Abstract
Although a number of studies have revealed the underlying mechanisms which regulate the development of colorectal cancer (CRC), we have not completely overcome this disease yet. Accumulating evidence has shown that the posttranscriptional regulation by the noncoding RNAs such as microRNAs plays an important role in the development or progression of CRC. Among a number of microRNAs, the let-7 microRNA family that was first discovered in C. elegans and conserved from worms to humans has been linked with the development of many types of cancers including CRC. The expression level of let-7 microRNAs is temporally low during the normal developmental processes, while elevated in the differentiated tissues. The let-7 microRNAs regulate the cell proliferation, cell cycle, apoptosis, metabolism, and stemness. In CRC, expressions of let-7 microRNAs have been reported to be reduced, and so let-7 microRNAs are considered to be a tumor suppressor. In this review, we discuss the mechanisms regulating the let-7 microRNA expression and the downstream targets of let-7 in the context of intestinal tumorigenesis. The application of let-7 mimics is also highlighted as a novel therapeutic agent.
1. Introduction
Colorectal cancer (CRC) is one of the most common causes of cancer deaths worldwide [1–3], and the number of CRC patients is increasing more and more [4]. Many studies have revealed the molecular mechanisms of the colonic tumorigenesis and have proposed various models such as the adenoma-carcinoma sequence (i.e., multistep carcinogenesis) and colitis-associated carcinogenesis observed in inflammatory bowel diseases (IBD). However, we have not yet completely understood the molecular basis of CRC tumorigenesis. One of the reasons which makes our understanding difficult is that not only genetic changes, such as mutation/deletion of oncogenes or tumor suppressor genes, but also the epigenetic modification or posttranscriptional regulation by RNA binding proteins and microRNAs is involved in the colonic tumorigenesis.
The let-7 microRNAs are noncoding RNAs which consist of ~22 nucleotides [5]. They are first discovered in C. elegans as a critical regulator of developmental process [6, 7]. Let-7 microRNAs have been experimentally confirmed to be conserved in a wide range of species including humans [8]. As the let-7 family, 9 members are identified so far: let-7a, let-7b, let-7c, let-7d, let-7e, let-7f, let-7g, let-7i, and miR-98. The mature let-7 negatively regulates the expression of target mRNAs at a posttranslational level via imperfect base pairing to the 3'-UTR of their target mRNAs [9]. During normal developmental process, the expression levels of let-7 microRNAs in stem and progenitor cells are maintained low. As progenitor cells differentiate, let-7 expression is increasing [10].
Let-7 microRNAs are downregulated in various types of cancers such as hepatocellular carcinoma (HCC), gastric adenocarcinoma, pancreatic cancer, ovarian cancer, prostate cancer, Burkitt lymphoma, renal cell carcinoma, breast cancer, and melanoma [11]. In the context of CRC, let-7 microRNAs are also downregulated, which affects the posttranscriptional regulation of target mRNAs resulting in the colonic tumorigenesis and progression.
In this review, we discuss the molecular mechanisms by which let-7 microRNAs regulate the colonic tumorigenesis, tumor progression, and chemotherapy resistance by a posttranscriptional regulation and also highlight the possibility of the application of let-7 targeting therapy against CRC.
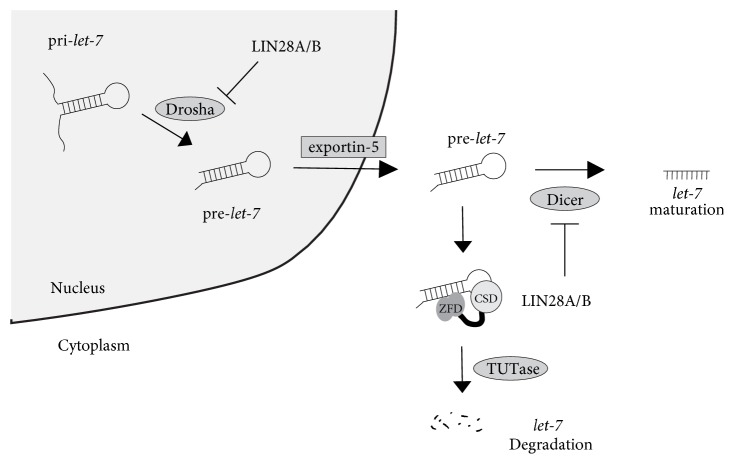
2. Biogenesis of Let-7 MicroRNAs (Figure 1)
Figure 1.
A schematic diagram of the let-7 biogenesis. Pri-let-7 microRNA precursors are processed in the nucleus by Drosha to pre-let-7. Pre-let-7 microRNA precursors are exported to the cytoplasm by exportin-5, where they are subsequently processed by Dicer resulting in the mature let-7. RNA binding protein LIN28A or LIN28B binds to either pri-let-7 or pre-let-7 using RNA binding domains, cold shock domain (CSD), and two zinc finger domains (ZFDs) and blocks the processing. Upon binding to pre-let-7, LIN28A or LIN28B recruits TUTase, which causes oligouridylation resulting in the degradation of pre-let-7.
