Phytoremediation of organic pollutant-contaminated sites using transgenic plants expressing bacterial enzyme has been well described. The major constraint of transgenic plants transferred with a single catabolic gene is that they can also accumulate/release intermediates, still causing phytotoxicity or additional environmental problems. On the other hand, bioaugmentation with degrading strains also has its drawbacks, including the instability of the inoculated strains and low bioavailability of pollutants. In this study, the synergistic relationship between a transgenic Arabidopsis plant expressing the bacterial N-demethylase PdmAB in the chloroplast and the inoculated intermediate-mineralizing bacterium Sphingobium sp. strain 1017-1 in the rhizosphere is used to develop an intriguing bioremediation method. The combinational transgenic plant-microbe remediation system shows a more efficient and complete removal of phenylurea herbicides from contaminated sites and can overcome the constraints of individual phytoremediation or bioaugmentation methods.
KEYWORDS: phenylurea herbicides, transgenic plant, plant-microbe remediation, rhizospheric microbes
ABSTRACT
The synergistic relationships between plants and their rhizospheric microbes can be used to develop a combinational bioremediation method, overcoming the constraints of individual phytoremediation or a bioaugmentation method. Here, we provide a combinational transgenic plant-microbe remediation system for a more efficient removal of phenylurea herbicides (PHs) from contaminated sites. The transgenic Arabidopsis thaliana plant synthesizing the bacterial N-demethylase PdmAB in the chloroplast was developed. The constructed transgenic Arabidopsis plant exhibited significant tolerance to isoproturon (IPU), a typical PH, and it took up the IPU through the roots and transported it to leaves, where the majority of the IPU was demethylated to 3-(4-isopropylphenyl)-1-methylurea (MDIPU). The produced intermediate was released outside the roots and further metabolized by the combinationally inoculated MDIPU-mineralizing bacterium Sphingobium sp. strain 1017-1 in the rhizosphere, resulting in an enhanced and complete removal of IPU from soil. Mutual benefits were built for both the transgenic Arabidopsis plant and strain 1017-1. The transgenic Arabidopsis plant offered strain 1017-1 a suitable accommodation, and in return, strain 1017-1 protected the plant from the phytotoxicity of MDIPU. The biomass of the transgenic Arabidopsis plant and the residence of the inoculated degrading microbes in the combinational treatment increased significantly compared to those in their respective individual transgenic plant treatment or bioaugmentation treatment. The influence of the structure of bacterial community by combinational treatment was between that of the two individual treatments. Overall, the combination of two approaches, phytoremediation by transgenic plants and bioaugmentation with intermediate-mineralizing microbes in the rhizosphere, represents an innovative strategy for the enhanced and complete remediation of pollutant-contaminated sites.
IMPORTANCE Phytoremediation of organic pollutant-contaminated sites using transgenic plants expressing bacterial enzyme has been well described. The major constraint of transgenic plants transferred with a single catabolic gene is that they can also accumulate/release intermediates, still causing phytotoxicity or additional environmental problems. On the other hand, bioaugmentation with degrading strains also has its drawbacks, including the instability of the inoculated strains and low bioavailability of pollutants. In this study, the synergistic relationship between a transgenic Arabidopsis plant expressing the bacterial N-demethylase PdmAB in the chloroplast and the inoculated intermediate-mineralizing bacterium Sphingobium sp. strain 1017-1 in the rhizosphere is used to develop an intriguing bioremediation method. The combinational transgenic plant-microbe remediation system shows a more efficient and complete removal of phenylurea herbicides from contaminated sites and can overcome the constraints of individual phytoremediation or bioaugmentation methods.
INTRODUCTION
Anthropogenic inputs of organic chemical compounds, such as polyaromatic hydrocarbons, polychlorinated biphenyls, and pesticides, into environments may lead to environmental pollution, exerting considerable adverse effects on human health and ecological security. Large-scale remediation of this kind of nonpoint pollution by a physical or chemical method is not feasible because of its high cost; in situ bioremediation is considered a cost-effective, less labor-intensive, safe, and environmentally friendly method (1–3).
Microbial remediation by inoculation of degrading microbes (also called bioaugmentation) has been widely used for the removal of various organic pollutants due to their versatile catabolic capacities. While microbial remediation might have its drawbacks (4–7), such as its instability due to the rapid decline in the inoculated cell amount during its competition with indigenous microorganisms and its low access to pollutants in relatively deep sites. Phytoremediation, the use of environmentally well-adapted and rapidly growing plants for removal of pollutants, is self-maintaining (i.e., autotrophic) and renewable (8). Plants can take up pollutants from relatively deep sites through their extensive root system and transport/translocate them to various plant tissues where they can be metabolized. Phytoremediation has additional benefits, including carbon sequestration, soil stabilization, biofuel or fiber production, and esthetic appearance (9). However, plants generally lack the versatile catabolic capacity for recalcitrant pollutants compared to microbes. Therefore, key genes for pollutant degradation are designed to be transferred from microbes to plants to enhance the catabolic ability of plants. For example, bacterial pentaerythritol tetranitrate reductase, nitroreductase, cytochrome P450, extradiol dioxygenase (DbfB), haloalkane dehalogenase (DhaA), and naphthalene dioxygenase systems have been successfully expressed in Arabidopsis plants, tobacco, and rice, for enhanced degradation, detoxification, and remediation of nitroglycerin (10), 2,4,6-trinitrotoluene (TNT) (11, 12), cyclotrimethylenetrinitramine (RDX) (13), 3-dihydroxybiphenyl (2,3-DHB), 1-chlorobutane (1-CB) (14), and aromatic hydrocarbons (15, 16). These studies demonstrated the usefulness of the phytoremediation of contaminated sites by transgenic plants expressing bacterial catabolic enzymes. However, it is usually difficult to transfer the complete catabolic gene cluster to plants for mineralizing the target pollutant, and plants transferred with a single catabolic gene can also accumulate/release intermediates, which can still cause phytotoxicity or additional environmental problems. To overcome these constraints, new strategies to improve the phytoremediation efficiency are needed.
Recently, the important role of rhizospheric microbes during phytoremediation to the removal of pollutants has been recognized (17–19). The plants and the rhizospheric microbes can establish synergistic relationships and build mutual benefits for both sides (20). Thus, the use of plants in combination with microbes has several advantages and could serve as an intriguing method to solve the problems encountered during the application of both individual phytoremediation and bioaugmentation techniques. In this study, a transgenic plant using the model plant Arabidopsis thaliana which expresses a bacterial N-demethylase (PdmAB) for N,N-dimethyl-substituted phenylurea herbicides (PHs) was constructed. Then, a new strategy for the efficient remediation of PHs in soil was established. In this strategy, the phytoremediation by the transgenic plant was combined with the bioaugmentation with Sphingobium sp. strain 1017-1, which is capable of mineralizing the intermediate of isoproturon (IPU; the typical PH) excreted from the transgenic plant in the rhizosphere. Furthermore, the mechanism underlying the enhanced removal rate of IPU was also revealed due to the synergistic relationship between the transgenic plant and the inoculated microbe. The combination of phytoremediation and bioaugmentation represents an innovative solution for the enhanced and complete removal of pollutants and can be a new strategy for the efficient bioremediation of organic chemical compound-contaminated sites.
RESULTS
Transgenic Arabidopsis plant expressing PdmAB in the chloroplast showed enhanced tolerance to IPU.
Sphingobium sp. strain YBL2 is able to mineralize IPU and can also degrade other PHs, like chlortoluron, metoxuron, monuron, diuron, fluometuron, and fenuron (21–23). Sphingobium sp. strain 1017-1 is the pdmAB-inactivated mutant of strain YBL2 (21). The initial degradation step of PHs in strain YBL2 is catalyzed by PdmAB. PdmAB is the terminal oxygenase component of the Rieske nonheme iron oxygenase (RO) system, which requires two additional components (ferredoxin and reductase) for electron transfer to perform its N-demethylase function (21). In the presence of proper electron transport components, PdmAB is able to catalyze the N-demethylation of IPU, generating 3-(4-isopropylphenyl)-1-methylurea (MDIPU). In addition, PdmAB also exhibits low activity toward MDIPU, producing 1-(4-isopropylphenyl) urea (DDIPU) (21, 22). Since PdmAB shows low specificity for electron transport components (21), the ferredoxin formed in plant chloroplast is assumed to shuttle electrons to PdmAB, eliminating the need for bacterial reductase and ferredoxin components. Based on this hypothesis, the chloroplast transit peptide-coding sequence of the Arabidopsis 5-enolpyruvylshikimate-3-phosphate (EPSP) synthase gene (AtCTP) was added to the 5′ ends of the pdmA and pdmB genes (Fig. 1a), and the expression cassettes for pdmAB genes were transformed into the genome of the Arabidopsis plant using Agrobacterium tumefaciens GV3101 (pDBN10938). Ten glufosinate ammonium-resistant lines were obtained and subjected to segregation analysis. After two rounds of selfing, three homozygous T3 lines (T3-2, T3-3, and T3-4) were selected for further analysis.
