Abstract
Background
HER2 over-expression is related with a poor prognosis in patients with invasive breast cancer tumors. Clinical associations have reported that somatic mutations of p53 more frequently detected in cases of sporadic breast cancer of the HER2 subtypes, besides a high percentage of HER2-amplifying tumors carry germline mutations of p53. The mechanisms responsible for the acquisition of oncogenic functions of p53 mutant proteins (mtp53), known as Gain of Function (GOF), over HER2 expression have not been reported. The objective of this study was to evaluate a possible relationship between p53 mutants and HER2 regulation.
Methods
HER2 expression (transcription and protein), as well as HER2 protein stabilization have been evaluated after inducing or silencing of p53 mutants’ expression in cell lines. Finally, we evaluated the interaction of the p53 mutants over the HER2 receptor promoter.
Results
Higher HER2 expression in cell lines harboring endogenous mtp53 compared with wt or null expression of p53 cell lines. Transfection of p53 mutants (R248Q and R273C) in cell lines increased the expression of HER2. Silencing of p53 mutants, decrease HER2 expression. The p53 mutants R248Q and R273C significantly increase the luciferase activity on the HER2 promoter, and both mutants also promote acetylation of H3 and H4 histones binding in it.
Conclusions
These findings show for the first time that p53 mutants induce over-expression of HER2 at transcriptional level of the HER2 protein. Our results could have clinical implications in breast cancer and other types of cancer where HER2 is over-expressed and used as a therapy target.
Electronic supplementary material
The online version of this article (10.1186/s12885-018-4613-1) contains supplementary material, which is available to authorized users.
Keywords: HER2, Mutant p53, Gain of function, Cancer
Background
The p53 tumor suppressor protein, which is encoded by TP53 gene, exerts its biological functions mainly by its transcriptional activity, although it is accepted that wild-type p53 (wtp53) has other biological activities that are transcription independent [1, 2]. Wild-type p53 levels are very low in normal cells; however, they rise rapidly in response to DNA damage, hypoxia, oxidant metabolism or oncogenic signaling, as well as in response to aging, maintaining genomic integrity and preventing tumor formation [3, 4]. TP53 is the most frequent target for mutations in human cancers, with more than half of all tumors exhibiting mutation at this locus. Unlike other tumor-suppressor genes, which typically are mutated by deletion or truncation, TP53 frequently undergoes missense mutations, resulting in single amino acid substitutions in the full length protein [3]. About a third of these missense mutations are located in six residues: R175, G245, R248, R249, R273, and R282, corresponding to the p53 DNA binding domain (DBD) and known as mutational hot-spots [5]. These p53 mutations can be roughly divided into two structural subgroups: DNA contact mutants (exemplified by R273 and R248 residues) directly involved in sequence-specific DNA contact; and conformational mutants (exemplified by the R175 residue), leading to a partial or complete abrogation of the conformational wtp53 DBD. The presence of these point mutations radically alters p53 function causing not only a simple loss of wild-type function, but also a dominant negative effect by binding and inhibiting wtp53 or a Gain of Function (GOF) acquiring novel activities independent of wtp53 [3]. p53 GOF mutations have been shown to result in oncogenic and a major proliferative processes such as increased tumorigenicity, anchorage independent cell growth and increased growth rate, increased metastasis and invasiveness, decreased sensitivity to chemotherapeutic drugs, disruption of the spindle checkpoint, activated topoisomerase I activity and induction of gene amplification, reviewed in [3]. Many of the GOF data come from p53-null systems where the expression levels of re-expressed mutant p53 were comparable to those observed in cancer cells. These results suggest a true patho-physiological role of the GOF of p53-mutants, which may lead to the development of a more aggressive cancer and poorer prognosis. The molecular mechanism of GOF’s phenotypes and up-regulation of gene expression by p53 mutants has not been clarified yet [6]. Among other important biomarkers implicated in several human cancers, there are the Human Epidermal growth factor Receptors (HERs), which regulate cell proliferation, angiogenesis, cell adhesion, cell motility, development and organogenesis, by activation of different downstream signaling pathways [7]. The HER family consists of four members (HER1–4) expressed in epithelial, mesenchymal, and neuronal cells, as well as in their cellular progenitors. Like all protein-tyrosine kinase receptors, the HER receptors exist as monomers on the cell surface and depend on their specific ligands for dimerization and trans-phosphorylation of their intracellular domains [8]. Although the product of the ERBB2 gene (HER2) has no known direct activating ligand due to its constitutively activated state, this receptor is known as the preferred dimerization partner for the other family members [9]. HER2 over-expression or gene amplification occurs in 20–30% of breast cancers with a correlation of poor prognosis prior to the advent of anti-HER2 therapies. Anti-HER2 therapy is currently approved for breast, gastric, and gastroesophageal cancers, when this receptor is over-expressed. However, HER2 over-expression have been reported in other malignancies, such as bladder, cervix, colon, endometrium, germ cell, glioblastoma, head and neck, liver, lung, ovarian, pancreas and salivary duct [10]. Elucidating the mechanism leading to HER2 gene up-regulation will be an important step to understand the pathogenesis of particularly aggressive subset of tumors over-expressing HER2 [11], as well as to find novel alternatives for therapy. Interestingly, Wilson et al. reported that patients carrying TP53 mutations (mtp53) show a significantly higher probability of developing breast cancer with HER2 gene amplification (83%) when compared to the cohort of early onset breast cancer cases (16%) [12]. A larger study that supports this association, found that patients with breast cancer harboring a germline TP53 mutation, have significantly higher HER2 positive tumors prevalence, compared to their counterparts who lack any mutation [13]. There are also studies where both somatic TP53 mutations and HER2 gene amplification are correlated with elevated risk of breast cancer recurrence and elevated overall mortality compared with patients with neither or only one alteration [14]. It is important to note that the studies mentioned above are just clinical associations and do not propose any mechanism responsible for this relation. In this work we found, for the first time, that p53 mutants can induce up-regulation of HER2 gene and favor the stabilization of the protein. Understanding the specific mechanisms for this relationship might reveal diagnostic and therapeutic insights in HER2 positive cancers.
Methods
Cell culture
Cancer cell lines: Saos-2, MCF-7, HeLa, C33A, OVCAR-3 and SKBR-3 were originally obtained from the American Type Culture Collection (ATCC). These cell lines were cultured in DMEM supplemented medium at 37 °C in a humidified 5% CO2 environment and were grown at 50 to 80% confluence before the next passage or further experiments.
Transient and stable transfections
Cells were plated (5 X 105 cells) in 60 mm × 15 mm culture dishes and transfected with: empty vector, p53R248Q, p53R273C [15], and wtp53 [16] plasmids; or with SureSilencing shRNA Plasmid Kit for TP53 (SABiosciences, Qiagen). Transfections were performed with Lipofectamine 2000 reagent (Invitrogen, CA) according to the manufacturer’s instructions. Transient transfections were performed for 48 h. Stable transfections were selected for 2 weeks in a growth media containing 1200 μg/ml of G-418 (Sigma-Aldrich, St Louis MO). To keep the clones selection, cells were grown continuously in media containing 800 μg/ml of G418.
Reverse transcription and quantitative polymerase chain reaction (RT-qPCR)
Total RNA was obtained using TRIZOL reagent (Invitrogen, CA) and cDNA was reverse transcribed from 3 μg of RNA using SuperScript II reverse transcriptase (Invitrogen, CA), following the manufacturer’s recommendations. Oligonucleotide primers were designed using the Primer Express Software and purchased from Sigma-Aldrich (St Louis MO). qPCR assays were performed by using SYBR green Master Mix (Thermo Scientific, Waltham MA) in an ABI 7300 Real Time PCR system (Applied Biosystems, California USA). The qPCR program was set as follow: 95 °C for 5 min, followed by 40 cycles at 95 °C for 30 s, 60 °C for 30 s, 72 °C for 30 s and a final elongation step of 72 °C for 7 min, with a final dissociation curve. Each reaction was carried out in triplicate and targeted genes were normalized to beta-2-microglobulin (B2M) gene. The threshold value (CT) for each sample was used to determine gene relative expression levels by the comparative CT method (2-ΔΔCT method) [17]. Detailed description of the primers used for HER2 and TP53 detection can be found in the Additional file 1: Table S1.
