Abstract
Mammalian genomes are folded in a hierarchy of compartments, topologically associating domains (TADs), subTADs and looping interactions. Here, we describe 3DNetMod, a graph theory-based method for sensitive and accurate detection of chromatin domains across length scales in Hi-C data. We identify nested, partially overlapping TADs and subTADs genome wide by optimizing network modularity and varying a single resolution parameter. 3DNetMod can be applied broadly to understand genome reconfiguration in development and disease.
Principles from the field of mathematics known as graph theory have emerged as powerful tools for quantifying connectivity patterns within complex systems1. Networks are graphs consisting of nodes connected by edges that can be used to represent the underlying structure of biological, social, physical and information systems. Complex networks often exhibit hierarchical patterns of connectivity across length scales2. Interacting nodes can form small subnetworks arranged in distinct recurrent configurations termed motifs. Subnetworks can in turn hierarchically aggregate into higher-order modules (or communities)2. Communities are important functionally because they can promote resilience to individual node failure within a network.
A complex system exhibiting hierarchical structure is the higher-order folding of chromatin in the 3D nucleus. The approximately two-meter-long genome is arranged in complex higher-order configurations to fit inside a mammalian nucleus that is 5–10 µm in diameter3,4. Individual chromosomes aggregate into higher-order ‘A’ and ‘B’ compartments5, which are further partitioned into Megabase (Mb)-sized TADs and smaller, nested subTADs6–10. Distal genomic segments connect to create long-range looping interactions within and between TADs and subTADs7. The functional role for TADs and subTADs, and the extent to which they change across biological conditions, remains poorly understood, in part because of the paucity of methods for sensitive and accurate detection of sub-Mb-scale domains. Two recent comparative analyses reported that existing domain-calling methods accurately identify TADs but show poor ability to capture the full nested hierarchy of partially overlapping subTADs11,12. Thus, there is a great need for computational tools that accurately and sensitively detect the full nested hierarchy of chromatin domains across length scales.
Here, we hypothesize that nested, partially overlapping TADs and subTADs can be identified with a community detection method based on the maximization of network modularity13. We abstracted Hi-C data as a square, symmetric adjacency matrix, A, of size N × N with N adjacent, equally spaced bins representing nodes and interaction frequencies between nodes representing edges (Supplementary Fig. 1). To identify communities, we maximized the modularity quality index, Q (equation (1)):
| (1) |
where Ai,j is the edge weight representing interaction frequency between nodes i and j, ki is the sum of all edge weights for node i, m is the sum of all nondiagonal edge weights in the network A, and γ is the resolution parameter (discussed below). Nodes i and j are assigned to communities gi and gj, respectively. The Kronecker delta, δ(gi,gj), is 1 if gi = gj and 0 otherwise. A modularity value close to 1 indicates strong community structure and a high-quality division of the network into communities, whereas a Q value close to 0 indicates that the strength of within-community connections is no higher than would be expected by chance.
To identify the optimal division of the network into communities, we employed a Louvain-like, locally greedy algorithm to maximize network modularity14 (3DNetMod-modularity maximization (MM); see Online Methods). The 3DNetMod-MM algorithm was chosen because it does not require a priori knowledge of the number of communities that will be detected15, and it offers the critical capability of identifying nested communities through the variation of a resolution parameter. The output from a single run of 3DNetMod-MM is an ‘optimal’ partition of nodes into communities.
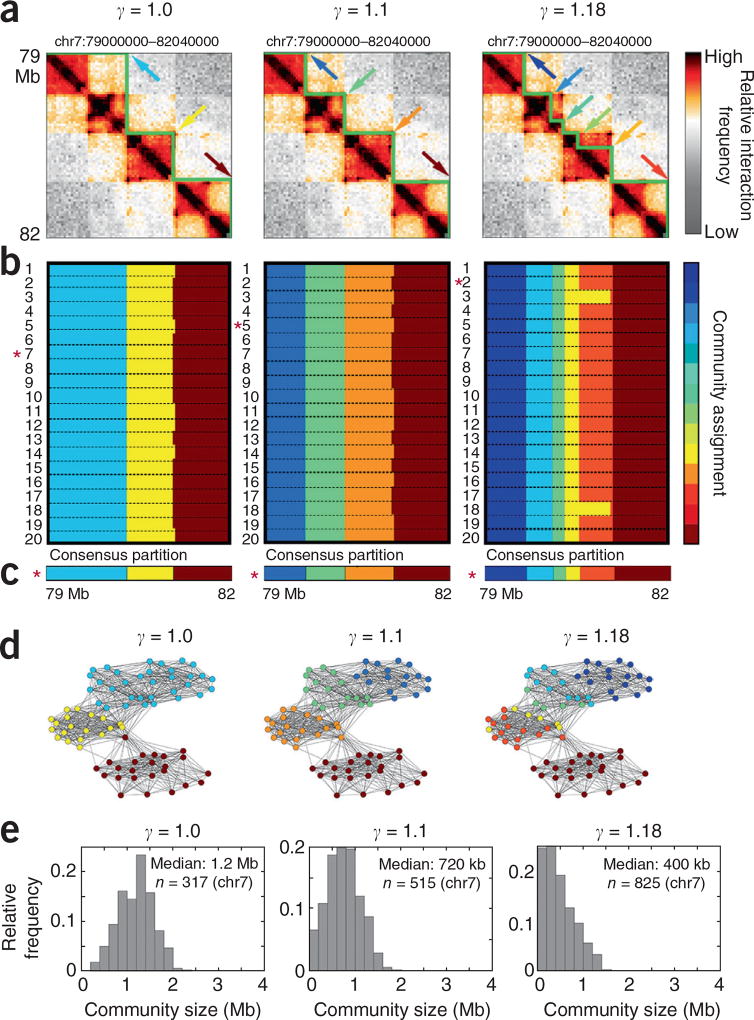
We first assessed the performance of 3DNetMod-MM in 40-kb-binned Hi-C data from human embryonic cortical plate tissue16 (Fig. 1a and Supplementary Table 1). To address boundary variation due to 3DNetMod-MM convergence on local maxima, we sampled the landscape of community partition solutions by applying 3DNetMod-MM 20 times (hereafter referred to as a ‘partition block’ with dimensions 20 × N) (Fig. 1b). We then identified the ‘consensus partition’ that is most similar to the other partitions (Fig. 1c; see Online Methods). We found that up to 1,000 partitions led to nearly the same consensus, which suggested that 20 partitions yields an acceptable balance of accuracy and speed (Supplementary Fig. 2). Thus, we address fluctuations in partitions due to algorithmic convergence on local versus global maxima by computing a consensus community partition. We hereafter refer to 3DNetMod-MM coupled with consensus community partitioning as ‘3DNetMod-MMCP’.
