Abstract
Human (h)4-1BB (TNFRSF9 or CD137) is an inducible tumor necrosis factor receptor (TNFR) superfamily member that interacts with its cognate ligand h4-1BBL to promote T lymphocyte activation and proliferation. h4-1BB is currently being targeted with agonists in cancer immunotherapy. Here, we determined the crystal structures of unbound h4-1BBL and both WT h4-1BB and a dimerization-deficient h4-1BB mutant (C121S) in complex with h4-1BBL at resolutions between 2.7 and 3.2 Å. We observed that the structural arrangement of 4-1BBL, both unbound and in the complex, represents the canonical bell shape as seen in other similar TNF proteins and differs from the previously reported three-bladed propeller structure of 4-1BBL. We also found that the binding site for the receptor is at the crevice formed between two protomers of h4-1BBL, but that h4-1BB interacts predominantly with only one ligand protomer. Moreover, h4-1BBL lacked the conserved tyrosine residue in the DE loop that forms canonical interactions between other TNFR family molecules and their ligands, suggesting h4-1BBL engages h4-1BB through a distinct mechanism. Of note, we discovered that h4-1BB forms a disulfide-linked dimer because of the presence of an additional cysteine residue found in its cysteine-rich domain 4 (CRD4). As a result, h4-1BB dimerization, in addition to trimerization via h4-1BBL binding, could result in cross-linking of individual ligand–receptor complexes to form a 2D network that stimulates strong h4-1BB signaling. This work provides critical insights into the structural and functional properties of both h4-1BB and h4-1BBL and reveals that covalent receptor dimerization amplifies h4-1BB signaling.
Keywords: tumor necrosis factor (TNF), protein-protein interaction, recombinant protein expression, cell surface receptor, protein structure, X-ray crystallography
Introduction
Human (h)4-1BB (TNFRSF9, CD137) is a tumor necrosis factor receptor (TNFR)2 superfamily (TNFRSF) member that contains four cysteine-rich domains (CRD) in the N-terminal extracellular region connected to a C-terminal cytoplasmic region that contains a TNF receptor-associated factor (TRAF)-binding motif to initiate subsequent signaling (1). 4-1BB acts as a co-stimulatory molecule on activated T cells to enhance their response to antigen and can also aid activation of other cells such as NK cells (2, 3). The ligand of h4-1BB, h4-1BBL (TNFSF9, CD137L), is a member of the TNF ligand superfamily expressed on activated antigen-presenting cells such as B cells, dendritic cells, and macrophages (4, 5). h4-1BBL can also be expressed extracellularly as a soluble homotrimer (6). Binding of 4-1BBL to 4-1BB results in aggregation of several receptors that allows for the efficient recruitment of TRAF adapter proteins such as TRAF1 and -2 to the cytoplasmic TRAF-binding motifs of separate 4-1BB receptors, ultimately initiating co-stimulatory signaling (2, 7). On T cells and NK cells, 4-1BB signaling can inhibit apoptosis while augmenting proliferation and effector functions such as cytokine production or CTL activity that can lead to eradication of established tumors (8, 9). Although the antitumor properties have made 4-1BB an ideal target for cancer immunotherapy (14, 15), 4-1BB signaling can also induce anti-inflammatory effects augmenting regulatory activity in T cells and dendritic cells, suggesting it is also a potential target for the treatment of autoimmune diseases (16–18).
Based on sequence variance and structural organization, TNF family ligands have been divided into conventional, EF disulfide, and divergent families (19, 20). The conventional family members (such as TNF, RANKL, and CD40L) share sequence identity and exist as trimers with a bell-shaped architecture. They bind three cognate monomeric receptors in a similar orientation via conserved hydrophobic interactions. The members of the EF-disulfide group (such as APRIL and TWEAK) contain disulfide bonds between the E and F strands resulting in a more globular shape and bind very small atypical TNFRs (BCMA, TACI, and Fn14). The third group contains TNF ligands (such as OX40L, GITRL, 4-1BBL, and CD30L) that exhibit greater sequence divergence and have a comparably shorter TNF homology domain (THD). Earlier crystallographic studies showed that two members of this group, OX40L and GITRL, exhibit an atypical expanded trimeric organization termed a blooming flower shape (21–23), and while similarly binding three cognate receptors these receptors are separated by a greater distance compared with the orientation seen in the conventional TNF ligand complexes. Although h4-1BBL has been thought to be similar to OX40L and GITRL, it has a considerably longer THD, and when crystallized was found in a novel more open trimeric arrangement, described as a three-bladed propeller architecture (24). Differences might also exist between mouse and human molecules. Human and mouse 4-1BBL are only 30% identical in sequence compared with ∼60% sequence identify found in other human and mouse TNF orthologs (22). Complicating making broad generalizations, our recent biochemical studies also demonstrated that murine 4-1BBL (m4-1BBL) is not naturally a trimer but exists as a covalently linked dimer (11). These data question whether the complexes of human 4-1BB and its ligand are analogous to mouse 4-1BB and its ligand, and whether they will be structurally related to other TNFRSF–TNFSF complexes.
Additionally, whereas 4-1BBL is the only recognized intercellular ligand for 4-1BB, other interactions have the potential to modify the complexes. Galectin-9 is a two-domain carbohydrate binding lectin family molecule with specificity for complex N-linked glycans that contain terminal galactose moieties (12, 13). Galectin-9 was found to bind to N-linked glycans on both mouse and human 4-1BB in a region distinct from the 4-1BBL–binding site, providing a means by which receptor clustering might be promoted when 4-1BB is engaged by 4-1BBL (10, 11). In mice, galectin-9 is required for 4-1BB to signal strongly and exert a more pronounced activity (10), perhaps related to m4-1BBL being a dimer and not efficiently clustering multiple 4-1BB receptors together. However, whether galectin-9 is required for, or can augment the ability of h4-1BB to signal when binding trimeric h4-1BBL has not been tested. Collectively, this suggests that a greater understanding of how 4-1BB forms a complex with 4-1BBL is needed.
In this study, we have determined the crystal structure of the WT h4-1BB–h4-1BBL complex, as well as the complex containing a 4-1BB C121S mutant. We have also crystallized unbound h4-1BBL for comparison. We have observed that in contrast to other TNFRs, human 4-1BB exists as a disulfide-linked dimer. Depending on the rate of disulfide-linked dimerization, 4-1BB can cross-link separate heterohexameric signaling complexes and assemble into a 2D lattice of signaling units, which would promote strong signaling. Our work provides unique insights into the structural and functional properties of the h4-1BBL/h4-1BB interaction and suggests that covalent receptor-dimerization, in addition to previously reported dimerization via the preligand assembly domain (PLAD), could be considered a tunable signal amplifier.
Results
Protein expression, characterization, and structure determination of the h4-1BB–4-1BBL complex
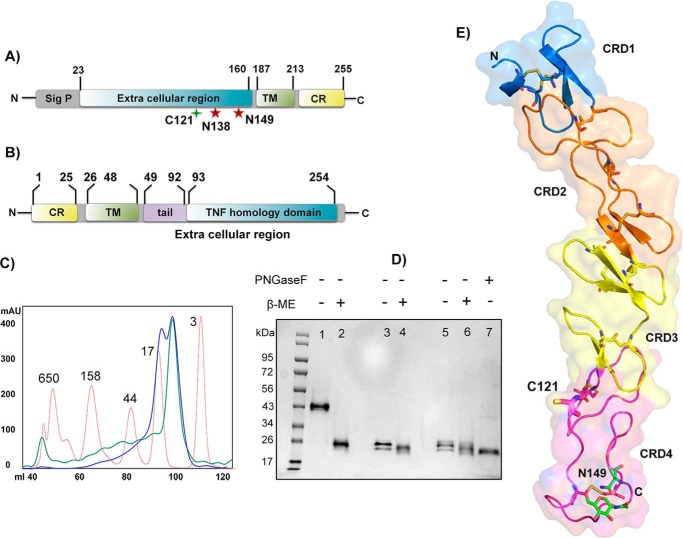
h4-1BB is a type I transmembrane receptor with four extracellular cysteine-rich domains followed by a short transmembrane domain and a C-terminal cytoplasmic region (Fig. 1A). 4-1BBL occurs both in soluble form, as well as a cell-bound type II transmembrane protein and is composed of a short N-terminal cytoplasmic region, followed by a transmembrane domain, and the extracellular TNF THD, which binds to 4-1BB (Fig. 1B). We expressed the extracellular region of 4-1BB (residues 24–160) in Sf9 insect cells and this was purified using ion metal affinity chromatography using the C-terminal His6 tag, followed by size exclusion chromatography (SEC). Biochemical analysis indicated that h4-1BB was expressed as a mixture of monomer and covalent dimer. The SEC profile of purified h4-1BB yielded two peaks of equal ratio (Fig. 1C). On nonreducing SDS-PAGE, the higher molecular mass peak migrated as two bands of equal intensity (as a result of differential glycosylation at Asn138 and Asn149 positions) with an apparent molecular mass of 43 kDa, whereas the lower molecular mass peak migrated at ∼24 kDa. Under reducing conditions, h4-1BB from both high and low Mr peaks migrated according to the lower molecular weight band, suggesting that h4-1BB contained in the high Mr peak was a covalent homodimer (Fig. 1D). Dimerization likely occurred through an unpaired cysteine (Cys121) found within CRD4 of h4-1BB. When we mutated Cys121 to Ser121 (C121S), h4-1BB eluted as a monodisperse peak that eluted comparably to the lower Mr peak on SEC and also migrated as a monomer on nonreducing SDS-PAGE (Fig. 1D). The unpaired cysteine within CRD4 is unique to h4-1BB, as all other known TNFRSF members lack any unpaired cysteines in their CRDs.
