ABSTRACT
Paracoccus denitrificans is a valuable model organism due to its versatile respiration capability and bioenergetic flexibility, both of which are critical to its survival in different environments. Quorum sensing (QS) plays a crucial role in the regulation of many cell functions; however, whether QS systems play a role in P. denitrificans is unknown. In this study, we demonstrated that iron uptake systems in P. denitrificans were directly regulated by a newly identified QS system. Genes coding for TonB-dependent systems, which transport chelated iron, were transcribed at higher levels in the QS-defective mutants. In contrast, genes coding for the Fbp system, which is TonB independent and transports unchelated ferric iron, were downregulated in the mutants. In brief, QS in P. denitrificans triggers a switch in iron uptake from TonB-dependent to TonB-independent transport during biofilm formation as higher concentrations of iron accumulate in the exopolysaccharide (EPS). Switching from TonB-dependent iron uptake systems to TonB-independent systems not only prevents cells from absorbing excess iron but also conserves energy. Our data suggest that iron uptake strategies are directly regulated by QS in Paracoccus denitrificans to support their survival in available ecological niches.
IMPORTANCE As iron is an important trace metal for most organisms, its absorption is highly regulated. Fur has been reported as a prevalent regulator of iron acquisition. In addition, there is a relationship between QS and iron acquisition in pathogenic microbes. However, there have been few studies on the iron uptake strategies of nonpathogenic bacteria. In this study, we demonstrated that iron uptake systems in Paracoccus denitrificans PD1222 were regulated by a newly identified PdeR/PdeI QS system during biofilm formation, and we put forward a hypothesis that QS-dependent iron uptake systems benefit the stability of biofilms. This report elaborates the correlation among QS, iron uptake, and biofilm formation and thus contributes to an understanding of the ecological behavior of environmental bacteria.
KEYWORDS: quorum sensing, iron transport system, biofilm formation, RNA-seq
INTRODUCTION
Iron is essential for a wide variety of biological functions from DNA synthesis to electron transport (1). Although iron is a key nutrient for most organisms, it is subject to biotic and abiotic redox reactions, typically forming insoluble Fe(III) oxyhydroxides under aerobic conditions (2). The bioavailability of such nutrients can thus be quite low regardless of local biological demand. To cope with iron limitation and to adapt to various environments, microbes have developed multiple iron uptake systems to absorb different forms of iron (3). These iron uptake systems are categorized into two classes according to the way iron passes through the outer membrane of cells. Class I systems (TonB-dependent systems) are responsible for chelated iron complex transport. Chelated iron is transported from the outer membrane into the periplasmic space by the energy transduction complex TonB-ExbB-ExbD (4). Class II systems (TonB-independent systems) are inner membrane systems that do not require TonB for iron acquisition; the diffusion of unchelated iron occurs through porins or similar proteins (5). Although iron is essential, high levels of iron are toxic due to its propensity to react with oxygen to generate reactive oxygen species (ROS) via Fenton and Haber-Weiss reactions (6). The expression of iron acquisition genes must thus be meticulously regulated. Fur is a prevalent ferric uptake regulator that regulates iron uptake genes and the biosynthesis of siderophores in response to the iron level in the cell. Usually, Fur indirectly regulates iron uptake systems by regulating two-component signal transduction systems, AraC-like regulators, and extracytoplasmic function (ECF) sigma factors (7).
Quorum sensing (QS) is a system by which a bacterial population monitors its cell density through the release of specific signaling molecules called autoinducers. The system has been reported to control a wide range of biological activities, including bioluminescence, sporulation, biofilm formation, conjugation, motility, and antibiotic production (8). QS also controls iron acquisition in several pathogens, such as Porphyromonas gingivalis (9) and Actinobacillus actinomycetemcomitans (10). The regulation of iron uptake systems by QS in pathogens contributes to the establishment and progression of infection (1). Moreover, several studies have reported a relationship among QS, iron uptake, and biofilm formation. A study reported by Banin et al. demonstrated that an exochelin siderophore uptake system is essential for biofilm formation under iron-limiting conditions in Pseudomonas aeruginosa (11). A Pseudomonas quinolone signal (PQS) was reported to act as an iron chelator, thus inducing iron starvation and upregulating a series of genes belonging to iron acquisition and the oxidative stress response (12). In addition, iron was reported to affect biofilm formation by interacting with the PQS systems (13).
P. denitrificans is a ubiquitous Gram-negative denitrifying bacterium originally isolated in 1910 by Martinus Beijerinck and renamed by D. H. Davis (14, 15). It has become a paradigm model organism for studies of prokaryotic respiration, specifically for studying the electron transfer chain and respiratory processes (16, 17). The P. denitrificans genome contains multiple homologs of iron acquisition systems, where each system transports a specific form of iron. These systems included TonB-dependent systems, such as Fec (18), Fhu (19), Ycl (20), Yfm (21), and Hmu (22), and TonB-independent systems, such as Fbp (23). However, although iron transport systems play an important role in P. denitrificans, whether iron acquisition is transcriptionally regulated through a signaling cascade is unclear. In a previous report, Schaefer et al. detected the production of a long-chain acyl-homoserine lactone (AHL), N-hexadecanoyl-homoserine lactone (C16-HSL) (24); however, the AHL synthase, LuxI/R-homologous QS system, and exact nature of downstream gene regulation in P. denitrificans have not been identified.
In this study, we chose P. denitrificans PD1222 as the research organism and identified a QS system, PdeI/PdeR, in P. denitrificans PD1222. RNA sequencing (RNA-Seq) was performed to examine the transcriptome of the wild type and QS-defective mutants (ΔpdeI and ΔpdeR) to provide a more comprehensive analysis of the regulation of P. denitrificans gene expression by the ΔpdeI and ΔpdeR mutants. Our results indicated that most genes related to iron uptake were regulated in the QS-defective mutants. Furthermore, we examined the relative mRNA transcriptional levels of the iron transport systems during biofilm formation and concluded that QS-dependent iron uptake was correlated with biofilm formation.
RESULTS
Identification of a LuxR/LuxI-type quorum-sensing regulatory system in P. denitrificans.
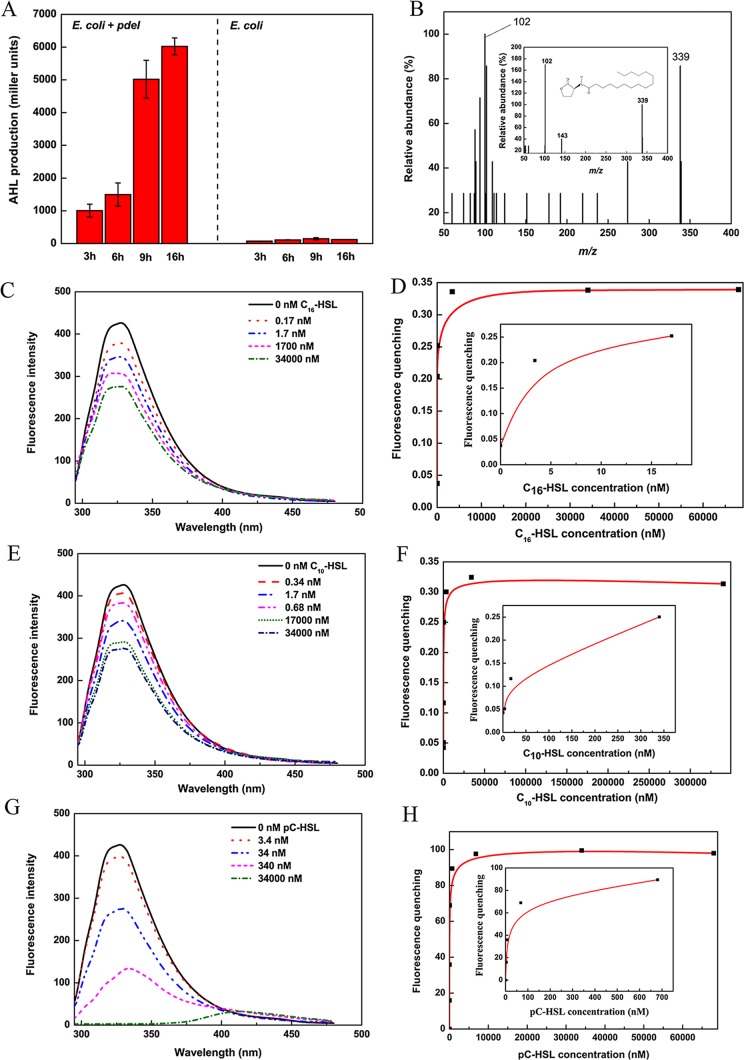
To investigate potential AHL synthases in P. denitrificans, we compared, cloned, and heterologously expressed their homologs from P. denitrificans. The sequence for the putative QS signal synthase pdeI from P. denitrificans was conserved among AHL synthase proteins. The open reading frame (ORF) pden_0787 encodes a putative protein of 199 amino acids whose sequence is 20% similar to that of the AHL synthase TraI from Agrobacterium tumefaciens. The pdeI gene was cloned into a pET-30(a) plasmid, which was then transformed into Escherichia coli BL21(DE3). The supernatant of the recombinant bacteria was added to the KYC55 medium. The AHL bioassay strain A. tumefaciens KYC55 uses a T7 expression system to strongly overexpress the regulator TraR. When AHL signals are present, TraR can activate a traI-lacZ reporter fusion protein, resulting in the production of β-galactosidase. The signals obtained from the recombinant sample turned the biosensor supernatant solution yellow, while no activity was observed in the control E. coli strain, which carried a pET-30(a) vector lacking the pdeI gene. The levels of lacZ induction by AHLs are reported in Miller units (Fig. 1A). These results suggest that pdeI from P. denitrificans encodes an AHL synthase. High-performance liquid chromatography-mass spectrometry (HPLC-MS) was used to identify the acyl chains of the AHLs. The extract of E. coli BL21(DE3) cells containing pET-pdeI was used for HPLC-MS analysis, and the retention times and mass spectra of the AHLs in the recombinant extract were compared with those of AHL standards reported by Schaefer et al. (24). The results showed that the features of an AHL with a molecular ion [M+H]+ of m/z 339, which had been extracted from the expression strain of BL21(DE3), were consistent with those of a C16-HSL standard (Fig. 1B).
