Abstract
To date, our knowledge of apoptosis regulation in insects comes almost exclusively from the model organism Drosophila melanogaster. In contrast, despite the identification of numerous genes that are presumed to regulate apoptosis in other insects based on sequence homology, little has been done to examine the molecular pathways that regulate apoptosis in other insects, including medically important disease vectors. In D. melanogaster, the core apoptosis pathway consists of the caspase negative regulator DIAP1, IAP antagonists, the initiator caspase Dronc and its activating protein Ark, and the effector caspase DrICE. Here we have studied the functions of several genes from the mosquito disease vector Aedes aegypti that share homology with the core apoptosis genes in D. melanogaster. Silencing of the iap1 gene in the A. aegypti cell line Aag2 caused spontaneous apoptosis, indicating that IAP1 plays a role in cell survival similar to that of DIAP1. Silencing A. aegypti ark or dronc completely inhibited apoptosis triggered by several different apoptotic stimuli. However, individual silencing of the effector caspases CASPS7 or CASPS8, which are the closest relatives to DrICE, only partially inhibited apoptosis, and silencing both CASPS7 and CASPS8 together did not have a significant additional effect. Our results suggest that the core pathway that regulates apoptosis in A. aegypti is similar to that of D. melanogaster, but that more than one effector caspase is involved in apoptosis in A. aegypti. This is interesting in light of the fact that the caspase family has expanded in mosquitoes compared to D. melanogaster.
Keywords: apoptosis, caspase, Aedes aegypti, inhibitor of apoptosis, DIAP1, Dronc, Ark
INTRODUCTION
Apoptosis is a type of programmed cell death that is important for many processes in metazoan organisms, including immunity, development, and tissue homeostasis. The molecular pathways that regulate apoptosis have been mainly studied in organisms that serve as genetic models: the nematode Caenorhabditis elegans, the insect D. melanogaster and the laboratory mouse. From studies done in these organisms, a conserved core apoptosis pathway has emerged [1]. Central to this core pathway are the caspases, a family of cysteine proteases that are present in most cells as inactive zymogens and become activated following a death signal. Activated caspases cleave cellular substrates, leading to the stereotypical morphological changes associated with apoptotic cell death, and so their activation by adaptor proteins must be carefully regulated. The first caspases that are activated following an apoptotic signal are initiator caspases, which auto-activate following binding to oligomerizing adaptor proteins, such as Apaf-1 in mammals or Ark in D. melanogaster. Once activated, initiator caspases cleave and activate effector caspases, which are responsible for cleaving numerous substrates in the cell, leading to apoptosis.
In flies and mammals, the main negative regulators of caspases are inhibitor of apoptosis (IAP) proteins, which inhibit caspases through direct binding and ubiquitination. Positive regulators of caspase regulation include the IAP antagonists, which bind to IAP proteins and destabilize IAP proteins, and also prevent them from binding to caspases. Together, caspases, IAPs, and IAP antagonists make up a core pathway in which the central players are largely conserved between flies and mammals.
In the 1990s, genetic studies in D. melanogaster began to define the molecules that were involved in the regulation of apoptosis in this model insect. Since then, work by many research groups has established the core apoptosis pathway in D. melanogaster (Fig 1). At the heart of this pathway is the initiator caspase Dronc, which is required for nearly all apoptotic cell deaths in the fly [2–4]. One of the notable exceptions where Dronc was not required was cell death in the midgut during the larval to pupal molt [3]; however, it was later reported that the cell death in this situation is due to autophagy, and is caspase-independent [5]. Dronc activation requires the adaptor protein Ark, the D. melanogaster ortholog of mammalian Apaf-1, which binds to Dronc and forms a complex known as the apoptosome which promotes Dronc dimerization and activation [6]. Unlike mammalian cells, which require release of cytochrome c from mitochondria in order for apoptosome assembly to occur, Ark-dependent Dronc activation appears to occur at a low constitutive level in D. melanogaster cells, since continuous expression of the IAP protein DIAP1 is required for cell survival in both the embryo and in S2 cells [7–10]. DIAP1, which contains a RING domain, functions as an E3 ubiquitin ligase and inactivates Dronc, as well as the effector caspase DrICE, by both degradative and non-degradative ubiquitylation [11]. A negative feedback mechanism appears to regulate the levels of both Dronc and Ark, wherein either protein can lower the levels of the other in a DIAP1-dependent fashion [12].
Fig 1. Genes that constitute the core apoptosis pathway in D. melanogaster, and their corresponding homologs in A. aegypti.
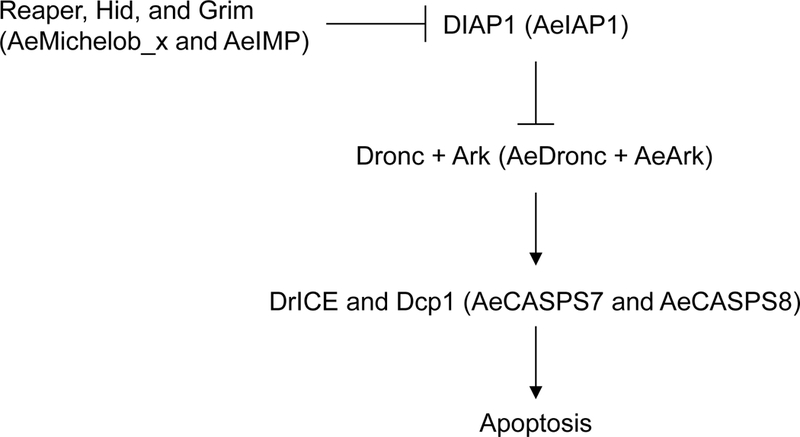
Genes that constitute the core apoptosis pathway in D. melanogaster and their corresponding homologs in A. aegypti (in parentheses) are shown. Negative regulation is illustrated by blunt symbols, while positive regulation is indicated by arrows.
