The α-amylase from Corallococcus sp. EGB, which was classified to the GH13_36 subfamily, can catalyze the conversion of maltooligosaccharides (≥G3) and soluble starch to maltose as the sole hydrolysate. An action mechanism for producing a high level of maltose without the attendant production of glucose has been proposed. Moreover, it also can hydrolyze γ-cyclodextrin and pullulan. Its biochemical characterization suggested that CoMA may be involved the accumulation of maltose in Corallococcus media.
KEYWORDS: Corallococcus, maltogenic α-amylase, transglycosylation, GH13, maltose
ABSTRACT
The gene encoding the novel amylolytic enzyme designated CoMA was cloned from Corallococcus sp. strain EGB. The deduced amino acid sequence contained a predicted lipoprotein signal peptide (residues 1 to 18) and a conserved glycoside hydrolase family 13 (GH13) module. The amino acid sequence of CoMA exhibits low sequence identity (10 to 19%) with cyclodextrin-hydrolyzing enzymes (GH13_20) and is assigned to GH13_36. The most outstanding feature of CoMA is its ability to catalyze the conversion of maltooligosaccharides (≥G3) and soluble starch to maltose as the sole hydrolysate. Moreover, it can hydrolyze γ-cyclodextrin and starch to maltose and hydrolyze pullulan exclusively to panose with relative activities of 0.2, 1, and 0.14, respectively. CoMA showed both hydrolysis and transglycosylation activities toward α-1,4-glycosidic bonds but not to α-1,6-linkages. Moreover, glucosyl transfer was postulated to be the major transglycosidation reaction for producing a high level of maltose without the attendant production of glucose. These results indicated that CoMA possesses some unusual properties that distinguish it from maltogenic amylases and typical α-amylases. Its physicochemical properties suggested that it has potential for commercial development.
IMPORTANCE The α-amylase from Corallococcus sp. EGB, which was classified to the GH13_36 subfamily, can catalyze the conversion of maltooligosaccharides (≥G3) and soluble starch to maltose as the sole hydrolysate. An action mechanism for producing a high level of maltose without the attendant production of glucose has been proposed. Moreover, it also can hydrolyze γ-cyclodextrin and pullulan. Its biochemical characterization suggested that CoMA may be involved the accumulation of maltose in Corallococcus media.
INTRODUCTION
Glycoside hydrolase family 13 (GH13), also known as the α-amylase family, is a large sequence-based family of glycoside hydrolases and includes a number of different enzyme activities and substrate specificities acting on α-glycosidic bonds. Based on the members' different substrate specificities, reaction mechanisms, and phylogenetic distributions, GH13 is currently divided into 42 subfamilies in the Carbohydrate-Active enZyme (CAZy) database (1). Maltogenic amylase (MAase; EC 3.2.1.133), neopullulanase (Npase; EC 3.2.1.135), and cyclomaltodextrinase (CDase; EC 3.2.1.54), belonging to subfamily GH13_20, are reported to be capable of hydrolyzing two or three of the following substrates: cyclomaltodextrins (CDs), pullulan, and starch (1–3). These three types of enzymes exhibit 40 to 86% amino acid sequence identity. They exhibit unique physicochemical and catalytic properties, including multisubstrate specificity and various catalytic capabilities for hydrolyzing α-1,4- and α-1,6-glycosidic linkages and transglycosylation of oligosaccharides to C-3, C-4, or C-6 hydroxyl groups of various acceptors (4). Although they belong to GH13, they are structurally different from typical α-amylases because they have an extra N-terminal domain that mediates their domain-swapping dimeric structure (3, 5). In addition, unlike other typical α-amylases, the three groups of enzymes are intracellular and have molecular masses that range from 62 to 90 kDa, which are slightly larger masses than those of most α-amylases. Maltogenic amylases prefer CDs to starch or pullulan as the substrates. Moreover, they hydrolyze CDs and starch to maltose and hydrolyze pullulan to panose by cleavage of α-1,4-glycosidic bonds, whereas α-amylases essentially lack activity against CDs and pullulan (6). A few of maltogenic amylases may have hydrolytic activity toward the acarbose, a competitive inhibitor of α-amylases (6–8).
There is considerable interest in the isolation of α-amylases that can produce high levels of specific maltooligosaccharides from the degradation of starch, as well as in the mechanism of action of these endo-acting enzymes (9). Maltose is one of these useful maltooligosaccharides, desired for its wide industrial applications in the food, pharmaceutical, biomedical, and fine chemicals industries (10). α-Amylases capable of producing high levels of maltose from the hydrolysis of starch have been isolated from fungal and bacterial sources, with maltose content ranging from 53% (wt/wt) to greater than 80% (wt/wt); these sources include Rhizopus oryzae (11), Penicillium expansum (12), Thermomonospora curvata (9), Bacillus megaterium G-2 (13), Streptomyces praecox (14), and Pyrococcus sp. strain ST04 (15). The mechanisms to produce high levels of maltose were postulated to involve these α-amylases exhibiting significant multimolecular reactions, including condensation and transglycosylation, in addition to their hydrolytic activity (14, 16–18) or exhibiting an exo-type maltose-forming α-amylase action pattern (13, 15). Saccharifying α-amylases, mainly α-amylase from Aspergillus oryzae, are used in the industrial production of maltose syrups with a 40 to 50% maltose content (19). However, starch degradation by almost all thermostable amylases involves the synchronous production of appreciable levels of glucose and other oligosaccharides (12). In addition, the α-amylase currently used in starch industries is active at pH 6.5 and requires Ca2+ for its activity and/or stability. Removal of Ca2+ and other oligosaccharides from the product streams by ion exchangers adds to the cost of the products (20).
Myxobacteria are a type of soil-dwelling and Gram-negative soil bacteria that are characterized by a multicellular stage in their life cycle that is induced by nutritional starvation and changes in the host environment (21). Starch degradation has been reported for every species of bacteriolytic myxobacteria, including the genera Myxococcus and, in particular, Corallococcus (22, 23). Corallococcus coralloides has an unusual pattern of carbohydrate utilization (24, 25). Corallococcus coralloides strain Cc c127 did not utilize mono- and disaccharides, but maltotriose and the polysaccharides starch, amylose, amylopectin, and pullulan stimulated its growth. However, only part of the starch has really been metabolized by the cells because a substantial amount of maltose and some glucose accumulated in the medium (24). This suggests that myxobacteria have an uncommon amylolytic system. Therefore, we were interested in understanding how these amylolytic enzymes of Corallococcus utilize carbohydrates. We previously isolated Corallococcus sp. EGB, which abundantly produces extracellular amylolytic enzymes (26) and also accumulates large amounts of maltose in the starch medium.
A gene encoding a putative GH13 amylase from the EGB strain was cloned and expressed in Escherichia coli. We classified it as a novel maltogenic α-amylase since it produces a high level of maltose from starch and γ-cyclodextrin and panose from pullulan. The maltogenic α-amylase also exhibited glycosyl transfer activity and Ca2+-independent properties. In the present study, the molecular cloning of the α-amylase gene and phylogenetic analysis, catalytic properties, and action mechanism of purified recombinant enzyme are described.
RESULTS AND DISCUSSION
Cloning of the α-amylase gene and sequence analysis.
To reveal the uncommon amylolytic system in Corallococcus, we sought to purify amylolytic enzymes and clone their encoding genes from Corallococcus sp. strain EGB. We isolated and characterized a novel liquefying maltohexaose-forming α-amylase (AmyM) and saccharifying α-amylase (AmyC) from the EGB strain (26, 27). We assumed that AmyM is the main extracellular liquefying amylase for utilizing starch in Corallococcus sp. EGB since AmyM showed a specific activity of 14,000 U/mg and produced 54% maltohexaose and other maltooligosaccharides from soluble starch. However, the enzymes which caused a substantial amount of maltose and some glucose to be accumulated in the medium have not been identified.