Western blot analysis
Equal amounts of whole-cell protein extracts were separated by size on discontinuous (8 and 10%) PAGE, and transferred to a 0.45 μm nitrocellulose membrane (Millipore, Billerica MA). After blocking, membranes were probed overnight with: HER2 monoclonal primary antibody (Cell signaling, Danvers, MA) and peroxidase-conjugated antibodies: p53-DO1 (Santa Cruz Biotechnology) and β-actin (Sigma-Aldrich, St Louis MO). Secondary rabbit antibody (Sigma-Aldrich, St Louis MO) required for HER2 was employed at 1:10000 dilution. Protein expression was detected by chemiluminiscence using Supersignal West Pico (Thermo Scientific, Waltham MA). The protein band intensity was analyzed from the Western blot images by using the Syngene software.
Promoter luciferase reporter assays
Stable transfected Saos-2 cells (empty vector, wtp53, p53R248Q, and p53R273C) were seeded at 1 × 105 cells in six-wells plates and then transiently transfected with HER2 promoter vector, pNeuLite (Addgene Inc., Cambridge MA), using Lipofectamine 2000 reagent (Invitrogen, CA) as described above. After 24 h, cells were lysed and samples probed with Dual-Glo®Luciferase Assay System (Promega, Madison WI) essentially as described in [18]. Assays were read with the Fluoroskan Ascent FL (Labsystems, Perú), 0.5 s integration/ well. Expression induction was calculated as the relative light units obtained for the Saos-2 cells expressing mtp53/average relative light units of the Saos-2 cells expressing empty vector, normalized with renilla relative light units. Results were plotted against the log of the compound concentration, using Prism 6 Software (GraphPad, San Diego CA).
ChIP promoter assay
Chromatin immunoprecipitation (ChIP) assay over HER2 promoter was performed in accordance to manufacturer’s kit protocol (ChIP One Day Kit, Qiagen). Briefly, cells were fixed at room temperature and washed with PBS. To obtain the cell insoluble material, cells were gently scraped with lysis buffer, collected and pelleted by centrifugation at 4 °C. DNA–protein complexes were sonicated to produce fragmented chromatin between 200 and 600 bp in length, determined by agarose gel electrophoresis. One-tenth of the fragmented chromatin was set aside as input control, and the remaining sample was pre-cleared with protein A/G. For each ChIP, either anti-p53 (DO-1) (Santa Cruz Biotechnology), anti-Histone 3 (Upstate, Millipore Merck) or anti-Histone 4 antibodies (Upstate, Millipore Merck), or non-specific control (IgG) was added and incubated overnight at 4 °C. Immune complexes were precipitated at 4 °C with magnetic beads and DNA-protein cross-links were reversed by adding NaCl. Samples were subsequently treated with proteinase K, and genomic DNA was recovered and purified by column affinity. Quantitation of DNA from p53, H3ac and H4ac ChIP, no antibody control, and input control was done with RT-qPCR as described above. Primers were designed by Primer Express Software (Applied Biosystems, Foster City, CA) and sequences are shown in Additional file 2: Table S2. Comparisons were normalized to input controls.
Statistical analysis
Each experiment was performed at least three times, the results were presented as mean ± Standard Error of the Mean (SEM). Statistical analysis was performed with 1-tailed paired Student’s t test for means between controls and experimental data. Histograms and statistical analysis were carried out using Prism (Graphpad). * p < 0.05, ns = non-significant statistical difference analysis.
Results
HER2 and p53 expression levels in cancer cell lines
As a first approach to the correlation that may exist between p53 mutant proteins and the HER2 receptor expression, we evaluated the mRNA and protein levels of p53 and HER2 in a panel of human cancer cells: MCF-7 and HeLa cells harboring wild-type p53 (wtp53), Saos-2 with null expression of p53 (p53-null), as well as, C33A, SKBR-3 and OVCAR-3 harboring three of the most frequent p53 hot-spot mutations (p53R273C, p53R175H and p53R248Q). For HER2 mRNA expression, quantitative PCR (RT-qPCR) assays revealed that the cell lines harboring mutant p53 (mtp53) had higher expression levels when compared with the cell lines with wild-type or null p53 expression (Fig. 1a). In agreement with the mRNA expression of HER2, we observed a higher HER2 protein expression in the cell lines harboring mutant p53 (Fig. 1b). In Additional file 3: Figure S1, (A) we showed that exogenous wtp53 expression in HeLa cell line, counteracting HPV-E6 effects, decrease HER2 mRNA expression; moreover, (B) wtp53 silencing in MCF-7 cell line increase HER2 mRNA expression; (C and D) both mRNA and protein HER2 levels are insignificantly decreased in Saos-2 cells transfected with wtp53.
Fig. 1.
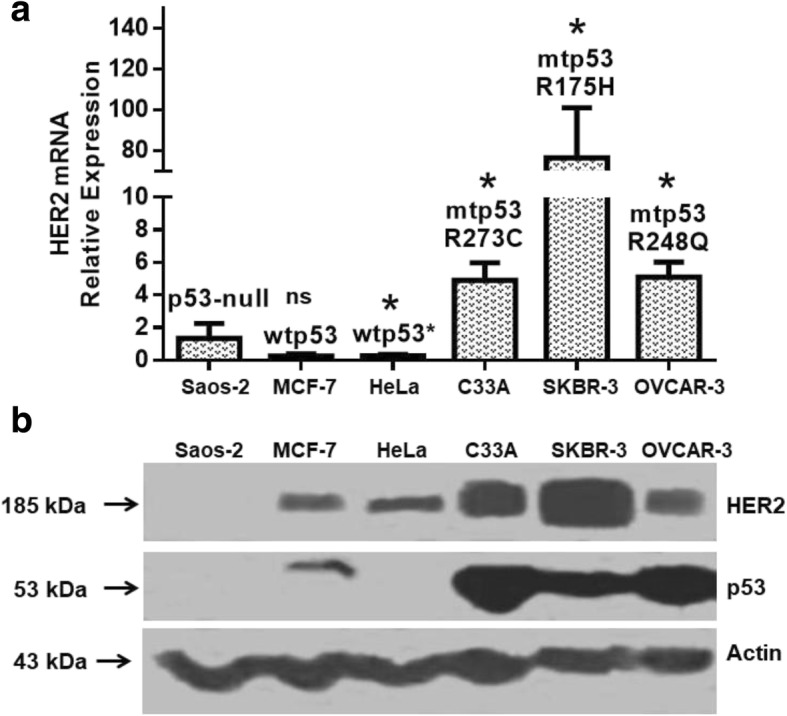
Cancer cell lines harboring p53 mutant proteins show elevated expression of both HER2 mRNA and protein. A panel of human cancer cell lines harboring p53R273C, p53R175H and p53R248Q mutant p53 proteins (C33A, SKBR-3 and OVCAR-3, respectively), as well as cells containing wild-type p53 (wtp53) protein (MCF-7 and HeLa), and null p53 expression (Saos-2) were analyzed to determine HER2 mRNA and protein expression. HeLa cell line expresses wtp53 but its expression is decreased by Human Papillomavirus 18 (wtp53•). a HER2 relative mRNA expression was measured by quantitative RT-PCR (RT-qPCR); final data were calculated by the comparative threshold cycle (CT) method (2-ΔΔCT), employing the β2-microglobulin (β2M) transcript as normalization control. The results are shown as the mean ± the standard error of the mean (SEM) of three independent assays. Statistical analysis was performed by comparing each cell line against Saos-2 cell line with t-student test, *p < 0.05; ns = non-significant statistical analysis. b HER2 protein levels were measured by Western Blot assay. Equal amounts of total cell lysates were blotted and proteins were identified as described in the Materials and Methods, with specific antibodies against HER2 and p53. Blots were stripped and re-probed for anti-β-actin as loading control. Image is representative of three independent experiments
