Abstract
Stage I colorectal carcinoma has excellent prognosis, with 5-year survival rate up to 95%. The occurrence of lymphovascular invasion, tumor budding, high number of PDC, or lymph node micrometastases is associated with tumor progression. The aim of this study was to evaluate the mutational status of 62 stage I colorectal carcinomas (CRC) (taken from 37 patients surviving more than five years since the initial diagnosis and from 25 patients who died of disease) and to correlate it with histopathological features and the clinical outcome. Mutations of KRAS, NRAS, BRAF, and PIK3CA genes were analyzed through Myriapod Colon Status Kit, using the high-throughput genotyping platform Sequenom MassARRAY System. Mutations in those genes were found in 31 cases (50%) and mainly in those with poor prognosis. The most frequent mutations occurred at codons 12 and 13 of the KRAS gene (40% of cases). We found concomitant PIK3CA mutations in 5 cases (8%). The presence of PIK3CA mutations was mainly observed in tumors with poor prognosis and with unfavorable histopathological prognostic features. High PDC grade (P = 0.0112), the presence of tumor budding (P = 0.0334), LVI (P < 0.0001), KRAS mutations (P = 0.0228), PIK3CA mutations (P = 0.0214), multiple genetic mutations in KRAS and PIK3CA genes (P = 0.039), and nodal micrometastases (P < 0.0001) were significant prognostic variables for CSS. The presence of LVI was the only independent and statistically significant prognostic variable for CSS in our cohort of pTNM stage I CRCs. The analysis of KRAS/PIK3CA mutational status may be used to identify patients with stage I CRC at high risk of bad outcome and who may need additional treatments, including biological therapies.
1. Introduction
It is well known that pTNM stage I (pT1N0M0 and pT2N0M0) colorectal carcinoma (CRCs) has an excellent prognosis, with 5-year survival rate up to 95% [1]. Therefore, patients with pTNM stage I CRC are not submitted to any adjuvant treatment after surgery [1]. However, in a small percentage of cases, they develop recurrences or metastases during the follow-up, and this event seems to be more frequent in cases showing lymphovascular invasion (LVI), tumor budding (TB), a high number of poorly differentiated clusters (PDC) in the primary tumor, or micrometastases in regional lymph nodes [2–5].
Up to now, only few studies have investigated the molecular profile of pTNM stage I CRC [6–8]. According to those studies, the percentage of mutated tumors ranges between 25% and 57%, and mutations mainly involve the KRAS gene in this subset of CRCs [6–8].
In this study, we aimed to investigate whether there is any correlation between the molecular profile and clinical outcome of pTNM stage I CRCs and whether mutations in KRAS, BRAF, NRAS, and PIK3CA genes are associated with histopathological features in this group of tumors.
2. Materials and Methods
2.1. Selection of Cases and Histological Examination
By a specialized Colorectal Cancer Registry instituted in Modena in 1984 [9], we identified all patients with stage I CRCs diagnosed between January 1984 and December 2004 (518 cases) and, among them, we selected those who died of disease (DOD) during the follow-up (37 cases). Paraffin blocks of the tumors and the relative haematoxylin and eosin- (H&E-) stained slides, stored in the archives of the Pathologic Anatomy of the University of Modena and Reggio Emilia, were available for only 25 of 32 patients (group A). This group of patients was matched with a group of 32 patients with stage I CRCs who were alive or who died of independent diseases (DOID) after a follow-up time longer than sixty months (group B). Cases in group B were consecutive stage I CRCs that fulfilled the inclusion criteria (at least 60-month follow-up) and with available paraffin blocks. All cases were anonymously collected.
Pathological features, including tumor size (maximum diameter in centimeters), tumor border configuration (expanding or infiltrating), WHO histological grade [10], pTNM stage [11], TB, LVI, grading based on the counting of PDC [12], and the presence of lymph node micrometastases (MM) [13], were available in all cases.
According to their location, the tumors were divided into 3 groups: (1) tumors located in the right colon, including the cecum and ascending and transverse colon; (2) tumors located in the left colon, including descending and sigmoid colon; and (3) tumors located in the rectum.
In accordance with the pathological staging at diagnosis (pTNM stage I) and the current oncological guidelines, none of the patients had been submitted to adjuvant chemoradiotherapy after surgical resection.