FIG 1.
Verification of pdeI and pdeR gene functions. (A) AHL production (in Miller units) of recombinant and control E. coli strains after being induced for 3, 6, 9, and 16 h was determined by A. tumefaciens KYC55(pJZ372)(pJZ384)(pJZ410) using a liquid assay. (B) LC-MS chromatograms of C16-HSL from E. coli(pET-pdeI) and the C16-HSL standard. The mass spectra reveal molecular ions [M+H] of m/z 339. (C) Interaction of PdeR with different concentrations of C16-AHL. The PdeR fluorescence was determined at a protein concentration of 34 μM. (D) Titrating PdeR with increasing amounts of C16-AHL. (E) Interaction of PdeR with different concentrations of C10-AHL. (F) Titrating PdeR with increasing amounts of C10-AHL. (G) Interaction of PdeR with different concentrations of pC-HSL. (H) PdeR titration with increasing amounts of pC-HSL.
An ORF upstream of pdeI encodes a putative protein of 203 amino acids that is similar to several LuxR-type transcriptional activators, such as TraR (23% amino acid identity). This ORF was therefore annotated pdeR. PdeR contains three tryptophan residues, two of which are predicted to be located in the N-terminal region (W44 and W72). LuxR homologs utilize N-terminal regions to dimerize and bind signal ligands; such intermolecular interactions generally invoke structural changes that then alter the intrinsic fluorescence intensity of the protein. Measurements taken at wavelengths between 300 and 400 nm showed that PdeR produced a fluorescence maximum at approximately 340 nm (Fig. 1C to H). This fluorescence decreased in intensity with increasing concentrations of AHLs. Maximal quenching was approximately 35% and resulted from reactions with N-decanoyl-l-homoserine lactone (C10-HSL) and C16-HSL; higher concentrations did not further quench the fluorescence. Maximal quenching reached 100% when the protein reacted with N-(p-coumaroyl)-l-homoserine lactone (pC-HSL) [AHLs with acyl groups contain aromatic functionality with electron-withdrawing groups; the pC-AHL used in this study was pC-HSL].
Profiling of ΔpdeI and ΔpdeR mutants and the autoregulation of PdeR/PdeI.
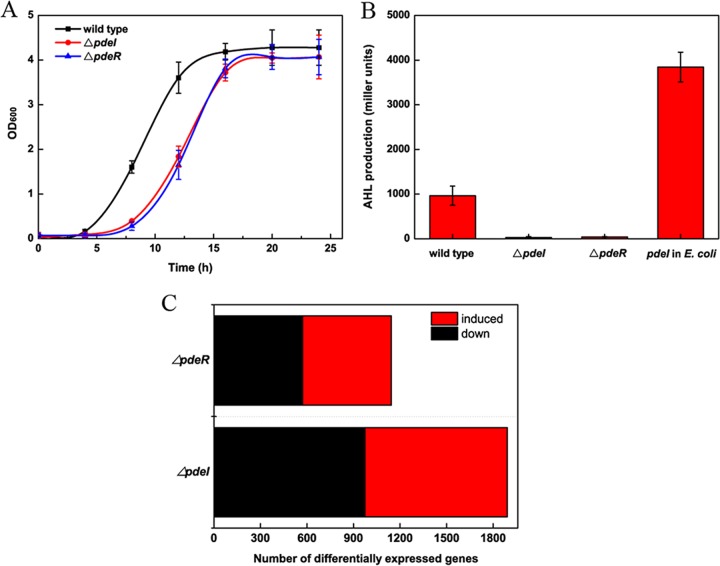
To investigate the function of the LuxR/LuxI homologs PdeR/PdeI in P. denitrificans, null mutations of pdeR and pdeI were generated by replacing each gene with a gfp cassette. Both the ΔpdeI and ΔpdeR mutants grew slower than the wild-type strain when cultured in LB medium (Fig. 2A), indicating that mutations in the quorum-sensing regulators affect the growth rate. Cell-free culture supernatants of these strains were assayed for AHL activity and AHL content. As noted above, the supernatant of the wild-type cultures exhibited high AHL activity; in contrast, no AHLs were detected in the supernatants of the ΔpdeI and ΔpdeR mutants via liquid assays (Fig. 2B).
FIG 2.
(A) Growth curves of wild-type and ΔpdeI and ΔpdeR mutant strains. (B) Effects of pdeR and pdeI genes on AHL production in P. denitrificans. Wild-type and ΔpdeI and ΔpdeR mutant cells were grown to a high cell density, and the cell-free supernatants were assayed for AHL activity. (C) Number of differentially expressed genes (DEGs) regulated by pdeI and pdeR.
A pdeI-lacZ transcriptional fusion was constructed in the wild-type and pdeR mutant strains of P. denitrificans PD1222 to examine the expression of the AHL synthase gene pdeI. This construct disrupted the pdeI locus, resulting in no AHL production in these strains (data not shown). Very low levels of β-galactosidase activity were detected against the pdeR mutant, even in the presence of AHLs (see Fig. S1A in the supplemental material, left). However, pdeI was fully induced in the wild type only when C16-HSL was added to the medium (Fig. S1A, right). These data indicate that pdeI expression requires both a functional PdeR and C16-HSL. To investigate pdeR expression, we created a pdeR-lacZ transcriptional fusion in the wild-type and pdeI mutant strains of P. denitrificans PD1222. Fig. S1B shows that pdeR expression is dependent on AHLs. The expression of pdeR was induced in the pdeI mutant by the addition of C16-HSL to the medium, while lacZ expression reached high levels at high cell densities in the wild type. These data collectively suggest that in P. denitrificans, the expression of both pdeR and pdeI genes is autoregulated, which is a hallmark of QS regulation.
QS regulates the iron acquisition systems of P. denitrificans.
To gain insight into the genome-scale gene expression patterns regulated by the QS system in P. denitrificans PD1222, we conducted RNA-Seq on the wild-type strain and the ΔpdeI and ΔpdeR mutants. The transcriptome of each strain was acquired during the stationary (S) phase (optical density at 600 nm [OD600], ∼3.0) under aerobic culture conditions. RNA-Seq was performed using an Illumina HiSeq 2000 system, and the samples yielded from 17,046,636 to 23,506,828 reads. To profile the gene expression, we mapped the total reads of the four libraries to the reference genome of P. denitrificans PD1222 by the BWA program. Approximately 9,098,930 to 14,448,966 reads were mapped. The gene expression levels were quantified based on their reads per kilobase of exon per million mapped sequence reads (RPKM) values. To identify genes regulated in a QS-dependent manner, we used the DEGseq package in R language, calculating the fold change (FC) from the ratio of the wild-type RPKM to mutant RPKM values (25). In total, 974 genes were upregulated in the ΔpdeI mutant from their wild-type values, and 918 genes were downregulated; 572 genes were upregulated in the ΔpdeR mutant, and 572 genes were downregulated (Fig. 2C). To validate the accuracy of the RNA-Seq results, the expression levels of six randomly selected genes for each group in wild-type samples relative to mutant samples were determined by quantitative real-time PCR (RT-qPCR) (Fig. S2B and D). This comparison of the RT-qPCR results and the RNA-Seq data showed that correlation coefficients were higher than 0.98 (Fig. S2A and C), indicating that our RNA-Seq data were reliable.