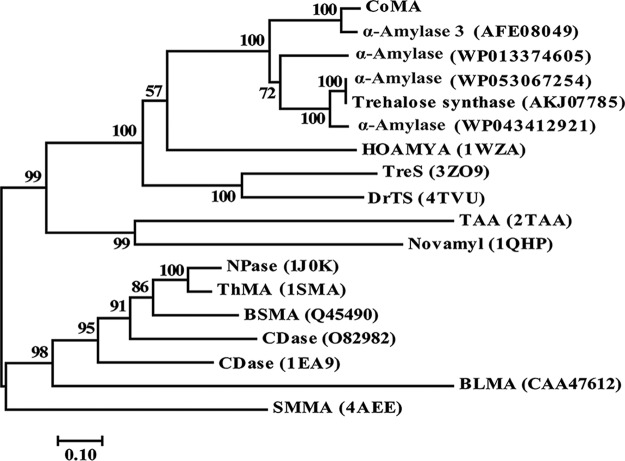
A novel amylase gene encoding the novel amylolytic enzyme designated CoMA was cloned from the chromosomal DNA of Corallococcus sp. EGB. The open reading frame that corresponds to CoMA consists of 1,665 nucleotides and encodes a protein of 554 amino acids with a predicted molecular mass of 59,946 Da and a calculated pI of 6.08. BLASTP analysis showed that CoMA shared its highest identity (93%) with the α-amylase 3 in the genome of C. coralloides DSM 2259 (28), followed by the trehalose synthase from Archangium gephyra (67%) and the α-amylase from Stigmatella aurantiaca (62%) (Fig. 1). However, all of these proteins have not been biochemically characterized. Among proteins with experimentally determined three-dimensional structures, CoMA showed the highest identity (34%) with the α-amylase HOAMYA (PDB 1WZA) from thermophilic halophile Halothermothrix orenii (29), 30% identity with the trehalose synthase TreS (PDB 3ZO9) from Mycobacterium smegmatis (30), and 29.7% identity with the trehalose synthase DrTS (PDB 4TVU) from Deinococcus radiodurans (31). The phylogenetic tree also revealed that CoMA exhibited low sequence identity (10 to 20%) with the multispecific enzymes of GH13_20, which includes neopullulanase (PDB 1J0K) from Bacillus stearothermophilus (32), maltogenic amylase ThMA (PDB 1SMA) of Thermus sp. IM6501 (5), and cyclomaltodextrinase (PDB 1EA9) from Bacillus sp. (3) (Fig. 1). Phylogenetic analysis showed that CoMA formed an independent branch apart from these proteins and may belong to the GH13_36 subfamily, which is most closely related to but distinct from GH13_16, whose members have trehalose synthase activity (see Fig. S1 in the supplemental material).
FIG 1.
Phylogenetic relationships between CoMA and other amylases from GH13. The phylogenetic tree was constructed by the neighbor-joining algorithm based on the amino acid sequence alignment in MEGA7. The amino acid sequence of CoMA was aligned with those of the following proteins: C. coralloides DSM 2259 α-amylase 3 (accession no. AFE08049), Stigmatella aurantiaca α-amylase (WP013374605), Archangium gephyra α-amylase (WP053067254), Archangium gephyra trehalose synthase (AKJ07785), Cystobacter violaceus α-amylase (WP043412921), Halothermothrix orenii α-amylase (HOAMYA; PDB 1WZA), Mycobacterium smegmatis trehalose synthase (TreS; PDB 3ZO9), Deinococcus radiodurans trehalose synthase (DrTS; PDB 4TVU), Aspergillus oryzae Taka-amylase A (PDB 2TAA), Bacillus stearothermophilus Novamy (PDB 1QHP), Bacillus stearothermophilus neopullulanase (NPase; PDB 1J0K), Thermus sp. IM6501 maltogenic amylase (ThMA; PDB 1SMA), Bacillus sp. A2-5a cyclomaltodextrinase (CDase; O82982), Bacillus sp. CDase (PDB 1EA9), Bacillus licheniformis maltogenic amylase (BLMA; CAA47612), and Staphylothermus marinus maltogenic amylase (SMMA; PDB 4AEE).
Multiple alignments of the primary sequences revealed that the N-terminal 130 amino acids in the former four enzymes are completely absent in TAKA-amylase A (TAA), HOAMYA, and CoMA (see Fig. S2 in the supplemental material). The N-terminal domains of MAase, Npase, and CDase have been shown to be involved in the oligomerization of these enzymes and the formation of the enzyme active site, together with the central (β/α)8 barrel of the adjacent subunit, forming a narrow and deep cleft suitable for binding cyclodextrins (33, 34). CoMA maintains highly conserved sequences at regions I to VII, and the three catalytic residues Asp-328, Glu-357, and Asp-424 (ThMA numbering) are invariant (5, 35). With regard to conserved sequence region V, there is a characteristic sequence change from QPDLN to MPKLN via MPDLN for the members of the α-amylase family (36–38). CoMA has intermediary sequence MPDLN in the fifth conserved sequence region (see Fig. S2 in the supplemental material), while the sequence QPDLN (or QxDLN) is typical for a large group of α-amylase family enzymes such as trehalose-6-phosphate hydrolase, amylosucrase, and trehalose synthase, and MPKLN is found in MAases, Npases, and CDases (38). Horvathova et al. concluded that the members of the α-amylase family that contained the intermediary sequence MPDLN may exhibit mixed activities of amylase, cyclodextrinase, and neopullulanase (38).
A lipoprotein signal peptide of initial 18 residues of CoMA was also found by using the LipoP 1.0 prediction program. The sequence Leu-Ser-Ala-Cys at positions 15 to 18 is strongly reminiscent of typical lipoprotein signal peptidase cleavage sites, which have the consensus sequence Leu/Val/Ile-Ala/Ser/Thr/Gly-Gly/Ala-Cys (39). Moreover, since cell cultures do not show enzyme activity in the culture filtrates, it is possible that the enzyme is secreted but remains associated with the membrane or cell wall (see Table S1 in the supplemental material). CoMA is proposed to be a lipoprotein with extracellular localization. This behavior is different from the three groups of these enzymes (MAases, NPases, and CDases), which are generally intracellular enzymes (3, 5).
Expression and purification of CoMA.
The structural gene of CoMA (with the signal peptide sequence) was successfully expressed in Escherichia coli BL21(DE3) at low expression levels of 3.3 mg liter−1. The recombinant CoMA with a C-terminal His6 tag was purified by immobilized metal-affinity chromatography using nickel-nitrilotriacetic acid (Ni2+-NTA) resin. Recombinant CoMA was purified 67-fold with a yield of 75%. The purified CoMA has amylase activity, as shown by zymogram analysis, and exhibits a molecular weight of ∼60.0 kDa on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels (Fig. 2). The purified CoMA had a specific activity of 980 U/mg toward soluble starch.
FIG 2.
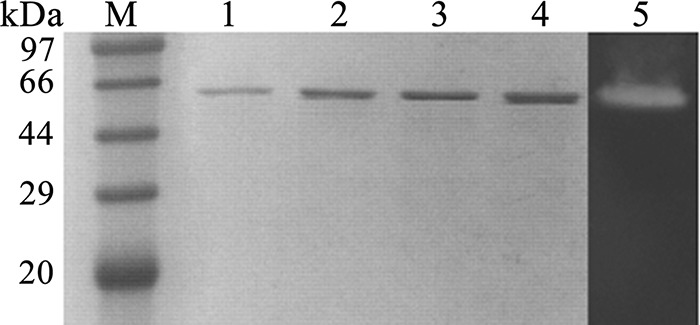
SDS-PAGE and zymogram analysis of the recombinant CoMA. The purity of the recombinant CoMA is shown on a 12% SDS-PAGE gel. Lane M, standard molecular mass markers. For lanes 1 to 4, 0.5, 1, 2, and 4 μg of purified CoMA were loaded into each lane, respectively, and for lane 5, enzyme zymogram analysis of CoMA (0.5 μg) was visualized by Lugol's iodine solution staining on soluble starch plate.
Biochemical properties of purified CoMA. (i) Effects of pH and temperature on enzyme activity and stability.
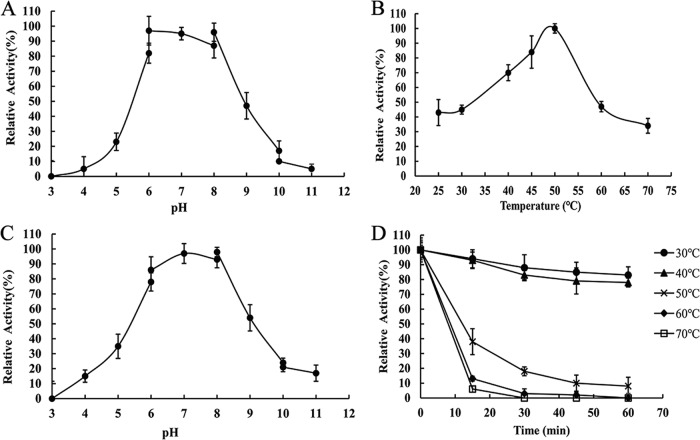
The enzyme activity at different pH and temperatures was determined with soluble starch as the substrate. As shown in Fig. 3A and C, the optimal working pH of CoMA was ∼7.0. In addition, CoMA retained more than 50% activity after being stored at pH 5.0 to 9.0 at 4°C for 24 h. The optimal temperature of the recombinant CoMA activity was 50°C (Fig. 3B). The specific activity of CoMA decreased significantly when the temperature was >60 or <30°C. Almost all of the enzyme activity remained after the enzyme was incubated for 60 min at temperatures of <40°C (Fig. 3D). However, a dramatic loss of enzymatic stability occurred in CoMA at temperatures higher than 50°C after 30 min of incubation. These results suggested that CoMA was a mesophilic enzyme.
FIG 3.
