Abstract
NMDA receptors are a diverse family of excitatory channels with critical roles in central synaptic transmission, development, and plasticity. Controlled expression of seven subunits and their combinatorial assembly into tetrameric receptors produces a range of molecularly distinct receptor subtypes. Despite relatively similar atomic structures, each subtype has input–output functions with unique biophysical and pharmacologic profiles. Here, we briefly summarize recent advances in understanding how gating and allosteric modulation are similar or distinct across NMDA receptor isoforms and identify open questions that will focus research in this area going forward.
Introduction
NMDA receptors mediate fundamental processes in the central nervous system (CNS). For this reason, they have garnered substantial attention since their discovery over 50 years ago. Notwithstanding significant progress in understanding their direct participation in health and disease, much remains unknown. In this review, we briefly summarize recent advances in delineating subtype-specific similarities and differences in the activation and modulatory mechanisms of NMDA receptors.
NMDA receptors belong to the ionotropic glutamate receptor (iGluR) family [1]. In this family, 18 separate genes encode subunits that segregate by homology into four functionally distinct classes: GluA (1–4) form AMPA receptors, GluK (1–5) form Kainate receptors, GluN (1, 2A–D, 3A–B) form NMDA receptors, and GluD (1–2) form delta receptors. Most, although not all, iGluRs function as glutamate-gated ion channels and mediate the bulk of excitatory neurotransmission in the mammalian CNS. Family members share transmembrane topology, modular tertiary architecture, and similar three-dimensional shapes [2,3].
Despite substantial structural similarity among iGluRs, NMDA receptors have a number of distinctive functional features, which relate directly to the receptor’s unique physiological roles (reviewed in [4]). First, NMDA receptors have characteristically large conductance, high Ca2+ permeability, and slow kinetics, which ensure substantial cationic influx, especially Ca2+ [5]. Second, due to their voltage-dependent block by physiological levels of Mg2+, membrane voltage affects the magnitude of their currents, which are largest when both the pre-synaptic and post-synaptic cells are concurrently active [6]. Third, NMDA receptor currents are sensitive to a large variety of endogenous and synthetic modulators, which expand the range of receptor behaviors and represent an important means for rapid, temporally, and regionally regulated control [7]. Because in most cases cells of the CNS co-express several iGluR subunits, these unique biophysical and pharmacologic features help to separate experimentally the current mediated specifically by NMDA receptors from the AMPA and Kainate receptor components of the glutamate-elicited response. However, even these features that distinguish NMDA receptors from other family members vary considerably across CNS regions and developmental stage, and with experience. In part, this functional heterogeneity reflects the expression of molecularly distinct NMDA receptor subtypes [8,9].
NMDA receptors assemble from seven gene products, GluN1, GluN2A–D, and GluN3A–B, each having distinct temporal and cell-specific expression patterns [9]. Unlike other iGluRs, NMDA receptors are obligate heterotetramers, and functional receptors must contain GluN1 and GluN2 and/or GluN3 subunits. The exact molecular composition of native NMDA receptors is still largely undetermined, mostly because several of the seven subunits co-localize and presently, experimental approaches cannot distinguish unambiguously responses from individual receptor types in mixtures. Therefore, it remains uncertain how NMDA receptor subunits combine to form working channels [8]. Functional characterization of recombinant receptors with defined molecular composition combined with in situ pharmacologic and genetic approaches support the view that NMDA receptors assemble as dimers of heterodimers [10,11]. As such they can be diheteromers, if they contain only two types of subunits, that is two GluN1 and the same two of GluN2 or GluN3 subunits; or they can be triheteromers, if they contain three types of subunits, that is two GluN1 and two different GluN2(A–D) and/or GluN3(A–B). To determine the molecular identity of NMDA receptors in neurons and glial cells, and to understand how each receptor subtype contributes to CNS physiology and pathology, it is necessary to delineate how NMDA receptor subtypes are similar and different functionally and structurally.
Activation of NMDA receptor subtypes
The most important known function of NMDA receptors is their ability to flux Na+ and Ca2+ across the plasma membrane, which produces simultaneously cellular depolarization and intracellular Ca2+ elevation. In turn, these effects change in real time the cell’s excitability and its intracellular signaling cascades. With this view, the information conveyed by NMDA receptor signals correlates directly with the time-dependent amplitude of the ensemble current and its ionic composition. The first depends on the receptor’s activation mechanism and the second on the receptor’s ionic permeability (reviewed by Wollmuth in this issue and in [5]). Here, we briefly summarize recent advances in understanding commonalities and differences in the activation and modulatory mechanisms of NMDA receptor subtypes.
At the simplest level, the activation of all iGluRs consists of two steps: glutamate binding and channel opening. The activation process begins when glutamate binds to resting receptors, which are impermeable or closed (C), and continues with an avalanche of structural changes that culminates with active receptors, which are permeable or open (O). Importantly, the conformational changes that gate the channel also prevent glutamate dissociation and, therefore, receptor deactivation necessarily consists of the reverse sequence: channel closing followed by glutamate dissociation.
Electrophysiological recordings largely agree with this bare-bone model and illustrate that when applying glutamate briefly (milliseconds) onto a cell expressing glutamatergic channels the current rises to a peak amplitude as dictated by binding and opening rates and then decays as dictated by closing and dissociation rates. Research over many years has added the necessary detail to explain additional features of the observed current. For example, glutamate applications elicit currents from NMDA receptors only when glycine is also present. In addition, when glutamate exposure is longer than milliseconds, after reaching an initial peak amplitude the current declines to a lower steady state level, illustrative of macroscopic desensitization. These and many other observations indicate that NMDA receptors activate along a more complex sequence of events and many more transition rates control the rise and fall of the synaptic current. Recently developed multi-step reaction mechanisms explain comprehensively receptor behaviors over several time domains, recording resolutions, stimulation patterns, and modulatory influences.