Figure 1.
Network modularity maximization and consensus partitioning (3DNetMod-MMCP) identifies nested, partially overlapping chromatin domains across length scales. (a) Hi-C heatmaps from human cortical plate tissue16. Domains identified with 3DNetMod-MMCP at γ = 1 (left), γ = 1.1 (center) and γ = 1.18 (right) are outlined in green. (b) Community partitions from 20 applications of 3DNetMod-MM at γ = 1 (left), γ = 1.1 (center) and γ = 1.18 (right). (c) Consensus partitions for each γ value from 20 partitions. (d) Spring-force diagrams representing network communities in Hi-C data. Node proximity corresponds to the relative interaction frequency between the two genomic bins. Node color indicates community assignment. (e) Chromosome-wide distribution (chr7) of domain sizes identified with γ = 1.0 (left), γ = 1.1 (center) and γ = 1.18 (right).
We next hypothesized that communities of different sizes can be identified with 3DNetMod-MMCP through modification of the resolution parameter. When γ > 1, the algorithm is biased toward detection of smaller communities, whereas the bias is toward detection of larger communities when γ < 1. By varying only a single resolution parameter, we detected a wide size range of hierarchically nested chromatin domains with 3DNetMod-MMCP (Fig. 1).
We next sought to comprehensively identify high-confidence domains genome wide. 3DNetMod-MM is estimated to run in time O(n)14, and the addition of 3DNetMod-MMCP and a sweep of γ values would make the method computationally intractable for identifying genome-wide domains. We devised a strategy in which we split the genome into overlapping regions and called nested domains on all regions in parallel (Supplementary Fig. 3a). Region size is provided as a user parameter and should be tuned to the resolution of the Hi-C library and the biological question of interest (Supplementary Fig. 4a–c). Thus, each genomic region can be run in parallel, significantly cutting down on the runtime and making it feasible to explore the range of γ values required to detect TADs and subTADs genome wide (Supplementary Table 2).
We devised a method, ‘gamma plateau sweep’ (3DNetMod-GPS), to select γ values that lead to high-confidence domains. To find the informative γ range, we generated random graphs with preserved weight, degree and strength distributions17 (Supplementary Figs. 3b and 5a,b). We noticed that modularity of the real network diverges from the random network at low γ and converges again at high γ (Supplementary Figs. 3c and 5c). We computed the 3DNetMod-GPS endpoint as the maximum γ across five representative regions where modularity converges between real and random networks. We also uncovered plateaus in which adjacent γ values gave rise to the same consensus partition and average number of communities (Supplementary Figs. 3d and 5d). Additionally, we noticed that some γ values did not belong to a plateau and gave rise to low-confidence partitions (Supplementary Fig. 6a–c). We observed that γ values resulting in a plateau size of 3 lead to robust domain calls in high-read-depth, 40-kb-binned Hi-C data (Supplementary Fig. 5d–f). Overall, 3DNetMod-MMCP and 3DNetMod-GPS coupled with our region-splitting strategy (Supplementary Fig. 3e–g) result in a large, diverse set of nested domains across length scales genome wide.
TADs and subTADs are known to interact in higher-order structures termed compartments5. Compartments appear in Hi-C maps as long-range, plaid patterns of alternating strong and weak interacting segments between groups of domains. By contrast, TADs and subTADs are thought to be structurally and functionally distinct from compartments, and defined as contiguous genomic intervals in which all pairs of loci exhibit elevated contact frequency7. To dissect higher-order compartments from domains, we devised a new ‘hierarchical spatial variance minimization’ (3DNetMod-HSVM) method (Supplementary Fig. 3h–l). We noticed that domains with continuously high interaction signal for the majority of bin pairs exhibit highly consistent partition blocks from 3DNetMod-MMCP (Supplementary Fig. 7a). By contrast, communities exhibiting a nonuniform, alternating interaction pattern characteristic of compartments tended to exhibit greater variability in boundary assignment across the 20 partitions (Supplementary Figs. 7b and 8a–e).
We posited that community assignment degeneracies in the partition blocks could represent (i) low-confidence boundaries indicative of ‘soft’ community structure18 and/or (ii) longer range compartments with nonuniform, alternating high/low signal. We computed a spatial variance for each domain boundary (see Online Methods). Spatial variance distributions were dependent on the length scale of the domain (Supplementary Fig. 8e). By instituting length-scale-specific spatial variance thresholds, we removed many communities exhibiting alternating interaction patterns indicative of compartments (Supplementary Fig. 8f–i). High-confidence domain boundaries passing the spatial variance thresholds exhibited enrichment for occupancy of the architectural protein CTCF, whereas communities with high spatial variance did not show enrichment (Supplementary Table 3 and Supplementary Fig. 9). Spatial variance distributions are widely variable among data sets (Supplementary Table 1); thus, we recommend tuning thresholds on a Hi-C-library-specific basis (Supplementary Figs. 10–12). These results indicate that 3DNetMod-HSVM has utility in dissecting high-confidence domains with corroborative enrichment of known epigenetic marks from low-confidence communities and higher-order compartments.