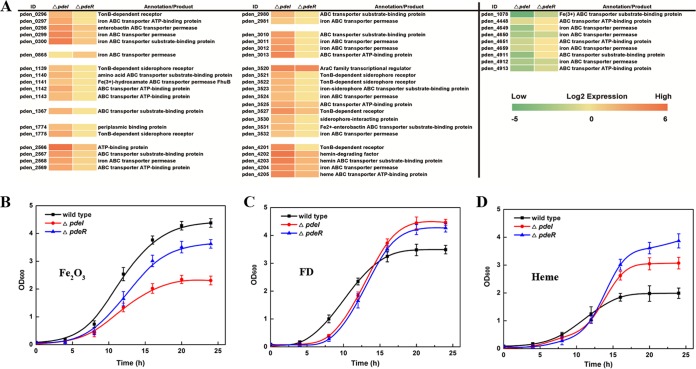
The gene expression patterns of iron transport systems were visualized by a heatmap, shown in Fig. 3A; 39 genes related to TonB-dependent iron transport and nine genes related to TonB-independent transport are listed. These data suggested that TonB-dependent systems were upregulated and that Ton-independent systems were downregulated in the QS-defective mutants. These findings were verified using physiological experiments in which chelated iron (heme and dicitrate chelated iron) stimulated the growth of pdeI and pdeR mutants better than that of the wild-type strain. In contrast, the two mutants grew less strongly in medium treated with ferric oxide than did the wild type (Fig. 3B to D).
FIG 3.
(A) Gene expression patterns of the iron uptake-related ABC transporter systems. The data were visualized as a heatmap. Genes differentially expressed between the ΔpdeI mutant and wild-type strain and between the ΔpdeR mutant and wild-type strain are listed in the map. Each column of the heatmap represents the log2 FC value in expression, with a green-yellow-red scheme in which the lower limit is −5 and the upper limit is 6. (B) Growth curves of the wild type and QS-defective mutants in medium with Fe2O3 only. (C) Growth curves of the wild type and QS-defective mutants in medium with dicitrate-chelated iron (ferric dicitrate [FD]) only. (D) Growth curves of the wild type and QS-defective mutants in medium with heme only.
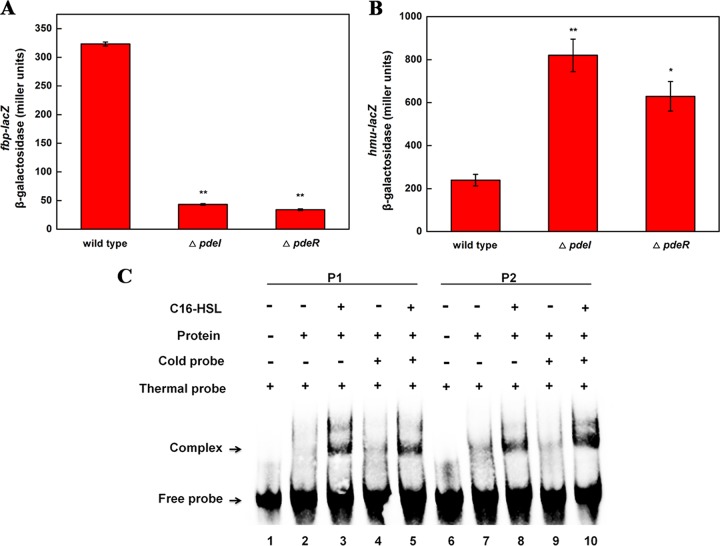
To further investigate the regulation of gene clusters related to iron transport by a QS system, we focused on two operons, one encoding a homolog of the fbp unchelated ferric transport system (pden_1077, pden_1078, and pden_1079) and one encoding a homolog of the hmu heme transport system (pden_4202, pden_4203, pden_4204, and pden_4205). To study the expression of these two operons, we constructed promoter-lacZ transcriptional fusions in the wild-type, ΔpdeI mutant, and ΔpdeR mutant strains. These constructs preserved a functional copy of the fbp or hmu operon. β-Galactosidase (in Miller units) was detected in the late-logarithmic-growth stage, and results indicated that the expression of the fbp-lacZ fusion was approximately 7-fold lower in both the ΔpdeI and ΔpdeR mutants than in the wild type, whereas the expression of the hmu-lacZ fusion was 3- to ∼4-fold higher in both the ΔpdeI and ΔpdeR mutants than in the wild type (Fig. 4A and B). To further understand the regulation system, we also measured the Miller units of lacZ fusions under different iron conditions (iron depletion and iron overdosage). As shown in Fig. S3, the results indicated that under iron-depleting conditions, the expression pattern of the fbp-lacZ fusion was slightly different from that cultured in medium with optimum iron concentration. The Miller units of hmu-lacZ fusion in all three strains were increased by almost 100%. Under iron overdosage conditions, the expression pattern of fbp-lacZ was similar to that cultured in medium with optimum iron concentration; however, the Miller units of the hmu-lacZ fusion in all three strains decreased by almost 30%. The different expression levels of the hmu-lacZ fusion under different iron conditions indicated the presence of other iron-induced regulatory mechanisms for heme uptake, such as Fur or sigma factors. Although the hmu operon was differentially expressed under different iron conditions, the QS regulatory mechanism was suitable for iron uptake systems under both iron-depleting and iron overdosage conditions.
FIG 4.
Expression of fbp and hmu operons is QS dependent. (A) fbp-lacZ expression in the wild type and the QS-defective mutants. (B) hmu-lacZ expression in the wild type and the QS-defective mutants. β-Galactosidase activity assays were performed as described previously, and data are presented in Miller units. The results are representative of three experiments. Error bars indicate standard deviations. Asterisks indicate a significant increase compared to the control in the same treatment period (*, P < 0.1; **, P < 0.05; ***, P < 0.01). (C) Gel mobility shift assays using purified PdeR-His and a DNA fragment containing the hmu promoter (P1) or fbp promoter (P2) regions. Lanes 1 and 6, a synthetic end-labeled 50-bp DNA fragment (thermal probe); lanes 2 and 7, PdeR protein did not form a complex with the labeled DNA fragment; lanes 3 and 8, the addition of C16-HSL resulted in complex formation; lanes 4 and 5 and 9 and 10, the binding specificity of PdeR for the probe DNA was not affected by unlabeled competitor DNA (cold probe).
To determine whether this QS regulatory mechanism is direct regulation or not, an electrophoretic mobility shift assay (EMSA) experiment was performed. The in vivo data of Fig. 4A and B indicate that PdeR regulates the promoter gene fusions of the hmu and fbp operons, presumably by binding to the PdeR box located in these regions. We therefore conducted DNA mobility shift assays to define the in vitro DNA-binding characteristics of PdeR. Figure 4C shows that PdeR bound the promoter region of the hmu and fbp operons but only when C16-HSL was present (lanes 1 to 3 and lanes 6 to 8). The addition of an unlabeled probe DNA did not affect complex formation (lanes 5 and 10). This result proves that fbp and hmu expression is directly activated by the QS regulator PdeR.
QS-dependent iron uptake systems were correlated with biofilm formation.
