ABSTRACT
Two Baeyer-Villiger monooxygenases (BVMOs), designated BoBVMO and AmBVMO, were discovered from Bradyrhizobium oligotrophicum and Aeromicrobium marinum, respectively. Both monooxygenases displayed novel features for catalyzing the asymmetric sulfoxidation of bulky and pharmaceutically relevant thioethers. Evolutionary relationship and sequence analysis revealed that the two BVMOs belong to the family of typical type I BVMOs and the subtype ethionamide monooxygenase. Both BVMOs are active toward medium- and long-chain aliphatic ketones as well as various thioether substrates but are ineffective toward cyclohexanone, aromatic ketones, and other typical BVMO substrates. BoBVMO and AmBVMO showed the highest activities (0.117 and 0.025 U/mg protein, respectively) toward thioanisole among the tested substrates. Furthermore, these BVMOs exhibited distinct activity and excellent stereoselectivity toward bulky and prochiral prazole thioethers, which is a unique feature of this family of BVMOs. No native enzyme has been reported for the asymmetric sulfoxidation of bulky prazole thioethers into chiral sulfoxides. The identification of BoBVMO and AmBVMO provides an important scaffold for discovering enzymes capable of asymmetrically oxidizing bulky thioether substrates by genome mining.
IMPORTANCE Baeyer-Villiger monooxygenases (BVMOs) are valuable enzyme catalysts that are an alternative to the chemical Baeyer-Villiger oxidation reaction. Although BVMOs display broad substrate ranges, no native enzymes were reported to have activity toward the asymmetric oxidation of bulky prazole-like thioether substrates. Herein, we report the discovery of two type I BVMOs from Bradyrhizobium oligotrophicum (BoBVMO) and Aeromicrobium marinum (AmBVMO) which are able to catalyze the asymmetric sulfoxidation of bulky prazole thioethers (proton pump inhibitors [PPIs], a group of drugs whose main action is a pronounced and long-lasting reduction of gastric acid production). Efficient catalysis of omeprazole oxidation by BoBVMO was developed, indicating that this enzyme is a promising biocatalyst for the synthesis of bulky and pharmaceutically relevant chiral sulfoxide drugs. These results demonstrate that the newly identified enzymes are suitable templates for the discovery of more and better thioether-converting BVMOs.
KEYWORDS: Baeyer-Villiger monooxygenase, ethionamide monooxygenase, stereoselectivity, asymmetric sulfoxidation, prazole thioether
INTRODUCTION
Baeyer-Villiger monooxygenases (BVMOs) catalyze the challenging Baeyer-Villiger oxidation by inserting a single oxygen atom adjacent to the carbonyl carbon to form an ester or lactone (1–3). Apart from the classical Baeyer-Villiger oxidation, other BVMO-mediated reactions have been reported, such as epoxidations and oxidations of nitrogen, boron, selenium, and sulfur compounds (4–6). Practical applications of BVMOs, including steroid transformations, terpenoid metabolism, degradation of linear, cyclic, and aromatic ketones, and prodrug activation, have been studied extensively (3). Compared with the chemical oxidation of a thioether, which suffers from the usage of hazardous oxidants, such as peracids or hydrogen peroxide, biooxidations catalyzed by enzymes have clear advantages, such as high regio-, chemo-, and stereoselectivity, prevention of undesirable formation of sulfones because of overoxidization, and very mild reaction conditions (7–11).
Optically active sulfoxides are important chiral intermediates for asymmetric synthesis, and these sulfoxides also constitute the structure of pharmaceuticals, such as prazoles, which are proton pump inhibitors (PPIs) (12–14). Although metal catalysts, such as titanate/(+)-(1R,2S)-cis-1-amino-2-indanol and vanadium/chitosan systems, have been used in the asymmetric oxidation of prazole thioethers (13, 15–20), the practical utilization of heavy metals, expensive chiral ligands, and compounds with relatively low stereo- and chemoselectivity is hampered because they are environmentally detrimental. In contrast, biooxidation approaches have rapidly been developed in recent years (Fig. 1). The earliest report was the oxidation of thioethers catalyzed by cyclohexanone monooxygenase (CHMO) from Acinetobacter sp. strain NCIMB 9871 (21). Subsequently, camphor-grown Pseudomonas putida NCIMB 10007 (22), CHMO from an Escherichia coli strain (23), 4-hydroxyacetophenone monooxygenase (HAPMO) from Pseudomonas fluorescens ACB (24–26), phenylacetone monooxygenase (PAMO) from Thermobifida fusca (27), BVMOAf1 from Aspergillus fumigatus Af293 (28), and Yarrowia monooxygenases A to H (YMOA-H) from the eukaryote Yarrowia lipolytica (29) have been successfully used for the enzymatic oxidation of various alkyl and aryl thioethers to the corresponding sulfoxides or sulfones. Unfortunately, these native BVMOs have failed to catalyze the oxidation of sterically bulky prazole-like thioether substrates.
FIG 1.
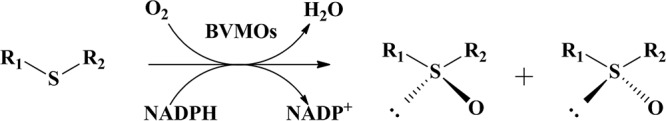
Asymmetric oxidation of thioether substrates by BVMOs for the production of chiral sulfoxides.
Therefore, biocatalytic monooxygenation or sulfoxidation of prazole-like thioethers by whole cells or engineered enzymes has received significant attention. For example, whole-cell-catalyzed oxidation of rabeprazole and omeprazole thioethers was achieved with Cunninghamella echinulata MK40 and Lysinibacillus sp. strain B71, respectively (30, 31). The products rabeprazole and omeprazole were accumulated to concentrations of up to 2.5 g/liter and 0.115 g/liter, respectively, with high enantiomeric excesses (ee) [>99% (S)-enantiomer]. Moreover, an engineered CHMO with more than 30 mutation sites from Acinetobacter sp. NCIMB 9871 selectively oxidized the omeprazole thioether to the desired (S)-omeprazole (32, 33). These results reveal the possible oxidation of bulky prazole-like thioethers by oxidases and inspired us to screen for such native enzymes in nature.
According to the literature, the BVMO EtaA (which belongs to the subtype ethionamide monooxygenase) from Mycobacterium tuberculosis oxidizes several bulky thioether substrates, including ethionamide, thiocarlide, and 3-(m-tolyl)-5-[(1-piperidinyl)carbonylmethyl]thio-1,2,4-thiadiazole (34–36). Preliminary experiments revealed that EtaA from M. tuberculosis reacted poorly with omeprazole thioether (Table 1). Thus, genome mining using the sequence of EtaA from M. tuberculosis as a template in a BLAST search of the UniProt/Swiss-Prot database was performed in an effort to exploit BVMOs with catalytic oxidation activity toward bulky prazole thioethers. Thirty enzyme genes from the search were identified, cloned, and assayed for their activity toward omeprazole thioether. Two BVMOs were identified, and their biochemical properties, substrate preference, and potential for application to the production of prazole-like compounds were investigated in detail.