Mature let-7 microRNAs are synthesized through the multiple processing by microprocessors [12]. Let-7 microRNAs are initially transcribed in long transcripts called primary let-7 (pri-let-7), which are processed in the nucleus by Drosha and Pasha to pre-let-7 with hairpin structures of about 70 nucleotides. Next, pre-let-7 are exported to the cytoplasm by exportin-5, where they are subsequently processed by the enzyme Dicer resulting in the mature let-7 [12]. Let-7 is known to be negatively regulated by the RNA binding proteins, LIN28A or LIN28B. LIN28A or LIN28B has two RNA binding domains, cold shock domain (CSD) and two Cys-Cys-His-Cys- (CCHC-) type zinc finger domains (ZFDs) [13–16], which bind to the conserved GGAG motif in the terminal loop of pri-let-7 or pre-let-7, respectively, resulting in the inhibition of the maturation of let-7 [17, 18]. The binding of LIN28A or LIN28B to either pri-let-7 or pre-let-7 inhibits the processing of let-7 precursors by Drosha and Dicer [19]. Upon binding to pre-let-7, LIN28A or LIN28B recruits TUT4/TUT7, which causes oligouridylation at the 3′ terminal of pre-let-7 [20–22]. Under normal conditions, Dicer recognizes the two nucleotides at the 3′ terminal via its PAZ domain; however, oligouridylation elongates the 3′ terminal resulting in the resistance to Dicer cleavage. Oligouridylated pre-let-7 can also be degenerated by the 3′-5′ exonuclease Dis312 [23, 24]. Thus, LIN28A or LIN28B not only inhibits the biogenesis of let-7 family miRNAs, but also induces their degradation. LIN28A and LIN28B are the negative regulators of let-7 microRNAs, and so the expression level of LIN28A is high in the developmental process, while that of LIN28B is low in the adult tissues [10]. LIN28B has two isoforms: LIN28B-long and LIN28B-short isoforms. Both isoforms preserve the binding ability to pre-let-7. However, LIN28B-short isoform lacks the inhibitory ability against the processing by microprocessors. Namely, LIN28B-short isoform functions as a competitor against the LIN28B-long isoform in the context of let-7 maturation [25].
3. Let-7 and Colon Cancer
Regarding the LIN28AB/let-7 axis, the downregulation of let-7 or upregulation of either LIN28A or LIN28B has been reported to be related to the worse prognosis in CRC patients. Stage I/II CRC patients with high expression of LIN28B exhibit poorer prognosis compared to those with no LIN28B expression [26]. King et al. reported that the expression level of LIN28B was inversely correlated to that of mature let-7a in human CRC [27]. Tu et al. examined the expression of LIN28A or LIN28B in the cohort of almost 600 CRC patients and found that LIN28A or LIN28B was highly expressed in 38 % of CRC patients [28]. Importantly, the overall survival was lower in the patients with high expression of LIN28A or LIN28B than in those with low expression of LIN28A or LIN28B. Madison et al. demonstrated that intestinal tumors were spontaneously developed in the transgenic mice with intestinal-specific LIN28B expression (Villin-Lin28b), which suggests that LIN28B functions as an oncogene [29]. Let-7b and let-7c can escape the downregulation by LIN28B in mice. Therefore, they crossed these mice with let-7b and let-7c knockout mice (let-7IEC-KO) and found that the additional downregulation of let-7b and let-7c resulted in the increase of tumor incidence [30]. These results suggest that let-7b or let-7c plays an important role in the intestinal tumorigenesis in mice.
4. Let-7 Target mRNAs in Intestine
As let-7 target mRNAs in the intestinal epithelium, Madison et al. revealed that HMGA1, HMGA2, IGF2BP2, HIF3A, ARID3A, and E2F5 were the most highly induced targets of let-7 microRNAs in the context of intestinal homeostasis [31]. Among these target molecules, the protein expression levels of HMGA1, HMGA2, ARID3A, and HIF3A were dramatically increased in the intestinal tumors. They also demonstrated that HMGA2 plays an important role in the downstream of let-7 in the context of colonic tumorigenesis. They found that the forced expression of HMGA2 could increase the colony forming capacity in intestinal organoids and that the haploinsufficiency of HMGA2 significantly decreased the tumor incidence in villin-Lin28b mice. HMGA2 is a member of the High Mobility Group A class of proteins which bind to AT-rich DNA stretches and modulate gene expression by introducing structural alterations in the chromatin landscape [32]. HMGA2-deficiency impaired the growth in mice, whereas the transgenic expression of HMGA2 variants enhanced the formation of benign tumors, indicating that HMGA2 confers a growth advantage and thus promotes tumorigenesis [33]. In fact, overexpression of HMGA2 was reported to promote metastasis and impact the survival of CRC patients [34]. RNA binding protein IGF2BP1 (IMP1) is also considered as a let-7 target molecule and highly expressed during developmental process, and prominent upregulation and/or de novo synthesis are observed in various tumors [33, 35]. Hamilton et al. demonstrated that enforced expression of IMP1 in SW480 cells could increase the growth of xenografts [36], which indicates that IMP1 plays an important role in the formation or dissemination of colon cancer.