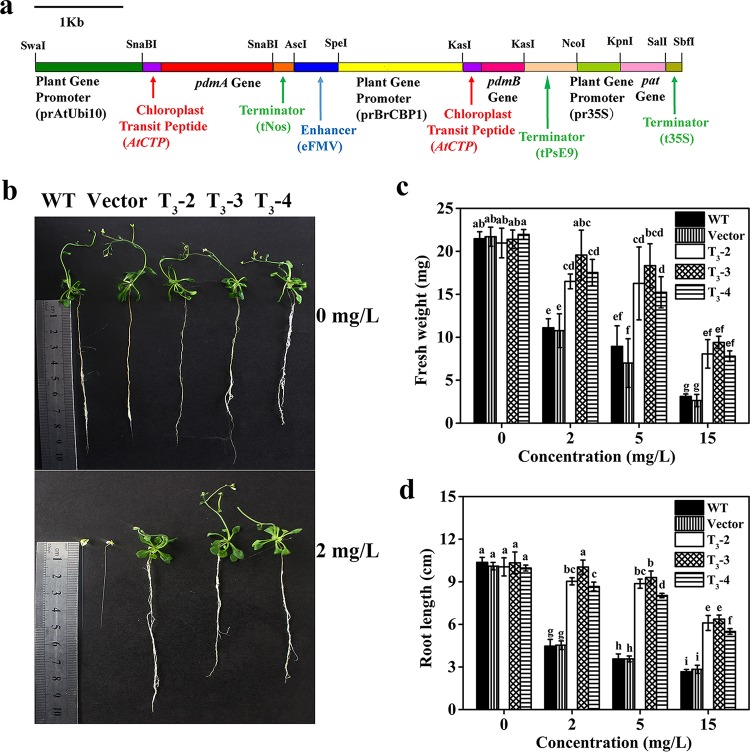
FIG 1.
Schematic diagram of the expression cassettes for pdmAB genes used for plant transformation, and the transgenic Arabidopsis plant shows tolerance to IPU. (a) The pdmA expression cassette contains the promoter of the Arabidopsis polyubiquitin 10 gene (prAtUbi10) (reference patent, CN201210570529), chloroplast transit peptide-coding sequence (AtCTP) (32), and a terminator of the tobacco nopaline synthase gene (tNos). The pdmB expression cassette contains the enhancer of the figwort mosaic virus 35S gene (eFMV) (48), the promoter of the Brassica CBP1 gene (prBrCBP) (reference patent, CN201310724357), the chloroplast transit peptide-coding sequence (AtCTP) (32), and a terminator of the pea rbcSE9 gene (tPsE9) (49). The phosphinothricin (glufosinate) N-acetyltransferase gene (pat) expression cassette for the selection of transgenic lines contains the promoter of the cauliflower mosaic virus 35S gene (pr35s), the pat gene from Streptomyces viridochromogenes, and the terminator of the cauliflower mosaic virus 35S gene (t35S). (b) Comparative root morphology and leaf surface between 30-day-old transgenic and nontransgenic Arabidopsis seedlings grown on 1/2 MS agar plates containing 0 to 2 mg/liter IPU. (c and d) Fresh weight (c) and root length (d) of 30-day-old transgenic and nontransgenic Arabidopsis seedlings grown on 1/2 MS agar plates containing 0 to 15 mg/liter IPU. The Arabidopsis seedlings used in the experiment include wild-type (WT) Arabidopsis seedlings, Arabidopsis seedlings transferred with an empty vector (vector control), and transgenic Arabidopsis seedlings (T3-2, T3-3, and T3-4). The data in panels c and d are derived from five independent measurements, and the error bars indicate standard deviations. Different lowercase letters above the bars indicate significant differences (P < 0.05).
Reverse transcription-PCR (RT-PCR) and quantitative RT-PCR (RT-qPCR) analyses showed that pdmA and pdmB transcripts accumulated in the roots, stems, and leaves of the transgenic lines instead of the wild-type (WT) and vector control lines (both called the nontransgenic lines) (see Fig. S1 in the supplemental material). Relatively higher transcription levels were observed in Arabidopsis leaves than in the roots and stems (Fig. S1a and b), which might be due to the stronger function of pdmAB promoters (prAtUbi10 and prBrCBP) in leaves than in the roots and stems. The transcription levels of pdmAB in the leaves of T3-3 were slightly higher (1.1- to ∼1.6-fold) than those in T3-2 and T3-4 (Fig. S1b), so the transgenic T3-3 line was selected for phytoremediation study.
No significant difference in the growth of transgenic and nontransgenic lines was observed in the absence of IPU. However, the transgenic Arabidopsis plant showed enhanced tolerance to 2 to 15 mg/liter IPU compared to the nontransgenic lines (Fig. S2). In the presence of IPU, damage symptoms were pronounced in the nontransgenic lines, including stunted root and shoot development, bleaching, and fresh weight (FW) decrease (48 to 88% decrease) (Fig. 1b and c). In contrast, most transgenic lines survived in the presence of 15 mg/liter IPU, and the FW and average root length of transgenic lines were approximately 2.5- to ∼3.8-fold and 1.9- to ∼2.4-fold those of the nontransgenic lines (Fig. 1c and d). A transgenic Arabidopsis plant without chloroplast transit peptide did not exhibit significant tolerance to IPU (data not shown). These results demonstrated that a transgenic Arabidopsis plant expressing PdmAB in the chloroplast destroyed the herbicidal activity of IPU before the herbicide could reach phytotoxic levels.
The action site of the PHs is the chloroplast photosynthesis system (24). The physiological and biochemical characteristics of the Arabidopsis plant also showed that the transformation of pdmAB alleviated the inhibition of IPU to Arabidopsis photosynthesis. Although the total chlorophyll contents decreased in all lines after IPU spraying, the chlorophyll content of the leaves in the transgenic lines (0.8 to ∼1.0 μg/mg FW) was approximately 1.2- to ∼1.5-fold that in the nontransgenic lines (Fig. S3a). The ratio of variable fluorescence to maximum chlorophyll fluorescence (Fv/Fm) of the nontransgenic lines decreased by 93.3 to 95.7% compared to that of the blank control and by 41.8 to 75.8% for the transgenic lines (Fig. S3b). The content of malondialdehyde (MDA) in the nontransgenic lines was 3.3- to ∼8.8-fold higher than that in the transgenic lines (Fig. S3c). The hydrogen peroxide content in the nontransgenic lines increased 4.0- to ∼4.8-fold compared to the blank control, while the hydrogen peroxide content in transgenic lines only increased 0.5- to ∼1.3-fold (Fig. S3d).
A transgenic Arabidopsis plant took up IPU and released its demethylated metabolite outside.
In the 30 ml of solid medium containing 15 mg/liter IPU, over 99% of IPU was removed by the transgenic lines, whereas less than 7.8% of the IPU was removed by the nontransgenic lines (Fig. 2a). Approximately 0.1 to 0.2 μg and 0.8 to 1.2 μg of IPU were detected in the roots and leaves of nontransgenic lines, respectively (Fig. 2b and c). In the transgenic lines, 0.2 μg of IPU was detected in the leaves, and no IPU was detected in the roots (Fig. 2b and c). Approximately 149.9 μg of MDIPU, the demethylated metabolite of IPU by PdmAB, was detected in the medium planted with transgenic lines, and small amounts of MDIPU (0.7 μg in leaves and 0.3 μg in roots) and DDIPU (0.2 μg in leaves and 0.1 μg in roots) were detected in the transgenic plant tissues. MDIPU was also detected in the growth medium, roots, and leaves of nontransgenic lines, but the concentrations were significantly lower than those in their counterparts in the transgenic lines. These data showed that IPU could be absorbed and demethylated efficiently by a transgenic Arabidopsis plant.
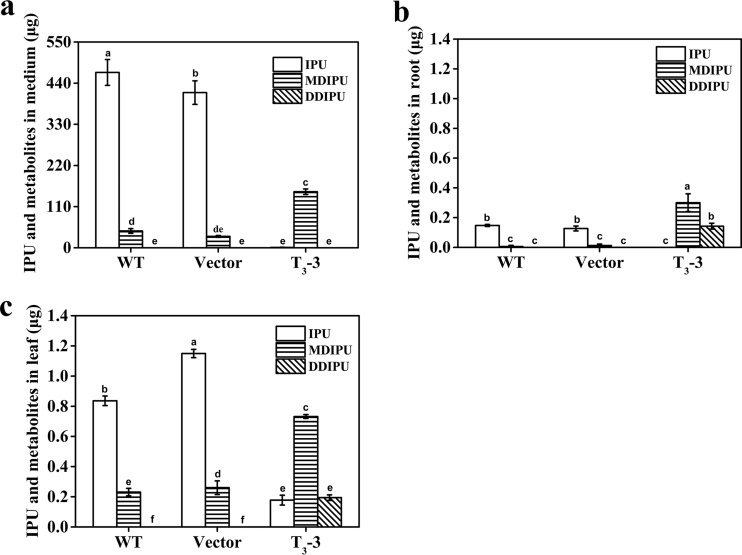
FIG 2.
Removal of 15 mg/liter IPU in 30 ml solid medium by a transgenic Arabidopsis plant. IPU and metabolites (MDIPU and DDIPU) in the medium (a), roots (b), and leaves (c) were determined by HPLC. The Arabidopsis seedlings used in the experiment include wild-type (WT) Arabidopsis seedlings, Arabidopsis seedlings transformed with an empty vector (vector), and transgenic Arabidopsis seedlings (T3-3). The results are the mean and standard deviation of the results from three replicates. Different lowercase letters above the bars indicate significant differences (P < 0.05).