The initiation of apoptosis involves an increase in either the expression level or the activity of IAP antagonists, such as Reaper, Hid, and Grim (reviewed in [11]. The IAP antagonists inactivate DIAP1 by two mechanisms: first, by displacing caspases from DIAP1 due to their ability to bind to the same sites on DIAP1 as caspases, and second, by inducing DIAP1 auto-ubiquitination. Following an apoptotic signal, IAP antagonists become activated through a combination of transcriptional and/or post-transcriptional mechanisms, and prevent DIAP1 from inhibiting Dronc. Activated Dronc cleaves and activates DrICE, which is the main effector caspase involved in apoptosis. Another closely related effector caspase, DCP-1, has an effect only in certain cell types and only when DrICE is mutated [13, 14]. The importance of DrICE was also supported by experiments utilizing in vivo RNAi to assess the contribution of effector caspases to suppressing death due to RNAi of DIAP1, in which co-silencing of DrICE, DCP-1, Decay and Damm was only slightly more effective at rescuing the phenotype due to DIAP1 silencing than silencing DrICE alone [15].
Unlike in mammals, where there are at least two distinct pathways of caspase activation, the intrinsic and the extrinsic pathways, a single core pathway appears to be involved in all apoptotic cell deaths in D. melanogaster. Although there is a role for the tumor necrosis factor homolog Eiger in certain apoptotic events, downstream apoptosis signaling by Eiger still occurs via the same core pathway as other death signals, involving Dronc and Ark [16].
The mosquito A. aegypti is the main vector involved in the transmission of yellow fever and Dengue fever. Sequencing of the A. aegypti genome allowed for the identification of a number of proteins which are predicted to play roles in regulating apoptosis, based on their homology to D. melanogaster proteins [17, 18]. These include A. aegypti IAP1, Ark, Dronc, the IAP antagonists Mx and IMP, a number of effector caspases, and several other proteins more peripheral to the core apoptosis pathway [17]. To date, functional studies of these proteins have been limited. Overexpression of Mx or IMP has been shown to induce apoptosis in heterologous species [17, 19]. Introduction of A. aegypti IAP1 dsRNA into adult mosquitoes results in death of the mosquito, but a role in apoptosis has not been directly established [20]. A. aegypti Dronc has been shown to be an active caspase, and to be regulated by hormonal signaling [21]. No studies have been done studying the overall apoptosis pathway in A. aegypti.
Here, we have undertaken a functional analysis of several of the A. aegypti genes that are predicted to be important in the core apoptosis pathway by examining the effects of silencing iap1, ark, dronc, and the effector caspases casps7 and casps8 in an A. aegypti cell line. A functional analysis of the IAP antagonist genes Imp and Mx is the subject of a separate study (H. Wang and R.J. Clem, in preparation). Our results demonstrate that the core apoptotic pathway is intact in A. aegypti and is regulated in a manner largely similar to that of D. melanogaster, but that at least two effector caspases appear to play non-redundant roles in apoptosis in A. aegypti.
MATERIALS AND METHODS
Cell culture
The A. aegypti cell line Aag2 [22] was obtained from Carol Blair (Colorado State University) and propagated at 27°C in Schneider’s medium (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Atlanta Biologicals).
Production of dsRNA
The open reading frames (ORFs) of A. aegypti ark, dronc, casps7, casps8, iap1 and the control baculovirus gene polyhedrin were cloned into the plasmid pCRII® (Invitrogen) and templates were amplified by PCR using primers bearing T7 polymerase binding sites both upstream (CTAATACGACTCACTATAGGGCAGGAAACAGCTATGAC) and downstream (CAGTCACGACGTTGTAAAACGACGGCC) of the ORFs of interest. The resulting PCR products were purified by phenol-chloroform-isoamyl alcohol extraction and were transcribed to synthesize complementary RNA strands using AmpliScribe™ T7 Transcription Kit (EPICENTRE Biotechnologies). The transcription products were treated with DNase I (37°C, 15 min) and then subjected to phenol-chloroform-isoamyl alcohol extraction. The purified complementary RNA strands were heated to 65°C for 15 min followed by cooling slowly to room temperature to obtain dsRNA which were then stored at −80°C.
RNAi
Aag2 cells were plated in 6 well plates (1×106 cells/well in 2 ml Schneider’s medium supplemented with 10% FBS) and incubated at 27°C overnight. The following day, the medium was replaced by 1 ml serum free medium and 40 μg dsRNA per gene being silenced was added into the medium and incubated at 27°C for 2 hours, followed by the addition of 1 ml medium supplemented with 20% FBS bringing the final concentration of FBS to 10%. Control Aag2 cells were subjected to the same procedure except that no dsRNA or control (baculovirus polyhedrin) dsRNA was added. The cells were incubated for the indicated time period to induce the RNAi effect.
Semi-quantitative RT-PCR
Total RNA was isolated from mock-treated or dsRNA-treated Aag2 cells using Trizol reagent (Invitrogen) as described in the protocol by the manufacturer. 1 μg of total RNA was used in the first strand cDNA synthesis by using ImProm-II™ Reverse Transcription System (Promega) according to the instructions provided by the manufacturer. Estimation of relative mRNA levels of specific genes was performed by PCR in the linear range of amplification using gene-specific primers. The number of PCR cycles was kept in the linear range by testing an increasing number of cycles (18, 21, 23, 25, 28, and 32) for each set of template and primers, and then choosing a cycle number where the amount of product was still increasing. The housekeeping gene used was the actin paralog AAEL001673.
Induction of apoptosis
To induce apoptosis, Aag2 cells with or without the indicated RNAi treatment were subjected to UV irradiation (254nm) by placing the plates on a transilluminator for 15 min, or by treating the cells with cycloheximide (CHX, at a final concentration of 100 ng/ml, Sigma), staurosporine (STS, at a final concentration of 1 μM, Sigma) or actinomycin D (Act D, at a final concentration of 50 ng/ml, Sigma) for the amount of time indicated.
