ABSTRACT
The recently revived Clostridium acetobutylicum-based acetone-butanol-ethanol (ABE) fermentation is widely celebrated and studied for its impact on industrial biotechnology. C. acetobutylicum has been studied and engineered extensively, yet critical areas of the molecular basis for how solvent formation is regulated remain unresolved. The core solventogenic genes (adhE1/aad, ctfA, ctfB, and adc) are carried on the sol locus of the pSOL1 megaplasmid, whose loss leads to asporogenous, “degenerate” cells. The sol locus includes a noncoding small RNA (sRNA), SolB, whose role is presumed to be critical for solventogenesis but has eluded resolution. In the present study, SolB overexpression downregulated the sol-locus genes at the transcript level, resulting in attenuated protein expression and a solvent-deficient phenotype, thus suggesting that SolB affects expression of all sol-locus transcripts and seemingly validating its hypothesized role as a repressor. However, deletion of solB resulted in a total loss of acetone production and severe attenuation of butanol formation, with complex effects on sol-locus genes and proteins: it had a small impact on adc mRNA or its corresponding protein (acetoacetate decarboxylase) expression level, somewhat reduced adhE1 and ctfA-ctfB mRNA levels, and abolished the ctfA-ctfB-encoded coenzyme A transferase (CoAT) activity. Computational predictions support a model whereby SolB expressed at low levels enables the stabilization and translation of sol-locus transcripts to facilitate tuning of the production of various solvents depending on the prevailing culture conditions. A key predicted SolB target is the ribosome binding site (RBS) of the ctfA transcript, and this was verified by expressing variants of the ctfA-ctfB genes to demonstrate the importance of SolB for acetone formation.
IMPORTANCE Small noncoding RNAs regulate many important metabolic and developmental programs in prokaryotes, but their role in anaerobes has been explored minimally. Regulation of solvent formation in the important industrial organism C. acetobutylicum remains incompletely understood. While the genes for solvent formation and their promoters are known, the means by which this organism tunes the ratios of key solvents, notably the butanol/acetone ratio to balance its electron resources, remains unknown. Significantly, the roles of several coding and noncoding genes in the sol locus in tuning the solvent formation ratios have not been explored. Here we show that the small RNA SolB fine-tunes the expression of solvents, with acetone formation being a key target, by regulating the translation of the acetone formation rate-limiting enzyme, the coenzyme A transferase (CoAT). It is notable that SolB expressed at very low levels enables CoAT translation, while at high, nonphysiological expression levels, it leads to degradation of the corresponding transcript.
KEYWORDS: SolB, small RNA, noncoding RNA, Clostridium acetobutylicum, acetone, butanol, ABE, sol locus, metabolic engineering, mRNA stability, solvent formation, RBS, translational regulation
INTRODUCTION
The year 2016 marked the 100th anniversary of the first industrial operation of the acetone-butanol-ethanol (ABE) fermentation, based on the Weizmann Clostridium acetobutylicum strain. C. acetobutylicum has been established as a model organism not only for clostridial genetics and physiology but also for the genetics and physiology of the industrially significant ABE fermentation (1–3). The last 15 years have seen great progress in understanding and engineering the regulation of the organism's metabolism, particularly that of solvent formation (4–8). Yet several important regulatory issues remain unresolved, including the role of small RNAs (sRNAs) in the regulation of the sol-locus genes responsible for solvent production (9). The sol locus is carried on the pSOL1 megaplasmid of C. acetobutylicum (10) and consists of the tricistronic sol operon (adhE1 [or aad]-ctfA-ctfB) (11, 12) on the positive strand and the convergent adc gene on the opposite strand. The bifunctional aldehyde-alcohol dehydrogenase (AdhE1; also known as AAD), encoded by the adhE1 (or aad) gene, is responsible for butanol production (13), while the ctfA and ctfB genes code for the two subunits of the coenzyme A transferase (CoAT) that catalyzes the conversion of acetoacetyl-CoA to acetoacetate while transferring the CoA moiety to either acetate or butyrate (14). Acetoacetate is converted to acetone upon the action of the adc-encoded acetoacetate decarboxylase (AADC) (15, 16).
The sol-locus genes are regulated by the master regulator of sporulation, Spo0A (17, 18). A 0A box (the DNA motif for binding of activated [phosphorylated] Spo0A) and two adjacent imperfect repeats found in the intergenic region upstream of adhE1 are essential for successful sol operon transcription (19, 20). Since inactivation of spo0A does not fully eliminate solvent formation and the expression of the sol-locus genes (18), it can be concluded that Spo0A is one of possibly many regulators of the sol-locus genes (5). These additional regulators would account for the fact that solventogenesis is controlled by several physiological conditions (i.e., stress, low pH, and source levels of carbon, nitrogen, and other nutrients) (21–24), for which molecular details are not yet known. Small, noncoding regulatory RNAs have been known to regulate the responses of metabolic and developmental programs in prokaryotes to various environmental signals, including stress, nutrient composition and availability, ion concentrations, pH, and oxygen tension, to name a few (9, 25–28). Many small RNAs in Clostridium organisms have been identified using genomic approaches (9, 26, 29), but with few exceptions, the roles and mechanistic actions of these small RNAs remain largely unexplored (9, 26, 30–32).
Here we explore the role of a small noncoding RNA, SolB, which was identified by Peter Dürre a few years ago, although there is no published work on it outside meeting presentations, a patent (33), and a Ph.D. thesis from his lab (34). We previously detected low levels of expression of SolB (annotated sCAP_176) in studies that aimed to identify and examine the expression of small RNAs in this organism under stress (9, 26). The 196-bp solB sequence lies upstream of the adhE1 gene of the sol operon. Zimmerman (34) identified a transcriptional start site and a putative promoter matching clostridial consensus promoters. Downregulation of SolB under butyrate stress and its upregulation under butanol stress have been demonstrated and are opposite the pattern observed for sol operon expression under the same conditions (5). These findings support the hypothesis, proposed by Dürre, that SolB is a repressor of solventogenesis, which was the starting hypothesis for the present study. Accordingly, we knocked out and overexpressed SolB in C. acetobutylicum ATCC 824 and examined the impacts on solventogenesis and the expression of the sol-locus genes/proteins.
RESULTS
Expression of SolB in ATCC 824 and generation of the solB deletion strain.
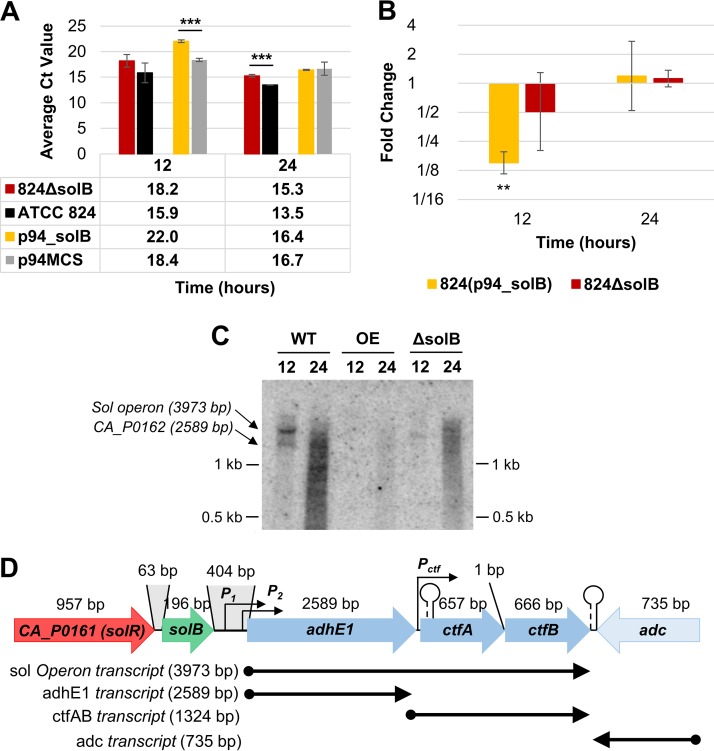
Overexpression of SolB from a strong clostridial promoter (Pptb) in C. acetobutylicum was achieved through cloning of a 234-bp sequence consisting of the complete 196-bp solB coding sequence (34) and the neighboring 18-bp upstream region and 20-bp downstream region into the p94MCS expression vector (Table 1) (see Materials and Methods). Transformation of this plasmid into wild-type ATCC 824 (ATCC 824 WT) generated the overexpression strain 824(p94_solB). A solB deletion strain, 824 ΔsolB, was generated using the pKO_mazF allelic exchange system (35, 36), which enables facile and precise deletion of small and large DNA sequences from the genome. Construction of the 824(p94_solB) strain was confirmed by Sanger sequencing (see Fig. S1 in the supplemental material); successful construction of the 824 ΔsolB strain was also confirmed by Sanger sequencing and PCR to identify both ends of the allelic exchange integration (Fig. 1). Loss of SolB expression in 824 ΔsolB was confirmed by quantitative reverse transcription-PCR (Q-RT-PCR) analysis (Fig. 2A and B). Northern blot analysis revealed significantly increased SolB expression in 824(p94_solB) (Fig. 2C). There were no detectable bands for SolB in 824 ΔsolB or the WT, the latter due to the very low expression level of SolB (9, 34). In agreement with the results of the Northern analysis, cycle threshold (CT) values (lower CT values indicate higher expression levels) and fold changes generated from Q-RT-PCR data substantiated the significant increase of SolB levels in 824(p94_solB), the absence of SolB expression in 824 ΔsolB (since the solB DNA was deleted), and the very low native SolB expression level in the WT strain, which has consistently been observed in prior transcriptome sequencing (RNA-seq) studies under normal and stress conditions (5).
TABLE 1.
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| E. coli strains | ||
| Turbo | lacIq endA1 Δ(hsdS-mcrB)5 | NEB |
| 10-beta competent strain | recA1 endA1 Δ(mrr-hsdRMS-mcrBC) | NEB |
| BL21(pAN3) | pAN3 Kmr ϕ3T I | 35 |
| C. acetobutylicum strains | ||
| ATCC 824 (WT) | Wild-type strain | ATCC |
| 824(p94MCS) (empty plasmid strain) | p94MCS; MLSr | 35 |
| 824(p94_solB) (solB overexpression strain) | p94_solB; MLSr | This study |
| 824(pKO_mazF::solB) | pKO_mazF::solB; Thr | This study |
| 824 ΔsolB | solB deletion; Thr | This study |
| Plasmids | ||
| p94MCS | Ampr MLSr; ptb promoter; MCS | 35 |
| p94MCS_solB | Ampr MLSr; ptb promoter; solB | This study |
| pKO_mazF | Thr-FRT; repL ori; bgaR and Pbgal upstream of mazF | 35 |
| pKO_mazF::solB | Thr-FRT; repL ori; bgaR and Pbgal upstream of mazF; Thr cassette flanked by ∼900-bp upstream and downstream solB regions of homology | This study |
| pAN3 | Kmr; ϕ3T I | 35 |
lacIq, lac repressor gene; endA1, nonspecific endonuclease gene deleted; Δ(hsdS-mcrB)5, host-specific restriction and methylcytosine-specific restriction genes deleted; recA1, homologous recombination gene deleted; Δ(mrr-hsdRMS-mcrBC), host-specific restriction and methylcytosine-specific restriction genes deleted; Kmr, kanamycin resistance; ϕ3T I, Bacillus subtilis phage ϕ3T I methyltransferase gene; MLSr, macrolide-lincosamide-streptogramin B resistance; Thr, thiamphenicol resistance; Ampr, ampicillin resistance; ptb, phosphotransbutyrylase gene; MCS, multiple cloning site; Thr-FRT, FRT-flanked thiamphenicol resistance gene; repL, pIM13 Gram-positive origin of replication; ori, ColE1 origin of replication; bgaR, beta-galactosidase regulator; Pbgal, promoter of beta-galactosidase; mazF, codon-optimized E. coli MazF toxin gene; pUC ori, pUC Gram-negative origin of replication.