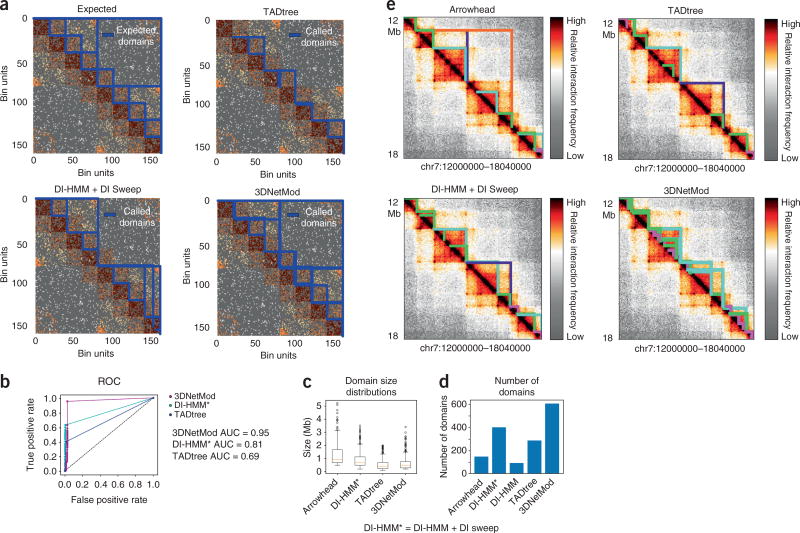
To compare the performance of 3DNetMod to other leading domain-calling methods, we generated TAD and subTAD simulations. We first simulated a network of nonoverlapping, nonhierarchical domains. We observed that 3DNetMod, TADtree19 and the Directionality Index-Hidden Markov Model (DI-HMM) method8 accurately identify nonoverlapping domains (Supplementary Fig. 13a,b). Next, to simulate a network with partially overlapping domains resembling subTADs, we created a construct of nested subtriangles derived from a Sierpinski triangle (Fig. 2a and Supplementary Fig. 13c,d). 3DNetMod outperformed DI-HMM and TADtree in capturing simulated domains with nested, partially overlapping structure, even when we added a full sweep of DI windows to the DI-HMM method (Fig. 2b). Finally, we compared the performance of 3DNetMod to Arrowhead7, TADtree and the full DI-HMM parameter sweep on Hi-C data from human cortical plate tissue (Fig. 2c–e). 3DNetMod outperformed current leading genome-wide domain-calling methods for sensitive and accurate detection of nested, partially-overlapping subTADs within TADs.
Figure 2.
3DNetMod outperforms leading domain-calling methods in real and simulated Hi-C data. (a) Simulated Hi-C data with nested, partially overlapping domain structure. Expected domains (top), DI-HMM + DI sweep domains (center top), TADtree domains (center bottom) and 3DNetMod domains (bottom) are shown. The maps shown are a zoomed-in view of the full simulations shown in Supplementary Figure 13d. (b) Receiver operating characteristic (ROC) curves showing the true positive rate and false positive rate of 3DNetMod (magenta), DI-HMM + DI Sweep (teal) and TADtree (blue) domain detection performance in the simulated Hi-C network with nested, partially overlapping domains. (c,d) Distribution of (c) domain sizes and (d) number of domains identified in chromosome 7 of human cortical plate tissue Hi-C16 by Arrowhead, DI-HMM + DI sweep, TADtree and 3DNetMod. (e) Heatmap of a 6-Mb region from human cortical plate tissue Hi-C16. Domains identified by Arrowhead (top), DI-HMM + DI sweep (center top), TADtree (center bottom) and 3DNetMod (bottom) are outlined in colors corresponding to their sizes: ≤400 kb (magenta), 401–800 kb (green), 801–1.6 Mb (cyan), 1.6–3 Mb (indigo), >3 Mb (orange).
3DNetMod’s ability to sensitively and accurately quantify chromatin domains may shed new light into how genome structure governs function across development and during the onset and progression of disease. Code, sample input data and usage instructions are freely available at https://bitbucket.org/creminslab/3dnetmod_method_v1.0_10_06_17.
ONLINE METHODS
Hi-C mapping, normalization and binning
Mapped, normalized and 40-kb-binned Hi-C matrices from human fetal cortical plate and germinal zone tissue generated in Won et al.16 were downloaded because raw reads were not readily available from GEO (Supplementary Table 1). Because the three central diagonals of the normalized data were all zero, we filled in these pixels with values drawn from the first off-diagonal of bins containing nonzero values. Mapped and 20-kb-binned Hi-C data from wild-type and Setdb1-deficient mouse neural cells generated in Jiang et al.20 were downloaded from GEO (Supplementary Table 1). Raw sequencing reads from two mouse embryonic stem (ES) cell replicates and two mouse cortical tissue replicates from Dixon et al.8 were downloaded from GEO (Supplementary Table 1). Dixon et al.8 paired-end reads were aligned independently to the mm9 mouse genome using bowtie2 (global parameters:–very-sensitive −L 30 −score-min L, −0.6, −0.2 −end-to-end–reorder; local parameters:–very-sensitive −L 20 −score-min L, −0.6, −0.2 −end-to-end–reorder) through HiC-Pro21. Unmapped reads, nonuniquely mapped reads and PCR duplicates were filtered, and uniquely aligned reads were paired. Hi-C maps were generated at 40 kb matrix resolution and balanced using the iterative correction and eigenvector decomposition (ICED) technique22. The resulting genome-wide Hi-C data is represented in a matrix of the format Aij, where the ijth element represents the interaction frequency between bins i and j. For all data, counts ≥1 were log transformed.
Hi-C data network construction
Hi-C data at 40 kb matrix resolution from mouse ES cells and cortical tissue8 and human cortical plate germinal zone tissue16 were parsed into 6-Mb regions with 4 Mb of overlap between adjacent regions. Hi-C data at 20 kb matrix resolution from Jiang et al.20 were parsed into 3-Mb regions with 2 Mb of overlap. Regions overlapping centromeres or telomeres were discarded. Additionally, low-information-content regions were discarded if they exhibited either (i) severe count sparsity, as indicated by a high percentage of pixels with zero values along the diagonal of the counts matrix (>5%) or (ii) >3 consecutive bins along the diagonal with zero counts.