Diheteromeric GluN1/GluN2 receptors activate with similar mechanisms but distinct rates
To date, comprehensive NMDA receptor reaction mechanisms have been developed and tested only for diheteromeric GluN1/GluN2 receptors. This is because the modeling process requires high quality single-channel recordings, which remain elusive for diheteromeric GluN1/GluN3 receptors and even more so for triheteromeric receptors. Generally, the activation sequence of diheteromeric GluN1/GluN2 receptors includes binding steps (two for glutamate and two for glycine), gating steps (C3–C2–C1–O1–O2), and desensitizing steps (D1 and D2) (Figure 1). In addition, all four GluN1/GluN2 receptors display modal behavior (reviewed in [12]).
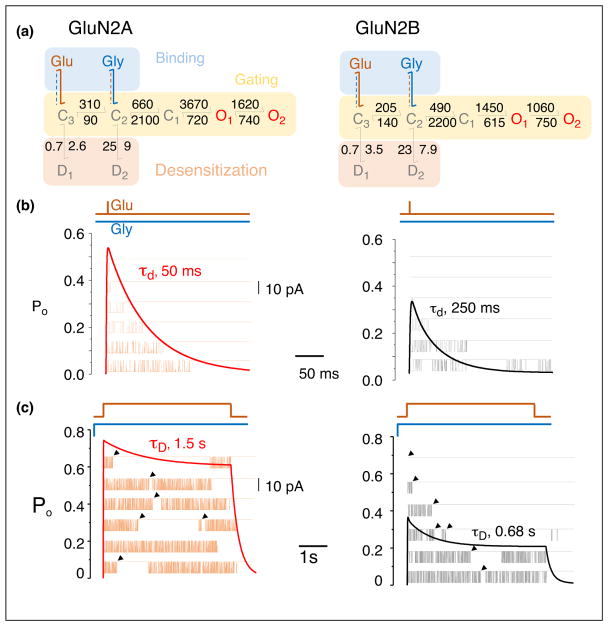
Figure 1.
NMDA receptor isoforms activate with similar steps but distinct transition rates, which dictate characteristic functional output. (a) Kinetic models derived from stationary single-channel recordings of GluN1-1a/GluN2A (left) and GluN1-1a/GluN2B receptors (right). After binding glutamate (Glu) and glycine (Gly), both receptor types cycle among five closed states, C1–3 and D1–2, and two open states, O1–2 (3C2O2D). Rate constants are in s−1. Agonist binding steps reflect mechanisms determined by separate experiments holding one ligand constant (dashed) while varying the concentration of the other ligand (solid). Gating and desensitization rates converged on similar values for both experiments. (b) Synaptic-like response to 1-ms pulse of glutamate (Glu, 1 mM) in the continued presence of glycine (Gly, 0.1 mM), simulated with the kinetic models in (a) as deterministic time-dependency of open state occupancies of several channels (thick line), or stochastic one-channel openings to sequential stimuli (thin line). The decay of the macroscopic synaptic-like current corresponds to the stochastic distribution of cluster durations, which is influenced in complex ways by Glu dissociation kinetics and all gating rate constants. (c) Extrasynaptic-like response predicted by the kinetic models in (a) to 5-s pulse of Glu in the continued presence of 0.1 mM glycine. The decay of the macroscopic current (desensitization, tauD) corresponds to the time-dependent increase in the frequency of long, closed durations (arrows), which reflect the increased occupancy of states D1 and D2.
More specifically, the activation reaction of NMDA receptors starts with two sequential glutamate binding reactions to resting (glycine-bound) receptors, which lead into a fully liganded closed state (C3), which initiates gating. The ensuing gating reaction consists of a linear series of steps (3C2O) (Figure 1a). The first, C3–C2, traps glutamate and permits glycine dissociation/association; the second, C2–C1, traps glycine and permits channel opening. Last, the channel opens (C1–O) and cycles between two kinetically distinct open states of similar conductance (O1–O2). All four diheteromeric GluN1/GluN2 receptors conform to this activation model [13–16]. Isoform-specific differences arise from the distinct values of rate constants for agonist binding and dissociation, and for each of the steps within the gating reaction [4]. This linear activation sequence depicts the most rapid pathway by which resting receptors can open (activation) and open receptors can deactivate (deactivation). Recently, single-molecule FRET data have supported this linear arrangement of states [17•]. Its transition rates set the rise and decay timecourse for synaptic NMDA receptor currents (Figure 1b) and the decay timecourse of the excitatory postsynaptic current (EPSC).
Two additional types of functional changes can occur, which take receptors away from this linear activation/deactivation sequence. The first allows receptors to transition into desensitized states, defined as off-path agonist-bound closed states; and the second takes receptors into a distinct gating mode, defined as an activation sequence with similar topology but altered kinetics [12]. GluN1/Glu2A and GluN1/GluN2B receptors can access two kinetically distinct desensitized states: D1 from C3 (C3–D1) and D2 from C2 (C2–D2) [14,15]. Models developed for GluN1/GluN2C receptors envision that for these receptor subtypes both desensitized states occur from C3 (C3–D1 and C3–D2), whereas models for heteromeric GluN1/GluN2D receptors lack desensitized states [13,16,18,19]. Transitions into and from desensitized states control the shape of the macroscopic response to prolonged agonist exposure and the steady-state current level [20] (Figure 1c), and may influence critically the physiological responses of extrasynaptic receptors [4].