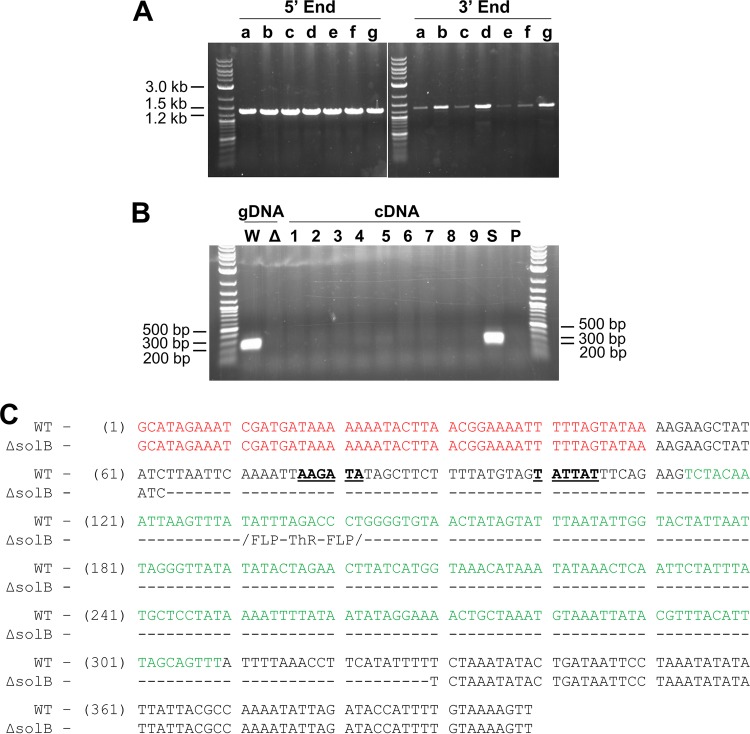
FIG 1.
PCR confirmation of 824 ΔsolB and sequencing of the solB region. (A) Amplification products of the Thr cassette 5′ and 3′ integration sites in genomic DNAs from 824 ΔsolB replicate strains a to g (strain b was chosen for further study). (B) Amplification products of the solB sequence from genomic DNAs (gDNA) of WT ATCC 824 (W) and 824 ΔsolB strain b (Δ) as well as from cDNAs from 3 replicate cultures of 824 ΔsolB (lanes 1, 4, and 7, 6 h; lanes 2, 5, and 8, 12 h; and lanes 3, 6, and 9, 24 h) and cDNAs from exponential-phase cultures (6 h) of the solB overexpression strain (S) and the empty plasmid strain (P). (C) Sequencing of the solB region in 824 ΔsolB, aligned with the ATCC 824 genome, showing deletion of solB (in green), together with 50 bp upstream (including its sigA promoter consensus sequence [34]) (bold and underlined) and 20 bp downstream, but no interference with the solR open reading frame (ORF) (in red) or any downstream sol operon regulatory sequences (not shown). FLP-ThR-FLP, 1,169-bp thiamphenicol resistance cassette containing flippase recognition sequences and inserted into the 824 ΔsolB genome.
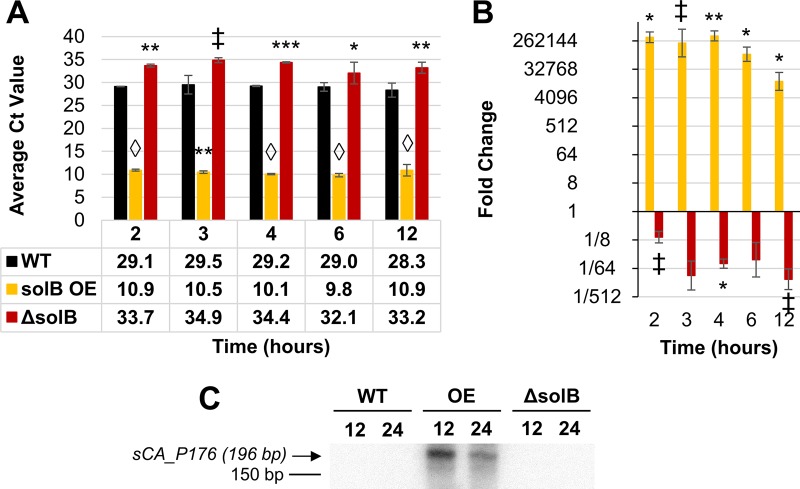
FIG 2.
Expression of solB in recombinant strains. (A) Raw CT values from Q-RT-PCR analysis of solB transcript levels (the average CT value for solB in nontemplate controls was 31.96 ± 0.41). (Note that CT values are inversely proportional to expression levels.) (B) Fold changes in solB overexpression (relative to the WT expression level), derived from Q-RT-PCR analysis. All error bars represent standard deviations (SD) (n = 2). ‡, P ≤ 0.07; *, P ≤ 0.05; **, P ≤ 0.01, ***, P ≤ 0.001, ♢, P ≤ 0.0001 (two-tailed, unpaired t test). (C) Northern blot analysis of solB expression in the WT strain (ATCC 824), the 824(p94_solB) solB overexpression strain (OE), and the 824 ΔsolB strain, using 5 μg total RNA.
SolB overexpression reduces solvent formation, but against expectations, solB deletion abolishes acetone formation and severely attenuates butanol formation.
Based on the hypothesis that SolB is a repressor of solventogenesis, it would be expected that its overexpression will result in a solvent-deficient phenotype and that its deletion will lead to higher solvent titers. Three biological replicates of static cultures of ATCC 824 WT, 824(p94MCS) (empty plasmid control), 824(p94_solB) (SolB overexpression strain), and 824 ΔsolB were grown for 72 h and assayed regularly for cell density, glucose consumption, and metabolite concentrations. The 824(p94_solB) strain exhibited significantly reduced production of butanol (19.8 mM), acetone (6.6 mM), and ethanol (3.7 mM) relative to that in the plasmid control strain and also that in the WT strain (Fig. 3 and Table 2). Acid production was sustained throughout the stationary phase, with butyrate and acetate concentrations peaking at ca. 53 mM and 43 mM, respectively, and remaining reasonably constant after approximately 24 h. That is, there was no acid reuptake, suggesting that the expression or activity of the CoA transferase (CoAT) enzyme was very low. (Note that the culture medium contained acetate at about 30 mM, which is the level of acetate at time zero in Fig. 3; Table 2 shows final net product titers.) The 824 ΔsolB deletion strain exhibited an even more severe solvent-deficient phenotype (Fig. 3 and Table 2), with 5.5 mM butanol, 2.7 mM ethanol, and no acetone that could be detected. This solvent-deficient phenotype was again associated with sustained acid production and high acid titers, with no acid uptake. This behavior of 824 ΔsolB appears to negate the hypothesized role of SolB as an sRNA repressor of the solventogenic genes.
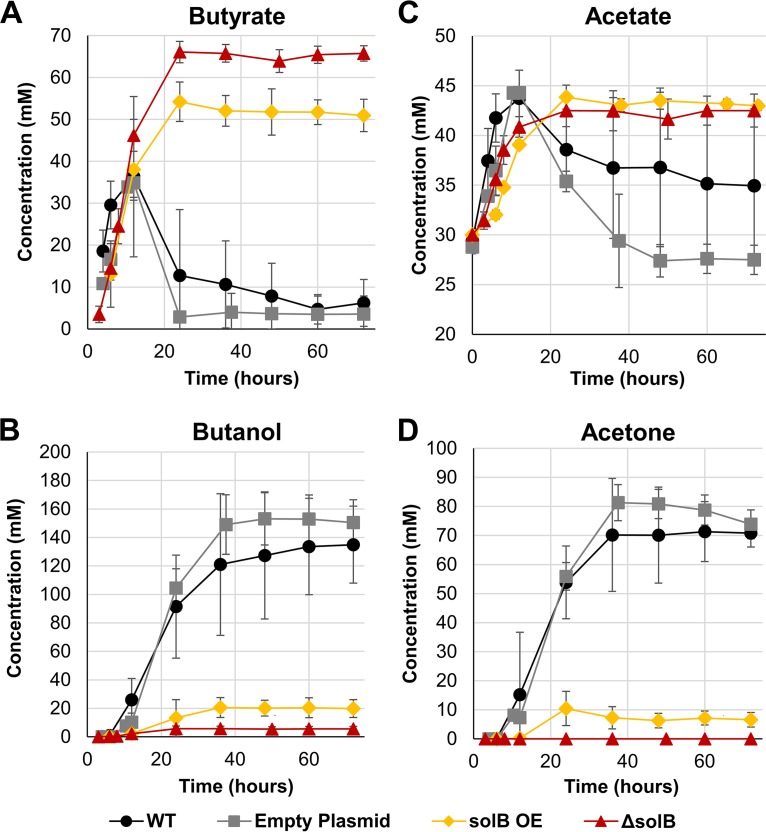
FIG 3.
Metabolite profiles of recombinant strains. Time profiles are shown for butyrate (A), butanol (B), acetate (C), and acetone (D) concentrations in the WT strain (ATCC 824), the empty plasmid strain [824(p94MCS)], the solB overexpression strain [824(p94_solB)] (solB OE), and the 824 ΔsolB strain over 72 h of static culture without pH adjustment. Error bars represent SD (n = 3). Endpoint concentrations (net product titers) are listed in Table 2; the time profiles shown here include starting medium concentrations, notably 30 mM acetate.
TABLE 2.
Endpoint metabolite concentrations after 72 h of static-flask growth
| Strain | Net product titer (mM)a |
|||||
|---|---|---|---|---|---|---|
| Butanol | Acetone | Ethanol | Acetate | Butyrate | Glucoseb | |
| WT | 135.0 ± 27.0 | 70.8 ± 4.8 | 19.2 ± 5.5 | 1.2 ± 6.6 | 6.2 ± 5.6 | 276.9 ± 52.1 |
| 824(p94MCS) | 150.4 ± 16.1 | 73.9 ± 4.9 | 29.6 ± 4.9 | −1.3 ± 1.9 | 3.6 ± 4.3 | 311.8 ± 24.2 |
| 824(p94_solB) | 19.8 ± 6.2 | 6.6 ± 2.5 | 3.7 ± 1.2 | 14.8 ± 3.2 | 50.9 ± 3.9 | 105.2 ± 5.3 |
| 824 ΔsolB | 5.5 ± 1.8 | 0.0 ± 0.0 | 2.7 ± 0.6 | 12.5 ± 1.6 | 65.7 ± 1.8 | 85.8 ± 10.4 |
Values are averages (n = 3) ± SD and are normalized to the level at the first time point. Metabolite profiles are shown in Fig. 3.