Figure 1.
Protein constructs, expression, and h4-1BB structure. A and B, domain architecture of h4-1BB (A) and h4-1BBL (B). Sig p, signal peptide; TM, transmembrane region; CR, cytoplasmic region. The unpaired cysteine 121 and two potential N-linked glycosylation sites of h4-1BB are indicated. C, size exclusion profile of purified wildtype (WT) (blue line) and C121S mutant (green line) of h4-1BB with reference to molecular mass marker proteins (red line) in kDa. D, SDS-PAGE analysis of purified WT and C121S mutant of h4-1BB under nonreducing (−β-ME (lanes 1, 3, and 5) and reducing conditions (+β-ME (lanes 2, 4, and 6). Lanes 1 and 2 contain dimeric WT 4-1BB (blue high Mr peak from C) and lanes 3 and 4 contain monomeric WT 4-1BB (blue low Mr peak from C). Lanes 5 and 6 contain the C121S mutant 4-1BB (green low Mr peak from C). Lane 7 represents deglycosylated (PNGase (peptide N-glycosidase F) treated) WT 4-1BB. E, crystal structure of h4-1BB (in the 4-1BB–4-1BBL complex) colored by the four cysteine-rich domains as cartoon overlaid onto transparent molecular surface representation. Unpaired Cys121 and disulfide bridges are shown as sticks in respective colors of CRDs. The N-linked glycosylation site and the N-glycans are represented as sticks with carbon atoms in green, oxygen in red, and nitrogen atoms in blue.
For structural studies, the TNF homology domain of h4-1BBL including the C-terminal end of the tail region (residues 80–244) was either co-expressed with the extracellular region of 4-1BB (residues 24–160) or expressed by itself with a C-terminal His6 tag using the baculovirus expression system. Unbound h4-1BBL and the complex was purified similar to h4-1BB as described above. We have obtained crystals of unbound 4-1BBL in the H32 space group, the WT h4-1BBL/h4-1BB complex in the C2 space group, and the complex containing the C121S mutant of h4-1BB in space group P21. The structures were determined by molecular replacement. Data collection and refinement statistics for the different crystals are listed in Table 1. The asymmetric unit (ASU) of the crystals belonging to the C2 space group contained the minimal signaling unit composed of three 4-1BB monomers bound to one trimeric 4-1BBL, whereas the crystals of the complex with the C121S mutant version of h4-1BB contained two heterohexameric complexes. The ASU of unbound h4-1BBL crystals contained one molecule of h4-1BBL.
Table 1.
Data collection and refinement statistics
| h4-1BBL | h4-1BB–4-1BBL complex | h4-1BB (C121S)–4-1BBL complex | |
|---|---|---|---|
| PDB ID | 6D3N | 6CPR | 6CU0 |
| Data collection statistics | |||
| Space group | H32 | C2 | P21 |
| Cell dimension | |||
| a, b, c, (Å) | 73.16, 73.16, 162.8 | 115.3, 66.5, 129.4 | 72.9, 114.7, 126.0 |
| α, β, γ (°) | 90.00, 90.00, 120.00 | 90.00, 103.09, 90.00 | 90.00, 101.24, 90.00 |
| Resolution range (Å) (outer shell) | 59.0–2.70 (2.83–2.70) | 38.0–2.70 (2.80–2.70) | 50.0–3.20 (3.31–3.20) |
| No. of unique reflections | 4,818 (630) | 25,892 (2,513) | 32,003 (3,027) |
| Rmeas (%) | 9.3 (83.2) | 14.3 (89.6) | 9.7 (62.8) |
| Rpim (%) | 3.3 (30.3) | 6.2 (42.3) | 4.5 (31.1) |
| Multiplicity | 9.2 (8.4) | 5.7 (5.0) | 4.2 (3.5) |
| Average I/σI | 11.6 (3.6) | 7.0 (1.7) | 20.3 (1.8) |
| Completeness (%) | 99.6 (99.4) | 97.8 (95.7) | 94.9 (89.5) |
| Refinement statistics | |||
| No. atoms | 1,017 | 6,257 | 11,476 |
| Protein | 1,001 | 6,022 | 11,404 |
| Ligand | 0 | 42 | 28 |
| Water | 10 | 148 | 44 |
| Glycerol/Na/Cl/sulfate | 6 | 45 | |
| Ramachandran plot (%) | |||
| Favored | 95.3 | 96.2 | 94.8 |
| Allowed | 3.8 | 3.7 | 5.1 |
| Outliers | 0.7 | 0.1 | 0.1 |
| R.m.s. deviations | |||
| Bonds (Å) | 0.008 | 0.009 | 0.009 |
| Angles (°) | 1.26 | 1.34 | 1.33 |
| B-factors (Å2) | |||
| Protein | 61.8 | 57.2 | 128 |
| Ligand | 47.2 | 186 | |
| Water | 56.7 | 53.3 | 30 |
| Glycerol/Na/Cl/sulfate | 18.3 | 66.4 | |
| R factor (%) | 20.7 | 24.8 | 24.7 |
| Rfree (%) | 25.2 | 27.9 | 29.5 |
Structure of h4-1BB
The interactions between 4-1BB and 4-1BBL are conserved in the different complexes and we will describe the structure for the complex containing WT h4-1BB, which has been refined to higher resolution. In the final structure, the N-terminal residues (Asp80–Gln89) of h4-1BBL were disordered. As expected, the three copies of 4-1BB and 4-1BBL were highly similar in structure and superimpose with root mean square deviation values of less than 0.5 Å.
Each h4-1BB monomer folded into a long and elongated molecule with the four CRDs arranged in a linear fashion. The ectodomain of h4-1BB contains a total of 21 cysteine residues in which 20 cysteine residues form 10 intra-disulfide linkages to maintain the structural and functional integrity of the protein (Fig. 1E). The additional cysteine at position 121 in CRD4 formed an inter-molecular disulfide bond between all three h4-1BB monomers of one ASU with the three symmetry-related molecules of another ASU, effectively cross-linking two different ASUs (Fig. S1, A and B). The complex containing the C121S mutant of h4-1BB lacks this intermolecular cross-linking of two heterohexameric units (Fig. S1C). The structural data of h4-1BB validated our biochemical data supporting the presence of covalent dimers of 4-1BB in solution. The structure also showed ordered electron density for N-linked glycans at position Asn149 of h4-1BB in all three monomers. The overall structure of h4-1BB in the complex is identical to that of the recently solved m4-1BB structure (11), except for CRD1 and part of CRD2, which deviate between both species (Fig. S2A). Although both h4-1BB and m4-1BB share 60% sequence similarity, CRD3 and CRD4 are almost identical in topology, whereas CRD1 is the most divergent (r.m.s. deviation of 1.49 Å between 115 Cα atoms from both receptors). However, all 10 disulfide linkages are conserved between m4-1BB and h4-1BB. The CRD1–CRD4 regions of h4-1BB adopt the so-called B2, A1–B2, A2, and A1–B1 modules, where A and B represent the type of module and the numbers denote the intradisulfide linkages in the module. Similar to m4-1BB, h4-1BB also contains a partial CRD1 and lacks the canonical disulfide linkage that is present in all other TNFR members. Additionally, analogous to m4-1BB, h4-1BB also has a bend in the central hinge region that connects CRD2 and CRD3 changing the relative orientation of its CRD3 and CRD4. Hence, h4-1BB is similar to other TNFR members only at its CRD2 region (Fig. S2B).