For the NMDA receptors investigated thus far, up to three distinct kinetic modes were identified. In kinetic schemes, modes are represented as parallel gating sequences, each with the same 3C2O2D arrangement, but with distinct rate constants, and receptors can transition stochastically between modes [21]. The rates with which receptors switch gating mode are slower than the activation/deactivation transitions such that receptors are unlikely to switch mode during a synaptic event. However, receptors in each mode deactivate with a distinct monophasic deactivation timecourse, suggesting that the well-known biphasic decay of the NMDA receptor synaptic current and the EPSC decay reflect the kinetic heterogeneity of synaptic NMDA receptors at the time of stimulation [22]. Not much is known of what causes modes or whether they are or can be controlled physiologically or experimentally. It is important to keep in mind that due to technical challenges in identifying and quantifying modes, the majority of investigators describe one-channel records with only one arm of the tiered model, and, therefore, report average values for transition rates, which in turn predict an average monophasic decay for the population response (Figure 1a).
The GluN1/GluN2D receptor is to date the only isoform reported for which differential splicing of the GluN1 subunit affects gating kinetics [18] and modal gating [16]. Whether splice-isoforms also differ in desensitization is unclear because the model lacks desensitization steps. GluN1/GluN2D receptors participate in synaptic transmission and control cellular excitability in hippocampal inhibitory interneurons [23]. Therefore, it will be important to develop a kinetic model that can describe the entirety of single-channel behaviors for this receptor subtype as well.
Among neurotransmitter-gated channels, NMDA receptors are unique in their functional requirement for glycine, a property arising from their structural requirement for the glycine-binding GluN1 subunits. The observation that macroscopic NMDA receptor currents desensitize faster and deeper when glycine is present in sub-saturating concentrations was referred to in the literature as ‘glycine-dependent desensitization’, and was thought to reflect negative cooperativity between the affinities of glutamate and glycine binding sites [24]. Recently, precise modeling of the glycine binding and dissociation reactions onto the kinetic model of GluN1/GluN2A receptors placed the glutamate and glycine binding reactions on separate kinetic steps within the activation sequence, namely on C3 and C2, respectively (Figure 1a). This expanded kinetic model predicted accurately all known glycine dependent current behaviors, without the need to postulate agonist-dependent changes in microscopic binding, dissociation, or desensitization rate constants [25]. Although the experimental data refer to GluN1/GluN2A receptors, given the overall conservation in activation mechanisms across GluN1/GluN2 subtypes, it is likely that for other receptor subtypes the conformations that most avidly bind glutamate and glycine are kinetically and structurally distinct.
Triheteromeric receptors
Historically, the majority of subtype-specific NMDA receptor properties were inferred from studies on recombinant diheteromeric receptors, and on synaptic currents recorded in the presence of GluN2-selective modulators. However, more recent studies suggest that triheteromeric GluN1/GluN2A/GluN2B receptors dominate at many adult synapses ([26] and reviewed by Paoletti in this volume). It had been assumed that triheteromeric channels have functional properties intermediate to GluN1/GluN2A and GluN1/GluN2B diheteromers. Yet, with new methodology that permits the expression of pure populations of recombinant triheteromeric receptors, it is evident that constituent GluN2 subunits have unequal impact on the current deactivation kinetics and its sensitivity to allosteric inhibitors [27•]. Specifically, these experiments showed a dominant functional role for the GluN2A subunit in GluN1/GluN2A/GluN2B triheteromers, consistent with results from constitutively active GluN2A or GluN2B subunits [28]. Recent observations of structurally asymmetric interactions formed by GluN2A relative to GluN2B subunits may represent the basis of such functional dominance [29•]. Therefore, it is likely that triheteromeric GluN1/GluN2A/GluN2B receptors gate with kinetic schemes similar in sequence to those of diheteromeric GluN1/GluN2 receptors, but with rate constants closer in value to those of GluN1/GluN2A receptors. Given their widespread expression, it will be important to develop quantitative reaction mechanisms for triheteromeric receptors, a task not yet attempted.
GluN1/GluN3 receptors as excitatory glycinergic receptors
Among, the iGluR family, GluN1 and GluN3 subunits are unique in that their agonist-binding site is specific to glycine (and D-serine). Therefore, diheteromeric GluN1/GluN3 channels are glycinergic and insensitive to glutamate or other GluN2-specific ligands. No kinetic model is yet available for GluN1/GluN3 receptors, although they play important roles in synaptogenesis and neurodegeneration [30,31]. Results reported to date indicate that their macroscopic behaviors and mechanisms are distinct in many aspects from their glutamatergic GluN1/GluN2 brethren and are likely more similar to those of AMPA and Kainate receptors [32].
For instance, unlike unitary currents recorded from glutamatergic GluN1/GluN2 receptors, those obtained from GluN1/GluN3 receptors have multiple conductance levels, a feature resembling AMPA and Kainate receptors [32,33]. Similarly, at the macroscopic level, GluN1/GluN3A currents desensitize much faster and more deeply than GluN1/GluN2 receptors [34]. Structurally as well, the roles of inter subunit interfaces influence function differently. In contrast to GluN1/GluN2 receptors, the efficacy of glycine at GluN1/GluN3 channels depends on allosteric interactions between the amino-terminal domains of the GluN3 subunits [35]; and an influential proton-binding site resides at the intersubunit interface of ligand-binding domains [36•]. Lastly, a strong dimer interface within the ligand-binding domain appears to increase activity by decreasing desensitization as in AMPA receptors [37], which is in contrast with glutamatergic GluN1/GluN2 receptors, where a flexible dimer interface is required for channel activation [38,39].
Given that astrocyte-released glycine is ambient in the extracellular milieu, it is likely that GluN1/GluN3 receptors operate in a tonic rather than phasic mode, being permanently glycine-bound and largely desensitized. In this scenario, the steady-state current, which has the potential to regulate excitatory tone, is controlled by factors that alter receptor desensitization [36•]. However, reaction schemes and structure–function relationships for these receptors, as well as the implications for synaptic physiology and pathology are presently lacking or incipient.