Total glucose consumption after 72 h of static growth.
The transition phase, typically occurring between 10 and 12 h and representing the metabolic shift from acid to solvent production, marks the point at which both 824(p94_solB) and 824 ΔsolB diverge from the WT culture (Fig. 3). Growth curves for all strains (Fig. S2) reveal that cell growth peaked at lower cell densities for 824(p94_solB) and 824 ΔsolB than for the WT and plasmid control strains; this stunted growth is not surprising given the abnormally high levels of acid metabolites and the corresponding low pH (due to a lack of pH control in these simple batch cultures), which is known to inhibit cell growth (27, 37). As a result, glucose consumption in both strains [824(p94_solB) and 824 ΔsolB] was reduced about 3-fold as well (Table 2). Given that SolB expression is downregulated under butyrate stress (9) and that there is large variability in the timing and levels of acid production in batch cultures of C. acetobutylicum, we were concerned that premature culture termination due to acid production and low pH might mask the physiological role of SolB in regulating solventogenesis. We thus carried out bioreactor experiments whereby the pH was prevented from dropping below 5.0 but was not controlled from rising to higher values. This is a method we have used extensively in the past for physiological and molecular strain characterization, and several detailed fermentation profiles of WT and engineered strains have been published (38–40). Two bioreactor experiments with 824 ΔsolB resulted in higher levels of butanol (ca. 30 to 35 mM) but still no acetone production (Fig. 4). We note the expected, much higher butyrate and acetate levels due to pH control. Thus, the phenotype of no acetone production of this strain is robust and reproducible and shows that the cells cannot produce acetone and thus cannot uptake the two acids via the CoAT reaction. These phenotypic data from the 824 ΔsolB bioreactor fermentations resemble those for the M5(pCAAD) strain (13). M5 is the 824 strain cured of the pSOL1 megaplasmid that carries the sol-locus genes. Plasmid pCAAD expresses the AdhE1/AAD protein from its natural sol operon promoter. Thus, there is no acetone production due to the absence of the ctfA-ctfB and adc genes. This reinforces the hypothesis that there is no expression of the CoAT protein in 824 ΔsolB, and also that the AdhE1/AAD protein is expressed and functional, thus enabling butanol (and ethanol) production.
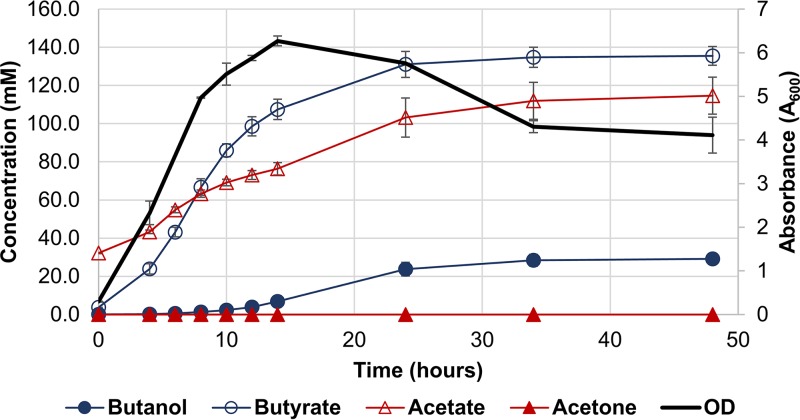
FIG 4.
Growth curve and metabolite profile for batch fermentation cultures of 824 ΔsolB. Cultures of 824 ΔsolB were grown in 4-liter bioreactors and maintained at a pH of ≥5.0 throughout growth. Error bars represent SD (n = 2). Metabolite profiles are not normalized and thus reflect starting medium concentrations, notably 30 mM acetate.
Taken together, the data from the SolB overexpression strain [824(p94_solB)] support the hypothesis that SolB is a transcriptional repressor of some or all sol-locus genes (sol operon and adc), but the data from the 824 ΔsolB strain paint a more complex picture.
Overexpression of SolB results in decreased transcript levels and severely attenuated protein levels of sol-locus genes/proteins.
We investigated the effects of SolB overexpression on solvent production (Fig. 3 and Table 2) by examining its impact on transcript levels of three primary sol-locus genes—adhE1/aad, ctfA, and adc—by Q-RT-PCR and Northern blot analysis. We also examined the impact of SolB overexpression on protein levels of CoAT and AADC, for which we had available polyclonal antibodies for Western analysis. Relative transcript levels of the sol-locus genes were measured by Northern analysis and Q-RT-PCR analysis of ATCC 824 WT, 824(p94MCS), 824(p94_solB), and 824 ΔsolB. Initial Northern blots revealed strong expression of all genes in both ATCC 824 WT and 824(p94MCS) samples (Fig. S3); thus, to conserve gel lanes, only ATCC 824 WT samples were included for comparison in all subsequent Northern blots.
The sol operon had heretofore been assumed to generate one long, 3,973-bp transcript (Fig. 5D). However, in recent RNA-seq studies, the junction between adhE1 and ctfA revealed a sustained low coverage of read counts (41) (Fig. S4), and upon further examination, a previously undiscovered putative terminator sequence was identified within the first 60 bp of the ctfA coding sequence (coordinates 178488 to 178525 on pSOL1) (41). This Rho-independent terminator, with a ΔG value of −9.6 kcal/mol, was not identified previously due to its location within a coding region, but the presence and relative sizes of the two bands seen in Northern blot analysis (Fig. 5C [discussed below] and the multiple bands in the adhE1 and ctfA Northern blots of Fig. S3) suggest that it is both present and active, generating the originally identified polycistronic transcript as well as a truncated transcript containing only adhE1. There is also a strong clostridial promoter, TTCATA(13)TATAAT, just upstream of the ribosome binding site (RBS) of the ctfA gene, suggesting that the ctfA-ctfB genes can be expressed independently from such a promoter. Thus, there are a total of three potential transcripts (Fig. 5D and data not shown). Since the transcript for ctfB remains on the same mRNA molecule as that with the ctfA transcript, expression of ctfA was deemed to be representative of both ctfA and ctfB expression.
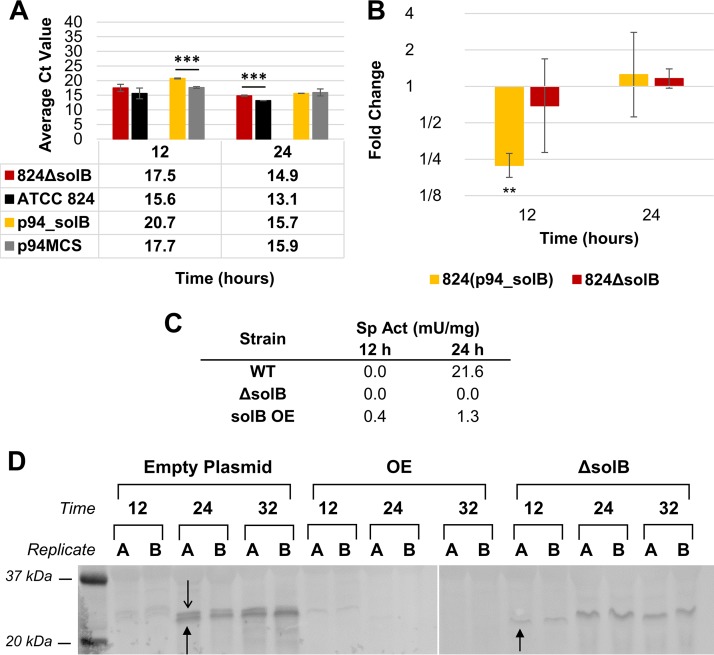
FIG 5.
Expression analysis of adhE1. (A) Average raw CT values (n = 3) from Q-RT-PCR analysis of adhE1 transcript levels (the average CT value for adhE1 in nontemplate controls was 29.58 ± 0.12). (B) Fold changes in adhE1 expression in 824(p94_solB) and 824 ΔsolB [relative to the expression levels in 824(p94) and the WT, respectively], derived from Q-RT-PCR analysis. All error bars represent SD (n = 3). **, P ≤ 0.01; ***, P ≤ 0.001 (two-tailed, unpaired t test). (C) Northern blot analysis of adhE1 expression at 12 and 24 h, revealing expression both as part of the polycistronic sol operon and as a single transcript, using 15 μg of total RNA. (D) Schematic of solB and the sol-locus genes, showing labels for the distal and proximal sol operon promoters (P1 and P2), the putative ctfA-ctfB promoter (Pctf), the hairpin Rho-independent terminator starting 36 bp into the ctfA coding sequence, and the strong bidirectional Rho-independent terminator between ctfB and the convergent adc gene. Also indicated are the proposed transcripts of sol operon genes (lengths listed; not drawn to scale), which are supported by RNA-seq data (9) (see Fig. S4 in the supplemental material).
Using a probe against adhE1, the SolB overexpression strain 824(p94_solB) showed very low to undetectable levels of the adhE1 transcript on Northern blots (Fig. 5C) as well as higher CT values and fold changes (Fig. 5A and B), indicating significantly reduced expression at 12 and 24 h, which represent the most meaningful points for expression of solventogenic genes, given that solvent production starts at a point between 10 and 12 h and has the highest rate between 12 and 24 h. Consistent with the delayed, low level of butanol production (Fig. 3 and Table 2), there was a faint, degraded adhE1 transcript at 24 h on the Northern blot and a concomitantly lower CT value for the SolB overexpression strain [824(p94_solB)]. Note in particular that for the samples exhibiting adhE1 expression, two bands appeared on the Northern blot, with both appearing larger than 1,000 bp. As discussed above, one represents the expected polycistronic sol operon transcript (3,973 bp), and the other, smaller band likely corresponds to a smaller transcript containing only adhE1 (2,589 bp) (Fig. 5C and D). Although mRNA degradation after the mid-exponential phase in simple batch cultures remains a serious problem (less so in pH-controlled cultures), one may perhaps conclude that the shorter, adhE1-containing transcript prevails over the longer transcript at 24 h.
Several efforts to generate high-quality Northern blots of ctfA and ctfB transcripts did not prove successful. While two transcripts could be seen (31) (Fig. S3), presumably corresponding to the 3,973-bp and 1,324-bp transcripts shown in Fig. 5D, the bands showed excessive degradation. Nevertheless, their patterns correspond qualitatively to those of the Q-RT-PCR data (Fig. 6A and B), which indicate substantial reductions in transcript levels only for the SolB overexpression strain [824(p94_solB)]. We note that although mRNA degradation was generally a problem, it somehow affected the transcripts probed with the ctfA probe more than those probed with the adhE1 probe, with very little degradation when the mRNA was probed with the adc probe (see below). These data thus suggest that ctfA-containing transcripts are less stable than the other sol-locus transcripts.