To investigate the distribution of functional PdmAB, the leaf, stem, and root pieces were harvested separately and used to transform IPU. The leaf, stem, and root pieces of the transgenic lines removed 79.8%, 69.7%, and 19.3% of the 15 μg of IPU added in the reaction system, respectively. In contrast, the leaf, stem, and root pieces of the nontransgenic lines removed 12.7%, 10.7%, and 11.3% of the 15 μg of IPU added in the reaction system, respectively. In addition, 4.4 μg, 3.0 μg, and 1.7 μg of MDIPU were detected in the reaction systems of the transgenic leaf, stem, and root pieces, respectively (Fig. S4a to c), while no MDIPU was detected in the reaction systems of the nontransgenic pieces. No demethylation activity of IPU was detected in the medium when the previously cultured transgenic lines had been removed, showing that no PdmAB was secreted outside the plant tissue and that the demethylation of IPU occurred inside transgenic plant tissue. These results showed that most functional PdmAB was located in the leaf and stem of the transgenic lines. The attempt to assay the activity of the crude PdmAB extracted from the tissues of the transgenic lines failed, even when NADH was added (data not shown). The reason underlying this might be that PdmAB, the multicomponent demethylase system, was damaged during the protein extraction procedure.
Removal of PHs by transgenic Arabidopsis spp. in water and further metabolism of the released intermediate by Sphingobium sp. strain 1017-1.
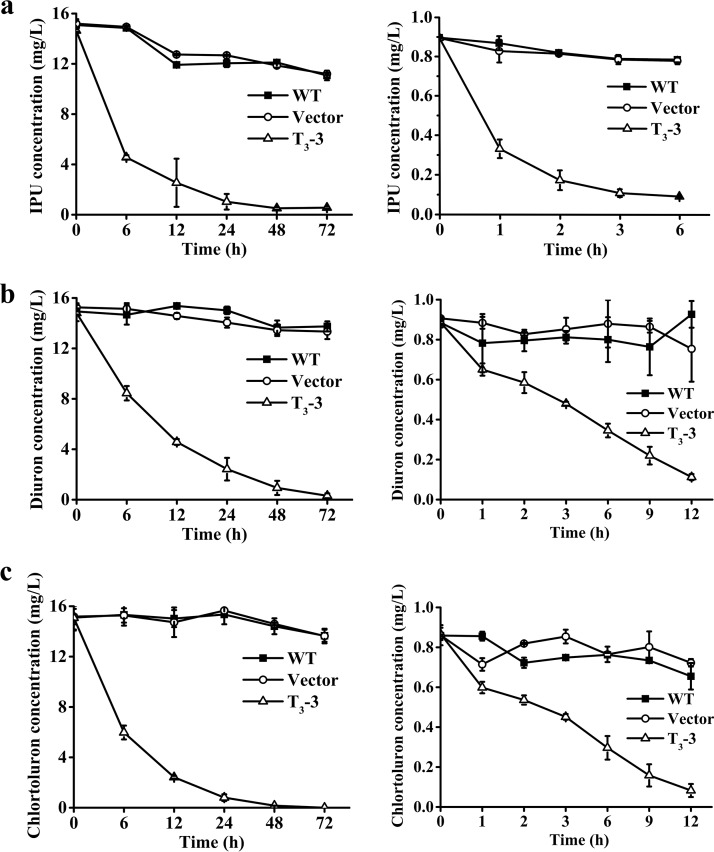
The transgenic lines (40 seedlings per treatment) showed excellent removal efficiency for low (0.9 mg/liter) and high (15 mg/liter) concentrations of IPU, diuron, or chlortoluron in the 1/2 Murashige and Skoog (MS) liquid medium (Fig. 3). The transgenic lines could also simultaneously remove 100% of the IPU, 89.8% of the diuron, and 97.6% of the chlortoluron within 72 h (Fig. S5a) from the medium containing a mixture of 6 mg/liter IPU, 6 mg/liter diuron, and 6 mg/liter chlortoluron. Furthermore, the transgenic lines could successively remove 93.4% of the total IPU (0.75 mg) within 48 h, which was added in four rounds at an interval of 12 h (Fig. S5b). In all treatments using the nontransgenic lines, negligible amounts of PHs were removed, and the growth of Arabidopsis spp. was severely stunted (Fig. S6).
FIG 3.
Removal of PHs in water by transgenic Arabidopsis plant. (a) IPU at 15 mg/liter (left) and 0.9 mg/liter (right). (b) Diuron at 15 mg/liter (left) and 0.9 mg/liter (right). (c) Chlortoluron at 15 mg/liter (left) and 0.9 mg/liter (right). The results are the mean and standard deviation of three replicates (40 seedlings per treatment).
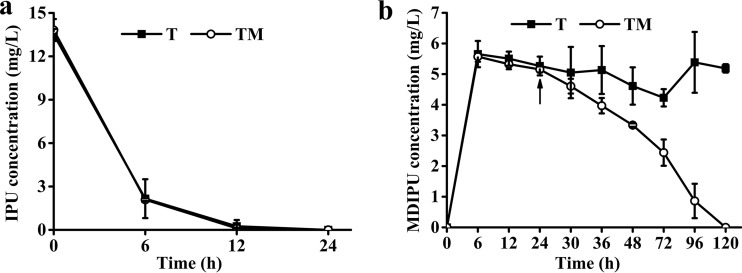
It was found that 5.5 mg/liter MDIPU was released into the medium at 6 h and kept unchanged until 120 h during the removal of 15 mg/liter of IPU by transgenic Arabidopsis seedlings (40 seedlings per flask) individually (Fig. 4a and b). When (2.46 ± 0.12) × 105 CFU/ml (values are means ± standard deviations calculated from the results from triplicate assays) of Sphingobium sp. strain 1017-1 was additionally inoculated at 24 h, the produced 5.5 mg/liter MDIPU decreased to a nondetectable level after 96 h. The results showed that the combination of a transgenic Arabidopsis plant and strain 1017-1 could completely remove IPU without accumulating intermediates (Fig. 4b).
FIG 4.
(a and b) Removal of 15 mg/liter IPU (a) and its metabolite MDIPU (b) in 1/2 MS liquid medium by a transgenic Arabidopsis plant (■, T) as well as by the combination of a transgenic Arabidopsis plant and Sphingobium sp. strain 1017-1 (○, TM). The arrow indicates the subsequent inoculation of Sphingobium sp. strain 1017-1 at the concentration of (2.46 ± 0.12) × 105 CFU/ml. The results are the mean and standard deviation of the results from three replicates (40 seedlings per treatment).
Enhanced and complete removal of IPU in soil by combination of a transgenic Arabidopsis plant and Sphingobium sp. strain 1017-1.
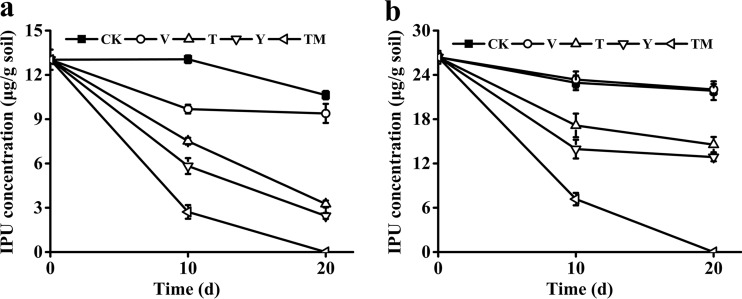
The conceptual framework and experimental design of the transgenic plant-microbe combined remediation system are illustrated in Fig. 5. For the individual phytoremediation by transgenic Arabidopsis seedlings (T) (10 seedlings per pot), 75% of the 15 μg/g IPU and 44.8% of the 30 μg/g IPU were removed from the soil within 20 days. For individual bioaugmentation by Sphingobium sp. strain YBL2 (Y), 81.2% of the 15 μg/g IPU and 51.2% of the 30 μg/g IPU were removed within 20 days, respectively. Interestingly, for both the low (15 μg/g) and high (30 μg/g) concentrations of IPU, the combinational remediation by transgenic Arabidopsis seedlings (10 seedlings per pot) and Sphingobium sp. strain 1017-1 (TM) completely removed IPU within 20 days (Fig. 6a and b). In the control (CK) and Arabidopsis (empty vector) (V) groups, no significant removal of IPU was observed, and the nontransgenic lines died in the soil spiked with 15 mg/kg IPU (Fig. 6a and 7a). These results demonstrated that combinational remediation had great potential to accelerate the bioremediation process of PH-contaminated soil.