Community detection with network modularity maximization and consensus partitioning
To partition genome-folding networks into communities, we employed a Louvain-like, locally greedy algorithm14 to maximize modularity. We first computed the modularity matrix, M, according to equation (2):
| (2) |
where M is a C × C matrix, C is the total number of communities, Mx,y is the normalized interaction frequency between x and y communities, kx is the sum of all edge weights for community x, γ is the resolution parameter (discussed below), and m is the total sum of all edge weights in network A (i.e., the sum of all interaction counts in the matrix excluding the i = j diagonal). For the first iteration of the algorithm (discussed in detail below), each individual node is an independent community; thus, indices for communities x and y correspond to the indices for nodes i and j. For any given γ, Axy/m is the normalized edge strength connecting communities x and y, and kxky /m2 is the expected normalized strength of edges at communities x and y if edges are placed at random. From the modularity matrix, M, the modularity, Q, is computed according to equation (3):
| (3) |
where Mx,y is the interaction frequency between x and y communities normalized to the expected interaction probability (equation (2)). To ensure that only edges within communities are added to the summation, δx,y is 1 if x and y are assigned to the same community (i.e., x = y) and 0 otherwise (i.e., x ≠ y).
The assumption of the Louvain-like algorithm is that the optimal community structure can be resolved by maximizing Q. The algorithm relies on an iterative, dynamic programming approach to rapidly converge on a local maximum in Q without comprehensively examining the entire search space. For each iteration t, individual nodes are given the opportunity to move into a new community placement that yields a locally maximal gain in Q, ΔQ, for the M network according to equation (4):
| (4) |
In the first iteration, t = 0, matrix M has the same dimensions as matrix A such that every node is assigned to its own community (i.e., C = N and x = i). Each node is given the chance to merge; and at the end of the iteration, if ΔQ ≤ 10−10, the algorithm terminates based on the assumption that no further moves will lead to a notable increase in Q. If ΔQ > 10−10, then the algorithm proceeds to the next iteration and resizes the modularity matrix, M, to C × C dimensions (where C is the number of communities computed in the previous t = t − 1 iteration, and (Mx,y)t=t is the sum of all previous (Mx,y)t=t−1 constituent edges that are merged into (Mx,y)t=t). The intuition for the iterations is that with increasing t the average size of communities increases with the gain in modularity. The algorithm terminates when no further single community merge leads to an improvement in modularity. Because modularity-maximization algorithms can converge on local maxima23, the Louvain-like algorithm was applied 20 times to sample the landscape of local maxima for each resolution parameter value (Supplementary Fig. 3e). To ensure random sampling of the landscape of possible maximum Q values, a randomized node selection with a unique seed value was used to run each of the 20 partitions.
To determine a consensus partition among the set of 20 partitions (Supplementary Fig. 3f), we identified the partition most similar to the 19 other partitions through use of a similarity metric: the adjusted RAND (aRAND) index24,25 from the sklearn toolbox sklearn.metrics.adjusted_rand_score. For perfectly matched partitions, aRAND is equal to unity. For partitions that are no more similar than would be expected by random chance, aRAND is close to 0 or slightly negative. The aRand was computed between all pairs of partitions within the set, and the partition with the highest average aRand, representing the partition that is most similar to all other partitions, was selected as the consensus partition26,27.
3DNetMod splitting and stitching approach to improve genome-wide Hi-C runtimes
A critical challenge in the application of 3DNetMod-MMCP to genome-wide Hi-C data is the runtimes. Although the Louvain-like locally greedy algorithm for modularity maximization scales linearly with network size14, computing the consensus partition with the aRAND index is a pairwise combinatorial algorithm that is nonpolynomial in scaling. Therefore, using chromosome-wide networks would give computationally intractable runtimes on Hi-C data binned at 40 kb matrix resolution in a genome-wide network of more than 75,000 nodes. High-resolution Hi-C data binned at 1–20 kb resolution would be computationally infeasible on a chromosome-wide network. Moreover, to find the optimal γ range (see below), 3DNetMod requires the random rewiring of genome-folding networks. The computational time to randomly rewire a network scales nonlinearly in time with network size, which also makes chromosome-wide 3DNetMod computationally intractable. We solved the time scaling issue by splitting the genome into defined 3-or-6 Mb regions and performing all domain-calling computations on each region in parallel.
Determining structural resolution parameter values with a γ plateau sweep
We developed 3DNetMod-GPS to determine the γ values that provide high-confidence domain calls across a range of length scales. To determine the maximum γ value for a given chromosome of a given Hi-C sample of interest, we first selected a representative subset of five evenly spaced regions of sufficient counts across a given chromosome. We constructed a randomly rewired network for each representative region by reshuffling the edge weights in a manner that preserves the symmetric nature of the adjacency matrix as well as preserving node degree while approximately preserving node strength17,28 (Supplementary Fig. 3b). To create randomly rewired networks, we implemented a python version of null_model_und_sign.m from the Brain Connectivity Toolbox (https://sites.google.com/site/bctnet/). Briefly, the expected weight, Ei,j, for all edges is computed as ki * kj, where ki is the degree of node i, and kj is the degree of node j. Observed (Ai,j) and expected (Ei,j) edge weights are then rank ordered in two independent lists. We then proceed with the following randomization steps: (i) a specific edge i,j (where i ≠ j) is randomly selected, (ii) the rank of that edge in the expected list is noted, (iii) the observed edge weight at the noted expected rank is then assigned to edge Ai,j, and (iv) the expected and observed values are then removed from their respective lists. New sets of remaining expected values and remaining observed values are computed and then ranked to reflect edge removal, and (v) steps i–iv are repeated until all observed edge values have been assigned. For each selected representative region and its accompanying randomly rewired network, modularity, Q, was computed beginning at γ = 0 and increasing in increments of 0.1 until the curves of Q versus γ for the real and randomly rewired networks converged (Supplementary Fig. 3a). The maximum γ value was selected as the largest γ value from the five regions at the convergence point between real and random networks. Max γ values were determined for all Hi-C data sets and all chromosomes.