In summary, kinetic modeling of one-channel current traces has been successful in producing reaction mechanisms for the four diheteromeric GluN1/GluN2 receptors. These cover receptor transitions over six orders of time magnitude (10−4–102 s) and reproduce faithfully population behaviors peculiar to NMDA receptors, such as biphasic deactivation kinetics, glycine-dependent desensitization, and frequency-dependent potentiation. Importantly, these models allow for the first time direct measurement of absolute open probabilities for individual receptor subtypes and account for the striking differences in their deactivation kinetics. Several physiologically important receptor subtypes remain uncharacterized.
Modifying the NMDA receptor response with modulators and blockers
Multiple physiological mechanisms regulate the amplitude and timecourse of NMDA receptor responses. Controlled expression of specific subunit types is only one of these. On a faster timescale, diffusible small molecules that bind reversibly to membrane embedded channels can shape NMDA receptor responses (Figure 2a). The effects of these reversible modulators are receptor subtype specific.
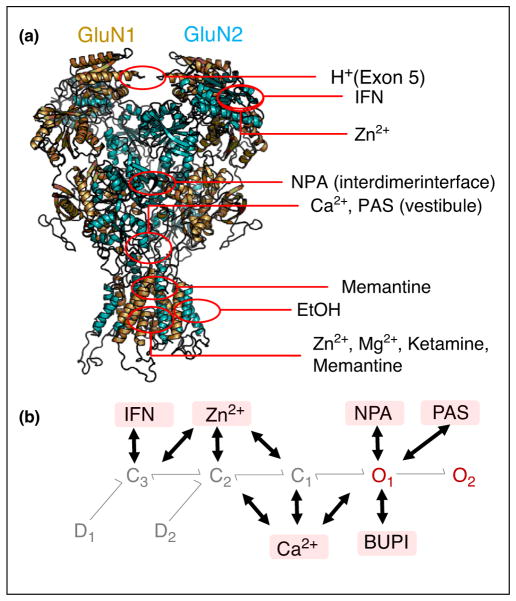
Figure 2.
Mechanism of NMDA receptor activity modulation by diffusible ligands. (a) Structure of tetrameric receptors (GluN1, gold; GluN2, teal; PDB 4PE5) with highlighted regions of modulator-binding sites defined by structural or functional studies. (b) For each modulator, a full reaction mechanism is described by the 3C2O2D sequence of modulator-free receptors as in Figure 1 and an additional 3C2O2D sequence of modulator-bound receptors depicted by the pink box, which has modulator-dependent rate constants. Thick arrows indicate the preferred transition states between the two 3C2O2D arms for each modulator. Proton (H+), ifenprodil (IFN), Ca2+, Zn2+, bupivacaine (BUPI), naphthoic acid (NPA), and pregnanolone sulfate (PAS).
It has been long observed that experimental conditions, including patch configuration and ionic composition of extra and intracellular compartments, especially H+, Ca2+, Zn2+ and Mg2+, influence the NMDA receptor macroscopic response and the pattern of single-channel activity. However, the mechanism by which this happens has been unclear and occasionally controversial.
Modulators change gating kinetics not mechanism
This question was recently addressed by leveraging the robust kinetic models developed for GluN1/GluN2 receptor subtypes to interpret the changes in opening pattern while bound to physiologic or synthetic modulators [40], or while carrying mutations introduced artificially [41] or occurring naturally in patients [42] (Figure 2a). Overall, these new reports show that modulator-bound and mutated receptors generally follow the same 3C2O2D gating sequence as modulator-free wild-type receptors but transitions within this sequence have altered rate constants; moreover, each modulator binds preferentially to separate receptor states and each perturbation, modulator or mutation, modifies a different collection of rate constants.
For example, Zn2+, ifenprodil (IFN), and H+ bind preferentially to closed states, which are accessed early during activation (C3, C2) [41,43]; whereas Mg2+, Ca2+ and pregnanolone sulfate (PAS) bind preferentially to open states (O1, O2), which are populated late in the activation sequence [15] (Figure 2b). The observation that overall, modulators with structurally overlapping or vicinal binding sites bind to the same kinetic state(s) has led to the exciting speculation that the sequence of kinetic states identified functionally may correspond to an ordered series of conformational changes that map vectorially onto the receptor’s atomic structure, top-down, or from the N-terminal toward the gate-hosting transmembrane domain.
Early evidence from measurements of intramolecular motions in functional receptors subject to allosteric inhibition [44,45] and from molecular dynamics simulations ([46] and see below) largely support this hypothesis. However, the information necessary to begin assigning even rough correspondence between kinetic states and families of structural conformations is currently insufficient. This area of investigation will benefit from ongoing sustained efforts to resolve atomic structures and kinetic reaction mechanisms for additional receptor subtypes, and mutants, with or without bound modulator.
Blockers
Channel blockers represent a special case of diffusible modulators, which bind within the membrane pore and obstruct ionic flux. The effect of blockers on NMDA receptor responses can differ widely depending on their association/dissociation kinetics, the physical pathway by which they access their binding site, and whether blocker-bound channels can continue to gate or must wait for the blocker to dissociate before closing. These characteristics can also be NMDA receptor subtype dependent. Given this complexity, the mechanisms by which diverse blockers affect the NMDA receptor activation reaction have been more difficult to reveal. However, NMDA receptor channel blockers hold enormous therapeutic potential and this area will continue to be the focus of intense research.
In particular, memantine and amantadine are NMDA receptor blockers that are clinically well tolerated and have demonstrated therapeutic effects in neurodegenerative disorders [47]. Similarly, ketamine, an anesthetic with long clinical history, has recently garnered renewed interest as a rapid and effective antidepressant [48]. Detailed kinetic models for the action of these therapeutically important blockers are not yet available.