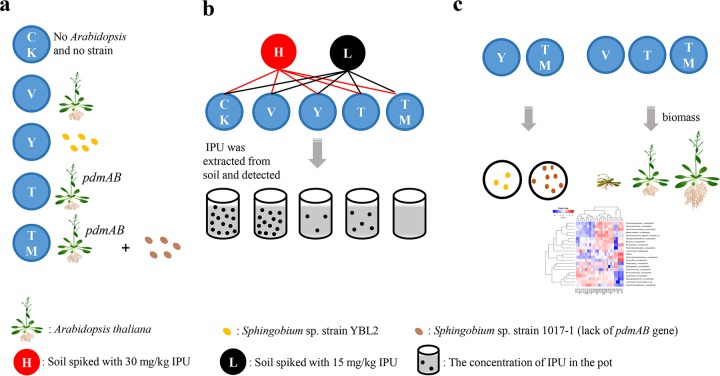
FIG 5.
Conceptual framework and experimental design of the transgenic plant-microbe combined remediation system. (a) Definitions of the five different treatments. Five treatments were set as follows: CK, neither strain inoculation nor Arabidopsis planting; V, planting of Arabidopsis seedlings (empty vector); T, planting of transgenic Arabidopsis seedlings (pdmAB); Y, inoculation with Sphingobium sp. strain YBL2; TM, inoculation with Sphingobium sp. strain 1017-1 together with planting of transgenic Arabidopsis plants (pdmAB). Strain 1017-1 is derived from strain YBL2, with the pdmAB genes deleted. (b) High (30 mg/kg) and low (15 mg/kg) concentrations of IPU were designed, and the concentration of IPU in each treatment was detected at 0, 10, and 20 days. (c) The cell amounts of the inoculated strains YBL2 and strain 1017-1 in the Y treatment and TM treatment, respectively, and the biomass (FW, root length and seedling length) of the transgenic Arabidopsis plant in the T treatment and the TM treatment, respectively, were determined. The bacterial community structure in each treatment was also analyzed at 0, 10, and 20 days.
FIG 6.
Removal of 15 μg/g (a) and 30 μg/g (b) IPU in soil by different treatments. The abbreviations for different treatments are the same as those in Fig. 5. The results are the mean and standard deviation of the results from three replicates.
FIG 7.
The growth status of transgenic Arabidopsis and the cell amounts of inoculated degrading strains during the removal of IPU in soil. The abbreviations for different treatments are the same as those in Fig. 5. (a) The growth status of Arabidopsis seedlings during the removal of low (L; 15 mg/kg) and high (H; 30 mg/kg) concentrations of IPU. (b) The cell amounts of inoculated strains of YBL2 and 1017-1 in the individual bioaugmentation treatment (T) and the combinational treatment (TM) by time, respectively. (c to e) The fresh weight (c), root length (d), and seedling length (e) of transgenic Arabidopsis seedlings in their respective treatments were measured after 20 days. The results are the mean and standard deviation of the results from three replicates. Different lowercase letters above the bars indicate significant differences (P < 0.05).
The transgenic Arabidopsis plant in the TM treatment grew more vigorously than that in the T treatment, especially in the soil treated with high concentrations of IPU (Fig. 7a). In the 15 mg/kg IPU-treated soil, the FW, root length, and seedling length of the transgenic Arabidopsis plant in the TM treatment were about 1.3-, 1.2-, and 1.0-fold those in the T treatment at 20 days, respectively (Fig. 7c to e). For the soil spiked with 30 mg/kg IPU, the FW, root length, and seedling length of the transgenic Arabidopsis plant in the TM treatment were approximately 2.5-, 2.1-, and 1.5-fold those in the T treatment, respectively (Fig. 7c to e). These results showed that the inoculated strain 1017-1 in the TM treatment could promote the growth of the transgenic plant, probably by metabolism of the released intermediate MDIPU in the rhizosphere, further relieving the phytotoxicity of the MDIPU.
The cell amounts of the initially inoculated Sphingobium sp. strains YBL2 and 1017-1 in their respective soil increased from (4.2 ± 0.05) × 106 CFU/g soil to (0.8 ± 0.13) to (1.4 ± 0.11) × 107 CFU/g soil at 10 days and decreased to (0.7 ± 0.01) to (5.6 ± 0.09) × 106 CFU/g soil at 20 days in both low and high concentrations of IPU-treated soils. Interestingly, the amount of strain 1017-1 cells in the TM treatment increased more significantly at 10 days and decreased less at 20 days compared to the amount of strain YBL2 cells in Y treatment (Fig. 7b). At 20 days, the amounts of strain 1017-1 cells were about 6.4-fold and 5.9-fold those of strain YBL2 in low and high concentrations of IPU-treated soils, respectively (Fig. 7b). These data showed that the transgenic Arabidopsis plant could provide a more suitable rhizospheric niche for the survivability of the inoculated strain.
Influence of different treatments on the soil bacterial community.
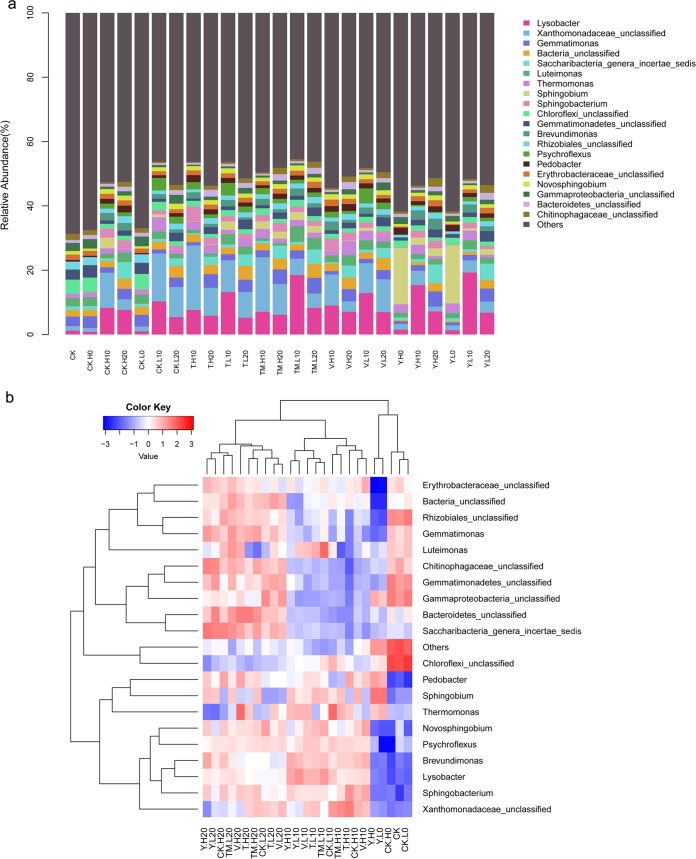
Alterations in the indigenous bacterial communities in the five soils of different treatments were investigated by MiSeq sequencing. A total of 1,709,256 (90%) tag sequences were obtained after filtration (Table S1). The dominant length of the tag sequences was more than 200 bp (86%). The classified sample sequences from the five differently treated soils were affiliated with 20 bacterial phyla (Fig. 8a and b). In soils sampled at 0 days (CK, CK.L/H0, and Y.L/H0), the content of actinobacteria was remarkably higher than in other samples. The decreased abundance of proteobacteria was detected in CK and CK.L/H0 but not in Y.L/H0, which might result from the inoculation of Sphingobium strains. The abundance of Sphingobium strains in Y.L/H0 was significantly higher than in other samples (Fig. S7). Although the relative contents of Sphingobium strains in soils with inoculation treatments at 10 and 20 days (Y.L/H10, Y.L/H20, TM.L/H10, and TM.L/H20) decreased, their abundance was higher than that in samples without inoculation treatments. The relative contents of Lysobacter, Sphingobacterium, Brevundimonas, Psychroflexus, and Novosphingobium spp. were much lower in soils sampled at 0 days, which increased significantly after treatments at 10 and 20 days (Fig. S7).
FIG 8.
The relative abundances of the bacterial phyla (a) and their variations (b) among samples from different treatments during the time. Red, increase in relative abundance; blue, decrease in relative abundance. CK, control; V, Arabidopsis plant transferred with an empty vector; T, transgenic A. thaliana T3-3; Y, strain YBL2; TM, combination of strain 1017-1 and transgenic A. thaliana T3-3; L, 15 mg/kg IPU applied; H, 30 mg/kg IPU applied. The numbers 0, 10, and 20 show that samples were collected 0, 10, and 20 days after inoculation, respectively. The experiment was performed in triplicate.
The CK, CK.L0, and C.H0 treatments showed the highest Shannon and Chao1 indices, while the lowest indices were found in Y.L0 and Y.H0 (Table S2). In the control soils with IPU application (CK.L/H), the richness estimators and diversity indices decreased significantly at 10 days and then increased at 20 days. Unlike the trend for control soils, the richness estimators and diversity indices increased continuously from 0 to 20 days with the Y treatment. The comparison between CK and Y treatments revealed that these indices increased in Y.L treatments at 20 days, which were neither detected at 10 days nor in the Y.H treatment at 20 days. The increased bacterial community richness and diversity might be due to the efficient elimination of low concentrations of IPU by bioaugmentation with strain YBL2. Compared to control samples with low IPU concentration at 10 days (CK.L.10), the individual transgenic plant treatment (T.L) and combinational remediation (TM.L) improved the bacterial community richness and diversity, while the nontransgenic line treatment (V.L) did not (Table S2). Similar results were detected in samples at 20 days. At a high concentration of IPU, the indices were only increased in TM.H treatments at 10 days. The results indicated that the bacterial community richness and diversity could be recovered by individual transgenic plant remediation only at low IPU concentrations, while the combinational remediation by the transgenic plant and strain 1071-1 could recover the bacterial community even at high IPU concentrations.
