernández-Sáiz V., van Bebber F., Garz A. K., Lemeer S., Rudelius M., Targosz B. S., Jacobs L., Knorn A. M., Slawska J., Platzbecker U., Germing U., Langer C., Knop S., et al. (2016) Immunomodulatory drugs disrupt the cereblon-CD147-MCT1 axis to exert antitumor activity and teratogenicity. Nat. Med. 22, 735–743 10.1038/nm.4128 [DOI] [PubMed] [Google Scholar]
- 51. Sawamura N., Wakabayashi S., Matsumoto K., Yamada H., and Asahi T. (2015) Cereblon is recruited to aggresome and shows cytoprotective effect against ubiquitin-proteasome system dysfunction. Biochem. Biophys. Res. Commun. 464, 1054–1059 10.1016/j.bbrc.2015.07.068 [DOI] [PubMed] [Google Scholar]
- 52. Gaestel M., Kotlyarov A., and Kracht M. (2009) Targeting innate immunity protein kinase signalling in inflammation. Nat. Rev. Drug. Discov. 8, 480–499 10.1038/nrd2829 [DOI] [PubMed] [Google Scholar]
- 53. Reinhard C., Shamoon B., Shyamala V., and Williams L. T. (1997) Tumor necrosis factor α-induced activation of c-jun N-terminal kinase is mediated by TRAF2. EMBO J. 16, 1080–1092 10.1093/emboj/16.5.1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Harrison L. M., Cherla R. P., van den Hoogen C., van Haaften W. C., Lee S. Y., and Tesh V. L. (2005) Comparative evaluation of apoptosis induced by Shiga toxin 1 and/or lipopolysaccharides in human monocytic and macrophage-like cells. Microb. Pathog. 38, 63–76 10.1016/j.micpath.2004.12.003 [DOI] [PubMed] [Google Scholar]
- 55. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., and Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell Proteomics 1, 376–386 10.1074/mcp.M200025-MCP200 [DOI] [PubMed] [Google Scholar]
- 56. Ong S. E., and Mann M. (2006) A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC). Nat. Protoc. 1, 2650–2660 [DOI] [PubMed] [Google Scholar]
- 57. Zhou Y., Xiong L., Zhang Y., Yu R., Jiang X., and Xu G. (2016) Quantitative proteomics identifies myoferlin as a novel regulator of a disintegrin and metalloproteinase 12 in HeLa cells. J. Proteomics 148, 94–104 10.1016/j.jprot.2016.07.015 [DOI] [PubMed] [Google Scholar]
- 58. Ren H., Fu K., Mu C., Zhen X., and Wang G. (2012) L166P mutant DJ-1 promotes cell death by dissociating Bax from mitochondrial Bcl-XL. Mol. Neurodegener. 7, 40 10.1186/1750-1326-7-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhang Y., Duan C., Yang J., Chen S., Liu Q., Zhou L., Huang Z., Xu Y., and Xu G. (2018) Deubiquitinating enzyme USP9X regulates cellular clock function by modulating the ubiquitination and degradation of a core circadian protein BMAL1. Biochem. J. 475, 1507–1522 10.1042/BCJ20180005 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.