The effect of bupivacaine, a NMDA receptor blocker widely used as local anesthetic, has been recently investigated in more depth [49]. In the presence of bupivacaine, NMDA receptor single-channel recordings revealed an additional, concentration-sensitive closed state reflective of blocker-bound receptors (Figure 2b). Consistent with an open channel block mechanism, this additional closed state was most likely accessed from open receptor states. In addition to rendering channels impermeable, the blocker changed transition rates in the activation sequence, indicative of a trapping block mechanism. This kinetic model identified blocker-sensitive transition rates, blocker concentration-dependence of receptor open probability, and stimulus-dependence of blocker efficacy thus supporting the premise that in addition to occluding permeation this ligand also alters activation kinetics [17•]. Delineating blocker mechanisms with similar detail will be necessary to understand why different blockers affect NMDA receptor responses differentially, a vexing unanswered question. Moreover, it will be necessary to delineate the subtype-specificity of NMDA receptor channel blockers as a mechanism for the distinct clinical effects of blockers.
Mutations
Similar to diffusible ligands, NMDA receptor mutations change the channel response by altering gating rate constants, and, therefore, the thermodynamic stability of kinetics states, rather than the sequence of conformational events. Even for mutations known for their drastic effects on permeation, gating effects are part of the overall change in response. For example, the single substitution GluN2A N615G located at the tip of the selectivity filter eliminates, as expected, voltage-dependent block by drastically reducing the binding of divalent cations (Zn2+, Mg2+) in the pore. However, even in the absence of divalent cation blockers, this mutation reduces the rate constants governing the activation reaction resulting in decreased occupancy of open states, and reduced macroscopic currents, which decay faster [41]. Given that NMDA receptor isoforms have gating landscapes that are thermodynamically distinct, it is highly likely that even mutations at conserved locations within NMDA receptors, such as this one, will have distinct effects across receptor subtypes. Therefore, when mechanistic detail is necessary, as for example when aspiring for structure-based drug design, extrapolating functional results from one receptor subtype to another is unhelpful.
Predicting structural correlates of kinetic states
Ligand-dependent channel activation involves coupling of conformational changes in the ligand-binding domain to pore opening. For this reason, the linkers connecting the extracellular ligand binding domains with the membrane-embedded helices (M1–M4) have received intense attention as physical conduits for ligand-dependent gating motions. Especially, because the top part of M3 helices form the ligand-sensitive gate, the linkers connecting the ligand-binding domain to the M3 helices have been studied in detail. When elongating these linkers by inserting glycine residues, the open probabilities of the resulting receptors were smaller, most likely due to reduced tension in these linkers and, therefore, looser coupling between ligand-binding domains and the gate [50]. Similarly, constraining movements of the peripheral M4 helices with disulfide bridges also decreased the open probability [51]. Even in advance of atomic resolution NMDA receptor structures, mathematical models envisioned gating trajectories that correlate with single-channel recordings to reproduce a number of kinetic features of channel function [52]. As more atomic-resolution structures become available, using mathematical modeling to predict the correspondence between kinetic states and discrete families of receptor conformations is the next frontier in understanding structure–function correlations in NMDA receptors.
To date, only three structural models for tetrameric NMDA receptors have been proposed [29•,53,54]; these are diheteromeric or triheteromeric GluN2B-containing receptors, each revealing a static snapshot of closed receptors. A cryo-EM study of GluN1/GluN2B receptors revealed a diversity of structural configurations adopted by channels in the presence of agonist, which included for the first time a portion of active receptor [55•]. These groundbreaking reports permit for the first time the use of molecular modeling approaches to infer structures of the resting, and agonist-bound closed, open, and desensitized states. Further, they represent springboards for inferring structures of all NMDA receptor subtypes to begin to identify the subtype-dependent differences that underlie subtype-dependent output and biological functions.
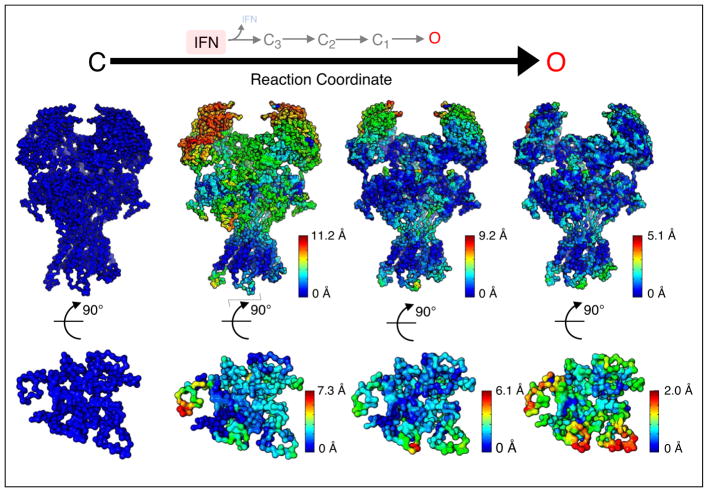
Of all NMDA receptor subtypes, the activation sequence of GluN1/GluN2A diheteromers is best understood to date, yet a structural model for this NMDA receptor subtype is not available. Homology modeling based on existing atomic-resolution structures offered a hypothetical arrangement for this subtype [56]. Further, using as initial and end-states homology models for the IFN-bound closed state and a predicted IFN-free open state, respectively, coarse-grained molecular dynamics simulations predicted a series of intermediate conformations. Importantly, the trajectory of structural change observed in these simulations followed a vectorial sequence starting in the N-terminal domain, continuing with the ligand-binding domain, and culminating with progressive movements within the transmembrane domain (Figure 3). This model is consistent with the pattern of change produced by modulators and blockers in the gating sequence. For example, N-terminal domain ligands (H+, Zn2+, IFN) influence steps that occur early in the activation reaction [41,43,57]; whereas open-channel blockers (Mg2+, bupivacaine) and modulators binding in or close to the transmembrane-domain (PAS) influence steps that occur later [15,49] (Figure 2). Initial validation for this model comes from considerable overlap between the location of newly identified disease-related mutations and functional hot spots predicted computationally by the model.
Figure 3.