5. Functions of Let-7 in the Colonic Tumorigenesis
5.1. Cell Cycle, Proliferation, and Apoptosis
The downregulation of let-7 promotes cell proliferation through the activation of a variety of cellular proliferation signaling and cell cycle signaling pathways. For instance, let-7 negatively regulates RAS expression through direct binding, which could downregulate MAPK pathway and PI3K/AKT pathway [11, 37]. Let-7 also directly downregulates some oncoproteins such as MYC, HMGA2, and IGF1 [38–41] that are known as critical regulators for the growth of CRC.
The downregulation of let-7 upregulates some cell cycle regulators such as cyclin A2, cyclin D1/2, CDK6, CDC34, CDC25A, Aurora A and B kinases, CDK8, PLAGL2, and TRIM71, which results in the activation of cell cycle [11, 42]. Let-7 also inhibits cell proliferation by regulating transcriptional factors such as STAT3, E2F5, E2F6, CBFB, PLAGL2, SOX9, GZF1, YAP1, GTF2I, and ARID3A [42, 43]. Let-7 microRNAs also suppress cell proliferation by regulating Wnt signaling. The inhibition of Wnt signaling by let-7 is reported in various types of cancers [44–47]. In the context of intestinal tumorigenesis, Tu et al. demonstrated that overexpressed LIN28 could accelerate the growth of intestinal tumors in ApcMin /+ mice and that this tumor-promoting effect was let-7 dependent [28].
The relationship between let-7 microRNAs and apoptosis in CRC is still controversial. Geng et al. reported that let-7 inhibited apoptosis by decreasing the expression of Fas through the direct inhibition against Fas mRNA in HT29 cells [48]. They demonstrated that let-7 inhibition increased Fas expression and the sensitivity to the FAS-related apoptosis. Conversely, Zhang et al. demonstrated that forced expression of let-7c promoted apoptosis in CRC cell lines at least by targeting BCL2L1 [49]. Mongroo et al. showed that the loss of IMP1, a let-7 target, promoted caspase- and lamin-mediated cell death through CYFIP2 by the cross-talk with KRAS in SW480 cells [50]. Another let-7 target in intestine, HMGA2, was reported to inhibit apoptosis [51]. The effect of let-7 microRNAs on apoptosis might be context-dependent and so further investigation will be needed.
5.2. Let-7 and Intestinal Stem Cells
Intestinal stem cell marker LGR5 is known to be upregulated in human colon cancers and sporadic adenomas [67]. The intestinal stem cell markers such as LGR5, ASCL2, SMOC2, Msi1, and Tert are also known to be increased in colitis-associated carcinogenesis [68]. Madison et al. examined the expression levels of let-7 microRNAs and intestinal stem cell markers (e.g., LGR5, EPHB2, and ASCL2) in human CRC tissues and normal adjacent tissues. Let-7a and let-7b were significantly downregulated in CRC specimens, while intestinal stem cell markers were significantly upregulated, which suggests that depletion of let-7a and let-7b may contribute to a stem cell phenotype in CRC [31]. They also demonstrated that tumors from Villin-Lin28b/let-7IEC-KO mice exhibited a significant upregulation of stem cell markers including Bmi1, Lrig1, Olfm4, ASCL2, Prom1, LGR5, Msi1, and SOX9, suggesting an expansion of CRC and +4 stem cell-like compartments. They evaluated the coexpression of let-7 target mRNAs and stem cell markers in mouse samples, and found that HMGA1 and HMGA2 were intensely correlated with stem cell markers. HMGA2 expression was also correlated with LGR5 expression in human CRC samples [31].
5.3. Tumor Progression and Let-7 MicroRNAs in CRC
The downregulation of let-7 promotes migration and invasion of normal intestinal epithelial cells and CRC cells [27]. Xenografts of the DLD-1 cells in which let-7 expression was decreased by LIN28B overexpression developed lung and liver metastases in a mouse model, which suggests that LIN28B/let-7 axis could affect the metastasis in CRC [26]. Some let-7 target oncogenes such as RAS and MYC might contribute to the tumor progression [69, 70]. HMGA2, one of the let-7 target molecules in intestine, is reported to promote epithelial-to-mesenchymal transition (EMT) through the induction of transcription factor Slug [71]. Forced expression of another let-7 target IMP1 in CRC cell line promoted the growth of xenograft tumors and dissemination into the blood in a mouse model. IMP1 overexpression decreased the expression of E-cadherin, suggesting that IMP1 contributes to the tumor progression through the loss of epithelial identity [36].
5.4. Let-7 and Chemoresistance
The downregulation of let-7 has been reported to contribute to the acquisition of resistance against chemotherapy in many types of cancers such as breast cancer [72], esophageal cancer [43], acute myeloid leukemia [73], pancreatic cancer [74], ovarian cancer [75], and HCC [76]. Although only a single paper reported that the high let-7g expression was significantly associated with the chemoresistance against S-1 in CRC patients [77], most of the studies have shown that the downregulation of let-7 contributes to the chemoresistance in the context of CRC. One of the underlying mechanisms by which the downregulated let-7 contributes to the chemoresistance is that the decreased let-7 induces the expression of DNA excision repair protein, excision repair cross-complementing group 1 (ERCC1), which contributes to the resistance against cisplatin or 5-FU [25].