Assessment of apoptosis
Observation of morphological change.
At the indicated time points after RNAi treatment or induction of apoptosis, cells and apoptotic bodies were photographed by bright field microscopy at 400X magnification.
DNA fragmentation assay.
Equal numbers (1×106) of Aag2 cells treated with iap1 or polyhedrin dsRNA for 24 hours were collected by centrifugation at 500 g for 5 min and apoptotic bodies were collected from the cell supernatant by centrifugation at 14,000 g for 10 min. Cells and apoptotic bodies were combined and suspended in 100 μl lysis buffer (10 mM Tris-HCl, pH 8.0, 150 mM NaCl, 10 mM EDTA, 0.4% SDS, 1 mg/ml proteinase K, 200 μg/ml RNase A) and incubated at 56°C for 3 hours. After phenol-chloroform-isoamyl alcohol extraction, total DNA was isolated by precipitation with 3M NaAc and ethanol and dissolved in TE buffer (10 mM Tris-HCl, 1 mM EDTA). Low molecular weight DNA was visualized under ultraviolet light after being subjected to electrophoresis on a 1.5% agarose gel in the presence of 0.5 μg/mL ethidium bromide.
Caspase activity assay.
Aag2 cells and apoptotic bodies were collected as described above for the DNA fragmentation assay and suspended in lysis buffer containing 10 mM HEPES, pH 7.0, 2 mM EDTA, 0.1% CHAPS and 5 mM dithiothreitol, supplemented with one complete mini EDTA-free protease inhibitor tablet (Roche Molecular Biochemicals) per 50 ml lysis buffer. Cells and apoptotic bodies were lysed by one cycle of freeze-thaw followed by centrifugation at 14,000 g for 15 min. Cell lysate (20 μg total protein/sample) was added to reaction mixture containing 25 mM HEPES, pH 7.5, 1 mM EDTA, 0.1% CHAPS, 10% sucrose, 10 mM dithiothreitol and 50 μM Ac-DEVD-AFC (MP Biochemicals) as substrate in a total volume of 100 μl. Fluorescence was measured in a Perkin Elmer Victor 3 multilabel counter at 37°C for 2 hours and final values of the measurements were used to evaluate the caspase activities of the according cell lysates.
RESULTS
Expression of apoptosis-related genes in the A. aegypti cell line Aag2
The core apoptosis pathway in D. melanogaster includes initiator and effector caspases, as well as other proteins which positively or negatively regulate caspase activation (Fig 1). A number of A. aegypti genes have been identified that share sequence homology with D. melanogaster apoptotic regulatory genes, and thus are predicted to participate in regulating apoptosis in A. aegypti [17, 18]. These include eleven caspases (dronc, dredd, casps7 – 8, and casps15 – 21); three IAPs (iap1, 2, and 5); two IAP antagonists (mx and imp); and ark, BG4 (fadd) and dnr1. To determine whether these genes are expressed in the A. aegypti cell line Aag2, RT-PCR was performed on total RNA from Aag2 cells. Transcripts for all 19 of the genes tested were amplified by RT-PCR (data not shown), indicating that all of these genes are expressed in Aag2 cells.
Aag2 cells are able to take up dsRNA and mount a specific RNAi response
When dsRNAs corresponding to iap1, dronc, ark, casps7, or casps8 were individually added to Aag2 cells, a dramatic decrease in steady-state level of transcripts was observed in each case (Fig 2 and Fig 3a). In the case of iap1, dronc, and casps8, transcripts were undetectable by RT-PCR as soon as 7 hours after addition of dsRNA, and the level of transcript remained undectable through at least 33 hrs (Fig 2 and Fig 3a). ark transcript levels were unchanged at 7 hrs, rose slightly at 11 hrs, but were greatly reduced after 27 and 33 hrs (Fig. 2). Although we observed in two independent experiments that ark transcript levels rose slightly at 11 hrs, this did not seem to affect the results in later experiments (see Fig 5 and Fig 6). casps7 transcript levels were dramatically lowered from 7 hrs till 33 hours but not completely silenced as seen with transcript levels of iap1, dronc and casps8 (Fig 2). In order to find out if higher amounts of dsRNA would completely silence casps7 transcription, we increased the amount of dsRNA used, but casps7 transcript levels were not completely silenced even when the amount of dsRNA was increased by 2-fold (80 μg) or 5-fold (200 μg) (data not shown). Treatment with any of the dsRNAs did not have any effect on levels of any of the other transcripts (Fig 2), suggesting that the silencing response was specific. These results demonstrate that Aag2 cells are able to take up dsRNA and mount a robust and specific RNAi response, which makes them useful for studying apoptosis regulation in A. aegypti.
Fig 2. Silencing of apoptosis-related genes in Aag2 cells.
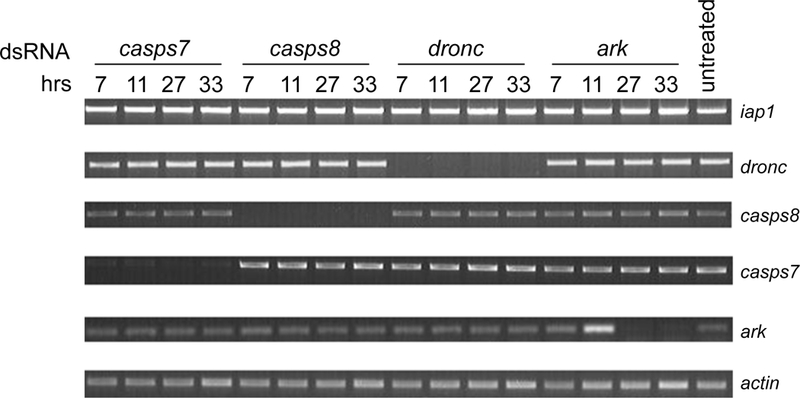
Total RNA was prepared from untreated Aag2 cells or Aag2 cells treated with the indicated dsRNAs for 7, 11, 27, or 33 hrs. Total RNA was subjected to RT-PCR using gene-specific primers for A. aegypti iap1, dronc, casps7, casps8, ark, or actin. The RT-PCR products were analyzed by agarose gel electrophoresis and visualized under UV light.