FIG 6.
Expression analysis of ctfA. (A) Average raw CT values (n = 3) from Q-RT-PCR analysis of ctfA transcript levels (the average CT value for ctfA in nontemplate controls was 30.85 ± 0.16). (B) Fold changes in ctfA expression in 824(p94_solB) and 824 ΔsolB [relative to the expression levels in 824(p94) and the WT, respectively], derived from Q-RT-PCR analysis. All error bars represent SD (n = 3). **, P ≤ 0.01; ***, P ≤ 0.001 (two-tailed, unpaired t test). (C) Specific activities of the CoAT enzyme from crude lysates of the WT (ATCC 824), 824 ΔsolB, and 824(p94_solB) (solB OE) strains. (D) Western blot analysis of CoAT protein expression (note bands for both subunits only for the wild type, with one band for 824 ΔsolB), using 50-μg samples, primary antisera diluted 1:500, and secondary antisera diluted 1:10,000.
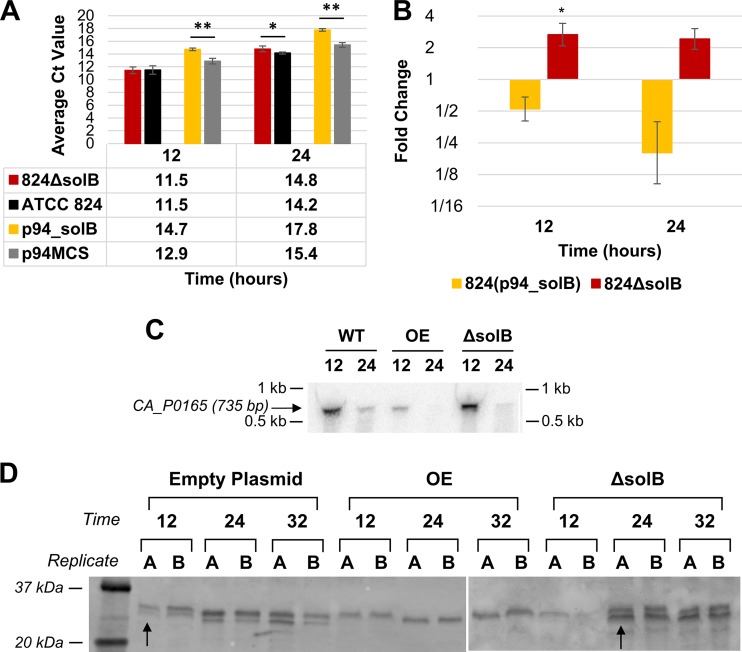
Transcription of adc peaks at approximately 12 h and then steadily decreases throughout stationary phase (15); this expression pattern was seen in Northern blot analyses of all samples (Fig. 7C). Both Northern blot and Q-RT-PCR data for the adc gene (Fig. 7) show that SolB overexpression leads to strong downregulation of, but does not abolish, the adc transcript. We note here that the stability of the adc mRNA enabled higher-quality Northern blots than those with the other sol operon transcripts, especially those with the ctfA-ctfB mRNAs.
FIG 7.
Expression analysis of adc. (A) Average raw CT values (n = 3) from Q-RT-PCR analysis of adc transcript levels (the average CT value for adc in nontemplate controls was 30.53 ± 0.17). (B) Fold changes in adc expression in 824(p94_solB) and 824 ΔsolB [relative to the expression levels in 824(p94MCS) and the WT, respectively], derived from Q-RT-PCR analysis. All error bars represent SD (n = 3). **, P ≤ 0.01; ***, P ≤ 0.001 (two-tailed, unpaired t test). (C) Northern blot analysis of adc expression at 12 and 24 h. (D) Western blot analysis of AADC protein expression (lower band, as previously verified [43]), using 50-μg samples, primary antisera diluted 1:250, and secondary antisera diluted 1:10,000. Panel D is a combination of images of two separate gels/membranes, since it was not possible to fit all lanes in one gel.
Next, we investigated the impact of SolB overexpression on the protein expression of CoAT and AADC by using Western analysis (we did not have an antibody for AdhE1, preventing its investigation at the protein level). As previously documented (42), two bands, nearly indistinguishably close together, representing the ∼26-kDa (CtfA) and ∼28-kDa (CtfB) CoAT subunits, were seen for the WT strain at 24 and 32 h, with faint bands at 12 h (Fig. 6D). The 824(p94_solB) strain displayed one or possibly two CoAT bands at 12 h only. Additionally, cell lysates displayed low levels of CoAT enzymatic activity for the SolB overexpression strain 824(p94_solB) compared to those for the WT strain (Fig. 6C). These data and the low levels of acetone produced (Fig. 3D) suggest that the cells of the 824(p94_solB) strain contain low levels of CoAT protein, which are apparently below the level of detection in our Western blots (Fig. 6D).
Two bands appeared on the AADC Western blots, near the expected protein size (Fig. 7D): the lower band, marked with an arrow in the figure, represents AADC, with the upper band showing a separate, unknown protein to which the antibody cross-reacts (43). The AADC band was present for all WT samples but none of the 824(p94_solB) strain samples (Fig. 7D), despite detectable (although low levels of) adc transcripts (Fig. 7C) and low levels of acetone formed by the strain. This suggests that AADC expression is low and was below the detectable level in our Western gels.
While translational repression by mRNA target degradation is a common regulatory mechanism for numerous prokaryotic sRNAs (44), transcriptional repression may also result indirectly from a trans-acting sRNA repressing the translation of a transcriptional activator to block the transcription of its target genes. As stated above, the transcription factor Spo0A, the master regulator of sporulation in Clostridium and all endospore formers, is an essential activator of the sol-locus genes (4, 18) and a possible target of SolB (34). Therefore, we were interested in investigating possible indirect repression of the sol-locus genes by SolB via its interaction with the spo0A mRNA. We thus assayed the spo0A transcript by Q-RT-PCR. CT values and fold change calculations show for both 824(p94_solB) and 824 ΔsolB that spo0A transcript levels remained nearly identical to those in the WT strain (Fig. S5), indicating that SolB does not affect the level of spo0A mRNA.
Collectively, these data show that in the SolB overexpression strain [824(p94_solB)], the sol operon and adc transcripts are severely downregulated but not completely absent and result in low levels of the corresponding proteins and low levels of butanol and acetone formation. These data support the role of SolB as a translational repressor that targets the mRNAs of the sol-locus genes, whereby the presumed binding leads to transcript degradation by endogenous ribonucleases recognizing and degrading the abnormal RNA duplexes (25, 42, 44). We note, however, that given the very low levels of SolB expression under normal growth conditions, this role of SolB as a translational repressor does not have physiological relevance. Its physiological role must therefore be different. The fact that SolB overexpression dramatically and specifically reduces the adhE1, ctfA-ctfB, and adc transcript levels supports computational models (see below) indicating that SolB interacts with these three transcripts. This is a critical hypothesis in deciphering the likely physiological role of SolB.
solB deletion abolishes any detectable CoAT protein expression and activity and attenuates adhE1 levels but leaves adc mRNA and AADC protein levels largely unaffected.
As with the WT strain, bands were present on the Northern blots for 824 ΔsolB samples, showing detectable but lower adhE1 mRNA expression, and this was corroborated by the Q-RT-PCR data (Fig. 5). Although the adhE1 bands were less strong than those of the WT strain, the transcript was nevertheless present. Q-RT-PCR data (Fig. 6A and B) showed that transcript levels of ctfA were lower for 824 ΔsolB samples than for the WT strain but were similar to those for the plasmid control strain, which produces solvents at high levels. In enzyme assays, we were not able to detect any CoAT activity (Fig. 6C), and the Western blot showed either the lower band (corresponding to CtfA) at 12 h or the upper band at 24 and 32 h, but never both; this is consistent with the lack of detectable enzyme activity. Bands of the adc mRNA with an intensity comparable to that for the WT strain were seen in 824 ΔsolB samples (Fig. 7C), and this was supported by the Q-RT-PCR data (Fig. 7A and B), collectively indicating that adc transcription under solB deletion conditions occurs at levels similar to those in the WT strain. The Western blot showed strong expression levels of the AADC protein at 24 and 32 h (Fig. 7D), yet no acetone was produced at those times or any time by the 824 ΔsolB strain (Fig. 1D), suggesting that the AADC level was not limiting acetone formation, a conclusion also supported by earlier work in our lab (42).
Taken together, these data show that solB deletion does not affect the transcript levels of ctfA-ctfB or adc significantly, yet it abolishes CoAT activity. The adhE1 mRNA levels in the deletion strain appeared to be lower than those in the WT strain but comparable to those in the plasmid control strain. AdhE1 protein expression or activity could not be assessed directly, but the metabolite data in Fig. 4 document butanol formation, albeit at a level lower than that of the WT strain, thus suggesting the presence of AdhE1 activity. It appears that only the ctfA-ctfB transcript cannot be translated in the absence of SolB, which suggests that SolB binding to ctfA-ctfB-containing mRNAs is necessary for their translation. As a result, there is no CoAT activity in the 824 ΔsolB strain, and thus no acetone production is possible (Fig. 3 and 4), as AADC does not limit acetone formation per the discussion above.
Secondary structures and SolB binding predictions indicate likely regulatory interactions between SolB and the sol-locus transcripts.
To elaborate on our experimental findings and consider potential mechanisms of molecular interaction between SolB and the sol-locus genes, we performed multiple computational analyses (see Materials and Methods). First, putative secondary structure folding was generated for the 5′ untranslated region (UTR) and the first 100 bp of the coding region for each sol-locus gene transcript, using the Vienna RNA Websuite (45). Second, predictions for binding between SolB and the 5′ UTR plus 100 bp of each transcript were generated using the IntaRNA online prediction program (46, 47) as well as the RNAup Web server (also part of the Vienna RNA Websuite). Given the variability that can exist among computational algorithms, both programs were utilized to more thoroughly examine binding potential.
The secondary structure folding predictions revealed significant stem-loop motifs in adhE1, ctfA, and adc transcripts (Fig. S6 to S8). Large, negative free energy values associated with the structure predictions (−23.4 kcal/mol for adhE1, −27.2 kcal/mol for adc, and −22.1 and −40 kcal/mol for ctfA) indicate strong probabilities of the natural occurrence of these folding motifs. Additionally, the predicted folding of each transcript occludes the ribosomal binding site, suggesting possible regulatory involvement of the secondary structures of these transcripts, particularly the potential need for translational activation, which can typically require the action of a regulatory sRNA (48).