Proposed structural model of NMDA receptor activation. Top, sequence of kinetic transitions initiated by IFN dissociation from IFN-bound closed conformations into IFN-free closed (C3, C2, C1) and open (O) states, according to kinetic models derived from single-channel electrical measurements. Middle, sequence of structural changes initiated by IFN dissociation from the IFN-bound closed structure (based on PDB 4PE5) into IFN-free closed and open conformations (based on PDB 5FXG), as proposed by coarse-grain and transition pathway modeling [56]. Spheres represent the alpha carbon of each residue; for each transition, structural motions were mapped onto the structure and color-coded according to their relative magnitude. Bottom, intracellular view of the transmembrane domain.
The first structural model of triheteromeric GluN1/GluN2A/GluN2B receptors based on cryo-EM data revealed subunit-specific interactions relative to the diheteromeric structures [29•]. In triheteromers, the GluN2A subunit participates in additional interactions with the GluN1 subunit, suggesting functional asymmetry for the two types of GluN2 subunits. Functional work with the triheteromeric receptor is consistent with a dominant role for the GluN2A, relative to the GluN2B, subunit [27•,28]. These pioneering studies pave the way for better understanding how subunit composition, through distinct intra-molecular interactions, supports kinetic transitions adapted to fulfill the biological roles of each NMDA receptor subtype.
Conclusions
Reaction mechanisms developed for the principal receptor subtypes connect accurately single-channel microscopic with macroscopic behaviors over a broad range of observation times and experimental conditions. Going forward these models will be increasingly useful in understanding how subtle differences in the structure of NMDA receptor subtypes produce biologically relevant functional differences. They will also assist in relating kinetic states with families of structural conformations and in uncovering possible signaling roles of electrically silent conformations [58]. Additional efforts in this area will have to expand the number of NMDA receptor subtypes for which appropriate models exist and to map the action of modulators and blockers onto these kinetic schemes. These quantitative models will help to understand in more detail how NMDA receptors contribute to physiological and pathological states and to envision rational approaches to modify their activities for therapeutic gain.
Acknowledgments
Grants from the NIH (R01NS09701602 and R21NS09838502 to GKP) supported this work.
Footnotes
Conflict of interest
None declared.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
- 1.Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK, Hansen KB, Yuan H, Myers SJ, Dingledine R. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev. 2010;62:405–496. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mayer ML. The challenge of interpreting glutamate-receptor ion-channel structures. Biophys J. 2017;113:2143–2151. doi: 10.1016/j.bpj.2017.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou HX, Wollmuth LP. Advancing NMDA receptor physiology by integrating multiple approaches. Trends Neurosci. 2017;40:129–137. doi: 10.1016/j.tins.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iacobucci GJ, Popescu GK. NMDA receptors: linking physiological output to biophysical operation. Nat Rev Neurosci. 2017;18:236–249. doi: 10.1038/nrn.2017.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glasgow NG, Siegler Retchless B, Johnson JW. Molecular bases of NMDA receptor subtype-dependent properties. J Physiol. 2015;593:83–95. doi: 10.1113/jphysiol.2014.273763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nowak L, Bregestovski P, Ascher P, Herbet A, Prochiantz A. Magnesium gates glutamate-activated channels in mouse central neurones. Nature. 1984;307:462–465. doi: 10.1038/307462a0. [DOI] [PubMed] [Google Scholar]
- 7.Hedegaard M, Hansen KB, Andersen KT, Bräuner-Osborne H, Traynelis SF. Molecular pharmacology of human NMDA receptors. Neurochem Int. 2012;61:601–609. doi: 10.1016/j.neuint.2011.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wyllie DJ, Livesey MR, Hardingham GE. Influence of GluN2 subunit identity on NMDA receptor function. Neuropharmacology. 2013;74:4–17. doi: 10.1016/j.neuropharm.2013.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paoletti P, Bellone C, Zhou Q. NMDA receptor subunit diversity: impact on receptor properties, synaptic plasticity and disease. Nat Rev Neurosci. 2013;14:383–400. doi: 10.1038/nrn3504. [DOI] [PubMed] [Google Scholar]
- 10.Schuler T, Mesic I, Madry C, Bartholomaus I, Laube B. Formation of NR1/NR2 and NR1/NR3 heterodimers constitutes the initial step in NMDA receptor assembly. J Biol Chem. 2008;283:37–46. doi: 10.1074/jbc.M703539200. [DOI] [PubMed] [Google Scholar]
- 11.Lee CH, Gouaux E. Amino terminal domains of the NMDA receptor are organized as local heterodimers. PLoS ONE. 2011;6:e19180. doi: 10.1371/journal.pone.0019180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Popescu GK. Modes of glutamate receptor gating. J Physiol. 2012;590:73–91. doi: 10.1113/jphysiol.2011.223750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dravid SM, Prakash A, Traynelis SF. Activation of recombinant NR1/NR2C NMDA receptors. J Physiol. 2008;586:4425–4439. doi: 10.1113/jphysiol.2008.158634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amico-Ruvio SA, Popescu GK. Stationary gating of GluN1/GluN2B receptors in intact membrane patches. Biophys J. 2010;98:1160–1169. doi: 10.1016/j.bpj.2009.12.4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kussius CL, Kaur N, Popescu GK. Pregnanolone sulfate promotes desensitization of activated NMDA receptors. J Neurosci. 