Structure of h4-1BBL
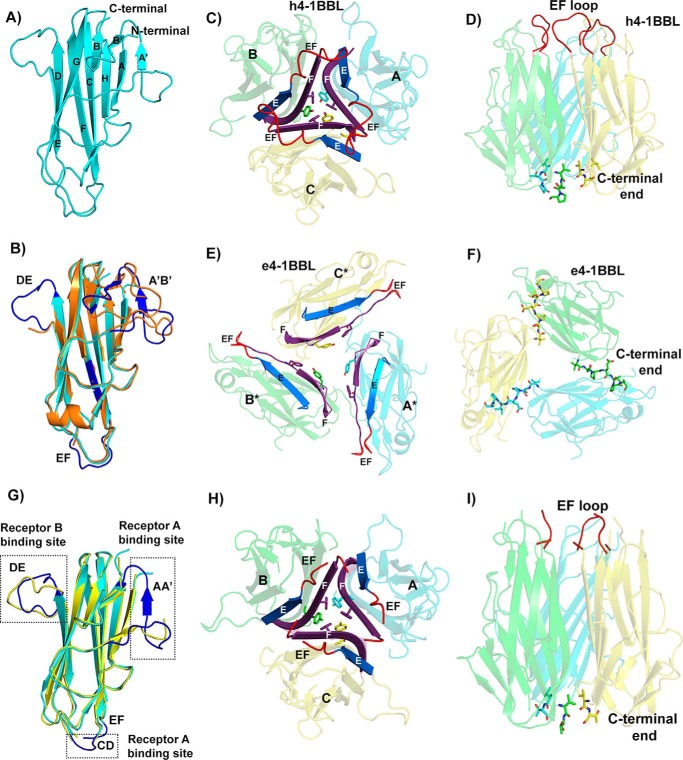
The THD region of h4-1BBL shares only 20–25% of sequence identity with other known TNF ligands, however, the overall structure of h4-1BBL is similar to other noncovalent homotrimeric TNF ligands. This is in contrast to murine 4-1BBL, which we previously reported to form a disulfide-linked homodimer (11). Each protomer of h4-1BBL adopts a jellyroll-fold composed of inner and outer β sheets. The inner sheet is formed by β strands A, A′, H, C, and F, whereas the outer sheet is formed by B′, B, G, D, and E strands (Fig. 2A). The core residues that form these β strands are conserved among TNF ligands, which is largely confined to internal aromatic residues of assembled monomers, however, they exhibit variation in the loops connecting the β strands. Superposition of a single h4-1BBL protomer with a protomer of the previously published crystal structure of unbound h4-1BBL (e4-1BBL; PDB 2X29) resulted in an r.m.s. deviation value of 0.56 Å between 103 Cα atoms. The core of h4-1BBL formed by inner and outer β sheets are very similar, however, the F strand, DE and EF loops are disordered in the e4-1BBL structure (24) (Fig. 2B). In addition, the two N-terminal strands A′ and B′ of h4-1BBL in the complex are replaced by a long AB loop in the e4-1BBL structure. Both the N-terminal region (AA′ loop) as well as the DE loop of h4-1BBL are in contact with the receptor, which likely limits their flexibility and gives rise to an ordered confirmation.
Figure 2.
Structure of h4-1BB ligand in the h4-1BB–4-1BBL complex. A, cartoon representation of the THD region of the human 4-1BBL monomer showing the classical jellyroll-fold with inner and outer β sheets labeled consecutively. B, superposition of h4-1BBL of the complex (cyan color) with e4-1BBL (orange color) showing significant structural ordering of DE, EF, and A′B′ loops (blue color) in h4-1BBL when bound to 4-1BB. C, h4-1BBL homotrimer composed of three protomers (A, B and C) is illustrated as transparent cartoon with cyan, green, and yellow colors, respectively. Trimerization interface at the middle portion of h4-1BBL, showing packing of E (blue) and F (purple) strands against each other in the homotrimer. The tyrosine and phenylalanine residues that form the inner hydrophobic core are represented as sticks. The ordered EF loop that connects strands E and F is shown in red color. D, stabilization of h4-1BBL trimer at the upper and lower regions by residues coming from the EF loop (red color) and C-terminal end (shown as sticks). E, cartoon rendering of trimeric e4-1BBL colored and labeled similarly to C. For clear distinction between h4-1BBL and e4-1BBL, the protomers of the later are marked with *. F, trimerization interface in e4-1BBL. C-terminal residues of one protomer interacting with adjacent protomer are indicated as sticks. G, superposition of h4-1BBL of the complex (cyan color) with unbound h4-1BBL produced in insect cells (yellow color). The structurally ordered loops in the h4-1BBL upon binding to the receptor are highlighted in blue color and the loops are indicated. H, cartoon representation of trimeric unbound h4-1BBL represented similarly to C. I, conventional bell-shaped arrangement of unbound h4-1BBL produced from insect cells, which are stabilized by residues from the N-terminal and C-terminal ends (shown as sticks) with EF loops colored in red.
Although the overall jellyroll-fold of an individual protomer of e4-1BBL appears similar to that of the h4-1BBL protomer in the complex, the superimposition of trimeric e4-1BBL with trimeric h4-1BBL results in large r.m.s. deviation values between Cα atoms (24 Å) (Fig. S3). The most striking feature is that the trimeric architecture of h4-1BBL in our complex emerges as a conventional bell-shaped arrangement rather than the extended three-bladed propeller structure observed in the e4-1BBL structure (Fig. 2, C–F, Fig. S3, A and B). In e4-1BBL, the individual protomers are spread out and do not form a typical trimerization interface but rather form a ring-like structure of three monomers, in which each protomer contacts the neighboring protomer linked as a daisy chain (Fig. 2, E and F).
Because these differences between both h4-1BBL structures were unexpected and difficult to rationalize, we determined the crystal structure of unbound h4-1BBL produced in the same insect cell expression system. As expected from our complex structure, insect cell-expressed h4-1BBL forms the identical bell-shaped trimeric arrangement found in the complex (Fig. 2, G–I, Fig. S3D). The only structural differences are localized in the EF loop region and the three receptor-binding sites, which are mostly disordered in the unbound h4-1BBL structure, likely due to the flexible nature of these loops (Fig. 2G).
Our unbound h4-1BBL structure is much more tightly packed with ∼45 residues involved in the interface rather than the smaller contact surface formed by 14 residues in e4-1BBL (24). Hence, the total buried surface area of h4-1BBL is now 4137 Å2 compared with 2934 Å2 in e4-1BBL. In our h4-1BBL structure, the trimer is arranged such that residues from the EF loop and the N-terminal and C-terminal portion stabilize the upper and lower part of the trimer, whereas the E and F strands of one protomer packs against the same of the adjacent protomers to stringently assemble the interior portion (Fig. 2, C, D, H, and I). Two hydrophobic residues, Tyr142 and Phe199, of strands C and F form a hydrophobic core that helps to maintain the core integrity of the trimer. Previous data showed that mutation of both of these residues to alanine and the removal of the C-terminal region of h4-1BBL impede the formation of trimers (24) signifying the importance of the hydrophobic core in upholding the bell-shaped architecture.