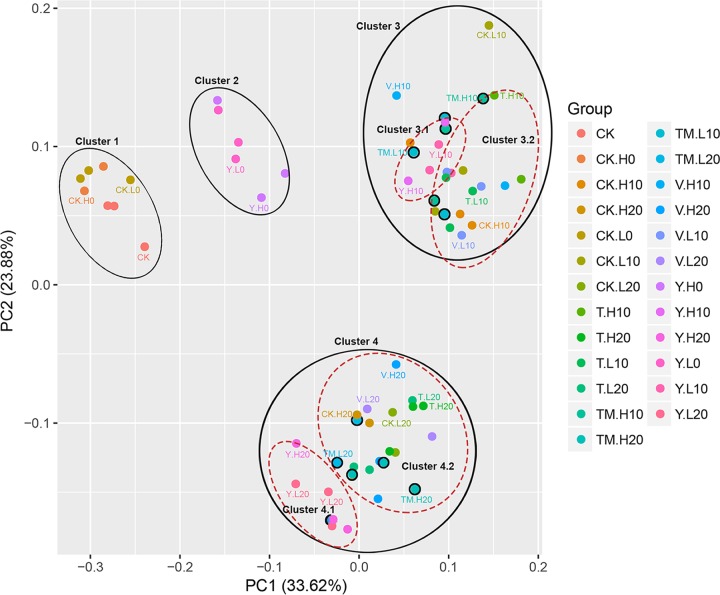
The principal-coordinate analysis (PCoA) plot separated bacterial communities into four distinct clusters (Fig. 9), showing that the communities with same sampling time clustered together tightly and differentiated with the sampling time. Consistently, the hierarchical clustering analysis also showed a similar separation (Fig. S8), suggesting that majority of the variance resulted from the sampling time. However, when each cluster was analyzed specifically, it was worth noticing that the communities in clusters 3 and 4 could be further divided into two subclusters (Fig. S8). Subclusters 4.1 and 4.2 separately contained bioaugmentation treatments (Y.L and Y.H) and Arabidopsis treatments (T/V/TM.L and T/V/TM.H). Similar results could be found in subclusters 3.1 and 3.2. Interestingly, the distribution of community in combinational treatments (TM.L and TM.H) changed in clusters 3 and 4. In cluster 3, the communities in combinational treatments could be found in both subclusters 3.1 and 3.2 but were detected in subcluster 4.2 and discriminated from subcluster 4.1, indicating that the influence of combinational treatment on bacterial community structure was dynamic along with the IPU elimination. These analyses indicated that the influence of the structure of bacterial community by combinational treatments (TM) was between that of individual bioaugmentation treatments (Y) and transgenic plant treatments (T).
FIG 9.
Principal-coordinate analysis (PCoA) of bacterial communities in soils with different treatments based on weighted Unifrac distances. The abbreviations for different treatments are the same as those in Fig. 8.
DISCUSSION
Bioaugmentation with degrading microbes has exhibited great potential for the cleanup of organic pollutants in many cases, while the remedial efficiencies in actual fields are sometimes not stable. The soil colonization ability of the inoculated microbes and the bioavailability of the tightly soil-bound pollutants (the ability of the microbes to spread through the soil and reach the pollutant) often are the limiting factors in successful bioaugmentation (25). Plants, stably present in the environment, can use their deep extensive root systems to take up pollutants from relatively deep soil and transport/translocate them to various plant tissues where they can be metabolized (9). The lack of a versatile catabolic capacity of plants can be remedied by transferring suitable genes from microbes into plants (11–13, 15, 16, 26, 27). However, plants transferred with key catabolic genes cannot achieve a complete removal of pollutants. The synergistic relationships between plants and microbes in the rhizosphere can be used to develop a new bioremediation strategy, overcoming the drawbacks of an individual bioaugmentation or phytoremediation method.
In this study, a transgenic Arabidopsis plant expressing the bacterial N-demethylase PdmAB, which is specially designed for the initial degradation of N,N-dimethyl-substituted PHs, was developed. Although the optimal bacterial ferredoxin of PdmAB may be the [3Fe-4S] type, PdmAB showed low specificity for electron transport components (21), making it possible to accept electrons from the electron transport components of the chloroplast (28). With the help of the chloroplast transit peptide-coding region, which targets PdmAB to the chloroplasts, the only expression of the terminal oxygenase PdmAB in the Arabidopsis plant endowed it with the N-demethylation function of N,N-dimethyl-substituted PHs, the rate-limiting step for PH mineralization (29). The importance of the chloroplast transit peptide was also confirmed by the fact that the Arabidopsis plant expressing PdmAB without transit peptide showed very low resistance levels to IPU (data not shown). These results indicate that the abundant ferredoxin formed in plant chloroplast can shuttle electrons to PdmAB. Additionally, why an Arabidopsis plant expressing PdmAB without transit peptide showed very low resistance levels to IPU may be explained by two reasons. First, cytosolic electron transport components of the Arabidopsis plant probably could not support the activity of PdmAB. Second, the action site of IPU is the chloroplast photosynthesis system, so compared to the PdmAB expressed in the cytoplasm, PdmAB located in the chloroplast can detoxify IPU more effectively.
It was found that a low concentration of IPU was detected in the roots and a relatively higher concentration of IPU was found in the leaves of the nontransgenic Arabidopsis plant. In addition, IPU was detected in the leaves instead of the roots in the transgenic Arabidopsis plant, although the leaves of the transgenic lines had higher demethylation activity than the roots. These results showed that IPU was adsorbed by the Arabidopsis plant through the roots and translocated to the leaves. The IPU taken up by the transgenic Arabidopsis plant was demethylated to MDIPU and small amounts of DDIPU, which were released into the environment through the roots. Although MDIPU and DDIPU showed lower phytotoxicity than IPU, their toxicity and recalcitrance in the environment remain unknown and need to be completely removed.
The inoculation of intermediate-degrading microbes together with transgenic plant can be an important additive to completely remove pollutants. The approach in this study provides a strong framework for producing a combinational transgenic plant-microbe system in which transgenic plant takes up IPU and initially catabolizes IPU to MDIPU efficiently, while the inoculated MDIPU-mineralizing strains completely mineralize the excreted MDIPU in the rhizosphere. The significantly enhanced removal of IPU from soils by the combinational remediation compared to individual phytoremediation or bioaugmentation is mainly due to the mutual benefits between the plants and microbes. The growing plants secrete a wide range of chemicals in root exudates and till the soil to improve aeration, providing a nutrient-rich and suitable microenvironment to prevent a rapid decline of the inoculated microbes and stimulating the action of the microbes in the rhizosphere. It was found that the amount of the inoculated strain 1017-1 cells in TM treatment increased more significantly at 10 days and decreased less at 20 days compared to the cell amounts of strain YBL2 in the Y treatment. The abundance of the Sphingobium strain in TM treatments was higher than that in the Y treatments at 20 days (Fig. S2). Furthermore, the root system of plants can act as an injection system to spread the microbes through the soil (30), establishing an increase in contact between the degrading microbes and the pollutants in the deeper soil layer. On the other hand, the inoculated degrading microbes efficiently catabolize the excreted intermediates from the transgenic plants, further releasing the phytotoxicity, enhancing the growth of the host transgenic plants. The transgenic Arabidopsis plant in the TM treatments grew more vigorously and had a higher biomass than in the T treatments, especially in the soil treated with high concentrations of IPU, which resulted in the more efficient removal of IPU from the soil. Overall, the combination of phytoremediation and bioaugmentation represents an innovative strategy for the enhanced and complete remediation of organic pollutant-contaminated sites, which warrants further verification with field experiments.
MATERIALS AND METHODS
Chemicals, bacterial strains, and culture conditions.
IPU, MDIPU, DDIPU, diuron, and chlortoluron (all >99% purity) were purchased from J&K Scientific Ltd. (Shanghai, China). Glufosinate-ammonium was purchased from Sigma-Aldrich (Shanghai, China). Murashige and Skoog medium (31) with vitamins (MS medium) was purchased from Beijing Seajet Scientific (Beijing, China). The IPU-mineralizing strain Sphingobium sp. YBL2 (= CCTCC AB2013269) (21, 22) and the pdmAB mutant strain Sphingobium sp. 1017-1 (21) were cultured in Luria-Bertani (LB) medium at 30°C. Antibiotics were added as follows: ampicillin (Amp), 100 mg/liter; spectinomycin (Spe), 100 mg/liter; and rifampin (Rif), 50 mg/liter.
Construction of transgenic Arabidopsis plants expressing the bacterial N-demethylase PdmAB.