2.2. Molecular Analysis
DNA was extracted from representative 10 μm-thick sections cut from formalin-fixed and paraffin-embedded blocks of each tumor sample containing at least 50% tumor cells. Extraction was performed with QIAamp DNA Mini Kit (Qiagen, Hilden, Germany), and DNA was quantified with Xpose-NGS (Trinean NV, Gentbrugge, Belgium). Mutations were detected in genome-amplified DNA using the high-throughput genotyping platform Sequenom MassARRAY System (Sequenom, San Diego, CA, USA) and the Myriapod Colon Status Kit (Diatech Pharmacogenetics, Italy) following the manufacturer's protocol. This molecular array allows to identify the most important mutations of KRAS (codons 12, 13, 59, 61, 117, and 146), NRAS (codons 12, 13, 18, 59, 61, 117, and 146), BRAF (codons 594, 600, and 601), and PIK3CA genes (codons 38, 81, 88, 93, 108, 118, 345, 420, 539, 542, 545, 546, 549, 1021, 1025, 1043, 1047, and 1049). In brief, 25 ng/μ DNA of each specific primitive tumor was amplified through multiplex PCR, and then unincorporated nucleotides were inactivated by shrimp alkaline phosphatase (SAP). A single-base extension reaction was performed using extension primers that hybridize immediately adjacent to the mutations and a custom mixture of nucleotides. Salts were removed by the addition of a cation exchange resin. Multiplexed reactions were spotted into SpectroCHIP II arrays, and DNA fragments were resolved by MALDI-TOF on the Compact Mass Spectrometer (Sequenom, San Diego, CA) with a limit of detection of 5%. Data were evaluated using MassARRAY Typer Analyser software 4.0, which allows to identify mutated alleles by comparing the ratio of the wild-type peak of all suspected mutants and to generate a specific report.
2.3. Statistical Analysis
Fisher exact and chi-squared tests were used to assess the statistical association between mutational status of the tumor and the clinicopathological parameters. Median age of the patients (68 years) and median size of the tumors (3 centimeters) were used as cutoff values for statistical analyses.
Cancer specific survival (CSS) was assessed by the Kaplan-Meier method, with the date of primary surgery as the entry date. CSS was characterized as the length of survival to death from CRC or to the last follow-up date.
The Mantel-Cox log-rank test was applied to assess the strength of association between CSS and each of the parameters (age and gender of the patient, size of the tumor, WHO histological grade, PDC grade, pT stage, tumor border configuration, TB, LVI, and MM) as a single variable.
Subsequently, a stepwise multivariate analysis (Cox regression model) was utilized to determine the independent effect of each variable on survival. Multivariate analysis was carried out by using stepwise method and including only clinicopathological variables with significant prognostic value at univariate analyses.
A probability (P) value less than 0.05 was considered statistically significant. Statistical analysis was done using MedCalc 12.1.4.0 statistical software (MedCalc Software, Mariakerke, Belgium).
2.4. Ethical Issues
All procedures were performed in accordance with the Helsinki Declaration. Ethical issues were discussed with the Local Ethics Committee. No formal approval was necessary to perform the histological review and molecular analyses.
3. Results
Clinical and pathological features of 62 tumors included in the study are summarized in Table 1. 31 tumors (50%) had mutations in KRAS, BRAF, and PI3KCA genes; details are listed in Table 2.
Table 1.
Clinicopathological characteristics of the 62 CRCs at pTNM stage I.
| Clinicopathological variables | Total patients (n = 62) | Alive/DOID patients (n = 37) | DOD patients (n = 25) | |
|---|---|---|---|---|
| M/F | 35/27 | 22/15 | 13/12 | |
| Mean age | 69.5 years | 68.7 years | 70.84 years | |
| Age range | 48–90 years | 48–85 years | 55–90 years | |
| Right colon | 11 | 8 | 3 | |
| Left colon | 15 | 11 | 4 | |
| Rectum | 26 | 18 | 8 | |
| Mean size of the tumor (cms) | 3.28 | 3.23 | 3.45 | |
| Size range of the tumor (cms) | 1–6.5 | 1–5.5 | 1.5–6.5 | |
| pT1/pT2 | 4/58 | 1/36 | 3/22 | |
| Micrometastases (present/absent) | 17/45 | 0/37 | 17/8 | |
| Tumor border configuration (expansive/infiltrative) | 18/45 | 14/23 | 4/21 | |
| WHO grading (G1/G2/G3) | 9/46/7 | 3/29/5 | 6/17/2 | |
| PDC grading (G1/G2/G3) | 26/23/13 | 17/14/6 | 9/9/7 | |
| Tumor budding (present/absent) | 32/30 | 15/22 | 17/8 | |
| LVI (present/absent) | 22/40 | 9/28 | 13/12 |
DOID: died of independent diseases; DOD: died of disease; M: male; F: female; LVI: lymphovascular invasion.
Table 2.