Fig 3. Silencing iap1 induces apoptosis in Aag2 cells.
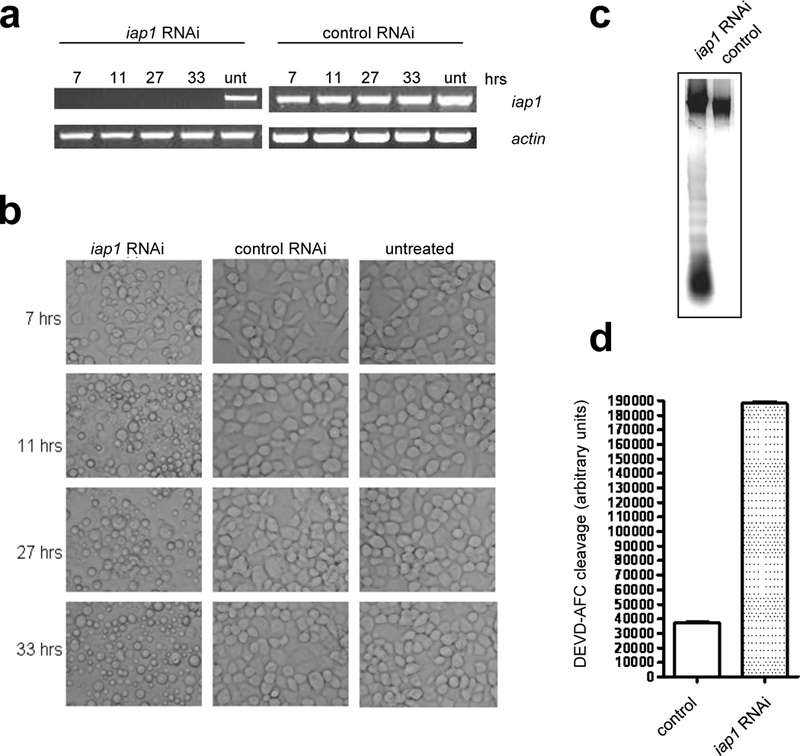
a) Total RNA was prepared from untreated Aag2 cells (unt) or Aag2 cells treated for 7, 11, 27 and 33 hrs with iap1 dsRNA or control polyhedrin dsRNA and then semi-quantitative RT-PCR was performed using gene-specific primers for iap1 or actin. b) Untreated Aag2 cells or cells treated with iap1 or polyhedrin dsRNA were photographed at the indicated hours after treatment. c-d) Aag2 cells were treated with iap1 dsRNA or polyhedrin dsRNA for 24 hrs, after which the cells were harvested and subjected to DNA fragmentation assay (c) or caspase activity assay (d). In (d), the values shown represent the mean +/− SE of three assays performed on each of two independent samples (six assays total).
Fig 5. Effect of silencing ark, dronc, casps7 or casps8 on caspase activity induced by Act D or UV treatment.
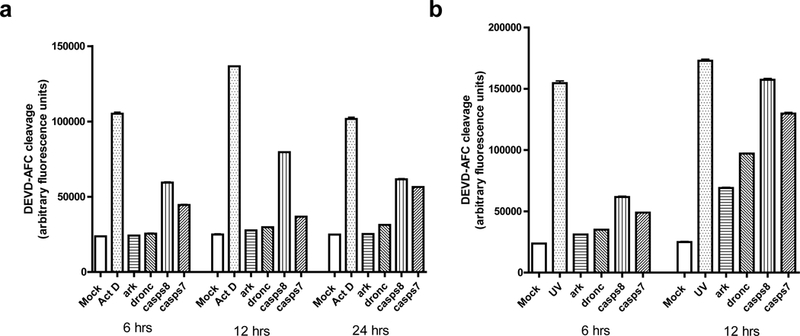
Aag2 cells were treated with the indicated dsRNAs for 24 hrs and then apoptosis was induced by Act D (a) or UV (b) treatment. At the indicated times after apoptosis induction, the cells were harvested and subjected to caspase activity assay. Mock-treated Aag2 cells (Mock) or Aag2 cells treated with Act D or UV were used as controls. The values shown represent the mean +/− SE of three assays performed on each of two independent samples (six assays total).
Fig 6. Effect of co-silencing casps7 and casps8 on caspase activity induced by Act D or UV treatment.