2009;29:6819–6827. doi: 10.1523/JNEUROSCI.0281-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vance KM, Hansen KB, Traynelis SF. Modal gating of GluN1/GluN2D NMDA receptors. Neuropharmacology. 2013;71:184–190. doi: 10.1016/j.neuropharm.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17•.Dolino DM, Chatterjee S, MacLean DM, Flatebo C, Bishop LDC, Shaikh SA, Landes CF, Jayaraman V. The structure-energy landscape of NMDA receptor gating. Nat Chem Biol. 2017;13:1232–1238. doi: 10.1038/nchembio.2487. This paper uses single molecule FRET to provide the first experimental evidence for a range of distinct and stable conformations adopted by the transmembrane domain of NMDA receptors while actively gating in biological membranes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vance KM, Hansen KB, Traynelis SF. GluN1 splice variant control of GluN1/GluN2D NMDA receptors. J Physiol. 2012;590:3857–3875. doi: 10.1113/jphysiol.2012.234062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wyllie DJ, Behe P, Colquhoun D. Single-channel activations and concentration jumps: comparison of recombinant NR1a/NR2A and NR1a/NR2D NMDA receptors. J Physiol. 1998;510(Pt 1):1–18. doi: 10.1111/j.1469-7793.1998.001bz.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Le Meur K, Galante M, Angulo MC, Audinat E. Tonic activation of NMDA receptors by ambient glutamate of non-synaptic origin in the rat hippocampus. J Physiol. 2007;580(Pt 2):373–383. doi: 10.1113/jphysiol.2006.123570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Popescu G, Auerbach A. Modal gating of NMDA receptors and the shape of their synaptic response. Nat Neurosci. 2003;6:476–483. doi: 10.1038/nn1044. [DOI] [PubMed] [Google Scholar]
- 22.Zhang W, Howe JR, Popescu GK. Distinct gating modes determine the biphasic relaxation of NMDA receptor currents. Nat Neurosci. 2008;11:1373–1375. doi: 10.1038/nn.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perszyk RE, DiRaddo JO, Strong KL, Low CM, Ogden KK, Khatri A, Vargish GA, Pelkey KA, Tricoire L, Liotta DC, Smith Y, et al. GluN2D-containing N-methyl-D-aspartate receptors mediate synaptic transmission in hippocampal interneurons and regulate interneuron activity. Mol Pharmacol. 2016;90:689–702. doi: 10.1124/mol.116.105130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benveniste M, Clements J, Vyklicky L, Jr, Mayer ML. A kinetic analysis of the modulation of N-methyl-D-aspartic acid receptors by glycine in mouse cultured hippocampal neurones. J Physiol. 1990;428:333–357. doi: 10.1113/jphysiol.1990.sp018215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cummings KA, Popescu GK. Glycine-dependent activation of NMDA receptors. J Gen Physiol. 2015;145:513–527. doi: 10.1085/jgp.201411302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rauner C, Kohr G. Triheteromeric NR1/NR2A/NR2B receptors constitute the major N-methyl-D-aspartate receptor population in adult hippocampal synapses. J Biol Chem. 2011;286:7558–7566. doi: 10.1074/jbc.M110.182600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27•.Hansen KB, Ogden KK, Yuan H, Traynelis SF. Distinct functional and pharmacological properties of triheteromeric GluN1/GluN2A/Glun2B NMDA receptors. Neuron. 2014;81:1084–1096. doi: 10.1016/j.neuron.2014.01.035. This study reports a method to express molecularly pure population of recombinant triheteromeric receptors and demonstrates that these receptors have distinct pharmacology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun W, Hansen KB, Jahr CE. Allosteric interactions between NMDA receptor subunits shape the developmental shift in channel properties. Neuron. 2017;94:58–64 e53. doi: 10.1016/j.neuron.2017.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29•.Lu W, Du J, Goehring A, Gouaux E. Cryo-EM structures of the triheteromeric NMDA receptor and its allosteric modulation. Science. 2017;355 doi: 10.1126/science.aal3729. This report is first to describe the atomic arrangement of a triheteromeric NMDA receptor. It reveals that GluN2A and GluN2B subunits have unique contacts with GluN1 subunits. This structural asymmetry likely represents the basis for the distinct functional properties of this channel type. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mahfooz K, Marco S, Martinez-Turrillas R, Raja MK, Perez-Otano I, Wesseling JF. GluN3A promotes NMDA spiking by enhancing synaptic transmission in Huntington’s disease models. Neurobiol Dis. 2016;93:47–56. doi: 10.1016/j.nbd.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 31.Rozeboom AM, Queenan BN, Partridge JG, Farnham C, Wu JY, Vicini S, Pak DT. Evidence for glycinergic GluN1/GluN3 NMDA receptors in hippocampal metaplasticity. Neurobiol Learn Mem. 2015;125:265–273. doi: 10.1016/j.nlm.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 32.Chatterton JE, Awobuluyi M, Premkumar LS, Takahashi H, Talantova M, Shin Y, Cui J, Tu S, Sevarino KA, Nakanishi N, Tong G, et al. Excitatory glycine receptors containing the NR3 family of NMDA receptor subunits. Nature. 2002;415:793–798. doi: 10.1038/nature715. [DOI] [PubMed] [Google Scholar]
- 33.Perez-Otano I, Schulteis CT, Contractor A, Lipton SA, Trimmer JS, Sucher NJ, Heinemann SF. Assembly with the NR1 subunit is required for surface expression of NR3A-containing NMDA receptors. J Neurosci. 2001;21:1228–1237. doi: 10.1523/JNEUROSCI.21-04-01228.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smothers CT, Woodward JJ. Expression of glycine-activated diheteromeric NR1/NR3 receptors in human embryonic kidney 293 cells is NR1 splice variant-dependent. J Pharmacol Exp Ther. 2009;331:975–984. doi: 10.1124/jpet.109.158493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mesic I, Madry C, Geider K, Bernhard M, Betz H, Laube B. The N-terminal domain of the GluN3A subunit determines the efficacy of glycine-activated NMDA receptors. Neuropharmacology. 2016;105:133–141. doi: 10.1016/j.neuropharm.2016.01.014. [DOI] [PubMed] [Google Scholar]