Let-7 microRNAs play an important role in regulating the response to antiepidermal growth factor receptor (EGFR) therapies in CRC patients. Cappuzzo et al. demonstrated that the KRAS wild-type metastatic CRC patients with high let-7c exhibited better response to EGFR monoclonal antibodies [78]. Although anti-EGFR treatment is not effective in CRC patients with mutated KRAS, Ruzzo et al. reported that the upregulation of let-7a may rescue the sensitivity of anti-EGFR therapies in such CRC patients [79]. A functional variant of a let-7 complementary site (LCS6) in the KRAS 3′UTR mRNA might be a useful biomarker to predict the sensitivity of anti-EGFR therapies in patients with metastatic CRC, and further prospective confirmation is needed [80].
6. Let-7 MicroRNAs as a Therapeutic Target
As discussed above, accumulating evidence has indicated that let-7 microRNAs function as a tumor suppressor in CRC. Therefore, gene replacement therapy which attempts to introduce the analogous let-7 molecules could be effective. Recent advances in genetic engineering have enabled us to make the microRNA mimics, artificial structures of RNA duplexes, that are identical to the mature microRNA sequence. An microRNA mimic is designed to have the function of the endogenous microRNA, attempting to restore its loss of function as a tumor suppressor [81]. However, systemic administration of microRNAs mimics might induce unexpected side effects because microRNAs including let-7 can be functional not only in cancer cells but also in normal cells (i.e., development, cell proliferation, apoptosis, cell cycle control, differentiation, migration, and metabolism [82, 83]). Therefore, alternative delivery systems functionalized with miRNA using nanoparticles have recently gained intense attention [53] and have been investigated in vitro/in vivo and also in clinical trials. Different types of microRNA mimics have been used in the medical field as therapeutic agents loaded on the surface of nanoparticles (Table 1). Let-7 mimics have been proved to have therapeutic efficacy in mouse models of lung and prostate cancers [55, 84]. These delivery systems have been tested on several animal models, and some disadvantages regarding toxicity, immune, and inflammatory responses were observed [85].
Table 1.
MicroRNA replacement therapy with mimics.
| microRNA | Cancer |
|---|---|
| microRNA-29 | Lung [52] |
| microRNA-31 | Neuroblastoma [53] |
| microRNA-34a | Neuroblastoma [54], Lung [55–57], HCC [58], Pancreas [59], Lymphoma [60], Prostate [61], Multiple Myeloma [62] |
| microRNA-113b | Lung [63] |
| microRNA-143 | CRC [64], Pancreas [59] |
| microRNA-145 | Pancreas [59], Lung [65] |
| microRNA-365 | CRC [66] |
| LET-7 | Lung [55] |
CRC: colorectal cancer; HCC: hepatocellular carcinoma.
For clinical application, gene replacement therapy with microRNA mimics met many obstacles: the instability of therapeutic molecules, nonspecific inflammation, controlled release of therapeutic molecules, specificity and efficiency of the delivery systems [53]. Advanced delivery strategies are still needed.
7. Conclusions
Accumulating evidence has showed that let-7 microRNAs function as tumor suppressor through the posttranscriptional regulation of target mRNAs in CRC. Let-7 microRNAs regulate various biological events such as cell proliferation, cell cycle, migration, progression, stem cell biology, metabolism, and chemoresistance. Gene replacement therapy with let-7 mimics which attempts to restore the tumor suppressive function of let-7 is tested and therapeutic efficacy has been proved in both in vitro experiments and in vivo animal models. However, many obstacles still remain to be overcome for the safe clinical application. Advanced drug delivery strategies are required.
Conflicts of Interest
The authors declare that there are no conflicts of interest regarding this paper.