TABLE 1.
Screening results of BVMOs for omeprazole thioether sulfoxidationa
| Substrate entry | NCBI accession no. | Microorganism | Sequence identity (%) | Conversion (%)b | % ee (configuration)b |
|---|---|---|---|---|---|
| S1 | WP_003899731.1 | Mycobacterium tuberculosis | 100 | <1.0 | 99 (R) |
| S2 | WP_015665598.1 | Bradyrhizobium oligotrophicum | 56 | 67 | 99 (R) |
| S3 | WP_091530404.1 | Fontimonas thermophila | 56 | 1.5 | 99 (R) |
| S4 | WP_050035958.1 | Rhodococcus aetherivorans | 55 | 1.8 | 99 (R) |
| S5 | WP_024102511.1 | Rhodococcus pyridinivorans | 53 | 1.8 | 99 (R) |
| S6 | WP_010953714.1 | Pseudomonas putida KT2440 | 50 | 2.9 | 99 (R) |
| S7 | WP_007076782.1 | Aeromicrobium marinum | 49 | 52 | 99 (R) |
| S8 | WP_005193356.1 | Gordonia amarae | 42 | 12 | 99 (R) |
The 500-μl reaction mixture contained diluted crude enzyme extracts, 1 mM NADPH, 1 mM omeprazole thioether, 2% (vol/vol) DMSO, and KPB (100 mM, pH 9.0) at 30°C.
Conversion and enantiomeric excess were determined by chiral HPLC.
RESULTS
Discovery of BVMOs.
To identify BVMOs with the desired activity toward prazole thioethers, EtaA from M. tuberculosis (NCBI accession no. WP_003899731.1) was selected as the template for genome mining. Thirty BVMOs with 35% to 91% sequence identities were cloned into the pET28a vector and overexpressed in E. coli BL21(DE3). Seven of these BVMOs were successfully expressed in a partially soluble form and displayed measurable activity (>1% conversion, using crude enzyme extracts as the catalysts) toward omeprazole thioether (Table 1). The enzymes from Bradyrhizobium oligotrophicum and Aeromicrobium marinum, which had the top two conversions, were chosen for further investigation and designated BoBVMO (gene NCBI accession number BAM88475.1; protein NCBI accession number WP_015665598.1) and AmBVMO (gene NCBI accession number EFQ82481.1; protein NCBI accession number WP_007076782.1), respectively. The sequence identity between BoBVMO and AmBVMO is 48%.
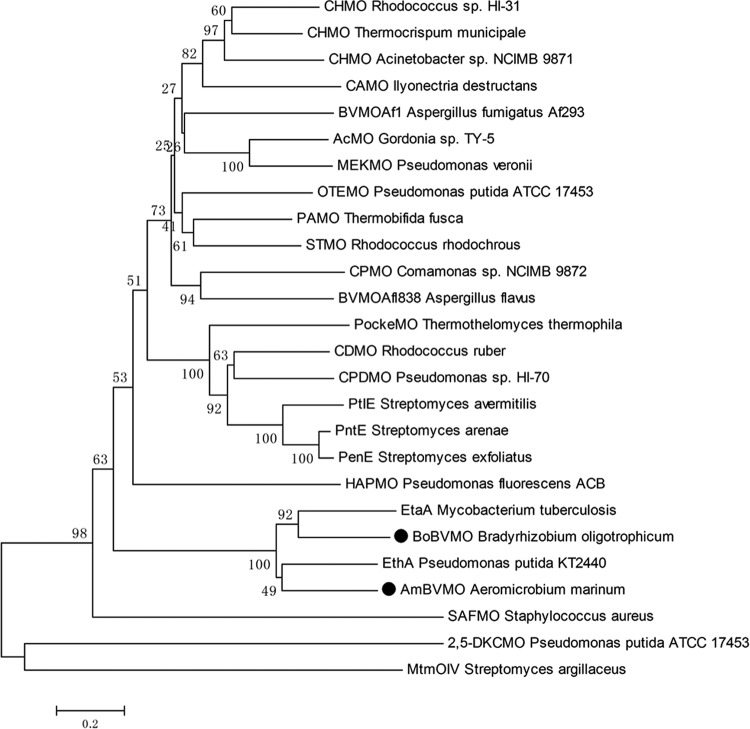
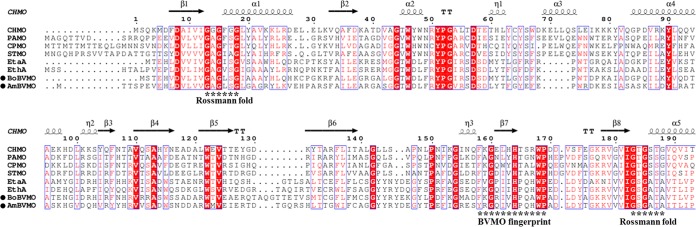
To establish the evolutionary relationship of BoBVMO and AmBVMO with other reported BVMOs, a phylogenetic tree (Fig. 2) based on their amino acid sequences was constructed. Twenty-six BVMOs with various catalytic functions (37, 38) were selected for this analysis. As illustrated in Fig. 2, the two BVMOs display high sequence identities with M. tuberculosis EtaA and with an EthA from Pseudomonas putida KT2440, a BVMO exhibiting high specificity toward short-chain aliphatic ketones (39). The sequence identities of BoBVMO and AmBVMO with EthA are 47% and 54%, respectively. A sequence alignment between the two BVMOs and other well-studied type I BVMOs was carried out (Fig. 3). Accordingly, two Rossmann fold GXGXX(G/A) sequences were identified to flank the fingerprint motif FXGXXXHXXXW(P/D) (40), which is the typical consensus sequence for type I BVMOs, in the two BVMOs. These results indicate that the two BVMOs are members of type I BVMOs, which are NADPH- and FAD-dependent enzymes (41).
FIG 2.