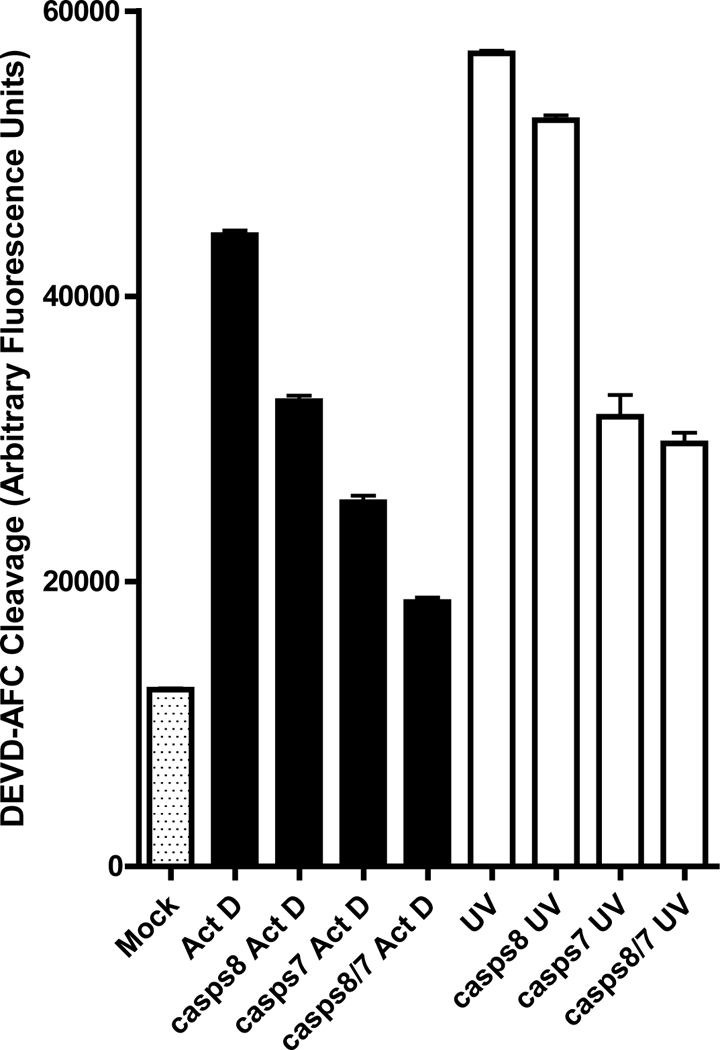
Aag2 cells were treated with dsRNA of casps8 or casps7 alone or together (CASPS8/7) for 24 hrs and then apoptosis was induced by Act D or UV treatment. 12 hrs after apoptosis induction, the cells were harvested and subjected to caspase activity assay. Mock-treated cells or cells treated with Act D or UV were used as controls. The values shown represent the mean +/− SE of three assays performed on each of two independent samples (six assays total).
Silencing of A. aegypti IAP1 induces rapid, spontaneous apoptosis in Aag2 cells
In cells from D. melanogaster and the lepidopteran insect Spodoptera frugiperda, RNAi of the IAP1 orthologs DIAP1 or Sf-IAP induces spontaneous apoptosis, indicating that continuous expression of IAP1 is necessary to maintain cell viability [8–10]. We therefore first examined the effect of silencing expression of A. aegypti iap1. Addition of iap1 dsRNA to Aag2 cells successfully silenced iap1 expression, as seen by a dramatic reduction in the level of iap1 transcripts within 7 hours (Fig 3a). Accordingly, we observed plasma membrane blebbing, indicative of apoptosis, in Aag2 cells treated with iap1 dsRNA for 7 hrs (Fig 3b). By 11 hrs, clear apoptosis was observed, including apoptotic body formation, and by 27 hrs, nearly all of the cells had undergone apoptosis (Fig 3b). Cells treated with control dsRNA remained unchanged (Fig 3b). In addition to the stereotypical morphological changes that accompany apoptosis, we also confirmed that the iap1 dsRNA-treated cells underwent DNA fragmentation into oligonucleosomal ladders (Fig 3c), and iap1 dsRNA-treated cells contained high levels of effector caspase activity as measured by cleavage of the effector caspase substrate DEVD-afc (Fig 3d). Thus, silencing of A. aegypti iap1 induced rapid, spontaneous apoptosis in Aag2 cells, similar to what has been previously observed in D. melanogaster S2 cells and S. frugiperda Sf21 cells.
Silencing Dronc or Ark completely blocks apoptosis, while silencing the effector caspases CASPS7 or CASPS8 partially blocks apoptosis induced by various apoptotic stimuli
A. aegypti contains clear one-to-one orthologs of D. melanogaster Ark and Dronc; however, there is no clear ortholog of DrICE, since DrICE and Dcp-1 resulted from duplication of an ancestral effector caspase after the divergence of mosquitoes and flies [23]. The two caspases most related to DrICE in A. aegypti are CASPS7 and CASPS8, which also arose by duplication after the divergence of mosquitoes and fruitflies [23]. We therefore tested whether silencing A. aegypti Ark, Dronc, CASPS7, or CASPS8 can rescue apoptosis induced by silencing A. aegypti IAP1. dsRNAs corresponding to these genes were added to Aag2 cells, followed 27 hrs later by addition of iap1 dsRNA. Silencing of each gene was effective, as judged by RT-PCR (Fig 4a). As expected, addition of ark, dronc, casps7, or casps8 dsRNA alone had no effect on cell viability (Fig 4b). However, when these genes were silenced prior to addition of iap1 dsRNA, apoptosis was either completely inhibited (in the case of silencing ark and dronc) or delayed and partially inhibited (in the case of silencing casps7 and casps8) (Fig 4c). Prior addition of control dsRNA had no effect on apoptosis induced by silencing of iap1 (data not shown). By 6 hrs after iap1 dsRNA addition, cell blebbing was evident in the cells treated only with iap1 dsRNA, but no blebbing was observed in cells that had been pre-treated with ark, dronc, casps7, or casps8 dsRNA (Fig 4c). By 20 hrs, extensive apoptosis was observed in the cells treated only with iap1 dsRNA, and no apoptosis was observed in the cells pre-treated with ark or dronc dsRNA, indicating that silencing these genes completely inhibited the apoptosis stimulated by loss of iap1 (Fig 4c). However, at 20 hrs after addition of iap1 dsRNA, some apoptosis was observed in cells pre-treated with casps7 or casp8 dsRNA, although less than in cells treated with iap1dsRNA alone (Fig 4c). Thus, CASPS7 and CASPS8 both appear to play a role in apoptosis stimulated by silencing IAP1.
Fig 4. Effect of silencing ark, dronc, casps7 or casps8 on apoptosis induced by silencing of iap1.