To determine the subset of γ values that lead to high-confidence domain calls in a region-specific manner, we developed a gamma plateau selection method. First, the number of communities detected with a single application of the Louvain-like locally greedy algorithm for each γ value between 0 and the computed maximum value in increments of 0.01 was determined (Supplementary Fig. 3d). Next, ≥ n consecutive γ values with the same number of communities detected were grouped into ‘plateaus’, where n is a user-defined parameter. Notably, n = 3 was used for Won et al.16 Hi-C (Supplementary Fig. 6), n = 3 was used for Jiang et al.20 Hi-C, and n = 8 was used for Dixon et al.8 with lower sequencing depth. The median γ value in each plateau was then used for all subsequent domain-calling computations. Finally, 3DNetMod-MMCP (Supplementary Fig. 3e,f) was then applied for each selected γ value for each region to detect the nested hierarchy of domains (Supplementary Fig. 3g).
Processing filters for initial domain call set from 3DNetMod-GPS and 3DNetMod-MMCP
The initial full set of domains detected through 3DNetMod-GPS and 3DNetMod-MMCP was then processed to remove small communities and edge cases. Domains smaller than a user-defined size based on the resolution limit of the HiC data were removed. For all data sets, domains smaller than five genomic bins were removed. Thus, for 40-kb-binned Dixon et al.8 and Won et al.16 data, the minimum domain size was 200 kb. For 20-kb-binned Jiang et al.20 data, minimum domain size was 100 kb. Second, domains with at least one boundary within four bins of the edge of the region were removed. Third, redundant domains with exact match start and stop coordinates that were identified at multiple γ values or in multiple adjacent overlapping regions were merged into one domain. Finally, we observed that lower sequencing depth Hi-C data from Dixon et al.8 was prone to egregiously wrong domain calls that were driven by the high spatial noise and high number of outlier pixels. Thus, we developed an optional ‘trash’ community filter that identifies and discards domain calls with very sparse counts (less than 80% of counts along the domain edge are greater than the lowest 1% of total counts for the region). This filter was only applied to the low-sequencing-depth Dixon et al.8 data.
Hierarchical spatial variance minimization to refine domain calls
To identify high-confidence domains across length scales, we developed and applied a new hierarchical spatial variance minimization method (3DNetMod-HSVM). We first stratified communities by size: (i) minimum domain size to 400 kb (Level 1), (ii) 401 to 800 kb (Level 2), (iii) 801 to 1,600 kb (Level 3), (iv) 1,601 kb to 3 Mb (Level 4) and (v) greater than 3 Mb (Level 5). We then quantified the variability of a community boundary call across the 20 partitions by computing a boundary spatial variance. Specifically, we computed the spatial variance per boundary across the 20 partitions according to equation (5):
| (5) |
where xi is the coordinate of the boundary in partition i (in units of nodes), x̅ is the coordinate of the consensus boundary, and n is the number of partitions in the set. Boundaries with perfect agreement across the set of 20 partitions will have a variance of zero, whereas boundaries with large fluctuations in position across the set of partitions will have a higher variance. Boundary spatial variance values were then pooled for all communities of a given size stratum across a given chromosome.
A size-stratum-specific variance threshold can be selected to minimize false positive domain calls and inadvertent detection of compartments as domains (Supplementary Fig. 3h–l). For Won et al.16 human cortical plate and human germinal zone tissue Hi-C, we selected thresholds of L1: 70% area under the curve (AUC), L2: 100% AUC, L3: 100% AUC, L4: 60% AUC, L5: 0 variance (Supplementary Fig. 10). For Dixon et al.8 mouse cortical tissue, we selected thresholds of L1: 0 variance, L2: 70% AUC, L3: 60% AUC, L4: 100% AUC, L5: 0 variance (Supplementary Fig. 11). For Jiang et al.20 wild-type mouse neural cells, we selected thresholds of L1: 0 variance, L2: 35% AUC, L3: 30% AUC, L4: 100% AUC, L5: 0 variance (Supplementary Fig. 12). Communities with a boundary that did not pass their size-stratum-specific variance threshold were considered low confidence and were removed from the list of domain calls. The resulting communities represent the high-confidence list of domains identified for a given cell type and replicate (Supplementary Fig. 3l).
Postprocessing chaos filter to remove domains called in regions with minimal substructure
To allow users to remove domains called in regions where there is minimal structure, we developed and implemented an optional ‘chaos filter’. This filter is based on the observation that segments of the genome with nested substructure have greater maximum counts and variance in the band 2–10 bins from the diagonal compared to regions with no nested substructure (or regions of ‘chaos’). To identify regions with minimal structure, each chromosome (excluding regions that were previously discarded for having low counts or being at centromeres or telomeres) is evaluated in a sliding window of 20 bins with a one-bin increment. For each 20-bin window, Wi, the maximum count and variance of each n off-diagonal, where n ranges from 2 to 10, is computed. These metrics are then compared to the corresponding off-diagonal chromosome-wide average of maximum and variance across all W. If none of the metrics for window Wi exceed a user-defined percentage of the corresponding chromosome-wide average, Wi is considered to have no nested substructure, and domains identified within Wi are discarded after variance thresholding. For Won et al.16 and Jiang et al.20 Hi-C data, we used a chaos filter percentage of 85%. For Dixon et al.8 Hi-C data, we used a more stringent chaos filter percentage of 92%. Domain calls post-chaos filter for Won et al.16 and Jiang et al.20 were used for comparison of methods in Figure 2 and Supplementary Figures 14 and 15.