Effects of temperature and pH on the activity and stability of CoMA. (A) Determination of the optimal pH. Assays were carried out by the addition of 0.3 μg of purified CoMA to 5 mg/ml soluble starch substrate at 50°C for 10 min in buffers of various pH values (pH 2.0 to 11.0). (B) Determination of the optimal temperature. The activity was measured by the addition of 0.3 μg of purified CoMA to 20 mM Tris-HCl (pH 7.0) buffer containing 5 mg/ml soluble starch substrate at 20 to 70°C for 10 min. (C) Stability of CoMA at different pH values. The residual enzyme activity was measured under optimal conditions after incubation of the 0.3 μg of purified enzyme with each buffer (various pH values) at 4°C for 24 h. (D) Thermostability of CoMA. The residual activity was measured under optimal conditions after incubation of 0.3 μg of purified enzyme at the indicated temperatures for up to 60 min. Mean values and standard deviations from three independent experiments are shown.
(ii) Effects of metal ions and chemical reagents on CoMA activity.
The residual activity was assayed by incubation of metal-free enzyme with 1 and 5 mM metal ion for 10 min. As shown in Table 1, the amylase activity was not significantly affected by K+, Ba2+, and Mg2+ ions at 1 and 5 mM. However, activity of CoMA was significantly inhibited by Cu2+, Cr3+, Zn2+, Fe3+, and Ca2+ ions. Interestingly, 1 mM Mn2+ ions is an effective activator of CoMA, since it was able to stimulate the activity by approximately 30%. In addition, SDS strongly inhibited CoMA enzyme activity. In contrast, dithiothreitol (DTT) and β-mercaptoethanol could significantly activate amylase activity, whereas dimethyl sulfoxide had no significant effect on the enzyme's activity (Table 2). Since the presence of 1 mM Ca2+ ions and 5 mM EGTA had no noticeable effect on enzyme activity, it was confirmed that the enzyme is Ca2+ independent. Traditionally, Ca2+ ions have been considered necessary for the maintenance of the structure of the active site and thus for the activity and stability of α-amylases (20).
TABLE 1.
Effect of metal ions on α-amylase activitya
| Metal ion | Mean relative activity (%) ± SD |
|
|---|---|---|
| 1 mM | 5 mM | |
| K+ (KCl) | 98.7 ± 3.0 | 104.4 ± 4.7 |
| Mg2+ (MgCl2) | 94.3 ± 2.0 | 92.5 ± 0.1 |
| Mn2+ (MnCl2) | 130.4 ± 2.7 | 118.6 ± 1.9 |
| Ba2+ (BaCl2) | 96.4 ± 1.0 | 101.6 ± 8.5 |
| Ca2+ (CaCl2) | 105.3 ± 3.2 | 78.0 ± 0.6 |
| Ni2+ (NiCl2) | 84.9 ± 1.7 | 33.3 ± 0.2 |
| Cu2+ (CuCl2) | 22.3 ± 2.4 | 1.1 ± 0.1 |
| Cr3+ (CrCl3) | 73.6 ± 0.6 | 12.5 ± 1.2 |
| Zn2+ (ZnCl2) | 7.1 ± 1.8 | 2.7 ± 0.2 |
| Fe3+ (FeCl3) | 85.5 ± 0.1 | 3.9 ± 1.3 |
Samples were preincubated with various metal ions (1 and 5 mM) for 10 min at 4°C, and the remaining activity was measured under standard assay conditions. Activity in the absence of any additives was taken as the 100% value. Mean values from three independent experiments are shown.
TABLE 2.
Effect of chemical agents on α-amylase activitya
| Chemical reagent | Concn | Mean relative activity (%) ± SD |
|---|---|---|
| Ethanol | 10.00% (vol/vol) | 66.0 ± 1.6 |
| Acetonitrile | 10.00% (vol/vol) | 27.1 ± 4.8 |
| Acetone | 10.00% (vol/vol) | 23.9 ± 5.6 |
| Tween 80 | 1.00% (vol/vol) | 57.4 ± 1.6 |
| SDS | 1.00% (wt/vol) | 4.9 ± 1.7 |
| Urea | 10 mM | 69.7 ± 8.6 |
| EDTA | 1 mM | 63.3 ± 1.5 |
| 5 mM | 44.1 ± 1.1 | |
| EGTA | 1 mM | 100 ± 0.8 |
| 5 mM | 103.2 ± 1.2 | |
| DTT | 1 mM | 124.7 ± 2.8 |
| 5 mM | 149.5 ± 1.6 | |
| PMSF | 1 mM | 75.6 ± 0.6 |
| 5 mM | 51.4 ± 1.8 | |
| β-Mercaptoethanol | 1 mM | 111.2 ± 4.9 |
| 5 mM | 151.6 ± 2.9 | |
| DMF | 1 mM | 86.6 ± 2.0 |
| 5 mM | 73.5 ± 0.6 |
The enzyme solution was incubated at 50°C in the presence of different concentrations of organic solvents for 10 min. Residual activity was measured under standard assay conditions. The activity in the absence of any additives was taken as the 100% value. Mean values from three independent experiments are shown.
Catalytic properties of CoMA.
The ability of recombinant CoMA to hydrolyze various carbohydrates under the standard assay conditions was examined (Table 3). Soluble starch was most effectively hydrolyzed by the enzyme. Moreover, it hydrolyzed γ-cyclodextrin and soluble starch to maltose and hydrolyzed pullulan to panose with relative activities of 0.2, 1, and 0.14, respectively, whereas CoMA had no activity toward maltose, α-cyclodextrin, β-cyclodextrin, panose, and dextran (Table 3). Furthermore, CoMA was active against maltotriose but not isomaltotriose (α-glucosyl-[1→6]-α-glucosyl-[1→6]-glucose). This finding revealed that CoMA harbored hydrolysis activities only toward α-1,4-glycosidic bonds and not α-1,6-linkages, which was consistent with the hydrolysis of pullulan to panose. The enzyme also did not hydrolyze acarbose and amylase activity of CoMA was completely inhibited by acarbose, a pseudotetrasaccharide competitive inhibitor of α-amylases (see Fig. S3 in the supplemental material). Recently, many GH13 enzymes that have an activity of cleaving acarbose have been reported, including neopullulanase from Bacillus stearothermophilus IMA6503 (40), maltogenic amylase ThMA from Thermus strain IM6501 (6), maltogenic amylase BSMA from Bacillus stearothermophilus (41), cyclodextrin-hydrolyzing enzyme PFTA from Pyrococcus furiosus (42), and cyclomaltodextrinase from Bacillus sp. I-5 (8).
TABLE 3.
Substrate specificity of CoMAa
| Substrateb | Solubility | Mean relative activity (%) ± SD |
|---|---|---|
| Starch from potato | Soluble | 46.8 ± 3.4 |
| Dextrin from corn | Soluble | 78.5 ± 7.4 |
| Soluble starch | Soluble | 100.00 |
| Amylose from potato | Insoluble | 35.1 ± 0.2 |
| Amylopectin from potato | Soluble | 12.5 ± 0.5 |
| Glycogen from bovine muscle | Soluble | 2.1 ± 0.3 |
| Pullulan | Soluble | 14.3 ± 1.5 |
| α-Cyclodextrin | Soluble | ND |
| β-Cyclodextrin | Soluble | ND |
| γ-Cyclodextrin | Soluble | 20.2 ± 2.1 |
| Dextran 64-74 | Soluble | ND |
| Dextran 10 | Soluble | ND |
| Panose* | Soluble | ND |
| Isomaltotriose* | Soluble | ND |
| Maltose* | Soluble | ND |
| Acarbose* | Soluble | ND |
For the determination of substrate specificity, the purified CoMA was incubated in 20 mM Tris-HCl buffer (pH 7.0) for 10 min at 50°C with 0.5% (wt/vol) concentrations of different substrates. Activity was measured using the DNS method (48). The activity against soluble starch was set as the 100% value by the addition of 0.3 μg of purified CoMA with 980 U/mg protein. ND, no activity detected. Mean values from three independent experiments are shown.
*, enzyme activity was measured by TLC or HPLC.
In addition, CoMA has a most remarkable feature: its ability to hydrolyze maltooligosaccharides (maltotriose to maltoheptaose) and starch to produce high levels of maltose without synchronous glucose production (Fig. 4). These results indicate that maltotriose is the smallest substrate, suggesting that CoMA has at least three glycosyl-binding subsites (−1 to +2). Although extracellular α-amylase TVA I has a similar substrate specificity, it hydrolyzed starch to produce 74.1% of maltose and 11.8% glucose (4, 34, 43). David et al. (58) reported a new Bacillus megaterium amylase displayed multispecific activity toward α-, β-, and γ-cyclodextrin, pullulan, and soluble starch and α-1,4-linkage transglycosylation activity. However, it hydrolyzed α-, β-, and γ-cyclodextrin to DP2 and DP3, soluble starch to low-molecular-weight oligosaccharides, and pullulan to panose, respectively. Thus, CoMA can be classified as a maltogenic α-amylase since it produces high levels of maltose from starch and γ-cyclodextrin, it produces exclusively panose from pullulan, and it displays transglycosylation activities toward α-1,4-glycosidic linkages. Those results also revealed that CoMA possesses some properties that distinguishes it from maltogenic amylases (GH13_20) and typical α-amylases, whereas maltogenic amylases, which are always intracellular enzymes, prefer cyclodextrin to starch or pullulan; in addition, cyclodextrin and pullulan are resistant to the hydrolytic activity of typical α-amylases.