Phylogenetic analysis of the two BVMOs and other BVMOs with various catalytic functions. All protein sequences were retrieved from the NCBI database. The proteins are as follows: CHMO from Rhodococcus sp. strain HI-31 (accession no. BAH56677.1) (47), CHMO from Thermocrispum municipale (accession no. 5M10_A) (48), CHMO from Acinetobacter sp. strain NCIMB 9871 (accession no. BAA86293.1) (49), cycloalkanone monooxygenase (CAMO) from Ilyonectria destructans (accession no. AET80001.1) (50), BVMOAf1 from Aspergillus fumigatus Af293 (accession no. XP_747160.1) (28), acetone monooxygenase (AcMO) from Gordonia sp. strain TY-5 (accession no. BAF43791.1) (51), methyl ketone monooxygenase (MEKMO) from Pseudomonas veronii (accession no. ABI15711.1) (52), 2-oxo-Δ3-4,5,5-trimethylcyclopentenylacetyl-coenzyme A monooxygenase (OTEMO) from Pseudomonas putida ATCC 17453 (accession no. H3JQW0.1) (53), PAMO from Thermobifida fusca (accession no. Q47PU3.1) (54), steroid monooxygenase (STMO) from Rhodococcus rhodochrous (accession no. BAA24454.1) (55), cyclopentanone monooxygenase (CPMO) from Comamonas sp. strain NCIMB 9872 (accession no. BAC22652.1) (56), BVMOAfl838 from Aspergillus flavus (accession no. 5J7X_A) (57), polycyclic ketone monooxygenase (PockeMO) from Thermothelomyces thermophila (accession no. 5MQ6_A) (38), cyclododecanone monooxygenase (CDMO) from Rhodococcus ruber (accession no. AAL14233.1) (58), cyclopentadecanone monooxygenase (CPDMO) from Pseudomonas sp. strain HI-70 (accession no. BAE93346.1) (59), PtIE from Streptomyces avermitilis (accession no. WP_010984425.1), PntE from Streptomyces arenae (accession no. E3VWI7.1) (60), PenE from Streptomyces exfoliatus (accession no. E3VWK3.1) (61), 4-hydroxyacetophenone monooxygenase (HAPMO) from Pseudomonas fluorescens ACB (accession no. Q93TJ5.1) (62), EtaA from Mycobacterium tuberculosis (accession no. WP_003899731.1) (34), BoBVMO from Bradyrhizobium oligotrophicum (accession no. WP_015665598.1), EthA from Pseudomonas putida KT2440 (accession no. WP_010953714.1) (39), AmBVMO from Aeromicrobium marinum (accession no. WP_007076782.1), flavin-containing monooxygenase from Staphylococcus aureus (SAFMO) (accession no. Q99R54.1) (63), 2,5-diketocamphane monooxygenase (2,5-DKCMO) from Pseudomonas putida ATCC 17453 (accession no. Q6STM1.1) (64), and mithramycin oxygenase IV (MtmOIV) from Streptomyces argillaceus (accession no. 4K5S_A) (65).
FIG 3.
Sequence alignment of the two BVMOs with other well-studied type I BVMOs. All protein sequences were retrieved from the NCBI database. The proteins are as follows: CHMO from Acinetobacter sp. strain NCIMB 9871 (accession no. BAA86293.1), PAMO from Thermobifida fusca (accession no. Q47PU3.1), CPMO from Comamonas sp. strain NCIMB 9872 (accession no. BAC22652.1), STMO from Rhodococcus rhodochrous (accession no. BAA24454.1), EtaA from Mycobacterium tuberculosis (accession no. WP_003899731.1), EthA from Pseudomonas putida KT2440 (accession no. WP_010953714.1), BoBVMO from Bradyrhizobium oligotrophicum (accession no. WP_015665598.1), and AmBVMO from Aeromicrobium marinum (accession no. WP_007076782.1). The two Rossmann folds (GXGXXG) and the BVMO fingerprint [FXGXXXHXXXW(P/D)] are marked with asterisks.
Substrate scope of the two BVMOs.
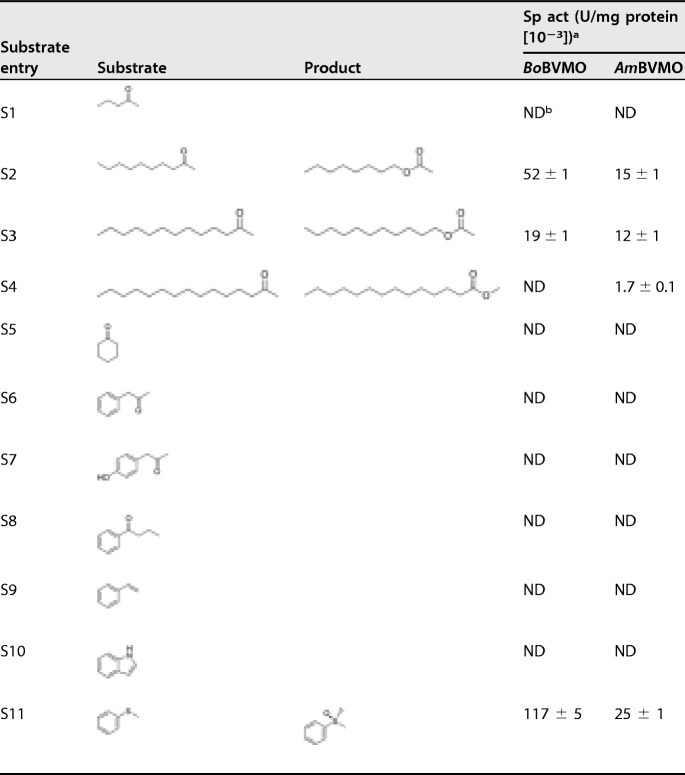
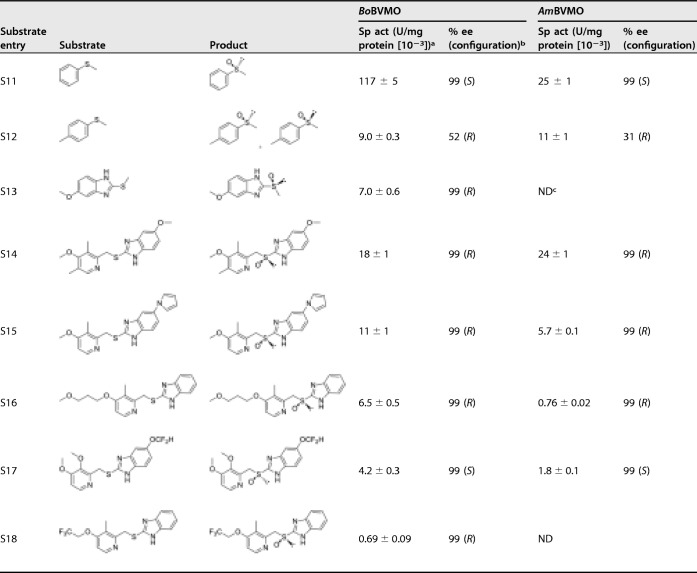
The substrate preferences of the two BVMOs toward 18 substrates, including aliphatic ketones, aromatic ketones, cyclic ketone, aromatic olefin, N-heterocycles, and thioethers, were explored with the purified enzymes. As listed in Table 2, the catalytic activity toward aliphatic ketones (substrates S2 to S4) decreased with increasing chain length of the fatty acid. No activity toward the short-chain aliphatic ketone (substrate S1), cyclohexanone (substrate S5), and aromatic ketones (substrates S6 to S8) was detected, indicating that the two BVMOs have high specificity toward medium- and long-chain aliphatic ketones. Styrene (substrate S9) and indole (substrate S10) were also not accepted. Thioanisole (substrate S11) was the best substrate among the tested compounds, giving specific activities of 0.117 U/mg protein (BoBVMO) and 0.025 U/mg protein (AmBVMO), respectively. The results of subsequent substrate scope extension experiments are shown in Table 3, which indicated that the ability of these two enzymes to convert bulky thioether substrates is similar to that of EtaA reported in the literature (34–36). Most of the thioethers tested were oxidized with good activities and excellent chemo- and stereoselectivities. Only 4-methylthioanisole (substrate S12) was converted with lower stereoselectivities (52% with BoBVMO and 31% with AmBVMO). Importantly, no sulfones were detected as by-products by overoxidation. The five bulky prazole thioethers (substrates S14 to S18) omeprazole, ilaprazole, rabeprazole, pantoprazole, and lansoprazole were converted by BoBVMO with high stereoselectivity. These prazole thioethers were also tested using AmBVMO, and four of these compounds were converted. Therefore, BoBVMO and AmBVMO are type I BVMOs with novel features because both of them catalyze the pharmaceutically relevant sulfoxidation of bulky prazole thioethers.