The binding predictions between SolB and the target transcripts reveal a strong putative interaction between SolB and each transcript, as indicated by significant negative free energy values. The generally accepted target model for such binding predictions is the 5′ UTR plus 100 bp of coding sequence (49); however, given the dynamic nature of such molecular interactions, as well as the changing conditions possible in vivo, we deemed it prudent to test multiple models to provide wider insight and a more comprehensive picture (see Materials and Methods). The resulting sets of free energy values and associated alignment predictions reveal the probability of each binding interaction. The binding predictions for SolB with the adhE1 transcript exhibited predicted free energies ranging from −3.88 to −4.62 kcal/mol, with each prediction exhibiting highly analogous base pairing (Fig. S6). Each predicted SolB binding region on adhE1 lay on a strongly predicted stem-loop, with one loop containing the ribosomal binding site and start codon. On the adc transcript UTR, two regions on opposite sides of the same bubble motif were predicted to interact with SolB, with nearly perfect alignment and with free energies of −4.63 and −6.15 kcal/mol (Fig. S8). For ctfA, strong binding with SolB was predicted for a single sequence within the ctfA coding region, 93 bp downstream of the ribosomal binding site (Fig. S7A). This was the sole sequence identified by both prediction algorithms and using each prediction model, showing a free energy of −6.23 kcal/mol. On the secondary structure for the ctfA transcript, this predicted binding sequence aligns neatly with the ribosomal binding site (Fig. S7C). Significantly, using every prediction parameter, all three transcript targets were predicted to bind with SolB at one of two strongly predicted central regions of its secondary structure (Fig. S9).
To determine the specificity/preference for the sol-locus genes as SolB regulatory targets, genome-wide target prediction computations were also conducted. Using the TargetRNA prediction algorithm and prediction criteria outlined in Materials and Methods and in Table S3, the most consistently predicted targets (i.e., appearing in at least 4 predictions) for SolB were ctfA and ctfB, along with thiL, the gene for the plasmid-borne acetyl-CoA acetyltransferase (thiolase), with highest rankings of 5, 19, and 1, respectively, across the 17 queries (Table S4). Computational predictions can be imperfect indicators of in vivo conditions, so multiple queries utilizing different parameters (see Materials and Methods and Table S3) were run to achieve a more comprehensive view and to account for variabilities. Although not predicted with the highest rank or as the sole target of SolB, the sol locus was frequently and consistently identified by the prediction algorithm, and we placed value in this consistency with which it appeared as we adjusted the query parameters, interpreting it as indicative of the strength of the prediction. Along with thiL, the sol locus was uniquely predicted to interact with SolB at the same sequence and with the same energy in all predictions.
Expression of ctfA with a mutated RBS restores acetone production in the SolB deletion strain.
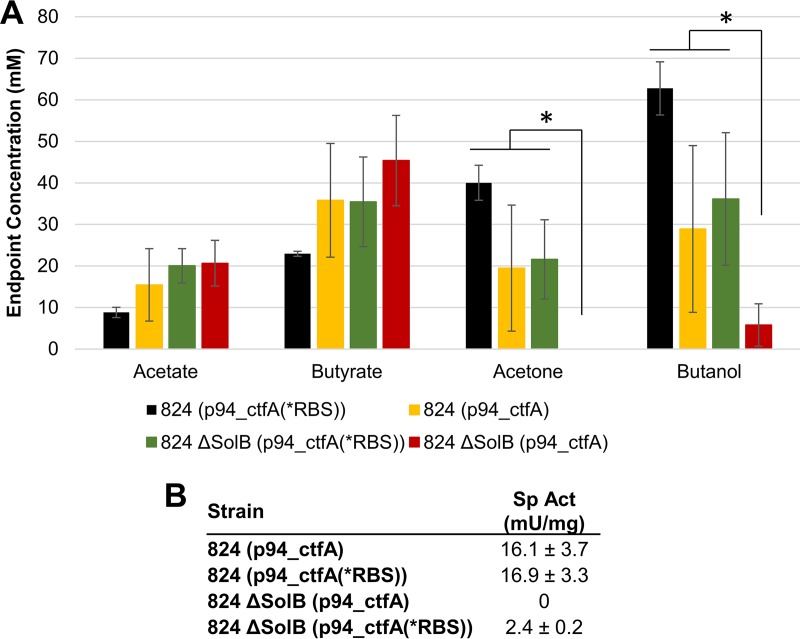
Our data described above suggest that SolB is an activator of ctfA translation or that it stabilizes the ctfA-ctfB transcript. This transcript folds back on itself, blocking the RBS; the predicted SolB binding site on ctfA is the exact sequence that interacts with the RBS during this folding. Since this occlusion of the RBS creates the proposed need for SolB binding and stabilization, mutagenesis that prevents the folding should allow for translation of ctfA-ctfB in the absence of SolB. Thus, to investigate the proposed interaction between SolB and the sol locus in vivo, site-directed mutagenesis of the ctfA RBS was performed (see Materials and Methods and Fig. S10). The RBS was changed into a different clostridial RBS sequence (the RBS for adhE1). We chose the adhE1 RBS because we wanted to use a related solvent formation RBS. The adhE1 RBS is not affected by any significant SolB-related regulation, since, as we discussed, AdhE1/AAD expression off its natural sol operon promoter from plasmid pCAAD results in good protein expression and butanol production in complementing the degenerate strain M5, which does not contain the solB gene (13). Secondary structure prediction (Fig. S10) indicates significant but not detrimental alteration of the ctfA transcript, which effectively interrupts the transcript's secondary structure to prevent its folding. Transformation of the mutated plasmid into the 824 ΔsolB strain generated the strain 824 ΔsolB(p94_ctfA(*RBS)). In addition, a plasmid containing the ctfA-ctfB transcript with its native RBS (p94_ctfA) was also transformed as a control. Both plasmids were also transformed into WT ATCC 824 as additional controls.
Small batch cultures without pH control of the 824 ΔsolB(p94_ctfA(*RBS)) strain exhibited restored acetone and butanol production, while the same solB deletion strain carrying the plasmid with the native RBS produced no acetone and diminished levels of butanol (P < 0.05), as seen before (Fig. 8A). Interestingly, statistically significantly higher acetone and butanol titers were observed in the WT 824 cultures transformed with p94_ctfA(RBS*) than in those transformed with the nonmutated control; this would be expected from the proposed model given the very low levels of SolB and the high expression of the ctfA-ctfB transcript from a multicopy plasmid. Note that the two plasmid-containing WT 824 strains showed lower levels of butanol and acetone than those typically observed in WT 824 fermentations. The interpretation here is that the larger number of ctfA RNA copies deriving from plasmid expression titrates SolB and alters its impact on the expression of AdhE1, resulting in lower butanol expression, which, in WT 824, is coupled to acid uptake and thus to acetone formation. To further support the proposed model, CoAT activities were measured in cell extracts prepared from batch cultures after 24 h. CoAT activity was detected in 824 ΔsolB(p94_ctfA(*RBS)), at 2.4 ± 0.2 mU/mg protein (Fig. 8B). No CoAT activity was observed in the plasmid control with the unmutated RBS. While the CoAT activity in 824 ΔsolB(p94_ctfA(*RBS)) is clearly sufficient to restore solvent formation, the measured activity is still lower than those of the WT 824 strains transformed with the mutated and unmutated ctfA plasmids, which showed activities of 16.9 ± 3.3 mU/mg and 16.1 ± 3.7 mU/mg, respectively. These results indicate that elimination of transcript folding on the RBS region in the absence of SolB restores CoAT translation and acetone production. As shown previously in this study, SolB deletion results in elimination of acetone production and the absence of CoAT translation (Fig. 3D and 6C and D). Restored enzyme and acetone production through a transcript that lacks the proposed need for SolB stabilization therefore indicates that it was the lack of SolB binding to the ctfA-ctfB transcript to enable translation that initially abolished CoAT translation and acetone production; therefore, per the proposed model, SolB binding is necessary for activation of the native transcript.
FIG 8.
Metabolite production and CoAT enzyme activities in C. acetobutylicum strains expressing ctfA-ctfB with the native RBS and a mutated RBS. (A) Endpoint concentrations of acetate, butyrate, acetone, and butanol (net product titers) from 824(p94_ctfA), 824(p94_ctfA(*RBS)), 824 ΔsolB(p94_ctfA), and 824 ΔsolB(p94_ctfA(*RBS)) after 72 h of static-flask culture without pH adjustment. Error bars represent SD (n = 3). *, P < 0.05. (B) Specific activities of the CoAT enzymes from 824(p94_ctfA), 824(p94_ctfA(*RBS)), 824 ΔsolB(p94_ctfA), and 824 ΔsolB(p94_ctfA(*RBS)).
DISCUSSION
Our data show that SolB plays a prominent role in solventogenesis that is more complex than the original hypothesis of SolB being a translational repressor of the solventogenic genes of the sol locus, whereby SolB presumably would bind the sol-locus transcripts and promote their degradation or prevent their translation. Our data here are consistent with this model for SolB overexpression: high levels of SolB expression lead to strong reductions in sol-locus transcripts (Fig. 5 to 7), which are accompanied by a commensurate reduction in (but not abolishment of) the production of butanol and acetone (Fig. 3). Computational predictions (Fig. S6 to S9) also support this model, whereby high levels of heteroduplexes of SolB with sol-locus transcripts would elicit the action of ribonucleases that would degrade such complexes (25, 44). However, given that SolB is expressed physiologically at low levels, this model cannot explain the physiological role of SolB. Significantly, we found that solB deletion attenuated butanol production and abolished acetone production, suggesting a more complex role for SolB. The fact that solB deletion leaves the adc transcript and AADC protein levels largely unaffected suggests that SolB does not have an impact on AADC expression under physiological conditions. However, solB deletion appears to reduce the levels of the adhE1 transcript and leads to significantly attenuated butanol production. This suggests that low, physiological levels of SolB are necessary for stabilizing the adhE1 transcript. Computational predictions (Fig. S6) may explain such a role, but this would be very difficult to interrogate experimentally given the difficulty of attaining in vitro the necessary physiological conditions and RNA stoichiometries that prevail in vivo, especially for the vary labile mRNAs of this organism. The impacts of SolB on the ctfA-ctfB transcript and on CoAT protein and activity are the most interesting and intriguing elements. solB deletion appears to reduce, but not severely so, the ctfA-ctfB transcript, but it abolishes CoAT translation (Fig. 6) and the production of acetone (Fig. 3 and 4). It thus appears that SolB is necessary for translation of the ctfA-ctfB transcript, and this is supported by computational predictions (Fig. S7). The hairpin Rho-independent terminator (Fig. 5C; Fig. S7) in the early portion of the ctfA-ctfB transcript apparently enables the generation of a monocistronic adhE1 transcript (Fig. 5). A similar hairpin engaging the RBS sequence on the separate ctfA-ctfB transcript would prevent translation of the ctfA-ctfB mRNA unless the hairpin structure is destabilized to free the RBS, and perhaps this is what SolB at low levels achieves.