Distribution of KRAS, PIK3CA, and BRAF mutations according to the status of patients.
| Mutations | Patients alive/DOID | Patients DOD |
|---|---|---|
| KRAS G12C | 1 | 0 |
| KRAS G12D | 2 | 4 |
| KRAS G12D + PIK3CA R108H | 0 | 1 |
| KRAS G12D + G13D + G117N | 0 | 1 |
| KRAS G12F | 0 | 1 |
| KRAS G12R + PIKRCA E545K | 0 | 1 |
| KRAS G12V | 2 | 3 |
| KRAS G13D | 5 | 2 |
| KRAS G13D + PIK3CA R88Q | 1 | 0 |
| KRAS G13D + PIK3CA E545K | 0 | 1 |
| KRAS Q61H | 1 | 0 |
| KRAS A146T | 0 | 1 |
| KRAS A 146T + PIK3CA R108H | 0 | 1 |
| BRAF V600E | 1 | 2 |
| Absent (WT) | 24 | 7 |
DOID: died of independent disease; DOD: died of disease.
3.1. KRAS Mutations
28/62 cases (45%) had mutations in the KRAS gene. Among those, 22 (78%) had a single KRAS mutation (G13D in 7 cases, G12D in 6 cases, G12V in 5 cases, G12C in 1 case, G12F in 1 case, A146T in 1, and Q61H in 1), while 6 showed multiple mutations. In detail, 5 cases had also PIK3CA mutations (R108H in 2 cases, E545K in 2 cases, and R88K in 1), and 1 had two additional KRAS mutations (G12D + G13D + G117N). KRAS mutations were significantly more frequent in the group of DOD patients (P = 0.019) (Table 3) Figure 1.
Table 3.
Statistical correlations between mutational status and the various clinicopathological parameters of the tumors.
| KRAS mutation | P | PIK3CA mutation | P | n mutations/case | P | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Absent | Present | Absent | Present | 0 | 1 | 2 | ||||
| Age | ||||||||||
| ≤68 years | 16 | 15 | 28 | 3 | 16 | 11 | 4 | |||
| >68 years | 18 | 13 | 0.798 | 29 | 2 | 1 | 15 | 14 | 2 | 0.588 |
| Gender | ||||||||||
| Male | 24 | 11 | 35 | 0 | 22 | 20 | 0 | |||
| Female | 10 | 17 | 0.02 | 22 | 5 | 0.012 | 9 | 12 | 6 | 0.004 |
| Size | ||||||||||
| ≤3 cms | 18 | 16 | 31 | 3 | 17 | 14 | 3 | |||
| >3 cms | 16 | 12 | 0.801 | 26 | 2 | 0.652 | 14 | 11 | 3 | 0.965 |
| Site | ||||||||||
| Right colon | 9 | 2 | 11 | 0 | 6 | 5 | 0 | |||
| Left colon | 9 | 6 | 15 | 0 | 9 | 5 | 1 | |||
| Rectum | 16 | 20 | 0.083 | 31 | 5 | 0.14 | 16 | 15 | 5 | 0.608 |
| pT | ||||||||||
| T1 | 1 | 3 | 4 | 0 | 1 | 3 | 0 | |||
| T2 | 33 | 25 | 0.319 | 53 | 5 | 1 | 30 | 22 | 6 | 0.329 |
| Nodal micrometastases | ||||||||||
| Absent | 28 | 17 | 44 | 1 | 27 | 17 | 1 | |||
| Present | 6 | 11 | 0.086 | 13 | 4 | 0.017 | 4 | 8 | 5 | 0.001 |
| Tumor border configuration | ||||||||||
| Expansive | 12 | 6 | 18 | 0 | 11 | 7 | 0 | |||
| Infiltrative | 22 | 22 | 0.271 | 39 | 5 | 0.309 | 20 | 18 | 6 | 0.212 |
| WHO grade | ||||||||||
| G1 | 4 | 5 | 7 | 2 | 4 | 3 | 2 | |||
| G2 | 24 | 22 | 43 | 3 | 24 | 18 | 4 | |||
| G3 | 6 | 1 | 0.2 | 7 | 0 | 0.202 | 3 | 4 | 0 | 0.562 |
| PDC grade | ||||||||||
| G1 | 23 | 13 | 35 | 1 | 23 | 11 | 2 | |||
| G2 | 7 | 7 | 14 | 0 | 6 | 8 | 0 | |||
| G3 | 4 | 8 | 0.168 | 8 | 4 | 0.001 | 2 | 6 | 4 | 0.004 |
| Tumor budding | ||||||||||
| Absent | 17 | 13 | 27 | 3 | 17 | 10 | 3 | |||
| Present | 17 | 15 | 0.803 | 30 | 2 | 0.666 | 14 | 15 | 3 | 0.541 |
| LVI | ||||||||||
| Absent | 21 | 12 | 32 | 1 | 20 | 12 | 1 | |||
| Present | 13 | 16 | 0.201 | 25 | 4 | 0.176 | 11 | 13 | 5 | 0.078 |
| Status | ||||||||||
| Alive, DOID | 25 | 12 | 36 | 1 | 24 | 12 | 1 | |||
| DOD | 9 | 16 | 0.019 | 21 | 4 | 0.147 | 7 | 13 | 5 | 0.006 |
DOID: died of independent diseases; DOD: died of disease; LVI: lymphovascular invasion.