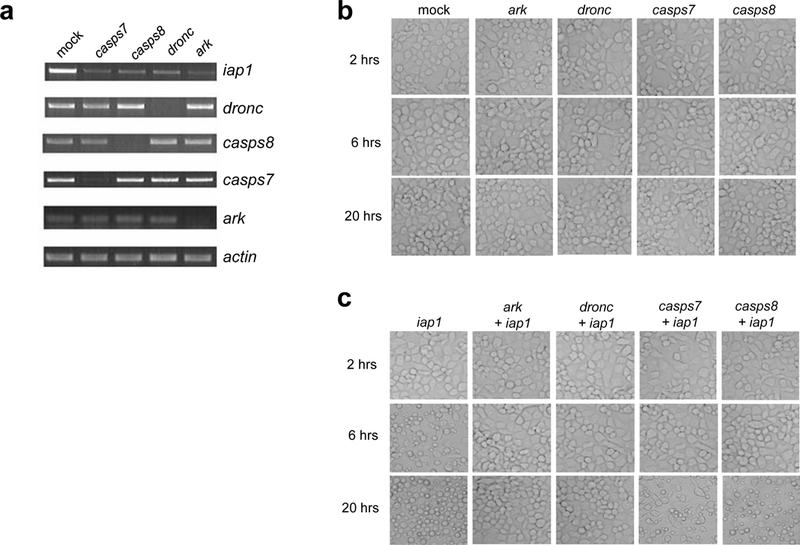
a) Aag2 cells were mock-treated or treated with dsRNAs corresponding to casps7, casps8, dronc, or ark for 24 hrs and then were treated with iap1 dsRNA. 20 hrs later, the cells were harvested and subjected to semi quantitative RT-PCR using gene specific primers for iap1, casps7, casps8, dronc, ark or actin. b-c) Aag2 cells were mock-treated or treated with the indicated dsRNAs for 24 hrs, and then the cells were photographed at the indicated times after being treated with control polyhedrin dsRNA (b) or iap1 dsRNA (c).
The roles of these apoptotic regulatory genes was also examined in response to several other treatments that induce apoptosis, namely ultraviolet radiation (UV) and the drugs cycloheximide (CHX), staurosporine (STS), and actinomycin D (Act D). Similar to the results with iap1 RNAi, silencing of ark or dronc resulted in excellent protection against all of these stimuli (Table 1). Also similar to iap1 RNAi, silencing of casps7 or casps8 provided only partial protection against CHX, Act D, and UV. casps7 RNAi provided partial protection against STS, but casps8 RNAi did not in the time frame examined (Table 1). Silencing of casps7 also seemed to provide more protection than silencing of casps8 against CHX-induced apoptosis (Table 1), suggesting that CASPS7 may play a somewhat more important role in apoptosis, at least in the case of certain stimuli. However, this would depend on the efficiency of silencing at the protein level, which is unknown in the current study.
Table 1.
Silencing of dronc, casps7, casps8 or ark delayed or inhibited apoptosis induced by multiple stimuli
| Gene silenced | CHX | STS | Act D | UV |
|---|---|---|---|---|
| dronc | +++ | +++ | +++ | +++ |
| casps7 | ++ | + | + | + |
| casps8 | + | - | + | + |
| ark | +++ | +++ | +++ |
+++ |
Aag2 cells were treated with dsRNAs corresponding to dronc, casps7, casps8 or ark for 24 hrs and then were treated with cycloheximide (CHX), staurosporine (STS), actinomycin D (Act D) or ultraviolet radiation (UV). 20 hrs later, cell viability was assessed by morphology. Symbols:
<5% viable;
5–25% viable;
25–50% viable;
>50% viable.
To verify these results and obtain more quantitative information, we examined effector caspase activity in cells subjected to RNAi and treated with Act D or UV. Consistent with what was observed by cell morphology (Table 1), RNAi of ark or dronc prevented effector caspase activation in response to Act D, with levels of caspase activity remaining similar to mock-treated cells through 24 hrs after Act D treatment (Fig 5a). RNAi of casps7 or casps8 partially inhibited effector caspase activity following treatment with Act D, with RNAi of casps7 having more of an effect than casps8 RNAi at 6 and 12 hrs, but the level of effector caspase activity was similar in the two treatments at 24 hrs.
With respect to UV treatment, RNAi of ark or dronc decreased effector caspase activity, although not as effectively as against Act D (Fig 5b). Silencing of casps8 or casps7 was again less effective than silencing ark or dronc, with casps8 RNAi being slightly less able to decrease effector caspase activity than casps7 RNAi (Fig 5b).
Both of the DrICE-related effector caspases, CASPS7 and CASPS8, are involved in apoptosis in Aag2 cells
Since silencing of casps7 or casps8 individually had only a partial inhibitory effect, we considered whether these two effector caspases exhibit functional redundancy. To test this, we silenced casps7 and casps8 either separately or together, and then induced apoptosis by treating the cells with either Act D or UV. Again, in response to either stimulus, silencing of casps7 had more effect on caspase activity than silencing casps8 (Fig 6). In the case of Act D, silencing both casps7 and casps8 reduced caspase activity further than casps7 alone, although not completely to background levels (Fig 6). In response to UV, silencing both caspases resulted in caspase activity that was similar to silencing casps7 alone, and still higher than background (Fig 6). The observation that silencing both casps7 and casps8 still did not completely eliminate caspase activity suggests that either silencing of at least one of these effector caspases was not completely effective, or there may be other effector caspases that are activated following treatment with Act D or UV. Regardless, the observation that silencing either casps7 or casps8 partially inhibits apoptosis demonstrates that there are at least two effector caspases involved in the core apoptosis pathway in Aag2 cells.
DISCUSSION
In this study we have performed the first comprehensive functional examination of the core apoptosis pathway in a disease vector, and indeed, to our knowledge the first in any insect other than D. melanogaster. Our results indicate that the core apoptosis pathway in A. aegypti functions in a manner that is overall similar to that in D. melanogaster but with some differences. As in D. melanogaster, continuous expression of A. aegypti IAP1 is required to prevent spontaneous apoptosis. Although we did not examine the roles of the IAP antagonists Mx and IMP in this study, results from an accompanying study indicate that these proteins also function similarly in the two species (H. Wang and R. J. Clem, in preparation). We also found that apoptosis in A. aegypti requires the initiator caspase Dronc, along with the activator protein Ark. Downstream of A. aegypti Dronc and Ark, CASPS7 and CASPS8 both appear to play roles in apoptosis. However, some questions remain concerning the effector caspases that are involved in apoptosis and their exact roles in A. aegypti.
