ABSTRACT
The concept of Helicobacter pylori biofilm formation is relatively new. To help provide a foundation for future biofilm studies, we characterized the biofilm formation ability of a common H. pylori lab strain, G27. The goal of this study was to evaluate biofilm formation by G27 in response to common culture conditions and to explore the biofilm matrix. Our results indicate that while various types of growth media did not dramatically affect biofilm formation, surface selection had a significant effect on the final biofilm mass. Furthermore, enzymatic assays and confocal microscopy revealed that proteins appear to be the primary structural component of the H. pylori extracellular matrix; extracellular DNA (eDNA) and polysaccharides were also present but appear to play a secondary role. Finally, we found that two well-characterized antibiofilm cationic peptides differentially affected early and late-stage biofilms. Together these results provide interesting avenues for future investigations that will seek to understand H. pylori biofilm formation.
IMPORTANCE The study of H. pylori biofilm formation is still in its infancy. As such, there is great variability in how biofilm assays are performed across labs. While several groups have begun to investigate factors that influence H. pylori biofilm formation, it is not yet understood how H. pylori biofilm formation may vary based on commonly used conditions. These inconsistencies lead to difficulties in interpretation and comparison between studies. Here, we set out to characterize biofilm formation by a commonly available lab strain, G27. Our findings provide novel insight into optimal biofilm conditions, the biofilm matrix, and possible mechanisms to block or disrupt biofilm formation.
KEYWORDS: Helicobacter pylori, antimicrobial peptides, biofilms
INTRODUCTION
Helicobacter pylori is a Gram-negative pathogen that colonizes the gastric mucosa of humans and nonhuman primates (1). An estimated 50% of the world's population is colonized (2), and unless treated, individuals remain colonized for life (3). Though infection with H. pylori is most often asymptomatic, H. pylori-infected individuals are at an increased risk for development of gastritis, peptic ulcers, and gastric cancer (4). Given that gastric cancer is the fourth leading cause of cancer-related death (5), research on this bacterium remains of significant interest.
Multiple research groups have recently shown that H. pylori has the ability to form biofilms (6–9). A biofilm is defined as a collection of cells that are adhered to a surface and encased by an extracellular matrix composed of polymeric substances created by the bacteria (10). The polymeric substances found in the matrix of various taxa include assorted proteins, polysaccharides, and extracellular DNA (eDNA) (11). The composition of the extracellular matrix is diverse, varying between species and even between strains (11). Though the individual cells might compete for resources like space, existence in a biofilm gives advantages for the bacteria found within; these advantages include protection from the stresses of the environment, access to nutrients, etc. (12). In particular, pathogenic bacteria found in biofilms display increased (adaptive) resistance to antibiotic treatment even if the bacteria do not possess traditional resistance mechanisms to the antibiotics being used (13). Equally important is that bacteria found in biofilms are often resistant to clearance by the immune system (14–17), leading to the establishment of chronic infections. Indeed, H. pylori biofilms have been found in biopsy specimens from patients for whom treatment has failed (18), suggesting that the ability to form biofilms might contribute to treatment failure. Thus, the study of H. pylori biofilms and the mechanisms of their formation and maintenance may lead to a better understanding of how to combat infections by this important pathogen.
Little is currently known about the molecular mechanisms that drive H. pylori biofilm formation. However, with increased awareness that H. pylori can form biofilms comes increased interest in the genes involved with biofilm formation and in their regulation. Indeed, recent studies have examined the genes that contribute to biofilm formation (19, 20). Given the relatively small genome of H. pylori (21, 22), there are probably many genes that have been investigated in other contexts that also play a role in biofilm formation. For example, the ArsRS two-component system was previously characterized in the context of its response to acidic pH (23, 24). This two-component system is also involved in biofilm formation; both deletion of arsS and the creation of a nonphosphorylatable form of ArsR (ArsR-D52N) result in a hyperbiofilm formation phenotype. Notably, each of these strains shows visually distinct differences in biofilm architecture when viewed by scanning electron microscopy (SEM), perhaps suggesting that the change in the composition of the extracellular matrix may not be uniform across the two mutant strains (9). Clearly, this two-component system appears to be important in the regulation of biofilm formation in H. pylori. Given the small regulatory network of H. pylori (25), there are likely additional regulatory factors that contribute to H. pylori biofilm formation.
As investigations into H. pylori's ability to live in a biofilm-associated state expand, one obstacle in comparing studies is the diversity in how assays are performed in different laboratories. In some instances, groups have focused on various clinical strains (26) or have altered medium additives, such as fetal bovine serum (FBS) (20). For many bacterial species that form biofilms, even small alterations in how the assay is performed can lead to large changes in biofilm formation (27–29). Therefore, the H. pylori field may benefit from an increased awareness of how differing conditions and assay variations affect H. pylori biofilm formation.
To help lay a foundation for future H. pylori biofilm investigations, we sought to characterize the biofilm-forming ability of a well-known H. pylori lab strain, G27, in standard rich laboratory media and on commonly used surfaces. Under these conditions, we characterized the extracellular matrix of the H. pylori biofilm via enzymatic assays and using confocal microscopy. Additionally, we tested the effects of two reported antibiofilm peptides during biofilm formation and on preformed biofilms. Together, these data help to build a more complete picture of H. pylori biofilm formation and help to expand the scientific communities' understanding of H. pylori biofilms.
RESULTS
Biofilm formation in G27 is unaffected by long-term serial passage or nutrient-rich medium type.
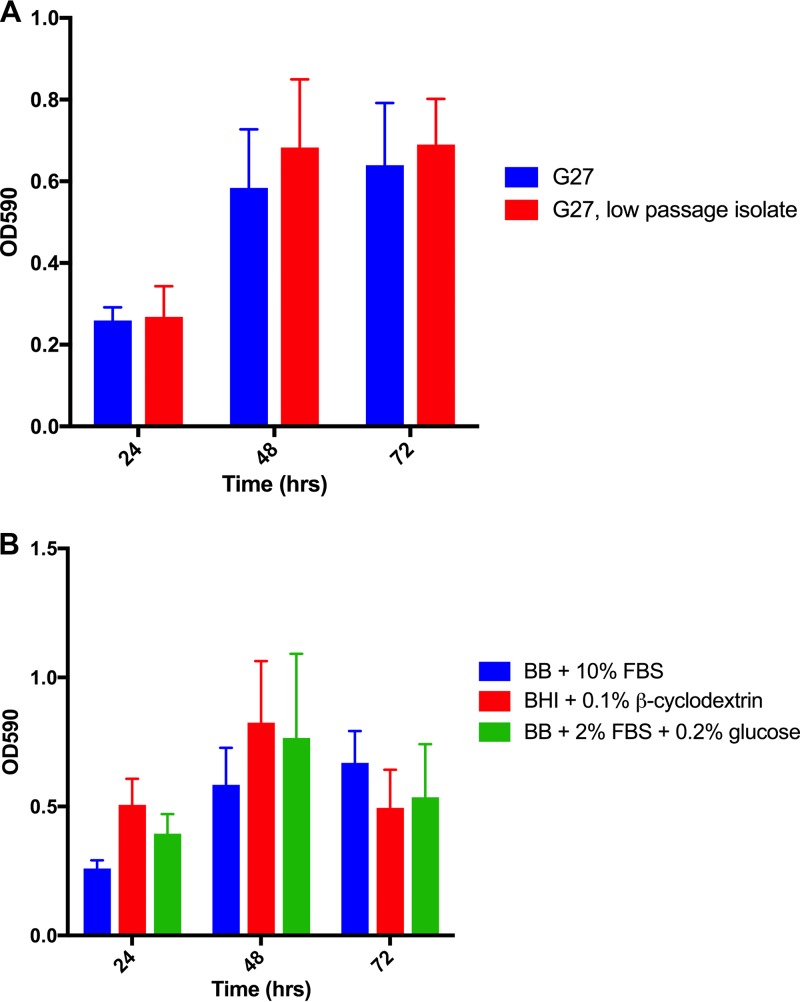
Previous work from our group demonstrated the ability of G27, a common laboratory strain, to form biofilm at the air-liquid interface under standard laboratory culture conditions (9). H. pylori is known for its frequent chromosomal rearrangement and high mutation rate (30), leading to the possibility that the biofilm-forming capability of the lab strain could have been altered due to long-term serial passage. To address this concern, we compared the biofilm-forming ability of our lab's particular strain, G27 (DSM 1), to that of a low-passage-number variant of strain G27 (DSM 359); DSM 359 represents the 12th passage of the original cultured G27 strain and represents one passage beyond the strain used for genomic sequencing of G27 (31). Despite the strain passages over the last 3 decades, we observed no appreciable difference in biofilm formation between DSM 1 and DSM 359 (Fig. 1A). This finding suggests that the biofilm phenotype observed with DSM 1 is not an artifact of long-term in vitro passage.
FIG 1.
Biofilm development in the strain G27 background. (A) Comparison between G27 (DSM 1) and an independent G27 low-passage-number isolate (DSM 359). (B) G27 (DSM 1) was grown in the indicated media. For each time point, each bar indicates the mean OD590 of solubilized crystal violet for three biological replicates at 24, 48, and 72 h; error bars display standard errors of the means.
Given the fastidious nature of H. pylori and the different media used for standard culture, we assessed biofilm formation using several of the most-common nutrient-rich H. pylori growth media (9, 26, 32). Despite variations in the medium compositions, DSM 1 formed similar levels of biofilm in each (Fig. 1B). Additionally, we examined the response to the addition of various H. pylori preferred carbon sources (33) to the media (see Fig. S1 in the supplemental material); again, H. pylori biofilm formation was similar in each supplemented medium. These data suggest that slight variations between standard H. pylori nutrient-rich growth media do not substantially affect H. pylori biofilm formation and indicate that our observation of biofilm formation is not an artifact of medium choice.
Substrate affects H. pylori biofilm formation.