Interface of h4-1BBL–h4-1BB complex
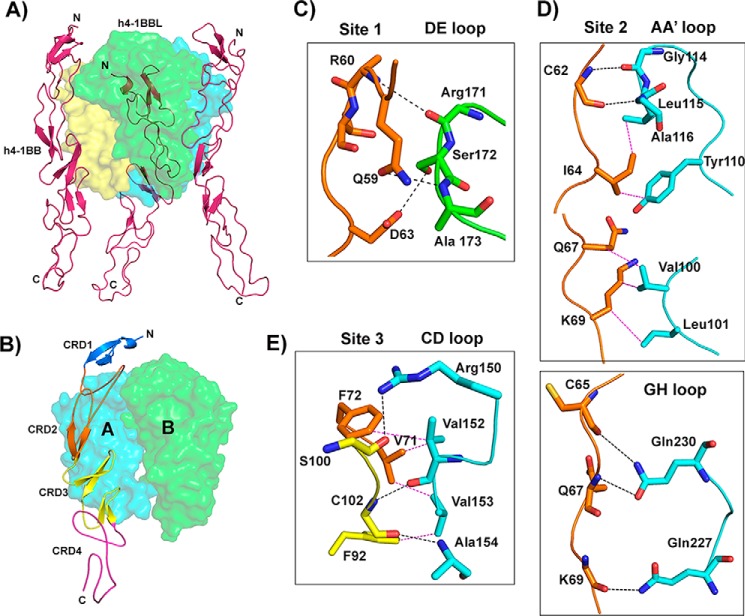
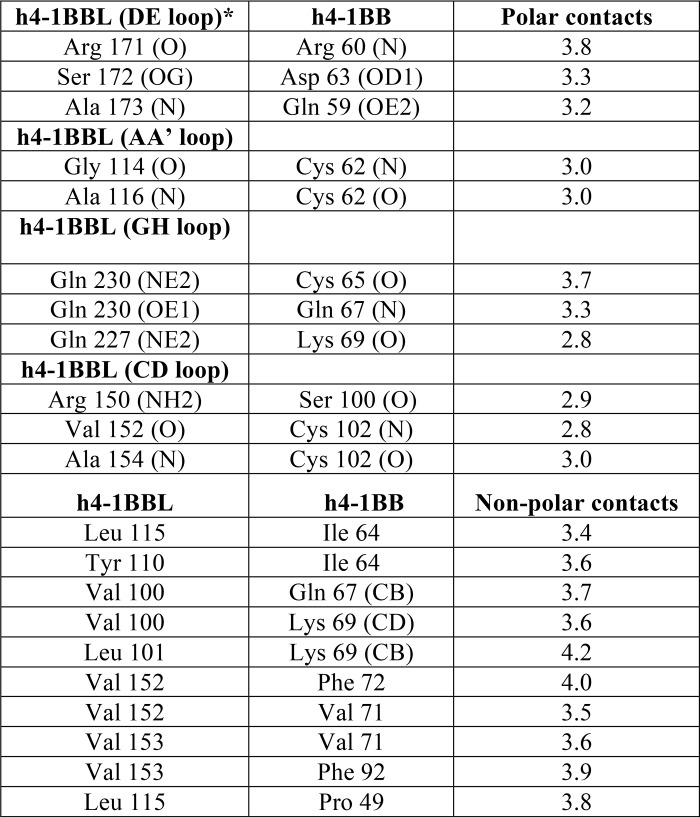
h4-1BBL forms a homotrimer around which three molecules of h4-1BB bind to form a hexameric complex similar to that observed in other TNF–TNFR complexes (Fig. 3A). The extracellular domain of h4-1BB binds at the interface formed by two adjacent protomers (protomer A and protomer B) of h4-1BBL and thereby interacts with two protomers simultaneously (Fig. 3B). Many of the TNF receptors e.g. TNFR1, TNFR2, RANK, and DR5, use CRD2 and CRD3 to interact with their respective ligands (25–28), whereas OX40 uses CRD1, CRD2, and CRD3 to contact OX40L (23). Our crystal structure showed that CRD1 of h4-1BB does not interact with its ligand and most of the contact area is covered by CRD2 and partially by CRD3. In addition, h4-1BBL uses its AA′, CD, DE, and GH loops to contact its cognate receptor rather than the β strands (Fig. S4). We divided the binding interface into three major binding sites. Site 1 corresponds to the interaction of the A1 module of CRD2 with the DE loop of one ligand protomer (protomer B); site 2 relates to the interactions of the long C loop of CRD2 containing the B2 module with the AA′, and GH loops of another ligand (protomer A); whereas site 3 describes the interactions between the A2 module of CRD3 with the CD loop of this second ligand protomer (protomer A) (Fig. 3, Table 2).
Figure 3.
Crystal structure of the h4-1BB–h4-1BBL complex and binding interface. A, asymmetric unit representing the functional hexameric complex with a h4-1BBL trimer surrounded by three h4-1BB receptors. h4-1BBL is illustrated as a transparent surface in green, cyan, and yellow colors; h4-1BB is shown as a magenta cartoon. B, binding site for each receptor formed by two adjacent protomers of h4-1BBL (surface). Receptor is shown as a cartoon and each CRD is colored separately and marked. C–E, interactions between: residues of the A1 module of h4-1BB CRD2 with the DE loop of h4-1BBL protomer B (C); the long C loop and B2 module of CRD2 (orange) with the AA′ and GH loop of h4-1BBL protomer A (D); and the A2 module of CRD3 with the CD loop of h4-1BBL protomer A (E). In all figures, protomer A is shown in cyan and protomer B in green. All interacting residues are shown as sticks, with CRD2 residues in orange and CRD3 residues in yellow. Hydrogen bonds are represented as black and van der Waals contacts as magenta dashed lines. In C, D, and E, residues of 4-1BB are labeled as single letter amino acids and those of 4-1BBL are marked as three-letter amino acids.
Table 2.
Interactions between h4–1BBL and h4–1BB
Binding of h4-1BB to 4-1BBL occurs predominantly through polar and hydrophobic interactions at all three sites that play a key role in the complex interface, whereas charged interactions (e.g. salt-bridges) are absent. The interface area is dominated by hydrogen bonding interactions between main chain atoms of most of the residues from both ligand and receptor. Only four residues from h4-1BBL (Arg150 of the CD loop, Ser172 of the DE loop, and Gln227 and Gln230 of the GH loop) and two residues from h4-1BB (Gln59 and Asp63 of CRD2) mediate hydrogen-bonding contacts using their side chain atoms (Fig. 3). Apart from these polar interactions, a hydrophobic core formed between residues Tyr110, Leu115, and Ala116 of the AA′ loop and Val152, and Val153 of the CD loop of h4-1BBL with Ile64, Val71, Phe72, and Phe92 from the central region of h4-1BB participates in the complex formation (Fig. 3E). At site 1, Ser172 of h4-1BBL appears to be important as it engages Asp63 of CRD2 of h4-1BB via side chain hydrogen bonding interactions (Fig. 3C). However, earlier mutational data showed that mutation of this Ser172 to Gly172 did not affect the binding affinity toward the receptor (24). Considering that interaction at sites 2 and 3 mostly form a wide polar and hydrophobic patch on protomer A, it can be rationalized that a single point mutation in h4-1BBL is not always sufficient to significantly affect the binding affinity.
Comparison to other TNF–TNFR complexes
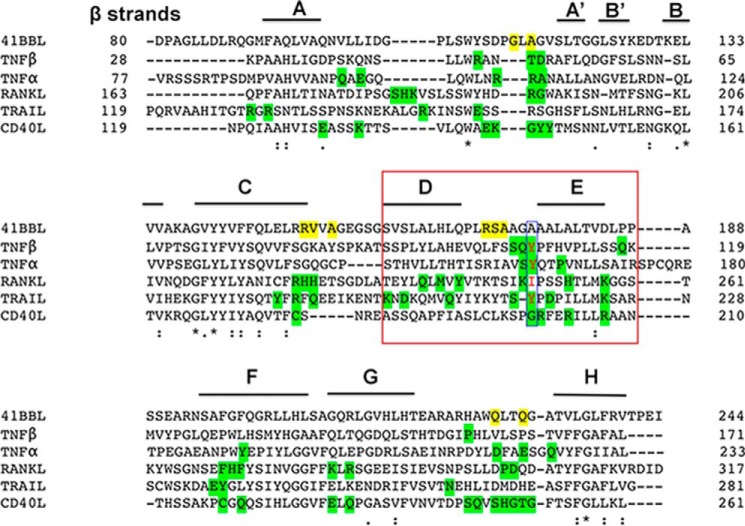
Until now the crystal structures of conventional family ligands (TNF, RANKL, and CD40L) revealed a compact bell-shaped trimeric architecture (26, 27, 29), whereas divergent family members (OX40L, GITRL, and 4-1BBL) display a more planar blooming flower-shaped structure (21, 23, 24). The bell-shaped trimers have very long CD, DE, and EF loops connecting the long C, D, E, and F strands, allowing one edge of each subunit to pack against an adjacent subunit to conserve the rigid pyramidal shape of the trimer (20). Based on its sequence (Fig. 4), h4-1BBL was proposed to belong to the divergent family (19, 20). However, it also possesses a significantly longer THD region (residues 93–254) than those found in divergent members (120 residues). This is similar to TNF, RANKL, and CD40L that also have ∼160 residues. Compared with OX40L and GITRL, which possess shorter β strands in both their inner (C and F strands) and outer (D and E strands) faces and shorter CD, DE, and EF loops (21, 23), h4-1BBL has relatively long loops connecting the long C-D, D-E, and E-F strands. Hence, the inter-protomer contacts that are missing in the upper region of the OX40L and GITRL trimers are restored in h4-1BB by long loops and β strands. Conversely, the h4-1BBL trimer adopts a bell-shaped architecture that can be comparable with conventional ligands rather than being in an extended planar shape as seen in the divergent members. Supporting this, the alignment of trimeric h4-1BBL with other conventional TNF ligands results in r.m.s. deviation of 2–3 Å between Cα atoms, with major conformational changes focused on the loops connecting the β strands while exhibiting enormous variation with trimeric OX40L and GITRL (r.m.s. deviation ∼ 20 Å) (Fig. S5).
Figure 4.
Sequence alignment of the THD domain of h4-1BBL and representatives of conventional TNFSF members. β Strands present in h4-1BBL are labeled. The residues that are forming hydrogen bond interactions with h4-1BB are shaded yellow for h4-1BBL and green for other conventional TNFSF members. The conserved hydrophobic residue (tyrosine) present in the DE loop of conventional members is highlighted with a blue box. The residues of the D and E strands of protomer B that are making contacts with h4-1BB are highlighted in the red box.