In D. melanogaster, DrICE is the predominant effector caspase involved in apoptosis, while DCP-1 plays only a minor role, which is only evident when DrICE is lacking [13, 14]. Phylogenetic analysis of caspases from twelve Drosophila and four mosquito species indicates that DrICE and DCP-1 arose by duplication after the divergence of fruitflies and mosquitoes [23]. This would seem to suggest that DrICE is the ancestral effector caspase responsible for carrying out apoptosis, and DCP-1 arose more recently and only plays a minor role.
In A. aegypti, CASPS7 is the closest relative to DrICE and DCP-1, while CASPS8 is in a closely related mosquito-specific clade that appears to have arisen by duplication after the divergence of fruitflies and mosquitoes [17, 23]. Our results suggest that CASPS7 may play a somewhat more important role than CASPS8, but both caspases appear to be involved in regulating apoptosis in A. aegypti. This is different from the situation in D. melanogaster, where a single effector caspase, DrICE, plays a predominant role in apoptosis. Interestingly, there have also been expansions in the numbers of other effector caspases in mosquitoes compared to D. melanogaster [17]. In addition, it is noteworthy that silencing both CASPS7 and CASPS8 did not completely protect Aag2 cells against apoptosis, as silencing Dronc or Ark did. At this time we cannot distinguish whether additional effector caspases also are involved in apoptosis in A. aegypti, or whether silencing of CASPS7 and/or CASPS8 was not completely effective at the protein level. The other effector-type (short prodomain) caspases in A. aegypti include the outlier CASPS20, and caspases that are closest phylogenetically to D. melanogaster Decay (CASPS18 and CASPS19) and Damm (CASPS15, 16, 17, and 21). In vivo RNAi of D. melanogaster Decay and Damm did not reveal a significant role in apoptosis [15], so these additional A. aegypti caspases would not be expected to have roles in apoptosis, but this possibility cannot be ruled out. Overexpression of CASPS19 in Aag2 cells results in high levels of caspase activity but no apoptosis is observed, suggesting that this caspase may serve a non-apoptotic function, while CASPS18 acts as a decoy caspase and enhances the activity of CASPS19, both in cells and in vitro [23].
For all of the genes examined, gene silencing was less effective at inhibiting apoptosis induced by UV than by Act D (compare Fig 5a and b). The short wavelength UV used in these experiments (254 nm) causes DNA damage through the formation of pyrimidine dimers and other photoproducts, and also causes the formation of reactive oxygen species. UV has a myriad of effects on cellular processes, since it inhibits both DNA replication and transcription, and also stimulates DNA repair pathways and cellular stress pathways, including JNK and p38 MAPK signaling [24]. Act D is an antibiotic and chemotherapeutic agent that intercalates into both single-stranded and double-stranded DNA and inhibits cellular transcription by RNA polymerases [25]. There is also evidence, however, suggesting that Act D causes DNA damage in cells [26]. While Act D is commonly used in experiments to inhibit transcription, it has the ability to induce apoptosis in many mammalian and insect cell lines. Inhibition of gene expression at either the transcriptional or translational levels induces apoptosis in many types of insect cells, presumably at least in part because of the requirement for expression of IAP1. However, the mechanisms involved in induction of apoptosis by either UV or Act D are likely to be complex, and are also likely to vary depending on the type of cell and dosage used, making it difficult to draw any conclusions about why gene silencing was less effective at inhibiting UV-induced apoptosis in these experiments.
It has been shown in vivo that topical application of A. aegypti IAP1 dsRNA causes lethality in adult mosquitoes, and although it was presumed that the lethality was due to induction of apoptosis, the mechanism was not explored [20]. Our results clearly indicate for the first time that silencing iap1 induces apoptosis in A. aegypti cells. There are advantages to using a cell line for these types of studies, namely ease of performing the experiments and the reduced complexity of assaying only a single cell type.
In Drosophila research, cell lines have been exploited in numerous studies that have contributed to our understanding the core apoptosis pathway in flies. However, there are of course also limitations to using cell lines, since cells may respond differently in the context of an intact tissue, and different cell types may utilize different pathways. It will be interesting, therefore, to determine the roles of these core pathway genes in regulating apoptosis in vivo, especially casps7 and casps8.
ACKNOWLEDGEMENTS
This work was supported by NIH grant R21 AI067642 (to R.J.C.) and by the Kansas Agricultural Experiment Station. This is contribution number 10–342-J from the Kansas Agricultural Experiment Station.
Footnotes
Conflict of Interest
The authors declare that they have no conflicts of interest.