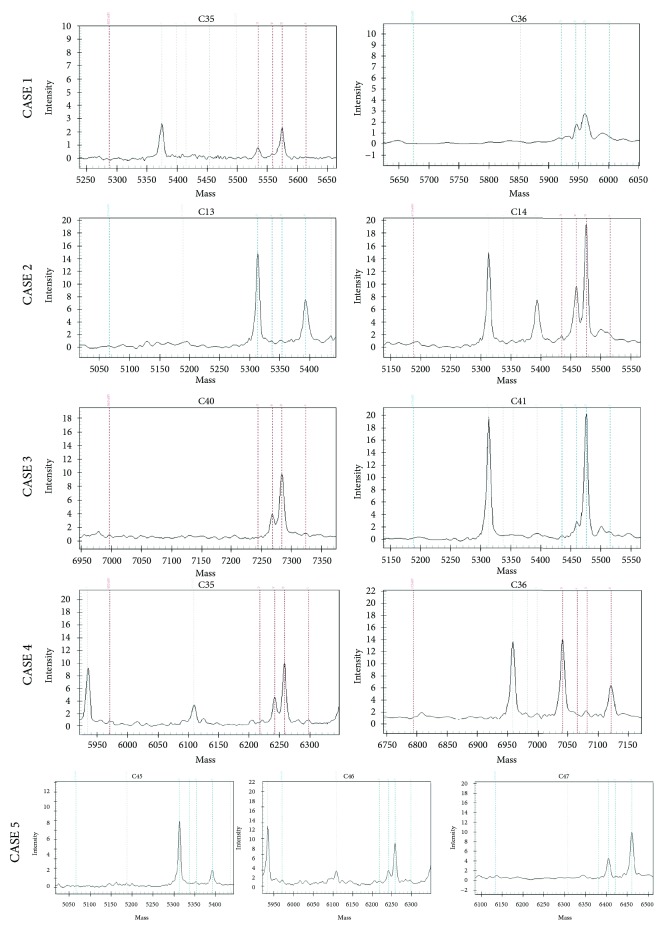
Figure 1.
KRAS and PIK3CA MassArray (Sequenom) graphic assay. pTNM stage I CRCs with unfavourable clinical outcome showed frequently multiple KRAS mutations involving unusual codons and KRAS mutations associated to PIRKCA mutations.
3.2. PIK3CA Mutations
PIK3CA mutations were found in a total of five cases (Table 2), all of which showed KRAS mutations as well. Mutations were E545K in 2 CRCs, R108K in 2, and R88Q in 1 (Table 2). The presence of PIK3CA mutations was significantly more frequent in female patients (P = 0.01) and in cases with nodal micrometastases (P = 0.017) and PDC G3 (P = 0.001) (Table 3).
All but one patient with PIK3CA-mutated tumor died of CRC; the only one who was alive 168 months since the initial diagnosis had PIK3CA R88Q mutation.
3.3. NRAS Mutations
NRAS mutations were not identified in any of the cases.
3.4. BRAF Mutations
BRAF V600E mutation was found in 3 cases: 2 were from DOD patients, while one was from a DOID patient, who was alive 167 months since the initial diagnosis. Due to the limited number of cases with BRAF mutations, statistical association with other parameters was not investigated. However, all 3 mutated cases were localized in the right colon (Table 3).
3.5. CRC with Multiple Mutations
CRCs with multiple mutations (KRAS + PIK3CA or multiple KRAS mutations) occurred mainly in female patients (P = 0.004) and in DOD patients (P = 0.006) and had significant association with lymph node micrometastases (P = 0.011) and high PDC grade (P = 0.004). Multiple mutations were more frequent in CRCs showing LVI, though statistical significance was not reached (Table 3).