The structure of h4-1BB is also slightly distorted from other TNFR members. Although the overall topology of h4-1BB is similar, superposition of h4-1BB from the complex with TNFR1, TNFR2, RANK, CD40, and OX40 result in r.m.s. deviation values ranging from 4 to 8 Å. (Fig. S2B). This large variation arises due to the bend in the beginning of CRD3 at the hinge region, thereby altering the orientation of CRD3 and CRD4 of h4-1BB compared with CRD1 and CRD2. The degree of bending deviates substantially compared with other TNFR structures. In many TNFRs, the hinge region affords freedom of rotation to CRD3 with respect to CRD2 and the central twisting here plays a significant role to promote the tight interaction with cognate ligands. For instance, in the RANKL–OPG complex, the central turn at the hinge region favors the CRD2 and CRD3 of OPG to reach the binding interface provided by two RANKL molecules thereby engaging with both simultaneously (26, 30).
Because the trimeric arrangement of h4-1BBL and the general topological fold of h4-1BB are analogous to conventional TNF/TNFR members, one might expect a similar mode of interaction between h4-1BBL and h4-1BB compared with other TNF–TNFR complexes. However, the considerable structural changes and sequence diversity of h4-1BBL at its loop regions appears to impact the overall shape of the interaction interface. Our structure shows that h4-1BBL employs a somewhat different approach to engage its receptor while maintaining the canonical mode of receptor recognition. First, the residues forming the interface area between ligand and receptor are different compared with existing TNF–TNFR complexes (25, 29). h4-1BBL employs only 14 residues, whereas other TNF ligands recruit ∼30 residues in the interface. The interaction surfaces of most TNF ligands (e.g. TNF, CD40L, and RANKL) and TNF receptors (e.g. TNFR2, CD40, and OPG) exhibit clear charge complementarity and hence the electrostatic interactions predominate to form the complex (Fig. 4). For instance, the positively charged residues of CD40L (His249, Arg203, and Arg207) and negatively charged residues of CD40 (Glu74, Asp84, and Glu117) interact to form the CD40–CD40L complex and mutation of these residues prevent the formation of the complex and receptor activation (29). Similarly, Arg113 and Arg77 of TNFR2 are important residues to form salt bridges with Asp219, Gln225, and Glu99 of TNF (25). In contrast, in the h4-1BBL–h4-1BB complex, most of the interface area is neutral and nonpolar, hence the hydrophobic and polar contacts drive the formation of this complex.
Another striking difference between h4-1BBL and other conventional ligands is among the residues of the DE loop region. In most of the conventional ligands (e.g. TNF, TRAIL, and TL1A) conserved hydrophobic residues (importantly tyrosine) in the DE loop (Fig. 4) are found to be energetically important for receptor binding (23, 27, 28, 31) and in the absence of tyrosine (as seen in RANKL and CD40L) its role was taken over by subsequent charged residues that form salt bridges with the receptor to provide sufficient binding energy (29, 30, 32). In the case of h4-1BBL, the DE loop lacks the conserved tyrosine residue and our interface analysis showed that Arg171, Ala173 (main chain atoms), and Ser172 (side chain) are making hydrogen-bonding contacts with the receptor (Fig. 3C). As mutation of both Arg171 and Ser172 did not affect the binding affinity, we can propose that the DE loop residues are not contributing energetically toward binding to the receptor. Similar to h4-1BBL, OX40L also lacks hydrophobic contacts contributed by the DE loop and only makes one contact with Glu123 that is shown dynamically not to be important for binding to its receptor (23).
Discussion
In this study, we have determined the crystal structure of unbound h4-1BBL and h4-1BB–4-1BBL complexes. The most surprising finding was that the structural arrangement of trimeric h4-1BBL in the complex as a conventional bell shape differs strongly compared with the previously published crystal structure of unbound e4-1BBL, which was described to resemble a three-bladed propeller and was somewhat similar to the type of blooming flower shape seen in divergent TNF ligands like OX40L and GITRL (24). However, the structure of unbound h4-1BBL is identical to that found in the complex, when both proteins are produced in the same expression system. Therefore, it is questionable whether a three-bladed propeller structure truly exists in naturally occurring TNF ligands.
The appearance of the planar blooming flower structure might be predictable. In trimeric hOX40L and hGITRL, both proteins have a short THD and short loops and β strands (20, 23). In contrast, h4-1BBL has an elongated THD region, similar to conventional TNF family members, which would favor a bell-shaped structure. Why unbound e4-1BBL then adopted the three-bladed propeller shape is not clear. It is possible that the manner by which e4-1BBL was expressed in Escherichia coli influenced its shape. Because the features that form the conventional bell-shaped trimer are disordered in the e4-1BBL structure, the E. coli-derived protein may not have been fully intact during crystallization or proteolytic cleavage of the protein could have contributed to the reported structure. Furthermore, the e4-1BBL construct used for crystallization was 20 residues longer than that used in this report and this may have formed a long disordered N-terminal tail upstream of the TNF homology domain that also could have affected the structure of e4-1BBL.
Previously published mutagenesis data of h4-1BBL identified possible key residues of h4-1BBL that control ligand trimerization or binding to h4-1BB (24). Leu115, which mediates contacts between e4-1BBL protomers is part of the h4-1BBL–h4-1BB binding interface and the mutation L115G indeed reduces binding to h4-1BB. Alanine scanning mutagenesis of Gln227 and Gln230 of h4-1BBL also decreased the binding affinity toward h4-1BB. In the complex structure both Gln227 and Gln230 of the GH loop use their side chains to form hydrogen bond interactions with h4-1BB (Fig. 3D) and hence disruption of this interaction reduces binding affinity.
Most of the TNF–TNFR complexes exhibit a 3:3 ratio of stoichiometric binding between ligand and receptor, in which each receptor binds at the crevice formed by two adjacent protomers of the ligand and the interaction patch of the receptor is almost uniformly distributed on both subunits. In contrast, the crystal structure of the h4-1BB–h4-1BBL complex revealed that each receptor predominantly binds to one ligand protomer (protomer A) with an extensive interface area (800 Å2) while contacting the adjacent subunit (protomer B) with an almost four times reduced interface area (200 Å2) (Fig. 5). The asymmetric nature of the binding of h4-1BB to predominantly one 4-1BBL protomer was also observed by EM using e4-1BBL (24). Our results show that CRD2 and CRD3 of h4-1BB interact with the ligand. A major role of CRD2 and CRD3 in binding to h4-1BBL has been reported previously using truncated h4-1BB molecules (35). The uneven contribution of both protomers of h4-1BBL in binding to 4-1BB mainly arises because the major interactions involving the second half of CRD2 and CRD3 are formed exclusively by protomer A, whereas interactions with the N-terminal part of CRD2 are shared between both protomers. We have noted structural differences in h4-1BB compared with other TNFRs that likely account for its lack of interactions with protomer B of h4-1BBL. In many TNFRs (e.g. TNFR1, TNFR2, and RANK), the twist in the central hinge region pulls the CRD3–4 region forward at an angle of ∼45 degrees with respect to the crystallographic axis so that CRD3 can come near to ligand subunit B. In contrast, in h4-1BB, these regions are pulled forward by only ∼20 degrees, hence making it difficult for CRD3 to reach the second ligand protomer (Fig. S6).
Figure 5.
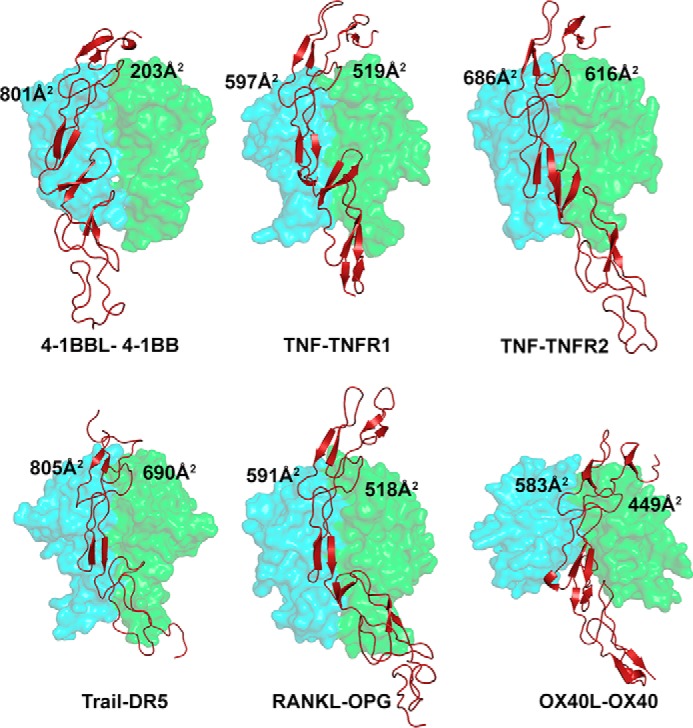
Binding site of various TNF receptors at the interface formed between two protomers of their cognate ligands. The receptor contact area with each protomer of each ligand is depicted in all the indicated complexes. The ligand protomers A and B are shown as a cyan and green transparent surface and the receptor as a brick red cartoon. PDB codes of crystal structures of TNF–TNFR complexes are as follows: TNF–TNFR1, 1TNR; TNF–TNFR2, 3ALQ; TRAIL–DR5, 1D4V; OX40L–OX40, 2HEV; RANKL–OPG, 3URF; all figures were made in PyMOL (33) (Schrödinger).