Assembly of final domain calls
To assemble the list of final domain calls, highly similar domains identified within the same sample were merged together. Two domains were considered ‘highly similar’ if the distance between their start coordinates was less than or equal to a user-defined allowance (typically equivalent to one genomic bin), and the distance between their stop coordinates also fell within the user-defined allowance. The new merged domain has boundary coordinates that encompass the constituent domains (i.e., the upstream boundary extends from the first upstream boundary coordinate to the last boundary coordinate of the constituent domains). For data sets without biological replicates for a given cellular condition (Won et al.16), the merged domain calls (post-chaos filter if chaos filter is used) represent the final 3DNetMod domain calls. For data sets with two biological replicates of a given cellular condition (Dixon et al.8, Jiang et al.20), an additional set of consistent domains was made containing calls that were identified in both biological replicates within the user-defined tolerance window (i.e., the distance between the start coordinate of a given domain in replicate one and replicate two must be less than or equal to the user-defined allowance, and the distance between their stop coordinates must also fall within the allowance).
Final 3DNetMod domain calls for human cortical plate Hi-C data16 as well as mouse neural cell Hi-C data20 can be found in the bitbucket repository. Genomic regions that were removed from consideration because of count sparsity or gaps in counts can also be found within the bitbucket repository.
3DNetMod runtimes
Using a local machine (2.5 GHz Intel Core i7) across four processors, the runtime for chromosome 7 of 40 kb binned data from Won et al.16 was 52 min (3,096 s). Genome-wide runtime was approximately 22 h (80,640 s) for a quadcore. With access to a high-performance computing cluster, all chromosomes and regions can be run in parallel, and genome-wide domain calls can be identified in ~1 h for 40-kb-binned data.
We found that average runtimes were consistent for regions with the same number of nodes but different matrix resolutions (data not shown) when using the same plateau size. For small region sizes (i.e., split regions up to 300 nodes instead of chromosome-wide networks), we observed that the runtime per region scales linearly with region size. For region sizes larger than 300 nodes, 3DNetMod has nonlinear scaling that makes the method computationally intractable. The critical aspects of 3DNetMod contributing to the nonlinear scaling are: (i) region size (i.e., number of nodes) during 3DNetMod-MMCP, (ii) region size during the random wiring step of 3DNetMod-GPS, and (iii) plateau size during 3DNetMod-GPS (smaller plateaus take significantly longer time to process because of more γ values). Thus, we recommend that users split the genome into regions of ~75–300 nodes.
Comparison of domain calls in 3-Mb, 6-Mb and 12-Mb regions
To assess the effect of region size on the quality of domains identified (Supplementary Fig. 4), Hi-C data from human cortical plate tissue16 were split into 3-Mb regions with 2 Mb overlap, 6-Mb regions with 4 Mb overlap and 12-Mb regions with 8 Mb overlap. 3DNetMod-GPS and 3DNetMod-MMCP were performed with a plateau size of 3 and all other default parameters. 3DNetMod-HSVM was performed with L1: 70% area under the curve (AUC), L2: 100% AUC, L3: 100% AUC, L4: 60% AUC, L5: 0 variance. Chaos filtering was not performed.
Benchmarking 3DNetMod against leading domain-calling methods in Hi-C
To qualitatively assess the performance of 3DNetMod compared with that of three leading domain-calling methods, we identified domains in one or more Hi-C data sets using DI-HMM8, Arrowhead7 and TADtree19. Across the methods, domains identified in regions with low counts or regions overlapping centromeres and telomeres (i.e., regions excluded from 3DNetMod domain calling) were excluded from further analysis.
To identify domains using the Directionality Index and Hidden Markov Model (DI-HMM) method detailed in Dixon et al.8, we created a python implementation of the method. Briefly, the DI test statistic used to measure upstream and downstream portions of a topological domain was computed according to equation (6):
| (6) |
where B is the total summation of counts within a 1D horizontal array of length L (a user-adjustable number of genomic bins) upstream of the diagonal, A is the total summation of counts within a 1D horizontal array of length L downstream of the diagonal, and E is the mean of A and B. For 40 kb Hi-C, the standard DI A/B length L is 50 bins (2 Mb). DI computation was then followed by a mixed Hidden Markov Model (mHMM) using Gaussian mixtures to predict ‘upstream bias’, ‘downstream bias’ and ‘no bias’ states using the procedure of Dixon et al.8. We used the Baum–Welch expectation-maximization algorithm and the Forward–Backward algorithm to estimate posterior marginals and compute the maximum likelihood estimate. The mixture with best goodness of fit was chosen by AIC criterion (equation (7)):
| (7) |
where k is the number of parameters in the model, and L is the maximum likelihood estimate. mHMM was then followed by a postprocessing step where only regions with median posterior probabilities ≥99% or a region comprised of at least 2 bins were selected. Domains were initiated at the beginning of a downstream-biased mHMM state and continued throughout consecutive downstream states. Domains ended when the last of the upstream-biased states was reached.
To identify a nested hierarchy of domains in human cortical plate cells16 using the DI-HMM method (Fig. 2e), we used a range of Directionality Index lengths8: 7 bins, 8 bins, 20 bins, 35 bins and 50 bins. We determined 7 bins to be the lower limit on account of the narrow distribution of DI values relative to the number of bins genome wide, resulting in rank deficiency during matrix inversion within HMM.
To call domains using the TADtree method19, we downloaded the TADtree python tool from http://compbio.cs.brown.edu/projects/tadtree. TADtree recursively maximizes a boundary index that looks for shifts in interaction frequency at TAD boundaries to form an optimal set of nested TAD trees19. We kept the default or recommended parameters to identify domains in Won et al.16 cortical plate chromosome 7 (Fig. 2e and Supplementary Fig. 14c). For 40 kb resolution data, these parameters are: s = 50, m = 10, P = 3, q = 12, gamma = 500 and n = 40019. The TADtree authors recommend 2 Mb as the maximal detected TAD size, which translates to s = 50 for 40-kb-binned data. The reported ~O(s5) scaling of the TADtree method19 leads to computationally intractable run times for Hi-C data binned lower than 40 kb matrix resolution. Because Jiang et al.20 wild-type neuron Hi-C is binned at 20 kb resolution, we could only run the method on a small individual chromosome (chr 18) with s = 50 (1 MB maximal detected TAD size) and default n, m, p, q and gamma values (Supplementary Fig. 15b). To detect domains with standard s = 100 (2 Mb maximal detected TAD size), we could only run half of chromosome 18 with n scaled to 200 and default m, p, q and gamma values (Supplementary Fig. 15c). Per the recommendation of TADtree authors, a final set of domain calls was chosen such that at most 2% of all outputted TADs are duplicates.