The p53R248Q and p53R273C mutants increase HER2 expression
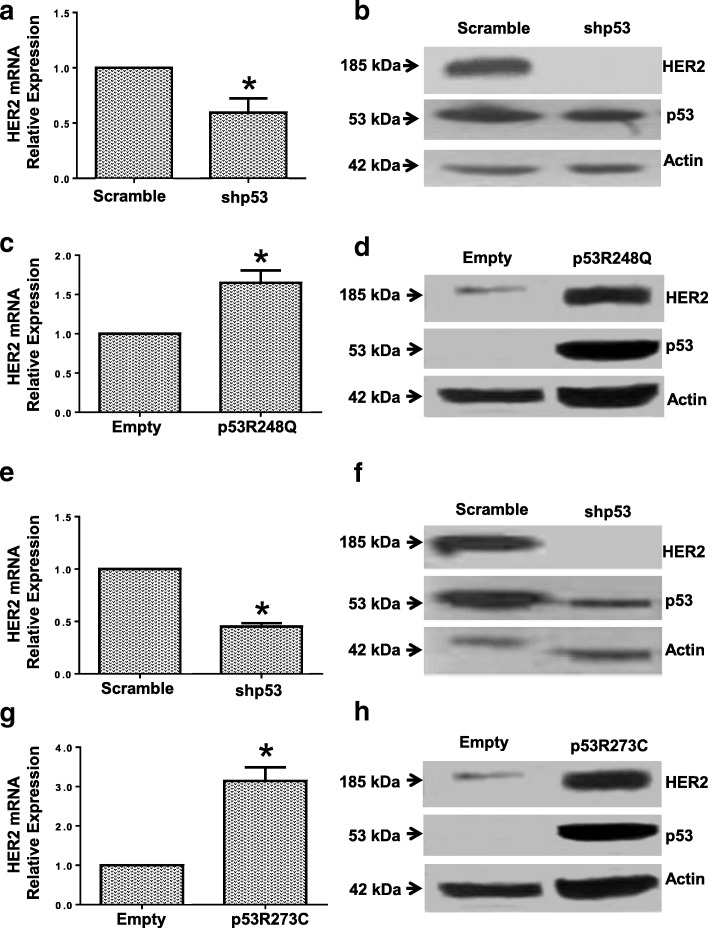
We then evaluated the effect of silencing or expressing exogenously two specific mutants (p53R248Q and p53R273C), on the expression of HER2. First, we silenced p53 expression in OVCAR-3 cell line which endogenously express p53R248Q mutant and observed a decreased expression of both the HER2 mRNA and protein, when compared to the shRNA negative control plasmid (Fig. 2a and b). On the contrary, when we expressed exogenously the mutant p53R248Q by stably transfection in the Saos-2 cell line (p53-null) (Fig. 2c and d), we observed that the p53R248Q mutant, lead to an elevated mRNA and protein expression of HER2. We also transfected transiently p53R248Q mutant in the HeLa cell line with similar results than in Saos-2 cell line (Data not shown). Next, also based on our previous results where we observed an elevated expression of HER2 in C33A, a cervical cancer cell line harboring the p53R273C mutant; we silenced the endogenous expression of the p53R273C mutant in C33A cell line, and this silencing lead to a decreased in the expression in both mRNA and protein levels of HER2 as well as in the case of p53R248Q mutant (Fig. 2e and f). In counterpart, stably transfection in the p53-null cell line (Saos-2) (Fig. 2g and h), lead to an increased HER2 protein expression. We also transiently transfected the p53R273C mutant, in the HeLa cell line (data not shown), in this case we also found an increase of HER2 protein level between HeLa and Saos-2 cells, but in HeLa we did not observe an increase of HER2 mRNA expression. These results suggest that the p53R248Q mutant, regulates positively HER2 expression at both transcriptional and protein level while regulation by p53R273C mutant could be cell context dependent.
Fig. 2.
p53R248Q and p53R273C mutants positively regulates the expression of both HER2 mRNA and protein. a-b The mutant p53R248Q harbored in the OVCAR-3 cell line was inhibited by specific shRNAs for p53 (shp53), as indicated in Materials and Methods. After 48 h, transfected OVCAR-3 cells were processed to determine a HER2 mRNA relative levels by RT-qPCR and b HER2 protein levels by Western Blot. c-d The Saos-2 cell line (p53-null) was stably transfected with mutant p53R248Q, or with the empty vector, as indicated in Materials and Methods. After stable selection, c HER2 mRNA expression assay was performed by RT-qPCR, and d protein expression was measured by Western Blot. The RT-qPCR results are the means of at least three independent experiments ± SEM and the image of the blot is representative of three independent experiments. Statistical analysis was performed with t-student test by comparing the results obtained in: shp53 vs scramble transfected OVCAR-3 cells; or p53R248Q vs empty vector transfected Saos-2 cells: * p < 0.05. e-f The expression of p53R273C mutant in C33A cell line was transiently inhibited by short-hairpin RNA against p53 (shp53), under the conditions indicated in Materials and Methods. After 48 h the cells were processed for: e HER2 mRNA expression assay by RT-qPCR, and f protein expression assay by Western Blot. g-h The Saos-2 cell line (null-p53) was stably transfected with mutant p53R273C, or with empty vector, as indicated in Materials and Methods. Transfected cells were processed after stable selection for: g HER2 mRNA expression assay by RT-qPCR, and h protein expression assay by Western Blot. The RT-qPCR results are the means of at least three independent experiments ± SEM and the image of the blot is representative of three independent experiments. Statistical analysis was performed with t-student test by comparing the results obtained in: shp53 vs scramble transfected C33A cells; or p53R273C vs empty vector transfected Saos-2 cells: * p < 0.05
The p53R248Q and p53R273C mutants induce transcriptional activation of HER2 promoter in Saos-2 cell line
Because we also observed that p53R248Q and p53R273C mutants induce an increase in the HER2 mRNA expression, we evaluated the effect of both mutants over the HER2 promoter using a luciferase transactivation assay in the HER2 promoter. We used the Saos-2 cells expressing stably either the p53R248Q or p53R273C mutants and we co-transfected the plasmid containing the HER2 promoter sequence in a luciferase reporter vector (pNeuLite) [19]. The luciferase activity was normalized with data obtained for renilla activity (Fig. 3). These results showed that, under our experimental conditions, p53R248Q and p53R273C significantly increased up to 50% the HER2 promoter activity, with respect to the control (empty vector). We also evaluated the transient ectopic expression of wtp53 in Saos-2 cell line and in this case, the results showed that exogenous wtp53 significantly diminished the HER2 promoter activity, in contrast to the results observed with the mutants of p53 analyzed (Fig. 3).
Fig. 3.