References
- 1.Malvezzi M., Bertuccio P., Levi F., La Vecchia C., Negri E. European cancer mortality predictions for the year 2014. Annals of Oncology. 2014 doi: 10.1093/annonc/mdu138. [DOI] [PubMed] [Google Scholar]
- 2.Hori M., Matsuda T., Shibata A., Katanoda K., Sobue T., Nishimoto H. Cancer incidence and incidence rates in Japan in 2009: a study of 32 population-based cancer registries for the Monitoring of Cancer Incidence in Japan (MCIJ) project. Japanese Journal of Clinical Oncology. 2015;45(9):884–891. doi: 10.1093/jjco/hyv088. [DOI] [PubMed] [Google Scholar]
- 3.Jemal A., Ward E. M., Johnson C. J., et al. Annual report to the nation on the status of cancer, 1975-2014, featuring survival. Journal of the National Cancer Institute. 2017;109(9) doi: 10.1093/jnci/djx030.djx030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arnold M., Sierra M. S., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2016;66(4):683–691. doi: 10.1136/gutjnl-2015-310912. [DOI] [PubMed] [Google Scholar]
- 5.Reinhart B. J., Slack F. J., Basson M., et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403(6772):901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 6.Rougvie A. E. Control of developmental timing in animals. Nature Reviews Genetics. 2001;2(9):690–701. doi: 10.1038/35088566. [DOI] [PubMed] [Google Scholar]
- 7.Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107(7):823–826. doi: 10.1016/S0092-8674(01)00616-X. [DOI] [PubMed] [Google Scholar]
- 8.Roush S., Slack F. J. The let-7 family of microRNAs. Trends in Cell Biology. 2008;18(10):505–516. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 9.Berindan-Neagoe I., Calin G. A. Molecular pathways: MicroRNAs, cancer cells, and microenvironment. Clinical Cancer Research. 2014;20(24):6247–6253. doi: 10.1158/1078-0432.CCR-13-2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Viswanathan S. R., Daley G. Q. Lin28: A MicroRNA Regulator with a Macro Role. Cell. 2010;140(4):445–449. doi: 10.1016/j.cell.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 11.Wang T., Wang G., Hao D., et al. Aberrant regulation of the LIN28A/LIN28B and let-7 loop in human malignant tumors and its effects on the hallmarks of cancer. Molecular Cancer. 2015;14(1, article no. 125) doi: 10.1186/s12943-015-0402-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee H., Han S., Kwon C. S., Lee D. Biogenesis and regulation of the let-7 miRNAs and their functional implications. Protein & Cell. 2016;7(2):100–113. doi: 10.1007/s13238-015-0212-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Balzer E., Moss E. G. Localization of the developmental timing regulator Lin28 to mRNP complexes, P-bodies and stress granules. RNA Biology. 2007;4(1):16–25. doi: 10.4161/rna.4.1.4364. [DOI] [PubMed] [Google Scholar]
- 14.Newman M. A., Thomson J. M., Hammond S. M. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14(8):1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Piskounova E., Polytarchou C., Thornton J. E., et al. Lin28A and Lin28B inhibit let-7 MicroRNA biogenesis by distinct mechanisms. Cell. 2011;147(5):1066–1079. doi: 10.1016/j.cell.2011.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mayr F., Heinemann U. Mechanisms of Lin28-mediated miRNA and mRNA regulation--a structural and functional perspective. International Journal of Molecular Sciences. 2013;14(8):16532–16553. doi: 10.3390/ijms140816532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nam Y., Chen C., Gregory R. I., Chou J. J., Sliz P. Molecular basis for interaction of let-7 microRNAs with Lin28. Cell. 2011;147(5):1080–1091. doi: 10.1016/j.cell.2011.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loughlin F. E., Gebert L. F. R., Towbin H., Brunschweiger A., Hall J., Allain F. H.-T. Structural basis of pre-let-7 miRNA recognition by the zinc knuckles of pluripotency factor Lin28. Nature Structural & Molecular Biology. 2012;19(1):84–91. doi: 10.1038/nsmb.2202. [DOI] [PubMed] [Google Scholar]
- 19.Piskounova E., Viswanathan S. R., Janas M., et al. Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. The Journal of Biological Chemistry. 2008;283(31):21310–21314. doi: 10.1074/jbc.c800108200. [DOI] [PubMed] [Google Scholar]
- 20.Hagan J. P., Piskounova E., Gregory R. I. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nature Structural & Molecular Biology. 2009;16(10):1021–1025. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heo I., Joo C., Cho J., Ha M., Han J., Kim V. N. Lin28 mediates the terminal uridylation of let-7 precursor microRNA. Molecular Cell. 2008;32(2):276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 22.Heo I., Joo C., Kim Y.-K., et al. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138(4):696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 23.Mullen T. E., Marzluff W. F. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes & Development. 2008;22(1):50–65. doi: 10.1101/gad.1622708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang H.-M., Triboulet R., Thornton J. E., Gregory R. I. A role for the Perlman syndrome exonuclease Dis3l2 in the Lin28-let-7 pathway. Nature. 