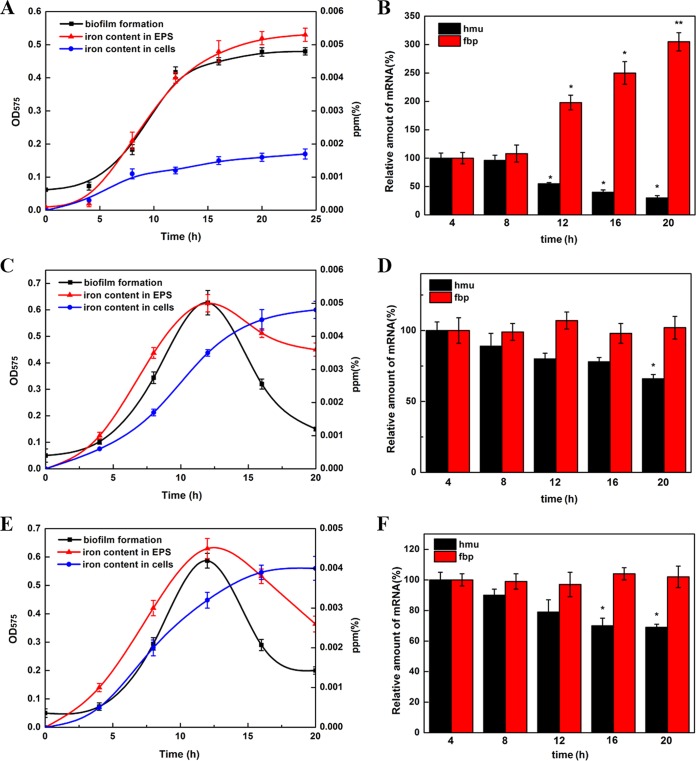
To reveal how the QS system regulates iron uptake during biofilm formation, we measured the iron content in EPS and cells enveloped in the EPS. The iron content is displayed in parts per million (the ratio of the mass of iron to the mass of all elements in biofilms). Meanwhile, the expression of the iron uptake systems of the wild-type strain and QS-defective strains during biofilm formation was also detected. As shown in Fig. 5, the majority of iron was accumulated in the EPS during biofilm formation of the wild-type strain. However, iron was absorbed in the QS-defective mutant cells continuously during biofilm formation. The increased intracellular iron concentration in the QS-defective mutants was correlated with the high expression levels of the hmu operon compared with the wild type. In addition, the decrease in iron mass in the EPS matrix was presumably due to the disintegrating biofilm surface during the late period.
FIG 5.
Relationship between QS-dependent iron uptake systems and biofilm formation. (A) Development of wild-type biofilm formation and the iron content in the EPS and cells enveloped in the EPS. Biofilms were stained with crystal violet and quantified by their absorbance at 575 nm. The iron concentration was determined during biofilm development and is shown in parts per million. (B) RT-qPCR of hmu and fbp operons during biofilm formation of the wild-type strain. Data were analyzed using the 2−ΔΔCT method. (C) Development of ΔpdeI mutant biofilm formation and the iron content in the EPS and cells enveloped in the EPS. (D) RT-qPCR of hmu and fbp operons during biofilm formation of the ΔpdeI mutant. (E) Development of ΔpdeR mutant biofilm formation and the iron content in the EPS and cells enveloped in the EPS. (F) RT-qPCR of hmu and fbp operons during biofilm formation of the ΔpdeR mutant. Asterisks indicate a significant increase compared to the control during the same treatment period (*, P < 0.1; **, P < 0.05; ***, P < 0.01).
DISCUSSION
Paracoccus denitrificans is a ubiquitous bacterium isolated from soil. In the reported P. denitrificans PD1222, DSM 413, and DSM 415 genomes (https://www.ncbi.nlm.nih.gov/genome/genomes/1658), many homologous genes of the iron transport systems have been identified. All of these strains contain homologs of the luxI and luxR genes, and the putative LuxR/LuxI systems showed 100% identity. In this study, we demonstrated that the iron acquisition systems were regulated by a newly identified PdeR/PdeI QS system.
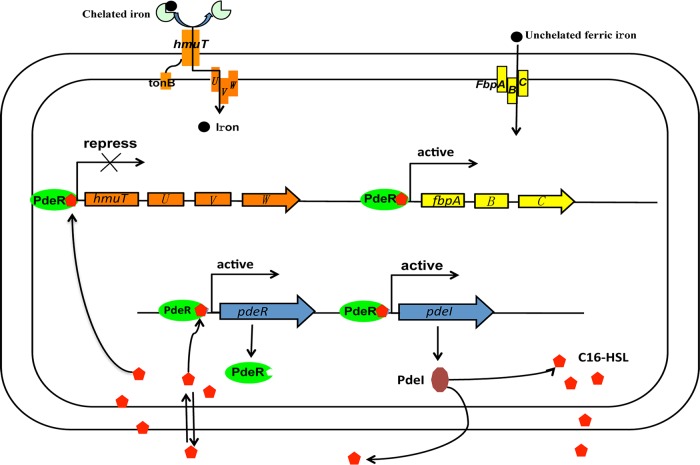
In the newly identified PdeR/PdeI QS system, PdeI was responsible for synthesis of the long-chain quorum-sensing signal molecule C16-HSL, and the pdeR gene, which encoded the regulatory protein PdeR, was located upstream of pdeI. Interestingly, unlike most reported LuxR-type proteins (26–29), the PdeR does not require cognate AHLs for correct folding but needs AHLs to regulate target gene expression. Moreover, in vivo fluorescence quenching experiments showed that PdeR binds not only its cognate, C16-HSL, but also C10-HSL and pC-HSL. P. denitrificans was thus assumed to respond not only to its own “language” but also to the “languages” of other populations (Fig. 1). By the promoter-lacZ fusions of pdeI and pdeR, constructed in the wild type and QS-defective mutants, the results show that very low levels of β-galactosidase activity were detected in the absence of AHLs or PdeR proteins (Fig. S2). The expression of pdeI and pdeR requires both a functional PdeR and autoinducers produced by PdeI, creating an autoinduction feedback loop (Fig. 6). QS feedback, a signature of QS regulation, has been observed in many bacteria with QS systems, such as Mesorhizobium tianshanense, Vibrio harveyi, and P. aeruginosa (30, 31).
FIG 6.
Schematic for regulation of iron acquisition by PdeR/PdeI and the autoregulation of PdeR/PdeI. hmuTUVW encodes the TonB-dependent Hmu transport system, while fbpABC encodes the TonB-independent Fbp transport system. In the presence of C16-HSL, PdeR actives the expression of fbpABC, pdeI, and pdeR but represses the expression of hmuTUVW.
By conducting RNA-Seq between the wild type and QS-defective mutants, we demonstrated that the QS system activated the expression of TonB-independent systems and repressed the expression of the TonB-dependent systems, thereby regulating the acquisition of iron from active transport to passive transport (Fig. 6). Furthermore, we demonstrated that PdeR bound to the promoters of fbp and hmu gene clusters in the presence of C16-HSL in vitro, indicating the direct regulation of iron acquisition systems by QS. The unique regulation of iron uptake systems in P. denitrificans raised a question: do QS-dependent iron uptake systems affect physiological changes in P. denitrificans? The QS systems reported in several pathogenic bacteria affect pathogenicity by regulating iron uptake systems (1). In P. aeruginosa, iron uptake systems affect biofilm formation by interacting with the PQS system (13). In this study, we demonstrated that QS-dependent iron uptake systems play an important role in maintaining biofilm integrity. In Fig. 5, we show that iron was enriched in the EPS of biofilms formed by P. denitrificans. This finding is consistent with the results reported by Friese et al., who demonstrated that biofilms bind significant quantities of metals under natural conditions and serve as matrices for the precipitation of insoluble mineral phases (32). To investigate the expression of iron transport systems during biofilm formation, the HmuSTUV and FbpABC systems were selected as representatives of TonB-dependent and TonB-independent systems, and the mRNA levels of the two iron transporters in biofilms were measured by an RT-qPCR method. In the wild-type strain, during biofilm formation, the expression of hmu was “turned off” by the QS system, and the less effective but more energy-conserving iron uptake system Fbp was “turned on.” The iron uptake pattern most probably switched from Hmu to Fbp during biofilm formation to protect cells from toxic iron concentration in the EPS and thus maintain biofilm integrity. In contrast, in the QS-defective mutants, the expression of fbp and hmu was not regulated by the QS system, causing iron accumulation in the cells enveloped by the EPS. The accumulation of iron in cells is likely to induce Fenton reactions within the cells and lead to the generation of reactive oxygen species (ROS), resulting in the rapid dissolution of the biofilm (33).
Based on the above-mentioned conclusions, we proposed the following in vivo model. (i) Under culture conditions where iron concentrations are sufficient to meet the rapid prebiofilm growth and reproduction needs, in the early stage of bacterial growth, before biofilm formation, the acquisition of iron is dominated mainly by the TonB-dependent transport in P. denitrificans. (ii) As cell density increases and biofilm formation begins, with its concurrent iron enrichment, iron acquisition switches from TonB-dependent Hmu-type transport to more energy-efficient TonB-independent Fbp-type transport to avoid excessive iron damage to cells.