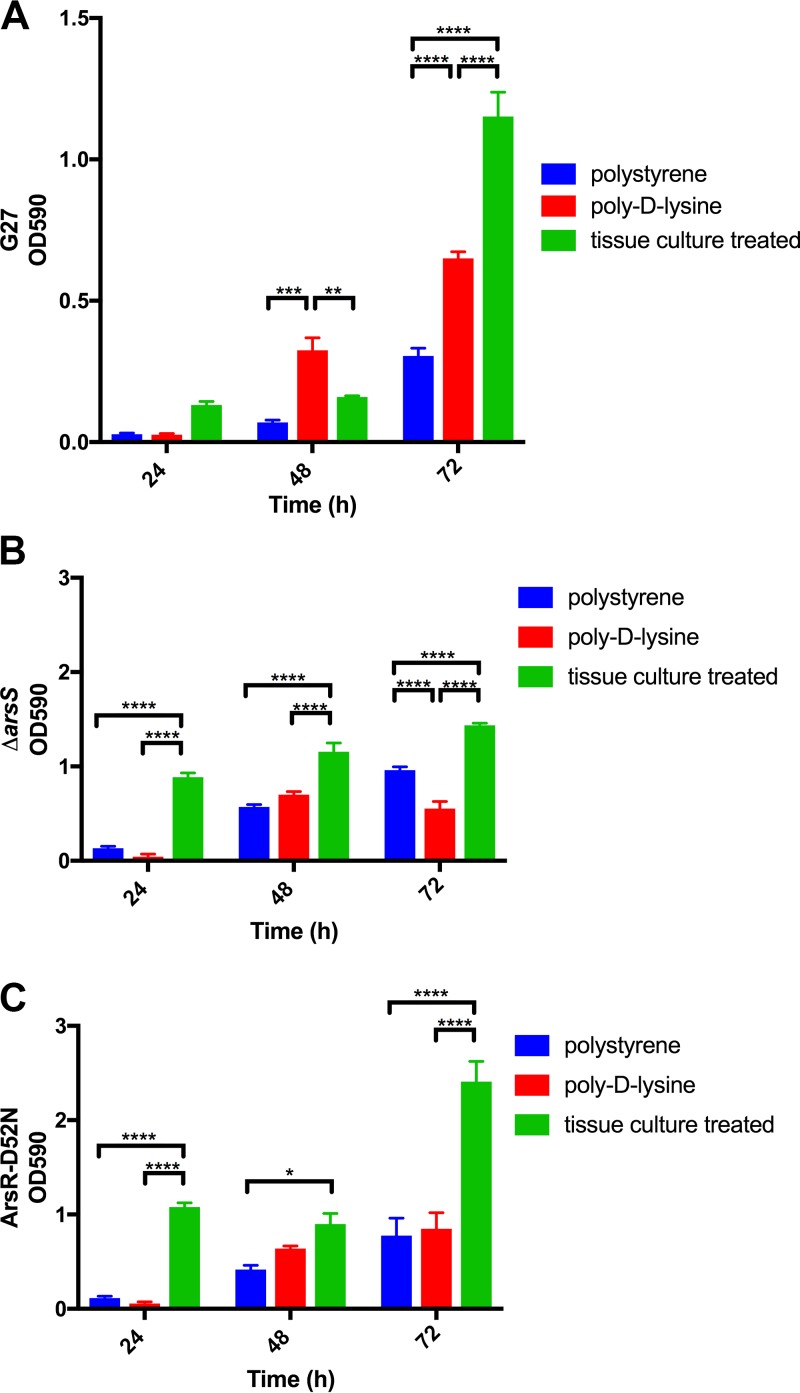
In addition to the medium selected, another commonly cited environmental condition that affects biofilm formation is the surface on which the biofilm is formed. In particular, precoating of a polystyrene plate with different substrates can affect how some bacterial biofilms adhere to the plate in an in vitro assay (34–36). However, it is unknown how H. pylori might change biofilm development in response to different surfaces. Thus, to assess the importance of surface to H. pylori biofilm development, we grew DSM 1 on three different substrates that each had distinct properties: untreated polystyrene (hydrophobic surface), poly-d-lysine-precoated polystyrene (hydrophilic, positive charge), or tissue culture-treated polystyrene (hydrophilic, negative charge) (Fig. 2). Additionally, since H. pylori strains containing mutations in the ArsRS two-component system have been shown to display a hyperbiofilm-forming phenotype (9), we also tested strains containing a ΔarsS mutation and an ArsR-D52N substitution in this assay to determine if biofilm formation by the hyperbiofilm-forming strains would be similarly affected by changes in the surface properties. We found that for all three tested strains, the most robust biofilms grew on tissue culture-treated plates (Fig. 2). In contrast, significantly less biofilm formation was observed for all three strains on either poly-d-lysine-precoated plates or untreated plates (Fig. 2). These data suggest that for G27, an ionic surface alone was not sufficient for maximal biofilm development. However, tissue culture treatment of the surface enhanced G27 biofilm development.
FIG 2.
Effects of alteration of the artificial substrate on H. pylori biofilm adherence. H. pylori strains G27 (A), PRINTER: this does not look like the standard uppercase Greek delta; please use standard delta as usual ΔarsS mutant (B), and ArsR-D52N mutant (C) were grown in 24-well plates with different surface properties: untreated polystyrene plate, poly-d-lysine-treated plate, and tissue culture-treated plate. The mean OD590 values for solubilized crystal violet, which are representative of the density of the biofilm, are shown; error bars show standard errors of the means. Each graph represents three biologically independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Composition of the H. pylori biofilm extracellular matrix.
The cells of a biofilm are encased within a matrix of their own creation; this keeps the cells adhered to each other and to the surface. Generally, the extracellular matrix of a biofilm formed by a particular species of bacteria is composed of an assortment of proteins, polysaccharides, and extracellular DNA (11). The composition of the extracellular matrix can be evaluated in several ways. For this study, we chose to take a two-pronged approach and used chemical/enzymatic disruption of the biofilm matrix as well as confocal microscopy combined with specific fluorescent probes to evaluate different components.
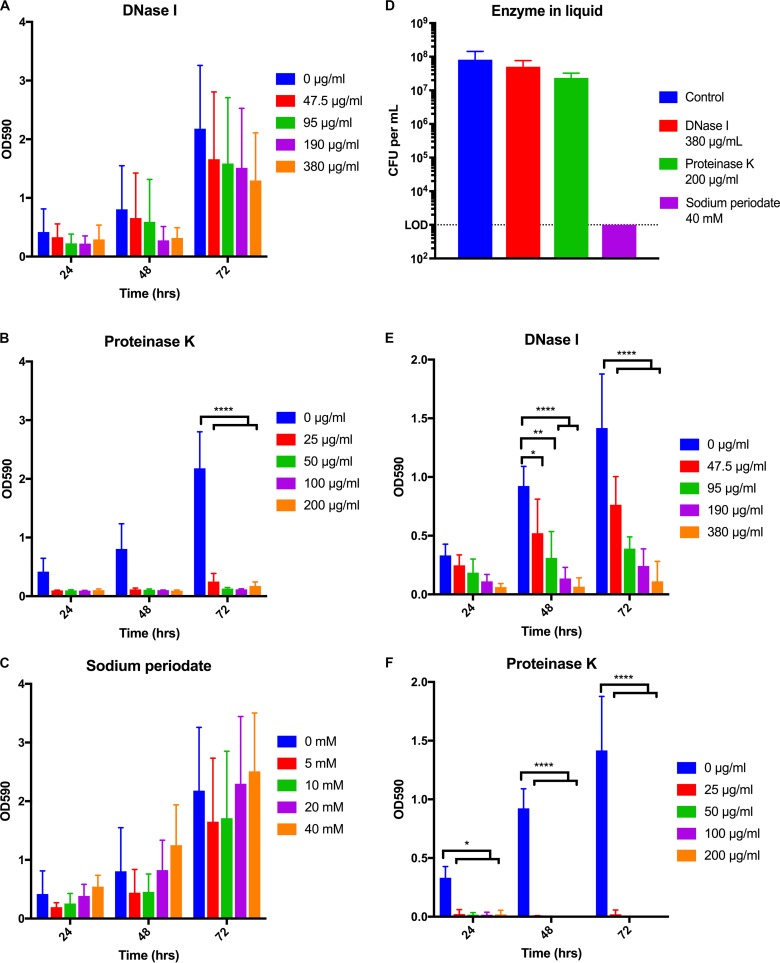
Identification of the components of the extracellular matrix of a biofilm via the addition of enzymes or chemicals is a well-known tactic (29, 37, 38). If incubation with a compound reduces the amount of biofilm in comparison to what is seen with the control, then the target of the utilized substance is likely an important part of the extracellular matrix. We individually assessed the effects of DNase I, proteinase K, and sodium periodate on preformed H. pylori biofilms using a range of concentrations for each of the substances (Fig. 3A to C). Though we observed a trend toward a dose-dependent decrease in total biofilm mass, incubation with DNase I did not cause a statistically significant reduction at any time or concentration tested (Fig. 3A). In contrast, treatment with proteinase K had the most-dramatic effect; biofilm was completely eliminated even at the lowest tested concentration (Fig. 3B). Conversely, incubation with sodium periodate to oxidize polysaccharides did not cause an observable reduction in biofilm mass at any of the time points or concentrations examined (Fig. 3C). Together, these data suggested that while eDNA may play a minor role, proteins constitute the integral structural component of the extracellular matrix of preformed H. pylori biofilms.
FIG 3.