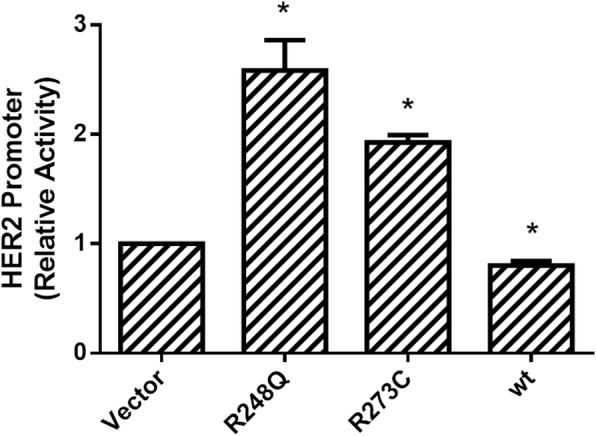
p53R248Q and p53R273C mutants but not p53 wild-type transactivate the HER2 promoter in stably transfected Saos-2 cells. Transcriptional activity of the HER2 promoter was assessed by a Luciferase reporter assay in Saos-2 cells stably transfected with R248Q and R273C p53 mutants, as well as with wild-type p53 plasmid and empty vector. Stably transfected Saos-2 cells were co-transfected with pNeuLite luciferase reporter plasmid for the HER2 promoter. After 48 h, luciferase activity was measured as described in the Materials and Methods. The results of the luciferase activity were normalized to renilla activity and are reported as the mean ± SEM of three independent experiments. Statistical analysis was performed with t-student test by comparing the results obtained for the Relative Activity in Saos-2 cells transfected with p53R48Q or p53R273C or wtp53 against empty vector: * p < 0.05
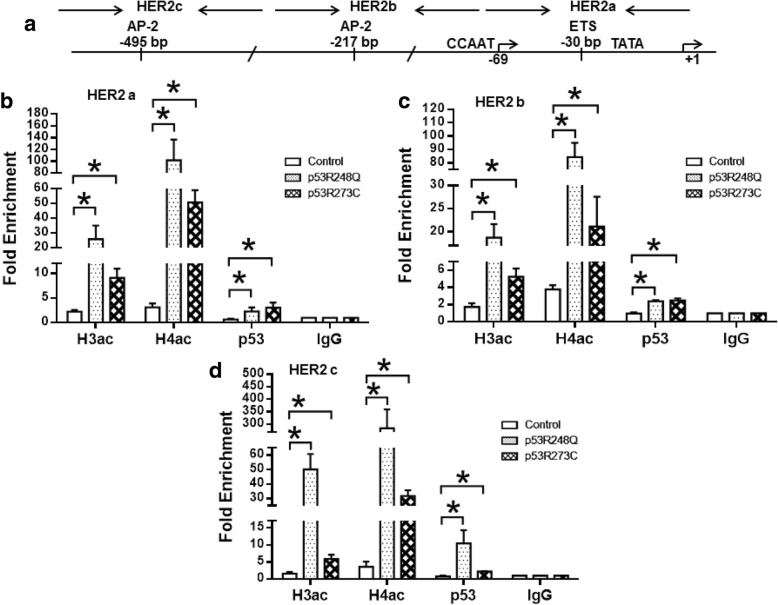
The p53R248Q and p53R273C mutants bind and induce histone acetylation on the HER2 promoter
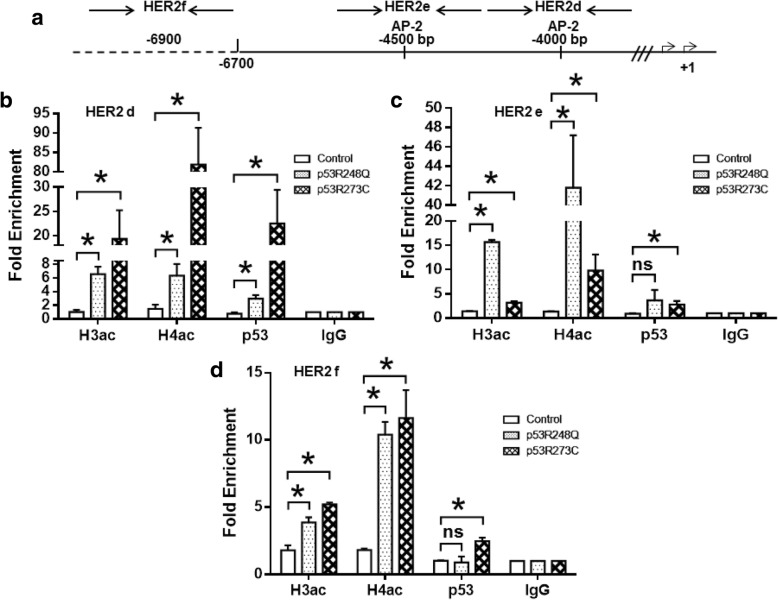
Due to our previous results indicating that p53 mutants up-regulate HER2 promoter activity, we decided to gain additional insight into the molecular mechanisms underlying a possible physical interaction between the HER2 promoter and the p53R248Q or p53R273C mutants. We performed chromatin immunoprecipitation (ChIP) assays, using antibodies directed against p53 and against acetylated forms of histones H3 and H4. These assays were performed with chromatin extracted from the Saos-2 cells stably expressing p53R248Q or p53R273C, as well as with the empty vector (control). We designed specific primers to dissect the HER2 proximal promoter region into three PCR products, denominated as HER2a, HER2b and HER2c, according to the description reviewed by Hurst in 2001 and Delacroix in 2005 [20, 21]. The HER2a region has a TATA box in position − 22 to − 26 bp, a binding site for transcriptional factors of the ETS family in the − 30 bp position, and a CAAT (CCAAT) box in position ranging from − 71 to − 75 bp. The HER2b region, include a sequence corresponding to AP-2 transcriptional binding factor at the − 217 position. The HER2c region, also include an AP-2 binding site at the − 495 bp (Fig. 4a). We found that in HER2a and HER2b regions, both p53R248Q and p53R273C mutants had a similar fold enrichment after p53 immunoprecipitation: HER2a (2.3 and 3.0 fold enrichment, respectively) and HER2b (2.4 and 2.5, respectively); moreover, in both regions there is an important increase in the acetylation marks of H3 and H4 histones (Fig. 4b and c). For HER2c we found that p53R248Q mutant has the highest enrichment, while p53R273C mutant had a similar enrichment both HER2a and HER2c (10.4 and 2.2 fold enrichment, respectively); the most important enrichment in acetylated H3 and H4 marks was also observed in this region (Fig. 4d). The Saos-2 cell line transfected with p53R248Q presented both the highest p53 protein level and acetylated histones (particularly H4) in the three proximal regions. For the HER2 distal promoter, we used the same set of primers reported by Delacroix et al., 2005 [22]. These primers were denominated HER2d and HER2e, which respectively cover two AP-2 binding sites located in the − 4000 bp and − 4500 bp of the HER2 promoter. Another region upstream of the HER2 promoter, with no HER2 activator binding sites, denominated as HER2f was included (Fig. 5a). We observed that HER2d has the major fold enrichment for the p53R273C mutant in the HER2 promoter. In general, there was a modest enrichment of acetylated histone marks for HER2d and HER2e (Fig. 5b and c). The region denominated HER2f had the lowest enrichment for acetylation marks in H3 and H4 histones, coinciding with the lowest p53 recruitment in the promoter (Fig. 5d). It is worth noticing that in distal regions (HER2d and HER2e) the enrichment of acetylated H3 and H4 have differences between the two evaluated mutants; while p53R273C induced a higher increase in the enrichment of acetylated histone marks for HER2d region, the p53R248Q favor a higher increase of these marks in the HER2e region. Taken together, these results suggest that both the p53R248Q and p53R273C mutants are directly involved in HER2 promoter activation. Moreover, both p53 mutants are contributing to the modification of the HER2 promoter towards a more activated chromatin by enriching H3 and H4 acetylation marks.
Fig. 4.
The p53R248Q and p53R273C mutants associate with chromatin and induce histone H3 and H4 acetylation in HER2 promoter proximal region. Saos-2 cells stably transfected with p53R248Q, p53R273C or empty vector were analyzed by Chromatin immunoprecipitation (ChIP) as described in the Materials and Methods. Antibodies against acetylated histone H3, H4, p53 protein and IgG as non-relevant antibody were used for the immunoprecipitation. a Shows a graphic representation of the regions encompassed by the primers used for ChIP assay. Precipitated DNA was amplified with primers described in Additional file 2: Table S2. HER2a (b) region from − 22 to − 75 bp; HER2b (c) -217 bp, and (d) region with not known HER2 regulatory sequences. Relative amounts of acetylated H3 and H4 histones, as well as p53 interaction with HER2 promoter regions were determined on the precipitated DNA by quantitative PCR (qPCR). Normalized data were calculated relative to each primer set (b-d), comparing p52R248Q or p53R273C vs IgG ChIP 1.0% input sample. The results of Fold Enrichment are reported as the mean ± SEM of three independent experiments. Statistical analysis was performed with t-student test by comparing the results obtained for the Fold Enrichment in Saos-2 cells transfected with p53R48Q or p53R273C against empty vector transfected cells: * p < 0.05
Fig. 5.