Our biochemical and structural analysis also revealed that the extracellular region of h4-1BB can form a covalent dimer in solution mediated by Cys121. h4-1BB was co-immunoprecipitated as a dimer and tetramer, as well as monomer, from a homogenous population of T cells (clone F-1), suggesting that the majority of 4-1BB on the cell surface may indeed exist as a dimer (3, 8). Dimerization is not uncommon among cell-surface receptors (e.g. insulin receptor, the B cell-surface Ig receptor, the T cell Ag receptor, the CD28 costimulatory receptor) and has been suggested to provide a mechanism to allow effective signaling (3, 37, 38), although the manner by which dimerization occurs may differ. h4-1BB has a unique cysteine in CRD4, but other TNFR, such as TRAIL-R1 or OPG have additional cysteines outside their CRDs, close to the transmembrane domain. As such, this mode of receptor dimerization may not be limited to h4-1BB. Our structure additionally showed that each monomer of dimeric 4-1BB could bind to two different 4-1BBL trimers. This implies that when 4-1BBL engages 4-1BB on a cells surface, it could result in cross-linking of individual ligand–receptor complexes together to form a 2D network, which would then induce multimerization of 4-1BB monomers and allow the generation of a strong intracellular signal. Other TNFRSF molecules may utilize other variants of this mechanism to generate strong signaling complexes. TNFR1 and -2, CD40, and the TRAIL receptors can form noncovalent dimers via a region termed the PLAD (8, 39), which is present within CRD1 of these receptors. This PLAD was described to drive the formation of homotypic, ligand-independent receptor complexes, and the appearance of higher order multimers was also suggested to occur after ligand binding resulting in enhanced downstream signaling. Analogous to this, Galectin-9 can additionally dimerize m4-1BB, via bridging CRD4 of two adjacent monomers, representing another way to facilitate the formation of pre-assembled 4-1BB complexes that further get cross-linked by their ligand (10).
In summary, the structure of 4-1BB–4-1BBL revealed several interesting features. First, that the trimeric arrangement of 4-1BBL is comparable with that seen for conventional TNF family ligands when complexed with their receptors. Second, that covalent dimerization of the receptor can readily occur and that this can result in the engagement of two 4-1BBL trimers. Our results provide significant insight into aggregation of 4-1BB signaling units and how signal strength can potentially be tuned in response to cellular stress or other factors to modulate the activity of this receptor.
Experimental procedures
Design of h4-1BB and h4-1BBL constructs
The h4-1BB extracellular region containing four cysteine-rich domains (CRD 1–4; amino acids 24–160) and the TNF homology domain of human 4-1BBL (including the C-terminal end of the tail region spanning amino acids 80–244) were produced using a Baculovirus expression system. The corresponding DNA fragments of h4-1BB and 4-1BBL were both amplified by PCR and cloned downstream of the gp67 secretion signal sequence into separate baculovirus transfer vectors (pAcGP67A). A C-terminal His6 tag was introduced downstream of h4-1BB to assist in its purification, whereas no tag was added to 4-1BBL. For structural studies of unbound h4-1BBL, a His6 tag was added at the C terminus and the protein was expressed by itself. To generate the h4-1BB C121S construct, the cysteine at position 121 of h4-1BB was exchanged for serine by site-directed mutagenesis using the Quick Change II Multi-site Mutagenesis Kit (Stratagene, La Jolla, CA). The identity and correct sequence of all the clones were confirmed by sequencing.
Preparation of recombinant baculovirus
The recombinant baculovirus transfer vector containing either h4-1BBL or WT or the C121S mutant of 4-1BB was transfected separately under aseptic conditions using BacPAK6DNA according to the manufacturer's protocol. To obtain recombinant virus, first 2 × 106 healthy dividing Spodoptera frugiperda (Sf) 9 cells were seeded into T-25 flasks and incubated for 15 min at 27 °C. In parallel, the transfection mixture was prepared by gently mixing 1 μg of transfer vector containing the target gene, 5 μl of BacPAK6DNA, 4 μl of Bacfectin reagent, in 100 μl of serum-free media without any antibiotics, and incubated for 15 min in the dark. As a control, the transfection mixture without the BacPAK6DNA was used. Both mixtures were added separately to the seeded Sf9 cells and grown for 7 days at 27 °C in serum-free medium containing antibiotics (100 units/ml of penicillin and 100 μg/ml of streptomycin). The recombinant virus was collected by centrifugation at 1000 × g for 10 min and then used for a first round of virus amplification. 500 μl of virus with a multiplicity of infection below 1 (m.o.i. <1) were used to infect 2 × 106 Sf9 cells in a total volume of 5 ml in T-25 flasks incubated at 27 °C. After 5 days, the virus was collected (m.o.i. < 1) and 1.5 ml was used to infect 14 × 106 Sf9 cells in T-175 flasks incubated for 5 days at 27 °C. The virus titer was determined by an end-point dilution assay. Prior to protein expression, 1 ml of low titer second round amplified recombinant virus (m.o.i. = 1) was infected in 14 × 106 Sf9 cells in T-175 flasks to obtain high titer virus over 5 days by incubating at 27 °C. For protein production, several individual 2-liter Erlenmeyer flasks seeded with 2 × 106 Sf9 cells/ml were infected with high titer recombinant virus stock having m.o.i. values ranging from 3 to 5. The flasks were incubated at 27 °C by shaking at 145 rpm for 72–84 h and the protein was collected from the supernatant by centrifugation for 10 min at 1000 × g.
Expression of h4-1BBL, h4-1BB–4-1BBL, and h4-1BB (C121S)–4-1BBL complex
For crystallization studies, the ectodomain of WT h4-1BB and h4-1BB (C121S) were separately co-expressed with the THD region of h4-1BBL in Sf9 insect cells. h4-1BBL was also expressed by itself. The recombinant viral stock for h4-1BB–h4-1BBL complexes was also prepared in a similar way to individual stocks mentioned above. Equal concentrations (2 μg) of WT h4-1BB or h4-1BB (C121S) containing transfer vector and h4-1BBL transfer vector (both in separate pAcGp67A vectors) were mixed with 0.5 μg of BacPAK6DNA and 5 μl of Bacfectin reagent in a total volume of 100 μl, and transfected as reported above. For protein expression of the h4-1BB–4-1BBL complex, 2.5 × 106 Sf9 cells/ml were seeded in 2-liter Erlenmeyer flasks and infected with high titer recombinant virus stock (made from low titer virus m.o.i. = 1 of second virus amplification) and grown as suspension cultures (135 rpm) at 27 °C for 3.5 days and collected from the supernatant by centrifugation.
Protein purification of h4-1BBL, h4-1BB, and h4-1BB–4-1BBL complex
For protein purification, the cell supernatants were further centrifuged at maximum speed to remove any cell debris and then buffer exchanged against 1× PBS by tangential flow filtration using 10-kDa molecular mass cut-off membranes (PALL). 5 ml of settled nickel-nitrilotriacetic acid resin was added to the ∼0.5-liter supernatants and slowly stirred overnight at 4 °C. Next, the beads were transferred to Econo columns (Bio-Rad) and washed with 20 mm imidazole (in 50 mm Tris-HCl, 300 mm NaCl, pH 8.0) to remove unwanted impurities. The individual h4-1BBL, h4-1BB, or h4-1BB–4-1BBL complexes were eluted with 250 mm imidazole (in 50 mm Tris-HCl, 300 mm NaCl, pH 8.0). The proteins were further purified by size exclusion chromatography using Superdex S200 columns. The native h4-1BB and h4-1BB (C121S) mutant proteins were used for biochemical analyses, whereas unbound h4-1BBL, and h4-1BB/4-1BBL and h4-1BB (C121S)–4-1BBL complexes were used for crystallization.