The N-demethylase PdmAB was identified from Sphingobium sp. strain YBL2 and can catalyze the N-demethylation of a number of N,N-dimethyl-substituted PHs, such as IPU, chlortoluron, metoxuron, monuron, diuron, fluometuron, and fenuron (21, 22). A strategy for expression of PdmAB in Arabidopsis plants with the help of the chloroplast transit peptide-coding region, which targets PdmAB to chloroplasts, was used in this study. The nucleotide sequences of the pdmA and pdmB genes were optimized using GenScript's OptimumGene codon optimization system according to the codon usage bias and GC content to make the genes well expressed in plants. The chloroplast transit peptide-coding sequence (AtCTP) (32) was fused to the 5′ ends of the pdmA and pdmB genes. AtCTP-pdmA was digested with SnaBI and then cloned into pGEM-T (Promega, Madison, WI, USA), while AtCTP-pdmB was digested with KasI and cloned into pGEM-T. Then, the gene expression cassettes for AtCTP-pdmA and AtCTP-pdmB were cut with SnaBI and KasI, respectively, and inserted into the corresponding sites of vector pDBNBC-02 (derived from pCAMBIA2301; Cambia) to produce pDBN10938. The plasmid pDBN10938 was introduced into Agrobacterium tumefaciens GV3101 using the liquid nitrogen method (33). Arabidopsis thaliana ecotype Columbia was transfected with A. tumefaciens cells harboring pDBN10938 using the floral dip method (34). The seeds of transgenic plants were screened on MS medium (31) containing 8 mg/liter glufosinate-ammonium, and the T1-resistant seedlings were transferred to soil. Finally, the homozygous genotypes of transgenic plants were obtained from self-fertilization, and homozygous lines were identified in the T3 generation via segregation analysis (35).
Analyses of the transcription level of pdmAB in a transgenic Arabidopsis plant by RT-PCR and RT-qPCR.
Total RNA was isolated from the roots, stems, and leaves of a 3-week-old Arabidopsis plant using RNAiso Plus (TaKaRa, Dalian, China). The isolated RNA was purified with the RT reagent PrimeScript kit with genomic DNA (gDNA) Eraser (TaKaRa) to remove DNA contamination, and then cDNA was synthesized according to the manufacturer's instructions. RT-PCR was performed as described previously with minor modifications (36). The primer pairs pdmAF/pdmAR (5′-GAGACTGAAATCCCTAAGAGCG-3′/5′-CTGACCGTGTGACTATAACCTG-3′), pdmBF/pdmBR (5′-CTTTCACACGAAGCCAAACTC-3′/5′-CTTCTCTGTCGAAATCCAGGG-3′), and AtAc2F/AtAc2R (5′-GCACCCTGTTCTTCTTACCGAG-3′/5′-AGTAAGGTCACGTCCAGCAAGG-3′) were used for the amplification of pdmA, pdmB, and AtAc2 (the reference gene in Arabidopsis), respectively. qRT-PCR was performed in the Applied Biosystems 7500 Fast real-time PCR system (Applied Biosystems, USA) with SYBR Premix Ex Taq II (Tli RNase H Plus; TaKaRa). All analyses were performed in triplicate, and the 2−ΔΔCT method was used for the quantitative analysis of the transcription of pdmAB genes in a wild-type Arabidopsis (WT) plant, an Arabidopsis plant transferred with an empty vector (vector control), and a transgenic Arabidopsis plant (T3-2, T3-3, and T3-4).
Assay of the resistance and physiological and biochemical characteristics of a transgenic Arabidopsis plant in response to IPU.
To study the IPU resistance of a transgenic Arabidopsis plant, Arabidopsis seeds were surface sterilized with 6% sodium hypochlorite for 15 min and stratified at 4°C for 2 days. Forty seeds were sown on solidified medium (pH 5.8, 1.5% agar) containing 30 ml of 1/2 MS (31), 1.5% sucrose, and different concentrations of IPU (0, 2, 5, or 15 mg/liter). The plates were grown vertically at 23°C/20°C with a 16-h light/8-h dark cycle in a growth chamber (Jiangnan, Ningbo, China). After 30 days, the root length, fresh weight (FW), and leaf surface area of the Arabidopsis plant were measured.
To study the effect of IPU on photosynthesis in Arabidopsis plants, 15-day-old plant seedlings of uniform size were transplanted to synthetic soil composed of a mixture of peat-vermiculite (1:3 [vol/vol]). The organic matter content in the soil was 3.5%, and the pH was 7.2. In accordance with the levels normally used for weed control in agricultural applications (1.05 to 1.2 kg/ha IPU dissolved in 750 to 900 kg/ha water), 0.09 g IPU dissolved in 33 g water was evenly sprayed on the leaves of 160 Arabidopsis seedlings (20 days old) in 40 pots (0.4 m2). After 7 days, 3 to 5 leaves were used for chlorophyll content analysis. The chlorophyll content was determined according to the Lichtenthaler method (37) by measuring the absorbance at 470 nm, 649 nm, and 665 nm. The photosynthetic parameters were determined using an Imaging-PAM (Heinz Walz Gmbh, Germany) photosynthesis system. Fv and Fm were determined after 30 min in the dark (38, 39). Hydrogen peroxide content was determined as described by Alexieva et al. (40), and MDA content was measured according to the method of Zhang et al. (41).
Analysis of IPU and its metabolites in plant tissue, soil, and water.
To detect IPU and its metabolites in plant tissues, the roots and leaves were washed with deionized water and 20% methanol three times to remove adherent IPU/metabolites from the surface. IPU and metabolites were extracted with acetonitrile containing 1% acetic acid, and chlorophyll was removed using a PSA/GCB/C18 (containing anhydrous magnesium sulfate, primary secondary amine [PSA], octadecyl-bonded silica [C18], and graphitized carbon black [GCB] sorbents) Clean Up tube (Anpel Laboratory Technologies, Shanghai, China). The IPU and its metabolites in soil and water were extracted using dichloromethane with a ratio of 10:1 (milliliters/gram) and 1:1 (milliliters/milliliter), respectively, and extraction was repeated three times. All of the extracts were dried over anhydrous Na2SO4 and evaporated using a vacuum rotary evaporator at room temperature. Then, the residual was dissolved in 100 μl methanol and analyzed using high-performance liquid chromatography (HPLC; UltiMate 3000 RSLC; Thermo Fisher Scientific, USA). For the HPLC analysis, a separation column (internal diameter, 4.6 mm; length, 250 mm) filled with Syncronis C18 (Thermo Fisher Scientific) was used. The mobile phase was acetonitrile-water (50:50 [vol/vol]), and the flow rate was 1.0 ml/min. The detection wavelength was 250 nm (21), and the injection volume was 20 μl. All experiments were performed in triplicate.
Transformation of IPU by root, stem, and leaf pieces of transgenic Arabidopsis plant.
The leaves, stems, and roots of a 21-day-old transgenic Arabidopsis plant were cut into 1-cm pieces, and 1 g of pieces of each part was placed into a reaction mixture containing 3 ml of 20 mM Tris-HCl (pH 7.0) and 5 mg/liter IPU. After incubation at 30°C for 48 h, the IPU was extracted using dichloromethane, and the concentration of IPU was detected by HPLC. Pieces of the WT and vector control were used as negative controls. All experiments were performed in triplicate.
Removal of PHs in water by transgenic Arabidopsis.
Forty seedlings of a 15-day-old transgenic Arabidopsis plant were transferred to a 250-ml conical flask containing 60 ml sterilized 1/2 MS liquid medium (0.5% sucrose [pH 5.8]) under sterile conditions, and the culture conditions were 23/20°C and 16-h light/8-h dark. PHs were filtered and added to the liquid medium after 10 days. For the removal of a high concentration of a single PH, a final concentration of 15 mg/liter of IPU, diuron, or chlortoluron was added. For the removal of a low concentration of a single PH, a final concentration of 0.9 mg/liter IPU, diuron, or chlortoluron was added. For the removal of mixed PHs, final concentrations of 6 mg/liter IPU, 6 mg/liter diuron, and 6 mg/liter chlortoluron were added. For the successive removal of IPU, 12 ml of 1/2 MS (0.5% sucrose [pH 5.8], containing 12.5 mg/liter IPU) was added three times every 12 h to the original 60 ml of medium (initial IPU concentration, 12.5 mg/liter). For the detection of the concentration of PHs, 3-ml samples were taken at intervals and detected by HPLC.
Removal of IPU in water by combination of transgenic Arabidopsis seedlings and Sphingobium sp. strain 1017-1.
Forty transgenic Arabidopsis (15-day-old) seedlings were cultured in 250-ml conical flasks as described above. Ten days later, IPU (15 mg/liter) was filtered and added, and 24 h later, Sphingobium sp. strain 1017-1 was additionally inoculated into the medium at a concentration of (2.46 ± 0.12) × 105 CFU/ml. At intervals, 2-ml samples of medium were taken, and the concentrations of IPU and its metabolite MDIPU were detected by HPLC, as described previously (21). Treatment without inoculation of strain 1017-1 was used as control.
Removal of IPU in soil by combination of a transgenic Arabidopsis plant and Sphingobium sp. strain 1017-1.