REFERENCES
- 1.Oberst A, Bender C, Green DR (2008) Living with death: The evolution of the mitochondrial pathway of apoptosis in animals. Cell Death Differ 15: 1139–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chew SK, Akdemir F, Chen P, Lu WJ, Mills K, Daish T, Kumar S, Rodriguez A, Abrams JM (2004) The apical caspase dronc governs programmed and unprogrammed cell death in Drosophila. Dev Cell 7: 897–907 [DOI] [PubMed] [Google Scholar]
- 3.Daish TJ, Mills K, Kumar S (2004) Drosophila caspase DRONC is required for specific developmental cell death pathways and stress-induced apoptosis. Dev Cell 7: 909–915 [DOI] [PubMed] [Google Scholar]
- 4.Xu D, Li Y, Arcaro M, Lackey M, Bergmann A (2005) The CARD-carrying caspase Dronc is essential for most, but not all, developmental cell death in Drosophila. Development 132: 2125–2134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Denton D, Shravage B, Simin R, Mills K, Berry DL, Baehrecke EH, Kumar S (2009) Autophagy, not apoptosis, is essential for midgut cell death in Drosophila. Curr Biol 19: 1741–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu X, Wang L, Acehan D, Wang X, Akey CW (2006) Three-dimensional structure of a double apoptosome formed by the Drosophila Apaf-1 related killer. J Mol Biol 355: 577–589 [DOI] [PubMed] [Google Scholar]
- 7.Wang SL, Hawkins CJ, Yoo SJ, Muller H-AJ, Hay BA (1999) The Drosophila caspase inhibitor DIAP1 is essential for cell survival and is negatively regulated by HID. Cell 98: 453–463 [DOI] [PubMed] [Google Scholar]
- 8.Muro I, Hay BA, Clem RJ (2002) The Drosophila DIAP1 protein is required to prevent accumulation of a continuously generated, processed form of the apical caspase DRONC. J Biol Chem 277: 49644–49650 [DOI] [PubMed] [Google Scholar]
- 9.Zimmermann KC, Ricci J-E, Droin NM, Green DR (2002) The role of ARK in stress-induced apoptosis in Drosophila cells. J Cell Biol 156: 1077–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Igaki T, Yamamoto-Goto Y, Tokushige N, Kanda H, Miura M (2002) Downregulation of DIAP1 triggers a novel Drosophila cell death pathway mediated by Dark and Dronc. J Biol Chem 277: 23103–23106 [DOI] [PubMed] [Google Scholar]
- 11.Orme M, Meier P (2009) Inhibitor of apoptosis proteins in Drosophila: gatekeepers of death. Apoptosis 14: 950–960 [DOI] [PubMed] [Google Scholar]
- 12.Shapiro PJ, Hsu HH, Jung HH, Robbins ES, Ryoo HD (2008) Regulation of the Drosophila apoptosome through feedback inhibition. Nat Cell Biol 10: 1387–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muro I, Berry DL, Huh JR, Chen CH, Huang H, Yoo SJ, Guo M, Baehrecke EH, Hay BA (2006) The Drosophila caspase Ice is important for many apoptotic cell deaths and for spermatid individualization, a nonapoptotic process. Development 133: 3305–3315 [DOI] [PubMed] [Google Scholar]
- 14.Xu D, Wang Y, Willecke R, Chen Z, Ding T, Bergmann A (2006) The effector caspases drICE and dcp-1 have partially overlapping functions in the apoptotic pathway in Drosophila. Cell Death Diff 13: 1697–1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leulier F, Ribeiro PS, Palmer E, Tenev T, Takahashi K, Robertson D, Zachariou A, Pichaud F, Ueda R, Meier P (2006) Systematic in vivo RNAi analysis of putative components of the Drosophila cell death machinery. Cell Death Diff 13: 1663–1674 [DOI] [PubMed] [Google Scholar]
- 16.Moreno E, Yan M, Basler K (2002) Evolution of TNF signaling mechanisms: JNK-dependent apoptosis triggered by Eiger, the Drosophila homolog of the TNF superfamily. Curr Biol 12: 1263–1268 [DOI] [PubMed] [Google Scholar]
- 17.Bryant B, Blair CD, Olson KE, Clem RJ (2008) Annotation and expression profiling of apoptosis-related genes in the yellow fever mosquito, Aedes aegypti. Insect Biochem Mol Biol 38: 331–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Waterhouse RM, Kriventseva EV, Meister S, Xi Z, Alvarez KS, Bartholomay LC, Barillas-Mury C, Bian G, Blandin S, Christensen BM, Dong Y, Jiang H, Kanost MR, Koutsos AC, Levashina EA, Li J, Ligoxygakis P, Maccallum RM, Mayhew GF, Mendes A, Michel K, Osta MA, Paskewitz S, Shin SW, Vlachou D, Wang L, Wei W, Zheng L, Zou Z, Severson DW, Raikhel AS, Kafatos FC, Dimopoulos G, Zdobnov EM, Christophides GK (2007) Evolutionary dynamics of immune-related genes and pathways in disease-vector mosquitoes. Science 316: 1738–1743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou L, Jiang G, Chan G, Santos CP, Severson DW, Xiao L (2005) Michelob_x is the missing inhibitor of apoptosis protein antagonist in mosquito genomes. EMBO Rep 6: 769–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pridgeon JW, Zhao L, Becnel JJ, Strickman DA, Clark GG, Linthicum KJ (2008) Topically applied AeIAP1 double-stranded RNA kills female adults of Aedes aegypti. J Med Entomol 45: 414–420 [DOI] [PubMed] [Google Scholar]
- 21.Cooper DM, Thi EP, Chamberlain CM, Pio F, Lowenberger C (2007) Aedes Dronc: a novel ecdysone-inducible caspase in the yellow fever mosquito, Aedes aegypti. Insect Mol Biol 16: 563–572 [DOI] [PubMed] [Google Scholar]
- 22.Lan Q, Fallon AM (1990) Small heat shock proteins distinguish between two mosquito species and confirm identity of their cell lines. Am J Trop Med Hyg 43: 669–676 [DOI] [PubMed] [Google Scholar]
- 23.Bryant B, Ungerer MC, Liu Q, Waterhouse RM, Clem RJ (2010) A caspase-like decoy molecule enhances the activity of a paralogous caspase in the yellow fever mosquito, Aedes aegypti. Insect Biochem Mol Biol 40: 516–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Batista LFZ, Kaina B, Meneghini R, Menck CFM (2009) How DNA lesions are turned into powerful killing structures: Insights from UV-induced apoptosis. Mutat Res 681: 197–208 [DOI] [PubMed] [Google Scholar]
- 25.Sobell HM (1985) Actinomycin and DNA transcription. Proc Natl Acad Sci U S A. 82: 5328–5331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mischo HE, Hemmerich P, Grosse F, Zhang S (2005) Actinomycin D induces histone γ-H2AX foci and complex formation of γ-H2Ax with Ku70 and nuclear DNA helicase II. J Biol Chem 280: 9586–9594 [DOI] [PubMed] [Google Scholar]