2013;497(7448):244–248. doi: 10.1038/nature12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mizuno R., Chatterji P., Andres S. Differential Regulation of LET-7 by LIN28B Isoform–Specific Functions. Mol Cancer Res. 2018;16(3):403–416. doi: 10.1158/1541-7786.MCR-17-0514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.King C. E., Cuatrecasas M., Castells A., Sepulveda A. R., Lee J.-S., Rustgi A. K. LIN28B promotes colon cancer progression and metastasis. Cancer Research. 2011;71(12):4260–4268. doi: 10.1158/0008-5472.CAN-10-4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.King C. E., Wang L., Winograd R., et al. LIN28B fosters colon cancer migration, invasion and transformation through let-7-dependent and-independent mechanisms. Oncogene. 2011;30(40):4185–4193. doi: 10.1038/onc.2011.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tu H.-C., Schwitalla S., Qian Z., et al. LIN28 cooperates with WNT signaling to drive invasive intestinal and colorectal adenocarcinoma in mice and humans. Genes & Development. 2015;29(10):1074–1086. doi: 10.1101/gad.256693.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Madison B. B., Liu Q., Zhong X., et al. LIN28B promotes growth and tumorigenesis of the intestinal epithelium via Let-7. Genes & Development. 2013;27(20):2233–2245. doi: 10.1101/gad.224659.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Triboulet R., Pirouz M., Gregory R. I. A Single Let-7 MicroRNA Bypasses LIN28-Mediated Repression. Cell Reports. 2015;13(2):260–266. doi: 10.1016/j.celrep.2015.08.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Madison B. B., Jeganathan A. N., Mizuno R., et al. Let-7 represses carcinogenesis and a stem cell phenotype in the intestine via regulation of Hmga2. PLoS Genetics. 2015;11(8) doi: 10.1371/journal.pgen.1005408.e1005408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yong S. L., Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes & Development. 2007;21(9):1025–1030. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Busch B., Bley N., Müller S., et al. The oncogenic triangle of HMGA2, LIN28B and IGF2BP1 antagonizes tumor-suppressive actions of the let-7 family. Nucleic Acids Research. 2016;44(8):3845–3864. doi: 10.1093/nar/gkw099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang X., Liu X., Li A. Y.-J., et al. Overexpression of HMGA2 promotes metastasis and impacts survival of colorectal cancers. Clinical Cancer Research. 2011;17(8):2570–2580. doi: 10.1158/1078-0432.CCR-10-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bell J. L., Wächter K., Mühleck B., et al. Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs): Post-transcriptional drivers of cancer progression? Cellular and Molecular Life Sciences. 2013;70(15):2657–2675. doi: 10.1007/s00018-012-1186-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hamilton K. E., Noubissi F. K., Katti P. S., et al. IMP1 promotes tumor growth, dissemination and a tumor-initiating cell phenotype in colorectal cancer cell xenografts. Carcinogenesis. 2013;34(11):2647–2654. doi: 10.1093/carcin/bgt217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson S. M., Grosshans H., Shingara J., et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120(5):635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 38.Jung H. J., Suh Y. Regulation of IGF -1 signaling by microRNAs. Frontiers in Genetics. 2014;5(472) doi: 10.3389/fgene.2014.00472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang K., Gao H., Wu X., et al. Frequent overexpression of HMGA2 in human atypical teratoid/rhabdoid tumor and its correlation with let-7a3/let-7b miRNA. Clinical Cancer Research. 2014;20(5):1179–1189. doi: 10.1158/1078-0432.ccr-13-1452. [DOI] [PubMed] [Google Scholar]
- 40.Chen K.-J., Hou Y., Wang K., et al. Reexpression of let-7g microRNA inhibits the proliferation and migration via K-Ras/HMGA2/snail axis in hepatocellular carcinoma. BioMed Research International. 2014;2014 doi: 10.1155/2014/742417.742417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Balzeau J., Menezes M. R., Cao S., Hagan J. P. The LIN28/let-7 pathway in cancer. Frontiers in Genetics. 2017;8(31) doi: 10.3389/fgene.2017.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barh D., Malhotra R., Ravi B., Sindhurani P. MicroRNA let-7: an emerging next-generation cancer therapeutic. Current Oncology. 2010;17(1):70–80. doi: 10.3747/co.v17i1.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sugimura K., Miyata H., Tanaka K., et al. Let-7 expression is a significant determinant of response to chemotherapy through the regulation of IL-6/STAT3 pathway in esophageal squamous cell carcinoma. Clinical Cancer Research. 2012;18(18):5144–5153. doi: 10.1158/1078-0432.CCR-12-0701. [DOI] [PubMed] [Google Scholar]
- 44.Jin B., Wang W., Meng X.-X., et al. Let-7 inhibits self-renewal of hepatocellular cancer stem-like cells through regulating the epithelial-mesenchymal transition and the Wnt signaling pathway. BMC Cancer. 2016;16(1, article no. 863) doi: 10.1186/s12885-016-2904-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cai H., Chen Y., Yang X., et al. Let7b modulates the Wnt/β-catenin pathway in liver cancer cells via downregulated Frizzled4. Tumor Biology. 2017;39(7):1–7. doi: 10.1177/1010428317716076. [DOI] [PubMed] [Google Scholar]