The p53R248Q and p53R273C mutants increase histone acetylation and p53 bind to HER2 promoter distal region. Chromatin immunoprecipitation (ChIP) assays were carried out in p53R248Q, p53R273C and empty vector stably transfected Saos-2 cells as described in the Material and Methods. Antibodies against acetylated histone H3, H4, p53 protein and IgG as non-relevant antibody were used for the immunoprecipitation. a Shows a graphic representation of the regions encompassed by the primers used for ChIP assay. Precipitated DNA was amplified with primers described in Additional file 2: Table S2. HER2d (b) -4000 bp; HER2e (c) -4500 bp, and HER2f (d) up-steam the HER2 promoter. Relative amounts of acetylated H3 and H4 histones, as well as p53 interaction with HER2 distal promoter regions were determined by quantitative PCR (qPCR). Normalized data were calculated relative to each primer set (b-d), comparing p52R248Q or p53R273C vs IgG ChIP 1.0% input sample. The results of Fold Enrichment are reported as the mean ± SEM of three independent experiments. Statistical analysis was performed with t-student test by comparing the results obtained for the Fold Enrichment in Saos-2 cells transfected with p53R48Q or p53R273C against empty vector transfected cells: *p < 0.05
Discussion
It is generally accepted that mutant p53 (mtp53) proteins promote carcinogenesis by exerting a dominant-negative effect over wtp53 protein, as well as by new oncogenic gain-of-function (GOF) activities independent of wtp53. However, the mechanisms of the GOF phenotype by p53 mutants, are only starting to be defined [23]. In previous works, it has been observed that p53 mutants are more frequently detected in a subset of breast and gastric cancer patients with poor prognosis, such as in those presenting the HER2 subtype [24, 25]. We choose the cell lines mentioned in the Material and Methods section, because Saos-2 is a cell line known for be p53-null, which is common used to study new roles for the wild-type and mutants of p53 [26]. In the case of MCF-7 and SKBR-3 breast cancer cell lines, Xiao et al., observed that MCF-7 cells have a typical number of gene copies (i.e., 2) and very low HER2 protein content, whereas SKBR-3 cells contain approximately 10-fold more copies of the HER2 gene and 15-fold higher HER2 protein content [27]. Consistent with Xiao’s results, we observed the most expression of mRNA and protein expression in SKBR-3 cell line. We decided not to use SKBR-3 cell line or p53R175H mutant in subsequent experiments indeed the amplification of HER2 in this cell line [28]. In case of cervical adenocarcinomas, there are reports that conclude that amplification of HER2 and mutations in TP53 are rare in cervical adenocarcinomas and that, low level chromosome 17q copy number gains are not associated with HER2 over-expression [29]. Beaufort et al., described several ovarian cancer cell lines and confirms the p53 mutation for the OVCAR-3 cell line but no HER2 amplification reported [30]. In the present study, we provided evidence to support the notion that mtp53 is implicated in the positive regulation of HER2 expression. We showed that in two different cell lines harboring wtp53 (HeLa and MCF-7), HER2 mRNA is expressed in very low level, which agrees with the results of Yang et al. (2006) indicating that wtp53 could negatively regulate HER2 [31]. In this regard, HeLa is a cervical cancer cell line containing the human papillomavirus-18 (HPV-18) genome that encodes for the E6 oncoprotein and causes p53 degradation [32]; which might explain the HER2 protein expression in HeLa cells. On the other hand, in spite that MCF-7 also harbors wtp53, the high basal HER2 protein levels observed in this cell line is probably inherent to the normal genetic background needed to maintain the function of breast cells; additionally, the p53 pathway has been suggested to be impaired in this tissue [33, 34]. In our work, we found that cell lines harboring three of the most common p53 mutants (p53R273C, p53R175H and p53R248Q) express higher levels of HER2 (mRNA and protein) compared to the cell lines with functional wtp53 (MCF-7), HPV-inactivated wtp53 (HeLa) or null-p53 (Saos-2). Our results are consistent with studies that suggest a clinical association between the presence of p53 mutations and HER2 over-expression [12–14]. We demonstrated that the exogenous expression of the p53R248Q or p53R273C mutants in two different cell types, HeLa and Saos-2 induces a significant increase in the levels of HER2 protein. On counterpart, we analyzed the effect of inhibiting the expression of the endogenous p53R248Q or p53R273C mutants harbored in the cell lines OVCAR-3 and C33A, respectively. We observed differences in the action efficiency of the shRNA for p53 silencing, which may be due the efficiency of transfection intrinsic to each cell line. The most p53 expression inhibition was for the p53R273C mutant expression compared with the expression for the p53R248Q mutant. Despite this, in both cases the inhibition of the p53 mutants’ expression was enough to decrease the expression of HER2, both at the level of mRNA and protein. The regulation of HER2 by p53 mutants, suggests important clinical implications in the diagnostic and therapy of HER2 positive breast cancer. To gain some insight on the molecular mechanisms by which p53 mutants positively regulate the expression of HER2, we evaluated the possibility of induction of HER2 promoter activity. In this sense, previous reports have demonstrated the promoter activation of genes such as MDR1, EGFR, NF-kB and Axl by mtp53 activity [6, 35]. Our results showed that both p53 mutants (R248Q and R273C) stably expressed in the Saos-2 cell line induced HER2 gene promoter activity employing a luciferase reporter assay. Up-regulation of the HER2 promoter activity could be the result of direct mutant p53 DNA-binding, induction or recruitment of other transcription factors and transcriptional activators such as histone acetyl transferases. Among known HER2 promoter activators are the transcription factors AP-2, YY1 and members of the ETS family [21, 36, 37]; there are also repressors such as FOXP3, PEA3 and GATA4 [19, 38]. According with the HER2 promoter described by Hurst in 2001, we evaluated if the observed up-regulation in HER2 is achieved through binding of mutant p53 to DNA fragments containing AP-2 or ETS sequences [21]. We demonstrated the specific recruitment of p53 mutants to both, proximal and distal regions of the promoter by ChIP assay but not all the regions were enriched at the same level. Interestingly, we observed that the highest enrichment for the p53R248Q mutant was in the HER2 proximal region while the mutant p53R273C shows its highest recruitment level in the distal HER2 promoter region even though both protein enrichments were located at regions with AP-2 binding sites. It is possible that the p53 mutants alone or bound with AP-2 proteins have different affinity or bind selectively to different sequences near AP-2 binding sites in the HER2 promoter. AP-2 factors have been demonstrated to bind to wtp53 in vitro and in vivo [39, 40]; for example, in a p53-dependent binding AP-2 can regulate the p21WAF1/CIP1 gene promoter [39, 41]. Recently it was also determined, using p53 point mutants, that the AP-2 binding region of p53 is located in amino acids 305–375 [42]. It is worth mentioning that the p53R248Q and p53R273C mutants are capable of activating AP-2 binding regions in other promoters, suggesting that these p53 mutants could cooperate with AP-2 to activate HER2 transcription. Other transcription factors have been shown to interact with p53 mutants, including SP1, ETS1, ETS2, NF-Y, and the Vitamin D receptor [43], altering their target genes expression. Mutant p53R273H might exert its oncogenic gain-of-function by binding to ETS human genomic sequences, both within proximal and distal promoters [44]. In agreement with these studies, in the HER2 proximal promoter there is an ETS sequence, and we observed that p53R248Q and p53R273C mutants bind to this region, suggesting that these p53 mutants interact with ETS transcription factors. On the other hand, we evaluated if these p53 mutants had any repercussion on H3 and H4 histone acetylation, enhancing the epigenetic marks and inducing directly or indirectly HER2 promoter activity [45, 46]. As expected, histone H3 and H4 acetylation enrichment induced by p53 mutants coincides with the level of HER2 promoter activation observed in the luciferase reporter assays. In general, the major increase in the level of the acetylated marks in H3 and particularly in H4 histones was due to p53R248Q mutant, and observed in the HER2 proximal promoter, indicating a more relevant participation of the proximal promoter for the HER2 activation. In agreement with our results, Zhu et al. showed that p53 GOF mutants bind to and up-regulate chromatin regulatory genes, such as the methyltransferases MLL1, MLL2 and acetyltransferase MOZ, resulting in genome-wide increases of histone methylation and acetylation. Analysis of The Cancer Genome Atlas shows a specific tendency of up-regulation of MLL1, MLL2 and MOZ in selected patient tumors that express GOF p53-mutants [47]. The MOZ gene, encodes an enzyme that adds an acetyl group to K9 of H3 histone, allowing increased gene expression [47]. In agreement with this report, we observed that p53 mutants, R248Q and R273C, increase the level of acetylated marks in H3 histone at the HER2 promoter. In addition, we observed that the acetylation level of histone H4 was higher than that for histone H3 in all the regions analyzed, coinciding with Sandip K et al., 2001 observation, using HDAC inhibitors, suggesting that H4 acetylation is more important for HER2 activation [46]. A schematic representation for the histone arrangement and p53 binding in the proximal and distal HER2 promoter regions is shown in Fig. 6.