Crystallization of h4-1BBL and h4-1BB–4-1BBL and h4-1BB (C121S)–4-1BBL complexes
Purified h4-1BBL and co-purified h4-1BB–4-1BBL and h4-1BB (C121S)–4-1BBL complexes were concentrated to 10 mg/ml and subjected to crystallization using the sitting drop vapor diffusion method. Initial crystallization trials were performed in a 96-well format using a nanoliter dispensing liquid handling robot (Phenix, Art Robbins Ltd.) while mixing 0.3 μl of protein with 0.3 μl of precipitant from different commercially available crystallization screens (JCSG core+ and JCSG core 1–4 screens). Over 800 crystallization conditions were tested and crystallization trails were carried out at both 4 and 22 °C. Optimization of all crystallization conditions was performed manually by both hanging drop and sitting drop methods. Crystals were obtained in various conditions containing ammonium sulfate or PEG 6000 as common precipitant and were grown over 7 days. Among all conditions, only two conditions yielded high quality-diffraction crystals. The h4-1BB/4-1BBL crystals used for the X-ray diffraction experiment were grown over 10 days at 4 °C by equilibrating 1 μl of protein complex (10 mg/ml of h4-1BB–4-1BBL complex in 50 mm HEPES and 150 mm NaCl at pH 7.5) and 1 μl of reservoir solution (containing 0.1 m sodium acetate, pH 4.6, 10% PEG 4000, and 0.2 m ammonium sulfate) against 1 ml of reservoir solution. Crystals of the h4-1BB (C121S)–4-1BBL complex grown over 4 days at 22 °C in high pH buffer containing 0.1 m Bicine and 30% PEG 6000. The h4-1BBL crystals were grown in 0.085 m trisodium citrate, pH 5.6, 25.5% PEG 4000, 0.17 m ammonium acetate, and 15% glycerol. All crystals were flash-cooled in liquid nitrogen in their crystallization buffer containing either 20% glycerol or in a mixture of paratone oil and paraffin oil in 1:1 ratio for subsequent data collection.
Data collection and refinement
Native diffraction data for different crystals was collected remotely at Stanford Synchrotron Radiation Light Source (SSRL) beamline 9-2 using a PILATUS 6M PAD detector at a wavelength of 0.97 Å and a temperature of 100 K. Each image was collected with 0.15 degree oscillation and a 1.25–5-s exposure time. The collected images were processed and scaled using XDS package (40, 41) as implemented in the AUTOXDS script at SSRL (Ana Gonzalez) to an overall resolution of 2.7 Å for h4-1BBL, 2.7 Å for h4-1BB–4-1BBL complex, and 3.2 Å for h4-1BB (C121S)–4-1BBL complex. The position of 4-1BBL in the asymmetric unit was determined by using molecular replacement method PHASER-MR (42) based on the structural model of human 4-1BBL (PDB 2X29). Similarly, subsequent model building of h4-1BB is performed by obtaining phase information from the m4-1BB model (PDB 5WI8). Starting with initial phases obtained by MR, 4-1BB was built gradually by cycles of iterative model building and consequently 4-1BB was manually built into the Fo − Fc electron density map using COOT and ARP/wARP function (43–45) as part of the CCP4 suite (36) to complete the model building. At the final stages of refinement, water molecules were added automatically using the Refmac program. The final model of h4-1BBL, h4-1BB–4-1BBL complex, and the h4-1BB (C121S)–4-1BBL complex were refined in PHENIX/REFMAC (34) with residual factors R/Rfree = 20.7/25.2, 24.8/27.9, and 24.7/29.5, respectively. All three structures have good geometry with 2 residues (0.1–0.7%) as outliers and 96 and 95% of residues in the favored region of the Ramachandran plot. The data collection and refinement statistics are summarized in Table 1. All figures were made in PyMOL (33).
Author contributions
A. B., M. C., and D. M. Z. conceptualization; A. B. and T. D. data curation; A. B. and D. M. Z. validation; A. B. investigation; A. B. and T. D. methodology; A. B. and D. M. Z. writing-original draft; T. D. software; M. C. and D. M. Z. funding acquisition; M. C. and D. M. Z. project administration; M. C. and D. M. Z. writing-review and editing; D. M. Z. supervision.
Supplementary Material
Acknowledgments
We thank the Stanford Synchrotron Lightsource (SSRL) for access to remote data collection and the SSRL beamline scientists for support. Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by United States Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research and National Institutes of Health, NIGMS (including Grant P41GM103393).
This work was supported by NIAID, National Institutes of Health Grant AI110929 (to M. C. and D. M. Z.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S6.
The atomic coordinates and structure factors (codes 6CPR, 6CU0, and 6D3N) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- TNFR
- tumor necrosis factor receptor
- TNFRSF
- tumor necrosis factor receptor superfamily
- 4-1BBL
- 4-1BB ligand
- CRD
- cysteine-rich domain
- TRAF
- TNF receptor-associated factor
- THD
- TNF homology domain
- PLAD
- preligand assembly domain
- SEC
- size exclusion chromatography
- ASU
- asymmetric unit
- r.m.s. deviation
- root mean square deviation
- RANKL
- receptor activator of nuclear factor κB ligand
- OPG
- osteoprotegerin
- m.o.i.
- multiplicity of infection
- Bicine
- N,N-bis(2-hydroxyethyl)glycine
- PDB
- Protein Data Bank.
References
- 1. Locksley R. M., Killeen N., and Lenardo M. J. (2001) The TNF and TNF receptor superfamilies: integrating mammalian biology. Cell 104, 487–501 10.1016/S0092-8674(01)00237-9 [DOI] [PubMed] [Google Scholar]
- 2. Lee S.-W., and Croft M. (2009) 4-1BB as a therapeutic target for human disease. in Therapeutic Targets of the TNF Superfamily (Grewal I. S., ed) pp. 120–129, Springer; New York: [DOI] [PubMed] [Google Scholar]
- 3. Pollok K. E., Kim Y. J., Zhou Z., Hurtado J., Kim K. K., Pickard R. T., and Kwon B. S. (1993) Inducible T cell antigen 4-1BB: analysis of expression and function. J. Immunol. 150, 771–781 [PubMed] [Google Scholar]
- 4. Pollok K. E., Kim Y.-J., Hurtado J., Zhou Z., Kim K. K., and Kwon B. S. (1994) 4-1BB T-cell antigen binds to mature B cells and macrophages, and costimulates anti-μ-primed splenic B cells. Eur. J. Immunol. 24, 367–374 10.1002/eji.1830240215 [DOI] [PubMed] [Google Scholar]
- 5. Vinay D. S., and Kwon B. S. (1998) Role of 4-1BB in immune responses. Semin. Immunol. 10, 481–489 10.1006/smim.1998.0157 [DOI] [PubMed] [Google Scholar]
- 6. Wyzgol A., Müller N., Fick A., Munkel S., Grigoleit G. U., Pfizenmaier K., and Wajant H. (2009) Trimer stabilization, oligomerization, and antibody-mediated cell surface immobilization improve the activity of soluble trimers of CD27L, CD40L, 41BBL, and glucocorticoid-induced TNF receptor ligand. J. Immunol. 183, 1851–1861 10.4049/jimmunol.0802597 [DOI] [PubMed] [Google Scholar]
- 7. Snell L. M., Lin G. H., McPherson A. J., Moraes T. J., and Watts T. H. (2011) T-cell intrinsic effects of GITR and 4-1BB during viral infection and cancer immunotherapy. Immunol. Rev. 244, 197–217 10.1111/j.1600-065X.2011.01063.x [DOI] [PubMed] [Google Scholar]
- 8. Sanchez-Paulete A. R., Labiano S., Rodriguez-Ruiz M. E., Azpilikueta A., Etxeberria I., Bolaños E., Lang V., Rodriguez M., Aznar M. A., Jure-Kunkel M., and Melero I. (2016) Deciphering CD137 (4-1BB) signaling in T-cell costimulation for translation into successful cancer immunotherapy. Eur. J. Immunol. 46, 513–522 10.1002/eji.201445388 [DOI] [PubMed] [Google Scholar]
- 9. Melero I., Shuford W. W., Newby S. A., Aruffo A., Ledbetter J. A., Hellström K. E., Mittler R. S., and Chen L. (1997) Monoclonal antibodies against the 4-1BB T-cell activation molecule eradicate established tumors. Nat. Med. 3, 682–685 10.1038/nm0697-682 [DOI] [PubMed] [Google Scholar]
- 10. Madireddi S., Eun S.-Y., Lee S.-W., Nemčovičová I., Mehta A. K., Zajonc D. M., Nishi N., Niki T., Hirashima M., and Croft M. (2014) Galectin-9 controls the therapeutic activity of 4-1BB–targeting antibodies. J. Exp. Med. 211, 1433–1448 10.1084/jem.20132687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bitra A., Doukov T., Wang J., Picarda G., Benedict C. A., Croft M., and Zajonc D. M. (2018) Crystal structure of murine 4-1BB and its interaction with 4-1BBL support a role for galectin-9 in 4-1BB signaling. J. Biol. Chem. 293, 1317–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wada J., and Kanwar Y. S. (1997) Identification and characterization of galectin-9, a novel β-galactoside-binding mammalian lectin. J. Biol. Chem. 272, 6078–6086 10.1074/jbc.272.9.6078 [DOI] [PubMed] [Google Scholar]