A recent study (50) elaborated on the crucial regulatory role of a stem-loop sequence in the 5′ UTR of adhE in Escherichia coli. The role of 5′ UTR loop structures in protein translation was recently demonstrated in C. acetobutylicum (51). Previous studies (19, 20) documented the regulatory importance of the adhE1 5′ UTR, including its secondary structure, in regulating solvent production in C. acetobutylicum. The sol locus apparently codes for multilayered regulation at both the transcriptional and translational levels, as several prior studies discussed here have reported, including the study by Thormann et al. (19), who reported that the adhE1 transcript gives rise to both a bifunctional AdhE1 protein and a separate butanol dehydrogenase. The present work adds another layer of complex regulation through which this organism is apparently able to adjust the levels of various solvents produced (e.g., the butanol-to-acetone ratio) according to the prevailing culture conditions. There are several well-documented cases where the culture conditions dictate butanol but no acetone production by this organism. Among them are the study of CO gassing that inhibits H2 production and leads to butanol formation without acetone production (52) and a study of growth on cheese whey that exhibited normal levels of butanol formation but virtually no acetone production (53). It is possible that SolB has a role in such fine-tuning of product formation by this organism. While SolB expression is upregulated under metabolite stress, it is possible that its expression is also modulated by other factors, such as the prevailing redox environment in the cells, which would explain how cells tune acetone production to the prevailing environmental conditions, as discussed above. This and other small RNAs are apparently important in the extensive posttranscriptional regulation of cell metabolism in this organism, as previously detailed by combining and integrating RNA-seq and proteomic data (54).
By catalyzing the transfer of CoA from acetoacetyl-CoA to either butyrate or acetate, CoAT facilitates the uptake of these acids, detoxifying their inhibitory effects and leading to the non-growth-associated solvent production of stationary phase. CoAT functions with AADC in the production of acetone, and the uptake of butyrate and acetate by CoAT also facilitates the production of butanol and ethanol by AdhE1. Antisense RNA knockdown of CoAT severely diminishes acetone titers, indicating that CoAT is the rate-limiting enzyme in bidirectional CoAT activity and acetone production (43). Expression of adhE1 from a multicopy plasmid in the M5 degenerate strain (discussed above) enables butanol formation without acetone production, but to levels significantly below those of the WT strain; this suggests that butanol production in solventogenic cultures originates primarily from AdhE1 and that the adhE1 mRNA apparently can be translated without the presence of SolB, albeit at significantly (>60%) reduced levels (compare Table 4 in reference 12 to Table 3 in reference 13). These findings are thus consistent with the presently reported data (Fig. 3 to 5) and proposed model for the impact of SolB on AdhE1 production. What is unusual and not previously well documented for an sRNA is that, according to the proposed model, the level of expression of SolB dramatically changes its effect from that of a translational activator (its proposed physiological role) at low levels to that of a translational repressor at high levels.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Relevant characteristics and sources of all bacterial strains and plasmids used in this study are listed in Table 1. E. coli strains were grown aerobically at 37°C and 220 rpm in liquid or solid LB medium supplemented with ampicillin (Amp) (50 μg/ml), kanamycin (Kan) (25 μg/liter), or chloramphenicol (Cm) (35 μg/ml) where necessary. Colonies of C. acetobutylicum were grown anaerobically at 37°C on solid 2×YTG medium (16 g/liter tryptone, 10 g/liter yeast extract, 5 g/liter NaCl, 10 g/liter glucose, 15 g/liter agar; pH 5.8) for at least 5 days before being transferred to 10 ml of liquid clostridial growth medium (CGM), with recombinant strains grown similarly, but in media supplemented with erythromycin (Em) (40 μg/ml in solid media and 50 μg/ml in liquid media) or thiamphenicol (Th) (5 μg/ml) where necessary. Expression of the lactose-inducible promoter of the pKO_mazF vector was induced by addition of 40 mM β-lactose (Sigma-Aldrich) to the culture media.
Static-flask cultures.
Strains were grown in replicate 40-ml static cultures in 50-ml conical tubes, with inoculation from 10-ml overnight cultures (A600 ≥ 0.5). All cultures were grown in CGM, supplemented with antibiotics where appropriate, with no pH adjustment.
Bioreactor studies.
The SolB deletion strain (824 ΔsolB) was grown in replicate 4-liter batch fermentations in a BioFlo 310 bioreactor (New Brunswick Scientific). Cultures were grown in undefined CGM supplemented with 5 μg/ml thiamphenicol. Bioreactor conditions were kept anaerobic through continuous sparging of N2 gas, and the pH was controlled (pH ≥ 5) by addition of 6 N ammonium hydroxide.
Construction of plasmids.
All primers and their sequences utilized in this study are listed in Table S1 in the supplemental material. Plasmid p94_solB, used for the overexpression of SolB, was constructed by PCR amplifying the 215-bp solB coding region by use of primers that appended BamHI and KasI restriction sites to the product's 5′ and 3′ ends, respectively. After double digestion with BamHI and KasI (New England BioLabs), the PCR product was ligated by use of Instant Sticky-End master mix (NEB) into the p94MCS expression vector, which had been linearized by digestion with BamHI and KasI and dephosphorylated with Antarctic phosphatase (NEB). Plasmid p94_ctfAB(*RBS), used for the expression of the mutated ctfA-ctfB transcript, was constructed using Gibson assembly to achieve mutagenesis of the ribosomal binding site (RBS) of the ctfA gene. Primers were used to separately amplify the ctfA gene and the ctfA promoter region while introducing the RBS mutation (a change from 5′-AGGAGG-3′ to 5′-AAGAAG-3′) into the PCR products (using primers listed in Table S1). The Gibson assembly, consisting of the two PCR products containing the native promoter, the new mutated RBS, and the ctfA-ctfB genes, was then cloned into the p94MCS vector, which had been digested with SalI. Plasmid p94_cftAB(WT RBS), which contains the native RBS in place of the mutated one, was constructed in a similar manner, by cloning the PCR product of the full native gene and promoter region into the SalI-digested p94MCS backbone.
Construction of the solB deletion plasmid pKO_mazF::solB followed a previously described procedure (35). Two regions of homology (RH) were amplified by standard PCR from genomic DNA of C. acetobutylicum ATCC 824. The 936-bp upstream RH corresponds to nucleotides −986 to −50 relative to the transcriptional start site of solB, and the 901-bp downstream RH corresponds to nucleotides +21 to +901 relative to the end of the solB sequence. The following restriction sites were added to the ends of each RH product: sites for XmaI (5′) and NotI (3′) for the upstream RH and sites for AvaII (5′) and SphI (3′) for the downstream RH. The PCR products were double digested with the appropriate restriction enzymes (NEB) and ligated into the pKO_mazF vector as described above. First, the downstream RH was ligated into pKO_mazF, which was linearized by AvaII and SphI double digestion and dephosphorylated. After transformation into chemically competent Turbo E. coli cells (NEB) and selection by EcoRI (NEB) confirmation digestion, successful constructs were linearized by XmaI and NotI digestion and dephosphorylated, and the upstream RH was cloned into the vector.
Plasmid transformations.
Newly constructed plasmids were transformed into chemically competent Turbo or 10-beta E. coli cells (both from NEB), harvested using a spin miniprep kit (Qiagen), and confirmed by 1-h restriction digestion and Sanger sequencing analysis. Plasmids were then transformed into electrocompetent E. coli BL21(pAN3) cells for DNA methylation (35, 55). Methylated plasmids were harvested and subsequently transformed via electroporation into electrocompetent C. acetobutylicum ATCC 824 as previously described (35, 56).
Isolation of solB deletion mutants.
Following transformation and 3 days of growth on 2×YTG solid medium supplemented with thiamphenicol (5 μg/ml), triplicate colonies were selected for growth in liquid 2×YTG. To facilitate allelic exchange of the Thr cassette, 10 subsequent vegetative transfers with fresh supplemented medium were conducted as described previously (35), with the final transfer grown with added 40 mM β-lactose to cure the cells of the pKO_mazF vector. Cultures were then serially diluted and grown on 2×YTG solid medium supplemented with thiamphenicol (5 μg/ml) and lactose (40 mM) to complete the curing of the remaining vector backbone, as described previously (35).
Confirmation of mutant strains by PCR and sequencing.
Genomic DNAs were extracted from deletion mutant cultures by use of a DNeasy blood and tissue kit (Qiagen) and were screened by PCR amplification of both crossover integration sites and the deleted solB sequence (Fig. 1A and B). PCR products were also purified using a PCR purification kit (Qiagen), adenylated using GoTaq hot start polymerase (Promega), and ligated into the pCR4-TOPO-TA sequencing vector (Life Technologies) for Sanger sequencing analysis (Fig. 1C).
RNA extraction and quantitative RT-PCR (Q-RT-PCR).
Cultures used for RNA extraction were inoculated with 10-ml overnight cultures (A600 ≥ 0.5) to a 1:16 dilution in 150 ml of CGM, supplemented with antibiotics where appropriate. In addition to regular sampling for optical density (A600) measurements and high-performance liquid chromatography (HPLC) analysis, 15-ml samples were collected at 6, 12, and 24 h (corresponding to the mid-exponential, transition, and stationary growth phases). These samples were immediately centrifuged for 10 min at 8,000 × g, and resultant cell pellets were stored at −80°C for up to 10 days. Isolation of total RNA, including small RNA, was performed using a Qiagen miRNeasy minikit as described previously (40), except that samples were not split and diluted after TRIzol addition and buffer RWT (optimized for microRNA [miRNA] and sRNA retention) was used in place of buffer RW1. All RNA samples were treated by use of a Turbo DNase kit (Ambion) according to the manufacturer's protocol to remove contaminant genomic DNA and were further purified by ethanol precipitation at −20°C before storage at −80°C. Generation of cDNA was performed using 2 μg of RNA template and a High-Capacity cDNA reverse transcription kit (Applied Biosystems).
All primer sets used for Q-RT-PCR analysis are listed in Table S1, and the efficiency of each primer set was validated on a test plate. Each cDNA sample was tested in triplicate using iTaq Universal SYBR green supermix (Bio-Rad). All cDNA samples intended for direct comparison were prepared from the same master mix preparation and run on the same plate to minimize variation in expression results. Primer efficiency was assumed to be 100% for calculation purposes, because the calculated efficiency for every primer set fell within the accepted range for the assumption of 100% efficiency (57).
Northern blotting.
Northern blotting was performed as described previously (5). Five- or 15-μg aliquots of total RNA samples were electrophoretically resolved in 5% precast Tris-borate-EDTA (TBE)–urea polyacrylamide Ready gels (Bio-Rad) in a Bio-Rad Mini Protean Tetra cell apparatus and then transferred to a positively charged nylon membrane (Ambion). Single-stranded oligonucleotide DNA probes used for hybridization (listed in Table S2) were labeled with [γ-32P]ATP by use of a phosphatase-negative T4 polynucleotide kinase (Affymetrix). Excess radioactive material was removed by use of NucAway spin columns (Ambion), and membrane prehybridization and hybridization with the radioactive probe were conducted with ultrasensitive hybridization buffer (Ambion), with samples incubated for 16 to 22 h at 42°C under gentle agitation in a hybridization incubator (Fisher Scientific).
Western blotting.