- 46.Sun X., Xu C., Tang S.-C., et al. Let-7c blocks estrogen-activated Wnt signaling in induction of self-renewal of breast cancer stem cells. Cancer Gene Therapy. 2016;23(4):83–89. doi: 10.1038/cgt.2016.3. [DOI] [PubMed] [Google Scholar]
- 47.Viñas J. L., Ventayol M., Brüne B., et al. miRNA let-7e modulates the wnt pathway and early nephrogenic markers in mouse embryonic stem cell differentiation. PLoS ONE. 2013;8(4) doi: 10.1371/journal.pone.0060937.e60937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Geng L., Zhu B., Dai B.-H., et al. A let-7/Fas double-negative feedback loop regulates human colon carcinoma cells sensitivity to Fas-related apoptosis. Biochemical and Biophysical Research Communications. 2011;408(3):494–499. doi: 10.1016/j.bbrc.2011.04.074. [DOI] [PubMed] [Google Scholar]
- 49.Zhang H., Zong Y., Qiu G., et al. Silencing Lin28 promotes apoptosis in colorectal cancer cells by upregulating let7c targeting of antiapoptotic BCL2L1. Molecular Medicine Reports. 2018;17(4):5143–5149. doi: 10.3892/mmr.2018.8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mongroo P. S., Noubissi F. K., Cuatrecasas M., et al. IMP-1 displays cross-talk with K-Ras and modulates colon cancer cell survival through the novel proapoptotic protein CYFIP2. Cancer Research. 2011;71(6):2172–2182. doi: 10.1158/0008-5472.CAN-10-3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nana A. W., Chin Y., Lin C., et al. Tetrac downregulates β-catenin and HMGA2 to promote the effect of resveratrol in colon cancer. Endocrine-Related Cancer. 2018;25(3):279–293. doi: 10.1530/ERC-17-0450. [DOI] [PubMed] [Google Scholar]
- 52.Wu Y., Crawford M., Mao Y., et al. Therapeutic delivery of MicroRNA-29b by cationic lipoplexes for lung cancer. Molecular Therapy—Nucleic Acids. 2013;2, article e84 doi: 10.1038/mtna.2013.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jurj A., Braicu C., Pop L.-A., Tomuleasa C., Gherman C. D., Berindan-Neagoe I. The new era of nanotechnology, an alternative to change cancer treatment. Drug Design, Development and Therapy. 2017;11:2871–2890. doi: 10.2147/DDDT.S142337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tivnan A., Orr W. S., Gubala V., et al. Inhibition of neuroblastoma tumor growth by targeted delivery of microRNA-34a using anti-disialoganglioside GD2 coated nanoparticles. PLoS ONE. 2012;7(5) doi: 10.1371/journal.pone.0038129.e38129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trang P., Wiggins J. F., Daige C. L., et al. Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Molecular Therapy. 2011;19(6):1116–1122. doi: 10.1038/mt.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hu Q. L., Jiang Q. Y., Jin X., et al. Cationic microRNA-delivering nanovectors with bifunctional peptides for efficient treatment of PANC-1 xenograft model. Biomaterials. 2013;34(9):2265–2276. doi: 10.1016/j.biomaterials.2012.12.016. [DOI] [PubMed] [Google Scholar]
- 57.Wiggins J. F., Ruffino L., Kelnar K., et al. Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34. Cancer Research. 2010;70(14):5923–5930. doi: 10.1158/0008-5472.CAN-10-0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bader A. G. MiR-34 - a microRNA replacement therapy is headed to the clinic. Frontiers in Genetics. 2012;3, article 120 doi: 10.3389/fgene.2012.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pramanik D., Campbell N. R., Karikari C., et al. Restitution of tumor suppressor microRNAs using a systemic nanovector inhibits pancreatic cancer growth in mice. Molecular Cancer Therapeutics. 2011;10(8):1470–1480. doi: 10.1158/1535-7163.MCT-11-0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Craig V. J., Tzankov A., Flori M., Schmid C. A., BaDer A. G., Müller A. Systemic microRNA-34a delivery induces apoptosis and abrogates growth of diffuse large B-cell lymphoma in vivo. Leukemia. 2012;26(11):2421–2424. doi: 10.1038/leu.2012.110. [DOI] [PubMed] [Google Scholar]
- 61.Liu C., Kelnar K., Liu B., et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nature Medicine. 2011;17(2):211–215. doi: 10.1038/nm.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cosco D., Cilurzo F., Maiuolo J., et al. Delivery of miR-34a by chitosan/PLGA nanoplexes for the anticancer treatment of multiple myeloma. Scientific Reports. 2015;5 doi: 10.1038/srep17579.17579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu Y., Crawford M., Yu B., Mao Y., Nana-Sinkam S. P., Lee L. J. MicroRNA delivery by cationic lipoplexes for lung cancer therapy. Molecular Pharmaceutics. 2011;8(4):1381–1389. doi: 10.1021/mp2002076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Borralho P. M., Kren B. T., Castro R. E., Moreira da Silva I. B., Steer C. J., Rodrigues C. M. P. MicroRNA-143 reduces viability and increases sensitivity to 5-fluorouracil in HCT116 human colorectal cancer cells. FEBS Journal. 2009;276(22):6689–6700. doi: 10.1111/j.1742-4658.2009.07383.x. [DOI] [PubMed] [Google Scholar]
- 65.Chiou G.-Y., Cherng J.-Y., Hsu H.-S., et al. Cationic polyurethanes-short branch PEI-mediated delivery of Mir145 inhibited epithelial-mesenchymal transdifferentiation and cancer stem-like properties and in lung adenocarcinoma. Journal of Controlled Release. 2012;159(2):240–250. doi: 10.1016/j.jconrel.2012.01.014. [DOI] [PubMed] [Google Scholar]