- 13. Wada J., Ota K., Kumar A., Wallner E. I., and Kanwar Y. S. (1997) Developmental regulation, expression, and apoptotic potential of galectin-9, a β-galactoside binding lectin. J. Clin. Investig. 99, 2452–2461 10.1172/JCI119429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sznol M., Hodi F., Margolin K., McDermott D., Ernstoff M., Kirkwood C., Wojtaszek C., Feltquate D., and Logan T. (2008) Phase I study of BMS-663513, a fully human anti-CD137 agonist monoclonal antibody, in patients (pts) with advanced cancer (CA). J. Clin. Oncol. 26, Abstr. 3007 [Google Scholar]
- 15. Fisher T. S., Kamperschroer C., Oliphant T., Love V. A., Lira P. D., Doyonnas R., Bergqvist S., Baxi S. M., Rohner A., Shen A. C., Huang C., Sokolowski S. A., and Sharp L. L. (2012) Targeting of 4-1BB by monoclonal antibody PF-05082566 enhances T-cell function and promotes anti-tumor activity. Cancer Immunol. Immunother. 61, 1721–1733 10.1007/s00262-012-1237-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Croft M. (2009) The role of TNF superfamily members in T-cell function and diseases. Nat. Rev. Immunol. 9, 271–285 10.1038/nri2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vinay D. S., Cha K., and Kwon B. S. (2006) Dual immunoregulatory pathways of 4-1BB signaling. J. Mol. Med. 84, 726–736 10.1007/s00109-006-0072-2 [DOI] [PubMed] [Google Scholar]
- 18. So T., Lee S. W., and Croft M. (2008) Immune regulation and control of regulatory T cells by OX40 and 4-1BB. Cytokine Growth Factor Rev. 19, 253–262 10.1016/j.cytogfr.2008.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bodmer J. L., Schneider P., and Tschopp J. (2002) The molecular architecture of the TNF superfamily. Trends Biochem. Sci. 27, 19–26 10.1016/S0968-0004(01)01995-8 [DOI] [PubMed] [Google Scholar]
- 20. Chattopadhyay K., Lazar-Molnar E., Yan Q., Rubinstein R., Zhan C., Vigdorovich V., Ramagopal U. A., Bonanno J., Nathenson S. G., and Almo S. C. (2009) Sequence, structure, function, immunity: structural genomics of costimulation. Immunol. Rev. 229, 356–386 10.1111/j.1600-065X.2009.00778.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chattopadhyay K., Ramagopal U. A., Mukhopadhaya A., Malashkevich V. N., DiLorenzo T. P., Brenowitz M., Nathenson S. G., and Almo S. C. (2007) Assembly and structural properties of glucocorticoid-induced TNF receptor ligand: implications for function. Proc. Natl. Acad. Sci. U.S.A. 104, 19452–19457 10.1073/pnas.0709264104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chattopadhyay K., Ramagopal U. A., Brenowitz M., Nathenson S. G., and Almo S. C. (2008) Evolution of GITRL immune function: murine GITRL exhibits unique structural and biochemical properties within the TNF superfamily. Proc. Natl. Acad. Sci. U.S.A. 105, 635–640 10.1073/pnas.0710529105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Compaan D. M., and Hymowitz S. G. (2006) The crystal structure of the costimulatory OX40-OX40L complex. Structure 14, 1321–1330 10.1016/j.str.2006.06.015 [DOI] [PubMed] [Google Scholar]
- 24. Won E.-Y., Cha K., Byun J.-S., Kim D.-U., Shin S., Ahn B., Kim Y. H., Rice A. J., Walz T., Kwon B. S., and Cho H.-S. (2010) The structure of the trimer of human 4-1BB ligand is unique among members of the tumor necrosis factor superfamily. J. Biol. Chem. 285, 9202–9210 10.1074/jbc.M109.084442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mukai Y., Nakamura T., Yoshikawa M., Yoshioka Y., Tsunoda S., Nakagawa S., Yamagata Y., and Tsutsumi Y. (2010) Solution of the structure of the TNF-TNFR2 complex. Sci. Signal. 3, ra83 [DOI] [PubMed] [Google Scholar]
- 26. Nelson C. A., Warren J. T., Wang M. W., Teitelbaum S. L., and Fremont D. H. (2012) RANKL employs distinct binding modes to engage RANK and the osteoprotegerin decoy receptor. Structure 20, 1971–1982 10.1016/j.str.2012.08.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Banner D. W., D'Arcy A., Janes W., Gentz R., Schoenfeld H.-J., Broger C., Loetscher H., and Lesslauer W. (1993) Crystal structure of the soluble human 55 kd TNF receptor-human TNFβ complex: implications for TNF receptor activation. Cell 73, 431–445 10.1016/0092-8674(93)90132-A [DOI] [PubMed] [Google Scholar]
- 28. Cha S.-S., Sung B.-J., Kim Y.-A., Song Y.-L., Kim H.-J., Kim S., Lee M.-S., and Oh B.-H. (2000) Crystal structure of TRAIL-DR5 complex identifies a critical role of the unique frame insertion in conferring recognition specificity. J. Biol. Chem. 275, 31171–31177 10.1074/jbc.M004414200 [DOI] [PubMed] [Google Scholar]
- 29. An H.-J., Kim Y. J., Song D. H., Park B. S., Kim H. M., Lee J. D., Paik S.-G., Lee J.-O., and Lee H. (2011) Crystallographic and mutational analysis of the CD40-CD154 complex and its implications for receptor activation. J. Biol. Chem. 286, 11226–11235 10.1074/jbc.M110.208215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Luan X., Lu Q., Jiang Y., Zhang S., Wang Q., Yuan H., Zhao W., Wang J., and Wang X. (2012) Crystal structure of human RANKL complexed with its decoy receptor osteoprotegerin. J. Immunol. 189, 245 10.4049/jimmunol.1103387 [DOI] [PubMed] [Google Scholar]
- 31. Zhan C., Patskovsky Y., Yan Q., Li Z., Ramagopal U., Cheng H., Brenowitz M., Hui X., Nathenson S. G., and Almo S. C. (2011) Decoy strategies: the structure of TL1A:DcR3 complex. Structure 19, 162–171 10.1016/j.str.2010.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu C., Walter T. S., Huang P., Zhang S., Zhu X., Wu Y., Wedderburn L. R., Tang P., Owens R. J., Stuart D. I., Ren J., and Gao B. (2010) Structural and functional insights of RANKL–RANK interaction and signaling. J. Immunol. 184, 6910 10.4049/jimmunol.0904033 [DOI] [PubMed] [Google Scholar]
- 33. DeLano W. (2002) The PyMOL Molecular Graphics System, Schrödinger, LLC, New York [Google Scholar]
- 34. Murshudov G. N., Vagin A. A., and Dodson E. J. (1997) Refinement of macromolecular structures by the maximum likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 10.1107/S0907444996012255 [DOI] [PubMed] [Google Scholar]
- 35. Yi L., Yan Z., Jia H., Wang X., Zhao Y., and Zhang H. (2017) CD137-CRDI is not necessary in the role of contacting its natural ligand. Immunol. Cell Biol. 95, 24–32 [DOI] [PubMed] [Google Scholar]
- 36. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., and Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 10.1107/S0907444910045749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. van Lier R. A., Borst J., Vroom T. M., Klein H., Van Mourik P., Zeijlemaker W. P., and Melief C. J. (1987) Tissue distribution and biochemical and functional properties of Tp55 (CD27), a novel T cell differentiation antigen. J. Immunol. 139, 1589 [PubMed] [Google Scholar]
- 38. Haskins K., Kubo R., White J., Pigeon M., Kappler J., and Marrack P. (1983) The major histocompatibility complex-restricted antigen receptor on T cells: I. isolation with a monoclonal antibody. J. Exp. Med. 157, 1149 10.1084/jem.157.4.1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chan F. K.-M. (2007) Three is better than one: pre-ligand receptor assembly in the regulation of TNF receptor signaling. Cytokine 37, 101–107 10.1016/j.cyto.2007.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kabsch W. (2010) Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D Biol. Crystallogr. 66, 133–144 10.1107/S0907444909047374,10.1107/S0108767310004551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. McCoy A. J., Grosse-Kunstleve R. W., Storoni L. C., and Read R. J. (2005) Likelihood-enhanced fast translation functions. Acta Crystallogr. D Biol. Crystallogr. 61, 458–464 10.1107/S0907444905001617 [DOI] [PubMed] [Google Scholar]
- 43. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 45. Morris R. J., Perrakis A., and Lamzin V. S. (2003) ARP/wARP and automatic interpretation of protein electron density maps. Methods Enzymol. 374, 229–244 10.1016/S0076-6879(03)74011-7 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.