In conclusion, the presence of multiple iron uptake systems is a manifestation of the adaptation of microbes to their environment. Our analyses based on related data suggest that switching between different iron uptake systems is a form of collective decision-making and confers benefits to biofilm formation in the environment, providing a new theoretical basis for studying the adaptability of nonpathogenic microorganisms.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The plasmids and bacterial strains used are listed in Table 1. P. denitrificans PD1222 was a gift from David Schleheck (34). Agrobacterium tumefaciens KYC55 was provided by Huiming Zheng (35). The suicide plasmids pJQ200SK and pVIK112 were gifts from Zhongli Cui (36). P. denitrificans PD1222 and the QS-defective ΔpdeI and ΔpdeR mutants were grown in LB medium, and E. coli strains were grown in LB medium at 37°C. Ampicillin, kanamycin, gentamicin, chloramphenicol, and rifampin were added to the medium at concentrations of 100, 50, 30, 30, and 100 μg/ml, respectively. A. tumefaciens KYC55(pJZ372)(pJZ384)(pJZ410) was cultivated at 28°C and 200 rpm in A. tumefaciens (AT) medium containing 100 μg/ml spectinomycin, 100 μg/ml gentamicin, and 5 μg/ml tetracycline.
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid | Characteristic(s)a | Source or reference |
|---|---|---|
| Strains | ||
| P. denitrificans | ||
| PD1222 | Wild type, G−, Rifr | David Schleheck |
| ΔpdeI mutant | pdeI deletion mutant of PD1222 | This study |
| ΔpdeR mutant | pdeR deletion mutant of PD1222 | This study |
| PD0786 | PD1222 derivative with PpdeR-lacZ | This study |
| PD0787 | PD1222 derivative with PpdeI-lacZ | This study |
| PD4202 | PD1222 derivative with Phmu-lacZ | This study |
| PD1077 | PD1222 derivative with Pfbp-lacZ | This study |
| E. coli | ||
| DH5α | Host strain for cloning vectors | Lab stock |
| SM10λpir | Conjugation strain | Lab stock |
| BL21(DE3) | F− ompT hsdS(rB− mB−) gal dcm lacY1 (DE3) | Lab stock |
| A. tumefaciens | ||
| KYC55 | AHL bioassay strain | Huiming Zheng |
| Plasmids | ||
| For gene expression | ||
| pMD19T | T-A cloning vector | Lab stock |
| pET-30(a) | Expression vector, Kanr | Lab stock |
| pET-pdeI | pET-30(a) derivative carrying pdeI | This study |
| pET-pdeR | pET-30(a) derivative carrying pdeR | This study |
| For gene knockout | ||
| pJQ200SK | Gmr, Mob+, lacZα, SacB | Zhongli Cui |
| pJQ-pdeI | pdeI deletion construct in pJQ200SK | This study |
| pJQ-pdeR | pdeR deletion construct in pJQ200SK | This study |
| pRK2013 | Helper plasmid, mob+ tra+, Kanr | Zhongli Cui |
| For lacZ fusion | ||
| pVIK112 | lacZ transcriptional fusion vector, R6K origin | Zhongli Cui |
| pPD0787 | pVIK112 derivative, PpdeI-lacZ | This study |
| pPD0786 | pVIK112 derivative, PpdeR-lacZ | This study |
| pPD4202 | pVIK112 derivative, Phmu-lacZ | This study |
| pPD1077 | pVIK112 derivative, Pfbp-lacZ | This study |
G−, Gram negative; Rifr, rifampin resistant; Kanr, kanamycin resistant; Gmr, gentamicin resistant.
DNA manipulations.
Expression strains were constructed by standard methods and verified by DNA sequencing. DNA isolated from P. denitrificans PD1222 was used as the template for pdeI and pdeR gene amplification. The primers 5′-GGAATTCCATATGCAGACCACCACACTTTC-3′ (5′ end) and 5′-CCGCTCGAGGTGCATCTTGGCCGCCAG-3′ (3′ end) were used for pdeI amplification. The primers 5′-GGAATTCCATATGATGTCGTCTCGCGCGGAAAT-3′ (5′ end) and 5′-CCGCTCGAGCAGCAAGCGGTAATCCTTGG-3′ (3′ end) were used for pdeR amplification. The 5′-end primer included an NdeI restriction site, and the 3′-end primer included an XhoI restriction site (both underlined). The PCR conditions for the two genes were 5 min at 95°C, followed by 30 cycles of 30 s at 95°C, 30 s at 55°C, and 1 min at 72°C, with a final step of 10 min at 72°C. Amplified pdeI and pdeR genes were cloned into pET-30(a), according to the manufacturer's instructions (EMD Biosciences [Novagen]). The plasmids were transformed into E. coli by heat shock.
To construct the QS-defective ΔpdeI mutant strain, the regions 500 bp upstream (pdeItyb1) and 500 bp downstream (pdeItyb2) of pdeI were amplified using the primer pairs pdeI-TYB1-F/pdeI-TYB1-R and pdeI-TYB2-F/pdeI-TYB2-R, respectively (Table 2). The green fluorescent protein (gfp) gene was amplified using the primers gfp-F and gfp-R. We cloned pdeItyb1, gfp, and pdeItyb2 in that order into the plasmid pMD19T. The inserted fragment was then excised with SacI and XhoI to obtain the pdeItyb1-gfp-pdeItyb2 fusion. We cloned the fusion into the suicide plasmid pJQ200SK, yielding pJQ-pdeI, and the plasmid was then transferred into P. denitrificans by triparental mating. Colonies with single-recombination events were selected based on their resistance to gentamicin and rifampin on LB plates. Double-recombination events were selected after growth on medium containing 10% (wt/vol) sucrose. The construction of the ΔpdeR mutant was similar to that of the ΔpdeI mutant. The primers used are listed in Table 2.
TABLE 2.
Primers used for construction of QS-defective mutations and promoter-lacZ fusion
| Primer name | Primer sequence (5′ to 3′) |
|---|---|
| pdeI-TYB1-F | AAGGAAAAAAGCGGCCGCCAACGCTACGACGGCTTCAC |
| pdeI-TYB1-R | GACTAGTTCAAGGAATACATGCCGCAGCGC |
| pdeI-TYB2-F | AAAACTGCAGGAACGCCGCCGTGTGCATTT |
| pdeI-TYB2-R | CCGCTCGAGCCCGAAAAGATCGCCGAGAT |
| pdeR-TYB1-F | AAGGAAAAAAGCGGCCGCTAAGCGATGGTATGGGCCCG |
| pdeR-TYB1-R | GACTAGTTCATCGAAAAGCCCCAGGCGAT |
| pdeR-TYB2-F | AAAACTGCAGTTCACGAGATCACGGAGCCG |
| pdeR-TYB2-R | CCGCTCGAGATAGGAATACATGCCGCAGC |
| gfp-F | GACTAGTACCAAGAACCCCAGGAGAAGACCATGAGTAAAGGAGAAGAACTTTTCAC |
| gfp-R | AAAACTGCAGCTATTTGTATAGTTCATCCATGCC |
| pdeI-F | GGAATTCCCACGGACGACATGGAGT |
| pdeI-R | GCTCTAGAGCGCGGTGTCGAAAAGGTCT |
| pdeR-F | GGAATTCGGCGGTTATGTTCGCATCAT |
| pdeR-R | GCTCTAGAGAAAAGCCCCAGGCGATGGT |
| hmu-R | GCTCTAGAATCTCGGGGTCCACGGTCAT |
| hmu-R | GCTCTAGAATCTCGGGGTCCACGGTCAT |
| fbp-F | GGAATTCGCATCGGGTGCCTGATCGGC |
| fbp-R | GCTCTAGATCAGCGATCAGCTCGCGCAA |
Fusions of the probable promoter regions for the P. denitrificans PD1222 homologs of hmu (pden_4202 to pden_4205), fbp (pden_1077 to pden_1079), pdeI (pden_0787), and pdeR (pden_0786) to a promoterless E. coli lacZ β-galactosidase gene were created in the plasmid pVIK112 (37). An intergenic region upstream of pden_4202 containing the 5′ end of the gene was PCR amplified using the primers hmu-F and hmu-R, given in Table 2. The hmu-F primer included an EcoRI restriction site, and the hmu-R primer included an XbaI restriction site. The PCR product was digested with EcoRI and XbaI, and the resulting fragment was ligated into a similarly digested pVIK112 plasmid to create pPD4202, which was confirmed by sequencing. The regions upstream of the fbp and pdeR coding sequences were similarly PCR amplified using the primer pairs fbp-F/fbp-R and pdeR-F/pdeR-R, respectively, and then subcloned into pVIK112, creating plasmids pPD1077 and pPD0786. The internal fragment of pdeI, excluding the 5′ end of the gene, was PCR amplified using the primer pair pdeI-F/pdeI-R (Table 2), and the resulting fragment was ligated into a similarly digested pVIK112 plasmid to create pPD0787. These pPD constructs were transformed into P. denitrificans PD1222 by triparental mating. Colonies with recombination events were selected based on their resistance to kanamycin and rifampin on LB plates, resulting in PD4202, PD1077, PD0786, and PD0787 mutants.