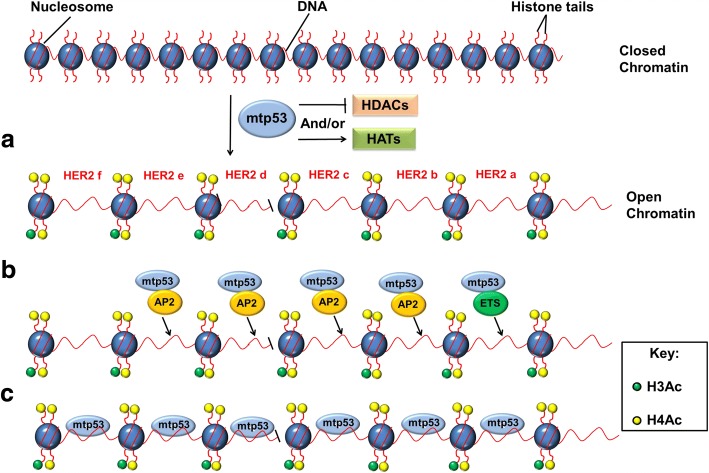
Fig. 6.
Model of HER2 up-regulation by p53 mutants. Three different molecular mechanisms of how mutant p53 may activate transcription from HER2 promoter are depicted. a Mutant p53 may induce histone (s) acetylation and/or inhibit histone deacetylase (s) causing enrichment of H3 and H4 acetylated forms and chromatin reorganization on the HER2 promoter; b Mutant p53 may induce interaction of one or more transcription factor (s), with the HER2 promoter, such as AP-2 and ETS; c Mutant p53 itself can interact directly on the HER2 promoter. These 3 possibilities are not mutually exclusive and would lead to the HER2 promoter activation with the consequent HER2 gene expression. In this representation, the histones are large blue spheres, the histone H3 acetylated marks are small green circles, the histone H4 acetylated marks are small yellow circles, the mutant p53 proteins are shown as light blue ovals, the AP-2 transcription factor are yellow ovals and ETS are green ovals. The DNA sequence is shown in red
On the other hand, mutant p53 also could increase the stability of HER2 protein, previous reports had demonstrated that p53R175H can regulate some genes like EGFR at the transcriptional level, and EGFR contributes to the HER2 protein stability by increasing its dimerization state [7, 28]. In addition, it has been previously reported that HER2 protein stabilization is also induced by Hsp90 [48]. The chaperone system controls the stability of the nascent forms of both EGF-R and HER2, but only the mature form of HER2. Interestingly, it has been also reported that Hsp90 also plays a key role in the folding of p53 mutants and stabilization, suggesting that a target therapy to Hsp90 could inhibit at the same time the HER2 signaling pathway and mutant p53 gain of function [49]. Recently, it was also demonstrated that mutants of p53 promote HSF1 phosphoactivation, protein stabilization and specific DNA-binding of this protein to its target gene promoters; HSF1 is the master transcriptional regulator of heat shock chaperones, including HSP90, suggesting a mtp53-HSF1-HSP90 oncogenic cooperation to stabilize the HER2 protein [50].
Conclusion
In conclusion, our study has demonstrated for the first time that HER2 over-expression is induced by p53 mutants through HER2 transcriptional activation. Acetylated H3 and H4 histones are associated to the HER2 proximal promoter in the presence of both p53R248Q and p53R273C mutants. However, more studies are required to completely elucidate the mechanism by which p53 mutants can induce HER2 mRNA up-regulation and its protein over-expression. Exploring the oncogenic gain of functions of p53 mutants may reveal new opportunities for diagnosis and therapy. In this regard, our results could have clinical implications in breast cancer and other cancer types where HER2 is over-expressed and used as a therapy target. As we mentioned in background section, Wilson et al. reported that patients carrying TP53 mutations show a significantly higher probability of developing breast cancer with ERBB2 gene amplification in human tumors. It will be necessary to demonstrate if the p53R248Q and p53R273C mutants are associated to HER2 over-expression in cancer or if there are specific mutations of p53 associated to overexpression of HER2.
Additional files
Table S1. The sequences of the primers used for HER2 and p53 analysis. (PDF 339 kb)
Table S2. The sequences of the primers used for HER2 promoter analysis. (PDF 336 kb)
Figure S1. Wild-type p53 expression inversely correlates with HER2 mRNA levels. HeLa (A) and Saos-2 (B) cell lines were transiently transfected with wtp53, while MCF-7 cell line (C) was transiently transfected with shp53, as indicated in Materials and Methods. Cells were processed to determine HER2 mRNA levels by RT-qPCR. These experiments were performed by triplicate and data are shown as the mean ± SEM. Statistical analysis was performed with t-student test by comparing the results obtained in: wtp53 vs empty vector transfected HeLa and Saos-2 cells; or shp53 vs scramble transfected MCF-7 cells: * p < 0.05, ns = non-significant statistical difference analysis. (D) HER2 Protein expression in Saos-2 transfected with wtp53 was measured by Western Blot and the image of the blot is representative of three independent experiments. (PDF 169 kb)
Acknowledgements
This study was performed in partial fulfillment of the requirement for the obtention of the Doctoral degree of Ana Alejandra Román Rosales (A.A.R.R.), student of the Programa de Doctorado en Ciencias Biomédicas, Instituto de Fisiología Celular, Universidad Nacional Autónoma de México (UNAM), Ciudad de México, México.
The authors would like to thank to Elizabeth Álvarez-Ríos, Rodolfo Ocádiz-Delgado and Lauro Macias-González for technical support.
Funding
Roman-Rosales AA was supported by grants from the CONACYT, México. Due to AARR’s role as first author, this implies a direct role of CONACYT in design, collection, analysis and interpretation of the data. Moreover, other grants obtained from CONACyT (168896) to Díaz-Chávez J and (236767) to Gariglio P were used for this work. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request.
Abbreviations
- ATTC
American Type Culture Collection
- B2M
Beta-2-microglobulin
- ChIP
Chromatin immunoprecipitation
- CT
Threshold Cycle value
- DBD
DNA Binding Domain
- DMEM
Dulbecco’s Modified Eagle Medium
- DNA
Deoxyribonucleic acid
- EGFR
Epidermal Growth Factor Receptor
- G245
Glycine in 245 codon of p53 protein
- GOF
Gain of Function
- HDAC
Histone Deacetylase
- HER2
Epidermal Growth Factor Receptor 2
- HER2
Epidermal Growth Factor Receptor 2 gene
- mRNA
Messenger Ribonucleic acid
- mtp53
Mutant p53
- p53
Tumor suppressor p53
- R175
Arginine in 175 codon of p53 protein
- R248
Arginine in 248 codon of p53 protein
- R249
Arginine in 249 codon of p53 protein
- R273
Arginine in 273 codon of p53 protein
- R282
Arginine in 282 codon of p53 protein
- RNA
Ribonucleic acid
- RT-qPCR
Reverse Transcription and quantitative Polymerase Chain Reaction
- SEM
Standard Error of the Mean
- shRNA
Short hairpin RNA
- TP53
Tumor suppressor p53 gene
- wtp53
Wild-type p53
Authors’ contributions
Conception and design: AARR, JDC. Development of methodology: AARR, EGV. Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): AARR, EGV, JDC. Writing, review, and/or revision of the manuscript: AARR, EGV, LAH, PG, JDC Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): EGV, PG, JDC. Study supervision: LAH, PG, JDC. All authors have read and approved the present manuscript in the current version.