FIG 4.
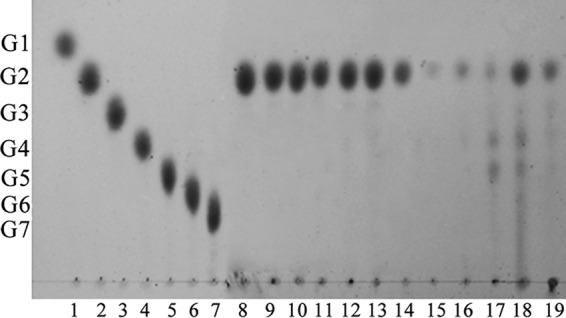
Hydrolysis products of CoMA on various substrates. Lanes 1 to 7, maltooligosaccharide standards (glucose to maltoheptaose); lanes 8 to 19, hydrolysis product of maltooligosaccharide (maltose to maltoheptaose), soluble starch, amylose, amylopectin, glycogen, starch from potato, and dextrin from corn. CoMA was incubated with 5 mg/ml of the various substrates at 30°C for 12 h. The end products were analyzed by TLC.
The kinetic parameters of the recombinant CoMA for soluble starch and amylopectin were determined with a Lineweaver-Burk plot. The kinetic parameters Km and Kcat were determined to be 2.9 mg ml−1 and 3,021.2 s−1 toward soluble starch and 10.3 mg ml−1 and 2,274.8 s−1 toward amylopectin (Table 4).
TABLE 4.
Apparent kinetic constants of CoMAa
| Substrate | Mean ± SD |
|||
|---|---|---|---|---|
| Sp act (U mg−1) | Km (mg ml−1) | Kcat (s−1) | Kcat/Km (ml mg−1 s−1) | |
| Soluble starch | 980.0 ± 7.1 | 2.9 ± 0.3 | 3,021.2 ± 10.1 | 1,019.6 ± 8.2 |
| Amylopectin from potato | 121.3 ± 5.3 | 10.3 ± 0.6 | 2,274.8 ± 9.6 | 220 ± 4.4 |
Mean values from three independent experiments are shown.
Hydrolytic mode of CoMA.
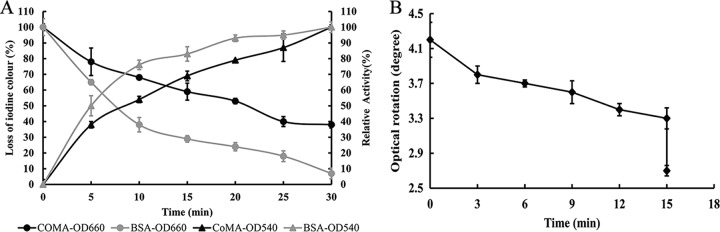
After adding CoMA, the blue value of the starch-iodine complex and reducing sugar were dramatically decreased and rapidly produced during the hydrolysis of soluble starch in a manner similar to the liquefying α-amylase from Bacillus subtilis (Sigma, St. Louis, MO) (Fig. 5A). The result indicated that the action mode of the CoMA is endolytic in nature, which agreed with its hydrolytic activity against γ-CD. The optical rotation of the soluble starch hydrolysis products was shifted downward (Fig. 5B), indicating that hydrolysis products have an α-anomeric configuration. Thus, CoMA could be identified as an α-amylase that yields α-maltose by splitting the α-1,4-glucosidic linkages in starch through an endo-type mechanism. Takasaki reported that the novel maltose-forming α-amylase PSMA exhibited a different exo-type mechanism; its enzymatic properties, including its action pattern, were very similar to those of β-amylase with exception of the configuration of the maltose that it produced (13). It produced only maltose from soluble starch, as CoMA did, but it did not act on pullulan or cyclodextrin.
FIG 5.
Hydrolytic mode of CoMA. (A) Determination of blue loss percentage and reducing sugar production of starch. Approximately 0.05 U of CoMA and liquefying α-amylase from Bacillus subtilis (BSA) were incubated with 1 ml of 5 mg/ml soluble starch at 40°C and pH 7.0. Hydrolysis of starch was estimated by assaying the decrease in the blue value from the starch-iodine complex (OD660, circle) and reducing sugar production (OD540, triangle). (B) Quantifying the anomeric form of the hydrolyzed products. A reaction mixture (1 ml) consisting of 5 mg/ml soluble starch in 20 mM Tris-HCl (pH 7.0) and 0.05 U of purified CoMA was added to the sample cell (cuvette). The optical rotation of the mixture was periodically measured in a polarimeter (WZZ-2S; Shenguang, China) using light-emitting diode. The mutarotation of the hydrolysate was determined by adding 25 μl of 4 M NaOH after the optical rotation became almost constant. Mean values and standard deviations from three independent experiments are shown.
Action pattern on soluble starch.
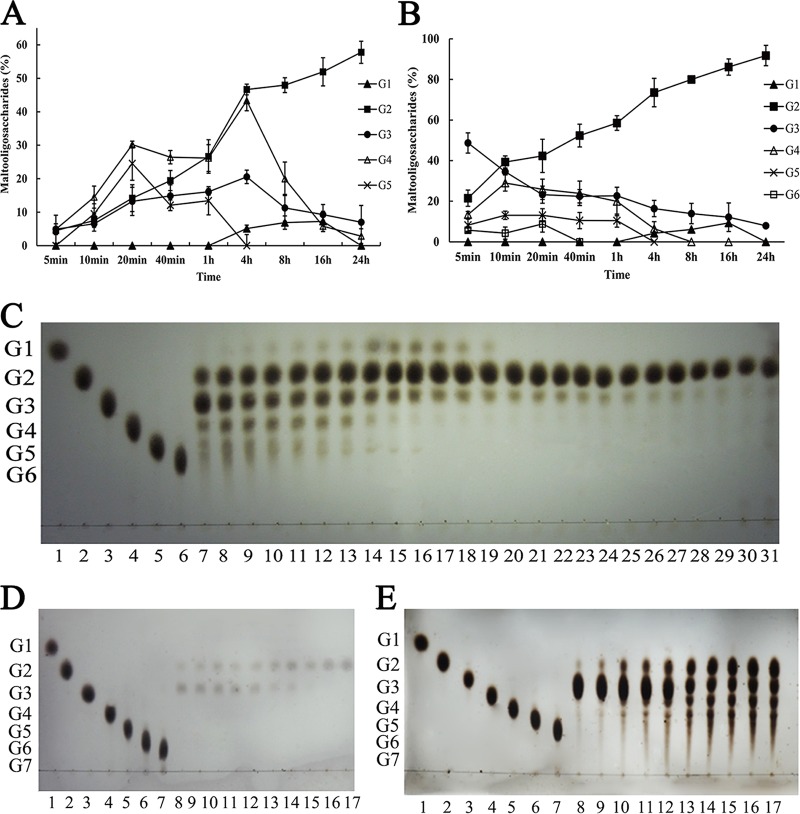
CoMA exhibited an interesting end product profile when it degraded starch (Fig. 6A). Degradation of 1% (wt/vol) starch resulted in the formation of oligosaccharides in the maltose to maltopentaose range at early time points, followed by an accumulation of maltose, maltotetraose, and small amounts of glucose after 1 h, and its final end products consisted of a high level of 58% (wt/wt) maltose without concomitant production of glucose. Although amylase enzymes producing larger amounts of maltose have been reported already, for example, the α-amylase of Thermomonospora curvata produced very high levels (73% [wt/wt]) of maltose from starch without synchronous glucose formation (9), the by-product maltotriose was simultaneously over 20%. The authors explained the thermophilic α-amylase producing high levels of maltose as being due to a combination of multimolecular reactions (transglycosylation and condensation) and the enzyme's low affinity for maltotriose. Thus, CoMA has potential value in the elimination of by-products for the production of higher-purity maltose.
FIG 6.
Analysis of end products formed by CoMA on starch and maltotriose. CoMA (10 U/ml) was incubated with 10 mg/ml starch (A) and 20 mM maltotriose (≥95% purity) (B) in 20 mM Tris-HCl (pH 7.0) at 30°C for up to 24 h, and the end products were quantitatively analyzed by HPLC. Constituents of end products (% [wt/wt]) are defined as (concentration of end product/original starch or maltotriose concentration) × 100. Mean values and standard deviations from three independent experiments are shown. (C) Time course of CoMA action on 20 mM maltotriose. Lanes 1 to 6, maltooligosaccharide standards (glucose to maltohexaose); lanes 7 to 31, hydrolysis product of 20 mM maltotriose (≥95% purity) at different reaction times (5, 10, 20, 30, 40, and 50 min and 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 24, 26, 28, and 30 h). (D and E) Time courses of CoMA action on 2 mM (D) and 200 mM (E) maltotriose. Lanes 1 to 6, maltooligosaccharide standards (glucose to maltohexaose); lanes 7 to 31, hydrolysis product of 20 mM maltotriose (≥95% purity) at different reaction times (5, 10, 20, 30, 40, and 50 min and 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 24, 26, 28, and 30 h). CoMA (10 U/ml) was incubated with the substrate at 30°C, and the end products were analyzed by TLC. G1 to G7 represent glucose to maltoheptaose.