Effects of DNase I, proteinase K, and sodium periodate on H. pylori biofilms. (A to C) Biofilms were grown for 24, 48, or 72 h. At the time points indicated on the x axis, biofilms were treated with DNase I (A), proteinase K (B), or sodium periodate (C) at the indicated concentrations. The biofilms were incubated with the enzymes/chemical for another 24 h at 37°C. (D) The viability of planktonic H. pylori was tested in response to the highest concentrations of DNase I, proteinase K, and sodium periodate utilized as shown in panels A to C. (E, F) DNase I (E) and proteinase K (F) were added to the growth media at the start of the H. pylori biofilm assays at the indicated concentrations. Biofilm development was monitored at 24, 48, or 72 h. The means for three biological replicates are indicated on the bar graph; error bars represent standard errors of the means. *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
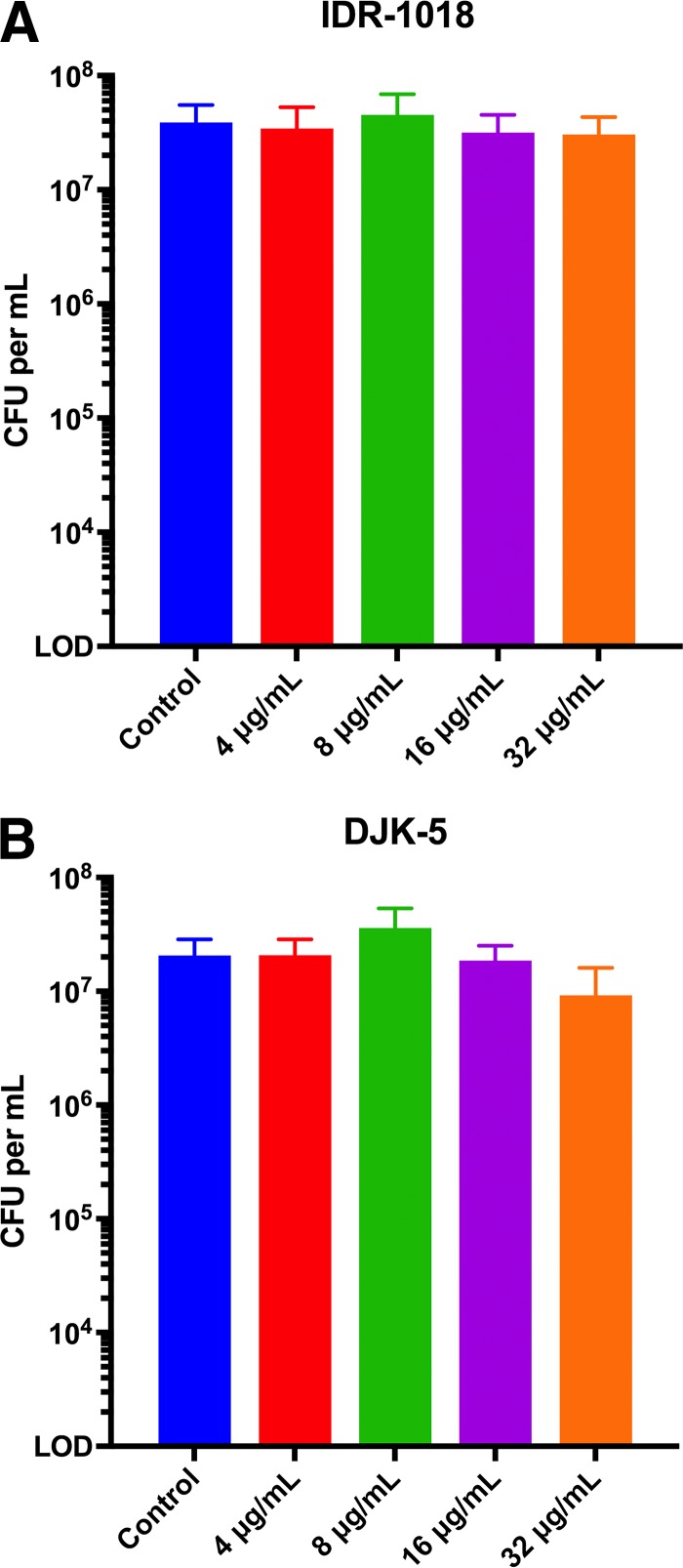
To ensure that observations regarding biofilm composition were not a result of cell death in the presence of the chosen additives, we also assessed the effects of each on the cell viability of planktonic H. pylori. We chose to test the highest concentrations used in the biofilm assays (Fig. 3D). Neither DNase I nor proteinase K altered H. pylori viability compared to an untreated control, demonstrating that the effects of the enzymes on biofilm disruption were not due to altered H. pylori cell viability. Conversely, treatment with sodium periodate reduced viability to below the limit of detection (Fig. 3D); this was true even when dramatically lower concentrations were assessed (see Fig. S2 in the supplemental material). Given that sodium periodate killed cells in liquid culture but did not affect the total mass of preformed biofilms, either H. pylori within the biofilm was protected against killing by sodium periodate, or the cells within the biofilm were killed but the biofilm remained intact.
It is commonly accepted that the composition of the biofilm extracellular matrix can change over time and that a component that is important at one stage of development might not be as necessary at other stages (39). Thus, we evaluated the effects of DNase I and proteinase K on H. pylori biofilm formation (Fig. 3E and F). We did not include sodium periodate in this analysis due to the exquisite sensitivity of H. pylori to this substance (Fig. 3D and S2). We found that the addition of DNase I at the initiation of biofilm formation decreased the final biofilm mass in a dose-dependent manner (Fig. 3E). Furthermore, this trend was noticeable by the first day and statistically significant at both 48 and 72 h. Similar to the effect found on preformed biofilms, the addition of proteinase K at time zero inhibited biofilm formation at all concentrations tested (Fig. 3F). The effect was so profound that the results were statistically significant by 24 h and across the duration of the experiment. These data suggest that both eDNA and protein were necessary in the early development of biofilms, as biofilm development was inhibited by their absence. However, eDNA did not appear to be as important in a preformed biofilm (Fig. 3A).
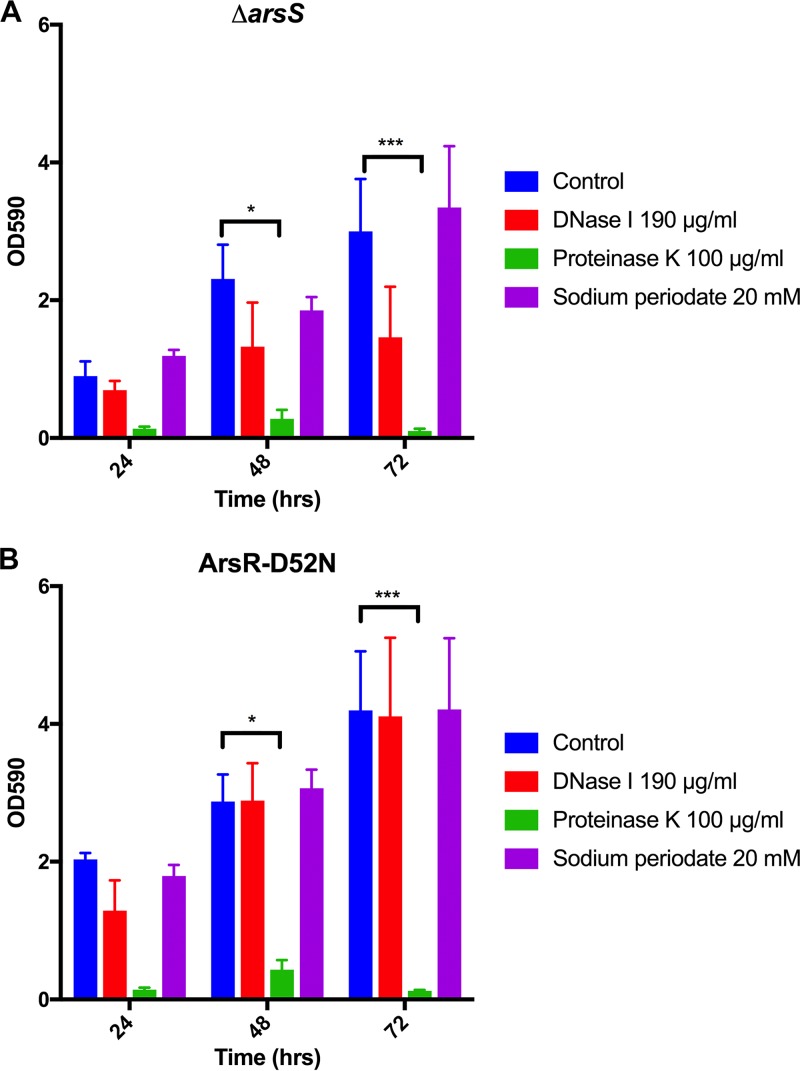
We previously found that the H. pylori ΔarsS and ArsR-D52N mutant strains displayed hyperbiofilm formation and increased aggregation (9). Furthermore, the visually different phenotypes displayed in SEM images of these strains (9) suggested that each might show distinct changes in the extracellular matrix. To determine if the increased biofilm formation by these strains was due to alterations in the presence of eDNA, protein, and/or polysaccharides in the extracellular matrix of the biofilm, we also performed enzymatic/chemical disruption assays on ΔarsS and ArsR-D52N biofilms; a single concentration for DNase I (190 μg/ml), proteinase K (100 μg/ml), and sodium periodate (20 mM) was used (Fig. 4). While a noticeable decrease in biofilm mass was observed after DNase I treatment of the ΔarsS mutant strain (Fig. 4A), this decrease was not statistically significant. No differences between untreated and DNase I-treated biofilms were observed for the ArsR-D52N strain (Fig. 4B). Similar to the effect on wild-type (WT) G27 biofilms, the addition of sodium periodate did little to affect the biofilms of either the ΔarsS or ArsR-D52N strain. Similar to the effect of proteinase K on wild-type strains, we found that proteinase K eliminated the biofilm for both the ΔarsS (Fig. 4A) and ArsR-D52N (Fig. 4B) mutant strains. En masse, these results lead us to conclude that the hyperbiofilm phenotype of the ΔarsS and ArsR-D52N strains was not due to alteration of the presence of eDNA or polysaccharides. Moreover, protein remained the most important constituent of the H. pylori biofilm extracellular matrix.