Ethics approval and consent to participate
‘Not applicable’ for that section.
Consent for publication
‘Not applicable’ for that section.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12885-018-4613-1) contains supplementary material, which is available to authorized users.
Contributor Information
A. A. Román-Rosales, Email: aleja_roman_r@hotmail.com
E. García-Villa, Email: liebre1963@yahoo.com
L. A. Herrera, Email: herreram@biomedicas.unam.mx
P. Gariglio, Phone: 52-(55)-57473337, Email: vidal@cinvestav.mx
J. Díaz-Chávez, Phone: 52-(55)-5628-0400, Email: jdiazchavez03@gmail.com, Email: josediaz030178@hotmail.com
References
- 1.Liu G, Chen X. Regulation of the p53 transcriptional activity. J Cell Biochem. 2006;97:448–458. doi: 10.1002/jcb.20700. [DOI] [PubMed] [Google Scholar]
- 2.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 3.Liu J, Zhang C, Feng Z. Tumor suppressor p53 and its gain-of-function mutants in cancer. Acta Biochim Biophys Sin Shanghai. 2014;46:170–179. doi: 10.1093/abbs/gmt144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 5.Olivier M, Hussain SP, Caron de Fromentel C, Hainaut P, Harris CC. TP53 mutation spectra and load: a tool for generating hypotheses on the etiology of cancer. IARC Sci Publ. 2004:247–70. http://www.ncbi.nlm.nih.gov/pubmed/15055300. [PubMed]
- 6.Vaughan C, Pearsall I, Yeudall A, Deb SP, Deb S. p53: its mutations and their impact on transcription. Subcell Biochem. 2014;85:71–90. doi: 10.1007/978-94-017-9211-0_4. [DOI] [PubMed] [Google Scholar]
- 7.Roskoski R., Jr The ErbB/HER family of protein-tyrosine kinases and cancer. Pharmacol Res. 2014;79:34–74. doi: 10.1016/j.phrs.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 8.Yarden Y, Pines G. The ERBB network: at last, cancer therapy meets systems biology. Nat Rev Cancer. 2012;12:553–563. doi: 10.1038/nrc3309. [DOI] [PubMed] [Google Scholar]
- 9.Graus-Porta D, Beerli RR, Daly JM, Hynes NE. ErbB-2, the preferred heterodimerization partner of all ErbB receptors, is a mediator of lateral signaling. EMBO J. 1997;16:1647–1655. doi: 10.1093/emboj/16.7.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan M, Parker BA, Schwab R, Kurzrock R. HER2 aberrations in cancer: implications for therapy. Cancer Treat Rev. 2014;40:770–780. doi: 10.1016/j.ctrv.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 11.Dillon RL, Brown ST, Ling C, Shioda T, Muller WJ. An EGR2/CITED1 transcription factor complex and the 14-3-3sigma tumor suppressor are involved in regulating ErbB2 expression in a transgenic-mouse model of human breast cancer. Mol Cell Biol. 2007;27:8648–8657. doi: 10.1128/MCB.00866-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson JR, Bateman AC, Hanson H, An Q, Evans G, Rahman N, et al. A novel HER2-positive breast cancer phenotype arising from germline TP53 mutations. J Med Genet. 2010;47:771–774. doi: 10.1136/jmg.2010.078113. [DOI] [PubMed] [Google Scholar]
- 13.Melhem-Bertrandt A, Bojadzieva J, Ready KJ, Obeid E, Liu DD, Gutierrez-Barrera AM, et al. Early onset HER2-positive breast cancer is associated with germline TP53 mutations. Cancer. 2012;118:908–913. doi: 10.1002/cncr.26377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bull SB, Ozcelik H, Pinnaduwage D, Blackstein ME, Sutherland DA, Pritchard KI, et al. The combination of p53 mutation and neu/erbB-2 amplification is associated with poor survival in node-negative breast cancer. J Clin Oncol. 2004;22:86–96. doi: 10.1200/JCO.2004.09.128. [DOI] [PubMed] [Google Scholar]
- 15.Chan KT, Lung ML. Mutant p53 expression enhances drug resistance in a hepatocellular carcinoma cell line. Cancer Chemother Pharmacol. 2004;53:519–526. doi: 10.1007/s00280-004-0767-4. [DOI] [PubMed] [Google Scholar]
- 16.Baker SJ, Markowitz S, Fearon ER, Willson JK, Vogelstein B. Suppression of human colorectal carcinoma cell growth by wild-type p53. Science (80- ) 1990;249:912–915. doi: 10.1126/science.2144057. [DOI] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Weisz L, Zalcenstein A, Stambolsky P, Cohen Y, Goldfinger N, Oren M, et al. Transactivation of the EGR1 gene contributes to mutant p53 gain of function. Cancer Res. 2004;64:8318–8327. doi: 10.1158/0008-5472.CAN-04-1145. [DOI] [PubMed] [Google Scholar]
- 19.Xing X, Wang SC, Xia W, Zou Y, Shao R, Kwong KY, et al. The ets protein PEA3 suppresses HER-2/neu overexpression and inhibits tumorigenesis. Nat Med. 2000;6:189–195. doi: 10.1038/72294. [DOI] [PubMed] [Google Scholar]
- 20.Delacroix L, Vernimmen D, Begon D, Jackers P, Winkler R. Mechanisms responsible for ERBB2 gene overexpression in human breast and non-breast cancer cells. The role of AP-2 transcription factors. Cancer Ther. 2005;3:14. [Google Scholar]
- 21.Hurst HC. Update on HER-2 as a target for cancer therapy: the ERBB2 promoter and its exploitation for cancer treatment. Breast Cancer Res. 2001;3:395–398. doi: 10.1186/bcr329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delacroix L, Begon D, Chatel G, Jackers P, Winkler R. Distal ERBB2 promoter fragment displays specific transcriptional and nuclear binding activities in ERBB2 overexpressing breast cancer cells. DNA Cell Biol. 2005;24:582–594. doi: 10.1089/dna.2005.24.582. [DOI] [PubMed] [Google Scholar]
- 23.Goldstein I, Marcel V, Olivier M, Oren M, Rotter V, Hainaut P. Understanding wild-type and mutant p53 activities in human cancer: new landmarks on the way to targeted therapies. Cancer Gene Ther. 2011;18:2–11. doi: 10.1038/cgt.2010.63. [DOI] [PubMed] [Google Scholar]
- 24.Jaehne J, Urmacher C, Thaler HT, Friedlander-Klar H, Cordon-Cardo C, Meyer HJ. Expression of Her2/neu oncogene product p185 in correlation to clinicopathological and prognostic factors of gastric carcinoma. J Cancer Res Clin Oncol. 1992;118:474–479. doi: 10.1007/BF01629433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langerod A, Zhao H, Borgan O, Nesland JM, Bukholm IR, Ikdahl T, et al. TP53 mutation status and gene expression profiles are powerful prognostic markers of breast cancer. Breast Cancer Res. 2007;9:R30. doi: 10.1186/bcr1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He H, Ni J, Huang J. Molecular mechanisms of chemoresistance in osteosarcoma (review) Oncol Lett. 2014;7:1352–1362. doi: 10.3892/ol.2014.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiao Y, Gao X, Maragh S, Telford WG, Tona A. Cell lines as candidate reference materials for quality control of ERBB2 amplification and expression assays in breast Cancer. Clin Chem. 2009;55:1307–1315. doi: 10.1373/clinchem.2008.120576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yallowitz AR, Li D, Lobko A, Mott D, Nemajerova A, Marchenko N. Mutant p53 amplifies epidermal growth factor receptor family signaling to promote mammary tumorigenesis. Mol Cancer Res. 2015;13:743–754. doi: 10.1158/1541-7786.MCR-14-0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chavez-Blanco A, Perez-Sanchez V, Gonzalez-Fierro A, Vela-Chavez T, Candelaria M, Cetina L, et al. HER2 expression in cervical cancer as a potential therapeutic target. BMC Cancer. 2004;4:59. doi: 10.1186/1471-2407-4-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beaufort CM, Helmijr JCA, Piskorz AM, Hoogstraat M, Ruigrok-Ritstier K, Besselink N, et al. Ovarian cancer cell line panel (OCCP): clinical importance of in vitro morphological subtypes. PLoS One. 2014;9:e103988. doi: 10.1371/journal.pone.0103988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang JW, Lee EY, Kang KW. ErbB2 overexpression in p53-inactivated mammary epithelial cells. FEBS Lett. 2006;580:6501–6508. doi: 10.1016/j.febslet.2006.10.059. [DOI] [PubMed] [Google Scholar]