TABLE 2.
Substrate spectrum of the two BVMOs discovered
aSpecific activity was determined at pH 9.0 and 25°C with 2 mM substrate (substrates S1 to S11) using purified enzyme.
bND, not detected.
TABLE 3.
Specific activity and stereoselectivity of the two BVMOs toward thioethers
aSpecific activity was determined at pH 9.0 and 25°C with 2 mM (substrates S11 and S12) or 0.2 mM (substrates S13 to S18) using purified enzyme.
bEnantiomeric excess was determined by chiral HPLC.
cND, not detected.
Biochemical properties of the two BVMOs.
The biochemical properties of the two BVMOs were characterized using the proteins purified by nickel affinity chromatography. The activities of purified BoBVMO and AmBVMO were measured at temperatures ranging from 20 to 50°C. The maximum activity was observed at 35°C for both BVMOs (see Fig. S1 in the supplemental material). The effect of pH on the activity of the two BVMOs was investigated at various pH values ranging from 6.0 to 11.0. The optimum pH was 9.0 for both BVMOs with different buffers, i.e., Tris-HCl buffer for BoBVMO and Gly-NaOH buffer for AmBVMO, and 60% of the maximum activity was still retained at pHs of between pH 8.5 and 9.5 (Fig. S2). The optimum pH of both BVMOs is slightly alkaline, which is general among BVMOs, as described in a review (3). Furthermore, the thermostability was examined over a temperature range of from 30 to 40°C (Fig. S3). According to thermal inactivation curves, BoBVMO had half-lives (t1/2) of 4.4 h and 0.024 h at 30 and 40°C, respectively, whereas AmBVMO had t1/2 values of 5.4 h and 0.097 h, respectively, indicating that these enzymes have lower stability at temperatures above 30°C.
Catalytic performance of BoBVMO for sulfoxidation.
The reaction conditions with respect to the presence of NADP+ or FAD were optimized using omeprazole thioether as the substrate and a crude enzyme extract of BoBVMO as the catalyst (Table S1). The conversion was 58% after 4 h in the absence of NADP+. The omeprazole thioether was completely converted to the corresponding (R) product within 2 h by BoBVMO when 0.2 mM NADP+ was added to the reaction mixture. The catalytic rate did not increase when the NADP+ concentration was increased to 0.5 mM. The cosolvent loading was also optimized. When the cosolvent was replaced by methanol, the reaction reached 99% conversion within 1 h. However, the presence of acetone reduced the conversion by BoBVMO to only 19% after 4 h. Addition of FAD did not increase the catalytic rate, indicating that the endogenous FAD in the E. coli cell lysate was sufficient to meet the catalytic requirements. Following optimization, the catalytic performance of BoBVMO toward the omeprazole thioether was examined with different substrate loads (1, 3, and 5 g/liter) using the same dosage of BoBVMO (Fig. 4). In the case of 3- or 5-g/liter substrate loading, 89 and 71% conversions were achieved in 6 h, respectively. However, in all cases, the conversion did not increase beyond 6 h, indicating that the activity of the enzyme may have been lost.
FIG 4.
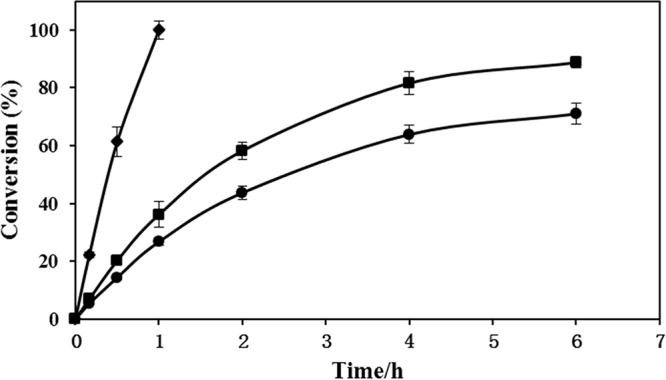
Progress curves of the BoBVMO-catalyzed sulfoxidation of omeprazole thioether performed under optimized conditions with different substrate loads. Symbols: ◆ 1.0 g/liter; ■, 3.0 g/liter; ●, 5.0 g/liter.
DISCUSSION
Biooxidations of bulky prazole thioethers by whole-cell catalysts or engineered CHMOs have been explored. However, the genetic information and the biochemical properties of the responsible enzymes were neither disclosed nor characterized. At the same time, low efficiency and the intricate evolutionary process inspired us to explore the possibility of finding oxidases in nature that convert bulky prazole thioethers. As a result, BoBVMO and AmBVMO with novel asymmetric sulfoxidation activity toward bulky prazole thioethers were discovered by a genome mining approach.
Even though the measured activities of BoBVMO and AmBVMO were lower than the ones determined with an engineered CHMO reported previously (32, 33) and the configuration of the product was opposite the desired one when the omeprazole thioether was used as a substrate, these newly identified native enzymes exhibited relatively high specific activities and excellent regio-, chemo-, and stereoselectivities toward various thioether substrates.
In summary, the ability of these two enzymes to catalyze asymmetric sulfoxidation of bulky prazole precursors is unique and provides an incentive to discover more powerful monooxygenases that catalyze the production of pharmaceutically relevant bulky thioethers. Besides, on the basis of the directed evolution technology for manipulating the selectivity and activity of BVMOs toward thioethers (42–44), further engineering of the two BVMOs is ongoing to improve their catalytic performance and to release their potential for chiral prazole synthesis in industry.
MATERIALS AND METHODS
General.