The synthetic soil was first sprayed with IPU that was dissolved in methanol. When the methanol evaporated, the polluted soil was mixed with unpolluted soil to obtain the final concentration of 15 mg/kg IPU (low concentration [L]) or 30 mg/kg IPU (high concentration [H]). The mixed soil (100 g) was packed into pots (top diameter, 10 cm; bottom diameter, 7 cm; height, 7.5 cm) and equilibrated in a glass greenhouse for 2 days. Five treatments were set as follows: (i) CK, neither strain inoculation nor Arabidopsis planting; (ii) V, planting of Arabidopsis seedlings (empty vector); (iii) T, planting of transgenic Arabidopsis seedlings containing pdmAB; (iv) Y, inoculation with Sphingobium sp. strain YBL2; and (v) TM, inoculation with Sphingobium sp. strain 1017-1 together with planting of transgenic Arabidopsis seedlings containing pdmAB. Each treatment was performed in triplicate. For the planting of Arabidopsis seedlings, 10 equivalently sized seedlings of a 15-day-old transgenic Arabidopsis plant or Arabidopsis plant with empty vector were transplanted into the soil in pots and placed at 23/20°C with a 16-h light/8-h dark cycle. For inoculation of the strains, Sphingobium sp. strain YBL2 or Sphingobium sp. strain 1017-1 was inoculated into the soil at the same concentration of (4.2 ± 0.05) × 106 CFU/g soil. The soil water content was controlled at approximately 40% to mitigate possible leaching of IPU.
The rhizospheric soil or respective bulk soil (3 g) was collected for each replicate at 0, 10, and 20 days after planting or inoculation. The concentration of IPU in the soil was measured as described previously to evaluate the effect of remediation (12). The amounts of inoculated Sphingobium sp. strain 1017-1 and Sphingobium sp. strain YBL2 were also determined. Sterile water (1.5 ml) was added to 0.4 g soil, mixed adequately, and plated on the LB medium with 100 mg/ml streptomycin. The grown colonies showing morphology similar to Sphingobium species on the plate were counted 3 days later, and the ddhA gene was amplified as a marker to confirm the authenticity of these colonies. The growth of the Arabidopsis plant was observed at intervals. The Arabidopsis seedlings in the soil were removed 20 days later, and the root, stem length, and fresh weight (biomass) of the Arabidopsis plant were measured.
Bacterial community analysis during remediation.
The total DNA of the rhizospheric soil or bulk soil (0.5 g; three replicates for each treatment) was extracted using an E.Z.N.A. soil DNA kit (Omega Bio-Tek, USA), according to the manufacturer's instructions. The specific primer set for bacteria, 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′), with the reverse primer containing a 6-bp barcode, was used to amplify the V3-V4 region of the 16S rRNA gene. PCR amplification was performed as described previously (42). The PCR products were purified by using AMPure XT beads (Beckman Coulter Genomics, Danvers, MA, USA) and quantified by a Qubit fluorometer (Invitrogen, USA). The purified amplicons were sequenced on a 300PE MiSeq platform (LC-Bio Technology, Hangzhou, China), according to standard protocols.
Pairs of reads from the original DNA fragments were merged by using FLASH (43). Reads were assigned to each sample according to the unique barcode of that sample. Sequences were quality filtered by QIIME pipeline using the criteria described previously (44, 45). The sequences were assigned to operational taxonomic units (OTUs) with a 97% similarity cutoff, and the OTUs were chosen using UPARSE (46). Representative sequences for each OTU were selected and assigned to taxonomic data using the RDP Classifier (47). Alpha diversity was applied in analyzing complexity of bacterial community diversity for a sample. In order to estimate the alpha diversity, the OTU table was rarified, and four metrics were calculated with QIIME pipeline, including Chao1 metric, observed OTU metric, Shannon index, and Simpson index. Beta diversity, used to evaluate differences in bacterial community structure among samples, was calculated by nonmetric multidimensional scaling (NMDS) and hierarchical clustering with the QIIME pipeline.
Accession number(s).
The sequence data of the 16S rRNA genes have been submitted to the GenBank database under accession numbers MH096057 to MH100661.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the National Key Research and Development Program of China (grant 2016YFD0800203), the Joint NSFC-ISF Research Program (grant 31461143009), and the National Natural Science Foundation of China (grant 31670111).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00273-18.
REFERENCES
- 1.Muud P, Hance R, Wright S. 1983. The persistence and metabolism of isoproturon in soil. Weed Res 23:239–246. doi: 10.1111/j.1365-3180.1983.tb00545.x. [DOI] [Google Scholar]
- 2.Gaillardon P, Sabar M. 1994. Changes in the concentrations of isoproturon and its degradation products in soil and soil solution during incubation at two temperatures. Weed Res 34:243–250. doi: 10.1111/j.1365-3180.1994.tb01992.x. [DOI] [Google Scholar]
- 3.Cox L, Walker A, Welch SJ. 1996. Evidence for the accelerated degradation of isoproturon in soils. Pest Manag Sci 48:253–260. doi:. [DOI] [Google Scholar]
- 4.Sun JQ, Huang X, Chen QL, Liang B, Qiu JG, Ali SW, Li SP. 2009. Isolation and characterization of three Sphingobium sp strains capable of degrading isoproturon and cloning of the catechol 1,2-dioxygenase gene from these strains. World J Microbiol Biotechnol 25:259–268. doi: 10.1007/s11274-008-9888-y. [DOI] [Google Scholar]
- 5.Bending GD, Lincoln SD, Sørensen SR, Morgan JAW, Aamand J, Walker A. 2003. In-field spatial variability in the degradation of the phenyl-urea herbicide isoproturon is the result of interactions between degradative Sphingomonas spp. and soil pH. Appl Environ Microbiol 69:827–834. doi: 10.1128/AEM.69.2.827-834.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elkhattabi K, Bouhaouss A, Scrano L, Lelario F, Bufo SA. 2007. Influence of humic fractions on retention of isoproturon residues in two Moroccan soils. J Environ Sci Health B 42:851–856. doi: 10.1080/03601230701555104. [DOI] [PubMed] [Google Scholar]
- 7.Hussain S, Devers-Lamrani M, Spor A, Rouard N, Porcherot M, Beguet J, Martin-Laurent F. 2013. Mapping field spatial distribution patterns of isoproturon-mineralizing activity over a three-year winter wheat/rape seed/barley rotation. Chemosphere 90:2499–2511. doi: 10.1016/j.chemosphere.2012.10.080. [DOI] [PubMed] [Google Scholar]
- 8.Abhilash PC, Jamil S, Singh N. 2009. Transgenic plants for enhanced biodegradation and phytoremediation of organic xenobiotics. Biotechnol Adv 27:474–488. doi: 10.1016/j.biotechadv.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 9.Doty SL, Shang TQ, Wilson AM, Tangen J, Westergreen AD, Newman LA, Strand SE, Gordon MP. 2000. Enhanced metabolism of halogenated hydrocarbons in transgenic plants containing mammalian cytochrome P450 2E1. Proc Natl Acad Sci U S A 97:6287–6291. doi: 10.1073/pnas.97.12.6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.French CE, Rosser SJ, Davies GJ, Nicklin S, Bruce NC. 1999. Biodegradation of explosives by transgenic plants expressing pentaerythritol tetranitrate reductase. Nat Biotechnol 17:491–494. doi: 10.1038/8673. [DOI] [PubMed] [Google Scholar]
- 11.Hannink N, Rosser SJ, French CE, Basran A, Murray JAH, Nicklin S, Bruce NC. 2001. Phytodetoxification of TNT by transgenic plants expressing a bacterial nitroreductase. Nat Biotechnol 19:1168–1172. doi: 10.1038/nbt1201-1168. [DOI] [PubMed] [Google Scholar]
- 12.van Dillewijn P, Couselo JL, Corredoira E, Delgado A, Wittich RM, Ballester A, Ramos JL. 2008. Bioremediation of 2,4,6-trinitrotoluene by bacterial nitroreductase expressing transgenic aspen. Environ Sci Technol 42:7405–7410. doi: 10.1021/es801231w. [DOI] [PubMed] [Google Scholar]
- 13.Rylott EL, Jackson RG, Edwards J, Womack GL, Seth-Smith HMB, Rathbone DA, Strand SE, Bruce NC. 2006. An explosive-degrading cytochrome P450 activity and its targeted application for the phytoremediation of RDX. Nat Biotechnol 24:216–219. doi: 10.1038/nbt1184. [DOI] [PubMed] [Google Scholar]
- 14.Uchida E, Ouchi T, Suzuki Y, Yoshida T, Habe H, Yamaguchi I, Omori T, Nojiri H. 2005. Secretion of bacterial xenobiotic-degrading enzymes from transgenic plants by an apoplastic expressional system: an applicability for phytoremediation. Environ Sci Technol 39:7671–7677. doi: 10.1021/es0506814. [DOI] [PubMed] [Google Scholar]