- 66.Nie J., Liu L., Zheng W., et al. microRNA-365, down-regulated in colon cancer, inhibits cell cycle progression and promotes apoptosis of colon cancer cells by probably targeting Cyclin D1 and Bcl-2. Carcinogenesis. 2012;33(1):220–225. doi: 10.1093/carcin/bgr245. [DOI] [PubMed] [Google Scholar]
- 67.Uchida H., Yamazaki K., Fukuma M., et al. Overexpression of leucine-rich repeat-containing G protein-coupled receptor 5 in colorectal cancer. Cancer Science. 2010;101(7):1731–1737. doi: 10.1111/j.1349-7006.2010.01571.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim H. S., Lee C., Kim W. H., Maeng Y. H., Jang B. G. Expression profile of intestinal stem cell markers in colitis-associated carcinogenesis. Scientific Reports. 2017;7(1) doi: 10.1038/s41598-017-06900-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Goel S., Huang J., Klampfer L. K-Ras, Intestinal homeostasis and colon cancer. Current Clinical Pharmacology. 2015;10(1):73–81. doi: 10.2174/1574884708666131111204440. [DOI] [PubMed] [Google Scholar]
- 70.Garte S. J. The c-myc oncogene in tumor progression. Critical Reviews in Oncogenesis. 1993;4(4):435–449. [PubMed] [Google Scholar]
- 71.Li Y., Zhao Z., Xu C., Zhou Z., Zhu Z., You T. HMGA2 induces transcription factor Slug expression to promote epithelial-to-mesenchymal transition and contributes to colon cancer progression. Cancer Letters. 2014;355(1):130–140. doi: 10.1016/j.canlet.2014.09.007. [DOI] [PubMed] [Google Scholar]
- 72.Lv K., Liu L., Wang L., et al. Lin28 mediates paclitaxel resistance by modulating p21, Rb and Let-7a miRNA in breast cancer cells. PLoS ONE. 2012;7(7) doi: 10.1371/journal.pone.0040008.e40008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen Y., Jacamo R., Konopleva M., Garzon R., Croce C., Andreeff M. CXCR4 downregulation of let-7a drives chemoresistance in acute myeloid leukemia. The Journal of Clinical Investigation. 2013;123(6):2395–2407. doi: 10.1172/JCI66553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bhutia Y. D., Hung S. W., Krentz M., et al. Differential processing of let-7a precursors influences RRM2 expression and chemosensitivity in pancreatic cancer: role of LIN-28 and SET oncoprotein. PLoS ONE. 2013;8(1) doi: 10.1371/journal.pone.0053436.e53436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhu H., Ng S.-C., Segr A. V., et al. The Lin28/let-7 axis regulates glucose metabolism. Cell. 2011;147(1):81–94. doi: 10.1016/j.cell.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ma X., Li C., Sun L., et al. Lin28/let-7 axis regulates aerobic glycolysis and cancer progression via PDK1. Nature Communications. 2014;5, article no. 5212 doi: 10.1038/ncomms6212. [DOI] [PubMed] [Google Scholar]
- 77.Nakajima G., Hayashi K., Xi Y., et al. Non-coding microRNAs hsa-let-7g and hsa-miR-181b are associated with chemoresponse to S-1 in colon cancer. Cancer Genomics & Proteomics. 2006;3(5):317–324. [PMC free article] [PubMed] [Google Scholar]
- 78.Cappuzzo F., Sacconi A., Landi L., et al. MicroRNA signature in metastatic colorectal cancer patients treated with anti-EGFR monoclonal antibodies. Clinical Colorectal Cancer. 2014;13(1):37–45.e4. doi: 10.1016/j.clcc.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 79.Ruzzo A., Graziano F., Vincenzi B., et al. High let-7a microRNA levels in KRAS-mutated colorectal carcinomas may rescue anti-EGFR therapy effects in patients with chemotherapy-refractory metastatic disease. The Oncologist. 2012;17(6):823–829. doi: 10.1634/theoncologist.2012-0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Saridaki Z., Weidhaas J. B., Lenz H.-J., et al. A let-7 microRNA-binding site polymorphism in KRAS predicts improved outcome in patients with metastatic colorectal cancer treated with salvage cetuximab/panitumumab monotherapy. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20(17):4499–4510. doi: 10.1158/1078-0432.CCR-14-0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Braicu C., Tomuleasa C., Monroig P., Cucuianu A., Berindan-Neagoe I., Calin G. A. Exosomes as divine messengers: are they the Hermes of modern molecular oncology? Cell Death & Differentiation. 2015;22(1):34–45. doi: 10.1038/cdd.2014.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jansson M. D., Lund A. H. MicroRNA and cancer. Molecular Oncology. 2012;6(6):590–610. doi: 10.1016/j.molonc.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ekin A., Karatas O. F., Culha M., Ozen M. Designing a gold nanoparticle-based nanocarrier for microRNA transfection into the prostate and breast cancer cells. The Journal of Gene Medicine. 2014;16(11-12):331–335. doi: 10.1002/jgm.2810. [DOI] [PubMed] [Google Scholar]
- 84.Hydbring P., Badalian-Very G. Clinical applications of microRNAs. F1000Research. 2013;2, article 136 doi: 10.12688/f1000research.2-136.v3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Guo W., Chen W., Yu W., Huang W., Deng W. Small interfering RNA-based molecular therapy of cancers. Chinese Journal of Cancer. 2013;32(9):488–493. doi: 10.5732/cjc.012.10280. [DOI] [PMC free article] [PubMed] [Google Scholar]