Commercial chemicals were purchased from TCI, Macklin, Aladdin, or Sigma-Aldrich. All prazole thioethers, sulfoxides, and sulfones were available from Aosaikang Pharmaceutical Co., Ltd. (Nanjing, China). The conversion and regio-, chemo-, and stereoselectivities of the reactions were determined by high-performance liquid chromatography (HPLC) or gas chromatography-mass spectrometry (GC-MS), as described in the supplemental material.
Genome data mining for BVMOs.
A library of putative Baeyer-Villiger monooxygenases was constructed by genome mining. A total of 30 monooxygenases, each with 35% to 91% amino acid sequence homology to the template BVMO EthA sequence, were selected from the UniProt/Swiss-Prot database.
Cloning, expression, and purification of BVMOs.
BVMO genes were amplified from the genomic DNA of the original strains and cloned into the pET28a vector under the control of the T7 promoter and then transformed into E. coli BL21(DE3) for overexpression. The positive transformants were grown at 37°C to an optical density at 600 nm of 0.6 to 0.8 in Luria broth (LB) medium containing 50 μg/ml kanamycin, protein production was induced with isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 0.2 mM, and the cells were cultured for a further 16 h at 16°C. Cells were harvested by centrifugation at 5,000 × g at 4°C for 10 min, washed twice with ice-chilled potassium phosphate buffer (KPB; 100 mM, pH 9.0), and then disrupted by ultrasonication. After centrifugation at 12,000 × g at 4°C for 30 min, the supernatant was loaded onto a His-Trap Ni-nitrilotriacetic acid FF column (5 ml; GE Healthcare Co.) that had been preequilibrated with buffer A (50 mM KPB, 500 mM NaCl, 10 mM imidazole, pH 8.0). The target protein was eluted using an increasing gradient of imidazole from 10 to 150 mM at a flow rate of 5 ml/min and detected by SDS-PAGE. The fraction containing the purified protein was collected, and this fraction was concentrated by ultrafiltration. After measuring the protein concentration, commercial FAD (>95% purity) from Sigma-Aldrich was added in excess (1.5-fold equivalent) to the purified enzyme solution, and the freshly purified enzyme was then used for further measurements.
Activity and selectivity assays.
The conversions of recombinant BVMOs toward omeprazole thioether were tested using crude enzyme extracts. In a 500-μl reaction mixture, 1 mM omeprazole thioether, 2% (vol/vol) dimethyl sulfoxide (DMSO), 1 mM NADPH, and diluted crude enzyme extracts were mixed in KPB (100 mM, pH 9.0). The reaction was performed at 30°C with mixing at 1,000 rpm in a Thermomixer (Eppendorf, Germany). After incubation for 3 h, the reaction mixture was extracted with an equal volume of ethyl acetate. The extract was dried over anhydrous Na2SO4 for at least 6 h. The conversions and stereoselectivities were determined by HPLC, as described in the supplemental material. The specific activities and the regio-, chemo-, and stereoselectivities of BoBVMO and AmBVMO toward various substrates were examined with the purified enzymes. In a 500-μl reaction mixture, 0.2 to 2 mM substrate, 2% (vol/vol) DMSO, 0.2 to 2 mM NADPH, and a diluted enzyme solution were mixed in KPB (100 mM, pH 9.0). The reaction mixture was incubated for 10 to 60 min at 25°C, and then the samples were treated as described above. The specific activities and stereoselectivities were determined by HPLC or GC-MS, as described in the supplemental material. One unit of activity was defined as the amount of enzyme required for the production of 1.0 μmol product per minute under the assay conditions. The mathematical formula ee = {([(R)] − [(S)])/([(R)] + [(S)])} × 100%, where [(R)] and [(S)] are the concentrations of the (R) and (S) enantiomers, respectively, was used for calculation of ee values. All the presented results are average values for the data from triplicate experiments.
Construction of a phylogenetic tree.
Phylogenetic analysis of the two BVMOs and other BVMOs with various catalytic functions was performed. The phylogenetic tree was constructed with the ClustalW alignment using the neighbor-joining method (45). The bootstrap values were based on 1,000 replicates. These analyses were carried out using MEGA6 software (46). All protein sequences were retrieved from the NCBI database.
Characterization of the two BVMOs.
The optimum temperature was determined by testing temperatures over the temperature range of 20 to 50°C. The optimum pH was determined by testing different pH values (6.0 to 11.0) in the following buffers (100 mM): potassium phosphate (pH 6.0 to 9.0), Tris-HCl (pH 8.0 to 9.0), and Gly-NaOH (pH 9.0 to 11.0). The highest activity was normalized as 100%. To investigate the thermostability of the two BVMOs, pure enzyme solutions (3.5 mg/ml) were incubated at different temperatures (30 and 40°C) in KPB for a set period, followed by measurement of the residual activity. The activity of the enzyme incubated for 0 h was normalized as 100%. The relative activities were determined by HPLC using omeprazole thioether as the substrate. Various substrates (aliphatic ketones, aromatic ketones, cyclic ketone, aromatic olefin, N-heterocycle, and thioethers [substrates S1 to S18]) were applied to explore the substrate preference of the two BVMOs. The activities were assayed with 0.2 to 2 mM substrates S1 to S18 (2% [vol/vol] DMSO) by using the above-mentioned method.
Reaction conditions for the oxidation of omeprazole thioether.
The full potency of the BVMOs was developed by optimizing the reaction with omeprazole thioether as the substrate with respect to the presence of NADP+ or FAD and the type of cosolvents. The 10-ml reaction mixture contained 1 g (wet weight) cells (resuspended in 9 ml Tris-HCl buffer and disrupted by ultrasonication), 10 mg omeprazole thioether, 20 mg Bacillus megaterium glucose dehydrogenase (cell extract), 20 mM glucose, 0.2 mM NADP+, 10% (vol/vol) cosolvent (DMSO, methanol, or acetone), and Tris-HCl buffer (100 mM, pH 9.0). The reaction mixture was shaken at 25°C and 180 rpm. Samples were intermittently removed and extracted for direct analysis of the conversion rate by HPLC. Under the optimal reaction conditions, the catalytic activity of BoBVMO toward omeprazole thioether was investigated with varied substrate loads (1 to 5 g/liter).
Supplementary Material
ACKNOWLEDGMENTS
We thank Liwen Bianji, Edanz Editing China, for editing the English text of a draft of the manuscript.
This work was financially supported by the National Natural Science Foundation of China (no. 21536004, 21672063, and 21776085), the Fundamental Research Funds for the Central Universities (no. 22221818014), and the Shanghai Science and Technology Program (no. 15JC1400403).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00638-18.