FIG 4.
Effects of DNase I, proteinase K, and sodium periodate on preformed ΔarsS and ArsR-D52N biofilms. DNase I, proteinase K, and sodium periodate were tested against preformed biofilms of ΔarsS (A) and ArsR-D52N (B) mutant strains. At the time points indicated, biofilms were treated with the indicated enzyme or chemical and incubated for another 24 h at 37°C. The means from the three biologically independent experiments are plotted; error bars indicate the standard errors of the means. *, P < 0.05; ***, P < 0.001.
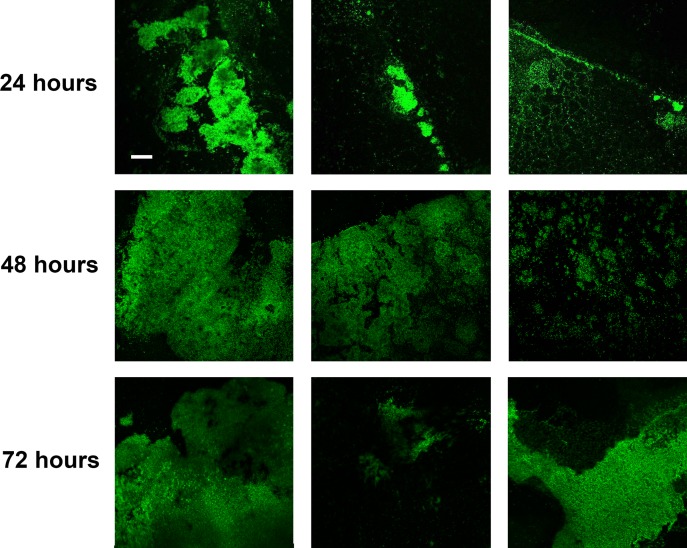
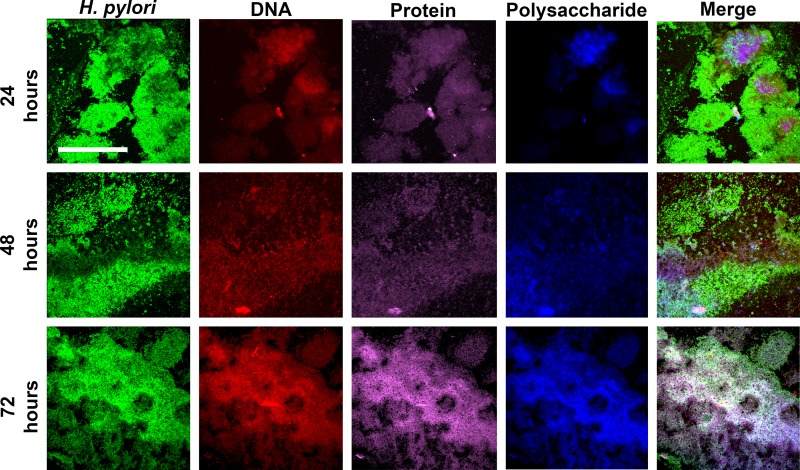
As the second part of our two-pronged approach, we next observed H. pylori biofilms by using confocal microscopy combined with fluorescent probes specific for protein, eDNA, and polysaccharides, as well as a fluorescently labeled antibody specific for H. pylori cells. We first simply observed H. pylori biofilm development (Fig. 5); G27 H. pylori biofilms were grown for 24, 48, and 72 h on glass coverslips before staining with H. pylori-specific antibody. The first prominent observation was regarding the location of the biofilm; the original position of the air-liquid interface could clearly be seen by the presence of large aggregates of H. pylori cells that attached in a distinct line in the 24-hour biofilm, though this line became less distinct as the biofilm increased in age (Fig. 5). Furthermore, the biofilms at this point were quite heterogeneous; though the majority of aggregates lay at the air-liquid interface, there were also many small aggregates and individual H. pylori cells that adhered to the surface below the air-liquid interface. This heterogeneity decreased as the biofilm grew older, leading to a more-uniform appearance that extended below the air-liquid interface. To properly visualize the extracellular matrix, we focused on the larger aggregates for additional staining (Fig. 6). DNA and protein were easily detectable. Moreover, even though incubation with sodium periodate suggested that polysaccharides were not an important structural component of the extracellular matrix (Fig. 3C), confocal microscopy revealed the presence of polysaccharides in the larger aggregates of H. pylori cells. The varied intensities of staining found within the biofilm likely suggested that eDNA, protein, and polysaccharides were unevenly distributed throughout the biofilm at the 24-hour time point. The distribution of these components did not seem to correspond to a recognizable position within the biofilm, nor did it appear to be related to the distance from the air-liquid interface. Rather, the extracellular matrix tended to occur in regions of the greatest cell density. Similar to how H. pylori biofilms grew more homogenous over time (Fig. 5), so too did the distribution of the extracellular matrix components. These data would suggest that eDNA, protein, and polysaccharides are all found in the extracellular matrix of H. pylori biofilms.
FIG 5.
H. pylori biofilm development. Immunofluorescent images of H. pylori biofilms were taken at 24, 48, and 72 h to visualize biofilm formation. Three representative images from different fields of a single coverslip per time point are shown at a magnification of ×10. Bar, 100 μm (scale is identical for all panels).
FIG 6.
Visualization of eDNA, protein, and polysaccharides in the extracellular matrix of H. pylori biofilms. Staining of the H. pylori biofilm matrix at 24, 48, and 72 h. Representative regions for areas with pronounced biofilm formation are shown. Bar, 100 μm (scale is identical for all panels). Each dye was captured on a single channel, and the images show a compressed z-stack; merged images at a magnification of ×40 are shown on the far right. Biofilms were stained with H. pylori-specific antibodies labeled with Alexa Fluor 488 (H. pylori), propidium iodide (DNA), FilmTracer Sypro Ruby biofilm matrix stain (protein), and calcofluor white (polysaccharides).
The antibiofilm peptides DJK-5 and IDR-1018 alter H. pylori biofilm formation.
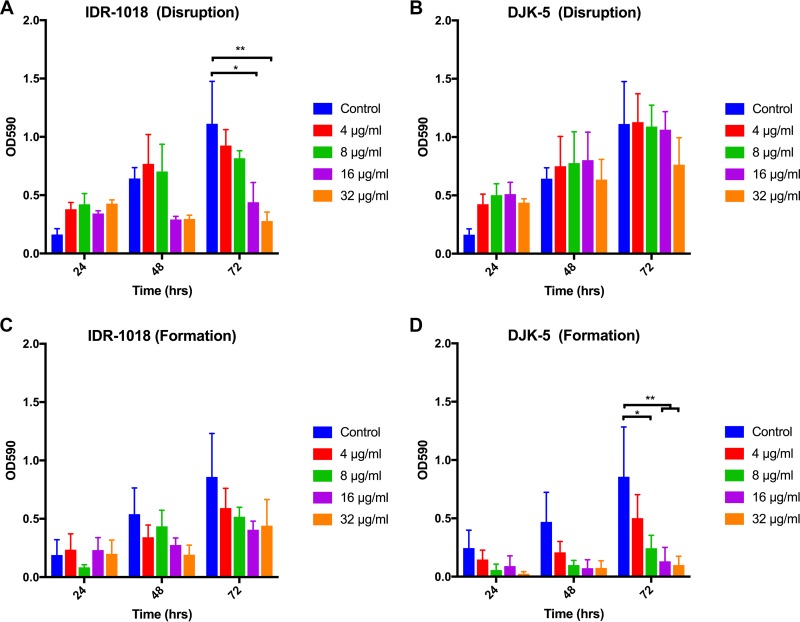
Due to the recalcitrant nature of biofilm-associated bacteria in response to antimicrobial therapy, identification of potential biofilm dispersal agents is of significant interest (40). The synthetic cationic peptides IDR-1018 and DJK-5 have each been shown to have a broad spectrum of activity against the biofilms of multiple species of bacteria (41, 42). We first assessed the effects of each peptide on the viability of H. pylori in liquid culture (Fig. 7). We found that after 24 h of growth, neither peptide demonstrated an appreciable effect on the viability of H. pylori at any concentration examined. We then evaluated the effects of the cationic peptides against preformed H. pylori G27 biofilms. We found that IDR-1018 decreased the final biofilm mass in relation to the control (Fig. 8A). Conversely, DJK-5 showed no appreciable effect on the preformed H. pylori biofilm (Fig. 8B). In addition to disruption of preformed biofilms, we also tested the effects of the two peptides on biofilm formation. When added at the start of the assay, IDR-1018 demonstrated a trend toward decreased biofilm formation, though this decrease was not statistically significant (Fig. 8C). In contrast, DJK-5 inhibited biofilm formation by wild-type G27 in a dose-dependent manner (Fig. 8D). Neither peptide had an effect either on preformed biofilms or on biofilm formation of the hyperbiofilm-forming strains (see Fig. S3 and S4 in the supplemental material). Together, these results suggested that H. pylori utilizes mechanisms of biofilm attachment and growth similar to those of other biofilm-forming species that can be targeted by antibiofilm peptides. However, mutations in ArsRS alter the effectiveness of these molecules.
FIG 7.
Effects of antibiofilm peptides on planktonic H. pylori. H. pylori viability after 24-hour incubation with IDR-1018 (A) and DJK-5 (B) was assayed. Bars represent the mean CFU from three biologically independent experiments; error bars display standard errors of the means. LOD, limit of detection.
FIG 8.
Effects of the antibiofilm peptides IDR-1018 and DJK-5 on G27 H. pylori biofilm formation. IDR-1018 and DJK-5 were added to preformed wild-type G27 biofilms at the time points indicated and incubated for another 24 h (Disruption) (A and B) or were added to the media when biofilm growth was started (Formation) (C and D). Biofilm formation was measured by crystal violet staining, and the mean OD590 values from three biologically independent replicates are shown; error bars display standard errors of the means. *, P < 0.05; **, P < 0.01.