- 36•.Cummings KA, Popescu GK. Protons potentiate GluN1/GluN3A currents by attenuating their desensitisation. Sci Rep. 2016;6:23344. doi: 10.1038/srep23344. This study revealed that, in contrast to their known inhibitory effect at all other NMDA receptor subtypes, protons potentiate GluN1/GluN3A channels. This observation provides a tool to identify pharmacologically responses from this receptor subtype in native preparations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Armstrong N, Jasti J, Beich-Frandsen M, Gouaux E. Measurement of conformational changes accompanying desensitization in an ionotropic glutamate receptor. Cell. 2006;127:85–97. doi: 10.1016/j.cell.2006.08.037. [DOI] [PubMed] [Google Scholar]
- 38.Borschel WF, Murthy SE, Kasperek EM, Popescu GK. NMDA receptor activation requires remodelling of intersubunit contacts within ligand-binding heterodimers. Nat Commun. 2011;2:498. doi: 10.1038/ncomms1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Borschel WF, Cummings KA, Tindell LK, Popescu GK. Kinetic contributions to gating by interactions unique to N-methyl-D-aspartate (NMDA) receptors. J Biol Chem. 2015;290:26846–26855. doi: 10.1074/jbc.M115.678656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ogden KK, Traynelis SF. New advances in NMDA receptor pharmacology. Trends Pharmacol Sci. 2011;32:726–733. doi: 10.1016/j.tips.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Amico-Ruvio SA, Murthy SE, Smith TP, Popescu GK. Zinc effects on NMDA receptor gating kinetics. Biophys J. 2011;100:1910–1918. doi: 10.1016/j.bpj.2011.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yuan H, Hansen KB, Zhang J, Pierson TM, Markello TC, Fajardo KV, Holloman CM, Golas G, Adams DR, Boerkoel CF, Gahl WA, et al. Functional analysis of a de novo GRIN2A missense mutation associated with early-onset epileptic encephalopathy. Nat Commun. 2014;5:3251. doi: 10.1038/ncomms4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Amico-Ruvio SA, Paganelli MA, Myers JM, Popescu GK. Ifenprodil effects on GluN2B-containing glutamate receptors. Mol Pharmacol. 2012;82:1074–1081. doi: 10.1124/mol.112.078998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sirrieh RE, MacLean DM, Jayaraman V. A conserved structural mechanism of NMDA receptor inhibition: a comparison of ifenprodil and zinc. J Gen Physiol. 2015;146:173–181. doi: 10.1085/jgp.201511422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sirrieh RE, MacLean DM, Jayaraman V. Subtype-dependent N-methyl-D-aspartate receptor amino-terminal domain conformations and modulation by spermine. J Biol Chem. 2015;290:12812–12820. doi: 10.1074/jbc.M115.649723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dai J, Zhou HX. Semiclosed conformations of the ligand-binding domains of NMDA receptors during stationary gating. Biophys J. 2016;111:1418–1428. doi: 10.1016/j.bpj.2016.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muir KW. Glutamate-based therapeutic approaches: clinical trials with NMDA antagonists. Curr Opin Pharmacol. 2006;6:53–60. doi: 10.1016/j.coph.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 48.Kavalali ET, Monteggia LM. Synaptic mechanisms underlying rapid antidepressant action of ketamine. Am J Psychiatry. 2012;169:1150–1156. doi: 10.1176/appi.ajp.2012.12040531. [DOI] [PubMed] [Google Scholar]
- 49.Paganelli MA, Popescu GK. Actions of bupivacaine, a widely used local anesthetic, on NMDA receptor responses. J Neurosci. 2015;35:831–842. doi: 10.1523/JNEUROSCI.3578-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kazi R, Dai J, Sweeney C, Zhou HX, Wollmuth LP. Mechanical coupling maintains the fidelity of NMDA receptor-mediated currents. Nat Neurosci. 2014;17:914–922. doi: 10.1038/nn.3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kazi R, Gan Q, Talukder I, Markowitz M, Salussolia CL, Wollmuth LP. Asynchronous movements prior to pore opening in NMDA receptors. J Neurosci. 2013;33:12052–12066. doi: 10.1523/JNEUROSCI.5780-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dai J, Wollmuth LP, Zhou HX. Mechanism-based mathematical model for gating of ionotropic glutamate receptors. J Phys Chem B. 2015;119:10934–10940. doi: 10.1021/acs.jpcb.5b00521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee CH, Lu W, Michel JC, Goehring A, Du J, Song X, Gouaux E. NMDA receptor structures reveal subunit arrangement and pore architecture. Nature. 2014;511:191–197. doi: 10.1038/nature13548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Karakas E, Furukawa H. Crystal structure of a heterotetrameric NMDA receptor ion channel. Science. 2014;344:992–997. doi: 10.1126/science.1251915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55•.Tajima N, Karakas E, Grant T, Simorowski N, Diaz-Avalos R, Grigorieff N, Furukawa H. Activation of NMDA receptors and the mechanism of inhibition by ifenprodil. Nature. 2016;534:63–68. doi: 10.1038/nature17679. This article describes the first tetrameric structure of a putative ‘active’ conformation. In addition, multiple closed, inactive conformations were observed providing structural support for the complex functional gating models developed from single-channel observations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zheng W, Wen H, Iacobucci GJ, Popescu GK. Probing the structural dynamics of the NMDA receptor activation by coarse-grained modeling. Biophys J. 2017;112:2589–2601. doi: 10.1016/j.bpj.2017.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Banke TG, Dravid SM, Traynelis SF. Protons trap NR1/NR2B NMDA receptors in a nonconducting state. J Neurosci. 2005;25:42–51. doi: 10.1523/JNEUROSCI.3154-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dore K, Aow J, Malinow R. Agonist binding to the NMDA receptor drives movement of its cytoplasmic domain without ion flow. Proc Natl Acad Sci U S A. 2015;112:14705–14710. doi: 10.1073/pnas.1520023112. [DOI] [PMC free article] [PubMed] [Google Scholar]