Action pattern on maltotriose.
The enzyme produces maltose as the sole product from maltooligosaccharides (maltotriose to maltoheptaose) (Fig. 4). This behavior is unusual since equimolar amounts of glucose and maltose should be produced when either glycosidic bond of maltotriose is hydrolyzed. The high-maltose-producing (74%) α-amylase of P. expansum appeared to hydrolyze maltotriose directly to maltose and glucose (12). In an attempt to gain some insight into the mechanism utilized by CoMA, maltotriose was used as a substrate, and the end product profile was examined.
As the time course of degrading 20 mM maltotriose showed (Fig. 6B and C), maltooligosaccharides in the range of maltose to maltohexaose were produced in the initial stage of the catalytic reaction, followed by an accumulation of maltose and formation of small amounts of glucose; at last, 92% (wt/wt) maltose was detected in the final hydrolysate of the maltotriose solution. Thus, the participation of some mechanism other than simple hydrolysis such as transglycosylation or condensation was strongly suggested. The action pattern on maltotriose is similar to that of the α-amylase of Streptomyces praecox NA-273 (14), which produces in excess of 80% (wt/wt) maltose as the sole end products. This unique mechanism of action resulted in the proposal of a transglycosylation mechanism to convert maltotriose to maltose via maltotetraose as an intermediate (2G3→G2+G4→3G2). Among enzymes of GH13_36, AmyM from uncultured bacterium also showed hydrolysis and transglycosylation properties and be regarded as an intermediate type of maltogenic amylase, α-amylase, and α-glucanotransferase (57). Moreover, the lipoprotein amylase AmyA from Anaerobranca gottschalkii displayed high transglycosylation activity on maltooligosaccharides and also had significant β-cyclodextrin glycosyltransferase (CGTase) activity. However, the main products of AmyM and AmyA hydrolysis were maltose and glucose (45).
The enzyme also produces maltose exclusively without appreciable formation of glucose when the initial solution contains 2 and 200 mM maltotriose (Fig. 6D and E). This hydrolytic profile indicates a mechanism of action that is distinct from the pattern of the α-amylase of T. curvata (9), in which unimolecular hydrolytic events predominate at a low concentration of maltotriose (2 mM) and multimolecular reactions shift the G1/G2 ratio away from unity with increasing substrate concentration (200 mM). These findings suggested that the transglycosylation activity of CoMA is higher than that of the T. curvata α-amylase in degradation of G3.
Model of CoMA action on maltotriose.
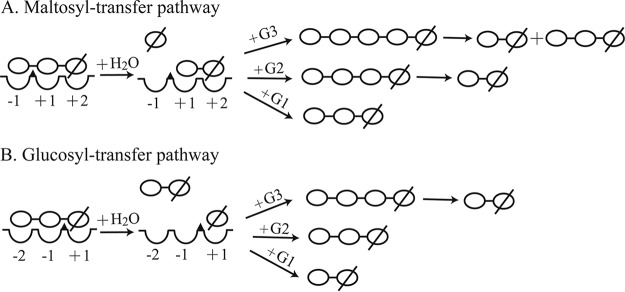
For multicatalytic reactions, condensation reactions would lead to the accumulation of maltohexaose whereas maltotetraose and maltopentaose are products of transfer reactions (9). Since a small quantity of maltohexaose was detected when CoMA reacted with maltotriose, transglycosylation is postulated to be the major multimolecular mechanism that contributes to the high yields of maltose and the absence of glucose (Fig. 6B). A model of the action of CoMA on maltotriose (Fig. 7) was proposed based on the results described above. Glucose was not detected in the initial stages of the hydrolysis of starch and maltotriose (Fig. 6A and B), and only maltose was detected throughout the process of hydrolyzing 2 mM maltotriose (Fig. 6D). Thus, glucosyl transfer (Fig. 7B), rather than maltosyl transfer was postulated to be the major transglycosidation reaction. It also means that CoMA prefers to release maltose from the nonreducing end and precludes glucose formation by a glucosyl transfer reaction. The reaction mechanism of the hydrolysis and transglycosylation by CoMA is considered to follow a double-displacement reaction (5, 44) that is similar to that of α-amylase, CGTase, and debranching enzymes. In addition, the detailed transglycosylation catalytic mechanism of CoMA was similar to that of a member of subfamily GH13_4, amylosucrase (EC 2.4.1.4), which catalyzes the synthesis of amylose-like polymers from sucrose (46). We speculated that maltotriose occupies binding sites −2 to +1 and a proton donor (Glu286) facilitates departure of the leaving group by donating a proton to the oxygen atom between glycosyl −1 and +1 in the first step; the nucleophile (Asp243) forms an enzyme-sequestered covalent intermediate with the +1 glucose residue and releases the maltose part of the maltotriose as the leaving group. In the second step, another oligosaccharide, as acceptor, occupies binding sites −2 and −1, and the deprotonated proton donor (Glu286) acts as a general base to activate the C-4 hydroxyl of the acceptor and leads to the formation of a new transglycosylation product (Fig. 7B). Those new transglycosylation products would be further hydrolyzed to maltose.
FIG 7.
Possible multimolecular maltosyl (A) and glucosyl (B) transfer pathways catalyzed by CoMA on maltotriose. The wedge (▲) represents the catalytic site of the enzyme. The concave features represent the glycosyl-binding subsites. A glucose residue is represented by ○; a reducing glucose residue is represented by ∅.
In brief, Corallococcus sp. EGB produces an unusual maltogenic amylase, CoMA, which produces high levels of maltose from maltooligosaccharides and starch without the attendant production of glucose as follows. First, CoMA hydrolyzes the α-1,4 linkages in starch by an endomechanism that yields sugars in the range of maltose to maltopentaose. Next, these intermediates are converted to maltose as a result of a multimolecular reaction that includes hydrolysis, condensation, and transglycosylation events. In addition, the glucosyl transfer events are postulated to be the major pathway. Finally, the overall result of these events is the accumulation of maltose in yields of 58% (wt/wt) (without synchronous glucose production).
Conclusion.
In the present study, the gene encoding the novel maltogenic α-amylase CoMA has been cloned, and its gene product has been expressed and characterized. CoMA (with a signal peptide sequence) can produce high levels of maltose from starch, maltooligosaccharides (≥G3), and γ-cyclodextrin and produce exclusively panose from pullulan. Based on the physicochemical properties of CoMA, we postulated that the maltogenic amylase from Corallococcus sp. EGB may be involved in maltose accumulation in the starch medium. A multimolecular reaction mechanism was responsible for producing a high level of maltose without the attendant production of glucose. In summary, CoMA has potential value in the elimination of by-products from the industrial production of high-maltose syrups and in increasing the level of maltose content.
MATERIALS AND METHODS
Chemicals and media.
Peptone and yeast extract were purchased from the Oxoid Co., Ltd. (Beijing, China). Substrates used for the determination of enzyme activity such as amylose, amylopectin, soluble starch, glycogen, isomaltotriose, panose, pullulan, and dextran were obtained from the Sigma Chemical Co. (St. Louis, MO). Cyclodextrins (CDs) such as α-CD, β-CD, and γ-CD and maltooligosaccharides (maltose [G2], maltotriose [G3], maltotetraose [G4], maltopentaose [G5], and maltohexaose [G6]) were acquired from Wako Pure Chemical (Osaka, Japan). All molecular reagents were purchased from TaKaRa (Dalian, China).
The lysogeny broth (LB) contained 10 g/liter tryptone, 5 g/liter yeast extract, and 5 g/liter NaCl. CTT medium (pH 7.6) consists of 10 g/liter casitone, 8 mM MgSO4, 10 mM Tris-HCl, and 1 mM potassium phosphate.
Strains, plasmids, and primers.
The strains, plasmids and primers used in this study are listed in Table 5. Corallococcus sp. EGB (CCTCC, catalog no. M2012528) was cultivated in CTT medium at 30°C and used as the source of the maltogenic amylase gene. E. coli strains were routinely cultivated aerobically at 37°C in LB medium. E. coli transformants were grown in LB medium containing ampicillin (100 mg/ml) or kanamycin (50 mg/ml).
TABLE 5.
Strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | ϕ80dlacZΔM15 Δ(lacZY-argF)U169 deoR recA1 endA1 hsdR17(rK− mK+) supE44 thi-1 gyrA96 relA1 | Invitrogen |
| E. coli BL21(DE3) | F− ompT hsdSB(rB− mB−) gal dcm (DE3) | Novagen |
| Corallococcus sp. EGB | Wild type, isolated from soil | This study |
| Plasmids | ||
| pMD19-T simple | E. coli cloning vector; Ampr | TaKaRa |
| pET29a (+) | E. coli expression vector, T7 RNA polymerase gene promoter and terminator; Kanr | Novagen |
| pMD19-coMA | pMD19-T simple derivate, containing the CoMA gene from Corallococcus sp. EGB | This study |
| pET29a-coMA | 7.1-kb pET29a derivate carrying the CoMA gene | This study |
| Primers | ||
| F1 | GAATTCCATATGCGCCCCCTCCGCGGACTCTC | |
| R1 | TCCGCTCGAGGCGCAGCCGCCAGATGCC |
Underlined sequences within the primers are the NdeI and XhoI restriction sites. Ampr, ampicillin resistance; Kanr, kanamycin resistance.
Gene cloning, sequencing, and construction of expression vector.
Genomic DNA was extracted from Corallococcus sp. EGB cells using the method described by Kaiser et al. (47). In accordance with the putative α-amylase 3 (GenBank accession no. AFE08049) gene sequence from C. coralloides DSM 2259 (28), the structural gene of α-amylase was PCR amplified from the chromosomal DNA of Corallococcus sp. EGB using the primers F1 and R1 (Table 5). The PCRs were performed for 32 cycles (95°C for 30 s, 60°C for 30 s, and 72°C for 90 s), followed by a 10-min extension at 72°C. The amplified PCR products were purified and ligated into the pMD19-T Simple vector and sequenced by Invitrogen Corporation (Shanghai, China). The positive recombinant plasmids were digested with NdeI and XhoI, and the gene was inserted into the pET29a(+) expression plasmid with a C-terminal His6 tag to generate the pET-coMA plasmid. The recombinant plasmid was transformed into competent E. coli BL21(DE3) for protein expression.
Expression and purification of the recombinant enzyme.
E. coli BL21(DE3) harboring the pET29a-coMA plasmid were cultured in 100 ml of LB broth that was supplemented with kanamycin at 200 rpm and 37°C until the optical density at 600 nm (OD600) reached 0.5 to 0.6. Protein expression was induced with a final concentration of 0.2 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and culturing continued at 150 rpm and 18°C for 16 h. The enzyme may be secreted but stays associated with the cells (based on preliminary data), which is why it is necessary to sonicate the cells for enzyme purification. The bacterial cells were harvested by centrifugation at 2,124 × g and 4°C for 10 min, washed twice with equilibration buffer (20 mM Tris-HCl buffer [pH 7.0], 300 mM NaCl) at 4°C and disrupted by ultrasonication (Insonator M201; Kubota, Japan). The cell debris was then removed by centrifugation at 12,580 × g and 4°C for 20 min, and the supernatant was retained as crude extract. The recombinant CoMA was purified by HisTrap HP column on an ÄKTA purifier 900 (GE Healthcare, USA). The target protein was eluted with elution buffer (20 mM Tris-HCl, 300 mM NaCl, 250 mM imidazole [pH 7.0]). The purified CoMA fractions were collected and dialyzed against 20 mM Tris-HCl buffer (pH 7.0) overnight at 4°C to remove the imidazole. At last, protein samples were concentrated using ultrafiltration columns (Millipore, USA) over a 10-kDa cut off. Samples containing various amounts of the purified CoMA were analyzed by SDS-PAGE using standard methods.
After electrophoresis, the gel was cut into two parts. One part was stained with 0.1% Coomassie brilliant blue R-250 and used to analyze the molecular weight of CoMA; the other was soaked in 2.5% (vol/vol) Triton X-100 for 30 min to remove the SDS. After washing the gel twice, the gel was incubated with 0.1 M Tris-HCl buffer (pH 8.0) for 1 h at 4°C to renature the CoMA and then stored in 20 mM Tris-HCl buffer (pH 7.0) at 4°C for amylase zymogram analysis (42). Gel sections with renatured CoMA were incubated on 20 mM Tris-HCl agar plates that contained 5 mg/ml soluble starch for 30 min at 37°C; the plate was then stained with Lugol's iodine. The locations where CoMA existed became visible as clear zones on a dark-blue background.
Enzyme assay.
The standard assay for CoMA activity was carried out at 50°C for 10 min by the addition of 0.3 μg of purified CoMA to 5 mg/ml soluble starch substrate in 20 mM Tris-HCl buffer (pH 7.0). The amount of reducing sugar was determined spectrophotometrically at 540 nm using the DNS method as previously described (48). One unit of enzyme activity was defined as the amount of enzyme that was required to release reducing sugars equivalent to 1 μmol of maltose per min under the test conditions. The protein concentration was photodensitometrically determined by the Bradford method using bovine serum albumin (BSA) as the standard.
Biochemical properties of the purified CoMA.
The optimal reaction pH was determined under standard conditions of 5 mg/ml soluble starch hydrolysis using several buffers with various pH values (20 mM citrate buffer [pH 3.0 to 6.0], 20 mM Tris-HCl [pH 6.0 to 8.0], 20 mM phosphate-buffered saline [pH 8.0 to 10.0], 20 mM glycine-NaOH buffer [pH 10.0 to 11.0]). The effects of temperature on CoMA activity were measured for 10 min under standard conditions with only the temperature changing (ranging from 20 to 70°C). To measure the pH stability, the 0.3 μg of purified enzyme was incubated at 4°C for 24 h in each buffer, and the residual activity was determined. The thermal stability assays were performed by incubating the 0.3 μg of purified enzyme at each temperature for 1 h in 20 mM Tris-HCl buffer (pH 7.0), followed by measuring its activity under the standard conditions at 50°C, and nonheated enzyme was used as the control (100%).
Then, 0.3 μg of purified CoMA was treated with 1 mM EDTA at 4°C for 5 h and dialyzed against 20 mM Tris-HCl buffer (pH 7.0) to remove the EDTA. For reactivation, the metal-free enzyme was incubated with appropriate metal salts (containing K+, Mg2+, Mn2+, Ba2+, Ca2+, Ni2+, Cu2+, Cr3+, Zn2+, and Fe3+) at final concentrations of 1 and 5 mM for 10 min, and the residual activity was then measured under standard conditions. Activity without any additives was taken as the 100% value.
To determine the effects of EDTA, EGTA, dimethylformamide (DMF), phenylmethylsulfonyl fluoride (PMSF), β-mercaptoethanol (at 1 and 5 mM), ethanol, acetonitrile, acetone (10%), and various concentrations of surfactants (Tween, SDS, and urea) on the activity, the procedure that was used for metal ions was used. Activity in the absence of any additives was taken as the 100% value.
The substrate specificity of CoMA was determined by measuring the enzyme's activity toward various carbohydrates (starch, amylose, amylopectin, glycogen, pullulan, dextran, acarbose, and α-, β-, and γ-cyclodextrin) at a concentration of 5 mg/ml in 20 mM Tris-HCl (pH 7.0). All reactions were monitored under the optimal condition, and the activity against soluble starch was set as the 100% value. Kinetics experiments were performed using soluble starch and amylopectin as the substrates, and the values of Km and Kcat were estimated by linear regression from double-reciprocal plots according to the method of Lineweaver (49).
Hydrolytic mode of CoMA.
The presence of endo- or exomodes of hydrolytic action were determined using a previously reported method (50) that employed an assay to determine of the amount of blue color from the presence of starch. In addition, the amount of the anomeric form of the hydrolyzed products was estimated by optical rotation measurement as described by Konsula and Liakopoulou-Kyriakides (51). Assays were performed by incubating approximately 0.05 U of CoMA or liquefying α-amylase from Bacillus subtilis (BSA) with 1 ml of 5 mg/ml soluble starch at 40°C and pH 7.0 for up to 30 min.
Action pattern of CoMA.
Sugars produced by enzymatic activity on soluble starch, maltotriose, pullulan, and γ-cyclodextrin were quantified, and the enzyme's hydrolytic pattern was analyzed by thin-layer chromatography (TLC) and high-performance liquid chromatography (HPLC). CoMA (10 U/ml) was incubated with starch or maltotriose (≥95% purity) in 20 mM Tris-HCl (pH 7.0) at 30°C for different times. For TLC analysis, the hydrolytic products were examined on silica gel 60 plates (Merck, Germany) using n-butanol/methanol/H2O (8:4:3, vol/vol/vol) as the solvent system (52). The reaction products were visualized by spraying a sulfuric acid-methanol (1:1, vol/vol) solution onto the plate, followed by baking at 95°C for 10 min. HPLC analysis was performed using a Cosmosil Sugar-D column (Nacalai Tesque, Kyoto, Japan) and a refractive index detector (RID-20A; Shimadzu, Kyoto, Japan) maintained at 30°C. The mobile phase was a mixture of acetonitrile and water (75:25, vol/vol) at a flow rate of 1 ml min−1 (9).
Sequence analysis.