Lastly, we investigated how the peptides affected the number of H. pylori cells found within the biofilm (see Fig. S5 in the supplemental material). While IDR-1018 disrupted biofilm mass in the crystal violet assay, it did not affect the total CFU found within the biofilm matrix (Fig. S5A). DJK-5 showed no effects on preformed biofilm mass or CFU (Fig. S5B). When H. pylori cells were enumerated from biofilms that had been incubated with the peptides from the start of the experiment, both IDR-1018 and DJK-5 resulted in a dose-dependent reduction in the total biofilm mass (Fig. S5C and D). However, only DJK-5 demonstrated a statistically significant reduction in the number of H. pylori cells recovered from the biofilm (Fig. S5D).
DISCUSSION
The growing interest in the ability of H. pylori to form a biofilm is reflected by the increased appearance of H. pylori biofilm-related studies in the literature. However, a hurdle faced in the comparison of the studies from different laboratories is the use of different H. pylori strains and the different conditions used to induce biofilm formation; there is currently little information on the conditions that affect H. pylori biofilm formation. To address this knowledge gap, we systemically analyzed the effects of commonly utilized rich media and surfaces on H. pylori biofilm formation. Additionally, we evaluated biofilm composition and the possible effects of two antibiofilm peptides.
Mutation of lab strains due to accidental selection imposed during in vitro serial passage is a common concern for the field of microbiology (43). This is particularly true for H. pylori, which has a tendency to mutate, change its chromosome, and acquire free DNA (30). Despite this, we found no appreciable difference in biofilm formation between low- and high-passage-number G27 isolates (Fig. 1A). Additionally, while it has been well established that alteration of the media can affect biofilm formation of many species (9, 26, 32), we found no significant effects in H. pylori biofilm formation in any of the commonly utilized rich media used for H. pylori culture (Fig. 1B); this was true even when additional preferred carbon sources were added (Fig. S1). While somewhat surprising, this lack of effect of carbon source might be due to a key difference between H. pylori and other bacterial species; often, when the addition of a preferred carbon source alters biofilm formation, carbon catabolite repression is involved (29, 44). However, no form of carbon catabolite repression has been identified in H. pylori. Indeed, investigation of the H. pylori genome suggests that the genes involved in carbon catabolite repression are absent (25). Thus, it is perhaps no surprise that H. pylori did not alter biofilm formation in response to the available carbon source. Despite this fact, it was clear that regulation in response to various stimuli does play a role in the biofilm formation of this organism. For example, we previously found that the ArsRS two-component system, which responds to acidic pH (24), also plays a role in regulating biofilm formation (9). Additionally, zinc depletion has been shown to promote H. pylori biofilm formation (45). While the ArsRS contribution appears to involve transcriptional regulation (9), the zinc effect appears to be posttranscriptional (45). Thus, regulation of biofilm formation in H. pylori is likely a complex process that will need to be investigated in greater detail in future studies.
Of the conducted characterization experiments, only modification of the properties of the surface had a significant impact on biofilm formation (Fig. 2); while there was little difference between biofilms grown in untreated polystyrene plates and those grown in plates coated with poly-d-lysine, tissue culture-treated plates supported the most-robust biofilm formation. This observation was consistent for G27 and the isogenic PRINTER: this does not look like the standard uppercase Greek delta; please use standard delta as usual ΔarsS and ArsR-D52N mutant strains, suggesting that surface properties are important even for the hyperbiofilm-forming strains (Fig. 2). Though not exhaustive, the tested surfaces were chosen because they possess different properties: untreated polystyrene wells are hydrophobic, poly-d-lysine wells are hydrophilic and positively charged, and tissue culture-treated surfaces are hydrophilic and negatively charged. Given the specific difference between the poly-d-lysine- and tissue culture-treated plates, the data suggested that H. pylori biofilm formation and adherence were mediated, at least in part, by surface charge in this assay; a negative charge appeared to promote optimal biofilm formation. This is in keeping with literature that suggests that negatively charged surfaces promote more biofilm accumulation than positively charged surfaces for Gram-negative bacteria (46).
Characterization of the biofilm matrix components revealed that the addition of proteinase K both disrupted preformed H. pylori biofilm (Fig. 3B) and inhibited H. pylori biofilm formation (Fig. 3F); these findings strongly suggest that proteins are a primary component of the H. pylori biofilm matrix. The same was true for biofilms formed by the hyperbiofilm-forming ΔarsS and ArsR-D52N strains; protein appears to be the primary constituent of the extracellular matrix (Fig. 4). While the identity of the protein(s) that is critical for biofilm formation remains to be elucidated, we note that the H. pylori genome encodes more than 60 outer membrane proteins (47) that could play a role in H. pylori aggregation or adherence to the surface. Of note, several of the genes encoding these proteins have been suggested to be regulated by the ArsRS system (24, 48). Thus, dysregulation of one or several of these outer membrane proteins could explain the hyperbiofilm-forming phenotype of strains bearing mutations in the ArsRS system. Further studies will be required to determine the identity of the proteins found within the extracellular matrix of H. pylori biofilms and to define the contribution of these proteins to biofilm formation.
Though our results suggested that protein is the most important component of the extracellular matrix of H. pylori biofilms, it is not the only component; eDNA is also present. Although incubation with DNase I did not result in a statistically significant disruption of preformed biofilms (Fig. 3A), there was a clear trend toward biofilm reduction. Furthermore, confocal microscopy revealed the presence of eDNA in H. pylori biofilms at 24, 48, and 72 h (Fig. 6). Additionally, the formation of total biofilm mass was significantly prevented when the effect of DNase I on biofilm formation was assessed (Fig. 3E). eDNA may also be structurally important for the biofilms of the ΔarsS mutant strain; though the decrease was not statistically significant, there was a noticeable decrease in biofilm mass after DNase I treatment (Fig. 4A). However, the ArsR-D52N mutant strain was unaffected by the addition of DNase I (Fig. 4B). Together, the data suggest that eDNA is important for early development of the H. pylori biofilm but does not play as great a structural role in older biofilms. A possible reason for the shift in the structural importance of eDNA may involve a change in function. For example, others have shown the presence of eDNA in the H. pylori biofilm (32) and have found that extensive recombination occurs within the biofilm (49). This led Grande et al. (49) to propose that the H. pylori biofilm acts as an environmental niche that promotes recombination. It follows then that perhaps H. pylori eDNA, repurposed as an adhesin and structural component of the extracellular matrix at early stages, is returned to the role of genetic material as the biofilm matures and other components take on the structural roles. Perhaps the ArsR-D52N mutant strain may have made this switch earlier than the ΔarsS mutant strain, thus explaining the differential sensitivity to DNase treatment. It is also worth noting that the biofilms formed by the ΔarsS and ArsR-D52N mutant strains are visually (9) and quantitatively (Fig. 4) different. Therefore, the differential effects of DNase treatment may be attributable simply to variations in the biofilms of these mutant strains.
The final potential component of the extracellular matrix that we investigated was polysaccharides. Confocal microscopy and calcofluor white staining revealed the presence of polysaccharides in the H. pylori biofilm (Fig. 6). This was an intriguing result, given that sodium periodate did not significantly alter H. pylori biofilm mass in strain G27 (Fig. 3C) or the ΔarsS and ArsR-D52N mutant strains (Fig. 4); loss of biofilm mass would be expected if the polysaccharides played a crucial structural role. Indeed, the data suggest either that polysaccharides do not play an important structural role in the extracellular matrix or that any polysaccharides that are present are somehow protected from the effects of sodium periodate. It is also possible that, like eDNA, polysaccharides are important for early biofilm development and then lose structural prominence as the biofilm matures. Unfortunately, we could not test this possibility given the exquisite sensitivity of H. pylori to sodium periodate (Fig. 3D). Future research using defined exoglycosidases or N- or O-linked glycosidases will likely help to more clearly define the constituency and contribution of polysaccharides to the H. pylori biofilm.
Targeted disruption of biofilms has important medical implications. Thus, we were interested to know whether the H. pylori biofilm would be affected by treatment with two synthetic antibiofilm peptides, IDR-1018 and DJK-5, each of which has been shown to be effective against the biofilms of a number of different bacterial species (50, 51). In these prior studies, it was noted that these peptides are effective in promoting biofilm disruption but do not affect planktonic bacteria (41, 42). Similarly, we saw no effect of either peptide on planktonic H. pylori (Fig. 7). Conversely, the two peptides displayed different effects on H. pylori biofilms. While IDR-1018 caused disruption of preformed biofilm (Fig. 8A) and trended toward inhibited biofilm formation (Fig. 8C), DJK-5 had little effect on preformed biofilm (Fig. 8B) but was highly effective at blocking H. pylori biofilm development (Fig. 8D). Unexpectedly, IDR-1018 did not alter the CFU that could be recovered from biofilms, either via disruption of a preformed biofilm (Fig. S5A) or when tested against biofilm formation (Fig. S5A). Since there was not a change in CFU, these data likely suggest that the reduction in biofilm demonstrated by IDR-1018 was likely due to a decrease in the extracellular matrix. Similar to the lack of effect of DJK-5 on preformed biofilm (Fig. 8B), no significant alteration in recovered CFU was detected (Fig. S5B). However, DJK-5 did decrease the CFU that could be recovered from the biofilm when tested against formation (Fig. S5D). Though the decrease appeared small when presented on a logarithmic scale, the decrease in CFU was 75% or greater for the concentrations of DJK-5 tested. While the exact reason that the two peptides displayed differential effects on H. pylori biofilms is currently unknown, recent studies have proposed that both IDR-1018 and DJK-5 promote degradation of the secondary messenger ppGpp, which is involved in the stringent stress response (50, 51). Indeed, the stringent response has been found to be important for biofilm formation in other bacterial species (52, 53). Whether IDR-1018 and DJK-5 were able to target ppGpp in H. pylori remains to be determined. However, given the effects of the two peptides in our assays, it is intriguing to speculate that the stringent response is important for H. pylori biofilm development. Regardless of the precise mechanism of action of these peptides, the data suggest that DJK-5 may be the most effective one to prevent H. pylori biofilm formation. This being said, we note that neither peptide was able to disrupt preformed biofilms formed by the ΔarsS and ArsR-D52N mutant strains or to prevent biofilm formation of these strains (Fig. S3 and S4). These results suggest that the ΔarsS and ArsR-D52N mutations modify H. pylori biofilm development in such a way that the antibiofilm peptides are less effective. Resistance to cationic peptides is often characterized by modifications that lead to a change in the surface charge of the bacterium (54). Like that of most bacteria, the surface of the H. pylori cell is negatively charged (55, 56). In fact, modification of H. pylori lipid A was shown to impart increased resistance to antimicrobial peptides (57). Thus, the lack of effect of IDR-1018 and DJK-5 against the mutant strains further supports the possibility that modulation of ArsRS regulation may lead to changes in outer membrane protein expression, which would result in changes to the surface charge of H. pylori. Alternatively, it is also possible that these strains have an altered stringent response. Both of these possibilities could lead to enhanced biofilm formation as well as reduced susceptibility to the antibiofilm peptides. Further studies will be required to fully understand the hyperbiofilm phenotype.