3.6. Correlations between CSS and Clinicopathological Parameters or Mutational Status
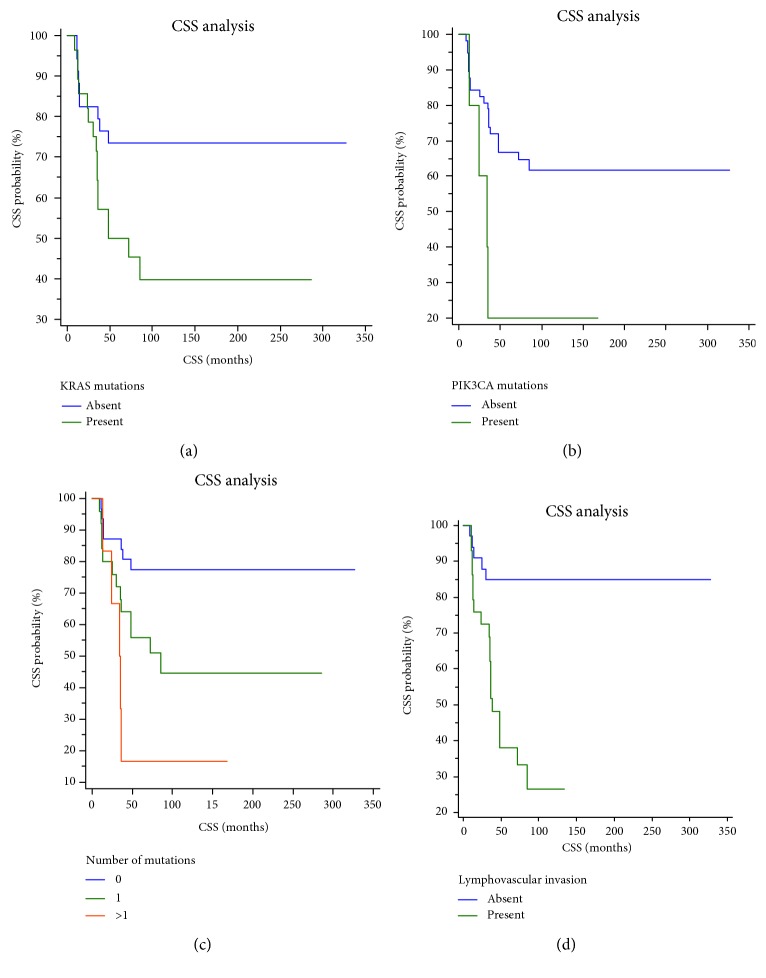
Univariate analyses showed that high PDC grade (P = 0.0112), the presence of tumor budding (P = 0.0334), LVI (P < 0.0001), KRAS mutations (P = 0.0228), PIK3CA mutations (P = 0.0214), multiple genetic mutations in KRAS and PIK3CA genes (P = 0.039) (Figure 2), and nodal micrometastases (P < 0.0001) (Figure 2(b)) were significant prognostic variables for CSS (Table 4). The presence of LVI was the only independent and statistically significant prognostic variable for CSS in our cohort of pTNM stage I CRCs (Table 4) (Figure 2(d)).
Figure 2.
(a–c) Kaplan-Meier curves showing CSS of patients with pTNM stage I CRC according to mutational status of KRAS, BRAF, and PIK3CA genes. Patients having CRC with (a) KRAS, (b) PIK3CA, or (c) 2 or 3 mutations had significantly lower CSS, compared to patients with CRC with absent or 1 mutation. (d) Kaplan-Meier curves showing CSS of patients with pTNM stage I CRC according to the presence of LVI. Patients having CRC with LVI had significantly lower CSS compared to patients with CRC not having LVI.
Table 4.
Univariate and multivariate analyses for CSS in fifty patients with stage I CRC.
| Parameter | Univariate analyses | Multivariate analyses | ||
|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | |
| Age | ||||
| ≤68 years | 1 | |||
| >68 years | 1.2 (0.5–2.6) | 0.642 | ||
| Gender | ||||
| Male | 1 | |||
| Female | 1.2 (0.5–2.8) | 0.537 | ||
| Size | ||||
| ≤3 cms | 1 | |||
| >3 cms | 0.8 (0.4–1.9) | 0.783 | ||
| Site | ||||
| Right colon | 1 | |||
| Left colon | 0.8 (0.2–2.9) | |||
| Rectum | 1.9 (0.6–5.5) | 0.241 | ||
| pT | ||||
| T1 | 1 | |||
| T2 | 0.4 (0.07–2.3) | 0.139 | ||
| Nodal micrometastases | ||||
| Absent | 1 | |||
| Present | 10.4 (3.6–29.8) | <0.0001 | ||
| Tumor border configuration | ||||
| Expansive | 1 | |||
| Infiltrative | 2.6 (1.1–6.1) | 0.0542 | ||
| WHO grade | ||||
| G1 | 1 | |||
| G2 | 0.5 (0.1–1.6) | |||
| G3 | 0.4 (0.08–2.1) | 0.322 | ||
| PDC grade | ||||
| G1 | 1 | |||
| G2 | 2.6 (1–7.2) | |||
| G3 | 3.6 (1.1–10.9) | 0.0112 | ||
| Tumor budding | ||||
| Absent | 1 | |||
| Present | 2.3 (1–5.2) | 0.0334 | ||
| LVI | ||||
| Absent | 1 | 1 | ||
| Present | 5.9 (2.6–13.2) | <0.0001 | 6.2 (2.3–16.6) | 0.0003 |
| KRAS mutations | ||||
| Absent | 1 | |||
| Present | 2.4 (1.1–5.4) | 0.0228 | ||
| PIK3CA mutations | ||||
| Absent | 1 | |||
| Present | 3.2 (0.5–17.7) | 0.0214 | ||
| Number of mutations/case | ||||
| 0 | 1 | |||
| 1 | 2.7 (1.1–6.2) | |||
| 2/3 | 5.6 (1.1–27) | 0.039 | ||
LVI: lymphovascular invasion.