The DNA and protein sequence alignments were performed at the National Center for Biotechnology Information (NCBI) using the BLASTN and BLASTP programs (http://www.ncbi.nlm.nih.gov/BLAST), respectively. Lipoprotein signal sequence prediction was carried out using the LipoP 1.0 server of the Center for Biological Sequence Analysis, Technical University of Denmark (http://www.cbs.dtu.dk) (39). The molecular mass and the isoelectric point (pI) were calculated using the ExPASy Proteomics server (53). The protein sequence alignments were generated with the MUSCLE alignment in MEGA 7.0 software (54). The phylogenetic tree was constructed using the neighbor-joining algorithm in MEGA 7.0 and assessed using 1,000 bootstrap replications (55). The conserved domains and the GH family classification were identified via the NCBI website (https://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml) (56).
Accession number(s).
The sequence for the gene encoding the novel amylolytic enzyme designated CoMA cloned from Corallococcus sp. strain EGB was deposited in the GenBank database under accession number MF069146.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the National Natural Science Foundation of China (31700054, 31560031, and 31400056), the Natural Science Foundation of Jiangsu Province (BK 20140687 and BK 20170997), and the China Postdoctoral Science Foundation (2016M591859).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00152-18.
REFERENCES
- 1.Stam MR, Danchin EG, Rancurel C, Coutinho PM, Henrissat B. 2006. Dividing the large glycoside hydrolase family 13 into subfamilies: towards improved functional annotations of α-amylase-related proteins. Protein Eng Des Sel 19:555–562. doi: 10.1093/protein/gzl044. [DOI] [PubMed] [Google Scholar]
- 2.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. 2009. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res 37:233–238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee H-S, Kim M-S, Cho H-S, Kim J-I, Kim T-J, Choi J-H, Park C, Lee H-S, Oh B-H, Park K-H. 2002. Cyclomaltodextrinase, neopullulanase, and maltogenic amylase are nearly indistinguishable from each other. J Biol Chem 277:21891–21897. doi: 10.1074/jbc.M201623200. [DOI] [PubMed] [Google Scholar]
- 4.Park K-H, Kim T-J, Cheong T-K, Kim J-W, Oh B-H, Svensson B. 2000. Structure, specificity and function of cyclomaltodextrinase, a multispecific enzyme of the α-amylase family. Biochim Biophys Acta 1478:165–185. doi: 10.1016/S0167-4838(00)00041-8. [DOI] [PubMed] [Google Scholar]
- 5.Kim J-S, Cha S-S, Kim H-J, Kim T-J, Ha N-C, Oh S-T, Cho H-S, Cho M-J, Kim M-J, Lee H-S. 1999. Crystal structure of a maltogenic amylase provides insights into a catalytic versatility. J Biol Chem 274:26279–26286. doi: 10.1074/jbc.274.37.26279. [DOI] [PubMed] [Google Scholar]
- 6.Kim T-J, Kim M-J, Kim B-C, Kim J-C, Cheong T-K, Kim J-W, Park K-H. 1999. Modes of action of acarbose hydrolysis and transglycosylation catalyzed by a thermostable maltogenic amylase, the gene for which was cloned from a Thermus strain. Appl Environ Microbiol 65:1644–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cha HJ, Yoon HG, Kim YW, Lee HS, Kim JW, Kweon KS, Oh BH, Park KH. 1998. Molecular and enzymatic characterization of a maltogenic amylase that hydrolyzes and transglycosylates acarbose. Eur J Biochem 253:251–262. doi: 10.1046/j.1432-1327.1998.2530251.x. [DOI] [PubMed] [Google Scholar]
- 8.Kim T-J, Shin J-H, Oh J-H, Kim M-J, Lee S-B, Ryu S, Kwon K, Kim J-W, Choi E-H, Robyt JF. 1998. Analysis of the gene encoding cyclomaltodextrinase from alkalophilic Bacillus sp. I-5 and characterization of enzymatic properties. Arch Biochem Biophys 353:221–227. [DOI] [PubMed] [Google Scholar]
- 9.Collins BS, Kelly CT, Fogarty WM, Doyle EM. 1993. The high maltose-producing α-amylase of the thermophilic actinomycete, Thermomonospora curvata. Appl Microbiol Biotechnol 39:31–35. doi: 10.1007/BF00166844. [DOI] [PubMed] [Google Scholar]
- 10.Fogarty WM, Kelly CT. 1990. Recent advances in microbial amylases, p 71–132. In Fogarty WM, Kelly CT (ed), Microbial enzymes and biotechnology, 2nd ed Elsevier Science Publishers, London, United Kingdom. [Google Scholar]
- 11.Li S, Zuo Z, Niu D, Singh S, Permaul K, Prior BA, Shi G, Wang Z. 2011. Gene cloning, heterologous expression, and characterization of a high maltose-producing α-amylase of Rhizopus oryzae. Appl Biochem Biotechnol 164:581–592. doi: 10.1007/s12010-011-9159-5. [DOI] [PubMed] [Google Scholar]
- 12.Doyle EM, Kelly CT, Fogarty WM. 1989. The high maltose-producing α-amylase of Penicillium expansum. Appl Microbiol Biotechnol 30:492–496. doi: 10.1007/BF00263854. [DOI] [PubMed] [Google Scholar]
- 13.Takasaki Y. 1989. Novel maltose-producing amylase from Bacillus megaterium G-2. Agric Biol Chem 53:341–347. doi: 10.1271/bbb1961.53.341. [DOI] [Google Scholar]
- 14.Suganuma T, Mizukami T, Moori K, Ohnishi M, Hiromi K. 1980. Studies of the action pattern of an α-amylase from Streptomyces praecox NA-273. J Biochem 88:131–138. [PubMed] [Google Scholar]
- 15.Jung J-H, Seo D-H, Holden JF, Park C-S. 2014. Maltose-forming α-amylase from the hyperthermophilic archaeon Pyrococcus sp. ST04. Appl Microbiol Biotechnol 98:2121–2131. doi: 10.1007/s00253-013-5068-6. [DOI] [PubMed] [Google Scholar]
- 16.Allen JD, Thoma JA. 1978. Multimolecular substrate reactions catalyzed by carbohydrases. Aspergillus oryzae alpha-amylase degradation of maltooligosaccharides. Biochemistry 17:2338–2344. [DOI] [PubMed] [Google Scholar]
- 17.Allen J, Thoma J. 1978. Model for carbohydrase action. Aspergillus oryzae alpha-amylase degradation of maltotriose. Biochemistry 17:2345–2350. [DOI] [PubMed] [Google Scholar]
- 18.Kelly CT, Collins BS, Fogarty WN, Doyle EM. 1993. Mechanisms of action of the α-amylase of Micromonospora melanosporea. Appl Microbiol Biotechnol 39:599–603. doi: 10.1007/BF00205059. [DOI] [PubMed] [Google Scholar]
- 19.Norman B. 1979. Application of polysaccharide degrading enzymes in the starch industry, p 339–376. In Berkley R. (ed), Microbial polysaccharides. Academic Press, New York, NY. [Google Scholar]
- 20.Sharma A, Satyanarayana T. 2012. Cloning and expression of acidstable, high maltose-forming, Ca2+-independent α-amylase from an acidophile Bacillus acidicola and its applicability in starch hydrolysis. Extremophiles 16:515–522. doi: 10.1007/s00792-012-0451-2. [DOI] [PubMed] [Google Scholar]
- 21.Reichenbach H. 1999. The ecology of the myxobacteria. Environ Microbiol 1:15–21. doi: 10.1046/j.1462-2920.1999.00016.x. [DOI] [PubMed] [Google Scholar]
- 22.Beebe J. 1943. Studies on the myxobacteria. 3. The utilization of carbohydrates. Iowa State Coll J Sci 17:227–240. [Google Scholar]
- 23.Norén B. 1955. Studies on myxobacteria. III. Organic factors in nutrition. Bot Not 108:81–134. [Google Scholar]
- 24.Irschik H, Reichenbach H. 1985. An unusual pattern of carbohydrate utilization in Corallococcus (Myxococcus) coralloides (Myxobacterales). Arch Microbiol 142:40–44. doi: 10.1007/BF00409234. [DOI] [Google Scholar]
- 25.Farez-Vidal ME, Fernandez-Vivas A, Arias J. 1990. Amylase programming during the life cycle of Myxococcus coralloides. J Appl Bacteriol 69:119–124. doi: 10.1111/j.1365-2672.1990.tb02919.x. [DOI] [Google Scholar]
- 26.Li Z, Wu J, Zhang B, Wang F, Ye X, Huang Y, Huang Q, Cui Z. 2015. AmyM, a novel maltohexaose-forming α-amylase from Corallococcus sp. strain EGB. Appl Environ Microbiol 81:1977–1987. doi: 10.1128/AEM.03934-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu J, Xia B, Li Z, Ye X, Chen Q, Dong W, Zhou J, Huang Y, Cui Z. 2015. Molecular cloning and characterization of a novel GH13 saccharifying α-amylase AmyC from Corallococcus sp. EGB. Starch 67:810–819. doi: 10.1002/star.201400258. [DOI] [Google Scholar]