Protein extraction and Western blotting were performed as previously described (58), with 40 μg of total protein loaded into a 4 to 20% PAGE gel (Genscript) and transferred to a supported nitrocellulose membrane (Bio-Rad). Membranes were washed with a blocking solution containing Tris-buffered saline plus Tween 20 (TBST) and 1% gelatin for 2 h and then hybridized for 2 h with either rabbit anti-AADC (20) or sheep anti-CoAT (43), diluted 1:250 and 1:500, respectively. Secondary antibodies were diluted 1:10,000 and hybridized for 1 h; mouse anti-rabbit–Alexa Fluor 647 and donkey anti-sheep–Alexa Fluor 647 (both from Life Technologies) were used for detection of AADC and CoAT, respectively. Fluorescence imaging was conducted with a Typhoon 9400 variable-mode imager (GE Healthcare).
CoAT assay.
Protein samples were quantified using the RC DC protein assay (Bio-Rad), with bovine serum albumin as the standard. CoAT activity was assayed at room temperature in the butyrate conversion direction as described previously (14). Combined in a cuvette were 100 mM Tris-HCl (pH 7.5), 100 mM potassium butyrate (pH 7.5), 20 mM MgCl2, 5% (vol/vol) glycerol, and 50 μl of crude lysate; the final volume was brought to 900 μl with deionized (DI) water. A control reaction mixture that omitted potassium butyrate (to measure the hydrolysis of acetoacetyl-CoA) was prepared for each sample. To initiate the reaction, 0.10 mM acetoacetyl-CoA was added, and the decrease in A310 was measured as an indication of the disappearance of the enolate form of acetoacetyl-CoA. The slopes obtained from control reactions were subtracted from those for their corresponding sample reactions (this hydrolysis was less than 10% of the uncorrected slope). One unit of activity is defined as the amount of enzyme which converts 1 μmol of acetoacetyl-CoA per min under these conditions. The extinction coefficient is 8.0 mM−1 · cm−1. The following equation was utilized to calculate the approximate specific activity of each crude lysate sample: specific activity (U/mg) = (ΔA310/min)/(8.0 × mass [mg]).
General analytical methods.
Cell density was determined by measuring the A600 by use of a Beckman Coulter DU 730 spectrophotometer (model A23616). Culture supernatants were assayed for glucose, acetate, ethanol, butyrate, acetone, and butanol concentrations in an Agilent Technologies 1200 series high-performance liquid chromatograph (HPLC), using an H2SO4 mobile phase and a 45-min separation method (59).
Standard PCRs utilized the Q5 Hot Start high-fidelity DNA polymerase (NEB), a 200 μM concentration of each deoxynucleoside triphosphate (dNTP), 0.5 μM (each) primers, and 50 to 300 ng of DNA template. Reactions were run in a Bio-Rad C1000 Touch thermal cycler; Q-RT-PCR included the addition of a CFX96 real-time system (Bio-Rad). Standard PCR conditions consisted of initial denaturation (98°C for 30 s) followed by 30 cycles consisting of 10 s of denaturation at 98°C, 45 s for annealing at a temperature specific to the primer set (typically 3°C higher than the lower of the two primers' annealing temperatures), and extension at 72°C for a variable time dependent on the size of the product, with a final extension step of 5 min at 72°C.
Computational methods.
Secondary structure folding was generated using the Vienna RNA Websuite (45). The 5′ UTR plus the first 100 bp of the coding region for each sol-locus gene transcript was utilized. The adhE1 UTR was generated using the proximal promoter for the sol operon, given that several studies have concluded that most transcriptional activity occurs from this promoter (11, 12, 20), and for ctfA predictions, the putative 5′ UTR for ctfA (based on the predicted promoter described in Results) plus the first 100 bp was used, as well as the terminal 100 bp of adhE1 and the first 100 bp of ctfA together with the intergenic region.
Binding predictions for SolB were generated using the IntaRNA online prediction program (46, 47) as well as the RNAup Web server (also part of the Vienna RNA Websuite). For the IntaRNA predictions, the full 196-bp solB sequence was queried against the full 5′ UTR plus the first 50 bp of coding sequence for both adhE1 and adc and against the 63-bp intergenic region between adhE1 and ctfA plus the first 100 bp of ctfA. For the RNAup predictions, the full solB sequence was queried against the following three models for each transcript: (i) the 5′ UTR plus 100 bp of coding sequence, (ii) the 5′ UTR plus 50 bp of coding sequence, and (iii) only the 5′ UTR.
Genome-wide target prediction for SolB was conducted using the TargetRNA prediction algorithm (v2.01) (60, 61). Seventeen prediction runs, reflecting combinations of different prediction parameters (Table S3), were conducted by querying of SolB against the C. acetobutylicum genome. Target results from all 17 predictions were screened for transcripts of characterized proteins that appeared in at least four predictions.
Supplementary Material
ACKNOWLEDGMENTS
A.J.J. thanks Keerthi Venkataramanan for his insight, training, and support for Northern blot analysis. We thank Matthew Ralston for sharing RNA-seq data and for his insights and support in bioinformatics analysis.
This work was supported by a genomic science grant from the U.S. Department of Energy (grant DE-SC0007092) and by the National Science Foundation (award number CBET-1511660).
The funding agencies had no role in the experimental design, data collection and interpretation, or decision to publish this research.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00597-18.
REFERENCES
- 1.Papoutsakis ET. 2008. Engineering solventogenic clostridia. Curr Opin Biotechnol 19:420–429. doi: 10.1016/j.copbio.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 2.Paredes CJ, Alsaker K, Papoutsakis ET. 2005. A comparative genomic view of clostridial sporulation and physiology. Nat Rev Microbiol 3:969–978. doi: 10.1038/nrmicro1288. [DOI] [PubMed] [Google Scholar]
- 3.Lee SY, Park JH, Jang SH, Nielsen LK, Kim J, Jung KS. 2008. Fermentative butanol production by clostridia. Biotechnol Bioeng 101:209–228. doi: 10.1002/bit.22003. [DOI] [PubMed] [Google Scholar]
- 4.Durre P, Bohringer M, Nakotte S, Schaffer S, Thormann K, Zickner B. 2002. Transcriptional regulation of solventogenesis in Clostridium acetobutylicum. J Mol Microbiol Biotechnol 4:295–300. [PubMed] [Google Scholar]
- 5.Wang QH, Venkataramanan KP, Huang HZ, Papoutsakis ET, Wu CH. 2013. Transcription factors and genetic circuits orchestrating the complex, multilayered response of Clostridium acetobutylicum to butanol and butyrate stress. BMC Syst Biol 7:120. doi: 10.1186/1752-0509-7-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lutke-Eversloh T, Bahl H. 2011. Metabolic engineering of Clostridium acetobutylicum: recent advances to improve butanol production. Curr Opin Biotechnol 22:634–647. doi: 10.1016/j.copbio.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 7.Tracy BP, Jones SW, Fast AG, Indurthi DC, Papoutsakis ET. 2012. Clostridia: the importance of their exceptional substrate and metabolite diversity for biofuel and biorefinery applications. Curr Opin Biotechnol 23:364–381. doi: 10.1016/j.copbio.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 8.Lutke-Eversloh T. 2014. Application of new metabolic engineering tools for Clostridium acetobutylicum. Appl Microbiol Biotechnol 98:5823–5837. doi: 10.1007/s00253-014-5785-5. [DOI] [PubMed] [Google Scholar]
- 9.Venkataramanan KP, Jones SW, McCormick KP, Kunjeti SG, Ralston MT, Meyers BC, Papoutsakis ET. 2013. The Clostridium small RNome that responds to stress: the paradigm and importance of toxic metabolite stress in C. acetobutylicum. BMC Genomics 14:849. doi: 10.1186/1471-2164-14-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cornillot E, Nair RV, Papoutsakis ET, Soucaille P. 1997. The genes for butanol and acetone formation in Clostridium acetobutylicum ATCC 824 reside on a large plasmid whose loss leads to degeneration of the strain. J Bacteriol 179:5442–5447. doi: 10.1128/jb.179.17.5442-5447.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischer RJ, Helms J, Dürre P. 1993. Cloning, sequencing, and molecular analysis of the sol operon of Clostridium acetobutylicum, a chromosomal locus involved in solventogenesis. J Bacteriol 175:6959–6969. doi: 10.1128/jb.175.21.6959-6969.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nair RV, Bennett GN, Papoutsakis ET. 1994. Molecular characterization of an aldehyde/alcohol dehydrogenase gene from Clostridium-acetobutylicum ATCC-824. J Bacteriol 176:871–885. doi: 10.1128/jb.176.3.871-885.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nair RV, Papoutsakis ET. 1994. Expression of plasmid-encoded aad in Clostridium acetobutylicum M5 restores vigorous butanol production. J Bacteriol 176:5843–5846. doi: 10.1128/jb.176.18.5843-5846.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wiesenborn DP, Rudolph FB, Papoutsakis ET. 1989. Coenzyme A transferase from Clostridium acetobutylicum ATCC 824 and its role in the uptake of acids. Appl Environ Microbiol 55:323–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerischer U, Dürre P. 1992. mRNA analysis of the adc gene region of Clostridium acetobutylicum during the shift to solventogenesis. J Bacteriol 174:426–433. doi: 10.1128/jb.174.2.426-433.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petersen DJ, Cary JW, Vanderleyden J, Bennett GN. 1993. Sequence and arrangement of genes encoding enzymes of the acetone-production pathway of Clostridium acetobutylicum ATCC824. Gene 123:93–97. doi: 10.1016/0378-1119(93)90545-E. [DOI] [PubMed] [Google Scholar]
- 17.Ravagnani A, Jennert KCB, Steiner E, Grunberg R, Jefferies JR, Wilkinson SR, Young DI, Tidswell EC, Brown DP, Youngman P, Morris JG, Young M. 2000. Spo0A directly controls the switch from acid to solvent production in solvent-forming clostridia. Mol Microbiol 37:1172–1185. doi: 10.1046/j.1365-2958.2000.02071.x. [DOI] [PubMed] [Google Scholar]
- 18.Harris LM, Welker NE, Papoutsakis ET. 2002. Northern, morphological, and fermentation analysis of spo0A inactivation and overexpression in Clostridium acetobutylicum ATCC 824. J Bacteriol 184:3586–3597. doi: 10.1128/JB.184.13.3586-3597.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thormann K, Feustel L, Lorenz K, Nakotte S, Durre P. 2002. Control of butanol formation in Clostridium acetobutylicum by transcriptional activation. J Bacteriol 184:1966–1973. doi: 10.1128/JB.184.7.1966-1973.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scotcher MC, Huang KX, Harrison ML, Rudolph FB, Bennett GN. 2003. Sequences affecting the regulation of solvent production in Clostridium acetobutylicum. J Ind Microbiol Biotechnol 30:414–420. doi: 10.1007/s10295-003-0057-x. [DOI] [PubMed] [Google Scholar]