4. Discussion
In this study, we investigated the molecular profile of a cohort of stage I CRCs and assessed its association with patient's prognosis and various clinicopathological variables. While the molecular profile of advanced CRC has been largely investigated to predict response to biological drugs [14–16], the mutational status of early CRC (stage I) has been rarely analyzed [6–8, 17, 18] and only in studies including stage I CRCs in heterogeneous cohorts of tumors at different pTNM stages [6–8, 17, 18]. Therefore, to the best of our knowledge, this represents the first study focused on the molecular profile of stage I CRC.
Our results can be summarized as follows: (1) mutations in the genes involved in RAS/MAPK and PI3K-PTEN-AKT signalling pathways were found in 50% of stage I CRC and mainly in cases with poor prognosis rather than in cases with favourable outcome (72% versus 40%), (2) the most frequent mutations observed in stage I CRC occurred at codons 12 and 13 of the KRAS gene (G13D, G12D, and G12V mutations), (3) PI3KCA mutations were strongly associated with KRAS mutations, and (4) synchronous KRAS and PIK3CA mutations or multiple KRAS mutations at codons 12 and 13 were mainly found in tumors with poor prognosis and with unfavorable histopathological prognostic features.
Hence, our findings confirm previous evidence that mutations at codons 12 and 13 (G13D, G12D, and G12V) of the KRAS gene are the most frequent mutations in stage I CRC [6–8, 17, 18], while NRAS, BRAF, and PIK3CA mutations are rare in this subgroup of CRC [6–8, 17].
The prognostic role of KRAS mutations in stage I CRC was previously investigated in two studies that showed that mutations in this gene are associated with shorter overall survival and recurrence-free survival [6, 18], while the specific prognostic role of PIK3CA mutations in stage I CRC had not been analyzed thus far. Our findings showed a significant association between KRAS or PIK3CA mutations and shorter CSS. Interestingly, the presence of multiple KRAS mutations and that of KRAS/PIK3CA bimutations were significant negative prognostic factors in our patients with stage I CRC. Li et al. [17] previously demonstrated that KRAS/PIK3CA bimutations are significantly more frequent in patients with stage IV CRC, compared to lower-stage carcinomas. KRAS/PIK3CA bimutations may induce metastasization through a synergic effect in the activation of PI3K-AKT pathway [17, 19, 20]. Since KRAS and PIK3CA mutations seem to be early events in colorectal carcinogenesis [21], we may hypothesize that early-stage CRCs with bimutations are more likely to behave as stage IV tumors and to develop distant metastases and worse outcome during the follow-up. Besides, in a previous study, we already showed the association between KRAS/PIK3CA bimutations and nodal metastases or lymphovascular invasion [22]. Further supporting the tendency of bimutated or trimutated stage I CRC to metastasization is that, in our cohort, this subgroup of tumors had significantly higher frequency of PDC G3 and nodal micrometastases. Indeed, several studies showed that PDC G3 CRCs have high metastatic potential at any pTNM stage [23]. In addition, the presence of nodal micrometastases in bi- or trimutated CRCs indicates disease diffusion in spite of pTNM stage I. Different from our previous study [24] on a cohort of CRCs at pTNM stages I, II, III, and IV, the presence of a single KRAS mutation was not associated with PDC grade or tumor budding in this group of pTNM stage I CRCs. Studies on a larger cohort are needed to clarify whether the association between KRAS mutations and PDC grade is stage-dependent or not.
Although further studies are necessary to corroborate these findings, patients with stage I CRC showing this biomolecular profile—duple or triple mutations—might be candidate to adjuvant treatments in the aim to prevent unfavorable outcome. The association between KRAS/PIK3CA multiple mutations and PDC G3 may be used as a screening tool to select cases with high probability to have KRAS/PIK3CA bimutations or KRAS multiple mutations and which may be referred to molecular analyses. Indeed, in order to avoid costs related to extensive molecular analyses on stage I CRC, evaluation of KRAS and PIK3CA mutational status may be limited only to cases showing PDC G3 at histological examination. Besides, the identification of PIK3CA mutations might also have therapeutic implications. Indeed, there is some evidence that CRC with PIK3CA mutations could be treated with PI3K/mTOR inhibitors [25].