- 15.Peng RH, Fu XY, Zhao W, Tian YS, Zhu B, Han HJ, Xu J, Yao QH. 2014. Phytoremediation of phenanthrene by transgenic plants transformed with a naphthalene dioxygenase system from Pseudomonas. Environ Sci Technol 48:12824–12832. doi: 10.1021/es5015357. [DOI] [PubMed] [Google Scholar]
- 16.Peng R, Fu X, Tian Y, Zhao W, Zhu B, Xu J, Wang B, Wang L, Yao Q. 2014. Metabolic engineering of Arabidopsis for remediation of different polycyclic aromatic hydrocarbons using a hybrid bacterial dioxygenase complex. Metab Eng 26:100–110. doi: 10.1016/j.ymben.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 17.Kuiper I, Lagendijk EL, Bloemberg GV, Lugtenberg BJJ. 2004. Rhizoremediation: a beneficial plant-microbe interaction. Mol Plant Microbe Interact 17:6–15. doi: 10.1094/MPMI.2004.17.1.6. [DOI] [PubMed] [Google Scholar]
- 18.Weyens N, van der Lelie D, Taghavi S, Newman L, Vangronsveld J. 2009. Exploiting plant-microbe partnerships to improve biomass production and remediation. Trends Biotechnol 27:591–598. doi: 10.1016/j.tibtech.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 19.Barac T, Taghavi S, Borremans B, Provoost A, Oeyen L, Colpaert JV, Vangronsveld J, van der Lelie D. 2004. Engineered endophytic bacteria improve phytoremediation of water-soluble, volatile, organic pollutants. Nat Biotechnol 22:583–588. doi: 10.1038/nbt960. [DOI] [PubMed] [Google Scholar]
- 20.Abhilash PC, Powell JR, Singh HB, Singh BK. 2012. Plant-microbe interactions: novel applications for exploitation in multipurpose remediation technologies. Trends Biotechnol 30:416–420. doi: 10.1016/j.tibtech.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 21.Gu T, Zhou CY, Sorensen SR, Zhang J, He J, Yu PW, Yan X, Li SP. 2013. The novel bacterial N-demethylase PdmAB is responsible for the initial step of N,N-dimethyl-substituted phenylurea herbicide degradation. Appl Environ Microbiol 79:7846–7856. doi: 10.1128/AEM.02478-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan X, Gu T, Yi ZQ, Huang JW, Liu XW, Zhang J, Xu XH, Xin ZH, Hong Q, He J, Spain JC, Li SP, Jiang JD. 2016. Comparative genomic analysis of isoproturon-mineralizing sphingomonads reveals the isoproturon catabolic mechanism. Environ Microbiol 18:4888–4906. doi: 10.1111/1462-2920.13413. [DOI] [PubMed] [Google Scholar]
- 23.Huang X, He J, Yan X, Hong Q, Chen K, He Q, Zhang L, Liu X, Chuang S, Li S, Jiang J. 2017. Microbial catabolism of chemical herbicides: Microbial resources, metabolic pathways and catabolic genes. Pestic Biochem Physiol 143:272–297. doi: 10.1016/j.pestbp.2016.11.010. [DOI] [PubMed] [Google Scholar]
- 24.Williams SL, Carranza A, Kunzelman J, Datta S, Kuivila KM. 2009. Effects of the herbicide diuron on cordgrass (Spartina foliosa) reflectance and photosynthetic parameters. Estuaries Coast 32:146–157. doi: 10.1007/s12237-008-9114-z. [DOI] [Google Scholar]
- 25.Chin-A-Woeng TFC, Bloemberg GV, Mulders IH, Dekkers LC, Lugtenberg BJ. 2000. Root colonization by phenazine-1-carboxamide-producing bacterium Pseudomonas chlororaphis PCL1391 is essential for biocontrol of tomato foot and root rot. Mol Plant Microbe Interact 13:1340–1345. doi: 10.1094/MPMI.2000.13.12.1340. [DOI] [PubMed] [Google Scholar]
- 26.Karavangeli M, Labrou NE, Clonis YD, Tsaftaris A. 2005. Development of transgenic tobacco plants overexpressing maize glutathione S-transferase I for chloroacetanilide herbicides phytoremediation. Biomol Eng 22:121–128. doi: 10.1016/j.bioeng.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 27.Kawahigashi H, Hirose S, Ohkawa H, Ohkawa Y. 2006. Phytoremediation of the herbicides atrazine and metolachlor by transgenic rice plants expressing human CYP1A1, CYP2B6, and CYP2C19. J Agric Food Chem 54:2985–2991. doi: 10.1021/jf052610u. [DOI] [PubMed] [Google Scholar]
- 28.Behrens MR, Mutlu N, Chakraborty S, Dumitru R, Jiang WZ, Lavallee BJ, Herman PL, Clemente TE, Weeks DP. 2007. Dicamba resistance: enlarging and preserving biotechnology-based weed management strategies. Science 316:1185–1188. doi: 10.1126/science.1141596. [DOI] [PubMed] [Google Scholar]
- 29.Scheunert I, Reuter S. 2000. Formation and release of residues of the 14C-labelled herbicide isoproturon and its metabolites bound in model polymers and in soil. Environ Pollut 108:61–68. doi: 10.1016/S0269-7491(99)00202-X. [DOI] [PubMed] [Google Scholar]
- 30.Kuiper I, Bloemberg GV, Lugtenberg BJJ. 2001. Selection of a plant-bacterium pair as a novel tool for rhizostimulation of polycyclic aromatic hydrocarbon-degrading bacteria. Mol Plant Microbe Interact 14:1197–1205. doi: 10.1094/MPMI.2001.14.10.1197. [DOI] [PubMed] [Google Scholar]
- 31.Murashige T, Skoog F. 1962. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15:473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x. [DOI] [Google Scholar]
- 32.Della-Cioppa G, Bauer SC, Klein BK, Shah DM, Fraley RT, Kishore GM. 1986. Translocation of the precursor of 5-enolpyruvylshikimate-3-phosphate synthase into chloroplasts of higher-plants in vitro. Proc Natl Acad Sci U S A 83:6873–6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zambryski P, Depicker A, Kruger K, Goodman HM. 1982. Tumor induction by Agrobacterium tumefaciens: analysis of the boundaries of T-DNA. J Mol Appl Genet 1:361–370. [PubMed] [Google Scholar]
- 34.Zhang XR, Henriques R, Lin SS, Niu QW, Chua NH. 2006. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat Protoc 1:641–646. doi: 10.1038/nprot.2006.97. [DOI] [PubMed] [Google Scholar]
- 35.Chen Y, Xu W, Shen H, Yan H, Xu W, He Z, Ma M. 2013. Engineering arsenic tolerance and hyperaccumulation in plants for phytoremediation by a PvACR3 transgenic approach. Environ Sci Technol 47:9355–9362. doi: 10.1021/es4012096. [DOI] [PubMed] [Google Scholar]
- 36.Su ZH, Xu ZS, Peng RH, Tian YS, Zhao W, Han HJ, Yao QH, Wu AZ. 2012. Phytoremediation of trichlorophenol by phase II metabolism in transgenic Arabidopsis overexpressing a Populus glucosyltransferase. Environ Sci Technol 46:4016–4024. doi: 10.1021/es203753b. [DOI] [PubMed] [Google Scholar]
- 37.Lichtenthaler HK. 1987. Chlorophylls and carotenoids–pigments of photosynthetic biomembranes. Methods Enzymol 148:350–382. doi: 10.1016/0076-6879(87)48036-1. [DOI] [Google Scholar]
- 38.Schreiber U, Schliwa U, Bilger W. 1986. Continuous recording of photochemical and non-photochemical chlorophyll fluorescence quenching with a new type of modulation fluorometer. Photosynth Res 10:51–62. doi: 10.1007/BF00024185. [DOI] [PubMed] [Google Scholar]
- 39.Heraud P, Beardall J. 2000. Changes in chlorophyll fluorescence during exposure of Dunaliella tertiolecta to UV radiation indicate a dynamic interaction between damage and repair processes. Photosynth Res 63:123–134. doi: 10.1023/A:1006319802047. [DOI] [PubMed] [Google Scholar]
- 40.Alexieva V, Sergiev I, Mapelli S, Karanov E. 2001. The effect of drought and ultraviolet radiation on growth and stress markers in pea and wheat. Plant Cell Environ 24:1337–1344. doi: 10.1046/j.1365-3040.2001.00778.x. [DOI] [Google Scholar]
- 41.Zhang L, Tian LH, Zhao JF, Song Y, Zhang CJ, Guo Y. 2009. Identification of an apoplastic protein involved in the initial phase of salt stress response in rice root by two-dimensional electrophoresis. Plant Physiol 149:916–928. doi: 10.1104/pp.108.131144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ovreås L, Forney L, Daae FL, Torsvik V. 1997. Distribution of bacterioplankton in meromictic Lake Saelenvannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl Environ Microbiol 63:3367–3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Magoč T, Salzberg SL. 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen C, Zhang JN, Lu M, Qin C, Chen YH, Yang L, Huang QW, Wang JC, Shen ZG, Shen QR. 2016. Microbial communities of an arable soil treated for 8 years with organic and inorganic fertilizers. Biol Fertil Soils 52:455–467. doi: 10.1007/s00374-016-1089-5. [DOI] [Google Scholar]
- 46.Edgar RC. 2013. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 47.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A 108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sanger M, Daubert S, Goodman RM. 1990. Characteristics of a strong promoter from figwort mosaic virus: comparison with the analogous 35S promoter from cauliflower mosaic virus and the regulated mannopine synthase promoter. Plant Mol Biol 14:433–443. doi: 10.1007/BF00028779. [DOI] [PubMed] [Google Scholar]
- 49.Sahoo DK, Dey N, Maiti IB. 2014. pSiM24 is a novel versatile gene expression vector for transient assays as well as stable expression of foreign genes in plants. PLoS One 9:e98988. doi: 10.1371/journal.pone.0098988. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.