REFERENCES
- 1.Huisgea VR. 1986. Adolf von Baeyers wissenschaftliches Werk-ein Vermächtnis. Angew Chem 98:297–311. doi: 10.1002/ange.19860980404. [DOI] [Google Scholar]
- 2.Renz M, Meunier B. 1999. 100 years of Baeyer-Villiger oxidations. Eur J Org Chem 1999:737–750. doi:. [DOI] [Google Scholar]
- 3.Leisch H, Morley K, Lau PCK. 2011. Baeyer-Villiger monooxygenases: more than just green chemistry. Chem Rev 111:4165–4222. doi: 10.1021/cr1003437. [DOI] [PubMed] [Google Scholar]
- 4.Colonna S, Gaggero N, Carrea G, Ottolina G, Pasta P, Zambianchi F. 2002. First asymmetric epoxidation catalysed by cyclohexanone monooxygenase. Tetrahedron Lett 43:1797–1799. doi: 10.1016/S0040-4039(02)00029-1. [DOI] [Google Scholar]
- 5.Branchaud BP, Walsh CT. 1985. Functional group diversity in enzymic oxygenation reactions catalyzed by bacterial flavin-containing cyclohexanone oxygenase. J Am Chem Soc 107:2153–2161. doi: 10.1021/ja00293a054. [DOI] [Google Scholar]
- 6.Ottolina G, Pasta P, Carrea G, Colonna S, Dallavalle S, Holland HL. 1995. A predictive active site model for the cyclohexanone monooxygenase catalyzed oxidation of sulfides to chiral sulfoxides. Tetrahedron Asymmetry 6:1375–1386. doi: 10.1016/0957-4166(95)00170-T. [DOI] [Google Scholar]
- 7.Hollmann F, Arends IWCE, Holtmann D. 2011. Enzymatic reductions for the chemist. Green Chem 13:2285–2314. doi: 10.1039/c1gc15424a. [DOI] [Google Scholar]
- 8.Bornscheuer UT, Huisman GW, Kazlauskas RJ, Lutz S, Moore JC, Robins K. 2012. Engineering the third wave of biocatalysis. Nature 485:185–194. doi: 10.1038/nature11117. [DOI] [PubMed] [Google Scholar]
- 9.Reetz MT. 2013. Biocatalysis in organic chemistry and biotechnology: past, present, and future. J Am Chem Soc 135:12480–12496. doi: 10.1021/ja405051f. [DOI] [PubMed] [Google Scholar]
- 10.van Beek HL, Romero E, Fraaije MW. 2017. Engineering cyclohexanone monooxygenase for the production of methyl propanoate. ACS Chem Biol 12:291–299. doi: 10.1021/acschembio.6b00965. [DOI] [PubMed] [Google Scholar]
- 11.Yachnin BJ, Lau PCK, Berghuis AM. 2016. The role of conformational flexibility in Baeyer-Villiger monooxygenase catalysis and structure. Biochim Biophys Acta 1864:1641–1648. doi: 10.1016/j.bbapap.2016.08.015. [DOI] [PubMed] [Google Scholar]
- 12.Carreno MC. 1995. Applications of sulfoxides to asymmetric synthesis of biologically active compounds. Chem Rev 95:1717–1760. doi: 10.1021/cr00038a002. [DOI] [Google Scholar]
- 13.Fernandez I, Khiar N. 2003. Recent developments in the synthesis and utilization of chiral sulfoxides. Chem Rev 103:3651–3705. doi: 10.1021/cr990372u. [DOI] [PubMed] [Google Scholar]
- 14.Pellissier H. 2006. Use of chiral sulfoxides in asymmetric synthesis. Tetrahedron 62:5559–5601. doi: 10.1016/j.tet.2006.03.093. [DOI] [Google Scholar]
- 15.Cotton H, Elebring T, Larsson M, Li L, Sorensen H, von Unge S. 2000. Asymmetric synthesis of esomeprazole. Tetrahedron Asymmetry 11:3819–3825. doi: 10.1016/S0957-4166(00)00352-9. [DOI] [Google Scholar]
- 16.Legros J, Dehli JR, Bolm C. 2005. Applications of catalytic asymmetric sulfide oxidations to the syntheses of biologically active sulfoxides. Adv Synth Catal 347:19–31. doi: 10.1002/adsc.200404206. [DOI] [Google Scholar]
- 17.Delamare M, Belot S, Caille J-C, Martinet F, Kagan HB, Henryon V. 2009. A new titanate/(+)-(1R,2S)-cis-1-amino-2-indanol system for the asymmetric synthesis of (S)-tenatoprazole. Tetrahedron Lett 50:1702–1704. doi: 10.1016/j.tetlet.2009.01.111. [DOI] [Google Scholar]
- 18.Mahale RD, Rajput MR, Maikap GC, Gurjar MK. 2010. Davis oxaziridine-mediated asymmetric synthesis of proton pump inhibitors using DBU salt of prochiral sulfide. Org Process Res Dev 14:1264–1268. doi: 10.1021/op100075v. [DOI] [Google Scholar]
- 19.Talsi EP, Rybalova TV, Bryliakov KP. 2015. Isoinversion behavior in the enantioselective oxidations of pyridylmethylthiobenzimidazoles to chiral proton pump inhibitors on titanium salalen complexes. ACS Catal 5:4673–4679. doi: 10.1021/acscatal.5b01212. [DOI] [Google Scholar]
- 20.Shen C, Qiao J, Zhao L, Zheng K, Jin J, Zhang P. 2017. An efficient silica supported chitosan@vanadium catalyst for asymmetric sulfoxidation and its application in the synthesis of esomeprazole. Catal Commun 92:114–118. doi: 10.1016/j.catcom.2017.01.018. [DOI] [Google Scholar]
- 21.Ryerson CC, Ballou DP, Walsh C. 1982. Mechanistic studies on cyclohexanone oxygenase. Biochemistry 21:2644–2655. doi: 10.1021/bi00540a011. [DOI] [PubMed] [Google Scholar]
- 22.Beecher J, Richardson P, Willetts A. 1994. Baeyer-Villiger monooxygenase-dependent biotransformations: stereospecific heteroatom oxidations by camphor-grown Pseudomonas putida to produce chiral sulfoxides. Biotechnol Lett 16:909–912. doi: 10.1007/BF00128623. [DOI] [Google Scholar]
- 23.Zambianchi F, Pasta P, Carrea G, Colonna S, Gaggero N, Woodley JM. 2002. Use of isolated cyclohexanone monooxygenase from recombinant Escherichia coli as a biocatalyst for Baeyer-Villiger and sulfide oxidations. Biotechnol Bioeng 78:489–496. doi: 10.1002/bit.10207. [DOI] [PubMed] [Google Scholar]
- 24.Kamerbeek NM, Olsthoorn AJJ, Fraaije MW, Janssen DB. 2003. Substrate specificity and enantioselectivity of 4-hydroxyacetophenone monooxygenase. Appl Environ Microbiol 69:419–426. doi: 10.1128/AEM.69.1.419-426.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kamerbeek NM, Janssen DB, van Berkel WJH, Fraaije MW. 2003. Baeyer-Villiger monooxygenases, an emerging family of flavin-dependent biocatalysts. Adv Synth Catal 345:667–678. doi: 10.1002/adsc.200303014. [DOI] [Google Scholar]