Interestingly, the occurrence of LVI emerged as the only significant and independent prognostic variable in our patients with stage I CRC, as we already found in a previous study [26]. Interestingly, two of the pT1 CRCs included in our cohort who died of disease during the follow-up exhibited multiple synchronous KRAS mutations. Therefore, the assessment of KRAS/PIK3CA mutational status may be particularly important to predict prognosis of patients with pT1 CRC submitted to endoscopic conservative treatments in the aim to identify patients at high risk of metastatic diffusion and worse outcome, who could be submitted to surgery and adjuvant treatments.
In conclusion, this is the first study specifically analyzing the molecular profile of stage I CRC. The presence of KRAS mutations, that of simultaneous mutations in PIK3CA gene, or that of multiple KRAS mutations was significantly associated with shorter CSS. PIK3CA or multiple KRAS mutations were associated with nodal micrometastases and PDC G3 as well. If our findings are confirmed in further studies, the analysis of KRAS/PIK3CA mutational status may be used to identify patients with stage I or pT1 CRCs at high risk of worse outcome, who may need additional treatments.
Acknowledgments
The study was supported by funds from “Programma di Ricerca Regione Univerisità 2010-2012”, funded by Regione Emilia-Romagna, entitled “Use of biomarkers and gene expression profiles to identify cancer patients with different prognosis and sensitivity to molecular targeted agents.”
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.
Conflicts of Interest
The authors have no conflict of interest to declare.
Authors' Contributions
Luca Reggiani Bonetti and Valeria Barresi contributed equally to this paper.
References
- 1.Ferlay J., Shin H. R., Bray F., Forman D., Mathers C., Parkin D. M. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. International Journal of Cancer. 2010;127(12):2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Barresi V., Reggiani-Bonetti L., di Gregorio C., Ponz de Leon M., Barresi G. Lymphatic vessel density and its prognostic value in stage I colorectal carcinoma. Journal of Clinical Pathology. 2011;64(1):6–12. doi: 10.1136/jcp.2010.083550. [DOI] [PubMed] [Google Scholar]
- 3.Ichimasa K., Kudo S. E., Miyachi H., et al. Patient gender as a factor associated with lymph node metastasis in T1 colorectal cancer: a systematic review and meta-analysis. Molecular and Clinical Oncology. 2017;6(4):517–524. doi: 10.3892/mco.2017.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bianco F., De Franciscis S., Belli A., et al. T1 colon cancer in the era of screening: risk factors and treatment. Techniques in Coloproctology. 2017;21(2):139–147. doi: 10.1007/s10151-017-1586-z. [DOI] [PubMed] [Google Scholar]
- 5.Yazilitas D., Özdemir N., Yazıcı O., Hocazade C., Demirci N. S., Zengin N. The clinical and pathological features affecting the time of relapse in patients with early stage colorectal cancer. Journal of Cancer Research and Therapeutics. 2016;12(4):1257–1260. doi: 10.4103/0973-1482.199527. [DOI] [PubMed] [Google Scholar]
- 6.Dobre M., Dinu D. E., Panaitescu E., et al. KRAS gene mutations - prognostic factor in colorectal cancer? Romanian Journal of Morphology and Embryology. 2015;56(Supplement 2):671–678. [PubMed] [Google Scholar]
- 7.Zhang J., Zheng J., Yang Y., et al. Molecular spectrum of KRAS, NRAS, BRAF and PIK3CA mutations in Chinese colorectal cancer patients: analysis of 1,110 cases. Scientific Reports. 2015;5(1, article 18678) doi: 10.1038/srep18678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Asaka S., Arai Y., Nishimura Y., et al. Microsatellite instability-low colorectal cancer acquires a KRAS mutation during the progression from Dukes’ A to Dukes’ B. Carcinogenesis. 2009;30(3):494–499. doi: 10.1093/carcin/bgp017. [DOI] [PubMed] [Google Scholar]
- 9.Losi L., Ponti G., Gregorio C. D., et al. Prognostic significance of histological features and biological parameters in stage I (pT1 and pT2) colorectal adenocarcinoma. Pathology, Research and Practice. 2006;202(9):663–670. doi: 10.1016/j.prp.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 10.Hamilton S. R., Bosman F. T., Boffetta P., et al. Carcinoma of the colon and rectum. In: Bosman T., Carneiro F., Hruban R. H., Theise N. D., editors. WHO Classification of Tumors of the Digestive System. Lyon: IARC Press; 2010. pp. 134–146. [Google Scholar]