To call domains using the Arrowhead method, Juicer_tools_0.7.0 was installed from https://github.com/theaidenlab/juicer/wiki/Download. Arrowhead calculates the likelihood that a given pixel corresponds to the corner of a domain and is capable of finding a nested hierarchy of domains7. For Won et al.16 data, the −n flag was used to indicate that data were already matrix balanced, whereas for Jiang et al.20 data, the −n flag was omitted, and default matrix normalization was performed. All other default parameters were used to identify domains in Won et al.16 (Fig. 2e and Supplementary Fig. 14a) and Jiang et al.20 Hi-C data (Supplementary Fig. 15a).
To more appropriately compare domains identified across the methods, domains identified in regions with low counts or regions overlapping centromeres and telomeres (i.e. regions already excluded from 3DNetMod analysis) were excluded from further analysis.
We compared the run times of 3DNetMod to TADtree on chromosome 18 of Jiang et al.20 wild-type neurons and chromosome 7 of Won et al.16 cortical plate. We used these single-chromosome runtimes in combination with the equations provided in Weinreb et al.19 to estimate a genome-wide runtime for TADtree (Supplementary Table 2) and compared this runtime to actual runtimes of 3DNetMod. We estimate that a genome-wide runtime of TADtree on 40-kb-binned data would take ~44.5 h of computation time, whereas a 3DNetMod takes ~22 h of computation time. Furthermore, we estimate that the genome-wide computation time of TADtree on 20-kb-binned data using all processors on a quad-core computer would take >63 d, whereas 3DNetMod takes ~2 d of computation time.
Benchmarking 3DNetMod against leading domain-calling methods in simulations
To quantitatively compare the performance of DI-HMM, 3DNetMod and TADtree in identifying domain structures, we constructed two different network simulations with known community structure: (i) binarized (Supplementary Fig. 13a) and (ii) nested overlapping (Fig. 2a and Supplementary Fig. 13c,d). A 400 × 400 bin binary network was constructed with three domain sizes of 100, 50 and 10 bins (14 in total). Each pixel within a domain was assigned a value of 1,000, while pixels not belonging to a domain were each assigned a value of zero. Additionally, we derived a nested overlapping 480 × 480 bin simulation from a well-ordered Sierpinski fractal pattern. In the nested construct, the simulated counts within the original layered triangles were randomly scrambled within the same layer to form square domains with the corners preserved (unscrambled). We constructed five nested layers of domains: the first inner layer consisted of 24 total 20-bin-sized domains. The second layer consisted of 22 total overlapping domains of size 40 bins offset midway at 20 bins. The third layer consisted of six 80-bin-sized domains, and the fourth and fifth layers contained three 160-bin domains and one 320-bin domain, respectively. The intensity of each layer was set to scale with the distance dependence of chromosome conformation capture data. The intensity of the domain corners was raised beyond the distance expectation to simulate loops anchoring chromatin-folding domains.
We applied DI-HMM, 3DNetMod and TADtree to find domains in our simulated Hi-C networks (Fig. 2 and Supplementary Fig. 13). DI A/B lengths L of 20, 30, 40, 60, 80, 120, 160, 240, 320 and 360 bins were chosen to capture as much nested structure as possible in the nested simulation, while L of 100 and 50 bins were chosen for the binary simulation. In 3DNetMod, we used a convergence threshold of 0.0035 for max γ value determination, 20 partitions, a plateau size of 1 and a minimal size threshold of two nodes. No variance thresholding at any length scale or filtering was applied.
To run TADtree on the nested overlapping simulations, an s value of 200 bins was used on account of limitations of O(s5) scaling (TADtree with s = 200 required 433,862 s to detect domains in the simulation). Thus, only one domain at the largest scale (320 bins) of the 56 known simulated domains could not be detected and was omitted from consideration. For the other TADtree parameters, m = 5, n = 56, P = 3, gamma = 500 and q = 12 were selected. For binary networks, an s value of 100 bins was used and other parameter choices included: m = 2, n = 14, P = 3, gamma = 500 and q = 12. We were unable to detect domains in our simulations using the Arrowhead method, as our simulation data structure was not readily transformed into the format required for the method’s software implementation.
A receiver operating characteristic (ROC) curve was constructed for DI sweep, 3DNetMod and TADtree (Fig. 2b and Supplementary Fig. 13b) by comparing a combined test array of all nested layers against a combined binary true array of all layers with one at a true domain boundary and zero elsewhere. The simulations shown in Supplementary Figure 13a were used to construct the ROC shown in Supplementary Figure 13b. The simulations shown in Supplementary Figure 13d were used to construct the ROC shown in Figure 2b. To score the confidence in the domain calls for the test array for DI sweep, boundaries of domains were assigned the domain average probability of each most likely hidden state. For 3DNetMod, boundaries of domains were assigned the boundary spatial variance. For TADtree, no metric is provided for the confidence in domain calls; thus, the boundaries of domains were assigned a confidence of 1. If called boundaries were within 3 bins of the actual boundary, and the domain sizes did not differ by more than 4 bins, the call was considered correct, and the boundary position in the test array was assigned to the same boundary position in the true array. Otherwise, the test boundary was tallied as a false positive.
Pileups of CTCF at domain boundaries
To assess the enrichment of chromatin marks at domain boundaries identified in wild-type mouse neurons at different length scales (Supplementary Fig. 9), we computed the average number of wild-type CTCF ChIP-seq peaks per 40-kb genomic interval at increasing distances from boundary coordinates.