- 26.de Gonzalo G, Torres Pazmino DE, Ottolina G, Fraaije MW, Carrea G. 2006. 4-Hydroxyacetophenone monooxygenase from Pseudomonas fluorescens ACB as an oxidative biocatalyst in the synthesis of optically active sulfoxides. Tetrahedron Asymmetry 17:130–135. doi: 10.1016/j.tetasy.2005.11.024. [DOI] [Google Scholar]
- 27.de Gonzalo G, Torres Pazmino DE, Ottolina G, Fraaije MW, Carrea G. 2005. Oxidations catalyzed by phenylacetone monooxygenase from Thermobifida fusca. Tetrahedron Asymmetry 16:3077–3083. doi: 10.1016/j.tetasy.2005.08.004. [DOI] [Google Scholar]
- 28.Mascotti ML, Juri AM, Dudek H, Sanz MK, Fraaije MW. 2013. Cloning, overexpression and biocatalytic exploration of a novel Baeyer-Villiger monooxygenase from Aspergillus fumigatus Af293. AMB Express 3:33. doi: 10.1186/2191-0855-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bordewick S, Beier A, Balke K, Bornscheuer UT. 2018. Baeyer-Villiger monooxygenases from Yarrowia lipolytica catalyze preferentially sulfoxidations. Enzyme Microb Technol 109:31–42. doi: 10.1016/j.enzmictec.2017.09.008. [DOI] [PubMed] [Google Scholar]
- 30.Yoshida T, Kito M, Tsujii M, Nagasawa T. 2001. Microbial synthesis of a proton pump inhibitor by enantioselective oxidation of a sulfide into its corresponding sulfoxide by Cunninghamella echinulata MK40. Biotechnol Lett 23:1217–1222. doi: 10.1023/A:1010521217954. [DOI] [Google Scholar]
- 31.Babiak P, Kyslikova E, Stepanek V, Valesova R, Palyzova A, Maresova H, Hajicek J, Kyslik P. 2011. Whole-cell oxidation of omeprazole sulfide to enantiopure esomeprazole with Lysinibacillus sp. B71. Bioresour Technol 102:7621–7626. doi: 10.1016/j.biortech.2011.05.052. [DOI] [PubMed] [Google Scholar]
- 32.Bong YK, Clay MD, Collier SJ, Mijts B, Vogel M, Zhang X, Zhu J, Nazor J, Smith D, Song S. June 2011. Engineered cylohexanone monooxygenases for synthesis of prazole compounds. Patent WO2011071982A2. [Google Scholar]
- 33.Bong YK, Clay MD, Collier SJ, Mijts B, Vogel M, Zhang X, Zhu J, Nazor J, Smith D, Song S. November 2014. Synthesis of prazole compounds. US patent 8895271B2.
- 34.Fraaije MW, Kamerbeek NM, Heidekamp AJ, Fortin R, Janssen DB. 2004. The prodrug activator EtaA from Mycobacterium tuberculosis is a Baeyer-Villiger monooxygenase. J Biol Chem 279:3354–3360. doi: 10.1074/jbc.M307770200. [DOI] [PubMed] [Google Scholar]
- 35.Grant SS, Wellington S, Kawate T, Desjardins CA, Silvis MR, Wivagg C, Thompson M, Gordon K, Kazyanskaya E, Nietupski R, Haseley N, Iwase N, Earl AM, Fitzgerald M, Hung DT. 2016. Baeyer-Villiger monooxygenases EthA and MymA are required for activation of replicating and non-replicating Mycobacterium tuberculosis inhibitors. Cell Chem Biol 23:666–677. doi: 10.1016/j.chembiol.2016.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang HN, Xu ZW, Jiang HW, Wu FL, He X, Liu Y, Guo SJ, Li Y, Bi LJ, Deng JY, Zhang XE, Tao SC. 2017. Cyclic di-GMP regulates Mycobacterium tuberculosis resistance to ethionamide. Sci Rep 7:5860. doi: 10.1038/s41598-017-06289-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beneventi E, Niero M, Motterle R, Fraaije M, Bergantino E. 2013. Discovery of Baeyer-Villiger monooxygenases from photosynthetic eukaryotes. J Mol Catal B Enzym 98:145–154. doi: 10.1016/j.molcatb.2013.10.006. [DOI] [Google Scholar]
- 38.Fuerst MJLJ, Savino S, Dudek HM, Gomez Castellanos JR, Gutierrez de Souza C, Rovida S, Fraaije MW, Mattevi A. 2017. Polycyclic ketone monooxygenase from the thermophilic fungus Thermothelomyces thermophila: a structurally distinct biocatalyst for bulky substrates. J Am Chem Soc 139:627–630. doi: 10.1021/jacs.6b12246. [DOI] [PubMed] [Google Scholar]
- 39.Rehdorf J, Kirschner A, Bornscheuer UT. 2007. Cloning, expression and characterization of a Baeyer-Villiger monooxygenase from Pseudomonas putida KT2440. Biotechnol Lett 29:1393–1398. doi: 10.1007/s10529-007-9401-y. [DOI] [PubMed] [Google Scholar]
- 40.Fraaije MW, Kamerbeek NM, van Berkel WJ, Janssen DB. 2002. Identification of a Baeyer-Villiger monooxygenase sequence motif. FEBS Lett 518:43–47. doi: 10.1016/S0014-5793(02)02623-6. [DOI] [PubMed] [Google Scholar]
- 41.Willetts A. 1997. Structural studies and synthetic applications of Baeyer-Villiger monooxygenases. Trends Biotechnol 15:55–62. doi: 10.1016/S0167-7799(97)84204-7. [DOI] [PubMed] [Google Scholar]
- 42.Reetz MT, Daligault F, Brunner B, Hinrichs H, Deege A. 2004. Directed evolution of cyclohexanone monooxygenases: enantioselective biocatalysts for the oxidation of prochiral thioethers. Angew Chem Int Ed Engl 43:4078–4081. doi: 10.1002/anie.200460311. [DOI] [PubMed] [Google Scholar]
- 43.Pazmino DET, Snajdrova R, Rial DV, Mihovilovic MD, Fraaije MW. 2007. Altering the substrate specificity and enantioselectivity of phenylacetone monooxygenase by structure-inspired enzyme redesign. Adv Synth Catal 349:1361–1368. doi: 10.1002/adsc.200700045. [DOI] [Google Scholar]
- 44.Balke K, Kadow M, Mallin H, Sass S, Bornscheuer UT. 2012. Discovery, application and protein engineering of Baeyer-Villiger monooxygenases for organic synthesis. Org Biomol Chem 10:6249–6265. doi: 10.1039/c2ob25704a. [DOI] [PubMed] [Google Scholar]
- 45.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. [DOI] [PubMed] [Google Scholar]
- 46.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mirza IA, Yachnin BJ, Wang S, Grosse S, Bergeron H, Imura A, Iwaki H, Hasegawa Y, Lau PCK, Berghuis AM. 2009. Crystal structures of cyclohexanone monooxygenase reveal complex domain movements and a sliding cofactor. J Am Chem Soc 131:8848–8854. doi: 10.1021/ja9010578. [DOI] [PubMed] [Google Scholar]