- 28.Huntley S, Zhang Y, Treuner-Lange A, Kneip S, Sensen CW, Søgaard-Andersen L. 2012. Complete genome sequence of the fruiting myxobacterium Corallococcus coralloides DSM. 2259. J Bacteriol 194:3012–3013. doi: 10.1128/JB.00397-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sivakumar N, Li N, Tang JW, Patel BK, Swaminathan K. 2006. Crystal structure of AmyA lacks acidic surface and provide insights into protein stability at poly-extreme condition. FEBS Lett 580:2646–2652. doi: 10.1016/j.febslet.2006.04.017. [DOI] [PubMed] [Google Scholar]
- 30.Caner S, Nguyen N, Aguda A, Zhang R, Pan YT, Withers SG, Brayer GD. 2013. The structure of the Mycobacterium smegmatis trehalose synthase reveals an unusual active site configuration and acarbose-binding mode. Glycobiology 23:1075–1083. doi: 10.1093/glycob/cwt044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y-L, Chow S-Y, Lin Y-T, Hsieh Y-C, Lee G-C, Liaw S-H. 2014. Structures of trehalose synthase from Deinococcus radiodurans reveal that a closed conformation is involved in catalysis of the intramolecular isomerization. Acta Crystallogr D Biol Crystallogr 70:3144–3154. doi: 10.1107/S1399004714022500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hondoh H, Kuriki T, Matsuura Y. 2003. Three-dimensional structure and substrate binding of Bacillus stearothermophilus neopullulanase. J Mol Biol 326:177–188. doi: 10.1016/S0022-2836(02)01402-X. [DOI] [PubMed] [Google Scholar]
- 33.Kim T-J, Nguyen VD, Lee H-S, Kim M-J, Cho H-Y, Kim Y-W, Moon T-W, Park CS, Kim J-W, Oh B-H. 2001. Modulation of the multisubstrate specificity of Thermus maltogenic amylase by truncation of the N-terminal domain and by a salt-induced shift of the monomer/dimer equilibrium. Biochemistry 40:14182–14190. doi: 10.1021/bi015531u. [DOI] [PubMed] [Google Scholar]
- 34.Abe A, Tonozuka T, Sakano Y, Kamitori S. 2004. Complex structures of Thermoactinomyces vulgaris R-47 α-amylase 1 with malto-oligosaccharides demonstrate the role of domain N acting as a starch-binding domain. J Mol Biol 335:811–822. doi: 10.1016/j.jmb.2003.10.078. [DOI] [PubMed] [Google Scholar]
- 35.Janeček Š. 2002. How many conserved sequence regions are there in the α-amylase family. Biologia 57:29–41. [Google Scholar]
- 36.Janeček Š. 1992. New conserved amino acid region of alpha-amylases in the third loop of their (β/α)8-barrel domains. Biochem J 288:1069. doi: 10.1042/bj2881069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Janeček Š. 1995. Close evolutionary relatedness among functionally distantly related members of the (β/α)8-barrel glycosyl hydrolases suggested by the similarity of their fifth conserved sequence region. FEBS Lett 377:6–8. doi: 10.1016/0014-5793(95)01309-1. [DOI] [PubMed] [Google Scholar]
- 38.Horvathova V, Janeček Š, Sturdik E. 2001. Amylolytic enzymes: molecular aspects of their properties. Gen Physiol Biophys 20:7–32. [PubMed] [Google Scholar]
- 39.Liebl W, Stemplinger I, Ruile P. 1997. Properties and gene structure of the Thermotoga maritima alpha-amylase AmyA, a putative lipoprotein of a hyperthermophilic bacterium. J Bacteriol 179:941–948. doi: 10.1128/jb.179.3.941-948.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheong KA, Kim TJ, Yoon JW, Park CS, Lee TS, Kim YB, Park KH, Kim JW. 2002. Catalytic activities of intracellular dimeric neopullulanase on cyclodextrin, acarbose and maltose. Biotechnol Appl Biochem 35:27–34. doi: 10.1042/BA20010052. [DOI] [PubMed] [Google Scholar]
- 41.Park KH, Kim MJ, Lee HS, Han NS, Kim D, Robyt JF. 1998. Transglycosylation reactions of Bacillus stearothermophilus maltogenic amylase with acarbose and various acceptors. Carbohydr Res 313:235–246. doi: 10.1016/S0008-6215(98)00276-6. [DOI] [PubMed] [Google Scholar]
- 42.Yang S-J, Lee H-S, Park C-S, Kim Y-R, Moon T-W, Park K-H. 2004. Enzymatic analysis of an amylolytic enzyme from the hyperthermophilic archaeon Pyrococcus furiosus reveals its novel catalytic properties as both an α-amylase and a cyclodextrin-hydrolyzing enzyme. Appl Environ Microbiol 70:5988–5995. doi: 10.1128/AEM.70.10.5988-5995.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tonozuka T, Mogi S-i, Shimura Y, Ibuka A, Sakai H, Matsuzawa H, Sakano Y, Ohta T. 1995. Comparison of primary structures and substrate specificities of two pullulan-hydrolyzing α-amylases, TVA I and TVA II, from Thermoactinomyces vulgaris R-47. Biochim Biophys Acta 1252:35–42. doi: 10.1016/0167-4838(95)00101-Y. [DOI] [PubMed] [Google Scholar]
- 44.Uitdehaag JC, Mosi R, Kalk KH, van der Veen BA, Dijkhuizen L, Withers SG, Dijkstra BW. 1999. X-ray structures along the reaction pathway of cyclodextrin glycosyltransferase elucidate catalysis in the α-amylase family. Nat Struct Mol Biol 6:432–436. doi: 10.1038/8235. [DOI] [PubMed] [Google Scholar]
- 45.Ballschmiter M, Armbrecht M, Ivanova K, Antranikian G, Liebl W. 2005. AmyA, an alpha-amylase with beta-cyclodextrin-forming activity, and AmyB from the thermoalkaliphilic organism Anaerobranca gottschalkii: two alpha-amylases adapted to their different cellular localizations. Appl Environ Microbiol 71:3709–3715. doi: 10.1128/AEM.71.7.3709-3715.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Skov LK, Mirza O, Henriksen A, De Montalk GP, Remaud-Simeon M, Sarçabal P, Willemot R-M, Monsan P, Gajhede M. 2001. Amylosucrase, a glucan-synthesizing enzyme from the α-amylase family. J Biol Chem 276:25273–25278. doi: 10.1074/jbc.M010998200. [DOI] [PubMed] [Google Scholar]
- 47.Kaiser D, Manoil C, Dworkin M. 1979. Myxobacteria: cell interactions, genetics, and development. Annu Rev Microbiol 33:595–639. doi: 10.1146/annurev.mi.33.100179.003115. [DOI] [PubMed] [Google Scholar]
- 48.Miller GL. 1959. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31:426–428. doi: 10.1021/ac60147a030. [DOI] [Google Scholar]
- 49.Dowd JE, Riggs DS. 1965. A comparison of estimates of Michaelis-Menten kinetic constants from various linear transformations. J Biol Chem 240:863–869. [PubMed] [Google Scholar]
- 50.Jana M, Maity C, Samanta S, Pati BR, Islam SS, Mohapatra PKD, Mondal KC. 2013. Salt-independent thermophilic α-amylase from Bacillus megaterium VUMB109: an efficacy testing for preparation of maltooligosaccharides. Ind Crops Prod 41:386–391. doi: 10.1016/j.indcrop.2012.04.048. [DOI] [Google Scholar]
- 51.Konsula Z, Liakopoulou-Kyriakides M. 2004. Hydrolysis of starches by the action of an α-amylase from Bacillus subtilis. Process Biochem 39:1745–1749. doi: 10.1016/j.procbio.2003.07.003. [DOI] [Google Scholar]
- 52.Hansen SA. 1975. Thin-layer chromatographic method for the identification of mono-, di-and trisaccharides. J Chromatogr A 107:224–226. doi: 10.1016/S0021-9673(00)82770-3. [DOI] [Google Scholar]
- 53.Gasteiger E, Hoogland C, Gattiker A, Duvaud Se Wilkins MR, Appel RD, Bairoch A. 2005. Protein identification and analysis tools on the ExPASy server, p 571–607. In Walker JM. (ed), The proteomics protocol handbook. Humana Press, Totowa, NJ. [Google Scholar]
- 54.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marchler-Bauer A, Zheng C, Chitsaz F, Derbyshire MK, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lanczycki CJ. 2012. CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res 41:348–352. doi: 10.1093/nar/gks1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yun J, Kang S, Park S, Yoon H, Kim MJ, Heu S, Ryu S. 2004. Characterization of a novel amylolytic enzyme encoded by a gene from a soil-derived metagenomic library. Appl Environ Microbiol 70:7229–7235. doi: 10.1128/AEM.70.12.7229-7235.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.David M-H, Günther H, Röper HH. 1987. Catalytic properties of Bacillus megaterium amylase. Starch 39:436–440. doi: 10.1002/star.19870391208. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.