Spring force diagram visualization
We visualized networks as spring-force diagrams using the MATLAB BGL toolbox (Fruchterman and Reingold, Code from MatlabBGL toolbox, David Gleich, Fig. 1). Networks were thresholded such that the top 15% of edge weights were visualized. A threshold of 15% was chosen because it was stringent enough to improve visualization, but it was lenient enough for the graph to remain fully connected. All analyses were performed on the fully weighted graphs; ad hoc thresholds were applied to facilitate visualization.
Life Sciences Reporting Summary
Further information regarding the experimental design may be found in the Life Sciences Reporting Summary.
Data availability
Code, sample input data and usage instructions are available at the following Bitbucket repository: https://bitbucket.org/creminslab/3dnetmod_method_v1.0_10_06_17. The data analyzed in this study are summarized in Supplementary Tables 1 and 3. Source data for Figures 1 and 2 are available online.
Supplementary Material
Acknowledgments
J.E.P.-C. is a New York Stem Cell Foundation (NYSCF) Robertson Investigator and an Alfred P. Sloan Foundation Fellow. This work was funded by The New York Stem Cell Foundation (J.E.P.-C.), the Alfred P. Sloan Foundation (J.E.P.-C.), the NIH Director’s New Innovator Award (1DP2MH11024701; J.E.P.-C.), a 4D Nucleome Common Fund grant (1U01HL12999801; J.E.P.-C.) and a joint NSF-NIGMS grant to support research at the interface of the biological and mathematical sciences (1562665; J.E.P.-C.). This material is based upon work supported by the National Science Foundation Graduate Research Fellowship under DGE-1321851 (H.N.). D.S.B. would also like to acknowledge support from the John D. and Catherine T. MacArthur Foundation.
Footnotes
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
J.E.P.-C., D.S.B. and H.K.N. conceived of the study. H.K.N., D.J.E., S.G., H.H., K.R.T. and J.K. implemented the computational pipeline. J.E.P.-C., H.K.N., D.J.E. and D.S.B. wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Bullmore E, Sporns O. Nat. Rev. Neurosci. 2009;10:186–198. doi: 10.1038/nrn2575. [DOI] [PubMed] [Google Scholar]
- 2.Girvan M, Newman ME. Proc. Natl. Acad. Sci. USA. 2002;99:7821–7826. doi: 10.1073/pnas.122653799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lanctôt C, Cheutin T, Cremer M, Cavalli G, Cremer T. Nat. Rev. Genet. 2007;8:104–115. doi: 10.1038/nrg2041. [DOI] [PubMed] [Google Scholar]
- 4.Dekker J, Marti-Renom MA, Mirny LA. Nat. Rev. Genet. 2013;14:390–403. doi: 10.1038/nrg3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lieberman-Aiden E, et al. Science. 2009;326:289–293. [Google Scholar]
- 6.Phillips-Cremins JE, et al. Cell. 2013;153:1281–1295. doi: 10.1016/j.cell.2013.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rao SS, et al. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dixon JR, et al. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nora EP, et al. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sexton T, et al. Cell. 2012;148:458–472. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 11.Dali R, Blanchette M. Nucleic Acids Res. 2017;45:2994–3005. doi: 10.1093/nar/gkx145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forcato M, et al. Nat. Methods. 2017;14:679–685. doi: 10.1038/nmeth.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Newman ME. Proc. Natl. Acad. Sci. USA. 2006;103:8577–8582. doi: 10.1073/pnas.0601602103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blondel VD, Guillaume J-L, Lambiotte R, Lefebvre E. J. Stat. Mech. Theory. E. 2008;2008:P10008. [Google Scholar]
- 15.Newman MEJ. Nat. Phys. 2012;8:25–31. [Google Scholar]
- 16.Won H, et al. Nature. 2016;538:523–527. doi: 10.1038/nature19847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rubinov M, Sporns O. Neuroimage. 2011;56:2068–2079. doi: 10.1016/j.neuroimage.2011.03.069. [DOI] [PubMed] [Google Scholar]
- 18.Ball B, Karrer B, Newman MEJ. Phys. Rev. E. 2011;84:036103. doi: 10.1103/PhysRevE.84.036103. [DOI] [PubMed] [Google Scholar]
- 19.Weinreb C, Raphael BJ. Bioinformatics. 2016;32:1601–1609. doi: 10.1093/bioinformatics/btv485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang Y, et al. Nat. Genet. 2017;49:1239–1250. doi: 10.1038/ng.3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Servant N, et al. Genome Biol. 2015;16:259. doi: 10.1186/s13059-015-0831-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Imakaev M, et al. Nat. Methods. 2012;9:999–1003. doi: 10.1038/nmeth.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Good BH, de Montjoye Y-A, Clauset A. Phys. Rev. E. 2010;81:046106. doi: 10.1103/PhysRevE.81.046106. [DOI] [PubMed] [Google Scholar]
- 24.Traud AL, Kelsic ED, Mucha PJ, Porter MA. SIAM Rev. 2011;53:526–543. [Google Scholar]
- 25.Hubert L, Arabie P. J. Classif. 1985;2:193–218. [Google Scholar]
- 26.Doron KW, Bassett DS, Gazzaniga MS. Proc. Natl. Acad. Sci. USA. 2012;109:18661–18668. doi: 10.1073/pnas.1216402109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lohse C, Bassett DS, Lim KO, Carlson JM. PLoS Comput. Biol. 2014;10:e1003712. doi: 10.1371/journal.pcbi.1003712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maslov S, Sneppen K. Science. 2002;296:910–913. doi: 10.1126/science.1065103. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Code, sample input data and usage instructions are available at the following Bitbucket repository: https://bitbucket.org/creminslab/3dnetmod_method_v1.0_10_06_17. The data analyzed in this study are summarized in Supplementary Tables 1 and 3. Source data for Figures 1 and 2 are available online.