- 21.Roos JW, McLaughlin JK, Papoutsakis ET. 1985. The effect of pH on nitrogen supply, cell-lysis, and solvent production in fermentations of Clostridium-acetobutylicum. Biotechnol Bioeng 27:681–694. doi: 10.1002/bit.260270518. [DOI] [PubMed] [Google Scholar]
- 22.Husemann MHW, Papoutsakis ET. 1988. Solventogenesis in Clostridium-acetobutylicum fermentations related to carboxylic-acid and proton concentrations. Biotechnol Bioeng 32:843–852. doi: 10.1002/bit.260320702. [DOI] [PubMed] [Google Scholar]
- 23.Tomas CA, Beamish J, Papoutsakis ET. 2004. Transcriptional analysis of butanol stress and tolerance in Clostridium acetobutylicum. J Bacteriol 186:2006–2018. doi: 10.1128/JB.186.7.2006-2018.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alsaker KV, Paredes C, Papoutsakis ET. 2010. Metabolite stress and tolerance in the production of biofuels and chemicals: gene-expression-based systems analysis of butanol, butyrate, and acetate stresses in the anaerobe Clostridium acetobutylicum. Biotechnol Bioeng 105:1131–1147. doi: 10.1002/bit.22628. [DOI] [PubMed] [Google Scholar]
- 25.Storz G, Vogel J, Wassarman KM. 2011. Regulation by small RNAs in bacteria: expanding frontiers. Mol Cell 43:880–891. doi: 10.1016/j.molcel.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen YL, Indurthi DC, Jones SW, Papoutsakis ET. 2011. Small RNAs in the genus Clostridium. mBio 2:e00340-10. doi: 10.1128/mBio.00340-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Borden JR, Jones SW, Indurthi D, Chen YL, Papoutsakis ET. 2010. A genomic-library based discovery of a novel, possibly synthetic, acid-tolerance mechanism in Clostridium acetobutylicum involving non-coding RNAs and ribosomal RNA processing. Metab Eng 12:268–281. doi: 10.1016/j.ymben.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gaida SM, Al-Hinai MA, Indurthi DC, Nicolaou SA, Papoutsakis ET. 2013. Synthetic tolerance: three noncoding small RNAs, DsrA, ArcZ and RprA, acting supra-additively against acid stress. Nucleic Acids Res 41:8726–8737. doi: 10.1093/nar/gkt651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soutourina OA, Monot M, Boudry P, Saujet L, Pichon C, Sismeiro O, Semenova E, Severinov K, Le Bouguenec C, Coppee JY, Dupuy B, Martin-Verstraete I. 2013. Genome-wide identification of regulatory RNAs in the human pathogen Clostridium difficile. Plos Genetics 9:e1003493. doi: 10.1371/journal.pgen.1003493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soutourina O. 2017. RNA-based control mechanisms of Clostridium difficile. Curr Opin Microbiol 36:62–68. doi: 10.1016/j.mib.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 31.Jones AJ. 2015. An investigation of the roles of small RNA on solvent tolerance and production in Clostridium acetobutylicum. MS thesis University of Delaware, Newark, DE. [Google Scholar]
- 32.Jones AJ, Venkataramanan KP, Papoutsakis T. 2016. Overexpression of two stress-responsive, small, non-coding RNAs, 6S and tmRNA, imparts butanol tolerance in Clostridium acetobutylicum. FEMS Microbiol Lett 363:fnw063. doi: 10.1093/femsle/fnw063. [DOI] [PubMed] [Google Scholar]
- 33.Wach W, Harms K, Klingeberg M, Duerre P, Nold N, Schiel B. March 2014. Production of acid and solvent in microorganisms. US patent 8679799 B2.
- 34.Zimmermann T. 2013. Untersuchungen zur Butanolbildung von Hyperthermus butylicus und Clostridium acetobutylicum. PhD thesis Universität Ulm, Ulm, Germany. [Google Scholar]
- 35.Al-Hinai MA, Fast AG, Papoutsakis ET. 2012. Novel system for efficient isolation of Clostridium double-crossover allelic exchange mutants enabling markerless chromosomal gene deletions and DNA integration. Appl Environ Microbiol 78:8112–8121. doi: 10.1128/AEM.02214-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Al-Hinai MA, Jones SW, Papoutsakis ET. 2014. sigma(K) of Clostridium acetobutylicum is the first known sporulation-specific sigma factor with two developmentally separated roles, one early and one late in sporulation. J Bacteriol 196:287–299. doi: 10.1128/JB.01103-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Papoutsakis ET, Bussineau CM, Chu IM, Diwan AR, Huesemann M. 1987. Transport of substrates and metabolites and their effect on cell metabolism (in butyric-acid and methylotrophic fermentations). Ann N Y Acad Sci 506:24–50. doi: 10.1111/j.1749-6632.1987.tb23808.x. [DOI] [PubMed] [Google Scholar]
- 38.Alsaker KV, Papoutsakis ET. 2005. Transcriptional program of early sporulation and stationary-phase events in Clostridium acetobutylicum. J Bacteriol 187:7103–7118. doi: 10.1128/JB.187.20.7103-7118.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tomas CA, Alsaker KV, Bonarius HPJ, Hendriksen WT, Yang H, Beamish JA, Paredes CJ, Papoutsakis ET. 2003. DNA array-based transcriptional analysis of asporogenous, nonsolventogenic Clostridium acetobutylicum strains SKO1 and M5. J Bacteriol 185:4539–4547. doi: 10.1128/JB.185.15.4539-4547.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones SW, Paredes CJ, Tracy B, Cheng N, Sillers R, Senger RS, Papoutsakis ET. 2008. The transcriptional program underlying the physiology of clostridial sporulation. Genome Biol 9:R114. doi: 10.1186/gb-2008-9-7-r114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ralston MT. 2015. Assembling improved gene annotations in Clostridium acetobutylicum with RNA sequencing. MS thesis University of Delaware, Newark, DE. [Google Scholar]
- 42.Tummala SB, Junne SG, Papoutsakis ET. 2003. Antisense RNA downregulation of coenzyme A transferase combined with alcohol-aldehyde dehydrogenase overexpression leads to predominantly alcohologenic Clostridium acetobutylicum fermentations. J Bacteriol 185:3644–3653. doi: 10.1128/JB.185.12.3644-3653.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tummala SB, Welker NE, Papoutsakis ET. 2003. Design of antisense RNA constructs for downregulation of the acetone formation pathway of Clostridium acetobutylicum. J Bacteriol 185:1923–1934. doi: 10.1128/JB.185.6.1923-1934.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Desnoyers G, Bouchard MP, Masse E. 2013. New insights into small RNA-dependent translational regulation in prokaryotes. Trends Genet 29:92–98. doi: 10.1016/j.tig.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 45.Gruber AR, Lorenz R, Bernhart SH, Neuböck R, Hofacker IL. 2008. The Vienna RNA Websuite. Nucleic Acids Res 36:W70–W74. doi: 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Busch A, Richter AS, Backofen R. 2008. IntaRNA: efficient prediction of bacterial sRNA targets incorporating target site accessibility and seed regions. Bioinformatics 24:2849–2856. doi: 10.1093/bioinformatics/btn544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wright PR, Georg J, Mann M, Sorescu DA, Richter AS, Lott S, Kleinkauf R, Hess WR, Backofen R. 2014. CopraRNA and IntaRNA: predicting small RNA targets, networks and interaction domains. Nucleic Acids Res 42:W119–W123. doi: 10.1093/nar/gku359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Papenfort K, Vanderpool CK. 2015. Target activation by regulatory RNAs in bacteria. FEMS Microbiol Rev 39:362–378. doi: 10.1093/femsre/fuv016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jain K, Updegrove TB, Wartell RM. 2011. A thermodynamic perspective of sRNA-mRNA interactions and the role of Hfq, p 111–131. In Sheardy RD, Winkle SA (ed), Frontiers in nucleic acids. American Chemical Society, Washington, DC. [Google Scholar]
- 50.Ito K, Hamasaki K, Kayamori A, Nguyen PAT, Amagai K, Wachi M. 2013. A secondary structure in the 5′ untranslated region of adhE mRNA required for RNase G-dependent regulation. Biosci Biotechnol Biochem 77:2473–2479. doi: 10.1271/bbb.130618. [DOI] [PubMed] [Google Scholar]
- 51.Lee J, Jang YS, Papoutsakis ET, Lee SY. 2016. Stable and enhanced gene expression in Clostridium acetobutylicum using synthetic untranslated regions with a stem-loop. J Biotechnol 230:40–43. doi: 10.1016/j.jbiotec.2016.05.020. [DOI] [PubMed] [Google Scholar]
- 52.Meyer CL, Roos JW, Papoutsakis ET. 1986. Carbon-monoxide gasing leads to alcohol production and butyrate uptake without acetone formation in continuous cultures of Clostridium-acetobutylicum. Appl Microbiol Biotechnol 24:159–167. [Google Scholar]
- 53.Bahl H, Gottwald M, Kuhn A, Rale V, Andersch W, Gottschalk G. 1986. Nutritional factors affecting the ratio of solvents produced by Clostridium-acetobutylicum. Appl Environ Microbiol 52:169–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Venkataramanan KP, Min L, Hou SY, Jones SW, Ralston MT, Lee KH, Papoutsakis ET. 2015. Complex and extensive post-transcriptional regulation revealed by integrative proteomic and transcriptomic analysis of metabolite stress response in Clostridium acetobutylicum. Biotechnol Biofuels 8:81. doi: 10.1186/s13068-015-0260-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mermelstein LD, Papoutsakis ET. 1993. In vivo methylation in Escherichia coli by the Bacillus subtilis phage phi 3T I methyltransferase to protect plasmids from restriction upon transformation of Clostridium acetobutylicum ATCC 824. Appl Environ Microbiol 59:1077–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mermelstein LD, Welker NE, Bennett GN, Papoutsakis ET. 1992. Expression of cloned homologous fermentative genes in Clostridium acetobutylicum ATCC 824. Biotechnology 10:190–195. [DOI] [PubMed] [Google Scholar]
- 57.Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 58.Tracy BP, Jones SW, Papoutsakis ET. 2011. Inactivation of sigma(E) and sigma(G) in Clostridium acetobutylicum illuminates their roles in clostridial-cell-form biogenesis, granulose synthesis, solventogenesis, and spore morphogenesis. J Bacteriol 193:1414–1426. doi: 10.1128/JB.01380-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tomas CA, Welker NE, Papoutsakis ET. 2003. Overexpression of groESL in Clostridium acetobutylicum results in increased solvent production and tolerance, prolonged metabolism, and changes in the cell's transcriptional program. Appl Environ Microbiol 69:4951–4965. doi: 10.1128/AEM.69.8.4951-4965.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kery MB, Feldman M, Livny J, Tjaden B. 2014. TargetRNA2: identifying targets of small regulatory RNAs in bacteria. Nucleic Acids Res 42:W124–W129. doi: 10.1093/nar/gku317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tjaden B. 2008. TargetRNA: a tool for predicting targets of small RNA action in bacteria. Nucleic Acids Res 36:W109–W113. doi: 10.1093/nar/gkn264. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.