There are clear limitations to our study. For example, there are distinct differences between the polystyrene plate assays (Fig. 3, 4, and 8) and the glass coverslip confocal assays (Fig. 5 and 6). Though the biofilms were grown under the same temperature and aeration conditions and in the same media, the target substrates differ; the tissue culture-treated polystyrene plates used in the crystal violet assays have been altered by gas plasma treatment, leaving the surface hydrophilic and negatively charged to better promote attachment. In contrast, the glass coverslips used in the confocal assays have been coated with poly-d-lysine, resulting in a positive surface charge. Since the negative charge of the tissue culture-treated plates was found to better promote H. pylori biofilm formation (Fig. 2), the biofilm seen in the confocal assays was likely not as robust as it could have been. Despite this limitation, the results are congruent; the biofilm formed at the air-liquid interface in both assays, and both assays demonstrated the importance of protein and eDNA in the extracellular matrix. En masse, these results provide novel information about the H. pylori biofilm that will likely help to better develop a robust future biofilm model.
Given the newness of H. pylori biofilm research, two major questions are (i) how life in a biofilm is compatible with what is currently understood about H. pylori infection and (ii) what the purpose of the biofilm is in the H. pylori life cycle. While these questions remain to be answered, it is possible that biofilm formation helps H. pylori to evade aspects of the immune system, facilitates genetic diversity via providing a niche for enhanced recombination, provides protection from the harsh environment of the gastric niche, etc. Indeed, H. pylori may form biofilms for any of these reasons or for all of them. Future research that is focused on identification of conditions, environmental stressors, metabolites, and regulatory elements that promote biofilm formation will help to answer key questions about the importance of H. pylori biofilms. Hopefully, the in vitro characterization studies presented here will provide a foundation for future mechanistic studies of H. pylori biofilm formation.
MATERIALS AND METHODS
Bacterial strains and growth.
The strains used in this study are listed in Table 1. H. pylori strains were grown as previously described (58). Briefly, all cultures were grown at 37°C in a microaerobic environment (5% O2, 10% CO2, and 85% N2) generated using the Anoxomat system (Advanced Instruments, Inc.). Strains grown on solid media were cultured on horse blood agar (HBA), composed of 4% Columbia agar (Neogen Corporation), 5% defibrinated horse blood (HemoStat Laboratories, Dixon, CA), 2 mg/ml β-cyclodextrin (Sigma-Aldrich), and an antibiotic-antifungal cocktail (10 mg/ml vancomycin [Amresco], 5 μg/ml cefsulodin [Sigma-Aldrich], 2.5 μg/ml polymyxin B [Sigma], 5 μg/ml trimethoprim [Sigma-Aldrich], and 8 μg/ml amphotericin B [Amresco]). H. pylori liquid cultures were grown in medium consisting of Brucella broth (BB) (Neogen Corporation) supplemented with 10% fetal bovine serum (FBS; Gibco) and 10 μg/ml vancomycin, unless otherwise noted. Liquid growth was carried out at 37°C with shaking (110 rpm). Stock cultures of H. pylori strains were maintained at −80°C in brain heart infusion broth (BD Biosciences) containing 10% FBS and 20% glycerol (EMD Chemicals, Inc.).
TABLE 1.
Bacterial strains used in this paper
Crystal violet biofilm quantification.
Biofilm quantification assays were performed as described previously (9). Briefly, 1 ml of BB–10% FBS per well was inoculated from an overnight liquid culture to an optical density at 600 nm (OD600) of 0.1. Three separate 24-well tissue culture-treated plates (Corning) were used for time points T24, T48, and T72. The plates were grown at 37°C in a microaerobic environment with shaking at 110 rpm. At each time point, the medium was aspirated, and wells were washed with phosphate-buffered saline (PBS; EMD chemicals) and then dried prior to methanol fixation (J. T. Baker). Gram's crystal violet solution (Sigma-Aldrich) diluted to one percent with water was then added to each well, including to one empty control well that served as a blank, and plates were incubated for 15 min. Following incubation, wells were washed with distilled water and air dried for 5 min at 37°C, and then crystal violet was solubilized with a differentiation solution (Sigma-Aldrich) for 15 min. To quantify biofilm formation, solubilized crystal violet solution was read at an absorbance of 590 (OD590) for each sample. The data shown represent results from three biologically independent experiments.
In addition to standard biofilm assays, some minor modifications were included to assay different media, carbon sources, or surface coatings. We evaluated standard rich H. pylori growth media: BB supplemented with 10% FBS and 10 μg/ml vancomycin, (9), brain heart infusion medium supplemented with 0.1% beta-cyclodextrin and 10 μg/ml vancomycin (26), and BB supplemented with 2% FBS, 10 μg/ml vancomycin, and 0.2% glucose (Sigma-Aldrich) (32). Additionally, we assessed the addition of various carbon sources (arabinose, sodium pyruvate, alanine, asparagine) to BB supplemented with 10% FBS and 10 μg/ml vancomycin. Overnight cultures were all grown in BB–10% FBS–vancomycin. Aliquots of the starter culture were centrifuged and resuspended in different media. The resuspended cells were then used to inoculate 1 ml of liquid medium per well to an OD600 of 0.1, and the experiment was conducted as described above. To test different surfaces, assays were performed as described above, except that the standard 24-well tissue culture-treated plates were replaced by either untreated (Corning) or poly-d-lysine (Corning)-treated 24-well plates. For these experiments, poly-d-lysine-coated plates were prepared per the protocol recommended by Corning.
Antibiofilm peptide, enzymatic, and chemical assays.
To evaluate the composition of the H. pylori biofilm extracellular matrix, we used treatments with different enzymes/chemicals to target matrix components. For these assays, we chose DNase I (Sigma-Aldrich), proteinase K (Sigma-Aldrich), and sodium periodate (Sigma-Aldrich); sodium periodate was chosen to test for polysaccharides because, unlike commonly used enzymes like dispersin B (59) and alginate lyase (60), the chemical is not specific to the linkages in polysaccharides. We also tested the effects of two previously described synthetic peptides, IDR-1018 and DJK-5 (41, 42), on H. pylori biofilms.
To determine the composition of biofilms over time, we assessed the effects of the above-mentioned compounds on preformed biofilms that were 24, 48, or 72 h old. Multiple concentrations of DNase I (47.5 μg/ml to 380 μg/ml), proteinase K (25 μg/ml to 200 μg/ml), sodium periodate (5 mM to 40 mM), IDR-1018 (4 μg/ml to 64 μg/ml), or DJK-5 (4 μg/ml to 64 μg/ml) were added to the media. Following treatment, the wells were incubated for a further 24 h before biofilm quantification by crystal violet staining as detailed above. Additionally, to determine if any of the compounds would affect biofilm formation, DNase I, proteinase K, IDR-1018, or DJK-5 was added to the media when the 24-well assays were initiated (T0). Biofilm development was monitored then over 24, 48, or 72 h by crystal violet staining. To verify that the compounds affected the biofilm and not cell viability, we assessed the viability of planktonic H. pylori cells exposed to either the antibiofilm peptides, the enzymes, or the chemicals at the concentrations listed above. Overnight starter cultures of H. pylori were used to inoculate a master culture adjusted to an OD600 of 0.05. The master culture was then split into 1-ml volumes in test tubes. Antibiofilm peptides or enzymes were added at the same concentrations as those used in the biofilm assays, and cultures were incubated at 37°C in gas evacuation jars under microaerobic conditions for 24 h, after which the CFU were enumerated.
To quantitate CFU for the peptide experiment (Fig. S5), two sets of wells were prepared. One set of wells was developed by crystal violet staining as detailed above. The second set of wells was aspirated and washed twice with 2 ml of warm BB–10% FBS to remove any cells left over from the liquid culture. To remove the cells from the surface of the polystyrene plate, the wells were then filled with 1 ml of BB–10% FBS supplemented with 200 μg/ml proteinase K and incubated for 1 h. To further break up cellular aggregates, the 1-ml cultures were then supplemented with 2 μM EDTA and sonicated for 10 min in a benchtop water bath (Branson 1510 ultrasonic cleaner). The cells were then diluted and plated onto HBA plates for later enumeration.
Confocal microscopy, immunofluorescence, and antibodies.