- 48.Romero E, Fraaije MW, Castellanos JRG, Mattevi A. 2016. Characterization and crystal structure of a robust cyclohexanone monooxygenase. Angew Chem Int Ed Engl 55:15852–15855. doi: 10.1002/anie.201608951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen YCJ, Peoples OP, Walsh CT. 1988. Acinetobacter cyclohexanone monooxygenase: gene cloning and sequence determination. J Bacteriol 170:781–789. doi: 10.1128/jb.170.2.781-789.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leipold F, Wardenga R, Bornscheuer UT. 2012. Cloning, expression and characterization of a eukaryotic cycloalkanone monooxygenase from Cylindrocarpon radicicola ATCC 11011. Appl Microbiol Biotechnol 94:705–717. doi: 10.1007/s00253-011-3670-z. [DOI] [PubMed] [Google Scholar]
- 51.Kotani T, Yurimoto H, Kato N, Sakai Y. 2007. Novel acetone metabolism in a propane-utilizing bacterium, Gordonia sp. strain TY-5. J Bacteriol 189:886–893. doi: 10.1128/JB.01054-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Onaca C, Kieninger M, Engesser K-H, Altenbuchner J. 2007. Degradation of alkyl methyl ketones by Pseudomonas veronii MEK700. J Bacteriol 189:3759–3767. doi: 10.1128/JB.01279-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leisch H, Shi R, Grosse S, Morley K, Bergeron H, Cygler M, Iwaki H, Hasegawa Y, Lau PCK. 2012. Cloning, Baeyer-Villiger biooxidations, and structures of the camphor pathway 2-oxo-Δ(3)-4,5,5-trimethylcyclopentenylacetyl-coenzyme A monooxygenase of Pseudomonas putida ATCC 17453. Appl Environ Microbiol 78:2200–2212. doi: 10.1128/AEM.07694-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fraaije MW, Wu J, Heuts DP, van Hellemond EW, Spelberg JH, Janssen DB. 2005. Discovery of a thermostable Baeyer-Villiger monooxygenase by genome mining. Appl Microbiol Biotechnol 66:393–400. doi: 10.1007/s00253-004-1749-5. [DOI] [PubMed] [Google Scholar]
- 55.Morii S, Sawamoto S, Yamauchi Y, Miyamoto M, Iwami M, Itagaki E. 1999. Steroid monooxygenase of Rhodococcus rhodochrous: sequencing of the genomic DNA, and hyperexpression, purification, and characterization of the recombinant enzyme. J Biochem 126:624–631. doi: 10.1093/oxfordjournals.jbchem.a022494. [DOI] [PubMed] [Google Scholar]
- 56.Iwaki H, Hasegawa Y, Wang S, Kayser MM, Lau PCK. 2002. Cloning and characterization of a gene cluster involved in cyclopentanol metabolism in Comamonas sp. strain NCIMB 9872 and biotransformations effected by Escherichia coli-expressed cyclopentanone 1,2-monooxygenase. Appl Environ Microbiol 68:5671–5684. doi: 10.1128/AEM.68.11.5671-5684.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ferroni FM, Tolmie C, Smit MS, Opperman DJ. 2016. Structural and catalytic characterization of a fungal Baeyer-Villiger monooxygenase. PLoS One 11:e0160186. doi: 10.1371/journal.pone.0160186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kostichka K, Thomas SM, Gibson KJ, Nagarajan V, Cheng Q. 2001. Cloning and characterization of a gene cluster for cyclododecanone oxidation in Rhodococcus ruber SC1. J Bacteriol 183:6478–6486. doi: 10.1128/JB.183.21.6478-6486.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Iwaki H, Wang S, Grosse S, Bergeron H, Nagahashi A, Lertvorachon J, Yang J, Konishi Y, Hasegawa Y, Lau PCK. 2006. Pseudomonad cyclopentadecanone monooxygenase displaying an uncommon spectrum of Baeyer-Villiger oxidations of cyclic ketones. Appl Environ Microbiol 72:2707–2720. doi: 10.1128/AEM.72.4.2707-2720.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Seo M-J, Zhu D, Endo S, Ikeda H, Cane DE. 2011. Genome mining in Streptomyces. Elucidation of the role of Baeyer-Villiger monooxygenases and non-heme iron-dependent dehydrogenase/oxygenases in the final steps of the biosynthesis of pentalenolactone and neopentalenolactone. Biochemistry 50:1739–1754. doi: 10.1021/bi1019786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jiang J, Tetzlaff CN, Takamatsu S, Iwatsuki M, Komatsu M, Ikeda H, Cane DE. 2009. Genome mining in Streptomyces avermitilis: a biochemical Baeyer-Villiger reaction and discovery of a new branch of the pentalenolactone family tree. Biochemistry 48:6431–6440. doi: 10.1021/bi900766w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kamerbeek NM, Moonen MJH, Van der Ven JGM, Van Berkel WJH, Fraaije MW, Janssen DB. 2001. 4-Hydroxyacetophenone monooxygenase from Pseudomonas fluorescens ACB. A novel flavoprotein catalyzing Baeyer-Villiger oxidation of aromatic compounds. Eur J Biochem 268:2547–2557. [DOI] [PubMed] [Google Scholar]
- 63.Hwang WC, Xu Q, Wu B, Godzik A. 2018. Crystal structure of a Baeyer-Villiger flavin-containing monooxygenase from Staphylococcus aureus MRSA strain MU50. Proteins 86:269. doi: 10.1002/prot.24661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Iwaki H, Grosse S, Bergeron H, Leisch H, Morley K, Hasegawa Y, Lau PC. 2013. Camphor pathway redux: functional recombinant expression of 2,5- and 3,6-diketocamphane monooxygenases of Pseudomonas putida ATCC 17453 with their cognate flavin reductase catalyzing Baeyer-Villiger reactions. Appl Environ Microbiol 79:3282–3293. doi: 10.1128/AEM.03958-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gibson M, Nur-e-alam M, Lipata F, Oliveira MA, Rohr J. 2005. Characterization of kinetics and products of the Baeyer-Villiger oxygenase MtmOIV, the key enzyme of the biosynthetic pathway toward the natural product anticancer drug mithramycin from Streptomyces argillaceus. J Am Chem Soc 127:17594–17595. doi: 10.1021/ja055750t. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.