Bioassays of AHLs.
AHL bioactivity was determined by measuring the β-galactosidase bioactivity of the ultrasensitive AHL biosensor strain A. tumefaciens KYC55 (38). Cultures of recombinant E. coli BL21(DE3)(pET-pdeI), wild-type PD1222, and ΔpdeI and ΔpdeR mutants were centrifuged at 5,000 × g for 5 min to pellet the cells. A 0.22-μm syringe filter was used to filter assay supernatants, and the cell-free culture supernatants were stored at −20°C. A. tumefaciens KYC55 (5 × 107) was inoculated into 2 ml of AT culture medium containing 200 μl of supernatant from the growth of recombinant E. coli. The supernatant of the E. coli strain carrying pET-30(a) without the pdeI gene was used as a negative control. The OD600 of each sample was recorded after approximately 10 h at 28°C, and 200 μl of the supernatant of the AT culture medium was then combined with 0.8 ml of Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, and 50 mM 2-mercaptoethanol [pH 7.0]). Two drops of 0.05% SDS solution and 3 drops of chloroform were added to this solution in 2-ml microcentrifuge tubes. The samples were vortexed vigorously for 10 s, 0.1 ml of 4 mg/ml ortho-nitrophenyl-β-d-galactopyranoside (ONPG) was added, and the samples were then placed into a 30°C water bath for 10 min. Reactions were stopped by the addition of 0.6 ml of 1 M Na2CO3. Cell debris were removed by centrifugation for 3 min at 16,000 × g and room temperature, and the OD420 of the supernatant was then measured. β-Galactosidase units were calculated according to the following equation: Miller units = (1,000 × OD420)/(OD600 × 10 × 0.2), where the OD420 was read from the supernatant of the reaction mixture.
LC-MS analysis of AHLs.
The supernatants of recombinant E. coli strains were extracted twice with equal volumes of acidified ethyl acetate (EtAc) containing 0.2% glacial acetic acid and finally dried by a rotary evaporator. The extracts and AHL standard were reconstituted in HPLC-grade acetonitrile. AHL profiling was confirmed by an HPLC-MS system using a C18 reverse-phase column (5 μm by 250 mm by 4.6 mm) coupled with positive-ion electrospray ionization (ESI)-mass spectrometry (MS), and compounds were eluted with a linear gradient of acetonitrile in water (10 to 70%) at a flow rate of 1 ml/min. The retention times and spectral properties of samples whose ESI spectra (m/z range, 50 to 400) exhibited a fragment product at m/z 102 were compared to those of the corresponding synthetic AHL standards (27).
Purification of PdeR protein.
E. coli BL21(DE3) cells containing the plasmid pET-pdeR were grown in 900 ml of LB broth with kanamycin at 37°C to an OD600 of 0.5. The temperature was lowered to 16°C, and 20 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) was added. After 16 h of agitation at 16°C, cells were harvested by centrifugation at 8,000 × g for 10 min at 4°C. The cells were suspended in 10 ml of binding buffer (20 mM Tris-HCl, 500 mM NaCl, and 5 mM imidazole [pH 7.9]) and lysed by sonication. The lysate was centrifuged at 13,000 × g for 30 min, and the resulting supernatant was fractionated by nickel-nitrilotriacetic acid (Ni-NTA) agarose column chromatography. The bound protein was washed with a buffer containing 20 mM Tris-HCl and 500 mM NaCl at pH 7.9. Proteins were eluted by increasing concentrations of imidazole. Fractions containing His-tagged PdeR were identified by SDS-PAGE.
Assay of PdeR-AHL interaction by fluorescence spectroscopy.
Fluorescence measurements were performed with a Shimadzu RF-5301PC spectrofluorophotometer. All experiments were conducted in 50 mM Tris (pH 7.5). Purified PdeR protein was used for all experiments. The PdeR protein and relevant AHLs were reacted for 30 min at 30°C before measurements were made. Spectra were recorded between 290 and 480 nm, with an excitation wavelength of 284 nm and a scan speed of 0.8 nm per second. To avoid errors caused by sample dilution, we performed autoinducer titrations by adding 1 μl of suitable stock solutions (39).
Transcriptional profiling.
P. denitrificans RNAs were isolated and purified with an RNeasy mini system (Qiagen, Hilden, Germany). Triplicate cultures of P. denitrificans PD1222, ΔpdeI mutant, and ΔpdeR mutant cultures were grown overnight in LB medium under aerobic conditions. When the OD600 reached ∼3.0, cells were harvested, and total RNAs were extracted. The quality of purified RNAs was analyzed by electrophoresis and quantified by a NanoDrop 1000 spectrophotometer (NanoDrop Technologies). The RNA was subjected to Solexa/Illumina sequencing at Beijing Auwigene Tech.
Raw data were generated and filtered by removing reads that contained possible sequencing errors. After RNA sequencing, raw reads were mapped to the downloaded reference genome sequences of P. denitrificans PD1222 using the BWA program. SAM files generated by mapping were converted into BAM files, which are in binary, and the BAM files were then sorted by chromosomal coordinates using the SAMtools program. Mapped reads per annotated gene (total, 5,134 genes) were counted by Bam-readcount. The relative transcript abundance was measured in reads per kilobase of exon per million mapped sequence reads (RPKM) (40).
Reverse transcriptase PCR and RT-qPCR analyses.
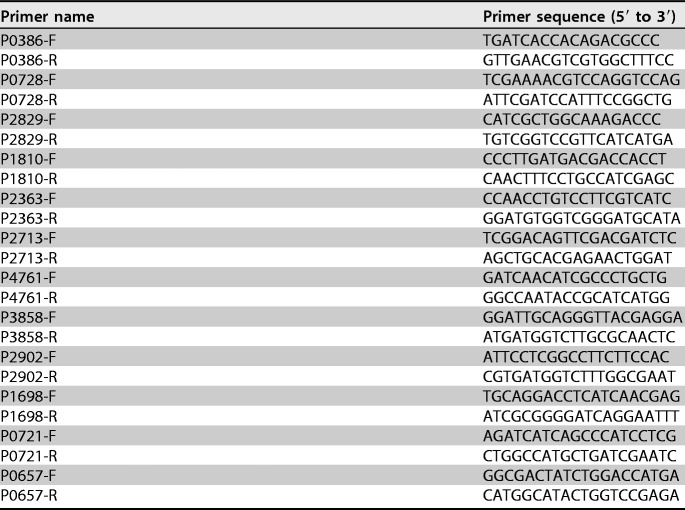
Twelve genes that had been identified as up- or downregulated by RNA-Seq analysis were selected, and quantitative real-time PCR (RT-qPCR) was carried out to confirm the gene expression changes of these 12 genes from the RNA-Seq results. PCR primers were designed using Primer 5.0 software and are listed in Table 3. Immunofluorescence analysis was performed with SYBR green master mix using a 7500 real-time PCR system, as previously described (41). Relative gene expression was calculated by the 2−ΔΔCT method. All reactions were carried out in triplicate.
TABLE 3.
Primers used for RT-qPCR
Physiological and biochemical iron uptake experiments.
The growth curves of the wild type and QS-defective mutants were detected in LB medium containing different types of iron. The original iron in the medium was removed by 2,2′-dipyridyl. By adding increasing concentrations of dipyridyl to the LB medium, we determined that the minimum concentration of dipyridyl that could remove iron from the LB medium was 150 nM (data not shown). Then, 50 nm iron dicitrate, Fe2O3, and heme were added to the medium separately, and the growth curves were determined by the OD600.