- 32.Lagunas-Martinez A, Madrid-Marina V, Gariglio P. Modulation of apoptosis by early human papillomavirus proteins in cervical cancer. Biochim Biophys Acta. 2010;1805:6–16. doi: 10.1016/j.bbcan.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 33.Lacroix M, Toillon RA, Leclercq G. p53 and breast cancer, an update. Endocr Relat Cancer. 2006;13:293–325. doi: 10.1677/erc.1.01172. [DOI] [PubMed] [Google Scholar]
- 34.Park JW, Neve RM, Szollosi J, Benz CC. Unraveling the biologic and clinical complexities of HER2. Clin Breast Cancer. 2008;8:392–401. doi: 10.3816/CBC.2008.n.047. [DOI] [PubMed] [Google Scholar]
- 35.Muller PA, Vousden KH. p53 mutations in cancer. Nat Cell Biol. 2013;15:2–8. doi: 10.1038/ncb2641. [DOI] [PubMed] [Google Scholar]
- 36.Allouche A, Nolens G, Tancredi A, Delacroix L, Mardaga J, Fridman V, et al. The combined immunodetection of AP-2alpha and YY1 transcription factors is associated with ERBB2 gene overexpression in primary breast tumors. Breast Cancer Res. 2008;10:R9. doi: 10.1186/bcr1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bosc DG, Janknecht R. Regulation of Her2/neu promoter activity by the ETS transcription factor, ER81. J Cell Biochem. 2002;86:174–183. doi: 10.1002/jcb.10205. [DOI] [PubMed] [Google Scholar]
- 38.Hua G, Zhu B, Rosa F, Deblon N, Adelaide J, Kahn-Perles B, et al. A negative feedback regulatory loop associates the tyrosine kinase receptor ERBB2 and the transcription factor GATA4 in breast cancer cells. Mol Cancer Res. 2009;7:402–414. doi: 10.1158/1541-7786.MCR-08-0175. [DOI] [PubMed] [Google Scholar]
- 39.McPherson LA, Loktev AV, Weigel RJ. Tumor suppressor activity of AP2alpha mediated through a direct interaction with p53. J Biol Chem. 2002;277:45028–45033. doi: 10.1074/jbc.M208924200. [DOI] [PubMed] [Google Scholar]
- 40.Modugno M, Tagliabue E, Ardini E, Berno V, Galmozzi E, De Bortoli M, et al. p53-dependent downregulation of metastasis-associated laminin receptor. Oncogene. 2002;21:7478–7487. doi: 10.1038/sj.onc.1205957. [DOI] [PubMed] [Google Scholar]
- 41.Wajapeyee N, Somasundaram K. Cell cycle arrest and apoptosis induction by activator protein 2alpha (AP-2alpha) and the role of p53 and p21WAF1/CIP1 in AP-2alpha-mediated growth inhibition. J Biol Chem. 2003;278:52093–52101. doi: 10.1074/jbc.M305624200. [DOI] [PubMed] [Google Scholar]
- 42.Stabach PR, Thiyagarajan MM, Woodfield GW, Weigel RJ. AP2alpha alters the transcriptional activity and stability of p53. Oncogene. 2006;25:2148–2159. doi: 10.1038/sj.onc.1209250. [DOI] [PubMed] [Google Scholar]
- 43.Santoro R, Strano S, Blandino G. Transcriptional regulation by mutant p53 and oncogenesis. Subcell Biochem. 2014;85:91–103. doi: 10.1007/978-94-017-9211-0_5. [DOI] [PubMed] [Google Scholar]
- 44.Vaughan CA, Deb SP, Deb S, Windle B. Preferred binding of gain-of-function mutant p53 to bidirectional promoters with coordinated binding of ETS1 and GABPA to multiple binding sites. Oncotarget. 2014;5:417–427. doi: 10.18632/oncotarget.1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Falahi F, Huisman C, Kazemier HG, van der Vlies P, Kok K, Hospers GA, et al. Towards sustained silencing of HER2/neu in cancer by epigenetic editing. Mol Cancer Res. 2013;11:1029–1039. doi: 10.1158/1541-7786.MCR-12-0567. [DOI] [PubMed] [Google Scholar]
- 46.Mishra SK, Mandal M, Mazumdar A, Kumar R. Dynamic chromatin remodeling on the HER2 promoter in human breast cancer cells. FEBS Lett. 2001;507:88–94. doi: 10.1016/S0014-5793(01)02951-9. [DOI] [PubMed] [Google Scholar]
- 47.Zhu J, Sammons MA, Donahue G, Dou Z, Vedadi M, Getlik M, et al. Gain-of-function p53 mutants co-opt chromatin pathways to drive cancer growth. Nature. 2015;525:206–211. doi: 10.1038/nature15251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Powers MV, Workman P. Targeting of multiple signalling pathways by heat shock protein 90 molecular chaperone inhibitors. Endocr Relat Cancer. 2006;13(Suppl 1):S125–S135. doi: 10.1677/erc.1.01324. [DOI] [PubMed] [Google Scholar]
- 49.Li D, Marchenko ND, Schulz R, Fischer V, Velasco-Hernandez T, Talos F, et al. Functional inactivation of endogenous MDM2 and CHIP by HSP90 causes aberrant stabilization of mutant p53 in human cancer cells. Mol Cancer Res. 2011;9:577–588. doi: 10.1158/1541-7786.MCR-10-0534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li D, Yallowitz A, Ozog L, Marchenko N. A gain-of-function mutant p53-HSF1 feed forward circuit governs adaptation of cancer cells to proteotoxic stress. Cell Death Dis. 2014;5:e1194. doi: 10.1038/cddis.2014.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. The sequences of the primers used for HER2 and p53 analysis. (PDF 339 kb)
Table S2. The sequences of the primers used for HER2 promoter analysis. (PDF 336 kb)
Figure S1. Wild-type p53 expression inversely correlates with HER2 mRNA levels. HeLa (A) and Saos-2 (B) cell lines were transiently transfected with wtp53, while MCF-7 cell line (C) was transiently transfected with shp53, as indicated in Materials and Methods. Cells were processed to determine HER2 mRNA levels by RT-qPCR. These experiments were performed by triplicate and data are shown as the mean ± SEM. Statistical analysis was performed with t-student test by comparing the results obtained in: wtp53 vs empty vector transfected HeLa and Saos-2 cells; or shp53 vs scramble transfected MCF-7 cells: * p < 0.05, ns = non-significant statistical difference analysis. (D) HER2 Protein expression in Saos-2 transfected with wtp53 was measured by Western Blot and the image of the blot is representative of three independent experiments. (PDF 169 kb)
Data Availability Statement
The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request.