Overnight cultures of DSM 1 (G27) were adjusted to a starting OD600 of 0.1 in a 16- by 100-mm glass tube to a total volume of 0.4 ml. Two circular glass coverslips (Corning poly-l-lysine) were placed back to back and suspended upright within the tube to allow for formation of a biofilm at the air-liquid interface of the exposed side of each coverslip. After 24, 48, and 72 h of growth, coverslips were carefully removed and placed into 500 μl of filter-sterilized PBS in a 12-well plate. Slips were rinsed 3 times in filtered PBS and then stained using the subsequent parameters. First, 500 μl of 1× FilmTracer Sypro Ruby biofilm matrix stain (Thermo Fisher Scientific) was added, and plates were incubated for 20 min. Sypro Ruby was removed, and wells were washed 2 times with filtered PBS. Next, blocking buffer (1× PBS, 3% bovine serum albumin [BSA], 0.05% sodium azide) was added for 45 min without rocking. Following blocking, coverslips were incubated with a polyclonal rabbit anti-H. pylori antibody (ab20459; Abcam) at a 1:400 dilution in blocking buffer for 45 min. After 45 min, cells were washed 3 times for 5 min each in blocking buffer and then incubated for 45 min with anti-rabbit Alexa Fluor 488 (1:800 dilution). After 40 min in the secondary antibody, 500 nM propidium iodide (Thermo Fisher Scientific) was added with the secondary antibody and incubated for the final 5 min. Cells were then washed 3 times with filtered PBS, and the final stain, 0.1% calcofluor white (fluorescent brightener 28; Sigma), was added for 30 min. After the final incubation, cells were washed 3 times with distilled water and then affixed to a slide with Vectashield mounting medium (Vector Laboratories). Confocal images were collected via a Zeiss 710 microscope (Carl Zeiss, Jena, Germany) and analyzed with Zen Black software (Zeiss).
Statistical analysis.
Statistical analyses were conducted using GraphPad Prism version 7 (Graph Pad Software, Inc.). A two-way analysis of variance (ANOVA) with Dunnett's correction for multiple comparisons was used to analyze increases in biofilm formation over time for individual strains and also to identify differences in biofilm formation between strains at each time point. Ordinary one-way ANOVA with Dunnett's correction was utilized for enumerating CFU within the biofilm (Fig. S5).
Supplementary Material
ACKNOWLEDGMENTS
We thank Cara Olsen for help with statistical analyses.
The work described here was supported by National Institutes of Health National Institute of Allergy and Infectious Disease (NIAID) grant R21AI121517 (to D.S.M.). R.E.W.H. is supported by Canadian Institutes from Health Research grant FDN-154287 and holds a Canada Research Chair and UBC Killam Professorship. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
The contents of this article are solely our responsibility and do not necessarily represent the official views of the Department of Defense, the Uniformed Services University, or the NIH.
Conflict of interest: The peptides described here have been filed for patent protection, assigned to R.E.W.H.'s employer, the University of British Columbia, and licensed to ABT Innovations Inc., in which the University of British Columbia and R.E.W.H. own shares.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00391-18.
REFERENCES
- 1.Marshall BJ, Warren JR. 1984. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet i:1311–1315. [DOI] [PubMed] [Google Scholar]
- 2.Moodley Y, Linz B, Bond RP, Nieuwoudt M, Soodyall H, Schlebusch CM, Bernhoft S, Hale J, Suerbaum S, Mugisha L, van der Merwe SW, Achtman M. 2012. Age of the association between Helicobacter pylori and man. PLoS Pathog 8:e1002693. doi: 10.1371/journal.ppat.1002693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blaser MJ. 1990. Epidemiology and pathophysiology of Campylobacter pylori infections. Rev Infect Dis 12(Suppl 1):S99–S106. doi: 10.1093/clinids/12.Supplement_1.S99. [DOI] [PubMed] [Google Scholar]
- 4.Yamamoto Y, Fujisaki J, Omae M, Hirasawa T, Igarashi M. 2015. Helicobacter pylori-negative gastric cancer: characteristics and endoscopic findings. Dig Endosc 27:551–561. doi: 10.1111/den.12471. [DOI] [PubMed] [Google Scholar]
- 5.WHO. 2017. Cancer. WHO, Geneva, Switzerland. http://www.who.int/mediacentre/factsheets/fs297/en/ Accessed 21 September 2017.
- 6.Carron MA, Tran VR, Sugawa C, Coticchia JM. 2006. Identification of Helicobacter pylori biofilms in human gastric mucosa. J Gastrointest Surg 10:712–717. doi: 10.1016/j.gassur.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 7.Coticchia JM, Sugawa C, Tran VR, Gurrola J, Kowalski E, Carron MA. 2006. Presence and density of Helicobacter pylori biofilms in human gastric mucosa in patients with peptic ulcer disease. J Gastrointest Surg 10:883–889. doi: 10.1016/j.gassur.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 8.Attaran B, Falsafi T, Moghaddam AN. 2016. Study of biofilm formation in C57Bl/6J mice by clinical isolates of Helicobacter pylori. Saudi J Gastroenterol 22:161–168. doi: 10.4103/1319-3767.178529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Servetas SL, Carpenter BM, Haley KP, Gilbreath JJ, Gaddy JA, Merrell DS. 2016. Characterization of key Helicobacter pylori regulators identifies a role for ArsRS in biofilm formation. J Bacteriol 198:2536–2548. doi: 10.1128/JB.00324-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. 1995. Microbial biofilms. Annu Rev Microbiol 49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- 11.Flemming HC, Wingender J. 2010. The biofilm matrix. Nat Rev Microbiol 8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 12.Hall-Stoodley L, Costerton JW, Stoodley P. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol 2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 13.Stewart PS, Costerton JW. 2001. Antibiotic resistance of bacteria in biofilms. Lancet 358:135–138. doi: 10.1016/S0140-6736(01)05321-1. [DOI] [PubMed] [Google Scholar]
- 14.Leid JG, Shirtliff ME, Costerton JW, Stoodley P. 2002. Human leukocytes adhere to, penetrate, and respond to Staphylococcus aureus biofilms. Infect Immun 70:6339–6345. doi: 10.1128/IAI.70.11.6339-6345.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Domenech M, Ramos-Sevillano E, Garcia E, Moscoso M, Yuste J. 2013. Biofilm formation avoids complement immunity and phagocytosis of Streptococcus pneumoniae. Infect Immun 81:2606–2615. doi: 10.1128/IAI.00491-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vuong C, Kocianova S, Voyich JM, Yao Y, Fischer ER, DeLeo FR, Otto M. 2004. A crucial role for exopolysaccharide modification in bacterial biofilm formation, immune evasion, and virulence. J Biol Chem 279:54881–54886. doi: 10.1074/jbc.M411374200. [DOI] [PubMed] [Google Scholar]
- 17.Mann EE, Wozniak DJ. 2012. Pseudomonas biofilm matrix composition and niche biology. FEMS Microbiol Rev 36:893–916. doi: 10.1111/j.1574-6976.2011.00322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cammarota G, Sanguinetti M, Gallo A, Posteraro B. 2012. Review article: biofilm formation by Helicobacter pylori as a target for eradication of resistant infection. Aliment Pharmacol Ther 36:222–230. doi: 10.1111/j.1365-2036.2012.05165.x. [DOI] [PubMed] [Google Scholar]
- 19.De la Cruz MA, Ares MA, von Bargen K, Panunzi LG, Martinez-Cruz J, Valdez-Salazar HA, Jimenez-Galicia C, Torres J. 2017. Gene expression profiling of transcription factors of Helicobacter pylori under different environmental conditions. Front Microbiol 8:615. doi: 10.3389/fmicb.2017.00615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shao C, Sun Y, Wang N, Yu H, Zhou Y, Chen C, Jia J. 2013. Changes of proteome components of Helicobacter pylori biofilms induced by serum starvation. Mol Med Rep 8:1761–1766. doi: 10.3892/mmr.2013.1712. [DOI] [PubMed] [Google Scholar]
- 21.Alm RA, Ling LS, Moir DT, King BL, Brown ED, Doig PC, Smith DR, Noonan B, Guild BC, deJonge BL, Carmel G, Tummino PJ, Caruso A, Uria-Nickelsen M, Mills DM, Ives C, Gibson R, Merberg D, Mills SD, Jiang Q, Taylor DE, Vovis GF, Trust TJ. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 22.Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA, Nelson K, Quackenbush J, Zhou L, Kirkness EF, Peterson S, Loftus B, Richardson D, Dodson R, Khalak HG, Glodek A, McKenney K, Fitzegerald LM, Lee N, Adams MD, Hickey EK, Berg DE, Gocayne JD, Utterback TR, Peterson JD, Kelley JM, Cotton MD, Weidman JM, Fujii C, Bowman C, Watthey L, Wallin E, Hayes WS, Borodovsky M, Karp PD, Smith HO, Fraser CM, Venter JC. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 23.Pflock M, Dietz P, Schar J, Beier D. 2004. Genetic evidence for histidine kinase HP165 being an acid sensor of Helicobacter pylori. FEMS Microbiol Lett 234:51–61. doi: 10.1111/j.1574-6968.2004.tb09512.x. [DOI] [PubMed] [Google Scholar]
- 24.Pflock M, Finsterer N, Joseph B, Mollenkopf H, Meyer TF, Beier D. 2006. Characterization of the ArsRS regulon of Helicobacter pylori, involved in acid adaptation. J Bacteriol 188:3449–3462. doi: 10.1128/JB.188.10.3449-3462.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Vries N, van Vliet AHM, Kusters JG. 2001. Gene regulation, p 321–334. In Mobley HLT, Mendz GL, Hazell SL (ed), Helicobacter pylori: physiology and genetics. American Society for Microbiology, Washington, DC. [Google Scholar]