- 11.Edge S. B., Byrd D. R., Compton C. C., Fritz A. G., Greene F. L., Trotti A. E. AJCC Cancer Staging Manual. 7th. New York, NY, USA: Springer; 2010. [Google Scholar]
- 12.Ueno H., Hase K., Hashiguchi Y., et al. Novel risk factors for lymph node metastasis in early invasive colorectal cancer: a multi-institution pathology review. Journal of Gastroenterology. 2014;49(9):1314–1323. doi: 10.1007/s00535-013-0881-3. [DOI] [PubMed] [Google Scholar]
- 13.Reggiani Bonetti L., di Gregorio C., de Gaetani C., et al. Lymph node micrometastasis and survival of patients with stage I (Dukes’ A) colorectal carcinoma. Scandinavian Journal of Gastroenterology. 2011;46(7-8):881–886. doi: 10.3109/00365521.2011.571708. [DOI] [PubMed] [Google Scholar]
- 14.Misale S., Yaeger R., Hobor S., et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486(7404):532–536. doi: 10.1038/nature11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Roock W., Claes B., Bernasconi D., et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. The Lancet Oncology. 2010;11(8):753–762. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- 16.Siena S., Sartore-Bianchi A., Di Nicolantonio F., Balfour J., Bardelli A. Biomarkers predicting clinical outcome of epidermal growth factor receptor-targeted therapy in metastatic colorectal cancer. Journal of the National Cancer Institute. 2009;101(19):1308–1324. doi: 10.1093/jnci/djp280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H. T., Lu Y. Y., An Y. X., Wang X., Zhao Q. C. KRAS, BRAF and PIK3CA mutations in human colorectal cancer: relationship with metastatic colorectal cancer. Oncology Reports. 2011;25(6):1691–1697. doi: 10.3892/or.2011.1217. [DOI] [PubMed] [Google Scholar]
- 18.Sideris M., Moorhead J., Diaz-Cano S., Haji A., Papagrigoriadis S. KRAS mutant status may be associated with distant recurrence in early-stage rectal cancer. Anticancer Research. 2017;37(3):1349–1358. doi: 10.21873/anticanres.11454. [DOI] [PubMed] [Google Scholar]
- 19.Rahbari N. N., Bork U., Motschall E., et al. Molecular detection of tumor cells in regional lymph nodes is associated with disease recurrence and poor survival in node-negative colorectal cancer: a systematic review and meta-analysis. Journal of Clinical Oncology. 2012;30(1):60–70. doi: 10.1200/JCO.2011.36.9504. [DOI] [PubMed] [Google Scholar]
- 20.Benvenuti S., Frattini M., Arena S., et al. PIK3CA cancer mutations display gender and tissue specificity patterns. Human Mutation. 2008;29(2):284–288. doi: 10.1002/humu.20648. [DOI] [PubMed] [Google Scholar]
- 21.Velho S., Moutinho C., Cirnes L., et al. BRAF, KRAS and PIK3CA mutations in colorectal serrated polyps and cancer: primary or secondary genetic events in colorectal carcinogenesis? BMC Cancer. 2008;8(1):p. 255. doi: 10.1186/1471-2407-8-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reggiani Bonetti L., Barresi V., Bettelli S., Caprera C., Manfredini S., Maiorana A. Analysis of KRAS, NRAS, PIK3CA, and BRAF mutational profile in poorly differentiated clusters of KRAS-mutated colon cancer. Human Pathology. 2017;62:91–98. doi: 10.1016/j.humpath.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 23.Barresi V., Reggiani Bonetti L., Ieni A., Caruso R. A., Tuccari G. Poorly differentiated clusters: clinical impact in colorectal cancer. Clinical Colorectal Cancer. 2017;16(1):9–15. doi: 10.1016/j.clcc.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 24.Barresi V., Reggiani Bonetti L., Bettelli S. KRAS, NRAS, BRAF mutations and high counts of poorly differentiated clusters of neoplastic cells in colorectal cancer: observational analysis of 175 cases. Pathology. 2015;47(6):551–556. doi: 10.1097/PAT.0000000000000300. [DOI] [PubMed] [Google Scholar]
- 25.Foley T. M., Payne S. N., Pasch C. A., et al. Dual PI3K/mTOR inhibition in colorectal cancers with APC and PIK3CA mutations. Molecular Cancer Research. 2017;15(3):317–327. doi: 10.1158/1541-7786.MCR-16-0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barresi V., Reggiani Bonetti L., Branca G., di Gregorio C., Ponz de Leon M., Tuccari G. Colorectal carcinoma grading by quantifying poorly differentiated cell clusters is more reproducible and provides more robust prognostic information than conventional grading. Virchows Archiv. 2012;461(6):621–628. doi: 10.1007/s00428-012-1326-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.