Gel electrophoresis mobility shift assay.
Two pairs of double-stranded nucleotide sequences, one from the promoter region of the hmu (pden_4202 to pden_4205) operon and one from that of the fbp (pden_1077 to pden_1079) operon, were synthesized and labeled with biotin for chemiluminescence detection. The two fragments contained regions 1 to 50 bp upstream of their respective methionine start codons. For specific competitor DNA, we used the 50-bp mutated probe. The sequences of the probes are listed in Table 4. Purified PdeR-His (100 nM) was incubated with 1 nM labeled DNA in binding buffer [10 mM Tris-HCl (pH 7.6), 10 mM KCl, 0.5 mM EDTA, 1 mM dithiothreitol, 5% (vol/vol) glycerol, and 1 μg/μl poly(dI-dC)] with or without C16-HSL (1 μM) for 20 min at 30°C. The mixtures were size fractionated in a nondenaturing 6% polyacrylamide gel and transferred to nitrocellulose membranes, at which point the biotin-labeled probes were detected by streptavidin-horseradish peroxidase chemiluminescence.
TABLE 4.
Sequence of the hot and cold probes used in the EMSA experiment
| Probe namea | Probe sequence (5′ to 3′)b |
|---|---|
| fbp-H | CAAGGCAGTGGCTGGCTGAATGCGCCAGCATCGTGCATCCGAACCGCAAG |
| fbp-C | CAAGGCAGTGGCTGGATTTTACAGAACGGCCCGTGCATCCGAACCGCAAG |
| hmu-H | CCTTGATGTCGATCAGCGGCGTGCCGTCCAGGCAGTCGAGCCCGCGCACG |
| hmu-C | CCTTGATGTCGATCTTGGGCCGCACGGGCGAGCGCAGGAGCCCGCGCACG |
H, hot probe; C, cold probe.
Underlining indicates mutated sequence in the cold probe.
Biofilm formation.
Biofilm formation was assessed as previously described (42). Cultures started at an initial OD660 of 0.05 and were incubated in polystyrene 96-well microtiter plates for 24 h at 30°C. To simulate the flow model under natural conditions, after 4 h of culturing (the lag phase), the culture fluid was replaced with sterilized fresh medium every half hour. Biofilms were stained with 0.1% (wt/vol) crystal violet solution for 30 min. Then, 20% (vol/vol) dimethyl sulfoxide in ethanol was added to each well and the absorbance at 575 nm measured to quantify biofilm formation.
Concentration measurements of iron in the EPS and cells enveloped in the EPS.
The EPS of the biofilms was extracted as described previously (43). Biofilms formed by P. denitrificans were centrifuged for 10 min at 4,000 × g, and the supernatant was collected and marked as the soluble EPS. The sediment was suspended in 1 ml of 0.85% NaCl and kept at 70°C for 30 min. The solution was centrifuged for 20 min at 20,000 × g, and the supernatant was collected and marked as the bond EPS. Soluble EPS and bond EPS were mixed together and filtered through a 0.22-μm-pore-size membrane. The extracted EPS and the cells enveloped within it were freeze-dried and weighed. The biosamples were then digested by the addition of 5 ml of nitric acid and subsequent heating at 300°C for 5 h, until the liquid became completely clear. The digested solution was transferred to a 50-ml volumetric flask and diluted with 5% HCl to a volume of 50 ml. Iron concentrations were determined using an inductively coupled plasma optical emission spectrometer (ICP-OES; Thermo iCAP 6000), and the relative content of iron in the biofilms was calculated (44).
Data analysis.
All data were analyzed and graphed by Origin 9.0 (OriginLab Corporation). Significance analysis was completed in SPSS (Statistical Product and Service Solutions) using one-way analysis of variance (ANOVA) (45).
This study does not involve human participants or animals.
Accession number(s).
The raw data were submitted to the National Center for Biotechnology Information Sequence Read Archive (NCBI SRA) database (https://www.ncbi.nlm.nih.gov/sra). The accession numbers are SRP128965 and SRP128852.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (grant XDB15030101), the National Natural Science Foundation of China (grants 41501250, 21377157, and 31670507), and the National Key Research and Development Program of China (grants 2016YFC0500401 and 2017YFC0505803-01).
We declare no competing interests.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00865-18.
REFERENCES
- 1.Oglesby AG, Farrow JM III, Lee J-H, Tomaras AP, Greenberg EP, Pesci EC, Vasil ML. 2008. The influence of iron on Pseudomonas aeruginosa physiology a regulatory link between iron and quorum sensing. J Biol Chem 283:15558–15567. doi: 10.1074/jbc.M707840200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Touati D. 2000. Iron and oxidative stress in bacteria. Arch Biochem Biophys 373:1–6. doi: 10.1006/abbi.1999.1518. [DOI] [PubMed] [Google Scholar]
- 3.Caza M, Kronstad JW. 2013. Shared and distinct mechanisms of iron acquisition by bacterial and fungal pathogens of humans. Front Cell Infect Microbiol 3:80. doi: 10.3389/fcimb.2013.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moeck GS, Coulton JW. 1998. TonB-dependent iron acquisition: mechanisms of siderophore-mediated active transport. Mol Microbiol 28:675–681. doi: 10.1046/j.1365-2958.1998.00817.x. [DOI] [PubMed] [Google Scholar]
- 5.Wyckoff EE, Mey AR, Leimbach A, Fisher CF, Payne SM. 2006. Characterization of ferric and ferrous iron transport systems in Vibrio cholerae. J Bacteriol 188:6515–6523. doi: 10.1128/JB.00626-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braun V. 1997. Avoidance of iron toxicity through regulation of bacterial iron transport. Biol Chem 378:779–786. [PubMed] [Google Scholar]
- 7.Cornelis P, Matthijs S, Van Oeffelen L. 2009. Iron uptake regulation in Pseudomonas aeruginosa. Biometals 22:15–22. doi: 10.1007/s10534-008-9193-0. [DOI] [PubMed] [Google Scholar]
- 8.Miller MB, Bassler BL. 2001. Quorum sensing in bacteria. Annu Rev Microbiol 55:165–199. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- 9.James CE, Hasegawa Y, Park Y, Yeung V, Tribble GD, Kuboniwa M, Demuth DR, Lamont RJ. 2006. LuxS involvement in the regulation of genes coding for hemin and iron acquisition systems in Porphyromonas gingivalis. Infect Immun 74:3834–3844. doi: 10.1128/IAI.01768-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fong KP, Gao L, Demuth DR. 2003. luxS and arcB control aerobic growth of Actinobacillus actinomycetemcomitans under iron limitation. Infect Immun 71:298–308. doi: 10.1128/IAI.71.1.298-308.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Banin E, Vasil ML, Greenberg EP. 2005. Iron and Pseudomonas aeruginosa biofilm formation. Proc Natl Acad Sci U S A 102:11076–11081. doi: 10.1073/pnas.0504266102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bredenbruch F, Geffers R, Nimtz M, Buer J, Häussler S. 2006. The Pseudomonas aeruginosa quinolone signal (PQS) has an iron-chelating activity. Environ Microbiol 8:1318–1329. doi: 10.1111/j.1462-2920.2006.01025.x. [DOI] [PubMed] [Google Scholar]
- 13.Yang L, Barken KB, Skindersoe ME, Christensen AB, Givskov M, Tolker-Nielsen T. 2007. Effects of iron on DNA release and biofilm development by Pseudomonas aeruginosa. Microbiology 153:1318–1328. doi: 10.1099/mic.0.2006/004911-0. [DOI] [PubMed] [Google Scholar]
- 14.Beijerinck M, Minkman DCJ. 1910. Bildung und Verbrauch von Stickoxydul durch Bakterien. Zentralbl Bakteriol Parasitenkd Abt II 25:30–63. [Google Scholar]
- 15.Kelly DP, Euzeby JP, Goodhew CF, Wood AP. 2006. Redefining Paracoccus denitrificans and Paracoccus pantotrophus and the case for a reassessment of the strains held by international culture collections. Int J Syst Evol Microbiol 56:2495–2500. doi: 10.1099/ijs.0.64401-0. [DOI] [PubMed] [Google Scholar]
- 16.John P, Whatley F. 1975. Paracoccus denitrificans and the evolutionary origin of the mitochondrion. Nature 254:495–498. doi: 10.1038/254495a0. [DOI] [PubMed] [Google Scholar]