- 26.Cole SP, Harwood J, Lee R, She R, Guiney DG. 2004. Characterization of monospecies biofilm formation by Helicobacter pylori. J Bacteriol 186:3124–3132. doi: 10.1128/JB.186.10.3124-3132.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stepanovic S, Cirkovic I, Ranin L, Svabic-Vlahovic M. 2004. Biofilm formation by Salmonella spp. and Listeria monocytogenes on plastic surface. Lett Appl Microbiol 38:428–432. doi: 10.1111/j.1472-765X.2004.01513.x. [DOI] [PubMed] [Google Scholar]
- 28.Kadam SR, den Besten HM, van der Veen S, Zwietering MH, Moezelaar R, Abee T. 2013. Diversity assessment of Listeria monocytogenes biofilm formation: impact of growth condition, serotype and strain origin. Int J Food Microbiol 165:259–264. doi: 10.1016/j.ijfoodmicro.2013.05.025. [DOI] [PubMed] [Google Scholar]
- 29.Seidl K, Goerke C, Wolz C, Mack D, Berger-Bachi B, Bischoff M. 2008. Staphylococcus aureus CcpA affects biofilm formation. Infect Immun 76:2044–2050. doi: 10.1128/IAI.00035-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Falush D, Kraft C, Taylor NS, Correa P, Fox JG, Achtman M, Suerbaum S. 2001. Recombination and mutation during long-term gastric colonization by Helicobacter pylori: estimates of clock rates, recombination size, and minimal age. Proc Natl Acad Sci U S A 98:15056–15061. doi: 10.1073/pnas.251396098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baltrus DA, Amieva MR, Covacci A, Lowe TM, Merrell DS, Ottemann KM, Stein M, Salama NR, Guillemin K. 2009. The complete genome sequence of Helicobacter pylori strain G27. J Bacteriol 191:447–448. doi: 10.1128/JB.01416-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grande R, Di Giulio M, Bessa LJ, Di Campli E, Baffoni M, Guarnieri S, Cellini L. 2011. Extracellular DNA in Helicobacter pylori biofilm: a backstairs rumour. J Appl Microbiol 110:490–498. doi: 10.1111/j.1365-2672.2010.04911.x. [DOI] [PubMed] [Google Scholar]
- 33.Lee WC, Goh KL, Loke MF, Vadivelu J. 2017. Elucidation of the metabolic network of Helicobacter pylori J99 and Malaysian clinical strains by phenotype microarray. Helicobacter 22:e12321. doi: 10.1111/hel.12321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Donlan RM. 2002. Biofilms: microbial life on surfaces. Emerg Infect Dis 8:881–890. doi: 10.3201/eid0809.020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beenken KE, Blevins JS, Smeltzer MS. 2003. Mutation of sarA in Staphylococcus aureus limits biofilm formation. Infect Immun 71:4206–4211. doi: 10.1128/IAI.71.7.4206-4211.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bonifait L, Grignon L, Grenier D. 2008. Fibrinogen induces biofilm formation by Streptococcus suis and enhances its antibiotic resistance. Appl Environ Microbiol 74:4969–4972. doi: 10.1128/AEM.00558-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tetz GV, Artemenko NK, Tetz VV. 2009. Effect of DNase and antibiotics on biofilm characteristics. Antimicrob Agents Chemother 53:1204–1209. doi: 10.1128/AAC.00471-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nguyen UT, Burrows LL. 2014. DNase I and proteinase K impair Listeria monocytogenes biofilm formation and induce dispersal of pre-existing biofilms. Int J Food Microbiol 187:26–32. doi: 10.1016/j.ijfoodmicro.2014.06.025. [DOI] [PubMed] [Google Scholar]
- 39.Sutherland IW. 2001. The biofilm matrix—an immobilized but dynamic microbial environment. Trends Microbiol 9:222–227. doi: 10.1016/S0966-842X(01)02012-1. [DOI] [PubMed] [Google Scholar]
- 40.Rabin N, Zheng Y, Opoku-Temeng C, Du Y, Bonsu E, Sintim HO. 2015. Agents that inhibit bacterial biofilm formation. Future Med Chem 7:647–671. doi: 10.4155/fmc.15.7. [DOI] [PubMed] [Google Scholar]
- 41.Mansour SC, de la Fuente-Nunez C, Hancock RE. 2015. Peptide IDR-1018: modulating the immune system and targeting bacterial biofilms to treat antibiotic-resistant bacterial infections. J Pept Sci 21:323–329. doi: 10.1002/psc.2708. [DOI] [PubMed] [Google Scholar]
- 42.Ribeiro SM, de la Fuente-Nunez C, Baquir B, Faria-Junior C, Franco OL, Hancock RE. 2015. Antibiofilm peptides increase the susceptibility of carbapenemase-producing Klebsiella pneumoniae clinical isolates to beta-lactam antibiotics. Antimicrob Agents Chemother 59:3906–3912. doi: 10.1128/AAC.00092-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fux CA, Shirtliff M, Stoodley P, Costerton JW. 2005. Can laboratory reference strains mirror “real-world” pathogenesis? Trends Microbiol 13:58–63. doi: 10.1016/j.tim.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 44.O'Toole GA, Gibbs KA, Hager PW, Phibbs PV Jr, Kolter R. 2000. The global carbon metabolism regulator Crc is a component of a signal transduction pathway required for biofilm development by Pseudomonas aeruginosa. J Bacteriol 182:425–431. doi: 10.1128/JB.182.2.425-431.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gaddy JA, Radin JN, Cullen TW, Chazin WJ, Skaar EP, Trent MS, Algood HM. 2015. Helicobacter pylori resists the antimicrobial activity of calprotectin via lipid A modification and associated biofilm formation. mBio 6:e01349-. doi: 10.1128/mBio.01349-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gottenbos B, Grijpma DW, van der Mei HC, Feijen J, Busscher HJ. 2001. Antimicrobial effects of positively charged surfaces on adhering Gram-positive and Gram-negative bacteria. J Antimicrob Chemother 48:7–13. doi: 10.1093/jac/48.1.7. [DOI] [PubMed] [Google Scholar]
- 47.Oleastro M, Menard A. 2013. The role of Helicobacter pylori outer membrane proteins in adherence and pathogenesis. Biology 2:1110–1134. doi: 10.3390/biology2031110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Loh JT, Gupta SS, Friedman DB, Krezel AM, Cover TL. 2010. Analysis of protein expression regulated by the Helicobacter pylori ArsRS two-component signal transduction system. J Bacteriol 192:2034–2043. doi: 10.1128/JB.01703-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grande R, Di Campli E, Di Bartolomeo S, Verginelli F, Di Giulio M, Baffoni M, Bessa LJ, Cellini L. 2012. Helicobacter pylori biofilm: a protective environment for bacterial recombination. J Appl Microbiol 113:669–676. doi: 10.1111/j.1365-2672.2012.05351.x. [DOI] [PubMed] [Google Scholar]
- 50.de la Fuente-Nunez C, Reffuveille F, Haney EF, Straus SK, Hancock RE. 2014. Broad-spectrum anti-biofilm peptide that targets a cellular stress response. PLoS Pathog 10:e1004152. doi: 10.1371/journal.ppat.1004152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de la Fuente-Nunez C, Reffuveille F, Mansour SC, Reckseidler-Zenteno SL, Hernandez D, Brackman G, Coenye T, Hancock RE. 2015. D-enantiomeric peptides that eradicate wild-type and multidrug-resistant biofilms and protect against lethal Pseudomonas aeruginosa infections. Chem Biol 22:196–205. doi: 10.1016/j.chembiol.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taylor CM, Beresford M, Epton HA, Sigee DC, Shama G, Andrew PW, Roberts IS. 2002. Listeria monocytogenes relA and hpt mutants are impaired in surface-attached growth and virulence. J Bacteriol 184:621–628. doi: 10.1128/JB.184.3.621-628.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.He H, Cooper JN, Mishra A, Raskin DM. 2012. Stringent response regulation of biofilm formation in Vibrio cholerae. J Bacteriol 194:2962–2972. doi: 10.1128/JB.00014-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Joo HS, Fu CI, Otto M. 2016. Bacterial strategies of resistance to antimicrobial peptides. Philos Trans R Soc Lond B Biol Sci 371(1695):20150292. doi: 10.1098/rstb.2015.0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smith JI, Drumm B, Neumann AW, Policova Z, Sherman PM. 1990. In vitro surface properties of the newly recognized gastric pathogen Helicobacter pylori. Infect Immun 58:3056–3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pruul H, Goodwin CS, McDonald PJ, Lewis G, Pankhurst D. 1990. Hydrophobic characterisation of Helicobacter (Campylobacter) pylori. J Med Microbiol 32:93–100. doi: 10.1099/00222615-32-2-93. [DOI] [PubMed] [Google Scholar]
- 57.Tran AX, Whittimore JD, Wyrick PB, McGrath SC, Cotter RJ, Trent MS. 2006. The lipid A 1-phosphatase of Helicobacter pylori is required for resistance to the antimicrobial peptide polymyxin. J Bacteriol 188:4531–4541. doi: 10.1128/JB.00146-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Carpenter BM, West AL, Gancz H, Servetas SL, Pich OQ, Gilbreath JJ, Hallinger DR, Forsyth MH, Merrell DS, Michel SL. 2015. Crosstalk between the HpArsRS two-component system and HpNikR is necessary for maximal activation of urease transcription. Front Microbiol 6:558. doi: 10.3389/fmicb.2015.00558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boles BR, Horswill AR. 2008. Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog 4:e1000052. doi: 10.1371/journal.ppat.1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alkawash MA, Soothill JS, Schiller NL. 2006. Alginate lyase enhances antibiotic killing of mucoid Pseudomonas aeruginosa in biofilms. APMIS 114:131–138. doi: 10.1111/j.1600-0463.2006.apm_356.x. [DOI] [PubMed] [Google Scholar]
- 61.Xiang Z, Censini S, Bayeli PF, Telford JL, Figura N, Rappuoli R, Covacci A. 1995. Analysis of expression of CagA and VacA virulence factors in 43 strains of Helicobacter pylori reveals that clinical isolates can be divided into two major types and that CagA is not necessary for expression of the vacuolating cytotoxin. Infect Immun 63:94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.