- 17.Stroh A, Anderka O, Pfeiffer K, Yagi T, Finel M, Ludwig B, Schägger H. 2004. Assembly of respiratory complexes I, III, and IV into NADH oxidase supercomplex stabilizes complex I in Paracoccus denitrificans. J Biol Chem 279:5000–5007. doi: 10.1074/jbc.M309505200. [DOI] [PubMed] [Google Scholar]
- 18.Luck SN, Turner SA, Rajakumar K, Sakellaris H, Adler B. 2001. Ferric dicitrate transport system (Fec) of Shigella flexneri 2a YSH6000 is encoded on a novel pathogenicity island carrying multiple antibiotic resistance genes. Infect Immun 69:6012–6021. doi: 10.1128/IAI.69.10.6012-6021.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fecker L, Braun V. 1983. Cloning and expression of the fhu genes involved in iron(III)-hydroxamate uptake by Escherichia coli. J Bacteriol 156:1301–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zawadzka AM, Kim Y, Maltseva N, Nichiporuk R, Fan Y, Joachimiak A, Raymond KN. 2009. Characterization of a Bacillus subtilis transporter for petrobactin, an anthrax stealth siderophore. Proc Natl Acad Sci U S A 106:21854–21859. doi: 10.1073/pnas.0904793106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamamoto H, Uchiyama S, Nugroho FA, Sekiguchi J. 1997. Cloning and sequencing of a 35.7 kb in the 70°–73° region of the Bacillus subtilis genome reveal genes for a new two-component system, three spore germination proteins, an iron uptake system and a general stress response protein. Gene 194:191–199. doi: 10.1016/S0378-1119(97)00130-3. [DOI] [PubMed] [Google Scholar]
- 22.Hornung JM, Jones HA, Perry RD. 1996. The hmu locus of Yersinia pestis is essential for utilization of free haemin and haem-protein complexes as iron sources. Mol Microbiol 20:725–739. doi: 10.1111/j.1365-2958.1996.tb02512.x. [DOI] [PubMed] [Google Scholar]
- 23.Adhikari P, Berish SA, Nowalk AJ, Veraldi KL, Morse SA, Mietzner TA. 1996. The fbpABC locus of Neisseria gonorrhoeae functions in the periplasm-to-cytosol transport of iron. J Bacteriol 178:2145–2149. doi: 10.1128/jb.178.7.2145-2149.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schaefer AL, Taylor TA, Beatty JT, Greenberg EP. 2002. Long-chain acyl-homoserine lactone quorum-sensing regulation of Rhodobacter capsulatus gene transfer agent production. J Bacteriol 184:6515–6521. doi: 10.1128/JB.184.23.6515-6521.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang L, Feng Z, Wang X, Wang X, Zhang X. 2010. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26:136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- 26.Bottomley MJ, Muraglia E, Bazzo R, Carfì A. 2007. Molecular insights into quorum sensing in the human pathogen Pseudomonas aeruginosa from the structure of the virulence regulator LasR bound to its autoinducer. J Biol Chem 282:13592–13600. doi: 10.1074/jbc.M700556200. [DOI] [PubMed] [Google Scholar]
- 27.Gao J, Ma A, Zhuang X, Zhuang G. 2014. An N-acyl homoserine lactone synthase in the ammonia-oxidizing bacterium Nitrosospira multiformis. Appl Environ Microbiol 80:951–958. doi: 10.1128/AEM.03361-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malott RJ, Baldwin A, Mahenthiralingam E, Sokol PA. 2005. Characterization of the cciIR quorum-sensing system in Burkholderia cenocepacia. Infect Immun 73:4982–4992. doi: 10.1128/IAI.73.8.4982-4992.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu J, Winans SC. 2001. The quorum-sensing transcriptional regulator TraR requires its cognate signaling ligand for protein folding, protease resistance, and dimerization. Proc Natl Acad Sci U S A 98:1507–1512. doi: 10.1073/pnas.98.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng H, Zhong Z, Lai X, Chen W-X, Li S, Zhu J. 2006. A LuxR/LuxI-type quorum-sensing system in a plant bacterium, Mesorhizobium tianshanense, controls symbiotic nodulation. J Bacteriol 188:1943–1949. doi: 10.1128/JB.188.5.1943-1949.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fuqua WC, Winans SC, Greenberg EP. 1994. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J Bacteriol 176:269–275. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Friese K, Mages M, Wendt-Potthoff K, Neu T. 1997. Determination of heavy metals in biofilms from the River Elbe by total-reflection X-ray fluorescence spectrometry. Spectrochim Acta B 52:1019–1025. doi: 10.1016/S0584-8547(96)01633-3. [DOI] [Google Scholar]
- 33.Gosselin F, Madeira LM, Juhna T, Block JC. 2013. Drinking water and biofilm disinfection by Fenton-like reaction. Water Res 47:5631–5638. doi: 10.1016/j.watres.2013.06.036. [DOI] [PubMed] [Google Scholar]
- 34.Felux A-K, Denger K, Weiss M, Cook AM, Schleheck D. 2013. Paracoccus denitrificans PD1222 utilizes hypotaurine via transamination followed by spontaneous desulfination to yield acetaldehyde and, finally, acetate for growth. J Bacteriol 195:2921–2930. doi: 10.1128/JB.00307-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng H, Mao Y, Zhu Q, Ling J, Zhang N, Naseer N, Zhong Z, Zhu J. 2015. The quorum sensing regulator CinR hierarchically regulates two other quorum sensing pathways in ligand-dependent and-independent fashions in Rhizobium etli. J Bacteriol 197:1573–1581. doi: 10.1128/JB.00003-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shen W, Liu W, Zhang J, Tao J, Deng H, Cao H, Cui Z. 2010. Cloning and characterization of a gene cluster involved in the catabolism of p-nitrophenol from Pseudomonas putida DLL-E4. Bioresour Technol 101:7516–7522. doi: 10.1016/j.biortech.2010.04.052. [DOI] [PubMed] [Google Scholar]
- 37.Kalogeraki VS, Winans SC. 1997. Suicide plasmids containing promoterless reporter genes can simultaneously disrupt and create fusions to target genes of diverse bacteria. Gene 188:69–75. doi: 10.1016/S0378-1119(96)00778-0. [DOI] [PubMed] [Google Scholar]
- 38.Zhu J, Chai Y, Zhong Z, Li S, Winans SC. 2003. Agrobacterium bioassay strain for ultrasensitive detection of N-acylhomoserine lactone-type quorum-sensing molecules: detection of autoinducers in Mesorhizobium huakuii. Appl Environ Microbiol 69:6949–6953. doi: 10.1128/AEM.69.11.6949-6953.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Minogue TD, Trebra MWv Bernhard F, von Bodman SB. 2002. The autoregulatory role of EsaR, a quorum-sensing regulator in Pantoea stewartii ssp. stewartii: evidence for a repressor function. Mol Microbiol 44:1625–1635. doi: 10.1046/j.1365-2958.2002.02987.x. [DOI] [PubMed] [Google Scholar]
- 40.Kim S, Park J, Choi O, Kim J, Seo Y-S. 2014. Investigation of quorum sensing-dependent gene expression in Burkholderia gladioli BSR3 through RNA-seq analyses. J Microbiol Biotechnol 24:1609–1621. doi: 10.4014/jmb.1408.08064. [DOI] [PubMed] [Google Scholar]
- 41.Dorak MT. 2006. Real-time PCR. Taylor & Francis Group, New York, NY. [Google Scholar]
- 42.O'Toole GA, Kolter R. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol 28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 43.D'Abzac P, Bordas F, Van Hullebusch E, Lens PN, Guibaud G. 2010. Extraction of extracellular polymeric substances (EPS) from anaerobic granular sludges: comparison of chemical and physical extraction protocols. Appl Microbiol Biotechnol 85:1589–1599. doi: 10.1007/s00253-009-2288-x. [DOI] [PubMed] [Google Scholar]
- 44.Michalak I, Chojnacka K, Marycz K. 2011. Using ICP-OES and SEM-EDX in biosorption studies. Mikrochim Acta 172:65–74. doi: 10.1007/s00604-010-0468-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.So YC, Sen PK. 1982. M-estimators based repeated significance tests for one-way ANOVA with adaptation to multiple comparisons. Seq Anal 1:101–119. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.